Abstract
The ongoing pandemic of the new severe acute respiratory syndrome coronavirus (SARS-CoV-2) has caused more than one million deaths, overwhelmed many public health systems, and led to a worldwide economic recession. This has raised an unprecedented need to develop antiviral drugs and vaccines, which requires profound knowledge of the fundamental pathology of the virus, including its entry, replication, and release from host cells. The genome of coronaviruses comprises around 30 kb of positive single-stranded RNA, representing one of the largest RNA genomes of viruses. The 5′ part of the genome encodes a large polyprotein, PP1ab, which gives rise to 16 non-structural proteins (nsp1– nsp16). Two proteases encoded in nsp3 and nsp5 cleave the polyprotein into individual proteins. Most nsps belong to the viral replicase complex that promotes replication of the viral genome and translation of structural proteins by producing subgenomic mRNAs. The replicase complexes are found on double-membrane vesicles (DMVs) that contain viral double-stranded RNA. Expression of a small subset of viral proteins, including nsp3 and nsp4, is sufficient to induce formation of these DMVs in human cells, suggesting that both proteins deform host membranes into such structures. We will discuss the formation of DMVs and provide an overview of other membrane remodeling processes that are induced by coronaviruses.
Keywords: SARS-CoV2, coronavirus, replication, autophagy
Introduction
Coronaviruses are enveloped viruses that belong to the order Nidovirales. They possess a positive-sense single-stranded RNA genome and are characterized by the production of subgenomic mRNAs during infection. The subfamily Orthocoronavirinae comprises alpha-, beta-, gamma-, and delta-coronaviruses that infect a broad range of amphibians, birds, and mammals. Alpha-coronaviruses comprise the human coronavirus NL63 that causes mild to moderate respiratory tract infections. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), together with SARS-CoV and MERS-CoV (Middle East respiratory syndrome–related coronavirus), belongs to the genus beta-coronavirus, which also comprises human coronavirus (HCoV)-OC43 and -HKU1. The latter cause mild upper respiratory tract infections such as the common cold, whereas MERS and SARS-CoV are responsible for severe forms of lower respiratory tract infections. The highest mortality rate was observed in patients infected with MERS-CoV (30%)1, followed by SARS-CoV2 (9%) and SARS-CoV-2 (<0.5%)3. However, the total number of deaths does not correlate with the mortality rate. For example, MERS-CoV and SARS-CoV-2 have caused 848 and 774 deaths, respectively, while SARS-CoV-2 has already killed more than 1,000,000 people4–6. The primary reason for the large number of Covid-19 cases and victims is related to the extremely rapid spread of the disease, which is triggered by a large proportion of asymptomatic infections and superspreading events7–9. Until an efficient treatment of the disease or a vaccine is found, social distancing and physical barriers are essential to control viral circulation and to limit the propagation of the virus.
Since the onset of the first coronavirus outbreak with SARS-CoV in 2002, our knowledge regarding the fundamental biology of coronaviruses has been tremendously advanced. However, many fundamental questions regarding the viral life cycle remain to be answered. In this review, we provide an overview of the molecular biology of SARS-CoV-2 and focus on membrane remodeling events that are induced by viral proteins to promote viral replication.
Uptake of severe acute respiratory syndrome coronavirus 2: a plan B to avoid endocytic compartments
Coronaviruses are enveloped by a membrane derived from the host cell during budding. Three viral transmembrane proteins are embedded in this membrane: the spike (S) protein, the envelope (E) protein, and the membrane (M) protein. Much effort has been dedicated to characterize the S-protein of SARS-CoV-2 since this protein is key to enter cells and a primary target of the adaptive immune system of the host. The S-protein belongs to the well-characterized class I fusion proteins and shares high similarity with the S-protein of SARS-CoV, to hemagglutinin of influenza A viruses, and to the envelop protein of HIV10. The human receptor and binding partner for the S-proteins of both SARS coronaviruses is the angiotensin-converting enzyme 2 (ACE2)11,12. The firm interaction of the receptor binding (S1) domain of the S-protein with ACE2 promotes tight adhesion of the virus to the host cell plasma membrane13. Entry of viruses into the host cells depends on the fusion of viral envelope and host cell membranes (Figure 1a). The fusion process is triggered by the S2 domain of the S-protein, which harbors a fusion peptide. In order to be activated, the S-protein needs to be cleaved at two proteolytic cleavage sites. The first cleavage site of the SARS-CoV-2 S-protein, which is located between the S1 and S2 domains, is cleaved by furin-like proteases probably during S-protein biosynthesis14. The second cleavage site in the S2 domain is cut by the host cell protease TMPRSS2 after the S-protein has bound to ACE2 on target cells11. The S1 subunit is released and the S2 subunit undergoes a first conformational rearrangement to expose the fusion peptide. This is followed by an insertion of this peptide into the host cell membrane. A second large conformational change of the S2 subunit triggers fusion of the viral envelope and host cell membranes15. Since TMPRSS2 is a plasma membrane protein, cleavage and activation of the SARS-CoV-2 S-protein take place at the host cell plasma membrane. A similar mechanism has been reported for the uptake of SARS-CoV16. However, SARS-CoV and MERS-CoV can also be taken up into cells that do not express TMPRSS2 via clathrin-dependent endocytosis followed by cleavage and activation by the pH-sensitive protease cathepsin L (Figure 1b)17–19. Establishing the entry pathway of SARS-CoV-2 is of tremendous importance to understand its pathogenicity and to identify vulnerable cell types20. ACE2 and TMPRSS2 are highly enriched in nasal epithelial cells which correlates with viral replication in the upper respiratory tract21. The efficient colonization of these cells promotes viral spreading and infection and renders SARS-CoV-2 more contagious than other related coronaviruses22. Epithelial cells of the lower respiratory tract, including bronchial and small airway tissues, express significantly less ACE2 and TMPRSS2, suggesting that they are less susceptible to viral infection. However, clathrin-dependent and -independent endocytosis followed by the fusion of viral envelope and host endolysosomal membranes provides a second access route to host cells23–25. Lysosomotropic agents such as chloroquine inhibit entry of SARS-CoV-2, suggesting that this coronavirus can also be taken up by endocytosis, which contributes to the colonization of cells lacking TMPRSS2 (Figure 1b)26. Although the specific contribution of the two alternative routes for the entry of SARS-CoV-2 into host cells remains to be established, recent evidence suggests that clinical symptoms of patients with Covid-19 correlate with expression levels of ACE2 and TMPRSS2. In children, known to be less susceptible to SARS-CoV-2 infections, strongly reduced expression levels of ACE2 and TMPRSS2 in upper and lower respiratory tract tissues have been observed27.
Figure 1. Infection cycle of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The infection cycle of SARS-CoV-2 is shown. (a) Viral entry by binding to the cell surface receptor angiotensin-converting enzyme 2 (ACE2) (red) and fusion of the spike protein after proteolytic processing at the plasma membrane by TMPRSS2. (b) Viral entry by taking advantage of the endocytic pathway involving clathrin-dependent or -independent endocytosis and proteolytic processing in lysogenic compartments involving cathepsin L. (c) Formation of double-membrane vesicles (DMVs) at the endoplasmic reticulum (ER). (d) DMVs serve as replication organelles to produce viral genomic RNA. The interaction of exported RNA and viral N-proteins gives rise to viral capsids (dark blue) that are delivered to budding compartments (light blue). Translation of structural proteins, including nucleocapsid (N), membrane (M), surface (S), and envelope (E) proteins, occurs from subgenomic mRNAs that are produced by the viral replication–translation complex. (e) Trafficking of viral structural proteins (dark blue) from the ER to the Golgi and the plasma membrane following the exocytotic pathway. Proteins accumulate in viral budding compartments that are derived from membranes of the exocytic pathway, where capsids bud into the lumen. The compartments fuse with the plasma membrane to release viruses. ERGIC, endoplasmic reticulum–Golgi intermediate compartment; rER, rough endoplasmic reticulum; sER, smooth endoplasmic reticulum; TGN, trans-Golgi network.
Critical functions but intrinsically disordered – viral non-structural proteins
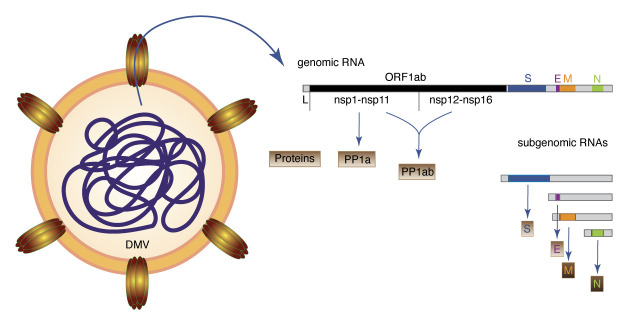
Whatever path is taken by viral particles to gain access into human cells, fusion of viral and host membranes releases viral capsids into the cytoplasm of host cells. The capsid contains the genome of CoVs, which is single-stranded positive-sense RNA that possesses a 5′ cap and a 3′ poly (A) tail. The genome thus serves as a template for translation. In the early phase of infection, the two partially overlapping open reading frames (ORFs) 1 and 2 of the genome are expressed which comprise two thirds of the total genome, giving rise to the two polyproteins (PPs) 1a and 1ab (Figure 2).
Figure 2. Replication and translation of viral genomic RNA.
Double-membrane vesicles (DMVs) contain double-stranded viral RNA, which is an intermediate of viral genomic RNA replication. Channels that span the double membrane allow viral RNAs, including positive-strand viral genomic RNA and subgenomic RNAs, to be exported. Translation of open reading frame 1a (ORF1a) gives rise to the polyprotein 1a (PP1a) comprising nsp1 to nsp11. A programmed ribosomal frameshift leads to transcription of PP1ab comprising nsp1 to nsp16. Subgenomic RNAs contain a common 5′ leader sequence and their translation gives rise to S (surface), E (envelope), M (membrane), and N (nucleocapsid) structural proteins. The 3′ part of the genome comprises additional ORFs (3a, 6, 7a, 7b, 8, and 10) that are not depicted. Their transcription leads to additional subgenomic RNAs (not shown). L, leader sequence.
PP1a comprises 11 non-structural proteins (nsps) that are cleaved into single nsps by the papain-like protease nsp3 and the 3C-like protease nsp5. A programmed ribosomal -1 frameshift upstream of the stop codon of nsp11 allows continuation of translation and gives rise to PP1ab that is cleaved into 15 polypeptides, comprising nsp1 to nsp1628. Proteins that are generated by cleavage of PP1a modulate cellular pathways and remodel endomembranes of host cells to generate compartments for viral replication. Proteins, including nsp12 to nsp16, are derived from PP1b and, owing to the lower frequency of the programmed frameshift, are less abundant29. However, these proteins assemble into a complex that associates with host endomembranes to coordinate viral replication and translation reactions.
Nsp1 of SARS-CoV-2 shuts down translation of host mRNAs and RIG-I–dependent innate immunity by binding to the mRNA entry tunnel of ribosomes30. Nsp2 is dispensable for viral replication in cell culture and no specific function has yet been revealed31. The multidomain protein nsp3 combines various different functions32. Its papain-like protease (PLpro) cleaves nsp1, nsp2, and nsp3. Moreover, nsp3 is a major interaction hub and integral member of the viral replication complex. The macrodomains of nsp3 suppress the host immune response by exerting several enzymatic activities, including ADP-ribose-1“-phosphate phosphatase, de-mono-ADP-ribosylation, and de-poly-ADP-ribosylation33–35. Another important function of nsp3 in remodeling host cell endomembranes is related to its two transmembrane helices and requires its interaction with the transmembrane protein nsp4 and nsp636. Moreover, nsp3 was recently shown to form part of a pore that spans double-membrane vesicles (DMVs)37. Such vesicles are generated from the endoplasmic reticulum (ER) and serve as viral replication organelles.
The two proteins nsp7 and nsp8 are co-factors of the main RNA-dependent RNA polymerase nsp1238 and their interaction enhances RNA binding and processivity of nsp1239–41. The two remaining proteins of PP1a—nsp9 and nsp10—are associated with the RNase complex through a direct interaction between nsp8 and nsp942. They bind single-stranded RNA and are supposed to regulate or modulate the activity of nsp12 during the viral replication cycle43.
The C-terminal part of PP1a contains nsp11, which is a small polypeptide that becomes part of nsp12 upon programmed ribosomal frameshift. In addition to nsp12, PP1b contains proteins that are essential for viral replication, including the helicase nsp13 that unwinds RNA44,45. Replication of the large and complex viral genome of SARS-CoV-2 depends also on nsp14 that proofreads RNA during replication and corrects errors made by nsp12. The corresponding 3′-to-5′ exoribonuclease activity of nsp14 from HCoV-229E has been shown to be essential for the production of viable virus46. Nsp14 is also involved in mRNA cap formation by methylating the inverted guanosine moiety at the N7 position. The second methylation of the cap at the 2′-O position of the first transcribed nucleotide is added by nsp1647. Cap methylation also depends on nsp10, which interacts with nsp14 and nsp16, suggesting that cap methylating enzymes assemble together with the RNA polymerase and associated proteins nsp8, nsp9, and nsp13 to form part of a bigger replication complex48. Recent studies have started to reveal the molecular composition of the replication machinery, providing insights into the function of the complex49–51. Nsp15 is a nidoviral RNA uridylate-specific endoribonuclease (NendoU) which is involved mainly in suppressing antiviral immune responses by degrading cytoplasmic viral RNA52,53.
Double is better than single – viral replication and double-membrane vesicles
Cells have evolved various antiviral strategies to counteract viral infection by detecting characteristic structures termed pathogen-associated molecular patterns (PAMPs). Double-stranded RNA (dsRNA) is a potent PAMP and is an intermediate that is produced during replication of RNA viruses, including SARS-CoV-254. Pattern recognition receptors such as Toll-like receptor 3 as well as RIG-I–like receptors RIG-1 and MDA-5 detect dsRNA and activate signaling cascades that induce the production of type 1 interferons (IFNs), including IFN-α and IFN-β55. These cytokines induce antiviral responses in neighboring cells and activate innate and adaptive immune responses. Coronaviruses, including SARS-CoV and SARS-CoV-2, shield their dsRNA intermediates in DMVs, probably to evade IFN-1 activation56. Together with other strategies, including RNA capping and interference with antiviral signaling pathways, coronaviruses severely delay IFN-1 induction and corresponding pro-inflammatory immune responses57,58.
DMVs are abundant structures induced by many +RNA viruses, including arteriviruses59,60, hepatitis C virus61, noro- and picornaviruses62,63, and coronaviruses64,65. The morphology of these vesicles shows remarkable similarities with autophagosomes, and for some viruses, including SARS-CoV, a direct link between formation of DMVs and autophagy has been proposed66. Interestingly, expression of nsp3 and nsp4 appears to be sufficient to induce DMVs67. However, how these structures are formed remains elusive. Insights into potential molecular mechanisms for DMV formation come from transmission electron microscopy studies of cells that express single nsps or combinations of them.
Individually expressed nsp3 and nsp4 co-localize with ER markers, consistent with their co-translational insertion into the ER67,68. A recent study demonstrated that nsp3 forms together with other, yet-to-be-identified viral or host proteins pores that span DMVs, allowing for exchange of luminal and cytoplasmic material and for export of viral RNA37. The pore possesses a sixfold symmetry, and nsp3 is the major constituent of its cytoplasmic portion that possesses a crown-like structure. This suggests that the transmembrane domains of nsp3 span the cytoplasmic membrane of DMVs. Furthermore, nsp4 has been shown to interact with nsp3 and this interaction is required and sufficient to induce pairing of ER membranes36. Given that DMVs originate from the ER, it is tempting to speculate that nsp3 and nsp4 engage each other to form the pore with nsp4 spanning the inner membrane of DMVs. Consistent with this hypothesis, intraluminal loops of nsp3 and nsp4 are involved in the interaction of both proteins36. Whether DMVs directly emerge from these paired ER structures remains to be established.
A recent study provided evidence that DMVs are the primary site for viral RNA synthesis56. Moreover, electron-tomography of cells infected with the murine hepatitis coronavirus (MHV) revealed channel-like structures in DMVs, connecting the lumen of DMVs with the cytoplasm37. Although replication organelles of Flock House nodavirus (FHV) differ from DMVs, they also contain a pore opening toward the cytoplasm which shares remarkable structural similarity to channels observed in DMVs induced by MHV69. Moreover, this pore contains the FHV protein A, which harbors RNA polymerase and RNA capping activities, suggesting that these pores are central organization platforms for the viral replication and transcription machinery.
The major component of the protein pore in DMVs of MHV is nsp3, which lacks similar enzymatic activities. However, nsp3 of SARS-CoV was found to interact with nsp7 and nsp12, both of which form part of the viral replication and transcription complex70. Furthermore, nsp3 interacts with the N-protein, which together with viral RNA forms the capsid, suggesting that nsp3 is a central organization hub for viral replication and capsid assembly71.
Double-membrane vesicles remain mysterious compartments
The generation of DMVs is not only a hallmark of cells infected by all coronaviruses. Similar structures are also observed in arteri-, noro-, and picorna-viruses and hepatitis C virus. Inhibiting their biogenesis can lead to potent antiviral therapies with a broad spectrum of action. This, however, requires that the molecular mechanism of DMV biogenesis is fully characterized and well understood. Unfortunately, our current knowledge is rather limited, and controversial observations regarding the biogenesis of DMVs have been made. This is particularly true for autophagy, one of the most versatile recycling pathways in eukaryotic cells.
Autophagy delivers damaged or superfluous cytoplasmic components, including organelles and protein aggregates, to lysosomes for degradation72. During this process, a double-membrane cup-like structure that engulfs autophagic cargo is formed. Sealing of this membrane gives rise to double-membrane limited autophagosomes. The obvious morphological similarity between such autophagosomes and viral DMVs suggests that viruses hijack autophagy-related (ATG) proteins to induce the formation of DMVs. Consistent with this hypothesis, ATG5, which is essential for canonical autophagy, was found to be required for MHV replication in embryonic stem cells73. By contrast, replication of MHV in mouse embryonic fibroblasts did not depend on ATG574 or ATG766, implying that the contribution of canonical autophagy to viral replication is cell type–specific. This, however, does not necessarily indicate that MHV replicates independently of autophagy in these cells because an ATG5/ATG7-independent autophagy pathway exists in embryonic tissues75.
Another potential connection between autophagy and viral replication involves LC3, an important autophagy protein and widely used cellular marker for autophagosomes. In canonical autophagy, LC3 is conjugated to the lipid phosphatidylethanolamine in autophagic membranes. The reaction is known as LC3 lipidation and involves conversion of LC3-I (non-lipidated) into LC3-II (lipidated) by a Ub-like conjugation system comprising E1 enzyme ATG7, the E2 enzyme ATG3, and the E3-ligase ATG12–ATG5-ATG16L176. LC3 was found to co-localize with MHV nsp8, indicating that components of the autophagy pathways are indeed hijacked by MHV73. Moreover, depletion of LC3 strongly impaired MHV replication, suggesting that LC3 is required for viral replication66. Similar observations were made in SARS-CoV–infected cells77. By contrast, lipidation of LC3 was not required for replication of MHV or SARS-CoV in bone marrow–derived macrophages or mouse embryonic fibroblasts, although LC3 was found to co-localize with viral nsps78.
LC3-II tethers cargo to autophagic membranes through its interaction with autophagy receptors such as p6279. Given that DMVs are devoid of such cargo, it is plausible that LC3-II is dispensable for viral replication. This implies that nsps co-localize with LC3-I instead of LC3-II. Although autophagy-independent functions of LC3-II have been discovered in the past, the molecular function of LC3-I remained less well understood80. However, LC3-I appears to be involved in ER-associated degradation (ERAD), which is an ER stress response and quality control pathway that ensures that unfolded proteins are removed from the ER81,82. In this pathway, LC3-I was found to co-localize with vesicles that remove ERAD regulators from the ER. Thus, it is possible that coronaviruses coopt ERAD-related pathways by recruiting LC3-I to induce the formation of DMVs81.
Without any doubt, the ER plays a central role in the replication of coronaviruses. Many electron microscopy studies of infected cells demonstrated that ER membranes are heavily deformed and remodeled56. As discussed in the previous section, the viral proteins nsp3, nsp4, and nsp6 are central coordinators of such rearrangements. Nsp6 is of particular interest regarding the relationship between ER membranes, viral replication, and autophagy. Autophagosomes are formed at distinct domains of the ER which are enriched in phosphatidylinositol-3-phosphate (PtdIns3P) by the action of an autophagy-specific PtdIns3–kinase complex. PtdIns3P binding proteins, including DFCP1, are recruited to these domains and induce the formation of omegasomes, which are cradle-like extensions of the ER (Figure 1c). Omegasomes serve as platforms for autophagosome biogenesis by coordinating the recruitment of autophagy proteins and lipids83,84. Nsp6 of the avian coronavirus IBV (infectious bronchitis virus) was shown to co-localize with the PtdIns (3)P binding proteins DFCP1 and WIPI2 at the ER, suggesting that nsp8 initiates the formation of autophagosomes85. Similar observations have been made for nsp6 from MHV and SARS-CoV. However, expansion of such autophagosomes was limited by nsp6. As a result, much smaller autophagosomes were formed in nsp6-expressing cells86. Although expression of nsp6 is sufficient to induce autophagy, the formation of DMVs depends on the co-expression of nsp3 and nsp4. This suggests that nsp6 initiates the formation of omegasomes but that nsp3 and nsp4 are required to prevent formation of canonical autophagosomes by inducing the formation of DMVs (Figure 1c).
Conclusions
Understanding membrane dynamics in cells is challenging. Even for fundamental transport processes between organelles, the biogenesis of organelles, and their degradation, many open questions remain. Revealing how complex viruses such as SARS-CoV-2 and other coronaviruses replicate and bud by manipulating cellular organelles and membrane trafficking pathways adds another layer of complexity. On the other hand, viral and bacterial pathogens have been excellent models to study fundamental cellular functions for a long time. Many insights into the dynamics of the actin cytoskeleton have been revealed by studying intracellular pathogens. Using coronavirus as models is a remarkable opportunity to reveal fundamental principles of membrane dynamics but also represents a challenge. This is particularly true for the budding process of coronaviruses, which is not at all understood at a molecular level54. Electron microscopy studies of cells infected with MHV revealed that virions bud into vacuolar structures that are derived from membranes of the secretory pathway, notably the ER–Golgi intermediate compartment and the Golgi87. However, these vacuoles possess remarkably different morphologies compared with canonical cellular organelles88. Furthermore, viral budding requires that viral RNA produced in DMVs and host membranes containing viral structural proteins converge in budding compartments (Figure 1e). This requires that structural proteins, including S- and M-protein of coronaviruses, are delivered to these structures. If the S-protein is expressed in human cells, it is transported by the canonical secretory pathway to the plasma membrane89. In infected cells, however, the S-protein needs to be diverted to budding compartments. How transport of viral capsid, donor membranes, and structural proteins is coordinated remains another open question and a major challenge for future research (Figure 1e).
The peer reviewers who approve this article are:
Montserrat Bárcena, Section Electron Microscopy, Department of Cell and Chemical Biology, Leiden University Medical Center, Leiden, Netherlands
Qing Zhong, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Department of Pathophysiology, Shanghai Jiao Tong University School of Medicine, Shanghai, 200025, China
Funding Statement
The authors declare that no grants were involved in supporting this work.
References
- 1. Who Mers-Cov Research Group: State of Knowledge and Data Gaps of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in Humans. PLoS Curr. 2013; 5: ecurrents.outbreaks.0bf719e352e7478f8ad85fa30127ddb8. 10.1371/currents.outbreaks.0bf719e352e7478f8ad85fa30127ddb8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sørensen MD, Sørensen B, Gonzalez-Dosal R, et al. : Severe acute respiratory syndrome (SARS): Development of diagnostics and antivirals. Ann N Y Acad Sci. 2006; 1067(1): 500–5. 10.1196/annals.1354.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toyoshima Y, Nemoto K, Matsumoto S, et al. : SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J Hum Genet. 2020; 65(12): 1075–82. 10.1038/s10038-020-0808-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Petrosillo N, Viceconte G, Ergonul O, et al. : COVID-19, SARS and MERS: Are they closely related? Clin Microbiol Infect. 2020; 26(6): 729–34. 10.1016/j.cmi.2020.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Azhar EI, Hui DSC, Memish ZA, et al. : The Middle East Respiratory Syndrome (MERS). Infect Dis Clin North Am. 2019; 33(4): 891–905. 10.1016/j.idc.2019.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hui DSC, Zumla A: Severe Acute Respiratory Syndrome: Historical, Epidemiologic, and Clinical Features. Infect Dis Clin North Am. 2019; 33(4): 869–89. 10.1016/j.idc.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li R, Pei S, Chen B, et al. : Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science. 2020; 368(6490): 489–93. 10.1126/science.abb3221 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 8. Rivett L, Sridhar S, Sparkes D, et al. : Screening of healthcare workers for SARS-CoV-2 highlights the role of asymptomatic carriage in COVID-19 transmission. eLife. 2020; 9: e58728. 10.7554/eLife.58728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Long QX, Tang XJ, Shi QL, et al. : Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020; 26(8): 1200–4. 10.1038/s41591-020-0965-6 [DOI] [PubMed] [Google Scholar]
- 10. Rey FA, Lok SM: Common Features of Enveloped Viruses and Implications for Immunogen Design for Next-Generation Vaccines. Cell. 2018; 172(6): 1319–34. 10.1016/j.cell.2018.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoffmann M, Kleine-Weber H, Schroeder S, et al. : SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020; 181(2): 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 12. Li W, Moore MJ, Vasilieva N, et al. : Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003; 426(6965): 450–4. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 13. Yan R, Zhang Y, Li Y, et al. : Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020; 367(6485): 1444–8. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 14. Hoffmann M, Kleine-Weber H, Pöhlmann S: A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol Cell. 2020; 78(4): 779–784.e5. 10.1016/j.molcel.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 15. Cai Y, Zhang J, Xiao T, et al. : Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020; 369(6511): 1586–92. 10.1126/science.abd4251 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 16. Heurich A, Hofmann-Winkler H, Gierer S, et al. : TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014; 88(2): 1293–307. 10.1128/JVI.02202-13 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 17. Kleine-Weber H, Elzayat MT, Hoffmann M, et al. : Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci Rep. 2018; 8(1): 16597. 10.1038/s41598-018-34859-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoffmann M, Hofmann-Winkler H, Pöhlmann S: Priming Time: How Cellular Proteases Arm Coronavirus Spike Proteins. Activation of Viruses by Host Proteases. 2018; 71–98. 10.1007/978-3-319-75474-1_4 [DOI] [Google Scholar]
- 19. Shirato K, Kawase M, Matsuyama S: Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018; 517: 9–15. 10.1016/j.virol.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ou X, Liu Y, Lei X, et al. : Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020; 11(1): 1620. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 21. Sungnak W, Huang N, Bécavin C, et al. : SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020; 26(5): 681–7. 10.1038/s41591-020-0868-6 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 22. Petersen E, Koopmans M, Go U, et al. : Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis. 2020; 20(9): e238–e244. 10.1016/S1473-3099(20)30484-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang H, Yang P, Liu K, et al. : SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008; 18(2): 290–301. 10.1038/cr.2008.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park JE, Li K, Barlan A, et al. : Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A. 2016; 113(43): 12262–7. 10.1073/pnas.1608147113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burkard C, Verheije MH, Wicht O, et al. : Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014; 10(11): e1004502. 10.1371/journal.ppat.1004502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang M, Cao R, Zhang L, et al. : Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020; 30(3): 269–71. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 27. Saheb Sharif-Askari N, Saheb Sharif-Askari F, Alabed M, et al. : Airways Expression of SARS-CoV-2 Receptor, ACE2, and TMPRSS2 Is Lower in Children Than Adults and Increases with Smoking and COPD. Mol Ther Methods Clin Dev. 2020; 18: 1–6. 10.1016/j.omtm.2020.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 28. Kelly JA, Olson AN, Neupane K, et al. : Structural and functional conservation of the programmed -1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J Biol Chem. 2020; 295(31): 10741–8. 10.1074/jbc.AC120.013449 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 29. Namy O, Moran SJ, Stuart DI, et al. : A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006; 441(7090): 244–7. 10.1038/nature04735 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 30. Thoms M, Buschauer R, Ameismeier M, et al. : Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020; 369(6508): 1249–55. 10.1126/science.abc8665 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 31. Graham RL, Sims AC, Baric RS, et al. : The nsp2 proteins of mouse hepatitis virus and SARS coronavirus are dispensable for viral replication. Adv Exp Med Biol. 2006; 581: 67–72. 10.1007/978-0-387-33012-9_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lei J, Kusov Y, Hilgenfeld R: Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018; 149: 58–74. 10.1016/j.antiviral.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fehr AR, Channappanavar R, Jankevicius G, et al. : The Conserved Coronavirus Macrodomain Promotes Virulence and Suppresses the Innate Immune Response during Severe Acute Respiratory Syndrome Coronavirus Infection. mBio. 2016; 7(6): e01721–16. 10.1128/mBio.01721-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Egloff MP, Malet H, Putics A, et al. : Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol. 2006; 80(17): 8493–502. 10.1128/JVI.00713-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fehr AR, Athmer J, Channappanavar R, et al. : The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol. 2015; 89(3): 1523–36. 10.1128/JVI.02596-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hagemeijer MC, Monastyrska I, Griffith J, et al. : Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014; 458–459: 125–35. 10.1016/j.virol.2014.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolff G, Limpens RWAL, Zevenhoven-Dobbe JC, et al. : A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020; 369(6509): 1395–8. 10.1126/science.abd3629 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 38. Imbert I, Guillemot JC, Bourhis JM, et al. : A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006; 25(20): 4933–42. 10.1038/sj.emboj.7601368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Q, Wu J, Wang H, et al. : Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell. 2020; 182(2): 417–428.e13. 10.1016/j.cell.2020.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 40. Gao Y, Yan L, Huang Y, et al. : Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020; 368(6492): 779–82. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 41. Hillen HS, Kokic G, Farnung L, et al. : Structure of replicating SARS-CoV-2 polymerase. Nature. 2020; 584(7819): 154–6. 10.1038/s41586-020-2368-8 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 42. Sutton G, Fry E, Carter L, et al. : The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004; 12(2): 341–53. 10.1016/j.str.2004.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Romano M, Ruggiero A, Squeglia F, et al. : A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells. 2020; 9(5): 1267. 10.3390/cells9051267 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 44. Tanner JA, Watt RM, Chai YB, et al. : The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases. J Biol Chem. 2003; 278(41): 39578–82. 10.1074/jbc.C300328200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jang KJ, Jeong S, Kang DY, et al. : A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci Rep. 2020; 10(1): 4481. 10.1038/s41598-020-61432-1 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 46. Minskaia E, Hertzig T, Gorbalenya AE, et al. : Discovery of an RNA virus 3'->5' exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A. 2006; 103(13): 5108–13. 10.1073/pnas.0508200103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bouvet M, Debarnot C, Imbert I, et al. : In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010; 6(4): e1000863. 10.1371/journal.ppat.1000863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Viswanathan T, Arya S, Chan SH, et al. : Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun. 2020; 11(1): 3718. 10.1038/s41467-020-17496-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 49. Yan L, Ge J, Zheng L, et al. : Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell. 2021; 184(1): 184–193.e10. 10.1016/j.cell.2020.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 50. Yan L, Zhang Y, Ge J, et al. : Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat Commun. 2020; 11(1): 5874. 10.1038/s41467-020-19770-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen J, Malone B, Llewellyn E, et al. : Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell. 2020; 182(6): 1560–1573.e13. 10.1016/j.cell.2020.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 52. Kim Y, Jedrzejczak R, Maltseva NI, et al. : Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020; 29(7): 1596–605. 10.1002/pro.3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deng X, Hackbart M, Mettelman RC, et al. : Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A. 2017; 114(21): E4251–E4260. 10.1073/pnas.1618310114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hartenian E, Nandakumar D, Lari A, et al. : The molecular virology of coronaviruses. J Biol Chem. 2020; 295(37): 12910–34. 10.1074/jbc.REV120.013930 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 55. Gantier MP, Williams BRG: The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 2007; 18(5–6): 363–71. 10.1016/j.cytogfr.2007.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Snijder EJ, Limpens RWAL, de Wilde AH, et al. : A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020; 18(6): e3000715. 10.1371/journal.pbio.3000715 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 57. Kindler E, Thiel V: SARS-CoV and IFN: Too Little, Too Late. Cell Host Microbe. 2016; 19(2): 139–41. 10.1016/j.chom.2016.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Channappanavar R, Fehr AR, Vijay R, et al. : Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016; 19(2): 181–93. 10.1016/j.chom.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pedersen KW, van der Meer Y, Roos N, et al. : Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999; 73(3): 2016–26. 10.1128/JVI.73.3.2016-2026.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Knoops K, Bárcena M, Limpens RWAL, et al. : Ultrastructural characterization of arterivirus replication structures: Reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol. 2012; 86(5): 2474–87. 10.1128/JVI.06677-11 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 61. Romero-Brey I, Merz A, Chiramel A, et al. : Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012; 8(12): e1003056. 10.1371/journal.ppat.1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Doerflinger SY, Cortese M, Romero-Brey I, et al. : Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog. 2017; 13(10): e1006705. 10.1371/journal.ppat.1006705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Limpens RWAL, van der Schaar HM, Kumar D, et al. : The transformation of enterovirus replication structures: A three-dimensional study of single- and double-membrane compartments. mBio. 2011; 2(5): e00166–11. 10.1128/mBio.00166-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Knoops K, Kikkert M, van den Worm SHE, et al. : SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008; 6(9): e226. 10.1371/journal.pbio.0060226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. de Wilde AH, Raj VS, Oudshoorn D, et al. : MERS-coronavirus replication induces severe In vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J Gen Virol. 2013; 94(Pt 8): 1749–60. 10.1099/vir.0.052910-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reggiori F, Monastyrska I, Verheije MH, et al. : Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010; 7(6): 500–8. 10.1016/j.chom.2010.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 67. Oudshoorn D, Rijs K, Limpens RWAL, et al. : Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio. 2017; 8(6): e01658–17. 10.1128/mBio.01658-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Angelini MM, Akhlaghpour M, Neuman BW, et al. : Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013; 4(4): e00524–13. 10.1128/mBio.00524-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ertel KJ, Benefield D, Castaño-Diez D, et al. : Cryo-electron tomography reveals novel features of a viral RNA replication compartment. eLife. 2017; 6: e25940. 10.7554/eLife.25940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Imbert I, Snijder EJ, Dimitrova M, et al. : The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008; 133(2): 136–48. 10.1016/j.virusres.2007.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hurst KR, Koetzner CA, Masters PS: Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol. 2013; 87(16): 9159–72. 10.1128/JVI.01275-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nishimura T, Tooze SA: Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discov. 2020; 6: 32. 10.1038/s41421-020-0161-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Prentice E, Jerome WG, Yoshimori T, et al. : Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004; 279(11): 10136–41. 10.1074/jbc.M306124200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhao Z, Thackray LB, Miller BC, et al. : Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007; 3(6): 581–5. 10.4161/auto.4782 [DOI] [PubMed] [Google Scholar]
- 75. Nishida Y, Arakawa S, Fujitani K, et al. : Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009; 461(7264): 654–8. 10.1038/nature08455 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 76. Mizushima N: The ATG conjugation systems in autophagy. Curr Opin Cell Biol. 2020; 63: 1–10. 10.1016/j.ceb.2019.12.001 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 77. Prentice E, McAuliffe J, Lu X, et al. : Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004; 78(18): 9977–86. 10.1128/JVI.78.18.9977-9986.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. de Haan CAM, Reggiori F: Are nidoviruses hijacking the autophagy machinery? Autophagy. 2008; 4(3): 276–9. 10.4161/auto.5241 [DOI] [PubMed] [Google Scholar]
- 79. Johansen T, Lamark T: Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J Mol Biol. 2020; 432(1): 80–103. 10.1016/j.jmb.2019.07.016 [DOI] [PubMed] [Google Scholar]
- 80. Fletcher K, Ulferts R, Jacquin E, et al. : The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018; 37(4): e97840. 10.15252/embj.201797840 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 81. Bernasconi R, Noack J, Molinari M: Unconventional roles of nonlipidated LC3 in ERAD tuning and coronavirus infection. Autophagy. 2012; 8(10): 1534–6. 10.4161/auto.21229 [DOI] [PubMed] [Google Scholar]
- 82. Bernasconi R, Molinari M: ERAD and ERAD tuning: Disposal of cargo and of ERAD regulators from the mammalian ER. Curr Opin Cell Biol. 2011; 23(2): 176–83. 10.1016/j.ceb.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hayashi-Nishino M, Fujita N, Noda T, et al. : A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009; 11(12): 1433–7. 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 84. Axe EL, Walker SA, Manifava M, et al. : Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008; 182(4): 685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 85. Cottam EM, Maier HJ, Manifava M, et al. : Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011; 7(11): 1335–47. 10.4161/auto.7.11.16642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cottam EM, Whelband MC, Wileman T: Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014; 10(8): 1426–41. 10.4161/auto.29309 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 87. Ulasli M, Verheije MH, de Haan CAM, et al. : Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010; 12(6): 844–61. 10.1111/j.1462-5822.2010.01437.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Stertz S, Reichelt M, Spiegel M, et al. : The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007; 361(2): 304–15. 10.1016/j.virol.2006.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Buchrieser J, Dufloo J, Hubert M, et al. : Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020; 39(23): e106267. 10.15252/embj.2020106267 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation