Abstract
A voltammetric genosensor has been developed for the early diagnosis of COVID-19 by determination of RNA-dependent RNA polymerase (RdRP) sequence as a specific target of novel coronavirus. The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) uses an RdRP for the replication of its genome and the transcription of its genes. Here, the silver ions (Ag+) in the hexathia-18-crown-6 (HT18C6) were used for the first time as a redox probe. Then, the HT18C6(Ag) incorporated carbon paste electrode (CPE) was further modified with chitosan and PAMAM dendrimer-coated silicon quantum dots (SiQDs@PAMAM) for immobilization of probe sequences (aminated oligonucleotides). The current intensity of differential pulse voltammetry using the redox probe was found to decrease with increasing the concentration of target sequence. Based on such signal-off trend, the proposed genosensor exhibited a good linear response to SARS-CoV-2 RdRP in the concentration range 1.0 pM–8.0 nM with a regression equation I (μA) = − 6.555 log [RdRP sequence] (pM) + 32.676 (R2 = 0.995) and a limit of detection (LOD) of 0.3 pM. The standard addition method with different spike concentrations of RdRP sequence in human sputum samples showed a good recovery for real sample analysis (> 95%). Therefore, the developed voltammetric genosensor can be used to determine SARS-CoV-2 RdRP sequence in sputum samples.
Graphical abstract
PAMAM-functionalized SiQDs were used as a versatile electrochemical platform for the SARS-CoV-2 RdRP detection based on a signal off sensing strategy. In this study, for the first time, the silver ions (Ag+) in the hexathia-18-crown-6 carrier were applied as an electrochemical probe.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00604-021-04773-6.
Keywords: Genosensor, SARS-CoV-2 RdRP sequence, PAMAM@SiQDs
Introduction
The world is currently witnessing the third large-scale pandemic in the last two decades after the severe acute respiratory syndrome (SARS) in 2003 and the Middle East respiratory syndrome (MERS) in 2012 [1]. The novel pandemic, known as COVID-19, is caused by the very contagious severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus. Due to the emergence and rapid global spread of COVID-19, the quick and accurate detection of virus or disease is increasingly vital to control the sources of infection and prevent the extensive spread of the disease. Despite all the efforts made, there is currently a global lack of reliable, fast, and ultrasensitive diagnostic tools for early detection of COVID-19.
Currently, the diagnostic tests for COVID-19 can be classified into two broad categories, namely molecular diagnostic tests and antibody or serology tests [2, 3]. The molecular diagnostic tests include nucleic acid amplification strategies, such as reverse transcription polymerase chain reaction (RT-PCR), reverse transcription loop-mediated amplification (RT-LAMP), and reverse transcription recombinase polymerase amplification (RT-RPA) [3, 4]. Although RT-PCR is considered the gold standard for COVID-19 diagnosis, it suffers from several limitations. An important issue with the RT-PCR test is the risk of eliciting false-negative and false-positive results [5]. The insufficient viral RNA at the point of detection may lead to false-negative results. Compared to RT-PCR, RT-LAMP is faster and does not require prior RNA isolation from the samples [6]. In addition, this isothermal amplification method requires six independent primers to recognize the target sequence and thus enables better specificity in the amplification [7]. Despite high specificity, RT-LAMP has some problems similar to other molecular detection techniques, such as unexpected cross-contamination due to the high amplification efficiency. This contamination may cause to false-positive results [8]. The amplification of nucleic acids using RT-RPA is faster than RT-LAMP, at 37 °C or less. Although this isothermal amplification method has been reported for the rapidly detection of SARS-CoV-2, there are still some deficiencies in the cost and operability. For example, the RT-RPA requires the participation of three enzymes [9].
The nucleic acid-based biosensors (genosensors) are being developed to improve the limitations of the conventional methods. Along with the advancement in medical diagnosis, the genosensors have become a sensitive and reliable technology for viral detection [10–12]. Among genosensors, electrochemical genosensors have attracted a lot of attention due to their fast response, sensitivity, economical properties, and compatibility with microfabrication technology and simple operation mode which makes them compatible with point-of-care (POC) testing.
The gene-based strategies for SARS-CoV-2 detection have some challenges, especially in terms of primer design [13]. Besides, there is a lack of information about the variety of genetic SARS-CoV-2 in humans and animals. Recently, the WHO recommended RNA-dependent RNA polymerase (RdRP) (oligonucleotide ID: RdRP_SARSr-P2 and RdRP_SARSr-P1), envelope (E), and nucleocapsid (N) genes for the detection of the novel coronavirus [14, 15]. Although these genes were reported as potential targets for the detection of coronavirus, Kakhki and co-workers [16] found out that only one of them (RdRP_SARSr-P2: 5′-CAGGTGGAACCTCATCAGGAGATGC-3′) was almost specific for the new coronavirus, and the other introduced probes would detect the other types of coronaviruses. Currently, the gene targets selected by different countries for RT-PCR molecular assays are genetically similar, including RdRP sequence and the open reading frame 1ab (ORF1ab) sequence [13, 16]. RdRP, a viral enzyme for viral RNA replication in host cells, plays an essential role in the RNA virus life cycle and has no host cell homolog. The SARS-CoV-2 RdRP sequence recommended by the WHO for detection of COVID-19 is very closely related to that of SARS. Specifically, in the selected gene sequences, only three fixed nucleotide bases were different between SARS-CoV-2 RdRP and SARS RdRP [17].
In this article, we focus on the label-free and nucleic acid amplification-free determination of SARS-CoV-2 RdRP sequence (RdRP_SARSr-P2) as a promising target for detection of COVID-19 using the silver ion-hexathia-18-crown-6 complex modified carbon paste electrode (CPE-HT18C6(Ag)). The chemically modified CPEs have received much attention in the electrochemical field [18]. Here, the silver ions in the HT18C6 were used for the first time as an electrochemical probe. HT18C6 is a macrocyclic thio crown ligand that is insoluble in water and can form a fairly stable complex with Ag+ ion in nonaqueous media [19]. It has been used as an excellent neutral carrier for the preparation of Ag-selective electrodes [20] and also for the selective extraction of silver ions into 1,2-dichloroethane phase [21]. As it was clearly pointed out in our previous study [20], this is likely due to the high selectivity of the ionophore for silver ion over other metal ions as well as the rapid exchange kinetics of the resulting complex. Chelating agents with soft coordination sites like sulfur seem to generate great affinity toward transition metal ions, such as Ag+. For the immobilization of probe sequences, CPE-HT18C6(Ag) was further modified with the chitosan and PAMAM dendrimer-coated silicon quantum dots (SiQDs@PAMAM). The SiQDs@PAMAM nanocomposite provides a large surface area and abundant amino groups for the immobilization of DNA probes. For the exploration of new sensitive platforms, a strong tendency toward biomolecules, high surface area, and good electrical conductivity is essential [22]. Generally, the signal amplification in the electrochemical biosensors can be achieved by using nanomaterials, such as quantum dots [23, 24], metal nanoparticles [2, 23], and graphene [2]. The conjugation of PAMAM functionalized nanomaterials with the DNA strands provides efficient nanocomposites for sensing purposes [25, 26]. Dendrimers are widely used as versatile platforms owing to their excellent properties, such as biocompatibility, uniformity, high-branched structure and large numbers of functional ending groups [27].
Experimental
Reagents and chemicals
Bovine serum albumin (BSA, 98%), (3-aminopropyl)trimethoxysilane (APTMS, 97%), potassium hexacyanoferrate(III) (K3Fe(CN)6, ≥ 99.0%), potassium hexacyanoferrate(II) trihydrate (K4Fe(CN)6 .3H2O, ≥ 99.95%), disodium hydrogen phosphate (99.95%), sodium dihydrogen phosphate (99%), potassium chloride (≥ 99%), sodium chloride (≥ 99%), methylacrylate (99%), ethylenediamine (≥ 99.5%), dimethyl sulfoxide (DMSO), maleic anhydride (99%), chitosan, acetonitrile, diethylether, glutaraldehyde solution (GA, 25% in water), N-hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), sodium hydroxide, and hexathia-18-crown-6 (HT18C6) were purchased from Sigma-Aldrich (Steinheim, Germany, https://www.sigmaaldrich.com). The extra pure graphite powder, paraffin oil, silver nitrate, methanol, hydrochloric acid, and 4-morpholineethane sulphonic acid sodium salt buffer (MES) were prepared from Merck (Darmstadt, Germany, https://www.merckmillipore.com). Phosphate buffer solution (PBS) was prepared using 0.1 M Na2HPO4 and 0.1 M NaH2PO4 (pH 7.2).
All selected oligonucleotides, including the SARS-CoV-2 RdRP, SARS RdRP, and E (envelope protein gene) sequences were provided by Faza Biotech Company (Iran) according to the previous reports [1, 16, 28] as follows:
SARS-CoV-2 RdRP (oligonucleotide ID: RdRP_SARSr-P2): 5′-CAGGT GGAAC CTCAT CAGGA GATGC-3′.
SARS RdRP: 5′-C CAGGT GGAAC ATCAT CCGGT GATGC-3′.
E: 5′-ACAC TAGCC ATCCT TACTG CGCTT CG-3′.
The probe sequence or receptor of SARS-CoV-2 RdRP was modified at the 5′-terminus with an NH2 group.
Instrumentation
Cyclic voltammetry (CV), differential pulse voltammetry (DPV), and electrochemical impedance spectroscopy (EIS) were performed using a PGSTAT 302N electrochemical workstation (Autolab, Netherlands, https://www.metrohm-autolab.com). The bias potential for EIS was 0.18 V. A conventional three-electrode cell, consisting of a Pt wire as the counter electrode, an Ag/AgCl electrode (KCl, 3 M) as the reference electrode, and a carbon paste electrode (CPE, i.d = 3.0 mm, Azar Electrode, Urmia, Iran) as the working electrode was utilized. Transmission electron microscopy (TEM) images were taken on an EM10C TEM from Zeiss (Germany, http://www.Zeiss.com) operating at 100 keV. Scanning electron microscopy (SEM) studies and energy dispersive X-ray spectroscopy (EDX) analysis were performed by a ZEISS EVO 18 analytic microscope (Germany, http://www.Zeiss.com) equipped with an energy dispersive X-ray analyzer. Spectrofluorimetric measurements were performed using an Agilent Cary Eclipse fluorescence spectrometer (Agilent Technologies, USA, https://www.agilent.com). The UV–Vis absorption spectra were recorded on a model Shimadzu UV 2550 (Shimadzu Corporation, Japan, https://www.shimadzu.com). A 780 pH Meter was used for measuring pH at 25 °C (Metrohm, Switzerland, https://www.metrohm.com).
Synthesis and characterization of SiQDs
Silicon QDs were synthesized through a one-step hydrothermal method which offers high temperature and pressure as the procedure reported by Feng and co-workers [29]. In brief, 0.368 g trisidium citrate was dissolved in 8 mL deionized water in a flask with sustainable aeration of nitrogen for 20 min to remove oxygen. Then, 2 mL APTMS was added into the above solution and then thoroughly stirred at room temperature and ambient pressure for 20 min. The as-prepared solution was loaded into a teflon-lined stainless steel autoclave, which was incubated at 200 °C for 2 h and cooled to room temperature. To remove byproducts, the resulting SiQDs colloidal solution was treated by dialysis tubing bag (1 kDa) against double distilled water for 12 h. After that, the colloidal solution was concentrated through a rotary evaporator followed by lyophilization to get brown-colored SiQD powder.
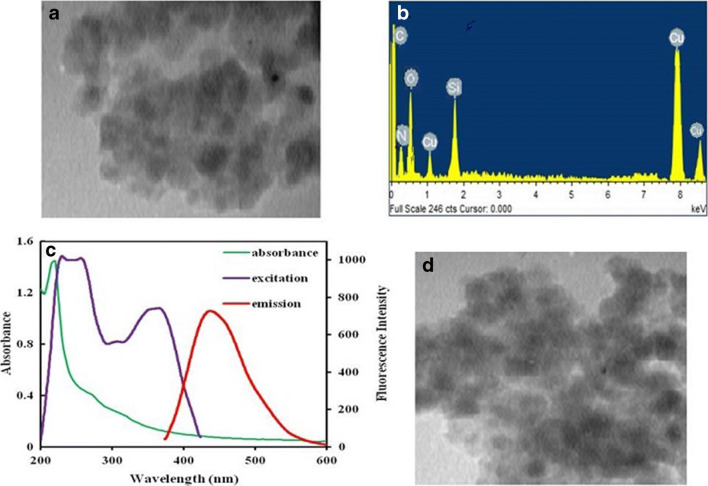
The synthesized SiQDs were characterized by TEM, fluorescence, and UV-vis absorption spectroscopy. The TEM image (Fig. 1a) confirms the formation of quasi-spherical SiQDs with an average size of ~ 10 nm. It should be mentioned that low contrast in the TEM image is due to the low atomic weight of silicon and nanoscale dimensions of the silicon dots, resulting in poor visualization [30]. The obtained image is similar to previously reported literature by Fan and co-workers [31].
Fig. 1.
a TEM image, b EDX spectrum, c UV-vis and fluorescence spectra of SiQDs, and d TEM image of SiQDs@PAMAM; the magnification of TEM images was adjusted in 100 KX
Elemental analysis of the SiQDs using EDX is shown in Fig. 1b, illustrating the presence of Si, N, and O. Peaks of C and Cu seen in this spectrum arose due to their integral presence in carbon-coated copper grids used for viewing the samples [30].
The SiQDs colloidal solution observed under a handheld UV lamp with an excitation wavelength of 360 nm shows intense blue color (Fig. 1c) suggesting the formation of blue-emitting SiQDs. Excitation and emission spectra of the resultant SiQDs indicates that the SiQDs possess good photoluminescence properties with symmetrical excitation and emission peaks. The emission maximum of the as-synthesized SiQDs is centered at 455 nm upon excitation at 360 nm.
Synthesis and characterization of PAMAM functionalized SiQDs
The preparation of amine-terminated generation 3 (G 3) PAMAM dendrimer by divergent technique starting from ethylenediamine core followed by repeated Michael type addition and amidation reaction was described in the ESM.
The carboxyl-containing SiQDs were functionalized with PAMAM by carbodiimide coupling reaction [32]. Briefly, 0.2 g of SiQDs was dissolved in 5 ml of 10 mM MES buffer (pH 6.5). One millimole of NHS and an equimolar amount of EDC were added to the SiQDs with constant stirring and continued for 1 h at room temperature to activate the carboxylic acid group of SiQDs. Then, 2 g of PAMAM dendrimer dissolved in 2 ml of 10 mM MES buffer solution was added slowly to the activated SiQDs with stirring condition. After 16 h of reaction at room temperature, the solution was purified using a dialysis tubing bag (3.5 kDa) against double distilled water for 3 days with change of water periodically to get PAMAM functionalized SiQDs (SiQDs@PAMAM).
The TEM image shows the morphology of SiQDs@PAMAM (Fig. 1d). The surface zeta potential of SiQDs and functionalized SiQDs was studied by Zetasizer. SiQDs show the negative zeta potential having a value of − 2.9 ± 1.1 mV. The negative zeta potential arises due to the presence of carboxylic acid (–COOH) groups on the surface of SiQDs. After conjugation of PAMAM dendrimers, the zeta potential of SiQDs changes to positive (10.7 ± 1.0 mV) and the magnitude of positive zeta potential gradually increases with an increase in the generation of PAMAM dendrimers.
Development of voltammetric genosensor
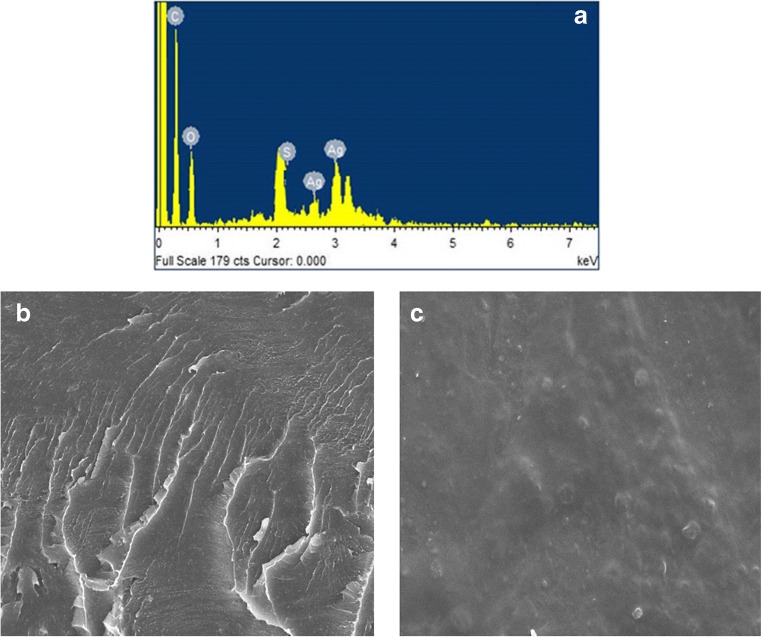
The synthesis of [Ag([18]aneS6)] ([18]aneS6 = 1,4,7,10,13,16-hexathiacyclooctadecane) was briefly described in the ESM. In continuation of our research, the CPE–HT18C6(Ag) was constructed under optimum conditions by mixing 55% w/w graphite powder, 15% w/w HT18C6(Ag), and 30% w/w of paraffin oil. For this purpose, 0.055 g of graphite powder and 0.015 g of HT18C6(Ag) were mixed in a mortar and homogenized. Then, 30 μL of paraffin oil was added and mixed well. The prepared paste was used to fill a Teflon holder. The microstructure of CPE–HT18C6(Ag) was studied by EDX (Fig. 2a). The peaks of S and Ag observed in this spectrum confirm the presence of HT18C6(Ag) in the composition of carbon paste.
Fig. 2.
a EDX spectrum of CPE-HT18C6(Ag); SEM images of b CPE-HT18C6(Ag) and c CPE-HT18C6(Ag)/chitosan with magnification of 2.00 KX
The surface of CPE–HT18C6(Ag) was polished on a clean paper until a smooth surface was observed. Then, 0.5 mL of 0.01% chitosan in 0.1 M acetic acid was dropcasted on the CPE–HT18C6(Ag) and dried at room temperature for 1 h. The surface morphology of CPE–HT18C6(Ag) (Fig. 2b) and CPE–HT18C6(Ag)/chitosan (Fig. 2c) were studied by SEM. It can be seen that surface of CPE–HT18C6(Ag) contains a relatively compact carbon paste matrix. After modification, a homogenous layer of chitosan was formed on the electrode surface.
The obtained CPE–HT18C6(Ag)/chitosan was dipped in 5% GA for 30 min to activate amine groups for the reaction with primary amines. The activated CPE–HT18C6(Ag)/chitosan was rinsed with distilled water repeatedly in order to remove unreacted GA from the modified electrode surface. For the immobilization of SiQDs@PAMAM on activated chitosan, the activated CPE–HT18C6(Ag)/chitosan was dipped in a 100 μL solution of as-synthesized SiQDs@PAMAM for 2 h. Then, the linking of the probe sequence to the amine functionalities of the immobilized dendrimer was performed in a buffered aqueous solution exploiting GA reagent. In the first stage, the CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM was dropcasted with 40 μL of an aqueous solution of 5% GA for 30 min. Subsequently, the so treated electrode was washed in MES solution and incubated for 1 h with 20 μL of 10 μM solution of the probe oligonucleotide, prepared properly diluting the standard solution in MES buffer. After removal of unreacted sequences by accurate rinsing with MES buffer, a blocking treatment aimed at preventing unspecific responses during the sample incubation was carried out by casting 25 μL of a 2.5 mg mL−1 solution of BSA dissolved in PBS on the modified electrode for an incubation time of 15 min.
It is important to mention that the stability of immobilized DNA probes on the modified electrode surface is one of the critical factors in the designing of genosensors. We observed the CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM/ssDNA is more stable than the CPE–HT18C6(Ag)/chitosan/carboxylated SiQDs/ssDNA. The functionalization of SiQDs with PAMAM can lead to more stability of interface compared to unfunctionalized SiQDs. It is because of the entanglement effects of PAMAM on the ssDNA and dsDNA [33]. Nandy and Maiti reported that DNA strands can be entangled on PAMAM dendrimers (G3-G5) [34]. They showed the amine-terminated PAMAM can form a stable complex with ssDNA and dsDNA. It is generally believed that the charge between the positively charged dendrimer and the negatively charged backbones of DNA strands plays a key role in the complex structure [33, 34]. In addition, abundant amino-terminated surface groups of PAMAM dendrimers can be used for the immobilization of aminated DNA probes on the modified electrode through the covalent bonding. Therefore, the electrostatic and covalent interactions of the dendrimer with probe sequences lead to more stability of the designed interface.
After the formation of interface, assays were carried out by incubating CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence with 20 μL of the target sequence for 25 min at room temperature. The electrode was then carefully washed to remove unspecifically bound oligonucleotides. All electrochemical measurements were carried out in pH 7.2 PBS.
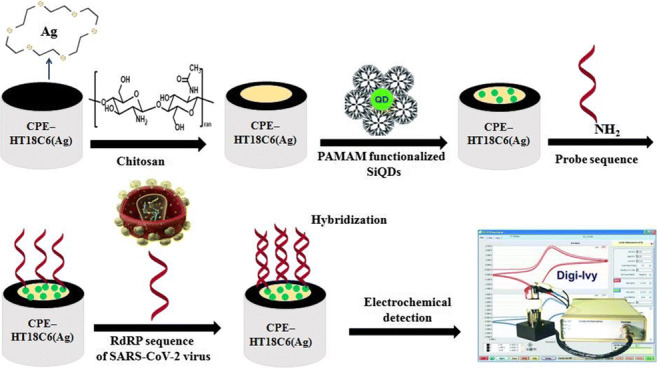
Figure 3 presents the schematic illustration of the genosensor fabrication process.
Fig. 3.
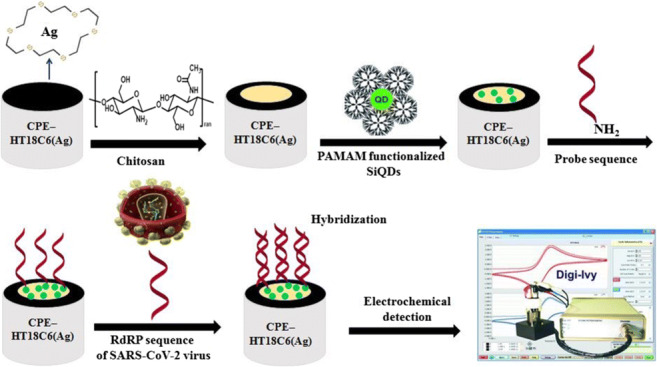
Schematic illustration of fabrication process of electrochemical genosensor
Results and discussion
Electrochemical characterizaction of step-wise electrode surface modification via CV
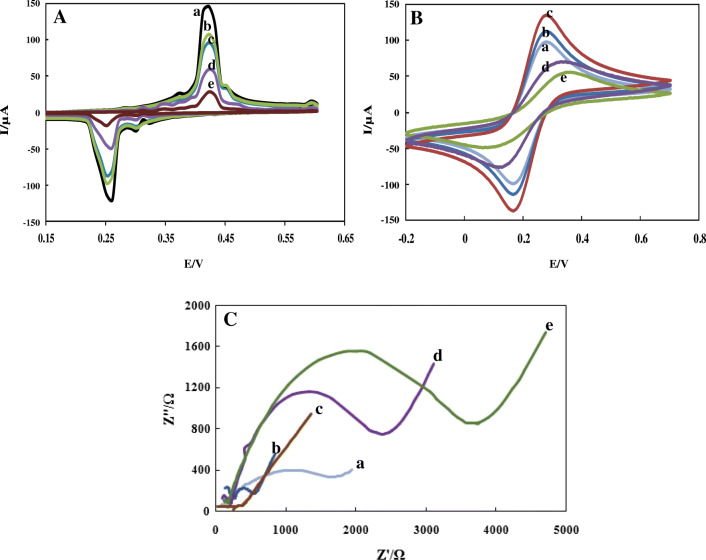
Cyclic voltammetry was recorded using the redox signal of silver ion in 0.1 M PBS (Fig. 4A). When the HT18C6(Ag) is mixed with carbon paste, a pair of well-defined redox peaks is observed at 0.42 and 0.26 V (vs. Ag/AgCl), as a result of oxidation of Ag to Ag + (curve a). While the modification of CPE–HT18C6(Ag) with chitosan blocks the electron transfer of silver and decreases redox signals (curve b). As can be seen, a furthermore decrease in the redox peak currents is recorded after modification of electrode surface with SiQDs@PAMAM (curve c). Besides, immobilization of probe sequences on the modified electrode surface leads to a remarkable decline of peak currents in curve d. Finally, as soon as the two sequences are hybridized, the redox signals of silver decrease further due to the thickening of the electron transfers (curve e). This observation was assigned to the hybridization of probe sequences with SARS-CoV-2 RdRP sequences at the electrode surface which inhibited the redox reaction of silver in CPE–HT18C6(Ag) structure.
Fig. 4.
CVs of CPE-HT18C6(Ag) (a), CPE-HT18C6(Ag)/chitosan (b), CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM (c), CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence (d), CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence/SARS-CoV-2 RdRP sequence (1 pM) (e) in A 0.1 M PBS (pH = 7.2) and B 0.1 M KCl containing 5 mM Fe(CN)63−/4−. C Nyquist plots of CPE-HT18C6(Ag) (a), CPE-HT18C6(Ag)/chitosan (b), CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM (c), CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence (d), CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence/SARS-CoV-2 RdRP sequence (1 pM) (e) in 0.1 M KCl containing 5 mM Fe(CN)63−/4 −
The stepwise modification of electrode surface was further studied using the CV method in the presence of a redox probe (0.1 M KCl containing 5 mM Fe(CN)63−/4−) (Fig. 4B). As can be seen, an increase in the redox peak currents is recorded for the electrode after modification with chitosan (curve b) compared to the CPE–HT18C6(Ag) (curva a), due to the positive charge of chitosan. Since the chitosan biopolymers carry the positive charges at the acidic conditions, there is an electrostatic attraction between Fe(CN)63−/4− and chitosan. Through immobilization of SiQDs@PAMAM on the surface of CPE–HT18C6(Ag)/chitosan, the peak currents are further enhanced due to the electrostatic attraction between positive-charged QDs and Fe(CN)63−/4− (curve c). After attachment of the probe sequence to the surface of CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM, the redox peak currents significantly decreased (curve d) related to the presence of phosphate groups in the DNA backbone that repelled the negatively charged probe from the electrode surface [10]. Similarly, the hybridization of SARS-CoV-2 RdRP sequence with the probe sequence led to decreasing the peak currents, owing to the further electrostatic repulsion between redox probe and target sequences (curve e).
EIS method was also carried out to characterize the fabrication process of the genosensor. It is a suitable method for monitoring the changes of the surface features during the fabrication process of electrochemical biosensors [35]. Figure 4 C shows the Nyquist plots of impedance spectra in the process of electrode modification in 0.1 M KCl containing 5 mM Fe(CN)63−/4−. The impedance of the CPE–HT18C6(Ag) was first obtained (curve a), indicating a very small semicircle at high frequency. When the chitosan solution was dropcasted on the surface of CPE–HT18C6(Ag), the Rct decreased due to the positive charge of chitosan (curve b). After the immobilization of SiQDs@PAMAM on the surface of CPE–HT18C6(Ag)/chitosan, the electrochemical semicircle became much smaller and close to a straight line (curve c). This is attributed to the electrostatic attraction between Fe(CN)63−/4− and positive-charged QDs. The assembling of ssDNA probes on the CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM led to a significant increase in Rct (curve d) due to electrostatic repulsion between Fe(CN)63−/4- anions and the negatively charged probe sequences. Finally, after the incubation of SARS-CoV-2 RdRP sequences with probe sequences, the Rct showed a further increase (curve e).
Calculation of the electroactive surface area
In order to increase the active surface area, the number of immobilized probe sequences and finally the sensitivity of the genosensor, the probe sequences were immobilized on the chitosan/SiQDs@PAMAM film. The electroactive surface area for CPE–HT18C6(Ag) and chitosan/SiQDs@PAMAM modified CPE–HT18C6(Ag) was determined from the cyclic voltammograms in solution of 5 mM of K3[Fe(CN)6] containing 0.1 M KCl at various sweep rates.
According to Eq. (1):
| 1 |
where ip is the peak current, n is the number of electrons in the reaction, A is the electrode area, D is the diffusion coefficient of the oxidized form hexacyanoferrate (III), C is the bulk concentration of the oxidized form and n is the scan rate. From the slope of the ip vs. ʋ1/2 line, the values of A were determined as 0.099 cm2 and 0.150 cm2 for the CPE–HT18C6(Ag) and chitosan/SiQDs@PAMAM-modified CPE–HT18C6(Ag), respectively. Therefore, after the electrode surface was modified with chitosan/SiQDs@PAMAM film, the electroactive surface area greatly increased, indicating that the introduction of interface provides more conduction pathways for the electron transfer of [Fe(CN)6]3−/4−. In addition, chitosan/SiQDs@PAMAM interface offers a large active surface area for immobilization of probe sequences.
Optimization of effective parameters on the genosensor response
The electrochemical responses of a genosensor can be influenced by several experimental parameters that should be optimized to obtain better performances. One of the effective factors in the electrode response is the amount of HT18C6(Ag) used in the paste composition. By increasing the amount of HT18C6(Ag) in the paste composition, the number of redox probes is increased. However, with a further increased HT18C6(Ag) amount, the electrical conductivity of the electrode is decreased [36]. To obtain the optimum value for HT18C6(Ag), the amount of paraffin oil was kept constant at a 30% level, while the amounts of HT18C6(Ag) and graphite powder were changed. Results showed the best response was obtained in 55% graphite powder, 15% HT18C6(Ag), and 30% paraffin oil (Fig. 1S).
Among the experimental parameters, the concentration of probe oligonucleotides is an important factor on the response of genosensors. As seen in Fig. 2S, the peak current of redox decreased with increasing concentration of loaded probe oligonucleotides on the surface of modified electrode up to 10 μM. However, the higher concentrations of probe sequences led to the saturation of electrode surface.
Since the hybridization time is a vital influencing factor on the response of genosensors, the effect of this parameter on the sensor response was also investigated (Fig. 3S). As is obvious, the current response decreased rapidly with increasing hybridization time up to 25 min and then leveled off at higher periods, suggesting that the formation of hybridized oligonucleotides at the electrode surface has reached a saturation level.
Analytical performance of signal-off voltammetric genosensor
The application of CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence for the detection of SARS-CoV-2 RdRP sequence was studied based on a signal-off strategy by the DPV technique in the potential range from 0 to 0.8 V (vs. Ag/AgCl). For this purpose, the prepared nanogenosensor was incubated with different concentrations of the target sequence, and the peak current of silver was measured as a redox probe. Obviously, the addition of target with different concentrations on the modified electrode surface induced different decreases in the peak current of the Ag probe. In fact, the oxidation peak current of silver decreased by adding more increments of SARS-CoV-2 RdRP sequence to the solution (Fig. 5a). Based on such signal-off trend, the genosensor exhibited a good linear response to target in the concentration range of 1.0 pM-8.0 nM with a regression equation of I (μA) = − 6.555 log [RdRP sequence] (pM) + 32.676 (R2 = 0.995) (n = 6) (Fig. 5b). The limit of detection (LOD) and limit of quantification (LOQ) evaluated as 3Sb/m and 10Sb/m were 0.3 pM and 1.0 pM respectively, where Sb is the blank standard deviation and m is the slope of the calibration curve.
Fig. 5.
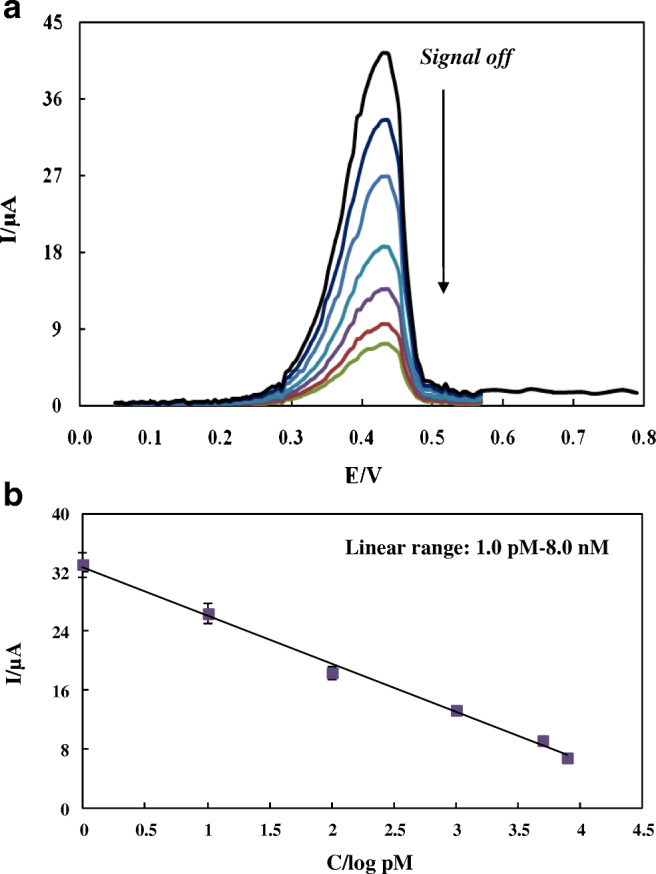
a DPVs of CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence in the presence of different concentrations of SARS-CoV-2 RdRP sequence (0, 1 pM, 10 pM, 100 pM, 1000 pM, 5000 pM, 8000 pM) in 0.1 M PBS of pH 7.2; b Calibration curve of Ip vs. log CRdRP
The selectivity of the detection largely reflects the performance of the designed genosensor. Thus, the specificity of the CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence toward different sequences including SARS-CoV-2 RdRP (10 pM), SARS RdRP (50 pM), and E (50 pM) sequences was investigated using DPV technique. The considerable decrease in the peak current of silver was only observed in the presence of the SARS-CoV-2 RdRP sequence, while negligible changes were observed with the other sequences tested (Fig. 6A). In addition, the bar chart of DPV response of CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence in the mixture of target gene (10 pM) and mentioned interferent genes (fivefold) showed a negligible decrease compared with that of modified electrode for target gene (Fig. 6B). Thus, it was concluded that the immobilized DNA probe selectively binds to the target sequence. These outcomes show the selectivity and specificity of the electrochemical genosensor toward the SARS-CoV-2 RdRP sequence.
Fig. 6.
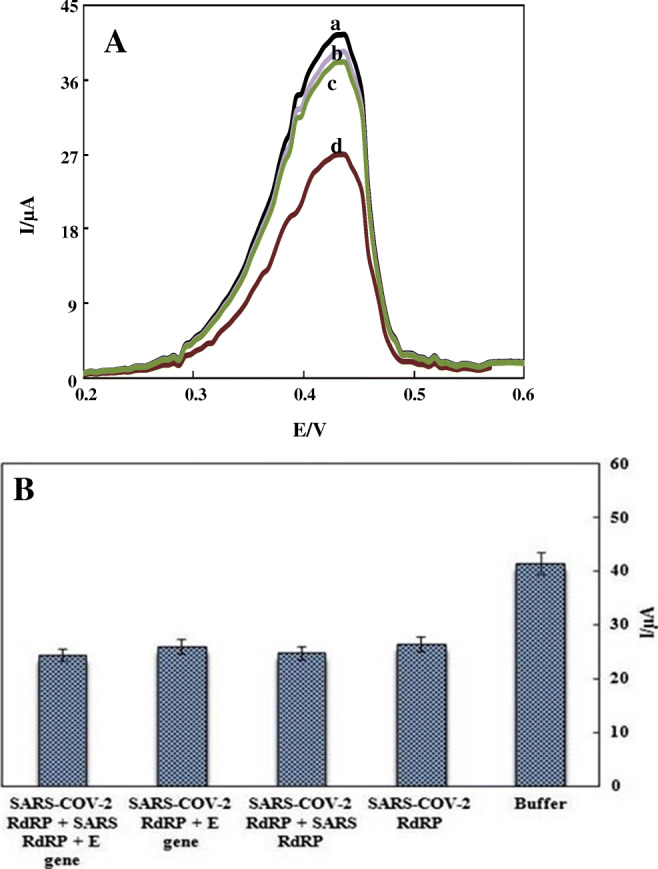
A DPVs of CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence in (a) PBS of pH 7.2, (b) E gene (50 pM), (c) SARS RdRP gene (50 pM), and (d) SARS-CoV-2 RdRP gene (10 pM). B Bar charts of DPV responses of CPE-HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence in PBS of pH 7.2, SARS-CoV-2 RdRP gene (10 pM), and the mixtures of SARS-COV-2 RdRP (10 pM) and other interferent genes (50 pM)
The relative standard deviations (RSDs) for single-electrode repeatability and electrode-to-electrode reproducibility were less than 2.7% and 6.6% (n = 5), respectively. Moreover, the stability was investigated by the electrode immersion in 0.1 M PBS (pH 7.2) for 15 days at the refrigerator (4 °C) and room temperature (25 °C). The constructed DPV aptasensor shows negligible current decay at 4 °C, with RSD of 1.94%; while RSD of 3.78% was found when the modified electrode was stored at room temperature. So, when not in use, the working modified electrode was stored at 4 °C.
To evaluate the practical application of this sensing strategy, the SARS-COV-2 RdRP gene was determined in the spiked sputum samples. For this purpose, sputum samples prepared from healthy volunteers (n = 3) were centrifuged at 5000 rpm (10 min) and then, diluted three times with MES buffer. The standard addition method and DPV technique at positive potential of ~ 0.42 V versus Ag/AgCl were used to measure target in the sputum samples. The samples were spiked with different concentrations of SARS-CoV-2 RdRP sequence (5.0 pM, 50.0 pM, and 5.0 nM). Each sample was analyzed four times, and the corresponding results are shown in Table 1, which discloses a good recovery in real serum samples (> 95%). Therefore, the voltammetric genosensor can be used to determine SARS-CoV-2 RdRP gene in sputum samples.
Table 1.
The obtained results from standard addition method with the spiked different concentrations of SARS-CoV-2 RdRP sequence in sputum samples prepared from healthy volunteers by electrochemical genosensor
| Sample | Added RdRP sequence | aFound RdRP sequence | RSD (%) | Recovery (%) |
|---|---|---|---|---|
| 1 | 5.0 pM | 4.8 | 1.9 | 96.0 |
| 2 | 50.0 pM | 50.4 | 1.0 | 100.8 |
| 3 | 5.0 nM | 5.1 | 1.6 | 102.0 |
aCalculated as a mean of four measurements
Comparison with the alternative biosensors for detection of COVID-19
The LOD of detection tools plays a major role in the early diagnosis of COVID-19. The low LOD of assay methods is desirable to prevent false-negative results. Table 1S presents several molecular diagnostic tests based on the nucleic acid amplification for SARS-CoV-2 detection. As mentioned, an important issue with the amplification strategies is the risk of eliciting false-positive results due to the unexpected cross-contamination. In addition to false-negative/positive results, the molecular diagnostics are not intended for POC diagnosis of COVID-19 [8].
Biosensors offer great potential to meet diagnostic requirements and can be used to detect COVID-19-related indicators. The advent of new technologies, such as nanotechnology, smartphones, and microfluidic technology has changed the world of sensors and provided portable biosensing systems for POC diagnostic applications. The biosensors also have the ability to simultaneously identify multiple markers. The simultaneous determination is an important need for the fast detection of multiple analytes present in a sample matrix. For example, areas with local infectious disease epidemics, such as Africa and the malaria epidemic have a high risk of simultaneous outbreaks together with COVID-19. The biosensors can simultaneously identify and screen COVID-19 and other local infectious diseases, serving as an early warning system in resource-poor areas [37]. In addition, these analytical devices can work with low sample volumes. More importantly, biosensors can be used for the label-free and nucleic acid amplification-free detection of DNA/RNA for the sensitive diagnosis of infection [38]. Therefore, ultralow LOD, ability for simultaneous detection, selectivity, and portability of biosensors make them a better choice for COVID-19 diagnosis.
In Table 2, the analytical characteristics of some previously reported biosensors for COVID-19 detection were compared with those of our proposed genosensor [2, 39–45]. The analytical figures of merit of the designed genosensor in this work are acceptable compared to the biosensors reported in the literature for COVID-19 detection. As is obvious, the electrode materials play a critical role in the construction of high-performance electrochemical sensing platforms for detecting ultralow levels of analytes. The PAMAM functionalized SiQDs provide a large surface area and abundant amino groups for the immobilization of DNA probes. In addition, the chitosan/SiQDs@PAMAM nanocomposite, as an efficient bio-interface film, possesses good biocompatibility. To the best of our knowledge, the silver ions in the HT18C6 were used for the first time as an electrochemical probe. The obtained complex was easily mixed with carbon paste. More importantly, no antibody or enzyme was used in our sensing method. Compared with other biosensors that require protein bioreceptors, such as antibodies, the present sensing method demonstrated a simplified need in label-free oligonucleotide probes. The use of nucleic acids can effectively reduce costs and increase the chemical or thermal stability of biosensors. The nucleic acids are in general more stable than antibodies and enzymes and have a longer shelf life.
Table 2.
Comparison of the analytical performance of the proposed electrochemical genosensor for SARS-CoV-2 detection with the other biosensors
| Detection method | Detection target | Type of affinity assay | LOD | DLR | References |
|---|---|---|---|---|---|
| Electrochemistry | N gene | GE/graphene/ssDNA-capped AuNPs | 6.9 copies μL−1 | 585.4–5.854 × 107 copies μL−1 | [2] |
| FET | S protein | SiO2/Si substrate/graphene/anti-S protein Ab |
1 fg mL−1 (in PBS) 100 fg mL−1 (in clinical samples) |
1 fg mL−1–1000 fg mL−1 (in PBS) | [39] |
| Colorimetry | N gene | ssDNA-capped AuNPs/RNaseH | 0.18 ng μL−1 | 0.2–3 ng μL−1 | [40] |
| LSPR | RdRP gene | ssDNA-functionalized AuNIs | 0.22 pM | – | [41] |
| Electrochemiluminescence | RdRP gene | GE/DT/DNA1/DNA2/Ru (bpy)32+ modified DNA3 | 2.67 fM | Up to 100 pM | [42] |
| Electrochemistry | S protein and N protein | SPE/MBs/anti-mouse IgG Ab/anti-S protein MAb (or anti-N protein MAb)/S protein (or N protein)/anti-S protein PAb (or anti-N protein PAb)/anti-rabbit IgG-AP |
19 ng mL−1 (S protein) 8 ng mL−1 (N protein) |
– | [43] |
| Electrochemistry | ncovNP gene | GE/4-ATP/DTSSP/ncovNP-MIP(PmPD) | 15 fM | Up to 111 fM | [44] |
| Electrochemistry | RNA | SPE/Au@Fe3O4-CP-HT/Target RNA/SCX8-RGO-Au-TB-LP-AP | 3 aM | 10 aM−1 pM | [45] |
| Electrochemistry | RdRP gene | CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM/ssDNA probe | 0.3 pM | 1.0 pM–8.0 nM | This work |
GE, gold electrode; N-gene, nucleocapsid phosphoprotein-gene; AuNPs, gold nanoparticles; S, spike protein; FET, field effect transistor; PBS, phosphate buffer solution; RNaseH, ribonuclease H; LSPR, localized surface plasmon resonance; AuNIs, gold nanoislands; DT, DNA tetrahedron; Ru (bpy)32+, tris(bipyridine)ruthenium(II) chloride; N protein, nucleocaspid protein; SPE, screen-printed electrode; MBs, magnetic beads; IgG, immunoglobulin G; MAb,monoclonal antibody; PAb, polyclonal antibody; AP, alkaline phosphatase; ncovNP, SARS-CoV-2 nucleoprotein; 4-ATP, 4-aminothiophenol; DTSSP, 3,3′-dithiobis [sulfosuccinimidyl propionate]; MIP, molecularly imprinted polymers; PmPD, poly-m-phenylenediamine; ncovNP-MIP(PmPD), ncovNP-MIP film generated from PmPD; CP, Capture probe; HT, hexane-1-thiol; SCX8-RGO, p-sulfocalix[8]arene (SCX8) functionalized graphene; TB, toluidine blue; LP, label probe; AP, auxiliary probe; CPE–HT18C6(Ag), HT18C6(Ag) incorporated carbon paste electrode; SiQDs@PAMAM, PAMAM dendrimer-coated silicon quantum dots
Conclusions
The COVID-19 pandemic has created huge damage to society and brought panic around the world. The accurate and reliable diagnosis followed by effective isolation and treatment of patients are pivotal at the early stage of virus breakouts. This is especially true when there is no cure or vaccine available for a transmissible disease, which is the case for the current COVID-19 pandemic. Up to now, more than 100 kits for COVID-19 diagnosis on the market have been proposed, while emerging sensing techniques for SARS-CoV-2 are also investigated. In this article, a signal-off sensing strategy was used for the voltammetric determination of SARS-CoV-2 RdRP sequence (RdRP_SARSr-P2) based on the CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe sequence. For the first time, the silver ions (Ag+) in the HT18C6 carrier were used as a redox probe. The modification of CPE–HT18C6(Ag) with nanolayers and then, hybridization of SARS-CoV-2 RdRP sequence with the probe sequence led to decreasing the peak currents of silver. The proposed genosensor exhibits a wide linear range of detection, low LOD, and good selectivity. Despite these advantages, the described sensing strategy is time and cost-consuming. In addition, such a design (electrochemical signal-off strategy) has a main limitation of negative signals. To overcome the aforementioned disadvantage, signal-on electrochemical biosensors were duch improved signaling; as the background current observed in the absence of a target is reduced, the gain of such a sensor, at least in theory, increases without limit [46, 47].
Supplementary Information
(DOCX 98 kb)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yang Y, Peng F, Wang R, Guan K, Jiang T, Xu G, Sun J, Chang C. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;109:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alafeef M, Dighe K, Moitra P, Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed eelectrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Zhang G, Liu L, Li Z. Emerging biosensing technologies for improved diagnostics of COVID-19 and future pandemics. Talnta. 2021;225:121986. doi: 10.1016/j.talanta.2020.121986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukas H, Xu C, Yu Y, Gao W. Emerging telemedicine tools for remote COVID-19 diagnosis, monitoring, and management. ACS Nano. 2020;14:16180–16193. doi: 10.1021/acsnano.0c08494. [DOI] [PubMed] [Google Scholar]
- 5.Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;2020:1–2. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu L, Li D, Ramadan S, Li Y, Klein N. Facile biosensors for rapid detection of COVID-19. Biosens Bioelectron. 2020;170:112673. doi: 10.1016/j.bios.2020.112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu X, Wang X, Han L, Chen T, Wang L, Li H, Li S, He L, Fu X, Chen S, Xing M, Chen H, Wang Y. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J Adv Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao J, Duan C, Xue L, Liu Y, Sun W, Xiang Y. DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens Bioelectron. 2020;167:112479. doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farzin L, Sadjadi S, Shamsipur M, Sheibani S. Electrochemical genosensor based on carbon nanotube/amine-ionic liquid functionalized reduced graphene oxide nanoplatform for detection of human papillomavirus (HPV16)-related head and neck cancer. J Pharm Biomed Anal. 2020;179:112989. doi: 10.1016/j.jpba.2019.112989. [DOI] [PubMed] [Google Scholar]
- 11.Farzin L, Shamsipur M, Samandari L, Sheibani S. HIV biosensors for early diagnosis of infection: the intertwine of nanotechnology with sensing strategies. Talanta. 2020;206:120201. doi: 10.1016/j.talanta.2019.120201. [DOI] [PubMed] [Google Scholar]
- 12.Grabowska I, Malecka K, Stachyra A, Gora-Sochacka A, Sirko A, Zagorski-Ostoja W, Radecka H, Radecki J. Single electrode genosensor for simultaneous determination of sequences encoding hemagglutinin and neuraminidase of avian influenza virus type H5N1. Anal Chem. 2013;85:10167–10173. doi: 10.1021/ac401547h. [DOI] [PubMed] [Google Scholar]
- 13.Li D, Zhang J, Li J. Primer design for quantitative real-time PCR for the emerging coronavirus SARS-CoV-2. Theranostics. 2020;10:7150–7162. doi: 10.7150/thno.47649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moitra P, Alafeef M, Dighe K, Frieman MB, Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakhki RK, Kakhki MK, Neshani A. COVID-19 target: a specific target for novel coronavirus detection. Gene Rep. 2020;20:100740. doi: 10.1016/j.genrep.2020.100740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 18.Roushani M, Valipour A, Saedi Z. Electroanalytical sensing of Cd2+ based on metal–organic frameworkmodified carbon paste electrode. Sens Actuator B-Chem. 2016;233:419–425. doi: 10.1016/j.snb.2016.04.106. [DOI] [Google Scholar]
- 19.Alberto R, Nef W, Smith A, Kaden TA, Neuburger M, Zehnder M, Frey A, Abram U, Schubiger PA. Silver(I) complexes of the derivatized crown thioether ligands 3,6,9,12,15,18-hexathianonadecanol and 3,6,9,13,16,19-hexathiaicosanol. Determination of stability constants and the crystal structures of [Ag(19-aneS6-OH)][CF3SO3] and [Ag(20-aneS6-OH)][BF4] Inorg Chem. 1996;35:3420. doi: 10.1021/ic951421y. [DOI] [PubMed] [Google Scholar]
- 20.Mashhadizadeh MH, Shamsipur M. Silver(I)-selective membrane electrode based on hexathia-18-crown-6. Anal Chim Acta. 1991;381:111–116. doi: 10.1016/S0003-2670(98)00672-2. [DOI] [Google Scholar]
- 21.Chayama K, Hara K, Tamari Y, Tsuji H, Kusaka Y, Sekido E, Mori Y. In: Solvent extraction in the process industries. Logsadil DH, Slater MJ, editors. New York: Elsevier; 1993. [Google Scholar]
- 22.Sarabaegi M, Roushani M, Hosseini H, Hoseini SJ, Bahrami M. Facile synthesis of a covalent organic framework (COF) based on the reaction of melamine and trimesic acid incorporated electrospun nanofiber and its application as an electrochemical tyrosinamide aptasensor. New J Chem. 2020;44:14922–14927. doi: 10.1039/D0NJ02837A. [DOI] [Google Scholar]
- 23.Valipour A, Roushani M. Using silver nanoparticle and thiol graphene quantum dots nanocomposite as a substratum to load antibody for detection of hepatitis C virus core antigen: electrochemical oxidation of riboflavin was used as redox probe. Biosens Bioelectron. 2017;89:946–951. doi: 10.1016/j.bios.2016.09.086. [DOI] [PubMed] [Google Scholar]
- 24.Farzin MA, Abdoos H. A critical review on quantum dots: from synthesis toward applications in electrochemical biosensors for determination of disease-related biomolecules. Talanta. 2021;224:121828. doi: 10.1016/j.talanta.2020.121828. [DOI] [PubMed] [Google Scholar]
- 25.Zhu N, Gu Y, Chang Z, He P, Fang Y. PAMAM dendrimers-based DNA biosensors for electrochemical detection of DNA hybridization. Electroanalysis. 2006;18:2107–2114. doi: 10.1002/elan.200603589. [DOI] [Google Scholar]
- 26.Niu X, Huang L, Zhao J, Yin M, Luo D, Yang Y. An ultrasensitive aptamer biosensor for the detection of codeine based on a Au nanoparticle/polyamidoamine dendrimer-modified screen-printed carbon electrode. Anal Methods. 2016;8:1091–1095. doi: 10.1039/C5AY01747E. [DOI] [Google Scholar]
- 27.Xie J, Wang J, Chen H, Shen W, Sinko PJ, Dong H, Zhao R, Lu Y, Zhu Y, Jia L. Multivalent conjugation of antibody to dendrimers for the enhanced capture and regulation on colon cancer cells. Sci Report. 2015;5:9445. doi: 10.1038/srep09445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuce M, Filiztekin E, Ozkaya KG. COVID-19 diagnosis —a review of current methods. Biosens Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y, Liu Y, Su C, Ji X, He Z. New fluorescent pH sensor based on label-free silicon nanodots. Sens Actuator B-Chem. 2014;203:795–801. doi: 10.1016/j.snb.2014.07.050. [DOI] [Google Scholar]
- 30.Wu J, Dai J, Shao Y, Sun Y. One-step synthesis of fluorescent silicon quantum dots (Si-QDs) and their application for cell imaging. RSC Adv. 2015;5:83581–83587. doi: 10.1039/C5RA13119G. [DOI] [Google Scholar]
- 31.Fan JW, Vankayala R, Chang CL, Chang CH, Chiang CS, Hwang KC. Preparation, cytotoxicity and in vivo bioimaging of highly luminescent water-soluble silicon quantum dots. Nanotechnology. 2015;26:215703. doi: 10.1088/0957-4484/26/21/215703. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh S, Ghosal K, Mohammad SA, Sarkar K. Dendrimer functionalized carbon quantum dot for selective detection of breast cancer and gene therapy. Chem Eng J. 2019;373:468–484. doi: 10.1016/j.cej.2019.05.023. [DOI] [Google Scholar]
- 33.Fatemi SM, Fatemi SJ, Abbasi Z. PAMAM dendrimer-based macromolecules and their potential applications: recent advances in theoretical studies. Polym Bull. 2020;77:6671–6669. doi: 10.1007/s00289-019-03076-4. [DOI] [Google Scholar]
- 34.Nandy B, Maiti PK. DNA compaction by a dendrimer. J Phys Chem B. 2010;115:217–230. doi: 10.1021/jp106776v. [DOI] [PubMed] [Google Scholar]
- 35.Valipour A, Roushani M. A glassy carbon immunoelectrode modified with vanadium oxide nanobelts for ultrasensitive voltammetric determination of the core antigen of hepatitis C virus. Microchim Acta. 2017;184:4477–4483. doi: 10.1007/s00604-017-2449-z. [DOI] [Google Scholar]
- 36.Khadem M, Faridbod F, Norouzi P, Foroushani AR, Ganjali MR, Shahtaheri SJ, Yarahmadi R. Modification of carbon paste electrode based on molecularly imprinted polymer for electrochemical determination of diazinon in biological and environmental samples. Electroanalysis. 2017;29:708–715. doi: 10.1002/elan.201600293. [DOI] [Google Scholar]
- 37.Mao K, Zhang H, Yang Z. An integrated biosensor system with mobile health and wastewater-based epidemiology (iBMW) for COVID-19 pandemic. Biosens Bioelectron. 2020;169:112617. doi: 10.1016/j.bios.2020.112617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu CC, Yen HY, Lai LT, Perng GC, Lee CR, Wu SJ. A label-free impedimetric genosensor for the nucleic acid amplification-free detection of extracted RNA of dengue virus. Sensors. 2020;20:3728. doi: 10.3390/s20133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, Kim SJ, Lee JO, Kim BT, Park EC, Kim SI. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 40.Moitra P, Alafeef A, Dighe K, Frieman MB, Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 42.Fan Z, Yao B, Ding Y, Zhao J, Xie M, Zhang K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens Bioelectron. 2021;178:113015. doi: 10.1016/j.bios.2021.113015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabiani L, Saroglia M, Galatà G, De Santis R, Fillo S, Luca V, Faggioni G, D’Amore N, Regalbuto E, Salvatori P, Terova G, Moscone D, Lista F, Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARSCoV-2 detection in saliva. Biosens Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raziq A, Kidakova A, Boroznjak R, Reut J, Opik A, Syritski V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens Bioelectron. 2021;178:113029. doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H, Liu F, Xie W, Zhou TC, Yang JO, Jin L, Li H, Zhao CY, Zhang L, Wei J, Zhang YP, Li CP. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens Actuator B-Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, Qian Y, Zhang Y, Meng S, Wang S, Li Y, Gao F. A novel label-free and signal-on electrochemical aptasensor based onthe autonomous assembly of hemin/G-quadruplex and direct electron transfer of hemin. Sens Actuator B-Chem. 2017;238:434–440. doi: 10.1016/j.snb.2016.07.086. [DOI] [Google Scholar]
- 47.Farzin L, Sadjadi S, Shamsipur M, Sheibani S, Mousazadeh MH. Employing AgNPs doped amidoxime-modified polyacrylonitrile (PANoxime) nanofibers for target induced strand displacement-based electrochemical aptasensing of CA125 in ovarian cancer patients. Mater Sci Eng C. 2019;97:679–687. doi: 10.1016/j.msec.2018.12.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 98 kb)