Abstract
Vitamin K antagonists are widely used anticoagulants targeting vitamin K epoxide reductases (VKOR), a unique family of integral membrane enzymes. To elucidate their catalytic cycle and inhibitory mechanism, here we report eleven x-ray crystal structures of human VKOR and a VKOR-like paralog with substrates and antagonists in different redox states. Substrates entering the active site in a partially oxidized state form a cysteine adduct that induces an open-to-closed conformational change, which triggers electron transfer to reduce the substrates. The reduction is facilitated by hydrogen-bonding interactions in a hydrophobic pocket. The antagonists bind specifically to the same hydrogen-bonding residues and induce a similar closed conformation. Thus, vitamin K antagonists act through mimicking the key interactions and conformational changes required for the VKOR catalytic cycle.
One-sentence summary:
Crystal structures reveal the catalytic cycle of integral membrane VKORs and the inhibition mechanism of vitamin K antagonists.
Vitamin K antagonists (VKAs) are oral anticoagulants used to treat and prevent thromboembolic diseases, including myocardial infarction and stroke, the two leading causes of human death and disability (1). Warfarin, a well-known VKA, is among the most commonly used drugs worldwide. However, warfarin overdose often causes major and fatal bleeding (2). In older adults, one third of hospitalizations for adverse drug reaction are due to warfarin use (3). Improving the safety of anticoagulation therapy requires the understanding of how VKAs inhibit their target, vitamin K epoxide reductase (VKOR).
VKOR is an endoplasmic membrane enzyme that sustains blood coagulation through the vitamin K cycle (4). This cycle begins with the epoxidation of vitamin K hydroquinone (KH2) that drives the γ-carboxylation of several coagulation factors, a posttranslational modification required for their activity. To regenerate this γ-carboxylase cofactor, VKOR reduces the vitamin K epoxide (KO) first to the quinone (K) and then to KH2. Each reduction step is coupled to the oxidation of two active-site cysteines in VKOR (fig. S1A). To return to their reactive state, these cysteines are reduced through an electron-transfer process, which is mediated by another pair of conserved cysteines in VKOR (5-8). This reductase activity is similar in a VKOR-like paralog, which functions to support bone mineralization and inhibit vascular calcification (9, 10). In addition, VKOR homologs catalyze disulfide-bond formation in many species, constituting a major family of integral membrane thiol oxidoreductases found from bacteria to humans (11).
VKAs inhibit VKOR and VKOR-like enzymes at both their epoxide and quinone reduction steps. The naphthoquinone ring of vitamin K is somewhat resembled by VKAs, which generally carry a pharmacophore of either 4-hydroxycoumarin or 1,3-indandione (fig. S1, B and C). Coumarin-based VKAs, including warfarin, phenprocoumon, and acenocoumarol, are the primary anticoagulants used in North American and many European countries. An indandione-based VKA, fluindione, is used mostly in France (12). Coumarin and indandione derivatives with large side groups, such as brodifacoum and chlorophacinone (fig. S1D), are known as superwarfarins that have a long-lasting effect of inducing severe bleeding. Owing to this high potency, brodifacoum is one of the most used rat poisons. After decades of VKA use, numerous mutations conferring resistance on VKOR have been identified in rodents and humans. Patients carrying such mutations require high warfarin dosage that varies by the type of mutations, adding another complication to the anticoagulation therapy.
To reveal the molecular basis of anticoagulation by VKAs, we determined crystal structures of human VKOR (HsVKOR) with representative VKAs and with the KO substrate captured in different functional states. We have also determined ligand-free, substrates-bound and warfarin-bound structures of a VKOR-like paralog. A total of eleven structures (Table S1 and fig. S2) illustrate almost the entire catalytic cycle of these integral membrane oxidoreductases and reveal the mechanism of vitamin K antagonism.
Overall structure of human VKOR with bound VKAs
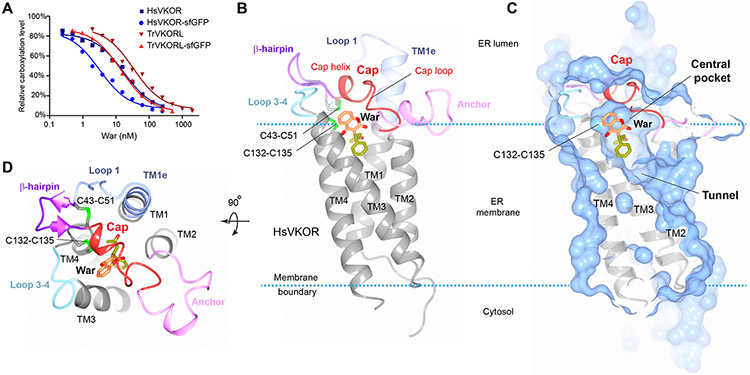
HsVKOR contains a large and flexible ER-luminal region and is a difficult target for structural characterization due to its in vitro instability (13). We found that pre-bound VKAs can stabilize the HsVKOR protein in a detergent solution (fig. S3, A and B; see Method). For additional stabilization in vitro, we fused the flexible N- and C-termini of HsVKOR to a split superfolder GFP (14), which also provides a scaffold for crystallization and facilitates structure determination (fig. S3, A, C and D). This termini-restrained construct is catalytically active and inhibitable by warfarin, both in cells (Fig. 1A) and after being purified with detergent (fig. S4, A and C); the increased warfarin sensitivity in cells and retained activity in detergent suggest that the protein fold of HsVKOR is stabilized after termini restraining. A combination of these strategies allowed the structure determination of HsVKOR at near-atomic resolution with warfarin, phenindione (a close analog of fluindione), brodifacoum and chlorophacinone in three different crystallographic space groups (Table S2).
Figure 1. Overall structure of HsVKOR with bound warfarin.
(A) Warfarin inhibition of epoxide reductase activity in cultured cells. The inhibition curves of untagged and sfGFP-fused HsVKOR and TrVKORL are compared. The activity assay has been repeated three times. (B) Structure of HsVKOR (side view) with warfarin (War). Secondary structure elements in the large ER luminal domain are named and presented in different colors. The four transmembrane helices (TM1–4) are shown in grey. The 4-hydroxycoumarin and side groups of warfarin are colored in orange and yellow green, respectively. The overall structure of HsVKOR with warfarin and with other vitamin K antagonists are very similar (fig. S3E). (C) Surface representation of the structure (exposed view). Warfarin is bound at the central pocket that contains the active site cysteines. The cap helix forms top part of the central pocket. The tunnel below is expected to bind the isoprenyl chain of K or KO substrate. (D) Top view of the structure.
HsVKOR with these four different VKAs bound adopts essentially the same overall structure (Fig. 1B and fig. S3E) under the different crystal packing conditions. The transmembrane helices (TM) form a four-helix bundle that creates a central pocket occupied by VKAs (Fig. 1C). This same pocket is designated as the active site of HsVKOR, where the catalytic cysteines, Cys132 and Cys135, are located. The coumarin or indandione ring of VKAs faces the ER luminal surface and their side group is buried in the transmembrane region of the protein. TM1 and TM2 are connected by the large ER-luminal region, which constitutes almost one-third of the protein sequence. This region starts from a helical extension of TM1 (named TM1e) and continues with a loop (loop 1) and a β-hairpin. The two ends of the β-hairpin are stapled together by a disulfide bond formed between Cys43 and Cys51 (Fig. 1D), which are the cysteine pair that mediates electron transfer during catalysis (5-8). Cys51 connects to a cap domain that covers the top of the central pocket. The cap domain consists of a short helix (cap helix) and a loop (cap loop), followed by an amphipathic anchor domain that is buried partially in the membrane. This anchor attaches the cap domain to the membrane and thereby stabilizes its covering of the central pocket. Residues whose mutation confers strong warfarin resistance are either located at this VKA-binding pocket or distributed in the other regions of the large luminal domain (fig. S5).
Binding interactions of VKAs
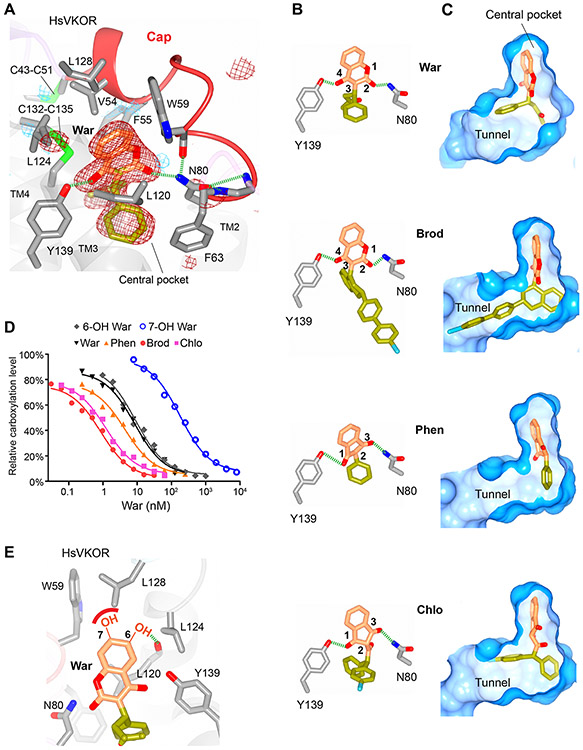
The structures show that all VKAs are hydrogen bonded to Asn80 on TM2 and Tyr139 on TM4 (Fig. 2, A and B). The Tyr139 hydroxyl forms a hydrogen bond with the 4-hydroxyl group of warfarin and brodifacoum. The importance of the 4-hydroxyl has been known since the discovery of vitamin K antagonization in 1939, as this coumarin modification in spoiled sweet clover caused fatal bleeding in cattle (15). Replacing the 4-hydroxyl of warfarin with chemical groups that disable hydrogen-bond formation results in almost no HsVKOR inhibition (16). Another key hydrogen bond is formed between the 2-ketone group of warfarin and the amide group of Asn80, whose side chain is well positioned through interacting with the protein backbone of the cap loop (Fig. 2A). Mutations disrupting either of these hydrogen bonds, such as Asn80Ala or Tyr139Phe, result in strong warfarin resistance (fig. S5A) (7). For phenindione and chlorophacinone, Asn80 and Tyr139 form hydrogen bonds with the 1,3-diketones of their indandione ring (Fig. 2B), which closely resemble the 2-ketone-4-hydoxyl of warfarin or the 2,4-diketones of its keto–enol tautomer (fig. S1B) (17). Thus, the hydrogen bonding with the meta-positioned (1,3- or 2,4-) diketones or ketone-hydroxyl, which provides the specificity of VKA recognition, is a shared key feature underlying the VKA inhibition of HsVKOR.
Figure 2. Critical molecular interactions between HsVKOR and vitamin K antagonists.
(A) The warfarin-binding pocket. The Fo-Fc omit map of warfarin is contoured at 3σ (red mesh) and −3σ (blue mesh). Hydrogen bonding interactions are indicated by dashed green lines. (B) Similar hydrogen-bonding patterns are observed for different VKAs (Brod: brodifacoum, Phen: phenindione, Chlo: chlorophacinone). Standard atomic numbers for the key positions of VKAs are indicated. (C) Side groups (yellow green) of VKAs bind differently to the isoprenyl-chain tunnel (surface view). (D) Inhibition curves of VKAs and hydroxywarfarins against HsVKOR using the cell-based assay. (E) Models of the 6- and 7-hydoxywarfarin in the warfarin-binding pocket. The dashed green line indicates a putative hydrogen bond to the 6-hydroxyl group, and the red curve represents a lack of favorable interaction between the polar 7-hydroxyl group and the hydrophobic protein side chains.
Except for these two hydrogen bonds, the VKAs are bound in a largely hydrophobic pocket that excludes water molecules (Fig. 2, A and C). The lowered dielectric constant in this central pocket may enhance the hydrogen bonding interactions. The coumarin or indandione rings of VKAs are stacked between Val54/Phe55 at the top and Leu120 from the bottom, and surrounded by Trp59, Phe63, Leu124 and Leu128 (Fig. 2A). All of these hydrophobic residues, when mutated, render HsVKOR strongly resistant to warfarin (fig. S5A) (7).
The large side groups of brodifacoum and chlorophacinone afford their high-potency inhibition of HsVKOR. The side group of brodifacoum occupies the entire central pocket, including a tunnel formed between TM2 and TM3 (Fig. 2C), where the isoprenyl chain of substrates should be bound (5). Hydrophobic residues aligning along this tunnel, such as Phe83, Phe87 and Tyr88, provide additional binding interactions to the side group of brodifacoum; these residues do not interact with warfarin, which only inhabits the inner end of the tunnel. The indandiones show a similar trend: the side group of chlorophacinone occupies half of this tunnel, whereas phenindione, with a nearly planar structure, cannot reach to this tunnel. These differences indicate that the large side groups of the superwarfarins interact with the isoprenyl-chain tunnel, thereby increasing the binding area and resulting in the strong inhibition (Fig. 2D).
Based on these structures, binding interactions of warfarin metabolites can be modeled. Warfarin is inactivated by cytochrome P450 2C9 (CYP2C9), whose genotype is a major predictor of warfarin dose (18). The CYP2C9 metabolism generates 7-hydroxywarfarin (71%) and 6-hydroxywarfarin (22%). If these metabolites bind in the same way as warfarin does, a polar 7-hydroxyl group would be energetically unfavorable in the hydrophobic pocket, whereas 6-hydroxyl substitution would be within the hydrogen bonding distance to the protein backbone, thereby saturating the polarity (Fig. 2E). Consistently, 6-hydroxywarfarin inhibits HsVKOR to a similar level as warfarin, whereas 7-hydroxywarfarin loses the inhibition efficacy (Fig. 2D) (16, 19). Thus, the different binding interactions of warfarin metabolites explain their relative activity.
Stabilization of the cap domain
The cap domain conformation and VKA binding are mutually stabilized. The association between the cap and TM domains requires VKA-mediated interactions (Fig. 2A and Movie S1), which explains the stabilization effect of bound VKAs during protein purification and crystallization. Consistently, footprinting of HsVKOR in cells by mass spectrometry detection showed that its structural flexibility is reduced with bound warfarin (7, 20). Neighboring protein regions also stabilize the cap domain conformation (Fig. 1D). At the horizontal direction, the cap domain is saddled between TM1/TM1e and a loop connecting TM3 and TM4 (loop 3–4). TM1/TM1e interacts with loop 1 primarily through Tyr25 and His28 (fig. S6A). Loop 1 and β-hairpin in turn stabilize the cap helix, and Asp44 is a central residue mediating such interactions (fig. S6B). The helical conformation of the cap helix is maintained by a hydrogen-bonding network involving Asp44 and several Ser residues (fig. S6C). In the vertical direction, the anchor domain positions the cap domain above the membrane surface, with Arg58 and Glu67 forming a salt bridge (fig. S6D). To anchor the cap loop in position, Asn80 from TM2 provides several hydrogen bond interactions (fig. S6E). Consistent with these structural observations, mutations such as Tyr25Ala, His28Ala and Asp44Ala show strong warfarin resistance (fig. S5A) (7); their mechanism had previously been enigmatic, because these residues did not appear to interact with warfarin (fig. S5B) (21, 22). The structures now reveal that such mutations would disrupt critical interactions that stabilize the cap domain. Taken together, warfarin resistance is caused by two distinct mechanisms: the resistant mutations can either directly affect the warfarin binding or destabilize the cap domain. This mechanistic understanding may improve the ability to predict warfarin dosage as a personalized medicine (23); the US Food and Drug Administration suggests pharmacogenomic tests when prescribing warfarin, but the clinical benefits of current dosing algorithms remain controversial (24).
Warfarin inhibits both fully oxidized and partially oxidized HsVKOR, which are the two major redox states of HsVKOR in a cellular environment (7). To mimic the partially oxidized state in vitro, we determined the warfarin-bound structure of a Cys43Ser mutant (fig. S7), which generates a free Cys135 (fig. S2). In this state, the Cys135 sulfhydryl is within hydrogen bonding distance to the 4-hydroxyl group of warfarin and the cap helix is covalently attached to TM4 through the Cys51–Cys132 disulfide, thereby stabilizing the binding of warfarin in the same orientation.
Warfarin-induced conformational changes
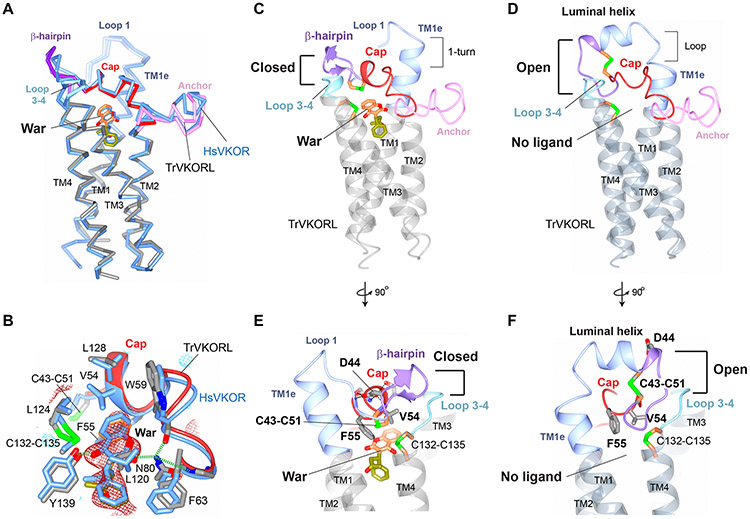
To understand the effect of warfarin binding, we determined the structures of a VKOR-like protein from Takifugu rubripes (TrVKORL) in both warfarin-bound and ligand-free states. TrVKORL is catalytically active and inhibitable by warfarin, both in cells and after being purified (Fig. 1A and fig. S4, B and C). This protein shares 73% sequence identity with human VKOR-like (HsVKORL) and 52% with HsVKOR (fig. S8); for simplicity of explanation, hereafter we number conserved residues in these proteins according to the HsVKOR sequence. TrVKORL was chosen for crystallization because it has higher protein expression level and better in vitro stability than HsVKORL (25). Compared to HsVKOR, the TrVKORL protein is stable in detergent micelles even without a bound ligand, enabling the capture of both the ligand-free and warfarin-bound states.
The conformations of warfarin-bound TrVKORL and HsVKOR in the fully oxidized state are remarkably similar (RMSD 1.1 Å) (Fig. 3, A and B). Both proteins adopt a closed conformation, with loop 1 and β-hairpin folded near the active site and the luminal domain forming extensive interactions with loop 3–4 (Fig. 3C). At the central pocket, warfarin interacts with surrounding residues in essentially the same way, and the hydrogen bonding of warfarin to Asn80 and Tyr139 is nearly identical (Fig. 3B). In fact, 90% of the residues critical to warfarin inhibition or catalysis are conserved between TrVKORL, HsVKORL and HsVKOR (fig. S8). The high sequence identity and structural similarity suggest that these VKOR enzymes share essentially the same catalytic and inhibitory mechanisms.
Figure 3. TrVKORL structures showing warfarin-induced conformational changes.
(A) Overall structure of TrVKORL with warfarin (colored by structure elements) is nearly the same as that of HsVKOR with warfarin (blue). (B) Similarity of their warfarin binding interactions. TrVKORL residues are colored in grey (carbon atoms) and HsVKOR residues in blue. The Fo-Fc omit map of warfarin is contoured at 3σ (red mash) and −3σ (blue mesh). C) TrVKORL with bound warfarin adopts a closed conformation, in which the cap helix and β-hairpin are formed and interact with loop 3–4. (D) The ligand-free TrVKORL adopts an open conformation. In the luminal helix, residues converted from the loop 1 and β-hairpin (in C) are colored in blue and purple, respectively. The TMs are colored in dark grey to indicate the open conformation. (E-F) Detailed view of the closed (E) and open (F) states. Warfarin binding induces the movements of Cys43-Cys51, Val54, and Phe55 and promotes the stabilizing interactions of Asp44 (dashed lines). See Movie S2 for modeled structural transition.
In the ligand-free state, the fully oxidized TrVKORL shows an open conformation, forming a luminal helix that is distant from the active site (Fig. 3D). This helix re-structures the residues of the loop 1 and β-hairpin found in the ligand-bound state. Moreover, the cap helix becomes a loop in absence of warfarin stabilization, and TM1e loses a helical turn. In the absence of these structural elements, few interactions are observed between loop 3–4 and the large luminal domain, a characteristic of the open conformation.
Modeling the structural transition between the ligand-free (open) and warfarin-bound (closed) states suggests that the induced fit prompted by warfarin brings Phe55 closer and reorients Val54 in the cap domain, thereby leading to the formation of the cap helix (Movie S2). This helical conformation brings the Cys43–Cys51 disulfide close to membrane surface to interact with Phe55 (Fig. 3E). The repositioning of Cys43–Cys51 induces the formation of β-hairpin and breaks up the luminal helix found in the open conformation. This change also reorients Asp44, which becomes a central residue holding the cap helix, β-hairpin and loop 1 together (fig. S6B). In contrast, in the open conformation, Asp44 is located on the luminal helix and does not participate in stabilizing interactions (Fig. 3F). Taken together, the local interactions with warfarin lead to global changes of protein conformation, a “domino effect” culminating into the closed conformation.
Substrate entrance
To understand the catalytic mechanism of VKORs, we first determined the structures of fully oxidized TrVKORL with KO and K. The protein conformations in these states are nearly identical to that in the ligand-free state. The Fo-Fc maps show weak electron densities (Fig. 4A and fig. S9A) of K or KO, indicating low-affinity binding of the substrates, probably because the oxidized enzyme is not at the state of reacting with the substrates. Furthermore, their naphthoquinone rings do not occupy the top of the central pocket (Fig. 4A and fig. S9B) but are located near the isoprenyl-chain tunnel, suggesting that the tunnel provides the path of substrate entrance and/or product release. The C2 atom of K or KO is distant (~7 Å) from the oxidized sulfur group of Cys135; the predicted reaction between these atoms (26, 27) therefore cannot occur. For the substrates to be bound with high affinity and positioned properly for catalysis, a reactive Cys135 is required.
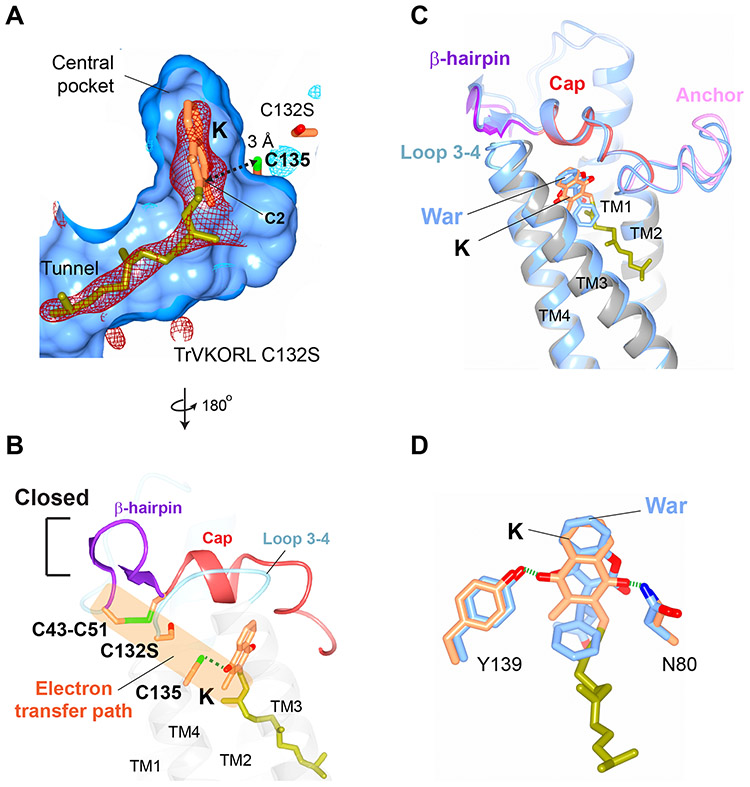
Figure 4. Structures of vitamin K-bound TrVKORL in noncatalytic and catalytic states.
(A) Fully oxidized TrVKORL with K. The central pocket, including the isoprenyl-chain tunnel, are shown in surface view. The Fo-Fc omit map of K is contoured at 3σ (red mash), −3σ (no visible density), and 2.5 σ (blue mash) due to the weak density of K in this redox state. The distance between the C2 atom of K and the Cys135 sulfur is indicated. (B) TrVKORL Cys132Ser mutant with K. The Fo-Fc omit map of K is contoured at 3σ (red mash) and −3σ (light blue mesh). The reduced Cys135 and K form a charge-transfer complex (dashed arrow). (C) Open conformation in the fully oxidized state. (D) Closed conformation with formation of the charge-transfer complex. The unimpeded electron-transfer path between the four cysteines and K is highlighted by the orange shading. (E) Superimposed structures of TrVKORL in the K-bound (as in (D); colored by structural elements) and warfarin-bound (as in Fig. 3C; blue) states. (F) Similar hydrogen-bonding interactions with K (orange and yellow green) and with warfarin (blue).
Substrate-induced electron transfer
To generate the reactive Cys135, we used a Cys132Ser mutant of TrVKORL (fig. S2) and determined a K-bound structure (Fig. 4B). In this catalytic state, Cys135 is close to the naphthoquinone ring of K, whose isoprenyl chain occupies the designated tunnel. The sulfhydryl group of Cys135 is ~3 Å to the C2 atom, consistent with the distance in a charge-transfer complex, in which the negative charge of Cys135 thiolate is partially transferred to the quinone ring; similar complexes have been found in other thiol oxidoreductases that use aromatic cofactors such as quinone and flavin (28, 29). Formation of this charge-transfer complex, namely a Cys135–K adduct, makes this substrate stably attached to the TrVKORL protein. The binding interactions in turn induce the protein transition from the open conformation in the noncatalytic state (Fig. 4C) to the closed conformation in the catalytic state (Fig. 4D). Importantly, this conformational change brings Cys43–Cys51 close to Cys132Ser and Cys135–K (Fig. 4D), a scenario occurring as these four cysteines transfer electrons in the wild-type protein (fig. S2). These residues are sequentially aligned within a short distance and thus are physically posed for electron transfer through thiol-disulfide exchanges. These unimpeded exchanges facilitate the passing of reducing equivalent to resolve the Cys135–K adduct, resulting in the full reduction of K.
The closed conformation induced by Cys135–K adduct is very similar to that induced by warfarin, with the cap domain, loop 1 and β-hairpin adopting essentially the same conformation in the two structures (Fig. 4E). Similar to warfarin binding interactions, Tyr139 and Asn80 form hydrogen bonds with the para-positioned 1,4-diketones of K (Fig. 4F). In addition, the isoprenyl chain of vitamin K interact with Phe83, Phe87 and Tyr88 along the tunnel, which are the same residues binding the side group of brodifacoum. These resemblances show that warfarin (and superwarfarin) exploits features that are tailored for the binding of substrates and/or transition states of the reaction to stably occupy the active site of VKORs.
Catalysis at the active site
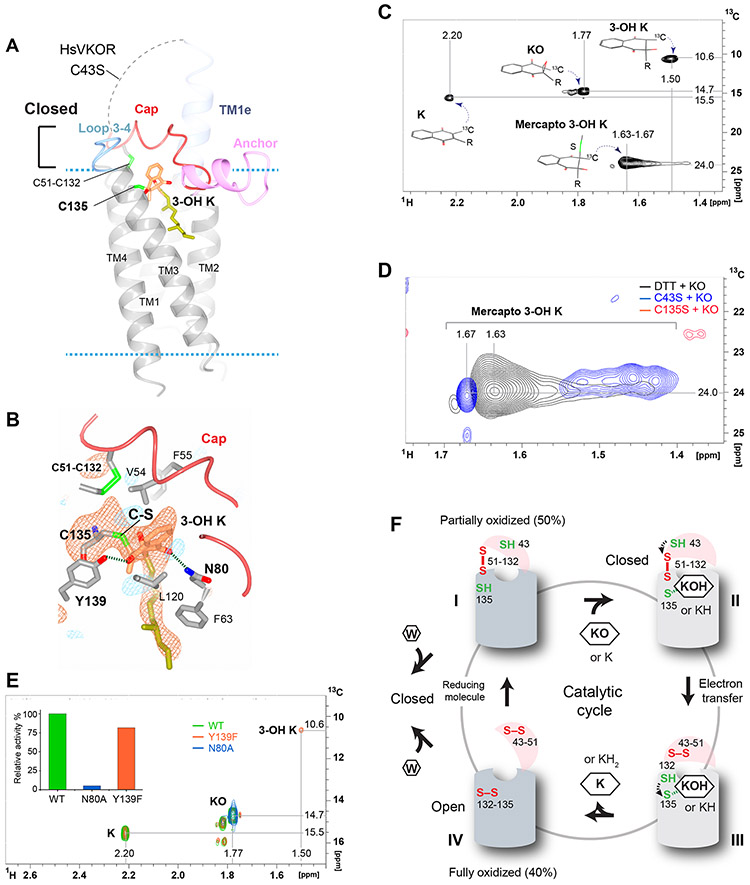
To further understand the catalytic mechanism, we used a similar strategy to determine the structure of HsVKOR with KO (Fig. 5A), this time employing a Cys43Ser mutation to generate the reduced Cys135 (fig. S2). This mutant mimics the state prior to attack of the Cys43 on the Cys51-Cys132 disulfide, as Cys135 is capable of attacking the substrate. The distance between the sulfur atom of Cys135 and the C2 atom of the naphthoquinone ring in the electron density map is ~2 Å (Fig. 5B), suggestive of a covalently bonded C–S adduct. To investigate this possibility, we incubated 13C-labeled KO with the Cys135Ser and Cys43Ser mutants of TrVKORL (chosen for their apo-protein stability). Cys135Ser lacks the reactive thiolate, whereas Cys43Ser is expected to carry out an initial covalent step of the reaction but not to proceed further without an externally supplied reducing agent (fig. S2). After proteolysis of the mutant proteins, different vitamin K derivatives with the 13C label show readily distinguishable positions in 2D 1H-13C correlated spectra (Fig. 5, C and D and fig. S10). Only the Cys43Ser mutant enzyme produces derivatives with signals (Fig. 5D) that are consistent with covalent adducts of 3-hydroxyl vitamin K (3-OH K) from nucleophilic attack of Cys135 on the epoxide (fig. S2). The crystal structure is consistent with this covalent adduct, which has been predicted from chemical modeling and quantum chemistry simulation (26, 27).
Figure 5. Structure of HsVKOR bound to a KO adduct.
(A) Overall structure of HsVKOR Cys43Ser mutant co-crystallized with KO. The dashed line indicates the disordered region between TM1e and cap domain. (B) Detailed view of the active site. The Fo-Fc omit map of C135-KOH is contoured at 3σ (red mash) and −3σ (blue mesh) ; 3-OH K is modeled into the density based on the geometric restraint and the catalytic chemistry (26, 27). The short distance between the C2 atom of 3-OH K and the sulfhydryl group of Cys135 suggests that they are connected by a C–S bond. (C) Two-dimensional 1H-13C HSQC spectrum (full spectrums in fig. S13) showing signals of 13C-labeled 2-methyl groups in four products of KO reduction with dithiothreitol (DTT) (52). Chemical structures of the products and the assigned chemical shifts are indicated. The chemical shifts of 13C 2-methyl group are strongly influenced by the ring current field of the naphthoquinone group (the 2-methyl is in plane with the naphthoquinone ring for K, in tetrahedral angle for 3-OH K and in other angles for KO and mercapto 3-OH K). (D) Superimposed regions of three 2D 1H-13C HSQC spectra (full spectra in fig. S13-S15) showing mercapto reduction products of 2-methyl-13C-labeled KO. Signals of products recovered from TrVKORL Cys43Ser catalyzed reaction are shown in blue and those from Cys135Ser in red. The non-enzymatic (DTT-reduced) products signals are shown in black. (E) Superimposed spectra (full spectra in fig. S16-S18) of reaction products obtained from wild-type HsVKOR (WT; green), Asn80Ala (blue) and Tyr139Phe mutants (orange) with KO and DTT. An area of 2D HSQC spectrum containing 1H-13C peaks of 2-methyl-13C-labeled K (2.20-15.5 ppm), KO (1.77-14.7 ppm) and 3-OH K (1.50-10.6 ppm) is shown. Inset, relative KO reduction activity of HsVKOR constructs in microsomes (see Materials and Methods). (F) The catalytic cycle and inhibition of HsVKOR is accompanied with redox-state and conformation changes. The large luminal domain is shown as a hemisphere (pink) and the transmembrane domain as a cylinder (light grey: closed conformation, dark grey: open conformation). State I, The partially oxidized state with free Cys43 and free Cys135. State II, Cys135 forms a stable adduct with 3-OH K (KOH) or K (KH), whose binding induces the closed conformation and the juxtaposition of Cys43 for electron transfer (dashed arrow). State III, The reduced Cys132 attacks Cys135–K-OH (or KH) to generate K (or KH2). State IV, The fully oxidized state is in an open conformation to release K (or KH2). Left, Warfarin (W) competes with the substrates for the partially oxidized enzyme. Unlike the substrates, warfarin binds also to the fully oxidized enzyme and removes it from the enzyme pool. The bound warfarin locks HsVKOR in both of the redox states into a closed conformation.
Similar to the K-bound state, the 1,4-diketones of 3-OH K retain hydrogen bonding to Tyr139 and Asn80 (Fig. 5B). Maintaining these hydrogen bonds is important for catalysis. In the in vitro assay using dithiothreitol (DTT) as the reductant, Asn80Ala shows nearly no enzymatic activity, whereas Tyr139Phe remains largely active but generates 3-OH K as a side product (Fig. 5E), consistent with previous reports (30, 31). In the cellular assay, Tyr139Phe and Asn80Ala also lower the HsVKOR activity (fig. S4, D and E). Taken together, Tyr139 and Asn80, which are the same residues recognized by VKAs (Fig. 2B), are also important for catalysis.
The HsVKOR structures with the 3-OH K adduct and with warfarin show interesting differences (fig. S11A). The naphthoquinone ring of 3-OH K rotates to an angle different from warfarin. The cap helix also rotates and is barely maintained in a helical conformation (fig. S11B). The cap loop and anchor domain interact differently, with Arg58 interacting with Glu67 in the 3-OH K state, whereas it inserts between the side chains of Glu67 and His68 when warfarin is bound. Warfarin was previously proposed to be a transition-state analog of 3-OH K because their chemical structures are similar (16, 30). This proposal, however, is not supported by the distinct structures in the warfarin and 3-OH K bound states. The polar 3-hydroxyl probably has prevented Phe55 from interacting with the naphthoquinone ring of 3-OH K (fig. S11B); Phe55, however, forms perpendicular π-π stacking interaction with the coumarin ring of warfarin (Fig. 2A). Moreover, this 3-hydroxyl does not participate in the hydrogen-bonding interaction as the 4-hydroxyl of warfarin does. On the other hand, the protein and ligand conformations in the warfarin-bound state are quite similar to those in the K-bound state (Fig. 4, E and F), suggesting that warfarin mimics a later step of the VKOR catalysis.
Discussion
Although extensive clinical experience with VKAs has been amassed over almost seven decades (32), there are still important questions as to how these anticoagulants inhibit VKOR, as well as the mechanism of this family of enzymes with natural substrates. Our structures reveal steps which may correspond to the catalytic cycle of VKOR (Fig. 5F and fig. S2) accompanied by protein conformational changes (Movie S3). In the cellular environment, ~50% of HsVKOR (and HsVKORL) is in the partially oxidized state (7, 21). At this state, we propose that the catalytic cycle begins with a reduced Cys135 (Fig. 5F, state I) that reacts with the substrates (K or KO), forming a charge-transfer or covalent complex that enables stable binding of the substrate (Fig. 5F, state II). Interactions of these adducts with the cap domain induce the closed conformation; the energy required for this conformational change probably originates from the binding and formation of the substrate adducts. In this closed conformation, Cys43 is juxtaposed with Cys51–Cys132 in an ideal position to undergo thiol-disulfide exchange (Fig. 4D). The resulting reduced Cys132 could in turn attack the Cys135–substrate adduct to generate the fully reduced reaction product (Fig. 5F, state III). Consequently, the active site becomes fully oxidized and the protein returns to its open conformation, resulting in a noncatalytic state that allows release of the reduced product (Fig. 5F, state IV). Notably, K is both a product and a substrate of VKOR catalysis (fig. S1A). The temporary binding of K at the low-affinity site (Fig. 4A) may allow K to quickly access Cys135 once it is reduced again, thereby expediting the reduction of KO to KH2 through the K intermediate. To return to the partially oxidized state, VKOR is likely reduced by partner proteins (33) or small molecules in the ER. Overall, the redox cycle of VKOR catalysis is associated with open and closed conformational changes. The closed conformation is induced by substrate binding and is conducive to later catalytic steps; this unique mechanism has not been observed before for the bacterial VKOR homolog or other membrane and soluble thiol oxidoreductases, such as DsbB and Ero1 (5, 34, 35).
The closed conformation induced by substrate binding closely resembles that induced by VKA binding (Fig. 4E). Comparison with the ligand-free structure show that the large luminal regions of these proteins adopt interconverting structures with remarkable plasticity and dynamics. The open and closed conformations are stabilized by electrostatic and hydrogen bonding interactions (fig. S6), which are relatively weak and may be readily biased by interaction with a ligand. Consequently, transitions between these conformations are utilized by substrates to trigger the catalytic cycle and by VKAs to achieve inhibition.
The binding of the VKAs and substate adducts both require hydrogen bonding to Asn80 and Tyr139. Tyr139 mutations result in the accumulation of a side-reaction product, 3-OH K, indicating that Tyr139 not only participates in substrate binding but is also essential to catalysis. Another important role of Tyr139 and Asn80 is that their hydrogen bonding should increase the redox potential of the naphthoquinone (36), thereby facilitating the substrate reduction. These hydrogen bonds may also provide particularly strong interactions to reaction transition states or intermediates and to VKAs owing to the hydrophobic environment at the active site in the closed conformation induced by bound substrates or VKAs. Overall, mimicking the hydrogen-bonding interactions required in the catalytic steps explains the very high affinity of VKA binding to VKOR, which is essentially irreversible (37).
VKAs simultaneously block two major steps of the catalytic cycle, the partially and fully oxidized states (Fig. 5F); these states together constitute ~90% of the HsVKOR cellular fraction (7) and both states are now captured in structures with warfarin bound (Fig. 2 and fig. S7). In contrast, only the partially oxidized enzyme is catalytically reactive to substrates, whereas the fully oxidized enzyme promotes product release (Fig. 4). VKAs and substrates occupy the same binding pocket and therefore should compete for the partially oxidized enzyme. On the other hand, the substrate reduction continuously generates the fully oxidized enzyme, which is removed by VKAs from further participating in the catalytic cycle (Fig. 5F). Inhibiting at both major states of the catalytic cycle enhances the potency of VKAs.
Overall, the crystal structures reported here reveal the conformational control and reaction chemistry of a family of integral membrane oxidoreductases and elucidate the multifaceted actions of their antagonists. The structures also explain the mechanism of warfarin resistance and the activity of warfarin metabolites, both of which contribute to warfarin dose variation (23); incorporating resistance profiles into warfarin dosage prediction may render better safety and accuracy. New therapeutic strategies of anticoagulation can be designed based on the different redox-state requirements between the VKA action and the substrate catalysis and based on the accompanied open and closed conformational changes.
Materials and Methods
Constructs and cloning
Codon optimized HsVKOR (accession code Q9BQB6; 3-155) and TrVKORL (NP_001027940; 6-175) were tagged at their two ends by the N- and C-halves of sfGFP (38). The DNAs encoding these constructs were cloned into a modified pPICZα expression vector (Invitrogen) by using a ligation-free PCR-based method (39). The Cys43Ser mutant of HsVKOR and Cys138Ser mutant of TrVKORL (corresponding to Cys132Ser in HsVKOR) were generated by site-directed mutagenesis (40). All nucleotide sequences were verified by DNA sequencing.
Protein expression and purification
The plasmids were linearized and transformed into Pichia pastoris by electroporation. Transformants were selected by Zeocin resistance on yeast extract peptone dextrose medium with sorbitol (YPDS) agar plates. The expression levels of resistant clones were compared through fluorescence-detection size-exclusion chromatography analysis (41). Clones with the highest expression levels were stored at −80°C.
For large-scale protein expression, 1 L culture was grown in the buffered minimal glycerol (BMG) media (1.2% glycerol, 0.34% yeast nitrogen base, 1% ammonium sulfate, 0.4 μg/mL biotin, and 100 mM potassium phosphate pH 6.0) at 30°C for 20 h. The growth media was then changed to 1 L buffered minimal methanol (BMM) media (0.34% yeast nitrogen base, 1% ammonium sulfate, 0.4 μg/mL biotin, and 200 mM potassium phosphate pH 6.0), and the protein expression were induced with 0.7% methanol. After 2 days at 25°C, the cells were harvested by centrifugation and flash frozen in liquid nitrogen.
Protein purification of wild-type TrVKORL and Cys138Ser mutant used the following procedure. Frozen cells (20 g) were broken by a ball mill (Retsch). The cell powder was resuspended in 40 mL lysis buffer (225 mM NaCl, 75 mM Tris-HCl pH 8.0, and 10 μg/mL DNase I). Subsequently, 1.2 g n-dodecyl-β-D-maltoside (DDM; 2% final concentration) was added to solubilize the membranes by stirring for 3 h at 4°C. The suspension was centrifuged at 130,000 g for 20 min. The supernatant was incubated with 3 mL TALON metal-affinity resin (Clontech) for 3 h at 4°C. The resin was subsequently collected on a gravity-flow column and washed with 60 mL wash buffer (10 mM imidazole, 150 mM NaCl, 0.2% DDM, and 20 mM Tris pH 8.0). The protein was eluted with 10 mL elution buffer (250 mM imidazole, 150 mM NaCl, 0.2% DDM, and 20 mM Tris pH 8.0). The eluted protein was concentrated and applied to Superdex 200 for size-exclusion chromatography (SEC) in a SEC buffer containing 0.05% lauryl maltose neopentyl glycol (LMNG, Anatrace), 150 mM NaCl, and 20 mM Tris pH 8.0. The peak fractions were collected and the TrVKORL protein was concentrated to 40 mg/mL and immediately used for crystallization.
This purification procedure was modified for the TrVKORL-warfarin complex in the following ways. The complex was generated by incubating the suspension of broken cells with 1 mM sodium salt of warfarin (Sigma) at 4°C for 20 min before the DDM solubilization. Warfarin at the 1 mM concentration was maintained throughout the membrane solubilization and immobilized metal-affinity chromatography (IMAC). However, the SEC buffer did not contain warfarin because excess amount of free warfarin interferes with crystallization.
To purify wild-type HsVKOR with VKAs, the suspension of broken cells was incubated with warfarin, phenindione, broadifacoum or chlorophacinone at 4°C for 20 min before the membrane solubilization. Warfarin was maintained at 1 mM concentration throughout the purification process, including the IMAC and SEC steps. The concentration of phenindione, broadifacoum, and chlorophacinone was 100 μM for IMAC and 500 μM or 1 mM for SEC. Purification of HsVKOR also required a lipid mixture, constituted of 0.1 mg/mL (final concentration) of 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (POPG) (Avanti) at a 3:1:1 molar ratio, in the IMAC and SEC buffers. The SEC buffer has the lipids and VKAs dissolved in 0.05% LMNG, 150 mM NaCl, and 20 mM Tris pH 8.0.
Purification of the Cys43Ser mutant of HsVKOR with warfarin used a similar protocol as that used for the wild-type protein with warfarin. To facilitate crystallization, the SEC buffer was changed to 200 mM potassium formate, 0.05% LMNG, 0.1 mg/mL lipids, and 200 μM warfarin.
Purification of the Cys43Ser mutant of HsVKOR with KO required the following modifications. The cell powder from 20 g cells was resuspended with 100 mL lysis buffer and sonicated to break the cell membranes. The membranes were collected by ultracentrifugation at 130,000 g for 45 min with a Beckman Ti45 rotor. The membrane pellet was homogenized into 40 mL lysis buffer and solubilized in 1% LMNG with 30 μM KO. KO at the same concentration was maintained in all the subsequent purification steps.
Crystallization and data collection
The wild-type and mutant HsVKOR and TrVKORL with different ligands were crystallized using the lipidic cubic phase (LCP) method (42). The protein and crystallization buffer conditions used for different constructs and different ligands are summarized in Table S3. For LCP crystallization, 40 mg/mL of protein was mixed with monoolein at a 2:3 ratio (v/v). For HsVKOR, all the ligands were added during purification, because this protein is unstable without a bound ligand. For TrVKORL, the ligands (except warfarin) were mixed with monoolein for co-crystallization with TrVKORL. The protein-monoolein mixture (0.1 μl) was covered by crystallization buffer (0.8 μl), and the LCP plates were incubated at 22°C. The crystals usually appeared within a week and grew to an optimal size after several weeks. The crystals were harvested from the mesophase bolus and flash-frozen by direct transfer to liquid nitrogen. Crystals embedded in the lipid bolus generally showed good diffraction under X-ray beams. The data were collected at the NECAT 24ID-C and 24ID-E beam lines at Advanced Photon Source, which are equipped with microdiffractometer-MD2 and Dectris's PILASTUS (24ID-C) and EIGER (24ID-E) detectors.
Structure determination
Molecular replacement with Phaser (43) used sfGFP (PDB code 2B3P) (14) as the search model. This initial sfGFP model was rigid-body refined with REFMAC (44). The partial phases were improved by solvent flattening, and in some cases by 2-fold non-crystallographic averaging, using the programs PARROT (45) and DM (46). The density modified maps were sufficient for automatic model building of the HsVKOR or TrVKORL region (~80–90% completeness) by the BUCCANEER software (45). The models were manually built to completion and refined against the data by using refinement programs installed in the PHENIX suites (47).
Activity assays
Cell-based activity assays were performed as previously described (48) using a HEK293 cell line that contains a chimeric FIXgla-Protein C gene and has endogenous VKOR and VKORL genes knocked out. Dual-expression plasmids containing HsVKOR, HsVKORL or TrVKORL constructs, along with a luciferase gene, were transfected into this double-knockout cell line. The carboxylation level of secreted FIXgla-PC in the cell-culture medium was measured by a sandwich ELISA, with luciferase activity serving as the control for transfection efficiency. The ELISA assay was conducted following a protocol previously reported (49). To obtain inhibition curves of VKAs and warfarin metabolites, the transfected cells were treated with 11 different concentrations of the compounds, with the concentration range optimized according to the dose response of each compound and each construct. The IC50s were analyzed using GraphPad Prism.
The activity assay using purified proteins followed a previously described protocol (8) with the following modifications. Purified HsVKOR, HsVKORL, and TrVKORL proteins (50 nM) were mixed with 50 μM K or 20 μM KO in a buffer containing 20 mM Tris-HCl pH 7.5, 0.1 M NaCl and 0.05% LMNG. The reaction was initiated by adding 5 mM DTT. The fluorescence of KH2 (excitation 250 nm and emission 430 nm) was detected using a plate reader (Molecular Devices).
Comparison of the cell-free activities of HsVKOR constructs (Fig. 5E, inset) used microsomes to provide a native-like membrane environment, thereby avoiding the use of detergent that may affect the enzymatic activity. Purification of microsomes followed a previous protocol (50) with the following modifications. Briefly, frozen Pichia cells were applied to a ball mill (Retsch) to break the cell walls. The cells were resuspended in 150 mM KCl and 50 mM HEPES pH 7.5, and sonicated to disrupt the cell membrane. After removal of cell debris, crude microsomes were collected by ultracentrifugation at 138,000 g for 1 h. The resuspended microsomes were purified through a step gradient of 35% and 60% sucrose (w/v), and the ER membranes were enriched after ultracentrifugation at 138,000 g for 12 h. Concentration of microsomal HsVKOR proteins was adjusted to the same level by their peak heights on FSEC, which used the fused GFP signal from solubilized microsomes. Catalysis of microsomal HsVKOR was initiated in a buffer containing 5 mM DTT, 5 μM KO, 150 mM KCl and 200 mM HEPES pH 7.5. The reactions were carried out at 30 °C for 2 h and subsequently analyzed by HPLC for the relative levels of KO and K (51).
NMR experiments
The selectively labeled (13C 2-methyl) vitamin K, purchased from Buchem BV, was used to synthesize (13C 2-methyl) KO and mercapto adducts (52). The 13C labeled 2-methyl KO was used as a substrate for all enzymatic and non-enzymatic reference reactions to facilitate the identification of different chemical species.
Purified TrVKORL Cys49Ser and Cys141Ser mutant proteins (corresponding to Cys43Ser and Cys135Ser in HsVKOR) at 30 μM concentration were incubated with 13C 2-methyl KO (150 μM) in a buffer containing 20 mM HEPES pH 7.5, 100 mM NaCl and 1 mM DDM at 4 °C overnight. Subsequently, 1 mM NEM (final concentration) was added to block free cysteines. For complete proteolysis of the mutant proteins, 5 % (wt/wt) protease K was added and the digestion was carried out at room temperature overnight. The vitamin K derivatives were extracted with isopropanol:hexane (3:2 v/v), and the upper hexane phase was collected and dried in argon. The residual was dissolved with deuterated chloroform and analyzed by NMR.
NMR experiments were carried out on a Bruker Avance III 600 MHz (14.09 T) spectrometer equipped with the cryoprobe. Deuterated chloroform (D-100%, CIL) was used as solvent for NMR experiments with all forms of vitamin K. The assignments of resonances that are available in previous literature (30, 52) were confirmed and extended to additional naphthoquinone protons and carbons using two-dimensional 1H-13C HSQC (53) and ADEQUATE experiments (54). The very limited quantities of derivatized forms of vitamin K were not separated into pure compounds. The mercapto and hydroxy modifications were distinguished based on their through-bond connectivities in ADEQUATE experiments, in which the C-2 carbon shifts were dominated by the changes in orbital hybridization of C-2 carbon. Consistently, in HSQC spectra 2-methyl group chemical shifts were strongly influenced by the ring current field of the naphthoquinone group.
The 1H and 13C resonances of 2-methyl group have readily distinguishable positions in different covalently modified forms of vitamin K (K, KO, 3-OH K and mercapto K), and the 13C label provided sufficient sensitivity in HSQC experiments. To observe a mercapto adduct (an analog of a cysteine adduct in VKOR reaction), we used a chemically synthesized racemic dithiothreitol (DTT) adduct of vitamin K (52).
Reactions with DTT and with VKOR mutants were carried out with a large excess of KO, and its signal dominated all resulting spectra; however, signals of derivative forms were readily observed. Compared to the mercapto adduct generated by using DTT, the enzymatic mercapto adduct isomers show additional signals, which result from heterogeneity of the proteolysis products.
Supplementary Material
Acknowledgment
We dedicate this paper to the memory of Dr. Evan Sadler, who has provided W.L. with generous support and invaluable advice as a mentor. We thank Dr. Daved Fremont for his critical reading of the manuscript.
Funding: W.L. is supported by NHLBI (R01 HL121718), W. M. Keck Foundation (Forefront of Science Award), Children’s Discovery Institute (MCII 2020-854), NEI (R21 EY028705), and NIGMS (R01 GM131008). The NECAT beamlines are funded by NIGMS (P30 GM124165), the Eiger detector is funded by a NIH-ORIP (HEI S10OD021527), and APS is supported by DOE (DE-AC02-06CH11357).
Footnotes
Competing interests: none.
Data and materials availability: Atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank under accession codes 6WV3 (HsVKOR-warfarin), 6WV6 (HsVKOR-phenindione), 6WVH (HsVKOR-Brodifacoum), 6WV7 (HsVKOR-Chlorophacinone), 6WV4 (HsVKOR Cys43Ser-warfarin), 6WV5 (HsVKOR Cys43Ser-KO), 6WVI (TrVKORL), 6WVB (TrVKORL-warfarin), 6WV9 (TrVKORL-K), 6WVA (TrVKORL-KO), and 6WV8 (TrVKORL ‘Cys132Ser’-K).
References and Notes
- 1.Goy J, Crowther M, Approaches to diagnosing and managing anticoagulant-related bleeding. Seminars in Thrombosis and Hemostasis 38, 702–710 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Schulman S, Beyth RJ, Kearon C, Levine MN, Hemorrhagic complications of anticoagulant and thrombolytic treatment: American College of Chest Physicians evidence-based clinical practice guidelines (8th edition). (2008), vol. 133. [DOI] [PubMed] [Google Scholar]
- 3.Budnitz DS, Lovegrove MC, Shehab N, Richards CL, Emergency hospitalizations for adverse drug events in older Americans. The New England journal of medicine 365, 2002–2012 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Stafford DW, The vitamin K cycle. J Thromb Haemost 3, 1873–1878 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Li W et al. , Structure of a bacterial homologue of vitamin K epoxide reductase. Nature 463, 507–512 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rishavy MA, Usubalieva A, Hallgren KW, Berkner KL, Novel insight into the mechanism of the vitamin K oxidoreductase (VKOR): electron relay through Cys43 and Cys51 reduces VKOR to allow vitamin K reduction and facilitation of vitamin K-dependent protein carboxylation. The Journal of biological chemistry 286, 7267–7278 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen G et al. , Warfarin traps human vitamin K epoxide reductase in an intermediate state during electron transfer. Nature Structural & Molecular Biology 24, 69–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Cheng W, Fowle Grider R, Shen G, Li W, Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer. Nature Communications 5, 3110–3110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammed A et al. , VKORC1L1, an enzyme rescuing the vitamin K 2,3-epoxide reductase activity in some extrahepatic tissues during anticoagulation therapy. Journal of Biological Chemistry 288, 28733–28742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeer C, Hamulyak K, Vitamin K: lessons from the past. Journal of Thrombosis and Haemostasis 2, 2115–2117 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Goodstadt L, Ponting CP, Vitamin K epoxide reductase: homology, active site and catalytic mechanism. Trends in biochemical sciences 29, 2002–2005 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Ansell J et al. , Descriptive analysis of the process and quality of oral anticoagulation management in real-life practice in patients with chronic non-valvular atrial fibrillation: the international study of anticoagulation management (ISAM). J Thromb Thrombolysis 23, 83–91 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Chu P-H, Huang T-Y, Williams J, Stafford DW, Purified vitamin K epoxide reductase alone is sufficient for conversion of vitamin K epoxide to vitamin K and vitamin K to vitamin KH2. Proceedings of the National Academy of Sciences of the United States of America 103, 19308–19313 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS, Engineering and characterization of a superfolder green fluorescent protein. Nature Biotechnology 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Link KP, The discovery of dicumarol and its sequels. Circulation 19, 97–107 (1959). [DOI] [PubMed] [Google Scholar]
- 16.Gebauer M, Synthesis and structure-activity relationships of novel warfarin derivatives. Bioorganic & medicinal chemistry 15, 2414–2420 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Porter WR, Warfarin: history, tautomerism and activity. J Comput Aided Mol Des 24, 553–573 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Cooper GM et al. , A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 112, 1022–1027 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haque JA, McDonald MG, Kulman JD, Rettie AE, A cellular system for quantitation of vitamin K cycle activity: Structure-activity effects on vitamin K antagonism by warfarin metabolites. Blood 123, 582–589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen G et al. , Membrane Protein Structure in Live Cells: Methodology for Studying Drug Interaction by Mass Spectrometry-Based Footprinting. Biochemistry 57, 286–294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen G et al. , Stabilization of warfarin-binding pocket of VKORC1 and VKORL1 by a peripheral region determines their different sensitivity to warfarin inhibition. Journal of Thrombosis and Haemostasis 16, 1164–1175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinhadri BCS, Jin DY, Stafford DW, Tie JK, Vitamin K epoxide reductase and its paralogous enzyme have different structures and functions. Scientific Reports 7, 17632–17632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Warfarin Pharmacogenetics C et al. , Estimation of the warfarin dose with clinical and pharmacogenetic data. The New England Journal of Medicine 360, 753–764 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wigle TJ, Jansen LE, Teft WA, Kim RB, Pharmacogenomics Guided-Personalization of Warfarin and Tamoxifen. J Pers Med 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Liu S, Yang Y, Li W, Characterization of Warfarin Inhibition Kinetics Requires Stabilization of Intramembrane Vitamin K Epoxide Reductases. J Mol Biol 432, 5197–5208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silverman RB, Chemical Model Studies for the Mechanism of Vitamin K Epoxide Reductase. J. Am. Chem. SOC 103, 5939–5941 (1981). [Google Scholar]
- 27.Davis CH et al. , A quantum chemical study of the mechanism of action of Vitamin K epoxide reductase ( VKOR ). Journal of Molecular Graphics and Modelling 26, 401–408 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Inaba K, Takahashi Y.-h., Ito K, Hayashi S, Critical role of a thiolate-quinone charge transfer complex and its adduct form in de novo disulfide bond generation by DsbB. Proceedings of the National Academy of Sciences of the United States of America 103, 287–292 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller SM et al. , Use of a Site-Directed Triple Mutant To Trap Intermediates: Demonstration That the Flavin C(4a)-Thiol Adduct and Reduced Flavin Are Kinetically Competent Intermediates in Mercuric Ion Reductase. Biochemistry 29, 2831–2841 (1990). [DOI] [PubMed] [Google Scholar]
- 30.Fasco MJ, Preusch PC, Hildebrandt E, Suttie JW, Formation of hydroxyvitamin K by vitamin K epoxide reductase of warfarin-resistant rats. The Journal of biological chemistry 258, 4372–4380 (1983). [PubMed] [Google Scholar]
- 31.Matagrin B et al. , New insights into the catalytic mechanism of vitamin K epoxide reductase (VKORC1) - The catalytic properties of the major mutations of rVKORC1 explain the biological cost associated to mutations. FEBS open bio 3, 144–150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim GB, Milestone 2: Warfarin: from rat poison to clinical use. Nat Rev Cardiol, (2017). [DOI] [PubMed] [Google Scholar]
- 33.Schulman S, Wang B, Li W, Rapoport T. a., Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proceedings of the National Academy of Sciences of the United States of America 107, 15027–15032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross E, Kastner DB, Kaiser CA, Fass D, Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell 117, 601–610 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Inaba K et al. , Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation. Cell 127, 789–801 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Hui Y, Chng EL, Chua LP, Liu WZ, Webster RD, Voltammetric method for determining the trace moisture content of organic solvents based on hydrogen-bonding interactions with quinones. Anal Chem 82, 1928–1934 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Fasco MJ, Principe LM, R- and S-warfarin inhibition of vitamin K and vitamin K 2,3-epoxide reductase activities in the rat. Journal of Biological Chemistry 257, 4894–4901 (1982). [PubMed] [Google Scholar]
- 38.Cabantous S, Terwilliger TC, Waldo GS, Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nature Biotechnology 23, 102–107 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Li C et al. , FastCloning: A highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnology 11, 92–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng L, Baumann U, Reymond JL, An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic acids research 32, e115–e115 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hattori M, Hibbs RE, Gouaux E, A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 20, 1293–1299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, Cherezov V, Crystallization of membrane proteins in lipidic mesophases. Journal of visualized experiments : JoVE 49, 2501–2501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy AJ et al. , Phaser crystallographic software. Journal of applied crystallography 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ, Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D Biological Crystallography 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Cowtan KD, Recent developments in classical density modification. Acta crystallographica. Section D, Biological crystallography 66, 470–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cowtan KD, 'dm': An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 (1994). [Google Scholar]
- 47.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica. Section D, Biological crystallography 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tie JK, Jin DY, Tie K, Stafford DW, Evaluation of warfarin resistance using transcription activator-like effector nucleases-mediated vitamin K epoxide reductase knockout HEK293 cells. Journal of Thrombosis and Haemostasis 11, 1556–1564 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tie J-K, Jin D-Y, Straight DL, Stafford DW, Functional study of the vitamin K cycle in mammalian cells. Blood 117, 2967–2974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bevans CG et al. , Determination of the warfarin inhibition constant Ki for vitamin K 2,3-epoxide reductase complex subunit-1 (VKORC1) using an in vitro DTT-driven assay. Biochimica et biophysica acta 1830, 4202–4210 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K, Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 427, 537–541 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Preusch PC, Suttie JW, A chemical model for the mechanism of vitamin K epoxide reductase. The Journal of Organic Chemistry 48, 3301–3305 (1983). [Google Scholar]
- 53.Schleucher J et al. , A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. Journal of Biomolecular NMR 4, 301–306 (1994). [DOI] [PubMed] [Google Scholar]
- 54.Martin GE, Williamson RT, Dormer PG, Bermel W, Inversion of 1J(CC) correlations in 1,n-ADEQUATE spectra. Magn Reson Chem 50, 563–568 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.