Abstract
Background:
Little is known about mechanisms of resistance to poly(adenosine diphosphate-ribose) polymerase inhibitors (PARPi) and platinum chemotherapy in patients with metastatic breast cancer and BRCA1/2 mutations. Further investigation of resistance in clinical cohorts may point to strategies to prevent or overcome treatment failure.
Patients and methods:
We obtained tumor biopsies from metastatic breast cancer patients with BRCA1/2 deficiency before and after acquired resistance to PARPi or platinum chemotherapy. Whole exome sequencing was carried out on each tumor, germline DNA, and circulating tumor DNA. Tumors underwent RNA sequencing, and immunohistochemical staining for RAD51 foci on tumor sections was carried out for functional assessment of intact homologous recombination (HR).
Results:
Pre- and post-resistance tumor samples were sequenced from eight patients (four with BRCA1 and four with BRCA2 mutation; four treated with PARPi and four with platinum). Following disease progression on DNA-damaging therapy, four patients (50%) acquired at least one somatic reversion alteration likely to result in functional BRCA1/2 protein detected by tumor or circulating tumor DNA sequencing. Two patients with germline BRCA1 deficiency acquired genomic alterations anticipated to restore HR through increased DNA end resection: loss of TP53BP1 in one patient and amplification of MRE11A in another. RAD51 foci were acquired post-resistance in all patients with genomic reversion, consistent with reconstitution of HR. All patients whose tumors demonstrated RAD51 foci post-resistance were intrinsically resistant to subsequent lines of DNA-damaging therapy.
Conclusions:
Genomic reversion in BRCA1/2 was the most commonly observed mechanism of resistance, occurring in four of eight patients. Novel sequence alterations leading to increased DNA end resection were seen in two patients, and may be targetable for therapeutic benefit. The presence of RAD51 foci by immunohistochemistry was consistent with BRCA1/2 protein functional status from genomic data and predicted response to later DNA-damaging therapy, supporting RAD51 focus formation as a clinically useful biomarker.
Keywords: BRCA1, BRCA2, breast cancer, PARP inhibitor, platinum
INTRODUCTION
Approximately 5% of breast cancer patients carry germline mutations in BRCA1 or BRCA2, tumor suppressor genes that function in the repair of DNA double-stranded breaks by homologous recombination (HR).1 BRCA1/2-deficient cancers are particularly sensitive to two classes of DNA-damaging therapies: platinum chemotherapy and poly (adenosine diphosphate-ribose) polymerase inhibitors (PARPi).1,2 Platinum agents are increasingly used in patients with metastatic breast cancer (MBC) and germline BRCA1/2 mutations, and two PARPi (olaparib and talazoparib) obtained US Food and Drug Administration approval for this indication in 2018. As such, it is important to understand how resistance occurs and to develop biomarkers predictive of response.
Previously described mechanisms of resistance to PARPi or platinum chemotherapy fall into two main categories: alteration of a protein in the HR pathway (including acquired re-expression of functional BRCA protein, known as reversion),3,4 and altered expression of a protein in the replication fork protection pathway.5 Though BRCA1 or BRCA2 reversions have been described in many clinical cohorts, non-reversion mechanisms of resistance are almost exclusively described in in vitro models, and much of the clinical work has been in ovarian and prostate cancer,4,6–8 with fewer investigations in breast cancer.9,10
The goal of this study was to use tumor sequencing to identify both reversion and non-reversion mechanisms of acquired resistance to PARPi or platinum chemotherapy in patients with BRCA1/2-deficient MBC, and to explore RAD51 focus formation (a marker of intact HR) as a clinically useful biomarker of resistance to PARPi and platinum chemotherapy.
METHODS
Cohort
All patients provided written informed consent for research biopsies and sequencing, as approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board (Protocol 05–246). We identified all patients who had germline or somatic deleterious BRCA1 or BRCA2 alteration and a tissue biopsy carried out following acquired resistance to PARPi or platinum therapy between July 2015 and July 2017. Acquired resistance was defined as >3 months of complete response, partial response, or stable disease, followed by disease progression on therapy. Patient/tumor characteristics and breast cancer treatment history were extracted from the medical record.
Tumor and blood sequencing
DNA extraction and construction of libraries for massively parallel sequencing were carried out as previously described.11 Cell-free DNA was isolated and circulating tumor DNA (ctDNA) was sequenced using the ichorCNA method, as previously described and noted in supplementary Methods, available at Annals of Oncology online.12 Analysis pipeline details follow in supplementary Methods, available at Annals of Oncology online.
Identification of reversions
Short frame-restoring indels of length <100 bp were identified using the 2-out-of-3 voting scheme described in the supplementary Methods, available at Annals of Oncology online, regarding somatic alterations. Longer deletions of length ≥100 bp were identified using SvABA, and the resulting variant call format was annotated using svaba-annotate. R and AnnotSV version 1.2.13 Long deletions identified in BRCA1 and BRCA2 were checked for validity and impact to the reading frame via manual inspection of the raw reads (*.alignments.txt.gz) aligned to contigs assembled by SvABA.
Immunohistochemical staining
For RAD51 staining, serial sections of formalin-fixed, paraffin-embedded tumor biopsies were stained as previously described14,15 using antibodies to RAD51 and Geminin independently. A sample was classified as HR-proficient if more than three RAD51 foci were present in a minimum of one cell in three ×40 fields. If RAD51 foci were absent, the sample was classified as HR-deficient if >3% of the cells were Geminin-positive. If there were no RAD51 foci and <3% of the cells were Geminin-positive, the proliferation rate of the tumor was classified as low and HR status could not be determined. Formalin-fixed, paraffin-embedded sections of a cell line block containing irradiated and unirradiated HR-proficient (HCC1569) and HR-deficient (MDA-MB-436) breast cancer cell lines were used as positive and negative controls.14,15
RESULTS
Patient and tumor characteristics
Eight patients with MBC, germline and/or somatic inactivating mutation in BRCA1 or BRCA2, and acquired resistance to any PARPi or platinum chemotherapy were identified (Table 1).
Table 1.
Clinico-pathologic data, reversion status, and RAD51 staining results
| Patient ID | Germline mutationa | Age at dx (years) | Stage at dx | Receptor status at dx | DNA-damaging tx received (line of MBC tx)c | Duration of response (months) | Best overall response | Reason for stopping tx | BRCA reversion status post-DNA- damaging tx | RAD51 foci status pre- DNA-damaging tx | RAD51 foci status post- DNA-damaging tx | Subsequent DNA-damaging tx exposure | Best response to subsequent DNA-damaging tx |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 292 | BRCA1 p.E23Vfs*17 | 44 | II | ER+/PR+/HER2− | Carboplatin (4th line) | 9 | SD | PD | No | UNK | UNK | None | NA |
| 303 | BRCA2 c.e9–1 (splice site, somatic) | 43 | III | ER+/PR+/HER2− | Cisplatin (5th line) | 15 | CR | PD | No | Absent | Absent | None | NA |
| 318 | BRCA2 p.E1493fs*10 | 51 | I | ER+/PR+/HER2− | Olaparib (5th line) | 11 | SD | PD | Yes, definite | Absent | Present | Carboplatin | Intrinsic resistance |
| 339 | BRCA2 p.L1908fs*2 | 30 | III | ER+/PR+/HER2− | Carboplatin (6th line) | 6 | UNK | PD | Yes, definite | Absent | Present | None | NA |
| 349 | BRCA1 p.S454* | 29 | I | ER+/PR+/HER2− | Olaparib (4th line) | 11 | PaR | PD | Yes, definite | Absent | Present | None | NA |
| 359 | BRCA1 p.T1677fs*2 | 53 | II | ER+/PR−/HER2− | Olaparib (2nd line) | 4 | SD | PD | No | Absent | Present | Carboplatin | Intrinsic resistance |
| 510 | BRCA2 p.S1720fs*7 | 58 | III | ER+/PR+/HER2− | Carboplatin ± veliparibb (6th line) | 9 | PaR | PD | Yes, putative | Absent | Present | None | NA |
| 565 | BRCA1 p.E23Vfs*16 | 42 | II | ER−/PR−/ HER2− | Veliparib (2nd line) | 20 | PaR | PD | No | Absent | Present | Carboplatin and olaparib (separate regimens) | Intrinsic resistance to both |
Anatomic stage is indicated. Best overall response was determined by RECIST (if patient on a clinical trial) or chart review by a breast medical oncologist (if patient not on a clinical trial). Intrinsic resistance is defined as progressive disease at first restaging or clinical progression before first restaging.
CR, complete response; dx, diagnosis; ER, estrogen receptor; fs, frameshift; MBC, metastatic breast cancer; NA, not applicable; ns, nonsense mutation; PD, progressive disease; PaR, partial response; PR, progesterone receptor; SD, stable disease; tx, treatment; UNK, unknown.
One patient (patient 303) had a somatic mutation in BRCA2.
This patient underwent blinded randomization to veliparib or placebo on a clinical trial.
Timelines indicating all treatments received by all patients, with to-scale durations, are shown in Figure 1, supplementary Figures S1 and S2, available at Annals of Oncology online, and Figure 3. Only DNA-damaging treatment associated with acquired resistance is indicated in the table (and with red arrows in treatment timelines). No patients received platinum or poly(adenosine diphosphate-ribose) polymerase inhibitors for non-metastatic disease.
BRCA1/2 reversions identified following platinum or PARPi therapy
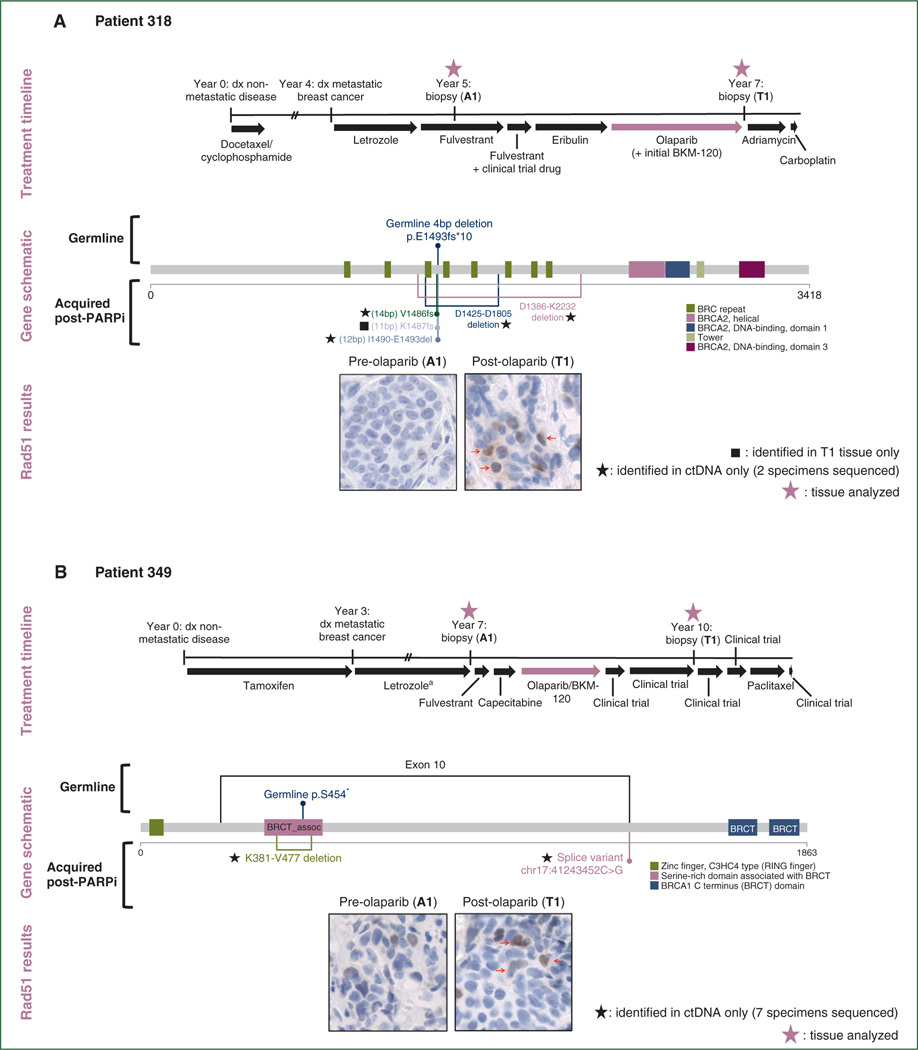
Four of eight patients demonstrated definite or putative reversion to a functional BRCA1 or BRCA2 open reading frame following acquired resistance (Figure 1, supplementary Figure S1, supplementary Table S1, available at Annals of Oncology online). These identified reversions suggested restoration of intact HR through reconstitution of functional BRCA1/2 protein as a likely mechanism of resistance to PARPi/platinum chemotherapy. Additionally, in all four patients, the pre-resistance tumors had no RAD51 foci whereas the post-resistance tumors did, indicative of HR restoration (Figure 1, supplementary Figure S1, available at Annals of Oncology online). We categorized reversion events as definite (patients 318, 339, and 349) if acquired restoration of BRCA1 or BRCA2 open reading frame could be concluded directly from gene sequence for one or more genomic event, and as putative (patient 510) if open reading frame sequence could not be directly concluded, but genomic events nearby the original inactivating event were newly acquired and accompanied by RAD51 foci post-resistance.
Figure 1. Reversions identified in BRCA1 and BRCA2 following exposure to PARP inhibitor or platinum.
(A) Following olaparib exposure, patient 318 acquired three short deletions immediately upstream of or encompassing the germline frameshift deletion in BRCA2, and two long in-frame deletions encompassing the germline frameshift deletion and contained within BRCA2 exon 11. Each of these acquired alterations is expected to restore BRCA2 open reading frame, re-establishing homologous recombination proficiency, and this is supported by the reacquisition of RAD51 foci in the post-resistance biopsy. (B) Patient 349 had a germline nonsense mutation in BRCA1 exon 10 and acquired an exon 10 splice site mutation following resistance to olaparib, with accompanying reacquisition of RAD51 foci. In addition, this patient acquired an in-frame deletion encompassing the germline frameshift deletion.
No clinical trials involving PARPi/platinum or other therapy specifically for BRCA1/2-mutant patients occurred between indicated sequenced biopsies. DNA-damaging treatment associated with acquired resistance is indicated by thick red arrow in the treatment timelines. Treatment timelines are to scale unless noted; double hash marks indicate treatment duration longer than diagrammed. Small red arrows identify cells with positive staining for RAD51 foci.
ctDNA, circulating tumor DNA; dx, diagnosis; PARP, poly(adenosine diphosphate-ribose) polymerase inhibitor.
a Exact treatment duration unknown.
Seven patients had successful sequencing of at least one ctDNA specimen (including all four patients with reversions) (supplementary Table S2, available at Annals of Oncology online). All ctDNA specimens were drawn at or after the post-resistance tumor sampling timepoint. In the four patients with reversions, there were 10 reversion events identified: 6 found in blood only, 3 in tumor only, and 1 in blood and tumor. Two patients had reversions identified only in ctDNA; in both cases, more samples were sequenced from ctDNA than from tumor (possibly explaining the discrepancy).
Genomic analysis of acquired resistance pathways
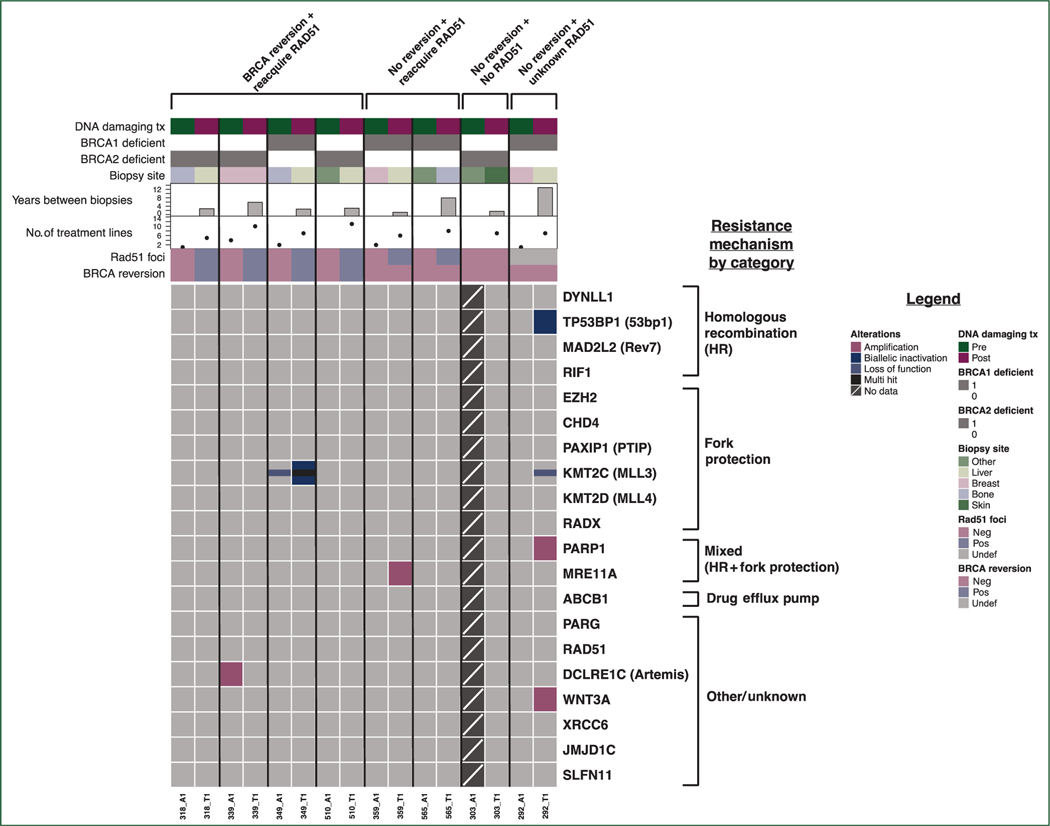
We compared tumor whole exome sequencing before and after acquired resistance to PARPi or platinum chemotherapy to identify potential non-reversion mechanisms of resistance to therapy. We analyzed single nucleotide variants and copy number variants in 20 genes from pathways previously linked to PARPi/platinum response or resistance in preclinical models and/or clinical specimens (Figure 2, supplementary Table S3, available at Annals of Oncology online).
Figure 2. Single nucleotide variant (SNV) and copy number variant (CNV) events in 20 genes from PARPi/platinum resistance pathways.
Co-mutation plot showing SNV and CNV events in 20 genes from pathways previously implicated in resistance to poly(adenosine diphosphate-ribose) polymerase inhibitors (PARPi) or platinum, across 15 metastatic breast cancer tumor samples obtained either before or after the acquisition of resistance to PARPi or platinum. Each column of data represents a unique tumor specimen; each row represents a gene of interest. No pre-resistance tumor sample was available for sequencing in patient 303. Horizontal tracks along the top of the plot indicate select clinical parameters for each specimen, presence or absence of detected BRCA reversion in either tumor tissue or circulating tumor DNA, and presence or absence of RAD51 foci staining.
Neg, negative; Pos, positive; Undef, undefined.
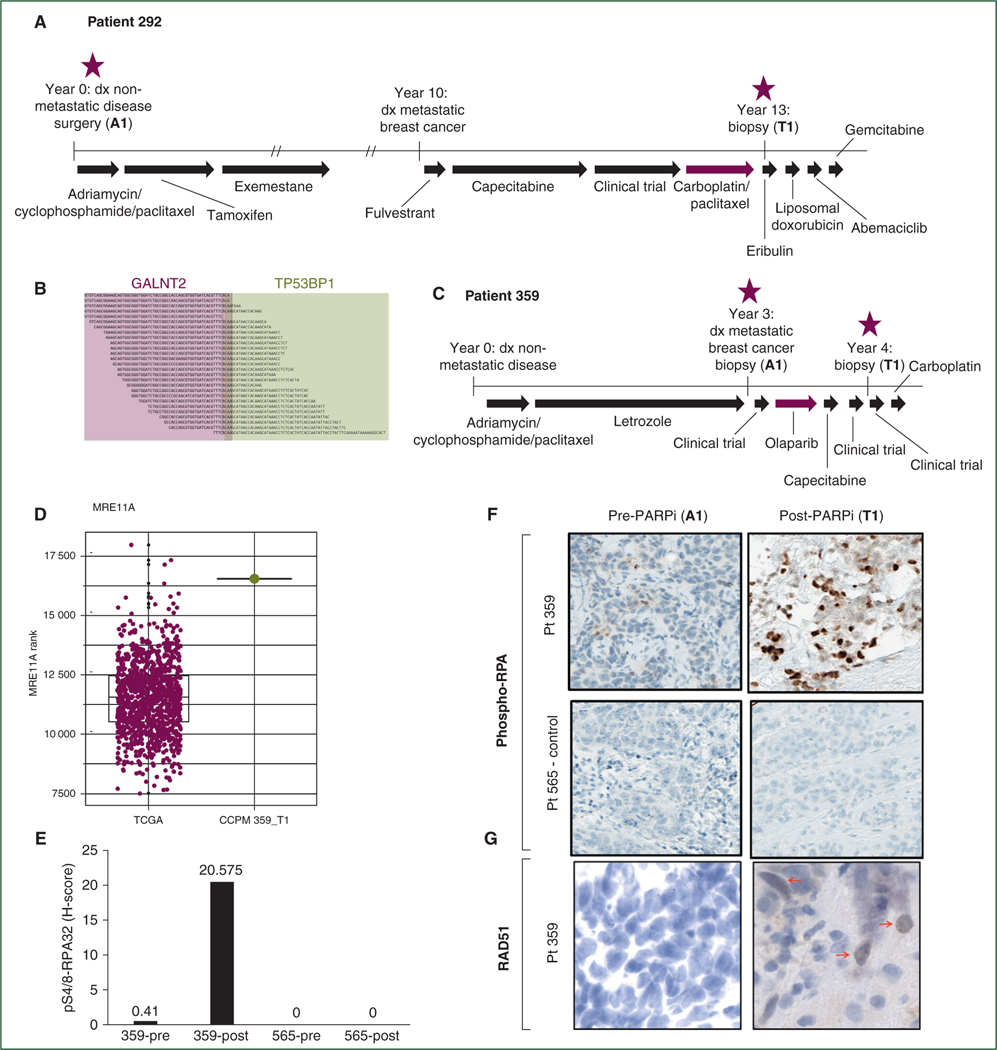
In two patients, we identified acquired genomic alterations anticipated to lead to HR restoration through increased DNA end resection. Patient 292 (pathogenic BRCA1 germline mutation and no reversion post-carboplatin; Figure 3A) acquired biallelic inactivation in TP53BP1 (Figure 3B). Low expression of TP53BP1 was also observed in the post-resistance tumor specimen. Loss of TP53BP1 is expected to facilitate BRCA1-independent end resection following double-stranded DNA breaks, since loss of 53BP1 in vitro restores HR in cells lacking BRCA1, leading to PARPi/platinum resistance despite maintained BRCA1 deficiency.16–18 Tissue was insufficient for RAD51 staining or 53BP1 protein staining.
Figure 3. Genomic alterations in TP53BP1 and MRE11A acquired in two patients with germline BRCA1 deficiency.
(A) Patient 292 had a germline deleterious BRCA1 mutation and acquired resistance to a carboplatin-containing regimen. (B) In a post-resistance tissue biopsy, patient 292 acquired biallelic inactivation of the gene TP53BP1 (loss of heterozygosity plus antisense fusion between TP53BP1 and GALNT2). (C) Patient 359 had a germline deleterious BRCA1 mutation and acquired resistance to olaparib. (D) In a post-resistance tissue biopsy, patient 359 showed very high RNA expression of MRE11A; figure shows comparison of MRE11A expression in all breast tumors from The Cancer Genome Atlas (N = 1093; burgundy dots) versus MRE11A expression in patient 359 post-resistance specimen (green dot; at 99.7th percentile of The Cancer Genome Atlas samples). (E) Pre- and post-resistance tumor biopsies from patient 359 show increase in phospho-RPA protein staining post-resistance; patient 565 (whose tumor showed no genomic evidence of acquired increased end resection post-resistance) is shown as a control. (F) Representative histology images of phospho-RPA stain for patient 359 pre- and post-resistance (patient 565 is again shown as a control). (G) Reacquisition of RAD51 foci following acquired resistance to olaparib in patient 359. Red arrows identify cells with positive staining for RAD51 foci. Treatment timelines are to scale unless noted; double hash marks indicate treatment duration longer than diagrammed.
dx, diagnosis; PARPi, poly(adenosine diphosphate-ribose) polymerase inhibitors.
Patient 359 (pathogenic BRCA1 germline mutation and no reversion post-olaparib; Figure 3C) acquired amplification of MRE11A, which encodes a DNA exonuclease that functions in end resection.19 MRE11A amplification could plausibly lead to PARPi resistance by increasing end resection at double-stranded DNA breaks, therefore restoring HR proficiency in tumor cells despite BRCA1 deficiency. Consistent with this, RNA expression of MRE11A was high in the post-resistance tumor specimen (Figure 3D), and tumor staining for phospho-RPA (a marker of DNA end resection) was substantially increased post-resistance (Figure 3E and F). Moreover, RAD51 foci were reacquired at the post-olaparib timepoint (Figure 3G). In this sample, we propose that the acquisition of MRE11A amplification is the likely biological mechanism of olaparib resistance.
One additional acquired genomic alteration that could potentially contribute to resistance was seen in the 20 genes analyzed. Patient 349 acquired biallelic inactivation of KMT2C, a histone methyltransferase necessary for the presence of MRE11 at replication forks,5,20 suggesting that a replication fork-stabilizing event may have occurred that conferred PARPi resistance.
Broader analysis of genomic alterations occurring in a larger set of 276 genes involved in all DNA damage repair processes in cancer21 did not reveal any mechanisms beyond those identified earlier. Given evidence that gene fusions driving overexpression of Abcb1 (drug efflux pump) can cause resistance to PARPi,4,9 we examined all gene fusion events; no relevant fusions were identified. Following all analyses, acquired resistance in patients 303 and 565 remained unexplained (supplementary Figure S2, available at Annals of Oncology online).
RAD51 foci and resistance to subsequent lines of platinum chemotherapy or PARPi
Of the six patients who acquired RAD51 foci following exposure to platinum chemotherapy or PARPi, three went on to have subsequent exposure to a different DNA-damaging therapy (Table 1), and all displayed intrinsic resistance to subsequent PARPi and/or platinum. By contrast, RAD51 foci were absent from all tested tumors before initial PARPi or platinum therapy (Figure 2), among patients selected for initial response to therapy. The data are consistent with the premise that intact or impaired HR, measured by the presence or absence of RAD51 foci, correlates with response to PARPi/platinum agents in BRCA1/2-deficient tumors, though the number of patients in this cohort is too small to allow definitive conclusions.
DISCUSSION
In this study, we used whole exome sequencing of tumor and blood, RNA sequencing of tumor, and formation of RAD51 foci by immunohistochemistry to interrogate resistance to PARPi or platinum chemotherapy and its correlation with tumor HR proficiency in a cohort of patients with MBC. Reversions were identified in one-half of patients, and sequencing data suggest additional biologically plausible non-reversion mechanisms of resistance, including amplification of MRE11A and biallelic inactivation of TP53BP1. The presence or absence of RAD51 foci correlated with resistance or response, respectively, to DNA-damaging therapy.
Reversion to protein-coding BRCA1 or BRCA2 transcript has been previously reported following exposure to platinum chemotherapy and/or PARPi, both in preclinical models of BRCA1/2-deficient tumor cells and in patients with BRCA1/2-mutated breast, ovarian, and prostate cancer.4,6,7,10,22,23 Prior evidence supports the biological plausibility of each revertant mechanism observed in this cohort.22–26 Of note, two patients with reversion received combined olaparib and phosphoinositide 3-kinase inhibitor, posited to synergize through a metabolic mechanism that is not expected to impact the chance of reversion as a specific mechanism of resistance.27
Despite co-sampling of tissue and ctDNA in seven of eight patients, and identification of reversions using both methods, the majority of reversion events were not shared between tumor and blood specimens. The preponderance of events identified in blood only may reflect the fact that multiple post-resistance blood specimens were sequenced in most patients (compared with only a single post-resistance tumor specimen sequenced in all patients). The discordance also highlights the limitations of a single tumor or blood sample in isolation to comprehensively capture heterogeneous genomics across multiple different metastatic lesions at distinct timepoints, and is consistent with a previous report showing incomplete overlap between reversions identified in blood versus tumor.6
Though genomic reversion is a frequently reported mechanism of clinical resistance to PARPi or platinum among patients with BRCA1/2-mutant tumors, as observed in our cohort, not all patients revert. At present, there are no known parameters to predict which patients will acquire somatic reversions and which will not. Work to compile and map reversions and associated germline mutations identified across tumor types may indicate whether the location and/or type of germline mutation in BRCA1/2 could assist in predicting which patients will experience reversion and which will not.
We identified two non-revertant patients in whom genomic evidence supports the acquisition of resistance through up-regulation of DNA end resection. Loss of 53BP1, a protein involved in DNA end resection, has been shown to restore HR functionality in BRCA1-deficient cells, and to eliminate the cells’ platinum/PARPi sensitivity.17,18 Reduced 53BP1 has also been described in platinum and PARPi-resistant ovarian cancer patient tumor specimens and patient-derived xenograft models, but to our knowledge has never been demonstrated as a mechanism of resistance in breast tumor specimens.28–30 53BP1 Normally inhibits the activity of MRE11 at DNA double-stranded breaks.5 MRE11A amplification (patient 359 post-olaparib) has not previously been reported as a mechanism of resistance to PARPi/platinum, but is plausible as an alternative means to promote DNA end resection. This biology is supported by increased phospho-RPA staining in our patient’s tumor, and represents a potential novel mechanism of resistance identified in this cohort. MRE11 also plays a role in replication fork degradation, and in this context, a theoretical consequence of its amplification could actually be increased sensitivity to PARPi/platinum.5,16 However, the broad evidence supporting 53BP1 loss as a resistance mechanism, the increase in phospho-RPA staining, and the fact that this patient’s tumor regained RAD51 foci, all contradict this as a predominant biological effect in this tumor. Overall, our results represent direct evidence of increased DNA end resection via 53BP1 loss or MRE11 up-regulation as clinically relevant mechanisms of resistance to PARPi/platinum in BRCA1-deficient breast tumors. Though numbers are too small to draw any conclusions about the broader prevalence of these mechanisms, further examination of the DNA end resection pathway—53BP1 and MRE11 in particular—is warranted in larger cohorts.
Our results suggest that immunohistochemical staining for RAD51 foci offers real-time assessment of a tumor’s HR proficiency, and correlates with response and resistance to PARPi/platinum therapy. The presence of RAD51 foci has been shown to correlate with decreased efficacy of PARP inhibition.30,31 We demonstrate that the presence or absence of RAD51 staining changes over time as predicted with HR-restoring mechanisms of resistance. RAD51 staining should be investigated as a predictive biomarker in larger cohorts of BRCA1/2-deficient patients treated with PARPi/platinum, as it has many potential clinical advantages. Rapidly available staining results could impact decisions about immediate next-line therapy, while offering a simple ‘on/off’ indicator of HR status—agnostic to specific underlying biology—to guide the use of any HR-disrupting treatment strategies in the clinic.
Our study has several limitations. Though the availability of paired tumor tissue offers a unique opportunity to examine resistance mechanisms, and, to our knowledge, this represents the largest such cohort of MBC patients reported to date, the cohort size is small. Due to the initial focus of the tissue collection protocol, most patients in the cohort have hormone receptor-positive breast cancer, which is not representative of the overall population of BRCA1/2 carriers with MBC. As this was not a treatment-based clinical trial, therapies received and biopsy timepoints are heterogeneous, and the biopsies carried out do not exactly bracket the PARPi/platinum treatments received. While it is not possible to conclude that the resistance mechanisms identified specifically resulted from selective pressure of PARPi/platinum, each mechanism highlighted has been previously reported to result from PARPi/platinum exposure, or, in the case of MRE11A amplification, is in a known resistance pathway. Though RAD51 staining is a proxy for overall HR function, there was insufficient tissue for specific BRCA1/2 functional assays which could indicate, for example, the presence of a functional hypomorph protein as a driver of resistance.
In this cohort of eight patients with MBC and BRCA1/2-deficient tumors who acquired resistance to PARPi or platinum therapy, we identified biologically plausible mechanisms of resistance in six patients. The fact that HR restoration explained resistance in the majority of this cohort suggests that HR-disrupting strategies (e.g. inhibition of phosphoinositide 3-kinase or cyclin-dependent kinases), or strategies disrupting both HR and replication fork stability (e.g. inhibition of ATR or CHK1)3 may represent the best opportunities to resensitize patients to PARPi or platinum therapies. Immunohistochemical assessment of RAD51 should be further explored as a predictive tool since, if validated, this biomarker could help clinicians to select optimal treatment regimens for patients with BRCA1/2-deficient tumors.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by an American Society of Clinical Oncology Young Investigator Award (AGW), the Breast Cancer Research Foundation (AGW, NUL), Specialized Program of Research Excellence (SPORE) in Breast Cancer National Institutes of Health [grant number P50 CA168504] (AGW, IEK, ADD, EPW, NUL, GIS, NW), a National Cancer Institute-Cancer Therapy Evaluation Program (CTEP) Biomarker Supplement to NIH [grant number UM1 CA186709] (BK, ADD, GIS), the Fashion Footwear Association of New York (to Dana-Farber Cancer Institute Breast Oncology Program), the National Comprehensive Cancer Network/Pfizer Collaborative Grant Program (NUL), Friends of Dana-Farber Cancer Institute (to NUL), Yale Cancer Center [grant number 1UM1CA86689–05] (PL), Department of Defense [grant number W81XWH-13–1-0032] (NW), AACR Landon Foundation [grant number 13–60-27-WAGL] (NW), Susan G. Komen [grant number CCR15333343] (NW), The V Foundation (NW), The Breast Cancer Alliance (NW), The Cancer Couch Foundation (NW), Twisted Pink (NW), Hope Scarves (NW), ACT NOW (to Dana-Farber Cancer Institute Breast Oncology Program). In addition, we thank Dana-Farber/Harvard Cancer Center in Boston, MA, for the use of the Specialized Histopathology Core, which provided immunohistochemical staining. Dana-Farber/Harvard Cancer Center is supported in part by an National Cancer Institute Cancer Center Support Grant [number NIH 5 P30 CA06516].
DISCLOSURE
AW receives institutional research funding from Genentech and MacroGenics. SW reports consulting/advisory board role for Foundation Medicine, InfiniteMD, Eli Lilly, and Puma Biotechnology; and equity in InfiniteMD. SSF filed a patent (WO2017161175A1) on methods applied in this study. SI receives research funding support from Genentech, PharmaMar, AstraZeneca, Merck (all to institution); and consulting fees from Genentech, Hengrui, Puma, Immunomedics, and Myriad. PL reports serving on advisory boards at AbbVie (2018–2019), Alexion (2016–2017), Ariad (2016–2017), GenMab (2016–2018), Glenmark (2016–2017), Menarini (2016–2017), Novartis (2016–2017), CytomX (2016–2019), Omniox (2016–2017), Ignyta (2016–2017), Genentech (2016–2019), Takeda (2017–2020), SOTIO Consultant (2018–2019), Cybrexa (2018–2019), Agenus (2018–2020), IQVIA (2019–2020), TRIGR (2019–2020), Pfizer (2019–2020), I-MAB (2019–2020), Immuno-Met (2018–2020), Black Diamond (2019–2020), Sartarius (2019–), and GlaxoSmithKline (2019–2020); data safety monitoring boards/committees at Agios (2016–2019), Five Prime (2017–2020), Halozyme (2016–2019), FivePrime (2017–2019), and Tyme (2018–2020); and reports work with the imCORE Alliance at Roche-Genentech (2016–2019). VAA filed a patent (WO2017161175A1) on methods applied in this study. SMT reports receiving institutional research support from Merck, Bristol-Myers Squibb, Exelixis, Eli Lilly, Pfizer, Novartis, AstraZeneca, Eisai, Nektar, Odonate, Sanofi, and Genentech and has served on advisory boards for Genentech, Eli Lilly, Novartis, Pfizer, Nektar, Immunomedics, Nanostring, Daiichi-Sankyo, Bristol-Meyers Squibb, Sanofi, Athenex, AstraZeneca, Eisai, Puma, and Merck. UM reports serving on advisory boards (paid) at Astrazeneca, Myriad Genetics, Clovis, Eli Lilly, Mersana, Geneos, Fuji Film, Cerulean; and consulting (paid) for Merck, 2X Oncology and Immunogen. IEK receives institutional research funding from Genentech/Roche, Pfizer, Daiichi-Sankyo; has advisory board (honoraria) roles at Genentech/Roche, Daiichi-Sankyo, Macrogenics, Context Therapeutics, Taiho Oncology; and reports DSMC (honoraria) at Merck; and reports DSMB (honoraria) at Novartis. EPW institutional research funding from Genentech/Roche and Merck; consultant/honoraria from Carrick Therapeutics, Genentech/Roche, Genomic Health, GSK, Jounce, Lilly, Merck, Seattle Genetics; advisory board/honoraria from Leap. ADD reports receiving commercial research grants from Eli Lilly & Company, Sierra Oncology, and EMD Serono and is a consultant/advisory board member for Eli Lilly & Company, Sierra Oncology, and EMD Serono. GIS has received research funding from Eli Lilly, Merck KGaA/EMD-Serono, Merck, and Sierra Oncology. He has served on advisory boards for Pfizer, Eli Lilly, G1 Therapeutics, Roche, Merck KGaA/EMD-Serono, Sierra Oncology, Bicycle Therapeutics, Fusion Pharmaceuticals, Cybrexa Therapeutics, Astex, Almac, Ipsen, Bayer, Angiex, and Daiichi Sankyo. NUL reports institutional research funding from Genentech, Merck, Pfizer, Seattle Genetics; and consulting/ad board roles at Puma, Daichii, Seattle Genetics. NW was previously a stockholder and consultant for Foundation Medicine; has been a consultant/advisor for Novartis and Eli Lilly; and has received sponsored research support from Novartis and Puma Biotechnology. None of these entities had any role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript. All other authors report no conflicts of interest.
REFERENCES
- 1.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6): 523–533. [DOI] [PubMed] [Google Scholar]
- 2.Tutt A, Tovey H, Cheang MCU, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med. 2018;24(5):628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5(11):1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patch AM, Christie EL, Etemadmoghadam D, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521(7553):489–494. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri AR, Callen E, Ding X, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535(7612): 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie EL, Fereday S, Doig K, Pattnaik S, Dawson SJ, Bowtell DDL. Reversion of BRCA1/2 germline mutations detected in circulating tumor DNA from patients with high-grade serous ovarian cancer. J Clin Oncol. 2017;35(12):1274–1280. [DOI] [PubMed] [Google Scholar]
- 7.Quigley D, Alumkal JJ, Wyatt AW, et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Disc. 2017;7(9):999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodall J, Mateo J, Yuan W, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Disc. 2017;7(9):1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brady SW, McQuerry JA, Qiao Y, et al. Combating subclonal evolution of resistant cancer phenotypes. Nat Commun. 2017;8(1):1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigelt B, Comino-Mendez I, de Bruijn I, et al. Diverse BRCA1 and BRCA2 Reversion mutations in circulating cell-free DNA of therapy-resistant breast or ovarian cancer. Clin Cancer Res. 2017;23(21):6708–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher S, Barry A, Abreu J, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12(1):R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8(1):1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geoffroy V, Herenger Y, Kress A, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018;34(20):3572–3574. [DOI] [PubMed] [Google Scholar]
- 14.Kochupurakkal BS, Parmar K, Lazaro J-B, et al. Abstract 2796: Development of a RAD51-based assay for determining homologous recombination proficiency and PARP inhibitor sensitivity. Cancer Res. 2017;77(13 Supplement):2796. [Google Scholar]
- 15.Hill SJ, Decker B, Roberts EA, et al. Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Disc. 2018;8(11):1404–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yazinski SA, Comaills V, Buisson R, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31(3):318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunting SF, Callen E, Wong N, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouwman P, Aly A, Escandell JM, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17(6):688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He YJ, Meghani K, Caron MC, et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature. 2018;563(7732):522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gala K, Li Q, Sinha A, et al. KMT2C mediates the estrogen dependence of breast cancer through regulation of ERalpha enhancer function. Oncogene. 2018;37(34):4692–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knijnenburg TA, Wang L, Zimmermann MT, et al. Genomic and molecular landscape of DNA damage repair deficiency across the Cancer Genome Atlas. Cell Rep. 2018;23(1):239–254.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451(7182):1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards SL, Brough R, Lord CJ, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451(7182):1111–1115. [DOI] [PubMed] [Google Scholar]
- 24.Saeki H, Siaud N, Christ N, et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci U S A. 2006;103(23):8768–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Bernhardy AJ, Cruz C, et al. The BRCA1-Delta11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 2016;76(9): 2778–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondrashova O, Nguyen M, Shield-Artin K, et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Disc. 2017;7(9):984–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matulonis UA, Wulf GM, Barry WT, et al. Phase I dose escalation study of the PI3kinase pathway inhibitor BKM120 and the oral poly (ADP ribose) polymerase (PARP) inhibitor olaparib for the treatment of high-grade serous ovarian and breast cancer. Ann Oncol. 2017;28(3):512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson N, Johnson SF, Yao W, et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc Natl Acad Sci U S A. 2013;110(42):17041–17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parmar K, Kochupurakkal BS, Lazaro J, et al. The CHK1 inhibitor prexasertib exhibits monotherapy activity in high-grade serous ovarian cancer models and sensitizes to PARP inhibition. Clin Cancer Res. 2019;25(20):6127–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz C, Castroviejo-Bermejo M, Gutierrez-Enriquez S, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol. 2018;29(5):1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castroviejo-Bermejo M, Cruz C, Llop-Guevara A, et al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med. 2018;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.