Abstract
Hck, a Src family nonreceptor tyrosine kinase (SFK), has recently been established as an attractive pharmacological target to improve pulmonary function in COVID-19 patients. Hck inhibitors are also well known for their regulatory role in various malignancies and autoimmune diseases. Curcumin has been previously identified as an excellent DYRK-2 inhibitor, but curcumin's fate is tainted by its instability in the cellular environment. Besides, small molecules targeting the inactive states of a kinase are desirable to reduce promiscuity. Here, we show that functionalization of the 4-arylidene position of the fluorescent curcumin scaffold with an aryl nitrogen mustard provides a stable Hck inhibitor (Kd = 50 ± 10 nM). The mustard curcumin derivative preferentially interacts with the inactive conformation of Hck, similar to type-II kinase inhibitors that are less promiscuous. Moreover, the lead compound showed no inhibitory effect on three other kinases (DYRK2, Src, and Abl). We demonstrate that the cytotoxicity may be mediated via inhibition of the SFK signaling pathway in triple-negative breast cancer and murine macrophage cells. Our data suggest that curcumin is a modifiable fluorescent scaffold to develop selective kinase inhibitors by remodeling its target affinity and cellular stability.
Keywords: curcumin derivative, cell signaling, kinase inhibition, Hck inhibitor, Src family kinase, enzyme kinetics
Abbreviations: Bcl-2, B-cell lymphoma 2; DLS, dynamic light scattering; DMEM, Dulbecco's modified Eagle's medium; ERKI1/2, extracellular signal-regulated kinase 1/2; FBS, foetal bovine serum; IISER, Indian Institute of Science Education and Research; PBS, phospahte buffered saline; PEP, phosphoenolpyruvate; SFK, Src family nonreceptor tyrosine kinase; STAT3, signal transducer and activator of transcription 3; TNF-α, tumor necrosis factor α
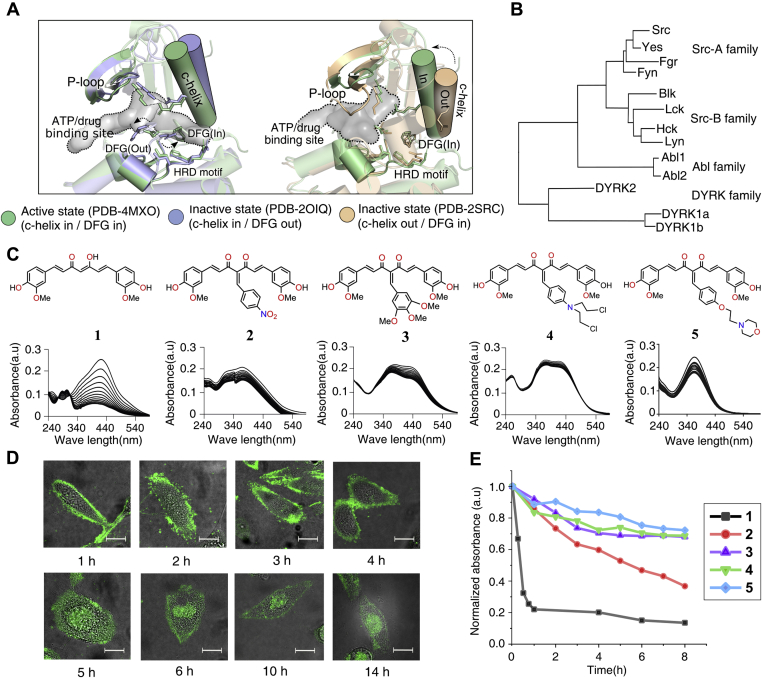
Curcumin, an active molecule against inflammation, microbial infections, cancer, and neurodegenerative diseases loses its drug-like potential due to its instability in the cellular environment (1). Besides, the planar symmetry makes curcumin less druggable toward asymmetric binding pockets such as kinases (2). Albeit, it is undeniable that pure curcumin shows excellent selectivity toward the DFG-Asp-in, active conformation of dual-specificity tyrosine-regulated kinase 2 (DYRK2) (3). The drugability of curcumin may be improved by nanoformulation and modification at relevant positions to enhance stability and specificity. A recent report suggests that curcumin self-assembles in the presence of Zn(II) and F-moc histidine to adopt nanoformulations enhancing stability (4). The functionalization of the methylenic position in the diketone motif of curcumin provides stable derivatives and introduces nonplanarity in the conformation (5, 6, 7). Analysis of the recent DYRK2 kinase domain structure in complex with curcumin suggests that the modification at the methylenic position in the diketone motif may prevent the docking of the curcumin derivatives to the ATP-binding pocket (Fig. 1A). We asked if the functionalization of the methylenic position will remodel the target preference of the curcumin compounds toward other kinases, namely Src, Hck, and Abl (Fig. 1B)? It may be noted that the clinical kinase inhibitors target one or more of the following conformations: the DFG-Asp-in active, the DFG-Asp-out Abl/cKit like inactive, or the DFG-Asp-in c-Src/CDK like inactive conformation (Fig. 1A) (8, 9, 10). Most type-I inhibitors targeting the active conformation are promiscuous as all kinases share more or less the same conformation in their active state. Type-II kinase inhibitors targeting the inactive conformations are desirable since the kinases adapt specific inactive conformation reducing the promiscuity.
Figure 1.
Design of curcumin derivatives, solution, and cellular stability.A, structural alignment of inactive (PDB: 2OIQ and 2SRC) and active (PDB: 4MXO) conformation of Src drug-binding pocket. The drug-binding pocket is shown in space filled model. B, phylogenetic tree constructed based on the kinase domain sequence of DYRK2, SFK, and Abl. C, chemical structure of compounds 1 to 5. Below the absorbance scan of the respective compound recorded for 1 h. D, confocal microscope images of live MDA-MB-231 cells treated with 4 at indicated time points. Scale bar 10 μm. E, a plot of normalized absorption versus time to demonstrate stability of 1 to 5 in pH 7.4 PBS containing 5% DMSO (see Figs. S1 and S2). PBS, phospahte buffered saline.
We targeted to design stable curcumin derivatives by modifying the methylenic position. The functionalization of the methylenic position in the diketone motif by 4-arylidene substituents would deter the free-radical-driven incorporation of oxygen in the curcumin scaffold, known to be occuring spontaneously in native curcumin at physiological pH (11, 12). The 4-aryl substituents (Fig. 1C) were chosen to promote noncovalent interactions with the ligand-binding pocket of the inactive form of the aforementioned kinases. Here in, we show that suitable functionalization of the methylenic position enhances the cellular stability and remodels the affinity toward DFG-in inactive conformation of Hck, instead of DYRK2. Hck is an attractive pharmacological target for its regulatory role in cancer development and virus infection (13, 14). Hck expresses explicitly in myeloid and B-lymphocyte lineage cells and regulates immune receptor signaling, cell migration, proliferation, and differentiation (15, 16). Recently, inhibition of Hck by ibrutinib in COVID-19 patients showed protection against lung injury and improved pulmonary function (17).
Results and discussion
We first made an in silico assessment of the ability of the compounds 1 to 5 to bind at the ligand-binding pocket of DYRK2 and different conformations of Abl and Src kinase domain (Fig. 1, A and C and Fig. S1A). The docking studies showed that the curcumin derivatives might have preserved the binding affinity toward DYRK2. Next, we probed, if the modified compounds have gained specificity toward the active or inactive conformations of the ligand-binding pocket. The curcumin did not distinguish between the active or inactive conformation of Src or Abl kinase domain (Fig. S1A). The plot of relative change in docking scores of 2 to 5 suggests that the functionalized curcumins bind preferentially to the DFG-Asp-in cSrc/CDK like inactive conformation (Fig. S1A). Kinase inhibitors targeted against the inactive state are desirable due to higher selectivity and potency (2, 18, 19). The four derivatives 2 to 5, of which two were taken from an earlier library of 4-arylidene functionalized curcumin derivatives (20, 21), were thus synthesized and characterized using standard analytical techniques. The synthetic method is a single-step Knoevenagel condensation of curcumin with the respective aldehydes (Fig. S2A). The curcumin (1) used in synthesis was purified by column chromatography from the crude commercial mixture.
We first evaluated the stability of 2 to 5 in comparison with curcumin in aqueous solution. The UV–visible data showed that the compounds 3 to 5 have greater solution half-lives (t1/2) in pH 7.4 (1× phospahte buffered saline [PBS] containing 5% DMSO) (Fig. 1E, Fig. S2, C–G and Table S1) compared with 1 and 2. Next, we studied the stability of 1 to 5 under the kinase assay condition (Fig. 1C). The bis(2-chloroethylamine) motif bearing compound 4 is the most stable under near-physiological conditions in the kinase assay buffer. Compound 4 also demonstrated to be stable for at least 24 h (observed period) in 8:2 v/v of 1x PBS and DMSO-d6 in 1H NMR (Fig. S18).
We determined the activity of 1 to 5, against DYRK2, Src, Hck, and Abl kinases (Fig. 2A) using coupled kinase assay. Curcumin inhibits DYRK2 kinase activity with Ki of 6 nM similar to the previous report (3) but compounds 2 to 5 do not (Fig. 2A and Fig. S3). Besides, 1 to 5 also display poor activity against Abl kinase (Ki > 3000 nM) in comparison with imatinib (Ki > 60 nM) (22) (Fig. 2A and Fig. S3). Compound 4 selectively inhibits Hck and Src kinase activity (Fig. 2A and Fig. S3) with Ki of 70 ± 11 nM and 460 ± 110 nM, respectively. Compound 1 to 3, 5, and imatinib showed a relatively weak inhibitory effect on Src and Hck (Ki more than tenfold compared to 4) (Fig. 2A and Fig. S3). We used imatinib as a control due to its specificity toward DFG-Asp-out Abl/cKit like inactive structure (Fig. 1A) (23, 24). Unlike Src, Abl kinase can adopt all three conformations of the kinase domain (Fig. 1A) (25). Thus, the majority of ATP-competitive, noncovalent Src inhibitors would be promiscuous to Abl. The specific inhibition of Src and Hck over Abl by 4 suggests that it is possibly binding to an inactive conformation of the Src kinase domain. We ruled out the interaction of 4 to the DFG-out inactive conformation of Src due to its relatively higher thermodynamic penalty. (22) It is exciting to find that 4 specifically inhibits Hck because of the importance of Hck in the regulation of cancer development and lung injury in COVID-19 patients (13, 14). Therefore, we further investigated the mechanism and mode of action of 4.
Figure 2.
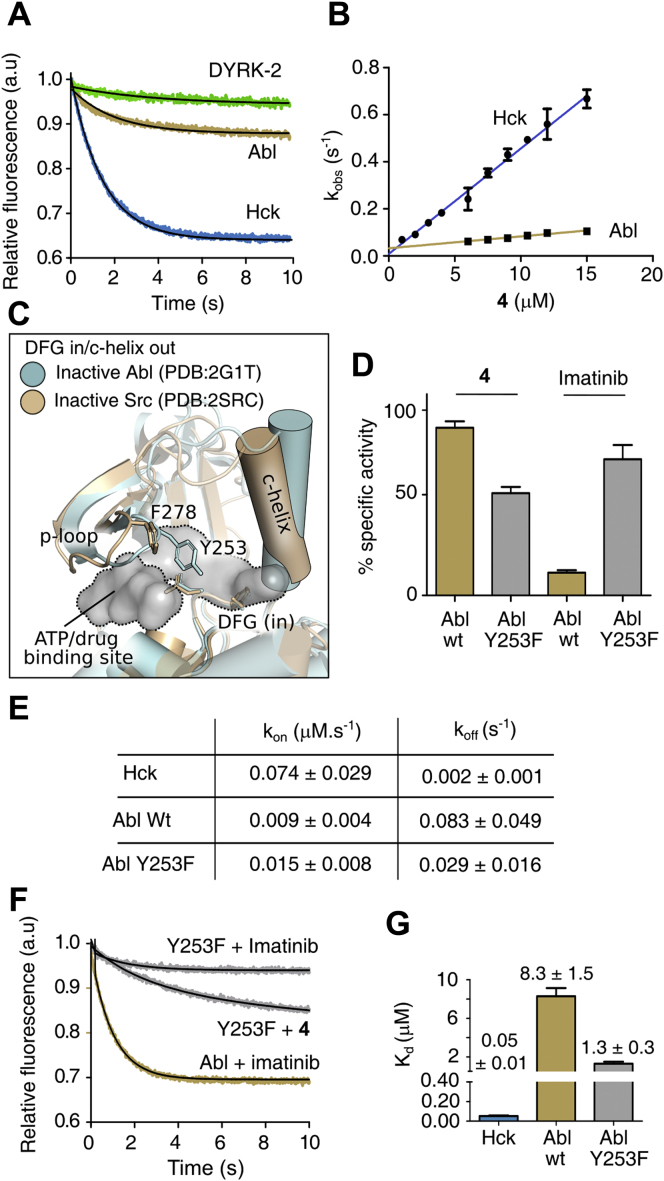
Compound 4 competitively binds with the inactive conformation of Hck kinase domainA, the plot of inhibitory constant (Ki) of 1 to 5 and imatinib for DYRK2 (100 nM), Abl (35 nM), Src (75 nM), and Hck (35 nM). B, % specific activity of Hck determined in the presence of 5 μM of 4 for the unphosphorylated (Y) and phosphorylated (pY) states. C, the plot of time-dependent decrease in tryptophan fluorescence upon binding of 4 to unphosphorylated and phosphorylated Hck. Each transient was fitted to single exponential kinetics. D, representative immunoblot showing the effect of 4 (15 μM) on Hck (250 nM) autophosphorylation at indicated time points (upper panel). The bottom panel is the plot of the relative rate of phosphorylation obtained from the densitometric analysis of the blot. E, Michaelis–Menten plot of Hck enzyme kinetics in the presence of 4 or DMSO as a control. F, tabulation of enzyme kinetic parameters of Hck at indicated concentration of 4. (n = 3 experiments; mean ± SD). All the kinetics data were recorded for 10 min (See Figs. S3–S5).
We first investigated if the oligomerization of 4 could explain the mode of kinase inhibition. The oligomeric state of 1 and 4 was determined from the hydrodynamic radius measured using the dynamic light scattering in an aqueous buffer (Fig. S2, H–K) and compared with imatinib and sucrose. Unlike curcumin (1), we observed that 4 did not aggregate in solution and behaved similar to imatinib.
Selective type-II inhibitors targeted against the inactive conformation of Abl are sensitive to the phosphorylation state of the activation loop (24, 26, 27, 28). The inhibitors that are nonselective for Src and Abl are often not sensitive to the phosphorylation of the kinase activation loop (29). Hence, we prepared and purified unphosphorylated Src and Hck, which was uniformly phosphorylated by treating with ATP and magnesium (Fig. S4B). Then the activity of the phosphorylated Hck (pY-Hck) and Src (pY-Src) was assayed in a time-dependent manner. The results show that 4 does not inhibit the pY-Hck and pY-Src (Fig. 2B and Fig. S4, C and D) emphasizing its preference toward the inactive conformation of Hck and Src.
A hallmark of Src activation is autophosphorylation of the activation loop (27, 28, 30), which shifts the equilibrium toward the active state (31, 32, 33). We determined the effect of 4 on the rate of autophosphorylation of Src and Hck and found that 4 reduces the rate of autophosphorylation by 2.5- and 500-fold, respectively (Fig. 2D and Fig. S4, E and F). We studied the interaction of 4 with Hck by measuring the kinetics of change in the intrinsic tryptophan fluorescence by stop-flow fluorimeter. The plot of the rate of change in tryptophan fluorescence (Fig. 2C) suggests that 4 preferentially interacts with the inactive unphosphorylated conformation of Hck; thus, further emphasizing that 4 shows preference to bind the inactive conformation and reduces the rate of autophosphorylation of the kinase domain.
The crystal structure of DYRK2 in complex with curcumin for the first time showed that curcumin occupies the ATP binding pocket potentially acting as a competitive inhibitor (9). To find out if 4 could inhibit Hck and Src as an ATP-competitive inhibitor, we determined the Km (Michaelis–Menten constant) for ATP and Vmax (Fig. 2, E and F and Fig. S4, F and G). Increasing concentration of 4 changes the Km without any significant change in the Vmax, indicating that 4 functions as a competitive inhibitor for the ATP-binding site.
The above data does not rule out the possibility of covalent modification of the ligand-binding site by the potential alkylating groups in 4, which may explain the specificity toward the SFK. The specificity of several compounds toward Src or Abl is attributed to the different sequence and structural dynamics of the P-loop (22, 34, 35). For example, in Src the Cys277 at the P-loop and Cys400 at the β7 could be covalently modified by alkylating agents, explaining the specificity of 4 toward Src (Fig. S5A). In Abl kinase domain, the amino acid residues at the corresponding positions are Gln and Val, respectively, providing no scope for 4 to covalently modify them, unlike a cysteine (36). Although the aromatic mustard motif is such that the alkylation ability would be low or absent, confirmation was required to exclude covalent modification as a possible mode of inhibition. To confirm the above we incubated the Src kinase domain and 4 in the kinase assay buffer for 1 h. The resultant sample was then analyzed by HPLC under denaturing condition to determine any covalent adduct formation. We observed that the 4-treated Src kinase elutes at the same resident time as untreated Src kinase with no coelution of 4 (Fig. S5C). The respective pure fraction of the protein sample from the HPLC was subjected to ESI-MS, which confirmed that the m/z envelope for the treated and untreated sample (Fig. S5D) corresponds to the unmodified protein. Thus, 4 is not covalently modifying the cysteine residues of Src kinase.
We studied the binding kinetics of 4 with DYRK2, Hck, and Abl to understand the mechanism of specificity toward Hck. We monitored the change in the intrinsic fluorescence of the protein corresponding to the pre-steady-state kinetics by titration (Fig. 3 and Fig. S6). The Hck binds to 4 with approximately tenfold faster kon rates than Abl and a Kd of 50 ± 10 nM, which matches with the Ki values from enzyme kinetics (Fig. 3, E and G). DYRK2 did not interact with 4 (Fig. 3A). The above result may be explained by comparing the structure of the ligand-binding-pocket in the Src DFG-Asp-in inactive structure (PDB ID: 2SRC) to the corresponding conformation of Abl kinase domain (PDB ID: 2G1T) (Fig. 3C). The P-loop of Abl takes a closed conformation in comparison to Src, which alters the conformational dynamics of the ligand-binding-pocket (37). The Y253 at the P-loop of Abl may sterically block the docking of 4 to the ligand-binding-pocket. In Src, the residue at the corresponding position is substituted by F278 (Fig. 3C and Fig. S5A). The Y253 is a critical residue in Abl, and its mutation to F253 causes imatinib resistance (38). Therefore, we speculate that the different structural dynamics of the P-loop residues in the Src and Abl kinase domain determine the specificity for 4. To find out if the Y253 is responsible for selectivity, we generated the Y253F mutant Abl to comparatively study imatinib and 4. The Abl Y253F bound weakly to imatinib and did not inhibit the kinase activity (Fig. 3, D, F and G). Whereas 4 binds with sevenfold higher affinity (Kd of 1.3 ± 0.3 μM) to the Abl Y253F and shows better inhibition compared with imatinib at a given dose (Fig. 3, D–G).
Figure 3.
Binding kinetics showing 4 specifically interacts with Hck.A, the plot of time-dependent decrease in tryptophan fluorescence of kinases domains upon binding to 4. Each transient was fit to a single exponential equation to obtain observed rate constants. B, the pseudo-first-order rate constants were plotted as a function of concentration of 4 to determine the kon and koff from the slope and intercept of linear fitting. C, drug-binding pocket of Src and Abl kinase domain in DFG-in/c-helix out inactive state. D, a plot of % activity of wildtype and Y253F mutant of Abl determined in the presence of 10 μM of 4 and imatinib. E, table of the rate constants (kon and koff) measured for binding of 4. F, the kinetic transient of imatinib or 4 binding to Abl or Y253F mutant. G, a plot of the dissociation constant (Kd) for the indicated kinase domain for 4 (n = 3 experiments; mean ± SD). (See Figs. S5 and S6)
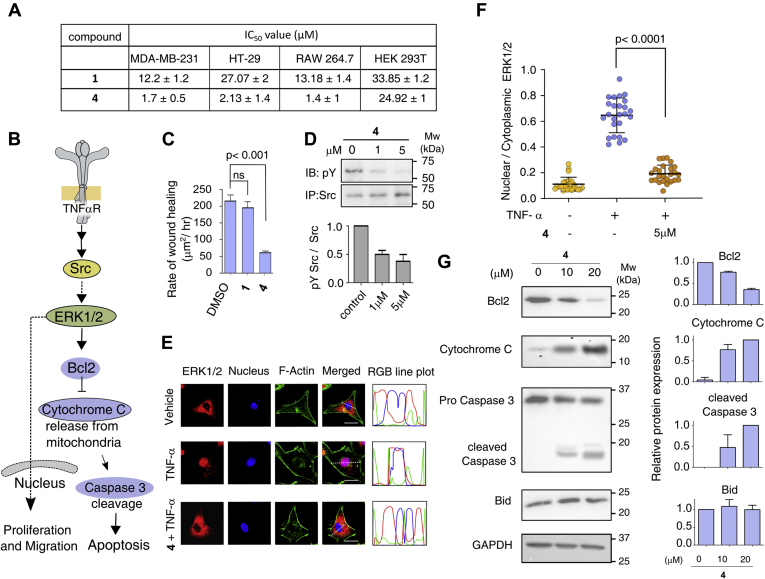
The functionalization of the methylenic position in the diketone motif of curcumin by the aryl nitrogen mustard thus altered the target specificity toward Hck over DYRK2, Abl, and Src. In spite of the encouraging results, the instability in the cellular environment may eclipse the potential of 4. We investigated the cellular stability exploiting the green fluorescence of 4, which allows to trace the molecule by live-cell imaging in the triple-negative-breast-cancer (TNBC) cell MDA-MB-231 (Fig. 1D). Curcumin, as expected (1), spontaneously degraded within 1 h inside the cell making it invisible (Fig. S1, B–D). Although the quantum yield (Table S1) of curcumin is higher than 4, the fluorescence of compound 4 was observed inside the MDA-MB-231 cell for 14 h, suggesting its higher stability (Fig. 1D and Fig. S1, E and F). To translate the inhibitory effect of 4 to cell-based experiments, we measured the cytotoxicity on the murine macrophage RAW 264.7, MDA-MB-231 (39), colon carcinoma (HT-29) (40), and HEK 293T cell line. The comparison of IC50 values (Fig. 4A and Fig. S7A) indicate that the 4 is ten times more potent than curcumin when tested against RAW 264.7, MDA-MB-231, and HT-29 cell lines, respectively. In contrary, 4 is tenfold less cytotoxic to HEK 293T cells (IC50 value of 24.92 ± 1 μM). This may be because among the above four cell lines HEK293 does not overexpress SFK. DNA cross-linking assay shows that 4 do not cross-link DNA in the in vitro assay (Fig. S7B). We next studied if 4 disrupts the plasma membrane integrity by live-cell imaging of MDA-MB-231 cell line treated with FM4-64-FX, a lipophilic probe used for staining plasma membrane (Fig. S7C) (41). The results support that 4 is not affecting the membrane integrity of the cells.
Figure 4.
Compound 4 inhibits Src kinase activation and ERK1/2 translocation in MDA-MB-231 cells.A, the table contains IC50 values of 1 and 4 in different cell lines. B, schematic representation of SFK signaling. Signaling modules probed are color-coded (KEGG pathway database). C, plot of the rate of wound healing measured for curcumin and 4-treated cells. D, western blot showing total Src kinase and the autophosphorylation level after treating cells with 4 followed by immunoprecipitation. Quantification of Src phosphorylation plotted as a bar diagram. E, representative images of MDA-MB-231 cells showing ERK1/2 localization after treating with 4 followed by activation with TNF-α, as indicated in each panel. F-actin and nucleus are stained with phalloidin (red) and DAPI (blue), respectively. Scale bar, 10 μm. F, quantification of nucleocytoplasmic distribution of ERK1/2 (n = 30 cells, from two independent experiments). G, representative western blot strips showing the cytoplasmic level of Bcl-2, Cytochrome C, caspase 3, and Bid in cells treated with 4. GAPDH was used as a loading control. At right panel, bar plot represents the quantification of expression level of each signaling module at indicated concentration of 4, (n = 3 experiments; mean ± SD). (See Figs. S7 and S8). Bcl-2, B-cell lymphoma 2; Bid, BH3 interacting domain death agonist; ERKI1/2, extracellular signal-regulated kinase 1/2; TNF-α, tumor necrosis factor α.
To evaluate if 4 inhibits Src-dependent cell migration, proliferation, and apoptosis, we performed a wound-healing assay with MDA-MB-231 and probed the signaling modules regulating the respective pathways (Fig. 4 and Fig. S8). The MDA-MB-231 cells rely on Src signaling for cell migration and metastasis (Fig. 4B) (42, 43). We observed that 4 inhibits the autophosphorylation of Src in the cell (Fig. 4D) and significantly impaired the wound-healing rates (IC10 and IC50 dose) (Fig. S8, B and C). The migration process involves the tumor necrosis factor α (TNF-α)-induced activation of Src signaling cascade and translocation of extracellular signal-regulated kinase 1/2 (ERK1/2) to the nucleus (44, 45). So, 4-treated MDA-MB-231 cells were activated with TNF-α and the localization of ERK1/2 to the nucleus was determined by immunofluorescence using confocal microscopy. In comparison to the untreated cells, treatment with 5 μM of 4 significantly increased the cytosolic fraction of ERK1/2 even after activation with TNF-α (Fig. 4, E and F). Comprehensively, our data suggest that 4 inhibits the Src-mediated ERK1/2 signaling necessary for cell migration and proliferation.
Activation of ERK1/2 promotes cell survival by enhancing the stability and activity of B-cell lymphoma 2 (Bcl-2), an antiapoptotic molecule that regulates the intrinsic apoptotic pathway (46, 47). Many oncogene-targeting molecules directly or indirectly inhibit ERK1/2 signaling to cause Bcl-2 degradation and cell death (47). Therefore, we examined if inhibition of ERK1/2 destabilizes Bcl-2 and activates intrinsic apoptotic pathway (Fig. 4B). Destabilization of Bcl-2 will release cytochrome c from mitochondria, subsequently activating Caspases3 causing apoptosis (48, 49, 50). We observed that 4 activates the mitochondria-mediated apoptotic pathway by decreasing Bcl-2 level in the cell, leading to release of cytochrome C from mitochondria and activation of Caspase3 (Fig. 4G). Compound 4 does not affect the BH3 interacting domain death agonist expression level suggesting that 4 induces apoptosis through an intrinsic apoptotic pathway. Together our data indicate that 4 interferes with multiple SFK-mediated signaling pathways regulating cell proliferation and apoptosis.
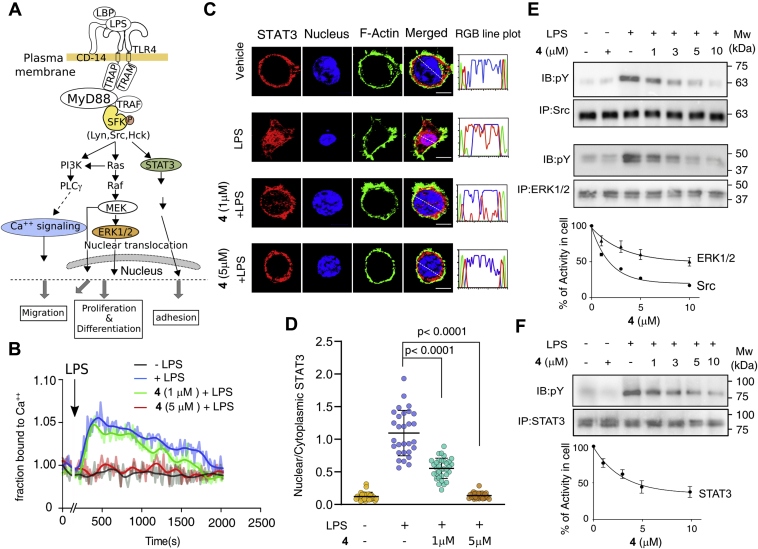
The upregulation of SFK in macrophage cells leads to tumorigenesis by activating diverse signaling modules, causing increase in angiogenesis, cell polarization, migration, and cell adhesion (Fig. 5A) (37, 51, 52). We investigated if inhibition of SFK by 4 in macrophages inhibits downstream signaling involved in tumorigenesis (51) following the activation of the macrophages with bacterial lipopolysaccharide (LPS). The cellular signaling changes were probed by measuring Ca2+ response, translocation of signal transducer and activator of transcription 3 (STAT3) to the nucleus, and phosphorylation of signaling proteins (ERK1/2 and STAT3) (Fig. 5A). Treatment with 5 μM of 4 shows a significant reduction in Ca2+ release and an increased cytosolic fraction of STAT3 (Fig. 5, B–D). The 4-treated cells showed concentration-dependent reduced phosphorylation of Src, ERK1/2, and STAT3 (Fig. 5, E and F) emphasizing that the cytotoxicity of 4 involves inhibition of SFK-mediated cell signaling. Compound 4 affects multiple SFK signaling pathways, yet given the diversity of the downstream signaling cascade, the SFKs may not be the sole endogenous targets.
Figure 5.
Compound 4 abolishes SFK signaling and STAT-3 nuclear translocation in macrophages: A, schematic of SFK signaling pathway. Signaling modules probed are color-coded (KEGG pathway database). B, Ca2+ flux as a function of time in RAW 264.7 cells measured after treating cells with indicated concentration of 4 followed by activation with LPS. C, representative images of RAW 264.7 cells showing STAT3 localization after treating with 4 following activation with LPS, as indicated in each panel. F-actin and the nucleus are stained with phalloidin (red) and DAPI (blue), respectively. Scale bar, 10 μm. D, quantification of nucleocytoplasmic distribution of STAT3 (n = 30 cells, from two independent experiments). E and F, western blot showing total protein and the autophosphorylation level of Src, Erk1/2, and STAT3 after treating cells with 4 following activation with LPS and immunoprecipitation. At the bottom of each panel, quantification of phosphorylation level is plotted as a function of 4 concentration. (n = 2 experiments; mean ± SD.). (See Fig. S9). ERKI1/2, extracellular signal-regulated kinase 1/2; STAT3, signal transducer and activator of transcription 3.
Experimental procedures
Materials and methods
All chemicals and solvents were purchased from commercial sources. Solvents were distilled and dried prior to use by standard procedures (53). Curcumin was purchased either from Sigma-Aldrich or Carbosynth Ltd. Curcumin obtained from commercial sources was further purified using column chromatography to get rid of curcuminoid mixtures. MTT [(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)] (USB), cell growth media, and their supplements were purchased from Gibco. Abl kinase domain was purchased from Addgene (Addgene plasmid # 79727) (54). The details for the list of antibodies used in this study are provided in Table S1. All the solvents used for spectroscopy and lipophilicity measurements are of spectroscopy grade and purchased from Merck. UV–visible measurements were done using either Agilent Technologies Cary 300 Bio or PerkinElmer Lambda 35 spectrophotometer. The fluorescence measurements were done using a Horiba-Yvon FluoroMax Plus spectrophotometer. The stopped-flow measurements were performed in na SFM2000 bioLogic spectrophotometer. FT-IR spectra were recorded using PerkinElmer SPECTRUM RX I spectrometer in KBr pellets. NMR spectra were recorded using either JEOL ECS 400 MHz or Bruker Avance III 500 MHz spectrometer at ambient temperature and calibrated using residual undeuterated solvents as internal reference. All 13C spectra reported are proton-decoupled. The chemical shifts (δ) are reported in parts per million (ppm). Elemental analyses were performed on a PerkinElmer CHN analyzer (Model 2400). Electrospray ionization mass spectra of compounds were recorded in positive-mode electrospray ionization using a Q-ToF Bruker maXis II instrument. All the compounds were kept in dark after purification and stored in refrigerator.
General synthetic method
Syntheses of the four curcumin derivatives (2–5) were conducted using scheme in Fig. S2A following modified procedure reported elsewhere (5). Briefly, 1.0 mmol of curcumin and 2 mmol of appropriate aromatic aldehyde were dissolved first in prewarmed toluene at 100 °C. Next into this hot mixture, catalytic amount of piperidine (0.05 mmol) and acetic acid (0.08 mmol) were added. The solution was then heated to reflux at 140 °C using a Dean–Stark apparatus. Resulting water as a by-product of reaction was eliminated by anhydrous Na2SO4 in the apparatus. The reaction was continued up to 18 to 20 h under dark. After completion of reaction, the whole solution was evaporated out under reduced pressure. Pure product was isolated using silica gel column chromatography (ethyl acetate–hexane mixture as eluent).
(1E,6E)-1,7-bis(4-hydroxy-3-methoxyphenyl)-4-(4-nitrobenzylidene)hepta-1,6-diene-3,5-dione (2)
Curcumin (1.0 mmol) and 4-nitro benzaldehyde (2.0 mmol) was used as reactant and a published protocol was followed (55). Pure product was isolated from silica gel chromatography by using 1:1 ethylacetate–hexane mixture as eluent. Yield: 51%, UV-vis [CH3OH, λmax, nm (ε/M−1 cm−1)]: 367 (27,900), 313 (24,100), 261 (18,100). FT-IR (KBr pellet, cm−1):3412(br), 2958(w), 2916(w), 2853(w), 1642(sh), 1596(s), 1573(m), 1514(sh), 1467(w), 1428(s), 1347(s), 1269(s), 1247(s), 1213(m), 1178(s), 1108(s), 1035(s), 977(m), 854(s), 810(w), 746(w), 718(w), 689(w). 1H NMR (500 MHz, CDCl3) δ 8.18 (d, 2H, J = 8.8 Hz), 7.80 (t, 2H, J = 9.0 Hz), 7.64 (d, 2H, J = 8.8 Hz), 7.45 (d, 1H, J = 16.2 Hz), 7.19 (m, 1H), 7.04 to 6.87 (m, 6H), 6.78 (d, 1H, J = 16.1 Hz), 5.94 (s, 2H), 3.93 (s, 3H), 3.90 (s, 3H). ESI-HRMS (Methanol) m/z (calc.): 524.1355 (524.1316) [C28H23NO8Na]+
(1E,6E)-1,7-bis(3-hydroxy-4-methoxyphenyl)-4-(3,4,5-trimethoxybenzylidene)hepta-1,6-diene-3,5-dione (3)
Curcumin (1.0 mmol) and 3,4,5-trimethoxybenzaldehyde (2.0 mmol) were used as reactants (20). Pure product was isolated from silica gel chromatography by using 2:1 ethylacetate-hexane mixture as eluent. Yield: 67%; UV-vis [CH3OH, λmax, nm (ε/M−1 cm−1)]: 364 (34,500), 250 (17,700). FT-IR (KBr pellet, cm−1): 3418(br), 2960(w), 2929(w), 2844(w), 1634(w), 1579(sh), 1512(sh), 1463(m), 1452(m), 1428(m), 1384(br), 1325(m), 1287(sh), 1270(w), 1244(s), 1210(s), 1180(s), 159(s), 1126(sh), 1031(s), 994(m), 982(w), 844(m), 816(w), 578(w), 550(w), 475(w). 1H NMR (400 MHz, CDCl3) δ 7.76 (t, 2H, J = 6.9 Hz), 7.52 (d, 1H, J = 16 Hz), 7.18 to 7.15 (m, 1H), 7.05 to 7.03 (m, 2H), 6.97 to 6.87 (m, 4H), 6.81 (d, 1H, J = 16 Hz), 5.91 (s, 2H), 3.93 (s, 3H), 3.90 (s, 3H), 3.84 (s, 3H), 3.80 (s, 6H). ESI-HRMS (Methanol) m/z (calc.): 569.1791 (569.1782) [C31H30O9Na]+
(1E,6E)-4-(4-(bis(2-chloroethyl)amino)benzylidene)-1,7-bis(4-hydroxy-3-methoxyphenyl)hepta-1,6-diene-3,5-dione (4)
Curcumin (1.0 mmol) and 4-(bis(2-chloroethyl)amino)benzaldehyde (2.0 mmol) were used as reactants. Pure product was isolated from silica gel chromatography by using 1:1 ethylacetate-hexane mixture as eluent. Yield: 40%. Anal. cal. for C32H31Cl2NO6 C, 64.43; H, 5.24; N, 2.35; found C, 62.51; H, 5.28; N, 2.41. UV-vis [CH3OH, λmax, nm (ε/M−1 cm−1)]: 418 (28,800), 375 (28,900), 257 (17,650). FT-IR (KBr pellet, cm−1): 3420(br), 3000(w), 2960(w), 2930(w), 2943(w), 1836(br), 1580(sh), 1511(sh), 1460(m), 1450(m), 1429(w), 1384(br), 1325(w), 1284(m), 1270(s), 1245(s), 1208(s), 1184(s), 1157(m), 1125(sh), 1030(m), 938(w), 848(m), 817(m), 730(w), 550(w), 455(br). 1H NMR (400 MHz, CDCl3) δ 7.76 (t, 2H, J = 6.9 Hz), 7.52 (d, 1H, J = 16 Hz), 7.18 to 7.15 (m, 1H), 7.05 to 7.03 (m, 2H), 6.97 to 6.87 (m, 4H), 6.81 (d, 1H, J = 16 Hz), 5.91 (s, 2H), 3.93 (s, 3H), 3.90 (s, 3H), 3.84 (s, 3H), 3.80 (s, 6H); 13C [1] NMR (125 MHz, CDCl3) δ 199.2, 186.6, 162.6, 148.8, 148.3, 147.9, 147.3, 146.8, 146.8, 144.6, 140.9, 136.7, 131.1, 127.5, 126.7, 125.6, 123.9, 123.4, 122.5, 120.0, 114.8, 111.7, 110.4, 110.1, 36.5, 31.5.ESI-HRMS (Methanol) m/z (calc.): 618.1411 (618.1421) [C32H31Cl2NO6Na]+ (Figs. S10–S13).
(1E,6E)-1,7-bis(4-hydroxy-3-methoxyphenyl)-4-(4-(2-morpholinoethoxy)benzylidene)hepta-1,6-diene-3,5-dione (5)
Curcumin (1.0 mmol) and 4-(2-morpholinoethoxy)benzaldehyde (2.0 mmol) were used as reactants. Pure product was isolated from silica gel chromatography by using 1:1 ethylacetate-hexane mixture as eluent. Yield: 42%.Anal. Cal. for C34H35NO8 C: 69.73; H: 6.02; N: 2.39; Found C: 69.68; H: 5.96; N: 2.41. UV-vis [CH3OH, λmax, nm (ε/M−1 cm−1)]: 364 (31,263); FT-IR (KBr pellet, cm−1): 3433(br), 2958(w), 2924(w), 2848(w), 1594(sh), 1511(sh), 1459(m), 1427(m), 1384(m)1266(s), 1167(s), 1115(s), 1029(m), 814(w); 1H NMR (500 MHz, CDCl3) δ 7.80 (s, 1H), 7.73 (d, J = 15.4 Hz, 1H), 7.45 (t, J = 12.2 Hz, 3H), 7.14 (d, J = 10 Hz, 1H), 7.01 (d, J = 9.1 Hz, 2H), 6.95 (d, J = 15.6 Hz, 2H), 6.91 to 6.85 (m, 2H), 6.82 (d, J = 8.8 Hz, 2H), 6.77 (s, 1H), 4.10 (t, J = 5.5 Hz, 2H), 3.89 (s, 3H), 3.86 (s, 3H), 3.74 to 3.70 (m, 4H), 2.80 (t, J = 5.5 Hz, 2H), 2.58 (s, 4H); 13C NMR (125 MHz, CDCl3) δ 198.66, 186.81, 160.38, 148.90, 148.52, 147.43, 146.84, 145.15, 140.35, 138.57, 132.42, 127.29, 126.60, 126.24, 125.31, 123.99, 123.56, 119.79, 114.88, 110.41, 110.00, 77.24, 76.99, 76.73, 66.61, 65.64, 57.34, 55.96, 53.92; ESI-HRMS (Methanol) m/z (calc.): 585.2452 (586.2435) [C34H35N1O8H+] (Figs. S14–S17).
Protein expression and purification
Kinase domain of Src (amino acid residues 254–535) and Hck (amino acid residues 166–445) were gifted from Markus Seeliger. The Abl (amino acid residues 229–460) and DYRK-2 (amino acid residues 146–540) plasmids were a gift from John Chodera, and Nicholas Levinson, and Markus Seeliger (Addgene plasmid # 79727). The Abl, Src, and Hck kinase domain were coexpressed with the YopH phosphatase and Trigger factor in E. coli BL21(DE3) and purified as described previously (54, 56). The DYRK2 was coexpressed with λ-phosphatase and Trigger factor and purified as described previously (3, 54). Briefly, the bacterial cells were grown to an OD600 of 1.2 at 37 °C before induction with 1 mM IPTG for 16 h at 18 °C. For purification of Src, Hck, and Abl kinase domain, cells were harvested and resuspended in buffer-A (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 25 mM imidazole, 5% glycerol). Cells expressing DYRK2 kinase domain were resuspended in lysis buffer (50 mM HEPES, pH 7.5, 500 mM NaCl, 10 mM imidazole, 5% glycerol and 5 mM β-mercaptoethanol). Cells were lysed by using cell homogenizer or sonication. Protein was purified by using nickel-affinity chromatography. The N-terminal His tag in Abl and Hck kinase domain was removed by dialyzing overnight in Buffer-C (20 mM Tris,100 mM NaCl, 1 mM DTT, 5% glycerol) containing Tev-protease. The YopH phosphatase and other contaminants were removed by anion-exchange chromatography followed by size-exclusion chromatography in the gel filtration buffer, (for Src, Hck, and Abl kinase, 50 mM Tris-HCl, pH8.0, 100 mM NaCl, 5% glycerol, 1 mM DTT were used) or (DYRK kinase, 25 mM HEPES, pH 7.5, 500 mM NaCl, 5 mM DTT were used). The purified protein was concentrated and stored at −80 °C.
Activity and IC50 measurement
Activity of Src, Hck, and Abl kinase was monitored by measuring the rate of phosphorylation of an Src-specific peptide (EAIYAAPFAKKK), and for DYRK2 Woodtide (KKISGRLSPIMTEQ) was used (3, 57). Reaction was continuously monitored by measuring the rate of oxidation of NADH to NAD in a buffer containing 10 mM MgCl2, 2.2 mM ATP, 100 mM Tris (pH 8.0), 1 mM Phosphoenolpyruvate (PEP), 0.6 mg/ml NADH, 75 U/ml Pyruvate Kinase, 105 U/ml lactate dehydrogenase, and 0.5 mM substrate peptide (58). For DYRK2, 1 mM ATP was used in each reaction. All the reactions were initiated by the addition of kinase at a final concentration of 35 to 100 nM. Decrease in NADH absorbance at 340 nM was monitored for 40 min at 25 °C. All the compounds were dissolved in DMSO and diluted in the gel filtration buffer. To determine the effect of curcumin and its derivatives, the enzyme and the compounds were incubated at 25 °C for 15 min before initiating the kinase reaction. Inhibitory constant (Ki) was calculated from IC50 according to the following relationship (59):
where [ATP] is the concentration of ATP in the assay and Km is the Michaelis constant for ATP. Km for ATP was determined for the Src kinase domains to be 99 μM and 210 μM in the absence and presence of 0.3% DMSO, respectively. The data analysis and curve fitting were done with GraphPad Prism 5 Ver 5.03. The Vmax and Km were determined by fitting Michaelis–Menten curve to the rate of phosphorylation measured at indicated ATP concentration.
The autophosphorylated Src or Hck kinase domain was prepared by incubating 10 μM the kinase with 1 mM ATP and 10 mM MgCl2 for 2 h at 25 °C, as described previously (42).
Autophosphorylation assays
The rate of autophosphorylation was analyzed by western blot using total antiphosphotyrosine antibody. In total, 250 nM of Src or Hck kinase domain was incubated with 15 μM Compound 4 or with DMSO at 25 °C for 15 min. The Src autophosphorylation was initiated by adding 1 mM ATP, 10 mM MgCl2, and 1 mM Na3VO4 at 25 °C (42). The reaction was stopped at indicated time points using 25 μl of 2X SDS-PAGE loading buffer. Level of phosphorylation and amount of Src Kinase domain loaded were detected by antiphosphotyrosine antibody (pY)(Abcam) and anti-His antibody (Biobharati), respectively. The amount of Hck loaded in each well was determined by Coomassie stain. Rate of phosphorylation was determined from the plot of normalized intensity of antiphosphotyrosine antibody (pY): amount of protein loaded in respective wells against time. The Image J software was used to determine the intensity from the blot (60). Rate of autophosphorylation was calculated from linear curve fitting.
Cytotoxicity by MTT assay
Cytotoxicity of 1 and 4 was determined against MDA-MB-231, HT-29 cells, RAW 264.7, and HEK 293T cell lines using MTT assay. In brief, cells were seeded in 96-well plate at a cell density of 6 × 106 cells per well in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% foetal bovine serum (FBS) (200 μl per well), antibiotics (100 units ml−1 penicillin and 100 μg ml−1 streptomycin) incubated at 37 °C and maintained in a 5% CO2 environment. RAW 264.7 cell line was grown in RPMI media supplemented with 10% FBS, antibiotics (100 units ml−1 penicillin and 100 μg ml−1 streptomycin). After 48 h of growth, the media was removed and replaced with fresh media. The compounds were dissolved in DMSO (molecular biology grade) and then diluted with growth media before adding to the cell culture at the desired concentration. In all the wells, final concentration of DMSO was within 0.2%. Cells were then incubated with 1 or 4 for another 48 h. After the incubation is over, the drug-containing media was removed and wells were replenished with fresh media (200 μl) followed by treatment with 20 μl of 1 mg/ml MTT dissolved in PBS (pH 7.2). After 3 h of incubation, media was removed and 200 μl of DMSO was added to each well. The inhibition of cell growth induced by the tested complexes was detected by measuring the absorbance of each well at 595 nm using SpectraMax M2e plate reader. The data was plotted and IC50 was calculated using GraphPad Prism 5 Ver 5.03. Each experiment was performed in triplicate, and average data with standard deviations are reported.
Wound-healing assay
MDA-MB-231 cells were grown to confluence in 6-well plates in DMEM medium supplemented with 10% FBS at 37 °C in a 5% CO2 atmosphere. A single wound was generated in each well by scratching gently with a sterile pipette tip. Then cells were washed twice with PBS (pH 7.2) followed by addition of fresh DMEM media containing 10% FBS. The respective IC10 (100 nM and 600 nM) and IC50 (1.3 μM and 13 μM) doses of 4 and 1 were used for the studies. The rate of wound healing was determined from the analysis of images of the inflicted wound recorded at different time points (ImageJ software). The percentage of wound closure was calculated by measuring the area unoccupied by cells at each time point and normalizing to the zero time point. Error bars represent the standard deviation from three independent experiments.
Compound stability
The compound stability was determined both in vitro and in a cell-based assay. The solution stability of 1 to 5 was determined by dissolving the compounds in 5% DMSO containing 1× PBS of pH 7.4 and measuring the absorbance for 24 h at 25 °C. The stability of 1 to 5 was also determined in the kinase assay buffer (50 mM Tris-HCl, pH 8.0/100 mM, NaCl/5%, glycerol/1 mM DTT) using 5% DMSO for the period of kinase assay (1 h).
The stability of the compound in cells was determined by recording the fluorescence images of 1 or 4-treated cells for 14 h. MDA-MB-231 cells were seeded in a glass-bottomed 35 mm Petri dish (Corning) at a density of 0.2 × 106 cells per plate. At 80% confluency, cells were treated with 10 μM of 1 or 4 as described previously in the cytotoxicity assay. Cells were allowed to grow for 1 h and washed with prewarmed 1X PBS and opti-MEM was added. Live-cell images of the cells were recorded at 1 h, 2 h, 3 h, 4 h, 5 h, 6 h, 10 h, and 14 h time point using a Leica confocal microscope. The fluorescence intensity for each cell was measured and analyzed by using the ImageJ software. For each time interval, background fluorescence of cells was subtracted from the control cells. The fluorescence intensity per μm2 for 15 cells was plotted against time; the error bar represents the standard deviation within the data set.
In-gel fluorescence assay was used to determine any covalent modification. In total, 10 μM Src-KD was incubated with 1 and 4 at 1:1 for 25 min at room temperature. Samples were run in 8% Native-PAGE at 4 °C. Gel fluorescence was checked at 400 to 500 nm and then stained with Coomassie brilliant blue.
HPLC and mass spectrometry
HPLC was done by using C18 reverse-phase HPLC column. Control sample was prepared by incubating 0.5 mg/ml Src-KD with 5 M Guanidinium chloride (Gu.HCl) for 10 min. To determine if 4 is covalently modifying the Src kinase domain, 0.5 mg/ml Src was incubated with an equimolar ratio of 4 for 1 h and then the sample was treated with 5 M Gu.HCl for 10 min. In total, 100 μl of each sample was injected into the column that was pre-equilibrated with 5% CH3CN + 95% water +0.1% TFA. Protein was eluted by a linear gradient of 10 to 90% CH3CN+0.1% TFA. The elution of protein and 4 was monitored by measuring the absorbance at 280 and 416 nm, respectively. The pure eluted fractions were used for ESI-MS mass spectrometry (Bruker maXis II instrument).
Ligand docking
Molecular docking was carried out using Autodock vina 4.0 (61).The structure of 4 was optimized in Gaussian09 software package (Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford, CT, 2016). Optimization of 4 was performed at the DFT level of theory with B3LYP function and 6-31G(d,p) basis set (62). Orbitals were defined as restricted during calculations. The conductor-like polarizable continuum model (CPCM) was used with water as the solvent during optimization (63, 64, 65). Structures of Src and Abl kinases were used from the protein data bank (PDB ID: 2SRC, 2OIQ, 4MXO, 1OPL, 1OPJ, 2G1T). Gaussian optimized structures of 1 to 5 were used as the ligand input files. Kinase structures were further optimized, and polar hydrogens were assigned by OPLS (optimized potential for liquid simulations) 3 force field to rectify the molecular geometries to get least energy conformations required for docking. All the water molecules (except those present in the binding site) were optimized for hydrogen-bonding network using OPLS3, corresponding to pH 7 (66). The receptor grid box was generated using autodock tools, surrounding ATP-binding domain of the respective kinase structures (Table 1). The vina output results along with possible interactions were visualized with PyMOL Molecular Graphics System, Version 2.0 (DeLano, W. L. (2009). The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, 2002).
Table 1.
The receptor grid boxes generated using autodock tools
| Conformation | PDB-ID | Grids used in docking study | |
|---|---|---|---|
| DFG-in inactive Src | 2SRC | center_x = 12.746 center_y = 19.425 center_z = 60.464 |
size_x = 24 size_y = 14 size_z = 12 |
| DFG-in active Src | 4MXO | center_x = 12.846 center_y = −36.933 center_z = −6.653 |
size_x = 17.5 size_y = 10 size_z = 13 |
| DFG-in inactive Abl | 2G1T | center_x = −1.239 center_y = 9.941 center_z = −40.033 |
size_x = 21.0 size_y = 19.5 size_z = 15.5 |
| DFG-out inactive Abl | 1OPJ | center_x = 16.917 center_y = 19.306 center_z = 52.058 |
size_x = 22 size_y = 12 size_z = 12 |
| DFG-in active Abl | 1OPL | center_x = 20.192 center_y = 50.378 center_z = 49.823 |
size_x = 14 size_y = 14 size_z = 20 |
Immunofluorescence microscopy
The nuclear translocation of ERK1/2 and STAT3 was determined by confocal microscopy of activated MDA-MB-231 and RAW 264.7 lines, respectively. The cells were grown on coverslip up to a confluency of 90% and treated with 1 μM and 5 μM 4 for 6 h. After incubation, cells were washed with PBS and activated. The MDA-MB-231 and RAW 264.7 cell lines were activated using 50 nM TNFα and 100 nM LPS for 30 min and 8 h, respectively. After activation, cell were washed three times with 1 PBS and fixed with 4% paraformaldehyde for 30 min. The cells were then permeabilized with 0.2% Triton X-100 for 20 min and blocked in 3% bovine serum albumin for 1 h. The cells were incubated with respective primary antibodies (STAT3 and ERK1/2) in blocking buffer for overnight. After washing three times with PBS containing 0.1% Tween-20, cells were incubated with secondary antibody conjugated with alexa fluor 562 and phalloidin (Invitrogen) for 2 h. Finally, coverslips were mounted with DAPI (4′,6-diamidino-2-phenylindole) and imaged under confocal laser scanning microscope (Leica TCS SP8).
Immunoprecipitation
In cell activity of Src kinase and its downstream signaling molecules such as ERK1/2 and STAT3 in response to 4 was determined by probing the phosphorylation level of respective immunoprecipitated proteins from MDA-MB-231 and RAW 264.7 cell line. MDA-MB-231 cells were grown up to 70% confluency and then treated with 1 to 10 μM of 4 or DMSO for 6 h. After incubation cells were activated as described previously. Activated cells were harvested and lysed with RIPA lysis buffer (25 mM Tris/HCl pH 7.4, 1 mM EDTA, 100 mM NaCl, 1% Nonidet P40, 1% Triton-x100 supplemented with 0.1 mM vanadate) and a protease-inhibitor cocktail (5 mg/l leupeptin, 0.1 mM phenylmethylsulphonyl fluoride, 2 mM Benzamidine). Cell lysate was sonicated and cleared by centrifuging at 15,000g for 10 min. The supernatant was incubated with anti-Src, anti-ERK1/2, or anti-STAT3 antibody for overnight at 4 °C. After overnight incubation, the antibody-bound protein solution was further incubated with 30 μl protein G beads for 2 h. The beads were washed four times with ice-cold PBS supplemented with 2 mM Na3VO4. The samples were boiled with Laemmli sample buffer and ran in SDS-PAGE. Following SDS-PAGE, lysates were transferred onto nitrocellulose paper and blocked with 5% skimmed milk in TBST (0.1% tween 20) for 2 h at room temperature. For detecting the level of phosphorylation, blocked membranes were incubated with total anti-pY antibody. To detect respective loading control, membranes were incubated with anti-Src, anti-ERK1/2, or anti-STAT3 for overnight at 4 °C followed by 2 h incubation with HRP-conjugated anti-mouse or anti-rabbit secondary antibody.
Western blot analysis of apoptosis makers
MDA-MB-231 cells were grown in DMEM/Ham’s F12 (50:50) media supplemented with 10% FBS and antibiotics at 37 °C in 5% CO2. To test the effect of 4 on the protein marker regulating the apoptotic pathway, the expression level of essential proteins such as Cytochrome C, caspase3, Bcl2, and BH3 interacting domain death agonist was observed. Cells were grown up to 90% confluency and then treated with 10 and 20 μM of 4 or DMSO vehicle for 12 h. The cells were lysed, and the protein samples were resolved by running through 6% SDS-PAGE. The protein was transferred onto nitrocellulose paper and blocked with 5% skimmed milk for 2 h at room temperature. The membrane was then incubated overnight at 4 °C with respective primary antibodies. The expression level of indicated proteins was detected with HRP-conjugated anti-rabbit secondary antibody. The normalized protein expression level was determined from the densitometric analysis of the respective blot using the program Image J (60).
Intracellular calcium detection
Time-dependent intracellular calcium content was measured with the help of the fluorescent dye, Indo-1 AM, using a multimode-plate reader (Synergy H1 Hybrid Multi-Mode Reader). RAW 264.7 cells were seeded in 96-well plates at 90% confluency before treating with 4 or DMSO for 3 h. The cells were washed twice with PBS and loaded with 1 μM of Indo1-AM for 60 min. Indo1-AM loaded cells were washed and activated with LPS (100 ng/ml). Real-time detection of change in fluorescence of Indo1-AM in live cells was measured every 15 s for 30 min at λex 355 nm and λem 400 nm and 475 nm. Fraction of Indo-1 AM bound to Ca2+ was obtained by the following equation and plotted as a function of time:
DNA cross-linking assay
The DNA cross-linking assay was performed by alkaline agarose gel electrophoresis. Purified pUC19 plasmid DNA (1500 ng) was incubated with various concentrations (0.1–20 μM) of melphalan (standard alkylating agent) or 4 in 40 μl binding buffer (3 mM sodium chloride/1 mM sodium phosphate, pH 7.4, and 1 mM EDTA) at 37 °C for 2 h. At the end of the reaction, the plasmid DNA was linearized by digestion with BamHI and precipitated with ethanol. The DNA was dissolved in alkaline buffer (0.5 N NaOH–10 mM EDTA) before resolving by 0.8% alkaline agarose gel electrophoresis run in the NaOH–EDTA buffer at 4 °C (67).
Pre-steady-state ligand-binding kinetics by stop-flow fluorescence spectroscopy
The interaction between the kinase domain and 4 was studied by measuring the kinetics of change in intrinsic tryptophan fluorescence of the kinase domain using a fluorimeter fitted with stop-flow attachment at room temperature. In all the experiments, λex and λem were fixed at 290 and 350 nm, respectively.
For the fluorescence kinetics measurements, the kinase domain at 100 nM concentration in 50 mM Tris (pH 8.0) buffer (containing 100 mM NaCl, 5% glycerol, 1 mM DTT) was titrated against indicated concentration of 4. To compare the kinetic transient of Abl-wt and Y253F-Abl in response to imatinib, 100 nM protein was mixed with 15 μM imatinib. Each transient was measured over 5 ms interval with 6000 to 12,000 time points and repeated four times. Data were fit to the following first-order kinetic equation:
The observed rate constants were plotted against the concentration of 4 and fitted to a linear function to determine the binding rate constant (kon) and dissociation rate constant (koff) from the slope and the intercept, respectively. Binding constant (Kd) was derived from the ratio of koff and kon.
Dynamic light scattering (DLS)
DLS measurements were performed at 30 °C using Malvern Zetasizer Nano ZS. Each measurement correlation time of 15 s per run was set, and ten runs per sample were collected. To check aggregation of 1 and 4, 15 μM of each compound was resuspended in assay buffer (50 mM Tris, 100 mM NaCl, 5% glycerol and 1 mM DTT) and incubated for 30 min at room temperature. Each sample was passed through 0.2 μm filters before measuring with DLS. Thirty percent (W/V) sucrose solution under the same condition was used as a monomer control (68). The imatinib was used as a reference sample for comparative study. The experimental results were analyzed using built-in Zetasizer software. During the calculation of hydrodynamic radius, a dispersant viscosity of 0.79 mPa and a refractive index of 1.33 were used.
Determination of plasma membrane integrity
MDA-MB-231 cells were seeded in a glass-bottomed 35 mm Petri dish at a density of 0.5 × 106 cells per plate. At 70 to 80% confluency, cells were treated with 15 μM of 1 and 4 as described previously in the cytotoxicity assay. After 6 h incubation, cells were washed with ice-cold Hanksʼ balanced salt solution (HBSS) without magnesium or calcium and immediately immersed in ice-cold FM4-64-FX staining solution (5 μg/ml) for 1 min on ice and immediately washed with HBSS (41). The live cells were imaged using LEICA TCS SP8 confocal microscope at λex/λem of 565 nm/744 and 480 nm/550 nm for the FM4-64-FX lipophilic dye and 4 (or 1), respectively.
Conclusion
In summary, we have designed a fluorescent, curcumin-based Hck inhibitor of nanomolar affinity and enhanced cellular stability. Hck inhibitors are shown to be useful in treating leukemia, management of viral-induced hyperimmune reactions (cytokine storm) and various autoimmune diseases. Almost all kinase inhibitors exhibit some degree of promiscuity. However, it is well understood that to reduce promiscuity, small molecules are designed to target the inactive conformations. Our biochemical studies suggest that the 4-aryl nitrogen-mustard curcumin derivative, 4, selectively targets the Hck over Abl, DYRK2, or Src kinase domain. Most importantly the selective targeting of the DFG-in inactive conformation of Hck prevents the structural transition of the kinase domain to the active state, which suggests that 4 may be less promiscuous. Cellular studies show that the cytotoxicity of 4 is predominantly due to inhibition of SFK-mediated cell signaling. This work widens the horizon of the fluorescent curcumin scaffold, through 4-arylidene modification at the methylenic position, to design kinase inhibitors with enhanced stability and remodeled target specificity.
Data availability
All the relevant data are contained within this article and in the supporting information.
Supporting information
This article contains supporting information (3, 5, 20, 42, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Professor John Kuriyan at the University of California, Berkeley, and Prof. Markus Seeliger at Stony Brook University for the Src, Yop, and Hck constructs. The authors thank the infrastructural facilities of Indian Institute of Science Education and Research (IISER) Kolkata. The authors are thankful to DST-FIST (Fund for Improvement of S&T infrastructure in universities & higher educational institutions, Department of Science and Technology) (SR/FST/LS-II/2017/93) for the analytical biology facility at IISER Kolkata.
Author contributions
The article was written through the contributions of all the authors. All the authors have approved the final version of the article. A. M. helped in the design, S. B. and S. R. synthesized and characterized the curcumin derivatives. S. B. and S. R. performed the stability studies. In vitro cytotoxicity was done by S. B., S. R., and M. P. C., R. D., M. P. C., and S. B. designed the biochemical and cell-based experiments. I. B. and M. P. C. performed the confocal microscopy.
Funding and additional information
The work presented in this article is extramurally funded by Science and Engineering Research Board (SERB), Government of India vide project no. EMR/2107/002324, ECR/2015/000142 and DBT (Department of Biotechnology) Ramalingaswami Fellowship (BT/RFF/Re-entry/14/2014) to R. D. We thank Dr Arnab Gupta for helping us with the confocal microscopy, a facility funded by DBT Wellcome Trust to A. G. Authors S. B. and M. P. C. thank CSIR (Council of Scientific and Industrial Research) and S. R. thanks INSPIRE for their respective research fellowships. We are thankful to IISER-K for the financial support.
Edited by Alex Toker
Contributor Information
Rahul Das, Email: rahul.das@iiserkol.ac.in.
Arindam Mukherjee, Email: a.mukherjee@iiserkol.ac.in.
Supporting information
References
- 1.Nelson Kathryn M., Walters Michael A., Dahlin Jayme L., Bisson J., Graham J., Pauli Guido F., Pauli Guido F. The essential medicinal chemistry of curcumin. J. Med. Chem. 2017;60:1620–1637. doi: 10.1021/acs.jmedchem.6b00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knight Z.A., Shokat K.M. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee S., Ji C., Mayfield J.E., Goel A., Xiao J., Dixon J.E., Guo X. Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2. Proc. Natl. Acad. Sci. U. S. A. 2018;115:8155–8160. doi: 10.1073/pnas.1806797115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Zou Q., Yuan C., Li S., Xing R., Yan X. Amino acid coordination driven self-assembly for enhancing both the biological stability and tumor accumulation of curcumin. Angew. Chem. Int. Ed. Engl. 2018;57:17084–17088. doi: 10.1002/anie.201810087. [DOI] [PubMed] [Google Scholar]
- 5.Qiu X., Du Y., Lou B., Zuo Y., Shao W., Huo Y., Huang J., Yu Y., Zhou B., Du J., Fu H., Bu X. Synthesis and identification of new 4-arylidene curcumin analogues as potential anticancer agents targeting nuclear factor-κB signaling pathway. J. Med. Chem. 2010;53:8260–8273. doi: 10.1021/jm1004545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J., Geng G., Shi Q., Sauriol F., Wu J.H. Design and synthesis of androgen receptor antagonists with bulky side chains for overcoming antiandrogen resistance. J. Med. Chem. 2009;52:5546–5550. doi: 10.1021/jm801218k. [DOI] [PubMed] [Google Scholar]
- 7.Lin L., Shi Q., Nyarko A.K., Bastow K.F., Wu C.-C., Su C.-Y., Shih C.C.Y., Lee K.-H. Antitumor agents. 250.Design and synthesis of new curcumin analogues as potential anti-prostate cancer agents. J. Med. Chem. 2006;49:3963–3972. doi: 10.1021/jm051043z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S., Huang W.-C., Li P., Guo H., Poh S.-B., Brady S.W., Xiong Y., Tseng L.-M., Li S.-H., Ding Z., Sahin A.A., Esteva F.J., Hortobagyi G.N., Yu D. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat. Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim L.C., Song L., Haura E.B. Src kinases as therapeutic targets for cancer. Nat. Rev. Clin. Oncol. 2009;6:587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 10.Belsches-Jablonski A.P., Biscardi J.S., Peavy D.R., Tice D.A., Romney D.A., Parsons S.J. Src family kinases and HER2 interactions in human breast cancer cell growth and survival. Oncogene. 2001;20:1465. doi: 10.1038/sj.onc.1204205. [DOI] [PubMed] [Google Scholar]
- 11.Gordon O.N., Schneider C. Vanillin and ferulic acid: Not the major degradation products of curcumin. Trends Mol. Med. 2012;18:361–363. doi: 10.1016/j.molmed.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider C., Gordon O.N., Edwards R.L., Luis P.B. Degradation of curcumin: From mechanism to biological implications. J. Agric. Food Chem. 2015;63:7606–7614. doi: 10.1021/acs.jafc.5b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poh Ashleigh R., O'Donoghue Robert J.J., Ernst M., O'Donoghue Robert J.J., Ernst M. Hematopoietic cell kinase (HCK) as a therapeutic target in immune and cancer cells. Oncotarget. 2015;6:15752–15771. doi: 10.18632/oncotarget.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito Y., Yuki H., Kuratani M., Hashizume Y., Takagi S., Honma T., Tanaka A., Shirouzu M., Mikuni J., Handa N., Ogahara I., Sone A., Najima Y., Tomabechi Y., Wakiyama M. A pyrrolo-pyrimidine derivative targets human primary AML stem cells in vivo. Sci. Transl. Med. 2013;5:116. doi: 10.1126/scitranslmed.3004387. [DOI] [PubMed] [Google Scholar]
- 15.Miranda M.B., Johnson D.E. Signal transduction pathways that contribute to myeloid differentiation. Leukemia. 2007;21:1363–1377. doi: 10.1038/sj.leu.2404690. [DOI] [PubMed] [Google Scholar]
- 16.Ernst M., Inglese M., Scholz G.M., Harder K.W., Clay F.J., Bozinovski S., Waring P., Darwiche R., Kay T., Sly P., Collins R., Turner D., Hibbs M.L., Anderson G.P., Dunn A.R. Constitutive activation of the Src family kinase Hck results in spontaneous pulmonary inflammation and an enhanced innate immune response. J. Exp. Med. 2002;196:589–604. doi: 10.1084/jem.20020873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treon S.P., Castillo J.J., Skarbnik A.P., Soumerai J.D., Ghobrial I.M., Guerrera M.L., Meid K., Yang G. The BTK inhibitor ibrutinib may protect against pulmonary injury in COVID-19-infected patients. Blood. 2020;135:1912–1915. doi: 10.1182/blood.2020006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornev A.P., Haste N.M., Taylor S.S., Ten Eyck L.F. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble M.E.M., Endicott J.A., Johnson L.N. Protein kinase inhibitors: Insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 20.Yue X., Zuo Y., Ke H., Luo J., Lou L., Qin W., Wang Y., Liu Z., Chen D., Sun H., Zheng W., Zhu C., Wang R., Wen G., Du J. Identification of 4-arylidene curcumin analogues as novel proteasome inhibitors for potential anticancer agents targeting 19S regulatory particle associated deubiquitinase. Biochem. Pharmacol. 2017;137:29–50. doi: 10.1016/j.bcp.2017.04.032. [DOI] [PubMed] [Google Scholar]
- 21.Chandrasekar T., Raman N. Exploration of cellular DNA lesion, DNA-binding and biocidal ordeal of novel curcumin based Knoevenagel Schiff base complexes incorporating tryptophan: Synthesis and structural validation. J. Mol. Struct. 2016;1116:146–154. [Google Scholar]
- 22.Seeliger M.A., Nagar B., Frank F., Cao X., Henderson M.N., Kuriyan J. c-Src binds to the cancer drug imatinib with an inactive Abl/c-kit conformation and a distributed thermodynamic penalty. Structure. 2007;15:299–311. doi: 10.1016/j.str.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Nagar B., Bornmann W.G., Pellicena P., Schindler T., Veach D.R., Miller W.T., Clarkson B., Kuriyan J. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 24.Schindler T., Bornmann W., Pellicena P., Miller W.T., Clarkson B., Kuriyan J. Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 25.Kwarcinski F.E., Brandvold K.R., Phadke S., Beleh O.M., Johnson T.K., Meagher J.L., Seeliger M.A., Stuckey J.A., Soellner M.B. Conformation-selective analogues of dasatinib reveal insight into kinase inhibitor binding and selectivity. ACS Chem. Biol. 2016;11:1296–1304. doi: 10.1021/acschembio.5b01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamontanara Allan J., Georgeon S., Tria G., Svergun Dmitri I., Hantschel O. The SH2 domain of Abl kinases regulates kinase autophosphorylation by controlling activation loop accessibility. Nat. Commun. 2014;5:5470. doi: 10.1038/ncomms6470. [DOI] [PubMed] [Google Scholar]
- 27.Osusky M., Taylor S.J., Shalloway D. Autophosphorylation of purified c-Src at its primary negative regulation site. J. Biol. Chem. 1995;270:25729–25732. doi: 10.1074/jbc.270.43.25729. [DOI] [PubMed] [Google Scholar]
- 28.Smart J.E., Oppermann H., Czernilofsky A.P., Purchio A.F., Erikson R.L., Bishop J.M. Characterization of sites for tyrosine phosphorylation in the transforming protein of Rous sarcoma virus (pp60v-src) and its normal cellular homolog (pp60c-src) Proc. Natl. Acad. Sci. U. S. A. 1981;78:6013–6017. doi: 10.1073/pnas.78.10.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hari S.B., Perera B.G.K., Ranjitkar P., Seeliger M.A., Maly D.J. Conformation-selective inhibitors reveal differences in the activation and phosphate-binding loops of the tyrosine kinases Abl and Src. ACS Chem. Biol. 2013;8:2734–2743. doi: 10.1021/cb400663k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter T. A tail of two src's: Mutatis mutandis. Cell. 1987;49:1–4. doi: 10.1016/0092-8674(87)90745-8. [DOI] [PubMed] [Google Scholar]
- 31.Schindler T., Sicheri F., Pico A., Gazit A., Levitzki A., Kuriyan J. Crystal structure of HcK in complex with a Src family-selective tyrosine kinase inhibitor. Mol. Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi H., Hendrickson W.A. Structural basis for activation of human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature. 1996;384:484–489. doi: 10.1038/384484a0. [DOI] [PubMed] [Google Scholar]
- 33.Johnson L.N., Noble M.E.M., Owen D.J. Active and inactive protein kinases: Structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 34.Lovera S., Sutto L., Boubeva R., Scapozza L., Dolker N., Gervasio F.L. The different flexibility of c-Src and c-Abl kinases regulates the accessibility of a druggable inactive conformation. J. Am. Chem. Soc. 2012;134:2496–2499. doi: 10.1021/ja210751t. [DOI] [PubMed] [Google Scholar]
- 35.Guimaraes C.R.W., Rai B.K., Munchhof M.J., Liu S., Wang J., Bhattacharya S.K., Buckbinder L. Understanding the impact of the P-loop conformation on kinase selectivity. J. Chem. Inf. Model. 2011;51:1199–1204. doi: 10.1021/ci200153c. [DOI] [PubMed] [Google Scholar]
- 36.Kwarcinski F.E., Fox C.C., Steffey M.E., Soellner M.B. Irreversible inhibitors of c-Src kinase that target a nonconserved cysteine. ACS Chem. Biol. 2012;7:1910–1917. doi: 10.1021/cb300337u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agafonov R.V., Wilson C., Otten R., Buosi V., Kern D. Energetic dissection of Gleevec's selectivity toward human tyrosine kinases. Nat. Struct. Mol. Biol. 2014;21:848–853. doi: 10.1038/nsmb.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roumiantsev S., Shah N.P., Gorre M.E., Nicoll J., Brasher B.B., Sawyers C.L., Van Etten R.A. Clinical resistance to the kinase inhibitor STI-571 in chronic myeloid leukemia by mutation of tyr-253 in the Abl kinase domain P-loop. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10700–10705. doi: 10.1073/pnas.162140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Z.-N., Zhang F., Tang P., Qi X.-W., Jiang J. RANKL-induced migration of MDA-MB-231 human breast cancer cells via Src and MAPK activation. Oncol. Rep. 2011;26:1243–1250. doi: 10.3892/or.2011.1368. [DOI] [PubMed] [Google Scholar]
- 40.Golubovskaya V.M., Gross S., Kaur A.S., Wilson R.I., Xu L.-H., Yang X.H., Cance W.G. Simultaneous inhibition of focal adhesion kinase and Src enhances detachment and apoptosis in colon cancer cell lines. Mol. Cancer Res. 2003;1:755–764. [PubMed] [Google Scholar]
- 41.Revelo N.H., Kamin D., Truckenbrodt S., Wong A.B., Reuter-Jessen K., Reisinger E., Moser T., Rizzoli S.O. A new probe for super-resolution imaging of membranes elucidates trafficking pathways. J. Cell Biol. 2014;205:591–606. doi: 10.1083/jcb.201402066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aleem S.U., Georghiou G., Kleiner R.E., Guja K.E., Craddock B.P., Lyczek A., Chan A.I., Garcia-Diaz M., Miller W.T., Liu D.R., Seeliger M.A. Structural and biochemical basis for intracellular kinase inhibition by Src-specific peptidic macrocycles. Cell Chem. Biol. 2016;23:1103–1112. doi: 10.1016/j.chembiol.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai P.-C., Chu C.-L., Chiu C.-C., Chang L.-S., Lin S.-R. Inhibition of Src activation with cardiotoxin III blocks migration and invasion of MDA-MB-231 cells. Toxicon. 2013;74:56–67. doi: 10.1016/j.toxicon.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 44.Wolczyk D., Zaremba-Czogalla M., Hryniewicz-Jankowska A., Tabola R., Grabowski K., Sikorski A.F., Augoff K. TNF-α promotes breast cancer cell migration and enhances the concentration of membrane-associated proteases in lipid rafts. Cell. Oncol. 2016;39:353–363. doi: 10.1007/s13402-016-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plotnikov A., Flores K., Maik-Rachline G., Zehorai E., Kapri-Pardes E., Berti D.A., Hanoch T., Besser M.J., Seger R. The nuclear translocation of ERK1/2 as an anticancer target. Nat. Commun. 2015;6:6685. doi: 10.1038/ncomms7685. [DOI] [PubMed] [Google Scholar]
- 46.Tamura Y., Simizu S., Osada H. The phosphorylation status and anti-apoptotic activity of Bcl-2 are regulated by ERK and protein phosphatase 2A on the mitochondria. FEBS Lett. 2004;569:249–255. doi: 10.1016/j.febslet.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Balmanno K., Cook S.J. Tumour cell survival signalling by the ERK1/2 pathway. Cell Death Differ. 2009;16:368–377. doi: 10.1038/cdd.2008.148. [DOI] [PubMed] [Google Scholar]
- 48.Rooswinkel R.W., van de Kooij B., de Vries E., Paauwe M., Braster R., Verheij M., Borst J. Antiapoptotic potency of Bcl-2 proteins primarily relies on their stability, not binding selectivity. Blood. 2014;123:2806–2815. doi: 10.1182/blood-2013-08-519470. [DOI] [PubMed] [Google Scholar]
- 49.Teijido O., Dejean L. Upregulation of Bcl2 inhibits apoptosis-driven BAX insertion but favors BAX relocalization in mitochondria. FEBS Lett. 2010;584:3305–3310. doi: 10.1016/j.febslet.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Cheng E.H.Y.A., Wei M.C., Weiler S., Flavell R.A., Mak T.W., Lindsten T., Korsmeyer S.J. BCL-2, BCL-XL sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 51.Poh A.R., Love C.G., Masson F., Preaudet A., Tsui C., Whitehead L., Monard S., Khakham Y., Burstroem L., Lessene G., Sieber O., Lowell C., Putoczki T.L., O'Donoghue R.J.J., Ernst M. Inhibition of hematopoietic cell kinase activity suppresses myeloid cell-mediated colon cancer progression. Cancer Cell. 2017;31:563–575.e565. doi: 10.1016/j.ccell.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suen P.W., Ilic D., Caveggion E., Berton G., Damsky C.H., Lowell C.A. Impaired integrin-mediated signal transduction, altered cytoskeletal structure and reduced motility in Hck/Fgr deficient macrophages. J. Cell Sci. 1999;112:4067–4078. doi: 10.1242/jcs.112.22.4067. [DOI] [PubMed] [Google Scholar]
- 53.Armarego W.L.F., Perrin D.D. 4th Ed. Elsevier; Burlington, MA: 1997. Purification of Laboratory Chemicals. [Google Scholar]
- 54.Albanese S.K., Parton D.L., Isik M., Rodriguez-Laureano L., Hanson S.M., Behr J.M., Gradia S., Jeans C., Levinson N.M., Seeliger M.A., Chodera J.D. An open library of human kinase domain constructs for automated bacterial expression. Biochemistry. 2018;57:4675–4689. doi: 10.1021/acs.biochem.7b01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandrasekar T., Pravin N., Raman N. DNA incision evaluation, binding investigation and biocidal screening of novel metallonucleases of 1,7-bis(4-hydroxy-3-methoxyphenyl)hepta-1,6-diene-3,5-dione based Knoevenagel condensate having methionine: Synthesis and structural validation. J. Mol. Struct. 2015;1081:477–485. [Google Scholar]
- 56.Seeliger M.A., Young M., Henderson M.N., Pellicena P., King D.S., Falick A.M., Kuriyan J. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foda Z.H., Shan Y., Kim E.T., Shaw D.E., Seeliger M.A. A dynamically coupled allosteric network underlies binding cooperativity in Src kinase. Nat. Commun. 2015;6:5939. doi: 10.1038/ncomms6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barker S., Kassel D.B., Weigl D., Huang X., Luther M., Knight W.B. Characterization of pp60c-Src tyrosine kinase activities using a continuous assay: Autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- 59.Cheng Y., Prusoff W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 60.Hartig Sean M. Basic image analysis and manipulation in ImageJ. Curr. Protoc. Mol. Biol. 2013 doi: 10.1002/0471142727.mb1415s102. Chapter 14:Unit14.15. [DOI] [PubMed] [Google Scholar]
- 61.Forli S., Huey R., Pique M.E., Sanner M.F., Goodsell D.S., Olson A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016;11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 63.Rashin A.A., Namboodiri K. A simple method for the calculation of hydration enthalpies of polar molecules with arbitrary shapes. J. Phys. Chem. 1987;91:6003–6012. [Google Scholar]
- 64.Stefanovich E.V., Truong T.N. Optimized atomic radii for quantum dielectric continuum solvation models. Chem. Phys. Lett. 1995;244:65–74. [Google Scholar]
- 65.Aguilar M.A., del Valle F.J.O. Solute-solvent interactions. A simple procedure for constructing the solvent cavity for retaining a molecular solute. Chem. Phys. 1989;129:439. [Google Scholar]
- 66.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., Wang L., Lupyan D., Dahlgren M.K., Knight J.L., Kaus J.W., Cerutti D.S., Krilov G., Jorgensen W.L., Abel R. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theor. Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 67.Hartley J.A., Berardini M.D., Souhami R.L. An agarose gel method for the determination of DNA interstrand crosslinking applicable to the measurement of the rate of total and “second-arm” crosslink reactions. Anal. Biochem. 1991;193:131–134. doi: 10.1016/0003-2697(91)90052-u. [DOI] [PubMed] [Google Scholar]
- 68.Kaszuba M., McKnight D., Connah M.T., McNeil-Watson F.K., Nobbmann U. Measuring sub nanometre sizes using dynamic light scattering. J. Nanoparticle Res. 2008;10:823–829. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the relevant data are contained within this article and in the supporting information.