Abstract
Upregulation of Angiotensin Converting Enzyme-2 (ACE2) was frequently observed in patients with lung cancer. Interestingly, our recent study revealed that the same ACE2 receptor was also strongly upregulated in lungs during SARS-CoV2 infection. Therefore, it is possible that the upregulated expression of ACE2 in lung tumors might increase the susceptibility to COVID-19 infection in lung cancer patients. However, the molecular mechanism for the regulation of ACE2 is known neither in lung tumors nor in COVID-19. Under this review, we attempt to identify transcription factors (TFs) in the promoter of ACE2 that promote the expression of ACE2 both in COVID-19 infection and lung cancer. This review would decipher the molecular role of ACE2 in the upscaled fatality of lung cancer patients suffering from COVID-19.
Abbreviations: ACE2, Angiotensin Converting Enzyme 2; RAS, Renin Angiotensin System; HBV, Hepatitis B virus; HCV, Hepatitis C virus; HSV, Herpes Simplex Virus; STAT, signal transducer and activator of transcription
Keywords: SARS-CoV2, ACE2, COVID-19, Lung cancer, STAT3
1. Physiological role of ACE2 in fluid balance, salt re-absorption, blood pressure maintenance and vasodilation
ACE2 is an important enzyme in renin-angiotensin system (RAS) of angiotensin metabolism (Burrell et al., 2004a). RAS is widely known endocrine pathway that regulates electrolyte balance, body fluid volume and cardiovascular control in peripheral circulation (Fountain and Lappin, 2020). RAS initiates with the reduction of renal blood flow and drop in fluid volume that directly stimulates the release of renin from kidney cortex (Handa and Johns, 1985) that in turn converts angiotensin precursor protein angiotensinogen to angiotensin-I (Reid and Moffat, 1978). Angiotensin converting enzyme (ACE), located at lungs, then enzymically transforms angiotensin-I to angiotensin-II (Erdos, 1976), which binds to the angiotensin1 subtype b (AT1b) receptor (Dabouras et al., 1973; Peach, 1977), triggers inward calcium current (Zhu et al., 1998), activates calcium-dependent CAM Kinase (CAMK) (Thomas et al., 1999), and phosphorylates Myosin light chain (MLC)(Anderson et al., 1981). Phosphorylation of MLC causes contraction of muscle that finally leads to the vasoconstriction (Zelis, 1983) resulting elevated blood pressure. In an indirect mechanism, angiotensin-1 also controls the reabsorption of minerals and water in kidney via synthesis of mineralocorticoid hormone in adrenal cortex (Brewster et al., 2003). Therefore, persistent activation of RAS pathway, upregulation of ACE, and elevated circulating levels of angiotensin- II directly lead to the development of hypertension and increased reabsorption in kidney. However, this physiological mechanism of water and salt balance by RAS has gained much importance after the discovery of angiotensin converting enzyme-2 in 2000 (Donoghue et al., 2000). Since then, ACE2 has been characterized as a ubiquitously-expressed, membrane-bound receptor that counterbalances the physiological action of ACE via enzymic conversion of angiotensin-II to Ang (1–7)(Allred et al., 2000). As a result, ACE2 facilitates the vasodilative response to lower blood pressure and maintain the fluid balance.
2. Pathological evidences support the beneficial role of ACE2 in preventing metabolic disorders
Besides its physiological roles, upregulation of ACE2 is also associated with the amelioration of pathogenesis of many metabolic disorders including liver fibrosis(Warner et al., 2007; Schrom et al., 2017), chronic kidney disease(Soler et al., 2013), heart failure (Crackower et al., 2002; Patel et al., 2017), and diabetes (Tikellis et al., 2003). Lipidoid nanoparticle-mediated delivery (Schrom et al., 2017), adeno-associated viral delivery (Mak et al., 2015) and pharmacological stimulation of ACE2 in hepatic stellate cells (Huang et al., 2010) was observed to attenuate the inflammation, reduce oxidative stress, and improve the morphological impairments in fibrotic livers. Similarly, adenoviral upregulation of ACE2 was found to improve glomerular filtration rate, lower systemic hypertension, and attenuate the expression of inflammatory genes (Lo et al., 2015) in mouse model of type-1 diabetes. Downregulation of ACE2 expression was also found to be correlated with kidney injury (Gupta et al., 2007; da Silveira et al., 2010; Velkoska et al., 2010) and inflammation in chronic kidney diseases such as glomerulopathy, diabetic nephropathy and hypertensive renal disease (Ye et al., 2006; Mizuiri et al., 2011; Burrell et al., 2012; Soler et al., 2013). Consequently, adenoviral overexpression of ACE2 gene through intravenous route was observed to completely reverse the glomerular injury in rat model of chronic kidney disease (Liu et al., 2011). Exogenous overexpression of recombinant ACE2 was found to be beneficial in preclinical model of heart failure in terms of improvement in endothelial dysfunction, suppression of tissue inflammation and myocardial fibrosis, correction of metabolic dysfunction, and reversal of pathological hypertrophy (Mori et al., 2014; Basu et al., 2017; Patel et al., 2017). These evidences collectively demonstrate how the activation of ACE2 directly improves the adverse pathological events in many metabolic disorders.
3. Upregulated expression of ACE2 is correlated with lung cancer
However, apart from all these beneficial roles, the expression of ACE2 was also found to be strongly upregulated in lung cancer tissue (Zhang et al., 2020). Upregulation of different epigenetic factors such as HAT-1, HDAC-2 and KDM5B are reported to trigger the transcription of ACE2 in patients with lung disease (Pinto et al., 2020). Although the signaling pathway behind the upregulated expression of ACE2 is not known yet, literatures (Burrell et al., 2004b; Uhal et al., 2012) suggest that the upregulation of ACE2 might contribute to an anti-tumorigenic response that catalyzes the synthesis of growth suppressive Ang1–7 peptide, which in turn slows down the tumor growth via mas receptor activation (Murphy et al., 2019) and subsequent inhibition of tumor-promoting MAP kinases (Lee et al., 2005). Therefore, the induced expression of ACE2 might be a compensatory mechanism to suppress the growth and progression of lung tumor. In lung cancer tissue, ACE2 expression was also tightly regulated with the strong attenuation of RAS pathway as supported by the significant reduction of ACE activity (Danilov et al., 2019). Combining all these evidences, upregulation of ACE2 is critical in the development of lung cancer pathogenesis, however, the mechanism involved in the expression of ACE2 is still unknown.
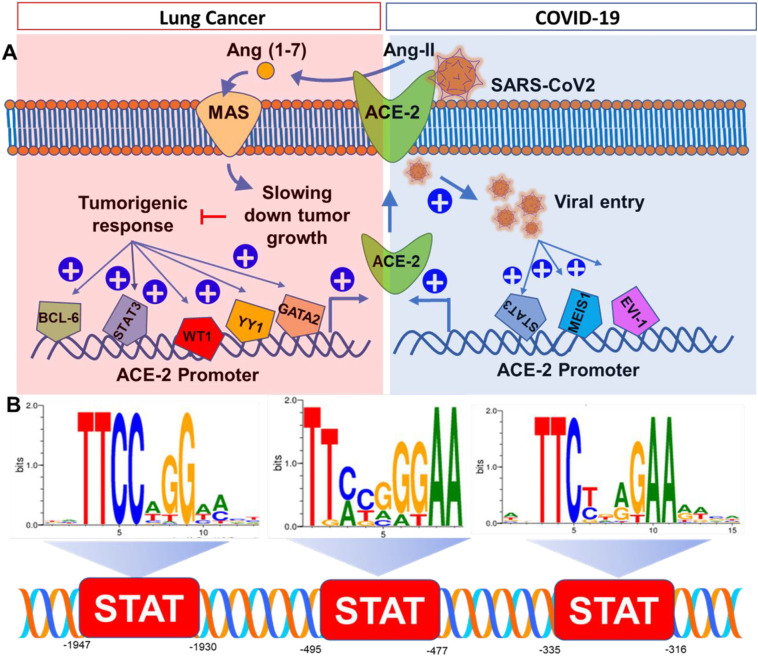
The most reliable strategy to understand the genetic regulation of ACE2 is to search its promoter for the transcription factor (TF) binding sites. Accordingly, MatInspector, a Genomatix software tool and a popular web-based program for predicting potential TF binding sites, was used to identify TFs in the ACE2 promoter. ACE2 promoter is located at chromosome X with 1947 nucleotide long sequence, which was extracted from PubMed (NCBI reference sequence ID: NG_068141), formatted in a FASTA format, loaded in the search box of MatInspector, and finally run through the search program. The possible TFs were summarized in a table (Table 1 ) with start and end position, core matching factors, and reported biological functions. TFs with core-match factor less than 0.95 were excluded from our study and those with more than 0.95 were selected as most suitable TFs. Since TFs were identified based on a predictive algorithms, to increase the confidence high cut-off value (0.95) was applied. Our analysis was further validated with TFs binding only to positive strand of ACE2 promoter. Total 51 TFs were identified on the promoter region of ACE2 (Table 1). Interestingly, many TFs with high-confidence binding sites (Match factor > 0.95) at ACE2 promoter are tumor-promoting transcription factors such as B-cell lymphoma 6 (BCL6), Wilms' tumor protein (WT1), Signal transducer and activator of transcription 3 (STAT3), Yin Yang-1 (YY1), AREB6, ERG, GKLF (or KLF4) and GATA TFs (Kumar et al., 2012; Hashiguchi et al., 2017). All these transcription factors are well-established contributing factors in the pathogenesis of lung and other tumors. Upregulation of BCL6 (Cardenas et al., 2017) and STAT3 (Tong et al., 2017) in bronchiolar epithelia are frequently observed in Non-small cell lung carcinoma tissue (NSCLC) (Deb et al., 2017). Transcriptional activation of Zinc finger containing transcription factor AREB6 also promoted migration and invasion of NSCLC (Guo et al., 2017). Upregulation of WT-1 was frequently detected in solid tumor tissue harvested from one of every 12 lung cancer patients (Menssen et al., 2000). Lung cancer patients with higher expression of YY1 had larger tumor size, poor differentiation, and higher rate of metastasis (Huang et al., 2017). Although, activation of ERG, an ETS gene family TF, contributes to the growth and spreading of prostate cancer (Sreenath et al., 2011), its activation was also noted in lung tumors (Scheble et al., 2010). Activation of GKLF or KLF-4 is frequently observed in stage III and IV lung tumors and considered as a potential prognostic biomarker in advanced lung cancer patients (Fadous-Khalife et al., 2016). GATA3 and GATA2 were reported to be a potential prognostic marker for lung adenocarcinoma (Hashiguchi et al., 2017) and NSCLC (Kumar et al., 2012) respectively. All these cancer-promoting transcription factors are highly expressed in lung tumors and could directly stimulate the transcription of ACE2 (Fig. 1A) during different stages of lung tumor progression.
Table 1.
Promoter analysis of ACE-2. Human Promoter sequence of ACE2 was derived from NCBI reference sequence ID: NG_068141. Promoter analysis was performed in MatInspector application of Genomatix gene analysis tool. Promoter length is 1947 base pairs (bp). Start and end numbers were calculated considering the start bp of open reading frame as 0. Core bases of the sequence were shown in block letters and possible biological functions were listed in the last column.
| Transcription factor | Start | End | Match factor | ResponseElement | Function |
|---|---|---|---|---|---|
| BCL6 | −1946 | −1930 | 0.969 | ggtTTCCtggaatgtgg | Tumorigenic |
| STAT3 | −1947 | −1930 | 0.968 | aggtTTCCtggaatgtggg | Tumorigenic |
| WT-1 | −1938 | −1919 | 0.967 | gaatgtgGGAGgagctttt | Tumorigenic |
| SMARCA-3 | −1852 | −1842 | 0.986 | tattACTTata | Lung cancer and tumorigenesis |
| GKLF | −1824 | −1806 | 0.965 | ttttttaAAAGgagagtat | Tumor suppressor (colorectal cancer) |
| MEIS-1 | −1752 | −1736 | 0.969 | actaaatcTGTCatctt | Myeloid Leukemia, viral integration |
| MIZ-1 | −1724 | −1710 | 1 | aaggcCCTCtg | Esophageal cancer |
| CDPCR3HD | −1634 | −1612 | 0.957 | aatacataGATCcatgttctgat | Cancer (?) |
| YY1 | −1627 | −1607 | 0.973 | atagatCCATgttctgattccat | Lung oncogene and tumorigenesis |
| AARE | −1613 | −1605 | 0.975 | aTTCCatca | Pulmonary obstruction and Lung cancer |
| CPHX | −1617 | −1605 | 0.977 | tcTGATtccatcatttgttagct | Development of Ovary |
| NKX2.5 | −1600 | −1582 | 0.984 | ttagcTGAGtgagatatag | Thyroid organogenesis and cancer |
| NKX2.5 | −1563 | −1546 | 0.951 | tttcaTAATtcataaaatg | Thyroid organogenesis and cancer |
| SOX6 | −1494 | −1472 | 0.986 | ataacACAAagcactttgaatag | Neurodevelopment and chondrogenesis |
| SMARCA-3 | −1473 | −1462 | 0.986 | agttACTTata | Chromatin remodeling and anti-depression |
| DLX-3 | −1473 | −1455 | 0,959 | agttacttaTAATtgtttt | Folliculogenesis |
| NOBOX | −1471 | −1452 | 0.978 | ttacttaTAATtgtttttt | Folliculogenesis |
| S8 | −1469 | −1450 | 0.995 | acttaTAATtgtttttttcct | Folliculogenesis |
| NFAT | −1422 | −1404 | 0.954 | gaataaGGAAaagcagtgg | T cell proliferatio and inflammation |
| GKLF | −1400 | −1382 | 0.97 | tttttttAAAGgcttgatt | Apoptotic |
| LHX6 | −1395 | −1373 | 0.975 | ttaaaggctTGATtattgcaatg | Neuro and lymphoid development |
| AREB6 | −1375 | −1363 | 0.978 | atgtCACCtgaac | Lung cancer and tumorigenesis |
| ZNF35 | −1382 | −1352 | 0.97 | tattgcaatgtcacctgaacctggAAGActt | Tumor suppressor |
| CEBPB | −1344 | −1329 | 0.952 | tgggtgaaGAAAtat | Macrophage function and inflammation |
| SMARCA3 | −1279 | −1269 | 0.985 | tgCCATttaaa | Chromatin remodeling and anti-depression |
| NKX3.1 | −1276 | −1258 | 0.955 | catttaaAGTGctcctctc | Prostate development and tumor suppressor |
| EVI1 | −1195 | −1179 | 1 | ttgacaAGATaaccact | Viral integration and myeloid leukemia |
| GATA1 | −1192 | −1180 | 0.957 | acaaGATAaccac | Erythroid development |
| CHR | −1173 | −1161 | 0.967 | ctctTTGAattct | RNA polymerase activation |
| RFX4 | −1126 | −1108 | 0.958 | gagttgacataGATActct | Spermatogenesis |
| SMARCA3 | −1097 | −1087 | 0.975 | taCCATgtgga | Chromatin remodeling and anti-depression |
| ARNT | −1078 | −1062 | 0.95 | tacttccaCGTGacctt | Toxin metabolism and hepatocellular carcinoma |
| MNT | −1077 | −1061 | 0.992 | acttcCACGtgaccttg | Transcriptional repressor of cell growth |
| DEC2 | −1076 | −1062 | 0.978 | cttccaCGTGacctt | Regulation of sleep and circadian rhythm |
| CARF | −1033 | −1022 | 0.97 | agaagGAGGca | Neuroprotection |
| ERG | −983 | −962 | 0.96 | gagaaataGGAAatgagcttt | Prostate tumor (fused with TMPRSS) |
| CRX | −853 | −837 | 0.963 | gcctgtaATCCtagcac | Photoreception and melatonin secretion |
| ESRRB | −821 | −799 | 0.978 | tgggcagatcacaAGGTcaggag | Neuroprotection, Stem cell pluripotency |
| SF1 | −814 | −799 | 0.996 | atcaCAAGgtcagga | Reproductive organ development |
| GATA1 | −803 | −791 | 0.961 | aggaGATAgagac | Blood cell maturation |
| GKLF | −739 | −721 | 0.988 | agctgggcGTGGtggtggg | Tumor Suppressor and cell reprogramming |
| AREB6 | −724 | −712 | 0.968 | tgggCACCtgtag | Lung cancer and tumorigenesis |
| ZNF750 | −500 | −486 | 1 | cgggaGGCTgaggca | Squamous epithelial cell development |
| IKZF3 | −521 | −509 | 0.995 | ggaagGGAAaatg | Lymphocyte development and differentiation |
| STAT3 | −495 | −477 | 0.98 | gagtttctgGGAAtatgat | Viral infection and tumor development |
| IK2 | −491 | −479 | 0.986 | ttctGGGAatatg | Embryonic development |
| SALL-1 | −472 | −460 | 0.977 | aaATAAaaataaa | Embryonic development of kidney and heart |
| GATA-4 | −458 | −446 | 0.965 | gtgaGATAaccta | Cardiac development |
| LMX1A | −453 | −431 | 0.969 | ataacctaTTAAtgaaattgtct | Insulin secretion |
| RU49 | −310 | −304 | 1 | aAGTAcc | Cerebellar development |
| GRHL | −309 | −297 | 0.973 | agtaccGGTTttg | Embryonic development |
| STAT3 | −335 | −316 | agctTTCTaggaaaatatt | Tumor formation and viral infection | |
| PDEF | −188 | −168 | 0.958 | cctctccaGGATgaactttat | Prostate-derived tumor suppressor |
| NRL | −169 | −145 | 1 | atattggctcAGCAgattgtttact | Rod photoreceptor development |
| TCF7L2 | −95 | −79 | 0.973 | ccaagttcAAAGgctga | HIV or HIV/HCV-coinfection and diabetes |
| GKLF or KLF4 | −94 | −76 | 0.98 | caagttcAAAGgctgataa | Tumor Suppressor and cell reprogramming |
| GATA | −84 | −72 | 0.995 | ggctGATAagaga | Growth, development and tumorigenesis |
Fig. 1.
ACE2 at the crossroad of lung cancer and COVID-19 infection. (A) Pathobiological roles of ACE2 during lung tumor progression (enclosed in red square) and COVID-19 (enclosed in blue square). (B) Promoter analysis of ACE2 for STAT3 binding. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. SARS-COV2 stimulated the expression of ACE2 in lung and kidney tissue
Recently, our study highlighted (Gottschalk et al., 2020) that the intranasal inoculation of Wuhan-standard SARS-CoV2 strongly upregulated the expression of ACE2 in both lungs and kidneys of K18-hACE2 mice. K18-hACE2 mice express human ACE2 receptor in lungs, kidney, and other tissue. Hence, these mice display human-like clinical manifestations of COVID-19 pathologies in lungs and kidney upon intranasal administration of SARS-CoV2 virus. Twenty-four hours after the viral inoculation, we observed that these mice experienced severe loss of oxygen saturation, drop in heart rate, hypothermia, and acute death response close to 30%. While investigating the mechanism, we observed strong upregulation of ACE2 in bronchiolar epithelium of lungs, glomerular epithelium, and renal tubular cells in kidney tissue of these animals. Moreover, that upregulation of ACE2 was positively correlated with the numbers of SARS-CoV2 virus particles entered within lung and kidney parenchyma suggesting that the stimulated expression of ACE2 was indeed involved in the entry of virus. Therefore, the cellular entry of SARS-CoV2 might operate through a feed-forward loop that stimulates the expression of ACE2 in order to facilitate the entry of more virus particles in pulmonary and renal epithelial cells. However, how the SARS-CoV2 infection triggers the expression of ACE2 is not known yet. While analyzing the promoter of ACE2, we detected multiple TFs that are known to be activated during viral infection. These include Myeloid ecotropic viral integration site 1 protein (MEIS1), ectopic viral integration transcription factor EVI1, cell cycle genes homology region protein CHR, STAT3, and Transcription factor 7-like 2 TCF7L2. All these nuclear factors are activated during the infection of positive-sense single-stranded RNA viruses. MEIS1 was originally discovered as a nuclear protein that was activated by the retroviral integration in the chromosome of myeloid leukemia tumor cells (Moskow et al., 1995). EVI1 is another TF that serves as common retroviral integration site in AKXD murine myeloid tumors (Metais and Dunbar, 2008). Constitutive activation of STAT3 exhibits a proviral function in several viral infections, including those of HBV, HCV, HSV-1, varicella zoster virus, human CMV and measles virus (Chang et al., 2018). Chronic hepatitis C, HIV, and HIV-HCV co-infection stimulates the polymorphism of TCF7L2 gene (Pineda-Tenor et al., 2015). Interestingly, promoter of ACE2 gene harbors high-confidence (>0.95) binding sites for all of these TFs indicating their potential involvement in the transcription of ACE2 gene upon infection of SARS-CoV2 (Fig. 1B).
5. Elevated expression of ACE2 was reported in lung cancer patients suffering from COVID-19
Patients with cancer are typically at higher risk of COVID-19 infection because of compromised host defenses and suppressed immunological responses due to strong side-effects of chemotherapeutic treatment (Ganatra et al., 2020). However, the molecular mechanism of upscaled COVID-19 infection in these patients was not known. One possible explanation might be the induced expression of ACE2 in these cancer patients promotes the viral infection. In fact, the expression of ACE2 was indeed found to be elevated in COVID-19 patients who has been suffering from chronic lung disease. Apart from cancer, Patients with pre-existing conditions such as diabetes, hypertensions and chronic obstructive lung disease also exhibited upregulated expression of ACE2 in lungs upon infection with SAR-CoV2 (Pinto et al., 2020).
Therefore, it is important to identify TFs, which might regulate the expression of ACE2 in COVID-19 patients with pre-existing condition of lung cancer. TFs that are possibly activated during retroviral infections such as STAT3, MEIS1, and EV-1 are also known to be involved in the pathogenesis of lung cancer. MEIS1 promotes the proliferation and migration of lung arterial cells suggesting its possible involvement in the chronic lung disease (Yao et al., 2020). STAT3 stimulates viral infection and infection-associated proliferation and migration of lung tumor cells (Lu et al., 2017). Transcriptional activation of EVI1 was reported to stimulate chromosomal rearrangements in solid lung tumor (Liang and Wang, 2020). Tumor microenvironment in lungs possibly induces the expressions of all these TFs that might promote the upregulation of ACE2 and subsequent infection of SARS-CoV2 virions through ACE2 receptors (Fig. 1).
6. Conclusion
In summary, we identified 7 TFs named as WT1, STAT3, YY1, AREB6, ERG, GKLF, and GATA2 as possible inducers of ACE2 transcription in Lung tumors, whereas 5 TFs including MEIS1, EVI1, CHR, STAT3, and TCF7L2 as potential stimulators of ACE2 during SARS-CoV2 infection. Combining all these predictions, MEIS1, EVI1, STAT3 are predicted TFs that serve as common TFs in stimulating the expression of ACE2 both in SARS-CoV2 infection and lung tumor pathogenesis. Among all these TFs, STAT-binding site was frequently observed in ACE2 promoter. Distal binding site is 1930 bp upstream (score = 0.968), proximal binding site is 495 bp upstream (score = 0.98) and the intermediate binding site is 335 bp upstream of start site (Fig. 1B). Therefore, based on our comprehensive promoter analysis of ACE-2, we conclude that STAT3-mediated upregulation of ACE2 might play critical roles both in lung tumor progression and COVID-19 infection. SARS-CoV2 infection might stimulate the binding of STAT3 in the promoter of ACE2 resulting the enhanced expression of ACE-2. That mechanism might turn on a feed-forward loop with upscaled entry of virus through ACE2 receptor. On the other hand, cancer pathologies in lung tissue also switches on the transcription of STAT3 leading to the upregulated expression of ACE2 in cancer cells.
Declaration of competing interest
Authors declare no conflict of interest.
Acknowledgments
Acknowledgement
This work is funded with the seed fund of Sotira LLC, AZ, USA and Simmaron Research Institute, NV, USA.
References
- Allred A.J., Diz D.I., Ferrario C.M., Chappell M.C. Pathways for angiotensin-(1—7) metabolism in pulmonary and renal tissues. American Journal of Physiology-Renal Physiology. 2000;279:F841–F850. doi: 10.1152/ajprenal.2000.279.5.F841. [DOI] [PubMed] [Google Scholar]
- Anderson J.M., Gimbrone M., Alexander R. Angiotensin II stimulates phosphorylation of the myosin light chain in cultured vascular smooth muscle cells. J. Biol. Chem. 1981;256:4693–4696. [PubMed] [Google Scholar]
- Basu R., Poglitsch M., Yogasundaram H., Thomas J., Rowe B.H., Oudit G.Y. Roles of angiotensin peptides and recombinant human ACE2 in heart failure. J. Am. Coll. Cardiol. 2017;69:805–819. doi: 10.1016/j.jacc.2016.11.064. [DOI] [PubMed] [Google Scholar]
- Brewster U.C., Setaro J.F., Perazella M.A. The renin-angiotensin-aldosterone system: cardiorenal effects and implications for renal and cardiovascular disease states. Am J Med Sci. 2003;326:15–24. doi: 10.1097/00000441-200307000-00003. [DOI] [PubMed] [Google Scholar]
- Burrell L.M., Johnston C.I., Tikellis C., Cooper M.E. ACE2, a new regulator of the renin-angiotensin system. Trends Endocrinol. Metab. 2004;15:166–169. doi: 10.1016/j.tem.2004.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell L.M., Johnston C.I., Tikellis C., Cooper M.E. ACE2, a new regulator of the renin–angiotensin system. Trends in Endocrinology & Metabolism. 2004;15:166–169. doi: 10.1016/j.tem.2004.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell L.M., Burchill L., Dean R.G., Griggs K., Patel S.K., Velkoska E. Chronic kidney disease: cardiac and renal angiotensin-converting enzyme (ACE) 2 expression in rats after subtotal nephrectomy and the effect of ACE inhibition. Exp. Physiol. 2012;97:477–485. doi: 10.1113/expphysiol.2011.063156. [DOI] [PubMed] [Google Scholar]
- Cardenas M.G., Oswald E., Yu W., Xue F., MacKerell A.D., Jr., Melnick A.M. The expanding role of the BCL6 oncoprotein as a cancer therapeutic target. Clin. Cancer Res. 2017;23:885–893. doi: 10.1158/1078-0432.CCR-16-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z., Wang Y., Zhou X., Long J.E. STAT3 roles in viral infection: antiviral or proviral? Future Virol. 2018;13:557–574. doi: 10.2217/fvl-2018-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E., Oliveira-dos-Santos A.J., da Costa J., Zhang L., Pei Y., Scholey J., Ferrario C.M., Manoukian A.S., Chappell M.C., Backx P.H., Yagil Y., Penninger J.M. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- Dabouras V., Eid S.R., Brändli A.W. 3.1. Identification of genes encoding Xenopus laevis angiotensin receptor type 1 and 2 and their expression in embryonic development. Genes and mechanisms regulating angiogenesis and kidney organogenesis-studies on ephrin-B and WT1 gene functions. 1973;81 [Google Scholar]
- Danilov S.M., Metzger R., Klieser E., Sotlar K., Trakht I.N., Garcia J.G. Tissue ACE phenotyping in lung cancer. PLoS One. 2019;14 doi: 10.1371/journal.pone.0226553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb D., Rajaram S., Larsen J.E., Dospoy P.D., Marullo R., Li L.S., Avila K., Xue F., Cerchietti L., Minna J.D., Altschuler S.J., Wu L.F. Combination therapy targeting BCL6 and Phospho-STAT3 defeats intratumor heterogeneity in a subset of non-small cell lung cancers. Cancer Res. 2017;77:3070–3081. doi: 10.1158/0008-5472.CAN-15-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Erdos E.G. Conversion of angiotensin I to angiotensin II. Am. J. Med. 1976;60:749–759. doi: 10.1016/0002-9343(76)90889-5. [DOI] [PubMed] [Google Scholar]
- Fadous-Khalife M.C., Aloulou N., Jalbout M., Hadchity J., Aftimos G., Paris F., Hadchity E. Kruppel-like factor 4: a new potential biomarker of lung cancer. Mol Clin Oncol. 2016;5:35–40. doi: 10.3892/mco.2016.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J.H., Lappin S.L. StatPearls; Treasure Island (FL): 2020. Physiology, Renin Angiotensin System. [PubMed] [Google Scholar]
- Ganatra S., Hammond S.P., Nohria A. The novel coronavirus disease (COVID-19) threat for patients with cardiovascular disease and cancer. JACC CardioOncol. 2020;2:350–355. doi: 10.1016/j.jaccao.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk G., Keating J.F., Kesler K., Knox K., Roy A. Intranasal administration of ACIS KEPTIDETM prevents SARS-CoV2-induced acute toxicity in K18-hACE2 humanized mouse model of COVID-19: a mechanistic insight for the prophylactic role of KEPTIDETM in COVID-19. bioRxiv. 2020 2020.11.13.378257. [Google Scholar]
- Guo X., Zhao L., Cheng D., Mu Q., Kuang H., Feng K. AKIP1 promoted epithelial-mesenchymal transition of non-small-cell lung cancer via transactivating ZEB1. Am. J. Cancer Res. 2017;7:2234–2244. [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Rhodes G.J., Berg D.T., Gerlitz B., Molitoris B.A., Grinnell B.W. Activated protein C ameliorates LPS-induced acute kidney injury and downregulates renal INOS and angiotensin 2. Am J Physiol Renal Physiol. 2007;293:F245–F254. doi: 10.1152/ajprenal.00477.2006. [DOI] [PubMed] [Google Scholar]
- Handa R.K., Johns E.J. Interaction of the renin-angiotensin system and the renal nerves in the regulation of rat kidney function. J. Physiol. 1985;369:311–321. doi: 10.1113/jphysiol.1985.sp015903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi T., Miyoshi H., Nakashima K., Yokoyama S., Matsumoto R., Murakami D., Mitsuoka M., Takamori S., Akagi Y., Ohshima K. Prognostic impact of GATA binding protein-3 expression in primary lung adenocarcinoma. Hum. Pathol. 2017;63:157–164. doi: 10.1016/j.humpath.2017.02.024. [DOI] [PubMed] [Google Scholar]
- Huang M.L., Li X., Meng Y., Xiao B., Ma Q., Ying S.S., Wu P.S., Zhang Z.S. Upregulation of angiotensin-converting enzyme (ACE) 2 in hepatic fibrosis by ACE inhibitors. Clin. Exp. Pharmacol. Physiol. 2010;37:e1–e6. doi: 10.1111/j.1440-1681.2009.05302.x. [DOI] [PubMed] [Google Scholar]
- Huang T., Wang G., Yang L., Peng B., Wen Y., Ding G., Wang Z. Transcription factor YY1 modulates lung cancer progression by activating lncRNA-PVT1. DNA Cell Biol. 2017;36:947–958. doi: 10.1089/dna.2017.3857. [DOI] [PubMed] [Google Scholar]
- Kumar M.S., Hancock D.C., Molina-Arcas M., Steckel M., East P., Diefenbacher M., Armenteros-Monterroso E., Lassailly F., Matthews N., Nye E., Stamp G., Behrens A., Downward J. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- Lee V.Y., Schroedl C., Brunelle J.K., Buccellato L.J., Akinci O.I., Kaneto H., Snyder C., Eisenbart J., Budinger G.S., Chandel N.S. Bleomycin induces alveolar epithelial cell death through JNK-dependent activation of the mitochondrial death pathway. Am. J. Phys. Lung Cell. Mol. Phys. 2005;289:L521–L528. doi: 10.1152/ajplung.00340.2004. [DOI] [PubMed] [Google Scholar]
- Liang B., Wang J. EVI1 in leukemia and solid tumors. Cancers (Basel) 2020;12 doi: 10.3390/cancers12092667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.X., Hu Q., Wang Y., Zhang W., Ma Z.Y., Feng J.B., Wang R., Wang X.P., Dong B., Gao F., Zhang M.X., Zhang Y. Angiotensin-converting enzyme (ACE) 2 overexpression ameliorates glomerular injury in a rat model of diabetic nephropathy: a comparison with ACE inhibition. Mol. Med. 2011;17:59–69. doi: 10.2119/molmed.2010.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C.S., Shi Y., Chang S.Y., Abdo S., Chenier I., Filep J.G., Ingelfinger J.R., Zhang S.L., Chan J.S. Overexpression of heterogeneous nuclear ribonucleoprotein F stimulates renal ace-2 gene expression and prevents TGF-beta1-induced kidney injury in a mouse model of diabetes. Diabetologia. 2015;58:2443–2454. doi: 10.1007/s00125-015-3700-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhang Y.G., Sun J. STAT3 activation in infection and infection-associated cancer. Mol. Cell. Endocrinol. 2017;451:80–87. doi: 10.1016/j.mce.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak K.Y., Chin R., Cunningham S.C., Habib M.R., Torresi J., Sharland A.F., Alexander I.E., Angus P.W., Herath C.B. ACE2 therapy using adeno-associated viral vector inhibits liver fibrosis in mice. Mol. Ther. 2015;23:1434–1443. doi: 10.1038/mt.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen H.D., Bertelmann E., Bartelt S., Schmidt R.A., Pecher G., Schramm K., Thiel E. Wilms’ tumor gene (WT1) expression in lung cancer, colon cancer and glioblastoma cell lines compared to freshly isolated tumor specimens. J. Cancer Res. Clin. Oncol. 2000;126:226–232. doi: 10.1007/s004320050037. [DOI] [PubMed] [Google Scholar]
- Metais J.Y., Dunbar C.E. The MDS1-EVI1 gene complex as a retrovirus integration site: impact on behavior of hematopoietic cells and implications for gene therapy. Mol. Ther. 2008;16:439–449. doi: 10.1038/sj.mt.6300372. [DOI] [PubMed] [Google Scholar]
- Mizuiri S., Hemmi H., Arita M., Aoki T., Ohashi Y., Miyagi M., Sakai K., Shibuya K., Hase H., Aikawa A. Increased ACE and decreased ACE2 expression in kidneys from patients with IgA nephropathy. Nephron Clin Pract. 2011;117:c57–c66. doi: 10.1159/000319648. [DOI] [PubMed] [Google Scholar]
- Mori J., Patel V.B., Abo Alrob O., Basu R., Altamimi T., Desaulniers J., Wagg C.S., Kassiri Z., Lopaschuk G.D., Oudit G.Y. Angiotensin 1-7 ameliorates diabetic cardiomyopathy and diastolic dysfunction in db/db mice by reducing lipotoxicity and inflammation. Circ Heart Fail. 2014;7:327–339. doi: 10.1161/CIRCHEARTFAILURE.113.000672. [DOI] [PubMed] [Google Scholar]
- Moskow J.J., Bullrich F., Huebner K., Daar I.O., Buchberg A.M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol. Cell. Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.T., Hossain M.I., Swiderski K., Chee A., Naim T., Trieu J., Haynes V., Read S.J., Stapleton D.I., Judge S.M. Mas receptor activation slows tumor growth and attenuates muscle wasting in cancer. Cancer Res. 2019;79:706–719. doi: 10.1158/0008-5472.CAN-18-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V.B., Lezutekong J.N., Chen X., Oudit G.Y. Recombinant human ACE2 and the angiotensin 1-7 Axis as potential new therapies for heart failure. Can J Cardiol. 2017;33:943–946. doi: 10.1016/j.cjca.2016.12.012. [DOI] [PubMed] [Google Scholar]
- Peach M.J. Renin-angiotensin system: biochemistry and mechanisms of action. Physiol. Rev. 1977;57:313–370. doi: 10.1152/physrev.1977.57.2.313. [DOI] [PubMed] [Google Scholar]
- Pineda-Tenor D., Berenguer J., Jimenez-Sousa M.A., Carrero A., Garcia-Alvarez M., Aldamiz-Echevarria T., Garcia-Broncano P., Diez C., Guzman-Fulgencio M., Fernandez-Rodriguez A., Resino S. rs7903146 polymorphism at transcription factor 7 like 2 gene is associated with total cholesterol and lipoprotein profile in HIV/hepatitis C virus-coinfected patients. AIDS Res. Hum. Retrovir. 2015;31:326–334. doi: 10.1089/aid.2014.0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto B.G.G., Oliveira A.E.R., Singh Y., Jimenez L., Gonçalves A.N.A., Ogava R.L.T., Creighton R., Schatzmann Peron J.P., Nakaya H.I. ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. medRxiv. 2020 doi: 10.1093/infdis/jiaa332. 2020.03.21.20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid I.A., Moffat B. Angiotensin II concentration in cerebrospinal fluid after intraventricular injection of angiotensinogen or renin. Endocrinology. 1978;103:1494–1498. doi: 10.1210/endo-103-4-1494. [DOI] [PubMed] [Google Scholar]
- Scheble V.J., Braun M., Wilbertz T., Stiedl A.C., Petersen K., Schilling D., Reischl M., Seitz G., Fend F., Kristiansen G., Perner S. ERG rearrangement in small cell prostatic and lung cancer. Histopathology. 2010;56:937–943. doi: 10.1111/j.1365-2559.2010.03564.x. [DOI] [PubMed] [Google Scholar]
- Schrom E., Huber M., Aneja M., Dohmen C., Emrich D., Geiger J., Hasenpusch G., Herrmann-Janson A., Kretzschmann V., Mykhailyk O., Pasewald T., Oak P., Hilgendorff A., Wohlleber D., Hoymann H.G., Schaudien D., Plank C., Rudolph C., Kubisch-Dohmen R. Translation of angiotensin-converting enzyme 2 upon liver- and lung-targeted delivery of optimized chemically modified mRNA. Mol Ther Nucleic Acids. 2017;7:350–365. doi: 10.1016/j.omtn.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira K.D., Pompermayer Bosco K.S., Diniz L.R., Carmona A.K., Cassali G.D., Bruna-Romero O., de Sousa L.P., Teixeira M.M., Santos R.A., Simoes e Silva A.C., Ribeiro Vieira M.A. ACE2-angiotensin-(1-7)-mas axis in renal ischaemia/reperfusion injury in rats. Clin Sci (Lond) 2010;119:385–394. doi: 10.1042/CS20090554. [DOI] [PubMed] [Google Scholar]
- Soler M.J., Wysocki J., Batlle D. ACE2 alterations in kidney disease. Nephrol. Dial. Transplant. 2013;28:2687–2697. doi: 10.1093/ndt/gft320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenath T.L., Dobi A., Petrovics G., Srivastava S. Oncogenic activation of ERG: a predominant mechanism in prostate cancer. J Carcinog. 2011;10:37. doi: 10.4103/1477-3163.91122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W.G., Pipolo L., Qian H. Identification of a Ca2+/calmodulin-binding domain within the carboxyl-terminus of the angiotensin II (AT1A) receptor. FEBS Lett. 1999;455:367–371. doi: 10.1016/s0014-5793(99)00904-7. [DOI] [PubMed] [Google Scholar]
- Tikellis C., Johnston C.I., Forbes J.M., Burns W.C., Burrell L.M., Risvanis J., Cooper M.E. Characterization of renal angiotensin-converting enzyme 2 in diabetic nephropathy. Hypertension. 2003;41:392–397. doi: 10.1161/01.HYP.0000060689.38912.CB. [DOI] [PubMed] [Google Scholar]
- Tong M., Wang J., Jiang N., Pan H., Li D. Correlation between p-STAT3 overexpression and prognosis in lung cancer: a systematic review and meta-analysis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0182282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhal B.D., Li X., Piasecki C.C., Molina-Molina M. Angiotensin signalling in pulmonary fibrosis. Int. J. Biochem. Cell Biol. 2012;44:465–468. doi: 10.1016/j.biocel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkoska E., Dean R.G., Burchill L., Levidiotis V., Burrell L.M. Reduction in renal ACE2 expression in subtotal nephrectomy in rats is ameliorated with ACE inhibition. Clin Sci (Lond) 2010;118:269–279. doi: 10.1042/CS20090318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner F.J., Lubel J.S., McCaughan G.W., Angus P.W. Liver fibrosis: a balance of ACEs? Clin Sci (Lond) 2007;113:109–118. doi: 10.1042/CS20070026. [DOI] [PubMed] [Google Scholar]
- Yao M.Z., Ge X.Y., Liu T., Huang N., Liu H., Chen Y., Zhang Z., Hu C.P. MEIS1 regulated proliferation and migration of pulmonary artery smooth muscle cells in hypoxia-induced pulmonary hypertension. Life Sci. 2020;255:117822. doi: 10.1016/j.lfs.2020.117822. [DOI] [PubMed] [Google Scholar]
- Ye M., Wysocki J., William J., Soler M.J., Cokic I., Batlle D. Glomerular localization and expression of angiotensin-converting enzyme 2 and angiotensin-converting enzyme: implications for albuminuria in diabetes. J. Am. Soc. Nephrol. 2006;17:3067–3075. doi: 10.1681/ASN.2006050423. [DOI] [PubMed] [Google Scholar]
- Zelis R. Mechanisms of vasodilation. Am. J. Med. 1983;74:3–12. doi: 10.1016/0002-9343(83)90848-3. [DOI] [PubMed] [Google Scholar]
- Zhang L., Han X., Shi Y. Comparative analysis of SARS-CoV-2 receptor ACE2 expression in multiple solid tumors and matched non-diseased tissues. Infect. Genet. Evol. 2020;85:104428. doi: 10.1016/j.meegid.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Zhang S.H., Wagner C., Kurtz A., Maeda N., Coffman T., Arendshorst W.J. Angiotensin AT1B receptor mediates calcium signaling in vascular smooth muscle cells of AT1A receptor–deficient mice. Hypertension. 1998;31:1171–1177. doi: 10.1161/01.hyp.31.5.1171. [DOI] [PubMed] [Google Scholar]