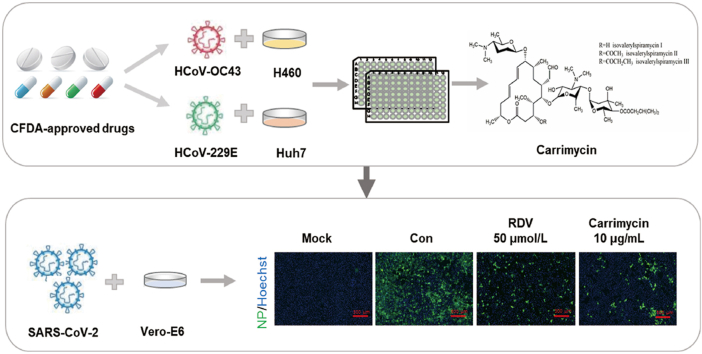
Abstract
COVID-19 pandemic caused by SARS-CoV-2 infection severely threatens global health and economic development. No effective antiviral drug is currently available to treat COVID-19 and any other human coronavirus infections. We report herein that a macrolide antibiotic, carrimycin, potently inhibited the cytopathic effects (CPE) and reduced the levels of viral protein and RNA in multiple cell types infected by human coronavirus 229E, OC43, and SARS-CoV-2. Time-of-addition and pseudotype virus infection studies indicated that carrimycin inhibited one or multiple post-entry replication events of human coronavirus infection. In support of this notion, metabolic labelling studies showed that carrimycin significantly inhibited the synthesis of viral RNA. Our studies thus strongly suggest that carrimycin is an antiviral agent against a broad-spectrum of human coronaviruses and its therapeutic efficacy to COVID-19 is currently under clinical investigation.
Key words: Coronavirus, SARS-CoV-2, HCoV-229E, HCoV-OC43, COVID-19, Carrimycin
Graphical abstract
CFDA-approved drug carrimycin is repurposed as an antiviral agent against human coronaviruses, including the currently pandemic SARS-CoV-2, through inhibition of post-entry replication events of human coronavirus infection.
1. Introduction
Coronaviruses (CoVs) are a large family of enveloped, positive-sense, single-stranded RNA viruses with broad host ranges1. Since the new millennium, cross species transmissions of CoVs from bats through intermediate mammalian hosts to humans have caused severe acute respiratory syndrome (SARS) in 2003, Middle East respiratory syndrome (MERS) in 2012, and current pandemic of coronavirus disease 2019 (COVID-19)2, 3, 4, 5. In addition, four human coronaviruses (HCoVs), including HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1, cause common cold and are speculated to be introduced into human population decades or even hundreds of years ago from unidentified animal hosts6. It is anticipated that emergence and re-emergence of CoV infections via cross species transmission will be a continuing challenge for human health and development of broad-spectrum antiviral agents against HCoVs are essential to cope with the current COVID-19 and future CoV epidemics.
Drug repurposing is an effective strategy for urgent treatment of emerging viral diseases7,8. In our efforts to search for the approved medicines that can suppress human CoV infections, an in-house collection of Chinese Food and Drug Administration (CFDA)-approved drugs including Chinese patent medicines, antibiotics, and antiviral agents were screened for their ability to protect the cytopathic effects (CPE) caused by HCoV-229E or HCoV-OC43 infection. We found a few macrolide antibiotics with antiviral activity against HCoV-229E and HCoV-OC43. Carrimycin, the most active one, was selected for further investigation of its antiviral activity against SARS-CoV-2 and determination of antiviral mechanism.
2. Materials and methods
2.1. Cells and viruses
Human hepatocellular carcinoma cell lines Huh7 and Huh7.5 and human lung cancer cell line H460 were kindly provided by Dr. Zonggen Peng and Dr. Zhen Wang, respectively, at Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College. Human hepatoblastoma cell line C3A was purchased from ATCC (Manassas, VA, USA). 293T-derived cell line expressing human recombinant angiotensin I converting enzyme 2 (293T-hACE2) was purchased from Delivectory Biosciences Inc. (Beijing, China). All cells cultured in Dulbecco's Modified Eagle Medium (DMEM, Invitrogen, Carlsbad, CA, USA) or Minimum Essential Media (MEM, Invitrogen) supplemented with 10% fetal bovine serum and antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin) at 37 °C in a 5% CO2 incubator.
HCoV-229E (strain VR740) was purchased from ATCC. HCoV-OC43 (strain VR1558) was a kind gift from Dr. Xuesen Zhao at Beijing Ditan Hospital, Capital Medical University (Beijing, China). SARS-CoV-2 (GenBank: MT123290) for immunofluorescence (IF) assay was isolated from a throat swab of a COVID-19 patient and stored in biosafety level-3 laboratory (Guangzhou Customs Technology Center, Guangzhou, China). The vesicular stomatitis virus (VSV) and SARS-CoV-2 pseudotyped viral particles were obtained from Delivectory Biosciences Inc. (Beijing, China).
2.2. Compounds
Carrimycin was provided by Shenyang Tonglian Group Co., Ltd. (Shenyang, China). Clarithromycin, midecamycin, erythromycin, roxithromycin, acetylspiramycin, azithromycin, clindamycin, remdesivir (RDV), and ammonium chloride (NH4Cl) were purchased from MedChemExpress (Monmouth Junction, NJ, USA). Ribavirin (RBV) and chloroquine (CQ) were purchased from Sigma–Aldrich (St. Louis, MO, USA).
2.3. Cell cytotoxicity assay
Cytotoxic effects of carrimycin on different cells were assayed by cell counting kit (CCK, TransGen Biotech, Beijing, China). Briefly, cells were seeded into 96-well culture plates and were incubated overnight. Then, the medium was removed and different concentrations of carrimycin were applied in triplicate. After 2 days’ incubation, the cytotoxicity of carrimycin was determined by CCK assay and then the 50% cytotoxic concentration (CC50) was calculated.
2.4. CPE inhibition assay
The anti-coronavirus activity of carrimycin was determined by a CPE inhibition assay. Briefly, cells were plated into 96-well culture plates and incubated for 24 h. The cells were infected with 100 times 50% tissue culture infective dose (TCID50) HCoV-229E or HCoV-OC43 and the indicated concentrations of compounds were added simultaneously. HCoV-229E infected Huh7 cells were treated for about 48 h and HCoV-OC43 infected H460 cells were treated for about 72 h. The 50% effective concentration (EC50) was determined by Reed & Muench method. The selectivity index (SI) was calculated as the ratio of CC50/EC50.
2.5. Immunofluorescence assay
C3A (2.0 × 105 cells/well), Huh7 (1.5 × 105 cells/well), or H460 (1.5 × 105 cells/well) cells grown on glass coverslips (Thermo Fisher Scientific, Waltham, MA, USA) were infected with coronavirus and treated with carrimycin at the same time of infection. At 48 h post infection, the culture medium was removed and the cells were washed and fixed. The cells were permeabilized in 0.5% Triton X-100 at room temperature for 15 min and blocked in phosphate buffer saline (PBS) containing 1% bovine serum albumin (BSA) for 60 min at room temperature. Cells were then incubated with an anti-coronavirus nucleoprotein (NP) antibody (Millipore, Billerica, MA, USA) or dsRNA antibody (SCICONS, Szirák, Hungary) at a dilution of 1: 200 for 2 h at room temperature. After washing three times with PBS, the samples were reacted with Alexa Fluor 488-labeled goat anti-mouse secondary antibody (Beyotime Institute of Biotechnology, China) for 1 h at room temperature. After washing with PBS, images were taken using a fluorescence microscope (Olympus IX71, Olympus, Japan).
2.6. Western blot assays
For analysis of proteins, the cellular proteins were extracted using M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific) with halt protease inhibitor single-use cocktail. Immunoblotting for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cell Signaling Technology, Boston, MA, USA, 1:1000) and coronavirus NP (Millipore, Billerica, MA, USA, 1:1000) was performed as described previously9.
2.7. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) assay
The total RNA of the infected cells was extracted using the RNeasy Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The one-step qRT-PCR was performed with TransScript Taqman One-Step qRT-PCR SuperMix (for HCoV-OC43 detection) and TransScript II Green One-Step qRT-PCR SuperMix (for HCoV-229E detection) (TransGen Biotech) using the ABI 7500 Fast Real-Time PCR system (Applied Biosystems)9. The applied primer sequences are shown in Table 1.
Table 1.
Primers used in qRT-PCR assay.
| Name | Primer | Sequence(5ʹ–3ʹ) |
|---|---|---|
| HCoV-OC43 NP | Sense | CGATGAGGCTATTCCGACTAGGT |
| Antisense | CCTTCCTGAGCCTTCAATATAGTAACC | |
| Probe | TAMRA-TCCGCCTGGCACGGTACTCCCT-BHQ2 | |
| GAPDH (human) | Sense | CGGAGTCAACGGATTTGGTCGTAT |
| Antisense | AGCCTTCTCCATGGTGGTGAAGAC | |
| Probe | TAMRA- CCGTCAAGGCTGAGAACGG -BHQ2 | |
| HCoV-229E NP | Sense | GACCRATCCTGTCACCTCTGAC |
| Antisense | GGGCATTYTGGACAAAKCGTCTACG | |
| GAPDH (human) | Sense | CTCTGGAAAGCTGTGGCGTGATG |
| Antisense | ATGCCAGTGAGCTTCCCGTTCAG | |
| 18S rRNA (human) | Sense | TGGAGGAGACGTTCCAGTGT |
| Antisense | GATCTGTCCAGGCAGTCCTT |
2.8. Time-of-addition assay
The viral replication step(s) targeted by carrimycin was mapped by determining the effect of sequentially delayed addition of the compounds on viral NP expression, i.e., time-of-addition experiment10. Briefly, C3A cells (3 × 105 cells/well) were infected with HCoV-OC43 at multiplicity of infection (MOI) of 0.5. Carrimycin (10 μg/mL) was added at the time of infection or at a different time post infection. All the cells were harvested at 24 h post infection and NP in the cell lysates were detected by Western blot assay.
2.9. Pseudovirus infection and luciferase assay
293T cells stably expressing human ACE2 were seeded into white wall and clear bottom 96-well plates and infected with lentiviruses pseudotyped with VSV glycoprotein (VSV-G) protein or SARS-CoV-2 spike protein in the absence or presence of carrimycin. NH4Cl was used as a positive control. At 24 h post infection, the media were removed and cells were lysed with 20 μL/well of cell lysis buffer (Promega, Madison, WI, USA) for 15 min, followed by adding 50 μL/well of luciferase substrate (Promega). The firefly luciferase activities were measured by luminometry with an EnSpire instrument (PerkinElmer, Waltham, MA, USA).
2.10. Click-iT nascent RNA capture assay
C3A cells (3 × 105 cells/well) were infected with HCoV-OC43 (MOI = 5) for 2 h. At 16 h post infection, the infected cells were mock-treated or treated with 10 μg/mL of carrimycin or 2 μmol/L of RDV for 3 h, and followed by continuing the treatment in the presence of 0.5 mmol/L 5-ethynyl uridine (EU) for 1 h. Total cellular RNA was extracted using the RNeasy Mini Kit (QIAGEN). EU-labeled nascent RNA was captured with Click-iT Nascent RNA Capture Kit according to the manufacturer's instructions (Thermo Fisher Scientific). Then, the captured RNA was detected by qRT-PCR assay with primers specified in Table 1 11.
2.11. Statistics analysis
Statistical analyses were performed using GraphPad Prism 7.0 software (GraphPad Software Inc., San Diego, CA, USA). Image J software (Rawak Software Inc., Stuttgart, Germany) was used for quantitative study on Immunofluorescence data. Results are expressed as mean ± standard deviation (SD). Data were analyzed by one-way ANOVA with Holm-Sidak's multiple comparisons test. P < 0.05 was considered significant.
3. Results
3.1. Carrimycin is a broad-spectrum antiviral agent against HCoVs
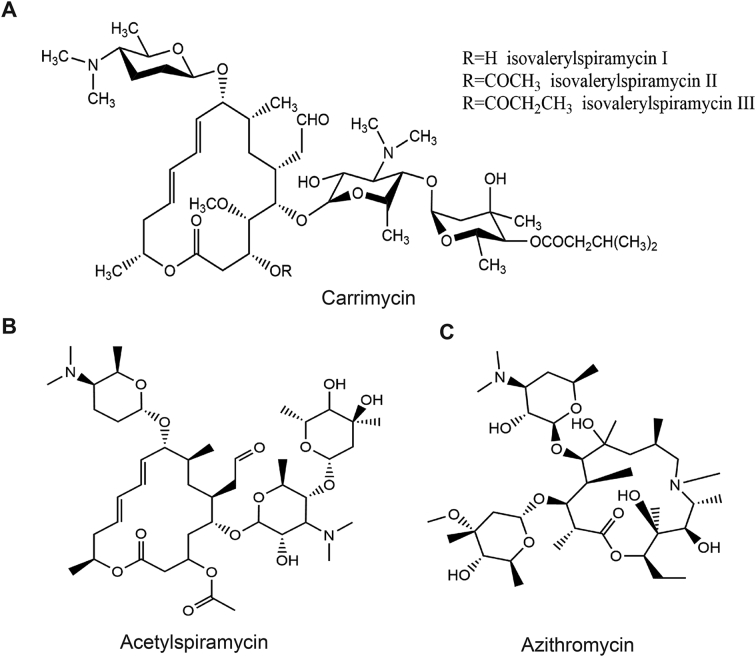
In order to identify the approved medicines that have a potential to be repurposed for the treatment of COVID-19, an in-house collection of more than 120 CFDA-approved drugs were screened for their ability to protect the CPE caused by HCoV-229E or HCoV-OC43 infection in Huh7 and H460 cells, respectively. Three macrolide antibiotics, acetylspiramycin, azithromycin, and carrimycin (Fig. 1), were found to inhibit the infection of both viruses with an SI higher than 5 (Table 2). In particular, carrimycin demonstrated the highest antiviral potency (EC50 value of 2.5 μg/mL) and selectivity (SI > 20) against both HCoV-229E and HCoV-OC43 (Table 2).
Figure 1.
The chemical structures of macrolide antibiotics. (A) Carrimycin. (B) Acetylspiramycin. (C) Azithromycin.
Table 2.
Antiviral activity of tested macrolide antibiotics against HCoV-229E and HCoV-OC43 in vitro.
| Drug | HCoV-229E |
HCoV-OC43 |
||||
|---|---|---|---|---|---|---|
| CC50 (μg/mL) | EC50 (μg/mL) | SI | CC50 (μg/mL) | EC50 (μg/mL) | SI | |
| Clarithromycin | >78.87 ± 29.88 | 21.18 ± 2.73 | >3.72 | >100 ± 0 | >100 ± 0 | – |
| Midecamycin | >100 ± 0 | 56.14 ± 18.66 | >1.78 | >100 ± 0 | 63.54 ± 8.20 | >1.57 |
| Erythromycin | >100 ± 0 | 39.17 ± 26.25 | >2.55 | >100 ± 0 | 63.54 ± 8.20 | >1.57 |
| Roxithromycin | >78.87 ± 29.88 | 25.43 ± 3.28 | >3.10 | >100 ± 0 | 40.70 ± 10.43 | >2.46 |
| Acetylspiramycin | >100 ± 0 | 18.71 ± 6.22 | >5.34 | >100 ± 0 | 13.57 ± 4.37 | >7.37 |
| Azithromycin | 63.54 ± 8.20 | 14.32 ± 0 | 4.44 | >100 ± 0 | 12.83 ± 9.07 | >7.79 |
| Ribavirin | >100 ± 0 | 3.77 ± 1.36 | >26.5 | >100 ± 0 | 7.06 ± 0.91 | >14.2 |
| Remdesivira | >5.0 ± 0 | 0.026 ± 0.003 | >192.3 | >5.0 ± 0 | 0.294 ± 0.053 | >17.0 |
| Carrimycin | 45.53 ± 17.25 | 2.35 ± 0.31 | 19.35 | 57.74 ± 0 | 2.51 ± 0.52 | 23.00 |
The cell cytotoxicity and antiviral activity assays presented in the table were tested by CPE assay.
"−" No antiviral activity at the maximal nontoxic concentration.
The unit of remdesivir concentration is μmol/L.
3.2. Carrimycin reduced the levels of HCoV RNA and protein in infected cultures
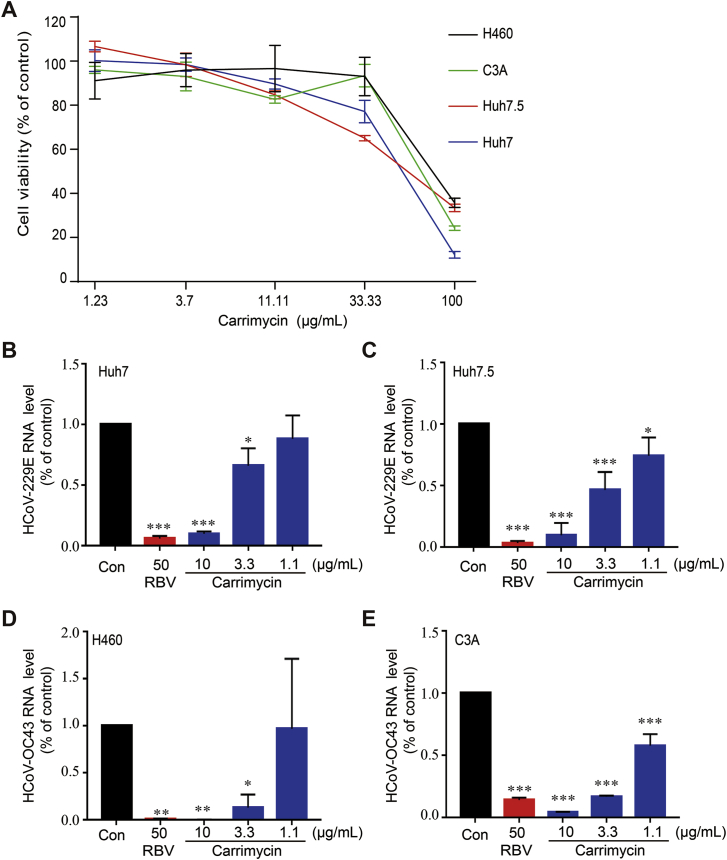
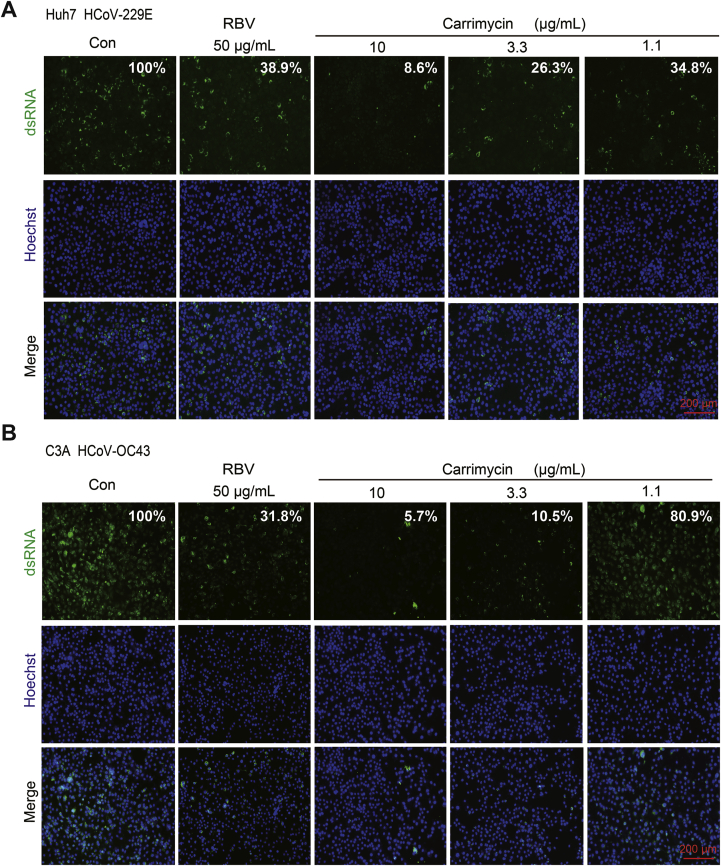
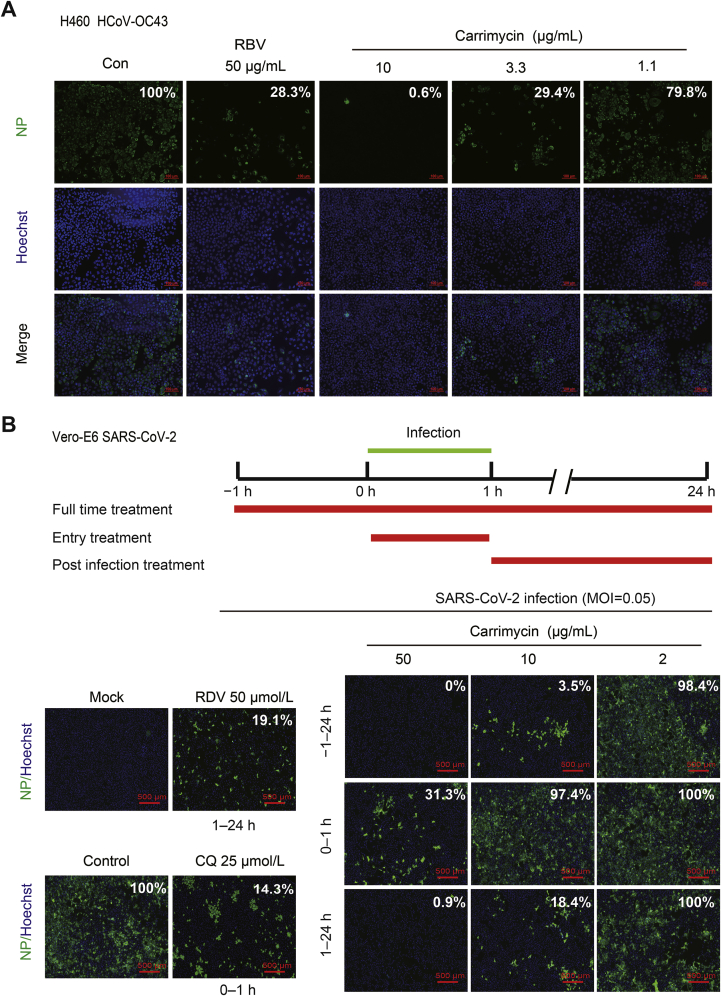
To ascertain the antiviral effects of carrimycin against HCoVs, we further examined the effects of carrimycin on the levels of viral nucleocapsid protein and RNA in infected cultures, with RBV as a positive control. As shown in Fig. 2, carrimycin reduced the levels of HCoV-229E and HCoV-OC43 RNA in multiple cells lines in a concentration dependent manner. As a positive-strand RNA virus, double stranded RNA is a key intermediate of viral RNA replication and can be visualized in the cytoplasm of HCoV infected cells. As shown in Fig. 3, carrimycin treatment significantly reduced the levels of dsRNA in HCoV-229E-infected Huh7 cells and HCoV-OC43-infected C3A cells. Moreover, immunofluorescent staining of virally infected cultures demonstrated that carrimycin treatment dose-dependently reduced the protein levels of HCoV-OC43 and SARS-CoV-2 NPs in H460 and Vero-E6 cells, respectively (Fig. 4).
Figure 2.
Carrimycin treatment significantly reduced HCoV RNA in multiple cell lines. (A) The cytotoxicity of carrimycin was determined by CCK assay. (B) and (C) Huh7 (2.5 × 105 cells/well) or Huh7.5 (2.5 × 105 cells/well) cells were plated into 12-well culture plates and incubated overnight. The cells were infected with HCoV-229E (MOI = 0.05) and various concentrations of carrimycin were added at the time of infection and treated for 24 h. The viral RNA levels were determined by a one-step qRT-PCR assay. (D) and (E) H460 (1.5 × 105 cells/well) or C3A (3 × 105 cells/well) cells were plated into 12-well culture plates and infected with HCoV-OC43 (MOI = 0.05) and various concentrations of carrimycin were added at the time of infection and treated for 48 h (H460) or 24 h (C3A). The viral RNA levels were determined by a one-step qRT-PCR assay. P values were calculated by one-way ANOVA (mean ± SD, n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. virus control (Con). RBV was used as the positive control.
Figure 3.
Carrimycin reduced the amounts of double-stranded RNA in HCoV infected cells. Huh7 (1.5 × 104 cells/well) or C3A (2.0 × 104 cells/well) cells were plated into 96-well culture plates and incubated overnight. The cells were infected with HCoV-229E (MOI = 0.005, A) or HCoV-OC43 (MOI = 0.05, B), and various concentrations of carrimycin were added at the time of infection and treated for 24 h. The dsRNA was visualized by immunofluorescent staining assay. Scale bar: 200 μm. The quantitative study on immunofluorescence was tested by Image J software.
Figure 4.
Carrimycin inhibited HCoVs infection as determined by immunofluorescent staining of viral NP proteins. (A) H460 cells (1.5 × 104 cells/well) were plated into 96-well culture plates and incubated overnight. The cells were infected with HCoV-OC43 (MOI = 0.05) and various concentrations of carrimycin were added at the same time for 48 h. Viral NP protein was visualized by immunofluorescence. Scale bar: 100 μm. (B) Vero-E6 cells (2.0 × 104 cells/well) were plated into 96-well culture plates and infected with SARS-CoV-2 (MOI = 0.05). Then, various concentrations of carrimycin and positive controls (RDV 50 μmol/L and CQ 25 μmol/L) were added at different times of infection. Viral NP protein was visualized by immunofluorescence. Scale bar: 500 μm. The quantitative study on immunofluorescence was tested by Image J software.
3.3. Carrimycin inhibited HCoV infection by targeting a post-entry replication event
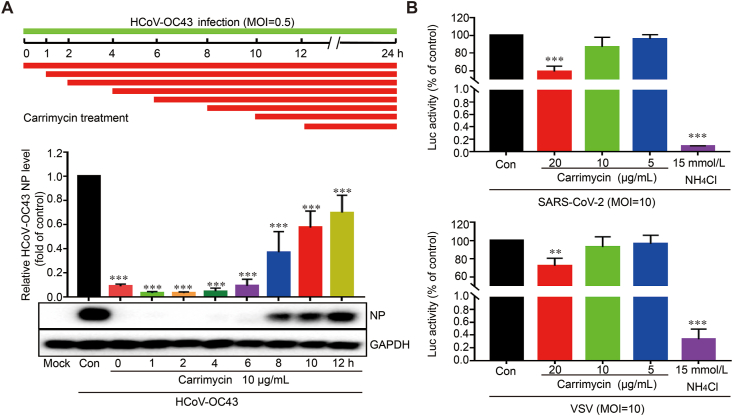
To determine the HCoV replication step(s) targeted by carrimycin, we took the advantage of robust infection of C3A cells by HCoV-OC43 to perform a time-of-addition experiment9. As shown in Fig. 5A, delayed addition of carrimycin at 6 h post infection still inhibited the expression of viral NP by approximate 90%, which is at the similar extent to that observed under the condition of treatment starting at 1 h before the infection (Fig. 4B). These results suggest that carrimycin most likely disrupted a post-entry replication step of the virus. In support of this notion, unlike NH4Cl that almost abolished the infection of lentiviral particles pseudotyped with SARS-CoV-2 envelope spike protein or VSV-G protein, carrimycin only modestly reduced the infection of both pseudoviruses at high concentration (Fig. 5B). Taken together, our results indicate that carrimycin efficiently inhibited the infection of multiple HCoVs by targeting one or multiple post-entry replication events.
Figure 5.
Carrimycin efficiently inhibited the infection of HCoVs by targeting a post-entry replication event. (A) Time-of-addition assay. C3A cells (3.0 × 105 cells/well) were plated into 12-well culture plates and infected with HCoV-OC43 (MOI = 0.5). Then, various concentrations of carrimycin were added at different times of infection and then the NP protein was analyzed using Western blot analysis. (B) 293T-hACE2 cells seeded in 96-well plates were infected with SARS-CoV-2 or VSV pseudovirus in the presence of the indicated concentrations of carrimycin and NH4Cl. At 24 h postinfection, the firefly luciferase activities were measured by microplate luminometry in a PerkinElmer EnSpire instrument. The luciferase activity was normalized to that of mock-treated control cells (mean ± SD, n = 3). ∗∗P < 0.01, ∗∗∗P < 0.001 vs. virus control (Con).
3.4. Carrimycin inhibited the synthesis of HCoV RNA
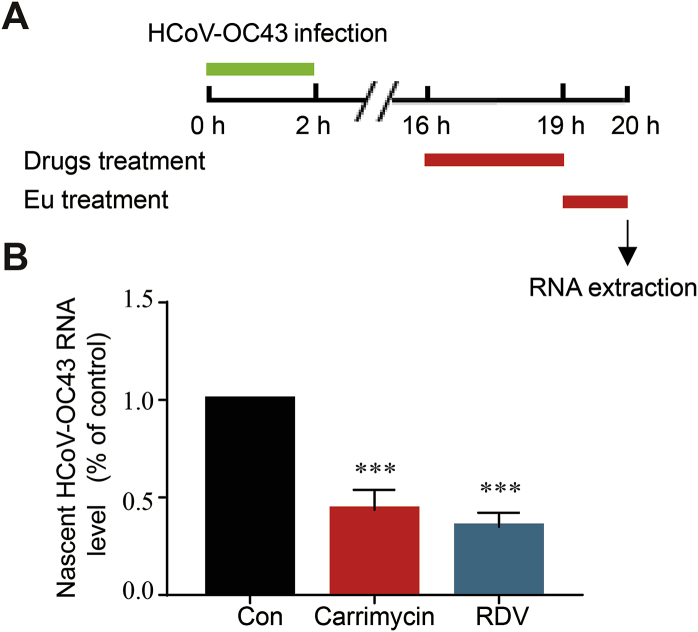
As carrimycin dose-dependently reduced the amounts of HCoV dsRNA, an intermediate of viral RNA replication, in infected cells, we further investigated whether the antibiotic inhibited the viral RNA synthesis by using a click chemistry method to detect the newly synthesized (nascent) viral RNA, with RDV, the HCoV RNA polymerase inhibitor, as a positive control. As depicted in Fig. 6A, HCoV-OC43 infected C3A cells were treated with carrimycin or RDV, starting at 16 h post infection for 3 h. The cells were then labeled with EU for 1 h. The EU-labeled nascent RNA was extracted from cell lysates by using a Click-iT Nascent RNA Capture Kit. HCoV-OC43 specific nascent RNA was quantified by a qRT-PCR assay. Similar to RDV, carrimycin also significantly reduced the amounts of HCoV-OC43 nascent RNA synthesis (Fig. 6B).
Figure 6.
Carrimycin inhibited the synthesis of viral RNA. (A) Experimental schedule. C3A cells (3.0 × 105 cells/well) were infected with HCoV-OC43 at an MOI of five for 2 h. The infected cells were mock-treated or treated with carrimycin (10 μg/mL) or RDV (2 μmol/L) at 16 h postinfection for 3 h and followed by continuing treatment in the presence of 0.5 mmol/L EU for 1 h. (B) Total EU-labeled RNA was extracted by using a Click-iT Nascent RNA Capture Kit as described in Materials and methods. HCoV-OC43 specific nascent RNA were determined by a one-step qRT-PCR assay and presented as the fraction of mock-treated control (mean ± SD, n = 4). ∗∗∗P < 0.001 vs. virus control (Con).
4. Discussion
Since the outbreak of COVID-19, there were no specific chemotherapeutic agents available to treat or prevent this disease. Currently, scientists around worldwide had focused on the repurposing of FDA approved drugs to treat COVID-19. Until now, more than 4000 clinical studies for COVID-19 were registered in the database of ClinicalTrials.gov (https://clinicaltrials.gov/ct2/results?cond=COVID-19). A review article systematically summarized some drugs repositioning for the control and treatment of COVID-1912. Although some drugs, such as remdesivir, can play a certain therapeutic effect in clinical treatment, the efficacy and adverse reactions of these drugs still need to be clarified13.
Carrimycin is a novel macrolide antibiotic produced by genetically engineered Streptomyces spiramyceticus harboring a 4″-O-isovaleryltransferase gene (ist) from Streptomyces thermotoleran. It mainly consists of isovalerylspiramycins (ISP) I–III, with trace amounts of other 4″-acylspiramycins14, 15, 16. Comparing with acylspiramycin, the longer alkyl chains at positions 4″ renders carrimycin more potent antibacterial activity, especially in vivo, as a result of higher lipophilicity17,18. Carrimycin was approved by CFDA for the treatment of acute tracheal-bronchitis caused by Haemophilus influenzae, Streptococcus pneumoniae, and for the treatment of acute sinusitis caused by Streptococcus pneumoniae, Haemophilus influenzae, Streptococcus pyogenes, Moraxella catarrhalis, and Staphylococcus. In this study, it was found that carrimycin exhibited broad-spectrum antiviral activity against HCoVs in multiple cells lines. As shown in Table 1, carrimycin showed the higher antiviral potency than acetylspiramycin. It remains to be determined whether the enhanced antiviral activity of carrimycin, as compared to acylspiramycin, is the result of higher lipophilicity and membrane permeability17,18.
Concerning the antiviral mechanism, the time-of-addition and pseudotyped lentiviral infection assays suggest that carrimycin efficiently inhibited the infection of multiple HCoVs by targeting one or multiple post-entry replication events. As positive strand RNA viruses, coronaviruses synthesize their RNA in the cytoplasmic replicase complexes consisting of viral nsp12–nsp7–nsp-8 core polymerase and cellular co-factors19,20. As shown in Fig. 6, similar to HCoV RNA polymerase inhibitor RDV, carrimycin significantly inhibited HCoV-OC43 RNA synthesis. Macrolide antibiotics inhibit bacterial protein synthesis by binding to the 50S ribosomal subunit21. As a macrolide antibiotic, it is not clear whether carrimycin hinders the viral protein synthesis, and it remains to be further investigated whether carrimycin directly inhibits the synthesis of viral RNA or regulates the synthesis of viral RNA by affecting host targets.
Pneumonia caused by viral infections usually has secondary bacterial infection22. COVID-19 may also be associated with secondary bacterial infections23,24. Now, the demonstrated antiviral activity to HCoVs, including SARS-CoV-2, as well as the preferential distribution in lungs via oral administration warranted the clinical trials of carrimycin for the treatment of COVID-19 in China (ChiCTR2000029867 and ChiCTR2000032242). The clinical trial results will be reported elsewhere.
Acknowledgments
We gratefully acknowledge Professor Ju-Tao Guo (Baruch S. Blumberg Institute, PA, USA) for helpful discussions and expert advice on the manuscript. The work was financially supported by CAMS Initiative for Innovative Medicine (2020-I2M-CoV19-008, China), the National Science and Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711003, China), the National Key Research and Development Program of China (2020YFC0844900, China), and Fundamental Research Funds for CAMS of China (2020HY320001, China).
Author contributions
Yuhuan Li and Jiandong Jiang designed experiments; Haiyan Yan, Jing Sun, Kun Wang, Shuo Wu, Linlin Bao, Airu Zhu, Tian Zhang, Rongmei Gao, Biao Dong, Jianrui Li, Qi Lv, Feifei Qin, Zhen Zhuang, and Xiaofang Huang carried out the experiments; Huiqiang Wang, Lu Yang, Ming Zhong, and Dong Wang analyzed the data and provided advice on the interpretation of data; Huiqiang Wang, Haiyan Yan, Jing Sun, Kun Wang, and Shuo Wu wrote the original draft with input from co-authors; Weiqing He and Xinyi Yang provided essential reagents; Yuhuan Li, Yongsheng Che, and Jiandong Jiang acquired funding; Yuhuan Li, Yongsheng Che, and Jiandong Jiang wrote the final draft; all authors approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Contributor Information
Yuhuan Li, Email: yuhuanlibj@126.com.
Yongsheng Che, Email: cheys@im.ac.cn.
Jiandong Jiang, Email: jiang.jdong@163.com.
References
- 1.Weiss S.R., Leibowitz J.L. Coronavirus pathogenesis. Adv Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Zaki A.M., Boheemen S.V., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol Sci. 2013;34:267–272. doi: 10.1016/j.tips.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X., Zheng S., Chen D., Zheng M., Li X., Li G. LY6E restricts entry of human coronaviruses, including currently pandemic SARS-CoV-2. J Virol. 2020;94:e00562-20. doi: 10.1128/JVI.00562-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y., Zhang J., Musharrafieh R., Hau R., Ma C., Wang J. Chemical genomics approach leads to the identification of hesperadin, an aurora B kinase inhibitor, as a broad-spectrum influenza antiviral. Int J Mol Sci. 2017;18:1929. doi: 10.3390/ijms18091929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagemeijer M.C., Vonk A.M., Monastyrska I., Rottier P.J., de Haan C.A. Visualizing coronavirus RNA synthesis in time by using click chemistry. J Virol. 2012;86:5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lima W.G., Brito J.C.M., Overhage J., Nizer W.S.D.C. The potential of drug repositioning as a short-term strategy for the control and treatment of COVID-19 (SARS-CoV-2): a systematic review. Arch Virol. 2020;165:1729–1737. doi: 10.1007/s00705-020-04693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beigel J.H., Tomashek K.M., Dodd L.E. Remdesivir for the treatment of covid-19 - final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano M., Sunazuka T., Tanaka H., Yamashita K., Okachi R., Omura S. Chemical modification of spiramycins. VI. Synthesis and antibacterial activities of 3,3ʹʹ-di-O-acyl-4ʹʹ-O-sulfonyl and 3,3ʹʹ-di-O-acyl-4ʹʹ-O-alkyl derivatives of spiramycin I. J Antibiot (Tokyo) 1985;38:1350–1358. doi: 10.7164/antibiotics.38.1350. [DOI] [PubMed] [Google Scholar]
- 15.Epp J.K., Huber M.L., Turner J.R., Goodson T., Schoner B.E. Production of a hybrid macrolide antibiotic in Streptomyces ambofaciens and Streptomyces lividans by introduction of a cloned carbomycin biosynthetic gene from Streptomyces thermotolerans. Gene. 1989;85:293–301. doi: 10.1016/0378-1119(89)90421-6. [DOI] [PubMed] [Google Scholar]
- 16.Shang G.D., Dai J.L., Wang Y.G. Construction and physiological studies on a stable bioengineered strain of shengjimycin. J Antibiot (Tokyo) 2001;54:66–73. doi: 10.7164/antibiotics.54.66. [DOI] [PubMed] [Google Scholar]
- 17.Shi X., Sun Y., Zhang Y., Zhong D. Tissue distribution of bitespiramycin and spiramycin in rats. Acta Pharmacol Sin. 2004;25:1396–1401. [PubMed] [Google Scholar]
- 18.Shi X., Fawcett J.P., Chen X., Zhong D. Structural identification of bitespiramycin metabolies in rat: a single oral dose study. Xenobiotica. 2005;35:343–358. doi: 10.1080/00498250500087580. [DOI] [PubMed] [Google Scholar]
- 19.Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J Biol Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brisson-Noël A., Trieu-Cuot P., Courvalin P. Mechanism of action of spiramycin and other macrolides. J Antimicrob Chemother. 1988;22:13–23. doi: 10.1093/jac/22.supplement_b.13. [DOI] [PubMed] [Google Scholar]
- 22.Manohar P., Loh B., Nachimuthu R., Hua X., Welburn S.C., Leptihn S. Secondary bacterial infections in patients with viral pneumonia. Front Med. 2020;7:420. doi: 10.3389/fmed.2020.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharov K.S. SARS-CoV-2-related pneumonia cases in pneumonia picture in Russia in March-May 2020: secondary bacterial pneumonia and viral co-infections. J Glob Health. 2020;10 doi: 10.7189/jogh.10.-020504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manna S., Baindara P., Mandal S.M. Molecular pathogenesis of secondary bacterial infection associated to viral infections including SARS-CoV-2. J Infect Public Health. 2020;13:1397–1404. doi: 10.1016/j.jiph.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]