Abstract
Computer-aided diagnosis for the reliable and fast detection of coronavirus disease (COVID-19) has become a necessity to prevent the spread of the virus during the pandemic to ease the burden on the healthcare system. Chest X-ray (CXR) imaging has several advantages over other imaging and detection techniques. Numerous works have been reported on COVID-19 detection from a smaller set of original X-ray images. However, the effect of image enhancement and lung segmentation of a large dataset in COVID-19 detection was not reported in the literature. We have compiled a large X-ray dataset (COVQU) consisting of 18,479 CXR images with 8851 normal, 6012 non-COVID lung infections, and 3616 COVID-19 CXR images and their corresponding ground truth lung masks. To the best of our knowledge, this is the largest public COVID positive database and the lung masks. Five different image enhancement techniques: histogram equalization (HE), contrast limited adaptive histogram equalization (CLAHE), image complement, gamma correction, and balance contrast enhancement technique (BCET) were used to investigate the effect of image enhancement techniques on COVID-19 detection. A novel U-Net model was proposed and compared with the standard U-Net model for lung segmentation. Six different pre-trained Convolutional Neural Networks (CNNs) (ResNet18, ResNet50, ResNet101, InceptionV3, DenseNet201, and ChexNet) and a shallow CNN model were investigated on the plain and segmented lung CXR images. The novel U-Net model showed an accuracy, Intersection over Union (IoU), and Dice coefficient of 98.63%, 94.3%, and 96.94%, respectively for lung segmentation. The gamma correction-based enhancement technique outperforms other techniques in detecting COVID-19 from the plain and the segmented lung CXR images. Classification performance from plain CXR images is slightly better than the segmented lung CXR images; however, the reliability of network performance is significantly improved for the segmented lung images, which was observed using the visualization technique. The accuracy, precision, sensitivity, F1-score, and specificity were 95.11%, 94.55%, 94.56%, 94.53%, and 95.59% respectively for the segmented lung images. The proposed approach with very reliable and comparable performance will boost the fast and robust COVID-19 detection using chest X-ray images.
Keywords: COVID-19, Image enhancement, Chest X-ray images, Convolutional neural networks, Lung segmentation
1. Introduction
Coronavirus Disease 2019 (COVID-19) pandemic with an exponential infection rate has overloaded worldwide healthcare systems [1]. There are more than twenty-one million active cases and more than two and half a million deaths in the world, as of March 2021 [2]. COVID-19 diagnosis is typically carried out by Reverse Transcription Polymerase Chain Reaction (RT-PCR), which suffers from low accuracy, delay, and low sensitivity [1,3,4]. Early diagnosis of disease increases the chances for successful treatment of infected patients and also reduces the chances of spreading in the community for a contagious disease like COVID-19. Wearable medical sensors and efficient artificial neural networks are used to classify COVID-19 patients based on physiological and questionnaire inputs [5]. Besides, different machine learning-based prediction models were implemented to predict the infection rate and probability of second and thirds waves of pandemic and risk of spreading associated to travel. Radiography images such as chest X-ray (CXR) or computed tomography (CT) are a routine technique for diagnosing lung-related diseases such as pneumonia [6], tuberculosis [7], and can be useful in COVID-19 detection as well [8,9]. One of the advantages of CXR is the ability to perform them easily using portable X-ray machines providing faster, and accurate COVID-19 diagnosis [8,[10], [11], [12], [13]]. CXRs are found to be potential for detecting COVID-19 with the help of artificial intelligence (AI) and are also less harmful to the human body compared to CT [8,[10], [11], [12]].
Recently, a large number of works have been carried out to detect COVID-19 using X-ray images with the help of different AI-based techniques. Different transfer learning techniques, novel network architectures, and ensemble solutions were proposed to improve the network performances to classify COVID-19, normal, and other lung diseases. Apostolopoulos et al. [6] reported 96.78% accuracy for COVID-19 detection from bacterial pneumonia and normal X-rays in a dataset of 1427 X-rays. Similarly, Abbas et al. [7] reported accuracy of 95.12% for COVID-19 classification from COVID-19, normal, and Severe Acute Respiratory Syndrome (SARS) CXR images using a pre-trained CNN model (DeTraC Decompose, Transfer and Compose) with a small database of 196 X-ray images. Minaee et al. [8] reported specificity and sensitivity of 90% and 97%, respectively using the ChexPert dataset [9]. Even though the results are promising, the dataset used for training machine learning (ML) models is small; however, it shows that deep ML models can be used for the COVID-19 detection. Khan et al. [10] explored a limited number of machine learning algorithms for a four-class classification problem (COVID-19, bacterial pneumonia, viral pneumonia, and normal) with a very small dataset. Goldstein et al. [11] built a classifier to detect COVID-19 using a pre-trained deep learning model (ResNet50) and enhanced by data augmentation and lung segmentation with the help of 2362 CXR images collected from four hospitals and achieved accuracy and sensitivity of 89.7% and 87.1%, respectively. Chowdhury et al. [12] proposed an ensemble of deep convolutional neural network (CNN) models named Efficient Convolutional Network (ECOVNet) to classify COVID-19, normal, and pneumonia using 16,493 CXR images using the transfer learning method and achieved an accuracy of 97%. Ashfar et al. [13] reported an accuracy of 95.7% using a Capsule Networks, called COVID-CAPS rather than a conventional CNN to deal with a smaller dataset. Yamac et al. [14] introduced a compact CNN architecture, Convolution Support Estimation Network (CSEN) that utilizes CheXNet as a feature extractor to classify the target CXR images as COVID-19, bacterial pneumonia, viral pneumonia, or normal. The network produced 98% COVID-19 detection sensitivity using a dataset of 462 COVID-19 CXR images. The same group of researchers has proposed a reliable warning system to diagnose early-stage COVID-19 cases using a compact CSEN network. Ahishali et al. [15] showed that CheXNet and CSEN have achieved a COVID-19 detection sensitivity of 97.1% and 98.5%, respectively on a smaller dataset. Wang and Wong [16] used around 14k CXR images but reported an accuracy of 83.5% using a deep CNN, called COVID-Net. Furthermore, Motamed et al. [17] proposed a randomized generative adversarial network (RANDGAN) that detects images of an unknown class (COVID-19) from known and labeled classes (normal and viral pneumonia) without the need for labels and training data from the unknown class of images (COVID-19) using 14,100 CXR images and attained an area under the curve (AUC) of 0.77. It was observed that combining different techniques can help to achieve better performance in COVID-19 detection. Angelica et al. [18] introduced a graph-based deep semi-supervised framework for classifying COVID-19 from CXR images using around 15,254 images and achieved an accuracy of 96.4%. Degerli et al. in Ref. [19] proposed a novel method for the joint localization, severity grading, and detection of COVID-19 from 15,495 CXR images by generating the so-called infection maps that can accurately localize and grade the severity of COVID-19 infection with 98.69% accuracy. Ahmed et al. in Ref. [20] proposed a novel CNN architecture, ReCoNet (residual image-based COVID-19 detection network) for COVID-19 detection using preprocessing steps, which was reported to be very useful for enhancing unique COVID-19 signature. The proposed modular architecture trained on 15,134 CXR images and achieved an accuracy, sensitivity, and specificity of 97.48%, 96.39%, and 97.53%, respectively. The machine learning model consists of a CNN-based multi-level preprocessing filter block in cascade with a multi-layer CNN-based feature extractor and a classification block. In the recent articles, transfer learning was very common approach to tackle such a problem and it showed very promising results.
The importance and the scientific motivations of the work are presented in this paragraph. There is a demand for medical image enhancement to help clinicians to make an accurate diagnosis of the diseases [6,8,12,32]. The image enhancement process consists of a collection of techniques that are used to improve the visual appearance of an image such as removing blur and noise of the image, which in turn increase the contrast and provides more details of an image. With the large number of X-ray images acquired every day in the hospitals for COVID-19 patients, the quality of the acquired images can vary due to several reasons: the condition of the patients, the breathing state of the patient, and human error. For front view X-ray, Posterior – Anterior (PA) and Anterior-posterior (AP) are the most common methods for chest X-ray images. Some differences between the procedure of both methods will reflect on the output image. The AP approach gives a less clear view of the heart and mediastinum because the patient is not instructed to take a few deep breaths and hold them for a couple of seconds. Thus, the output image is less clear compared to the PA approach. Besides, an AP image can also be taken with the patient sitting or at a supine state on the bed, which can affect the image quality. Secondly, many images are converted from DICOM (Digital Imaging and Communications in Medicine) to other image formats such as PNG (Portable Network Graphics) and JPG/JPEG (Joint Photographic Experts Group). This conversation can lead to lower-quality images. It is required to improve the overall quality of the image, which improves the spatial features of the image. The main purpose of image enhancement is to improve the interpretability or perception of information contained in the image for human viewers or feature extraction and creates an image that is subjectively better than the original image by changing the pixel intensity of the input image. A major concern is not to alter the information during the image enhancement process. Various image enhancement techniques such as de-noising algorithms, filtering, interpolation, wavelets, etc. [[34], [35], [36]] are applied for this purpose. Many functions are available to enhance the geometric features such as edges, corners, and ridges of the medical images. These techniques and approaches can enhance the classification performance of the machine learning models for medical images. Several local image enhancement algorithms have been introduced in the last two decades to improve the image quality to boost machine learning models’ performance [37,38]. Arun et al. [39] proposed the adaptive histogram equalization technique, which can help in image enhancement; however, this made the image appears fuzzy. This approach was further improved by Hasikin et al. [40], where they proposed the use of fuzzy set theory. It not only produces better quality images but also requires minimum processing time. Selvi et al. [41] proposed a method for enhancing the fingerprint images. A four-step image enhancement technique, i.e. preprocessing, fuzzy-based filtering, adaptive thresholding, and morphological operation, was utilized for producing noise-free fingerprint images. This technique produced better peak signal-to-noise (PSNR) values than many previous techniques. Mohammad et al. [42] presented Bi-and Multi-histogram methods to enhance the contrast while preserving the brightness and natural appearance of the images. This technique has been useful in many applications that require image enhancement [43]. Several other popular histogram techniques can be explored for CXR images to investigate whether they can help ML models in various image classification techniques or not [8,24,[44], [45], [46], [47], [48], [49], [50], [51]]. In our previous study [8], we have discussed four different pre-processing schemes that were tested for detecting COVID-19 from other coronavirus family diseases (SARS and Middle East Respiratory Syndrome (MERS)) using CXR images. It was observed that the 3-channel approach (a combination of original, Contrast Limited Adaptive Histogram Equalization (CLAHE) and image complement) outperforms other enhancement techniques and achieves sensitivities of 99.5%, 93.1%, and 97% for classifying COVID-19, MERS, and SARS images. Yujin et al. [49] used Histogram Equalization (HE) and gamma correction enhancement techniques for detecting COVID-19 from CXR images. They proposed a patch-based CNN approach with a relatively small number of trainable parameters for COVID-19 diagnosis and it showed 92.5% sensitivity for COVID-19. Heidari et al. [28] proposed a pre-trained VGG-19 network using histogram equalization and a new three-channel approach using 8474 CXR images consisting of COVID-19, community-acquired pneumonia, and normal cases. The three-channel approach used the two sets of filtered images from the enhanced CXR and the original images, which achieved 94.5% accuracy in classifying COVID-19 images. It can be summarized that the above-stated studies along with many others are relying on a limited dataset for developing and validating machine learning models.
Since the reported articles used a dataset containing a very small number of COVID-19 CXR images, it makes them difficult to generalize their findings, and cannot guarantee to reproduce the results when these models are evaluated on a larger dataset. Therefore, the investigation of different CXR image enhancement techniques investigated on a large dataset comprising normal (healthy class), non-COVID (other lung infections), and COVID-19 infected patients’ CXR images will be very useful. The contributions of this paper can be explicitly stated below:
-
•
To the best of the authors' knowledge, this is the first work where the effects of various CXR image enhancement techniques were extensively studied on plain and segmented CXR image classification.
-
•
This is the largest CXR and lung segmented image dataset comprising COVID-19, normal (healthy), and non-COVID (different lung infections) CXR images were gathered and used for classification.
-
•
A modified version of the U-Net model is proposed in this manuscript, which outperforms the standard U-Net architecture for the lung segmentation of CXR images.
-
•
The outcome of this study was verified by image visualization technique to confirm the findings of the deep networks.
-
•
The use of CXR image enhancement techniques, transfer learning, and segmentation of lung resulted in benchmark results of COVID-19 detection outperforming the state-of-the-art methods.
The remaining part of this paper is divided into the following sections: Section 2 provides the details of the various pre-trained classification models, lung segmentation models, different image enhancement, and visualization techniques. Section 3 describes the methodology followed in this study, and the results of the classification performance using the original and segmented CXR images enhanced using different techniques along with the visualization heat map in Section 4. The paper is then concluded in Section 5.
2. Background
2.1. Deep Convolutional Neural Networks
Deep CNNs have been popularly used in image classification due to their superior performance compared to other machine learning paradigm. The networks automatically extract the spatial and temporal features of an image. The approach of transfer learning has been successfully incorporated in many applications. [[21], [22], [23]], especially where a large dataset can be hard to find. Thus, it opens the opportunity of utilizing smaller datasets and also reduces the time required to develop a deep learning algorithm from scratch [24,25]. For COVID-19 detection, nine pre-trained deep learning CNNs such as ResNet18, ResNet50, ResNet101 [26], DenseNet201 [27], ChexNet [28], and InceptionV3 [29] were predominantly used in the literature. ChexNet is the only network that is trained on CXR images, unlike the other networks that are initially trained on the ImageNet database. Residual Network (in short ResNet) with several variants, solve vanishing gradient and degradation problem [26] and learn from residuals instead of features [30]. Dense Convolutional Network (in brief DenseNet) needs a smaller number of parameters than a conventional CNN, as it does not learn redundant feature maps. The DenseNet has layers with direct access to the original input image and loss function gradients. Another variant of DenseNet, ChexNet is trained and validated using a large number of CXR images [28]. Inception-v3 is a CNN architecture from the inception family that makes several improvements including using label smoothing, factorized 7 × 7 convolutions, and the use of an auxiliary classifier to propagate label information lower down the network (along with the use of batch normalization for layers in the side head). This network scales in ways that strive to use the added computation as effectively as possible through correctly factorized convolutions and aggressive regularization [29].
2.2. Image enhancement techniques
Image enhancement is an important image-processing technique, which highlights key information in an image and reduces or removes certain secondary information to improve the identification quality in the process. The aim is to make the objective images more suitable for a specific application than the original images. We employ five different enhancement techniques in this research. In the following section, these image enhancement techniques will be briefly introduced:
2.2.1. Histogram equalization (HE)
The histogram equalization (HE) technique aims to distribute the gray levels within an image. Each gray level is therefore has equal chances to occur. HE changes the brightness and contrast of the dark and low contrast images to enhance image quality [31]. The histogram would be skewed towards the lower end of the grayscale for a dark image, and the image information would be squeezed into the dark end of the histogram. In order to create a more evenly distributed histogram, the gray levels can be re-distributed at the dark end, which can make the image clear. The histogram of a digital image with intensity levels in the range [0, L-1] is a discrete function represented as follows:
| (1) |
Where, is kth intensity value, is the number of pixels in the image with intensity, . Histograms are frequently normalized by the total number of pixels in the image. Assuming an M x N image, a normalized histogram is related to the probability of occurrence of in the image as shown in equation (2).
| (2) |
2.2.2. Contrast limited adaptive histogram equalization (CLAHE)
An improved HE variant is called Adaptive Histogram Equalization (AHE). AHE performs histogram equalization over small regions (i.e., patches) in the image, and thus, AHE enhances the contrast of each region individually. Therefore, it improves local contrast and edges adaptively in each region of the image to the local distribution of pixel intensities instead of the global information of the image. However, AHE could over amplify the noise component in the image [32]. However, images enhanced with CLAHE are more natural in appearance than those produced by HE. When the HE technique was applied to the X-ray images, it was observed that it saturated certain regions. To address this issue, CLAHE uses the same approach as AHE but the amount of contrast enhancement that can be produced within the selected region is limited by a threshold parameter. Firstly, the original image is converted from RGB (red, green, and blue) color space to HSV (hue, saturation, and value) color space as a human sense color similar to the HSV version. Secondly, the value component of HSV is processed by CLAHE without affecting the hue and saturation. The initial histogram is cropped and each gray-level is redistributed to the cropped pixels. The value of each pixel is reduced to a user-selectable limit. Finally, the HSV processed image is re-transformed to RGB color space.
2.2.3. Image invert/complement
The image inversion or complement is a technique where the zeros become ones and ones become zeros so black and white are reversed in a binary image. For an 8-bit grayscale image, the original pixel is subtracted from the highest intensity value (255), the difference is considered as pixel values for the new image. For X-ray images, the dark spots turn lighter and light spots become darker. The mathematical expression is simply as follows:
| (3) |
Where, x and y are the intensity values of the original and the transformed (new) images. This technique shows the lungs area (i.e., the region of interest) lighter and the bones are darker. As this is a standard procedure, which was used widely by radiologists, it may equally help deep networks for a better classification. It can be noted that the histogram for the complemented image is a flipped copy of the original image.
2.2.4. Gamma correction
Typically, linear operations are performed on individual pixels in image normalization, such as scalar multiplication, addition, and subtraction. Gamma correction performs a non-linear operation on source image pixels. Gamma correction alternates the pixel value to improve the image using the projection relationship between the value of the pixel and the value of the gamma according to the internal map. If P represents the pixel value inside the range [0,255], Ω represents the angle value, Ґ is the symbol of the gamma value set, x is the grayscale value of the pixel (x ε P). Let be range midpoint [0, 255]. The linear map from group P to group Ω is defined as:
| (4) |
The mapping from Ω to Ґ is defined as:
| (5) |
| (6) |
where a ε [0, 1] denotes a weighted factor.
Based on this map, group P can be related to Ґ group pixel values. The arbitrary pixel value is calculated in relation to a given Gamma number. Let (x) = h(x), and the Gamma correction function is as follows
| (7) |
where g(x) represents the output pixel correction value in grayscale.
2.2.5. Balance contrast enhancement technique (BCET)
BCET represents an approach for improving balance contrast by stretching or compressing the contrast of the image without altering the histogram pattern of the image data. The solution is based on the parabolic function acquired from the image data. The general parabolic functional form using y coordinate and x coordinate in an XY plane is defined as
| (8) |
The three coefficients a, b and c are determined from the following equations using the minimum, the maximum, and the mean of the input and output image values.
| (9) |
| (10) |
| (11) |
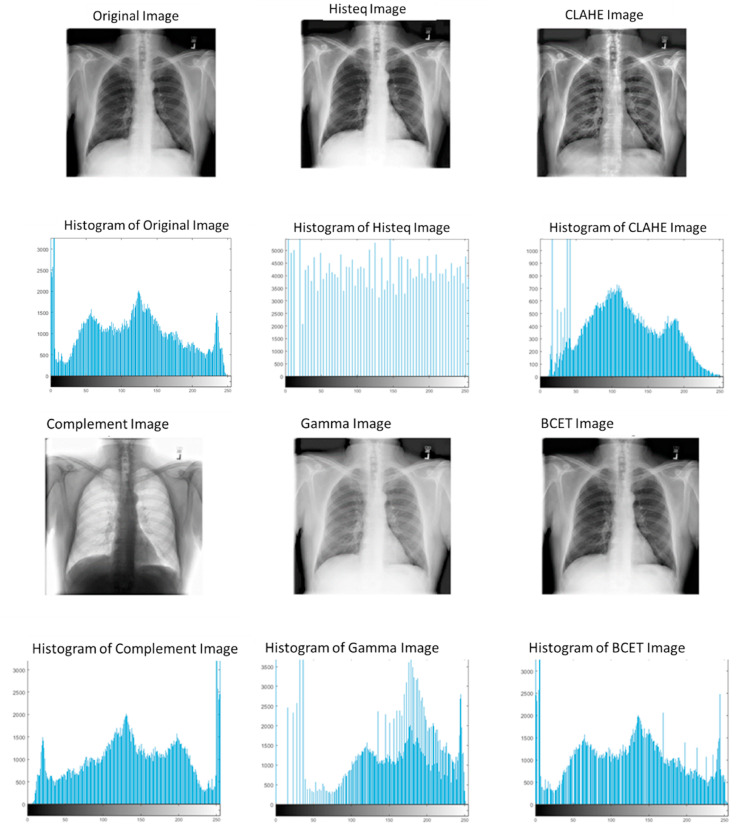
Where 'l' represents the minimum value of the input image, 'h' denotes the maximum value of the input image, 'e' denotes the mean value of the input image, 'L' the minimum value of the output image, 'H' denotes the maximum value of the output image and 'E' denotes the mean value of the output image. Fig. 1 shows the difference between the different image enhancement techniques.
Fig. 1.
Histogram for original X-ray image and the images undergo different enhancement techniques.
2.3. Visualization techniques
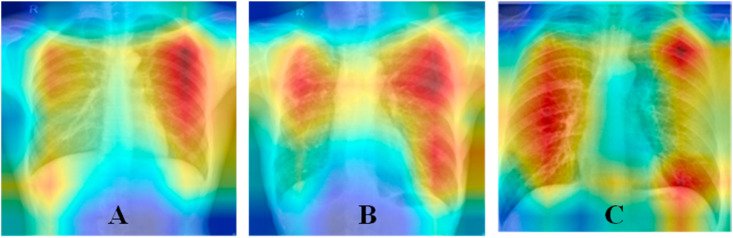
The emergence of visualization tools has led to growing interest in how CNN works and the logic behind the making of particular decisions by a network. In order to view the decision-making process of CNNs, visualization methods lead to better visual representation. These also improve the transparency of the model by visualizing the reasoning behind the inference that can be interpreted in a way that can be easily understood by humans, thereby increasing trust in the results of the CNNs. There are many popular visualization techniques such as SmoothGrad [33], Grad-CAM [34], Grad-CAM++ [35], Score-CAM [26]. But in this study, Score-CAM was used due to its promising performance [36]. Fig. 2 provides a sample Score-CAM visualization, where it highlights the regions used by CNN in making decisions. This visualization helps in increasing the confidence of the reliability of the deep layer networks, by confirming the decision making from the relevant region of the images.
Fig. 2.
Score-CAM visualization of A) COVID-19 CXR, B) Normal CXR, C) Non-COVID Lung Opacity CXR, to show the location of CNN model's learning.
3. Methodology
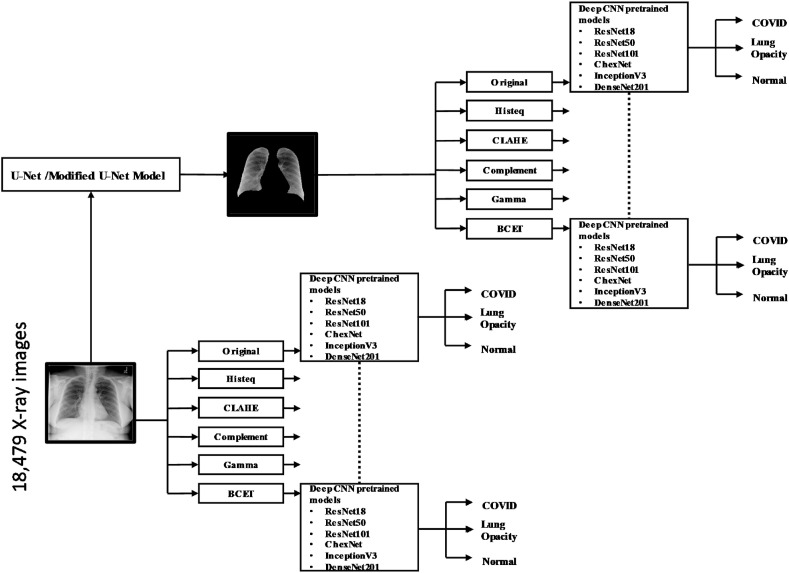
The block diagram of the methodology adopted in the study is shown in Fig. 3 . The study used two different CXR image databases: 1) lung segmentation and 2) classification databases. The major experiments that are carried out in this study: 1) Two U-Net models (original and modified U-Net model, which is proposed by the authors) to segment lungs from the CXR images, 2) Evaluation of original and five different enhanced plain CXR images for classification using seven different pre-trained deep-learning networks, and 3) Evaluation of segmented lungs CXR images (original and five different enhanced) for classification using pre-mentioned networks. Along with the calculation of different performance matrics to evaluate the network performance, the elapsed time per image for the best performing network in each image enhancement technique for plain and segmented lung X-ray images were calculated. The reliability of the last two experiments was verified using the Score-CAM visualization technique.
Fig. 3.
Block diagram of the system methodology.
The details of the study, i.e. dataset details, pre-processing and augmentation techniques adopted in this study, performance matrices utilized in the study, are discussed below.
3.1. Datasets description
In this study, the authors have used a large dataset compiled by the team and referred to the COVQU dataset, which is comprised of 18,479 CXR images across 15,000 patient cases.
3.1.1. Datasets -lung segmentation
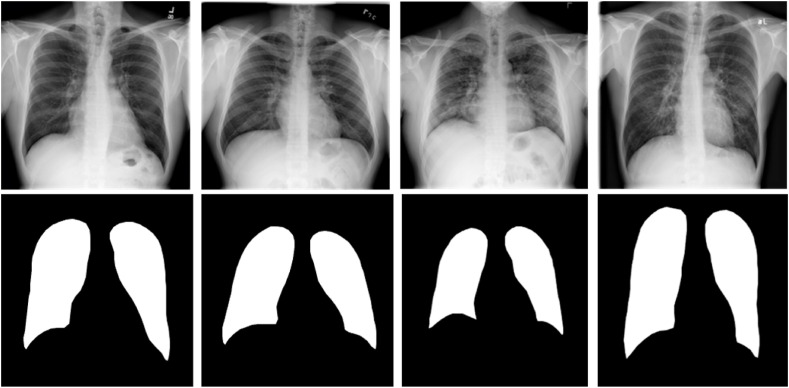
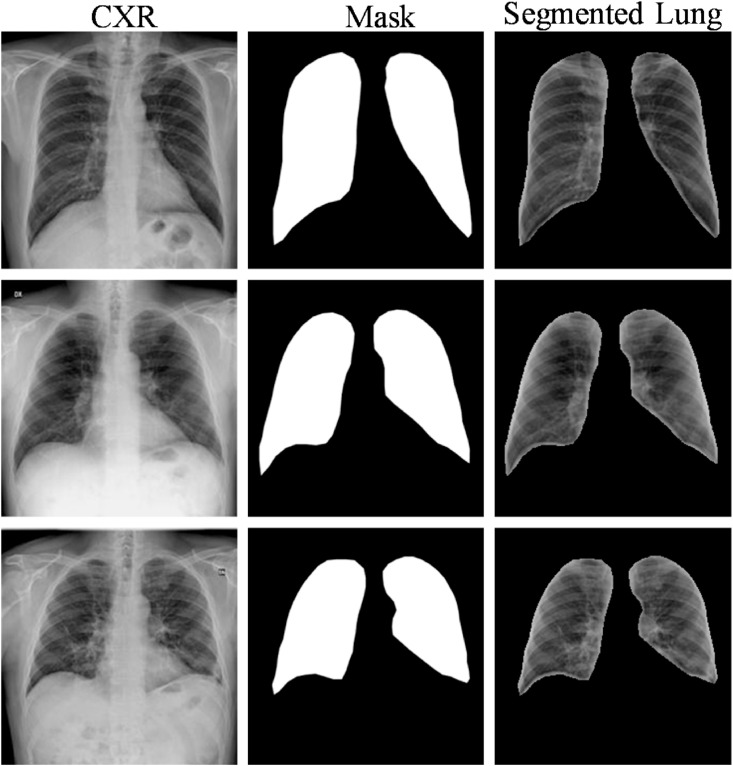
To investigate lung segmentation models, the authors have created ground truth lung masks for 18,479 CXR images, which are verified by expert radiologists as a part of a separate study (which is not reported in this study). Sample CXR images and masks are shown in Fig. 4 . The original and the modified U-Net networks were trained and tested with CXR images and their respective ground truth masks.
Fig. 4.
Samples of CXR images and their ground truth masks of the dataset.
3.1.2. Datasets -image classification
The dataset used to train and evaluate the proposed study is comprised of 18,479 CXR images across 15,000 patient cases. This COVQU dataset is the largest public COVID-19 positive dataset, according to the best of the authors’ knowledge. To generate this dataset, the authors have used and modified different open-access databases for three different types (COVID-19, normal (healthy), and non-COVID lung infections) CXR images. COVQU dataset combined the Radiological Society of North America (RSNA) CXR dataset [37] and COVID-19 dataset, details below.
3.1.3. RSNA CXR dataset
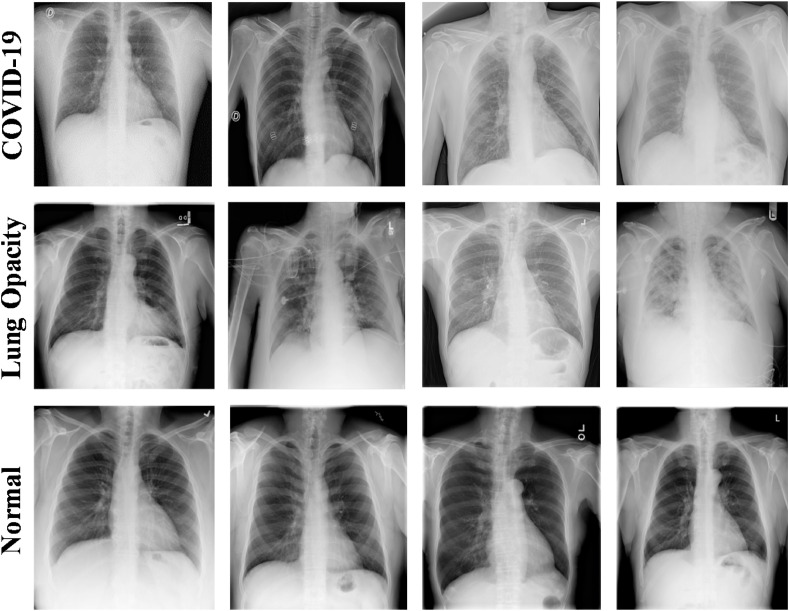
RSNA pneumonia detection challenge dataset [37], consists of about 26,684 CXR DICOM images, where 8851 images are normal, 11,821 images are with different lung abnormalities and 6012 are non-COVID ground grass lung opacity X-ray images. In this study, we have used 8851 normal (healthy) and 6012 non-COVID (lung infection) X-ray images. The CXR's in the dataset were reviewed by trained radiologists and the condition was confirmed through clinical history, vital signs, and laboratory exams. The dataset comprises several other health conditions in the lungs such as fluid overload (pulmonary edema), bleeding, volume loss (atelectasis or collapse), lung cancer, or post-radiation or surgical changes of lungs. The CXR images were correlated with clinical symptoms and history in classifying them to healthy control (normal) (without any underlying medical condition) and non-COVID lung infections. Sample X-ray images used in the study are shown in Fig. 5 .
Fig. 5.
CXR image samples from different datasets: (A) COVID-19, (B) non-COVID Lung Opacity, (C) and Normal (healthy).
3.1.4. COVID-19 dataset
COVID-19 dataset is comprised of 3616 positive COVID-19 CXR images, which are collected from different publicly available datasets, online sources, and published articles. Out of 3616 X-ray images, 2473 images are collected from the BIMCV-COVID19+ dataset [38], 183 images from a German medical school [39], 559 X-ray images are from the Italian Society of Medical Radiology (SIRM), GitHub, Kaggle & Twitter [[40], [41], [42], [43]], and 400 X-ray images from another COVID-19 CXR repository [44]. BIMCV-COVID19+ dataset is the single largest public dataset with 2473 CXR images of COVID-19 patients acquired from digital X-ray (DX) and computerized X-ray (CX) machines. The major difference between non-COVID and COVID categories is the lung opacity in the CXR images due to other lung-related diseases and COVID-19, respectively.
3.2. Preprocessing and data augmentation
The datasets were preprocessed to resize the X-Ray images to fit the input image-size requirements of different CNN models such as 256 × 256 pixels for the U-Net models, 299 × 299 pixels for InceptionV3, and 224 × 224 pixels for all other networks. Using the mean and standard deviation of the images, Z-score normalization was carried out.
3.2.1. Data augmentation
It is reported that data augmentation can improve the classification accuracy of the deep learning algorithms by augmenting the existing data rather than collecting new data [45]. Data augmentation can significantly increase the diversity of data available for the training models. Image augmentation is crucial when the dataset is imbalanced. In this study, the number of normal images was 8,851, which is more than twice the size of COVID-19 CXR images. Therefore, it was important to augment COVID-19 CXR images two-times to make the database balance. Some of the deep learning frameworks have built-in data augmentation facility within the algorithms, however, in this study, an image rotation based augmentation technique was utilized to generate training images of COVID-19 before applying to the CNN models for training.
3.3. Details of the experiments
As explained earlier in the methodology section, three major sets of experiments (Lung Segmentation, Classification without and with segmented lung CXR images) were carried out in this study. In these experiments, five-fold cross-validation was used, where 80% of the total images were used for training and 20% for testing. Out of the training dataset, a subset of 20% was utilized for validation to avoid overfitting [46]. Table 1 shows the details of the number of training, validation, and test CXR images used in the two experiments of plain and segmented lung X-ray images using five different image enhancement techniques. Finally, the weighted average of the five folds was calculated.
Table 1.
Details of the dataset used for training, validation, and testing.
| Database | Types | Count. of CXR's/class | Training Dataset |
|||
|---|---|---|---|---|---|---|
| Training image/fold | Augmented train image/fold | Validation image/fold | Test image/fold | |||
| COVID-19 dataset | COVID-19 | 3616 | 2314 | 4628 | 578 | 724 |
| RSNA CXR dataset | Normal | 8851 | 5664 | 5664 | 1416 | 1771 |
| Non-COVID | 6012 | 3847 | 3847 | 962 | 1203 | |
As mentioned earlier, different experiments conducted in this study were carried out using PyTorch library with Python 3.7 on Intel® Xeon® CPU E5-2697 v4 @ 2.30 GHz and 64 GB RAM, with a 16 GB NVIDIA GeForce GTX 1080 GPU. In the following section, each of them will be discussed separately:
3.3.1. Experiments on lung segmentation
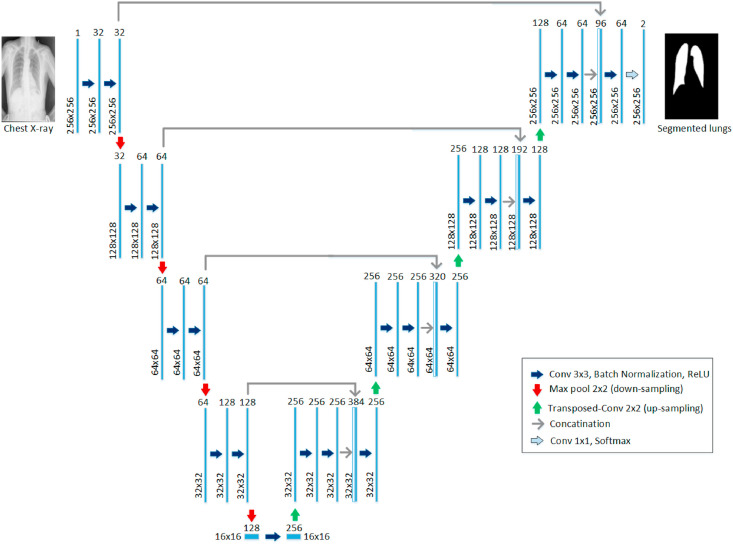
Recently, U-Net architecture has gained increasing popularity in different biomedical image segmentation applications [6, 62]. U-Net is one of the most important semantic segmentation frameworks for a convolutional neural network (CNN), which is widely used in the medical image analysis domain for lesion segmentation, anatomical segmentation, and classification. The advantage of this network framework is that it can accurately segment the desired feature target and effectively process and objectively evaluate medical images [63]. In this study, we have investigated the original U-Net architecture and proposed a modified version of the U-Net model (Fig. 6 ).
Fig. 6.
Modified U-Net model architecture proposed for lung segmentation.
The original U-Net model consists of a contracting path with 4 encoding blocks, followed by an expanding path with 4 decoding blocks. Each encoding block consists of two consecutive 3 × 3 convolutional layers followed by a max-pooling layer with a stride of 2 for downsampling. The decoding blocks consist of a transposed convolutional layer for upsampling, a concatenation with the corresponding feature map from the contracting path, and two 3 × 3 convolutional layers. While the decoding block has a variation in the modified U-Net architecture, where three convolutional layers are used instead of two. The modified decoder starts with an upsampling layer, followed by two 3 × 3 convolutional layers, a concatenation layer, and a 3 × 3 convolutional layer. All convolutional layers are followed by Batch normalization and ReLU activation. At the final layer, 1 × 1 convolution is utilized to map the output from the last decoding block to 2 channel feature maps, where a pixel-wise SoftMax is applied to map each pixel into a binary class of background or lung.
A standard U-Net and the proposed modified U-Net models were trained and validated to create lung segmentation using five-fold cross-validation. Out of 18,479 CXR images and ground truth lung masks, 80% of images and the corresponding masks were used for training and 20% images for testing. The training was done using a batch size of 4, the learning rate of 0.001 for a maximum of 20 epochs using Adam optimizer. The learning rate was decreased if no improvement was observed for consecutive 3 epochs and stopped if there was no improvement for consecutive 6 epochs.
3.3.2. Experiments on COVID-19 classification using plain and segmented lung CXRs
In two different classification experiments (classification using plain and segmented lung CXRs), five-fold cross-validation was used with 80% of 18,479 CXR images, and segmented lungs were used for training and 20% for testing. To avoid the overfitting issue, 20% of the training set was used for validation. Seven different CNN models were compared separately using non-segmented (plain) and segmented (lung) X-ray images using five different image enhancement techniques (HE, CLAHE, Complement, Gamma Correction, and BCET) for the classification of COVID-19, non-COVID lung opacity, and normal images to investigate the effect of image enhancement and lung segmentation on COVID-19 detection. The training was done using a batch size of 32 CXR images, a learning rate of 0.001, for a maximum of 20 epochs using Adam optimizer. As mentioned earlier, the learning rate was decreased with no improvement for consecutive 3 epochs and stopped with no improvement for consecutive 6 epochs.
Five deep pre-trained networks (Inceptionv3, ResNet50, ResNet101, ChexNet, and DenseNet201) and one comparatively shallow pre-trained network (ResNet18) and a shallow CNN were evaluated. Six pre-trained networks were trained from their ImageNet initial weights and a shallow CNN model was trained from scratch to see the comparative performance of transfer learning and learning from scratch. An eleven-layer CNN architecture was trained from scratch where the input to the cov1 layer is a 224 × 224 RGB image of a fixed size. The image is passed through a stack of convolutional (Conv.) layers where the filters have been used with a very limited receptive field: 3 to 3 (which is the smallest size to capture the left/right, up/down, center concept). It also utilizes 1 to 1 convolution filters in one of the setups, which can be seen as a linear transformation of the input channels (followed by non-linearity). The convolution stride is 1 pixel fixed; convolution's spatial padding. The layer input is such that after convolution, the spatial resolution is retained, i.e. the padding for 3-3 convolution is 1-pixel. All the convolution layers are not followed by max-pooling. Five max-pooling layers, which follow some of the convolution layers, perform spatial pooling. Max-pooling, with stride 2, is performed over a 2 to 2-pixel window. A stack of convolutional layers (which has a different depth in different architectures) follows three Fully-Connected (FC) layers: the first two have 4096 neurons each, the third performs 3-class classification and thus includes 3 neurons (one for each class). The soft-max layer is the final layer as shown in Supplementary Table 1.
3.4. Performance evaluation matrix
3.4.1. Evaluation matrix - lung segmentation
The performance of different networks in image segmentation for the testing dataset was evaluated after the completion of the training and validation phase and was compared using three performance metrics: accuracy, Intersection-Over-Union (IoU), and Dice. The equations used to calculate accuracy, IoU or Jaccard Index, and Dice coefficient (or F1-score) are shown in equation (13), (14), (15).
| (12) |
| (13) |
| (14) |
3.4.2. Evaluation matrix - COVID classification
The different CNNs’ performance in classification was evaluated using six performance metrics: overall accuracy, weighted sensitivity or recall, weighted specificity, weighted precision, and weighted F1 score using Eqs. (16), (17), (18), (19), (20). Since different classes had a different number of images, per class weighted performance metrics and overall accuracy were used to compare the networks. The performance was also evaluated using the area under the curve (AUC):
| (15) |
| (16) |
| (17) |
| (18) |
| (19) |
Here, true positive (TP), true negative (TN), false positive (FP), and false-negative (FN) were used to denote the number of COVID-19 CXR images were identified as COVID-19, the number of normal and non-COVID lung opacity CXRs were identified as normal and non-COVID CXRs, the number of normal and non-COVID CXRs incorrectly identified as COVID-19 CXRs and the number of COVID-19 CXRs incorrectly identified as normal and non-COVID, respectively.
In addition to the metrics stated above, the various classification networks were also compared in terms of the elapsed time per image, i.e. time taken by each network to classify an input image, represented in Eq. (20).
| (20) |
Where is the starting time for a network to classify an image, I and is the end time when the network has classified the same image, I.
4. Results and discussions
This section describes the performance of the lung segmentation models and classification networks’ performance on the plain X-ray images and segmented lung X-ray images.
4.1. Lung segmentation
Table 2 shows the overall accuracy and weighted IoU and Dice of the test-folds for the U-Net and modified U-Net model for lung segmentation.
Table 2.
Performance of segmentation networks.
| Network | Accuracy (A) | IoU | Dice |
|---|---|---|---|
| U-Net | 98.21 | 93.04 | 96.3 |
| Modified U-Net (Proposed) | 98.63 | 94.3 | 96.94 |
The modified U-Net model was used to segment the classification database (8851 normal, 6012 lung opacity, and 3616 COVID images), which was used for the classification of COVID, non-COVID lung opacity, and normal cases. It is important to see on a completely unseen image-set with a lung infection (COVID and non-COVID) and normal healthy images how well the trained segmentation model works. It can be seen from Fig. 7 that the modified U-Net model trained on the segmented CXR dataset can segment the lung areas of the X-ray images of the classification database very reliably. A qualitative evaluation was done to confirm that each X-ray image was segmented correctly.
Fig. 7.
CXR sample images (left), generated masks by the network (middle) and resulting segmented lung (right).
4.2. COVID-19 classification
As mentioned earlier, there are two different experiments (on plain and segmented lungs X-ray images) were conducted for the classification of COVID-19, non-COVID lung opacity and normal. The comparative performance of the best performing model for different enhancement techniques for classification between the three different classes of plain images is shown in Table 3 . Table 4 shows the comparative performance of the different CNN models in classifying COVID-19 using original and Gamma corrected X-ray images. It can be noted that the shallow CNN model was not performing better for original and enhanced images. The pre-trained models after re-training on CXR images can perform better than a shallow CNN model trained from scratch on CXR images. Moreover, it was observed that deep networks perform better than shallow networks.
Table 3.
Comparison of the best network for the COVID-19 classification using CXR images for different enhancement techniques.
| Different Enhancement | Model | Overall |
Weighted |
||||
|---|---|---|---|---|---|---|---|
| A | P | R | F | S | |||
| Original | InceptionV3 | 93.46 | 93.49 | 93.47 | 93.47 | 95.48 | 0.98 |
| Complement | DenseNet201 | 94.19 | 94.21 | 94.19 | 94.19 | 95.78 | 0.72 |
| Histeq | ChexNet | 94.34 | 94.17 | 94.14 | 94.14 | 95.98 | 0.62 |
| CLAHE | DenseNet201 | 94.08 | 94.09 | 94.08 | 94.07 | 95.77 | 0.75 |
| Gamma | ChexNet | 96.29 | 96.28 | 96.29 | 96.28 | 97.27 | 0.6 |
| BCET | DenseNet201 | 94.5 | 94.5 | 94.5 | 94.49 | 96.25 | 0.8 |
Table 4.
Comparison of different models for Covid-19 classification using original and Gamma corrected CXR images.
| Technique | Model | Overall |
Weighted |
|||
|---|---|---|---|---|---|---|
| A | P | R | F | S | ||
| Original | Resnet18 | 93.43 | 93.43 | 93.43 | 93.42 | 95.49 |
| Resnet50 | 93.01 | 93.12 | 93.02 | 93.04 | 95.5 | |
| Resnet101 | 93.01 | 93.04 | 93.01 | 93 | 95.11 | |
| ChexNet | 93.21 | 93.28 | 93.21 | 93.2 | 95.54 | |
| DenseNet201 | 92.7 | 92.78 | 92.7 | 92.72 | 95.35 | |
| InceptionV3 | 93.46 | 93.49 | 93.47 | 93.47 | 95.48 | |
| CNN | 91.3 | 91.28 | 91.2 | 91.28 | 91.4 | |
| Gamma correction | Resnet18 | 94.63 | 94.64 | 94.62 | 94.6 | 95.92 |
| Resnet50 | 94.56 | 94.58 | 94.56 | 94.53 | 95.81 | |
| Resnet101 | 94.93 | 94.94 | 94.93 | 94.92 | 96.2 | |
| ChexNet | 96.29 | 96.28 | 96.29 | 96.28 | 97.27 | |
| DenseNet201 | 95.05 | 95.06 | 95.05 | 95.05 | 96.55 | |
| InceptionV3 | 94.95 | 94.95 | 94.95 | 94.93 | 96.24 | |
| CNN | 92.2 | 92.18 | 92.2 | 92.18 | 92.34 | |
The best performing network has been reported for the different enhancement techniques in Table 3. It is evident that the Gamma correction image enhancement technique was the best performing technique not only for classification performance but also in terms of the time required to process a single image as shown in Table 3. It was further verified in Table 4 that the Gamma correction technique has consistently improved the performance for different networks over the original X-ray images. Finally, it was seen that ChexNet was the best performing networking on gamma-corrected CXR images for COVID-19 detection. The CheXNet showed an accuracy, precision, recall, F1-score, and specificity of 96.29%, 96.28%, 96.29%, 96.28%, and 96.27%, respectively. The superior performance of CheXNet in comparison to DenseNet201 is exhibiting that the deeper layer does not always perform better, rather CheXNet is the only DenseNet variant that was initially trained on chest X-ray images. Therefore, the CNN model initially trained on X-ray images can perform better on another CXR image problem. Similar performance was observed by the authors in their other COVID-19 detection experiments [47]. However, ResNet18, 50, and 101 showed increasingly better performance for the classification of images without segmentation. Fig. 8 (A) shows that the ROC curves for the best performing networks for the different image enhancement techniques. It is evident that the gamma correction is helping the network in discriminating different image classes better. The comparative performance of the different image enhancement techniques for the different CNN models is shown in Table 5 . Table 6 shows the comparative performance of the different CNN models for the classification of the three-class problem using original and Gamma corrected lung segmented X-ray images.
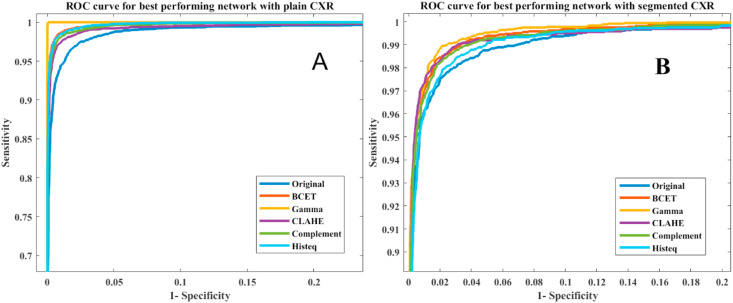
Fig. 8.
ROC curves for the best performing network in each image enhancement technique for plain CXR images (A) and segmented lung CXR images (B).
Table 5.
Comparison of the best networks for the COVID-19 classification using lung segmented CXR images for different image enhancement techniques.
| Enhancement techniques | Model | Overall |
Weighted |
||||
|---|---|---|---|---|---|---|---|
| A | P | R | F | S | |||
| Original | ChexNet | 93.22 | 93.22 | 93.22 | 93.22 | 95.51 | 0.65 |
| Complement | InceptionV3 | 93.46 | 93.49 | 93.47 | 93.47 | 95.48 | 1.2 |
| Histeq | DenseNet201 | 93.44 | 93.43 | 93.44 | 93.42 | 95.55 | 0.78 |
| CLAHE | ChexNet | 93.9 | 93.91 | 93.9 | 93.89 | 95.77 | 0.7 |
| Gamma | DenseNet201 | 95.11 | 94.55 | 94.56 | 94.53 | 95.59 | 0.72 |
| BCET | DenseNet201 | 94.12 | 94.17 | 94.14 | 94.14 | 95.98 | 0.85 |
Table 6.
Comparison of different models for COVID-19 classification using original and Gamma Corrected lung segmented CXR images.
| Technique | Model | Overall |
Weighted |
|||
|---|---|---|---|---|---|---|
| A | P | R | F | S | ||
| Original | Resnet18 | 92.23 | 92.23 | 92.22 | 92.21 | 94.66 |
| Resnet50 | 92.51 | 92.5 | 92.51 | 92.5 | 95.38 | |
| Resnet101 | 93.14 | 93.14 | 93.15 | 93.12 | 95.41 | |
| ChexNet | 93.22 | 93.22 | 93.22 | 93.22 | 95.51 | |
| DenseNet201 | 92.79 | 92.87 | 92.79 | 92.77 | 94.62 | |
| InceptionV3 | 92.43 | 92.44 | 92.43 | 92.4 | 94.81 | |
| CNN | 87.02 | 87.18 | 87.02 | 87.06 | 92.94 | |
| Gamma | Resnet18 | 93.31 | 93.3 | 93.31 | 93.3 | 95.82 |
| Resnet50 | 93.24 | 93.24 | 93.24 | 93.22 | 95.23 | |
| Resnet101 | 93.13 | 92.92 | 92.94 | 92.92 | 95.25 | |
| ChexNet | 93.63 | 93.67 | 93.63 | 93.59 | 95.34 | |
| DenseNet201 | 95.11 | 94.55 | 94.56 | 94.53 | 95.59 | |
| InceptionV3 | 93.92 | 93.92 | 93.92 | 93.9 | 95.7 | |
| CNN | 88.79 | 89.36 | 88.79 | 88.7 | 91.12 | |
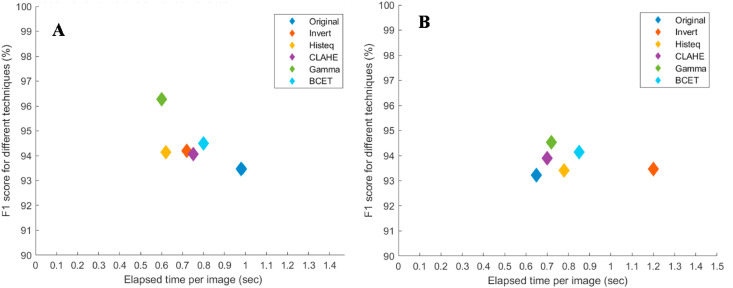
Table 5 shows that the Gamma correction technique was the best performing image enhancement technique for the segmented lung X-ray images. Fig. 8(B) shows the ROC curves for the different image enhancement techniques for the best performing network, where the image enhancement techniques had consistently improved the performance for different networks in comparison to the original X-ray images. It can also be seen that the Gamma correction technique is the best performing technique with a comparable elapsed time per image ( as shown in Fig. 9 . Finally, it was observed that DenseNet201 was the best performing network for the segmented lung CXR images in COVID-19 detection using gamma-corrected lungs. The network achieves accuracy, precision, recall, F1-score, and specificity of 95.11%, 94.55%, 94.56%, 94.53%, and 95.59%, respectively.
Fig. 9.
Comparison of F1 Score versus the elapsed time per image for the best performing network in each image enhancement technique for plain X-ray images (A) and segmented lung X-ray images (B).
4.3. Visualization using score-Cam
As mentioned earlier, it is important to see where the network is learning from the CXR images. It can be learning from relevant and non-relevant areas of the CXR images for classification, which can be verified using Score-CAM-based heat maps generated for original (non-segmented) and segmented CXR images.
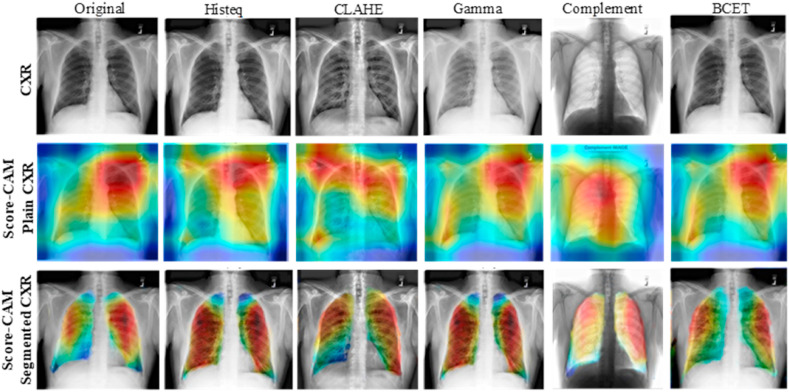
It is evident from the heat map of both the original and segmented lungs using the Score-CAM technique is that the decision-making by the CNN models in original CXR is not always coming from the lung areas (Fig. 10 ). The areas of CXR images that are mostly contributing to CNN's decisions are not always the lungs when the plain X-ray images are used for classification. It can be noted from Table 4, Table 6 that there is no performance improvement observed by the use of segmented lungs for this classification problem rather a small reduction in performance was observed. However, it evident from Fig. 10 that the plain X-ray images can produce a non-reliable performance of the CNN models, which is not desirable for such critical bio-medical application. On the other hand, a reliable classification of the diseases using chest X-ray images can be achieved using segmented CXR images, which is more useful for computer-aided diagnosis.
Fig. 10.
Score-CAM visualization of correctly classified COVID-19 X-ray images using the different enhancement techniques: CXR (top row), Score-CAM heat map on original CXR (middle row), and segmented lungs CXR (bottom row).
It was reported in the recent literature that the deep learning models can learn from the irrelevant areas and can take a decision from the irrelevant information [[48], [49], [50], [51]] and therefore even though high performing network's performance cannot be generalized in real-world applications. Therefore, the segmented lungs helped the CNN to decide on the main region of interest compared to the plain X-rays. In other words, the reliability of how the network is taking decisions for classification is important to increase the confidence of the end-user in the AI performance. As decisions for lung diseases should be done using the lung region in the CXR images. However, due to the limited availability of ground-truth lung masks in recent works, the results are reported on plain X-rays, which can partially or completely fail in a real-world application. This work has become possible by the authors due to the benchmark lung masks created by the authors with the help of a pool of radiologists. The benchmark COVQU dataset and corresponding lung masks will be made publicly available as a part of another scientific data article.
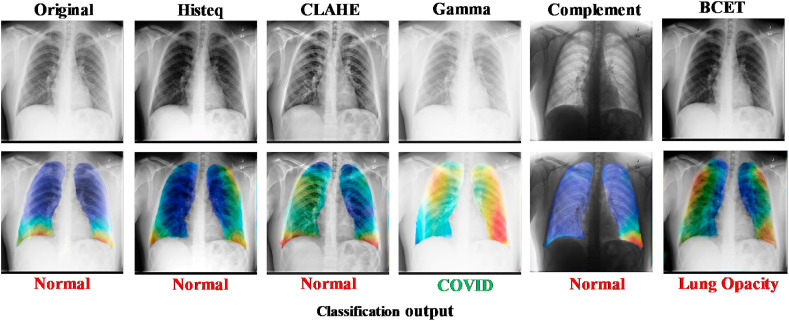
It is also interesting to see how the Gamma enhancement technique is outperforming other enhancement techniques for a sample case where almost all the techniques have misclassified COVID-19 X-rays to either normal or non-COVID lung opacity but the Gamma correction based image enhancement technique has correctly classified the image. It can also be seen from Fig. 11 that the Gamma correction technique on the segmented lungs is taking decisions from the region of interest, i.e. lungs, to correctly classify the image. In summary, it can be said that the performance reported in the recent literature in COVID-19 and other lung infection detection is comparable to the performance reported in this study (Table 7 ). However, three important aspects are reported in this study that is missing in other recent works. Firstly, in most of the works, a small number of COVID-19 CXR images were used for training and testing the CNN models. Secondly, a detailed comprehensive investigation of the image enhancement techniques on COVID-19 detection was not reported in the literature, and finally, no article has reported results on such large CXR images and corresponding ground truth lung masks to investigate the effect of reliable lung segmentation on COVID-19 detection. Therefore, the results reported in this study are not only comparable to the state-of-the-art results but also reliable and generalizable as it trained and validated on a large dataset.
Fig. 11.
Gamma Enhancement on segmented lungs correctly classifies COVID-19 x-ray while others miss classifying the sample X-ray images.
Table 7.
Comparison with the current state-of-art/relevant studies.
| Articles | Techniques | Dataset | Performance |
|---|---|---|---|
| Tsung et al. [52] | CNN (ResNet50) | 15478 chest X-ray images (473 COVID) | accuracy, sensitivity, and specifcity obtained is 93%, 90.1%, and 89.6% |
| Abbas et al. [7] | CNN (DeTraC) | 1768 chest X-ray images (949 COVID) | Accuracy-93.1% |
| Jain et al. [53] | CNN (Inception V3, Xception, and ResNet) | 6432 chest X-ray images (490 COVID) | Accuracy-96% and Recall-92% |
| Ohata et al. [54] | Transfer learning + machine learning method (DenseNet201 + MLP) | 388 chest X-ray images (194 COVID) | Acc: 95.641%, F1-score: 95.633%, FPR: 4.103% |
| Ioannis et al. [6] | CNN | 1427 chest X-ray images (224 COVID) | accuracy, sensitivity, and specifcity obtained is 96%, 96.66%, and 96.46% |
| Zulfaezal et al. [55] | CNN (ResNet101) | 5982 chest X-ray images (1765 COVID) | sensitivity, specificity, and accuracy of 77.3%, 71.8%, and 71.9%, respectively |
| Proposed study | Seven different deep CNN networks for classification and modified Unet network for segmentation | 18479 chest x-ray images (3616 COVID) | accuracy of 96.29%, the sensitivity of 97.28%, and the F1-score of 96.28%. In segmentation, Accuracy of 98.63%, and Dice score of 96.94% |
5. Conclusion
The immediate and accurate detection of highly infectious COVID-19 plays a vital role in preventing the spread of the virus. In this study, we have used CXR images since X-ray imaging is cheaper, easily accessible, and faster than the conventional methods commonly used, such as RT-PCR and CT. As an important contribution, the largest CXR dataset, COVQU, which consists of 3616 COVID-19, 6012 non-COVID lung opacity, and 8851 normal X-ray images, has been compiled and will be shared publicly as the benchmark dataset. Moreover, for the first time in the literature, we explored the effect of different image enhancement techniques in the automatic detection of COVID-19 from the CXR images using deep Convolutional Neural Networks. Furthermore, we proposed a novel variant of U-Net architecture for the lung segmentation from the X-ray images, which have outperformed the state-of-the-art U-Net model. This work explores the effect of different image enhancement techniques in the automatic detection of COVID-19 from the CXR images using deep Convolutional Neural Networks. In this study, six different deep learning pre-trained CNN models were trained on imageNet weights and a shallow CNN model was trained from scratch. The performance of seven CNN models for five different image enhancement techniques was evaluated for the classification of COVID-19, non-COVID lung infection, and normal CXR images. Our extensive experiments on image enhancement techniques show that a reliable COVID-19 diagnosis can be achieved with the accuracy, precision, and recall of 96.29%, 96.28%, and 96.28% without segmentation and 95.11%, 94.55%, and 94.56% with segmentation, respectively. ChexNet model with gamma enhancement technique provided the best performance without image segmentation whereas DenseNet201 with gamma enhancement technique outperforms for the segmented lungs. The detection accuracy of COVID-19 was found to be comparable to the state-of-the-art results reported in the recent literature even though the reported articles used a limited number of COVID-19 images. The Score-CAM visualization technique confirms the reliability of the trained models as the decision was made from the lung regions in the segmented CXR images. Thus, the results reaffirm the importance of accurate segmentation of lungs from the CXR images, which can assist machine learning models in diagnostic decisions. Considering the performance improvement observed using the proposed modified U-Net architecture over the standard U-Net model, the authors are planning to utilize different variants of U-Net (with residual connections, UNet+ [56], dense connections, UNet ++ [57]) along with the different encoder techniques that can help in improving the performance even further. This deep AI-based system can be useful as a fast screening tool that can save lives or prevent casualties, especially during the pandemic period when casualties can happen due to delay or miss-diagnosis.
Author contribution
Experiments were designed by TR and MEHC. Experiments were performed by TR, AK, AT, YQ and SBAK. Results were analyzed by MEHC, SK, MSK, MTI, SAM, and SMZ. The project is supervised by MEHC and SAM. All the authors were involved in the interpretation of data and paper writing and revision of the article.
Funding
COVID19 Emergency Response Grant #QUERG-CENG-2020-1 from Qatar University, Doha, Qatar provided the support for the work and the claims made herein are solely the responsibility of the authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Biographies

Tawsifur Rahman received his B.Sc (Eng.) degree from the department of Electrical and Electronic Engineering at the University of Chittagong, Bangladesh and M.Sc. degree from the department of Biomedical Physics and Technology (BPT) at the University of Dhaka, Bangladesh. He is currently working as a research student at the University of Dhaka, Bangladesh. His current research interest includes biomedical image and signal processing, machine learning, computer vision and data science. He has expertise on designing and developing Nerve stimulator for measuring conduction velocity in human body, developing electrocardiogram (ECG), electromyogram (EMG) circuit, developing Howland constant current source and Instrumentation amplifier to measure tetra polar bio-impedance, Detection of different stage of brain activity by analyzing various EEG wave. He has received ICT fellowship 2019–2020 from ICT ministry of Bangladesh for research entitled “Driver drowsiness detection from HRV and computer vision using machine learning”. He placed 3rd position in the 1st Bangladesh Electronics Olympiad at 2015.

Amith Khandakar (IEEE Senior Member) received the B.Sc. degree in electronics and telecommunication engineering from North South University, Bangladesh, and the master's degree in computing (networking concentration) from Qatar University, in 2014. He graduated as the Valedictorian (President Gold Medal Recipient) of North South University. He is currently the General Secretary of the IEEE Qatar Section and also the Qatar University IEEE Student Branch Coordinator and an Adviser (Faculty). He is also a certified Project Management Professional and the Cisco Certified Network Administrator. He was a Teaching Assistant and Lab Instructor for two years for courses, such as mobile and wireless communication systems, principle of digital communications, introduction to communication, calculus and analytical geometry, and Verilog HDL: modeling, simulation and synthesis. Simultaneously, he was a Lab Instructor for the following courses: programming course “C,” Verilog HDL, and general physics course. He has been with Qatar University, since 2010. After graduation, he was a Consultant in a reputed insurance company in Qatar and in a private company that is a sub-contractor to National Telecom Service Provider in Qatar.

Yazan Qiblawey (M′19) received the B.Sc in electrical engineering from Qatar University, Qatar in 2017 and M.Sc in electrical and electronics engineering from University of Nottingham, U.K in 2019. He is currently a Research Assistant at College of Engineering, Qatar University, Qatar. His publication has been selected for the top 5 papers in IEEE R8 Student Paper Contest in 2018 for the paper “Design and Implementation of Autonomous Surface Vehicle”. His research interests include signal and image processing, machine learning and fault detection.

ANAS M. TAHIR (Student Member, IEEE) received the B.S. degree in electrical engineering from Qatar University, Qatar, in 2018. He is currently pursuing the M.S. degree in electrical engineering with Qatar University, where he is currently working as a Research Assistant. His current research interests are machine learning and artificial intelligence application in biomedical engineering research field.

Serkan Kiranyaz (Senior Member, IEEE) is currently a Professor with Qatar University, Doha, Qatar. He published two books, five book chapters, more than 80 journal articles in high impact journals, and 100 papers in international conferences. He made contributions on evolutionary optimization, machine learning, bio-signal analysis, computer vision with applications to recognition, classification, and signal processing. He has coauthored the papers which have nominated or received the Best Paper Award in ICIP 2013, ICPR 2014, ICIP 2015, and IEEE TSP 2018. He had the most-popular articles in the years 2010 and 2016, and most-cited article, in 2018, in the IEEE Transactions on Biomedical Engineering. From 2010 to 2015, he authored the fourth most-cited article of Neural Networks journal. His theoretical contributions to advance the current state of the art in modeling and representation, targeting high long-term impact, while algorithmic, system level design and implementation issues target medium and long-term challenges for the next five to ten years. He is in particular aims at investigating scientific questions and inventing cutting edge solutions in personalized biomedicine which is in one of the most dynamic areas where science combines with technology to produce efficient signal and information processing systems. His research team has received the 2nd and 1st places in PhysioNet Grand Challenges 2016 and 2017, among 48 and 75 international teams, respectively. He received the Research Excellence Award and the Merit Award of Qatar University, in 2019.

Saad Bin Abul Kashem received his Ph.D. degree in Robotics and Mechatronics from Swinburne University of Technology (SUT), Melbourne, Australia, in 2013. He received his B.Sc. in Electrical and Electronic Engineering from East West University, Bangladesh, back in 2008. At present, he is working as a Robotics and Advanced Computer Skills Faculty at Qatar Armed Forces – Academic Bridge Program, Qatar Foundation in Qatar. He is also a Visiting Research Fellow in Electrical & Computer Engineering at Texas A&M University and a Visiting Assistant Professor at Presidency University. As a Lecturer in the Electrical Engineering department (Robotics and Mechatronics Engineering panel) at Swinburne University of Technology (October 1, 2014 to July 27, 2017), he had been convener, moderator, and instructor of different courses. He has over eleven years' experience in both industry and academia. Dr. Saad has already published 51 articles in the form of Book, Book Chapter, Journal and Conference Proceedings. He is a professional member of Institution of Engineering and Technology, UK (IET), Institute of Electrical and Electronics Engineers (IEEE), IEEE Robotics and Automation Society, and International Association of Engineers (IAENG). He is an editorial board member and reviewer of many international reputed Journals such as Journal of Electrical and Electronic Engineering (USA), IEEE Transactions on Control Systems Technology (USA), Vehicle System Dynamics, Taylor and Francis Ltd, (UK) etc. His research interests include vehicle dynamics, electric vehicle, renewable energy systems, intelligent and autonomous control, robotics, nonlinear control theory, and applications.

Mohammad Tariqul Islam (Senior Member, IEEE) is currently a Professor with the Department of Electrical, Electronic and Systems Engineering, Universiti Kebangsaan Malaysia (UKM), and a Visiting Professor with the Kyushu Institute of Technology, Japan. He is the author or coauthor of about 500 research journal articles, nearly 175 conference papers, and a few book chapters on various topics related to antennas, microwaves, and electromagnetic radiation analysis with 20 inventory patents filed. Thus far, his publications have been cited 5641 times and his H-index is 38 (Source: Scopus). His Google scholar citation is 8200 and H-index is 42. His research interests include communication antenna design, satellite antennas, and electromagnetic radiation analysis. He is a Chartered Professional Engineer (C.Eng.), a member of IET, U.K., and a Senior Member of IEICE, Japan. He was a recipient of the 2018 IEEE AP/MTT/EMC Excellent Award, the Publication Award from the Malaysian Space Agency, in 2014, 2013, 2010, and 2009, and the Best Paper Presentation Award from the International Symposium on Antennas and Propagation (ISAP 2012), Nagoya, Japan, in 2012, and IconSpace, in 2015. He received several international gold medal awards, including the Best Invention in Telecommunication Award, the Special Award from Vietnam for his research and innovation, and best researcher awards at UKM, in 2010 and 2011. He also won the Best Innovation Award, in 2011, and the Best Research Group in ICT Niche by UKM, in 2014. He was a recipient of more than 40 research grants from the Malaysian Ministry of Science, Technology and Innovation, the Ministry of Education, the UKM Research Grant, and international research grants from Japan and Saudi Arabia. He serves as a Guest Editor for Sensors Journal and an Associate Editor for IEEE Access and Electronics Letters (IET).

Somaya Al Maadeed(IEEE Senior Member) holds a PhD degree in computer science from Nottingham University. She supervised students through research projects related to computer vision and biomedical image applications. She is currently a professor at Computer Science Department, Qatar University. She is the Coordinator of the Computer Vision Research Group, Qatar University. She enjoys excellent collaboration with national and international institutions, and industry. She is a principal investigator of several funded research projects related to Medical Imaging. She published extensively in computer vision and pattern recognition and delivered workshops on teaching programming. In 2015, she was elected as the IEEE Chair of the Qatar Section. She filled IPs related to cancer detection from images and light.

Susu M. Zughaier, MSc, PhD is an Associate Professor of Microbiology and Immunology. Trained as a clinical microbiologist, earned a Diploma in Clinical Microbiology, University College London in 1991; MSc in Medical Microbiology in 1994 and PhD in Microbiology and Immunology from Cardiff University, UK in 1999. She did her postdoctoral training at Harvard Medical School in Boston, USA before joining Emory University in 2001. Dr. Zughaier was an Assistant Professor at the Department of Microbiology and Immunology, Emory University School of Medicine in Atlanta, USA before joining Qatar University College of Medicine in Fall 2017. Her research interests are focused on host-pathogen interactions and vaccine development. She investigates antibiotic resistance mechanisms and implements nanotechnology for rapid detection of bacterial infections. Her translational research is focused on vitamin D immune modulatory effects. To date, Dr. Zughaier published more than 50 scientific research papers and awarded two patents on her discoveries. Her H-index is 29 (Google Scholar) and listed among the top 2% of highly cited authors with more than 9000 citations. She is an active member of the American society of Microbiology (ASM) and serves as the Scientific Microbiology Councilor for the International Endotoxin and Innate Immunity Society. Dr Zughaier also serves on Women, Diversity and Equity Committee of the International Society of Leukocyte Biology. She is an expert in Meningococcal and Gonococcal nanovaccines and therefore served as consulted for Pfizer and the World Health Organization (WHO). In 2019 she joined Bacterial Vaccines Experts Network (BactiVac), University of Birmingham UK. She serves as Guest Editor, editorial board member and ad-hoc reviewer for multiple international journals.

Muhammad Salman Khan received the B.S. (Hons.), M.S., and Ph.D. degrees all in electrical engineering from the University of Engineering and Technology (UET), Peshawar, Pakistan, in 2007, the George Washington University, Washington, D.C., USA, in 2010, and Loughborough University, England, U.K., in 2013, respectively. He was a Postdoctoral Fellow within the Department of Electrical Engineering, Universidad de Chile, Santiago, Chile between 2013 and 2015. Since 2015 he is an Assistant Professor within the Department of Electrical Engineering, Jalozai campus, University of Engineering and Technology Peshawar, Pakistan, where he is also serving as the Head of Department since 2017. He is Principal Investigator of the Higher Education Commission Pakistan funded projects within Artificial Intelligence in Healthcare, Intelligent Information Processing Lab, National Center of Artificial Intelligence, UET Peshawar. In 2019 he was included as a member of the World Health Organization (WHO) Roster of Experts on Digital Health. His research interests include information processing, pattern recognition, machine learning and artificial intelligence.

Muhammad E. H. Chowdhury (Senior Member, IEEE) received the B.Sc. and M.Sc. degrees with record marks from the Department of Electrical and Electronics Engineering, University of Dhaka, Bangladesh, and the Ph.D. degree from the University of Nottingham, U.K., in 2014. He worked as a Postdoctoral Research Fellow and a Hermes Fellow at the Sir Peter Mansfield Imaging Centre, University of Nottingham. He is currently working as an Assistant Professor with the Department of Electrical Engineering, Qatar University. Before joining Qatar University, he worked in several universities of Bangladesh. He has two patents and published around 80 peer-reviewed journal articles, conference papers, and four book chapters. His current research interests include biomedical instrumentation, signal processing, wearable sensors, medical image analysis, machine learning, embedded system design, and simultaneous EEG/fMRI. He is also running several NPRP and UREP grants from QNRF and internal grants from Qatar University along with academic and government projects. He has been involved in EPSRC, ISIF, and EPSRC-ACC grants along with different national and international projects. He has worked as a Consultant for the projects entitled Driver Distraction Management Using Sensor Data Cloud (2013–14, Information Society Innovation Fund (ISIF) Asia). He is an Active Member of British Radiology, the Institute of Physics, ISMRM, and HBM. He received the ISIF Asia Community Choice Award 2013 for a project entitled Design and Development of Precision Agriculture Information System for Bangladesh. He has recently won the COVID-19 Dataset Award for his contribution to the fight against COVID-19. He is serving as an Associate Editor for IEEE Access and a Topic Editor and Review Editor for Frontiers in Neuroscience.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104319.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.S. A. Harmon, T. H. Sanford, S. Xu, E. B. Turkbey, H. Roth, Z. Xu, et al., "Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets," Nat. Commun., vol. 11 (1), p. 4080(2020/08/14 2020). [DOI] [PMC free article] [PubMed]
- 2.EIA U. US Energy Information Administration; Washington, DC: 2011. International Energy Outlook. [Google Scholar]
- 3.Yang Y., Yang M., Shen C., Wang F., Yuan J., Li J., et al. medRxiv; 2020. Evaluating the Accuracy of Different Respiratory Specimens in the Laboratory Diagnosis and Monitoring the Viral Shedding of 2019-nCoV Infections. [Google Scholar]
- 4.Yu M., Xu D., Lan L., Tu M., Liao R., Cai S., et al. Thin-section chest CT imaging of coronavirus disease 2019 pneumonia: comparison between patients with mild and severe disease. Radiology: Cardiothoracic Imaging. 2020;2(2) doi: 10.1148/ryct.2020200126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassantabar S., Stefano N., Ghanakota V., Ferrari A., Nicola G.N., Bruno R., et al. 2020. Coviddeep: Sars-Cov-2/covid-19 Test Based on Wearable Medical Sensors and Efficient Neural Networks. arXiv preprint arXiv:2007.10497. [Google Scholar]
- 6.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021;51:854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., et al. Proceedings of the AAAI Conference on Artificial Intelligence. 2019. Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison; pp. 590–597. [Google Scholar]
- 10.Khan A.I., Shah J.L., Bhat M.M. Computer Methods and Programs in Biomedicine. 2020. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images; p. 105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein E., Keidar D., Yaron D., Shachar Y., Blass A., Charbinsky L., et al. 2020. COVID-19 Classification of X-Ray Images Using Deep Neural Networks. arXiv preprint arXiv:2010.01362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury N.K., Rahman M., Rezoana N., Kabir M.A. 2020. ECOVNet: an Ensemble of Deep Convolutional Neural Networks Based on EfficientNet to Detect COVID-19 from Chest X-Rays. arXiv preprint arXiv:2009.11850. [Google Scholar]
- 13.P. Afshar, S. Heidarian, F. Naderkhani, A. Oikonomou, K. N. Plataniotis, and A. Mohammadi, "COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from X-ray images," Pattern Recogn. Lett., vol. 138 pp. 638-643(2020/10/01/2020). [DOI] [PMC free article] [PubMed]
- 14.Yamac M., Ahishali M., Degerli A., Kiranyaz S., Chowdhury M.E., Gabbouj M. 2020. Convolutional Sparse Support Estimator Based Covid-19 Recognition from X-Ray Images. arXiv preprint arXiv:2005.04014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahishali M., Degerli A., Yamac M., Kiranyaz S., Chowdhury M.E., Hameed K., et al. 2020. A Comparative Study on Early Detection of COVID-19 from Chest X-Ray Images. arXiv preprint arXiv:2006.05332. [Google Scholar]
- 16.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10(1):19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motamed S., Rogalla P., Khalvati F. 2020. RANDGAN: Randomized Generative Adversarial Network for Detection of COVID-19 in Chest X-Ray. arXiv preprint arXiv:2010.06418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aviles-Rivero A.I., Sellars P., Schönlieb C.-B., Papadakis N. 2020. GraphXCOVID: Explainable Deep Graph Diffusion Pseudo-labelling for Identifying COVID-19 on Chest X-Rays. arXiv preprint arXiv:2010.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Degerli A., Ahishali M., Yamac M., Kiranyaz S., Chowdhury M.E., Hameed K., et al. 2020. COVID-19 Infection Map Generation and Detection from Chest X-Ray Images. arXiv preprint arXiv:2009.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed S., Yap M.H., Tan M., Hasan M.K. medRxiv; 2020. Reconet: Multi-Level Preprocessing of Chest X-Rays for Covid-19 Detection Using Convolutional Neural Networks. [Google Scholar]
- 21.Christodoulidis S., Anthimopoulos M., Ebner L., Christe A., Mougiakakou S. Multisource transfer learning with convolutional neural networks for lung pattern analysis. IEEE J. Biomed, Health Informatics. 2016;21(1):76–84. doi: 10.1109/JBHI.2016.2636929. [DOI] [PubMed] [Google Scholar]
- 22.Yang H., Mei S., Song K., Tao B., Yin Z. Transfer-learning-based online Mura defect classification. IEEE Trans. Semicond. Manuf. 2017;31(1):116–123. [Google Scholar]
- 23.Akçay S., Kundegorski M.E., Devereux M., Breckon T.P. 2016 IEEE International Conference on Image Processing (ICIP) 2016. Transfer learning using convolutional neural networks for object classification within x-ray baggage security imagery; pp. 1057–1061. [Google Scholar]
- 24.Tajbakhsh N., Shin J.Y., Gurudu S.R., Hurst R.T., Kendall C.B., Gotway M.B., et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imag. 2016;35(5):1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 25.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009;22(10):1345–1359. [Google Scholar]
- 26.Wang H., Wang Z., Du M., Yang F., Zhang Z., Ding S., et al. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops. 2020. Score-CAM: score-weighted visual explanations for convolutional neural networks; pp. 24–25. [Google Scholar]
- 27.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 28.Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., et al. 2017. Chexnet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv preprint arXiv:1711.05225. [Google Scholar]
- 29.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 30.LeCun Y., Kavukcuoglu K., Farabet C. Proceedings of 2010 IEEE International Symposium on Circuits and Systems. 2010. Convolutional networks and applications in vision; pp. 253–256. [Google Scholar]
- 31.Veluchamy M., Subramani B. Image contrast and color enhancement using adaptive gamma correction and histogram equalization. Optik. 2019;183:329–337. [Google Scholar]
- 32.Zimmerman J.B., Pizer S.M., Staab E.V., Perry J.R., McCartney W., Brenton B.C. An evaluation of the effectiveness of adaptive histogram equalization for contrast enhancement. IEEE Trans. Med. Imag. 1988;7(4):304–312. doi: 10.1109/42.14513. [DOI] [PubMed] [Google Scholar]
- 33.Smilkov D., Thorat N., Kim B., Viégas F., Wattenberg M. 2017. Smoothgrad: Removing Noise by Adding Noise. arXiv preprint arXiv:1706.03825. [Google Scholar]
- 34.Selvaraju R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-CAM: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 35.Chattopadhay A., Sarkar A., Howlader P., Balasubramanian V.N. 2018 IEEE Winter Conference on Applications of Computer Vision. WACV); 2018. Grad-cam++: generalized gradient-based visual explanations for deep convolutional networks; pp. 839–847. [Google Scholar]
- 36.Rahman T., Khandakar A., Kadir M.A., Islam K.R., Islam K.F., Mazhar R., et al. Reliable tuberculosis detection using chest X-ray with deep learning, segmentation and visualization. IEEE Access. 2020;8:191586–191601. [Google Scholar]
- 37.kaggle. RSNA pneumonia detection challenge [Online]. Available: https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data. [Accessed on 09-June-2020].
- 38.BIMCV-COVID19 Datasets related to COVID19's pathology course. 2020. https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/#1590858128006-9e640421-6711 [Online]. Available:
- 39.covid-19-image-repository. 2020. https://github.com/ml-workgroup/covid-19-image-repository/tree/master/png [Online]. Available:
- 40.Chen R., Liang W., Jiang M., Guan W., Zhan C., Wang T., et al. Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 from a nationwide analysis in China. Chest. 2020;158(1):97–105. doi: 10.1016/j.chest.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weng Z., Chen Q., Li S., Li H., Zhang Q., Lu S., et al. 2020. ANDC: an Early Warning Score to Predict Mortality Risk for Patients with Coronavirus Disease 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., et al. MedRxiv; 2020. Neutrophil-to-lymphocyte Ratio Predicts Severe Illness Patients with 2019 Novel Coronavirus in the Early Stage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang I., Pranata R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): systematic review and meta-analysis. J. Intens. Care. 2020;8(1):1–10. doi: 10.1186/s40560-020-00453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.COVID-CXNet. 2020. https://github.com/armiro/COVID-CXNet [Online]. Available:
- 45.Mikołajczyk A., Grochowski M. 2018 International Interdisciplinary PhD Workshop (IIPhDW) 2018. Data augmentation for improving deep learning in image classification problem; pp. 117–122. [Google Scholar]
- 46.E. D. SCIENCE Overfitting in machine learning: what it is and how to prevent it. https://elitedatascience.com/overfitting-in-machine-learning [Online]. Available:
- 47.Chowdhury M.E.H., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 48.Schlemper J., Oktay O., Schaap M., Heinrich M., Kainz B., Glocker B., et al. Attention gated networks: learning to leverage salient regions in medical images. Med. Image Anal. 2019;53:197–207. doi: 10.1016/j.media.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahman T., Chowdhury M.E., Khandakar A., Islam K.R., Islam K.F., Mahbub Z.B., et al. Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest X-ray. Appl. Sci. 2020;10(9):3233. [Google Scholar]
- 50.Sengupta S., Basak S., Saikia P., Paul S., Tsalavoutis V., Atiah F., et al. A review of deep learning with special emphasis on architectures, applications and recent trends. Knowl. Base Syst. 2020:105596. [Google Scholar]
- 51.Hohman F., Kahng M., Pienta R., Chau D.H. Visual analytics in deep learning: an interrogative survey for the next frontiers. IEEE Trans. Visual. Comput. Graph. 2018;25(8):2674–2693. doi: 10.1109/TVCG.2018.2843369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin T.-C., Lee H.-C. Presented at the Proceedings of the 4th International Conference on Medical and Health Informatics. 2020. Covid-19 chest radiography images analysis based on integration of image preprocess, guided grad-CAM, machine learning and risk management. [Google Scholar]
- 53.Jain R., Gupta M., Taneja S., Hemanth D.J. Applied Intelligence; 2020/10/09 2020. Deep learning based detection and analysis of COVID-19 on chest X-ray images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohata E.F., Bezerra G.M., Chagas J.V. S.d., Neto A.V.L., Albuquerque A.B., Albuquerque V.H. C.d., et al. Automatic detection of COVID-19 infection using chest X-ray images through transfer learning. IEEE/CAA J. Autom. Sin. 2021;8(1):239–248. [Google Scholar]
- 55.M. Z. Che Azemin, R. Hassan, M. I. Mohd Tamrin, and M. A. Md Ali, "COVID-19 deep learning prediction model using publicly available radiologist-adjudicated chest X-ray images as training data: preliminary findings," Int. J. Biomed. Imag., vol. 2020 p. 8828855(2020/08/18 2020). [DOI] [PMC free article] [PubMed]
- 56.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 57.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. Unet++: a nested u-net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.