Abstract
Eosinophilic esophagitis (EoE) is an allergic inflammatory disease of the esophagus that occurs in both children and adults. Previous studies of affected tissue from pediatric cohorts have identified prominent signatures of eosinophilia and type 2 inflammation. However, the details of the immune response in adults with EoE are still being elucidated. To determine whether EoE in adults shares inflammatory profiles with those observed in children, we performed RNA-sequencing of paired human esophageal biopsies and blood samples from adults with EoE or gastroesophageal reflux disease (GERD). Unbiased analysis of differentially expressed genes in tissue revealed a strong interferon signature that was significantly enriched in EoE patients as compared to patients with GERD. Both type I and type II interferon responsive genes were upregulated in adult biopsies, but not in blood. A similar increase in expression of interferon gene sets was observed in pediatric EoE biopsies as compared to non-EoE samples, and in public pediatric and adult RNA-sequencing data. Finally, we found that human peripheral CD4+ T cells from children with EoE produce IFNγ upon activation with EoE-causal allergens. Together, this work identifies a conserved interferon signature in pediatric and adult EoE, highlighting a role for non-type 2 inflammatory networks in the disease process in humans.
Keywords: Human, Eosinophilic esophagitis, Interferon, T cells
Introduction
Eosinophilic esophagitis (EoE) is a chronic allergic inflammatory disease that is triggered by specific foods, and characterized by Th2 cell-mediated inflammation and esophageal eosinophilia (1). Affecting approximately 1 in 2500 (150,000) children and adults in the United States, EoE symptoms are debilitating and range from reflux, chest pain, and poor growth in children to esophageal stricture formation and food impaction in adolescents and adults (2). EoE is a chronic disease that affects both children and adults. However, the extent of pathophysiologic similarities and differences between pediatric and adult EoE are unknown.
EoE is an emerging disease, and our understanding of its pathophysiology is still evolving. The sine qua non of EoE is the presence of infiltrating eosinophils on esophageal biopsy and clinically there are several features that link EoE to an allergic pathophysiology including increased prevalence in individuals with a history of atopy (3), being triggered by exposure to food and aeroallergens, and therapeutic efforts focused on allergen avoidance and/or use of corticosteroids. These initial observations have led to an early recognition of the critical role for type 2 inflammatory networks in EoE immunopathology (4). Indeed, experimental evidence supporting type 2 inflammation in EoE includes more than two decades of work profiling the transcriptional, cellular, and inflammatory features of inflamed esophagus (5), as well as more recent efforts that have focused on understanding the contribution of systemic and local T cell responses (3, 6).
Despite the strong link to type 2 inflammation within the esophageal mucosa, therapeutic attempts to block allergic pathways have had limited success in treating EoE (7, 8). Simultaneously, there has been a recent recognition of a role for non-type 2 inflammatory mediators in EoE pathophysiology. For example, TGFβ is now recognized as playing a critical role in EoE-related esophageal remodeling (9-11). Further, there is evidence that cytokines and chemokines more often associated with type 1 inflammation (including IL-1 and IL-6) are elevated in the esophagus of individuals with EoE (12). Together, these observations suggest that although type 2 inflammatory networks represent an important subset of the relevant immune pathways activated during EoE, a deeper understanding of the mucosal inflammatory milieu is needed to advance diagnostic and therapeutic options for this disorder.
Here we utilize adult and pediatric patient cohorts to profile peripheral and esophageal EoE inflammatory networks in a minimally-biased manner. We observe conserved IFN gene signatures in the esophagus of adult and pediatric EoE patients. We further identify circulating T cells from pediatric EoE patients that produce IFN gamma (IFNγ) when stimulated with disease-causal allergens. Together, these results identify IFN-mediated inflammatory signaling as a feature of EoE immunopathology that is conserved between children and adults, and raise the possibility that circulating T cells are one source of IFN cytokines in EoE.
Materials and Methods
Adult Subjects.
Nine adult patients receiving endoscopy for suspected EoE were enrolled at the Gastroenterology Department at Virginia Mason Clinic. A diagnosis of EoE was confirmed in five patients by the presence of >15 eosinophils/hpf in biopsy sections. Four control patients underwent endoscopy for clinical indications but were found to have minimal or no eosinophil infiltrate in esophageal biopsies. None of the patients had received oral steroids before endoscopy, and all were on an open diet at the time of enrollment. Patient characteristics are summarized in Table I. Biopsies were collected in RNAlater (Thermo Fisher Scientific) and paired blood samples were collected in Tempus Blood RNA Tubes (Thermo Fisher) and stored at −80°C until use. The study was approved by the Institutional Review Board (IRB) at Benaroya Research Institute, protocol 7109-477.03. All subjects provided informed consent.
Table I.
Subject characteristics RNA expression studies
| Subject ID |
Diagnosis | Age | Sex | Eosinophils/hpf1, endoscopic appearance |
Medications | Biopsy- proven eliciting food trigger, if known |
Peanut IgE |
Milk IgE |
Soy IgE |
Wheat IgE |
Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adult tissue, blood | |||||||||||
| 1 | EoE | 75 | M | >80 proximal, >50 distal, edema and rings, EREFS2=3 | PPI3 | NA4 | <0.01 | <0.01 | <0.01 | ||
| 2 | EoE | 34 | F | 40-50, proximal and distal, edema, rings, exudate, furrows, stricture, EREFS=8 | Histamine-2 blocker, anti-histamine | peanut | 37 | 0.39 | 0.84 | 0.16 | History of IgE food allergy to peanut; allergic rhinitis |
| 3 | EoE | 51 | F | 120 proximal, 42 distal, edema, rings, exudate, EREFS=3 | Histamine-2 blocker, antibiotic | 0.17 | 0.16 | 0.21 | 0.27 | Asthma, allergic rhinitis, Eczema | |
| 4 | EoE | 32 | M | 25-50 proximal and distal, edema, rings, exudate, furrows, stricture, EREFS=6 | NA | 0.31 | <0.10 | <0.10 | Allergic rhinitis | ||
| 5 | EoE | 25 | M | 85 proximal, 122 distal, edema, rings, EREFS=3 | 4.84 | 0.14 | 1.12 | 4.04 | Allergic rhinitis, possible pollen related EoE (alder IgE 23.4) | ||
| 7 | Control | 39 | M | Negative | NA | <0.10 | <0.10 | <0.10 | Symptoms consistent with GERD5 | ||
| 8 | Control | 60 | F | Negative | PPI | NA | <0.10 | <0.10 | <0.10 | Symptoms consistent with GERD | |
| 9 | Control | 65 | F | Negative | PPI, anti-histamine | <0.01 | <0.10 | <0.10 | <0.10 | Symptoms consistent with GERD | |
| 10 | Control | 38 | M | 10-20, mid and distal | <0.01 | <0.10 | <0.10 | 0.24 | Symptoms consistent with GERD | ||
| Pediatric tissue | |||||||||||
| 11 | EoE | 16 | M | 25, gross furrowing | PPI | NA | SPT6(−) | SPT(−) | SPT(−) | History of food impaction | |
| 12 | EoE | 8 | F | 20 | PPI | NA | <0.01 | <0.01 | <0.01 | ||
| 13 | EoE | 12 | M | 50, reactive epithelial changes | NA | NA | NA | NA | |||
| 15 | EoE | 10 | M | 35, eosinophilic microabscesses and basal hyperplasia | PPI | peanut, tree nut | SPT(+) | SPT(−) | SPT(−) | NA | History of IgE food allergy to egg, had reincorporated into diet following food challenge |
| 16 | Control | 5 | M | Negative | NA | NA | NA | NA | |||
| 17 | Control | 5 | F | Negative | polyethylene glycol 3350 | NA | NA | NA | NA | ||
| 18 | Control | 6 | M | Negative | NA | NA | NA | NA | |||
| 19 | Control | 17 | M | Negative | NA | NA | NA | NA | |||
| 20 | Control | 16 | M | Negative | NA | NA | NA | NA | |||
| 21 | Control | 13 | F | Negative | PPI, dicyclomine | NA | NA | NA | NA | ||
HPF, high power field
EREFS score, edema, rings, exudate, furrows, and stricture, each scored on a scale of 0-2 and then summed across features
PPI, protein pump inhibitor
NA, not available
GERD, gastric esophageal reflux disease
SPT, skin prick test
Pediatric Subjects.
A total of 17 pediatric subjects with symptoms consistent with EoE and undergoing endoscopy were enrolled at Children’s Hospital of Philadelphia (CHOP). Esophageal biopsy and/or peripheral blood samples were obtained under CHOP IRB-approved protocols 08-005998 or 18-015524 with subject consent. All pediatric patients in the biopsy cohort were on an open diet at the time of biopsy. The results shown in Tables I and II represent their initial biopsy findings prior to therapy. For peripheral blood T cell analyses, EoE subjects with confirmed, milk-triggered EoE, or non-EoE controls, were recruited.
Table II.
Subject characteristics T cell studies
| Subject ID |
Diagnosis | Age | Sex | Eosinophils/hpf1, endoscopic appearance |
Medications | Biopsy- proven eliciting food trigger, if known |
Peanut IgE |
Milk IgE |
Soy IgE |
Wheat IgE |
Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pediatric blood | |||||||||||
| 22 | EoE | 11 | M | 18 | PPI2 | milk | SPT3(−) | SPT(−) | SPT(−) | SPT(−) | |
| 23 | EoE | 9 | F | up to 25 | PPI | milk, egg, soy, wheat, beef | NA4 | NA | NA | NA | Asthma, Allergic Rhinitis |
| 24 | EoE | 8 | M | 8 | PPI | milk | NA | NA | NA | NA | IgE food allergy, Allergic Rhinitis, Eczema, Adhesive allergy |
| 25 | EoE | 2 | M | 55 | PPI | milk | NA | NA | NA | NA | |
| 26 | EoE | 14 | M | 78 | PPI | milk | SPT(−) | SPT(−) | SPT(−) | SPT(−) | |
| 27 | EoE | 6 | M | 20 | PPI | milk, wheat, sesame, soy, tree nut, peanut | NA | NA | NA | NA | Allergic Rhinitis |
| 28 | EoE | 8 | M | 0-8 | PPI | milk, egg, tree nut | NA | NA | NA | NA | Adhesive allergy |
| 29 | EoE | 10 | M | Remission | PPI | milk | SPT(−) | SPT(+) | SPT(−) | SPT(−) | Consumes baked milk |
| 30 | EoE | 8 | F | Remission | PPI, Dupixent | milk | NA | NA | SPT(+) | SPT(−) | on Dupixent for eczema |
| 31 | Control | 18 | F | Negative | NA | NA | NA | NA | |||
| 32 | Control | 10 | F | Negative | NA | NA | NA | NA | Drug Allergy | ||
| 33 | Control | 12 | F | Negative | PPI | NA | NA | NA | NA | ||
| 34 | Control | 27 | F | NA | NA | NA | NA | NA | |||
| 35 | Control | 11 | F | 0-3 | PPI | NA | NA | NA | NA | ||
| 36 | Control | 12 | M | 1-2 | PPI | NA | NA | NA | NA | ||
| 37 | Control | 38 | F | NA | NA | NA | NA | NA | |||
| 38 | Control | 17 | F | Negative | PPI | NA | NA | NA | NA | Drug Allergy | |
| 39 | Control | 32 | F | NA | NA | NA | NA | NA | |||
HPF, high power field
PPI, protein pump inhibitor
SPT, skin prick test
NA, not available
RNA sequencing.
Total RNA was isolated from adult esophageal biopsies (1-2 per subject) by homogenizing tissue in QIAzol lysis reagent (Qiagen) followed by column purification using the miRNeasy kit with on column DNA digestion (Qiagen). Whole blood RNA was extracted using MagMax for Stabilized Blood Tubes RNA Isolation Kit, followed by globin reduction using GlobinClear Human (Thermo Fisher). Sequencing libraries were constructed from total RNA using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara) and NexteraXT DNA sample preparation kit (Illumina) to generate Illumina-compatible barcoded libraries. Dual-index, single-read sequencing of pooled libraries was carried out on a HiSeq2500 sequencer (Illumina) with 58-base reads, using HiSeq v4 Cluster and SBS kits (Illumina) with a target depth of 5 million reads per sample. Base calls were processed to FASTQs on BaseSpace (Illumina). Reads were processed using workflows managed on the Galaxy platform; reads were trimmed by 1 base at the 3’ end, and then trimmed from both ends until base calls had a minimum quality score of at least 30 (Galaxy FASTQ Trimmer tool v1.0.0) (13). FastqMcf (v1.1.2, https://benthamopen.com/ABSTRACT/TOBIOIJ-7-1) was used to remove any remaining adapter sequence. Reads were aligned using the TopHat aligner (v1.4.1) (14) with the GRCh38 reference genome and gene annotations from ensembl release 77. Gene counts were generated using HTSeq-count (v0.4.1) (15). Quality metrics were compiled from PICARD (v1.134, http://broadinstitute.github.io/picard/), FASTQC (v0.11.3, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), Samtools (v1.2) (16), and HTSeq-count (v0.4.1). RNA sequencing data from the adult biopsies and whole blood samples is deposited in the GEO Repository (GSE156651, https://www.ncbi.nlm.nih.gov/gds/).
NanoString gene expression analysis.
Total RNA was isolated from pediatric biopsies (Table I) and analyzed with a NanoString Human Immunology panel (NanoString Technologies, Seattle WA) consisting of 594 genes, including 15 internal reference genes. Immunology probe sets detect gene expression of a broad range of immune mediated molecules, including type I and type II IFNs and receptors. RNA was hybridized with unique barcoded probe sets overnight and digital images were processed with the nCounter digital analyzer to tabulate reporter probe counts. Gene expression was analyzed using the nSolver Analysis software (NanoString) by subtracting the geometric mean of the negative background probes, and normalization to the geometric mean of the housekeeping gene panel (ABCF1, EEF1G, G6PD, PPIA, RPL19, and TBP). Genes with expression below 13.19 reads across all subjects were excluded from analysis due to minimal expression.
Transcript analysis.
Aligned gene counts from RNA-sequencing were TMM normalized with the package edgeR (17). Median coefficient of variation coverage (<0.5) and percent alignment (>85%) was used to eliminate libraries with poor quality (no libraries met this criteria). Patient sex check was used to exclude sample swaps. Differential gene expression was modeled separately for tissue and blood samples by the LIMMA R package (18). A two-group comparison between adult EoE and non-EoE samples was performed. Differentially expressed genes were used to query STRING-db for interactions and visualized by Cytoscape (19). Network visualizations utilized genes differentially expressed at ≤0.01 false discovery rate (FDR) and genes connected to those with a cutoff of ≤0.05 FDR. Gene ontology enrichment analysis was performed using the GOana function in the LIMMA R package (20). Hallmark gene sets (refined by algorithmic and expert curation) were downloaded from MSigDb (19), and gene set enrichment was computed by the ROAST package (21). Heatmaps displaying enriched genes were filtered at an FDR of 0.05 and were subjected to unsupervised hierarchical clustering. Gene expression data from the Sherrill et al. study (22) was downloaded from the NCBI GEO database with accession number GSE58640 and gene set enrichment performed as described. Gene set enrichment analysis for the Hallmark IFNα and IFNγ response gene sets was performed in the same manner on the normalized pediatric EoE and non-EoE biopsy gene counts generated with the NanoString Human Immunology panel, without TMM normalization.
Quantitative PCR.
Tissue and globin reduced whole blood RNA (100ng) from adult EoE and GERD patients was reverse transcribed into cDNA using SuperScriptIV reverse transcriptase (Life Technologies). Gene expression was detected using Taqman gene expression assays specific for the STAT1, STAT2, ADAR, IFIT1, GBP2, and RPL36A genes (Applied Biosystems) on a 7500 Fast Real-Time PCR System (Applied Biosystems). Samples were run in triplicate in a single experiment. Mean Ct values were normalized to expression of the housekeeping gene (RPL36A), and then expressed relative to a technical positive control sample included on each plate.
Antigen-activated T cell responses.
The study protocol was approved by the IRB of CHOP, and informed consent was obtained from all participating donors (n=18, 9 per experimental arm). The donor characteristics are summarized in Table II. 5-15 mL of whole blood was obtained by venipuncture using sodium heparin collection tubes. In order to achieve processing within 5 hours, tubes were transported at room temperature by foot to the Hill Laboratory at CHOP. PBMCs were isolated by Ficoll gradient using standard protocols and following the manufacturer’s instructions.
The median PBMC number obtained was 4.29E7 (range 1.16E7-9.42E7), and PBMCs were washed, counted, and cultured at 1 million cells/mL at 37°C, 5% CO2 in a 96 well round-bottom plate (Corning) in CTS OpTimizer T cell expansion serum-free media (Gibco) in the presence or absence of 0.625 μg/well of Tetanus Toxoid, or 6.25 μg/well each of five commonly allergenic milk proteins (α-lactalbumin, β-lactoglobulin, and α/β/κ-casein) cleaned of endotoxin. After six days, control and experimental T cells were treated with PMA/ionomycin/brefeldin A for a period of four hours at 37°C, 5% CO2 to amplify the endogenous cytokine signal, washed, and surface stained with LiveDead Fixable Dye (Invitrogen, 1%), CD8 (Clone SK1, BV510, BioLegend, 1:100 dilution), CD19 (Clone HIB19, CD19, BioLegend, 5:100 dilution), CD3 (Clone SK7, APC780, BioLegend, 1:100 dilution), and CD4 (Clone OKT4, BV605, BioLegend, 2.5:100 dilution). Cells were washed followed by fixation and permeabilization (Invitrogen), and intracellular staining for IFNγ (Clone 4S.B3, PE/Cy7, BioLegend, 1:100 dilution) using standard protocols.
Samples were acquired using the BD FACSFortessa, using DIVA. Photo Multiplier Tube (PMT) voltages were set using CS&T beads. Compensation was calculated using single stained cells and/or compensation beads (eBioscience). A representative data set demonstrating the gating strategy is shown in Supplemental Figure 3, and the results were audited. The median background reactivity for IFNγ observed in this study was 0.75% of the parent gate (range 0.01-2.69) in CD4+ T cells. Culture supernatant IFNγ levels were assayed by high-sensitivity Human T cell Cytokine Luminex assay (Millipore). Levels below the detectable limit were considered zero.
Statistics.
Transcript analysis: A two-group comparison was performed to identify differentially expressed genes between adult EoE and non-EoE samples, with p-values computed by LIMMA via a modified t-statistic (2-sided). Gene ontology enrichment analysis was performed with the GOana function which utilized a Wallenius non-central hypergeometric test, based on an extension of the hypergeometric distribution. Gene set enrichment computed by the ROAST package used two-sided rotation tests, analogous to permutation tests, to test whether the genes within a gene set all change in the same direction. P-values resulting from all three analyses were adjusted for multiple comparisons using the Benjamini-Hochberg procedure. Quantitative PCR: A Mann Whitney U test was performed to determine significant differences in gene expression between EoE and non-EoE samples. Flow cytometry and Luminex: Significant outliers were identified and excluded by Grubbs' test, and significant differences between the experimental groups were determined by one-sided student’s t test.
Study approval.
All clinical investigation was conducted according to Declaration of Helsinki principles, following approval from the appropriate IRBs as detailed above. Written informed consent was obtained prior to patient participation in these studies.
Results
Tissue, but not blood RNA sequencing profiles distinguish adult EoE subjects from non-EoE patients.
Esophageal biopsies and paired blood samples were obtained from five adult patients with EoE and four control patients with normal endoscopies and histology who had symptoms consistent with GERD (Table I). Four of the EoE patients had a history of allergy and/or asthma, including one individual who developed EoE symptoms in response to peanut oral immunotherapy. No other EoE patients had a history of food-induced allergy. Non-EoE patients had no known history of allergy. None of the subjects had received oral glucocorticoids before endoscopy.
Bulk RNA-sequencing data from esophageal biopsy and whole blood RNA revealed a clear distinction in principal component (PC) 1 between the biopsy and blood transcript profiles, while PC2 distinguished profiles from EoE and non-EoE biopsies (Fig. 1A). In contrast, there was no distinction of transcript profiles from unstimulated whole blood samples between EoE and non-EoE patients.
Figure 1. Differential gene expression differentiates sample origin and disease type in adults.
Bulk RNA-sequencing was performed on whole esophageal biopsies or paired whole blood from five adult EoE patients and four non-EoE control patients with symptoms consistent with GERD. A) Principal component (PC) analysis distinguished tissue from blood samples (PC1), and EoE from non-EoE (PC2) in tissue samples but not blood. B) Differential gene expression analysis. A two-group comparison was performed to identify differentially expressed genes between adult EoE vs. non-EoE samples, with p-values computed by LIMMA via a modified t-statistic and adjusted using the Benjamini-Hochberg procedure. 677 genes were significantly differentially expressed in biopsies from adult EoE (n=476, red symbols) or non-EoE subjects (n=201, gold symbols) at FDR ≤0.05. C) Heatmap of differentially expressed genes subjected to unsupervised hierarchical clustering. Differential gene expression distinguished EoE from non-EoE biopsy samples. Scaled gene expression is shown as blue (low) to red (high). D) Enrichment of Gene Ontology (GO) gene sets in the differentially expressed genes in (B) was performed with the GOanna function in LIMMA using a Wallenius non-central hypergeometric test. Gene sets were filtered at FDR≤0.05 and plotted in a volcano plot as gene sets significantly enriched in EoE (red n=200) versus gene sets significantly enriched in non-EoE (gold n=19). The top 15 gene sets are annotated.
Differential gene expression analysis was performed to identify transcript signatures differentiating adult EoE and GERD samples. We found a total of 677 differentially expressed genes between EoE and GERD biopsies (false discovery rate (FDR) ≤0.05), 476 of which were upregulated in EoE and 201 upregulated in GERD (Fig. 1B, 1C and Supplemental Table I). There were no significantly differentially expressed genes between EoE and GERD whole blood samples (FDR≥0.071, data not shown), consistent with no difference in PC2 in Fig. 1A. Thus, an EoE-specific transcript signature is not detectable in unfractionated unstimulated blood samples in adult patients.
An IFN signature is enriched in esophageal tissue from adult and pediatric EoE patients.
Differentially expressed genes upregulated in EoE versus GERD biopsies were significantly enriched for gene sets (defined by the Gene Ontology consortium (20)) involving processes in the immune system, response to stress, defense response, response to stimulus, and innate immune response (FDR≤6.19e-8, Fig. 1D, Supplemental Table II). Likewise, gene set enrichment analysis (GSEA) identified genes involved in inflammation, signaling, and angiogenesis (FDR=0.0125, Supplemental Table III). STRING-db network interactions showed several nodes of interacting genes (Fig. 2A). Two nodes included genes related to type 2 immune responses, including CCR3 (eotaxin receptor involved in eosinophil recruitment), PTGDR2 (prostaglandin D2 receptor 2, the canonical Th2 cell marker CRTH2), and ALOX15 (arachidonate 15-lipoxigenase, which is highly expressed in Th2A cells (23)). As expected, there was expression of genes from a human eosinophil gene set (24) in adult EoE tissue, as well as genes from a previously identified EoE gene signature (Supplemental Fig. 1) (25). In contrast, genes enriched in non-EoE biopsies were enriched for gene sets related to keratinization, skin development, and epidermal cell differentiation (FDR≤7.02e-11, Fig. 1D, Supplemental Table 2)
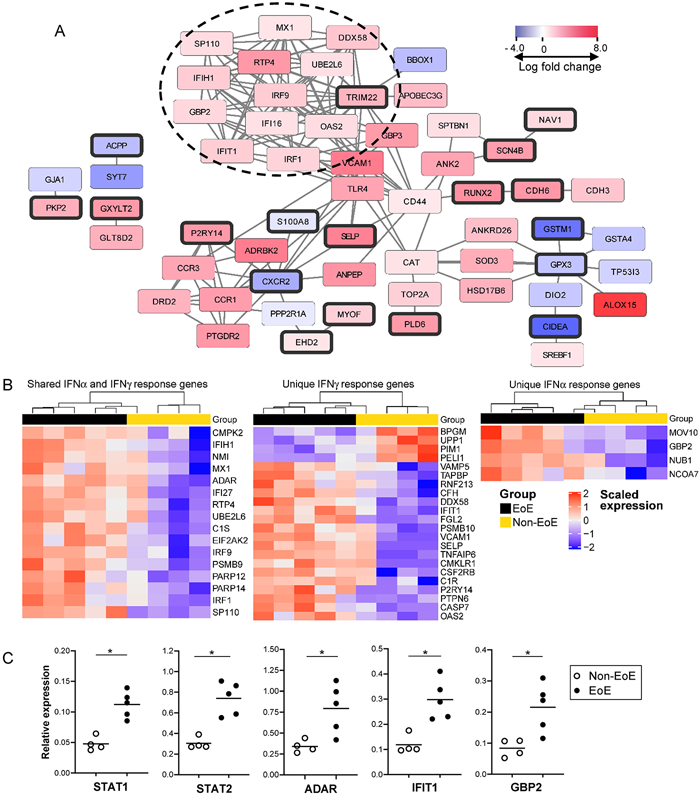
Figure 2. An IFN response signature is enriched in adult EoE tissue.
A) Differentially expressed genes (n=677) between five EoE and four non-EoE biopsy samples were analyzed for protein interactions using STRING-db. The network visualization shows genes at FDR ≤0.01 (bold borders) and connected genes at FDR ≤0.05 (no borders). The color of the rectangle indicates the log fold change in gene expression in EoE vs. non-EoE, and ranges from blue (up in non-EoE) to red (up in EoE). A prominent node of IFN related genes is indicated with the dotted circle. B) Gene set enrichment analysis of the Hallmark IFNα and IFNγ gene sets was computed on the biopsy RNA-sequencing data with the ROAST package, using two-sided rotation tests as described in the methods section. P-values were adjusted using the Benjamini-Hochberg procedure. Heatmaps display enriched genes filtered at FDR ≤0.05 and were subjected to unsupervised hierarchical clustering. Scaled gene expression is shown as blue (low) to red (high). Shown are heatmaps for genes unique to each IFN gene set and a common set of IFN response genes. C) Upregulation of select IFN response genes in EoE tissue compared with non-EoE biopsies was confirmed by quantitative PCR. Samples were run in triplicate in a single experiment. Group differences in gene expression were significant at p<0.05 indicated by an asterisk using a Mann Whitney U test.
Surprisingly, GSEA also revealed significant enrichment of IFN alpha (IFNα, FDR=0.0125) and IFNγ response genes (FDR=0.0141) in the differentially expressed genes from the EoE and non-EoE biopsies (Supplemental Table III). Further, there was a prominent node of upregulated IFN response genes related to TRIM22 in the STRING-db interaction network (Fig. 2A). To explore this further, we examined expression of genes in the Hallmark IFNα (n=97) and IFNγ (n=200) response gene sets in the biopsy RNA-sequencing data. Expression of 42 IFN response genes was enriched in the biopsies (FDR≤0.05), including 4 genes unique to IFNα, 22 unique to IFNγ, and 16 genes shared between the IFNα and γ response. Unsupervised clustering of the enriched genes differentiated EoE from non-EoE biopsies with the exception of one non-EoE subject who had a partial IFN signature (Fig. 2B). Heatmaps of the expression of the full set of IFNα and IFNγ response genes are shown in Supplemental Fig. 2.
We confirmed increased expression of select interferon response genes in EoE vs. non-EoE biopsies by quantitative PCR (Fig. 2C). Both STAT1 and STAT2 transcript levels were significantly higher in EoE biopsies consistent with IFN-mediated signaling; increased transcript levels were also detected for ADAR (induced by both IFNα and IFNγ), IFIT1 (IFNγ-induced), and GBP2 (IFNα-induced) in EoE vs. non-EoE tissue. There was no significant difference in expression of these genes in whole blood RNA from the same subjects (data not shown). These results highlight that IFN responses mediated by both IFNα and IFNγ are detected in EoE tissue. Likewise, there was no significant difference in gene enrichment between IFNα and IFNγ responsive genes in EoE biopsies relative to the total number of genes in the respective gene sets, indicating that both type I and type II IFNs are expressed in esophageal tissue from adult EoE patients.
To determine if upregulation of IFN response genes is a common finding in EoE, we validated our findings using a second publicly available tissue RNA-sequencing data set from a mixed age cohort of 10 EoE and six non-EoE patients (GSE58640)(22). The demographics of this cohort are listed in Supplemental Table IV, and included primarily pediatric patients in addition to three adults. Consistent with our data, GSEA demonstrated differential expression of IFNα and IFNγ response genes (FDR≤0.05) in the EoE biopsies compared to non-EoE biopsies (Fig. 3). We also noted that a small set of IFN response genes unique to IFNγ or shared between IFNγ and IFNα were enriched in the non-EoE biopsies in both the original and the validation dataset (Supplemental Fig. 2, Fig. 3), however the majority of IFN response genes were upregulated in the EoE tissue.
Figure 3. Enrichment of IFN signatures in tissue from a validation set of mixed age EoE patients.
Publically available RNA-sequencing data from pediatric (n=7) and adult (n=3) EoE biopsies and pediatric non-EoE (n=6) biopsy samples was downloaded from NCBI GEO database (accession number GSE58640) (22). Gene set enrichment analysis using the Hallmark interferon-α (IFNα) (n=97) and IFNγ (n=200) gene sets was computed with the ROAST package, using two-sided rotation tests as described in the methods section. P-values were adjusted using the Benjamini-Hochberg procedure. Heatmaps display enriched genes filtered at FDR ≤0.05 and were subjected to unsupervised hierarchical clustering. Scaled gene expression is shown as blue (low) to red (high). Shown are heatmaps for genes unique to each IFN gene set and a common set of IFN response genes.
An additional cohort of four pediatric EoE patients and six controls were studied using a more targeted approach (Table I). Non-EoE control pediatric patients were undergoing diagnostic evaluation for gastrointestinal symptoms and reported symptoms warranting esophagogastroduodenoscopy, but had no prior diagnosis of EoE and had biopsies which excluded histopathologic abnormalities. Half of these control patients had a prior history of atopic dermatitis, IgE-mediated food allergy, allergic rhinitis, or asthma. In contrast, all of the EoE patients in this pediatric cohort had a prior personal history of atopy at the time of presentation with EoE. One of these individuals had a history of developing EoE subsequent to reintroduction of an IgE-mediated food allergen into the diet (egg). None of these patients had received oral or topical glucocorticoids before endoscopy (Table I). Total RNA from whole biopsies was analyzed using the NanoString Human Immunology Panel consisting of 579 immune related genes. GSEA showed upregulation of gene expression for IFNγ response-specific genes, as well as shared IFNα and IFNγ response genes in the pediatric EoE tissue samples compared with the control samples (FDR<0.1, Fig. 4). Two of the six non-EoE patients expressed higher levels of a small subset of the enriched IFN response genes. Together, this work indicates that an IFN response signature characterizes EoE tissue from patients over a range of ages.
Figure 4. IFN gene expression is upregulated in pediatric EoE biopsies.
Gene expression in esophageal biopsies from four pediatric EoE patients (active) and six non-EoE patients (control) was analyzed using the NanoString Human Immunology panel. Gene set enrichment analysis was performed on the normalized read counts using the Hallmark IFNα (n=97) and IFNγ (n=200) response gene sets, of which 63 genes were included on the NanoString panel. Gene set enrichment was computed with the ROAST package, using two-sided rotation tests as described in the methods section. P-values were adjusted using the Benjamini-Hochberg procedure. Heatmaps display enriched genes filtered at an FDR <0.1 and were subjected to unsupervised hierarchical clustering. Scaled log gene expression is shown as blue (low) to red (high). Genes unique to the IFNγ response gene set are labeled light blue in the sidebar; genes in common between the IFNα and IFNγ gene sets are labeled pink. No genes unique to the IFNα gene set were enriched at an FDR<0.1.
Circulating T cells from EoE patients produce interferon upon allergen stimulation.
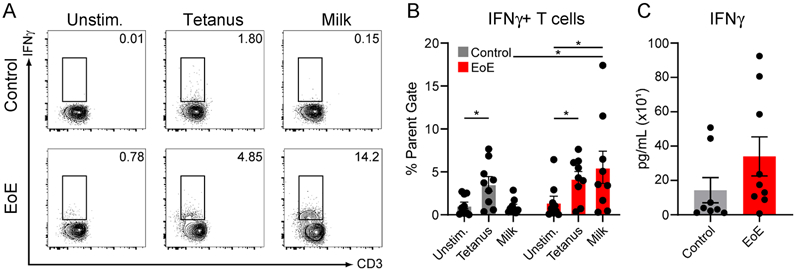
Previous studies have detected IFNγ expression in CD8+ T cells in EoE biopsy tissues, suggesting circulating T cells may be one potential source of IFN in esophageal tissue (26). In this regard, it is notable that we did not detect an IFN signature in whole blood. However, we hypothesized that circulating T cells may not express IFN when not actively responding to food antigens. To test this, we isolated PBMCs from the blood of patients with milk-triggered EoE or non-EoE controls (Table II), and cultured them in vitro for 6 days in the presence or absence of five allergenic milk proteins (α-lactalbumin, β-lactoglobulin, and α/β/κ-casein). We then assayed CD4+ T cell IFNγ production by intracellular flow cytometry (Supplemental Fig. 3). Compared with stimulated T cells isolated from controls, or unstimulated EoE T cells, EoE CD4+ T cells stimulated with purified milk proteins expressed significant amounts of IFNγ by intracellular cytokine staining (Fig. 5A, 5B). As compared with control PBMCs, there was a trend towards increased IFNγ levels in culture superannuates of milk-stimulated EoE PBMCs (Fig. 5C). We did not see a statistically significant increase in IFNγ expression in CD8+ T cells from EoE patients stimulated with milk peptides (not shown). Together, these results indicate that antigen-activated CD4 T cells may be a source of IFNγ in EoE patients.
Figure 5. IFNγ is produced by circulating, milk-activated pediatric EoE T cells.
A) Flow-cytometric analysis of CD4+ T cells from peripheral blood of non-EoE (control) or milk-allergic EoE (EoE) pediatric subjects. PBMCs were cultured in vitro in the absence (Unstimulated; Unstim.) or presence of tetanus toxoid or milk proteins (α-lactalbumin, β-lactoglobulin, and α/β/κ-casein) for 6 days. Gated on Live, CD8−, CD19−, CD4+. Numbers indicate percentage of parent gate. B) IFNγ production by peripheral CD4+ T cells across multiple non-EoE or milk-allergic EoE subjects. C) IFNγ amounts in culture supernatants of peripheral CD4+ T cells from non-EoE or milk-allergic EoE subjects cultured in vitro in the presence of milk proteins. N=8-9 subjects per experimental arm; *P≤0.05.
Discussion
Although it is known that the mucosal inflammation in EoE is heterogenous, study thus far has focused on the contribution of type 2 inflammation to the disease process. Further, studies to date have predominantly been performed in children leaving fundamental questions as to the extent to which adult EoE is a pathologic continuation of pediatric disease. Here, we characterized the inflammatory response in pediatric and adult EoE using transcriptional profiling of esophageal biopsies in an effort to identify inflammatory characteristics that are conserved across age-groups. Unbiased analysis of adult biopsies identified 677 differentially expressed genes compared with those from non-EoE patients. While genes related to type 2 immunity were also upregulated in EoE tissue, we demonstrate there is a strong type I and type II IFN response gene expression signature that is conserved in both pediatric and adult populations. One strength of our analyses is that we replicate this finding in three cohorts: an adult biopsy tissue RNA-seq discovery cohort, a second mixed-age biopsy tissue RNA-seq EoE and control cohort, and using a digital multiplex gene analysis of biopsy tissue gene expression in a pediatric cohort. Notably, we did not observe significantly differentially expressed genes in unstimulated, unfractionated adult whole blood. This could be due to a technological limit of detection, or the requirement for antigen activation of circulating, food-specific T cells. Consistent with the latter hypothesis, we found that peripheral CD4+ T cells from children with EoE produce IFNγ upon antigen activation. Together, these data reveal a conserved IFN signature in esophageal tissue of adults and children with EoE, that is also reflected in circulating antigen-activated T cells.
Previous profiling studies of affected tissue in EoE patients have identified signatures of eosinophilia, inflammation, and immune activation (22, 27). These studies led to the development of a molecular diagnostic gene expression panel that can distinguish EoE tissue from non-EoE biopsies in both pediatric and adult cohorts (25, 28). We found similar gene sets were enriched in adult EoE tissue, and observed several network clusters that included type 2 immune response genes. Accordingly, there was enrichment of eosinophil related genes and differential expression of the above gene expression panel, consistent with type 2 mediated disease in our adult cohort.
Aspects of an IFN response have been previously detected in EoE, though not thoroughly studied. For example, prior studies have found upregulated IFNγ expression in inflamed EoE biopsy tissue (29), and increased IFNγ has been detected in the supernatant of cultured EoE biopsy tissue using ELISA (30). Our study adds to these observations by comprehensively describing differential expression of both IFNα and IFNγ response genes in esophageal tissue from both adult and pediatric EoE patients in a minimally-biased effort across three independent patient cohorts and datasets. There was no preferential enrichment of IFNα or IFNγ response genes relative to the total numbers of genes in each Hallmark gene set and we confirmed increased expression of genes unique to the IFNα or IFNγ response by quantitative PCR, indicating that both type I and type II IFNs contribute to the IFN signature in EoE. The magnitude of the IFN signature differed amongst EoE patients, and a small number of IFN response genes were preferentially expressed in non-EoE patients, perhaps reflecting general effects of tissue damage. This finding may be related to the use of biopsies from patients with GERD as non-EoE controls for our adult cohort, rather than biopsies from population based controls with normal esophageal tissue. Notably, the EoE-associated gene C11orf30/EMSY is a repressor of IFN response gene expression, providing a plausible link to an increased IFN gene signature in EoE (31). One strength of our study is that our data derive from multiple medical centers and from patients across varying age groups, yet seem to consistently point toward enrichment of type I and type II IFN response genes in EoE biopsy tissue.
Bulk RNAseq of esophageal biopsies does not allow us to distinguish the cells producing IFN and responding to IFN in our study. However, we demonstrate that peripheral blood CD4+ cells stimulated with milk antigens produce IFNγ, raising the possibility that circulating T cells partly contribute to known IFNγ-producing T cells in the inflamed EoE mucosa (25). Indeed, Wen and colleagues have published single cell RNA-sequencing data of tissue infiltrating T cells and detected IFNγ RNA in over 84% of T cells in EoE biopsies, although there was no difference in the level of IFNγ expression between cells from active EoE versus controls (26). Sayej and colleagues examined lymphocytes from EoE biopsy tissue using flow cytometry and found significant elevation in TNFα and IFNγ production from the CD3+CD8+ T cell population but not from the CD3+CD4+ T cell population (32). We also detected enrichment of several genes unique to the IFNα response in two of the datasets analyzed in our study, although data regarding the cellular source of IFNα in EoE is lacking. A report that eosinophils in EoE tissue form extracellular traps that can elicit a type I IFN response from plasmacytoid dendritic cells (pDCs) suggests an additional plausible source of IFN (33). In support of this, we analyzed immune cell infiltrates in additional banked biopsies from a subset of the adult EoE and GERD patients in this study by flow cytometry and detected pDCs in EoE tissue but not GERD biopsies (data not shown). Regardless, additional studies in mice and humans are needed to clarify the cellular participants in the IFN response in EoE.
Does IFN play a role in the pathology of EoE? This cross-sectional study does not allow us to definitively determine if the IFN response gene signature correlates with disease course. Our data correlate an IFN gene response signature with the presence of EoE mucosal disease. However, additional prospective studies of EoE patients before and after remission are required to establish a causal link between IFN and pathophysiologic features in EoE. From a mechanistic standpoint, IFNγ has well known effects on HLA gene expression, which can enhance antigen presentation of allergens in esophageal tissue in EoE (34). IFNγ also potentiates release of the IL-1 cytokine family member IL-33 from esophageal epithelial cells, which can then act as a ligand for ST2 on innate lymphoid cells type 2 cells, Th2 cells, mast cells, and basophils, thereby contributing to the type 2 immune response in the tissue (35). IFNγ is known to enhance aspects of human eosinophil effector functions including superoxide anion generation, degranulation, adhesion, expression of multiple signaling and effector molecules (36), while both IFNγ and IFNα-induced necrosis may contribute to tissue destruction in the esophagus (37). Finally, there is mounting interest in the role of IgG4 in EoE immunopathology (38-40). This is of particular relevance as IFNα signaling has been shown to contribute to IgG4 production in other disease contexts (41). Future studies should focus on the mechanistic relationship between IFN and IgG4 in the context of EoE.
In summary, we show for the first time a conserved IFN gene expression signature in EoE biopsy tissue that is present in both pediatric and adult EoE patients. We conclude that IFN responsive genes are an additional feature within the biopsy gene expression signature in EoE patients. This study extends the current knowledge of the mucosal inflammatory response in EoE, and specifically points to type I and type II IFN responses as a form of non-Th2 mediated inflammation that may be relevant to EoE immunopathology.
Supplementary Material
Key points.
An IFN gene signature is present in the inflamed mucosa of individuals with EoE
This IFN signature is conserved between adults and children
IFNγ is produced by circulating EoE T cells activated with EoE-causal allergens
Acknowledgements
The authors acknowledge the Genomics Core laboratory at Benaroya Research Institute for the RNA-sequencing of adult biopsies and blood samples, Dr. Mario Rosasco for transcript alignment and processing, as well as Drs. Amanda Muir and Alain Benitez from the Children’s Hospital of Philadelphia Department of Gastroenterology for providing pediatric biopsy tissue for these studies. We also acknowledge Dr. Anne Hocking at Benaroya Research Institute for manuscript preparation.
Grant Support
This work was supported by a supplement to grant 1 R01 AI124220-01A (Ziegler), K08AI48456 (Ruffner), and K08DK116668 (Hill) from the National Institutes of Health, the Children’s Hospital of Philadelphia Food Allergy Frontier Grant (Spergel), and by a Hope Research Grant from the American Partnership for Eosinophilic Disorders and the American Academy of Allergy, Asthma & Immunology (Hill). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- CHOP
Children’s Hospital of Pennsylvania
- EoE
eosinophilic esophagitis
- FDR
false discovery rate
- GERD
gastroesophageal reflux disease
- GSEA
gene set enrichment analysis
- IRB
institutional review board
- PC
principal component
Footnotes
Disclosures
The authors have no conflicts of interests to report.
Data availability:
RNA-sequencing data from the adult biopsy and blood samples is publically available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156651).
References
- 1.Simon D, Cianferoni A, Spergel JM, Aceves S, Holbreich M, Venter C, Rothenberg ME, Terreehorst I, Muraro A, Lucendo AJ, Schoepfer A, Straumann A, and Simon HU. 2016. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy 71: 611–620. [DOI] [PubMed] [Google Scholar]
- 2.Dellon ES, Liacouras CA, Molina-Infante J, Furuta GT, Spergel JM, Zevit N, Spechler SJ, Attwood SE, Straumann A, Aceves SS, Alexander JA, Atkins D, Arva NC, Blanchard C, Bonis PA, Book WM, Capocelli KE, Chehade M, Cheng E, Collins MH, Davis CM, Dias JA, Di Lorenzo C, Dohil R, Dupont C, Falk GW, Ferreira CT, Fox A, Gonsalves NP, Gupta SK, Katzka DA, Kinoshita Y, Menard-Katcher C, Kodroff E, Metz DC, Miehlke S, Muir AB, Mukkada VA, Murch S, Nurko S, Ohtsuka Y, Orel R, Papadopoulou A, Peterson KA, Philpott H, Putnam PE, Richter JE, Rosen R, Rothenberg ME, Schoepfer A, Scott MM, Shah N, Sheikh J, Souza RF, Strobel MJ, Talley NJ, Vaezi MF, Vandenplas Y, Vieira MC, Walker MM, Wechsler JB, Wershil BK, Wen T, Yang GY, Hirano I, and Bredenoord AJ. 2018. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference. Gastroenterology 155: 1022–1033 e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capucilli P, and Hill DA. 2019. Allergic Comorbidity in Eosinophilic Esophagitis: Mechanistic Relevance and Clinical Implications. Clin Rev Allergy Immunol 57: 111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wechsler JB, and Bryce PJ. 2014. Allergic mechanisms in eosinophilic esophagitis. Gastroenterol Clin North Am 43: 281–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell JM, Paul M, and Rothenberg ME. 2017. Novel immunologic mechanisms in eosinophilic esophagitis. Curr Opin Immunol 48: 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cianferoni A, Ruffner MA, Guzek R, Guan S, Brown-Whitehorn T, Muir A, and Spergel JM. 2018. Elevated expression of activated TH2 cells and milk-specific TH2 cells in milk-induced eosinophilic esophagitis. Ann Allergy Asthma Immunol 120: 177–183 e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothenberg ME 2015. Molecular, genetic, and cellular bases for treating eosinophilic esophagitis. Gastroenterology 148: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wechsler JB, and Hirano I. 2018. Biological therapies for eosinophilic gastrointestinal diseases. J Allergy Clin Immunol 142: 24–31 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawson R, Yang T, Newbury RO, Aquino M, Doshi A, Bell B, Broide DH, Dohil R, Kurten R, and Aceves SS. 2016. TGF-beta1-induced PAI-1 contributes to a profibrotic network in patients with eosinophilic esophagitis. J Allergy Clin Immunol 138: 791–800 e794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen N, Fernando SD, Biette KA, Hammer JA, Capocelli KE, Kitzenberg DA, Glover LE, Colgan SP, Furuta GT, and Masterson JC. 2018. TGF-beta1 alters esophageal epithelial barrier function by attenuation of claudin-7 in eosinophilic esophagitis. Mucosal Immunol 11: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawson R, Anilkumar A, Newbury RO, Bafna V, Aquino M, Palmquist J, Hoffman HM, Mueller JL, Dohil R, Broide DH, and Aceves SS. 2015. The TGFbeta1 Promoter SNP C-509T and Food Sensitization Promote Esophageal Remodeling in Pediatric Eosinophilic Esophagitis. PLoS One 10: e0144651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, Alexander ES, Butz BK, Jameson SC, Kaul A, Franciosi JP, Kushner JP, Putnam PE, Abonia JP, and Rothenberg ME. 2011. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol 127: 208–217, 217 e201-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, and Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46: W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C, Pachter L, and Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anders S, Pyl PT, and Huber W. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome S Project Data Processing. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, and Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young MD, Wakefield MJ, Smyth GK, and Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, and Smyth GK. 2010. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26: 2176–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherrill JD, Kiran KC, Blanchard C, Stucke EM, Kemme KA, Collins MH, Abonia JP, Putnam PE, Mukkada VA, Kaul A, Kocoshis SA, Kushner JP, Plassard AJ, Karns RA, Dexheimer PJ, Aronow BJ, and Rothenberg ME. 2014. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun 15: 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wambre E, Bajzik V, DeLong JH, O'Brien K, Nguyen QA, Speake C, Gersuk VH, DeBerg HA, Whalen E, Ni C, Farrington M, Jeong D, Robinson D, Linsley PS, Vickery BP, and Kwok WW. 2017. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pages F, Speicher MR, Trajanoski Z, and Galon J. 2013. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39: 782–795. [DOI] [PubMed] [Google Scholar]
- 25.Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, Franciosi JP, Garza JM, Kaul A, King EC, Collins MH, Kushner JP, and Rothenberg ME. 2013. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology 145: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ, Putnam P, Mukkada V, Foote H, Rehn K, Darko S, Douek D, and Rothenberg ME. 2019. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest 129: 2014–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, Jameson SC, Kirby C, Konikoff MR, Collins MH, Cohen MB, Akers R, Hogan SP, Assa'ad AH, Putnam PE, Aronow BJ, and Rothenberg ME. 2006. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest 116: 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dellon ES, Veerappan R, Selitsky SR, Parker JS, Higgins LL, Beitia R, Genta RM, and Lash RH. 2017. A Gene Expression Panel is Accurate for Diagnosis and Monitoring Treatment of Eosinophilic Esophagitis in Adults. Clin Transl Gastroenterol 8: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta SK, Fitzgerald JF, Kondratyuk T, and HogenEsch H. 2006. Cytokine expression in normal and inflamed esophageal mucosa: a study into the pathogenesis of allergic eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 42: 22–26. [DOI] [PubMed] [Google Scholar]
- 30.Mulder DJ, Pooni A, Mak N, Hurlbut DJ, Basta S, and Justinich CJ. 2011. Antigen presentation and MHC class II expression by human esophageal epithelial cells: role in eosinophilic esophagitis. Am J Pathol 178: 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ezell SA, Polytarchou C, Hatziapostolou M, Guo A, Sanidas I, Bihani T, Comb MJ, Sourvinos G, and Tsichlis PN. 2012. The protein kinase Akt1 regulates the interferon response through phosphorylation of the transcriptional repressor EMSY. Proc Natl Acad Sci U S A 109: E613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sayej WN, Menoret A, Maharjan AS, Fernandez M, Wang Z, Balarezo F, Hyams JS, Sylvester FA, and Vella AT. 2016. Characterizing the inflammatory response in esophageal mucosal biopsies in children with eosinophilic esophagitis. Clin Transl Immunology 5: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon D, Radonjic-Hosli S, Straumann A, Yousefi S, and Simon HU. 2015. Active eosinophilic esophagitis is characterized by epithelial barrier defects and eosinophil extracellular trap formation. Allergy 70: 443–452. [DOI] [PubMed] [Google Scholar]
- 34.Hokland M, Basse P, Justesen J, and Hokland P. 1988. IFN-induced modulation of histocompatibility antigens on human cells. Background, mechanisms and perspectives. Cancer Metastasis Rev 7: 193–207. [DOI] [PubMed] [Google Scholar]
- 35.Shan J, Oshima T, Wu L, Fukui H, Watari J, and Miwa H. 2016. Interferon gamma-Induced Nuclear Interleukin-33 Potentiates the Release of Esophageal Epithelial Derived Cytokines. PLoS One 11: e0151701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi T, Kimura H, Kurabayashi M, Kozawa K, and Kato M. 2008. Interferon-gamma enhances human eosinophil effector functions induced by granulocyte-macrophage colony-stimulating factor or interleukin-5. Immunol Lett 118: 88–95. [DOI] [PubMed] [Google Scholar]
- 37.Thapa RJ, Nogusa S, Chen P, Maki JL, Lerro A, Andrake M, Rall GF, Degterev A, and Balachandran S. 2013. Interferon-induced RIP1/RIP3-mediated necrosis requires PKR and is licensed by FADD and caspases. Proc Natl Acad Sci U S A 110: E3109–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright BL, Kulis M, Guo R, Orgel KA, Wolf WA, Burks AW, Vickery BP, and Dellon ES. 2016. Food-specific IgG4 is associated with eosinophilic esophagitis. J Allergy Clin Immunol 138: 1190–1192 e1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuyler AJ, Wilson JM, Tripathi A, Commins SP, Ogbogu PU, Kruzsewski PG, Barnes BH, McGowan EC, Workman LJ, Lidholm J, Rifas-Shiman SL, Oken E, Gold DR, Platts-Mills TAE, and Erwin EA. 2018. Specific IgG4 antibodies to cow's milk proteins in pediatric patients with eosinophilic esophagitis. J Allergy Clin Immunol 142: 139–148 e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGowan EC, Platts-Mills TAE, and Wilson JM. 2019. Food allergy, eosinophilic esophagitis, and the enigma of IgG4. Ann Allergy Asthma Immunol 122: 563–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bozzalla Cassione E, and Stone JH. 2017. IgG4-related disease. Curr Opin Rheumatol 29: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data from the adult biopsy and blood samples is publically available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156651).