Abstract
The goal of this study was to determine the expression and distribution of the host restriction factors (RFs) TRIM5α and TRIM11 in non-human primate (NHP) neural retina tissue and the human Muller cell line MIO-M1. In addition, experiments were performed to determine the effect of TRIM5α and TRIM11 knockdown on FIVGFP transduction of MIO-M1 cells with the goal of devising strategies to increase the efficiency of lentiviral (LV) gene delivery vectors. Immunofluorescence (IF) studies indicated that TRIM5α and TRIM11 were localized predominantly in nuclei within the outer nuclear layer (ONL) and inner nuclear layer (INL) of NHP retina tissue. Double label IF indicated that TRIM5α and TRIM11 were localized to some of the retinal Muller cell nuclei. MIO-M1 cells expressed TRIM5α predominantly in the nucleus and TRIM11 primarily in the cytosol. FIVGFP transduction efficiency was significantly increased, at 4 and 7 days post transduction, in TRIM5α and TRIM11 knockdown clones (KD) compared to WT MIO-M1 cells. In addition, pretreatment with the proteasome inhibitor MG132 increased the transduction efficiency of FIVGFP in WT MIO-M1 cells. The nuclear translocation of NF-κB (p65), at 72 hours post FIVGFP transduction, was enhanced in TRIM5α and TRIM11 KD clones. The expression of TRIM5α and TRIM11 in macaque neural retina tissue and MIO-M1 cells indicate the presence of these RFs in NHP retina and human Muller cells. Our data indicate that even partial knockdown of TRIM5α or TRIM11, or a short proteasome inhibitor pretreatment, can increase the transduction efficiency of a LV vector.
Keywords: Retina, Muller cells, TRIM5α, TRIM11, Gene therapy, Viral vectors, Proteasome Inhibitor
1.0. Introduction:
The retina is an ideal target organ for gene therapy for several reasons including the compartmentalization of intraocular tissues, accessibility for microsurgical delivery of viral vectors, and immune privilege due to the intraocular environment (Kumaran et al., 2018). Gene therapy holds great promise for the treatment of inherited retinal dystrophies (IRD), a group of clinically and genetically heterogeneous degenerative disorders (Bennett 2017,Kumaran et al., 2018,Ludwig et al., 2019,Ong et al., 2019,Trapani and Auricchio 2018,Ziccardi et al., 2019). Adeno-associated virus (AAV) has emerged as the vector of choice for many gene delivery studies, although its limited transgene capacity of approximately 4.8 kilobases (kb) may require dual vector strategies (Bennett 2017,Hori et al., 2019,Kumaran et al., 2018,Trapani et al., 2014). Recently, the FDA approved the first gene therapy for an IRD using an AAV Vector (Georgiadis et al., 2016). For larger transgenes, lentiviral vectors (LVs), including non-integrating variants, can be utilized (Annoni et al., 2019,Balaggan and Ali 2012). The desire to avoid HIV-based vectors has led to the use of non-primate vectors including equine infectious anemia virus (EIAV) and feline immunodeficiency virus (FIV) for gene delivery (Balaggan et al., 2006,Cavalieri et al., 2018,Saenz et al., 2012). To date, sub-retinal administration has achieved high efficiency lentiviral transduction only in the retina pigment epithelium (RPE) (Balaggan and Ali 2012,Ziccardi et al., 2019), but EIAV may also transduce photoreceptor cells (Balaggan et al., 2006).
The efficacy of LV gene therapy can be influenced by factors including innate immune responses and cellular restriction factors (RFs), which block key steps in the viral life cycle, often in a species–specific manner (Annoni et al., 2019,Borsotti et al., 2016,Jia et al., 2015,Kajaste-Rudnitski and Naldini 2015,van Tol et al., 2017). RFs including APOBEC3G, tripartite motif (TRIM) proteins, tetherin, SAMHD1, and Mx2 can disrupt LV replication in mammalian cells (Borsotti et al., 2016,Kajaste-Rudnitski and Naldini 2015,Uchil et al., 2008,Yuan et al., 2016). The effect of these restriction factors must be considered when designing vectors for viral gene delivery, regardless of the intended cellular target.
The TRIM protein family is a group of type I interferon-inducible E3 ligases which regulate a variety of cellular functions including cell cycle progression, signal transduction, and transcription (van Gent et al., 2018). A large number of the approximately 80 human TRIM proteins are induced after viral infection or by interferon stimulation as part of the host antiviral response (Koepke et al., 2020). The antiviral activities of TRIM proteins include direct targeting of viral components, regulation of the innate immune response, and modulation of autophagy-mediated defenses (Koepke et al., 2020). A minimum of seven TRIM proteins, including TRIM5α, TRIM11, TRIM19, TRIM22, TRIM33, TRIM34 and TRIM37 can restrict LV replication (Koepke et al., 2020,van Tol et al., 2017).
The best studied of the TRIM proteins, TRIM5α, binds to and destabilizes the viral capsid via K63-linked ubiquitination (Imam et al., 2019), leading to premature disassembly and a reduction in reverse transcription (Stremlau et al., 2004). TRIM5α also plays a role in the innate immune response by promoting AP-1 and NF-κB signaling, through K63-ubiquitination of the TAK1 kinase complex, which leads to the transcription of inflammatory cytokines (Grutter and Luban 2012,Hage and Rajsbaum 2019,Imam et al., 2019,Lascano et al., 2016,Pertel et al., 2011). Another TRIM protein, TRIM11, interacts with the incoming LV capsid to accelerate uncoating and block reverse transcription, as well as inhibiting the release of viral particles from cells (Koepke et al., 2020,Yuan et al., 2016).
The majority of TRIM proteins are expressed ubiquitously, but their subcellular localization can vary (Reymond et al., 2001). In HeLa and U2OS cells, TRIM5α was located in cytoplasmic bodies while TRIM11 was distributed more evenly throughout the nucleus and cytoplasm (Reymond et al., 2001,Wu et al., 2006). TRIM function may be linked to intracellular distribution, as alteration of TRIM5α localization by the proteasome inhibitor MG132 disrupted targeting of the HIV-1 reverse transcription complex to the proteasome (Danielson et al., 2012,Wu et al., 2006). Interestingly, MG132 treatment increased the transduction efficiency of FIVGFP in human trabecular meshwork cells and monkey organ-cultured anterior segments, indicating a role for the proteasome in retroviral restriction in these cells (Aktas et al., 2018). Targeted LNEIE mutations in the HIV-1 capsid protein were able to overcome rhesus TRIM5α-mediated capsid degradation, which resulted in higher transduction efficiency in both rhesus and human primary T cells (He et al., 2017). These experiments indicate that reducing host cell TRIM restriction can significantly improve the efficiency of LV gene delivery vectors.
In this study we determined that the restriction factors TRIM5α and TRIM11 are expressed in NHP retina Muller cell nuclei. TRIM5α was predominantly expressed in MIO-M1 cell nuclei, while TRIM11 was mainly expressed in the cytosol of this cell type. We demonstrated that knockdown of TRIM5α or TRIM11 in MIO-M1 cells, or pretreatment with a proteasome inhibitor, significantly increased the transduction efficiency of the LV vector FIVGFP.
2.0. Methods:
2.1. Macaque retina tissue
Eyes from euthanized rhesus macaques (Macaca mulatta) were obtained as they became available from the Wisconsin National Primate Research Center of the University of Wisconsin-Madison. Animals were free of infectious agents at the time of sacrifice and no animals were deliberately sacrificed for these studies. Macaque eyes were kept on ice and dissected within one hour of sacrifice. Posterior eye cups were incubated with phosphate-buffered saline (PBS)/1 mM EDTA for 30 minutes at 37°C to loosen neural retina tissue and separate the retina from RPE cells. Neural retina tissue was rinsed in PBS before proceeding with further studies. All experiments adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
2.2. Cell Culture
The human Muller cell line MIO-M1 (Limb et al., 2002) and the human retinal pigment epithelial cell line ARPE-19 (ATCC CRL-2302) were maintained in DMEM/F12 (Cellgro/Mediatech, Manassas, VA, 10–090-CV)/10% FBS supplemented with a 1:100 dilution of L-glutamine-penicillin-streptomycin (SIGMA, St. Louis, MO, G1146) at 37°C in 5% CO2. HEK293 TN cells were maintained in DMEM /10% FBS supplemented with a 1:100 dilution of L-glutamine-penicillin-streptomycin at 37°C in 5% CO2.
2.3. CRISPR knockdown
MIO-M1 cells were transfected with equal amounts of TRIM5α CRISPR/Cas9 KO Plasmid (h) and TRIM5α HDR Plasmid (h) (Santa Cruz Biotechnology (SCBT), Dallas, TX, sc-409348, sc-409348-HDR) or TRIM11 CRISPR/Cas9 KO Plasmid (h) and TRIM11 HDR Plasmid (h) (SCBT, sc-408290, sc-408290-HDR) following the Lipofectamine 3000 reagent protocol (Invitrogen, Carlsbad, CA, L300000). Cells were maintained in DMEM/F12/10% FBS with a 1:100 dilution of L-glutamine-penicillin-streptomycin. Puromycin (SCBT, sc-108071A, 1 μg/ml) was added to the culture media at 3 days post transduction to select for transduced cells. Puromycin resistant cells were cloned by limiting dilution. Equal microgram amounts of wildtype (WT) Muller and TRIM5α, or WT Muller and TRIM11 KD lysates, were screened for protein knockdown by immunoblotting with TRIM5α or TRIM11 antibodies. Quantification of knockdown was performed on ECL images from three separate immunoblots utilizing ImageJ ((http://rsb.info.nih.gov/ij/) and the mean percent WT expression was graphed.
2.4. shRNA knockdown
ARPE-19 cells were transduced with TRIM5 shRNA lentiviral particles (Santa Cruz Biotechnology (SCBT), sc-61718-V) or TRIM11 shRNA lentiviral particles (SCBT, sc-76734-V) at an MOI of 2 in the presence of 10 mg/ml Polybrene (SCBT, sc-134220). Cells were maintained in DMEM/F12/10% FBS with the addition of Primocin (InvivoGen, San Diego, CA, ant-pm-1, 1:500). Puromycin (SCBT, sc-108071A 1, μg/ml) was added to the culture media at 3 days post transduction to select for transduced cells. Puromycin resistant cells were cloned by limiting dilution and screened for protein knockdown by immunoblotting with TRIM5α or TRIM11 antibodies.
2.5. Immunoblotting
Lysates were prepared from macaque neural retina tissue immediately post-sacrifice (Gerhardinger et al., 2001). MIO-M1 cell lysates were prepared in Laemmli’s sample buffer (BIORAD, Hercules, CA, 1610737) with added protease inhibitor (SIGMA P8340, 1:100) and phosphatase inhibitor (MilliporeSigma, Burlington, MA, 524633, 1:50) cocktails. Protein concentrations were determined by Pierce 660nm assay (Thermo Scientific, Rockford, IL, 2260). Lysates were electrophoresed on 4–15% Mini-PROTEAN TGX precast gels (BIORAD, 4568084S) with BenchMark Pre-stained Protein Standard (Invitrogen, Carlsbad, CA 10748–010) as a molecular weight marker (MWM). Proteins were electrophoretically transferred to nitrocellulose prior to blocking with 5% non-fat dry milk in Genius Buffer I (100mM maleic acid, 150mM NaCl, pH 7.5) containing 0.3% v/v Tween-20. The primary antibodies were diluted, as indicated, in 5% non-fat dry milk in Genius Buffer I, and incubated ON at 4°C followed by washing in Genius Buffer I containing 0.3% v/v Tween-20. HRP-conjugated secondary antibodies were diluted 1:5000 and applied for 1 hour at RT. After washing, the blots were developed using WesternSure PREMIUM chemilluminescent Substrate (LI-COR, Lincoln, NE, 926–95000).
The following primary antibodies were used for immunoblotting: rabbit anti-TRIM5α (Leinco Technologies, Fenton, MO, T331, 1:500), mouse anti-TRIM5 (SCBT, sc-373864), and rabbit anti-TRIM11 (Abcam, Cambridge, MA, ab111694, 1:1000). The secondary antibodies used for immunoblotting were mouse anti-rabbit HRP (SCBT, sc-2357, 1:5000) and goat anti-mouse HRP (Novus Biologicals, Centennial, CO, NBP1–75144, 1:5000).
2.6. Immunofluorescence
Neural retina tissue obtained from euthanized macaques was rinsed in PBS and fixed in 10% neutral buffered formalin (Thermo Fisher, Kalamazoo, MI, 305–510) for 24 hours before paraffin embedding and sectioning. Tissue sections were de-paraffinized and antigen retrieval was performed via heat treatment with pH 6 citrate buffer (10mM citric acid/ 0.05% Tween, pH 6, 20 minutes 92°C). Slides were blocked with 5% normal donkey serum (NDS) (ImmunoReagents Inc., Raleigh, NC, SP-072–020113) before incubation with the primary antibody diluted in 2.5% NDS/PBS overnight (ON) at 4°C.
MIO-M1 cells were plated on poly-L-lysine (SIGMA, P4707) coated chamber slides (Millipore, PEZGS0896). Cells were fixed in 4% paraformaldehyde/PBS and permeabilized for 10 minutes in 0.1%TritonX-100/PBS. Slides were then blocked with 5% NDS before incubation with the primary antibody diluted in 2.5% NDS/PBS for 1 hour at RT.
The secondary antibodies, diluted 1:400 in PBS containing 2.5% NDS, were applied for 1 hour at RT. Slides were stained with 1 μg/ml Hoechst (Invitrogen, 33342) to visualize nuclei before mounting with Immu-mount (Thermo Scientific, 9990402). Fluorescence images (40x objective) were taken with a Zeiss Axiocam 702 mono camera on a Zeiss Axio Imager.Z2 microscope with Zeiss ZEN 2.3 pro software (Zeiss Microimaging, Oberkochen, Germany). Scale bars represent 20 micrometers (μm).
The primary antibodies utilized for IF included: rabbit anti-TRIM5α (Leinco Technologies, T331, 1:100), rabbit anti-TRIM11 (Abcam, ab111694, 1:100), mouse anti-vimentin (SCBT, sc-373717, 1:200), mouse anti-calretinin (SCBT, sc-365956, 1:50), mouse anti-PKCα (SCBT, sc-8393, 1:50), rabbit anti-FLAG Tag (SIGMA, SAB4301135, 1:140), rabbit anti-TurboGFP (Evrogen, Moscow, Russia, AB513, 1:1000), and goat anti-NF-κB (p65) (SCBT, sc-372, 1:50). The secondary antibodies included: goat anti-mouse Alexa Fluor 594 (Life Technologies, Carlsbad, CA, A11032), goat anti-rabbit Alexa Fluor 488 (Life Technologies, A11008), and donkey anti-goat Alexa Fluor 594 (Invitrogen, A11058).
2.7. FIVGFP production
Packaging of the FIVGFP vector was described previously (Slauson et al., 2015). Briefly, the FIV-based vector pCDF1-MCS2-EF1α-copGFP (1.74mg, System Biosciences, Palo Alto, CA, CD111B-1) was co-transfected into HEK293TN cells with pFIV-34-N (326 μg) and pVSVG (408 μg) in eight 500 cm2 culture plates. Culture supernatants were collected after 48 hours and centrifuged at 24,000g for 30 minutes to pellet viral particles. The pellets were resuspended in Hanks balanced salt solution (HBSS) (Mediatech, 21–023-CM) and centrifuged through a 20% sucrose cushion in PBS. Viral pellets were resuspended in HBSS and stored at −80°C. Viral titers were determined by quantitation of fluorescent cells following serial dilution and transduction of Crandell-Rees feline kidney cells (CRFK).
2.8. FIVGFP Transduction
Wild type MIO-M1, TRIM5α KD clones 13 and 14, and TRIM11 KD clones 5 and 6 were transduced ON at 37°with FIVGFP (Multiplicity of infection (MOI) of 95). The following day, cells were trypsinized and plated onto poly-L-lysine coated glass chamber slides. At four or seven days post transduction, cells were fixed, permeabilized, blocked, and stained with anti-TurboGFP and goat anti-rabbit AlexaFluor 488 as described above. Cells were also stained with 1μg/ml Hoechst to visualize nuclei. DAPI positive nuclei and GFP positive cells were counted in five fields for each cell type and data was expressed as mean percent GFP positive cells. p values were determined by two-sample t-test. Sample size (N)=5. Similar experiments were performed with the ARPE-19 TRIM5α or TRIM11 KD clones.
2.9. MG132 assay
MG132 cytotoxicity was determined using a commercially available MTS assay (Cell Titer 96 Aqueous One Solution, Promega, Madison, WI, G3580). WT MIO-MI, TRIM5α−13 KD, or TRIM11–6 KD cells were seeded in a collagen treated (Advanced BioMatrix, Carlsbad, CA, PureCol, 5005) 96-well plate at 6 × 104 cells/well and grown overnight in complete medium (DMEM/F12/10% FBS supplemented with a 1:100 dilution of L-glutamine-penicillin-streptomycin) at 37°C in 5% CO2. The next day, the media was aspirated and replaced with 100 μl of complete media containing increasing concentrations of MG132 (SIGMA, Z-Leu-Leu-Leu-al, C2211) with an adjusted final concentration of 0.05% DMSO (SIGMA, D2650). After a 1 hour incubation at 37°C, the media was replaced with 100 μl complete media, and 20 μl Cell Titer 96 Aqueous solution was added to each well. The plate was incubated at 37°C for 1 hour and the absorbance was measured at 490 nm using a Biotek Neo2 plate reader (Biotek, Winooski, VT). OD values were blanked using the absorbance from media only control wells. Samples were run in duplicate and the percent cell viability was calculated by setting the mean blanked absorbance value of the 0 μM MG132 wells at 100 percent. The data was graphed with error bars representing SEM.
WT MIO-M1, TRIM5α−13 KD, and TRIM11–6 KD cells were pretreated with media (DMEM/F12/10% FBS supplemented with a 1:100 dilution of L-glutamine-penicillin-streptomycin), media plus 0.05% DMSO, or media plus 20 μM MG132/0.05% DMSO for 1 hour at 37°C. Cells were washed 3 times in PBS, media was added to wells, and cells were transduced ON at 37° with FIVGFP (MOI of 20). The following day, the cells were trypsinized and plated onto poly-L-lysine coated glass chamber slides. At six days post transduction, cells were fixed, permeabilized, blocked, and stained with anti-TurboGFP and goat anti-rabbit AlexaFLuor 488 as described above. Cells were also stained with 1μg/ml Hoechst to visualize nuclei. DAPI positive nuclei and GFP positive cells were counted in five fields for each cell type and data was expressed as mean percent GFP positive cells. p values were determined by two-sample t-test. Sample size (N)=5. The experiment was run in duplicate with similar results.
2.10. NFκB (p65) Activation
MIO-M1 WT, TRIM5α KD, and TRIM11 KD cells were plated in duplicate on poly-L-lysine coated chamber slides and transduced with FIVGFP (MOI of 12) at 37°C for 72 hours. Cells were fixed, permeabilized, and blocked as described above. Slides were stained with goat anti-NFκB(p65), followed by donkey anti-goat AlexaFluor 594. Nuclei were stained with Hoechst (1 μg/ml) before mounting with Immu-mount. The total cell number (DAPI positive nuclei) and p65 positive nuclei were determined in five random fields per well and data was expressed as mean percent nuclear p65. p values were determined by two-sample t-test. Sample size (N)=10.
3.0. Results:
3.1. TRIM5α and TRIM11 are expressed in NHP retina tissue
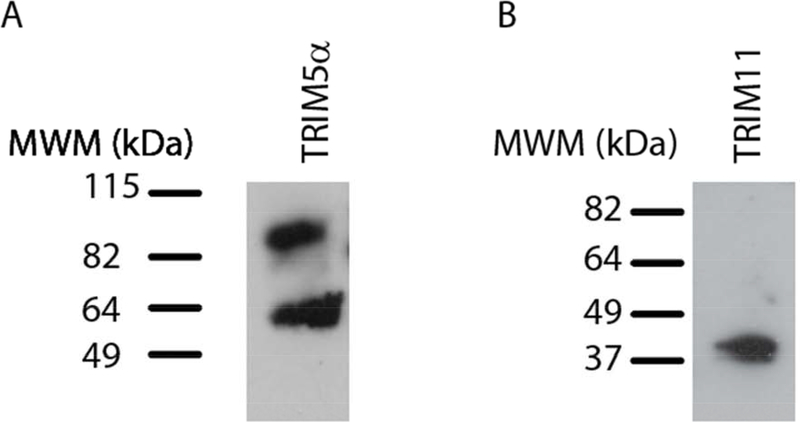
To determine whether the restriction factors TRIM5α and TRIM11 were expressed in NHP neural retina tissue, immunoblots were performed. Blotting of retina tissue lysate with a rabbit anti-TRIM5α antibody detected a doublet near the expected molecular weight (MW) of TRIM5α (Figure 1A). A polyclonal antibody against TRIM11 recognized a single band near the expected MW of TRIM11 (Figure 1B) indicating that both TRIM5α and TRIM11 proteins are produced in NHP retina tissue.
Figure 1: Expression of TRIM5α and TRIM11 proteins in NHP retina tissue.
Immunoblotting of macaque retina tissue lysates was performed with A) rabbit anti-TRIM5α or B) rabbit anti-TRIM11 primary antibodies. Goat anti-rabbit HRP-conjugated secondary antibodies were applied prior to ECL.
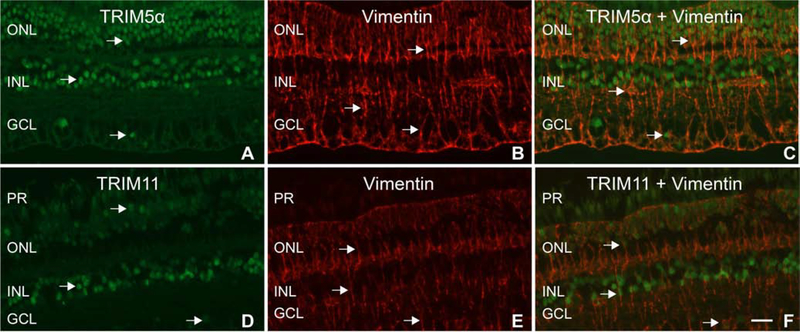
In HeLa cells, TRIM5α proteins localize mainly to cytoplasmic bodies, whereas TRIM11 is expressed in a diffuse pattern in both the cytosol and nucleus (Reymond et al., 2001). To our knowledge, the expression pattern of these restriction factors has not been determined in macaque neural retina tissue. To examine the cellular distribution of TRIM5α and TRIM11 within the neural retina, IF was performed on paraffin embedded macaque retina tissue. The TRIM5α antibody detected protein expression predominantly in nuclei of the outer nuclear layer (ONL) and inner nuclear layer (INL), as well as some scattered nuclei in the ganglion cell layer (GCL) (Figure 2A). TRIM11 expression was mainly detected in nuclei of the INL, with fainter staining in ONL nuclei, and some scattered expression in GCL nuclei (Figure 2D).
Figure 2: Expression patterns of TRIM5α and TRIM11 in macaque neural retina tissue:
A) Arrows indicate TRIM5α staining. B and E) Arrows indicate vimentin staining of Muller cells. C) Merged image with arrows indicating localization of TRIM5α staining to Muller cell nuclei. D) Arrows indicate TRIM11 staining. F) Merged image indicating localization of TRIM11 staining to Muller cell nuclei. PR= photoreceptor layer, ONL= outer nuclear layer, INL= inner nuclear layer, GCL= ganglion cell layer. Scale bar= 20μm.
To identify which retinal cells expressed TRIM5α and TRIM11 in their nuclei, double label IF was performed with the TRIM5α or TRIM11 antibody and cell type specific marker antibodies including vimentin (Muller), calretinin (amacrine), and PKCα (rod bipolar) (Sauter et al., 2018). Staining with the anti-vimentin antibody identified long, filamentous Muller cells which extended throughout the retina (Figure 2B, E). When the TRIM5α or TRIM11 images were merged with the vimentin stains, it became apparent that some of the vimentin-labeled cells expressed TRIM5α or TRIM11 in their nuclei (Figure 2C, F). These images indicate that Muller cells in the macaque retina express TRIM5α and TRIM11 proteins in their nuclei. Double label IF with calretinin and PKCα antibodies did not reveal TRIM5α or TRIM11 expression localizing to amacrine or rod bipolar cells in macaque neural retina tissue (data not shown).
3.2. Expression patterns of TRIM5α and TRIM11 in a human Muller cell line
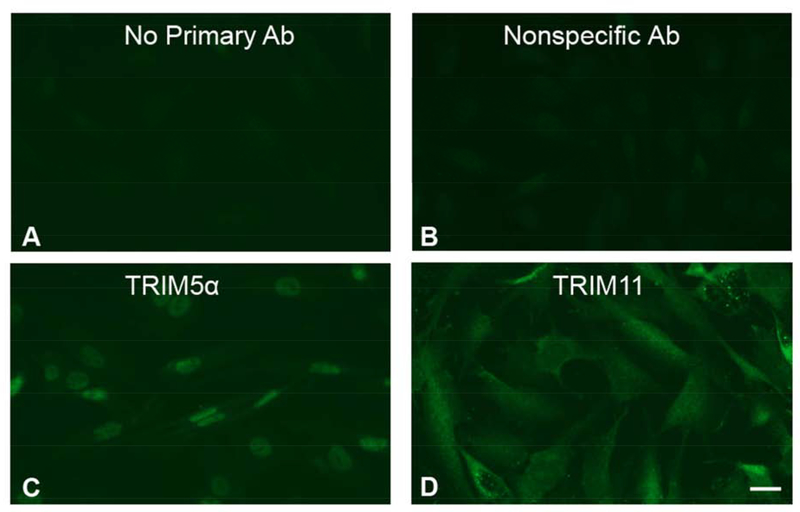
Since macaque Muller cell nuclei were shown to express both TRIM5α and TRIM11, we wanted to determine the cellular distribution pattern of these restriction factors in the human Muller cell line MIO-M1. MIO-M1 cells stained with the TRIM5α antibody demonstrated bright nuclear staining, and faint cytoplasmic staining (Figure 3C) compared to a no primary antibody control (Figure 3A) or a nonspecific rabbit polyclonal antibody control (Figure 3B). TRIM11 staining was cytosolic and diffuse, although some MIO-M1 cells also appeared to expressTRIM11 in their nuclei (Figure 3D). These images indicate that TRIM5α protein was primarily expressed in the nuclei, while TRIM11 expression appeared to be mainly cytoplasmic, in MIO-M1 cells.
Figure 3: Expression patterns of TRIM5α and TRIM11 in MIO-M1 cells.
MIO-M1 cells were fixed, permeabilized, and stained with A) no primary antibody, B) a nonspecific rabbit anti-FLAG Tag antibody, C) rabbit anti-TRIM5α, or D) rabbit anti-TRIM11 followed by a goat anti-rabbit Alexa Fluor 488 secondary antibody. All polyclonal antibodies were diluted to a final concentration of 10 μg/ml.
3.3. Knockdown of TRIM5α or TRIM11 increased FIVGFP transduction
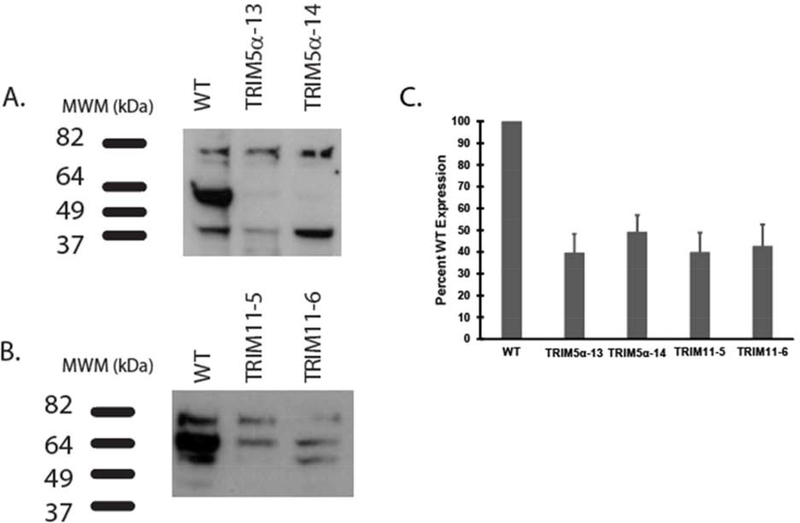
CRISPR technology was utilized to knock down TRIM5α or TRIM11 proteins in MIO-M1 cells and puromycin resistant clones were screened by immunoblotting with TRIM5α and TRIM11 antibodies. We obtained two clones with decreased levels of TRIM5α (Figure 4A) and two clones with decreased levels of TRIM11 (Figure 4B). ImageJ analysis of three separate immunoblots indicated that TRIM5α KD clones 13 and 14 had a mean of 39.6% or 49.2%, respectively, of WT levels of TRIM5α (Figure 4C). Analysis of TRIM11 clones 5 and 6 indicated that they expressed a mean of 39.9% or 42.7%, respectively, of WT levels of TRIM11 (Figure 4C). Total knockdown was not achieved in MIO-M1 cells for either TRIM5α or TRIM11, suggesting that these proteins are necessary for cell survival. We were also not able to generate a viable double KD of TRIM5α and TRIM11 in this cell type.
Figure 4: Immunoblots of TRIM5α and TRIM11 knockdown clones.
Equal micrograms of MIO-M1 WT and TRIM5α, or MIO-M1 WT and TRIM11 knockdown cell lysates, were electrophoresed and blotted with: A) mouse anti-TRIM5 or B) rabbit anti-TRIM11 antibodies. HRP-conjugated secondary antibodies were applied prior to ECL. C) ImageJ analysis was performed on three separate blots for each antibody and the mean percent WT expression was graphed with error bars representing standard error of the mean (SEM).
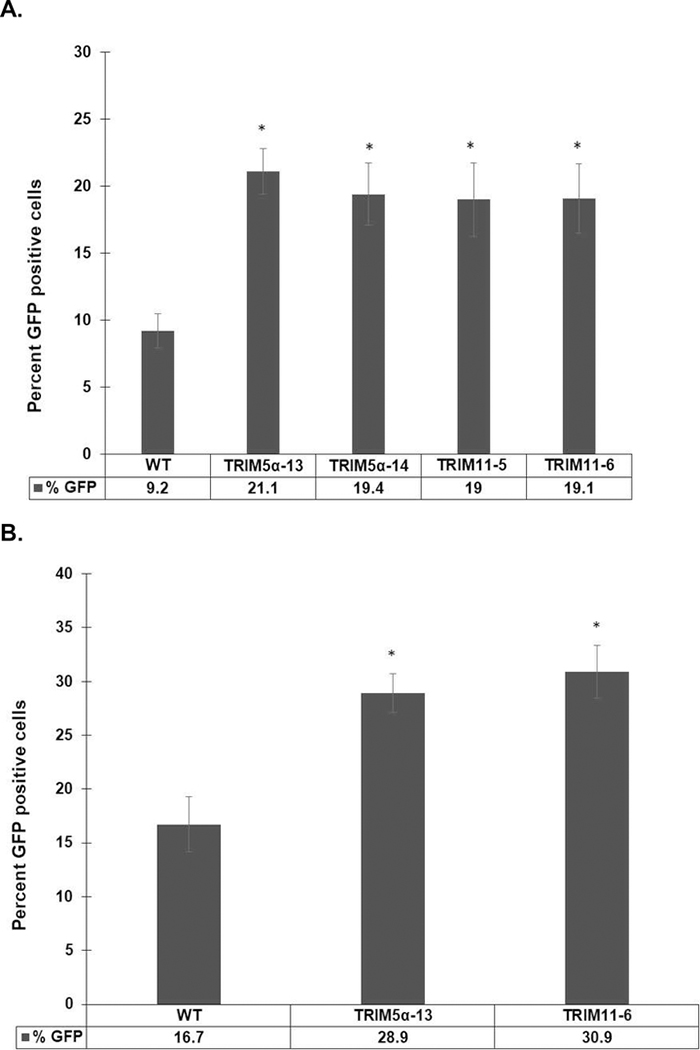
To determine if partial knockdown of TRIM5α or TRIM11 affected the transduction efficiency of a non-human lentiviral gene delivery vector in human cells, we exposed WT MIO-M1 cells, TRIM5α KD clones, and TRIM11 KD clones to FIVGFP. At 4 or 7 days post transduction, cells were fixed and stained with an anti-TurboGFP antibody to visualize GFP positive cells. The transduction efficiency of FIVGFP was very low in WT MIO-M1 cells, as expected for a feline retrovirus in human cells, with only 16.7% of cells expressing GFP at seven days post transduction (Figure 5B). In contrast, there was a roughly two-fold increase in the number of GFP positive cells in the two TRIM5α KD clones and the two TRIM11 KD clones, compared to WT MIO-M1 cells, at four days post transduction (Figure 5A). By seven days post transduction, the overall number of GFP positive cells had increased, and TRIM5α−13 KD and TRIM11–6 KD clones still had a significantly higher transduction efficiency compared to WT MIO-M1 cells (Figure 5B). These results, which were consistent in four replicate experiments, indicate that a decrease in the amount of TRIM5α or TRIM11 protein allowed for enhanced FIVGFP transduction of the human Muller cell line MIO-M1.
Figure 5: TRIM5α or TRIM11 knockdown increased FIVGFP transduction efficiency in a human Muller cell line.
WT MIO-M1, TRIM5α clones 13 and 14, and TRIM11 clones 5 and 6 were challenged with FIVGFP. Four days (A) or seven days (B) post transduction, cells were fixed and stained with an anti-TurboGFP antibody followed by goat anti-rabbit Alexa Fluor 488. Cells were stained with Hoechst to visualize nuclei. The percent GFP positive cells were determined in five fields for each cell type. Error bars represent SEM. p values were determined by two-sample t-test. Sample size (N) = 5. (*): p<0.02.
To study the effect of TRIM5α or TRIM11 knockdown in a different retinal cell line, we utilized shRNA technology to isolate stable knockdown clones in the human retinal pigment epithelial cell line ARPE-19. As with the MIO-M1 cells, we were not able to obtain total knockout of either TRIM5α or TRIM11 in ARPE-19 cells, suggesting that these proteins are vital for cellular function. FIVGFP transduction efficiency experiments with the ARPE-19 TRIM5α or TRIM11 KD clones yielded inconsistent results for reasons that are not clear at this time (data not shown).
3.4. Proteasome inhibition increased transduction of MIO-MI cells
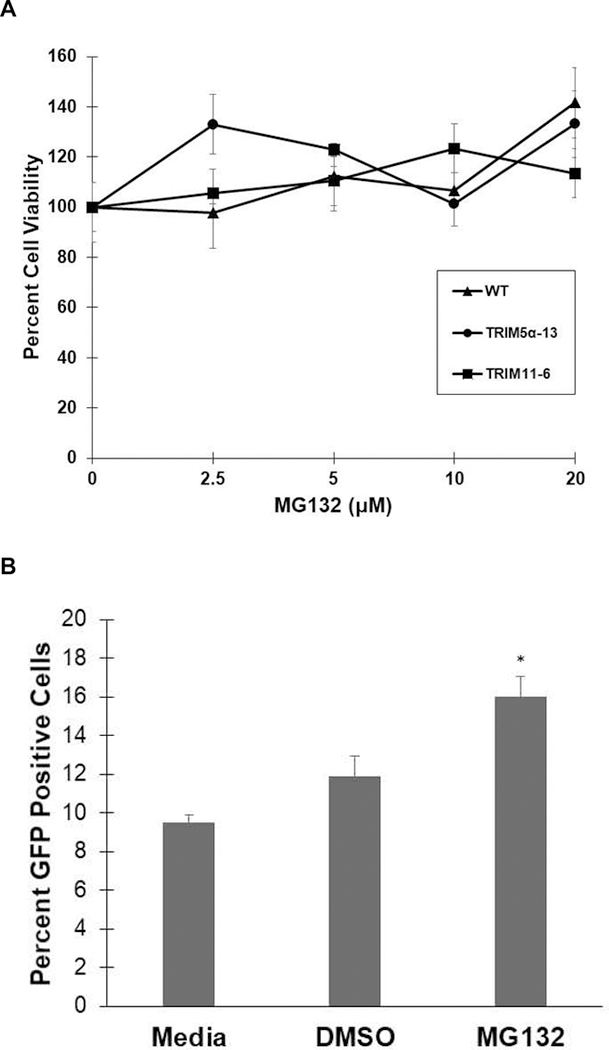
Prior to testing the effect of the proteasome inhibitor MG132 on transduction efficiency in MIO-M1 cells, the toxicity of the DMSO solvent and MG132 was determined. WT MIO-MI, TRIM5α−13 KD, or TRIM11–6 KD cells were exposed to increasing concentrations of MG132 in 0.05% DMSO for 1 hour at 37°C. A MTS assay indicated that no toxicity was observed for either 0.05% DMSO alone or concentrations of up to 20 μM MG132 for any of the Muller cells tested (Fig 6A).
Figure 6: Proteasome inhibitor pretreatment increased FIVGFP transduction efficiency in MIO-M1 cells.
A) WT MIO-M1, TRIM5α−13 KD cells, and TRIM11–6 KD cells were incubated with increasing concentrations of MG132 for 1 hour at 37°C, followed by MTS assay. The mean percent viability was calculated and graphed with error bars representing the SEM. B) WT MIO-M1 cells were pretreated with media, 0.05% DMSO, or 20 μM MG132/0.05% DMSO for 1 hour at 37°C prior to FIVGFP transduction (MOI 20). Six days post transduction, the cells were fixed and stained with an anti-TurboGFP antibody followed by goat anti-rabbit AlexaFluor 488. Cells were stained with Hoechst to visualize nuclei. The percent GFP positive cells were determined in five fields for each condition and the mean percent was graphed with error bars representing the SEM. p values were determined by two-sample t-test. Sample size (N)= 5. ( *): p=0.002.
To determine the effect of proteasome inhibitor pretreatment on FIVGFP transduction efficiency, we incubated WT MIO-M1 cells, TRIM5α−13 KD, and TRIM11–6 KD cells with 20 μM MG132 for 1 hour at 37°C. After washing with PBS, the cells were challenged with FIVGFP (MOI 20) for 6 days at 37°C. MG132 pretreatment had a significant effect on the transduction efficiency of FIVGFP in WT MIO-M1 cells with GFP positive cells increasing from 9.5% in media treated cells to 16% in MG132 treated cells (Fig 6B). DMSO alone did not have a significant effect on transduction efficiency (Fig 6B) and these results were consistent in duplicate experiments. We did not observe an increase in FIVGFP transduction efficiency after MG132 pretreatment in either TRIM5α−13 KD or TRIM11–6 KD cells (data not shown).
3.5. NF-κB (p65) activation is increased in transduced TRIM5α and TRIM11 knockdown cells
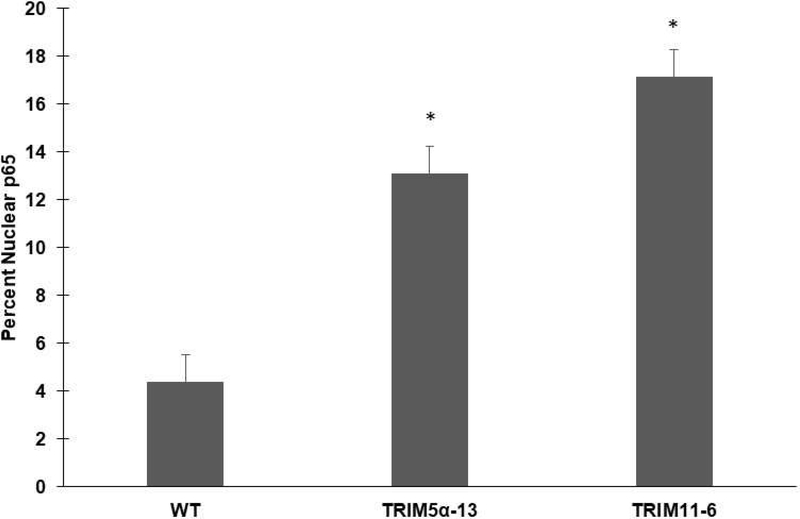
To determine whether TRIM5α or TRIM11 knockdown affect NF-κB activation, WT MIO-M1, TRIM5α KD, or TRIM11 KD cells were challenged with FIVGFP. Seventy-two hours post transduction, cells were fixed and stained with an antibody to NF-κB (p65). The percent nuclear p65 was calculated to serve as an indicator of NF-κB (p65) activation. An average of 4.4% of WT MIO-M1 cells exhibited p65 staining in their nuclei after a 72-hour FIVGFP challenge (Figure 7). In contrast, TRIM5α KD cells exhibited an approximate three-fold increase (13.1%) in the percent of cells with nuclear p65 staining following FIVGFP exposure (Fig 7). TRIM11 KD cells had an even greater increase in nuclear p65 staining (17.2%), compared to WT MIO-M1 cells, following FIVGFP challenge (Fig 7). These experiments indicate that KD of the restriction factors TRIM5α or TRIM11 in the human Muller cell line MIO-M1 resulted in significantly higher levels of NFκB (p65) activation at later time points post FIVGFP transduction.
Figure 7: NF-κB (p65) activation following FIVGFP transduction.
Duplicate wells of WT, TRIM5α−13 KD, and TRIM11–6 KD MIO-M1 cells were challenged with FIVGFP for 72 hours. Cells were fixed, permeabilized, and stained with an anti-NF-κB (p65) antibody followed by donkey anti-goat Alexa Fluor 594. Cells were stained with Hoechst to visualize nuclei. The total number of cells and p65 positive nuclei were determined in five random fields per well and data was graphed as mean percent nuclear p65. Error bars represent SEM. p values were determined by two-sample t-test. (*): p < 0.001. Sample size (N)=10
4.0. Discussion:
Although the expression of some TRIM proteins, including TRIM11, have been characterized in the developing mouse retina (Chowdhury et al., 2018), researchers have not yet explored the distribution of TRIM proteins in the primate retina. Given our interest in factors that reduce the efficiency of lentiviral gene delivery vectors, we focused on TRIM5α and TRIM11 in this study. Our data show that both proteins are expressed in NHP retina and that TRIM5α and TRIM11 were localized to some Muller cell nuclei. Reports of TRIM localization in HeLa and U2OS cells indicated cytoplasmic localization of TRIM5α and diffuse localization of TRIM11, suggesting that localization may be cell-type dependent (Reymond et. al., 2001; Wu et. al., 2006). TRIM5α is also expressed in neuroblastoma cell lines but the localization was not reported (Singh et. al., 2014). The nuclear distribution of TRIM11 in NHP retina was consistent with reports of TRIM11 localization in multiple cell types (Darshit and Ramanathan, 2016; Jabbari et.al., 2018; Rathinam and Fitzgerald, 2011; Reymond et. al., 2001).
Analysis of the human Muller cell line MIO-M1 indicated that TRIM5α was expressed predominantly in the nuclei while TRIM11 was primarily cytoplasmic. Expression of both proteins in Muller cells would be consistent with the role of Muller cells in responses to microbial infection and innate immune response signaling (Kumar et al., 2013, Kumar and Shamsuddin 2012). The differences in localization of these proteins in NHP retina and the MIO-M1 cells could be due to several factors including species-specific differences or differences due to tissue and cell processing. Finally, the differences may be related to the fact that we were comparing cells in situ with immortalized cells in tissue culture. Environmental factors are known to regulate expression and localization of TRIM proteins (Singh et. al., 2014) and the tissue and cell culture environments are quite different. The NHP tissue was obtained from pathogen-free animals and the MIO-M1 cells were not exposed to virus prior to analysis so infection related differences should not be relevant. It will be interesting to examine the localization of TRIM proteins in the context of an infection.
TRIM5α and TRIM11 bind to HIV-1 capsid complexes and accelerate uncoating (Yuan et al., 2016) despite low sequence homology (Li et al., 2007). Our results indicate that knockdown of TRIM5α in MIO-M1 cells resulted in a significant increase in transduction efficiency of the FIVGFP vector, suggesting that TRIM5α is involved in FIV restriction. Previously a negative correlation between TRIM5α expression and HIV-1 vector transduction efficiency in human and rhesus CD34+ cells was reported (Evans et al., 2014). Transient knockdown of TRIM5α in CD4+ lymphocytes or a human neuroblastoma cell line (SK-N-AS) also increased HIV-1 infection (Singh et al., 2014). These results indicate that even a partial decrease in TRIM5α protein can enhance lentiviral transduction in multiple cell types.
Our data show that knockdown of TRIM11 in MIO-M1 cells also increased FIVGFP transduction efficiency. TRIM11 was initially identified as a RF that inhibited HIV entry (Uchil et. al., 2008). Yuan et. al. (2016) found that the inhibition involved direct binding of TRIM11 to the capsid resulting in accelerated uncoating, decreased capsid stability, and inhibition of early stages of infection. TRIM11 also disrupts nuclear import of viral DNA (Yuan et. al., 2014; Yuan et. al., 2016), activates the proteasome (Chen et. al., 2018), and suppresses AIM2 inflammasomes (Liu et. al., 2016). This study supports a role for TRIM11 in restriction of lentiviral gdvs in human Muller cells and demonstrates that transduction efficiency can be increased by partial knockdown of TRIM11.
Other methods to overcome TRIM mediated restriction may include alteration of viral capsid proteins (He et al., 2017), the use of linkage-specific deubiquitinating enzymes (Imam et al., 2019), or proteasome inhibitor treatment (Aktas et al., 2018,Danielson et al., 2012,Wu et al., 2006). Our data indicate that pretreatment with the proteasome inhibitor MG132 significantly increased the transduction efficiency of a lentiviral vector in human Muller cells. These results are consistent with our previous studies on human trabecular meshwork cells and support a role for the use of proteasome inhibitors prior to ocular gene therapy (Aktas et al., 2018). Previous studies have linked the association of rhesus TRIM5α and proteasomes to restriction of HIV-1 in HeLa cells (Wu et al., 2006, Danielson et al., 2012). Our experiments reinforce the importance of proteasome function in lentiviral restriction and demonstrate this link for the first time in a human Muller cell line. Interestingly, TRIM 11 restriction on HIV-1 was not sensitive to proteasome inhibitors in HEK293 cells, revealing that although there is some functional overlap in terms of capsid binding, there are differences in effector functions of the TRIM5α and TRIM11 proteins (Tareen et al., 2011,Yuan et al., 2016). We did not observe an increase in FIVGFP transduction efficiency in MG132 treated TRIM5α or TRIM11 knockdown clones and future studies will be needed to explain this finding.
The early induction of NF-κB is essential for efficient lentiviral gene expression, yet it also triggers an antiviral innate immune response (Heusinger and Kirchhoff 2017). Lentiviruses cope with this by enhancing NF-κB activity early in infection and suppressing it at later stages. Our data show that the increase in FIVGFP transduction efficiency in TRIM5α or TRIM11 KD MIO-M1 cells was accompanied by increased nuclear translocation of NF-κB (p65) at 3 days post transduction. Since the FIVGFP gdv used in our studies is not replication competent, later stages of the lentiviral life cycle, in which NF-κB is usually suppressed, are not present in our system. This observation provides another way to quantitate transduction efficiency in cell types that are difficult to transduce, and suggests a role for NF-κB activation in efficient gene delivery by lentiviral vectors.
In summary, we have identified TRIM5α and TRIM11 expression in Muller cell nuclei of NHP retina, suggesting that these restriction factors may play a role in reducing the transduction efficiency of lentiviral gene delivery vectors in this tissue. We have also shown in a cultured Muller cell line that knockdown of either TRIM5α or TRIM11, or pretreatment with a proteasome inhibitor, increased the transduction efficiency of a lentiviral vector. These results suggest that transient inhibition of host restriction factors, just prior to vector delivery, could improve transduction efficiency of retinal cells and result in vector sparing which is especially important in ocular gene delivery due to the small volume of vector that can be administered (Aktas et al., 2018). Future experiments will determine if transient knockdown of TRIM5α or TRIM11 in primary NHP or human Muller cells increases transduction efficiency of lentiviral vectors.
Highlights.
NHP retina tissue expresses TRIM5α and TRIM11 in Muller cell nuclei
Cultured human Muller cells express TRIM5α in nuclei and TRIM11 in the cytosol
Knockdown of TRIM5α or TRIM11 increased transduction efficiency of FIVGFP
Proteasome inhibitor pretreatment increased transduction efficiency of FIVGFP
NF-κB (p65) activation was increased in TRIM5α and TRIM11 knockdown clones
Acknowledgements:
Inna Larsen for manuscript and figure preparation. Hongyu Rao, Aaron Kolb, and Sarah Ferguson for FIVGFP production.
Funding: The Retina Research Foundation, Houston TX, Core Grant for Vision Research NIH P30 EY016665 to Curtis Brandt, NIH P51OD011106 to the Wisconsin National Primate Research Center, University of Wisconsin-Madison, and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aktas Z, Rao H, Slauson SR, Gabelt BT, Larsen IV, Sheridan RTC, Herrnberger L, Tamm ER, Kaufman PL, Brandt CR: Proteasome inhibition increases the efficiency of lentiviral vector-mediated transduction of trabecular meshwork. Invest. Ophthalmol. Vis. Sci, 59 (2018), pp. 298–310, 10.1167/iovs.17-22074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annoni A, Gregori S, Naldini L, Cantore A: Modulation of immune responses in lentiviral vector-mediated gene transfer. Cell. Immunol, 342 (2019), pp. 103802, 10.1016/j.cellimm.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaggan KS, Ali RR: Ocular gene delivery using lentiviral vectors. Gene Ther, 19 (2012), pp. 145–153, 10.1038/gt.2011.153. [DOI] [PubMed] [Google Scholar]
- Balaggan KS, Binley K, Esapa M, Iqball S, Askham Z, Kan O, Tschernutter M, Bainbridge JW, Naylor S, Ali RR: Stable and efficient intraocular gene transfer using pseudotyped EIAV lentiviral vectors. J. Gene Med, 8 (2006), pp. 275–285, 10.1002/jgm.845. [DOI] [PubMed] [Google Scholar]
- Bennett J: Taking stock of retinal gene therapy: looking back and moving forward. Mol. Ther, 25 (2017), pp. 1076–1094, 10.1016/j.ymthe.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsotti C, Borroni E, Follenzi A: Lentiviral vector interactions with the host cell. Curr. Opin. Virol, 21 (2016), pp. 102–108, 10.1016/j.coviro.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Cavalieri V, Baiamonte E, Lo Iacono M: Non-primate lentiviral vectors and their applications in gene therapy for ocular disorders. Viruses, 10 (2018), pp. 10.3390/v10060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhu G, Johns EM, Yang X: TRIM11 activates the proteasome and promotes overall protein degradation by regulating USP14. Nat. Commun, 9 (2018), pp. 1223, 10.1038/s41467-018-03499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Laboissonniere LA, Wester AK, Muller M, Trimarchi JM: The Trim family of genes and the retina: Expression and functional characterization. PLoS One, 13 (2018), pp. e0202867, 10.1371/journal.pone.0202867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson CM, Cianci GC, Hope TJ: Recruitment and dynamics of proteasome association with rhTRIM5alpha cytoplasmic complexes during HIV-1 infection. Traffic, 13 (2012), pp. 1206–1217, 10.1111/j.1600-0854.2012.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshit BS, Ramanathan M: Activation of AKT1/GSK-3beta/beta-Catenin-TRIM11/Survivin Pathway by Novel GSK-3beta Inhibitor Promotes Neuron Cell Survival: Study in Differentiated SH-SY5Y Cells in OGD Model. Mol. Neurobiol, 53 (2016), pp. 6716–6729, 10.1007/s12035-015-9598-z. [DOI] [PubMed] [Google Scholar]
- Evans ME, Kumkhaek C, Hsieh MM, Donahue RE, Tisdale JF, Uchida N: TRIM5alpha variations influence transduction efficiency with lentiviral vectors in both human and rhesus CD34(+) cells in vitro and in vivo. Mol. Ther, 22 (2014), pp. 348–358, 10.1038/mt.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis A, Duran Y, Ribeiro J, Abelleira-Hervas L, Robbie SJ, Sunkel-Laing B, Fourali S, Gonzalez-Cordero A, Cristante E, Michaelides M, et al. : Development of an optimized AAV2/5 gene therapy vector for Leber congenital amaurosis owing to defects in RPE65. Gene Ther, 23 (2016), pp. 857–862, 10.1038/gt.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardinger C, McClure KD, Romeo G, Podesta F, Lorenzi M: IGF-I mRNA and signaling in the diabetic retina. Diabetes, 50 (2001), pp. 175–183, [DOI] [PubMed] [Google Scholar]
- Grutter MG, Luban J: TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr. Opin. Virol, 2 (2012), pp. 142–150, 10.1016/j.coviro.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage A, Rajsbaum R: To TRIM or not to TRIM: the balance of host-virus interactions mediated by the ubiquitin system. J. Gen. Virol, 100 (2019), pp. 1641–1662, 10.1099/jgv.0.001341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Xue J, Wang W, Liu L, Ye C, Cong Z, Kimata JT, Qin C, Zhou P: Efficient transduction of human and rhesus macaque primary T cells by a modified human immunodeficiency virus type 1-based lentiviral vector. Hum. Gene Ther, 28 (2017), pp. 271–285, 10.1089/hum.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusinger E, Kirchhoff F: Primate lentiviruses modulate NF-kappaB activity by multiple mechanisms to fine-tune viral and cellular gene expression. Front. Microbiol, 8 (2017), pp. 198, 10.3389/fmicb.2017.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Fukutome M, Koike C: Adeno associated virus (AAV) as a tool for clinical and experimental delivery of target genes into the mammalian retina. Biol. Pharm. Bull, 42 (2019), pp. 343–347, 10.1248/bpb.b18-00913. [DOI] [PubMed] [Google Scholar]
- Imam S, Komurlu S, Mattick J, Selyutina A, Talley S, Eddins A, Diaz-Griffero F, Campbell EM: K63-linked ubiquitin is required for restriction of HIV-1 reverse transcription and capsid destabilization by rhesus TRIM5alpha. J. Virol, 93 (2019), pp. 10.1128/JVI.00558-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari E, Woodside J, Tan MMX, Shoai M, Pittman A, Ferrari R, Mok KY, Zhang D, Reynolds RH, de Silva R, et al. : Variation at the TRIM11 locus modifies progressive supranuclear palsy phenotype. Ann. Neurol, 84 (2018), pp. 485–496, 10.1002/ana.25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Zhao Q, Xiong Y: HIV suppression by host restriction factors and viral immune evasion. Curr. Opin. Struct. Biol, 31 (2015), pp. 106–114, 10.1016/j.sbi.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajaste-Rudnitski A, Naldini L: Cellular innate immunity and restriction of viral infection: implications for lentiviral gene therapy in human hematopoietic cells. Hum. Gene Ther, 26 (2015), pp. 201–209, 10.1089/hum.2015.036. [DOI] [PubMed] [Google Scholar]
- Koepke L, Gack MU, Sparrer KM: The antiviral activities of TRIM proteins. Curr. Opin. Microbiol, 59 (2020), pp. 50–57, 10.1016/j.mib.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Pandey RK, Miller LJ, Singh PK, Kanwar M: Muller glia in retinal innate immunity: a perspective on their roles in endophthalmitis. Crit. Rev. Immunol, 33 (2013), pp. 119–135, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Shamsuddin N: Retinal Muller glia initiate innate response to infectious stimuli via toll-like receptor signaling. PLoS One, 7 (2012), pp. e29830, 10.1371/journal.pone.0029830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran N, Michaelides M, Smith AJ, Ali RR, Bainbridge JWB: Retinal gene therapy. Br. Med. Bull, 126 (2018), pp. 13–25, 10.1093/bmb/ldy005. [DOI] [PubMed] [Google Scholar]
- Lascano J, Uchil PD, Mothes W, Luban J: TRIM5 retroviral restriction activity correlates with the ability to induce innate immune signaling. J. Virol, 90 (2016), pp. 308–316, 10.1128/JVI.02496-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gold B, O’HUigin C, Diaz-Griffero F, Song B, Si Z, Li Y, Yuan W, Stremlau M, Mische C, et al. : Unique features of TRIM5alpha among closely related human TRIM family members. Virology, 360 (2007), pp. 419–433, 10.1016/j.virol.2006.10.035. [DOI] [PubMed] [Google Scholar]
- Limb GA, Salt TE, Munro PM, Moss SE, Khaw PT: In vitro characterization of a spontaneously immortalized human Muller cell line (MIO-M1). Invest. Ophthalmol. Vis. Sci, 43 (2002), pp. 864–869, [PubMed] [Google Scholar]
- Liu T, Tang Q, Liu K, Xie W, Liu X, Wang H, Wang RF, Cui J: TRIM11 suppresses AIM2 inflammasome by degrading AIM2 via p62-Dependent selective autophagy. Cell. Rep, 16 (2016), pp. 1988–2002, 10.1016/j.celrep.2016.07.019. [DOI] [PubMed] [Google Scholar]
- Ludwig PE, Freeman SC, Janot AC: Novel stem cell and gene therapy in diabetic retinopathy, age related macular degeneration, and retinitis pigmentosa. Int J. Retina Vitreous, 5 (2019), pp. 7, 10.1186/s40942-019-0158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong T, Pennesi ME, Birch DG, Lam BL, Tsang SH: Adeno-associated viral gene therapy for inherited retinal disease. Pharm. Res, 36 (2019), pp. 34, 10.1007/s11095-018-2564-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, et al. : TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature, 472 (2011), pp. 361–365, 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VAK, Fitzgerald KA: Cytosolic surveillance and antiviral immunity. Curr. Opin. Virol, 1 (2011), pp. 455–462, 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. : The tripartite motif family identifies cell compartments. EMBO J, 20 (2001), pp. 2140–2151, 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz DT, Barraza R, Loewen N, Teo W, Poeschla EM: Feline immunodeficiency virus-based lentiviral vectors. Cold Spring Harb. Protoc, 2012 (2012), pp. 71–76, 10.1101/pdb.ip067579. [DOI] [PubMed] [Google Scholar]
- Sauter MM, Kolb AW, Brandt CR: Toll-like receptors 4, 5, 6 and 7 are constitutively expressed in non-human primate retinal neurons. J. Neuroimmunol, 322 (2018), pp. 26–35, 10.1016/j.jneuroim.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Patel V, Mureithi MW, Naranbhai V, Ramsuran D, Tulsi S, Hiramen K, Werner L, Mlisana K, Altfeld M, et al. : TRIM5alpha and TRIM22 are differentially regulated according to HIV-1 infection phase and compartment. J. Virol, 88 (2014), pp. 4291–4303, 10.1128/JVI.03603-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauson SR, Peters DM, Schwinn MK, Kaufman PL, Gabelt BT, Brandt CR: Viral vector effects on exoenzyme C3 transferase-mediated actin disruption and on outflow facility. Invest. Ophthalmol. Vis. Sci, 56 (2015), pp. 2431–2438, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J: The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature, 427 (2004), pp. 848–853, 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Tareen SU, Emerman M: Human Trim5alpha has additional activities that are uncoupled from retroviral capsid recognition. Virology, 409 (2011), pp. 113–120, 10.1016/j.virol.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani I, Auricchio A: Seeing the light after 25 years of retinal gene therapy. Trends Mol. Med, 24 (2018), pp. 669–681, 10.1016/j.molmed.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Trapani I, Colella P, Sommella A, Iodice C, Cesi G, de Simone S, Marrocco E, Rossi S, Giunti M, Palfi A, et al. : Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med, 6 (2014), pp. 194–211, 10.1002/emmm.201302948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W: TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog., 4 (2008), pp. e16, 10.1371/journal.ppat.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent M, Sparrer KMJ, Gack MU: TRIM proteins and their roles in antiviral host defenses. Annu. Rev. Virol, 5 (2018), pp. 385–405, 10.1146/annurev-virology-092917-043323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tol S, Hage A, Giraldo MI, Bharaj P, Rajsbaum R: The TRIMendous role of TRIMs in virus-host interactions. Vaccines (Basel), 5 (2017), pp. 10.3390/vaccines5030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XL, Anderson JL, Campbell EM, Joseph AM, Hope TJ: Proteasome inhibitors uncouple rhesus TRIM5 alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. U. S. A, 103 (2006), pp. 7465–7470, 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Yao W, Huang F, Sun B, Yang R: The human antiviral factor TRIM11 is under the regulation of HIV-1 Vpr. PLoS One, 9 (2014), pp. e104269, 10.1371/journal.pone.0104269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Yao W, Tokunaga K, Yang R, Sun B: An HIV-1 capsid binding protein TRIM11 accelerates viral uncoating. Retrovirology, 13 (2016), pp. 72, 10.1186/s12977-016-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziccardi L, Cordeddu V, Gaddini L, Matteucci A, Parravano M, Malchiodi-Albedi F, Varano M: Gene therapy in retinal dystrophies. Int. J. Mol. Sci, 20 (2019), pp. 10.3390/ijms20225722. [DOI] [PMC free article] [PubMed] [Google Scholar]