Summary
We present a proteogenomic study of 108 human papilloma virus (HPV)-negative head and neck squamous cell carcinomas (HNSCCs). Proteomic analysis systematically catalogs HNSCC-associated proteins and phosphosites, prioritizes copy number drivers and highlights an oncogenic role for RNA processing genes. Proteomic investigation of mutual exclusivity between FAT1 truncating mutations and 11q13.3 amplifications reveals dysregulated actin dynamics as a common functional consequence. Phosphoproteomics characterizes two modes of EGFR activation, suggesting a new strategy to stratify HNSCCs based on EGFR ligand abundance for effective treatment with inhibitory EGFR monoclonal antibodies. Widespread deletion of immune modulatory genes accounts for low immune infiltration in immune-cold tumors, whereas concordant upregulation of multiple immune checkpoint proteins may underlie resistance to anti-PD-1 monotherapy in immune-hot tumors. Multi-omic analysis identifies three molecular subtypes with high potential for treatment with CDK inhibitors, anti-EGFR antibody therapy, and immunotherapy, respectively. Altogether, proteogenomics provides a systematic framework to inform HNSCC biology and treatment.
Graphical Abstract

eTOC Blurb
Huang et al. report a proteogenomic study on 108 HPV-negative head and neck squamous cell carcinomas (HNSCCs). In addition to creating a comprehensive resource for pathogenic insights, multi-omic analysis identifies therapeutic hypotheses that may inform more precise approaches to treatment.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common epithelial malignancy worldwide (Bray et al., 2018) and can be broadly classified into human papillomavirus (HPV)-associated (HPVpos) and HPV-negative (HPVneg) subtypes. Most HNSCC patients are treated with surgery, chemotherapy, and radiotherapy. Targeted agents, including an EGFR monoclonal antibody (mAb) inhibitor and two PD-1 inhibitors, have been approved by FDA for HNSCC treatment, but overall response rates have been moderate (Baselga et al., 2005; Burtness et al., 2005; Herbst et al., 2005; Seiwert et al., 2016; Vermorken et al., 2008; Vermorken et al., 2007). Recently, the Cancer Genome Atlas (TCGA) and other studies have defined the genomic landscape and transcriptomic subtypes of HNSCC (Cancer Genome Atlas, 2015; Chung et al., 2004; Keck et al., 2015; Walter et al., 2013). However, a complete understanding of how genetic aberrations drive tumor phenotypes remains elusive, and translation of genomic and transcriptomic findings into improved HNSCC treatment has been limited.
By integrating mass spectrometry (MS)-based proteomics with genomics and transcriptomics, we performed an integrated proteogenomic characterization of 108 HPVneg HNSCCs. We focused on HPVneg HNSCCs because they account for 75% of all HNSCCs and have distinct molecular profiles and significantly worse prognosis compared to HPVpos tumors (Kreimer et al., 2005). Our study systematically catalogs HPVneg HNSCC-associated proteins, phosphosites, and signaling pathways. Proteogenomic integration provides functional insights into genomic aberrations, with practical implications for precision treatment of patients with HPVneg HNSCC.
Results
Proteogenomic Profiling
We prospectively collected 110 treatment-naïve primary HNSCC tumors and matched blood samples (Table S1), and 66 tumors had matched normal adjacent tissues (NATs). Homogenized samples were aliquoted for molecular profiling using whole-exome sequencing (WES), whole-genome sequencing (WGS), methylation array, RNA sequencing (RNA-seq), microRNA sequencing (miRNA-seq), and isobaric tandem mass tag (TMT) labeling-based global and phosphoproteomics (Figure 1A). One sample with evidence of HPV infection by RNA-seq was removed from downstream analysis (Figure S1A). The cohort was 87% male and tumor sites were predominantly from the oral cavity and larynx (44.5% each). Consistent with self-reporting, genomics-based smoking inference associated 70% of the patients with strong evidence of smoking (Figure S1B-C).
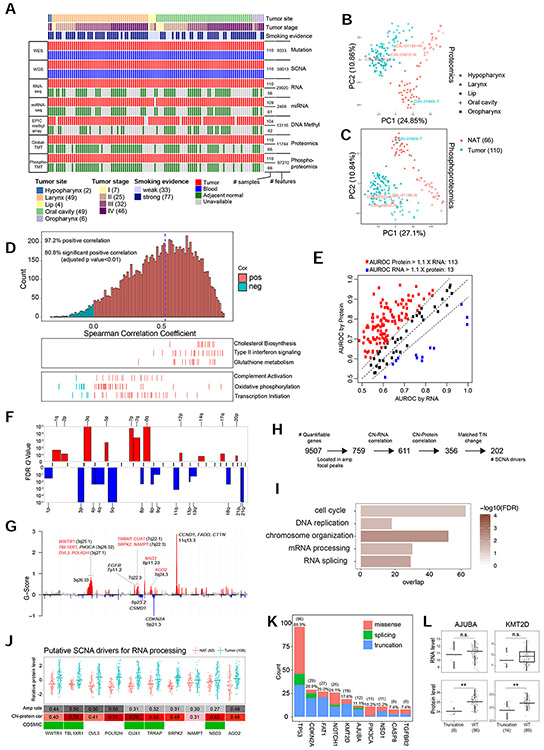
Figure 1. Proteogenomic profiling and impact of genetic aberrations on proteins.
(A) Cohort clinical features and omic data generation. (B) Global proteomics and (C) peptide-level phosphoproteomics PCA plots. (D) Gene-wise mRNA-protein correlation and pathway enrichment. (E) Area under the receiver operating characteristic curve (AUROC) for KEGG pathway membership prediction using RNA and protein data. Red and blue indicate pathways with >10% difference between the two. (F) Arm-level SCNAs. (G) Focal-level SCNAs with known drivers and RNA processing genes (red) annotated. (H) Prioritization of genes in focal amplification peaks. (I) GO terms enriched for prioritized SCNA drivers (Fisher’s exact test). (J) Protein abundance of RNA processing genes in tumors and NATs, annotated with amplification rate, copy number-protein correlation (Pearson’s correlation), and presence (green) in the COSMIC Cancer Gene Census. (K) Mutation frequency and type for the most frequently mutated genes. (L) Comparisons of RNA and protein levels for AJUBA and KMT2D between samples with truncating mutations and wild-type (WT) samples. **p<0.01, Student’s t-test. n.s., not significant. Numbers in parentheses represent the sample sizes for the involved groups. See also Figure S1 and Table S1-2.
Proteomic analysis identified 11,744 proteins. Phosphoproteomic analysis identified 97,210 phosphopeptides, covering 56,959 confidently localized phosphosites from 8,133 genes, including 81% on serine, 16% on threonine, and 3% on tyrosine. Replicate samples showed high measurement reproducibility across the TMT plexes, and there were no observable batch effects by TMT plex (Figure S1D-G). Unsupervised principal component analysis (PCA) of both proteomic and phosphoproteomic data separated tumor samples from NATs (Figure 1B-C). One tumor and three NAT samples with questionable tumor/NAT identity in data quality control (labeled in Figure 1B-C) were confirmed by pathological inspection and removed from downstream analysis.
For the 108 tumors, the median gene-wise Spearman’s correlation between protein and RNA abundance was 0.52, and the median sample-wise correlation was 0.43. Genes involved in forming large protein complexes, such as those related to complement activation, oxidative phosphorylation, and transcription initiation, showed lower protein-RNA correlation (Figure 1D). Protein data substantially outperformed RNA data in co-expression-based gene function prediction (Figure 1E), suggesting a critical role for protein-level regulation in determining gene functions.
Impact of Genetic Aberrations on Cognate Proteins
Somatic copy number alteration (SCNA) analysis identified arm level amplifications and deletions (Figure 1F). Focal peaks included amplifications of 3q26.33, 7p11.2, 7q22.3, 8p11.23, and 11q13.3 and deletions of 8p23.2 and 9p21.3, among others (Figure 1G). The strongest focal alteration was observed at 11q13.3. Some of these focal SCNA hotspots were also associated with structural variation events (Figure S1H and Table S2).
By filtering for correlated copy number (CN), mRNA, and protein levels across tumor samples and concordant protein level changes between paired tumor and NAT samples, we prioritized 202 putative SCNA drivers, including well-established PIK3CA, EGFR, CCND1, and CTTN, from a total of 759 quantifiable genes in the focal amplicons (Figure 1H-G, Table S2). The prioritized genes showed higher essentiality in HPVneg HNSCC cell lines in a genome-wide genetic perturbation screen (Tsherniak et al., 2017) (Figure S1I). Enrichment analysis associated the prioritized genes with cell cycle, DNA replication, chromosome organization, mRNA processing, and RNA splicing (Figure 1I). Amplification of the RNA processing genes (Figure 1G) has not been linked to HNSCC tumorigenesis previously, but some (WWTR1, TBL1XR1, CUX1, TRRAP, NSD3) are known cancer genes (Figure 1J).
Frequently mutated genes in this cohort (Figure 1K) were consistent with those in TCGA HPVneg HNSCCs, and we assessed the impact of mutations on cognate gene products. Missense mutations in TP53 were associated with increased p53 mRNA and protein abundance (Figure S1J), suggesting that certain TP53 mutations might endow oncogenic gain-of-function to this protein. Several frequently mutated genes had a substantial proportion of truncating mutations (Figure 1K), which typically induce nonsense-mediated decay (NMD) of cognate transcripts. Interestingly, truncating mutations in KMT2D and AJUBA were associated with reduced protein but not mRNA abundance (Figure 1L). Most of the truncating variants in these two genes may escape NMD according to the NMD rules (Lindeboom et al., 2016) (Table S2). Thus, proteomic data were crucial to support the tumor suppressor role of these genes.
DNA methylation of 91 genes was associated with both reduced mRNA and protein abundance in tumors (Table S2). These genes included several putative tumor suppressor genes whose expression was reported to be regulated by promoter methylation in other cancer types, such as NEFM (Calmon et al., 2015), MGMT (Rivera et al., 2010), GLDC (Min et al., 2016), and CHFR (Brandes et al., 2005).
Proteomic Alterations Associated with Tumorigenesis and Prognosis
We compared 63 tumors vs paired NATs to identify HNSCC-associated alterations in transcripts (mRNA, miRNA, and circRNA), proteins, and phosphosites (Table S3). Here we focus on results from the analysis of proteins and phosphosites quantified in ≥50% of the pairs. For proteins, 3,355 (35%) were significantly increased and 3,163 (33%) were significantly decreased in tumors (adjusted p<0.01, Wilcoxon signed-rank test, Figure 2A). The 104 proteins increased by >2-fold were enriched in biological processes such as protein hydroxylation, leukocyte migration, cell chemotaxis, and angiogenesis, whereas the 488 decreased proteins were enriched in acute inflammatory response, platelet degranulation, muscle system process, and fatty acid metabolic process (adjusted p<0.01, Fisher’s exact test, Table S3). After controlling for epithelium content in a multivariate model, 63 out of the 104 remained significantly elevated by >2-fold (Figure S2A, Table S3), and over two-thirds showed above average abundance of all proteins (Figure S2B). Among the 63, 22 are secretable and could serve as putative salivary biomarkers, seven can be targeted by FDA-approved drugs, and one is a cancer/testis (C/T) antigen (Figure 2B, Table S3). Notably, KIT, CAMP, and other highly increased proteins such as DEFA3, DEFA1B, CRTAP, and CLCNKA had decreased mRNA in tumors (Figure 2C, Figure S2C). Elevated tumor expression was supported by data-independent acquisition (DIA) proteomics (Figure S2D. I). Immunohistochemistry (IHC) data from the Human Protein Atlas (HPA) provided further independent validation for a subset of proteins (Figure S2E).
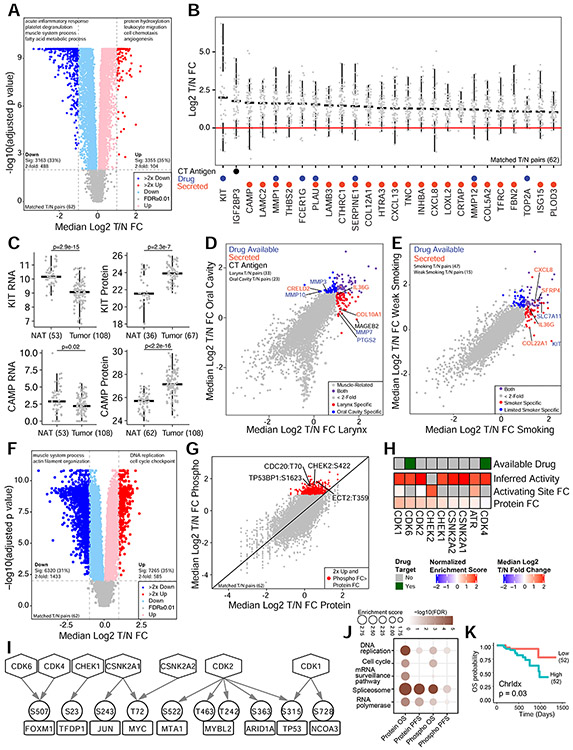
Figure 2. Proteomic alterations associated with tumorigenesis and prognosis.
(A) Protein abundance differences between tumors and NATs (Wilcoxon signed-rank test). Representative GO terms for 2-fold increased and decreased proteins are listed. (B) Abundance fold changes (FC) for selected highly elevated proteins annotated with potential clinical utilities. (C) Comparisons of RNA and protein levels for KIT and CAMP between tumors and NATs, Student’s t-test. (D) Comparison of protein changes in two anatomic sites. Dot colors indicate shared or site-specific elevations, and font colors indicate different types of clinical utilities. (E) Comparison of protein changes in tumors with strong and weak smoking evidence, colored as panel D. (F) Phosphosite abundance differences between tumors and NATs (Wilcoxon signed-rank test). Representative GO terms for proteins with 2-fold increased or decreased phosphosites are listed. (G) Comparison of abundance changes between phosphosites and their corresponding proteins. (H) Kinases with increased activity inferred from phosphorylation of its substrates (normalized enrichment score) or increased phosphorylation of its activating site. (I) Increased phosphorylation (circle) on transcription factor substrates (rectangle) of kinases (hexagon) with increased activity. All transcription factors had increased inferred activity from the RNA targets. (J) The common pathways enriched with proteins or phosphoproteins associated with OS or PFS (Fisher’s exact test). (K) Kaplan-Meier plot comparing OS for patients stratified by the median Chrldx score, logrank test. Numbers in parentheses represent the sample sizes for the involved groups. See also Figure S2 and Table S3.
Most proteins behaved similarly in the tumor vs NAT comparison whether derived from larynx or oral cavity (Figure 2D). However, 261 proteins, many muscle-related, were decreased specifically in oral cavity tumors, likely due to higher levels of muscle tissue in oral cavity NATs compared to larynx NATs. Several proteins with potential clinical utility were increased in a particular site (Figure 2D), including MAGEB2, PTGS2, MMP7, COL10A1, and IL36G in larynx (blue dots), and MMP3, MMP10, and CRELD2 in oral cavity (red dots). When grouped based on smoking evidence, tumors with strong smoking evidence showed specific increase of four secretable proteins (CXCL8, SFRP4, IL36G, COL22A1) and two targets of approved drugs (KIT, SLC7A11) (Figure 2E). The highly specific overexpression of KIT in tumors with strong smoking evidence explained the large variation of protein fold change observed for KIT across all tumors (Figure 2B).
For phosphosites, 7,265 (35%) were significantly increased and 6,320 (31%) were significantly decreased in tumors vs NATs (adjusted p<0.01, Wilcoxon signed-rank test, Figure 2F). Proteins with a phosphosite change >2-fold were enriched in biological processes such as DNA replication and cell cycle checkpoint, whereas proteins with a phosphosite change decreased by >2-fold were enriched in muscle system process and actin filament organization. Among the 559 phosphosites increased by >2-fold with stronger changes than in the corresponding protein (Figure 2G), only 8% had known functional annotations. Of these, 30% were involved in cell cycle regulation, including a site on the essential mitotic regulator CDC20 (Hein et al., 2017; Wang et al., 2015). An additional 12% were involved in cytoskeleton reorganization, including ECT2 T359, which contributes to tumor cell invasion (de Cárcer et al., 2017; Justilien et al., 2011). TP53BP1 S1623, which inhibits DNA repair, was also highly phosphorylated (Benada et al., 2015). Differential phosphosite analysis identified only one hyperphosphorylated activating site on a kinase, CHEK2 (Lovly et al., 2008). Kinase activities were also inferred based on the levels of substrate phosphorylation. A total of nine kinases had significantly increased activity, including targets of approved inhibitors (CDK4 and CDK6) (Figure 2H). Using predicted substrates for kinases (Linding et al., 2007), we identified 12 more kinases with increased activity in tumors, including CHUK and IKBKB, which are targets of approved inhibitors (Figure S2F). Increased kinase activity was supported by phosphorylation of corresponding transcription factor substrates and, in turn, increased transcription factor activity inferred from their mRNA targets (Figure 2I, Table S3).
Survival analysis identified 263 proteins and 173 phosphoproteins significantly associated with overall survival (OS), and 162 proteins and 164 phosphoproteins significantly associated with progression-free survival (PFS) (nominal p<0.01, Table S3). OXSR1 and GPALPP1 remained significant for both after multiple test adjustment, and 8 phosphoproteins remained significant for OS (Figure S2G-H, adjusted p<0.2). Strikingly, poor prognosis associated proteins/phosphoproteins were enriched in pathways reported in Figure 1I for SCNA drivers, including DNA replication, cell cycle, and RNA processing (Figure 2J), suggesting a potential relationship between SCNA and adverse clinical outcome. Indeed, tumors with higher chromosome instability scores (ChrIdx score) tended to have shorter OS (Figure 2K) and PFS (Figure S2I).
Mutually Exclusive FAT1 Truncating Mutations and 11q13.3 Amplification Converge to Protein-level Actin Dysregulation
FAT1 encodes an atypical cadherin and is one of the most frequently mutated genes in this cohort (Figure 1K) and the TCGA cohort. Truncating mutations account for >70% of all FAT1 mutations in both cohorts, in sharp contrast to other cancer types (Figure S3A). By integrating copy number data, we further divided FAT1 mutations into four groups (Figure S3B, Table S4). Compared to WT FAT1, samples with FAT1 biallelic truncations showed reduced FAT1 protein and mRNA levels (p=3.4e-6 and p=1.5e-3, Student’s t-test, Figure S3C-D). Since samples with other types of FAT1 mutations showed intermediate and more variable mRNA and protein levels, we excluded them from downstream analyses to focus on the most frequent mutation type with the strongest cis effects.
Mutual exclusivity was observed (p=6.0e-3, Fisher’s exact test) between FAT1 truncating mutation and 11q13.3 amplification (Figure 3A), the strongest focal SCNA in our cohort (Figure 1G). This was confirmed in TCGA HPVneg HNSCCs (p<0.001, Figure S3E). For all 9 protein coding genes in 11q13.3, amplification resulted in concordantly increased mRNA and protein abundance (Figure 3A).
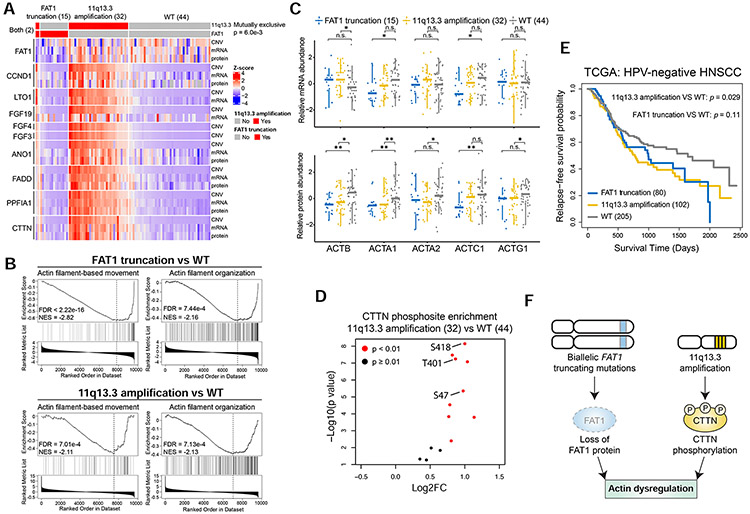
Figure 3. Mutually exclusive FAT1 truncating mutations and 11q13.3 amplification converge to protein-level actin dysregulation.
(A) Heatmap visualizing multi-omic profiles of FAT1 and the nine coding genes in 11q13.3. (B) GSEA plots for actin-related pathways in FAT1 truncation or 11q13.3 amplification vs WT comparisons. (C) Relative mRNA and protein abundance in the FAT1 truncation, 11q13.3 amplification, and WT groups for five actin isoforms. *p<0.05, **p<0.01, Student’s t-test. n.s., not significant. (D) CTTN phosphosite abundance differences between the 11q13.3 amplification and WT groups (Student’s t-test). (E) Relapse-free survival in HPVneg HNSCC TCGA patients with FAT1 truncation or 11q13.3 amplification compared to WT (logrank test). (F) Proposed model explaining the mutual exclusivity between FAT1 truncating mutations and 11q13.3 amplification. Numbers in parentheses represent the sample sizes for the involved groups. See also Figure S3 and Table S4.
Mutual exclusivity may arise when two aberrations are functionally equivalent (Ciriello et al., 2012). Pathway enrichment analysis using proteomic data showed downregulation of proteins involved in actin dynamics in both FAT1 truncated and 11q13.3 amplified groups compared to the WT group (adjusted p<0.05, gene set enrichment analysis [GSEA], Figure 3B). Despite varying mRNA abundance of actin genes between groups, protein abundance for five actin genes was higher in the WT group (p<0.05, Student’s t-test, Figure 3C). In particular, beta-actin (encoded by ACTB), a non-muscle actin implicated in cell motility, structure and integrity (Drazic et al., 2018), was significantly downregulated at the protein level in both FAT1 truncated and 11q13.3 amplified groups despite upregulated mRNA. This finding was verified using DIA proteomic data (Figure S3F). These data suggest that FAT1 truncation and 11q13.3 amplification converge on regulating actin dynamics at the protein level.
Depletion of FAT1 has been causally linked to dysregulated actin organization at the cell periphery, looser cell association, and abrogated cell polarity (Tanoue and Takeichi, 2004). Phosphoproteomic data showed significantly elevated levels of CTTN phosphosites in 11q13.3 amplified samples compared to WT (Figure 3D). The most elevated phosphosite was CTTN S418, reported to alter cell motility and cytoskeletal rearrangement to promote tumor progression (MacGrath and Koleske, 2012). Although small sample size and short follow-up time precluded prognostic association in our cohort, patients harboring FAT1 truncation or 11q13.3 amplification had worse survival than those without either alteration in TCGA HPVneg HNSCCs (Figure 3E). These results suggest that both FAT1 truncation and 11q13.3 amplification drive poor prognosis, possibly through a common mechanism of modulating actin dynamics (Figure 3F), which provides an explanation for the mutual exclusivity between these frequent genomic aberrations in HPVneg HNSCC.
Proteogenomic Delineation of the Rb Pathway
The most common genetic aberrations affected the cyclin D-CDK4/6-Rb pathway, including CDKN2A deletions (57%) and mutations (27%) and CCND1 amplifications (32%) (Figure 4A), all of which had comparable frequencies in the TCGA HPVneg HNSCCs. CDKN2A was hypermethylated in 13 tumors (12%), and hypermethylation was mutually exclusive with other loss of function alterations (Figures 4A and S4A-B, Table S4). Mutations in CDK6 (5%) and RB1 (2%) were also observed (Figure 4A).
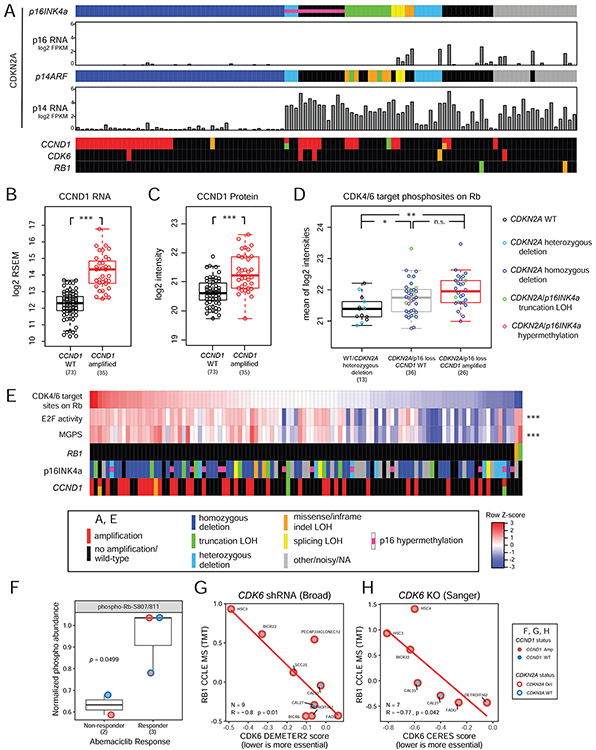
Figure 4. Proteogenomic delineation of the cyclin D-CDK4/6-Rb pathway.
(A) Genetic and epigenetic aberrations in pathway genes. Impact of CDKN2A aberrations for two major isoforms, p16INK4a (p16) and p14ARF (p14), on respective transcript mRNA levels are shown separately. LOH: loss of heterozygosity. (B-C) Cis-effects of CCND1 amplification on RNA (B) and protein abundance (C). ***p<1e-4, Wilcoxon rank sum test, n=108. (D) Comparison of Rb phosphorylation levels (average of all CDK4/6 target sites) among three tumor groups. *p<0.05. **p<0.001, Wilcoxon rank sum test. (E) Heatmap comparing Rb phosphorylation, E2F activity, and the mean of cell cycle regulated genes (MGPS), with genomic aberrations annotated. ***p<1e-4, Pearson’s correlation with Rb phosphorylation. (F) Comparison of phospho-Rb-S807/811 in non-responsive and responsive HPVneg HNSCC PDX models to abemaciclib, Student’s t-test. (G-H) Associations between mass spectrometry (MS)-based Rb abundance and CDK6 essentiality scores derived from shRNA (DEMETER2, G) or CRISPR (CERES, H)-based genetic perturbations, respectively, in seven HPVneg HNSCC cell lines. R: Pearson’s correlation coefficient. Numbers in parentheses represent the sample sizes for the involved groups. See also Figure S4 and Table S4.
Homozygous deletion of CDKN2A led to loss of mRNA expression of both major isoforms, p16INK4a (p16) and p14ARF (p14), but other aberrations such as promoter hypermethylation primarily affected p16 (Figure 4A and S4B). Five CDKN2A mutations associated with loss of heterozygosity (LOH) altered p16 but not p14, and an additional six mutations resulted in truncation of p16 but only missense or in-frame indel changes in p14 (Figure S4A). Altogether, 68 tumors (63%) had genetic or epigenetic events predicted to disrupt p16 expression (tumors with homozygous deletion, p16 hypermethylation, or p16 truncation LOH in Figure 4A), which may explain widespread missing proteomic measurements for p16. CCND1 amplification was associated with increased levels of CCND1 RNA and protein in general, but not in all tumors as evidenced by overlap between the distributions of the amplified and WT groups (Figure 4B-C). To assess the impact of CDKN2A aberrations and CCND1 amplifications on CDK4/6-Rb signaling, we defined three groups: samples that were WT for pathway genes, including those with CDKN2A heterozygous deletion (n = 13), samples with p16 aberration affecting RNA expression but no CCND1 amplification (n = 36), and samples with both p16 aberration and CCND1 amplification (n = 26). Mean phosphorylation levels of the CDK4/6 target sites on Rb protein (Rb phosphosite score) were significantly higher in the 2nd and 3rd groups than in the 1st group (Figure 4D). However, many of the samples from the 2nd and 3rd groups had Rb phosphosite scores that were well within the range of the 1st group. The observation that CDKN2A and CCND1 aberrations did not always result in increased CCND1 protein and CDK4/6 activity was also seen in data from TCGA HPVneg HNSCCs (Figure S4C-E).
Rb phosphosite scores were significantly correlated with both E2F activity scores and multi-gene proliferation scores (MGPS) inferred from the RNA data (Pearson’s correlation = 0.50 and 0.47, p=4e-8 and 4e-7, respectively, Figure 4E). However, several samples with low Rb phosphosite scores had high E2F activity and MGPS, indicating cell cycle activation through other mechanisms besides CDK4/6 mediated phosphorylation. For example, the two samples with the lowest Rb phosphosite scores had high E2F activity and MGPS. These samples harbored RB1 mutations, potentially bypassing the need for CDK4/6 to phosphorylate Rb. While nearly all samples with high cell cycle activity had CDKN2A or CCND1 aberrations, a number of samples with these aberrations had low cell cycle activity. These observations suggest that Rb status is an effective and necessary indicator of CDK4/6-dependent cell cycle activity, which cannot be accurately predicted using genomic or transcriptomic markers.
To test the clinical relevance of Rb for CDK4/6 targeting, we analyzed data from HPVneg HNSCC PDXs treated with abemaciclib (Karamboulas et al., 2018), a CDK4/6 inhibitor in phase II clinical trials for HNSCC. While CCND1 and/or CDKN2A status did not separate response to abemaciclib, treatment responsive PDXs had elevated phospho-Rb-S807/811 signal (p=0.05, Student’s t-test) (Figure 4F). Moreover, CDK6 dependency was examined in HPVneg HNSCC cell lines from two independent genetic perturbation screens with associated molecular profiles from the cancer cell line encyclopedia (CCLE) (Behan et al., 2019; McFarland et al., 2018; Nusinow et al., 2020). Cell lines with higher levels of Rb protein, which was highly correlated with the Rb phosphosite score in our dataset (Spearman rho = 0.89, p<0.0001), were more sensitive to genetic depletion of CDK6 (p<0.05, Pearson’s correlation, Figure 4G-H). Taken together, these results support the hypothesis that phospho- or total Rb may serve as markers for CDK4/6 inhibitors in HPVneg HNSCC.
Two Modes of EGFR Activation
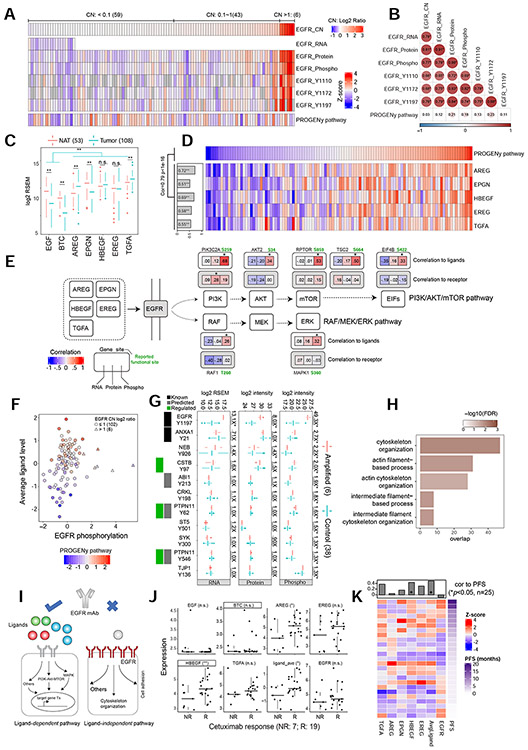
We analyzed our data to gain insights into the poor response of HNSCC patients to EGFR inhibition. EGFR mutations were identified in only three tumors, and none were hotspot mutations. Moreover, no samples harbored the EGFR VIII fusion variant. However, 49 samples showed EGFR amplification (CN log2 ratio > 0.1), and six had high amplification (CN log2 ratio > 1) (Figure 5A). EGFR CN was significantly associated with mRNA and protein abundance, overall phosphorylation level of EGFR, and phosphorylation levels of activation sites Y1110, Y1172, and Y1197 (Figure 5A-B). Thus, EGFR amplification is associated with EGFR activation.
Figure 5. Proteogenomic characterization of EGFR ligand-dependent and -independent pathways.
(A) Heatmap comparing EGFR multi-omics profiles and the inferred PROGENy EGFR pathway activity and (B) their Pearson’s correlation coefficients. *p<0.01. (C) EGFR ligand mRNA abundance in tumors and NATs. **p < 0.001, Student’s t-test. (D) Pearson’s correlation between EGFR pathway activity and mRNA abundance of individual ligands. (E) For genes in the ligand-dependent pathways downstream of EGFR, the Pearson’s correlations between each omics feature and average ligand abundance (correlation to ligands) or EGFR abundance (correlation to receptor) are shown. *p < 0.01. Reported functional sites are colored green. (F) Relationship between PROGENy EGFR pathway activity (color gradients) and average ligand abundance or EGFR phosphorylation level. The six triangles represent samples with the high EGFR amplification. (G) Abundance comparisons between amplified samples and controls for 11 tyrosine phosphosites and cognate mRNA and proteins. Green box indicates known regulation by EGFR, black and gray indicate known and predicted EGFR substrates, respectively. Numbers on the side indicate fold changes. *p<0.01, Student’s t-test. (H) GO biological processes enriched with proteins with EGFR CN-associated phosphorylation (Fisher’s exact test). (I) Diagram depicting two modes of EGFR activation with implications for EGFR mAb therapies. (J) Comparisons between non-responsive (NR) and responsive (R) HPVneg HNSCC PDX models to Cetuximab treatment for average ligand (ligand_ave), individual ligands, and EGFR mRNA abundance. *p<0.05; **p<0.01 Student’s t-test. (K) Spearman’s correlations between mRNA abundance and PFS using data from a clinical trial testing panitumumab in HNSCC patients. Numbers in parentheses represent the sample sizes for the involved groups. See also Figure S5 and Table S5.
We inferred EGFR pathway activity based on mRNA expression data using PROGENy (Schubert et al., 2018). Unexpectedly, the inferred pathway activity showed no or weak correlations with EGFR alterations (Pearson’s correlation = 0.03 - 0.23, Figure 5A-B). In contrast, except for the two with very low mRNA abundance in tumors (Figure 5C), all EGFR ligands (Singh et al., 2016) showed strong correlations with inferred pathway activity (Pearson’s correlation = 0.51 – 0.72, Figure 5D). These observations were fully recapitulated in data from TCGA HPVneg HNSCCs (Figure S5A-D). Moreover, phosphoproteomics quantified several phosphosites on proteins involved in the PI3K/Akt/mTOR and the RAF/MEK/ERK pathways, two primary downstream pathways of EGFR (Wee and Wang, 2017). These phosphosites, including several key functional sites such as PIK3C2A S259 (Margaria et al., 2019), RPTOR S859 (Wang et al., 2009) and EIF4B S422 (Shahbazian et al., 2006), showed strong correlations with EGFR ligands, independent of the mRNA and protein expression of the host genes (Figure 5E). Conversely, none were significantly correlated with EGFR protein abundance. Thus, both transcriptomic and phosphoproteomic data suggest that the EGFR ligands, instead of the receptor, are the rate-limiting factors for EGFR pathway activity.
To identify signaling changes associated with EGFR amplification, which did not result in increased EGFR pathway activity but seemed to enhance EGFR phosphorylation in a ligand-independent manner (Figure 5F), we compared phosphoproteomic profiles between the six samples with high EGFR amplification and 38 other samples with similar chromosomal instability (ChrIdx score >3) (Figure S5E). This analysis identified 297 phosphosites with significantly higher phosphorylation in the EGFR amplification group (p<0.01, Student’s t-test), and 212 phosphosites showed stronger changes than at the RNA or protein level, suggesting these are bona fide phosphorylation changes and not due to differential gene expression or cell type composition (Table S5). The 11 tyrosine sites with significantly increased phosphorylation (Figure 5G) included five known or predicted EGFR substrates (EGFR Y1197, ANXA1 Y21, PTPN11 Y546, PTPN11 Y62, and ABI1 Y213). Additionally, PTPN11 Y546, PTPN11 Y62, and CSTB Y97 are reported to be regulated by EGFR in the PhosphoSitePlus database. Proteins harboring the 212 sites were enriched in cytoskeleton organization, actin filament, and intermediate filament junction related pathways (adjusted p<0.01, Fisher’s exact test, Figure 5H), suggesting a role for EGFR in modulating intercellular junctions and cell motility, as previously reported (Klymkowsky and Parr, 1995; Stallaert et al., 2018).
Since our data suggest that EGFR amplification activates EGFR in a ligand-independent manner, and EGFR mAbs function primarily by binding to the EGFR extracellular domain to prevent ligand-induced pathway activity (Harding and Burtness, 2005; Messersmith and Hidalgo, 2007), EGFR ligand abundance, rather than EGFR amplification or overexpression, should be used to select HNSCC patients for treatment with anti-EGFR mAbs (Figure 5I). Indeed, utilizing data from an HNSCC PDX study with treatment response to an EGFR mAb, cetuximab (Klinghammer et al., 2017), we found that EGFR ligands, not the receptor, showed significantly higher expression in responders than non-responders (Figure 5J). Furthermore, in a clinical trial testing another EGFR mAb, panitumumab, in HNSCC patients (Siano et al., 2018), we found that EGFR ligand abundance, but not receptor abundance, significantly correlated with PFS (Figure 5K).
Immuno-proteogenomic Analysis Reveals Immunosuppressive SCNAs
To gain a deeper understanding of immune evasion and resistance to PD-1 inhibitors in HNSCC, we performed an immuno-proteogenomic analysis. Tumors showed a wide range of immune cell infiltration levels as consistently quantified by ESTIMATE immune score (Yoshihara et al., 2013), CD3 IHC staining, and CD3 proteomic data (Figure 6A). Higher immune cell infiltration was not linked to any anatomic sites but was associated with lower clinical stage, less smoking, and better prognosis (Figure 6B, Figure S6A-B). In silico deconvolution using xCell (Aran et al., 2017) showed that both cytotoxic immune cells (e.g., CD8 T cells and M1 macrophages) and immunosuppressive cells (e.g., Tregs and M2 macrophages) were enriched in tumors with high levels of immune cell infiltration. In these immune-hot tumors, both cytotoxic immune enzymes and immunosuppressive proteins were overexpressed at the protein and/or mRNA levels (Figure 6A), with high correlations observed across immune inhibitory genes (Figure 6C). These data may explain the moderate response rate to single-agent pembrolizumab treatment in PD-L1 positive HNSCCs (Seiwert et al., 2016) and suggest combinatorial checkpoint inhibition as a logical proposition to increase treatment efficacy.
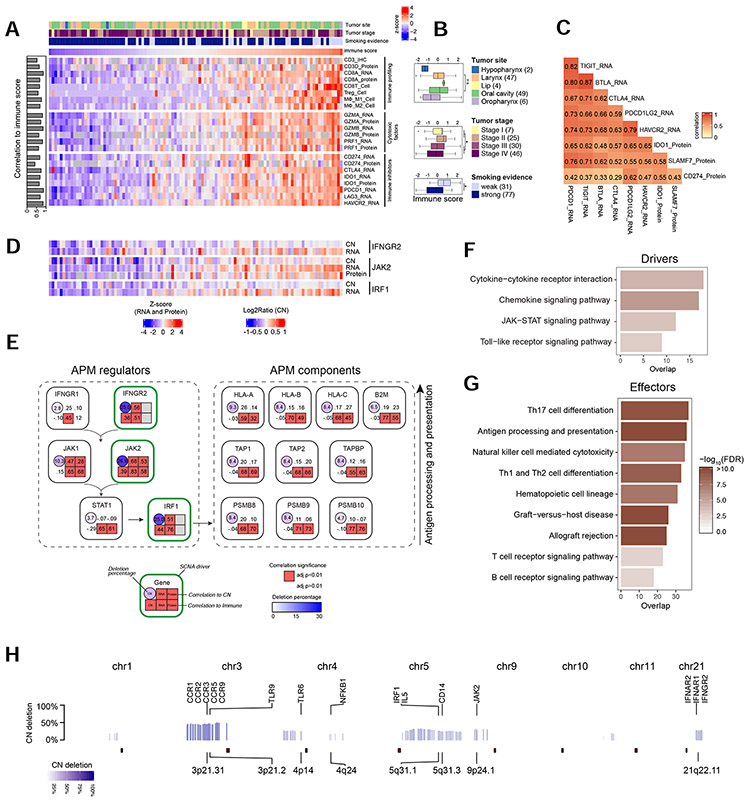
Figure 6. Immuno-proteogenomic analysis reveals immunosuppressive SCNA drivers.
(A) Pearson’s correlations between ESTIMATE immune score and proteogenomic profiles of immune infiltration, cytotoxic factors, and immune inhibitors. (B) Comparisons of the immune score across clinical attributes (*p<0.01, Student’s t-test). (C) Correlations among immune checkpoints and suppressors. (D) Copy number (CN), mRNA abundance, and protein abundance of three SCNA driver genes. (E) Diagram showing the information flow from antigen processing and presenting machinery (APM) regulators to APM components. The top row for each gene shows the cis-effect of CN on RNA and protein abundance, and the bottom row shows the correlation between immune score and each omics type. (F) Pathways enriched for immune-associated genes whose expression was suppressed by SCNA (i.e., immunosuppressive SCNA drivers). (G) Pathways enriched for immune-associated genes whose expression was not associated with SCNA (i.e., effectors of the immune-suppressive CN deletions). (H) The distribution of immunosuppressive SCNAs across the genome. Selected immune genes are highlighted. Numbers in parentheses represent the sample sizes for the involved groups. See also Figure S6 and Table S6.
Next, we sought to identify tumor intrinsic determinants of low immune infiltration in immune-cold tumors. We observed negative correlations between immune cell infiltration and either tumor mutational burden or protein abundance of quantified C/T antigens (Figure S6C). Moreover, proteomics-supported neoantigens (Wen et al., 2020) (Table S6) showed little correlation to immune cell infiltration (Figure S6C). Thus, the low immune infiltration was not driven by a lack of tumor antigen sources. Instead, we found significantly reduced expression of multiple components and regulators of the antigen presentation machinery (APM) pathway at both mRNA and protein levels in immune-cold tumors (Figure S6D-E). Few mutations in APM genes and their regulators were identified in our cohort (Figure S6F), but frequent somatic CN deletions (>25%) were found in APM regulators IFNGR2, JAK2, and IRF1 (Figure S6G). CN levels of these genes correlated strongly with mRNA and protein levels when data were available, and these genes further showed strong correlation with immune infiltration at all molecular levels (Figure 6D-E), suggesting a causal contribution of these gene deletions to APM deficiency and low immune infiltration (i.e., ‘SCNA drivers’). In comparison, APM components showed significant correlation with immune infiltration at the gene expression, but not CN level, indicating that the changes occurred in trans (i.e., “SCNA effectors”). Consistent with the model depicted in Figure 6E, gene regulatory network analysis using VIPER (Alvarez et al., 2016) identified STAT1 as the central transcription factor regulating immune activity.
Expanding the APM-focused analysis to a genome-wide search identified 294 putative SCNA drivers (Table S6) and 2,058 putative SCNA effectors. SCNA drivers were enriched in cytokine/chemokine receptor, JAK-STAT, and TLR pathways, all of which regulate immunogenicity or immune response within tumor cells (Figures 6F and S6H). By contrast, SCNA effectors were mostly involved in immune cytotoxicity, especially adaptive immune cell activation and function (Figure 6G). SCNA drivers were distributed widely across the genome (Figure 6H), including the most frequently deleted 3p region, which encodes chemokine/cytokine receptors and TLRs, and 9p24.1, which encodes JAK2, a key component of the JAK-STAT pathway. Genes deleted in 9p24.1 also include CD274 that encodes PD-L1, suggesting that PD-L1-mediated immune checkpoint is not needed in immune-cold HNSCCs. Our observations for both immune-cold and immune-hot tumors were supported by our reanalysis of transcriptomic data from TCGA HPVneg HNSCCs (Figure S6I-L), albeit at a lower sensitivity for detecting the driver CN pathway signals, suggesting that integrating proteomic data helpedprioritize driver CN events involved in immunogenicity.
Multi-omics Subtypes and Targeted Therapies
By integrating CN, RNA, miRNA, protein, and phosphopeptide data, an unsupervised clustering analysis grouped tumors into three clusters. Clusters I, II, and III were significantly associated with previously established classical, basal, and mesenchymal RNA subtypes (Walter et al., 2013), respectively (p<0.05, Fisher’s exact test, Figure 7A-B).
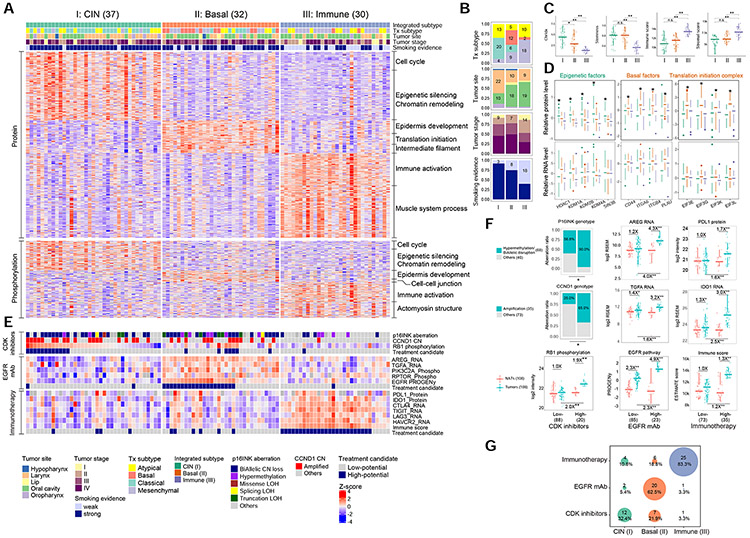
Figure 7. Integrated multi-omics subtypes and subtype-specific targeted therapies.
(A) Proteomic and phosphoproteomic profiles of the signature proteins and the enriched biological processes of the three integrated subtypes. (B) Sample distribution across different clinical attributes. (C) Comparisons of the three subtypes for four molecular phenotypes. *p<0.01, **p<0.001, Student’s t-test. (D) mRNA and protein levels of protein-specific gene signatures related to epigenetic, basal, and translation initiation factors for different subtypes. Each feature was tested for its differential abundance between the given subtype and the other two subtypes. *: adjusted p<0.01 for both comparisons, Student’s t-test. (E) Heatmap visualizing proteogenomic measurements of the suggested biomarkers for targeted therapies and candidacy for treatment with CDK inhibitors (upper), EGFR mAb (middle), and immune checkpoint blockade (bottom). (F) Comparisons of the proposed biomarkers between high-potential and low-potential tumors, and between each group of tumors and NATs. Numbers at the top denote fold changes. *p<0.01 **p<0.001. Student’s t-test. (G) The proportions of high-potential candidates for each target therapy in the three subtypes. Numbers in parentheses represent the sample sizes for the involved groups. See also Figure S7 and Table S7.
The classical, basal, and mesenchymal RNA subtypes have been characterized by overexpression of genes related to cell proliferation, epidermal development, and stromal infiltration, respectively (Walter et al., 2013). Proteomic and phosphoproteomic data not only confirmed these features but also provided new insights (Figures 7A and S7A-C, Table S7). Cluster I was associated with the larynx, strong smoking (Figure 7B, p<0.01, Fisher’s exact test), and high chromosome instability (CIN) (Figures 7C and S7D). Proteomic data, but not RNA-seq data, showed increased levels of multiple epigenetic regulators in this cluster (Figure 7D), supporting a suggested linkage between aberrant epigenetic activities and smoking and CIN in HNSCC (Ghantous et al., 2018; Papillon-Cavanagh et al., 2017). This cluster showed the worst prognosis (Figure S7E-F), which is consistent with the observation for the classical RNA subtype of HNSCC (Keck et al., 2015). Cluster II showed protein-level elevation of several basal factors (Figure 7D). Moreover, protein- and phosphorylation-specific elevation of eukaryotic translation initiation (EIF) complex members indicated higher translational activity in these tumors (Figures 7D and S7G). Both clusters I and II were associated with higher stemness (Figure 7C), likely due to aberrant epigenetic activity and basal-like factor activation, respectively (Malta et al., 2018). Cluster III was enriched with tumors with weak smoking evidence (p<0.01, Fisher’s exact test, Figure 7B). It was also associated with higher immune scores and, to a lesser degree, higher stromal scores (Figure 7C). Consistent with our observation, a more recent HNSCC subtyping study annotated the mesenchymal RNA subtype as “inflamed” (Keck et al., 2015). Notably, the atypical RNA subtype, which was enriched with HPVpos samples in the original study, collapsed to clusters I and III (Figure 7B), which was supported by their associations with relatively higher stemness cores and higher immune/stromal scores, respectively (Figure S7H). Taken together, our multi-omics subtyping identified three subtypes of HPVneg HNSCCs, which we named CIN, Basal, and Immune, respectively.
To examine the utility of these subtypes in guiding treatment selection, we evaluated their associations with our proposed biomarkers for targeted therapies in HNSCC (Figure 7E). The CIN subtype was associated with frequent genetic aberrations of CCND1 and CDKN2A and high CDK4/6 activity as indicated by Rb hyperphosphorylation, suggesting potential response to CDK4/6 inhibitors. The Basal subtype was characterized by high EGFR ligand expression (AREG and TGFA) and high EGFR pathway activity, suggesting potential response to EGFR mAb. The Immune subtype showed high expression of multiple immune checkpoint proteins, and thus may benefit from checkpoint blockade. For each treatment option, the high-potential tumors showed significantly higher levels of the biomarkers than the low-potential tumors and the matched NATs (p<0.01, Student’s t-test), whereas the latter two showed no or a less significant difference (Figure 7F). In total, 32% of the CIN tumors, 62% of the Basal tumors, and 83% of the Immune tumors had high potential for treatment with CDK inhibitors, EGFR mAb, and immunotherapy, respectively (Figure 7G).
Discussion
With several targeted therapies approved for the treatment of HNSCC and many more in development, the identification of accurate biomarkers to guide treatment selection is a major research priority (Santuray et al., 2018). Our study demonstrates the promise of proteogenomics in addressing this challenge. For EGFR targeted therapy, it is well acknowledged that EGFR amplification or overexpression cannot be used to predict response to EGFR mAbs in HNSCC (Ang et al., 2014; Burtness et al., 2005; Crombet et al., 2004; Psyrri et al., 2014). Our data suggest a new strategy of using EGFR ligand abundance to stratify patients for effective treatment with EGFR mAb. In addition, some tumors with EGFR ligand overexpression also harbor CCND1 and CDKN2A aberrations, which may render them resistant to anti-EGFR mAb monotherapy. Tumors with high EGFR amplification do not necessarily have high levels of EGFR ligands and may not respond to EGFR mAbs. However, these tumors show strong EGFR phosphorylation and thus could respond to small-molecule EGFR tyrosine kinase inhibitors (TKIs). Consistent with this hypothesis, the combination of p16-negativity and EGFR amplification identified HNSCC patients that achieved a clinically meaningful benefit from afatinib, an EGFR TKI, in a phase III trial (Santuray et al., 2018).
For immunotherapy, immune-hot tumors concordantly overexpress multiple checkpoints and other immunosuppressive genes, which may partially explain the moderate response rate in PD-L1 positive HNSCCs to a single-agent pembrolizumab treatment (Seiwert et al., 2016). Moreover, there was no clinical improvement from combining durvalumab (PD-L1 antibody) and tremelimumab (CTLA-4 antibody) in unselected patients with relapsed/metastatic HNSCC in a phase III trial (Ferris et al., 2020). Profiling of multiple immune checkpoint proteins may allow more precise personalization of combination immunotherapy regimens, potentially leading to improved outcomes through accurate patient selection.
Multiple clinical trials are evaluating CDK4/6 inhibitors in HNSCC, but there are no established biomarkers to guide patient selection (Adkins et al., 2019). Rb phosphorylation status could be considered as a biomarker together with CCND1/CDKN2A genomic aberrations for future clinical trials of CDK4/6 inhibitors, whereas these genomic markers alone, or transcriptomic markers of E2F activity, may not accurately reflect CDK4/6 activity.
We identified new targets for therapeutic development, such as KIT, FCER1G, PLAU, SERPINE1, TOP2A, several MMPs, and several cell cycle and DNA damage-related kinases. In addition, multiple C/T antigens are recurrently overexpressed in tumors compared to NATs, including IGF2BP3, MAGEB2, KIF2C, CEP55, and NUF2 (Figure 2, Figure S6), and proteomics-supported neoantigens were predicted for 20.4% of the patients (Table S6). Both C/T antigens and neoantigens are promising immunotherapy targets.
We also generated new knowledge concerning HNSCC biology. Proteomics data prioritized copy number drivers and highlighted an oncogenic role for RNA processing factors in HNSCC tumorigenesis. Widespread deletion of immune modulatory genes may account for loss of immunogenicity and low immune infiltration in HNSCC. FAT1 was among the most frequently mutated genes in HNSCC. Previous studies have linked FAT1 mutations to the WNT and HIPPO pathways (Ciriello et al., 2012; Martin et al., 2018) or apoptosis (Kranz and Boutros, 2014), but none of these theories were supported by our data (Figure S3G). Instead, proteomic investigation of the mutually exclusive relationship between FAT1 truncating mutations and 11q13.3 amplifications revealed their functional convergence on dysregulated actin dynamics, which may underlie poor prognosis of tumors with these genetic aberrations.
In summary, this study extends our biological understanding of HPVneg HNSCC and generates therapeutic hypotheses that may serve as the basis for future preclinical studies and clinical trials toward molecularly guided precision treatment of this aggressive cancer type. Meanwhile, we have made the primary and processed datasets available in publicly accessible data repositories and portals, which will allow full investigation of this extensively characterized cohort by both the HNSCC and broader scientific communities. We also expect wide application of the demonstrated proteogenomics framework to future studies of HNSCC and other cancer types.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bing Zhang (bing.zhang@bcm.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Raw proteomics data files are hosted by the CPTAC Data Portal and can be accessed at: https://proteomics.cancer.gov/data-portal. Genomic and transcriptomic data files can be accessed via the Genomic Data Commons (GDC) Data Portal: https://portal.gdc.cancer.gov. Processed data utilized for this publication can be accessed via LinkedOmics: http://www.linkedomics.org.
Several customized coding software packages were generated as part of this study and have been referenced in the corresponding STAR Methods section and listed with links to the coding script in the Key Resources Table: software codes generated by the Cieslik laboratory for genomic analyses (CNVEX), by the Nesvizhskii laboratory for proteomic data processing (Philosopher and TMT-Integrator), and by the Zhang lab for data processing and neoantigen detection (NeoFlow and PepQuery).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid | Alfa Aesar | Catalog: J63218 |
| Acetonitrile, Optima LC/MS | Fisher Chemical | A955-4 |
| Ammonium Hydroxide solution | Sigma | Catalog: 338818 |
| Aprotinin | Sigma | Catalog: A6103 |
| CD3 antibody (polyclonal) | Agilent - Dako | #A0452 |
| DAB | Agilent - Dako | #K3468 |
| Dithiothreitol | Thermo Fisher Scientific | Catalog: 20291 |
| Envision+ System HRP labelled polymer, anti-Rabbit | Agilent - Dako | #K4002 |
| Ethylenediaminetetraacetic acid | Sigma | Catalog: E7889 |
| Formic acid | Fisher Chemical | Catalog: A117-50 |
| Hydroxylamine solution | Aldrich | Catalog: 467804 |
| Iodoacetamide | Thermo Fisher Scientific | Catalog: A3221 |
| Iron (III) chloride | Sigma | Catalog:451649 |
| Leupeptin | Roche | Catalog: 11017101001 |
| Lysyl endopeptidase, aass spectrometry grade | Wako Chemicals | Catalog: 125-05061 |
| Ni-NTA agarose beads | QIAGEN | Catalog: 30410 |
| PUGNAc | Sigma | Catalog: A7229 |
| Phenylmethylsulfonyl fluoride | Sigma | Catalog: 93482 |
| Phosphatase Inhibitor Cocktail 2 | Sigma | Catalog: P5726 |
| Phosphatase Inhibitor Cocktail 3 | Sigma | Catalog: P0044 |
| Reversed-phase C18 SepPak | Waters | Catalog: WAT054925 |
| Sequencing grade modified trypsin | Promega | Catalog: V511X |
| Sodium chloride | Santa Cruz Biotechnology | Catalog: sc-295833 |
| Sodium fluoride | Sigma | Catalog: S7920 |
| TMT11-131C label reagent | Thermo Fisher Scientific | Catalog: A34807 |
| Tandem mass tags – 10plex | Thermo Fisher Scientific | Catalog: 90406 |
| Trifluoroacetic acid | Sigma | Catalog: 302031 |
| Tris(hydroxymethyl)amino methane | Invitrogen | Catalog: AM9855G |
| Urea | Sigma | Catalog: U0631 |
| Water, Optima LC/MS | Fisher Chemical | W6-4 |
| Critical Commercial Assays | ||
| TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold | Illumina | Catalog: RS-122-2301 |
| Infinium MethylationEPIC Kit | Illumina | Catalog: WG-317-1003 |
| Nextera DNA Exosome Kit | Illumina | Catalog: 20020617 |
| KAPA Hyper Prep Kit, PCR-free | Roche | Catalog: 07962371001 |
| BCA Protein Assay Kit | ThermoFisher Scientific | Catalog: 23225 |
| Software and Algorithms | ||
| Software | Source | Identifier (i.e. links) |
| Bowtie (v1.1.1) | (Langmead et al., 2009) | https://sourceforge.net/projects/bowtie-bio/files/bowtie/ |
| Bowtie2 (v2.3.3) | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| BWA (v0.7.17-r1188) | (Li and Durbin, 2009) | http://bio-bwa.sourceforge.net/ |
| cBioPortal | (Cerami et al., 2012; Gao et al., 2013) | https://www.cbioportal.org |
| CIRI (v2.0.6) | (Gao et al., 2015) | https://sourceforge.net/projects/ciri/ |
| CNVEX | Marcin Cieslik Lab | https://github.com/mctp/cnvex |
| Customprodbj | (Wang and Zhang, 2013) | https://github.com/bzhanglab/customprodbj |
| EricScript v0.5.5 | (Benelli et al., 2012) | https://sites.google.com/site/bioericscript/ |
| ESTIMATE | (Yoshihara et al., 2013) | https://bioinformatics.mdanderson.org/public-software/estimate/ |
| germlinewrapper v1.1 | Li Ding Lab | https://github.com/ding-lab/germlinewrapper |
| GISTIC2 | (Mermel et al., 2011) | https://www.genepattern.org/modules/docs/GISTIC_2.0 |
| GSVA | (Hänzelmann et al., 2013) | https://bioconductor.org/packages/release/bioc/html/GSVA.html |
| lmerTest | (Kuznetsova et al., 2017) | https://cran.r-project.org/web/packages/lmerTest/index.html |
| INTEGRATE v0.2.6 | (Zhang et al., 2016) | https://sourceforge.net/projects/integrate-fusion/ |
| iProFUN | (Song et al., 2019) | https://github.com/WangLab-MSSM/iProFun |
| LinkedOmics | (Vasaikar et al., 2018) | http://linkedomics.org |
| Manta v1.6.0 | (Chen et al., 2016) | https://github.com/Illumina/manta |
| MoonlightR | (Colaprico et al., 2020) | http://bioconductor.org/packages/MoonlightR/ |
| MSFragger-20190628 | (Kong et al., 2017) | http://msfragger.nesvilab.org/ |
| MS-GF+ | (Kim and Pevzner, 2014) | https://github.com/MSGFPlus/msgfplus/ |
| MuTect v1.1.7 | (Cibulskis et al., 2013) | https://github.com/broadinstitute/mutect |
| NeoFlow | (Wen et al., 2020) | https://github.com/bzhanglab/neoflow |
| NetworKIN | (Linding et al., 2008) | https://networkin.info |
| OmicsEV | Bing Zhang lab | https://github.com/bzhanglab/OmicsEV |
| OmicsOne | Hui Zhang Lab | https://github.com/huizhanglab-jhu/OmicsOne |
| PepQuery | (Wen et al., 2019) | http://pepquery.org/ |
| PeptideProphet | (Keller et al., 2002) | http://tools.proteomecenter.org/wiki/index.php?title=Main_Page |
| PDV | (Li et al., 2019) | https://github.com/wenbostar/PDV |
| Philosopher-v1.6.0 | Alexey Nesvizhskii lab | https://philosopher.nesvilab.org/ |
| Pindel v0.2.5 | (Ye et al., 2009) | https://github.com/genome/pindel |
| ProteinProphet | (Nesvizhskii et al., 2003) | http://tools.proteomecenter.org/wiki/index.php?title=Main_Page |
| PTMProphet | (Shteynberg et al., 2019) | http://tools.proteomecenter.org/wiki/index.php?title=Main_Page |
| QuickMIRSeq | (Zhao et al., 2017) | https://sourceforge.net/projects/quickmirseq/ |
| RSEM (v1.3.1) | (Li and Dewey, 2011) | https://deweylab.github.io/RSEM/ |
| Samtools (V1.1.0) | (Li et al., 2009) | https://github.com/samtools/samtools |
| SignatureAnalyzer | (Kim et al., 2016) | https://software.broadinstitute.org/cancer/cga/msp |
| somaticwrapper v1.5 | Li Ding Lab | https://github.com/ding-lab/somaticwrapper |
| STAR-Fusion v1.5.0 | (Haas et al., 2019) | https://github.com/STAR-Fusion/STAR-Fusion |
| Strelka v2.9.2 | (Kim et al., 2018) | https://github.com/Illumina/strelka |
| Sumer | (Savage et al., 2019) | https://github.com/bzhanglab/sumer |
| TCGAbiolinks | (Colaprico et al., 2016) | http://bioconductor.org/packages/TCGAbiolinks/ |
| TMT-Integrator-v1.0.9 | Alexey Nesvizhskii lab | http://tmt-integrator.nesvilab.org/ |
| VarScan v2.3.8 | (Koboldt et al., 2012) | https://dkoboldt.github.io/varscan/ |
| VIPER | (Alvarez et al., 2016) | http://califano.c2b2.columbia.edu/viper |
| VirusScan | (Cao et al., 2016) | https://github.com/ding-lab/VirusScan/tree/simplified |
| WebgestaltR | (Liao et al., 2019) | http://www.webgestalt.org/ |
| xCell | (Aran et al., 2017) | http://xcell.ucsf.edu/ |
| Deposited Data | ||
| cBioPortal | (Cerami et al., 2012) | https://www.cbioportal.org/ |
| CTDatabase | (Almeida et al., 2009) | http://www.cta.lncc.br |
| DoRothEA | (Garcia-Alonso et al., 2019) | https://github.com/saezlab/DoRothEA |
| Human Protein Atlas | (Uhlén et al., 2015) | https://www.proteinatlas.org |
| MSigDBv7.0 Hallmark gene sets | (Liberzon et al., 2015) | https://www.gsea-msigdb.org |
| PhosphoSitePlus | (Hornbeck et al., 2015) | https://www.phosphosite.org |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Specimens and Clinical Data
Tumor and germline blood samples from 110 qualified cases were collected from 6 tissue source sites in strict accordance to the CPTAC-3 protocol. All patients provided written informed consent. Institutional review boards at tissue source sites reviewed protocols and consent documentation adhering to the CPTAC guidelines. Normal adjacent tissues were collected from 83 cases. This study contained predominantly males (87%) and the cases were collected from 7 different countries. Histopathologically-defined squamous cell carcinomas were considered for analysis, with an age range of 23-81. A complete set of clinical data were obtained from the tissue source sites and reviewed for correctness and completeness.
Sample Processing
The CPTAC Biospecimen Core Resource (BCR) at the Pathology and Biorepository Core of the Van Andel Research Institute in Grand Rapids, Michigan manufactured and distributed biospecimen kits to the Tissue Source Sites (TSS) located in the US, Europe, and Asia. Each kit contains a set of pre-manufactured labels unique for each specimen respective to TSS location, disease, and sample type that is used to track the specimens through the BCR to the CPTAC proteomic and genomic characterization centers.
Tissue specimens averaging 200 mg were snap-frozen by the TSS within a 30-minute cold ischemic time (CIT) (CIT average = 13 minutes) and an adjacent segment was formalin-fixed paraffin-embedded (FFPE) and H&E stained by the TSS for quality assessment to meet the CPTAC HNSCC requirements. Routinely, several tissue segments for each case were collected. Tissues were flash frozen in liquid nitrogen (LN2) then transferred to a LN2 freezer for storage until approval for shipment to the BCR.
Specimens were shipped using a cryoport that maintained an average temperature of under −140°C to the BCR with a time and temperature tracker to monitor the shipment. Receipt of specimens at the BCR included a physical inspection and review of the time and temperature tracker data for specimen integrity, followed by barcode entry into a biospecimen tracking database. Specimens were again placed in LN2 storage until further processing. Acceptable HNSCC tumor tissue segments were determined by TSS pathologists based on the percent viable tumor nuclei (> 80%), total cellularity (> 50%), and necrosis (< 20%). Segments received at the BCR were verified by BCR and Leidos Biomedical Research (LBR) pathologists and the percent of total area of tumor in the segment was also documented. Additionally, disease-specific working group pathology experts reviewed the morphology to clarify or standardize specific disease classifications and correlation to the proteomic and genomic data.
Specimens selected for this study were determined on the maximal percent in the pathology criteria and best weight. Specimens were pulled from the biorepository using an LN2 cryocart to maintain specimen integrity and then cryopulverized. The cryopulverized specimen was divided into aliquots for DNA (30 mg) and RNA (30 mg) isolation and proteomics (50 mg) for molecular characterization. Nucleic acids were isolated and stored at −80°C until further processing and distribution; cryopulverized protein material was returned to the LN2 freezer until distribution. Shipment of the cryopulverized segments used cryoports for distribution to the proteomic characterization centers and shipment of the nucleic acids used dry ice shippers for distribution to the genomic characterization centers; a shipment manifest accompanied all distributions for the receipt and integrity inspection of the specimens at the destination. The DNA sequencing was performed at the Broad Institute, Cambridge, MA and RNA sequencing was performed at the University of North Carolina, Chapel Hill, NC. Material for proteomic analyses was sent to the Proteomic Characterization Center at Johns Hopkins University, Maryland, USA.
METHOD DETAILS
Genomics and Transcriptomics Profiling Experiments
Sample Processing for Genomic DNA and Total RNA Extraction
Our study sampled a single site of the primary tumor from surgical resections, due to the internal requirement to process a minimum of 125 mg of tumor issue and 50 mg of adjacent normal tissue. DNA and RNA were extracted from tumor and normal specimens in a co-isolation protocol using Qiagen’s QIAsymphony DNA Mini Kit and QIAsymphony RNA Kit. Genomic DNA was also isolated from peripheral blood (3-5 mL) to serve as matched germline reference material. The Qubit™ dsDNA BR Assay Kit was used with the Qubit® 2.0 Fluorometer to determine the concentration of dsDNA in an aqueous solution. Any sample that passed quality control and produced enough DNA yield to go through various genomic assays was sent for genomic characterization. RNA quality was quantified using the NanoDrop 8000 and quality was assessed using the Agilent Bioanalyzer. A sample that passed RNA quality control and had a minimum RIN (RNA integrity number) score of 7 was subjected to RNA sequencing. Identity match for germline, normal adjacent tissue, and tumor tissue was assayed at the BCR using the Illumina Infinium QC array. This beadchip contains 15,949 markers designed to prioritize sample tracking, quality control, and stratification.
Whole Exome Sequencing (WES)
Library Construction
Library construction was performed as described in (Fisher et al., 2011), with the following modifications: initial genomic DNA input into shearing was reduced from 3 μg to 20-250 ng in 50 μL of solution. For adapter ligation, Illumina paired-end adapters were replaced with palindromic forked adapters, purchased from Integrated DNA Technologies, with unique dual-indexed molecular barcode sequences to facilitate downstream pooling. Kapa HyperPrep reagents in 96-reaction kit format were used for end repair/A-tailing, adapter ligation, and library enrichment PCR. In addition, during the post-enrichment SPRI cleanup, elution volume was reduced to 30 μL to maximize library concentration, and a vortexing step was added to maximize the amount of template eluted.
In-solution Hybrid Selection
After library construction, libraries were pooled into groups of up to 96 samples. Hybridization and capture were performed using the relevant components of Illumina's Nextera Exome Kit and followed the manufacturer’s suggested protocol, with the following exceptions. First, all libraries within a library construction plate were pooled prior to hybridization. Second, the Midi plate from Illumina’s Nextera Exome Kit was replaced with a skirted PCR plate to facilitate automation. All hybridization and capture steps were automated on the Agilent Bravo liquid handling system.
Preparation of Libraries for Cluster Amplification and Sequencing
After post-capture enrichment, library pools were quantified using qPCR (automated assay on the Agilent Bravo) using a kit purchased from KAPA Biosystems with probes specific to the ends of the adapters. Based on qPCR quantification, libraries were normalized to 2 nM.
Cluster Amplification and Sequencing
Cluster amplification of DNA libraries was performed according to the manufacturer’s protocol (Illumina) using exclusion amplification chemistry and flowcells. Flowcells were sequenced utilizing sequencing-by-synthesis chemistry. The flowcells were then analyzed using RTA v.2.7.3 or later. Each pool of whole exome libraries was sequenced on paired 76 cycle runs with two 8 cycle index reads across the number of lanes needed to meet coverage for all libraries in the pool. Pooled libraries were run on HiSeq 4000 paired-end runs to achieve a minimum of 150x on target coverage per sample library. The raw Illumina sequence data were demultiplexed and converted to fastq files; adapter and low-quality sequences were trimmed. The raw reads were mapped to the hg38 human reference genome and the validated BAMs were used for downstream analysis and variant calling.
PCR-Free Whole Genome Sequencing
Preparation of Libraries for Cluster Amplification and Sequencing
An aliquot of genomic DNA (350 ng in 50 μL) was used as the input into DNA fragmentation (aka shearing). Shearing was performed acoustically using a Covaris focused-ultrasonicator, targeting 385bp fragments. Following fragmentation, additional size selection was performed using a SPRI cleanup. Library preparation was performed using a commercially available kit provided by KAPA Biosystems (KAPA Hyper Prep without amplification module) and with palindromic forked adapters with unique 8-base index sequences embedded within the adapter (purchased from IDT). Following sample preparation, libraries were quantified using quantitative PCR (kit purchased from KAPA Biosystems), with probes specific to the ends of the adapters. This assay was automated using Agilent’s Bravo liquid handling platform. Based on qPCR quantification, libraries were normalized to 1.7 nM and pooled into 24-plexes.
Cluster Amplification and Sequencing (HiSeq X)
Sample pools were combined with HiSeq X Cluster Amp Reagents EPX1, EPX2, and EPX3 into single wells on a strip tube using the Hamilton Starlet Liquid Handling system. Cluster amplification of the templates was performed according to the manufacturer’s protocol (Illumina) with the Illumina cBot. Flowcells were sequenced to a minimum of 15x on HiSeq X utilizing sequencing-by-synthesis kits to produce 151bp paired-end reads. Output from Illumina software was processed by the Picard data processing pipeline to yield BAMs containing demultiplexed, aggregated, aligned reads. All sample information tracking was performed by automated LIMS messaging.
Illumina Infinium MethylationEPIC BeadChip Array
The MethylationEPIC array uses an 8-sample version of the Illumina Beadchip capturing > 850,000 DNA methylation sites per sample. 250 ng of DNA was used for the bisulfite conversion using Infinium MethylationEPIC BeadChip Kit. The EPIC array includes sample plating, bisulfite conversion, and methylation array processing. After scanning, the data were processed through an automated genotype calling pipeline. Data generated consisted of raw IDAT files and a sample sheet.
RNA Sequencing
Quality Assurance and Quality Control of RNA Analytes
All RNA analytes were assayed for RNA integrity, concentration, and fragment size. Samples for total RNA-seq were quantified on a TapeStation system (Agilent, Inc. Santa Clara, CA). Samples with RINs > 8.0 were considered high quality.
Total RNA-seq Library Construction
Total RNA-seq library construction was performed from the RNA samples using the TruSeq Stranded RNA Sample Preparation Kit and bar-coded with individual tags following the manufacturer’s instructions (Illumina, Inc. San Diego, CA). Libraries were prepared on an Agilent Bravo Automated Liquid Handling System. Quality control was performed at every step and the libraries were quantified using the TapeStation system.
Total RNA Sequencing
Indexed libraries were prepared and run on HiSeq 4000 paired-end 75 base pairs to generate a minimum of 120 million reads per sample library with a target of greater than 90% mapped reads. Typically, these were pools of four samples. The raw Illumina sequence data were demultiplexed and converted to FASTQ files, and adapter and low-quality sequences were quantified. Samples were then assessed for quality by mapping reads to the hg38 human genome reference, estimating the total number of reads that mapped, amount of RNA mapping to coding regions, amount of rRNA in the sample, number of genes expressed, and relative expression of housekeeping genes. Samples passing this QA/QC were then clustered with other expression data from similar and distinct tumor types to confirm expected expression patterns. Atypical samples were then SNP typed from the RNA data to confirm source analyte. FASTQ files of all reads were then uploaded to the GDC repository.
miRNA-seq Library Construction
miRNA-seq library construction was performed from the RNA samples using the NEXTflex Small RNA-Seq Kit (v3, PerkinElmer, Waltham, MA) and bar-coded with individual tags following the manufacturer’s instructions. Libraries were prepared on the Sciclone Liquid Handling Workstation. Quality control was performed at every step, and the libraries were quantified using a TapeStation system and an Agilent Bioanalyzer using the Small RNA analysis kit. Pooled libraries were then size selected according to NEXTflex Kit specifications using a Pippin Prep system (Sage Science, Beverly, MA).
miRNA Sequencing
Indexed libraries were loaded on the Hiseq 4000 to generate a minimum of 10 million reads per library with a minimum of 90% reads mapped. The raw Illumina sequence data were demultiplexed and converted to FASTQ files for downstream analysis. Resultant data were analyzed using a variant of the small RNA quantification pipeline developed for TCGA (Chu et al., 2016). Samples were assessed for the number of miRNAs called, species diversity, and total abundance. Samples passing quality control were uploaded to the GDC repository.
Genomics and Transcriptomics Data Processing
Somatic Mutation Calling
Somatic variants were called by the Somaticwrapper pipeline, which includes four different callers, i.e., Strelka v.2 (Saunders et al., 2012), MUTECT v1.7 (Cibulskis et al., 2013), VarScan v.2.3.8 (Koboldt et al., 2012), and Pindel v.0.2.5 (Ye et al., 2009) from WES. We kept the exonic SNVs called by any 2 callers among MUTECT v1.7, VarScan v.2.3.8, and Strelka v.2 and indels called by any 2 callers among VarScan v.2.3.8, Strelka v.2, and Pindel v.0.2.5. For the merged SNVs and indels, we applied a 14X and 8X coverage cutoff for tumor and normal, separately. We also filtered SNVs and indels by a minimal variant allele frequency (VAF) of 0.05 in tumors and a maximal VAF of 0.02 in normal samples. Finally, we filtered any SNV which was within 10bp of an indel found in the same tumor sample.
Germline Variant Calling
Germline Variant Calling was performed using germlinewrapper v1.1, which implements multiple tools for the detection of germline INDELs and SNVs. Germline SNVs were identified using VarScan v2.3.8 (with parameters: --min-var-freq 0.10 --p-value 0.10, --min-coverage 3 --strand-filter 1) operating on a mpileup stream produced by samtools v1.2 (with parameters: -q 1 -Q 13) and GATK v4.0.0.0 (McKenna et al., 2010) using its haplotype caller in single-sample mode with duplicate and unmapped reads removed and retaining calls with a minimum quality threshold of 10. All resulting variants were limited to the coding region of the full-length transcripts obtained from Ensembl release 95 plus additional two base pairs flanking each exon to cover splice donor/acceptor sites. We required variants to have allelic depth ≥ 5 reads for the alternative allele. We filtered large INDELs that were longer than 100 bps.
DNA Methylation Array
The raw data from Illumina's EPIC methylation arrays were available as IDAT files from the CPTAC consortium. The methylation analysis was performed using the cross-package workflow methylationArrayAnalysis available on Bioconductor. In brief, the raw data IDAT files were processed to obtain the methylated (M) and unmethylated (U) signal intensities for each locus. The processing step included an unsupervised normalization step called functional normalization that has been previously implemented for Illumina 450K methylation arrays (Fortin et al., 2014). A detection p value was also calculated for each locus, and this p value captured the quality of detection at the locus with respect to negative control background probes included in the array. Loci having common SNPs (with MAF > 0.01), as per dbSNP build 132 through 147 via the UCSC “snp132common” track through “snp147common” track, were removed from further analysis. Beta values were calculated as M/(M+U), that is equal to the fraction methylated for each locus. Beta values of loci whose detection p values were > 0.01 were assigned NA in the output file. All loci were annotated with the EPIC Manifest from MethylationEPIC_v-1-0_B2.csv from the zip archive infinium-methylationepic-v1-0-b2-manifest-file-csv.zip from Illumina through the “IlluminaHumanMethylationEPICanno.ilm10b2.hg19” package on Bioconductor. To map EPIC arrays to the GRCh38 assembly, all probes were reannotated by annotation information from InfiniumAnnotation.
Copy-Number Analysis
Copy-number analysis was performed jointly leveraging both whole-genome sequencing (WGS) and whole-exome sequencing data of the tumor and germline DNA, using CNVEX (https://github.com/mctp/cnvex). CNVEX uses whole-genome aligned reads to estimate coverage within fixed genomic intervals, and whole-genome and whole-exome variant calls to compute B-allele frequencies at variable positions (we used TNScope germline calls). Coverages were computed in 10kb bins, and the resulting log coverage ratios between tumor and normal samples were adjusted for GC bias using weighted LOESS smoothing across mappable and non-blacklisted genomic intervals within the GC range 0.3-0.7, with a span of 0.5 (the target, blacklist, and configuration files are provided with CNVEX). The adjusted log coverage ratios (LR) and B-allele frequencies (BAF) were jointly segmented by a custom algorithm based on Circular Binary Segmentation (CBS). Alternative probabilistic algorithms were implemented in CNVEX, including algorithms based on recursive binary segmentation (RBS) (Gey and Lebarbier, 2008), and dynamic programming (Bellman, 1961), as implemented in the R package jointseg (Pierre-Jean et al., 2015). For the CBS-based algorithm, first LR and mirrored BAF were independently segmented using CBS (parameters alpha=0.01, trim=0.025) and all candidate breakpoints collected. The resulting segmentation track was iteratively “pruned” by merging segments that had similar LR, BAFs, and short lengths. For the RBS- and DP-based algorithms, joint-breakpoints were “pruned” using a statistical model selection method (Lebarbier, 2005). For the final set of CNV segments, we chose the CBS-based results as they did not require specifying a prior on the number of expected segments (K) per chromosome arm, were robust to unequal variances between the LR and BAF tracks, and provided empirically the best fit to the underlying data.
RNA Quantification and Circular RNA Prediction
The hg38 reference genome and RefSeq annotations were used for the RNA-seq data analysis and were downloaded from the UCSC table browser. First, CIRI (v2.0.6) was used to call circular RNA with default parameters and BWA (version 0.7.17-r1188) was used as the mapping tool. The cutoff of supporting reads for circRNAs was set to 10. Then we used a pseudo-linear transcript strategy to quantify gene and circular RNA expression (Li et al., 2017). In brief, for each sample, linear transcripts of circular RNAs were extracted and 75bp (read length) from the 3’ end was copied to the 5’ end. The modified transcripts were called pseudo-linear transcripts. Transcripts of linear genes were also extracted and mixed with pseudo-linear transcripts. RSEM (version 1.3.1) with Bowtie2 (version 2.3.3) as the mapping tool was used to quantify gene and circular RNA expression based on the mixed transcripts. After quantification, the upper quantile method was applied for normalization. The normalized matrix was log2-transformed and separated into gene and circular RNA expression matrices.
miRNA-Seq Data Analysis
Processed miRNA bam files were downloaded from GDC and transferred to fastq format using samtools (version 1.10). QuickMIRSeq (Zhao et al., 2017) with bowtie as a mapping tool (version 1.1.1) was used to quantify miRNA expression. The following parameters were used for QuickMIRSeq: 1) 2 bp extension / shorten were allowed in both upstream and downstream regions of mature miRNA; 2) The minimum and maximum length of miRNA reads were set to 16 and 28, respectively. Then RPM (reads per million) values were used to quantify miRNA expression levels.
HPV Virus Identification
The unmapped RNA-seq reads to the human reference genome were extracted and mapped to the virus reference by VirusScan (Cao et al., 2016). The reference contained the known HPV genotypes including the "high-risk" genotypes such as HPV 16 and HPV 18. Samples were classified as HPV positive using an empiric definition of detection of > 1,000 mapped RNA-seq reads.
Mutational Signature Analysis
Non-negative matrix factorization algorithm (NMF) was used in deciphering mutation signatures in cancer somatic mutations stratified by 96 base substitutions in trinucleotide sequence contexts. To obtain a reliable signature profile, we used the Somaticwrapper pipeline to call mutations from WES and WGS data. SignatureAnalyzer exploited the Bayesian variant of the NMF algorithm and enabled an inference for the optimal number of signatures from data itself at a balance between the data fidelity (likelihood) and the model complexity (regularization) (Kasar et al., 2015; Kim et al., 2016; Tan and Févotte, 2013). Signatures were compared against known signatures derived from COSMIC (Tate et al., 2019) and cosine similarity was calculated to identify the best match.
Gene Fusion Detection
Fusions in RNA-seq samples were called using three callers: STAR-Fusion v1.5.0 (Haas et al., 2019), EricScript v0.5.5 (Benelli et al., 2012), and INTEGRATE v0.2.6 (Zhang et al., 2016). As STAR-Fusion has higher sensitivity, calls made by this tool with higher supporting evidence (defined by fusion fragments per million total reads, or FFPM > 0.1) is required, or a given fusion must be reported by at least 2 callers being retained. Fusions present in the following databases were then excluded: 1) uncharacterized genes, immunoglobulin genes, mitochondrial genes, etc.; 2) fusions from the same gene or paralog genes; and 3) fusions reported in TCGA normal samples (Gao et al., 2018), GTEx tissues (reported in the STAR-Fusion output), and non-cancer cell studies (Babiceanu et al., 2016). Finally, normal fusions were filtered out from the tumor fusions.
Structural Variant Analysis
Structural variants in WGS samples were called with Manta 1.3.2 (Chen et al., 2016), retaining variants where sample site depth is less than 3x the median chromosome depth near one or both variant breakends, somatic score is greater than 30, and for small variants (<1000 bases) in the normal sample, the fraction of reads with MAPQ0 around either breakend does not exceed 0.4. It is optimized for the analysis of somatic variation in tumor/normal sample pairs. The paired and split-read evidence were combined during the SV discovery and scoring to improve accuracy. We prioritized the variants by the number of spanning read pairs which strongly (Q30) support the variants (>5 as high confidence level).
SCNA Arm and Focal Significance
From the segment-level SCNA data, we used GISTIC2 (Mermel et al., 2011) to assess the arm- and focal-level SCNA significance using the default parameters except for increased threshold of significance (i.e., -ta and -td parameters of GISTIC2) to 0.3 based on the distribution of germline copy number variants.
Proteomic and Phosphoproteomic Profiling Experiments
Sample Processing for Protein Extraction and Tryptic Digestion
All samples for the current study were prospectively collected for the Clinical Proteomic Tumor Analysis Consortium (CPTAC) project as described above and processed for mass spectrometric (MS) analysis at Johns Hopkins University (JHU). Tissue lysis and downstream sample preparation for global proteomic and phosphoproteomic analysis was carried out as previously described (Clark et al., 2018; Mertins et al., 2018). Approximately 25-120 mg of each cryo-pulverized head and neck squamous cell carcinoma (HNSCC) tissue or normal adjacent tissue (NAT) was resuspended in lysis buffer (8 M urea, 75 mM NaCl, 50 mM Tris, pH 8.0, 1 mM EDTA, 2 μg/mL aprotinin, 10 μg/mL leupeptin, 1 mM PMSF, 10 mM NaF, Phosphatase Inhibitor Cocktail 2 and Phosphatase Inhibitor Cocktail 3 [1:100 dilution], and 20 μM PUGNAc) by repeated vortexing. Lysates were clarified by centrifugation at 20,000 x g for 10 min at 4°C, and protein concentrations were determined by BCA assay (Pierce). Proteins were diluted to a final concentration of 8 mg/mL with lysis buffer, and 800 mg of protein was reduced with 5 mM dithiothreitol (DTT, ThermoFisher) for 1h at 37°C, and subsequently alkylated with 10 mM iodoacetamide (Sigma) for 45 min at room temperature (RT) in the dark. Samples were diluted 1:4 with 50 mM Tris-HCl (pH 8.0) and subjected to proteolytic digestion with LysC (Wako Chemicals) at 1 mAU:50 mg enzyme-to-substrate ratio for 2h at RT, followed by the addition of sequencing grade modified trypsin (Promega) at 1:50 enzyme-to-substrate ratio and overnight incubation at RT. The digested samples were then acidified with 50% Formic acid (FA, Fisher Chemicals) to pH 2. Tryptic peptides were desalted on reversed phase C18 SPE columns (Waters) and dried using Speed-Vac (Thermo Scientific).
TMT Labeling of Peptides
Dried peptides from each sample were labeled with 11-plex TMT (Tandem Mass Tag) reagents (Thermo Fisher Scientific). Peptides (300 μg) from each of the HNSCC and NAT samples were dissolved in 60 μL of 50 mM HEPES, pH 8.5 solution. An internal quality control (QC) sample, NCI-7 Cell Line (Clark et al., 2018), was interspersed among all TMT 11-plex sets. HNSCC and NAT samples with NCI-7 QC aliquots were co-randomized to 19 TMT sets. A reference sample was created by pooling an aliquot from 87 HNSCC tissues and 50 NAT tissues (representing ~80% of the sample cohort), and included in all TMT 11-plex sets as a pooled reference channel. Five mg of TMT reagent was dissolved in 250 μL of anhydrous acetonitrile, and then 20 μL of each TMT reagent was added to the corresponding aliquot of peptides. After 1h incubation at RT, the reaction was quenched by incubation with 5% NH2OH for 15 min at RT. Following labeling, peptides were desalted on reversed phase C18 SPE columns (Waters) and dried using Speed-Vac (Thermo Scientific).
Peptide Fractionation by Basic Reversed-phase Liquid Chromatography (bRPLC)
To reduce the likelihood of peptides co-isolating and co-fragmenting due to high sample complexity, we employed extensive, high-resolution fractionation via basic reversed phase liquid chromatography (bRPLC). Previous reports have indicated this approach can reduce the incidence of isobaric reporter ion ratio distortion effects, which would impact downstream quantitation (Ow et al., 2011; Rauniyar and Yates, 2014). For each TMT set, about 3.3 mg desalted peptides were reconstituted in 900 μL of 5 mM ammonium formate (pH 10) and 2% acetonitrile (ACN) and loaded onto a 4.6 mm x 250 mm RP Zorbax 300 A Extend-C18 column with 3.5 mm size beads (Agilent). Peptides were separated using an Agilent 1200 Series HPLC instrument using basic reversed-phase chromatography with Solvent A (2% ACN, 5 mM ammonium formate, pH 10) and a non-linear gradient of Solvent B (90% ACN, 5 mM ammonium formate, pH 10) at 1 mL/min as follows: 0% Solvent B (7 min), 0% to 16% Solvent B, (6 min), 16% to 40% Solvent B 60 min), 40% to 44% Solvent B (4 min), 44% to 60% Solvent B (5 min) and then held at 60% Solvent B for 14 min. Collected fractions were concatenated into 24 fractions as described previously (Mertins et al., 2018); 5% of each of the 24 fractions was aliquoted for global proteomic analysis, dried down in a Speed-Vac, and resuspended in 3% ACN, 0.1% formic acid prior to ESI-LC-MS/MS analysis. The remaining sample was utilized for phosphopeptide enrichment.
Enrichment of Phosphopeptides by Fe-IMAC
The remaining 95% of the sample were further concatenated into 12 fractions prior to phosphopeptide enrichment using immobilized metal affinity chromatography (IMAC) as previously described (Mertins et al., 2013). In brief, Ni-NTA agarose beads were utilized to prepare Fe3+-NTA agarose beads, and then about 250 μg peptides of each fraction reconstituted in 80% ACN/0.1% trifluoroacetic acid were incubated with 10 μL of the Fe3+-IMAC beads for 30 mins. Samples were then spun down and the supernatant containing unbound peptides was removed. The beads were brought up in 80% ACN, 0.1% trifluoroacetic acid and then loaded onto equilibrated C-18 Stage Tips, and washed by 80% ACN, 0.1% trifluoroacetic acid. Tips were rinsed twice with 1% formic acid, followed by sample elution off the Fe3+-IMAC beads and onto the C-18 State Tips with 70 μL of 500 mM dibasic potassium phosphate, pH 7.0 three times. C-18 Stage Tips were washed twice with 1% formic acid, followed by elution of the phosphopeptides from the C-18 Stage Tips with 50% ACN, 0.1% formic acid twice. Samples were dried down and resuspended in 3% ACN, 0.1% formic acid prior to ESI-LC-MS/MS analysis.
ESI-LC-MS/MS for Global Proteome and Phosphoproteome Analysis
The global proteome and phosphoproteome fractions were analyzed using the same instrumentation and methodology as described in a previous study (Clark et al., 2019). Peptides (~ 0.8 μg) were separated on an Easy nLC 1200 UHPLC system (Thermo Scientific) on an in-house packed 20 cm x 75 mm diameter C18 column (1.9 mm Reprosil-Pur C18-AQ beads (Dr. Maisch GmbH); Picofrit 10 mm opening (New Objective). The column was heated to 50°C using a column heater (Phoenix-ST). The flow rate was 0.200 μl/min with 0.1% formic acid and 2% acetonitrile in water (A) and 0.1% formic acid, 90% acetonitrile (B). The peptides were separated with a 6–30% B gradient in 84 mins and analyzed using the Thermo Fusion Lumos mass spectrometer (Thermo Scientific). Parameters were as follows MS1: resolution – 60,000, mass range – 350 to 1800 m/z, RF Lens – 30%, AGC Target 4.0e5, Max IT – 50 ms, charge state include - 2-6, dynamic exclusion – 45 s, top 20 ions selected for MS2; MS2: resolution – 50,000, high-energy collision dissociation activation energy (HCD) – 37, isolation width (m/z) – 0.7, AGC Target – 2.0e5, Max IT – 105 ms.
Proteomics and Phosphoproteomics Data Processing
Protein Database Searching and Quantification of Global and Phosphoproteomic Data
MS/MS spectra were searched using the MSFragger version 20190628 (Kong et al., 2017) against a CPTAC harmonized RefSeq protein sequence database appended with an equal number of decoy sequences. For the analysis of whole proteome data, MS/MS spectra were searched using a precursor-ion mass tolerance of 20 ppm, fragment mass tolerance of 20 ppm, and allowing C12/C13 isotope errors (1/0/1/2/3). Cysteine carbamidomethylation (+57.0215) and lysine TMT labeling (+229.1629) were specified as fixed modifications, and methionine oxidation (+15.9949), N-terminal protein acetylation (+42.0106), and TMT labeling of peptide N terminus and serine residues (to account for any over-labeling) were specified as variable modifications (Clark et al., 2019; Zecha et al., 2019). The search was restricted to fully tryptic peptides, allowing up to two missed cleavage sites. For the analysis of phosphopeptide enriched data, the set of variable modifications also included phosphorylation (+79.9663) of serine, threonine, and tyrosine residues. The post-processing of the search results was done using the Philosopher toolkit version v1.6.0 (https://philosopher.nesvilab.org). MSFragger output files (in pepXML format) were processed using PeptideProphet (Keller et al., 2002) (with the high–mass accuracy binning and semi-parametric mixture modeling options) to compute the posterior probability of correct identification for each peptide to spectrum match (PSM). In the case of the phosphopeptide-enriched dataset, PeptideProphet files were additionally processed using PTMProphet (Deutsch et al., 2015) to localize the phosphorylation sites. The resulting pepXML files from PeptideProphet (or PTMProphet) from all 20 TMT 11-plex experiments were then processed together to assemble peptides into proteins (protein inference) and to create a combined file (in protXML format) of high confidence protein groups. The combined protXML file and the individual PSM lists for each TMT 11-plex were further processed using the Philosopher filter command as follows. Each peptide was assigned either as a unique peptide to a particular protein group or assigned as a razor peptide to a single protein group that had the most peptide evidence. The protein groups assembled by ProteinProphet (Nesvizhskii et al., 2003) were filtered to 1% protein-level False Discovery Rate (FDR) using the chosen FDR target-decoy strategy and the best peptide approach (allowing both unique and razor peptides) and applying the picked FDR strategy (Savitski et al., 2015). In each TMT 11-plex, the PSM lists were filtered using a stringent, sequential FDR strategy, retaining only those PSMs with PeptideProphet probability of 0.9 or higher (which in these data corresponded to less than 1% PSM-level FDR) and mapped to proteins that also passed the global 1% protein-level FDR filter.
For each PSM that passed these filters, MS1 intensity of the corresponding precursor-ion was extracted using the Philosopher label-free quantification module based on the moFF method (Argentini et al., 2016) (using 10 p.p.m mass tolerance and 0.4 min retention time window for extracted ion chromatogram peak tracing). In addition, for all PSMs corresponding to a TMT-labeled peptide, eleven TMT reporter ion intensities were extracted from the MS/MS scans (using 0.002 Da window) and the precursor ion purity scores were calculated using the intensity of the sequenced precursor ion and that of other interfering ions observed in MS1 data (within a 0.7 Da isolation window). All supporting information for each PSM, including the accession numbers and names of the protein/gene selected based on the protein inference approach with razor peptide assignment and quantification information (MS1 precursor-ion intensity and the TMT reporter ion intensities) was summarized in the output PSM.tsv files, one file for each TMT 11-plex experiment.
To generate summary reports in different levels (gene, peptide, and protein for global and phosphopeptide enriched data; additional modification site report for phosphopeptide data), we processed the PSM.tsv files using TMT-Integrator. Each PSM in a PSM.tsv file passing the following criteria were kept for creating integrated reports, including (1) having TMT label, (2) having intensity in the reference channel, (2) precursor-ion purity above 50%, (3) summed reported ion intensity (across all channels) not in the lower 5% of all PSMs (2.5% for phosphopeptide enriched data), (4) peptide with phosphorylation (for phosphopeptide enriched data). For a peptide with redundant PSMs, only the PSM with the highest summed TMT intensity was kept for later analysis. PSMs mapping to common external contaminant proteins were excluded, and unique and razor peptides were both used for quantification. Next, the report ion intensities of each PSM were log2 transformed and normalized by the reference channel intensity (i.e., subtracted log2 reference intensity from those log2 report ion intensities), therefore the intensities were converted into log2-based ratio (denoted as ‘ratios’ in the following paragraphs). After converting the intensities to ratios, the PSMs were grouped based on the predefined level (i.e., gene, protein, peptide, and phosphopeptide-level). The interquartile range (IQR) algorithm was then applied to remove the outliers in each PSM group and the remaining ratios were normalized using the median centering. Finally, the normalized ratios were converted back to abundances using the weighted sum of the MS1 intensities of the top three most intense peptide ions. The details of the IQR, median centering, and abundance conversion algorithms are described in detail in (Clark et al., 2019). For phosphosite quantification, a phosphopeptide was assigned to a single protein based on razor assignment. Specifically, each phosphopeptide was assigned to the protein with most evidence when it could be mapped to multiple proteins. If all mapped proteins for a given phosphopeptide are indistinguishable (i.e. the same level of evidence), then one is selected alphabetically. The ratio for each site was calculated from the ratios of all phosphopeptides (including those with multiple phosphorylations) containing this site by median. Then the ratios of sites were converted to intensities using the way similar to that for protein quantification.
Data Harmonization
mRNA Data
Gene-level mRNA data (RSEM) were upper-quantile normalized and log2 transformed. Genes with missing values (i.e., raw RSEM equal to zero) in less than half of the samples were regarded as quantifiable (N = 29020) and used for downstream analyses (e.g., protein-RNA correlation). Another version of the mRNA data, which were represented as FPKM, was used in certain analyses that benefit from normalization by gene length (specified in the Integrated Analysis section).
Methylation Data
The probe-level methylation data (represented as beta-values) were aggregated to gene-level by extracting probes located in the CpG island of the promoter region of a given gene. The mean beta-value of these probes was used to represent gene-level methylation.
Proteomics Data
Gene-level proteomics data, which were represented as normalized MS intensity, were log2 transformed. Proteins with missing values in less than half of the samples were regarded as quantifiable (N = 9657) and used for downstream analyses.
Phosphoproteomics Data
Peptide-level, single site-level, and gene-level phosphoproteomics data, which were represented as normalized MS intensity, were log2 transformed. The peptide-level data contained both localized and non-localized phosphorylation events and the peptides could harbor single, double, or triple phosphorylations. This was utilized by the quality control analyses and a localized version was used for multi-omics subtyping (further described in Unsupervised Subtyping Using Non-negative Matrix Factorization (NMF)). Site-level phosphorylation data consisted of single confidently localized sites and were used for the majority of analyses. Gene-level data were used for survival association. Phosphopeptides, phosphosites, and genes with missing values in less than half of the samples were regarded as quantifiable unless otherwise described.
Handling of Missing Values in Proteomics and Phosphoproteomics Data
The missing values were handled in a few different ways for different analyses. 1) For PCA analysis, because it does not allow missing values, we removed any proteins and phosphopeptides missed in more than 50% of the samples and then did missing value imputation using the K nearest neighbors (KNN) algorithm for the remaining proteins and phosphopeptides. 2) For differential abundance analysis across tumors, non-missing values were required for 50% of the samples for each protein and phosphopeptide. 3) In the tumor versus normal analysis, multiplex bias is mitigated by the requirement that the protein or phosphopeptide needed to be quantified in both the tumor and paired normal sample for a patient in the paired statistical test. The paired samples for each patient were assigned to the same TMT plex by experimental design. 4) For the multi-omics subtyping analysis, all features with missing values were excluded.
Data Quality Control
Quality Control of Proteomic and Phosphoproteomic Data Generation
During the mass spectrometric analysis of global peptides and phosphopeptides, we established a quality control procedure to assess the data quality and instrument performance. For quality control of the HNSCC proteomic and phosphoproteomic raw data, we processed the raw data files through the MS-PyCloud proteomics pipeline (Chen et al., 2018) to generate a quality control table. The quality control table includes the number of PSMs, peptides, and proteins identified in each LC-MS/MS analysis and in each TMT set. Other reported quality control parameters include the total MS2 count; percent of peptides identified in 1, 2, 3, or 4+ charge states; and the minimum and maximum TMT channel intensity ratios relative to the median channel intensity for each TMT set, as well as the injection time for MS/MS analysis.
We calculated the number of unique and shared peptides/proteins in each fraction of a TMT set. A peptide/protein was considered shared if it was identified in more than one fraction. Each fraction contributed unique peptides/proteins relative to the other fractions. This ensured that no fraction was a duplicate of another fraction and each fraction generated quality data.
Correlation-based Checking for Potential Sample Mislabeling
In a large-scale multi-omics study, it is critical to validate the sample labels in experiments as well as in data analysis to avoid mislabeling. The cross-omics correlation (e.g., RNA and protein) of the same sample is usually highest among the correlation values of different samples. Thus, after calculating all pairs of sample correlation values among different omics levels, we could determine whether the highest correlated pairs between two levels have the same sample labels. Here, we applied the sample correlation method implemented in OmicsOne (https://github.com/huizhanglab-jhu/OmicsOne) to the abundance matrices of RNA-seq, proteomics, and phosphoproteomics data to check the sample labels of all 110 samples that passed pathological quality control. The three normalized gene level expression matrices of RNA-seq, proteomics, and phosphoproteomics data were firstly z-score transformed for each gene across all samples. In correlating RNA-seq with proteomics data, only the top 500 most correlated genes were considered for sample correlation based on the gene-wise correlation of the two levels. All pair-wise correlation values were calculated using Spearman’s rank correlation. All the samples were highly correlated with samples having the identical label, and 96% of them were the best ranked correlations. We also did the sample correlation tests on “RNA-seq and phosphoproteomics” and “proteomics and phosphoproteomics” comparisons. Based on all the correlation results, there were no mislabeled samples observed in the RNA-seq, proteomics, and phosphoproteomics data sets.
Machine Learning-Based Checking for Potential Sample Mislabeling
We use our omics data to build machine learning models to detect possible sample mislabeling. Specifically, we built Random Forest classification models using molecular data (RNA-seq, protein) as features to predict patient gender and disease status (tumor vs. normal). When predicting the gender, we only used known sex genes as input features. Leave-one-out cross-validation was used to predict the labels (gender, disease status respectively) for each sample. Samples with low predicted probability (<0.15) for the provided class for either gender or disease status are flagged for manual check. Following this procedure, two samples were identified for potential gender mislabeling. These two samples are a pair of tumor and NAT from the same patient. This was later confirmed as a data entry error for the patient and was corrected in the released data. Additionally, three NAT samples (C3L-01138-N, C3N-03612-N, and C3N-02275-N) and one tumor sample (C3N-01643-T) were predicted as tumor and normal samples, respectively, by both mRNA and protein models. These results were consistent with the unsupervised PCA analysis (Figure 1B-C). These samples with questionable tumor/NAT identity were confirmed by follow-up pathological inspection and then removed from all downstream analyses.
Function Prediction Based on Gene Co-expression
Co-expression network construction using mRNA and protein expression data and network-based gene function prediction for KEGG pathways were performed as previously described (Wang et al., 2017) using OmicsEV (https://github.com/bzhanglab/OmicsEV).
Data Quality Evaluation Using Quality Control Samples
As one of our quality control (QC) strategies, we included the reference sample (i.e., pooled HNSCC samples) in a non-reference channel in three different TMTs. For these QC samples, the R-squared value between each pair of replicate samples was calculated on log2 transformed protein expression data at gene level or phosphopeptide data. Then the R-squared values were used to evaluate the quality (reproducibility) of the data. In addition, a new quantification strategy was used to process both global proteomics and phosphoproteomics data, in which a virtual reference channel was built for each TMT experiment to serve as “common reference” rather than using the reference sample in the reference channel. The virtual reference was computed by taking the average of all channels in each TMT experiment. In this case, all the reference samples from the reference channels could be used as QC samples and they were treated equally as the samples in non-reference channels during the quantification. Then the R-squared values between each pair of the reference samples were calculated using the same way to evaluate the quality (reproducibility) of the data.
Batch Effect Evaluation
The batch effect was evaluated for both global proteome and phosphoproteome data using principal component regression analysis (Büttner et al., 2019). Specifically, we first removed any proteins missing in more than 50% samples followed by missing value imputation using KNN before performing PCA analysis. Next, for each PC, Pearson’s correlation coefficient with batch covariate was calculated and the significance of the correlation coefficient was estimated using one-way ANOVA. A p value less than 0.05 is considered significant. The analysis was performed using OmicsEV (https://github.com/bzhanglab/OmicsEV).
Immunohistochemistry (IHC)
IHC analysis of CD3 Protein
Cut tissue sections (5 μm) on charged glass slides were baked for 10-12 hours at 58°C in a dry slide incubator, deparaffinized in xylene and rehydrated via an ethanol step gradient. Heat-induced antigen retrieval steps were performed at pH 9.0, with the primary antibody incubated at room temperature for 1 hour (CD3, polyclonal, Dako, 1:100) followed by standard chromogenic staining protocol with the Envision Polymer-HRP/3,3’diaminobenzidine (DAB, Dako) process. Slides were counterstained in Harris hematoxylin. Immunohistochemistry scoring was performed using the percentage of stromal CD3-positive tumor infiltrating lymphocytes (TILS). All IHC results were evaluated against positive and negative tissue controls.
Data-Independent Acquisition (DIA) Analysis
ESI-LC-MS/MS
Unlabeled, digested peptide material from individual tissue samples (HNSCC and NAT) was spiked with index Retention Time (iRT) peptides (Biognosys) and subjected to data-independent acquisition (DIA) analysis. Peptides (~ 0.8 μg) were separated on an Easy nLC 1200 UHPLC system (Thermo Scientific) on an in-house packed 20 cm x 75 μm diameter C18 column (1.9 μm Reprosil-Pur C18-AQ beads (Dr. Maisch GmbH); Picofrit 10 μm opening (New Objective)). The column was heated to 50°C using a column heater (Phoenix-ST). The flow rate was 0.200 μl/min with 0.1% formic acid and 3% acetonitrile in water (A) and 0.1% formic acid, 90% acetonitrile (B). The peptides were separated with a 7–30% B gradient in 84 mins and analyzed using the Thermo Fusion Lumos mass spectrometer (Thermo Scientific). The DIA segment consisted of one MS1 scan (350-1650 m/z range, 120K resolution) followed by 30 MS2 scans (variable m/z range, 30K resolution). Additional parameters were as follows: MS1: RF Lens – 30%, AGC Target 4.0e5, Max IT – 50 ms, charge state include - 2-6; MS2: isolation width (m/z) – 0.7, AGC Target – 3.0e6, Max IT – 120 ms.
Spectral Library Generation
For spectral library generation, an aliquot (2 μg) of unlabeled, digested peptide material from individual tissue samples (HNSCC and NAT) was pooled and subjected to bRPLC as previously described. Collected fractions were concatenated into eight fractions by combining twelve fractions that are eight fractions apart (i.e., combining fractions #1, #9, #17, #25, #33, #41, #49, #57, #65, #73, #81, and #89; #2, #10, #18, #26, #34, #42, #50, #58, #66, #74, #82, and #90; and so on); dried down in a Speed-Vac, resuspended in 3% ACN, 0.1% formic acid, and was spiked with index Retention Time (iRT) peptides (Biognosys) prior to ESI-LC-MS/MS analysis. Parameters were the same as previously described for ESI-LC-MS/MS with a high-energy collision dissociation activation energy (HCD) – 34.
Protein Database Searching and Quantification
The spectral library file was generated based on PulSar search engine (SpectroNaut 13, Biognosys) against combined search results derived from DDA (n = 8) and DIA (n=203). In brief, the DDA files were searched with BGS factory search setting (default) against human database (uniprot released; 20,380 entries). MS1 and MS2 tolerance was set as “dynamic” and only tryptic peptides were allowed with two missed cleavages. Allowable modification on amino acid set to: oxidation (M, variable), acetylation (protein N-terminus, variable), and carbamidomethylation (C, fixed). The DIA files were searched using the PulSar search engine based on DIA umpire algorithm (Tsou et al., 2015) with default parameter as like DDA. To get homogenous protein inference and protein/peptide FDR of 1% in the integrated library, “generate library from search achieve” option was used on SpectroNaut. The spectral library precursor filter was set as follows: amino acid length > 3, relative intensity > 5%, m/z range between 300-1800, and best N fragment ions per peptide = 3~6. The retention times of these filtered PSMs were further transformed to indexed retention time (iRT) scale based on the standard peptides (iRT peptide kit, Biognosys) spiked into samples.
The integrated spectral library was loaded onto SpectroNaut (version13), and then targeted quantification was performed using default settings, as described in the previous report (Bruderer et al., 2017). The retention time of a peptide feature was transformed into iRT scale using the “local regression” option and the extracted ion chromatogram (XIC) retention time window was set to “dynamic” with 1 for the correction factor. The identified precursors and proteins were filtered at 1% of q-value was considered for the identification of precursor and protein (Reiter et al., 2011). Interference correction on corrections at MS1 and MS2 level was levels were enabled, removing fragments/isotopes from quantification based on the presence of interfering signals but keeping at least three for quantification. All abundances were calculated based on the area under the extracted ion chromatogram (XIC) of all assigned fragments that passed filtering.
Integrated Analysis
Protein-RNA Correlation
The gene-wise protein-RNA correlation for all genes quantifiable in both omics data types (N = 9579) was computed using Spearman’s correlation. The correlation significance was set at Benjamini-Hochberg adjusted p value < 0.01. Signed −log10 p value was used as the ranking metric for GSEA analysis using Webgestalt (Liao et al., 2019) to identify GO biological processes and KEGG pathways enriched for genes with low and high protein-RNA correlations, respectively.
Estimate Score
The ESTIMATE scores reflecting the overall immune and stromal infiltration were calculated by the R package ESTIMATE (Yoshihara et al., 2013) using the normalized RNA expression data (RSEM).
Differential Abundance Analysis
Differential analysis was performed for paired tumor and NAT samples using the Wilcoxon signed-rank test. Each feature was required to be non-missing in at least 50% of the paired samples. P values were adjusted using the Benjamini-Hochberg method and features were considered significant with an adjusted p value < 0.01. Proteomic and transcriptomic features with at least a median 2-fold increase in tumors were considered to be tumor-associated proteins. Over-representation analysis with the GO Biological Process (BP) gene sets was performed separately for proteins either increased or decreased >2-fold using WebGestaltR (Liao et al., 2019). The reference set was the proteins identified in at least 50% of the paired tumor and NAT samples. Over-representation analysis with the GO Biological Process (BP) gene sets was also performed separately for proteins with at least one phosphosite either increased or decreased >2-fold. The reference set was the proteins with an identified phosphosite in at least 50% of the paired tumor and NAT samples. GO BP terms were considered significant with a Benjamini-Hochberg (BH) adjusted p value < 0.01.
A linear mixed model (Kuznetsova et al., 2017) was used to identify differential proteins while controlling for non-epithelial content difference. Specifically, the z score-transformed ESTIMATE score (i.e., the sum of the ESTIMATE immune score and stromal score) was used as a fixed effect. Proteins with a significant (Benjamini-Hochberg adjusted p < 0.01) coefficient for tumor vs NAT expression change that remained greater than 1 after accounting for non-epithelial content were retained as tumor-associated proteins. Proteins were annotated as C/T Antigens (Almeida et al., 2009), secretable (Uhlén et al., 2015), and as the targets of FDA approved drugs (Human Protein Atlas, DrugBank). Phosphosites were annotated with the “ON_FUNCTION” and “ON_PROCESS” from PhosphoSitePlus.
Differential analysis was repeated for samples within a subgroup (e.g. larynx tumors, oral cavity tumors). Proteins were required to be non-missing in at least 50% of the paired tumor and NAT samples within the subgroup. Proteins increased 2-fold in one group but not the other were considered specific markers for that group.
Kinase Activity Inference
Activated kinases were first inferred from significantly increased phosphorylation of their sites annotated as ‘enzymatic activity, induced’ from PhosphoSitePlus. We also required these sites to have a median phosphorylation fold change greater than the median protein fold change in tumor compared to paired NAT. Kinase activity was additionally inferred from the phosphorylation of its substrates. Pre-ranked GSEA in WebGestaltR was performed on the signed −log10 p values from the differential abundance analysis using site-level substrates annotated in PhosphoSitePlus and UniProt. At least 10 substrates were required for each kinase. Kinases were considered significantly activated with a Benjamini-Hochberg adjusted p value < 0.05.
Prediction of Kinase Substrates
NetworKIN (Linding et al., 2007) was used to predict kinases for every identified phosphosite in the phosphoproteomics data. Substrate sets were generated using the combined set of known substrates from PhosphoSitePlus and UniProt used in Kinase Activity Inference and the predicted substrates from NetworKIN with a NetworKIN score ≥ 5. Kinase Activity Inference was performed as above with the new combined set of substrates.
Transcription Factor Activity Inference
Transcription factor activity for each sample was inferred using the VIPER package (Alvarez et al., 2016) on z-score transformed RNA data. The transcription factor targets were collected from DoRothEA (Garcia-Alonso et al., 2019) and the medium confidence targets were used for analysis. Activity scores for tumor and normal samples were compared using Student’s t-test and the p values were adjusted using the Benjamini-Hochberg method. Transcription factors with an adjusted p value < 0.05 were considered significant.
We also used the ARACNe algorithm (Lachmann et al., 2016) in VIPER to construct gene regulatory networks and infer transcription factor targets based on correlation to the transcription factor protein abundance. This allows for cancer-specific transcription factor gene regulation. We correlated the normalized enrichment protein activity scores with immune scores.
Stemness Score Inference
Stemness scores were calculated as previously described (Malta et al., 2018). We used MoonlightR (Colaprico et al., 2020) to query, download, and preprocess the pluripotent stem cell samples (ESC and iPSC) from the Progenitor Cell Biology Consortium (PCBC) dataset (Daily et al., 2017; Salomonis et al., 2016). To calculate the stemness scores based on mRNA expression, we built a predictive model using one-class logistic regression (OCLR) (Sokolov et al., 2016) on the PCBC dataset.
For mRNA expression-based signatures, to ensure compatibility with the CPTAC HNSCC cohort, we first mapped the gene names from Ensembl IDs to Human Genome Organization (HUGO), dropping any genes that had no such mapping. The resulting training matrix contained 12,955 mRNA expression values measured across all available PCBC samples. To calculate mRNA based stemness index (mRNASi) we used the FPKM (Fragments Per Kilobase Million) mRNA expression values for all the 161 HNSCC samples (108 tumor samples and 53 NAT samples).
We used the function TCGAanalyze_Stemness from the package TCGAbiolinks (Colaprico et al., 2016) and followed our previously-described workflow (Mounir et al., 2019)) with “stemSig” argument set to PCBC_stemSig.
MSigDB Hallmark Pathway single sample Gene Set Enrichment Analysis (ssGSEA)
ssGSEA was performed for each sample using gene-wise Z-scores of log2 upper-quartile normalized RNA-seq data for the MSigDB Hallmark gene sets (Liberzon et al., 2015) via WebGestaltR (Liao et al., 2019). For this analysis, read counts of 0 were treated as NAs prior to gene-wise normalization. Pathway activity scores are enrichment scores from ssGSEA.
Multi-Gene Proliferation Scores (MGPS)
MGPS were calculated as described previously (Ellis et al., 2017). Specifically, the scores are the mean of gene-wise Z-scores for log2 upper-quartile normalized RSEM data for all cell cycle-regulated genes identified by Whitfield et al. in each sample (Whitfield et al., 2002).
Curation of FAT1 and CDKN2A Genetic Aberration
In order to facilitate a granular analysis between tumor groups with distinct types of molecular loss of the tumor suppressor genes FAT1 and CDKN2A, a comprehensive tumor annotation was carried out using multiple molecular features for each patient. For example, for annotating CDKN2A, a gene largely affected by copy number events in HNSCC, we considered the following: 1) mutation types (Missense (including in-frame insertion and deletions (indels)), Truncation (stop gain, frameshift indels) and Splice site mutations) as separate categories. For cases with mutation, we next looked at both variant allele frequency of the mutation as adjudged from whole exome sequencing, estimated tumor purity, and copy number data (log ratio, absolute copy number, and B-allele frequency). Mutated samples with either lower tumor purity (as estimated from WGS) or with subclonal events based on mutation VAF, were excluded. Subsequently, cases with Truncation mutation + one copy loss were annotated as Bi-allelic truncation loss or truncation loss of heterozygosity (Biallelic. Trunc or truncation LOH). Cases with Splicing mutation with associated one copy loss were classified as Bi-allelic splicing or splicing LOH, and finally tumors with Missense mutations with associated one copy loss were annotated as missense LOH. While these tumors were classified as LOH mutants, the remaining mutant copy of CDKN2A frequently showed evidence for copy number amplification of the mutant alleles. 2) Based on CDKN2A copy number data, we next annotated tumors as Homozygous deletion (Homozyg. del.), one copy loss (Heterozyg. del) or no loss (Wild-type). Mutations affecting CDKN2A were assessed separately for both splice isoforms of the gene, p16INK4a and p14ARF.
Prioritizing Putative SCNA Drivers
The workflow to prioritize putative SCNA drivers was shown in Figure 1H. First, all the genes with quantifiable copy number, RNA expression, and proteomics (N = 9507) were filtered to keep the focal amplified genes, which were located in the segments identified by GISTIC2 with Q value < 0.25 (N = 759). These focal amplified genes were further filtered by their CN-mRNA correlation and next CN-protein correlation to keep the genes with significant CN cis-effect (BH adjusted p value < 0.01, Spearman’s correlation). Finally, the remaining genes (N = 356) were further filtered to keep the ones with significant higher protein levels in tumors than NATs (BH adjusted p value < 0.01, Student’s t-test), which generated a list of 202 putative SCNA driver genes. These genes were used to perform over-representation analysis against all quantifiable genes in the three omics to identify enriched GO biological processes.
cis-effect of DNA Methylations
To study the cis-effect of DNA methylation on mRNA and protein expression, we performed a multivariate correlation analysis that included SCNA and mutation effects as confounding variables using the software iProFun (Song et al., 2019). The DNA methylation levels were averaged from the CpG islands located in the upstream and nearby transcription start site (TSS) regions, including 5’UTR, 1st exon, and upstream TSS. Somatic mutations were considered if their mutation rate was >5%. In the analysis, we controlled age, gender, immune score, stromal score, inferred smoking status, and tumor location as covariates.
A significant association is identified if it passes three criteria: (1) biological filtering procedure highlighting significant methylations that are hyper- or hypo-methylated in tumor vs NAT, (2) posterior probabilities of associating to an outcome > 75%, and (3) empirical false discovery rate (eFDR) <10% from 100 permutations.
FAT1 Mutation and 11q13.3 Amplification Analysis
TCGA FAT1 mutation, CNV, and patient survival data were downloaded from cBioPortal (Gao et al., 2013). HPV+ samples were excluded from downstream analysis. 11q13.3 amplification was defined by the mean CNV values of the 9 genes in the focal region (mean CNVs > 1). These genes include FAT1, CCND1, LTO1, FGF19, FGF4, FGF3, ANO1, FADD, PPFIA1, and CTTN. Samples with both FAT1 truncation mutation and 11q13.3 amplification were randomly assigned to FAT1 truncation or 11q13.3 amplification groups. The R package “survival” (Therneau, 2020) was used to compare patient survival between groups.
Inferred HIPPO, WNT, and Apoptotic Activity Scores
All scores were inferred using protein abundance by the ssGSEA method from the GSVA R package (Barbie et al., 2009; Hänzelmann et al., 2013). The gene sets HIPPO pathway, WNT pathway, and apoptotic were from the KEGG pathway, Reactome pathway, and the MSigDB hallmark gene sets, respectively.
CDK4/6 Pathway Analysis
CCND1 amplified samples are those with copy-number log2 ratios >= 1 for the CCND1 gene. CDKN2A groups were defined as described above (Curation of FAT1 and CDKN2A Genetic Aberration). For cis analysis, wild-type samples are those without mutations, amplifications, or deletions of each respective gene, while wild-type samples for downstream analysis are those that are wild-type for both genes. For cis analysis of the effects of CDKN2A aberrations, transcript level log2 RSEM for the p16INK4a and p14ARF isoforms were evaluated separately, considering aberrations that affect the respective isoform. Scores for Hallmark E2F targets (ssGSEA) and multi-gene proliferation (MGPS) were calculated as described above. Rb phosphorylation is the mean of the 5 sites with complete data for all tumors that were annotated as CDK4 or CDK6 target sites in PhosphoSitePlus (pT252, pT356, pT373, pS780, and pS795). For the analysis presented in Figure 4D, the first group included tumors that were wild-type for all pathway genes (CDKN2A, CCND1, CDK6, and RB1; heterozygous deletions of CDKN2A were included if the retained allele was WT), while the second group included tumors with loss of expression aberrations affecting only CDKN2A/p16INK4a (CDKN2A homozygous deletion, p16 promoter hypermethylation, and p16 truncation LOH mutations), and the third group included tumors with CDKN2A/p16INK4a aberrations in combination with CCND1 amplifications.
EGFR Pathway Analysis
We inferred signaling activity of the EGFR pathway based on tumor mRNA expression data using PROGENy (Schubert et al., 2018), which computes pathway activity on the basis of mRNA abundance of a responsive gene set identified from a large compendium of publicly available cell line perturbation experiments. To identify ligand-depend EGFR pathway phosphorylation cascade, three omics measurements (RNA, protein, and site-level phosphorylation) for the curated pathway component genes (https://www.wikipathways.org/index.php/Pathway:WP437 and also from relevant literatures (Sigismund et al., 2018; Wee and Wang, 2017) were correlated to the average RNA level of five EGFR ligands (i.e., AREG, TGFA, EREG, EPGN, and HBEGF) using Pearson’s correlation. Only the phosphosites with both significant correlation (p < 0.01) and higher correlation coefficient than those from RNA and protein at the gene level were considered as evidence for phosphorylation-level regulation. Similar correlation analysis was performed to examine whether these pathway components were associated with the EGFR receptor protein level.
The pathway activity primarily driven by EGFR-amplification was investigated by comparing the highest six EGFR-amplified samples to the rest of the samples with high CN instability (ChrIdx > 3). Three omics measurements for (RNA, protein, and site-level phosphorylation) of all quantifiable genes/sites were used to perform Student’s t-test between the two groups. Only the phosphosites with both significant difference (p < 0.01) and higher fold-change than those from RNA and protein at gene level were considered as evidence for phosphorylation-level regulation. Over-representation analysis was performed on genes containing these qualified sites against all genes quantifiable in RNA, protein and phosphorylation to identify function enrichment of GO biological processes (BH adjusted p <0.05).
Driver Copy Number Deletions Associated with Low Immune Infiltration
The CN log2 ratio, RNA expression, and protein expression of quantifiable genes were correlated to the immune score, which was inferred by ESTIMATE (Yoshihara et al., 2013). For putative driver copy number deletions contributing to immune suppression, we required significant correlation for all three measurements to the immune score (Spearman’s correlation, BH adjusted p < 0.01). Meanwhile, the copy number was also required to have significant cis-effect (i.e., significant correlation from CN to both protein and RNA). SCNA effector genes were defined as those with significant correlation to the immune score at gene expression (both mRNA and protein) but not copy number level, indicating their changes occurred in trans. Over-representation analysis was performed using these qualified genes to identify pathway enrichment (adjusted p < 0.05).
Immune Cell Type Composition
The abundances of 64 different cell types in 162 HNSCC samples (108 tumor samples and 53 NAT samples) were computed via xCell (Aran et al., 2017). For this analysis, FPKM (Fragments Per Kilobase Million) mRNA expression values were utilized.
Neoantigen Identification
We used NeoFlow (Wen et al., 2020)(https://github.com/bzhanglab/neoflow) for neoantigen prediction. Specifically, Optitype (Szolek et al., 2014) was used to find human leukocyte antigens (HLA) in the WES data. Then we used netMHCpan 4.0 (Jurtz et al., 2017) to predict HLA peptide binding affinity for somatic mutation-derived variant peptides with a length between 8-11 amino acids. The IC50 binding affinity cutoff was set to 150 nM. HLA peptides with binding affinity higher than 150 nM were removed. Variant identification was also performed at both mRNA and protein levels using RNA-seq data and MS/MS data, respectively. To identify variant peptides, we used a customized protein sequence database approach (Wang et al., 2012). We derived customized protein sequence databases from matched WES data and then performed database searching using the customized databases for individual TMT experiments. We built a customized database for each TMT experiment based on somatic variants from WES data. We used Customprodbj (https://github.com/bzhanglab/customprodbj) for customized database construction. MS-GF+ was used for variant peptide identification for all global proteome and phosphorylation data. Results from MS-GF+ were filtered with 1% FDR at the PSM level. Remaining variant peptides were further filtered using PepQuery (http://www.pepquery.org) (Wen et al., 2019) with the p-value cutoff <= 0.01. The spectra of variant peptides were annotated using PDV (https://github.com/wenbostar/PDV) (Li et al., 2019).
Transcriptomics Subtypes
The transcriptomics-based subtyping was performed using the centroid-based method with previously established signature genes (Walter et al., 2013). Specifically, the RNA-seq matrix, represented as RSEM, were aggregated together with the TCGA RSEM matrix (downloaded from Broad GDAC Firehose data portal: https://gdac.broadinstitute.org/) and upper-quartile normalized. The RNA expression matrix was median-centered in the gene-wise manner, and each sample was correlated to each of the four centroid vectors representing the average signature gene expression for the four subtypes. The samples were assigned to different transcriptomics subtypes according to the highest correlations. Samples with an insignificant correlation to all subtypes (p > 0.01) were marked as ‘undecided’.
Unsupervised Subtyping Using Non-negative Matrix Factorization (NMF)
Non-negative matrix factorization (NMF) implemented in the NMF R package (Gaujoux and Seoighe, 2010) was used to perform unsupervised clustering of tumor samples and to identify proteogenomic features (proteins, phosphopeptide, mRNA transcripts, miRNAs, and somatic copy number alterations) that showed characteristic abundance patterns for each cluster. Briefly, given a factorization rank k (where k is the number of clusters), NMF decomposes a p x n data matrix V into two matrices W and H such that multiplication of W and H approximates V. Matrix H is a k x n matrix whose entries represent weights for each sample (1 to N) to contribute to each cluster (1 to k), whereas matrix W is a p x k matrix representing weights for each feature (1 to p) to contribute to each cluster (1 to k). Matrix H was used to assign samples to clusters by choosing the k with maximum score in each column of H. For each sample, we calculated a cluster membership score as the maximal fractional score of the corresponding column in matrix H. We defined a ”cluster core” as the set of samples with cluster membership score > 0.5. Matrix W containing the weights of each feature in a certain cluster was used to derive a list of representative features separating the clusters using the method proposed in (Kim and Park, 2007). Cluster-specific features were further subjected to a 2-sample moderated t-test (Ritchie et al., 2015) comparing the feature abundance between the respective cluster and all other clusters. Derived p-values were adjusted for multiple hypothesis testing using the methods proposed in (Benjamini and Hochberg, 1995).
Preprocessing of Data Tables
To enable integrative multi-omics clustering, we required all data types (and converted if necessary) to represent ratios to either a common reference measured in each TMT plex (proteome, phosphoproteome) or an in-silico common reference calculated as the median abundance across all samples (mRNA, mi-RNA). The phosphoproteome data consisted of phosphopeptides containing confidently localized sites. All data tables were then concatenated and only features non-missing in all tumors were used for subsequent analysis. Features with the lowest standard deviation (bottom 5th percentile) across all samples were deemed uninformative and were removed from the dataset. Each row in the data matrix was further scaled and standardized such that all features from different data types were represented as z-scores.
Since NMF requires a non-negative input matrix, the data matrix of z-scores was further converted into a non-negative matrix as follows:
Create one data matrix with all negative numbers zeroed.
Create another data matrix with all positive numbers zeroed and the signs of all negative numbers removed.
Concatenate both matrices resulting in a data matrix twice as large as the original, but with positive values only and zeros and hence appropriate for NMF.
Determination of Factorization Rank
The resulting matrix was then subjected to NMF analysis leveraging the NMF R package (Gaujoux and Seoighe, 2010) and using the factorization method described in (Brunet et al., 2004). To determine the optimal factorization rank k (number of clusters) for the multi-omic data matrix, a range of clusters between k=2 and 8 was tested. For each k we factorized matrix V using 50 iterations with random initializations of W and H. To determine the optimal factorization rank we calculated cophenetic correlation coefficients measuring how well the intrinsic structure of the data is recapitulated after clustering and chose the k with maximal cophenetic correlation for cluster numbers between k=3 and 8.
NMF Clustering
Having determined the optimal factorization rank k, and in order to achieve robust factorization of the multi-omics data matrix V, the NMF analysis was repeated using 500 iterations with random initializations of W and H and partitioning of samples into clusters as described above. Due to the non-negative transformation applied to the z-scored data matrix as described above, matrix W of feature weights contained two separate weights for positive and negative z-scores of each feature, respectively. In order to reverse the non-negative transformation and to derive a single signed weight for each feature, each row in matrix W was first normalized by dividing by the sum of feature weights in each row. Weights per feature and cluster were then aggregated by keeping the maximal normalized weight and multiplying with the sign of the z-score from the initial data matrix. Thus, the resulting transformed version of matrix Wsigned contained signed cluster weights for each feature present in the input matrix.
Subtype Signature Gene and Phosphosite Identifications
For each of the three subtypes, the subtype signature genes and phosphosites were defined as those significantly more abundant in that subtype compared to both of the other two subtypes (BH adjusted p value < 0.01, Student’s t-test). The subtype signature genes were identified at the RNA, protein, and phosphosite levels. All the subtype-significant genes and phosphosites (collapsed into gene-level) were used to perform over-representation analysis to identify enriched GO biological processes.
Multi-omics Gene Set Enrichment Analysis
GO biological processes enriched for each data type for a subtype were ranked by the signed −log10 p value and submitted to sumer (Savage et al., 2019). Briefly, sumer uses weighted set cover to remove redundant processes within a data type and clusters remaining pathways using affinity propagation (Frey and Dueck, 2007) based on the Simpson similarity index.
Chromosome Instability (CIN) Score
The CIN score (ChrIdx) reflects the overall copy number aberration across the whole genome. From the segmentation result, we used a weighted-sum approach to summarize the chromosome instability for each sample (Vasaikar et al., 2019). The absolute segment-level log2 ratios of all segments (indicating the copy number aberration of these segments) within a chromosome were weighted by the segment length and summed up to derive the instability score for the chromosome. The genome-wide chromosome instability index was calculated by summing up the instability score of all 22 autosomes.
Identification of High-potential Candidate Samples for Targeted Therapies
For each of the three targeted therapies, i.e., CDK4/6 inhibitors, EGFR mAb, and combinatorial immunotherapy, we proposed samples to be high-potential candidates for response to these three therapeutic strategies if they have both high level of the pathway activity targetable by the drug and high level of candidates reflecting the pathway activity. For CDK4/6 inhibitors, the pathway and candidates are CDK4/6-Rb1 phosphorylation-cell cycle (represented by phosphorylation of CDK4/6 substrates on Rb) and any of CCND1/CDKN2A genetic aberrations (including high CCND1 CN amplification, with CN log2 ratio > 1, CDKN2A bi-allelic loss or mutation), respectively. For EGFR mAb, the pathway and candidates are EGFR PROGENy pathway score and AREG or TGFA RNA expression, respectively. For combinatorial immunotherapy, the pathway and molecular signatures are ESTIMATE immune score and protein or RNA expression of immune checkpoint or suppressor molecules (e.g., PDL1). We required that the level of both pathways and molecular signatures from high-potential samples to be significantly higher than the rest of tumor samples (i.e., low-potential candidates) and also higher than the matched NAT samples. For each feature in the molecular signatures and pathways mentioned above, we performed univariate k-means clustering to cluster samples into low, medium, and high groups (k=3) using the Ckmeans.1d.dp R package (Wang and Song, 2011). Samples were assigned to the ‘high potential’ group if they belonged to the ‘high’ group based on the pathway clustering and simultaneously belonged to the ‘high’ group based on the clustering of at least one of the molecule signatures. The enrichment of each target therapies to the integrated subtypes was performed using Fisher’s exact test.
External Data Collection
For analyses using datasets from previously published, publicly available studies, we collected high throughput transcriptomic profiling and associated response data from NCBI GEO: GSE102995 (Siano et al., 2018) and GSE84713 (Klinghammer et al., 2017). 26 HPV-negative HNSCC PDX models were analyzed from GSE102995. For GSE84713, 1 out of 25 tumors was HPV-positive but not annotated in the sequencing files, therefore all 25 tumors were analyzed. In vitro genome-wide perturbation datasets of HNSCC cell lines using shRNA (McFarland et al., 2018) and CRISPR (Behan et al., 2019) (Wang and Song, 2011) were downloaded from DepMap (https://depmap.org/portal/). Of note, all of the HNSCC cell lines included here have CCND1 amplification and/or CDKN2A deletion, so genomic aberrations could not be used to predict response (Figure 4G, H). Phospho-Rb (Ser-807/811) and GAPDH loading control signals from untreated (control) HPV-negative HNSCC PDXs were quantified by densitometry analysis from western blotting data (Karamboulas et al., 2018) using Image Studio Lite Software (LI-COR). Normalized phospho-Rb abundance from each PDX was calculated by dividing phospho-Rb signals by their corresponding GAPDH signals. The TCGA data were downloaded from the Broad Firehose data portal (https://gdac.broadinstitute.org/). CDKN2A mutated gene matrix, CCND1 and CDKN2A copy number (GISTIC), CCND1 RSEM data, and CCND1 and Rb pS807/811 RPPA data for HPVneg TCGA tumors were downloaded from LinkedOmics (http://linkedomics.org).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using R unless explained otherwise. Multiple comparisons were adjusted by the Benjamini-Hochberg correction (Benjamini and Hochberg, 1995). Details can be found in Results and figure legends.
Supplementary Material
Table S1. Clinical information and molecular phenotypes of the samples analyzed in this study (related to Figure 1).
Table S2. Summary of significant genomic aberrations (related to Figure 1).
Table S3. Tumor-associated molecular features and protein changes and putative drug targets (related to Figure 2).
Table S5. Phosphosites associated with EGFR amplification (related to Figure 5).
Table S6. Proteomics supported putative neoantigens and putative immunosuppressive SCNAs (related to Figure 6).
Table S7. Subtype-specific molecular features and treatment opportunities (related to Figure 7).
Highlights.
A systematic inventory of HNSCC-associated proteins, phosphosites, and pathways
Three multi-omic subtypes linked to targeted treatment approaches and immunotherapy
Widespread deletion of immune modulatory genes accounts for loss of immunogenicity
Two modes of EGFR activation inform response to anti-EGFR monoclonal antibodies
Acknowledgments
This work was supported by grants U24 CA210954, U24 CA210985, U24 CA210972, U24 CA210979, U24 CA210986, U24 CA214125, U24 CA210967, and U24 CA210993 from the National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium (CPTAC), by a Cancer Prevention Institute of Texas (CPRIT) award RR160027, by grant T32 CA203690 from the Translational Breast Cancer Research Training Program, and by funding from the McNair Foundation. B.Z. is a CPRIT Scholar in Cancer Research and McNair Medical Institute Scholar. We thank Christopher J. Rickets (NCI) for helpful discussions.
Appendix
Secondary Authors
Anupriya Agarwal, Matthew L. Anderson, Shayan C. Avanessian, Dmitry Avtonomov, Oliver F. Bathe, Chet Birger, Michael J. Birrer, Lili Blumenberg, William E. Bocik, Uma Borate, Melissa Borucki, Meghan C. Burke, Shuang Cai, Anna Pamela Calinawan, Sandra Cerda, Alyssa Charamut, Lin S. Chen, Shrabanti Chowdhury, Karl R. Clauser, Houston Culpepper, Tomasz Czernicki, Fulvio D'Angelo, Jacob Day, Stephanie De Young, Emek Demir, Fei Ding, Marcin J. Domagalski, Joseph C. Dort, Brian Druker, Elizabeth Duffy, Maureen Dyer, Nathan J. Edwards, Kimberly Elburn, Tatiana S. Ermakova, David Fenyo, Renata Ferrarotto, Alicia Francis, Stacey Gabriel, Luciano Garofano, Yifat Geffen, Gad Getz, Linda I. Hannick, Pushpa Hariharan, David N. Hayes, David Heiman, Barbara Hindenach, Katherine A. Hoadley, Galen Hostetter, Martin Hyrcza, Scott D. Jewell, Corbin D. Jones, M. Harry Kane, Alicia Karz, Ramani B. Kothadia, Azra Krek, Chandan Kumar-Sinha, Tao Liu, Hongwei Liu, Jan Lubínski, Weiping Ma, Ewa Malc, Anna Malovannaya, Sailaja Mareedu, Sanford P. Markey, Annette Marrero-Oliveras, Nicollette Maunganidze, Jason E. McDermott, Peter B. McGarvey, John McGee, Piotr Mieczkowski, Simona Migliozzi, Rebecca Montgomery, Chelsea J. Newton, Umut Ozbek, Amanda G. Paulovich, Samuel H. Payne, Dimitar Dimitrov Pazardzhikliev, Amy M. Perou, Francesca Petralia, Lyudmila Petrenko, Paul D. Piehowski, Dmitris Placantonakis, Larisa Polonskaya, Elena V. Ponomareva, Olga Potapova, Liqun Qi, Ning Qu, Shakti Ramkissoon, Boris Reva, Shannon Richey, Karna Robinson, Nancy Roche, Karin Rodland, Daniel C. Rohrer, Dmitry Rykunov, Eric E. Schadt, Yan Shi, Yvonne Shutack, Shilpi Singh, Tara Skelly, Richard Smith, Lori J. Sokoll, Jakub Stawicki, Stephen E. Stein, James Suh, Wojciech Szopa, Dave Tabor, Donghui Tan, Darlene Tansil, Guo Ci Teo, Ratna R. Thangudu, Cristina Tognon, Elie Traer, Shirley Tsang, Jeffrey Tyner, Ki Sung Um, Dana R. Valley, Lyubomir Valkov Vasilev, Negin Vatanian, Uma Velvulou, Michael Vernon, Thomas F Westbrook, Jeffrey R. Whiteaker, Yige Wu, Midie Xu, Lijun Yao, Xinpei Yi, Fengchao Yu, Kakhaber Zaalishvili, Yuriy Zakhartsev, Robert Zelt, Grace Zhao, Jun Zhu
Consortia
Anupriya Agarwal, Eunkyung An, Matthew L. Anderson, Meenakshi Anurag, Shayan C. Avanessian, Dmitry Avtonomov, Oliver F. Bathe, Chet Birger, Michael J. Birrer, Lili Blumenberg, William E. Bocik, Emily S. Boja, Uma Borate, Melissa Borucki, Meghan C. Burke, Shuang Cai, Anna Pamela Calinawan, Liwei Cao, Steven A. Carr, Sandra Cerda, Daniel W. Chan, Hui-Yin Chang, Alyssa Charamut, Lijun Chen, Lin S. Chen, Xi S. Chen, Arul M. Chinnaiyan, Shrabanti Chowdhury, Marcin Cieślik, David J. Clark, Karl R. Clauser, Antonio Colaprico, Daniel Cui Zhou, Houston Culpepper, Tomasz Czernicki, Fulvio D'Angelo, Felipe da Veiga Leprevost, Jacob Day, Stephanie De Young, Emek Demir, Saravana M. Dhanasekaran, Fei Ding, Li Ding, Marcin J. Domagalski, Joseph C. Dort, Yongchao Dou, Brian Druker, Elizabeth Duffy, Maureen Dyer, Nathan J. Edwards, Adel K. El-Naggar, Kimberly Elburn, Matthew J. Ellis, Tatiana S. Ermakova, David Fenyo, Alicia Francis, Stacey Gabriel, Luciano Garofano, Yifat Geffen, Gad Getz, Michael A. Gillette, Charles A. Goldthwaite, Linda I. Hannick, Pushpa Hariharan, David N. Hayes, David Heiman, Tara Hiltke, Barbara Hindenach, Katherine A. Hoadley, Galen Hostetter, Yingwei Hu, Chen Huang, Martin Hyrcza, Eric J. Jaehnig, Scott D. Jewell, Jiayi Ji, Wen Jiang, Corbin D. Jones, M. Harry Kane, Alicia Karz, Karen A. Ketchum, Christopher R. Kinsinger, Ramani B. Kothadia, Azra Krek, Karsten Krug, Chandan Kumar-Sinha, Jonathan T. Lei, Kai Li, Qing Kay Li, Yize Li, Wen-Wei Liang, Yuxing Liao, Hongwei Liu, Tao Liu, Jan Lubínski, Weiping Ma, Ewa Malc, Anna Malovannaya, D. R. Mani, Sailaja Mareedu, Sanford P. Markey, Annette Marrero-Oliveras, Nicollette Maunganidze, Jason E. McDermott, Peter B. McGarvey, John McGee, Mehdi Mesri, Piotr Mieczkowski, Simona Migliozzi, George Miles, Rebecca Montgomery, Alexey I. Nesvizhskii, Chelsea J. Newton, Gilbert S. Omenn, Umut Ozbek, Jianbo Pan, Amanda G. Paulovich, Samuel H. Payne, Dimitar Dimitrov Pazardzhikliev, Amy M. Perou, Francesca Petralia, Lyudmila Petrenko, Alexander R. Pico, Paul D. Piehowski, Dmitris Placantonakis, Larisa Polonskaya, Elena V. Ponomareva, Olga Potapova, Liqun Qi, Jiang Qian, Ning Qu, Shakti Ramkissoon, Boris Reva, Shannon Richey, Karna Robinson, Ana I. Robles, Nancy Roche, Karin Rodland, Henry Rodriguez, Daniel C. Rohrer, Dmitry Rykunov, Shankha Satpathy, Sara R. Savage, Eric E. Schadt, Michael Schnaubelt, Yan Shi, Zhiao Shi, Yvonne Shutack, Andrew Sikora, Shilpi Singh, Tara Skelly, Richard Smith, Lori J. Sokoll, Xiaoyu Song, Jakub Stawicki, Stephen E. Stein, James Suh, Wojciech Szopa, Dave Tabor, Donghui Tan, Darlene Tansil, Guo Ci Teo, Ratna R. Thangudu, Mathangi Thiagarajan, Cristina Tognon, Elie Traer, Shirley Tsang, Jeffrey Tyner, Ki Sung Um, Dana R. Valley, Rodrigo Vargas Eguez, Lyubomir Valkov Vasilev, Negin Vatanian, Pankaj Vats, Uma Velvulou, Michael Vernon, Pei Wang, Yuefan Wang, Alissa M. Weaver, Bo Wen, Michael Wendl, Thomas F Westbrook, Jeffrey R. Whiteaker, Maciej Wiznerowicz, Yige Wu, Matthew A. Wyczalkowski, Midie Xu, Lijun Yao, Xinpei Yi, Seungyeul Yoo, Fengchao Yu, Kakhaber Zaalishvili, Yuriy Zakhartsev, Robert Zelt, Bing Zhang, Hui Zhang, Zhen Zhang, Grace Zhao, Jun Zhu
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
SAC is a member of the scientific advisory boards of Kymera, PTM BioLabs and Seer and a scientific advisor to Pfizer and Biogen. Other authors have no conflict of interests to declare.
References
- Adkins D, Ley J, Neupane P, Worden F, Sacco AG, Palka K, Grilley-Olson JE, Maggiore R, Salama NN, Trinkaus K, et al. (2019). Palbociclib and cetuximab in platinum-resistant and in cetuximab-resistant human papillomavirus-unrelated head and neck cancer: a multicentre, multigroup, phase 2 trial. Lancet Oncol 20, 1295–1305. [DOI] [PubMed] [Google Scholar]
- Almeida LG, Sakabe NJ, deOliveira AR, Silva MCC, Mundstein AS, Cohen T, Chen Y-T, Chua R, Gurung S, Gnjatic S, et al. (2009). CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res 37, D816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH, and Califano A (2016). Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet 48, 838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang KK, Zhang Q, Rosenthal DI, Nguyen-Tan PF, Sherman EJ, Weber RS, Galvin JM, Bonner JA, Harris J, El-Naggar AK, et al. (2014). Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol 32, 2940–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Hu Z, and Butte AJ (2017). xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 18, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argentini A, Goeminne LJE, Verheggen K, Hulstaert N, Staes A, Clement L, and Martens L (2016). moFF: a robust and automated approach to extract peptide ion intensities. Nat Methods 13, 964–966. [DOI] [PubMed] [Google Scholar]
- Babiceanu M, Qin F, Xie Z, Jia Y, Lopez K, Janus N, Facemire L, Kumar S, Pang Y, Qi Y, et al. (2016). Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res 44, 2859–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. (2009). Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Trigo JM, Bourhis J, Tortochaux J, Cortés-Funes H, Hitt R, Gascón P, Amellal N, Harstrick A, and Eckardt A (2005). Phase II multicenter study of the antiepidermal growth factor receptor monoclonal antibody cetuximab in combination with platinum-based chemotherapy in patients with platinum-refractory metastatic and/or recurrent squamous cell carcinoma of the head and neck. J Clin Oncol 23, 5568–5577. [DOI] [PubMed] [Google Scholar]
- Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, et al. (2019). Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 568, 511–516. [DOI] [PubMed] [Google Scholar]
- Bellman R (1961). On the approximation of curves by line segments using dynamic programming. Commun ACM 4, 284. [Google Scholar]
- Benada J, Burdová K, Lidak T, von Morgen P, and Macurek L (2015). Polo-like kinase 1 inhibits DNA damage response during mitosis. Cell Cycle 14, 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benelli M, Pescucci C, Marseglia G, Severgnini M, Torricelli F, and Magi A (2012). Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics 28, 3232–3239. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300. [Google Scholar]
- Brandes JC, van Engeland M, Wouters KAD, Weijenberg MP, and Herman JG (2005). CHFR promoter hypermethylation in colon cancer correlates with the microsatellite instability phenotype. Carcinogenesis 26, 1152–1156. [DOI] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, and Jemal A (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424. [DOI] [PubMed] [Google Scholar]
- Bruderer R, Bernhardt OM, Gandhi T, Xuan Y, Sondermann J, Schmidt M, Gomez-Varela D, and Reiter L (2017). Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility of Results. Molecular & cellular proteomics: MCP 16, 2296–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet J-P, Tamayo P, Golub TR, and Mesirov JP (2004). Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA 101, 4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtness B, Goldwasser MA, Flood W, Mattar B, Forastiere AA, and Eastern Cooperative Oncology G (2005). Phase III randomized trial of cisplatin plus placebo compared with cisplatin plus cetuximab in metastatic/recurrent head and neck cancer: an Eastern Cooperative Oncology Group study. J Clin Oncol 23, 8646–8654. [DOI] [PubMed] [Google Scholar]
- Büttner M, Miao Z, Wolf FA, Teichmann SA, and Theis FJ (2019). A test metric for assessing single-cell RNA-seq batch correction. Nat Methods 16, 43–49. [DOI] [PubMed] [Google Scholar]
- Calmon MF, Jeschke J, Zhang W, Dhir M, Siebenkäs C, Herrera A, Tsai H-C, O'Hagan HM, Pappou EP, Hooker CM, et al. (2015). Epigenetic silencing of neurofilament genes promotes an aggressive phenotype in breast cancer. Epigenetics 10, 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N (2015). Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Wendl MC, Wyczalkowski MA, Wylie K, Ye K, Jayasinghe R, Xie M, Wu S, Niu B, Grubb R, et al. (2016). Divergent viral presentation among human tumors and adjacent normal tissues. Sci Rep 6, 28294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang B, Schnaubelt M, Shah P, Aiyetan P, Chan D, Zhang H, and Zhang Z (2018). MS-PyCloud: An open-source, cloud computing-based pipeline for LC-MS/MS data analysis. bioRxiv, 320887. [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, and Saunders CT (2016). Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222. [DOI] [PubMed] [Google Scholar]
- Chu A, Robertson G, Brooks D, Mungall AJ, Birol I, Coope R, Ma Y, Jones S, and Marra MA (2016). Large-scale profiling of microRNAs for The Cancer Genome Atlas. Nucleic Acids Res 44, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, et al. (2004). Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 5, 489–500. [DOI] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, and Getz G (2013). Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Cerami E, Sander C, and Schultz N (2012). Mutual exclusivity analysis identifies oncogenic network modules. Genome Res 22, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DJ, Dhanasekaran SM, Petralia F, Pan J, Song X, Hu Y, da Veiga Leprevost F, Reva B, Lih T-SM, Chang H-Y, et al. (2019). Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell 179, 964–983.e931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DJ, Hu Y, Bocik W, Chen L, Schnaubelt M, Roberts R, Shah P, Whiteley G, and Zhang H (2018). Evaluation of NCI-7 Cell Line Panel as a Reference Material for Clinical Proteomics. J Proteome Res 17, 2205–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A, Olsen C, Bailey MH, Odom GJ, Terkelsen T, Silva TC, Olsen AV, Cantini L, Zinovyev A, Barillot E, et al. (2020). Interpreting pathways to discover cancer driver genes with Moonlight. Nat Commun 11, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. (2016). TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 44, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombet T, Osorio M, Cruz T, Roca C, del Castillo R, Mon R, Iznaga-Escobar N, Figueredo R, Koropatnick J, Renginfo E, et al. (2004). Use of the humanized anti-epidermal growth factor receptor monoclonal antibody h-R3 in combination with radiotherapy in the treatment of locally advanced head and neck cancer patients. J Clin Oncol 22, 1646–1654. [DOI] [PubMed] [Google Scholar]
- Daily K, Ho Sui SJ, Schriml LM, Dexheimer PJ, Salomonis N, Schroll R, Bush S, Keddache M, Mayhew C, Lotia S, et al. (2017). Molecular, phenotypic, and sample-associated data to describe pluripotent stem cell lines and derivatives. Sci Data 4, 170030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cárcer G, Wachowicz P, Martínez-Martínez S, Oller J, Méndez-Barbero N, Escobar B, González-Loyola A, Takaki T, El Bakkali A, Cámara JA, et al. (2017). Plk1 regulates contraction of postmitotic smooth muscle cells and is required for vascular homeostasis. Nat Med 23, 964–974. [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Mendoza L, Shteynberg D, Slagel J, Sun Z, and Moritz RL (2015). Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clin Appl 9, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Aksnes H, Marie M, Boczkowska M, Varland S, Timmerman E, Foyn H, Glomnes N, Rebowski G, Impens F, et al. (2018). NAA80 is actin's N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc Natl Acad Sci USA 115, 4399–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Suman VJ, Hoog J, Goncalves R, Sanati S, Creighton CJ, DeSchryver K, Crouch E, Brink A, Watson M, et al. (2017). Ki67 Proliferation Index as a Tool for Chemotherapy Decisions During and After Neoadjuvant Aromatase Inhibitor Treatment of Breast Cancer: Results From the American College of Surgeons Oncology Group Z1031 Trial (Alliance). Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology 35, 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris RL, Haddad R, Even C, Tahara M, Dvorkin M, Ciuleanu TE, Clement PM, Mesia R, Kutukova S, Zholudeva L, et al. (2020). Durvalumab with or without tremelimumab in patients with recurrent or metastatic head and neck squamous cell carcinoma: EAGLE, a randomized, open-label phase III study. Ann Oncol 31, 942–950. [DOI] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. (2011). A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 12, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J-P, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, and Hansen KD (2014). Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 15, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey BJ, and Dueck D (2007). Clustering by passing messages between data points. Science (New York, NY) 315, 972–976. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Liang W-W, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, Liao W-W, Reynolds SM, Wyczalkowski MA, Yao L, et al. (2018). Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep 23, 227–238.e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang J, and Zhao F (2015). CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 16, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alonso L, Holland CH, Ibrahim MM, Turei D, and Saez-Rodriguez J (2019). Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 29, 1363–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, and Seoighe C (2010). A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey S, and Lebarbier E (2008). Using CART to Detect Multiple Change Points in the Mean for Large Sample). [Google Scholar]
- Ghantous Y, Schussel JL, and Brait M (2018). Tobacco and alcohol-induced epigenetic changes in oral carcinoma. Curr Opin Oncol 30, 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Dobin A, Li B, Stransky N, Pochet N, and Regev A (2019). Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol 20, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, and Guinney J (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding J, and Burtness B (2005). Cetuximab: an epidermal growth factor receptor chemeric human-murine monoclonal antibody. Drugs Today 41, 107–127. [DOI] [PubMed] [Google Scholar]
- Hein JB, Hertz EPT, Garvanska DH, Kruse T, and Nilsson J (2017). Distinct kinetics of serine and threonine dephosphorylation are essential for mitosis. Nat Cell Biol 19, 1433–1440. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Arquette M, Shin DM, Dicke K, Vokes EE, Azarnia N, Hong WK, and Kies MS (2005). Phase II multicenter study of the epidermal growth factor receptor antibody cetuximab and cisplatin for recurrent and refractory squamous cell carcinoma of the head and neck. J Clin Oncol 23, 5578–5587. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, and Skrzypek E (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43, D512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, and Nielsen M (2017). NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol 199, 3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justilien V, Jameison L, Der CJ, Rossman KL, and Fields AP (2011). Oncogenic activity of Ect2 is regulated through protein kinase C iota-mediated phosphorylation. J Biol Chem 286, 8149–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamboulas C, Bruce JP, Hope AJ, Meens J, Huang SH, Erdmann N, Hyatt E, Pereira K, Goldstein DP, Weinreb I, et al. (2018). Patient-Derived Xenografts for Prognostication and Personalized Treatment for Head and Neck Squamous Cell Carcinoma. Cell Rep 25, 1318–1331.e1314. [DOI] [PubMed] [Google Scholar]
- Kasar S, Kim J, Improgo R, Tiao G, Polak P, Haradhvala N, Lawrence MS, Kiezun A, Fernandes SM, Bahl S, et al. (2015). Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat Commun 6, 8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M, Rieke D, Endhardt K, Fang P, Brägelmann J, et al. (2015). Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res 21, 870–881. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, and Aebersold R (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74, 5383–5392. [DOI] [PubMed] [Google Scholar]
- Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, Rosenberg JE, Van Allen EM, D'Andrea A, and Getz G (2016). Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet 48, 600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, and Pevzner PA (2014). MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Källberg M, Chen X, Kim Y, Beyter D, Krusche P, and Saunders CT (2018). Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 15, 591–594. [DOI] [PubMed] [Google Scholar]
- Klinghammer K, Otto R, Raguse J-D, Albers AE, Tinhofer I, Fichtner I, Leser U, Keilholz U, and Hoffmann J (2017). Basal subtype is predictive for response to cetuximab treatment in patient-derived xenografts of squamous cell head and neck cancer. Int J Cancer 141, 1215–1221. [DOI] [PubMed] [Google Scholar]
- Klymkowsky MW, and Parr B (1995). The body language of cells: the intimate connection between cell adhesion and behavior. Cell 83, 5–8. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, and Wilson RK (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, and Nesvizhskii AI (2017). MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods 14, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz D, and Boutros M (2014). A synthetic lethal screen identifies FAT1 as an antagonist of caspase-8 in extrinsic apoptosis. EMBO J 33, 181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer AR, Clifford GM, Boyle P, and Franceschi S (2005). Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev 14, 467–475. [DOI] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, and Christensen RHB (2017). lmerTest Package: Tests in Linear Mixed Effects Models. Journal of Statistical Software 82, 1–26. [Google Scholar]
- Lachmann A, Giorgi FM, Lopez G, and Califano A (2016). ARACNe-AP: gene network reverse engineering through adaptive partitioning inference of mutual information. Bioinformatics 32, 2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebarbier E (2005). Detecting multiple change-points in the mean of Gaussian process by model selection. Signal Processing 85, 717–736. [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Vaudel M, Zhang B, Ren Y, and Wen B (2019). PDV: an integrative proteomics data viewer. Bioinformatics 35, 1249–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xie X, Zhou J, Sheng M, Yin X, Ko E-A, Zhou T, and Gu W (2017). Quantifying circular RNA expression from RNA-seq data using model-based framework. Bioinformatics 33, 2131–2139. [DOI] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, and Zhang B (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47, W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RGH, Supek F, and Lehner B (2016). The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet 48, 1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Ostheimer GJ, van Vugt MATM, Jørgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. (2007). Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, Bork P, Yaffe MB, and Pawson T (2008). NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res 36, D695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovly CM, Yan L, Ryan CE, Takada S, and Piwnica-Worms H (2008). Regulation of Chk2 ubiquitination and signaling through autophosphorylation of serine 379. Mol Cell Biol 28, 5874–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGrath SM, and Koleske AJ (2012). Cortactin in cell migration and cancer at a glance. J Cell Sci 125, 1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J, Omberg L, Gevaert O, et al. (2018). Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 173, 338–354.e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaria JP, Ratto E, Gozzelino L, Li H, and Hirsch E (2019). Class II PI3Ks at the Intersection between Signal Transduction and Membrane Trafficking. Biomolecules 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Degese MS, Vitale-Cross L, Iglesias-Bartolome R, Valera JLC, Wang Z, Feng X, Yeerna H, Vadmal V, Moroishi T, et al. (2018). Assembly and activation of the Hippo signalome by FAT1 tumor suppressor. Nat Commun 9, 2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, Krill-Burger JM, Green TM, Vazquez F, Boehm JS, et al. (2018). Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun 9, 4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, and DePristo MA (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, and Getz G (2011). GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, and Carr SA (2013). Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods 10, 634–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, Clauser KR, Clauss TR, Shah P, Gillette MA, et al. (2018). Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc 13, 1632–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messersmith WA, and Hidalgo M (2007). Panitumumab, a monoclonal anti epidermal growth factor receptor antibody in colorectal cancer: another one or the one? Clin Cancer Res 13, 4664–4666. [DOI] [PubMed] [Google Scholar]
- Min HL, Kim J, Kim WH, Jang BG, and Kim MA (2016). Epigenetic silencing of the putative tumor suppressor gene GLDC (glycine dehydrogenase) in gastric carcinoma. Anticancer Res 36, 179–187. [PubMed] [Google Scholar]
- Mounir M, Lucchetta M, Silva TC, Olsen C, Bontempi G, Chen X, Noushmehr H, Colaprico A, and Papaleo E (2019). New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput Biol 15, e1006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, and Aebersold R (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, et al. (2020). Quantitative proteomics of the cancer cell line encyclopedia. Cell 180, 387–402.e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow SY, Salim M, Noirel J, Evans C, and Wright PC (2011). Minimising iTRAQ ratio compression through understanding LC-MS elution dependence and high-resolution HILIC fractionation. Proteomics 11, 2341–2346. [DOI] [PubMed] [Google Scholar]
- Papillon-Cavanagh S, Lu C, Gayden T, Mikael LG, Bechet D, Karamboulas C, Ailles L, Karamchandani J, Marchione DM, Garcia BA, et al. (2017). Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet 49, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jean M, Rigaill G, and Neuvial P (2015). Performance evaluation of DNA copy number segmentation methods. Brief Bioinform 16, 600–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psyrri A, Lee J-W, Pectasides E, Vassilakopoulou M, Kosmidis EK, Burtness BA, Rimm DL, Wanebo HJ, and Forastiere AA (2014). Prognostic biomarkers in phase II trial of cetuximab-containing induction and chemoradiation in resectable HNSCC: Eastern cooperative oncology group E2303. Clin Cancer Res 20, 3023–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauniyar N, and Yates JR (2014). Isobaric labeling-based relative quantification in shotgun proteomics. J Proteome Res 13, 5293–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter L, Rinner O, Picotti P, Hüttenhain R, Beck M, Brusniak M-Y, Hengartner MO, and Aebersold R (2011). mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods 8, 430–435. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, Bekele BN, and Aldape KD (2010). MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol 12, 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Dexheimer PJ, Omberg L, Schroll R, Bush S, Huo J, Schriml L, Ho Sui S, Keddache M, Mayhew C, et al. (2016). Integrated Genomic Analysis of Diverse Induced Pluripotent Stem Cells from the Progenitor Cell Biology Consortium. Stem Cell Reports 7, 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuray RT, Johnson DE, and Grandis JR (2018). New therapies in head and neck cancer. Trends Cancer 4, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, and Cheetham RK (2012). Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817. [DOI] [PubMed] [Google Scholar]
- Savage SR, Shi Z, Liao Y, and Zhang B (2019). Graph Algorithms for Condensing and Consolidating Gene Set Analysis Results. Molecular & cellular proteomics: MCP 18, S141–S152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski MM, Wilhelm M, Hahne H, Kuster B, and Bantscheff M (2015). A Scalable Approach for Protein False Discovery Rate Estimation in Large Proteomic Data Sets. Molecular & cellular proteomics: MCP 14, 2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Klinger B, Klünemann M, Sieber A, Uhlitz F, Sauer S, Garnett MJ, Blüthgen N, and Saez-Rodriguez J (2018). Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat Commun 9, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, Heath K, McClanahan T, Lunceford J, Gause C, et al. (2016). Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol 17, 956–965. [DOI] [PubMed] [Google Scholar]
- Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JWB, Blenis J, Pende M, and Sonenberg N (2006). The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J 25, 2781–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shteynberg DD, Deutsch EW, Campbell DS, Hoopmann MR, Kusebauch U, Lee D, Mendoza L, Midha MK, Sun Z, Whetton AD, and Moritz RL (2019). PTMProphet: Fast and Accurate Mass Modification Localization for the Trans-Proteomic Pipeline. J Proteome Res 18, 4262–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siano M, Espeli V, Mach N, Bossi P, Licitra L, Ghielmini M, Frattini M, Canevari S, and De Cecco L (2018). Gene signatures and expression of miRNAs associated with efficacy of panitumumab in a head and neck cancer phase II trial. Oral Oncol 82, 144–151. [DOI] [PubMed] [Google Scholar]
- Sigismund S, Avanzato D, and Lanzetti L (2018). Emerging functions of the EGFR in cancer. Mol Oncol 12, 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov A, Paull EO, and Stuart JM (2016). ONE-CLASS DETECTION OF CELL STATES IN TUMOR SUBTYPES. Pac Symp Biocomput 21, 405–416. [PMC free article] [PubMed] [Google Scholar]
- Song X, Ji J, Gleason KJ, Yang F, Martignetti JA, Chen LS, and Wang P (2019). Insights into Impact of DNA Copy Number Alteration and Methylation on the Proteogenomic Landscape of Human Ovarian Cancer via a Multi-omics Integrative Analysis. Molecular & cellular proteomics: MCP 18, S52–S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallaert W, Brüggemann Y, Sabet O, Baak L, Gattiglio M, and Bastiaens PIH (2018). Contact inhibitory Eph signaling suppresses EGF-promoted cell migration by decoupling EGFR activity from vesicular recycling. Sci Signal 11. [DOI] [PubMed] [Google Scholar]
- Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, and Kohlbacher O (2014). OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan VYF, and Févotte C (2013). Automatic relevance determination in nonnegative matrix factorization with the β-divergence. IEEE Trans Pattern Anal Mach Intell 35, 1592–1605. [DOI] [PubMed] [Google Scholar]
- Tanoue T, and Takeichi M (2004). Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J Cell Biol 165, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. (2019). COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 47, D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM (2020). A Package for Survival Analysis in R. In. [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. (2017). Defining a cancer dependency map. Cell 170, 564–576.e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou C-C, Avtonomov D, Larsen B, Tucholska M, Choi H, Gingras A-C, and Nesvizhskii AI (2015). DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Methods 12, 258–264, 257 p following 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science (New York, NY) 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Vasaikar S, Huang C, Wang X, Petyuk VA, Savage SR, Wen B, Dou Y, Zhang Y, Shi Z, Arshad OA, et al. (2019). Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell 177, 1035–1049.e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasaikar SV, Straub P, Wang J, and Zhang B (2018). LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 46, D956–D963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer H-R, Cupissol D, et al. (2008). Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med 359, 1116–1127. [DOI] [PubMed] [Google Scholar]
- Vermorken JB, Trigo J, Hitt R, Koralewski P, Diaz-Rubio E, Rolland F, Knecht R, Amellal N, Schueler A, and Baselga J (2007). Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. J Clin Oncol 25, 2171–2177. [DOI] [PubMed] [Google Scholar]
- Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, Ang MK, Hayward MC, Salazar AH, Hoadley KA, et al. (2013). Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS ONE 8, e56823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, and Song M (2011). Ckmeans.1d.dp: Optimal k-means Clustering in One Dimension by Dynamic Programming. R J 3, 29–33. [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma Z, Carr SA, Mertins P, Zhang H, Zhang Z, Chan DW, Ellis MJC, Townsend RR, Smith RD, et al. (2017). Proteome Profiling Outperforms Transcriptome Profiling for Coexpression Based Gene Function Prediction. Molecular & cellular proteomics: MCP 16, 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence JC, Sturgill TW, and Harris TE (2009). Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J Biol Chem 284, 14693–14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang J, Wan L, Zhou X, Wang Z, and Wei W (2015). Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol Ther 151, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Slebos RJC, Wang D, Halvey PJ, Tabb DL, Liebler DC, and Zhang B (2012). Protein identification using customized protein sequence databases derived from RNA-Seq data. J Proteome Res 11, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, and Zhang B (2013). customProDB: an R package to generate customized protein databases from RNA-Seq data for proteomics search. Bioinformatics 29, 3235–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee P, and Wang Z (2017). Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Li K, Zhang Y, and Zhang B (2020). Cancer neoantigen prioritization through sensitive and reliable proteogenomics analysis. Nat Commun 11, 1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wang X, and Zhang B (2019). PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations. Genome Res 29, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, and Botstein D (2002). Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, and Ning Z (2009). Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4, 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecha J, Satpathy S, Kanashova T, Avanessian SC, Kane MH, Clauser KR, Mertins P, Carr SA, and Kuster B (2019). TMT Labeling for the Masses: A Robust and Cost-efficient, In-solution Labeling Approach. Molecular & cellular proteomics: MCP 18, 1468–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, White NM, Schmidt HK, Fulton RS, Tomlinson C, Warren WC, Wilson RK, and Maher CA (2016). INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res 26, 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Gordon W, Du S, Zhang C, He W, Xi L, Mathur S, Agostino M, Paradis T, von Schack D, et al. (2017). QuickMIRSeq: a pipeline for quick and accurate quantification of both known miRNAs and isomiRs by jointly processing multiple samples from microRNA sequencing. BMC Bioinformatics 18, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinical information and molecular phenotypes of the samples analyzed in this study (related to Figure 1).
Table S2. Summary of significant genomic aberrations (related to Figure 1).
Table S3. Tumor-associated molecular features and protein changes and putative drug targets (related to Figure 2).
Table S5. Phosphosites associated with EGFR amplification (related to Figure 5).
Table S6. Proteomics supported putative neoantigens and putative immunosuppressive SCNAs (related to Figure 6).
Table S7. Subtype-specific molecular features and treatment opportunities (related to Figure 7).
Data Availability Statement
Raw proteomics data files are hosted by the CPTAC Data Portal and can be accessed at: https://proteomics.cancer.gov/data-portal. Genomic and transcriptomic data files can be accessed via the Genomic Data Commons (GDC) Data Portal: https://portal.gdc.cancer.gov. Processed data utilized for this publication can be accessed via LinkedOmics: http://www.linkedomics.org.
Several customized coding software packages were generated as part of this study and have been referenced in the corresponding STAR Methods section and listed with links to the coding script in the Key Resources Table: software codes generated by the Cieslik laboratory for genomic analyses (CNVEX), by the Nesvizhskii laboratory for proteomic data processing (Philosopher and TMT-Integrator), and by the Zhang lab for data processing and neoantigen detection (NeoFlow and PepQuery).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid | Alfa Aesar | Catalog: J63218 |
| Acetonitrile, Optima LC/MS | Fisher Chemical | A955-4 |
| Ammonium Hydroxide solution | Sigma | Catalog: 338818 |
| Aprotinin | Sigma | Catalog: A6103 |
| CD3 antibody (polyclonal) | Agilent - Dako | #A0452 |
| DAB | Agilent - Dako | #K3468 |
| Dithiothreitol | Thermo Fisher Scientific | Catalog: 20291 |
| Envision+ System HRP labelled polymer, anti-Rabbit | Agilent - Dako | #K4002 |
| Ethylenediaminetetraacetic acid | Sigma | Catalog: E7889 |
| Formic acid | Fisher Chemical | Catalog: A117-50 |
| Hydroxylamine solution | Aldrich | Catalog: 467804 |
| Iodoacetamide | Thermo Fisher Scientific | Catalog: A3221 |
| Iron (III) chloride | Sigma | Catalog:451649 |
| Leupeptin | Roche | Catalog: 11017101001 |
| Lysyl endopeptidase, aass spectrometry grade | Wako Chemicals | Catalog: 125-05061 |
| Ni-NTA agarose beads | QIAGEN | Catalog: 30410 |
| PUGNAc | Sigma | Catalog: A7229 |
| Phenylmethylsulfonyl fluoride | Sigma | Catalog: 93482 |
| Phosphatase Inhibitor Cocktail 2 | Sigma | Catalog: P5726 |
| Phosphatase Inhibitor Cocktail 3 | Sigma | Catalog: P0044 |
| Reversed-phase C18 SepPak | Waters | Catalog: WAT054925 |
| Sequencing grade modified trypsin | Promega | Catalog: V511X |
| Sodium chloride | Santa Cruz Biotechnology | Catalog: sc-295833 |
| Sodium fluoride | Sigma | Catalog: S7920 |
| TMT11-131C label reagent | Thermo Fisher Scientific | Catalog: A34807 |
| Tandem mass tags – 10plex | Thermo Fisher Scientific | Catalog: 90406 |
| Trifluoroacetic acid | Sigma | Catalog: 302031 |
| Tris(hydroxymethyl)amino methane | Invitrogen | Catalog: AM9855G |
| Urea | Sigma | Catalog: U0631 |
| Water, Optima LC/MS | Fisher Chemical | W6-4 |
| Critical Commercial Assays | ||
| TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold | Illumina | Catalog: RS-122-2301 |
| Infinium MethylationEPIC Kit | Illumina | Catalog: WG-317-1003 |
| Nextera DNA Exosome Kit | Illumina | Catalog: 20020617 |
| KAPA Hyper Prep Kit, PCR-free | Roche | Catalog: 07962371001 |
| BCA Protein Assay Kit | ThermoFisher Scientific | Catalog: 23225 |
| Software and Algorithms | ||
| Software | Source | Identifier (i.e. links) |
| Bowtie (v1.1.1) | (Langmead et al., 2009) | https://sourceforge.net/projects/bowtie-bio/files/bowtie/ |
| Bowtie2 (v2.3.3) | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| BWA (v0.7.17-r1188) | (Li and Durbin, 2009) | http://bio-bwa.sourceforge.net/ |
| cBioPortal | (Cerami et al., 2012; Gao et al., 2013) | https://www.cbioportal.org |
| CIRI (v2.0.6) | (Gao et al., 2015) | https://sourceforge.net/projects/ciri/ |
| CNVEX | Marcin Cieslik Lab | https://github.com/mctp/cnvex |
| Customprodbj | (Wang and Zhang, 2013) | https://github.com/bzhanglab/customprodbj |
| EricScript v0.5.5 | (Benelli et al., 2012) | https://sites.google.com/site/bioericscript/ |
| ESTIMATE | (Yoshihara et al., 2013) | https://bioinformatics.mdanderson.org/public-software/estimate/ |
| germlinewrapper v1.1 | Li Ding Lab | https://github.com/ding-lab/germlinewrapper |
| GISTIC2 | (Mermel et al., 2011) | https://www.genepattern.org/modules/docs/GISTIC_2.0 |
| GSVA | (Hänzelmann et al., 2013) | https://bioconductor.org/packages/release/bioc/html/GSVA.html |
| lmerTest | (Kuznetsova et al., 2017) | https://cran.r-project.org/web/packages/lmerTest/index.html |
| INTEGRATE v0.2.6 | (Zhang et al., 2016) | https://sourceforge.net/projects/integrate-fusion/ |
| iProFUN | (Song et al., 2019) | https://github.com/WangLab-MSSM/iProFun |
| LinkedOmics | (Vasaikar et al., 2018) | http://linkedomics.org |
| Manta v1.6.0 | (Chen et al., 2016) | https://github.com/Illumina/manta |
| MoonlightR | (Colaprico et al., 2020) | http://bioconductor.org/packages/MoonlightR/ |
| MSFragger-20190628 | (Kong et al., 2017) | http://msfragger.nesvilab.org/ |
| MS-GF+ | (Kim and Pevzner, 2014) | https://github.com/MSGFPlus/msgfplus/ |
| MuTect v1.1.7 | (Cibulskis et al., 2013) | https://github.com/broadinstitute/mutect |
| NeoFlow | (Wen et al., 2020) | https://github.com/bzhanglab/neoflow |
| NetworKIN | (Linding et al., 2008) | https://networkin.info |
| OmicsEV | Bing Zhang lab | https://github.com/bzhanglab/OmicsEV |
| OmicsOne | Hui Zhang Lab | https://github.com/huizhanglab-jhu/OmicsOne |
| PepQuery | (Wen et al., 2019) | http://pepquery.org/ |
| PeptideProphet | (Keller et al., 2002) | http://tools.proteomecenter.org/wiki/index.php?title=Main_Page |
| PDV | (Li et al., 2019) | https://github.com/wenbostar/PDV |
| Philosopher-v1.6.0 | Alexey Nesvizhskii lab | https://philosopher.nesvilab.org/ |
| Pindel v0.2.5 | (Ye et al., 2009) | https://github.com/genome/pindel |
| ProteinProphet | (Nesvizhskii et al., 2003) | http://tools.proteomecenter.org/wiki/index.php?title=Main_Page |
| PTMProphet | (Shteynberg et al., 2019) | http://tools.proteomecenter.org/wiki/index.php?title=Main_Page |
| QuickMIRSeq | (Zhao et al., 2017) | https://sourceforge.net/projects/quickmirseq/ |
| RSEM (v1.3.1) | (Li and Dewey, 2011) | https://deweylab.github.io/RSEM/ |
| Samtools (V1.1.0) | (Li et al., 2009) | https://github.com/samtools/samtools |
| SignatureAnalyzer | (Kim et al., 2016) | https://software.broadinstitute.org/cancer/cga/msp |
| somaticwrapper v1.5 | Li Ding Lab | https://github.com/ding-lab/somaticwrapper |
| STAR-Fusion v1.5.0 | (Haas et al., 2019) | https://github.com/STAR-Fusion/STAR-Fusion |
| Strelka v2.9.2 | (Kim et al., 2018) | https://github.com/Illumina/strelka |
| Sumer | (Savage et al., 2019) | https://github.com/bzhanglab/sumer |
| TCGAbiolinks | (Colaprico et al., 2016) | http://bioconductor.org/packages/TCGAbiolinks/ |
| TMT-Integrator-v1.0.9 | Alexey Nesvizhskii lab | http://tmt-integrator.nesvilab.org/ |
| VarScan v2.3.8 | (Koboldt et al., 2012) | https://dkoboldt.github.io/varscan/ |
| VIPER | (Alvarez et al., 2016) | http://califano.c2b2.columbia.edu/viper |
| VirusScan | (Cao et al., 2016) | https://github.com/ding-lab/VirusScan/tree/simplified |
| WebgestaltR | (Liao et al., 2019) | http://www.webgestalt.org/ |
| xCell | (Aran et al., 2017) | http://xcell.ucsf.edu/ |
| Deposited Data | ||
| cBioPortal | (Cerami et al., 2012) | https://www.cbioportal.org/ |
| CTDatabase | (Almeida et al., 2009) | http://www.cta.lncc.br |
| DoRothEA | (Garcia-Alonso et al., 2019) | https://github.com/saezlab/DoRothEA |
| Human Protein Atlas | (Uhlén et al., 2015) | https://www.proteinatlas.org |
| MSigDBv7.0 Hallmark gene sets | (Liberzon et al., 2015) | https://www.gsea-msigdb.org |
| PhosphoSitePlus | (Hornbeck et al., 2015) | https://www.phosphosite.org |