Abstract
Borrelia (Borreliella) burgdorferi, along with closely related species, is the etiologic agent of Lyme disease. The spirochete subsists in an enzootic cycle that encompasses acquisition from a vertebrate host to a tick vector and transmission from a tick vector to a vertebrate host. To adapt to its environment and persist in each phase of its enzootic cycle, B. burgdorferi wields three systems to regulate the expression of genes: the RpoN-RpoS alternative sigma (σ) factor cascade, the Hk1/Rrp1 two-component system and its product c-di-GMP, and the stringent response mediated by RelBbu and DksA. These regulatory systems respond to enzootic phase-specific signals and are controlled or fine-tuned by transcription factors, including BosR and BadR, as well as small RNAs, including DsrABb and Bb6S RNA. In addition, several other DNA-binding and RNA-binding proteins have been identified, although their functions have not all been defined. Global changes in gene expression revealed by high-throughput transcriptomic studies have elucidated various regulons, albeit technical obstacles have mostly limited this experimental approach to cultivated spirochetes. Regardless, we know that the spirochete, which carries a relatively small genome, regulates the expression of a considerable number of genes required for the transitions between the tick vector and the vertebrate host as well as the adaptation to each.
Introduction
The Lyme disease spirochete Borrelia (Borreliella) burgdorferi, and closely related species B. afzelii, B. garinii, B. bissettii, and B. mayonii, are maintained in an enzootic cycle involving a tick vector and a vertebrate host (Figure 1) (Lane et al., 1991; Radolf et al., 2012; Caimano et al., 2016). The spirochetes regulate the expression of their genes in a phase-specific fashion as they traverse this cycle in nature, moving back and forth between the tick and a vertebrate. Ixodes larvae feed on an infected host and acquire the spirochetes, which take up residence in the midgut. Following the molt, nymphs feed on a vertebrate and the spirochetes exit the midgut, migrate through the hemocoel, reach the salivary glands, and transmit to the naïve host (Ribeiro et al., 1987; Dunham-Ems et al., 2009). The spirochete survives in these two disparate habitats, and moves between them at the proper time, by sensing biological cues that alter the expression of the requisite collection of genes. Different ‘modulons’ are expressed as B. burgdorferi cycles from tick to vertebrate and vertebrate to tick, and adapts to these different environments (Iyer et al., 2015). The physiological signposts of the enzootic cycle, molecular cues prompting the spirochete to regulate its gene expression, have not been fully delineated nor have the mechanisms of signal integration and transduction to the regulatory machinery been elucidated. Schwan et al. (1995) uncovered temperature as an environmental signal; these seminal findings suggested that the increased temperature of the warm blood meal regulates the levels of outer membrane lipoproteins during tick feeding. B. burgdorferi is exposed to a variety of other factors that vary during the enzootic cycle and have been shown to affect gene expression, including pH (Carroll et al., 1999; Yang et al., 2000), dissolved oxygen (Seshu et al., 2004a) and carbon dioxide (Hyde et al., 2007), transition metals (Troxell et al., 2013), nutrients (Bugrysheva et al., 2015; Drecktrah et al., 2015), osmolarity (Bontemps-Gallo et al., 2016), weak organic acids (Dulebohn et al., 2017), and short-chain fatty acids (Lin et al., 2018). Nevertheless, uncertainty remains regarding the relative roles of each and their interactions.
Figure 1.
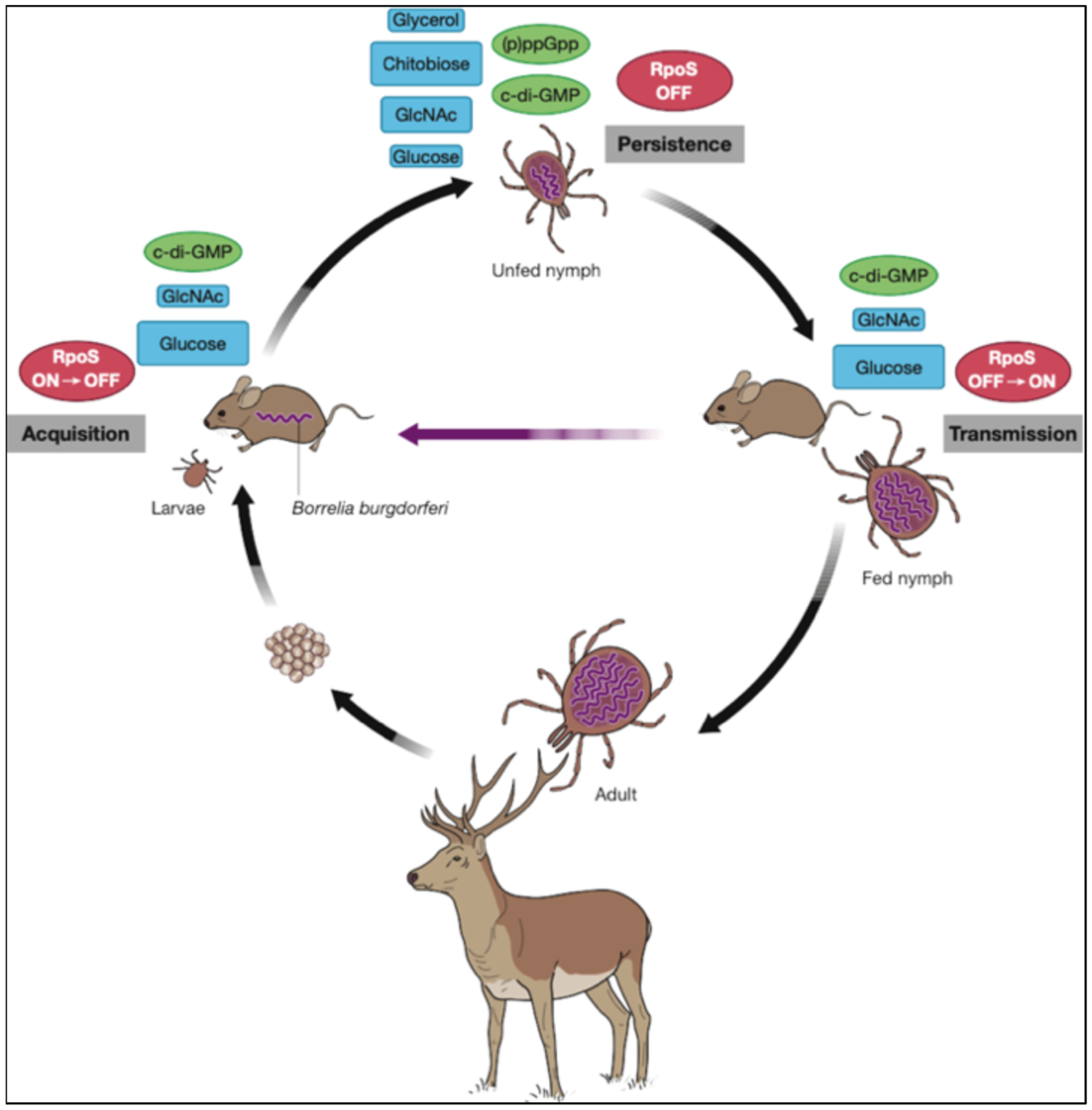
The enzootic cycle of B. burgdorferi showing milestones indicating the action of gene regulatory systems. Acquisition: larval ticks acquire B. burgdorferi by feeding on an infected vertebrate. Persistence: intracellular second messengers (p)ppGpp and c-di-GMP regulate persistence in the tick by various mechanisms, including control of genes encoding carbohydrate utilization, while the molecular gatekeeper RpoS is absent in unfed ticks. Transmission: following the molt into nymphs, infected ticks can transmit B. burgdorferi to uninfected vertebrates, completing the cycle. Reprinted with permission from Caimano et al. (2016). GlcNAc, N-acetyl glucosamine.
This review covers our current understanding of gene regulation during the enzootic cycle; since the original chapter was written (Skare et al., 2010), a number of recommended reviews have been published (Samuels, 2011; Radolf et al., 2012; Kung et al., 2013; Troxell and Yang, 2013; Groshong and Blevins, 2014; Caimano et al., 2016; Iyer and Schwartz, 2016; Samuels and Samuels, 2016; Ye et al., 2016; Lybecker and Samuels, 2017; Stevenson and Seshu, 2018). We begin the review by introducing the enzyme catalyzing transcription, and invoke the maxim spirochetes do it differently, as the B. burgdorferi RNA polymerase is unusually, albeit not uniquely, dependent on manganese (Boyle et al., 2020). We know of three main gene regulatory systems that are in play throughout the enzootic cycle: the RpoN-RpoS alternative sigma (σ) factor cascade; the Hk1/Rrp1 two-component system and its second messenger c-di-GMP; and stringent response regulators, RelBbu and DksA, and the alarmone (p)ppGpp. Orthologs of several transcriptional regulators that have been thoroughly characterized in model microorganisms are involved in modulating these gene regulatory networks, although in ways that, more often than not, are utterly idiosyncratic for this deeply branching group of bacteria (Samuels and Radolf, 2009). We discuss nucleic-acid binding proteins and mention a relatively recently described signaling molecule. We then take up post-transcriptional mechanisms, focusing on small regulatory RNAs, before turning our attention to what has been learned via high-throughput transcriptomic approaches.
RNA polymerase
The transcription of genes in B. burgdorferi is carried out by the RNA polymerase holoenzyme consisting of subunits encoded by rpoA (α subunit), rpoB (β subunit), rpoC (β′ subunit), and rpoZ (ω subunit), which constitute the core RNA polymerase, along with one of three σ factors encoded by rpoD (housekeeping sigma factor σ70 or σD), rpoN (σN or σ54), or rpoS (σS or σ38). It has the same α2ββ′ω plus σ architecture as other bacterial RNA polymerases. The σ subunit is required for recognition of gene promoters and separation of the two strands of the double helix (Marchetti et al., 2017). The σ subunit associates with the α2ββ′ω core to initiate transcription and then elongation proceeds, often after the σ subunit dissociates from the holoenzyme. The largest subunit, β′, houses the active site that catalyzes RNA synthesis. The ω subunit promotes RNA polymerase activity in other bacteria (Mathew and Chatterji, 2006; Kurkela et al., 2020); however, its role in the B. burgdorferi RNA polymerase has yet to be characterized.
Unlike prototypical bacterial RNA polymerases that require magnesium for activity (Sosunov et al., 2003), the enzymatic activity of the B. burgdorferi RNA polymerase is manganese-dependent (Boyle et al., 2020). Manganese-dependent RNA polymerases have been found in other bacteria, including Clostridium acetobutylicum, Bacillus subtilis, and Lactobacillus curvatus (Rutberg and Armentrout, 1972; Stetter and Zillig, 1974; Pich and Bahl, 1991). The molecular basis of the manganese requirement for the B. burgdorferi RNA polymerase is unclear, as the conserved catalytic site amino acids within the β′ subunit are identical to those found in both magnesium- and manganese-utilizing bacteria. The spirochete regulates the cellular concentrations of manganese through the BmtA transporter (Ouyang et al., 2009a; Wagh et al., 2015), and alterations in extracellular levels of manganese affect the expression of a variety of virulence genes (Troxell and Yang, 2013; Troxell et al., 2013). Changes in B. burgdorferi gene expression in response to lower concentrations of manganese may be the consequence of reducing the bioavailability of manganese required for RNA polymerase activity, resulting in transcriptional stalling as described in other bacteria (Rutberg and Armentrout, 1972; Stetter and Zillig, 1974; Borbely and Schneider, 1988; Pich and Bahl, 1991; Sosunov et al., 2003; Poranen et al., 2008; Agapov et al., 2017).
The enzymatic activity of the B. burgdorferi RNA polymerase also responds to temperature and pH (Boyle et al., 2020), which is notable because the environmental temperature and pH vary as B. burgdorferi traverses its enzootic cycle (Carroll et al., 1999; Schwan and Piesman, 2000; Yang et al., 2000). In vitro transcription assays initiated from RpoD (σ70)-dependent promoters using the B. burgdorferi RNA polymerase core and recombinant RpoD σ factor showed RNA polymerase activity is affected by temperature with maximal activity at 37°C. Similarly, RNA polymerase activity is pH-dependent within the physiological range experienced by B. burgdorferi with the lowest activity observed at pH 6.8 and the highest activity at pH 7.5 (Boyle et al., 2020). These in vitro experiments suggest regulation of B. burgdorferi gene expression in response to vector and host environments might be a consequence, at least partially, of biochemical changes in RNA polymerase activity.
During growth of B. burgdorferi in vitro, RpoD (σ70) is the primary σ factor responsible for the initiation of transcription, as the alternative σ factors, RpoN (σN) and RpoS (σS), are either inactive or present at low levels in the cell during logarithmic growth of B. burgdorferi (Hübner et al., 2001; Burtnick et al., 2007; Bontemps-Gallo et al., 2016). Sites of RpoD-dependent transcriptional initiation were mapped by in vitro transcription reactions initiated by recombinant RpoD (Adams et al., 2017; Boyle et al., 2020). The ability of B. burgdorferi to complete its enzootic cycle is dependent on RNA polymerase utilization of RpoN and RpoS (see the next section on the RpoN-RpoS alternative σ factor cascade). The expression of rpoN and rpoS, along with corresponding changes in RpoN- and RpoS-dependent gene expression, occur in response to environmental signals. While much of the work on gene regulation has focused on the impact of transcription factors, alternative σ factors and small RNAs in B. burgdorferi, the intracellular environment, including pH, temperature, and manganese availability, may directly alter RNA polymerase function along with the interactions of gene regulatory systems with RNA polymerase.
RpoN-RpoS alternative σ factor cascade
The RpoN-RpoS alternative σ factor cascade has undoubtedly been the most studied gene regulatory pathway in B. burgdorferi (Samuels, 2011; Radolf et al., 2012; Samuels and Samuels, 2016; Ye et al., 2016; Stevenson and Seshu, 2018) (Figure 2). It plays a vital role in the enzootic cycle of B. burgdorferi by activating transcription of genes that are essential for transmission and vertebrate host infection, while repressing genes that are required for spirochete survival in the tick vector (Hübner et al., 2001; Caimano et al., 2004; Caimano et al., 2005; Fisher et al., 2005; Caimano et al., 2007; Ouyang et al., 2008; Dunham-Ems et al., 2012; Grove et al., 2017; Arnold et al., 2018). The housekeeping σ factor (e.g., σ70 in E. coli) is responsible for transcription of most genes (see the section on RNA polymerase above). On the other hand, alternative σ factors recognize different promoters and allow for transcription of a subpopulation of genes in response to environmental or developmental signals (Kazmierczak et al., 2005; Österberg et al., 2011). RpoN (σN or σ54; bb0450) and RpoS (σS or σ38; bb0771) are the only two alternative σ factors encoded in the B. burgdorferi genome (Fraser et al., 1997). RpoN directly controls rpoS expression, which constitutes the RpoN-RpoS σ factor cascade (or RpoN-RpoS pathway).
Figure 2.
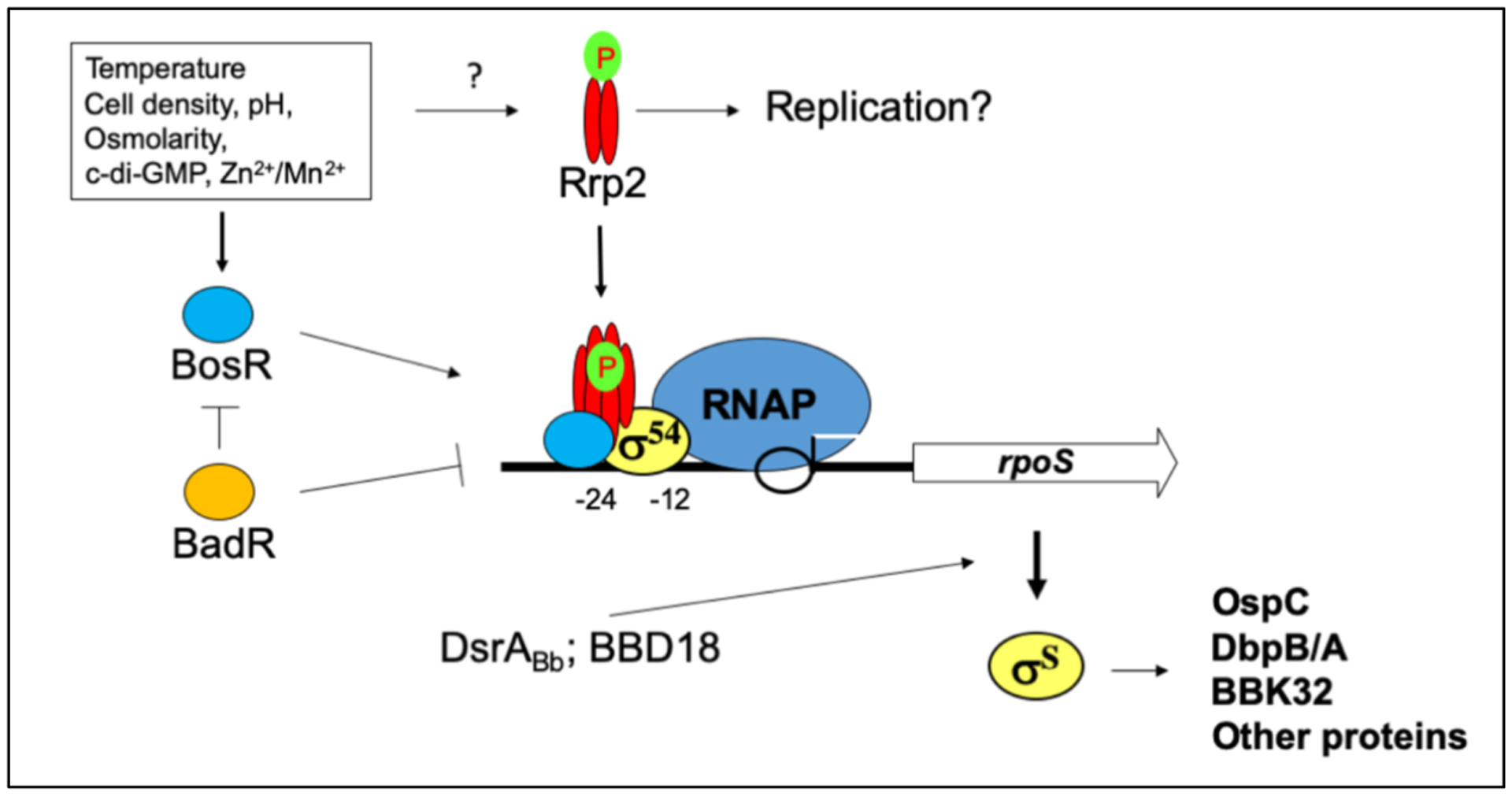
A model of the RpoN-RpoS σ factor cascade. During tick feeding and mammalian infection, environmental and host signals activate the RpoN-RpoS σ factor cascade. Activation of RpoN requires phosphorylation of Rrp2 and accumulation of BosR. Rrp2 is the sole prokaryotic enhancer-binding protein present in B. burgdorferi that is required for RpoN (σ54) activation. Phosphorylation of Rrp2 not only is required for RpoN-RpoS activation, but also is indispensable for cell survival, presumably replication. Levels of BosR respond to environmental signals and accumulation of BosR activates rpoS at its RpoN-dependent promoter via an unknown mechanism. BadR represses rpoS transcription by directly binding near the RpoN-dependent promoter region. In addition to the major rpoS mRNA species transcribed from the RpoN-dependent promoter, a longer rpoS transcript is produced at low cell density from an RpoN-independent promoter located within the upstream flgJ gene. The sRNA DsrABb regulates the efficiency of long rpoS mRNA species translation in response to temperature. DDB18 can regulate RpoS (σS) levels at the post-transcriptional level. Accumulation of rpoS transcript leads to the production of OspC, DbpA, DbpB, BBK32, and other mammalian infection-associated proteins.
RpoN is a unique σ factor that does not belong to the σ70 (RpoD) family (Reitzer and Schneider, 2001; Wigneshweraraj et al., 2008). RpoN-dependent gene expression possesses several distinctive features. First, RpoN-holoenzyme (Eσ54) exclusively requires a transcriptional activator, called bacterial enhancer binding protein (bEBP) (Bush and Dixon, 2012). B. burgdorferi has a single bEBP, Rrp2 (Yang et al., 2003). Second, RpoN recognizes a highly conserved −24/−12 promoter sequence that differs dramatically from the −35/−10 promoter sequence recognized by σ70 family σ factors (Studholme and Buck, 2000). Third, RpoN-dependent gene activation requires ATP binding (although not hydrolysis). As such, RpoN controls the expression of only a few genes in most bacteria (Reitzer and Schneider, 2001; Kazmierczak et al., 2005). In B. burgdorferi, the only gene with an RpoN-type promoter identified is rpoS, which has a perfect −24/−12 promoter sequence (Hübner et al., 2001; Smith et al., 2007).
RpoS from B. burgdorferi is evolutionarily distinct from the prototypical RpoS of ɣ-proteobacteria (Chiang and Schellhorn, 2010). However, it retains key features of its namesake, including the presence of a region σ1.2 and promoter recognition via an extended −10 (Eggers et al., 2006; Caimano et al., 2019). In most bacteria, RpoS serves to globally regulate gene expression to adapt to environmental stresses, such as entering stationary growth phase (reviewed in Hengge-Aronis, 2002a). RpoS accumulates following low or high temperature, low pH, high osmolarity, oxidative stress, UV exposure, and carbon starvation, resulting in regulation of genes to address the stressful conditions and prevent cellular damage. Yet, in B. burgdorferi, RpoS controls expression of enzootic phase-specific genes rather than the general stress response, and is required for infection of the vertebrate host (Caimano et al., 2004; Caimano et al., 2019) as well as transmission during tick feeding (Fisher et al., 2005; Dunham-Ems et al., 2012). RpoS in B. burgdorferi induces vertebrate host-specific genes and represses tick vector-specific genes (Hübner et al., 2001; Caimano et al., 2004; Caimano et al., 2005; Fisher et al., 2005; Caimano et al., 2007; Ouyang et al., 2008; Grove et al., 2017; Arnold et al., 2018) and, thus, RpoS has been designated the “gatekeeper” of the enzootic cycle (Figure 1) (Caimano et al., 2007; Mulay et al., 2009; Caimano et al., 2019).
RpoN and RpoS constitute a σ factor cascade
The discovery of the RpoN-RpoS σ factor cascade by Hübner et al. (2001) was made possible by inactivation of the rpoS and rpoN genes in an infectious B. burgdorferi strain. Both rpoS and rpoN mutants showed a striking phenotype: loss of production of OspC and DbpB/A, two of the abundant surface antigens. Abrogation of OspC and DbpB/A in the rpoN mutant was due to abolished production of RpoS, and constitutive expression of rpoS in the rpoN mutant rescued OspC and DbpB/A production. RpoN controlling rpoS activation appeared to be direct, as the rpoS gene has a perfect consensus −24/−12 RpoN-dependent promoter sequence (Hübner et al., 2001; Burtnick et al., 2007; Lybecker and Samuels, 2007; Smith et al., 2007). Further biochemical and genetic analyses indicated that the −24/−12 RpoN-dependent promoter is the major promoter responsible for cell density-dependent rpoS expression (Burtnick et al., 2007; Lybecker and Samuels, 2007; Smith et al., 2007).
To date, rpoS is the only gene identified whose transcription has been demonstrated to be directly controlled by RpoN in B. burgdorferi. Virtually all differentially expressed genes in the rpoS and rpoN mutants are overlapping (Ouyang et al., 2008), supporting the hypothesis that RpoN modulates B. burgdorferi genes mainly through RpoS; however, other data suggest that RpoN and RpoS can regulate different groups of genes under certain conditions (Fisher et al., 2005; Arnold et al., 2018). In addition, a second, RpoD (σ70)-dependent, promoter (−35/−10) has been reported upstream that transcribes a longer rpoS mRNA (Lybecker and Samuels, 2007). Translation of this long rpoS mRNA is regulated by the small RNA (sRNA) DsrABb (Lybecker and Samuels, 2007) and may explain the controversy regarding the RpoN-dependent and RpoS-dependent transcriptomes (see section below on Small regulatory RNAs and RNA chaperones). There is evidence that RpoS is post-transcriptionally regulated by BBD18 encoded on the linear plasmid lp17, although the mechanistic details are unknown (Dulebohn et al., 2014) (Figure 2).
Genes controlled by RpoN-RpoS σ factor cascade
The first genes identified as being under the control of RpoN-RpoS σ factor cascade were ospC and dbpBA (Hübner et al., 2001); microarray and RNA-seq analyses have revealed that the RpoN-RpoS σ factor cascade influences the expression of more than 100 genes (Fisher et al., 2005; Caimano et al., 2007; Boardman et al., 2008; Ouyang et al., 2008; Arnold et al., 2018; Caimano et al., 2019) (see the section on Transcriptomics below). Many of the vertebrate phase-specific are under the control of the RpoN-RpoS σ factor cascade and include, in addition to ospC and dbpBA, bbk32, oppA5, bba64, bba66, ospF and mlp (Fischer et al., 2003; Eggers et al., 2004; Grimm et al., 2004; Clifton et al., 2006; Fischer et al., 2006; Seshu et al., 2006; Gilmore et al., 2007; Medrano et al., 2007; Gautam et al., 2008; Maruskova et al., 2008; Shi et al., 2008). Many of these genes are differentially regulated under a variety of environmental conditions including temperature, pH, blood, and host adaptation (Revel et al., 2002; Brooks et al., 2003; Ojaimi et al., 2003; Tokarz et al., 2004; Caimano et al., 2007). These genes often encode surface lipoproteins, but many are hypothetical genes with no known function. Several chromosomal genes, including genes involved in chemotaxis and metabolism are also controlled by the RpoN-RpoS σ factor cascade (Fisher et al., 2005; Caimano et al., 2007; Ouyang et al., 2008). Recently, Caimano et al. (2019) compared the RpoS regulons of two B. burgdorferi strains and postulated that the RpoN-RpoS σ factor cascade controls two cohorts of genes: a core group that is required for maintenance of B. burgdorferi in the enzootic cycle and a group of plasmid-encoded genes encoding variable surface lipoproteins important for adaptation to diverse vertebrate reservoir hosts.
RpoS controls B. burgdorferi gene expression both directly and indirectly. Genetic evidence indicates that the −35/−10 promoters of ospC, ospF and several other RpoS-regulated genes are recognized exclusively by RpoS; in other words, RpoS directly governs ospC expression by binding to the ospC promoter (Eggers et al., 2004; Yang et al., 2005). RpoD (σ70) and RpoS (σS) are closely related and recognize similar core promoter elements (Gaal et al., 2001; Hengge-Aronis, 2002b). Promoter selectivity by the two σ factors in E. coli is complicated, involving minor sequence differences, architectural DNA-binding proteins, or DNA supercoiling (Kusano et al., 1996; Colland et al., 2000; Becker and Hengge-Aronis, 2001; Gaal et al., 2001; Hengge-Aronis, 2002b; Typas et al., 2007). Alternative σ factor utilization by RNA polymerase is also influenced by 6S RNA (Wassarman and Storz, 2000; Cavanagh and Wassarman, 2014; Steuten et al., 2014a; Steuten et al., 2014b; Wassarman, 2018) (see section below on Small regulatory RNAs and RNA chaperones). In B. burgdorferi, the sequence within the extended −10 region of the promoter contributes to RpoS recognition and a point mutation of C to G at position −13 abolished RpoS-dependent expression of ospC (Eggers et al., 2004; Yang et al., 2005; Caimano et al., 2019). Replacing the −10 region of the RpoD-dependent promoter of the ospE gene with the −10 region of the RpoS-dependent promoter of ospF converted ospE to an RpoS-dependent gene (Eggers et al., 2006). A putative RpoS-dependent consensus extended −10 region sequence of TG(G/A)(G/A)ATA(T/A)ATT has been proposed based on analyzing the promoters of ten RpoS-dependent genes of B. burgdorferi (Caimano et al., 2007). Although ospC transcription is chiefly controlled by RpoS, low levels of ospC mRNA have been detected in the absence of RpoS: He et al. (2008) reported that ospC expression was greatly reduced, but not completely eliminated in a rpoS ospAB double mutant. A recent transcriptome study detected ospC expression in the rpoN and rpoS mutants (Arnold et al., 2018). Given that RpoD and RpoS recognize similar promoter sequences, this ospC mRNA is likely due to leaky transcription mediated by RpoD.
RpoS appears also to regulate B. burgdorferi gene expression in an indirect manner. The bba66 genes has a 29-bp inverted repeat (IR) element upstream of the promoter that is required for transcription, suggesting that RpoS may influence bba66 indirectly via an unidentified factor that binds to the IR motif (Clifton et al., 2006). In addition, some genes are only partially controlled by RpoS, such as gltP (encoding a glutamate transporter) and cdr (encoding NAD-dependent coenzyme A disulphide reductase). These genes likely have a promoter that is recognized by both RpoS and RpoD (Caimano et al., 2007; Eggers et al., 2011).
In addition to activating transcription, RpoS represses expression of some B. burgdorferi genes (Caimano et al., 2005; Caimano et al., 2007; Grove et al., 2017; Arnold et al., 2018). Several of these RpoS-repressed genes are important for tick colonization and are downregulated following transmission, including ospAB, lp6.6, bba74, and bb0365 (de Silva et al., 1996; Schwan and Piesman, 2000; Pal et al., 2008; Mulay et al., 2009). Some bdr paralogs, which encode members of the Borrelia direct repeat protein family with no known function (Zückert et al., 1999), are also repressed by RpoS (Roberts et al., 2002). While the mechanism of RpoS-dependent repression of this regulon remains unclear, the T-rich track upstream of the promoter was proposed to be involved (Sohaskey et al., 1999; Brooks et al., 2003; Caimano et al., 2007). Recently, using a GFP reporter and the dialysis membrane chambers (DMC) model, Grove et al. (2017) uncovered two distinct mechanisms of RpoN-dependent gene repression: one is direct repression (ospA, glp) in which RpoS competes with RpoD at the −35/−10 sequence for promoter binding and the other is indirect repression (bba74) via an unknown trans-acting repressor whose expression is controlled by RpoS. However, other data suggest that BosR, a transcriptional activator of rpoS, not RpoS, directly represses ospAB and other lipoprotein genes (Wang et al., 2013; Shi et al., 2014) (see next section on Transcription factors BosR and BadR).
Factors regulating the RpoN-RpoS σ factor cascade
Rrp2, encoded by bb0763, is the sole bEBP present in B. burgdorferi (Yang et al., 2003). Rrp2 belongs to the well-studied NtrC protein family (Studholme and Buck, 2000). Members of this family contain three putative functional domains: an N-terminal response regulator domain, a central RpoN-activation domain, and a C-terminal helix-turn-helix (HTH) DNA-binding domain. The central domain becomes activated upon phosphorylation at a conserved Asp residue within the N-terminal domain. The activated central domain then contacts the RpoN-holoenzyme through DNA looping, hydrolyzes ATP, and promotes open promoter complex formation for transcriptional initiation. Yang et al. (2003) first demonstrated that Rrp2 is the upstream activator for the RpoN-RpoS alternative σ factor cascade (Figure 2): a single G239C point mutation within the central activation domain of Rrp2 abolished expression of rpoS, ospC, and other RpoN-RpoS pathway-dependent genes. The G239C mutation did not abolish the ATPase activity of Rrp2, suggesting that the mutation affects interaction of Rrp2 with RpoN necessary for initiation of open complex formation (Ouyang and Zhou, 2017). Artificial induction of an inducible rrp2 construct also resulted in rpoS transcription (Ouyang et al., 2014c). Interestingly, Rrp2 activates rpoS transcription without binding to a specific enhancer binding site (Burtnick et al., 2007; Blevins et al., 2009), despite having a predicted HTH-containing C-terminal domain and being capable of non-specific binding to DNA (Burtnick et al., 2007; Blevins et al., 2009). Surprisingly, rpoN is not required for B. burgdorferi growth and is readily inactivated, but rrp2 is essential (Yang et al., 2003; Burtnick et al., 2007; Blevins et al., 2009; Groshong et al., 2012; Yin et al., 2016). This suggests that Rrp2 has an additional role besides serving as the activator for the RpoN-RpoS σ factor cascade. Phosphorylation, ATPase activity, and the C-terminal-mediated DNA-binding are all required for the essential function of Rrp2, but the mechanistic details remain unclear (Yin et al., 2016; Ouyang and Zhou, 2017). Further work, such as chromatin immunoprecipitation (ChIP), will be required to identify Rrp2-dependent but RpoN-independent genes to gain insight the essential nature of Rrp2.
Given that Rrp2 is a two-component response regulator, its cognate histidine kinase Hk2 (BB0763) predicted in the genome was hypothesized to receive upstream signals to phosphorylate and activate Rrp2, which, in turn, activates the RpoN-RpoS σ factor cascade (Yang et al., 2003; Burtnick et al., 2007; Liu et al., 2020). However, unlike rrp2, hk2 is not essential for cell survival and, unexpectedly, disruption of hk2 does not affect the expression of rpoS and ospC, suggesting that Hk2 is not required for Rrp2 phosphorylation. One the other hand, Hk2 can function as a phosphatase to dephosphorylate Rrp2 and a strain that overexpresses Hk2 has reduced level of phosphorylated Rrp2 and impaired growth (Liu et al., 2020). Further study indicated that acetyl phosphate, a high-energy metabolic intermediate that can donate phosphoryl or acetyl groups, activates Rrp2 via phosphorylation (Xu et al., 2010); the hypothesis was compelling as B. burgdorferi senses the blood meal, begins to replicate and activates the RpoN-RpoS σ factor cascade upon tick feeding, so acetyl phosphate would serve to connect metabolic status and enzootic phase-specific gene expression. However, inactivation of the AckA-Pta pathway that produces acetyl phosphate proved to be lethal to spirochete survival but did not affect the expression of rpoS and ospC (Richards et al., 2015), arguing against a role for acetyl phosphate in Rrp2 phosphorylation. Therefore, the source of phosphate and the mechanism of Rrp2 phosphorylation are enigmatic, although acetyl phosphate, Hk2 and other small phosphate donors might have overlapping and complementary functions.
Another two-component system present in the B. burgdorferi genome, Hk1-Rrp1, appears to engage in cross-talk with the Rrp2-controlled RpoN-RpoS σ factor cascade (Rogers et al., 2009; He et al., 2011; Sze et al., 2013; He et al., 2014; Caimano et al., 2015) (see section below on Hk1/Rrp1 two-component system and c-di-GMP). The interplay between the Hk1-Rrp1 and Hk2-Rrp2 signaling systems has only begun to be revealed. Sze et al. (2013) reported that Rrp1 affects rpoS via regulation of bosR. c-di-GMP, synthesized by Rrp1, controls rpoS expression through the c-di-GMP binding protein PlzA and PlzA links the two sets of two-component systems (He et al., 2014; Caimano et al., 2015; Zhang et al., 2018).
Transcription factors BosR and BadR
BosR
BosR (BB0647; Borrelia oxidative stress regulator) is a transcriptional regulator that belongs to the ferric uptake regulator (Fur) family (Fraser et al., 1997; Boylan et al., 2003; Katona et al., 2004; Samuels and Radolf, 2009). It shares 22.5% and 21.8% identity, and 43% and 49% similarity, respectively, with the Fur protein in Vibrio cholerae and the stress-responsive Fur family protein PerR in Bacillus subtilis. B. burgdorferi does not rely on iron (Posey and Gherardini, 2000; Wang et al., 2012b), so BosR seemed unlikely to regulate iron homeostasis in the spirochete. Instead, BosR is analogous to PerR and was first reported to regulate the expression of dps/napA/bicA as well as other genes involved in oxidative stress response in B. burgdorferi (Boylan et al., 2003), but BosR was eventually shown to activate rpoS transcription and, thus, the RpoS-dependent regulon (Hyde et al., 2009; Ouyang et al., 2009b; Hyde et al., 2010; Ouyang et al., 2011; Ouyang et al., 2014a; Katona, 2015).
Expression and regulation of bosR
Transcription of bosR is driven by two promoters: a strong RpoD-dependent promoter located directly upstream of bosR (Katona et al., 2004; Hyde et al., 2010; Katona, 2015; Ouyang et al., 2016) and a second RpoD-dependent promoter upstream of an operon in which bosR is co-transcribed with bb0648 (encoding a putative serine/threonine kinase) and bb0646 (encoding a putative lipase) (Ouyang et al., 2009b; Ouyang et al., 2016). While bosR is expressed throughout the entire enzootic cycle (Medrano et al., 2007; Ouyang et al., 2016), environmental factors, such as temperature and pH, modulate its transcription (Katona et al., 2004; Ouyang et al., 2016). The intracellular second messengers c-di-GMP (Sze et al., 2013; He et al., 2014; Novak et al., 2014; Ye et al., 2014) and the alarmones (p)ppGpp (Bugrysheva et al., 2015; Drecktrah et al., 2015; Drecktrah et al., 2018) induce bosR expression at both transcriptional and post-transcriptional levels. Dissolved carbon dioxide (Hyde et al., 2007) and the transition metals manganese, zinc and copper (Troxell and Yang, 2013; Troxell et al., 2013; Wang et al., 2017) influence bosR expression at the post-transcriptional level. BadR, a ROK repressor, negatively affects bosR expression at both transcriptional and post-transcriptional levels (Miller et al., 2013; Ouyang and Zhou, 2015). Transcription of bosR also is activated by the BosR protein, indicating that BosR is an auto-activator (Ouyang et al., 2016), which is different from classical Fur family proteins as auto-repressors. The expression and regulatory patterns of bosR highlight the crucial role of BosR in the enzootic cycle of B. burgdorferi.
BosR is an atypical Fur family member
Fur family proteins consist of two structural domains including a N-terminal winged helix-turn-helix DNA-binding domain and a C-terminal dimerization domain, connected by a flexible hinge region. Structural analyses of numerous Fur proteins have revealed two sites involved in metal binding (Helmann, 2014; Pinochet-Barros and Helmann, 2018; Sarvan et al., 2018; Nader et al., 2019). The first site (S1), located in the dimerization domain, is usually occupied by a zinc ion and is crucial for protein dimerization, so it is called the structural site. The second metal binding site (S2) is located in the hinge region between the DNA-binding domain and the dimerization domain. The metal ion (e.g., iron, manganese, zinc, or nickel) occupying the S2 site determines the specific regulatory function of the Fur protein; thus, it is called the regulatory site. Occasionally, Fur proteins such as those in Helicobacter pylori and Campylobacter jejuni contain a third metal binding site (S3) in the dimerization domain, which is also important for the regulatory function of the protein (Dian et al., 2011; Butcher et al., 2012). The regulatory functions of Fur family members are responsive to the availability of transition metal ions such as iron, manganese, zinc, and nickel (Hantke, 1981, 2001; Carpenter et al., 2009; Fillat, 2014). In the absence of metal cofactors, the DNA-binding domain of Fur typically has an “open” conformation that does not bind DNA. Occupancy of S1 with a zinc ion induces Fur dimerization, which brings the DNA-binding domains of two monomers into closer proximity. Binding of metal ions to S2 triggers additional conformational changes, which in turn stimulates the formation of a “closed” DNA-binding domain. As a result, the “closed” Fur binds to promoter DNA with high affinity and represses gene expression. In addition, metal-dependent Fur family proteins also sense non-metal signals. For example, PerR contains the zinc-binding structural S1 site and a manganese- or iron-coordinating regulatory S2 site (Traoré et al., 2006; Jacquamet et al., 2009; Traoré et al., 2009). Manganese or iron occupying S2 triggers the formation of a “closed” PerR (i.e., PerR-Zn-Mn and PerR-Zn-Fe) that binds to the Per box and blocks gene transcription. When iron in the S2 site detects peroxide, such as H2O2, histidine residues of the PerR-Zn-Fe are oxidized. This leads to the release of iron and changes PerR to the “open” conformation. The “open” PerR then dissociates from the Per box, which results in the derepression of genes involved in the peroxide stress response (Traoré et al., 2006; Jacquamet et al., 2009; Traoré et al., 2009).
Recombinant BosR contains zinc, but no other metal ions (including iron, copper, manganese, chromium, cadmium, nickel, magnesium, or lead) (Ouyang et al., 2011; Wang et al., 2012b; Mason et al., 2019). Two CXXC motifs were identified in the dimerization domain of BosR that constitute the zinc-coordination structural S1 site of BosR (Mason et al., 2019). This site is similar to the S1 site in many Fur family proteins including PerR (Figure 3). The ability of BosR to bind Zn was dramatically reduced when the CXXC motifs were mutated (Mason et al., 2019). The CXXC motifs also contribute to the ability of BosR to form dimers and are crucial for the regulatory function of BosR. However, BosR lacks a recognizable regulatory S2 site. Of the residues constituting the iron, manganese, zinc, or nickel-coordinating regulatory S2 site of Fur proteins, BosR retains only a single conserved residue (His111) in the hinge region (Figure 3). The vicinity surrounding His111 lacks metal coordinating residues other than the known CXXC motifs. Therefore, the DNA-binding conformation of BosR may not be induced by metal binding, other than at the S1 site. Instead, the His111 may be modified through an as yet unknown mechanism to facilitate DNA binding, which is consistent with the presence of only zinc in recombinant BosR (Ouyang et al., 2011; Wang et al., 2012b; Mason et al., 2019). Alternatively, BosR may use a noncanonical S2 site for signal sensing. Recombinant BosR is able to bind copper, zinc and iron when incubated with unusually high amounts of these metal ions, so perhaps BosR contains two regulatory sites coordinating zinc, iron, or copper in addition to the structural S1 site (Wang et al., 2017). Surprisingly, adding zinc or copper, but not iron, inhibited the binding of BosR to the ospA promoter region (Wang et al., 2017). However, we do not know which metal is bound to BosR in vivo.
Figure 3.

Alignment of BosR and Fur proteins using Clustal Omega. Bs, B. subtilis; Ec, E. coli; Vc, V. cholera; Cj, C. jejuni; Hp, H. pylori. The residues constituting the S1, S2 and S3 metal binding sites are indicated in red, green, and blue, respectively. Residues potentially important for DNA binding are indicated in purple.
There remain considerable knowledge gaps regarding BosR. The role of this regulatory protein in the oxidative stress response and transition metal homeostasis is, frankly, uncertain. In addition, the contributions of the BosR regulon to the enzootic cycle need to be addressed. The binding of Fur protein from other bacteria to DNA results in gene repression; however, the binding of BosR to the promoter region of rpoS and bosR is known to activate transcription, so the mechanistic basis for this unusual activity should be explored. Furthermore, whether BosR has a second regulatory metal-binding site like other Fur proteins is unknown. Dissecting the unusual DNA binding activity and regulatory function of BosR is fundamental toward determining the pathway by which BosR senses the host- and vector-specific signals to control gene expression during the enzootic cycle.
BosR function
Seshu et al. (2004b) constructed the first bosR mutant in a high-passage, non-infectious strain of B. burgdorferi. However, the bosR allele (bosRR39K) in the parental high-passage strain possessed a point mutation (R39K) relative to the wild-type bosR in infectious strains (Seshu et al., 2004a; Hyde et al., 2006); the R39 residue of BosR is essential for protein function (Seshu et al., 2004a; Katona, 2015). bosR deletion mutants were then generated in low-passage, infectious B. burgdorferi strains. Hyde et al. (2009, 2010) reported that bosR contributes to the in vitro growth of B. burgdorferi, and the levels of Dps and SodA were decreased in the bosR mutant. However, neither the growth defect nor the change in Dps synthesis were observed in the bosR mutant generated by Ouyang et al (2009b; 2011). Despite these and other discrepancies, the strategic bosR mutants yielded breakthroughs in our understanding of BosR function. First, BosR is essential for tick to vertebrate transmission and vertebrate infection by B. burgdorferi. Second, BosR is required for the transcriptional activation of rpoS. The expression of rpoS and RpoS-dependent OspC and DbpA was abolished in bosR mutants. BosR directly activates transcription of rpoS by binding to the rpoS promoter region (Ouyang et al., 2011; Ouyang et al., 2014a; Katona, 2015). Specifically, BosR binds to three sites in the rpoS promoter region as a homodimer, including a site overlapping the −24/−12 RpoN-dependent promoter (Ouyang et al., 2011). This suggests that BosR interacts with RpoN (σ54) to facilitate the recruitment of RpoN-RNA polymerase holoenzyme to the rpoS promoter to activate gene expression. Further analyses revealed that BosR recognizes the rpoS promoter region via a core sequence called the BosR box, a novel DNA element consisting of a 13-bp palindromic sequence (ATTTAANTTAAAT) with dyad symmetry (Ouyang et al., 2014a). Different from the 19-bp Fur binding consensus (i.e., Fur box, GATAATGATAATCATTATC) and the 15-bp PerR binding consensus (i.e., Per box, TTATAATTATTATAA), the BosR box probably comprises a 6–1–6 inverted repeat composed of two hexamers (ATTTAA) in a head-to-tail orientation. The requirement of BosR in the activation of RpoN-dependent rpoS transcription was unexpected, as activation of bacterial RpoN (σ54) systems generally only requires the bacterial enhancer binding protein (bEBP) (Bush and Dixon, 2012), and suggests that BosR and Rrp2 together mimic the bEBP.
The BosR regulon
Recombinant BosR binds to many DNA sequences including the Per box (Katona et al., 2004; Ouyang et al., 2014a) and the promoter regions of dps (Boylan et al., 2003; Katona et al., 2004; Ouyang et al., 2011), sodA (Seshu et al., 2004a), cdr (Boylan et al., 2006), oppA4 (Medrano et al., 2007), ospA (Wang et al., 2013; Shi et al., 2014; Wang et al., 2017), ospD (Wang et al., 2013), and bosR itself (Katona et al., 2004; Ouyang et al., 2011). Such versatile DNA recognition capacity is not typical of canonical Fur family proteins. Contradictory results were also obtained regarding the DNA binding activity of BosR. Boylan et al. (2003) reported that the binding of BosR to the dps promoter was inhibited by manganese and required zinc as well as the reduction with DTT. Katona et al. (2004), however, found that binding of BosR to the Per box and the dps promoter was not affected by manganese, but was inhibited by zinc. Wang et al. (2017) showed that the binding of BosR to the ospA promoter was not affected by manganese or iron but was inhibited by copper and zinc. These discrepancies may be due to proteins with different conformations used in the in vitro binding studies. Analytical ultracentrifugation analyses revealed a monomer-dimer equilibrium of recombinant BosR in free solution (Ouyang et al., 2014a). Metal ions and oxidizing reagents may induce conformational changes, and thus affect the DNA binding activity of recombinant protein. In an attempt to discern the global regulatory effects of BosR, Katona et al. (2004) searched the B. burgdorferi genome for the Per box by using the BlastN program and found 26 genes possibly regulated by BosR. Ouyang et al. (2014a) searched the genome (Fraser et al., 1997) for putative BosR boxes using the Regulatory Sequence Analysis Tools, and found 47 genes carrying one or more BosR box sequences in their promoter regions. However, identification of a BosR binding site in a promoter does not mean that BosR regulates the gene. Microarray-based transcriptomic analysis of the bosR mutant identified 137 and 62 genes that were upregulated or downregulated, respectively, by BosR, although the function of most them is unknown (Ouyang et al., 2009b). A comparison of the bosR and rpoS microarray data (Ouyang et al., 2008) found that 50 and 57 genes were upregulated or downregulated, respectively, only by BosR, which suggests that BosR is a dual-functional global regulator in B. burgdorferi.
BadR
BadR (BB0693; Borrelia host adaption regulator) is a DNA-binding transcriptional regulator that represses rpoS transcription in B. burgdorferi (Fraser et al., 1997; Miller et al., 2013; Ouyang and Zhou, 2015). BadR synthesis is induced at low temperature, mimicking the tick environment (Miller et al., 2013). badR is expressed throughout the entire enzootic cycle of B. burgdorferi, reaching a maximal level in intermolt larvae (Ouyang and Zhou, 2015). BadR is required by B. burgdorferi to infect vertebrates; badR mutants are incapable of infecting mice through needle injection (Miller et al., 2013; Ouyang and Zhou, 2015). In addition, BadR is crucial for optimal growth of B. burgdorferi in vitro; badR mutants display a marked growth defect in culture compared to the wild-type parental strain. Expression of rpoS is enhanced in badR mutants, indicating that BadR is a repressor of rpoS transcription. Recombinant BadR binds to multiple sites in the rpoS promoter region; several residues in the putative sugar-binding domain of BadR were crucial for binding. A merodiploid with badR expression controlled by an inducible promoter was constructed to overcome the inability to complement the badR null mutant: induction of badR inhibited transcription of rpoS (Ouyang and Zhou, 2015). Some differences exist between the two genetic studies of badR (Miller et al., 2013; Ouyang and Zhou, 2015) including whether expression of rpoS in the badR mutants is growth-phase dependent. In addition, Miller et al. (2013) reported that binding of recombinant BadR to the rpoS promoter region was inhibited by the addition of phosphorylated sugars, while this inhibition was not observed by Ouyang et al (2015), which may be due to the presence of the maltose-binding protein tag on the recombinant BadR in the first study.
BadR inhibits expression of bosR at both transcriptional and post-transcriptional levels. Binding sites for BadR have been identified in the sequences upstream of bosR (Ouyang and Zhou, 2015). Both BadR and BosR bind to the promoter regions of rpoS and bosR, although they recognize different core sequences. BadR recognizes DNA at TAAAATAT motifs. Expression of rpoS is abolished in a bosR-badR double mutant (Ouyang and Zhou, 2015), which suggests that BosR functions to activate rpoS transcription rather than to derepress BadR. BadR represses or induces the expression of 127 or 80 genes, respectively (Miller et al., 2013). Subsequent RNA-seq analyses showed that expression of 134 and 100 genes were decreased and increased, respectively, in the badR mutant (Arnold et al., 2018). Many of these genes are associated with sugar transport and utilization.
How BadR modulates gene expression in B. burgdorferi remains elusive. BadR was originally annotated as a xylose operon regulatory protein XylR-1 involved in the utilization and degradation of D-xylose (Fraser et al., 1997). BadR, however, likely does not function as a xylose-responsive transcriptional regulator like XylR, as B. burgdorferi does not use xylose as a carbon source (von Lackum and Stevenson, 2005) and its genome does not encode proteins for xylose transport and catabolism (Fraser et al., 1997). Rather, BadR has ~21% amino acid sequence identity with the E. coli transcriptional regulator NagC. Three-dimensional protein modeling indicated that BadR had a structure similar to the Mlc protein in V. cholerae and E. coli (Ouyang and Zhou, 2015). Mlc is a homolog of NagC and both proteins belong to the ROK (Repressor, ORF, Kinase) protein family. ROK repressors contain an N-terminal helix-turn-helix (HTH) DNA-binding motif and a C-terminal sugar-binding domain. The DNA-binding activity and regulatory function of ROK repressors are responsive to the availability of specific sugars. Despite the homology to NagC and Mlc, BadR likely functions differently from other ROK repressors. Typically, ROK repressors bind a specific sugar or effector through specificity-determining residues (SDRs) (Titgemeyer et al., 1994; Kazanov et al., 2013). However, BadR lacks the SDRs found in NagC, Mlc, and other ROK repressors, consistent with the observation that the DNA-binding of recombinant BadR was not affected by phosphorylated sugars (Ouyang and Zhou, 2015), and the expression of genes involved in GlcNAc transport and utilization were not affected in the badR mutant (Miller et al., 2013), supporting that BadR does not function analogously to NagC or Mlc and is yet another example of an atypical transcriptional regulator influencing the expression of enzootic cycle-specific genes (Samuels and Radolf, 2009).
YebC
YebC (BB0025) is a transcription factor that belongs to a putative YebC/PmpR family of DNA-binding transcriptional regulators (TACO1 family, pfam PF01709) (Shin et al., 2002; Zhang et al., 2012) and controls vlsE expression (Zhang et al., 2020). VlsE undergoes antigenic variation in the vertebrate host and vlsE expression increases concomitantly with the decrease in ospC expression in response to the host adaptive immune response, but there are few mechanistic details on the regulation of vlsE expression (Norris, 2014; Chaconas et al., 2020). Zhang et al. (2020) very recently demonstrated that deletion of yebC dramatically reduced vlsE mRNA levels in vitro. Expression of yebC and vlsE is co-regulated in response to culture temperature, and is inversely regulated with expression of ospC during vertebrate infection. The yebC mutant has a similar phenotype as the vlsE mutant: both are infectious in immunocompromised mice but unable to infect immunocompetent mice. YebC binds to the vlsE promoter, suggesting that YebC directly regulates vlsE transcription; however, how YebC is activated during the vertebrate phase remains unknown.
Hk1/Rrp1 two-component system and c-di-GMP
Since its discovery as a novel allosteric activator of cellulose synthase in Komagataeibacter xylinus (formerly Acetobacter xylinus) more than 30 years ago (Ross et al., 1987), bis-(3′−5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) has emerged as a ubiquitous global secondary messenger associated with a wide range of lifestyle control networks, including the transition from planktonic to sessile states, biofilm formation, cell cycle progression, and virulence (Cotter and Stibitz, 2007; Wolfe and Visick, 2008; Hengge, 2009; Römling et al., 2013; Valentini and Filloux, 2019). Diguanylate cyclases (DGCs), the enzymes responsible for synthesis of c-di-GMP from two GTP molecules, are readily identified by the presence of an ~170 amino acid GGDEF (Gly-Gly-Asp-Glu-Phe) conserved domain (Pfam PF00990) (Pei and Grishin, 2001; Schirmer and Jenal, 2009). GGDEF domain-containing proteins are omnipresent in bacteria (Galperin, 2004; Ryjenkov et al., 2005), with most bacterial genomes encoding multiple DGCs. The potential relevance of c-di-GMP for environmental signaling in Lyme disease spirochetes first came to light in a bioinformatics study by Galperin et al. (2001). Using purified recombinant protein, Ryjenkov et al. (2005) provided direct biochemical evidence that Rrp1 (BB0419), the only identifiable GGDEF domain-containing protein encoded by Borrelia spp. (Galperin et al., 2001), functions as a diguanylate cyclase. Intracellular concentrations of c-di-GMP in B. burgdorferi are controlled by the activity of two evolutionarily distinct phosphodiesterases (PDEs), PdeA and PdeB, belonging to the EAL and HD-GYP families of non-specific PDEs, respectively (Sultan et al., 2010; Sultan et al., 2011; Novak et al., 2014). While both borrelial PDEs hydrolyze c-di-GMP with high affinity (Sultan et al., 2010; Sultan et al., 2011; Novak et al., 2014), PdeA- and PdeB-deficient spirochetes display different motility- and virulence-related phenotypes (Sultan et al., 2010; Sultan et al., 2011), suggesting that these PDEs are part of a complex regulatory system that involves multiple effectors and control mechanisms.
Rrp1 along with Hk1 form a prototypical bacterial sensory two-component signal transduction system (TCS). Hk1, the sensory component, is composed of four modular domains: a periplasmic sensor domain of three ligand-binding subdomains, each with homology to bacterial periplasmic solute-binding proteins (PBPs), a histidine kinase domain, a phosphoacceptor receiver (REC) domain, and a histidine phosphotransferase (Hpt) domain (Bauer et al., 2015). Ligand binding by Hk1 initiates a signal transduction cascade that results in phosphorylation of the Rrp1 REC domain and activation of c-di-GMP synthesis (Caimano et al., 2011; Bauer et al., 2015). Although the small molecule(s) responsible for activating Hk1 in nature have yet to be identified, bioinformatics and structural analyses of the Hk1 sensory PBP domains point to exogenously-derived amino acids and/or their derivatives as potential activating ligands (Caimano et al., 2011; Caimano et al., 2015). Consistent with this prediction, the Hk1/Rrp1 TCS is active when spirochetes are grown in BSK II (Rogers et al., 2009; Caimano et al., 2011), a semi-defined medium that contains abundant free amino acids (Barbour, 1984). The genes encoding Hk1 and Rrp1 (bb0420 and bb0419, respectively) are co-transcribed as a bicistronic message (Caimano et al., 2011). hk1/rrp1 is expressed more or less constitutively in vitro and throughout the enzootic cycle (Rogers et al., 2009; Caimano et al., 2011).
Phenotypic characterization of hk1 and rrp1 mutants demonstrated that this TCS functions exclusively during the tick phase of the enzootic cycle (Caimano et al., 2011; He et al., 2011; Kostick et al., 2011). Hk1- and Rrp1-deficient organisms display wild-type burdens in murine tissues following needle inoculation but are rapidly destroyed in the midguts of feeding larvae and nymphs as early as 36 hours into the acquisition blood meal (Caimano et al., 2011; He et al., 2011; Kostick et al., 2011). The phenotypic similarities between hk1 and rrp1 mutants in larvae and nymphs suggest that the ligand(s) responsible for activating c-di-GMP signaling are not tick life-stage-specific (Caimano et al., 2011; He et al., 2011; Kostick et al., 2011; Caimano et al., 2015). Little to no killing was observed in the absence of tick feeding, indicating that the destruction of both mutants is mediated by the feeding process. An important clue regarding this unusual tick phenotype was provided by Bontemps-Gallo et al. (2016), who found that hk1 and rrp1 mutant B. burgdorferi grown at 34°C in BSK II were substantially more osmo-sensitive than their wild-type isogenic parent under conditions that mimic the high osmolality (~550–650 mOsm) within fed midguts. Additional insight into the contribution of the Hk1/Rrp1 TCS to tick adaptation comes from genome-wide transcriptomics analyses by microarray (Rogers et al., 2009; He et al., 2011) and RNA-seq (Caimano et al., 2015). Results from these analyses revealed two broad categories of c-di-GMP-regulated genes that could contribute to survival within tick midguts during the blood meal. The first includes genes involved in utilization of alternate carbon sources for energy generation (glycolysis) and biosynthesis of phospholipids and/or peptidoglycan. In addition to the glycerol utilization (Glp) system, c-di-GMP upregulates expression of PTS transporters (malX2 and chbC) for N-acetylglucosamine (GlcNAc) and chitobiose (a GlcNAc dimer derived from chitin, the major component of tick cuticle), respectively (Tilly et al., 2001; Tilly et al., 2004; Rhodes et al., 2009; Gherardini et al., 2010; Pappas et al., 2011; Sze et al., 2013; Caimano et al., 2015; Corona and Schwartz, 2015; Caimano et al., 2016; Zhang et al., 2018). The phenotypes for hk1 and rrp1 mutants in feeding ticks, however, are more pronounced than those of either glp or chbC mutants. Presumably, utilization of multiple alternative carbon sources enables spirochetes to expand exponentially and maintain cell envelope homeostasis as blood meal-derived glucose becomes increasingly limited due to competition from midgut epithelial cells. Additional c-di-GMP-regulated genes contribute to maintenance of the cell envelope: the Rrp1 regulon includes ackA, encoding the acetate kinase required for generation of acetyl phosphate, which in turn gives rise to acetyl-CoA, the starting point for synthesis of peptidoglycan via the mevalonate pathway (Xu et al., 2010; Van Laar et al., 2012). The second category of c-di-GMP regulated genes includes differentially-expressed lipoproteins, including the cp32-encoded OspE/BbCRASP lipoproteins, which have been shown to inhibit complement-mediated lysis by binding complement factor H (CFH) and CFH-related proteins (Radolf et al., 2012; Bhattacharjee et al., 2013; Kraiczy and Stevenson, 2013). Other lipoproteins regulated by c-di-GMP (e.g., Mlps) also may protect B. burgdorferi from hazards encountered in the fed midgut (e.g., antimicrobial peptides and reactive oxygen species).
c-di-GMP modulates cellular processes in bacteria by a variety of mechanisms, including transcriptional, translational, allosteric inhibition of enzymatic activity (I-sites), protein-protein interactions, protein secretion, and protein stability (Cotter and Stibitz, 2007; Wolfe and Visick, 2008; Römling et al., 2013; Valentini and Filloux, 2019). A wide range of effector molecules involved in c-di-GMP signaling have been identified, including two types of riboswitches (Sudarsan et al., 2008; Lee et al., 2010; Smith et al., 2011) and numerous c-di-GMP receptor proteins (Schirmer and Jenal, 2009; Dahlstrom and O’Toole, 2017; Valentini and Filloux, 2019). One of the most well-studied group of c-di-GMP effector proteins are those containing an ~110 amino acid PilZ domain (PF07238) (Amikam and Galperin, 2006; Schirmer and Jenal, 2009), first identified at the C-terminus of the cellulose synthase catalytic subunit of BcsA from K. xylinum (Ross et al., 1987). To date, two c-di-GMP effector PilZ domain proteins, PlzA (BB0733) and PlzB, have been described in B. burgdorferi (Freedman et al., 2010; Pitzer et al., 2011; He et al., 2014; Mallory et al., 2016). A third PilZ-domain protein, PlzC, is found only in relapsing fever spirochetes (Mallory et al., 2016). PlzA, encoded on the chromosome, and PlzB, encoded on a linear plasmid, share only 63% amino acid identity and appear to carry out distinct functions (Mallory et al., 2016). The majority of Lyme disease spirochetes encode only PlzA. c-di-GMP binding by borrelial Plz proteins is mediated by a signature PilZ domain motif: RIHER and DzSYGG (z=hydrophobic residue) (Amikam and Galperin, 2006; Ryjenkov et al., 2006; Freedman et al., 2010; Pitzer et al., 2011; Mallory et al., 2016). In independent studies, Motaleb, Marconi and colleagues demonstrated that recombinant PlzA binds c-di-GMP at 1:1 stoichiometry with relatively high affinity (~1 – 6 μM) (Freedman et al., 2010; Pitzer et al., 2011; Mallory et al., 2016). Binding of c-di-GMP by PlzA is thought to induce a conformational change enabling a ‘switch mechanism’ (Mallory et al., 2016). Interestingly, the PilZ motifs in borrelial Plz proteins are separated by a variable 32 amino acid spacer that is significantly longer than the 19 to 23 residues typically found in prototypical PilZ domains, perhaps allowing spirochetal Plz proteins to engage a more extensive collection of downstream effector molecules (Mallory et al., 2016). Consistent with its c-di-GMP effector function, expression of plzA is increased during tick feeding (Freedman et al., 2010; Pitzer et al., 2011). However, there are conflicting reports regarding the extent to which PlzA is required for survival during tick feeding. Pitzer et al. (2011) reported that plzA mutants survive poorly in naturally-infected ticks during the acquisition and transmission blood meals, while Kostick-Dunn et al. (2018) found that PlzA was not strictly required to establish infection in ticks. Nonetheless, both groups demonstrated that plzA-infected ticks are unable to transmit spirochetes to naïve mice. Moreover, overexpression of PlzA is able to partially compensate for the loss of Rrp1 in feeding ticks (Zhang et al., 2018). This phenotype stems, in part, from the requirement for both c-di-GMP and PlzA for transcription of the glp operon (bb0240-bb0243) (Caimano et al., 2015; Zhang et al., 2018), which encodes a system for the uptake and utilization of glycerol (Pappas et al., 2011). However, unlike rrp1 mutants, which are infectious in mice, spirochetes lacking PlzA are substantially less virulent by needle-inoculation (Pitzer et al., 2011; He et al., 2014; Kostick-Dunn et al., 2018; Zhang et al., 2018). Decreased virulence of plzA mutants has been attributed, at least in part, to aberrant motility (i.e., decreased swarming, reduced translational motility and flex frequency) (Pitzer et al., 2011; Kostick-Dunn et al., 2018) and/or dysregulation of RpoS-dependent gene regulation (Sze et al., 2013; He et al., 2014). He et al. (2014) reported that PlzA influences the RpoS pathway via its effects on BosR, a Fur family protein required for expression of rpoS (see the section above on BosR and BadR transcription factors) (Samuels and Radolf, 2009); plzA mutants express two- to threefold lower levels of bosR mRNA compared to wild type and little to no BosR protein in vitro (He et al., 2014). Given that the Hk1/Rrp1 TCS is not required and presumably ‘OFF’ in vertebrates, any virulence-related aspects of the plzA mutant phenotype, including dysregulation of BosR and/or RpoS, are likely mediated by apo-PlzA (Pitzer et al., 2011; Zhang et al., 2018).
Genome-wide comparison of the RpoS and Hk1/Rrp1 regulons identified only a handful of genes that are regulated by both RpoS and c-di-GMP (Caimano et al., 2015). The expression profiles of these co-regulated genes also were strikingly different from those of the prototypical RpoS-regulated genes ospC and dbpA in fed nymphs (i.e., when both pathways are active) (Dunham-Ems et al., 2012; Caimano et al., 2019). Thus, while c-di-GMP may enhance expression of some RpoS-upregulated genes, these two pathways likely function in parallel rather than cooperatively. As part of its gatekeeper function, RpoS also represses the expression of RpoD (σ70)-dependent tick-phase genes, including ospA, bba62, and the glp operon, within the vertebrate (Caimano et al., 2005; Caimano et al., 2007; Dunham-Ems et al., 2012; Grove et al., 2017; Caimano et al., 2019) (see the section above on the RpoN-RpoS σ factor cascade). Although transcription of RpoS-upregulated genes begins during the transmission blood meal (Schwan et al., 1995; Dunham-Ems et al., 2012; Ouyang et al., 2012; Caimano et al., 2019), RpoS-mediated repression does not occur to any appreciable extent until spirochetes have entered the vertebrate host (Dunham-Ems et al., 2012; Caimano et al., 2019). Overlap between the Hk1/Rrp1 and RpoS regulons includes a number of tick phase-specific genes that are repressed by RpoS during infection. These data led Caimano et al. (2015) to propose that c-di-GMP antagonizes RpoS-mediated repression either directly or indirectly in feeding ticks. Once B. burgdorferi is in a vertebrate, the absence of c-di-GMP would allow unfettered repression of these tick phase-specific genes.
Stringent response and (p)ppGpp
RelBbu and DksA mediate adaptation to nutrient stress
B. burgdorferi adapts to environmental nutritional changes encountered during its enzootic cycle by globally altering gene expression through the stringent response, which is particularly pivotal during persistence in the tick vector between blood meals. The stringent response steels the spirochete by regulating the expression of numerous pathways, including replication, motility, morphology, stress responses, and virulence factors. These changes are driven by modulating the levels of two intracellular alarmones, guanosine pentaphosphate and guanosine tetraphosphate, collectively known as (p)ppGpp, which directly bind RNA polymerase and indirectly affect numerous regulatory systems to alter transcription (Potrykus and Cashel, 2008; Dalebroux and Swanson, 2012; Gaca et al., 2015; Hauryliuk et al., 2015; Liu et al., 2015; Steinchen and Bange, 2016; Gourse et al., 2018). In B. burgdorferi, (p)ppGpp levels are regulated by a single gene product, RelBbu, containing both a canonical synthetase (RelA) domain induced by amino acid starvation and a bifunctional synthetase/hydrolase (SpoT) domain (Concepcion and Nelson, 2003; Bugrysheva et al., 2005; Drecktrah et al., 2015). In addition to the catalytic domains, RelBbu also contains the regulatory TGS (threonyl-tRNA synthetases, GTPases and SpoT) and DC (aspartate-cysteine) oligomerization domains, but lacks the ACT regulatory domain in the C-terminal region (Grant, 2006; Hauryliuk et al., 2015; Ronneau and Hallez, 2019). (p)ppGpp levels rise during nutrient deprivation and fall upon return to nutrient-rich conditions in a RelBbu-dependent manner (Concepcion and Nelson, 2003; Bugrysheva et al., 2005; Drecktrah et al., 2015). The specific triggers of the stringent response in B. burgdorferi remain unknown, but amino acid starvation may not serve as the classic signal since the B. burgdorferi genome is predicted to contain few amino acid transporters and may rely more on peptide transport and proteolysis (Fraser et al., 1997; Groshong et al., 2017); instead, the spirochete may sense changes in levels of lipids or phosphate.
The RelBbu-mediated transcriptional changes of the stringent response play an important role in survival during nutrient stress both in culture and within the nymphal tick midgut during the transmission blood meal (Drecktrah et al., 2015). Survival in the midgut and transmission to mice via tick bite are not abolished in a relBbu mutant incapable of synthesizing (p)ppGpp, but are significantly curtailed (Drecktrah et al., 2015). Growth in nutrient-rich media at 23°C, the temperature used to represent the tick environment, is also RelBbu-dependent (Bugrysheva et al., 2015). The stringent response also regulates B. burgdorferi morphology as the relBbu mutant is more likely to form round bodies, a spherical condensed form within an outer membrane observed in vitro and within ticks between blood meals, in response to nutrient stress (Brorson and Brorson, 1997, 1998; Alban et al., 2000; Dunham-Ems et al., 2012; Drecktrah et al., 2015). These studies illustrate an important role for RelBbu and the stringent response in persistence in the tick and subsequent transmission to the host.
The transcription factor DksA (DnaK suppressor), which potentiates the transcriptional effects of (p)ppGpp on RNA polymerase activity (Gourse et al., 2018), also globally reprograms the B. burgdorferi transcriptome in response to nutrient stress (Boyle et al., 2019; Mason et al., 2020). There is considerable overlap between the DksA and RelBbu regulons, but differences exist (see below), illustrating that DksA is a cooperating partner in the stringent response but also exerts an independent influence. Recent studies have found that, similar to RelBbu, DksA regulates genes that affect B. burgdorferi survival under nutrient and osmotic stresses (Boyle et al., 2019). Surprisingly, DksA is essential for B. burgdorferi infectivity in the murine model of Lyme disease, at least by needle inoculation (Mason et al., 2020), while RelBbu is dispensable for murine infectivity (Drecktrah et al., 2015). Tick transmission studies await further investigation as do experiments examining the role of DksA in persistence in the tick.
The RelBbu regulon
The RelBbu-dependent regulon is characterized by broad patterns of gene regulation typical of bacteria adapting to nutrient stress by reducing growth, including repression of many genes involved in metabolism (glycolysis and mevalonate biosynthesis), translation (ribosomal proteins) and transcription (RNA polymerase subunits), as revealed by RNA-seq and microarray analyses (Bugrysheva et al., 2015; Drecktrah et al., 2015). RelBbu-dependent regulation of gene products whose function is not fully understood may provide clues to their role in the enzootic cycle. For example, the expression of the genes encoding the oligopeptide binding proteins (oppA1, oppA2, oppA3 and oppA5) are RelBbu-upregulated suggesting that peptide uptake functions during persistence of the spirochete in the tick, as this is the phase where survival of the relBbu mutant is compromised (Drecktrah et al., 2015). RelBbu also regulates the glycerol metabolic (glp) operon (Bugrysheva et al., 2015; Drecktrah et al., 2015). The glp operon contributes to persistence of B. burgdorferi in the tick where the spirochete is thought to utilize glycerol as a carbon source and environmental signal, as has been demonstrated in vitro (He et al., 2011; Pappas et al., 2011; Bugrysheva et al., 2015). The defect in persistence in the tick of the relBbu mutant is likely due, at least in part, to compromised glycerol metabolism. The metabolic fate of glycerol may be directed by the stringent response as well: RelBbu upregulates expression of genes encoding the glycerol uptake facilitator (glpF) and glycerol kinase (glpK), but studies disagree on the effect on the last gene in the operon, glycerol 3-phosphate dehydrogenase (glpD) (Bugrysheva et al., 2015; Drecktrah et al., 2015). If GlpD is upregulated then glycerol 3-phosphate would be trafficked to the glycolytic pathway, while downregulation of GlpD would instead shuttle phosphorylated glycerol to membrane and lipoprotein biosynthesis. This suggests that the stringent response coordinates transcriptional changes to affect metabolism to adapt the spirochete to nutritional flux in the enzootic cycle.
These transcriptomic studies also illuminate potential functions for RelBbu in processes not traditionally thought to be regulated by the stringent response. Expression of vlsE, a Vmp-like surface lipoprotein used to evade the adaptive host response (Norris, 2014; Chaconas et al., 2020) (see Radolf and Samuels, 2021), is RelBbu-upregulated (Drecktrah et al., 2015). Although vlsE recombination only occurs in the vertebrate host, the stringent response is the first signaling system identified that regulates vlsE expression. In addition, RelBbu regulates expression of the bosR gene (Drecktrah et al., 2015) (see section above on Transcription factors BosR and BadR). The reduction in bosR expression is not enough to render the relBbu mutant non-infectious by needle inoculation, but could play a role in the tick transmission defect. RelBbu also upregulates a number of genes encoding extracellular matrix-binding proteins important in the vertebrate host, such as dbpBA and bbk32 (Drecktrah et al., 2015). These findings suggest that B. burgdorferi may encounter nutrient-limited niches in the vertebrate, such as the synovial regions in joints, that trigger the stringent response or there may be subtler, as of yet unidentified, signals inducing (p)ppGpp production in the spirochete to modulate gene expression in the vertebrate host.
RelBbu also seems to coordinate the life cycle of B. burgdorferi bacteriophages by controlling numerous genes located on the cp32 prophages (Eggers and Samuels, 1999; Zhang and Marconi, 2005). RelBbu represses expression of many more cp32 genes than it activates, with most located in the putative late operon (Drecktrah et al., 2015). Furthermore, expression of the essential gene ftsH (Chu et al., 2016) was also RelBbu-downregulated (Bugrysheva et al., 2015; Drecktrah et al., 2015); the ftsH homolog in E. coli encodes a protease that regulates the λ phage life cycle (Kihara et al., 1997; Shotland et al., 1997). Thus, changes in nutrient availability in the tick midgut or vertebrate may signal, through the stringent response, conditions for prophage induction.
RelBbu regulates sRNAs
The stringent response regulates expression of a third of all identified small RNAs (sRNAs) in B. burgdorferi, including intragenic sRNAs, antisense sRNAs and 5′ UTR sRNAs (Popitsch et al., 2017; Drecktrah et al., 2018) (see the section below on Small regulatory RNAs and RNA chaperones). Threefold more sRNAs were RelBbu-upregulated than downregulated and were mapped throughout the genome (Drecktrah et al., 2015). Antisense sRNAs and 5′ UTR sRNAs associated with genes of known or predicted function mostly targeted genes encoding translation, transporter and cell envelope functions. One intriguing example is that RelBbu downregulates an antisense sRNA within the glpF ORF (SR0186) while concurrently upregulating the glpF mRNA (Drecktrah et al., 2018). The sense:antisense pairing may favor degradation of the glpF transcript in response to vertebrate-specific signals (Drecktrah et al., 2015). Thus, one mechanism B. burgdorferi may employ to survive in the tick involves sensing nutrient stress between blood meals to induce the stringent response that represses the glpF antisense sRNA, which, in turn, stabilizes the glpF mRNA and increases GlpF production to enhance transport and utilization of glycerol.
RelBbu may also indirectly modulate RNA polymerase activity to alter the global transcriptome through the Bb6S RNA (see the section below on Small regulatory RNAs and RNA chaperones). Bb6S RNA is a highly expressed sRNA that binds directly to the RpoD (σ70)-RNA polymerase (Drecktrah et al., 2020). RelBbu upregulates Bb6S RNA, potentially providing another mechanism to shape the transcriptional landscape in addition to the direct interaction of (p)ppGpp with RNA polymerase (Drecktrah et al., 2018). The vast contingent of sRNAs controlled by the stringent response represent multiple pathways for regulating gene expression in response to nutrient stress.
The DksA regulon
The transcription factor DksA collaborates with (p)ppGpp to globally regulate gene expression (Gourse et al., 2018). DksA targets many of the same genes as RelBbu, but also affects a distinct set (Boyle et al., 2019), a phenomenon that has been reported in other bacteria (Potrykus and Cashel, 2008; Dalebroux et al., 2010). Downregulation of genes encoding ribosomal proteins, DNA replication proteins, and transcriptional and translational machinery was DksA-dependent in addition to RelBbu-dependent (Bugrysheva et al., 2015; Drecktrah et al., 2015; Boyle et al., 2019). DksA-dependent genes were similarly found throughout the genome and spanned all functional categories, reinforcing the extensive genomic reach of this stringent response-associated transcription factor. As with RelBbu, DksA upregulated expression of factors important for persistence in the tick, such as the glycerol metabolic genes glpF and glpK, the bacterioferritin ortholog gene dps/napA/bicA, as well as the tick phase-specific lipoprotein-encoding genes ospA and lp6.6 (Boyle et al., 2019). Further transcriptional overlap with the RelBbu regulon includes the repression of many cp32 genes in the late phage operon and upregulation of a number of genes encoding lipoproteins associated with the vertebrate phase of the enzootic cycle, including dbpA and ospC (Drecktrah et al., 2015; Boyle et al., 2019).
Interplay between (p)ppGpp and DksA is evident as levels of dksA transcript increase in the relBbu null mutant (Bugrysheva et al., 2015; Drecktrah et al., 2015) while (p)ppGpp levels are increased in a dksA null mutant (Boyle et al., 2019). Thus, the spirochete may be compensating for the lack of one component of the stringent response by increasing the levels of its partner, illustrating the cooperative and complex relationship of (p)ppGpp and DksA in remodeling the transcriptome to adapt to nutrient flux in the dynamic environments of the enzootic cycle.
That both RelBbu and DksA upregulate expression of gene products controlled by the RpoN-RpoS σ factor cascade mediating vertebrate phase-specific gene expression and host infectivity was unexpected, and suggests a more intricate role for these factors than merely mediating the response to nutrient limitations. DksA, as well as RelBbu, does not alter the level of rpoS transcript during nutrient stress, but rather appears to affect RpoS levels post-transcriptionally (Drecktrah et al., 2015; Boyle et al., 2019; Mason et al., 2020), a function that has been previously reported in E. coli (Brown et al., 2002). The mechanism for post-transcriptional RpoS regulation in these mutants has not been determined but may be driven, at least in part, by RelBbu-upregulation of DsrABb, an sRNA known to post-transcriptionally increase RpoS levels (Lybecker and Samuels, 2007; Drecktrah et al., 2018). Thus, reduction of ospC and dbpA transcripts is likely due to the lack of RpoS protein in the DksA mutant and may account for the non-infectious phenotype observed when mice are challenged by needle inoculation (Mason et al., 2020). The finding that the RelBbu mutant is infectious indicates a marked divergence of the two regulators that have similar regulons and are thought to work hand-in-hand. Future studies are required to discern the subtle functional differences between RelBbu and DksA, and elucidate the induction mechanisms of genes encoding lipoproteins involved in B. burgdorferi infectivity as well as the complexities of the stringent response.
Small regulatory RNAs and RNA chaperones
Small non-coding RNAs have been described as the “dark matter of the cell” (Riddihough, 2005) because, despite their plethora, they remained mostly undiscovered in the pre-genomic world. Of course, many of these small non-coding RNAs were known since the early days of molecular biology, such as 6S RNA, the ubiquitous and, in certain situations, predominant global regulator of bacterial transcription whose function was outright mysterious for over 30 years (Wassarman and Storz, 2000). Aside from a few functional small RNAs (sRNAs), such as tmRNA (SsrA), annotated in the genome sequence (Fraser et al., 1997), the first small RNA identified in B. burgdorferi was DsrABb, a post-transcriptional regulator of the RpoN-RpoS alternative σ factor cascade (Lybecker and Samuels, 2007). The constellation of non-coding sRNAs in B. burgdorferi exponentially expanded a decade later via high-throughput transcriptome studies (Arnold et al., 2016; Adams et al., 2017; Popitsch et al., 2017; Drecktrah et al., 2018). The bevy of sRNAs was classified based on their genomic location: intergenic sRNAs, intragenic sRNAs, antisense sRNAs, and 5′ UTR-associated sRNAs (Figure 4) (Popitsch et al., 2017).
Figure 4.
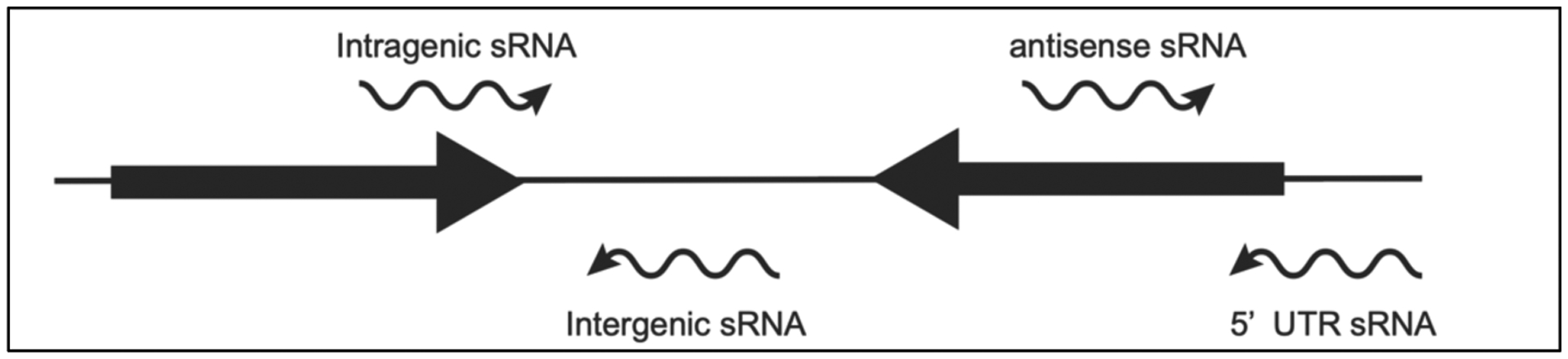
Classification of sRNAs based on their genomic location. sRNAs (black wavy arrows) were categorized based on their relation to annotated open reading frames (black solid arrows).
Several mechanisms of riboregulation by sRNAs have been defined, including those employed by pathogenic bacteria to modulate virulence strategies (Fröhlich and Vogel, 2009; Papenfort and Vanderpool, 2015; Svensson and Sharma, 2016; Hör et al., 2018; Lejars et al., 2019; Adams and Storz, 2020; González Plaza, 2020). Many sRNAs regulate gene expression via base-pairing with target mRNAs, affecting their stability, translation, transcription, or processing: trans-acting sRNAs base-pair with their target mRNAs and cis-acting antisense sRNAs are completely complementarity to their cognate mRNA (Waters and Storz, 2009; Storz et al., 2011; Caldelari et al., 2013). Other sRNAs interact with proteins, titrating their levels or otherwise affecting their activities. As of 2020, only three sRNAs have been functionally characterized in B. burgdorferi. (i) DsrABb is a trans-acting sRNA that base-pairs with rpoS mRNA regulating its translation (Lybecker and Samuels, 2007). (ii) Bb6S RNA is a protein-interacting sRNA that directly influences σ factor selectivity by RNA polymerase and globally regulates gene expression (Drecktrah et al., 2020). (iii) ittA is an sRNA that affects expression of several genes and is required for dissemination in the vertebrate host via an unknown molecular mechanism (Medina-Pérez et al., 2020). To date, there is no experimental evidence in B. burgdorferi for riboswitches, cis-acting regulatory RNAs usually found in the 5′ UTR of the mRNA they regulate that function by altering their secondary structure in response to binding intracellular metabolites (Roth and Breaker, 2009; Serganov and Nudler, 2013; Sherwood and Henkin, 2016; McCown et al., 2017); however, the class of 5′ UTR-associated sRNAs (Popitsch et al., 2017) may represent heretofore unrecognized riboswitches.
sRNAs
DsrABb
In Escherichia coli, DsrA is one of three regulatory sRNAs that affect levels of the σ factor RpoS (Hengge-Aronis, 2002a; Repoila et al., 2003; Majdalani et al., 2005; Narberhaus et al., 2006; Waters and Storz, 2009). The sRNA disrupts a hairpin stem-loop that blocks access of the ribosome to the Shine-Dalgarno sequence on the rpoS mRNA and enhances its translation at low temperature (Hengge-Aronis, 2002a; Repoila et al., 2003). DsrA is considered to be “a thermometer for the RpoS regulon” in E. coli (Repoila et al., 2003) because both its transcription and stability increase at low temperature, causing an accumulation of RpoS protein during the cold stress response (Repoila and Gottesman, 2001).
An sRNA was discovered in B. burgdorferi that has extensive complementarity to the upstream region of the rpoS mRNA, which appears to assume a hairpin stem-loop structure akin to that of E. coli, implying a similar mode of regulation (Lybecker and Samuels, 2007). Several experiments suggested that the sRNA functioned in post-transcriptional temperature-mediated regulation of RpoS, so it was called DsrABb; however, there are substantial differences between the E. coli and B. burgdorferi DsrA analogs (Lybecker and Samuels, 2007). Notably, DsrABb senses an increase in temperature, presumably when warm vertebrate blood enters the ambient feeding tick, while DsrA in E. coli responds to a decrease in temperature. In addition, DsrABb, which is about four times longer than DsrA from E. coli, has more extensive base-pairing potential (31 of 34 nucleotides, or up to 51 of 68 nucleotides, compared to 26 of 38 nucleotides) with its target, the rpoS mRNA in B. burgdorferi (Lybecker and Samuels, 2007). Furthermore, via an as yet unknown mechanism, DsrABb requires translation of an upstream and overlapping open reading frame encoded by bb0577 (Lybecker and Samuels, 2007). Lastly, the steady-state levels of DsrA in E. coli are influenced by temperature, while DsrABb is unaffected by temperature (Lybecker and Samuels, 2007) and instead is upregulated by the stringent response (Drecktrah et al., 2018) (see the section above on the Stringent response and (p)ppGpp). Other than the potential crosstalk between the stringent response (which regulates gene expression in response to starvation in the unfed tick midgut) and the RpoN-RpoS alternative σ factor cascade (which activates gene expression required in the vertebrate host and represses gene expression required in the tick vector), almost nothing is known about the temperature-sensing modus operandi of DsrABb.
DsrABb is hypothesized to serve as a molecular thermometer regulating gene expression during the enzootic cycle of B. burgdorferi by modulating the RpoN-RpoS σ factor cascade. Yet, the increase in rpoS mRNA levels (Caimano et al., 2004) does not require DsrABb (Lybecker and Samuels, 2007), which suggests the presence of two independent mechanisms that regulate RpoS. Two rpoS mRNAs have been identified (Lybecker and Samuels, 2007; Smith et al., 2007): a short rpoS mRNA that is transcribed from an RpoN-dependent promoter (Smith et al., 2007) at high cell density (Lybecker and Samuels, 2007) and a long rpoS mRNA that is transcribed from an RpoD-dependent promoter at low cell density (Lybecker and Samuels, 2007). Of note, DsrABb stimulates the translation of rpoS mRNA exclusively at low cell density in culture (Lybecker and Samuels, 2007). Only the long rpoS mRNA contains the sequence complementary to DsrABb (Lybecker and Samuels, 2007) and, therefore, DsrABb regulates translation of the RpoD-dependent long rpoS mRNA, but not translation of the RpoN-dependent short rpoS mRNA. The seminal studies (Hübner et al., 2001; Smith et al., 2007) on rpoS regulation missed the long rpoS mRNA presumably because the spirochetes were grown to a high cell density, culture conditions in which the short rpoS mRNA is transcribed (Lybecker and Samuels, 2007). The long rpoS transcript has been observed in both an rpoN null mutant and in a strain with the RpoN-dependent rpoS promoter mutated (Hall et al., 2021). What, then, is the regulatory role of the two rpoS mRNA species, and the DsrABb sRNA, in the enzootic cycle? The proposed model is that the pathway controlling RpoS and, subsequently, enzootic cycle phase-specific gene expression, bifurcates: the DsrABb-RpoS branch activates RpoS during transmission from the tick to the vertebrate and the RpoN-RpoS branch maintains RpoS levels, and RpoS-dependent gene expression, in the vertebrate (Hall et al., 2021). This accounts for the apparently discrepant observations that rpoN mutants migrate from the midgut to the salivary glands (Fisher et al., 2005), but rpoS mutants cannot exit the midgut (Dunham-Ems et al., 2012).
Bb6S RNA
6S RNA, encoded by ssrS, directly interacts with RpoD-holoenzyme (E), the RNA polymerase carrying the RpoD (Eσ70) σ factor (Wassarman and Storz, 2000; Cavanagh and Wassarman, 2014; Steuten et al., 2014a; Steuten et al., 2014b; Burenina et al., 2015; Wassarman, 2018), and serves as a global regulator of transcription (Waters and Storz, 2009; Storz et al., 2011). The primary sequences of 6S RNAs from different bacterial species are not conserved, but rather share a conserved secondary structure that imitates the open promoter complex (Wassarman and Storz, 2000; Barrick et al., 2005; Trotochaud and Wassarman, 2005; Chen et al., 2017), and 6S RNA prevents binding of Eσ70 to RpoD-dependent promoters, resulting in decreased transcription of RpoD-dependent genes and increased transcription of many RpoS-dependent genes (Wassarman and Storz, 2000; Trotochaud and Wassarman, 2004, 2006; Cavanagh et al., 2008; Cavanagh et al., 2010; Neusser et al., 2010). In general, the function of 6S RNA is to shift expression from the RpoD-dependent regulon to the RpoS-dependent regulon (Cavanagh et al., 2008). In E. coli and other bacteria, 6S RNA has been associated with adaptation to environmental stresses, but 6S RNA also affects bacterial virulence and infectivity (Faucher et al., 2010; Yan et al., 2013; Warrier et al., 2014; Ren et al., 2017).
Bb6S RNA, the canonical 6S RNA in B. burgdorferi, was first predicted by computational genome mining (Barrick et al., 2005) and was then found in the noncoding sRNA transcriptomes of B. burgdorferi (Arnold et al., 2016; Popitsch et al., 2017; Drecktrah et al., 2018). Recently, Bb6S RNA was shown to bind RNA polymerase in vivo, be processed by RNase Y, regulate expression of lipoprotein genes, and play a role in infectivity in the murine model (Drecktrah et al., 2020). Mapping and modeling of Bb6S RNA suggest that the secondary structure is conserved with a single-stranded central bubble surrounded by double-stranded stem regions (Barrick et al., 2005; Trotochaud and Wassarman, 2005; Chen et al., 2017; Drecktrah et al., 2020).
Unlike 6S RNA from E. coli and some other bacteria (Wassarman and Storz, 2000; Cavanagh and Wassarman, 2014; Steuten et al., 2014b), steady-state levels of Bb6S RNA were not affected in cultured B. burgdorferi by nutrient stress or growth phase (Drecktrah et al., 2020), which is consistent with RNA-seq studies showing temperature and growth phase had no effect on Bb6S RNA levels (Arnold et al., 2016; Popitsch et al., 2017). However, the amount of Bb6S RNA increased as fed larvae molted into nymphs and decreased after nymphs fed, indicating a function for Bb6S RNA during spirochete persistence in the tick vector (Drecktrah et al., 2020). Bb6S RNA also is upregulated by the stringent response (Drecktrah et al., 2018), which facilitates survival of B. burgdorferi between the larval and nymph blood meals (Drecktrah et al., 2015) (see section above on the Stringent response and (p)ppGpp). However, Bb6S RNA was not required for spirochetes to persist through the larval and nymph stages (Drecktrah et al., 2020). Note, though, that in other bacteria, the phenotypes of 6S RNA mutants are typically subtle (Wassarman and Storz, 2000; Trotochaud and Wassarman, 2004, 2006; Cavanagh et al., 2012; Cavanagh and Wassarman, 2013) and Bb6S RNA was required for full infectivity of mice by needle inoculation, although, the infectivity defect of the Bb6S RNA mutant was overcome by either an increased inoculum or tick transmission (Drecktrah et al., 2020). Bb6S RNA regulates expression of the RpoS-dependent genes ospC and dbpBA without significantly changing RpoS protein levels, results consistent with Bb6S RNA sequestering Eσ70 and promoting transcription by the RpoS-holoenzyme (EσS) as found in model microorganisms (Drecktrah et al., 2020).
ittA
The intergenic sRNA SR0736, encoded between bbd18 and bbd21 on lp17, was recently identified as attenuating vertebrate infectivity and dissemination of B. burgdorferi by filtering data from a global transposon insertion (Lin et al., 2012) sequencing screen with the output of an sRNA transcriptome (Popitsch et al., 2017). This sRNA was renamed ittA, for infectivity-associated and tissue-tropic sRNA locus A (Medina-Pérez et al., 2020). An ittA mutant strain differentially expresses 19 genes compared to the wild-type parental strain, including increased levels of bba66 and vra, as well as 11 other mRNAs, and decreased levels of ospD, ospA and bba74/oms28, as well as three other mRNAs (Medina-Pérez et al., 2020). The molecular mechanism(s) ittA employs to regulate gene expression are currently unknown. Curiously, despite restoring the in vivo infectivity phenotype, the ittA complement did not rescue the gene expression defect of many of the validated differentially regulated genes.
RNA chaperones
Hfq
Hfq is an RNA chaperone that facilitates the interaction between sRNAs and their targets (Gottesman, 2004; Valentin-Hansen et al., 2004; Majdalani et al., 2005; Brennan and Link, 2007; Waters and Storz, 2009). The Hfq of B. burgdorferi was difficult to identify because it has limited homology with the Hfq from E. coli and other bacteria, but it is able to heterologously complement an hfq mutant of E. coli (Lybecker et al., 2010). Conversely, the E. coli hfq gene heterologously complements several phenotypes of the pleiotropic B. burgdorferi hfq mutant in vitro, albeit not the vertebrate infectivity defect (Lybecker et al., 2010). B. burgdorferi Hfq is required for infectivity in mice, as well as induction of RpoS and its regulon (Lybecker et al., 2010); therefore, the phenotype is not due solely to a defect in facilitating the DsrABb interaction with rpoS mRNA, as RpoS can be induced at high cell density in the dsrABb mutant (Lybecker and Samuels, 2007), suggesting an Hfq-dependent sRNA network in the spirochete that regulates infectivity (Lybecker et al., 2010).
CsrA
CsrA is an RNA-binding protein that inhibits translation of several genes by binding the ribosome-binding sites of their mRNAs and blocking the ribosome; CsrB and CsrC are sRNAs that bind to and sequester CsrA, inhibiting its activity on mRNAs (Romeo, 1998; Majdalani et al., 2005; Hör et al., 2018; Romeo and Babitzke, 2018). CsrA regulates a variety of physiological processes in E. coli and other bacteria, including carbon metabolism and motility (Romeo, 1998; Majdalani et al., 2005; Hör et al., 2018; Romeo and Babitzke, 2018), as well as bacterial infectivity (Lucchetti-Miganeh et al., 2008; Vakulskas et al., 2015). Levels of CsrA in B. burgdorferi increase in response to elevated temperature and decreased pH (Sanjuan et al., 2009). Initial reports suggested that CsrA affects the expression of ospC as well as other RpoS-dependent genes and is required for murine infectivity in B. burgdorferi (Sanjuan et al., 2009; Karna et al., 2011; Sze and Li, 2011; Sze et al., 2011; Karna et al., 2013; Arnold et al., 2018; Lin et al., 2018); however, a subsequent study reported that CsrA does not activate RpoS or its regulon and that csrA mutants are infectious in the murine model (Ouyang et al., 2014b). As of 2020, the role of CsrA in the physiology of B. burgdorferi remains enigmatic and its RNA targets remain undefined.
RNA turnover and ribonucleases
The steady-state levels of RNAs are controlled not just by the rate of their transcription, but also by the rate of their degradation, which is catalyzed by a variety of ribonucleases (Mohanty and Kushner, 2016; Bechhofer and Deutscher, 2019; Trinquier et al., 2020). Compared to other bacteria, B. burgdorferi encodes a sparse quiver of ribonucleases, which includes the endoribonucleases RNase III (BB0705), RNase P (BB0441), RNase Z (BB0755), YbeY (BB0060), RNase Y (BB0504), RNase M5 (BB0626), and RNase HII (BB0046) and the exoribonuclease PNPase (BB0805) (Fraser et al., 1997; Archambault et al., 2013; Anacker et al., 2018; Drecktrah et al., 2020). Lyme disease spirochetes lack detectable orthologs of RNase E/G and RNase J1/J2; RNase Y functions in RNA processing and decay in Bacillus subtilis, Staphylococcus aureus, and Streptococcus pyogenes in an analogous fashion as RNase E or G in E. coli, so B. burgdorferi appears to be more similar in its suite of ribonucleases to the gram-positive bacteria (Shahbabian et al., 2009; Yao and Bechhofer, 2010; Bechhofer, 2011; Lehnik-Habrink et al., 2011; Marincola et al., 2012; Chen et al., 2013; Bechhofer and Deutscher, 2019). Two ribonucleases have been genetically characterized in B. burgdorferi: RNase III, encoded by the rnc gene (Anacker et al., 2018) and RNase Y, encoded by the essential rny gene (Drecktrah et al., 2020). RNase III, which recognizes and cleaves double-stranded RNA, is required for proper rRNA processing (Anacker et al., 2018) and for mRNA decay, at least for several transcripts (Snow et al., 2020), in the spirochete. RNase Y is involved in the biogenesis of 6S RNA in B. burgdorferi (Drecktrah et al., 2020), which has not been shown in any other bacterium, but the ribonuclease surely has additional roles in the cell, likely in the turnover of mRNA based on precedence in other bacteria (Lehnik-Habrink et al., 2011; Durand et al., 2012; Chen et al., 2013), as Bb6S RNA null mutants are viable but rny is essential for growth of the spirochete (Drecktrah et al., 2020).
Schlax and colleagues have assayed mRNA turnover kinetics in vitro and demonstrated that half-lives vary from a minute to almost an hour, which is considerably longer than the half-lives measured for the majority of mRNAs in most bacteria (Archambault et al., 2013). Note that these studies are more complicated in B. burgdorferi because mRNA turnover in bacteria is generally quantified following inhibition of transcription with rifampicin, but the unusual RNA polymerase of B. burgdorferi is resistant to this antibiotic, so, instead, actinomycin D is used to arrest transcription (Archambault et al., 2013). The half-lives and steady-state levels of several mRNAs, as well as the steady-state levels of the stable rRNAs, were significantly increased in an rnc mutant, indicating a role for RNase III in RNA degradation (Snow et al., 2020). However, the steady-state levels of a couple mRNAs decreased in the rnc mutant, suggesting a more nuanced regulatory scheme involving RNase III. At this point, the mechanism by which RNA degradation and RNase activity is regulated to effect post-transcriptional levels of gene expression in B. burgdorferi is unknown; however, RNase III is known to regulate the interaction between antisense sRNAs and their target mRNAs in several bacteria, including E. coli, B. subtilis, S. aureus, S. pyogenes and Streptomyces antibioticus (Lasa et al., 2011; Lasa et al., 2012; Lioliou et al., 2012; Lee et al., 2013; Lybecker et al., 2014a; Lybecker et al., 2014b; Le Rhun et al., 2016), and a similar mechanism may be employed by the spirochete.
Nucleic acid-binding proteins
The genome of B. burgdorferi encodes several proteins that resemble known nucleic acid-binding proteins of other bacteria, such as BosR and BadR (see the section above). However, most of those predicted nucleic acid-binding proteins have yet to be characterized. In addition, novel proteins have been identified that do not possess structures previously associated with nucleic acid interactions. Two of the most notable of these borrelial proteins, EbfC and SpoVG, are also encoded by a wide spectrum of other bacterial species. Thus, studies of B. burgdorferi are providing insights on the physiology of bacteria ranging from E. coli to S. aureus.
At least two novel B. burgdorferi proteins, SpoVG and BpuR, initially discovered due to their DNA-binding activities, have since been found to have greater affinities for RNA than for DNA. Combined with data from other species, this finding raises the possibility that many other “DNA-binding” proteins might also preferentially bind to RNAs. Additionally, the known activities of BpuR and SpoVG support a paradigm that RNA-binding proteins can have functions beyond serving as RNA chaperones, such as operating solo to directly affect mRNA translation.
EbfC
This 99-amino acid protein forms a homodimer in solution; the alpha helical N- and C-termini of both subunits form a “pincer” that fits over the DNA double helix (Lim et al., 2003; Riley et al., 2009). Consistent with binding by a homodimer, EbfC preferentially interacts with a palindromic sequence, 5′-GTnAC-3′. This sequence occurs approximately every 1.1 kb throughout the B. burgdorferi genome. EbfC also forms higher-order oligomers, including tetramers and octamers, an activity that could serve to bridge separate DNA strands (Riley et al., 2009). Microscopic analyses of GFP-tagged EbfC revealed that it associates with the borrelial nucleoid (Jutras et al., 2012a). These are all characteristics of nucleoid-associated proteins, leading to a model that a major function of EbfC is to organize the structure of borrelial DNA. Moreover, the ebfC gene is coexpressed with dnaX, which encodes a subunit of DNA polymerase, suggesting that DNA-binding by EbfC plays an indirect role in replication (Jutras et al., 2012a). Almost all known species of bacteria also encode an EbfC homolog (variously named YbaB, ORF12, and other terms.), implying a conserved function (Cooley et al., 2009; Wang et al., 2012a).
EbfC initially was identified due to its high affinity binding to the upstream region of B. burgdorferi erp operons (Babb et al., 2006). The majority of erp operators contain two to three complete EbfC-binding sites, a density that has not been identified elsewhere in the B. burgdorferi genome. EbfC acts as an anti-repressor of erp transcription by competing with the BpaB repressor for binding to the operator (Jutras et al., 2012b). Erp protein expression is repressed during colonization of an unfed tick, but is induced when the tick begins to feed, and those outer surface lipoproteins are expressed during transmission and throughout the vertebrate phase of the enzootic cycle. Studies of B. burgdorferi cultivated in vitro found that erp transcript levels increase as bacterial replication rates increase, a phenomenon that also occurs as a tick begins to feed and the spirochetes rapidly multiply due to nutrients in the blood (Jutras et al., 2013c). The current model for erp regulation posits that the rapid bacterial replication rate in feeding ticks necessitates DnaX and EbfC for DNA replication, and that erp operons have evolved to recognize elevated EbfC levels as a signal that the tick is feeding and the time is appropriate to produce the Erp proteins (Jutras et al., 2012a; Jutras et al., 2013c).
BpaB
All of the plasmids in the segmented genome of B. burgdorferi encode a unique BpaB protein. BpaB and two other ORFs are essential for plasmid maintenance and partitioning, with BpaB apparently performing a function comparable to the ParB proteins of other bacterial plasmids. The unique sequences of BpaB from each plasmid appear to be involved with compatibility (Zückert and Meyer, 1996; Casjens et al., 1997; Fraser et al., 1997; Casjens et al., 2000; Stewart et al., 2001; Eggers et al., 2002; Stewart et al., 2003; Brisson et al., 2013). The BpaB proteins of the cp32 family of prophages are the best studied to date. While BpaB presumably binds DNA near the origin of replication, as do ParB proteins from other species, three other binding sites have been identified on cp32s: one each in the erp operator, adjacent to the nucP promoter, and adjacent to the ssbP promoter (Burns et al., 2010; Chenail et al., 2012). In erp operators, BpaB binds to a specific sequence 5′ of the promoter, then oligomerizes along the DNA, stabilized by protein-protein interactions (Burns et al., 2010). Occlusion of the promoter by BpaB prevents RNA polymerase binding, thereby inhibiting transcription (Jutras et al., 2012b). BpaB appears to enhance transcription of nucP and ssbP, through unknown mechanisms (Chenail et al., 2012).
BpuR
BpuR is a homodimer of two 122-amino acid subunits, which fold together into a “PUR” domain (Graebsch et al., 2010; Jutras et al., 2013a). This domain is named because it preferentially binds to purine-rich stretches of nucleic acids. PUR domains are found in a few other bacterial genera, such as Treponema and Bacteroides, and in all multicellular eukaryotes. This curious distribution suggests that there was an exchange of a PUR motif between Domains over the course of evolution.
PUR domain proteins bind to double-stranded and single-stranded DNAs and to RNA, with BpuR exhibiting its greatest affinity for RNA. A specific nucleotide sequence has yet to be identified for BpuR binding, a feature shared with eukaryotic PUR domain proteins. This suggests that PUR domain proteins interact with higher-order structural determinants.
BpuR was first identified due to its ability to bind DNA, as it has a high affinity for sequences in erp operator sites. It acts as a co-repressor of erp transcription, enhancing the repressor activity of BpaB through an undefined mechanism. BpuR has also been found to bind several borrelial mRNAs, including bpuR itself, where binding to the 5′ end inhibits translation (Jutras et al., 2013d). Comparisons of transcriptomic and proteomic data indicate that BpuR is also likely to affect translation of numerous mRNAs (Jutras et al., 2019).
SpoVG
Homologs of SpoVG are produced by many species of spirochetes and firmicutes. Its name comes from early studies of Bacillus subtilis, where spoVG mutants are defective in stage V of sporulation (Rosenbluh et al., 1981). SpoVG proteins do not contain previously known nucleic acid-binding motifs, and their functions were a mystery until investigations of a DNA-binding protein of B. burgdorferi resulted in fishing out SpoVG (Jutras et al., 2013b). That initial study further demonstrated that the SpoVG proteins of Listeria monocytogenes and Staphylococcus aureus also bind to DNA, and mapped nucleic acid-binding specificity to the C-terminal alpha helix. Subsequent studies found that the L. monocytogenes and B. burgdorferi SpoVG proteins exhibit higher affinities for RNA than for DNA (Burke and Portnoy, 2016; Savage et al., 2018). A defined nucleotide sequence for SpoVG-binding has yet to be identified, suggesting that it interacts with conformational aspects of the nucleic acid sugar-phosphate backbone. B. burgdorferi spoVG mutants exhibit pleotropic defects, evidently due to both transcriptional and post-transcriptional regulatory activities (Savage et al., 2018).
Hbb
Integration host factor (IHF) was discovered on the basis of its role in the integration of bacteriophage λ into the E. coli chromosome (Drlica and Rouviere-Yaniv, 1987; Friedman, 1988; Nash, 1996). IHF binds to DNA at specific sites and induces bending. The origin of replication on the E. coli chromosome contains an IHF-binding site, which is conserved in several bacteria, and IHF is essential for replication (Nash, 1996). Another small DNA-binding protein in E. coli is HU, which is homologous to IHF (Drlica and Rouviere-Yaniv, 1987; Schmid, 1990; Nash, 1996). HU also bends DNA, but it lacks sequence specificity. Many bacteria have homologs of both IHF and HU, although several species have only a single homolog. Tilly et al. (1996) isolated a gene encoding an IHF/HU homolog, termed Hbb, from B. burgdorferi in a genetic screen based on complementation of the λ DNA packaging defect of E. coli IHF/HU mutants. Hbb is most similar to homologs from bacteria that encode a single IHF/HU homolog and is 27 to 30% identical to IHF and HU from E. coli (Tilly et al., 1996). Hbb is unable to complement an E. coli HU mutant for the defect in bacteriophage Mu growth (Tilly et al., 1996) and it binds DNA in a sequence-specific fashion (Kobryn et al., 2000; Mouw and Rice, 2007), suggesting that Hbb is more like IHF than HU. Kobryn et al. (2000) demonstrated that Hbb binds to a specific sequence that is related to the IHF consensus binding site. In addition, IHF binds to this Hbb-binding site with high affinity and Hbb will bind to an IHF consensus site (Mouw and Rice, 2007). However, Hbb affects DNA topology, which is a property of HU, so Hbb has a substantial non-specific DNA-binding activity and is not a straightforward IHF homolog (Kobryn et al., 2000).
Mouw and Rice (2007) solved the structure of an Hbb-DNA complex. The structure and mechanism of DNA bending is similar to that of IHF and HU proteins. Hbb binds DNA as a homodimer with two β-ribbon “arms” that wrap in the minor groove, with a pair of conserved prolines intercalating between adjacent base pairs (Mouw and Rice, 2007), which results in a DNA bend of more than 180° (Kobryn et al., 2000; Mouw and Rice, 2007). The binding site identified by Kobryn et al. (2000) is near the origin of replication on the chromosome (between dnaA and dnaN). Thus, Hbb may be involved in replication initiation (Chaconas, 2005), although results in an E. coli surrogate system show that recombinant Hbb increases transcription of oppA2 and oppA4 promoter fusions (Medrano et al., 2007).
Gac
Knight and Samuels (1999) identified Gac in a biochemical screen for proteins that bound to a sub-telomeric sequence from linear plasmid lp17. Gac has HU-like non-specific DNA-binding activity (Knight and Samuels, 1999; Corbett et al., 2004), but it is a novel protein that completely lacks sequence similarity to HU or other members of the architectural DNA-binding protein family: it has only been identified to date in B. burgdorferi. Gac is the C-terminal domain of GyrA, the A subunit of DNA gyrase, and it is produced as an independent protein by a mechanism unprecedented in bacteria. B. burgdorferi transcribes two mRNAs carrying gac sequences: the larger one encodes the entire gyrBA operon that presumably generates DNA gyrase, the essential type II DNA topoisomerase in bacteria that introduces negative DNA supercoiling (Reece and Maxwell, 1991; Corbett and Berger, 2004), and the smaller, more abundant one, encodes Gac. The gac mRNA is predicted to be transcribed from its own promoter embedded in the gyrA gene (Knight and Samuels, 1999). This unusual architectural DNA-binding protein was termed Gac (for GyrA C-terminal domain), and its gene gac, because, although it is translated from a subset of the GyrA ORF, it is a different gene product of a unique genetic locus (Knight et al., 2000). In other words, mutations can disrupt the synthesis of Gac, but have no phenotypic effect on GyrA. In fact, genetic disruption of Gac was accomplished by mutating the start codon and Met 5 codon (to Leu and Ile, respectively) (Knight et al., 2000). Disappointingly, the gac mutant had no detectable phenotype. The phenotype was hypothesized to be suppressed by the coumermycin A1 resistance marker in the upstream gyrB gene used to introduce the site-specific mutations (Samuels, 2006). Although reverse genetics has so far failed to reveal the function of Gac in B. burgdorferi, other experiments suggest that Gac is an architectural DNA-binding protein with HU activity. HU is essential for replicative transposition of bacteriophage Mu in E. coli: generating the Type I complex, an early intermediate, requires HU (Craigie et al., 1985). Gac was able to substitute for HU, albeit not as efficiently, in promoting formation of the Type I complex in an in vitro donor-cleavage reaction (Knight and Samuels, 1999). Notably, Gac was not able to substitute in the assay for IHF. In addition, Gac, expressed from its own promoter, was able to complement the defect in Mu growth, as assayed by efficiency of plating, in an E. coli HU mutant (Knight and Samuels, 1999). Like HU, IHF, and Hbb, Gac bends DNA, which was demonstrated by FRET measurements (Corbett et al., 2004). However, the mechanism by which Gac bends DNA is completely different than the other architectural DNA-binding proteins that involve a β-sheet in the minor groove. Instead, Gac has a novel structure, determined by Corbett et al. (2004) and termed a β-pinwheel, which is similar to a β-propeller; the DNA wraps around the perimeter of four of the six blades of the pinwheel, resulting in a profound bend. This structure also provided insight into the enzymatic mechanism of DNA gyrase (Corbett et al., 2004), as Gac, and the C-terminal domains of DNA gyrases from other bacteria, impart positive supercoiling on the wrapped DNA, which is converted to negative supercoiling following the strand-passage reaction (Ruthenburg et al., 2005; Huang et al., 2006; Kramlinger and Hiasa, 2006).
DNA supercoiling and cis-acting sequences
DNA supercoiling, which is known to regulate gene expression in other bacteria (López-García and Forterre, 2000; Cheung et al., 2003; Rui and Tse-Dinh, 2003; Peter et al., 2004), has been hypothesized to serve as a molecular thermometer for ospC expression (Alverson et al., 2003). There are several mechanisms by which DNA supercoiling affects gene expression, including the regulation of promoter recognition by RpoS. E. coli RpoS preferentially recognizes promoters on a relaxed template (Kusano et al., 1996) and DNA relaxation in E. coli is required for transcription by RNA polymerase holoenzyme containing RpoS during the cellular response to osmotic stress (Bordes et al., 2003). In fact, DNA supercoiling has been proposed to be a post-translational regulator of RpoS activity (Hengge-Aronis, 2002b; Bordes et al., 2003).
In B. burgdorferi, ospC gene is transcribed from an RpoS-dependent promoter (see the section above on the RpoN-RpoS alternative σ factor cascade), but transcription is repressed early during the infection because OspC elicits a potent immune response (Liang et al., 2002a; Liang et al., 2002b; Crother et al., 2004; Liang et al., 2004; Tilly et al., 2006). Two overlapping inverted repeats (IRs) are present upstream of the promoter (Margolis et al., 1994) that serve as an operator to mediate repression (Xu et al., 2007). The inverted repeats probably either bind a trans-acting repressor or alter secondary structure in response to changes in DNA supercoiling, or both. Surprisingly, mutating the inverted repeats has little effect on ospC transcription in trans from a shuttle vector (Eggers et al., 2004; Yang et al., 2005). Drecktrah et al. (2013) obtained different results using site-directed mutagenesis of the inverted repeats in cis on cp26: induction of ospC by DNA supercoiling, temperature, and pH required the distal inverted repeat, but not the proximal inverted repeat. In addition, the sequence of the IRs was less important for the regulation of ospC expression than the base-pairing potential of the two halves of the IR element (Drecktrah et al., 2013).
Cyclic-di-AMP
Cyclic-di-AMP is a recently identified second messenger in bacteria (Römling, 2008; Corrigan and Gründling, 2013; Commichau et al., 2019). It is synthesized from two molecules of ATP by diadenylate cyclase (DAC), and is hydrolyzed by phosphodiesterases (PDEs). Virtually all organisms identified to date that are endowed with c-di-AMP are gram-positive bacteria. B. burgdorferi is one of a few exceptions (Barker et al., 2013; Ye et al., 2014; Savage et al., 2015; Rubin et al., 2018). B. burgdorferi encodes a diadenylate cyclase, CdaA (BB0008) (Savage et al., 2015), and a phosphodiesterase, DhhP (BB0619) (Ye et al., 2014). In gram-positive bacteria, genes encoding c-di-AMP phosphodiesterase are often non-essential: mutants lacking phosphodiesterase were readily obtained and those mutants have been key to our understanding of c-di-AMP signaling (Römling, 2008; Corrigan and Gründling, 2013; Commichau et al., 2019). On the other hand, genes encoding c-di-AMP cyclase appear to be essential in most bacteria. In B. burgdorferi, the phenotypes of c-di-AMP mutants are unique. Inactivation of dhhP in B. burgdorferi could only be achieved in the presence of a plasmid encoded inducible dhhP gene (Ye et al., 2014). The conditional dhhP mutant failed to grow in the absence of the inducer. This mutant had a decreased, not increased, resistance to β-lactamase antibiotics, which also differs from gram-positive bacteria.
Wild-type B. burgdorferi cultured in vitro has very low levels of intracellular c-di-AMP (~9 nM), and overproduction of CdaA does not increase c-di-AMP levels (Ye et al., 2014; Savage et al., 2015), suggesting that CdaA may not be active under standard in vitro culture conditions. However, this may not be true as inactivation of dhhP resulted in over fiftyfold increase of intracellular c-di-AMP levels, suggesting that CdaA is active to produce c-di-AMP in cultured spirochetes. c-di-AMP appears to be involved in several cellular processes in other bacteria, including potassium transport, cell wall and membrane homeostasis, DNA damage repair, sporulation, and osmoregulation (Römling, 2008; Corrigan and Gründling, 2013; Commichau et al., 2019). In B. burgdorferi, the essential nature of the dhhP mutant strongly indicates that c-di-AMP plays an important role that is yet to be elucidated.
Transcriptomics
Transcriptomics, or genome-wide expression profiling, is a powerful tool that comparatively quantifies RNAs produced from the genome. Methods of transcriptome analyses are continuously evolving, but two of the most commonly used approaches to date are microarrays and RNA-sequencing (RNA-seq) technologies. Microarray-based studies quantify transcript levels of predetermined RNAs, while RNA-seq can identify all expressed RNAs. Microarray-based transcriptomic data from multiple B. burgdorferi studies was recently reviewed by Iyer et al. (2016). RNA-seq has more recently been utilized for differential gene expression analysis, and to globally identify transcription start sites (TSSs) and small RNAs (sRNAs).
The vast majority of transcriptomic studies have been performed on spirochetes cultivated in vitro under conditions attempting, perhaps not successfully, to mimic either the vertebrate host or tick vector (Table 1). Performing transcriptomics on spirochetes within infected ticks or vertebrate tissues is challenging due to the paucity of organisms present within these host environments. Using an enrichment-based microarray approach, Iyer et al. (2015) identified an in vivo transcriptome during acquisition (fed larvae) and transmission (fed nymphs), while Narasimhan et al. (2003) examined in vivo gene expression in the central nervous system (CNS) and heart tissues of non-human primates (NHP). Notably, the genome coverage is low in both the tick and NHP transcriptomes, perhaps because the transcriptome is indeed reduced in vivo compared to in vitro, or because of limitations of the microarray technology to detect low-level transcripts. In addition, differentially-expressed genes may have been missed due to the limited dynamic range of microarray technology. Several groups have taken advantage of a model developed by Akins et al. (1998) in which spirochetes are cultivated within dialysis membrane chambers (DMCs) implanted into the peritoneal cavity of a rat or rabbit to generated large numbers of spirochetes in a mammalian ‘host-adapted’ state. These ‘host-adapted’ organisms undergo many, but not all, of the transcriptomic and proteomic changes associated with vertebrate infection, including repression of tick-phase genes, such as ospA and the glp operon.
Table 1.
Transcriptome studies in B. burgdorferi.
| Experimental Condition | Strain | Platform | Reference |
|---|---|---|---|
| Increased temperature | B31 | Membrane microarray | Ojaimi et al., Infect. Immun., 2003 |
| Comparative analysis of clinical isolates | B31 | Membrane microarray | Ojaimi et al., Infect. Immun., 2005 |
| Temperature-shift in vitro vs. host adapted DMC | B31 | Glass slide microarray | Revel et al., PNAS, 2002 |
| In vitro vs. host adapted DMC | B31 | Membrane microarray | Brooks et al. Infect. Immun., 2003 |
| Host mimic: Influence of whole blood in vitro | B31 | Membrane microarray | Tokarz et al., Infect. Immun., 2004 |
| Monoclonal OspB antibody co-cultivation | B31 | Membrane microarray | Anderton et al., Infect. Immun., 2004 |
| Neurological cell co-cultivation | B31 | Affymetrix slide microarray | Livengood et al., Infect. Immun., 2008 |
| RpoS regulon (in vitro vs. DMC) | 297 | 70 m oligo glass slide microarray | Caimano et al., Mol. Microbiol., 2007 |
| RpoS regulon (in vitro vs. DMC) | B31, 297 | Illumina RNA sequencing | Caimano et al., Front Microbiol., 2019 |
| RpoN/RpoS regulon (in vitro) | B31 | 70 m oligo glass slide microarray | Fisher et al., PNAS, 2005 |
| BosR regulon (in vitro) | B31 | Membrane microarray | Hyde et al., Microbiology, 2006 |
| BosR regulon (in vitro) | B31 | 70 m oligo glass slide microarray | Ouyang et al., Mol. Microbiol., 2009 |
| Rrp2 regulon (in vilro) | B31 | 70 m oligo glass slide microarray | Boardman et al., Infect. Immun.,2008 |
| Rrp2/RpoN/RpoS regulon (in vitro) | B31 | 70 m oligo glass slide microarray | Ouyang et al., Microbiology, 2008 |
| CsrA, BadR, RpoN and RpoS | B31 | Illumina RNA sequencing | Arnold et al., PLOS One, 2018 |
| Rrp1 regulon (in vilro) | B31 | 70 m oligo glass slide microarray | Rogers et al. Mol. Microbiol., 2009 |
| Rrp1 regulon (in vilro) | B31 | 70 m oligo glass slide microarray | He et al., PLOS Pathog., 2011 |
| Rrp1 regulon (in vitro) | B31 | Illumina RNA sequencing | Caimano et al., Infect. Immun., 2015 |
| RelBbu regulon (in vitro) | 297 | 70 m oligo glass slide microarray | Bugrysheva et al., PLOS One, 2015 |
| BadR regulon (in vitro) | B31 | Nimblegen | Miller et al., Mol. Microbiol., 2013 |
| HrpA regulon (in vitro) | B31 | Nimblegen | Salman-Dilgimen et al., PLOS Pathog., 2013 |
| Non-human primate tissues | N40, JD1 | Glass slide microarray | Narasimhan et al., PNAS, 2003 |
| Fed nymphs | N40 | Glass slide microarray | Narasimhan et al., J. Bacteriol., 2002 |
| Infected murine tissues | B31 | 70 m oligo glass slide microarray | Pal et al., J. Infect. Dis., 2008 |
| Fed larvae and nymphs vs DMCs and in vitro | B31 | 70 m oligo glass slide microarray | Iyer et al., Mol. Microbiol., 2015 |
| GlcNAc starvation | B31 | Illumina RNA sequencing | Schneider et al., FEMS Microbiol Lett., 2018 |
| Global 5’ end identification | B31 | Illumina RNA sequencing | Adams et al., Nucleic Acid Res, 2016 |
| In vitro growth phases | B31 | Illumina RNA sequencing | Arnold et al., PLOS One, 2016 |
| Temperature response small RNA regulon | B31 | Illumina RNA sequencing | Popitsch et al., BMC genomics, 2016 |
| DNA methylation | B31 | Illumina RNA sequencing | Casselli et al., J. Bacteriol., 2018 |
| RelBbu regulon and starvation | B31 | Illumina RNA sequencing | Drecktrah et al., PLOS Pathogens, 2015 |
| RelBbu small RNA regulon | B31 | Illumina RNA sequencing | Drecktrah et al., Front. Cell. Infect. Microbiol., 2018 |
| Doxycycline treatment | B31 | Ion Torrent RNA sequencing | Caskey et al., Front Microbiol., 2019 |
| EbfC regulon | B31 | Illumina RNA sequencing | Jutras et al., J. Bacteriol., 2012 |
| Outer membrane vesicle transcriptome | B31 | Illumina RNA sequencing | Malge et al., FEMS Microbiol. Lett., 2018 |
| ittA regulon | B31 | Illumina RNA sequencing | Medina et al., PLOS Pathogens, 2020 |
| DksA and starvation regulon | B31 | Affymetrix slide microarray | Boyle et al., J. Bacteriol., 2019 |
Comparative transcriptome analyses using either microarray or RNA-seq have identified cohorts of genes regulated by specific environmental conditions as well as important regulatory networks in B. burgdorferi. Strikingly, there is often limited overlap between published data sets for seemingly identical strains (i.e., wild type) grown under ostensibly the same environmental condition(s). The lack of concordance between different data sets is likely due to a combination of biological and technical factors, including variations in growth media components, phase of growth at the time of harvest, methods used for RNA extraction, cDNA synthesis and library preparation, approach used to identify differentially expressed genes (i.e., microarray or RNA-seq), and data analyses. Population and cell-to-cell heterogeneity likely account for some of the differential gene expression observed in transcriptome studies, which is further confounded by the complex plasmid profiles of B. burgdorferi. Plasmid content and the repertoires of plasmid-encoded genes vary between strains. In addition, plasmids frequently are lost during in vitro cultivation. Several studies have compared gene expression profiles of different strains of B. burgdorferi, revealing strain-specific gene regulation (Ojaimi et al., 2005; Caimano et al., 2019).
Despite the lack of concordance among transcriptome studies in B. burgdorferi, several trends have emerged. Genes encoding outer surface proteins induced during transmission and vertebrate infection (OspC-like) often are induced by a temperature upshift, cultivation in DMCs and the RpoN/RpoS alternative σ factor cascade (Revel et al., 2002; Brooks et al., 2003; Ojaimi et al., 2003; Tokarz et al., 2004; Fisher et al., 2005; Hyde et al., 2006; Caimano et al., 2007; Boardman et al., 2008; Livengood et al., 2008; Ouyang et al., 2008; Ouyang et al., 2011; Caimano et al., 2019). Starvation, and the signaling molecules (p)ppGpp and c-di-GMP regulate genes encoding outer surface proteins, chemotactic proteins and transporters and enzymes required to utilize different carbon sources. Strikingly, differential expression of the glp operon in many of the transcriptome studies substantiate its role in the tick. This operon encodes three genes, glpF, glpK, and glpD, required for uptake and utilization of glycerol as an alternative carbon source in ticks; glp-deficient spirochetes survive at much lower levels during the tick blood meal and intermolt (He et al., 2011; Pappas et al., 2011). The glp operon or specific genes in the operon are upregulated by tick-associated regulators Rrp1, RelABbu, and DksA as well as starvation, while it is repressed by a vertebrate phase-specific environmental cues (temperature upshift, recovery from starvation, co-cultivation with neuroglial cells, and cultivation within DMCs) along with vertebrate phase regulators BosR, BadR and CsrA (Revel et al., 2002; Fisher et al., 2005; Hyde et al., 2006; Livengood et al., 2008; Ouyang et al., 2009b; Rogers et al., 2009; He et al., 2011; Miller et al., 2013; Ouyang et al., 2014b; Bugrysheva et al., 2015; Caimano et al., 2015; Drecktrah et al., 2015; Iyer et al., 2015; Ouyang and Zhou, 2015; Iyer and Schwartz, 2016; Schneider and Rhodes, 2018; Boyle et al., 2019).
Temperature response
Growth at different temperatures is often employed to mimic the environmental change spirochetes undergo as B. burgdorferi transitions between the tick vector and vertebrate host (23°C and 34–37°C, respectively). Revel et al. (2002) altered both the pH and temperature of in vitro B. burgdorferi cultures to simulate the unfed and fed tick environments (23°C/pH 7.5 and 37°C/pH 6.8, respectively). Ojaimi et al. (2003) and Iyer et al. (2015) also compared differential gene expression of cells grown at 23°C and 35°C, but did not alter the pH. To simulate nymphal transmission, Tokarz et al. (2004) compared the transcriptomes of spirochetes grown at 35°C in BSK II medium with and without the addition of 6% human blood.
Starvation response
In order to persist in the tick midgut in between blood meals, B. burgdorferi must adapt to and survive nutrient deprivation. Three transcriptomic analyses of B. burgdorferi cultured in nutrient-limited media provide insight into gene expression during spirochete persistence (Drecktrah et al., 2015; Schneider and Rhodes, 2018; Boyle et al., 2019). Drecktrah et al. (2015) shifted B. burgdorferi cultures growing in BSK II with rabbit serum (BSK II + RS) at stationary phase to RPMI without rabbit serum for 6 hours to represent nutrient limitation and starvation conditions in the tick. RPMI lacks many of the nutrients present in BSK II including neopeptone, yeastolate, N-acetylglucosamine and bovine serum albumin. To mimic the influx of nutrients that accompanies an incoming blood meal during transmission, cultures were subsequently shifted back to BSK II + RS for 2 hours. Boyle et al. (2019) also examined differential gene expression in response to starvation in RPMI media using microarray analyses instead of RNA-seq.
N-acetylglucosamine (GlcNAc) starvation also has been used to imitate tick overwintering in vitro (Schneider and Rhodes, 2018). GlcNAc is required for cell wall synthesis and, once phosphorylated, the sugar can be shuttled into the glycolytic pathway. B. burgdorferi is not able to synthesize GlcNAc from glucose but acquires it from the breakdown of chitobiose, a major component of tick cuticle, via the phosphoenolpyruvate-phosphotransferase systems (PEP-PTS) (Tilly et al., 2001; Corona and Schwartz, 2015; Caimano et al., 2016; Troy et al., 2016). Schneider and Rhodes (2018) used RNA-seq to compare the gene expression profiles of wild-type B. burgdorferi cultivated in the presence or absence of GlcNAc.
In vivo tick transcriptomics
As previously discussed, in vivo transcriptomic analyses are challenging due to the low numbers of spirochetes that can be recovered from the tick vector or infected host tissues. To date, the only genome-wide transcriptomic study of B. burgdorferi within ticks compared fed larvae (acquisition) and fed nymphs (transmission). Using an RNA enrichment pre-amplification step prior to microarray analysis, Iyer et al. (2015) detected ~500 transcripts in the fed nymphs and larvae. The transcriptomes of B. burgdorferi in both tick life stages were also compared to B. burgdorferi grown in vitro following temperature-shift or cultivated within DMCs. Hierarchal clustering revealed that the expression profile of temperature-shifted B. burgdorferi was more similar to that of fed larvae than fed nymphs, although the in vivo transcriptomes were very different than the in vitro transcriptome.
Host-adapted transcriptomics
Several studies also sought to demonstrate the transcriptomic expression profile of mammalian host-adapted organisms. Narasimhan et al. (2003) examined in vivo gene expression in the central nervous system (CNS) and heart tissues of non-human primates (NHP), which included a comparison between steroid treated and immunocompromised NHPs. An enrichment technique to selectively amplify specific bacterial transcriptomes was used to enhance detection of spirochetal mRNA in NHP tissues. While the detectable transcriptome was limited (~500 transcripts), the authors identified over 150 differentially expressed genes in the heart compared to CNS tissue. To date, this is the only in vivo transcriptome analysis performed using infected vertebrate tissues. However, multiple groups have attempted to recapitulate the tissue environment by cultivating spirochetes within DMCs or by co-cultivating spirochetes with human neuroglial cells (Revel et al., 2002; Brooks et al., 2003; Tokarz et al., 2004; Caimano et al., 2007; Livengood et al., 2008; Iyer et al., 2015; Caimano et al., 2019). Three microarray analyses have directly compared the transcriptomes of host-adapted B. burgdorferi within DMCs compared to in vitro cultured spirochetes at 37°C. These studies have demonstrated that gene expression varies substantially between the two growth environments (Revel et al., 2002; Brooks et al., 2003; Iyer et al., 2015). Livengood et al. (2008) identified genes related to spirochete-host interactions by comparing the transcriptomes of B. burgdorferi co-cultivated with human neuroglial cells to those cultured in cell-free medium.
Regulators of gene expression
The RpoN/RpoS alternative σ factor cascade (see section above) is activated within ticks during the nymphal blood meal resulting in a sea change of gene expression to allow for transmission and vertebrate infection. RpoS, an alternate σ factor, induces expression of genes required for transmission and maintenance of B. burgdorferi in the vertebrate host and represses many tick phase-specific genes. Transcription of rpoS is induced during the nymphal blood meal by RpoN, as part of a complex that also includes Rrp2 and the Fur family regulator BosR (see sections above on the RpoN/RpoS alternative σ factor cascade and the Transcription factors BosR and BadR).
The BosR regulon has been examined by microarray by two groups (Hyde et al., 2006; Ouyang et al., 2009b). Hyde et al. (2006) compared the transcriptomes of a bosR point mutant sensitive to oxidative stress and corresponding complemented strain. Ouyang et al. (2009b) analyzed the BosR-dependent transcriptome by comparing the transcriptomes of a wild-type strain and bosR null mutant.
Fisher et al. (2005) was first to define the RpoN- and RpoS-dependent transcriptome. Their comparative transcriptomics revealed a set of genes regulated through the RpoN/RpoS alternative σ factor cascade as well as subsets of genes that are uniquely regulated by either RpoN or RpoS alone. Ouyang et al. (2008) examined the Rrp2-, RpoN- and RpoS-dependent transcriptomes utilizing microarray analyses. They also identified a large number of genes coordinately regulated by all three factors suggesting a hierarchical coordination of Rrp2, RpoN and RpoS. In contrast to the Fisher et al. (2005) study, however, Ouyang et al. (2008) found only a limited number of genes regulated by RpoN or RpoS alone. In the latter study, the Rrp2 regulon contained >100 genes that were differentially expressed outside of the RpoN-RpoS pathway. Another Rrp2-dependent regulon was identified by Boardman et al. (2008).
Caimano et al. (2007) initially described the RpoS regulon from B. burgdorferi strain 297 grown in vitro at 37°C and in DMCs, by comparing the transcriptomes of wild type and a rpoS mutant via microarray analysis. The in vitro and DMC RpoS regulons overlapped significantly with many genes being induced by RpoS under both growth conditions. However, they also identified a large cohort of genes that were repressed by RpoS only in DMC, demonstrating that mammalian host-specific signals are necessary for RpoS repression. More recently, Caimano et al. (2019) compared the RpoS regulons of two phylogenetically distinct B. burgdorferi strains, 297 and B31, with different origins (human and tick, respectively): both in vitro temperature-shifted, and DMC-cultivated spirochetes were used for transcriptome analyses. These studies identified a core set of genes that were differentially regulated in both strains in vitro and/or in DMCs as well as RpoS-regulated genes that were unique to each strain. Notably, the core set of genes was primarily encoded on the chromosome, while the strain-specific genes were predominately plasmid-borne. Importantly, RpoS-mediated repression of tick phase-specific genes in both strains occurred only in response to mammalian host-specific signals.
Arnold et al. (2018) employed RNA-seq to examine the RpoN and RpoS regulons and, unexpectedly, detected only one differentially expressed transcript in the rpoS mutant, and two in the rpoN mutant compared to the wild type. The spirochetes were cultured under conditions that do not induce synthesis of RpoS and therefore levels of the alternate σ factor were assumed to be low in the wild-type strain. This study underscores the impact of growth phase and culture conditions on comparative transcriptome analyses.
The two-component system Hk1/Rrp1 appears to function primarily in ticks during transmission and acquisition (see section above on the Hk1/Rrp1 two-component system and c-di-GMP). In fact, B. burgdorferi lacking either Hk1 or Rrp1 do not survive within tick midguts during the blood meal (Caimano et al., 2011; He et al., 2011; Kostick et al., 2011; Caimano et al., 2015). The response regulator Rrp1 is a diguanylate cyclase that synthesizes c-di-GMP, a small signaling molecule that regulates many cellular processes in bacteria (Cotter and Stibitz, 2007; Wolfe and Visick, 2008; Hengge, 2009; Römling et al., 2013; Valentini and Filloux, 2019). Three groups have examined the Rrp1 regulon using both microarray and RNA-seq (Rogers et al., 2009; He et al., 2011; Caimano et al., 2015). Rogers et al. (2009) first identified the Rrp1 regulon using microarray technology and a deletion mutant of rrp1 in a non-infectious B. burgdorferi strain (B31 5A13). He et al. (2011) identified Rrp1-dependent differentially expressed transcripts also using microarray technology but in an infectious wild-type strain. The Rrp1 regulon was also examined by RNA-seq (Caimano et al., 2015).
B. burgdorferi also utilizes the stringent response mechanism to sense and adapt to nutrient limitation by adjusting gene expression that modifies their growth and physiology (Potrykus and Cashel, 2008; Dalebroux and Swanson, 2012; Gaca et al., 2015; Hauryliuk et al., 2015; Liu et al., 2015; Steinchen and Bange, 2016; Gourse et al., 2018) (see section above on the Stringent response and (p)ppGpp). The stringent response is mediated by levels of the alarmones, guanosine pentaphosphate and guanosine tetraphosphate, abbreviated (p)ppGpp. In B. burgdorferi, (p)ppGpp levels are controlled by RelBbu, a bifunctional synthetase/hydrolase containing domains homologous to RelA and SpoT (Concepcion and Nelson, 2003; Bugrysheva et al., 2005; Drecktrah et al., 2015). RelA typically synthesizes (p)ppGpp in response to amino acid starvation, while SpoT induces accumulation of (p)ppGpp in response to limiting fatty acids, phosphate, carbon, or iron. The RelBbu regulon has been characterized by RNA sequencing and microarray analyses (Bugrysheva et al., 2015; Drecktrah et al., 2015). Bugrysheva et al. (2015) used microarray analyses to characterize the RelBbu transcriptome from in vitro cultivated B. burgdorferi at exponential and stationary growth phase. Drecktrah et al. (2015) used RNA sequencing analyses to describe the RelBbu regulon at stationary growth phase, starvation conditions and recovery from starvation.
DksA works in conjunction with (p)ppGpp to affect RNA polymerase activity in E. coli (Gourse et al., 2018) (see section above on the Stringent response and (p)ppGpp). Boyle et al. (2019) identified the DksA regulon at mid-logarithmic growth phase and under starvation conditions using microarray analyses in B. burgdorferi. The dksA mutant has a significant increase in (p)ppGpp at both mid-logarithmic and starvation cultivation conditions, which thwarted an attempt to delineate the role of dksA changes in gene expression, independent of (p)ppGpp concentration.
Borrelia host adapted regulator (BadR) is a DNA-binding protein that is necessary for B. burgdorferi to colonize mice (Miller et al., 2013; Ouyang and Zhou, 2015) (see section above on the Transcription factors BosR and BadR). BadR binds the DNA upstream of both the rpoS and bosR genes (Miller et al., 2013; Ouyang and Zhou, 2015). Two transcriptomic studies have examined the BadR regulon. Miller et al. (2013) utilized microarray analyses, while Arnold et al. (2018) performed RNA-seq to compare wild-type and mutant badR strains.
The function of the carbon storage regulator A (csrA) homolog in B. burgdorferi remains unknown and there are contradictory reports as to its influence on the RpoS-RpoN σ factor cascade (Sanjuan et al., 2009; Karna et al., 2011; Sze and Li, 2011; Sze et al., 2011; Karna et al., 2013; Ouyang et al., 2014b; Lin et al., 2018) (see section above on Small regulatory RNAs and RNA chaperones). In other organisms, CsrA is an RNA-binding protein that regulates carbon storage, virulence factors, and other cellular processes (Romeo, 1998; Majdalani et al., 2005; Lucchetti-Miganeh et al., 2008; Vakulskas et al., 2015; Hör et al., 2018; Romeo and Babitzke, 2018). Arnold et al. (2018) utilized RNA-seq to assay the CsrA regulon and compared it to the BadR regulon revealing a cohort of genes regulated by both factors.
Differential gene expression on plasmids
Several plasmids appear to be hotbeds of differential gene expression. Genes on lp54 have been implicated in transmission and borrelial survival in the vertebrate host. Strikingly, many genes on lp54 are reported as differentially expressed in nearly all transcriptome studies. Many genes on lp28–2 are upregulated by the RpoN/RpoS alternative σ factor cascade and downregulated by RelABbu in response to starvation (Caimano et al., 2007; Boardman et al., 2008; Ouyang et al., 2008; Ouyang et al., 2009b; Drecktrah et al., 2015; Caimano et al., 2019). These data indicate that induction of genes on lp28–2 may be important for transmission and vertebrate survival or repression of these genes may be needed for adaptation to the tick vector. However, lp28–2 is not necessary for the murine infection via needle inoculation (Purser and Norris, 2000). The cp32 family of circular plasmids also harbor many genes that are differentially expressed under several environmental conditions and by several regulators, including the RpoN/RpoS alternative σ factor cascade, RelABbu and Rrp1 (Caimano et al., 2005; Caimano et al., 2007; Boardman et al., 2008; Ouyang et al., 2008; Ouyang et al., 2009b; Rogers et al., 2009; He et al., 2011; Drecktrah et al., 2015; Caimano et al., 2019). Genes on lp36 are downregulated by RpoS in the DMC and are primarily upregulated by RelABbu, suggesting phase-specific expression may be important as spirochetes traverse their enzootic cycle (Caimano et al., 2007; Drecktrah et al., 2015; Caimano et al., 2019). Jewett et al. (2007) reported that the lp36 plasmid is not required for survival in the tick but does play a role in mammalian infectivity. As discussed above, the complex plasmid profiles of B. burgdorferi pose a significant challenge to identifying bona fide differentially expressed genes harbored on plasmids. In comparative transcriptomic analyses, if all genes on a plasmid are either upregulated or downregulated, then the cause may be plasmid loss in one of the strains.
Identification of sRNAs
The advent of RNA-seq allowed researchers to globally identify sRNAs in B. burgdorferi (Arnold et al., 2016; Popitsch et al., 2017) (see section above on Small regulatory RNAs and RNA chaperones). Popitsch et al. (2017) size-selected for sRNAs 50–500 nucleotides in length, and sequenced the sRNA transcriptome from B. burgdorferi cultivated in vitro at 23°C and after a temperature shift to 37°C, identifying 1005 sRNAs. Arnold et al. (2016) sequenced total RNA from three phases of growth in vitro at 37°C and found putative sRNAs from short predicted transcriptional units (<425 nucleotide in length) in non-coding regions of the genome, or antisense to annotated protein coding genes. The Arnold et al. (2016) data set identified 351 sRNAs, but the type of RNA-seq and subsequent analyses do not allow for the identification of sRNAs that overlap an annotated open reading frame. As such, the data set excludes intragenic sRNAs and 5′ UTR-associated sRNAs. In addition, the size selection was performed in silico and included predicted transcriptional units. If the intragenic sRNAs and 5′ UTR-associated sRNAs are excluded, then the Popitsch et al. (2017) data set identifies 550 intergenic sRNAs and antisense sRNAs, similar to the findings of Arnold et al. (2016). Further comparison and overlap of the two data sets is difficult due to differences in the sRNA annotations and reference annotations for the genes. In addition, the spirochetes were cultivated in different media and at different growth phases, as well as the two studies used different cDNA library types, construction, and analyses.
The RelBbu-dependent sRNA transcriptome revealed that the stringent response regulates the expression of a large number of sRNAs (Drecktrah et al., 2018). Approximately one third of the annotated sRNAs, sans the intragenic sRNA class, were differentially expressed during starvation of in vitro cultivated B. burgdorferi. 187 sRNAs are upregulated by RelBbu, while 54 are downregulated. The non-coding RNA (ncRNA) regulons of CsrA and BadR were also identified using the predicted ncRNAs annotations from Arnold et al. (2016). Antisense RNAs associated with glpF and rpoN were differentially regulated by both regulators.
Identification of transcriptional start sites (TSSs) and 5′ ends
5′ end-specific RNA-sequencing (5′ RNA-seq) can globally identify the 5′ ends and transcriptional start sites (TSSs) of transcripts. This technique exploits some of the biochemical properties of transcription (Adams et al., 2017), namely that the first nucleotide transcribed in an RNA contains a triphosphate at the 5′ end, while processed or degraded 5′ ends of RNAs contain a monophosphate or a hydroxyl group. Adams et al. (2017) used 5′ RNA-seq on in vitro cultivated B. burgdorferi and identified 6042 TSSs. 63% of the TSSs were termed ‘internal’, denoting that the promoter and 5′ end is within an annotated gene in the same orientation, while 13% were antisense TSSs with the 5′ ends of RNAs encoded opposite to annotated genes. 22% of the TSSs were within 300 nucleotides of the start codon of an annotated open reading frame. Arnold et al. (2016) predicted 5′ ends of transcripts by analyzing total RNA sequencing data from B. burgdorferi cultivated in vitro at three different growth phases. A custom script was used to determine putative transcriptional units and their transcript boundaries. 600 to 800 putative 5′ ends were reported in each of the growth phases examined. The predicted 5′ ends identified could be associated with TSSs or processed and/or degraded RNAs.
Future perspectives
Transcriptomic studies have significantly contributed toward understanding global gene regulation in B. burgdorferi. The accumulated data from studies performed over the last twenty years are laudable, but many challenges still remain to more accurately understand global gene expression during the enzootic cycle of B. burgdorferi. More robust in vivo transcriptome analyses with higher genomic coverage in both the tick victor and the vertebrate host would paint a more accurate big picture of the changes in gene expression. Specifically, tick tissue-specific (salivary gland and midguts) and mouse tissue-specific (joint, heart, bladder, and others) transcriptomes will shed light on gene expression patterns necessary for B. burgdorferi to transmit and persist in the tick and vertebrate. RNA-sequencing technologies continue to evolve with increased sensitivity and coverage yields from significantly less input RNA. Single-cell RNA-seq is an advancing technology that will uncover cell-to-cell heterogeneity in populations of spirochetes that may determine infection and persistence outcomes. Dual RNA-seq analyzes host or vector transcriptomes simultaneously with the bacterial transcriptome to reveal correlations between bacterial and host-specific responses in gene expression. RNA-seq can be coupled to other biochemical approaches to identify RNA targets of RNA-binding proteins (via CLIP-seq, RIP-seq, RIL-seq, and CLASH and Grad-seq), novel ribo-regulators (via Term-seq) and functional antisense RNAs (via dsRNA-seq) (Lybecker et al., 2014b; Lybecker and Samuels, 2017; Saliba et al., 2017). Using and adapting these ‘-seq’ methodologies to in vitro and in vivo models of infection should rapidly advance our understanding of global gene expression patterns in the Lyme disease spirochete.
Concluding remarks
Gene expression is regulated in B. burgdorferi and related species as the spirochetes traverse their natural enzootic cycles, which requires acquisition by a naïve tick from an infected vertebrate, adaptation to and persistence in the tick, transmission from an infected tick to a naïve host, and adaptation to and survival in the vertebrate. Many of the molecular mechanisms controlling gene expression as the spirochete transitions between and adapts to these phases of the enzootic cycle have been experimentally uncovered and genetically dissected, albeit the majority of investigative approaches are restricted to cultured organisms or an emulation of the genuine environments B. burgdorferi experiences. The spirochete employs three regulatory systems to control gene expression, which include a sequential cascade of a pair of alternative σ factors, a couple of two-component systems, and, at the most recent count, three purine second messengers. A trio of transcription factors and three sRNAs are known to activate or fine-tune gene expression at the transcriptional and post-transcriptional levels. High-throughput transcriptomic approaches have identified the genes and defined the phase-specific regulons that are activated or repressed by environmental signals, although their precise roles have yet to be fully elucidated. Transcriptomic studies have revealed a surprising swath of previously unrecognized small RNAs, the vast majority of which have no known function, but are likely responsible for fine-tuning the expression of specific genes throughout the enzootic cycle of the spirochete. Many of the gene products controlled by the RpoN-RpoS σ factor cascade and the Hk1-Rrp1 two-component system function to move the spirochete between the vector and host or to facilitate survival by altering the metabolism of the spirochete or its surface that interfaces directly with the vector or host; however, many of these regulated genes have no homology to genes outside of the deeply branching spirochete phylum (or even genus), so their physiological functions remain a mystery. B. burgdorferi is renowned for doing things its own way, but, curiously, the spirochete wields the canonical bacterial response to nutrient deprivation, the stringent response, to survive in the tick between blood meals, which entails shifting the expression of genes encoding various metabolic functions, carbohydrate utilization and surface lipoproteins. Our hope is that this review has shed light into the current state-of-the-art and built a conceptual framework for continuing studies on gene regulation and transcriptomics in the Lyme disease spirochetes.
Acknowledgements
This review is a reboot of the Gene Regulation, Transcriptomics, and Proteomics chapter written by Jon Skare, Jay Carroll, Frank Yang, Scott Samuels, and Darrin Akins (Skare et al., 2010). DSS thanks JDR for his passion, mentorship and forbearance… and apologizes for the agita.
Research in our laboratories on gene regulation and transcriptomics is supported by Public Health Service grants R01 AI051486 to DSS, R01 AI130247 to DSS, DD, and ML, R21 AI133334 to DD and DSS, R03 AI146909 to ZO, R01 AI083640 to XFY, R21 AI128379 to MJC, R01 AI029735 to MJC, R21 AI126146 to MJC, R21 AI139940 to MJC, R01 AI144126 to BS, R21 AI147139 to BS, and R21 AI139956 to BS as well as Connecticut Children’s Medical Center to MJC.
References
- Adams PP, Flores Avile C, Popitsch N, Bilusic I, Schroeder R, Lybecker M, and Jewett MW (2017). In vivo expression technology and 5′ end mapping of the Borrelia burgdorferi transcriptome identify novel RNAs expressed during mammalian infection. Nucleic Acids Res 45, 775–792. 10.1093/nar/gkw1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PP, and Storz G (2020). Prevalence of small base-pairing RNAs derived from diverse genomic loci. Biochim Biophys Acta Gene Regul Mech 1863, 194524. 10.1016/j.bbagrm.2020.194524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agapov A, Olina A, Esyunina D, and Kulbachinskiy A (2017). Gfh factors and NusA cooperate to stimulate transcriptional pausing and termination. FEBS Lett 591, 946–953. 10.1002/1873-3468.12609 [DOI] [PubMed] [Google Scholar]
- Akins DR, Bourell KW, Caimano MJ, Norgard MV, and Radolf JD (1998). A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Invest 101, 2240–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alban PS, Johnson PW, and Nelson DR (2000). Serum-starvation-induced changes in protein synthesis and morphology of Borrelia burgdorferi. Microbiology 146, 119–127. [DOI] [PubMed] [Google Scholar]
- Alverson J, Bundle SF, Sohaskey CD, Lybecker MC, and Samuels DS (2003). Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi. Mol Microbiol 48, 1665–1677. 10.1046/j.1365-2958.2003.03537.x [DOI] [PubMed] [Google Scholar]
- Amikam D, and Galperin MY (2006). PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22, 3–6. 10.1093/bioinformatics/bti739 [DOI] [PubMed] [Google Scholar]
- Anacker ML, Drecktrah D, LeCoultre RD, Lybecker M, and Samuels DS (2018). RNase III processing of rRNA in the Lyme disease spirochete Borrelia burgdorferi. J Bacteriol 200, e00035–18. 10.1128/JB.00035-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault L, Borchert JS, Bergeron J, Snow S, and Schlax PJ (2013). Measurements of mRNA degradation in Borrelia burgdorferi. J Bacteriol 195, 4879–4887. 10.1128/JB.00659-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold WK, Savage CR, Brissette CA, Seshu J, Livny J, and Stevenson B (2016). RNA-seq of Borrelia burgdorferi in multiple phases of growth reveals insights into the dynamics of gene expression, transcriptome architecture, and noncoding RNAs. PLoS One 11, e0164165. 10.1371/journal.pone.0164165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold WK, Savage CR, Lethbridge KG, Smith TC 2nd, Brissette CA, Seshu J, and Stevenson B (2018). Transcriptomic insights on the virulence-controlling CsrA, BadR, RpoN, and RpoS regulatory networks in the Lyme disease spirochete. PLoS One 13, e0203286. 10.1371/journal.pone.0203286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb K, Bykowski T, Riley SP, Miller MC, Demoll E, and Stevenson B (2006). Borrelia burgdorferi EbfC, a novel, chromosomally encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete’s resident cp32 prophages. J Bacteriol 188, 4331–4339. 10.1128/JB.00005-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG (1984). Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57, 521–525. [PMC free article] [PubMed] [Google Scholar]
- Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, and Valdivia RH (2013). STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio 4, e00018–13. 10.1128/mBio.00018-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, and Breaker RR (2005). 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11, 774–784. 10.1261/rna.7286705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer WJ, Luthra A, Zhu G, Radolf JD, Malkowski MG, and Caimano MJ (2015). Structural characterization and modeling of the Borrelia burgdorferi hybrid histidine kinase Hk1 periplasmic sensor: a system for sensing small molecules associated with tick feeding. J Struct Biol 192, 48–58. 10.1016/j.jsb.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer DH (2011). Bacillus subtilis mRNA decay: new parts in the toolkit. Wiley Interdiscip Rev RNA 2, 387–394. 10.1002/wrna.66 [DOI] [PubMed] [Google Scholar]
- Bechhofer DH, and Deutscher MP (2019). Bacterial ribonucleases and their roles in RNA metabolism. Crit Rev Biochem Mol Biol 54, 242–300. 10.1080/10409238.2019.1651816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker G, and Hengge-Aronis R (2001). What makes an Escherichia coli promoter σS dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of σS. Mol Microbiol 39, 1153–1165. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Oeemig JS, Kolodziejczyk R, Meri T, Kajander T, Lehtinen MJ, Iwai H, Jokiranta TS, and Goldman A (2013). Structural basis for complement evasion by Lyme disease pathogen Borrelia burgdorferi. J Biol Chem 288, 18685–18695. 10.1074/jbc.M113.459040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins JS, Xu H, He M, Norgard MV, Reitzer L, and Yang XF (2009). Rrp2, a σ54-dependent transcriptional activator of Borrelia burgdorferi, activates rpoS in an enhancer-independent manner. J Bacteriol 191, 2902–2905. 10.1128/JB.01721-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman BK, He M, Ouyang Z, Xu H, Pang X, and Yang XF (2008). Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun 76, 3844–3853. 10.1128/IAI.00467-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontemps-Gallo S, Lawrence K, and Gherardini FC (2016). Two different virulence-related regulatory pathways in Borrelia burgdorferi are directly affected by osmotic fluxes in the blood meal of feeding Ixodes ticks. PLoS Pathog 12, e1005791. 10.1371/journal.ppat.1005791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbely G, and Schneider GJ (1988). Cyanobacterial DNA-dependent RNA polymerase. Methods Enzymol 167, 592–599. 10.1016/0076-6879(88)67067-4 [DOI] [Google Scholar]
- Bordes P, Conter A, Morales V, Bouvier J, Kolb A, and Gutierrez C (2003). DNA supercoiling contributes to disconnect σS accumulation from σS-dependent transcription in Escherichia coli. Mol Microbiol 48, 561–571. [DOI] [PubMed] [Google Scholar]
- Boylan JA, Hummel CS, Benoit S, Garcia-Lara J, Treglown-Downey J, Crane EJ III, and Gherardini FC (2006). Borrelia burgdorferi bb0728 encodes a coenzyme A disulphide reductase whose function suggests a role in intracellular redox and the oxidative stress response. Mol Microbiol 59, 475–486. 10.1111/j.1365-2958.2005.04963.x [DOI] [PubMed] [Google Scholar]
- Boylan JA, Posey JE, and Gherardini FC (2003). Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc Natl Acad Sci USA 100, 11684–11689. 10.1073/pnas.2032956100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WK, Groshong AM, Drecktrah D, Boylan JA, Gherardini FC, Blevins JS, Samuels DS, and Bourret TJ (2019). DksA controls the response of the Lyme disease spirochete Borrelia burgdorferi to starvation. J Bacteriol 201, e00582–18. 10.1128/JB.00582-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WK, Hall LS, Armstrong AA, Dulebohn DP, Samuels DS, Gherardini FC, and Bourret TJ (2020). Establishment of an in vitro RNA polymerase transcription system: a new tool to study transcriptional activation in Borrelia burgdorferi. Sci Rep 10, 8246. 10.1038/s41598-020-65104-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RG, and Link TM (2007). Hfq structure, function and ligand binding. Curr Opin Microbiol 10, 125–133. 10.1016/j.mib.2007.03.015 [DOI] [PubMed] [Google Scholar]
- Brisson D, Zhou W, Jutras BL, Casjens S, and Stevenson B (2013). Distribution of cp32 prophages among Lyme disease-causing spirochetes and natural diversity of their lipoprotein-encoding erp loci. Appl Environ Microbiol 79, 4115–4128. 10.1128/AEM.00817-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CS, Hefty PS, Jolliff SE, and Akins DR (2003). Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun 71, 3371–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brorson Ø, and Brorson S-H (1997). Transformation of cystic forms of Borrelia burgdorferi to normal, mobile spirochetes. Infection 25, 240–246. [DOI] [PubMed] [Google Scholar]
- Brorson Ø, and Brorson S-H (1998). A rapid method for generating cystic forms of Borrelia burgdorferi, and their reversal to mobile spirochetes. APMIS 106, 1131–41. [DOI] [PubMed] [Google Scholar]
- Brown L, Gentry D, Elliott T, and Cashel M (2002). DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol 184, 4455–4465. 10.1128/jb.184.16.4455-4465.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva JV, Bryksin AV, Godfrey HP, and Cabello FC (2005). Borrelia burgdorferi rel is responsible for generation of guanosine-3′-diphosphate-5′-triphosphate and growth control. Infect Immun 73, 4972–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva JV, Pappas CJ, Terekhova DA, Iyer R, Godfrey HP, Schwartz I, and Cabello FC (2015). Characterization of the RelBbu regulon in Borrelia burgdorferi reveals modulation of glycerol metabolism by (p)ppGpp. PLoS One 10, e0118063. 10.1371/journal.pone.0118063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burenina OY, Elkina DA, Hartmann RK, Oretskaya TS, and Kubareva EA (2015). Small noncoding 6S RNAs of bacteria. Biochemistry (Mosc) 80, 1429–1446. 10.1134/S0006297915110048 [DOI] [PubMed] [Google Scholar]
- Burke TP, and Portnoy DA (2016). SpoVG Is a conserved RNA-binding protein that regulates Listeria monocytogenes lysozyme resistance, virulence, and swarming motility. mBio 7, e00240. 10.1128/mBio.00240-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns LH, Adams CA, Riley SP, Jutras BL, Bowman A, Chenail AM, Cooley AE, Haselhorst LA, Moore AM, Babb K, et al. (2010). BpaB, a novel protein encoded by the Lyme disease spirochete’s cp32 prophages, binds to erp operator 2 DNA. Nucleic Acids Res 38, 5443–5455. 10.1093/nar/gkq284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, Hoover TR, and Gherardini FC (2007). Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol Microbiol 65, 277–293. 10.1111/j.1365-2958.2007.05813.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M, and Dixon R (2012). The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev 76, 497–529. 10.1128/MMBR.00006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher J, Sarvan S, Brunzelle JS, Couture J-F, and Stintzi A (2012). Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci USA 109, 10047–10052. 10.1073/pnas.1118321109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Drecktrah D, Kung F, and Samuels DS (2016). Interaction of the Lyme disease spirochete with its tick vector. Cell Microbiol 18, 919–927. 10.1111/cmi.12609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Dunham-Ems S, Allard AM, Cassera MB, Kenedy M, and Radolf JD (2015). Cyclic di-GMP modulates gene expression in Lyme disease spirochetes at the tick-mammal interface to promote spirochete survival during the blood meal and tick-to-mammal transmission. Infect Immun 83, 3043–3060. 10.1128/IAI.00315-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Gonzalez CA, and Radolf JD (2005). Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J Bacteriol 187, 7845–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Hazlett KRO, and Radolf JD (2004). RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect Immun 72, 6433–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Groshong AM, Belperron A, Mao J, Hawley KL, Luthra A, Graham DE, Earnhart CG, Marconi RT, Bockenstedt LK, et al. (2019). The RpoS gatekeeper in Borrelia burgdorferi: an invariant regulatory scheme that promotes spirochete persistence in reservoir hosts and niche diversity. Front Microbiol 10, 1923. 10.3389/fmicb.2019.01923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, and Radolf JD (2007). Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol 65, 1193–1217. 10.1111/j.1365-2958.2007.05860.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Kenedy MR, Kairu T, Desrosiers DC, Harman M, Dunham-Ems S, Akins DR, Pal U, and Radolf JD (2011). The hybrid histidine kinase Hk1 is part of a two-component system that is essential for survival of Borrelia burgdorferi in feeding Ixodes scapularis ticks. Infect Immun 79, 3117–3130. 10.1128/IAI.05136-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldelari I, Chao Y, Romby P, and Vogel J (2013). RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb Perspect Med 3, a010298. 10.1101/cshperspect.a010298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter BM, Whitmire JM, and Merrell DS (2009). This is not your mother’s repressor: the complex role of Fur in pathogenesis. Infect Immun 77, 2590–2601. 10.1128/IAI.00116-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JA, Garon CF, and Schwan TG (1999). Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect Immun 67, 3181–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, et al. (2000). A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35, 490–516. [DOI] [PubMed] [Google Scholar]
- Casjens S, van Vugt R, Tilly K, Rosa PA, and Stevenson B (1997). Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol 179, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh AT, Chandrangsu P, and Wassarman KM (2010). 6S RNA regulation of relA alters ppGpp levels in early stationary phase. Microbiology 156, 3791–3800. 10.1099/mic.0.043992-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh AT, Klocko AD, Liu X, and Wassarman KM (2008). Promoter specificity for 6S RNA regulation of transcription is determined by core promoter sequences and competition for region 4.2 of σ70. Mol Microbiol 67, 1242–1256. 10.1111/j.1365-2958.2008.06117.x [DOI] [PubMed] [Google Scholar]
- Cavanagh AT, Sperger JM, and Wassarman KM (2012). Regulation of 6S RNA by pRNA synthesis is required for efficient recovery from stationary phase in E. coli and B. subtilis. Nucleic Acids Res 40, 2234–2246. 10.1093/nar/gkr1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh AT, and Wassarman KM (2013). 6S-1 RNA function leads to a delay in sporulation in Bacillus subtilis. J Bacteriol 195, 2079–2086. 10.1128/JB.00050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh AT, and Wassarman KM (2014). 6S RNA, a global regulator of transcription in Escherichia coli, Bacillus subtilis, and beyond. Annu Rev Microbiol 68, 45–60. 10.1146/annurev-micro-092611-150135 [DOI] [PubMed] [Google Scholar]
- Chaconas G (2005). Hairpin telomeres and genome plasticity in Borrelia: all mixed up in the end. Mol Microbiol 58, 625–635. 10.1111/j.1365-2958.2005.04872.x [DOI] [PubMed] [Google Scholar]
- Chaconas G, Castellanos M, and Verhey TB (2020). Changing of the guard: How the Lyme disease spirochete subverts the host immune response. J Biol Chem 295, 301–313. 10.1074/jbc.REV119.008583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wassarman KM, Feng S, Leon K, Feklistov A, Winkelman JT, Li Z, Walz T, Campbell EA, and Darst SA (2017). 6S RNA mimics B-form DNA to regulate Escherichia coli RNA polymerase. Mol Cell 68, 388–397. 10.1016/j.molcel.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Itzek A, Malke H, Ferretti JJ, and Kreth J (2013). Multiple roles of RNase Y in Streptococcus pyogenes mRNA processing and degradation. J Bacteriol 195, 2585–2594. 10.1128/JB.00097-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenail AM, Jutras BL, Adams CA, Burns LH, Bowman A, Verma A, and Stevenson B (2012). Borrelia burgdorferi cp32 BpaB modulates expression of the prophage NucP nuclease and SsbP single-stranded DNA-binding protein. J Bacteriol 194, 4570–4578. 10.1128/JB.00661-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KJ, Badarinarayana V, Selinger DW, Janse D, and Church GM (2003). A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res 13, 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SM, and Schellhorn HE (2010). Evolution of the RpoS regulon: origin of RpoS and the conservation of RpoS-dependent regulation in bacteria. J Mol Evol 70, 557–571. 10.1007/s00239-010-9352-0 [DOI] [PubMed] [Google Scholar]
- Chu C-Y, Stewart PE, Bestor A, Hansen B, Lin T, Gao L, Norris SJ, and Rosa PA (2016). Function of the Borrelia burgdorferi FtsH homolog is essential for viability both in vitro and in vivo and independent of HflK/C. mBio 7, e00404–16. 10.1128/mBio.00404-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Nolder CL, Hughes JL, Nowalk AJ, and Carroll JA (2006). Regulation and expression of bba66 encoding an immunogenic infection-associated lipoprotein in Borrelia burgdorferi. Mol Microbiol 61, 243–258. 10.1111/j.1365-2958.2006.05224.x [DOI] [PubMed] [Google Scholar]
- Colland F, Barth M, Hengge-Aronis R, and Kolb A (2000). σ factor selectivity of Escherichia coli RNA polymerase: role for CRP, IHF and Lrp transcription factors. EMBO J 19, 3028–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Heidemann JL, Ficner R, and Stülke J (2019). Making and breaking of an essential poison: the cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di-AMP in bacteria. J Bacteriol 201, e00462–18. 10.1128/JB.00462-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion MB, and Nelson DR (2003). Expression of spoT in Borrelia burgdorferi during serum starvation. J Bacteriol 185, 444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley AE, Riley SP, Kral K, Miller MC, DeMoll E, Fried MG, and Stevenson B (2009). DNA-binding by Haemophilus influenzae and Escherichia coli YbaB, members of a widely-distributed bacterial protein family. BMC Microbiol 9, 137. 10.1186/1471-2180-9-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, and Berger JM (2004). Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct 33, 95–118. [DOI] [PubMed] [Google Scholar]
- Corbett KD, Shultzaberger RK, and Berger JM (2004). The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc Natl Acad Sci USA 101, 7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona A, and Schwartz I (2015). Borrelia burgdorferi: carbon metabolism and the tick-mammal enzootic cycle. Microbiol Spectr 3, MBP-0011–2014. 10.1128/microbiolspec.MBP-0011-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, and Gründling A (2013). Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11, 513–524. 10.1038/nrmicro3069 [DOI] [PubMed] [Google Scholar]
- Cotter PA, and Stibitz S (2007). c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol 10, 17–23. 10.1016/j.mib.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Craigie R, Arndt-Jovin DJ, and Mizuuchi K (1985). A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci USA 82, 7570–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crother TR, Champion CI, Whitelegge JP, Aguilera R, Wu X-Y, Blanco DR, Miller JN, and Lovett MA (2004). Temporal analysis of the antigenic composition of Borrelia burgdorferi during infection in rabbit skin. Infect Immun 72, 5063–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom KM, and O’Toole GA (2017). A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu Rev Microbiol 71, 179–195. 10.1146/annurev-micro-090816-093325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, and Swanson MS (2012). ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10, 203–212. 10.1038/nrmicro2720 [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, and Swanson MS (2010). Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol 76, 200–219. 10.1111/j.1365-2958.2010.07094.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva AM, Telford SR 3rd, Brunet LR, Barthold SW, and Fikrig E (1996). Borrelia burgdorferi OspA is an arthropod-specific transmission-blocking Lyme disease vaccine. J Exp Med 183, 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian C, Vitale S, Leonard GA, Bahlawane C, Fauquant C, Leduc D, Muller C, de Reuse H, Michaud-Soret I, and Terradot L (2011). The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol Microbiol 79, 1260–1275. 10.1111/j.1365-2958.2010.07517.x [DOI] [PubMed] [Google Scholar]
- Drecktrah D, Hall LS, Brinkworth AJ, Comstock JR, Wassarman KM, and Samuels DS (2020). Characterization of 6S RNA in the Lyme disease spirochete. Mol Microbiol 113, 399–417. 10.1111/mmi.14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Hall LS, Hoon-Hanks LL, and Samuels DS (2013). An inverted repeat in the ospC operator is required for induction in Borrelia burgdorferi. PLoS One 8, e68799. 10.1371/journal.pone.0068799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Hall LS, Rescheneder P, Lybecker M, and Samuels DS (2018). The stringent response-regulated sRNA transcriptome of Borrelia burgdorferi. Front Cell Infect Microbiol 8, 231. 10.3389/fcimb.2018.00231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Lybecker M, Popitsch N, Rescheneder P, Hall LS, and Samuels DS (2015). The Borrelia burgdorferi RelA/SpoT homolog and stringent response regulate survival in the tick vector and global gene expression during starvation. PLoS Pathog 11, e1005160. 10.1371/journal.ppat.1005160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K, and Rouviere-Yaniv J (1987). Histonelike proteins of bacteria. Microbiol Rev 51, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulebohn DP, Hayes BM, and Rosa PA (2014). Global repression of host-associated genes of the Lyme disease spirochete through post-transcriptional modulation of the alternative sigma factor RpoS. PLoS One 9, e93141. 10.1371/journal.pone.0093141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulebohn DP, Richards CL, Su H, Lawrence KA, and Gherardini FC (2017). Weak organic acids decrease Borrelia burgdorferi cytoplasmic pH, eliciting an acid stress response and impacting RpoN- and RpoS-dependent gene expression. Front Microbiol 8, 1734. 10.3389/fmicb.2017.01734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Ems SM, Caimano MJ, Eggers CH, and Radolf JD (2012). Borrelia burgdorferi requires the alternative sigma factor RpoS for dissemination within the vector during tick-to-mammal transmission. PLoS Pathog 8, e1002532. 10.1371/journal.ppat.1002532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Ems SM, Caimano MJ, Pal U, Wolgemuth CW, Eggers CH, Balic A, and Radolf JD (2009). Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J Clin Invest 119, 3652–3665. 10.1172/JCI39401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Gilet L, and Condon C (2012). The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet 8, e1003181. 10.1371/journal.pgen.1003181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, and Radolf JD (2002). Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for expression of fluorescent reporters in the Lyme disease spirochaete. Mol Microbiol 43, 281–296. [DOI] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Malizia RA, Kariu T, Cusack B, Desrosiers DC, Hazlett KRO, Claiborne A, Pal U, and Radolf JD (2011). The coenzyme A disulphide reductase of Borrelia burgdorferi is important for rapid growth throughout the enzootic cycle and essential for infection of the mammalian host. Mol Microbiol 82, 679–697. 10.1111/j.1365-2958.2011.07845.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, and Radolf JD (2004). Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J Bacteriol 186, 7390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, and Radolf JD (2006). Sigma factor selectivity in Borrelia burgdorferi: RpoS recognition of the ospE/ospF/elp promoters is dependent on the sequence of the −10 region. Mol Microbiol 59, 1859–1875. [DOI] [PubMed] [Google Scholar]
- Eggers CH, and Samuels DS (1999). Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J Bacteriol 181, 7308–7313. 10.1128/JB.181.23.7308-7313.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher SP, Friedlander G, Livny J, Margalit H, and Shuman HA (2010). Legionella pneumophila 6S RNA optimizes intracellular multiplication. Proc Natl Acad Sci USA 107, 7533–7538. 10.1073/pnas.0911764107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillat MF (2014). The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 546, 41–52. 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- Fischer JR, LeBlanc KT, and Leong JM (2006). Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect Immun 74, 435–441. 10.1128/IAI.74.1.435-441.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JR, Parveen N, Magoun L, and Leong JM (2003). Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci USA 100, 7307–7312. 10.1073/pnas.1231043100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, and Gherardini FC (2005). Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci USA 102, 5162–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. (1997). Genomic sequence of a Lyme disease spirochete, Borrelia burgdorferi. Nature 390, 580–586. [DOI] [PubMed] [Google Scholar]
- Freedman JC, Rogers EA, Kostick JL, Zhang H, Iyer R, Schwartz I, and Marconi RT (2010). Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol Med Microbiol 58, 285–294. 10.1111/j.1574-695X.2009.00635.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DI (1988). Integration host factor: a protein for all reasons. Cell 55, 545–554. [DOI] [PubMed] [Google Scholar]
- Fröhlich KS, and Vogel J (2009). Activation of gene expression by small RNA. Curr Opin Microbiol 12, 674–682. 10.1016/j.mib.2009.09.009 [DOI] [PubMed] [Google Scholar]
- Gaal T, Ross W, Estrem ST, Nguyen LH, Burgess RR, and Gourse RL (2001). Promoter recognition and discrimination by EσS RNA polymerase. Mol Microbiol 42, 939–954. [DOI] [PubMed] [Google Scholar]
- Gaca AO, Colomer-Winter C, and Lemos JA (2015). Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol, 1146–1156. 10.1128/JB.02577-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY (2004). Bacterial signal transduction network in a genomic perspective. Environ Microbiol 6, 552–567. 10.1111/j.1462-2920.2004.00633.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Nikolskaya AN, and Koonin EV (2001). Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203, 11–21. 10.1111/j.1574-6968.2001.tb10814.x [DOI] [PubMed] [Google Scholar]
- Gautam A, Hathaway M, McClain N, Ramesh G, and Ramamoorthy R (2008). Analysis of the determinants of bba64 (P35) gene expression in Borrelia burgdorferi using a gfp reporter. Microbiology 154, 275–285. 10.1099/mic.0.2007/011676-0 [DOI] [PubMed] [Google Scholar]
- Gherardini F, Boylan J, Lawrence K, and Skare J (2010). Metabolism and physiology of Borrelia. In Borrelia: Molecular Biology, Host Interaction and Pathogenesis, Samuels DS, and Radolf JD, eds. (Norfolk, UK: Caister Academic Press; ), pp. 103–138. [Google Scholar]
- Gilmore RD Jr., Howison RR, Schmit VL, Nowalk AJ, Clifton DR, Nolder C, Hughes JL, and Carroll JA (2007). Temporal expression analysis of the Borrelia burgdorferi paralogous gene family 54 genes BBA64, BBA65, and BBA66 during persistent infection in mice. Infect Immun 75, 2753–2764. 10.1128/IAI.00037-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Plaza JJ (2020). Small RNAs as fundamental players in the transference of information during bacterial infectious diseases. Front Mol Biosci 7, 101. 10.3389/fmolb.2020.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S (2004). The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol 58, 303–328. [DOI] [PubMed] [Google Scholar]
- Gourse RL, Chen AY, Gopalkrishnan S, Sanchez-Vazquez P, Myers A, and Ross W (2018). Transcriptional responses to ppGpp and DksA. Annu Rev Microbiol 72, 163–184. 10.1146/annurev-micro-090817-062444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graebsch A, Roche S, Kostrewa D, Söding J, and Niessing D (2010). Of bits and bugs–on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure. PLoS One 5, e13402. 10.1371/journal.pone.0013402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GA (2006). The ACT domain: a small molecule binding domain and its role as a common regulatory element. J Biol Chem 281, 33825–33829. 10.1074/jbc.R600024200 [DOI] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, Schwan TG, Policastro PF, Elias AF, and Rosa PA (2004). Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc Natl Acad Sci USA 101, 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groshong AM, and Blevins JS (2014). Insights into the biology of Borrelia burgdorferi gained through the application of molecular genetics. Adv Appl Microbiol 86, 41–143. 10.1016/B978-0-12-800262-9.00002-0 [DOI] [PubMed] [Google Scholar]
- Groshong AM, Dey A, Bezsonova I, Caimano MJ, and Radolf JD (2017). Peptide uptake is essential for Borrelia burgdorferi viability and involves structural and regulatory complexity of its oligopeptide transporter. mBio 8, e02047–17. 10.1128/mBio.02047-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groshong AM, Gibbons NE, Yang XF, and Blevins JS (2012). Rrp2, a prokaryotic enhancer-like binding protein, is essential for viability of Borrelia burgdorferi. J Bacteriol 194, 3336–3342. 10.1128/JB.00253-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove AP, Liveris D, Iyer R, Petzke M, Rudman J, Caimano MJ, Radolf JD, and Schwartz I (2017). Two distinct mechanisms govern RpoS-mediated repression of tick-phase genes during mammalian host adaptation by Borrelia burgdorferi, the Lyme disease spirochete. mBio 8, e01204–17. 10.1128/mBio.01204-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LS, Simonich CA, Brinkworth AJ, Lybecker MC, Drecktrah D, and Samuels DS (2021). A novel RpoN-independent rpoS transcript in Borrelia burgdorferi. Manuscript in preparation.
- Hantke K (1981). Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet 182, 288–292. 10.1007/BF00269672 [DOI] [PubMed] [Google Scholar]
- Hantke K (2001). Iron and metal regulation in bacteria. Curr Opin Microbiol 4, 172–177. 10.1016/s1369-5274(00)00184-3 [DOI] [PubMed] [Google Scholar]
- Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, and Gerdes K (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13, 298–309. 10.1038/nrmicro3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Oman T, Xu H, Blevins J, Norgard MV, and Yang XF (2008). Abrogation of ospAB constitutively activates the Rrp2-RpoN-RpoS pathway (sigmaN-sigmaS cascade) in Borrelia burgdorferi. Mol Microbiol 70, 1453–1464. 10.1111/j.1365-2958.2008.06491.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Ouyang Z, Troxell B, Xu H, Moh A, Piesman J, Norgard MV, Gomelsky M, and Yang XF (2011). Cyclic di-GMP is essential for the survival of the Lyme disease spirochete in ticks. PLoS Pathog 7, e1002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Zhang J-J, Ye M, Lou Y, and Yang XF (2014). Cyclic di-GMP receptor PlzA controls virulence gene expression through RpoS in Borrelia burgdorferi. Infect Immun 82, 445–452. 10.1128/IAI.01238-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD (2014). Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J Biol Chem 289, 28112–28120. 10.1074/jbc.R114.587071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R (2009). Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7, 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R (2002a). Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev 66, 373–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R (2002b). Stationary phase gene regulation: what makes an Escherichia coli promoter σS-selective? Curr Opin Microbiol 5, 591–595. [DOI] [PubMed] [Google Scholar]
- Hör J, Gorski SA, and Vogel J (2018). Bacterial RNA biology on a genome scale. Mol Cell 70, 785–799. 10.1016/j.molcel.2017.12.023 [DOI] [PubMed] [Google Scholar]
- Huang Y-Y, Deng J-Y, Gu J, Zhang Z-P, Maxwell A, Bi L-J, Chen Y-Y, Zhou Y-F, Yu Z-N, and Zhang X-E (2006). The key DNA-binding residues in the C-terminal domain of Mycobacterium tuberculosis DNA gyrase A subunit (GyrA). Nucleic Acids Res 34, 5650–5659. 10.1093/nar/gkl695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner A, Yang X, Nolen DM, Popova TG, Cabello FC, and Norgard MV (2001). Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci USA 98, 12724–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Seshu J, and Skare JT (2006). Transcriptional profiling of Borrelia burgdorferi containing a unique bosR allele identifies a putative oxidative stress regulon. Microbiology 152, 2599–2609. 10.1099/mic.0.28996-0 [DOI] [PubMed] [Google Scholar]
- Hyde JA, Shaw DK, Smith R III, Trzeciakowski JP, and Skare JT (2009). The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both homeostatic and pathogenic properties of the Lyme disease spirochete. Mol Microbiol 74, 1344–1355. 10.1111/j.1365-2958.2009.06951.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Shaw DK, Smith R III, Trzeciakowski JP, and Skare JT (2010). Characterization of a conditional bosR mutant in Borrelia burgdorferi. Infect Immun 78, 265–274. 10.1128/IAI.01018-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Trzeciakowski JP, and Skare JT (2007). Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J Bacteriol 189, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Caimano MJ, Luthra A, Axline D Jr., Corona A, Iacobas DA, Radolf JD, and Schwartz I (2015). Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Mol Microbiol 95, 509–538. 10.1111/mmi.12882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, and Schwartz I (2016). Microarray-based comparative genomic and transcriptome analysis of Borrelia burgdorferi. Microarrays 5, 9. 10.3390/microarrays5020009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquamet L, Traoré DAK, Ferrer J-L, Proux O, Testemale D, Hazemann J-L, Nazarenko E, El Ghazouani A, Caux-Thang C, Duarte V, et al. (2009). Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol Microbiol 73, 20–31. 10.1111/j.1365-2958.2009.06753.x [DOI] [PubMed] [Google Scholar]
- Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, VanRaden M, Gherardini F, and Rosa PA (2007). The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol Microbiol 64, 1358–1374. 10.1111/j.1365-2958.2007.05746.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Bowman A, Brissette CA, Adams CA, Verma A, Chenail AM, and Stevenson B (2012a). EbfC (YbaB) is a new type of bacterial nucleoid-associated protein and a global regulator of gene expression in the Lyme disease spirochete. J Bacteriol 194, 3395–3406. 10.1128/JB.00252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Chenail AM, Carroll DW, Miller MC, Zhu H, Bowman A, and Stevenson B (2013a). Bpur, the Lyme disease spirochete’s PUR domain protein: identification as a transcriptional modulator and characterization of nucleic acid interactions. J Biol Chem 288, 26220–26234. 10.1074/jbc.M113.491357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Chenail AM, Rowland CL, Carroll D, Miller MC, Bykowski T, and Stevenson B (2013b). Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins. PLoS One 8, e66683. 10.1371/journal.pone.0066683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Chenail AM, and Stevenson B (2013c). Changes in bacterial growth rate govern expression of the Borrelia burgdorferi OspC and Erp infection-associated surface proteins. J Bacteriol 195, 757–764. 10.1128/JB.01956-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Jones GS, Verma A, Brown NA, Antonicello AD, Chenail AM, and Stevenson B (2013d). Posttranscriptional self-regulation by the Lyme disease bacterium’s BpuR DNA/RNA-binding protein. J Bacteriol 195, 4915–4923. 10.1128/JB.00819-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Savage CR, Arnold WK, Lethbridge KG, Carroll DW, Tilly K, Bestor A, Zhu H, Seshu J, Zückert WR, et al. (2019). The Lyme disease spirochete’s BpuR DNA/RNA-binding protein is differentially expressed during the mammal-tick infectious cycle, which affects translation of the SodA superoxide dismutase. Mol Microbiol 112, 973–991. 10.1111/mmi.14336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Verma A, Adams CA, Brissette CA, Burns LH, Whetstine CR, Bowman A, Chenail AM, Zückert WR, and Stevenson B (2012b). BpaB and EbfC DNA-binding proteins regulate production of the Lyme disease spirochete’s infection-associated Erp surface proteins. J Bacteriol 194, 778–786. 10.1128/JB.06394-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karna SLR, Prabhu RG, Lin Y-H, Miller CL, and Seshu J (2013). Contributions of environmental signals and conserved residues to the functions of carbon storage regulator A of Borrelia burgdorferi. Infect Immun 81, 2972–2985. 10.1128/IAI.00494-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karna SLR, Sanjuan E, Esteve-Gassent MD, Miller CL, Maruskova M, and Seshu J (2011). CsrA modulates levels of lipoproteins and key regulators of gene expression critical for pathogenic mechanisms of Borrelia burgdorferi. Infect Immun 79, 732–744. 10.1128/IAI.00882-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona LI (2015). The Fur homologue BosR requires Arg39 to activate rpoS transcription in Borrelia burgdorferi and thereby direct spirochaete infection in mice. Microbiology 161, 2243–2255. 10.1099/mic.0.000166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona LI, Tokarz R, Kuhlow CJ, Benach J, and Benach JL (2004). The Fur homologue in Borrelia burgdorferi. J Bacteriol 186, 6443–6456. 10.1128/JB.186.19.6443-6456.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazanov MD, Li X, Gelfand MS, Osterman AL, and Rodionov DA (2013). Functional diversification of ROK-family transcriptional regulators of sugar catabolism in the Thermotogae phylum. Nucleic Acids Res 41, 790–803. 10.1093/nar/gks1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak MJ, Wiedmann M, and Boor KJ (2005). Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol Rev 69, 527–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Akiyama Y, and Ito K (1997). Host regulation of lysogenic decision in bacteriophage λ: transmembrane modulation of FtsH (HflB), the cII degrading protease, by HflKC (HflA). Proc Natl Acad Sci USA 94, 5544–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Kimmel BJ, Eggers CH, and Samuels DS (2000). Disruption of the Borrelia burgdorferi gac gene, encoding the naturally synthesized GyrA C-terminal domain. J Bacteriol 182, 2048–2051. 10.1128/jb.182.7.2048-2051.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, and Samuels DS (1999). Natural synthesis of a DNA-binding protein from the C-terminal domain of DNA gyrase A in Borrelia burgdorferi. EMBO J 18, 4875–4881. 10.1093/emboj/18.17.4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobryn K, Naigamwalla DZ, and Chaconas G (2000). Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol Microbiol 37, 145–155. [DOI] [PubMed] [Google Scholar]
- Kostick JL, Szkotnicki LT, Rogers EA, Bocci P, Raffaelli N, and Marconi RT (2011). The diguanylate cyclase, Rrp1, regulates critical steps in the enzootic cycle of the Lyme disease spirochetes. Mol Microbiol 81, 219–231. 10.1111/j.1365-2958.2011.07687.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostick-Dunn JL, Izac JR, Freedman JC, Szkotnicki LT, Oliver LD Jr., and Marconi RT (2018). The Borrelia burgdorferi c-di-GMP binding receptors, PlzA and PlzB, are functionally distinct. Front Cell Infect Microbiol 8, 213. 10.3389/fcimb.2018.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, and Stevenson B (2013). Complement regulator-acquiring surface proteins of Borrelia burgdorferi: Structure, function and regulation of gene expression. Ticks Tick Borne Dis 4, 26–34. 10.1016/j.ttbdis.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramlinger VM, and Hiasa H (2006). The “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J Biol Chem 281, 3738–3742. 10.1074/jbc.M511160200 [DOI] [PubMed] [Google Scholar]
- Kung F, Anguita J, and Pal U (2013). Borrelia burgdorferi and tick proteins supporting pathogen persistence in the vector. Future Microbiol 8, 41–56. 10.2217/fmb.12.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela J, Fredman J, Salminen TA, and Tyystjärvi T (2020). Revealing secrets of the enigmatic omega subunit of bacterial RNA polymerase. Mol Microbiol. 10.1111/mmi.14603 [DOI] [PubMed] [Google Scholar]
- Kusano S, Ding Q, Fujita N, and Ishihama A (1996). Promoter selectivity of Escherichia coli RNA polymerase Eσ70 and Eσ38 holoenzymes: effect of DNA supercoiling. J Biol Chem 271, 1998–2004. [DOI] [PubMed] [Google Scholar]
- Lane RS, Piesman J, and Burgdorfer W (1991). Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu Rev Entomol 36, 587–609. [DOI] [PubMed] [Google Scholar]
- Lasa I, Toledo-Arana A, Dobin A, Villanueva M, Ruiz de los Mozos I, Vergara-Irigaray M, Segura V, Fagegaltier D, Penadés JR, Valle J, et al. (2011). Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci USA 108, 20172–20177. 10.1073/pnas.1113521108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I, Toledo-Arana A, and Gingeras TR (2012). An effort to make sense of antisense transcription in bacteria. RNA Biol 9, 1039–1044. 10.4161/rna.21167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rhun A, Beer YY, Reimegård J, Chylinski K, and Charpentier E (2016). RNA sequencing uncovers antisense RNAs and novel small RNAs in Streptococcus pyogenes. RNA Biol 13, 177–195. 10.1080/15476286.2015.1110674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, and Breaker RR (2010). An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329, 845–848. 10.1126/science.1190713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Gatewood ML, and Jones GH (2013). RNase III is required for actinomycin production in Streptomyces antibioticus. Appl Environ Microbiol 79, 6447–6451. 10.1128/AEM.02272-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, and Stülke J (2011). RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol 81, 1459–1473. 10.1111/j.1365-2958.2011.07777.x [DOI] [PubMed] [Google Scholar]
- Lejars M, Kobayashi A, and Hajnsdorf E (2019). Physiological roles of antisense RNAs in prokaryotes. Biochimie 164, 3–16. 10.1016/j.biochi.2019.04.015 [DOI] [PubMed] [Google Scholar]
- Liang FT, Jacobs MB, Bowers LC, and Philipp MT (2002a). An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J Exp Med 195, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FT, Nelson FK, and Fikrig E (2002b). Molecular adaptation of Borrelia burgdorferi in the murine host. J Exp Med 196, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FT, Yan J, Mbow ML, Sviat SL, Gilmore RD, Mamula M, and Fikrig E (2004). Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect Immun 72, 5759–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K, Tempczyk A, Parsons JF, Bonander N, Toedt J, Kelman Z, Howard A, Eisenstein E, and Herzberg O (2003). Crystal structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function coexpressed with the recombinational DNA repair protein RecR. Proteins 50, 375–379. 10.1002/prot.10297 [DOI] [PubMed] [Google Scholar]
- Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, Chaconas G, Philipp MT, and Norris SJ (2012). Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PLoS One 7, e47532. 10.1371/journal.pone.0047532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Chen Y, Smith TC II, Karna SLR, and Seshu J (2018). Short-chain fatty acids alter metabolic and virulence attributes of Borrelia burgdorferi. Infect Immun 86, e00217–18. 10.1128/IAI.00217-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioliou E, Sharma CM, Caldelari I, Helfer A-C, Fechter P, Vandenesch F, Vogel J, and Romby P (2012). Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet 8, e1002782. 10.1371/journal.pgen.1002782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Bittner AN, and Wang JD (2015). Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol 24, 72–79. 10.1016/j.mib.2015.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Xu H, Zhang Y, Yang J, Du J, Zhou Y, Yang XF, and Lou Y (2020). Role of Hk2 in the enzootic cycle of Borrelia burgdorferi. Front Med 7, 652. 10.3389/fmed.2020.573648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livengood JA, Schmit VL, and Gilmore RD Jr. (2008). Global transcriptome analysis of Borrelia burgdorferi during association with human neuroglial cells. Infect Immun 76, 298–307. 10.1128/IAI.00866-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, and Forterre P (2000). DNA topology and the thermal stress response, a tale from mesophiles and hyperthermophiles. Bioessays 22, 738–746. [DOI] [PubMed] [Google Scholar]
- Lucchetti-Miganeh C, Burrowes E, Baysse C, and Ermel G (2008). The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 154, 16–29. 10.1099/mic.0.2007/012286-0 [DOI] [PubMed] [Google Scholar]
- Lybecker M, Bilusic I, and Raghavan R (2014a). Pervasive transcription: detecting functional RNAs in bacteria. Transcription 5, e944039. 10.4161/21541272.2014.944039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker M, Zimmermann B, Bilusic I, Tukhtubaeva N, and Schroeder R (2014b). The double-stranded transcriptome of Escherichia coli. Proc Natl Acad Sci USA 111, 3134–3139. 10.1073/pnas.1315974111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, Abel CA, Feig AL, and Samuels DS (2010). Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 78, 622–635. 10.1111/j.1365-2958.2010.07374.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, and Samuels DS (2007). Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol 64, 1075–1089. 10.1111/j.1365-2958.2007.05716.x [DOI] [PubMed] [Google Scholar]
- Lybecker MC, and Samuels DS (2017). Small RNAs of Borrelia burgdorferi: characterizing functional regulators in a sea of sRNAs. Yale J Biol Med 90, 317–323. [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Vanderpool CK, and Gottesman S (2005). Bacterial small RNA regulators. Crit Rev Biochem Mol Biol 40, 93–113. [DOI] [PubMed] [Google Scholar]
- Mallory KL, Miller DP, Oliver LD Jr., Freedman JC, Kostick-Dunn JL, Carlyon JA, Marion JD, Bell JK, and Marconi RT (2016). Cyclic-di-GMP binding induces structural rearrangements in the PlzA and PlzC proteins of the Lyme disease and relapsing fever spirochetes: a possible switch mechanism for c-di-GMP-mediated effector functions. Pathog Dis 74, ftw105. 10.1093/femspd/ftw105 [DOI] [PubMed] [Google Scholar]
- Marchetti M, Malinowska A, Heller I, and Wuite GJL (2017). How to switch the motor on: RNA polymerase initiation steps at the single-molecule level. Protein Sci 26, 1303–1313. 10.1002/pro.3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis N, Hogan D, Tilly K, and Rosa PA (1994). Plasmid location of Borrelia purine biosynthesis gene homologs. J Bacteriol 176, 6427–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marincola G, Schäfer T, Behler J, Bernhardt J, Ohlsen K, Goerke C, and Wolz C (2012). RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol Microbiol 85, 817–832. 10.1111/j.1365-2958.2012.08144.x [DOI] [PubMed] [Google Scholar]
- Maruskova M, Esteve-Gassent MD, Sexton VL, and Seshu J (2008). Role of the bbA64 locus of Borrelia burgdorferi in early stages of infectivity in a murine model of Lyme disease. Infect Immun 76, 391–402. 10.1128/IAI.01118-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason C, Liu X, Prabhudeva S, and Ouyang Z (2019). The CXXC motifs are essential for the function of BosR in Borrelia burgdorferi. Front Cell Infect Microbiol 9, 109. 10.3389/fcimb.2019.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason C, Thompson C, and Ouyang Z (2020). DksA plays an essential role in regulating the virulence of Borrelia burgdorferi. Mol Microbiol 114, 172–183. 10.1111/mmi.14504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, and Chatterji D (2006). The evolving story of the omega subunit of bacterial RNA polymerase. Trends Microbiol 14, 450–455. 10.1016/j.tim.2006.08.002 [DOI] [PubMed] [Google Scholar]
- McCown PJ, Corbino KA, Stav S, Sherlock ME, and Breaker RR (2017). Riboswitch diversity and distribution. RNA 23, 995–1011. 10.1261/rna.061234.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Pérez DN, Wager B, Troy E, Gao L, Norris SJ, Lin T, Hu L, Hyde JA, Lybecker M, and Skare JT (2020). The intergenic small non-coding RNA ittA is required for optimal infectivity and tissue tropism in Borrelia burgdorferi. PLoS Pathog 16, e1008423. 10.1371/journal.ppat.1008423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano M, Ding Y, Wang X-G, Lu P, Coburn J, and Hu LT (2007). Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J Bacteriol 189, 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Karna SLR, and Seshu J (2013). Borrelia host adaptation Regulator (BadR) regulates rpoS to modulate host adaptation and virulence factors in Borrelia burgdorferi. Mol Microbiol 88, 105–124. 10.1111/mmi.12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, and Kushner SR (2016). Regulation of mRNA decay in bacteria. Annu Rev Microbiol 70, 25–44. 10.1146/annurev-micro-091014-104515 [DOI] [PubMed] [Google Scholar]
- Mouw KW, and Rice PA (2007). Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb. Mol Microbiol 63, 1319–1330. 10.1111/j.1365-2958.2007.05586.x [DOI] [PubMed] [Google Scholar]
- Mulay VB, Caimano MJ, Iyer R, Dunham-Ems S, Liveris D, Petzke MM, Schwartz I, and Radolf JD (2009). Borrelia burgdorferi bba74 is expressed exclusively during tick feeding and is regulated by both arthropod- and mammalian host-specific signals. J Bacteriol 191, 2783–2794. 10.1128/JB.01802-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader S, Pérard J, Carpentier P, Arnaud L, Crouzy S, and Michaud-Soret I (2019). New insights into the tetrameric family of the Fur metalloregulators. Biometals 32, 501–519. 10.1007/s10534-019-00201-8 [DOI] [PubMed] [Google Scholar]
- Narasimhan S, Caimano MJ, Liang FT, Santiago F, Laskowski M, Philipp MT, Pachner AR, Radolf JD, and Fikrig E (2003). Borrelia burgdorferi transcriptome in the central nervous system of non-human primates. Proc Natl Acad Sci USA 100, 15953–15958. 10.1073/pnas.2432412100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F, Waldminghaus T, and Chowdhury S (2006). RNA thermometers. FEMS Microbiol Rev 30, 3–16. [DOI] [PubMed] [Google Scholar]
- Nash HA (1996). The HU and IHF proteins: accessory factors for complex protein-DNA assemblies. In Regulation of Gene Expression in Escherichia coli, Lin ECC, and Lynch AS, eds. (Austin, Texas: R. G. Landes Co; ), pp. 149–179. [Google Scholar]
- Neusser T, Polen T, Geissen R, and Wagner R (2010). Depletion of the non-coding regulatory 6S RNA in E. coli causes a surprising reduction in the expression of the translation machinery. BMC Genomics 11, 165. 10.1186/1471-2164-11-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SJ (2014). vls antigenic variation systems of Lyme disease Borrelia: eluding host immunity through both random, segmental gene conversion and framework heterogeneity. Microbiol Spectr 2, MDNA3–0038–2014. 10.1128/microbiolspec.MDNA3-0038-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak EA, Sultan SZ, and Motaleb MA (2014). The cyclic-di-GMP signaling pathway in the Lyme disease spirochete, Borrelia burgdorferi. Front Cell Infect Microbiol 4, 56. 10.3389/fcimb.2014.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, et al. (2003). Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun 71, 1689–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C, Mulay V, Liveris D, Iyer R, and Schwartz I (2005). Comparative transcriptional profiling of Borrelia burgdorferi clinical isolates differing in capacities for hematogenous dissemination. Infect Immun 73, 6791–6802. 10.1128/IAI.73.10.6791-6802.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Österberg S, del Peso-Santos T, and Shingler V (2011). Regulation of alternative sigma factor use. Annu Rev Microbiol 65, 37–55. 10.1146/annurev.micro.112408.134219 [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Blevins JS, and Norgard MV (2008). Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology 154, 2641–2658. 10.1099/mic.0.2008/019992-0 [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Deka RK, and Norgard MV (2011). BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog 7, e1001272. 10.1371/journal.ppat.1001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, He M, Oman T, Yang XF, and Norgard MV (2009a). A manganese transporter, BB0219 (BmtA), is required for virulence by the Lyme disease spirochete, Borrelia burgdorferi. Proc Natl Acad Sci USA 106, 3449–3454. 10.1073/pnas.0812999106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, and Norgard MV (2009b). BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol Microbiol 74, 1331–1343. 10.1111/j.1365-2958.2009.06945.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Narasimhan S, Neelakanta G, Kumar M, Pal U, Fikrig E, and Norgard MV (2012). Activation of the RpoN-RpoS regulatory pathway during the enzootic life cycle of Borrelia burgdorferi. BMC Microbiol 12, 44. 10.1186/1471-2180-12-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, and Zhou J (2015). BadR (BB0693) controls growth phase-dependent induction of rpoS and bosR in Borrelia burgdorferi via recognizing TAAAATAT motifs. Mol Microbiol 98, 1147–1167. 10.1111/mmi.13206 [DOI] [PubMed] [Google Scholar]
- Ouyang Z, and Zhou J (2017). The putative Walker A and Walker B motifs of Rrp2 are required for the growth of Borrelia burgdorferi. Mol Microbiol 103, 86–98. 10.1111/mmi.13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Zhou J, Brautigam CA, Deka R, and Norgard MV (2014a). Identification of a core sequence for the binding of BosR to the rpoS promoter region in Borrelia burgdorferi. Microbiology 160, 851–862. 10.1099/mic.0.075655-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Zhou J, and Norgard MV (2014b). CsrA (BB0184) is not involved in activation of the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. Infect Immun 82, 1511–1522. 10.1128/IAI.01555-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Zhou J, and Norgard MV (2014c). Synthesis of RpoS is dependent on a putative enhancer binding protein Rrp2 in Borrelia burgdorferi. PLoS One 9, e96917. 10.1371/journal.pone.0096917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Zhou J, and Norgard MV (2016). Evidence that BosR (BB0647) Is a positive autoregulator in Borrelia burgdorferi. Infect Immun 84, 2566–2574. 10.1128/IAI.00297-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, Dai J, Li X, Neelakanta G, Luo P, Kumar M, Wang P, Yang X, Anderson JF, and Fikrig E (2008). A differential role for BB0365 in the persistence of Borrelia burgdorferi in mice and ticks. J Infect Dis 197, 148–155. 10.1086/523764 [DOI] [PubMed] [Google Scholar]
- Papenfort K, and Vanderpool CK (2015). Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev 39, 362–378. 10.1093/femsre/fuv016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas CJ, Iyer R, Petzke MM, Caimano MJ, Radolf JD, and Schwartz I (2011). Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle. PLoS Pathog 7, e1002102. 10.1371/journal.ppat.1002102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, and Grishin NV (2001). GGDEF domain is homologous to adenylyl cyclase. Proteins 42, 210–216. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, and Cozzarelli NR (2004). Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol 5, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pich A, and Bahl H (1991). Purification and characterization of the DNA-dependent RNA polymerase from Clostridium acetobutylicum. J Bacteriol 173, 2120–2124. 10.1128/jb.173.6.2120-2124.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinochet-Barros A, and Helmann JD (2018). Redox Sensing by Fe2+ in Bacterial Fur Family Metalloregulators. Antioxid Redox Signal 29, 1858–1871. 10.1089/ars.2017.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR, and Motaleb MA (2011). Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infect Immun 79, 1815–1825. 10.1128/IAI.00075-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popitsch N, Bilusic I, Rescheneder P, Schroeder R, and Lybecker M (2017). Temperature-dependent sRNA transcriptome of the Lyme disease spirochete. BMC Genomics 18, 28. 10.1186/s12864-016-3398-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen MM, Salgado PS, Koivunen MR, Wright S, Bamford DH, Stuart DI, and Grimes JM (2008). Structural explanation for the role of Mn2+ in the activity of ϕ6 RNA-dependent RNA polymerase. Nucleic Acids Res 36, 6633–6644. 10.1093/nar/gkn632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey JE, and Gherardini FC (2000). Lack of a role for iron in the Lyme disease pathogen. Science 288, 1651–1653. 10.1126/science.288.5471.1651 [DOI] [PubMed] [Google Scholar]
- Potrykus K, and Cashel M (2008). (p)ppGpp: still magical? Annu Rev Microbiol 62, 35–51. 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- Purser JE, and Norris SJ (2000). Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci USA 97, 13865–13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD, Caimano MJ, Stevenson B, and Hu LT (2012). Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10, 87–99. 10.1038/nrmicro2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD and Samuels DS (2021). Lyme Disease and Relapsing Fever Spirochetes: Genomics, Molecular Biology, Host Interactions and Disease Pathogenesis (Norfolk, UK: Caister Academic Press; ). 10.21775/9781913652616 [DOI] [Google Scholar]
- Reece RJ, and Maxwell A (1991). DNA gyrase: structure and function. Crit Rev Biochem Mol Biol 26, 335–375. [DOI] [PubMed] [Google Scholar]
- Reitzer L, and Schneider BL (2001). Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65, 422–444. 10.1128/MMBR.65.3.422-444.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Sang Y, Qin R, Cui Z, and Yao Y-F (2017). 6S RNA is involved in acid resistance and invasion of epithelial cells in Salmonella enterica serovar Typhimurium. Future Microbiol 12, 1045–1057. 10.2217/fmb-2017-0055 [DOI] [PubMed] [Google Scholar]
- Repoila F, and Gottesman S (2001). Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J Bacteriol 183, 4012–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repoila F, Majdalani N, and Gottesman S (2003). Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol Microbiol 48, 855–861. [DOI] [PubMed] [Google Scholar]
- Revel AT, Talaat AM, and Norgard MV (2002). DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci USA 99, 1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes RG, Coy W, and Nelson DR (2009). Chitobiose utilization in Borrelia burgdorferi is dually regulated by RpoD and RpoS. BMC Microbiol 9, 108. 10.1186/1471-2180-9-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC, Mather TN, Piesman J, and Spielman A (1987). Dissemination and salivary delivery of Lyme disease spirochetes in vector ticks (Acari: Ixodidae). J Med Entomol 24, 201–205. 10.1093/jmedent/24.2.201 [DOI] [PubMed] [Google Scholar]
- Richards CL, Lawrence KA, Su H, Yang Y, Yang XF, Dulebohn DP, and Gherardini FC (2015). Acetyl-phosphate is not a global regulatory bridge between virulence and central metabolism in Borrelia burgdorferi. PLoS One 10, e0144472. 10.1371/journal.pone.0144472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddihough G (2005). In the forests of RNA dark matter. Science 309, 1507–1507. 10.1126/science.309.5740.1507 [DOI] [Google Scholar]
- Riley SP, Bykowski T, Cooley AE, Burns LH, Babb K, Brissette CA, Bowman A, Rotondi M, Miller MC, DeMoll E, et al. (2009). Borrelia burgdorferi EbfC defines a newly-identified, widespread family of bacterial DNA-binding proteins. Nucleic Acids Res 37, 1973–1983. 10.1093/nar/gkp027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DM, Caimano M, McDowell J, Theisen M, Holm A, Orff E, Nelson D, Wikel S, Radolf J, and Marconi RT (2002). Environmental regulation and differential production of members of the Bdr protein family of Borrelia burgdorferi. Infect Immun 70, 7033–7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Terekhova D, Zhang H-M, Hovis KM, Schwartz I, and Marconi RT (2009). Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol Microbiol 71, 1551–1573. 10.1111/j.1365-2958.2009.06621.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T (1998). Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol 29, 1321–1330. [DOI] [PubMed] [Google Scholar]
- Romeo T, and Babitzke P (2018). Global regulation by CsrA and its RNA antagonists. Microbiol Spectr 6, RWR-0009–2017. 10.1128/microbiolspec.RWR-0009-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U (2008). Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci Signal 1, pe39. 10.1126/scisignal.133pe39 [DOI] [PubMed] [Google Scholar]
- Römling U, Galperin MY, and Gomelsky M (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77, 1–52. 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneau S, and Hallez R (2019). Make and break the alarmone: regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol Rev 43, 389–400. 10.1093/femsre/fuz009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh A, Banner CD, Losick R, and Fitz-James PC (1981). Identification of a new developmental locus in Bacillus subtilis by construction of a deletion mutation in a cloned gene under sporulation control. J Bacteriol 148, 341–351. 10.1128/JB.148.1.341-351.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, et al. (1987). Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325, 279–281. 10.1038/325279a0 [DOI] [PubMed] [Google Scholar]
- Roth A, and Breaker RR (2009). The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78, 305–334. 10.1146/annurev.biochem.78.070507.135656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BE, Huynh TN, Welkie DG, Diamond S, Simkovsky R, Pierce EC, Taton A, Lowe LC, Lee JJ, Rifkin SA, et al. (2018). High-throughput interaction screens illuminate the role of c-di-AMP in cyanobacterial nighttime survival. PLoS Genet 14, e1007301. 10.1371/journal.pgen.1007301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui S, and Tse-Dinh Y-C (2003). Topoisomerase function during bacterial responses to environmental challenge. Front Biosci 8, d256–263. [DOI] [PubMed] [Google Scholar]
- Rutberg L, and Armentrout RW (1972). Deoxyribonucleic acid polymerase activity in a deoxyribonucleic acid polymerase I-deficient mutant of Bacillus subtilis infected with temperature bacteriophage SPO2. J Virol 10, 658–660. 10.1128/JVI.10.4.658-660.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Graybosch DM, Huetsch JC, and Verdine GL (2005). A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J Biol Chem 280, 26177–26184. 10.1074/jbc.M502838200 [DOI] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Römling U, and Gomelsky M (2006). The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281, 30310–30314. 10.1074/jbc.C600179200 [DOI] [PubMed] [Google Scholar]
- Ryjenkov DA, Tarutina M, Moskvin OV, and Gomelsky M (2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187, 1792–1798. 10.1128/JB.187.5.1792-1798.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba A-E, Santos SC, and Vogel J (2017). New RNA-seq approaches for the study of bacterial pathogens. Curr Opin Microbiol 35, 78–87. 10.1016/j.mib.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Samuels DS (2006). Antibiotic resistance in Borrelia burgdorferi: applications for genetic manipulation and implications for evolution. In Molecular Biology of Spirochetes, Cabello FC, Hulinska D, and Godfrey HP, eds. (Amsterdam, Netherlands: IOS Press; ), pp. 56–70. [Google Scholar]
- Samuels DS (2011). Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol 65, 479–499. 10.1146/annurev.micro.112408.134040 [DOI] [PubMed] [Google Scholar]
- Samuels DS, and Radolf JD (2009). Who is the BosR around here anyway? Mol Microbiol 74, 1295–1299. 10.1111/j.1365-2958.2009.06971.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS, and Samuels LRN (2016). Gene regulation during the enzootic cycle of the Lyme disease spirochete. For Immunopathol Dis Therap 7, 205–212. 10.1615/ForumImmunDisTher.2017019469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan E, Esteve-Gassent MD, Maruskova M, and Seshu J (2009). Overexpression of CsrA (BB0184) alters the morphology and antigen profiles of Borrelia burgdorferi. Infect Immun 77, 5149–5162. 10.1128/IAI.00673-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvan S, Butcher J, Stintzi A, and Couture J-F (2018). Variation on a theme: investigating the structural repertoires used by ferric uptake regulators to control gene expression. Biometals 31, 681–704. 10.1007/s10534-018-0120-8 [DOI] [PubMed] [Google Scholar]
- Savage CR, Arnold WK, Gjevre-Nail A, Koestler BJ, Bruger EL, Barker JR, Waters CM, and Stevenson B (2015). Intracellular concentrations of Borrelia burgdorferi cyclic di-AMP are not changed by altered expression of the CdaA synthase. PLoS One 10, e0125440. 10.1371/journal.pone.0125440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage CR, Jutras BL, Bestor A, Tilly K, Rosa PA, Tourand Y, Stewart PE, Brissette CA, and Stevenson B (2018). Borrelia burgdorferi SpoVG DNA- and RNA-binding protein modulates the physiology of the Lyme disease spirochete. J Bacteriol 200, e00033–18. 10.1128/JB.00033-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer T, and Jenal U (2009). Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7, 724–735. 10.1038/nrmicro2203 [DOI] [PubMed] [Google Scholar]
- Schmid MB (1990). More than just “histone-like” proteins. Cell 63, 451–453. [DOI] [PubMed] [Google Scholar]
- Schneider EM, and Rhodes RG (2018). N-acetylmannosamine (ManNAc) supports the growth of Borrelia burgdorferi in the absence of N-acetylglucosamine (GlcNAc). FEMS Microbiol Lett 365, fny243. 10.1093/femsle/fny243 [DOI] [PubMed] [Google Scholar]
- Schwan TG, and Piesman J (2000). Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol 38, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J, Golde WT, Dolan MC, and Rosa PA (1995). Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA 92, 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, and Nudler E (2013). A decade of riboswitches. Cell 152, 17–24. 10.1016/j.cell.2012.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Boylan JA, Gherardini FC, and Skare JT (2004a). Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect Immun 72, 1580–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Boylan JA, Hyde JA, Swingle KL, Gherardini FC, and Skare JT (2004b). A conservative amino acid change alters the function of BosR, the redox regulator of Borrelia burgdorferi. Mol Microbiol 54, 1352–1363. [DOI] [PubMed] [Google Scholar]
- Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, Höök M, and Skare JT (2006). Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol Microbiol 59, 1591–1601. [DOI] [PubMed] [Google Scholar]
- Shahbabian K, Jamalli A, Zig L, and Putzer H (2009). RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J 28, 3523–3533. 10.1038/emboj.2009.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood AV, and Henkin TM (2016). Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu Rev Microbiol 70, 361–374. 10.1146/annurev-micro-091014-104306 [DOI] [PubMed] [Google Scholar]
- Shi Y, Dadhwal P, Li X, and Liang FT (2014). BosR functions as a repressor of the ospAB operon in Borrelia burgdorferi. PLoS One 9, e109307. 10.1371/journal.pone.0109307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Xu Q, McShan K, and Liang FT (2008). Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect Immun 76, 1239–1246. 10.1128/IAI.00897-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Yokota H, Kim R, and Kim S-H (2002). Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus. Proc Natl Acad Sci USA 99, 7980–7985. 10.1073/pnas.132241399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotland Y, Koby S, Teff D, Mansur N, Oren DA, Tatematsu K, Tomoyasu T, Kessel M, Bukau B, Ogura T, et al. (1997). Proteolysis of the phage λ CII regulatory protein by FtsH (HflB) of Escherichia coli. Mol Microbiol 24, 1303–1310. [DOI] [PubMed] [Google Scholar]
- Skare JT, Carroll JA, Yang XF, Samuels DS, and Akins DR (2010). Gene regulation, transcriptomics and proteomics. In Borrelia: Molecular Biology, Host Interaction and Pathogenesis, Samuels DS, and Radolf JD, eds. (Norfolk, UK: Caister Academic Press; ), pp. 67–101. [Google Scholar]
- Smith AH, Blevins JS, Bachlani GN, Yang XF, and Norgard MV (2007). Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN). J Bacteriol 189, 2139–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Shanahan CA, Moore EL, Simon AC, and Strobel SA (2011). Structural basis of differential ligand recognition by two classes of bis-(3′−5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc Natl Acad Sci USA 108, 7757–7762. 10.1073/pnas.1018857108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow S, Bacon E, Bergeron J, Katzman D, Wilhelm A, Lewis O, Syangtan D, Calkins A, Archambault L, Anacker ML, et al. (2020). Transcript decay mediated by RNase III in Borrelia burgdorferi. Biochem Biophys Res Commun 529, 386–391. 10.1016/j.bbrc.2020.05.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey CD, Zückert WR, and Barbour AG (1999). The extended promoters for two outer membrane lipoprotein genes of Borrelia spp. uniquely include a T-rich region. Mol Microbiol 33, 41–51. [DOI] [PubMed] [Google Scholar]
- Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, and Goldfarb A (2003). Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J 22, 2234–2244. 10.1093/emboj/cdg193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinchen W, and Bange G (2016). The magic dance of the alarmones (p)ppGpp. Mol Microbiol 101, 531–544. 10.1111/mmi.13412 [DOI] [PubMed] [Google Scholar]
- Stetter KO, and Zillig W (1974). Transcription in Lactobacillaceae: DNA-dependent RNA polymerase from Lactobacillus curvatus. Eur J Biochem 48, 527–540. 10.1111/j.1432-1033.1974.tb03794.x [DOI] [PubMed] [Google Scholar]
- Steuten B, Hoch PG, Damm K, Schneider S, Köhler K, Wagner R, and Hartmann RK (2014a). Regulation of transcription by 6S RNAs: insights from the Escherichia coli and Bacillus subtilis model systems. RNA Biol 11, 508–521. 10.4161/rna.28827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuten B, Schneider S, and Wagner R (2014b). 6S RNA: recent answers–future questions. Mol Microbiol 91, 641–648. 10.1111/mmi.12484 [DOI] [PubMed] [Google Scholar]
- Stevenson B, and Seshu J (2018). Regulation of gene and protein expression in the Lyme disease spirochete. In Spirochete Biology: The Post Genomic Era, Adler B, ed. (Cham, Switzerland: Springer; ), pp. 83–112. 10.1007/82_2017_49 [DOI] [PubMed] [Google Scholar]
- Stewart P, Thalken R, Bono J, and Rosa P (2001). Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol 39, 714–721. [DOI] [PubMed] [Google Scholar]
- Stewart PE, Chaconas G, and Rosa P (2003). Conservation of plasmid maintenance functions between linear and circular plasmids in Borrelia burgdorferi. J Bacteriol 185, 3202–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J, and Wassarman KM (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43, 880–891. 10.1016/j.molcel.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme DJ, and Buck M (2000). The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol Lett 186, 1–9. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, and Breaker RR (2008). Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321, 411–3. 10.1126/science.1159519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Pitzer JE, Boquoi T, Hobbs G, Miller MR, and Motaleb MA (2011). Analysis of the HD-GYP domain cyclic-di-GMP phosphodiesterase reveals a role in motility and enzootic life cycle of Borrelia burgdorferi. Infect Immun 79, 3273–3283. 10.1128/IAI.05153-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Pitzer JE, Miller MR, and Motaleb MA (2010). Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol Microbiol 77, 128–142. 10.1111/j.1365-2958.2010.07191.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson SL, and Sharma CM (2016). Small RNAs in bacterial virulence and communication. Microbiol Spectr 4. 10.1128/microbiolspec.VMBF-0028-2015 [DOI] [PubMed] [Google Scholar]
- Sze CW, and Li C (2011). Inactivation of bb0184, which encodes carbon storage regulator A, represses the infectivity of Borrelia burgdorferi. Infect Immun 79, 1270–1279. 10.1128/IAI.00871-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Morado DR, Liu J, Charon NW, Xu H, and Li C (2011). Carbon storage regulator A (CsrABb) is a repressor of Borrelia burgdorferi flagellin protein FlaB. Mol Microbiol 82, 851–864. 10.1111/j.1365-2958.2011.07853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Smith A, Choi YH, Yang X, Pal U, Yu A, and Li C (2013). Study of the response regulator Rrp1 reveals its regulatory role in chitobiose utilization and virulence of Borrelia burgdorferi. Infect Immun 81, 1775–1787. 10.1128/IAI.00050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Elias AF, Errett J, Fischer E, Iyer R, Schwartz I, Bono JL, and Rosa P (2001). Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J Bacteriol 183, 5544–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Fuhrman J, Campbell J, and Samuels DS (1996). Isolation of Borrelia burgdorferi genes encoding homologues of DNA-binding protein HU and ribosomal protein S20. Microbiology 142, 2471–2479. 10.1099/00221287-142-9-2471 [DOI] [PubMed] [Google Scholar]
- Tilly K, Grimm D, Bueschel DM, Krum JG, and Rosa P (2004). Infectious cycle analysis of a Borrelia burgdorferi mutant defective in transport of chitobiose, a tick cuticle component. Vector Borne Zoonotic Dis 4, 159–168. 10.1089/1530366041210738 [DOI] [PubMed] [Google Scholar]
- Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, Byram R, Dorward D, Vanraden MJ, Stewart P, et al. (2006). Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun 74, 3554–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titgemeyer F, Reizer J, Reizer A, and Saier MH Jr. (1994). Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140 (Pt 9), 2349–2354. 10.1099/13500872-140-9-2349 [DOI] [PubMed] [Google Scholar]
- Tokarz R, Anderton JM, Katona LI, and Benach JL (2004). Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect Immun 72, 5419–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traoré DAK, El Ghazouani A, Ilango S, Dupuy J, Jacquamet L, Ferrer J-L, Caux-Thang C, Duarte V, and Latour J-M (2006). Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol Microbiol 61, 1211–1219. 10.1111/j.1365-2958.2006.05313.x [DOI] [PubMed] [Google Scholar]
- Traoré DAK, El Ghazouani A, Jacquamet L, Borel F, Ferrer J-L, Lascoux D, Ravanat J-L, Jaquinod M, Blondin G, Caux-Thang C, et al. (2009). Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat Chem Biol 5, 53–59. 10.1038/nchembio.133 [DOI] [PubMed] [Google Scholar]
- Trinquier A, Durand S, Braun F, and Condon C (2020). Regulation of RNA processing and degradation in bacteria. Biochim Biophys Acta Gene Regul Mech 1863, 194505. 10.1016/j.bbagrm.2020.194505 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, and Wassarman KM (2004). 6S RNA function enhances long-term cell survival. J Bacteriol 186, 4978–4985. 10.1128/JB.186.15.4978-4985.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, and Wassarman KM (2005). A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol 12, 313–319. 10.1038/nsmb917 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, and Wassarman KM (2006). 6S RNA regulation of pspF transcription leads to altered cell survival at high pH. J Bacteriol 188, 3936–3943. 10.1128/JB.00079-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B, and Yang XF (2013). Metal-dependent gene regulation in the causative agent of Lyme disease. Front Cell Infect Microbiol 3, 79. 10.3389/fcimb.2013.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B, Ye M, Yang Y, Carrasco SE, Lou Y, and Yang XF (2013). Manganese and zinc regulate virulence determinants in Borrelia burgdorferi. Infect Immun 81, 2743–2752. 10.1128/IAI.00507-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy EB, Lin T, Gao L, Lazinski DW, Lundt M, Camilli A, Norris SJ, and Hu LT (2016). Global Tn-seq analysis of carbohydrate utilization and vertebrate infectivity of Borrelia burgdorferi. Mol Microbiol 101, 1003–1023. 10.1111/mmi.13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Becker G, and Hengge R (2007). The molecular basis of selective promoter activation by the σS subunit of RNA polymerase. Mol Microbiol 63, 1296–1306. [DOI] [PubMed] [Google Scholar]
- Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, and Romeo T (2015). Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 79, 193–224. 10.1128/MMBR.00052-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, and Udesen C (2004). The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol 51, 1525–1533. [DOI] [PubMed] [Google Scholar]
- Valentini M, and Filloux A (2019). Multiple roles of c-di-GMP sgnaling in bacterial pathogenesis. Annu Rev Microbiol 73, 387–406. 10.1146/annurev-micro-020518-115555 [DOI] [PubMed] [Google Scholar]
- Van Laar TA, Lin Y-H, Miller CL, Karna SLR, Chambers JP, and Seshu J (2012). Effect of levels of acetate on the mevalonate pathway of Borrelia burgdorferi. PLoS One 7, e38171. 10.1371/journal.pone.0038171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lackum K, and Stevenson B (2005). Carbohydrate utilization by the Lyme borreliosis spirochete, Borrelia burgdorferi. FEMS Microbiol Lett 243, 173–179. [DOI] [PubMed] [Google Scholar]
- Wagh D, Pothineni VR, Inayathullah M, Liu S, Kim K-M, and Rajadas J (2015). Borreliacidal activity of Borrelia metal transporter A (BmtA) binding small molecules by manganese transport inhibition. Drug Des Devel Ther 9, 805–816. 10.2147/DDDT.S77063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang F, Hua X, Ma T, Chen J, Xu X, Wang L, Tian B, and Hua Y (2012a). Genetic and biochemical characteristics of the histone-like protein DR0199 in Deinococcus radiodurans. Microbiology 158, 936–943. 10.1099/mic.0.053702-0 [DOI] [PubMed] [Google Scholar]
- Wang P, Dadhwal P, Cheng Z, Zianni MR, Rikihisa Y, Liang FT, and Li X (2013). Borrelia burgdorferi oxidative stress regulator BosR directly represses lipoproteins primarily expressed in the tick during mammalian infection. Mol Microbiol 89, 1140–1153. 10.1111/mmi.12337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lutton A, Olesik J, Vali H, and Li X (2012b). A novel iron- and copper-binding protein in the Lyme disease spirochaete. Mol Microbiol 86, 1441–1451. 10.1111/mmi.12068 [DOI] [PubMed] [Google Scholar]
- Wang P, Yu Z, Santangelo TJ, Olesik J, Wang Y, Heldwein E, and Li X (2017). BosR is a novel Fur family member responsive to copper and regulating copper homeostasis in Borrelia burgdorferi. J Bacteriol 199, e00276–17. 10.1128/JB.00276-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrier I, Hicks LD, Battisti JM, Raghavan R, and Minnick MF (2014). Identification of novel small RNAs and characterization of the 6S RNA of Coxiella burnetii. PLoS One 9, e100147. 10.1371/journal.pone.0100147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM (2018). 6S RNA, a global regulator of transcription. Microbiol Spectr 6, RWR-0019–2018. 10.1128/microbiolspec.RWR-0019-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, and Storz G (2000). 6S RNA regulates E. coli RNA polymerase activity. Cell 101, 613–623. [DOI] [PubMed] [Google Scholar]
- Waters LS, and Storz G (2009). Regulatory RNAs in bacteria. Cell 136, 615–628. 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, et al. (2008). Modus operandi of the bacterial RNA polymerase containing the σ54 promoter-specificity factor. Mol Microbiol 68, 538–546. 10.1111/j.1365-2958.2008.06181.x [DOI] [PubMed] [Google Scholar]
- Wolfe AJ, and Visick KL (2008). Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol 190, 463–475. 10.1128/JB.01418-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Caimano MJ, Lin T, He M, Radolf JD, Norris SJ, Gheradini F, Wolfe AJ, and Yang XF (2010). Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi. PLoS Pathog 6, e1001104. 10.1371/journal.ppat.1001104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, McShan K, and Liang FT (2007). Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi. Mol Microbiol 64, 220–231. 10.1111/j.1365-2958.2007.05636.x [DOI] [PubMed] [Google Scholar]
- Yan Y, Su S, Meng X, Ji X, Qu Y, Liu Z, Wang X, Cui Y, Deng Z, Zhou D, et al. (2013). Determination of sRNA expressions by RNA-seq in Yersinia pestis grown in vitro and during infection. PLoS One 8, e74495. 10.1371/journal.pone.0074495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, and Norgard MV (2000). Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol Microbiol 37, 1470–1479. [DOI] [PubMed] [Google Scholar]
- Yang XF, Alani SM, and Norgard MV (2003). The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci USA 100, 11001–11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Lybecker MC, Pal U, Alani SM, Blevins J, Revel AT, Samuels DS, and Norgard MV (2005). Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J Bacteriol 187, 4822–4829. 10.1128/JB.187.14.4822-4829.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, and Bechhofer DH (2010). Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y. J Bacteriol 192, 3279–3286. 10.1128/JB.00230-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Zhang J-J, Fang X, Lawlis GB, Troxell B, Zhou Y, Gomelsky M, Lou Y, and Yang XF (2014). DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect Immun 82, 1840–1849. 10.1128/IAI.00030-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Zhou Y, Lou Y, and Yang XF (2016). Genome reduction of Borrelia burgdorferi: two TCS signaling pathways for two distinct host habitats. Sci China Life Sci 59, 19–21. 10.1007/s11427-015-4996-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Yang Y, Xiang X, Wang Q, Yang Z-N, Blevins J, Lou Y, and Yang XF (2016). Insight into the dual functions of bacterial enhancer-binding protein Rrp2 of Borrelia burgdorferi. J Bacteriol 198, 1543–1552. 10.1128/JB.01010-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, and Marconi RT (2005). Demonstration of cotranscription and 1-methyl-3-nitroso-nitroguanidine induction of a 30-gene operon of Borrelia burgdorferi: evidence that the 32-kilobase circular plasmids are prophages. J Bacteriol 187, 7985–7995. 10.1128/JB.187.23.7985-7995.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-J, Chen T, Yang Y, Du J, Li H, Troxell B, He M, Carrasco SE, Gomelsky M, and Yang XF (2018). Positive and negative regulation of glycerol utilization by the c-di-GMP binding protein PlzA in Borrelia burgdorferi. J Bacteriol 200, e00243–18. 10.1128/JB.00243-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen T, Raghunandanan S, Xiang X, Yang J, Liu Q, Edmondson DG, Norris S, Yang XF, and Lou Y (2020). YebC regulates variable surface antigen VlsE expression and is required for host immune evasion in Borrelia burgdorferi. PLoS Pathog 16, e1008953. 10.1371/journal.ppat.1008953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lin J, and Gao Y (2012). In silico identification of a multi-functional regulatory protein involved in Holliday junction resolution in bacteria. BMC Syst Biol 6 Suppl 1, S20. 10.1186/1752-0509-6-S1-S20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zückert WR, and Meyer J (1996). Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J Bacteriol 178, 2287–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zückert WR, Meyer J, and Barbour AG (1999). Comparative analysis and immunological characterization of the Borrelia Bdr protein family. Infect Immun 67, 3257–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]


