Arising from Lina Sieverling et al. Nature Communications 10.1038/s41467-019-13824-9 (2020)
The PCAWG Consortium has recently released an integrative re-analysis of a large set of tumor whole genome sequence (WGS) data from 2658 cancer patients across 38 different primary tumor sites1. In a companion paper, Sieverling et al. built a random forest classifier for the telomere maintenance mechanism (TMM) by regarding truncating ATRX or DAXX alterations, referred to as ATRX/DAXXtrunc, vs. TERT modifications (TERTmod; i.e., promoter mutations ± amplifications ± structural variations), as indicators of alternative lengthening of telomeres (ALT) vs. telomerase2. We show here that equating ATRX/DAXXtrunc and TERTmod with ALT and telomerase, respectively, results in TMM predictions which do not correlate well with TMM assay data. Although ATRX/DAXXtrunc mutations are associated with TMM, most tumors do not harbor them and they are heterogeneously distributed in ALT-positive (ALT + ) tumors of different types, as are TERTmod in telomerase-positive tumors3–6, making these mutations an insufficient basis for building a classifier in a large-scale pan-cancer study.
Here, we provide a new analysis of the PCAWG data, based on C-circle assay (CCA)7 data that are available for a subset of these tumors3. We show that the Sieverling et al. score overestimates the proportion of ALT associated with ATRX/DAXXtrunc and misclassifies ALT tumors when these mutations are absent. We also show some telomere variant repeats (TVR) correlate with ATRX/DAXXtrunc mutations, regardless of TMM. Finally, we propose a new classifier to identify ALT tumors in the PCAWG cohort.
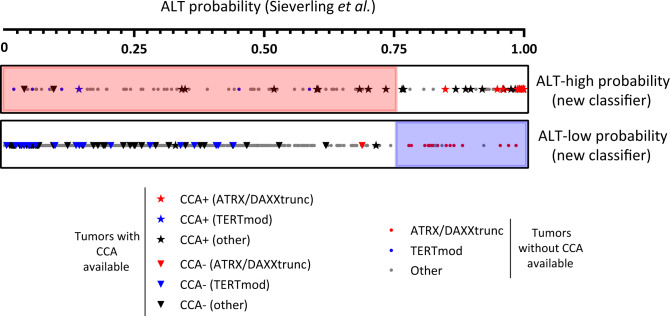
WGS data have been used as a means to assess telomere content and analyze TMM3. To our knowledge, only one previous genomic study generated an ALT-probability score based on ALT assay data. The CCA appears to be a reliable marker of the presence of ALT activity in cancers7, notwithstanding cell line studies showing that quantitative CCA data do not always correlate with the amount of ALT activity (e.g., ref. 8), highlighting the need for further biological studies to decipher the origin of C-circles and their functional relationship with the ALT mechanism. Lee et al.3 used the CCA to determine ALT status of 167 pancreatic neuroendocrine tumors (PaNET) and melanomas, and then applied machine learning to features including total telomeric and TVR content to develop an ALT classifier with an accuracy of 91.6%. The classifier was then applied to WGS data from 908 additional tumors, mostly from The Cancer Genome Atlas (TCGA) dataset. Of the total of 1075 tumors studied by Lee et al.3, 703 were included in the PCAWG study, and CCA data are available for 114 of these (melanoma, n = 46 and PanNET, n = 68). A comparison of the Sieverling and Lee scores revealed a poor correlation (r = 0.101 [95% CI 0.025–0.17], Spearman correlation; Supplementary Fig. 1). We then compared the CCA data with Sieverling’s score. The latter identified only 64.5% of CCA-positive tumors as ALT-high probability and misidentified CCA-negative ATRX/DAXXtrunc tumors (Fig. 1). Also, the only CCA-positive TERTmod tumor was identified as ALT-low probability by the Sieverling score.
Fig. 1. Comparison of the ALT-probability score of Sieverling et al.2, with the new classifier proposed in this article.
The 2497 patients described in Sieverling et al. are divided here into two groups: ALT-high probability (upper bar) and ALT-low probability (lower bar) according to the new classifier. Within the two bars, each patient is represented by a symbol placed on a scale from 0 to 1 according to its Sieverling ALT-probability score. Star and inverted triangle symbols represent patients whose tumors were C-circle assay (CCA) positive or negative, respectively3, and the dots represent patients for whom CCA data are unavailable. Red and blue symbols correspond to patients with ATRX/DAXXtrunc and TERTmod alterations, respectively2. The pink and blue shading highlights the tumors for which the two scores were discordant: the pink shading in the upper box indicates tumors with ALT-high probability (new classifier), that were ALT-probability <0.75 (ALT-low) by Sieverling’s score; the blue shading in the lower box indicates tumors with ALT-low probability (new classifier), that were ALT-probability >0.75 (ALT-high) by Sieverling’s score.
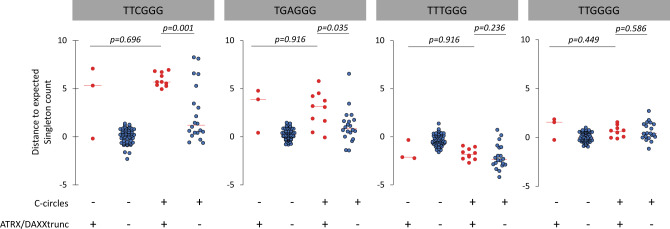
Sieverling et al.’s analysis of the PCAWG data identified an association between prevalence of TTCGGG singletons and ALT, and a more pronounced enrichment of the TGAGGG TVR than previously noted. Our re-analysis showed that TTCGGG, TGAGGG, TTTGGG, and TTGGGG singleton distributions are similar among ATRX/DAXXtrunc tumors, regardless of CCA result (Fig. 2). In contrast, in CCA-positive tumors, there were statistically significant differences in TTCGGG and TGAGGG distributions between tumors with and without ATRX/DAXXtrunc.
Fig. 2. Distance to the expected singleton repeat count in tumors with negative C-circle assay, with (n = 3), or without (n = 58) ATRX/DAXXtrunc, and in tumors with positive C-circle assay with (n = 10), or without (n = 20) ATRX/DAXXtrunc.
The center (horizontal) red line of the scattergrams is the median. p < 0.05 was considered as significant; Kolmogorov–Smirnov tests, two-tailed.
ATRX is a SWI/SNF-like chromatin remodeling protein that binds to G-rich tandem repeats9. Once recruited, ATRX cooperates with histone chaperone DAXX in replication-independent deposition of the histone variant H3.3. Loss of ATRX/DAXX function causes defects in multiple cellular processes, including defective sister chromatid cohesion and telomere dysfunction but in some cellular contexts is not sufficient by itself to induce ALT10. Our data suggest that ATRX/DAXXtrunc (rather than the ALT mechanism per se) could also play a role in TVR distribution. This observation could provide additional insights regarding the mechanisms involved in ALT promotion in the context of ATRX/DAXXtrunc in specific tumor types. TVR are associated with genomic instability, and could facilitate telomere spatial reconfiguration and affect telomere binding affinity11,12. Although histones are non-specific DNA binding proteins, nucleosome formation could potentially be influenced by telomeric DNA sequences and their structural properties13.
The international effort that enabled release of the PCAWG data set has provided a major new resource for seeking new insights into cancer, including the ALT pathway, which represents a potentially valuable, currently unexploited, target for anti-cancer therapies. Understanding more precisely how telomeres are maintained in cancer will shed light on replicative immortality and telomeric DNA damage response mechanisms. To develop a classifier, based on CCA data as an indicator of ALT, we searched for a signature associated with CCA status within the genomic features provided by the Telomere Hunter WGS tool2. Of the eight features used by Sieverling et al. in their random forest classifier, five were retained after Akaike information criterion stepwise regression analysis: telomere content (tumor/control log2 ratio) and the distance of TTTGGG, TTCGGG, TTGGGG, and GTAGGG singletons from their expected occurrence. A classifier was then built from these five features, which permitted definition of two groups as “high-probability” and “low probability”, using different combinations of the PanNET and melanoma datasets for training and testing. A 100% success rate was achievable when considering only one tumor type (i.e., using PanNET or melanoma for both the learning and the validation sets). Our final classifier, built using the whole cohort of patients with CCA data, has an accuracy of 93.86% (93.55% specificity and 93.98% sensitivity). This underlines the need for caution when extrapolating a classifier built on a specific tumor type to others. When applied to the 2497 PCAWG patients, our classifier identified 200 tumors with high probability of ALT (Supplementary data 1), and their reported distribution across different histological types (leiomyosarcomas 73%, osteosarcomas 63%, liposarcomas 53%, and low grade gliomas 29%) is consistent with previous reports5. In contrast, 27% of ATRX/DAXXtrunc tumors (n = 17/64) were classified as ALT-low probability, and 6% of TERTmod tumors (n = 15/269) as ALT-high probability (Fig. 1).
In conclusion, the Sieverling et al. score is an ATRX/DAXXtrunc vs. TERTmod classifier rather than a predictor of ALT. The view that ATRX/DAXX loss is essentially equivalent to the presence of ALT activity may apply only to specific types of tumors, in specific genomic or epigenetic contexts, as suggested by recent cell line based studies14,15. Adoption of this view as a generalization across cancer types may divert attention from the need to identify alternative molecular actors involved in TMM. The classifier based on telomeric content and TVR distributions we provide here is more accurate as judged by C-circles, a hallmark of ALT, for PanNETs and melanomas, the tumor types on which it was trained. Although its predictions for other tumor types are consistent with the reported prevalence of ALT, this classifier should be applied with caution, given the apparent tumor type specificity of TVR distribution.
Methods
Sieverling et al.2 and Lee et al.3 ALT-probability scores were compared using Spearman’s correlation. Distances to the expected singleton repeat count distributions2 in tumors with negative CCA3, with or without ATRX/DAXXtrunc2, and in tumors with positive CCA3 with, or without ATRX/DAXXtrunc2 were compared using two-tailed Kolmogorov–Smirnov tests at the 5% level of significance. To develop a classifier, based on CCA data3 as an indicator of ALT, we searched for the best combination signature associated with CCA status3 within the genomic features and telomere content determined by Sieverling et al. with the Telomere Hunter WGS tool2 (telomere content tumor/control log2 ratio, telomere insertion count, breakpoint count, and singleton distributions), using Akaike information criterion stepwise regression analysis. Statistical analyses were carried out using R version 3.6.2 (R Foundation for Statistical Computing).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Lars Feuerbach and colleagues for their positive approach and constructive comments regarding this Matters Arising, and acknowledge their response that the CCA data we refer to here were released after they submitted their study to the internal revision process of the PCAWG consortium, and thus could not be included in their work. We thank Hilda Pickett, Karen MacKenzie and Yangxiu Wu for helpful comments and critical reading of the manuscript. A.N. is supported by a Doc.Mobility fellowship from the Swiss National Science Foundation.
Author contributions
A.N. and R.R.R. designed the study; A.N. performed the analysis; A.N. and R.R.R. wrote the manuscript.
Data availability
This Matters Arising is a non-interventional, retrospective analysis and commentary of published data, publicly available within the Nature Communications paper “Sieverling et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat. Commun. 11, 1–13 (2020).”2, and the Nucleic Acids Research paper “Lee et al. Telomere sequence content can be used to determine ALT activity in tumors. Nucleic Acids Res. 46, 4903–4918 (2018).”3
Genomic features and telomere content determined by Sieverling et al., and the Sieverling et al. score, are available in the Sieverling et al.2 supplementary data 1 file. C-circle data, referenced by Lee et al.3, and the Lee et al. score, are available in the Lee et al.3 supplementary data file. Aligned PCAWG read data in BAM format are also available at the European Genome Phenome Archive (EGA; https://www.ebi.ac.uk/ega/search/site/pcawg under accession number EGAS00001001692). In addition, all open-tier PCAWG genomics data, as well as datasets used for PCAWG analysis, can be downloaded from the ICGC Data Portal at http://docs.icgc.org/pcawg/data/. The software TelomereHunter used by Sieverling et al.2 for telomeric in silico analysis is available from https://www.dkfz.de/en/applied-bioinformatics/telomerehunter/telomerehunter.html. The core computational pipelines used by the PCAWG Consortium for alignment, quality control, and variant calling are available to the public at https://dockstore.org/search?search=pcawg under the GNU General Public License v3.0, which allows for reuse and distribution.
Competing interests
R.R.R. is listed as a joint inventor on a patent regarding the C-circle assay: European Patent 10818148.8, US Patent US 08999643, Chinese Patent 201080048175.X: “Methods and assays for the detection of Alternative Lengthening of Telomeres (ALT) activity in cells”; Children’s Medical Research Institute, Inventors: Jeremy D. Henson and Roger R. Reddel.
Ethical approval
This Matters Arising comment is a non-interventional, retrospective analysis and commentary on published data, publicly available within the Nature Communications paper “Sieverling, L. et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat. Commun. 11, 1–13 (2020).” and its supplementary information files. Therefore, no ethics board/committee’s approval or formal patient consent was required. As described in Sieverling et al., the Ethics oversight for the PCAWG protocol was undertaken by the TCGA Program Office and the Ethics and Governance Committee of the ICGC; each individual ICGC and TCGA project that contributed data to PCAWG had its own local arrangements for ethics oversight and regulatory alignment.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alexandre de Nonneville, Email: adenonneville@cmri.org.au.
Roger R. Reddel, Email: rreddel@cmri.org.au
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-21794-0.
References
- 1.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sieverling L, et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-019-13824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee M, et al. Telomere sequence content can be used to determine ALT activity in tumours. Nucleic Acids Res. 2018;46:4903–4918. doi: 10.1093/nar/gky297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chudasama P, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat. Commun. 2018;9:144. doi: 10.1038/s41467-017-02602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthel FP, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017;49:349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayward NK, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 7.Henson JD, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J-M, Yadav T, Ouyang J, Lan L, Zou L. Alternative lengthening of telomeres through two distinct break-induced replication pathways. Cell Rep. 2019;26:955–968.e3. doi: 10.1016/j.celrep.2018.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyer, M. A., Qadeer, Z. A., Valle-Garcia, D. & Bernstein, E. ATRX and DAXX: mechanisms and mutations. Cold Spring Harb. Perspect. Med. 7, a026567 (2017). [DOI] [PMC free article] [PubMed]
- 10.Clynes D, et al. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 2015;6:7538. doi: 10.1038/ncomms8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conomos D, et al. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol. 2012;199:893–906. doi: 10.1083/jcb.201207189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzec P, et al. Nuclear-receptor-mediated telomere insertion leads to genome instability in ALT cancers. Cell. 2015;160:913–927. doi: 10.1016/j.cell.2015.01.044. [DOI] [PubMed] [Google Scholar]
- 13.Lai WKM, Pugh BF. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 2017;18:548–562. doi: 10.1038/nrm.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brosnan-Cashman JA, et al. ATRX loss induces multiple hallmarks of the alternative lengthening of telomeres (ALT) phenotype in human glioma cell lines in a cell line-specific manner. PLoS ONE. 2018;13:e0204159. doi: 10.1371/journal.pone.0204159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, M. K. et al. Functional loss of ATRX and TERC activates alternative lengthening of telomeres (ALT) in LAPC4 prostate cancer cells. Mol. Cancer Res. 10.1158/1541-7786.MCR-19-0654 (2019). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This Matters Arising is a non-interventional, retrospective analysis and commentary of published data, publicly available within the Nature Communications paper “Sieverling et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat. Commun. 11, 1–13 (2020).”2, and the Nucleic Acids Research paper “Lee et al. Telomere sequence content can be used to determine ALT activity in tumors. Nucleic Acids Res. 46, 4903–4918 (2018).”3
Genomic features and telomere content determined by Sieverling et al., and the Sieverling et al. score, are available in the Sieverling et al.2 supplementary data 1 file. C-circle data, referenced by Lee et al.3, and the Lee et al. score, are available in the Lee et al.3 supplementary data file. Aligned PCAWG read data in BAM format are also available at the European Genome Phenome Archive (EGA; https://www.ebi.ac.uk/ega/search/site/pcawg under accession number EGAS00001001692). In addition, all open-tier PCAWG genomics data, as well as datasets used for PCAWG analysis, can be downloaded from the ICGC Data Portal at http://docs.icgc.org/pcawg/data/. The software TelomereHunter used by Sieverling et al.2 for telomeric in silico analysis is available from https://www.dkfz.de/en/applied-bioinformatics/telomerehunter/telomerehunter.html. The core computational pipelines used by the PCAWG Consortium for alignment, quality control, and variant calling are available to the public at https://dockstore.org/search?search=pcawg under the GNU General Public License v3.0, which allows for reuse and distribution.