Abstract
Inflammatory bowel disease (IBD), clinically defined as Crohn’s disease (CD), ulcerative colitis (UC), or IBD-unclassified, results in chronic inflammation of the gastrointestinal tract in genetically susceptible hosts. Pediatric onset IBD represents ≥ 25% of all IBD diagnoses and often presents with intestinal stricturing, perianal disease, and failed response to conventional treatments. NOD2 was the first and is the most replicated locus associated with adult IBD, to date. However, its role in pediatric onset IBD is not well understood. We performed whole-exome sequencing on a cohort of 1,183 patients with pediatric onset IBD (ages 0–18.5 years). We identified 92 probands with biallelic rare and low frequency NOD2 variants accounting for approximately 8% of our cohort, suggesting a Mendelian inheritance pattern of disease. Additionally, we investigated the contribution of recessive inheritance of NOD2 alleles in adult IBD patients from a large clinical population cohort. We found that recessive inheritance of NOD2 variants explains ~ 7% of cases in this adult IBD cohort, including ~ 10% of CD cases, confirming the observations from our pediatric IBD cohort. Exploration of EHR data showed that several of these adult IBD patients obtained their initial IBD diagnosis before 18 years of age, consistent with early onset disease. While it has been previously reported that carriers of more than one NOD2 risk alleles have increased susceptibility to Crohn’s Disease (CD), our data formally demonstrate that recessive inheritance of NOD2 alleles is a mechanistic driver of early onset IBD, specifically CD, likely due to loss of NOD2 protein function. Collectively, our findings show that recessive inheritance of rare and low frequency deleterious NOD2 variants account for 7–10% of CD cases and implicate NOD2 as a Mendelian disease gene for early onset Crohn’s Disease.
Subject terms: Medical genomics, Immunogenetics, Sequencing
Introduction
Inflammatory bowel disease (IBD) is a chronic inflammatory condition of the gastrointestinal (GI) tract that arises as part of an inappropriate response to commensal or pathogenic microbiota in a genetically susceptible individual1–4. IBD encompasses Crohn’s Disease (CD); ulcerative colitis (UC); and IBD unclassified (IBDU). The etiology of IBD is complex and has been attributed to defects in a number of cellular pathways including pathogen sensing, autophagy, maintenance of immune homeostasis, and intestinal barrier function, among other processes3–18.
Great effort has been invested into defining the genetic factors that confer IBD susceptibility. To date, > 200 unique loci have been associated with IBD through genome-wide association studies (GWAS), primarily in adult populations19,20. Nearly all the identified susceptibility loci exhibit low effect sizes (ORs ~ 1.0–1.5) individually19, and collectively account for less than 20% of the heritable risk for IBD19–21. These observations support a complex disease model in which common variants of modest effect sizes interact with environmental factors including diet, smoking, and the intestinal microbiome22,23 to give rise to IBD susceptibility.
The earliest and most replicated genetic associations with IBD24–26 correspond to a locus on chromosome 16 that encompasses the nucleotide-binding and oligomerization domain-containing 2 (NOD2) gene, with an average allelic odds ratio across multiple studies of 3.119,20. NOD2 encodes an intracellular microbial sensor that recognizes muramyl dipeptide (MDP) motifs found on bacterial peptidoglycans27,28. Upon activation, NOD2 protein signals through the NF-κB family of proteins29 to modulate transcription of genes encoding pro-inflammatory cytokines IL-8, TNF-α, and IL-1β30–32, among others. Variation in NOD2 accounts for approximately 20% of the genetic risk among CD cases, with three variants—p.R702W (ExAC MAF = 0.0227 across all populations), p.G908R (ExAC MAF = 0.0099), and p.L1007fs (ExAC MAF = 0.0131)—accounting for over 80% of the disease-causing mutations in NOD2 associated with adult CD33, albeit not with UC; and particularly ileal versus colonic CD34. These three “common” risk variants, typically observed in a heterozygous state, are predicted to be loss-of-function alleles that impair NF-κB activation in response to MDP ligands, in vitro28,35–37.
With the assumption that genetic risk has a disproportionate effect over environmental risk in early onset disease, recent studies have focused on pediatric IBD cases (diagnosed < 18y)38–40. Pediatric IBD patients comprise 20–25% of all IBD cases and are typically more clinically severe than adult-onset patients, often exhibiting disease of the upper GI tract, small bowel inflammation, and perianal disease as well as failure to thrive and poor clinical response4,40. Results from GWAS conducted in this group of severely affected patients indicate that associated loci in early onset IBD significantly overlap with adult IBD loci, including both the NOD2 locus and an additional 28 CD-specific loci previously implicated in adult-onset IBD41–43. As the mechanism for these “common” IBD susceptibility loci in the pathogenesis of early onset IBD remains unclear44, we performed whole-exome sequencing and rare variant analysis on a cohort of 1,183 pediatric onset IBD patients to elucidate the role of rare protein coding variation in IBD-associated genes, specifically NOD2, in this disease.
Subjects and methods
Samples
We obtained informed consent for all individuals included in this study or parental informed consent was obtained for minors under 18 years of age. For pediatric IBD, we studied a cohort of 1,183 probands with pediatric onset IBD (ages 0–18.5 years), including their affected and unaffected parents and siblings, where available (total samples = 2,704). Individuals were consented for genetic studies under an IRB-approved protocol by the Toronto Hospital for Sick Children, Canada as part of the NEOPICS initiative (https://www.neopics.org/).
DiscovEHR participants are a subset of the Geisinger MyCode Community Health Initiative. The MyCode Community Health Initiative is a repository of blood, serum, and DNA samples from Geisinger patients that have been consented to participate in research and donate samples for broad research use, including genomic analyses that can be linked to de-identified electronic health record (EHR) information. DiscovEHR participants were consented in accordance with the Geisinger Institutional Review Board approved protocol, Study number 2006–0258.
Helsinki guidelines
All human experiments followed relevant guidelines and regulations according to the Declaration of Helsinki.
Exome sequencing
Sample preparation, whole exome sequencing, and sequence data production for both the pediatric IBD cohort and the DiscovEHR cohort were performed at the Regeneron Genetics Center (RGC) as previously described45. In brief, 1ug of high-quality genomic DNA was used for exome capture utilizing the NimbleGen VCRome 2.1 design. Captured libraries were sequenced on the Illumina HiSeq 2500 platform with v4 chemistry using paired-end 75 bp reads. Exome sequencing was performed such that > 85% of the bases were covered at 20 × or greater. Raw sequence reads were mapped and aligned to the GRCh37/hg19 human genome reference assembly, and called variants were annotated and analyzed using an RGC implemented cloud-based pipeline. Briefly, variants were filtered based on their observed minor allele frequencies at a < 2% cutoff using the internal RGC database and other public population control databases to filter out common polymorphisms and high frequency, likely benign variants in consideration of disease prevalence.
DiscovEHR statistical analyses
For NOD2 locus-specific statistical analyses in the DiscovEHR cohort, individuals with ICD diagnoses in their EHR consistent with IBD and carriers of NOD2 variants were annotated and filtered using the same pipeline as for the pediatric IBD cases. Odds ratios for all genetic models (additive, recessive and genotypic) were calculated using Fisher’s exact test with no covariates.
For large-scale association analyses, variants were annotated with snpEff using Ensembl 85 gene definitions46. Gene definitions were restricted to transcripts with annotated start and stop codons, totaling 19,467 protein-coding genes. Predicted loss-of-function (pLoF) variants were defined as any of the following: variants leading to a premature stop codon, loss of a start codon, or loss of a stop codon; single-nucleotide variants or indels disrupting canonical splice donor or acceptor sites; and frame-shifting indels predicted to result in premature stop codons. Phasing of putative compound heterozygotes was performed as previously described for this cohort47 using a combination of familial relationship based phasing48 and population allele frequency based phasing with EAGLE49. Biallelic pLoF and predicted deleterious missense variants with a MAF < 5% in the discovery set of 58,138 European ancestry individuals were aggregated at a gene level. Variants were aggregated for gene burden tests in two ways as previously described45,50: pLoFs only and pLoFs plus missense variants (M3) predicted to be deleterious (pdNS) by five different bioinformatic prediction algorithms for functional effects, namely SIFT51, LRT52, MutationTaster53, PolyPhen2 HumDiv, and PolyPhen2 HumVar54. Genotypes were coded as follows: homozygous reference as 0, heterozygotes as 1, and homozygous alternative or compound heterozygous as 2. PLINK 1.955 was used to run Firth logistic regression under both additive and recessive models using the ICD10 K50 phenotype, which is the ICD10 diagnosis code for Crohn’s disease [regional enteritis], (Ncases = 613 versus Ncontrols = 54,802). Phenome-wide associations were performed using all ICD-10 disease diagnosis codes available for the DiscovEHR dataset.
Results
An initial analysis of the exome sequencing data for pathogenic and expected pathogenic variants in genes known to cause monogenic forms of IBD in all probands from our pediatric IBD cohort identified 40 rare variants in 31 probands56. Additionally, we performed trio-based analysis of 492 complete trios using a proband-based analytical pipeline to identify all recessive (compound heterozygous and homozygous), X-linked, and de novo variants of interest in the affected probands. In our initial analyses, we identified 10 families with recessive (compound heterozygous or homozygous), rare variants (2% ≤ MAF) in NOD2, all with a diagnosis of CD. We observed that some of the rare variants in these probands were inherited in trans from previously-reported CD risk alleles, mainly the p.G908R missense variant. We identified two individuals who are compound heterozygous for the p.G908R risk allele in trans with a less common NOD2 CD risk variant (p.N852S) in one case and a novel truncating indel (p.S506Vfs*73) in the second case (Supplementary Table 1, Fam008 and Fam009). The observation of a CD-associated NOD2 risk allele in trans from other rare or novel alleles led us to survey the rest of the probands, including singletons and those part of incomplete trios, for recessive inheritance, either in a homozygous or compound heterozygous manner, of NOD2 variants, but expanding our allelic range to low-frequency variants (2% ≤ MAF ≤ 5%). Through this approach we identified 108 probands with putative recessive NOD2 variants. Visual inspection of sequence reads and orthogonal confirmation through Sanger sequencing excluded 13 probands with variants inherited in cis from an unaffected parent or heterozygous variants that were initially called as homozygous due to low coverage of the region and skewed allelic balance. Of note, we identified 5 probands carrying p.L1007fs and p.M863V risk variants, 4 of which were confirmed to occur in cis and were inherited from an unaffected parent. The remaining case with p.L1007fs and p.M863V was a singleton and thus phase could not be determined. These two variants segregate in cis within the same haplotype, as confirmed by segregation within the trios and as previously observed57. Therefore, we excluded these 4 probands from our final count of recessively-inherited NOD2 variants. Similarly, we identified 3 probands from 3 complete trios segregating the p.S431L and p.V793M reported risk variants in cis inherited from an unaffected carrier parent; these probands were also excluded. Three additional probands were excluded on the basis of a re-evaluation of the phenotype that excluded a clinical diagnosis of IBD.
Thus, we identified 92 probands with confirmed recessive NOD2 variants within our pediatric onset IBD cohort, none of which had variants of interest in known monogenic IBD associated genes. These included: 25 probands carrying homozygous variants, 41 probands with confirmed compound heterozygous variants, and an additional 26 singleton probands with putative compound heterozygous variants where phasing could not be performed (Supplementary Table 1, Supplementary Fig. 1). The majority of the compound heterozygous individuals (65/67) carry a known NOD2 CD-risk allele in addition to either another known NOD2 CD-risk allele or a novel NOD2 variant, including some truncating loss-of-function variants supporting loss or impaired function of NOD2 in the pathophysiology of CD6. In total, 92 of 1,183 (7.8%) of the probands in our pediatric onset IBD cohort conformed to a recessive, Mendelian inheritance mode for NOD2 rare and low frequency (MAF ≤ 5%) deleterious variants (Table 1, Fig. 1, Supplementary Table 1, and Supplementary Table 4).
Table 1.
Mutation spectrum of recessive NOD2 variants in an EO-IBD cohort.
| NOD2 variant | # EO-IBD probands | Mean age (range) | % CD Dx | Tissue involvement | ||
|---|---|---|---|---|---|---|
| Colon (%) | Ileum (%) | Perianal (%) | ||||
| Compound heterozygous | ||||||
| Rare/rare | 4 T; 1S | 12.5 (9.0–14.6) | 88.9 | 80.0 | 60.0 | 40.0 |
| Common/rare | 1Q; 9 T; 6D; 14S | 12.6 (5.5–16.5) | 93.1 | 57.1 | 82.1 | 25.0 |
| Common/common | 17 T; 5D; 10S | 11.8 (2.1–18.5) | 90.6 | 56.3 | 90.6 | 25.0 |
| Homozygous | ||||||
| Rare | 1D; 2S | 9.6 (4.2–15.1) | 66.7 | 33.3 | 33.3 | 33.3 |
| p.R702W | 1Q; 2 T; 3D; 1S | 11.7 (5.8–13.8) | 100 | 85.7 | 57.1 | 42.9 |
| p.G908R | 2 T; 1D; 2S | 10.6 (7.7–13.7 | 80.0 | 40.0 | 40.0 | 0.0 |
| p.L1007fs | 4 T; 2D; 4S | 12.1 (5.9–13.4) | 100 | 60.0 | 90.0 | 20.0 |
Common NOD2 variants refer to the three main low-frequency Crohn’s Disease risk variants p.R702W, p.G908R, and p.L1007fs; Rare NOD2 variants refer to other low-frequency variants (MAF ≤ 5%).
Q, quartet; T, trio; D, duo; S, singleton; Dx, diagnosis.
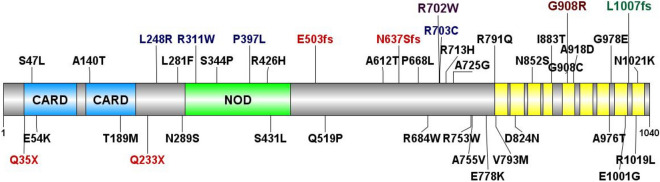
Figure 1.
Mutation spectrum of NOD2 in inflammatory bowel disease (IBD) patients. NOD2 variation identified in patients with pediatric early onset IBD (upper) and adult IBD cohort from the RGC-GHS DiscovEHR collaboration (lower). Variants in blue were observed in both cohorts; variants in red are predicted loss-of-function that result in nonsense mediated decay. The three “common” low-frequency Crohn’s Disease risk variants are highlighted: R702W (purple), G908R (brown), and L1007fs (green). Also depicted are the NOD2 protein structural domains: two caspase activation and recruitment domains (CARD), a nucleotide binding and oligomerization (NOD) domain, and leucine rich repeat domains in yellow.
The 92 pediatric patients homozygous for NOD2 mutations were predominantly male (71%) with a median age at diagnosis of 12.5 years (Supplementary Table 1). At diagnosis, 83% displayed diagnostic features of Crohn’s disease. 23% of the cohort displayed a constellation of extra-intestinal manifestations, mainly large joint arthritis, chronic recurrent multifocal osteomyelitis, recurrent fevers, erythema nodosum, and pyoderma gangrenosum. Only 6% of the cohort showed significant perianal disease (namely fistulae and abscesses; skin tags and fissures were not considered as perianal disease) (Supplementary Table 1). Per the Montreal classification of IBD58, 44% of the overall cohort of patients presented with ileal disease at diagnosis (L1). 25% presented with ileocolonic disease (L3) and 10% displayed features of colonic inflammation only (L2). Isolated upper disease was only present in 2% of the cohort (L4). We observed a progression of ileal disease in 21% (18.5% stricturing; 2.5% penetrating) with 21% requiring a resection. On review of the Crohn’s disease patients only, 50% displayed L1 disease (terminal ileal +/− limited cecal disease), 32.9% L3 (ileocolonic) disease; 86.8% B1 (non-stricturing, non-penetrating) disease, 10.5% B2 (stricturing) disease. Of the 92 patients with biallelic variants in NOD2, 42.4% (n = 39) had ileal disease, 27.17% (n = 25) had ileocolonic disease, and 11.95% (n = 11) had colonic inflammation only (Supplementary Table 1).
Given the substantial contribution of recessive NOD2 variants to CD in our pediatric onset IBD cohort and the known contribution of NOD2 to adult CD, we next investigated the contribution of NOD2 recessivity in a large clinical population. For this, we examined a cohort of adult IBD patients from the Geisinger-Regeneron DiscovEHR collaboration45. A key feature of the DiscovEHR study is the ability to link genomic sequence data to de-identified electronic health records (EHRs). Within this cohort, we identified 984 patients (of 51,289 total sequenced DiscovEHR patient-participants) with a diagnosis of IBD, defined as having a problem list entry or an encounter diagnosis entered for two separate clinical encounters on separate calendar days for the ICD-9 codes 555* (Regional enteritis) or 556* (Ulcerative enterocolitis) or ICD-10 K50* (Crohn's disease [regional enteritis]) or K51* (Ulcerative colitis). For our analysis, we surveyed all instances of homozygous NOD2 rare and low frequency variants (MAF ≤ 5%); the same parameters applied to our pediatric IBD probands. Among patients with an IBD diagnosis, we identified 18 individuals who are either homozygous for the p.R702W risk allele (N = 10) or homozygous for the p.L1007fs allele (N = 8) (Table 2, Supplementary Fig. 2). We did not identify any p.G908R homozygous individuals with an IBD diagnosis in this cohort. Next, we looked for instances of putative compound heterozygosity among these adult IBD DiscovEHR patients. First, we searched for occurrences of two or more of the three most prevalent NOD2 risk alleles (p.R702W, p.G908R, or p.L1007fs) in these individuals. We identified putative compound heterozygosity for the three main CD risk alleles, p.R702W/p.G908R (N = 6), p.G908R/p.L1007fs (N = 5), and p.R702W/p.L1007fs (N = 11) (Table 2). We also observed instances of putative compound heterozygosity for each of the three main CD risk alleles along with either a rarer CD risk allele or a novel allele or two rare alleles in trans (N = 24), parallel to the findings in our pediatric IBD cohort. Using familial relationships and pedigree reconstruction47, we were able to confirm appropriate segregation for 32 of the 64 DiscovEHR recessive NOD2 variant carriers with IBD, including trans inheritance in 13 putative compound heterozygotes (Supplementary Fig. 2). The other 32 were singleton cases where phase could not be confirmed. Overall, we identified 64 homozygous or putative compound heterozygous NOD2 variant carriers in the DiscovEHR IBD cohort, accounting for 6.5% of patients with an IBD diagnosis in this clinical population (Fig. 1, Supplementary Table 4).
Table 2.
Mutation spectrum of recessive NOD2 variants in the RGC-GHS DiscovEHR adult IBD cohort.
| NOD2 variants | # IBD patients | Mean age (range) | # CD Dx | % CD Dx | # Male | % Male | # Perianal | % Perianal |
|---|---|---|---|---|---|---|---|---|
| Homozygous | ||||||||
| p.R702W | 10 | 47.3 (16.0–76.3) | 10 | 100 | 5 | 50.0 | 1 | 10.0 |
| p.G908R | 0 | – | – | – | – | – | – | – |
| p.L1007fs | 8 | 40.25 (11.0–75.0) | 8 | 100 | 4 | 50.0 | 3 | 37.5 |
| Compound heterozygous | ||||||||
| Common/common | ||||||||
| p.R702W/p.G908R | 6 | 48.5 (11.4–54.2) | 3 | 50.0 | 5 | 83.3 | 1 | 16.7 |
| p.R702W/p.L1007fs | 11 | 38.5 (21.5–69.2) | 8 | 72.7 | 3 | 27.3 | 2 | 18.2 |
| p.G908R/p.L1007fs | 5 | 35.2 (20.0–52.4) | 4 | 80.0 | 2 | 40.0 | 0 | 0.0 |
| Common/rare | 16 | 40.3 (10.8–66.1) | 8 | 50.0 | 8 | 50.0 | 6 | 37.5 |
| Rare/rare | 8 | 60.9 (30.8–78.7) | 4 | 50.0 | 3 | 37.5 | 1 | 12.5 |
Common NOD2 variants refer to the three main low-frequency Crohn’s Disease risk variants p.R702W, p.G908R, and p.L1007fs; Rare NOD2 variants refer to other low-frequency variants (MAF ≤ 5%).
Dx, diagnosis.
We were also able to evaluate longitudinal de-identified medical records for all patients within the DiscovEHR IBD cohort. According to their EHR data, 21 patients received diagnoses of both UC and CD. To clarify these diagnoses, we performed manual evaluation of EHR information (which includes demographics, encounter and problem list diagnosis codes, procedure codes, and medications) for all 64 homozygous or compound heterozygous NOD2 patients with an IBD diagnosis. Through this review, 6 homozygotes exhibited a conflicting diagnosis of CD, of which 5 were resolved as CD and 1 could not be defined; 16 compound heterozygotes exhibited a conflicting diagnosis of CD of which 6 were resolved as CD and 10 were resolved as UC (Supplementary Table 3). In total, we found that 17/18 (94.4%) of homozygous NOD2 individuals and 33/46 (71.7%) compound heterozygous had a diagnosis of CD and that 9.9% of all CD cases in this cohort could be attributed to homozygous or compound heterozygous variants in NOD2. We next investigated age of disease onset using the first recorded date of an IBD diagnosis in the EHR. We identified 6 carriers of recessive NOD2 variants (9.4% of our recessive NOD2 patients with IBD) who were diagnosed with IBD prior to 18 years of age. We also identified additional 11 carriers of recessive NOD2 variants diagnosed with IBD prior to age 30 years, which is at or below the average age of IBD diagnosis59 and is consistent with earlier disease onset (Supplementary Table 3). Of note, our DiscovEHR cohort data extends to a median of 14 years (and maximum of 25 years) of electronically recorded medical information, concurrent with the adoption of the EHR by the Geisinger Health System. Since 72.4% of our cohort is currently over the age of 50 years, we cannot determine whether the age of onset for IBD occurred prior to the first electronically recorded date of an IBD diagnosis for many recessive NOD2 patients; thus it is possible that other individuals with homozygous or compound heterozygous variants in NOD2 might have had pediatric-onset disease that was not captured in the EHR.
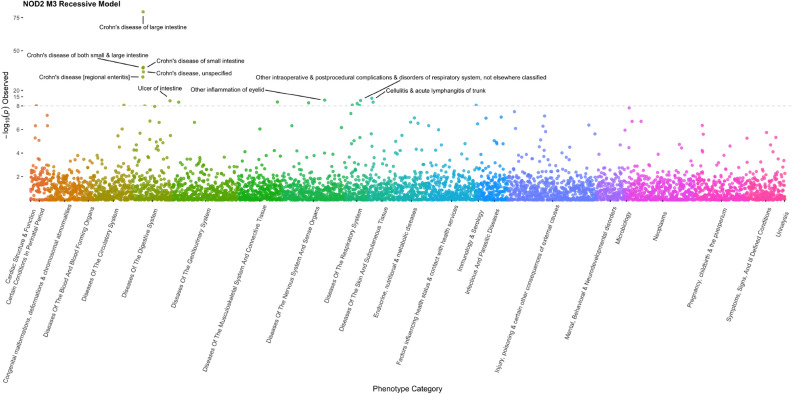
Incidentally, our manual evaluation of the EHR data for these individuals also revealed that 75% of the IBD patients had a diagnosis record of anemia in their history. In about 58% of these cases the anemia diagnosis was given concurrent or before the first recorded diagnosis of IBD, with an average of 2.26 years prior. This observation is consistent with previous reports of anemia as an important yet underappreciated and undertreated comorbidity in IBD60,61, but also suggests that anemia may be an early indicator of IBD onset. Interestingly, 16 of 48 individuals homozygous for the p.L1007fs variant that do not have a diagnosis of IBD and for which we were able to review their EHR information had a diagnosis of anemia in their chart and 11 of them had diagnosis codes related to gastrointestinal complaints. To further assess whether NOD2 genotype status associated with other phenotypes, we performed a PheWAS analysis using all ICD codes recorded in the EHR of NOD2 homozygous and compound heterozygous individuals. This analysis showed that NOD2 recessivity significantly and specifically associates with Crohn’s disease (Fig. 2).
Figure 2.
Phenome wide association analysis (PheWAS) of ICD diagnostic codes with biallelic recessive genotypes of NOD2. Analysis shows that NOD2 recessive status significantly associates with Crohn’s disease and related diagnoses.
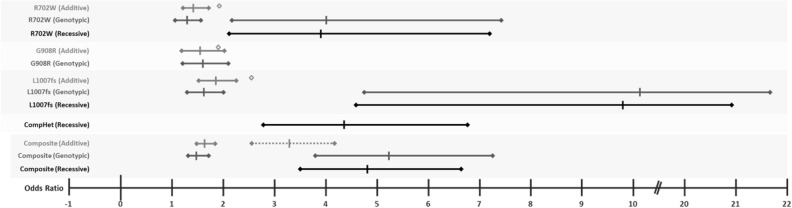
Next, given the recessive inheritance of NOD2 variants observed in both our pediatric onset and adult IBD cohorts, we estimated the disease risk for the three main known CD risk alleles (p.R702W, p.G908R, and p.L1007fs) in our adult IBD case cohort and their effect sizes using additive, genotypic, and recessive genetic models. Under an additive model, we observed similar effect sizes for each of the 3 variants [OR = 1.43 (1.20–1.71 95%CI, P-value 4.63 × 10–5) for p.R702W; OR = 1.56 (1.18–2.06 95%CI, P-value 1.54 × 10–3) for p.G908R; and OR = 1.84 (1.50–2.26 95%CI, P-value 1.69 × 10–9) for p.L1007fs], consistent with previously reported low to moderate effect sizes for each allele by GWAS20 and data available in the IBD Exomes Portal62 (Table 3, Fig. 3). However, for the two risk alleles with homozygous cases, in the genotypic model – which estimates distinct effect sizes for heterozygous and homozygous carriers – we observe substantially larger effects in homozygotes versus heterozygotes for the p.R702W variant (Het OR = 1.30 [1.06–1.58 95%CI], P-value 8.77 × 10–3, versus Hom OR = 4.02 [2.17–7.45 95% CI], P-value 6.86 × 10–6) and the p.L1007fs variant (Het OR = 1.63 [1.30–2.04 95%CI], P-value 1.67 × 10–5, versus Hom OR = 10.15 [4.75–21.69 95% CI], P-value 1.38 × 10–12). We also calculated the effect sizes using a recessive model for these two variants and the 22 compound heterozygotes carrying any combination of the 3 CD risk alleles. We found that recessive effect sizes for the p.R702W and p.L1007fs variants were similar to those observed under the homozygous genotypic model (OR = 3.91 [2.11–7.24 95% CI], P-value 2.86 × 10–6, and OR = 9.81 [4.59–20.94 95% CI], P-value 3.80 × 10–13, respectively) (Table 3, Fig. 2). Additionally, we calculated the relative risk for the identified putative compound heterozygous (pCHET) individuals under a recessive model. We observed that the effect size for the compound heterozygotes was also significant (OR = 4.35 [2.80–6.75 95% CI], P-value = 8.14 × 10–13), consistent with our previous observations (Table 3, Fig. 2). The calculated combined contribution of the 3 CD risk alleles under the different genetic models was as follows: additive (OR = 1.64 [1.45–1.86 95%CI], P-value 4.58 × 10–15), genotypic (Het OR = 1.49 [1.28–1.73 95%CI], P-value 2.75 × 10–7, versus Hom OR = 5.24 [3.77–7.27 95% CI], P-value 4.31 × 10–22), and recessive (OR = 4.81 [3.47–6.67 95% CI], P-value = 1.63 × 10–25) (Table 3, Fig. 3).
Table 3.
OR calculations for 3 NOD2 CD risk alleles (p.R702W, p.G908R, p.L1007fs), composite, and compound heterozygous combinations in the DiscovEHR cohort.
| NOD2 variant | DiscovEHR MAF | DiscovEHR controls (N = 50,305) | DiscovEHR IBD cases (N = 984) | Additive model OR [95% CI] (P-value) | Genotypic model (heterozygous) OR [95% CI] (P-value) | Genotypic model (homozygous) OR [95% CI] (P-value) | Recessive model OR [95% CI] (P-value) | ExAC MAF | IBD exomes OR (P-value) |
|---|---|---|---|---|---|---|---|---|---|
| p.R702W | 0.050 | Het = 4727; Hom = 145 | Het = 116; Hom = 11 | 1.43 [1.20–1.71] (4.63 × 10–5) | 1.30 [1.06–1.58] (0.008765) | 4.02 [2.17–7.45] (6.86 × 10–6) | 3.91 [2.11–7.24] (2.86 × 10–6) | 0.035 | 1.92 (< 1 × 10–16) |
| p.G908R | 0.017 | Het = 1683; Hom = 16 | Het = 52; Hom = 0 | 1.56 [1.18–2.06] (0.001544) | 1.61 [1.21–2.13] (0.00087) | NA | NA | 0.012 | 1.91 (< 1 × 10–16) |
| p.L1007fs | 0.029 | Het = 2808; Hom = 42 | Het = 86; Hom = 8 | 1.84 [1.50–2.26] (1.69 × 10–9) | 1.63 [1.30–2.04] (1.67 × 10–5) | 10.15 [4.75–21.69] (1.38 × 10–12) | 9.80 [4.59–20.94] (3.80 × 10–13) | 0.018 | 2.57 (< 1 × 10–16) |
| Compound Het | N = 263 | N = 22 | 4.35 [2.80–6.75] (8.14 × 10–13) | ||||||
| Composite NOD2 | Het = 8955; Rec = 450 | Het = 232; Rec = 41 | 1.64 [1.45–1.86] (4.58 × 10–15) | 1.49 [1.28–1.73] (2.75 × 10–7) | 5.24 [3.77–7.27] (4.31 × 10–22) | 4.81 [3.47–6.67] (1.63 × 10–25) | |||
| 3.29 [2.56–4.23] (2 alleles predicted) |
There were no homozygotes for the p.G908R variant affected with IBD in our cohort; therefore, no genotypic homozygous and recessive ORs could be calculated. The ‘Composite NOD2′ calculations account for all alleles and genotypes for the 3 CD risk variants in the different genetic models.
Figure 3.
Graphical representation of Odds Ratio (OR) point estimates and 95% confidence intervals (CI) for the three main CD risk alleles (p.R702W, p.G908R, p.L1007fs) under additive, genotypic, and recessive genetic models (corresponding to values in Table 3). The dotted line in the Composite panel depicts the calculated CI with corresponding calculated OR for 2 alleles under an additive genetic model; of note the point estimate (2xOR) is outside of the 95% CI for the Composite genotypic homozygous and recessive models. Diamonds correspond to estimated OR values for these same variants in the IBD Exomes Browser49; no confidence intervals are provided.
Subsequently, we combined all heterozygous, homozygous, and phased compound heterozygous predicted loss-of-function (pLoF) and predicted deleterious missense variants in NOD2 with a MAF ≤ 5% including the 3 risk alleles to calculate the CD risk using a burden test under additive and recessive models. The pLoF only burden analysis was significant under both the additive (P-value 5.5X10–20) and recessive (P-value = 2.67 × 10–19) models, however the risk was much higher under the recessive model (OR = 20.74 [10.70 – 40.20 95%CI] compared to the additive model (OR = 2.69 [2.18–3.32 95%CI]) (Table 4). The pLoF and predicted deleterious missense variant burden analysis showed similar results with a significantly higher risk under the recessive model: additive (OR = 2.38 [2.01–2.82]95%CI, P-value 4.60 × 10–24) and recessive (OR = 13.15 [8.50–20.37]95%CI, P-value 7.21 × 10–31) (Table 4). Collectively, these analyses show substantially larger effects for NOD2 homozygotes and compound heterozygotes than heterozygotes only and indicate that the genetic contribution of NOD2 alleles, in a subset of Crohn’s disease patients, is consistent with a recessive disease model.
Table 4.
Comparison of additive and recessive models for heterozygous, homozygous, and phased compound heterozygous pLoF and predicted deleterious missense variants for NOD2 variants (MAF < 5%) for Crohn’s Disease Risk in DiscovEHR.
| NOD2 gene burden variant class | Additive OR [95% CI] | Additive P-value | Recessive OR [95% CI] | Recessive P-value | Controls (ref/het/Hom) | Cases (ref/het/Hom) |
|---|---|---|---|---|---|---|
| pLoF only | 2.69 [2.18–3.32] | 5.50 × 10–20 | 20.74 [10.70–40.20] | 2.67 × 10–19 | 51,501/3254/47 | 529/73/11 |
| pLoF and predicted deleterious missense | 2.38 [2.01–2.82] | 4.60 × 10–24 | 13.15 [8.50–20.37] | 7.21 × 10–31 | 48,253/6388/161 | 474/115/24 |
Discussion
We use the term inflammatory bowel disease (IBD) throughout to encompass diagnoses of both Ulcerative Colitis and Crohn’s disease in the DiscovEHR cohort, which is similar to the referral diagnosis of the pediatric patients where some had diagnoses of ulcerative colitis, Crohn’s disease, or IBD unspecified (Table S1). Furthermore, prior to the release of ICD-10 codes, there was no specific diagnosis code for Crohn’s disease, as it was coded as ‘regional enteritis’ (ICD-9 555), lending itself to confusion and misdiagnoses. The DiscovEHR IBD cohort is not intended to be a ‘pure’ Crohn’s disease cohort but rather a representative sample of the adult population that is diagnosed with IBD. Both, the pediatric and adult cohorts reflect the clinical heterogeneity of patients diagnosed with IBD and the challenges of the clinical and molecular diagnosis of this disease.
Our observations are in line with previous analyses and meta-analyses of CD cohorts where individuals carrying any one of the main three CD associated risk alleles (p.R702W, p.G908R, or p.L1007fs) have 2–fourfold increased risk for developing CD63, whereas carriers of two or more of the same NOD2 variants have a 15–40 fold increased risk for developing CD33,64,65, exhibiting disease of the terminal ileum34, and earlier diagnosis (by an average of 3 years)33. Our observations support these studies but highlight a subset of IBD cases molecularly defined by recessive inheritance of NOD2 alleles that exhibit markedly increased risk for CD with significantly earlier age of onset (mean age of onset among recessive NOD2 carriers in the DiscovEHR IBD cohort: 43.4y; mean age of onset in the DiscovEHR IBD cohort: 51.5y; P-value: 4.0X10–4 by unpaired t test).
Further, while we observe a low effect size for single allele carriers, based on our allelic effect size calculations for each of the 3 main CD risk alleles in our DiscovEHR cohort (Table 3, Fig. 3), we hypothesize that homozygous and compound heterozygous NOD2 individuals included in large IBD GWAS cohorts have likely contributed to a large proportion of the relative risk calculations for IBD, specifically for CD, under additive models, and that homozygous effect sizes have been largely underappreciated or underreported. It is possible that stratification or conditional statistical analysis of these large and heterogeneous cohorts based on NOD2 genotypes may increase power to detect other loci that contribute to IBD.
While our observations strongly support recessive inheritance of NOD2 variants as a driver of early onset Crohn’s disease, we observed incomplete penetrance, as evidenced by homozygous or compound heterozygous NOD2 variant carriers that do not have a clinical presentation of IBD65–67. Penetrance and expressivity are two major genetic concepts that play into the onset of the phenotype and the clinical presentation of monogenic diseases68. In the case of IBD, penetrance is known to be incomplete and clinical presentation is extremely variable. Further, the contribution of additional environmental triggers that may enhance disease onset and/or severity in an already genetically-compromised individual should not be underestimated, especially considering that the loss of epithelial barrier function occurring during IBD allows for host exposure to up to 1014 gut microbiota69,70. Even in cases of monogenic IBD, such as IL-10 receptor deficiency71–73, intestinal flora are required for disease presentation in murine disease models74–76. Furthermore, variation in genes involved in NOD2-dependent signaling pathways, including XIAP77–79 and TRIM2280, result in Mendelian forms of IBD. For XIAP, and most likely TRIM22, viral triggers are required for disease onset and progression, and XIAP mutations have variable penetrance, with only a small percentage of XIAP-deficiency patients developing CD (age of onset between 3 months and 40 years64). As NOD2-deficient hosts are more susceptible to the pathogenic effects of a changing intestinal microenvironment81, the contribution of either discrete or continuous gene-environment exposures may further explain heterogeneity in onset and presentation of disease for genetically-sensitized recessive NOD2 carriers.
Given the wide variability in clinical presentation of IBD, we cannot exclude the possibility that recessive NOD2 carriers exhibit subclinical phenotypes not formally diagnosed as IBD or that they may eventually develop IBD. It is additionally possible that recessive NOD2 carriers in the DiscovEHR cohort have a diagnosis of IBD that has not been captured in the EHR. Detailed investigation into the medical histories of recessive NOD2 carriers may shed light on this variable expressivity or incomplete capture of medical information. We also cannot exclude the possibility that recessive NOD2 carriers possess additional genes or alleles that either contribute to disease onset and severity or, alternatively, provide protection or reduced expressivity of the phenotype. Identification of these genetic modifiers warrants future investigation both to unveil additional IBD-risk associated loci for early onset UC and CD cases and to identify protective genes and alleles that can be used to derive therapeutic avenues for IBD treatment and management.
In summary, in a cohort of 1,183 pediatric and early onset IBD patients, we report recessive inheritance of rare and low frequency variants in NOD2 accounting for about 8% of probands. We assessed the contribution of NOD2 recessive inheritance in a broader, heterogeneous cohort of adult IBD patients, similar to those recruited for GWAS, and found that recessive inheritance of variants in NOD2 account for 6.5% of these IBD patients, including 9.9% of CD cases. Thus, recessive inheritance of rare and low frequency NOD2 variants explain a substantial proportion of CD cases in a pediatric cohort and a large clinical population, with significantly earlier age of disease onset. Consistently, both pediatric and adult CD exhibit a broad spectrum of clinical presentation, suggesting a shared etiology across age groups, at least in the subgroup defined by recessive NOD2-driven CD. Our findings indicate that deleterious NOD2 variants should be considered as strong predictors of IBD-CD onset and implicate NOD2 as a Mendelian disease gene for early onset IBD, specifically for a molecularly defined subset of Crohn’s disease patients.
Supplementary Information
Acknowledgements
We are thankful to the patients and families who participated in this study. A.M.M. is funded by a CIHR – Operating Grant (MOP119457) and the Leona M. and Harry B. Helmsley Charitable Trust to study VEOIBD.
Author contributions
J.E.H., N.W., J.S., E.C., N.G., R.M., C.V.H., G.W., and C.G.-J. performed data analyses and interpretation. K.F., A.K.K., and A.B. provided project support. J.G.R. and J.D.O. contributed with sequence data generation. A.R.S., A.G., O.G, A.M.M., and C.G.-J. contributed to study design and execution. C.G-J. wrote the main manuscript text and elaborated Figs. 1 and 3. N.G. prepared Fig. 2. A.R.S., A.G., A.M.M., and C.G.-J. contributed to manuscript revision and editing. All authors reviewed the manuscript.
Competing interests
J.E.H., J.S., C.V.H., A.K.K., J.G.R., J.D.O., A.R.S., A.B., and C.G-J are full-time employees of the Regeneron Genetics Center from Regeneron Pharmaceuticals, Inc. and receive stock options as part of compensation. All other authors have no conflicts to disclose.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aleixo M. Muise, Email: aleixo.muise@utoronto.ca
Claudia Gonzaga-Jauregui, Email: clau.gonzagajauregui@regeneron.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-84938-8.
References
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N. Engl. J. Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Limbergen J, Wilson DC, Satsangi J. The genetics of Crohn's disease. Annu. Rev. Genomics Hum. Genet. 2009;10:89–116. doi: 10.1146/annurev-genom-082908-150013. [DOI] [PubMed] [Google Scholar]
- 3.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 4.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu. Rev. Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho JH, Abraham C. Inflammatory bowel disease genetics: Nod2. Annu. Rev. Med. 2007;58:401–416. doi: 10.1146/annurev.med.58.061705.145024. [DOI] [PubMed] [Google Scholar]
- 7.Abraham C, Cho JH. Functional consequences of NOD2 (CARD15) mutations. Inflamm. Bowel Dis. 2006;12:641–650. doi: 10.1097/01.MIB.0000225332.83861.5f. [DOI] [PubMed] [Google Scholar]
- 8.Chu H, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 10.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abraham C, Cho JH. IL-23 and autoimmunity: new insights into the pathogenesis of inflammatory bowel disease. Annu Rev Med. 2009;60:97–110. doi: 10.1146/annurev.med.60.051407.123757. [DOI] [PubMed] [Google Scholar]
- 13.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cattin AL, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol. Cell Biol. 2009;29:6294–6308. doi: 10.1128/MCB.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muise AM, et al. Polymorphisms in E-cadherin (CDH1) result in a mis-localised cytoplasmic protein that is associated with Crohn's disease. Gut. 2009;58:1121–1127. doi: 10.1136/gut.2008.175117. [DOI] [PubMed] [Google Scholar]
- 17.Scharl M, et al. Protection of epithelial barrier function by the Crohn's disease associated gene protein tyrosine phosphatase n2. Gastroenterology. 2009;137:2030–2040. doi: 10.1053/j.gastro.2009.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGovern DP, Kugathasan S, Cho JH. Genetics of Inflammatory Bowel Diseases. Gastroenterology. 2015;149:1163–1176. doi: 10.1053/j.gastro.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 23.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 25.Hugot JP, et al. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996;379:821–823. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 26.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 27.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 28.Inohara, N. et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 278, 5509–5512 (2003). [DOI] [PubMed]
- 29.Van Limbergen J, Radford-Smith G, Satsangi J. Advances in IBD genetics. Nat. Rev. Gastroenterol. Hepatol. 2014;11:372–385. doi: 10.1038/nrgastro.2014.27. [DOI] [PubMed] [Google Scholar]
- 30.Inohara, C., McDonald, C., & Nunez, G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem.74, 355–383 (2005). [DOI] [PubMed]
- 31.Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat. Rev. Immunol. 2006;6:9–20. doi: 10.1038/nri1747. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe T, Kitani A, Strober W. NOD2 regulation of Toll-like receptor responses and the pathogenesis of Crohn's disease. Gut. 2005;54:1515–1518. doi: 10.1136/gut.2005.071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesage S, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am. J. Hum. Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleynen I, et al. Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387:156–167. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonen DK, et al. Crohn's disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology. 2003;124:140–146. doi: 10.1053/gast.2003.50019. [DOI] [PubMed] [Google Scholar]
- 36.Chamaillard M, et al. Gene-environment interaction modulated by allelic heterogeneity in inflammatory diseases. Proc. Natl. Acad. Sci. USA. 2003;100:3455–3460. doi: 10.1073/pnas.0530276100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Heel DA, et al. Muramyl dipeptide and toll-like receptor sensitivity in NOD2-associated Crohn's disease. Lancet. 2005;365:1794–1796. doi: 10.1016/S0140-6736(05)66582-8. [DOI] [PubMed] [Google Scholar]
- 38.Moran CJ, Klein C, Muise AM, Snapper SB. Very early-onset inflammatory bowel disease: gaining insight through focused discovery. Inflamm. Bowel Dis. 2015;21:1166–1175. doi: 10.1097/MIB.0000000000000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosen MJ, Dhawan A, Saeed SA. Inflammatory bowel disease in children and adolescents. JAMA Pediatr. 2015;169:1053–1060. doi: 10.1001/jamapediatrics.2015.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crowley E, Muise A. Inflammatory bowel disease: what very early onset disease teaches us. Gastroenterol. Clin. N. Am. 2018;47:755–772. doi: 10.1016/j.gtc.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Cutler, D. J. et al. Dissecting allele architecture of early onset IBD using high-density genotyping. PLoS One. 10, e0128074: 10.1371/journal.pone.0128074 (2015). [DOI] [PMC free article] [PubMed]
- 42.Imielinski M, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kugathasan S, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genetics. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul T, et al. Distinct phenotype of early childhood inflammatory bowel disease. J. Clin. Gastroenterol. 2006;40:583–586. doi: 10.1097/00004836-200608000-00004. [DOI] [PubMed] [Google Scholar]
- 45.Dewey FE, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016 doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 46.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staples J, et al. Profiling and leveraging relatedness in a precision medicine cohort of 92,455 Exomes. Am. J. Hum. Genet. 2018;102:874–889. doi: 10.1016/j.ajhg.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staples J, et al. PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loh PR, et al. Reference-based phasing using the haplotype reference consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 52.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 54.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang, C. C., et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 4, 7. 10.1186/s13742-015-0047-8 (2015). [DOI] [PMC free article] [PubMed]
- 56.Crowley E, et al. Prevalence and clinical features of inflammatory bowel diseases associated with monogenic variants, identified by whole-exome sequencing in 1000 children at a single center. Gastroenterology. 2020;158:2208–2220. doi: 10.1053/j.gastro.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silverberg MS, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a working party of the 2005 montreal world congress of gastroenterology. Can. J. Gastroenterol. 2005;19(Suppl A):5A–36A. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 59.Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 60.Mücke V, Mücke MM, Raine T, Bettenworth D. Diagnosis and treatment of anemia in patients with inflammatory bowel disease. Ann. Gastroenterol. 2017;30:15–22. doi: 10.20524/aog.2016.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jimenez KM, Gasche C. Management of iron deficiency anaemia in inflammatory bowel disease. Acta Haematol. 2019;142:30–36. doi: 10.1159/000496728. [DOI] [PubMed] [Google Scholar]
- 62.IBD Exomes Portal, Cambridge, MA. http://ibd.broadinstitute.org/. Last accessed: December 2019.
- 63.Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am. J. Gastroenterol. 2004;99:2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 64.Hugot JP, et al. Prevalence of CARD15/NOD2 mutations in Caucasian healthy people. Am. J. Gastroenterol. 2007;102:1259–1267. doi: 10.1111/j.1572-0241.2007.01149.x. [DOI] [PubMed] [Google Scholar]
- 65.Uhlig HH, et al. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology. 2014;147:990–1007. doi: 10.1053/j.gastro.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kammermeier J, et al. Phenotypic and genotypic characterisation of inflammatory bowel disease presenting before the age of two years. J. Crohns Colitis. 2016;11:60–69. doi: 10.1093/ecco-jcc/jjw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ray A, Dittel BN. Interrelatedness between dysbiosis in the gut microbiota due to immunodeficiency and disease penetrance of colitis. Immunology. 2015;146:359–368. doi: 10.1111/imm.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 2013;132:1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mankertz J, Schulzke JD. Altered permeability in inflammatory bowel disease: pathophysiology and clinical implications. Curr. Opin. Gastroenterol. 2007;23:379–383. doi: 10.1097/MOG.0b013e32816aa392. [DOI] [PubMed] [Google Scholar]
- 70.Guinane, C. M., & Cotter, P. D. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap. Adv. Gastroenterol. 6, 295–308 (2013). [DOI] [PMC free article] [PubMed]
- 71.Glocker EO, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kotlarz D, et al. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology. 2012;143:347–355. doi: 10.1053/j.gastro.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 73.Muise AM, Snapper SB, Kugathasan S. The age of gene discovery in very early onset inflammatory bowel disease. Gastroenterology. 2012;143:285–288. doi: 10.1053/j.gastro.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 74.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-P. [DOI] [PubMed] [Google Scholar]
- 75.Sellon RK, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immunol. 1998;66:5224–5231. doi: 10.1128/IAI.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, I., et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS One.8, e70783. 10.1371/journal.pone.0070783 (2013). [DOI] [PMC free article] [PubMed]
- 77.Abbott DW, et al. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol. Cell Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Damgaard RB, et al. Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Mol. Med. 2013;5:1278–1295. doi: 10.1002/emmm.201303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krieg, A. et al. XIAP mediates NOD signaling via interaction with RIP2. Proc. Natl. Acad. Sci. USA.106, 14524–14529 (2009). [DOI] [PMC free article] [PubMed]
- 80.Li Q, et al. Variants in TRIM22 that affect NOD2 signaling are associated with very-early-onset inflammatory bowel disease. Gastroenterology. 2016;150:1196–1207. doi: 10.1053/j.gastro.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramanan D, et al. Helminth infection promotes colonization resistance via type 2 immunity. Science. 2016;352:608–612. doi: 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.