Abstract
Genomic DNA in eukaryotes is organized into chromatin through association with core histone proteins to form nucleosomes. To understand the structure and function of chromatin, we must determine the structures of nucleosomes containing native DNA sequences. However, to date, our knowledge of nucleosome structures is mainly based on the crystallographic studies of the nucleosomes containing non-native DNA sequences. Here, we discuss the technical issues related to the determination of the nucleosome structures and review the few structural studies on native-like nucleosomes. We show how an antibody fragment-aided single-particle cryo-EM can be a useful method to determine the structures of nucleosomes containing genomic DNA. Finally, we provide a perspective for future structural studies of some native-like nucleosomes that play critical roles in chromatin functions.
Genomic DNA in eukaryotes is organized into chromatin through association with core histone proteins to form repetitive nucleosomes [1, 2], the structural units of chromatin. Nucleosomes are the substrates of protein machinery responsible for essential processes of DNA replication, recombination, transcription, repair, and chromosome segregation. They differ from each other in their DNA sequences, histone components (variants), and post-translational modifications. The simplest form of the nucleosome is the nucleosome core particle (NCP). It comprises an octamer of two copies of each of the four histones (H2A, H2B, H3 and H4) wrapped with 145–147 bp of DNA [3–5]. To date, our knowledge of the interactions of core histones and DNA is almost entirely based on the crystal structures of NCPs containing either of the two types of DNA fragments: a palindromic one half of the human α-satellite DNA and the Widom 601 DNA [6, 7].
However, there are well-known uncertainties in these crystal structures likely caused by crystal packing [8]. In the crystals, one type of the interactions between neighboring NCPs involves packing of the ends of the DNA that could affect the DNA conformation within the nucleosome core (Figure 1). The NCPs containing 145 bp and 147 bp DNA have the same physical length in the crystal structures, leading to differences in the DNA conformations at regions that are the binding sites of the ATPase domains of chromatin remodelers [9–14]. Also, obtaining well-diffracting crystals of nucleosome core particles is strongly dependent on the DNA fragment used for NCP assembly, which also suggests sequence-dependent DNA conformational changes at other packing locations. In the literature, NCP is more often cited to consist of 147 bp DNA. However, direct experimental evidence has not been provided. The first structure of a native-like NCP is solved at 2.6 Å resolution using X-ray crystallography, which consists of the 147 bp 3’-LTR of the mouse mammary tumor virus promotor for nucleosome A (MMTV-A) DNA [15]. In contrast, 145 bp DNA is chosen for the structural determination of the NCP containing the human telomeric repeat DNA sequence [16]. Another limitation of the X-ray crystallographic approach is that the DNA length that is determined by biochemical methods may not be suitable for crystallization, and systematic screening of DNA lengths may be required to obtain crystals.
Figure 1. Illustration of crystal packing of DNA ends and its effects on DNA conformation within the nucleosome core particle.
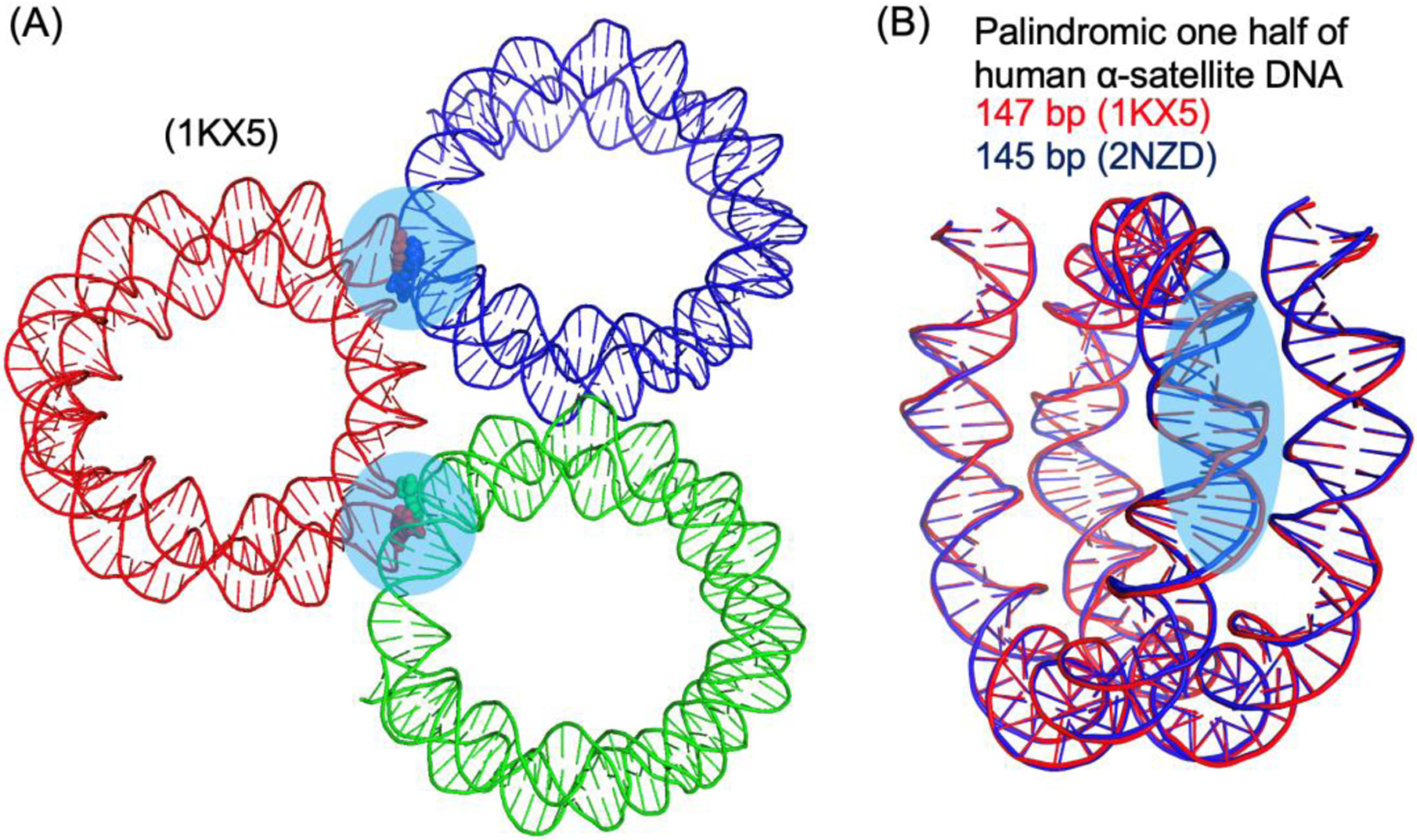
(A) Highlights of DNA ends packing between neighboring nucleosome core particles (transparent circles in cyan). The base pairs at the ends of DNA are shown in spheres. (B) Comparison of the crystal structures of nucleosome core particles with 145 bp and 147 bp DNA, showing that they have the same physical length. The region with significant conformational difference in DNA is highlighted using the transparent oval (cyan). PDB IDs are shown in the parentheses.
Single-particle cryo-EM provides an alternative way to determine the structure of nucleosomes at an atomic resolution without the need for crystals and the exact length of the DNA. The major obstacle for obtaining high-resolution structures of nucleosomes by cryo-EM is that nucleosomes tend to dissociate during the process of cryogenic sample preparation, which leads to free DNA that reduces the contrast difference between the nucleosome and the surrounding background [17]. Thus, most of the structural studies of nucleosomes by cryo-EM use Windom 601 DNA. Chemical cross-linking may help to prevent nucleosome dissociation but often leads to a decrease in resolution due to sample inhomogeneity.
In one study, the cryo-EM density map of the native human nucleosome bound to the intasome of the prototype foamy virus is determined at a 7.8 Å resolution [18]. Substantial conformation changes occur to the histones and DNA. But the low resolution of the density map excludes the analysis of detailed interactions between the DNA and the integrase. Cryo-EM, coupled with chemical cross-linking, has also been used to study the structures of the nucleosomes containing the 186 bp human ABL1 enhancer N1 DNA and the 162 bp human LIN28B DNA [19, 20]. The nucleosomes containing the above DNAs are the targets of DNA-sequence dependent recognition by the pioneer transcription factors of FoxA1 and Oct4, respectively. In these two cases, the density maps obtained have a resolution of 4.5 Å and 3.6 Å, which does not allow the nucleotides in the nucleosomes to be explicitly defined. Biochemical methods are used to determine the dyad of the nucleosomes in these cases. The ABL enhancer N1 DNA includes two target sites (eG and eH) for FoxA1. The cryo-EM density map suggests that the eH site is near the dyad region while the eG site is at the region opposite to the dyad in the nucleosome. In the case of LIN28B DNA, it contains three recognition sites for Oct4. The density map shows that one site is located near the linker DNA region while the other two sites are located within the nucleosome core particle. It is suggested that the site near the linker DNA region is the functional site.
Recently, it is demonstrated that an engineered single-chain antibody fragment (scFv) that binds to the acidic patch of core histones H2A-H2B on the surface of the nucleosome can help prevent the dissociation of nucleosomes on the cryo-EM grids. The scFv is derived from a mouse antibody (PL2–6), which binds to the free nucleosome released from apoptotic cells, serving as antigens to elicit antibodies causing systemic lupus erythematosous (SLE) autoimmune diseases in human and mouse [21, 22]. The scFv-NCP complex distributed homogenously in the vitrified ice as intact particles without significant dissociation [17]. Using the scFv cryo-EM approach, the structures of the human centromeric (CENP-A) NCP containing either 145 bp human α-satellite DNA or 147 bp Widom 601 DNA are determined at 2.6 Å (Fig. 3A). With such resolution, the nucleotides can be assigned specifically using the density map alone. Interestingly, the DNA structures at the SHL2 to SHL3 region also show a significant difference between the NCPs with α-satellite and Widom 601 DNAs. In the same study, the structure of H3 NCP containing the Widom 601 147 bp DNA is also solved at high-resolution.
Figure 3. scFv-assisted cryo-EM structure determination of nucleosomes.
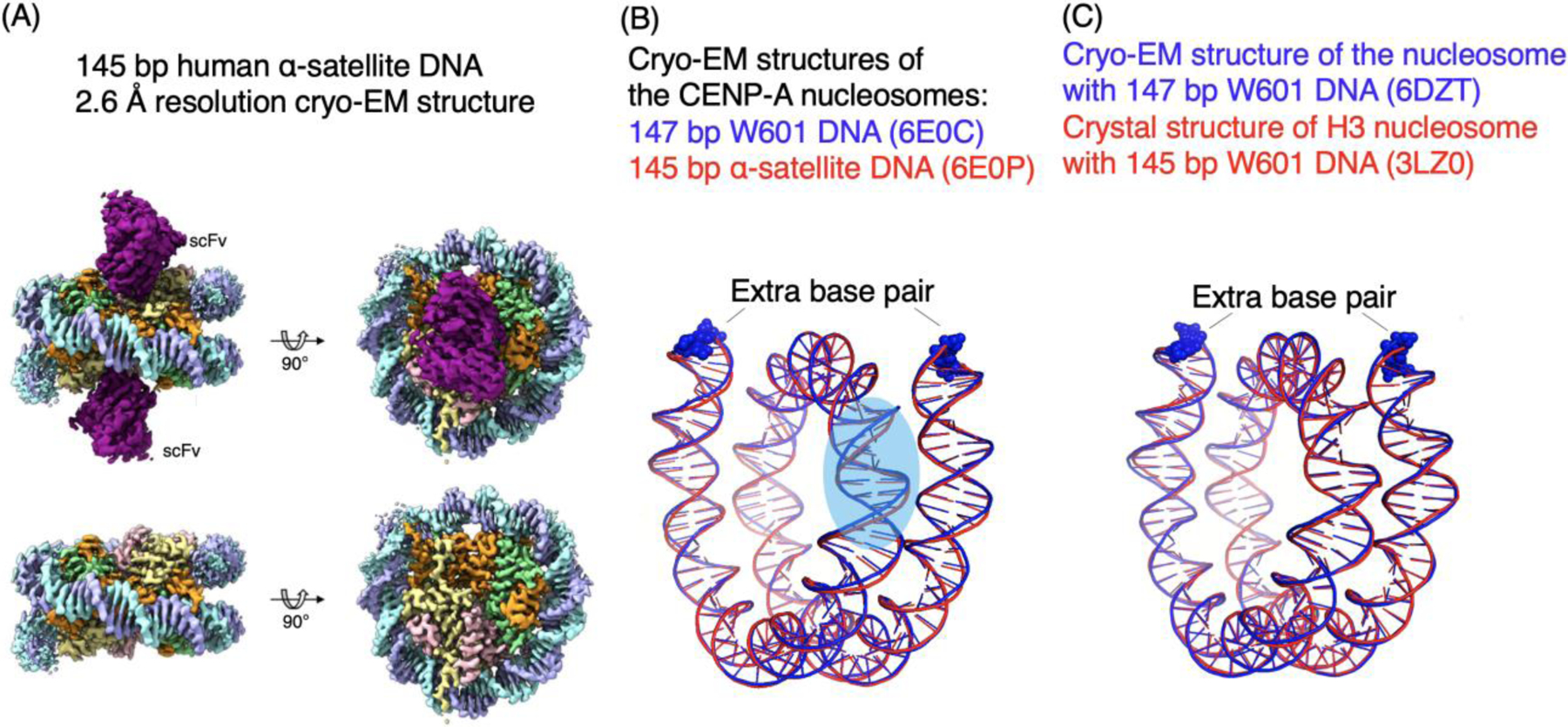
(A) Illustration of the density maps of the scFv-nucleosome complex at 2.6 Å resolution [27]. (B) Comparison of the cryo-EM structures of CENP-A nucleosomes with 147 bp Widom 601 DNA and 145 bp α-satellite DNA. The base pair at each end of the nucleosome in the 147 bp are shown in spheres. (C) Comparison of the cryo-EM structure of the nucleosome containing 147 bp Widom 601 DNA and the crystal structure of 145 bp Widom 601 nucleosome. The extra base pairs at the ends of the 147 bp DNA are shown in spheres.
From these studies, it is found that there is one additional base pair on each of the two DNA ends in the 147 bp Widom 601 CENP-A nucleosome structure in comparison with that of the CENP-A NCP containing the 145 bp α-satellite DNA. Similarly, there is one additional base pair DNA at each of the two ends of the 147 bp DNA in the cryo-EM structure of the H3 NCP compared with that of the crystal structure of the H3 NCP containing 145 bp Widom 601 DNA. Since the crystal structures of NCPs containing 145 bp and 147 bp have the same physical length, these cryo-EM structures suggest that crystal packing squeezes two base pairs of 147 bp DNA into the nucleosome core region that normally accommodate 145 bp in the absence of crystal packing. Therefore, the NCP in solution is likely to include 145 bp DNA. Since the 2-bp difference in NCP can lead to a substantial change in the DNA conformation and the relative positions/orientations of the two neighboring nucleosomes connected by the linker DNA, the determination of the precise number of base pairs in the NCP DNA is critical for understanding the function of the nucleosome and high-order structures of chromatin, in particular, considering the fact that 147 bp NCP is commonly used to analyze chromatin structure in genome-wide studies and in the theoretical modeling of nucleosomes and high-order structures of chromatin.
Figure 2. Examples of native-like nucleosomes studied by X-ray crystallography and cryo-EM.
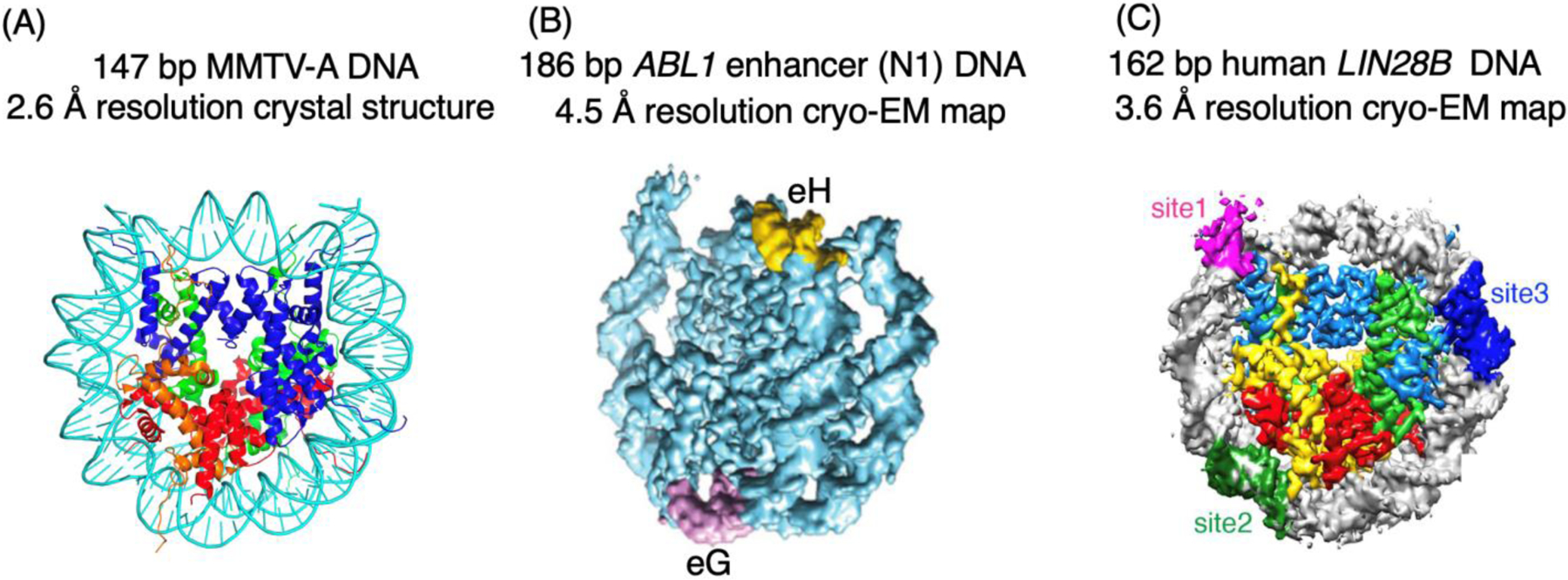
(A) The crystal structure of 147 bp MMTV-A nucleosome core particle [15]. (B) Cryo-EM reconstruction of 186 bp ABL1 enhance (N1) nucleosome. The FoxA1 binding sites (eG and eH) are shown in magenta and yellow colors [19]. (C) Cryo-EM density map for the 162 bp human LIN28B DNA and DNA motifs for binding of transcription factor Oct4 [20].
Perspective.
We anticipate the immediate use of the scFv-assisted cryo-EM approach to determine the structures of several native-like nucleosomes that are known to play important functional roles. These include the nucleosomes containing the DNAs with virus integration sites and targets of pioneer transcription factors, FoxA1 and Oct4, discussed above. There are also recently identified nucleosomes that are targets of various other pioneer transcription factors (ASCL1, BRN2, PU1, BRN2, CEBPa, and CEBPb) [23]. Also, 16 budding yeast centromeric nucleosomes contain unique AT-rich DNA [24], which are recognized by the centromere protein CBF3 in a sequence-dependent manner. Similarly, the human centromeric nucleosome contains the α-satellite DNA with the 17 bp CENP-B box, the target site of the centromere protein CENP-B. One can also use the scFv-aided cryo-EM approach to determine the structures of nucleosomes in complex with the proteins that do not bind to the acidic patch regions, for example, the native-like chromatosomes containing linker histone isoforms [25, 26]. Moreover, scFv-aided cryo-EM approach could be used to determine the intrinsic positioning of nucleosomes in genome-wide studies, which may require a community effort. It is likely that for weaker positioning sequences, the nucleosome may display multiple positions. High-resolution structures will likely provide distributions of nucleosome positions at a single-molecule level.
Highlights.
Nucleosome core particles consist of 145 bp DNA
scFv-aided cryo-EM approach allows structural determination of native-like nucleosomes
Acknowledgement
This work is supported by the intramural research program of National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Olins AL, Olins DE. Spheroid chromatin units (v bodies). Science. 1974;183:330–2. [DOI] [PubMed] [Google Scholar]
- [2].Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–71. [DOI] [PubMed] [Google Scholar]
- [3].Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. [DOI] [PubMed] [Google Scholar]
- [4].Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–50. [DOI] [PubMed] [Google Scholar]
- [5].Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J Mol Biol. 2007;368:1067–74. [DOI] [PubMed] [Google Scholar]
- [6].Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. [DOI] [PubMed] [Google Scholar]
- [7].Harp JM, Uberbacher EC, Roberson AE, Palmer EL, Gewiess A, Bunick GJ. X-ray diffraction analysis of crystals containing twofold symmetric nucleosome core particles. Acta Crystallogr D Biol Crystallogr. 1996;52:283–8. [DOI] [PubMed] [Google Scholar]
- [8].Tan S, Davey CA. Nucleosome structural studies. Curr Opin Struct Biol. 2011;21:128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chittori S, Hong J, Bai Y, Subramaniam S. Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome. Nucleic Acids Res. 2019;47:9400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yan L, Wu H, Li X, Gao N, Chen Z. Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat Struct Mol Biol. 2019;26:258–66. [DOI] [PubMed] [Google Scholar]
- [11].Liu X, Li M, Xia X, Li X, Chen Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature. 2017;544:440–5. [DOI] [PubMed] [Google Scholar]
- [12].Nodelman IM, Bleichert F, Patel A, Ren R, Horvath KC, Berger JM, et al. Interdomain Communication of the Chd1 Chromatin Remodeler across the DNA Gyres of the Nucleosome. Mol Cell. 2017;65:447–59 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Farnung L, Vos SM, Wigge C, Cramer P. Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature. 2017;550:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol. 2007;27:8306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Frouws TD, Duda SC, Richmond TJ. X-ray structure of the MMTV-A nucleosome core. Proc Natl Acad Sci U S A. 2016;113:1214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Soman A, Liew CW, Teo HL, Berezhnoy NV, Olieric V, Korolev N, et al. The human telomeric nucleosome displays distinct structural and dynamic properties. Nucleic Acids Res. 2020;48:5383–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chua EY, Vogirala VK, Inian O, Wong AS, Nordenskiold L, Plitzko JM, et al. 3.9 A structure of the nucleosome core particle determined by phase-plate cryo-EM. Nucleic Acids Res. 2016;44:8013–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, et al. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Takizawa Y, Tanaka H, Machida S, Koyama M, Maehara K, Ohkawa Y, et al. Cryo-EM structure of the nucleosome containing the ALB1 enhancer DNA sequence. Open Biol. 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Echigoya K, Koyama M, Negishi L, Takizawa Y, Mizukami Y, Shimabayashi H, et al. Nucleosome binding by the pioneer transcription factor OCT4. Sci Rep. 2020;10:11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kramers K, Stemmer C, Monestier M, van Bruggen MC, Rijke-Schilder TP, Hylkema MN, et al. Specificity of monoclonal anti-nucleosome auto-antibodies derived from lupus mice. J Autoimmun. 1996;9:723–9. [DOI] [PubMed] [Google Scholar]
- [22].Olins AL, Langhans M, Monestier M, Schlotterer A, Robinson DG, Viotti C, et al. An epichromatin epitope: persistence in the cell cycle and conservation in evolution. Nucleus. 2011;2:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xiao H, Wang F, Wisniewski J, Shaytan AK, Ghirlando R, FitzGerald PC, et al. Molecular basis of CENP-C association with the CENP-A nucleosome at yeast centromeres. Genes Dev. 2017;31:1958–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhou BR, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y. Structural Mechanisms of Nucleosome Recognition by Linker Histones. Mol Cell. 2015;59:628–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bednar J, Garcia-Saez I, Boopathi R, Cutter AR, Papai G, Reymer A, et al. Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Mol Cell. 2017;66:384–97 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhou BR, Yadav KNS, Borgnia M, Hong J, Cao B, Olins AL, et al. Atomic resolution cryo-EM structure of a native-like CENP-A nucleosome aided by an antibody fragment. Nat Commun. 2019;10:2301. [DOI] [PMC free article] [PubMed] [Google Scholar]


