Abstract
We have carried out meta-analyses of genome-wide association studies (GWAS) (n = 23 784) of the first two principal components (PCs) that group together cortical regions with shared variance in their surface area. PC1 (global) captured variations of most regions, whereas PC2 (visual) was specific to the primary and secondary visual cortices. We identified a total of 18 (PC1) and 17 (PC2) independent loci, which were replicated in another 25 746 individuals. The loci of the global PC1 included those associated previously with intracranial volume and/or general cognitive function, such as MAPT and IGF2BP1. The loci of the visual PC2 included DAAM1, a key player in the planar-cell-polarity pathway. We then tested associations with occupational aptitudes and, as predicted, found that the global PC1 was associated with General Learning Ability, and the visual PC2 was associated with the Form Perception aptitude. These results suggest that interindividual variations in global and regional development of the human cerebral cortex (and its molecular architecture) cascade—albeit in a very limited manner—to behaviors as complex as the choice of one’s occupation.
Keywords: brain development, cortical surface area, cortical thickness, genome-wide association study, occupational aptitude
Introduction
The radial unit hypothesis provides a framework for global and regional expansion of the primate cerebral cortex (Rakic 1988). Using magnetic resonance imaging (MRI), one can derive a number of metrics informative with regard to development and aging of the human cerebral cortex, including cortical surface area and cortical thickness. The two measures provide insights into different developmental processes, each with a different timeline. Cortical surface area reflects primarily the tangential growth of the cerebral cortex during prenatal development; the phase of “symmetric division of progenitor cells” in the proliferative zones during the first trimester is particularly important for the tangential growth through additions of ontogenetic columns (Rakic 1988). The subsequent phase of “asymmetric division” continues to increase the number of ontogenetic columns (and thus surface area), but it also begins to contribute to the thickness of cerebral cortex formed by postmitotic neurons migrating from the proliferative zones to the cortical plate in the inside-out manner (Rakic 1988). Ionizing radiation of the (monkey) fetus during early gestation reduces surface area (sparing cortical thickness), while the same radiation applied in midgestation affects both the surface area and cortical thickness (Selemon et al. 2013). While surface area remains stable after early childhood, cortical thickness continues to change, in particular during puberty and aging. Furthermore, both surface area and cortical thickness vary across individuals in global and regional manners.
In recent years, substantial progress has been made in revealing molecular architecture of a number of brain phenotypes, including those captured by global (e.g., intracranial volume (Adams et al. 2016)) and regional (e.g., hippocampal volume (Stein et al. 2012)) measures derived from T1-weighted MR images, as well as other MRI-derived brain phenotypes (e.g., interhemispheric connectivity (Mollink et al. 2019)). Recent efforts of the CHARGE (Hofer 2019) and ENIGMA (Grasby 2019) Consortia have focused on identifying genetic variations associated with values of surface area and thickness at each of the 34 cortical regions delineated by FreeSurfer. Given the previously observed genetic influences on both surface area (Chen et al. 2012) and thickness (Chen et al. 2013) across multiple regions of the human cerebral cortex, we took a different approach and performed, first, principal component analysis of the surface area and thickness of the 34 cortical regions and, then, executed genome-wide association studies (GWAS) of these derived cortical phenotypes, namely, the first two principal components of surface area and thickness, respectively. We meta-analyzed these in a total of 23 784 individuals (with replications in another 25 746 individuals). Finally, as a proof of concept, we asked whether interindividual differences in the derived cortical phenotypes relate to behaviors as complex as the choice of one’s occupation. To achieve this, we took advantage of a unique resource, the Dictionary of Occupational Titles (DOT) (National Research Council 1980), and related both the brain phenotypes and their genetic determinants to relevant occupational aptitudes in up to 220 000 individuals.
Materials and Methods
Participants
For the meta analyses of GWASs of principal components of cortical surface area and thickness across the 34 regions of interest, we analyzed data of the 23 784 participants from 19 cohort studies collaborating in the Cohorts of Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium (Psaty et al. 2009) and the UK Biobank (UKBB) (Sudlow et al. 2015). All the individuals, aged between 12 and 97 years, were stroke- and dementia-free and of European ancestry. Supplementary Table E1 provides the information on demographic characteristics, brain imaging measures and acquisition, and genotyping for each cohort study. Each study secured approval from institutional review boards or equivalent organizations, and all participants provided written informed consent. Our meta-GWAS results were replicated in participants from ENIGMA consortium and an independent subset of UKBB participants who were not included in our meta-GWAS analyses (total N = 25 746). For the association analyses of occupational aptitudes vs. brain phenotypes and genetic variants, we analyzed up to 220 000 UKBB participants.
Principal Component Analysis of Cortical Surface Area and Cortical Thickness
Each cohort estimated the surface area (left and right hemispheres summed) and the thickness (left and right hemispheres averaged) of the 34 cortical regions, using FreeSurfer (Fischl and Dale 2000) or FSL FIRST (Patenaude et al. 2011) and carried out principal component analyses to obtain the loadings of the first two leading components PC1 and PC2 (see Supplementary Figures E1 and E2 for cohort-specific loadings for the surface area and thickness, respectively). Then, for each cortical region, a median value of loadings was obtained across the cohorts. The median loading values (Supplementary Table E2) were then used to derive the “general” PC score for each individual (script available at https://doi.org/10.6084/m9.figshare.11619879) and later used as the outcome variable in the multiple linear regression models fitted for the GWAS analyses.
Genome-Wide Association Studies and Meta-analyses
The GWAS and meta-analyses were carried out as follows. Using the median PC loadings across the CHARGE consortium cohorts (Supplementary Figures E1, E2 and Supplementary Table E2), each cohort derived the general PC1 and PC2 scores for the surface area and the thickness (PC1-SA, PC2-SA, PC1-TH and PC2-TH) to be used as phenotypes in the genome-wide association tests in order to ensure “homogeneity” in phenotype derivation. All the association tests were adjusted for age, sex, and other cohort-specific confounding variables, such as study site and/or family structure. The cohort-specific GWAS results were then examined for quality control with Easy QC software (Winkler et al. 2014) and meta-analyzed with METAL (Willer et al. 2010) using fixed effects models. Quantile-quantile plots of associations yielded by the genome-wide association study meta-analyses are shown in Supplementary Figure E3. Results were inspected for inflation due to confounders such as population stratification or sample overlap, by calculating the inflation factor (λ) as well as the LD score intercept using LD score regression (Bulik-Sullivan et al. 2015b). An inflation factor λ > 1 suggests an inflation of the association statistics, which can be due to both spurious and genuine effects. It is likely to increase with sample size and degree of polygenicity of the phenotype, as the distribution of effect sizes begins to differ substantially from a null distribution when more variants have true associations. An LD score intercept >1 suggests that there is spurious association, but an intercept <1.10 is generally considered to suggest that the signal is mostly due to genuine association effects. LD score regression intercepts were 1.05 (SE = 0.0079), 1.03 (SE = 0.0076), 1.03 (SE = 0.0065), and 1.02 (SE = 0.007), for PC1-SA, PC2-SA, PC1-TH, and PC2-TH, respectively.
Replication analyses were carried out in an independent subset of 6234 UKBB participants, who were not included in the meta-GWAS, and in silico based on the ENIGMA3-GWAS summary statistics for the surface area of the 34 FreeSurfer-cortical regions of interest in 19 152 participants of European ancestry from 34 cohorts (Grasby 2019). The in silico replication analysis was done using the genome-wide inferred statistics (GWIS) approach (Nieuwboer et al. 2016). In particular, the ENIGMA3 meta-GWAS summary statistics and the median PC loadings used to derive the PC scores in our meta-GWAS were used to derive the coefficient estimates and their corresponding standard errors, using the delta method. We then performed a look-up of the SNPs associated with PC1- and PC2-SA in those summary GWIS statistics.
Gene Expression in the Human Cerebral Cortex
We examined temporal and spatial pattern of gene expression using the BrainSpan atlas (www.brainspan.org). The mRNA expression levels were measured by RNA sequencing in 607 brain tissues from 18 female and 23 male donors. Supplementary Table E4 in Extended Data provides details about the donor brains and the sample brain regions. Authors of the original report describing these data surveyed 10 potential confounders of the variation in gene expression using the top 10 components yielded by a multidimensional scaling analysis, namely, age, brain region, sequence depth, pH, RNA integrity number (RIN), sex, ethnicity, hemisphere, sample dissection score, and sequencing processing site; as reported in Fig. S11B of the original report, “only age and brain region correlated strongly with specific dimensions” (Li et al. 2018).
Genetic Correlations
Genetic correlations were conducted between PC1-SA and PC2-SA (surface area PC scores) and complex traits (growth, brain function and brain disorders) using LD score regression method for genetic correlation estimation using summary GWAS summary statistics and LD structure (Bulik-Sullivan et al. 2015a) implemented in LDSC v1.0.0 (https://github.com/bulik/ldsc). Single-nucleotide polymorphisms (SNPs) were filtered using the available HapMap3 SNPs, and genetic correlations were conducted using LDSC’s 1000 Genomes European LD scores. Genetic correlations were restricted to European GWAS samples (Supplementary Table E10). A Bonferroni correction was applied to correct for the 55 genetic correlations. The required LDSC files were obtained from the LDSC GitHub repository (https://data.broadinstitute.org/alkesgroup/LDSCORE/).
Partitioned Heritability
This analysis was conducted using LD score regression method for partitioaned heritability estimation (Finucane et al. 2015) implemented in LDSC v1.0.0 (https://github.com/bulik/ldsc) in order to assess the enrichment of PC1-SA and PC2-SA GWAS variants in chromatin regions with differential accessibility during cortical neurogenesis between the germinal zone (GZ) and cortical plate (CP), as reported previously for other phenotypes (de la Torre-Ubieta et al. 2018). Enrichment of heritability represents the ratio between (1) the proportion of heritability explained by a particular annotation and (2) the proportion of SNPs in the annotation (Finucane et al. 2015; de la Torre-Ubieta et al. 2018). An annotation file was generated for regions in which chromatin was more accessible in GZ, compared to CP (GZ > CP; 19 260) and vice versa (CP > GZ; 17 803), using the available supplementary data table (de la Torre-Ubieta et al. 2018). Next, LD scores for the chromatin annotation file were computed following the LDSC guidelines (https://github.com/bulik/ldsc/wiki/LD-Score-Estimation-Tutorial), using the available HapMap3 SNPs, a 1 cM window, and the Phase 3 1000 Genomes European genotype files. Finally, partitioned heritability was run combining the chromatin annotation file with the LDSC baseline annotation file. The required LDSC files were obtained from the LDSC GitHub repository (https://data.broadinstitute.org/alkesgroup/LDSCORE/).
Occupational Aptitudes
We used the DOT (National Research Council 1980) that contains 28 801 titles of which 12 099 are so-called “base” titles (corresponding to occupations). Each base title is associated with scores capturing, among other things, aptitudes defined as “the capacities or abilities required of a worker to facilitate the learning of job tasks” (National Research Council 1980, p. 29); DOT contains scores for a total of 11 aptitudes, including General Learning Ability and Form Perception. In DOT, General Learning Ability was defined as “The ability to ‘catch on’ or understand instructions and underlying principles; the ability to reason and make judgments. Closely related to doing well in school” (US Department of Labor 1991, p. 9–3). Form Perception aptitude was defined as “The ability to perceive pertinent details in objects or in pictorial or graphic material. Ability to make visual comparisons and discrimination and see slight differences in shapes and shadings of figures and widths and lengths of lines” (US Department of Labor 1991, p. 9–17)). To test these predictions, we used the data on occupations of the UK Biobank participants with available MRI data (n = 21 609). The UK BioBank encodes occupation using the UK 2000 Standard Occupation Classification (SOC2000UK) (Office for National Statistics 2000), while aptitude scores are associated with DOT (National Research Council 1980). A crosswalk, or mapping, from the SOC2000UK codes to the DOT codes was required in order to assign occupational aptitude scores to each participant. As a direct mapping was not available, we combined several available mappings: SOC2000UK to the 2010 version (SOC2010UK) (Office for National Statistics 2010b, 2012), SOC2010UK to the 2008 International Standard Classification of Occupations (ISCO2008) (Office for National Statistics 2010a; International Labour Office 2012), ISCO2008 to the US 2010 Standard Occupational Classification (SOC2010US) (Cosca and Emmel 2010; Emmel and Cosca 2010; Labor US Department, Statistics BoL (n.d.)), and SOC2010US to DOT (http://www.widcenter.org/document/legacy-crosswalks/). For a given SOC2000UK code, all of the matching SOC2010UK codes were found, and for each of these, all of the matching ISCO2008 codes were found; this procedure was repeated for each subsequent crosswalk. Thus, each SOC2000UK code could be associated with many DOT codes. The aptitude scores associated with each SOC2000UK code were the average of the aptitude scores associated with each corresponding DOT code (DOT codes without an associated aptitude were ignored).
Associations with Polygenic Scores for Surface Area PC1 and PC2
Polygenic scores for surface area PC1 and PC2 in the UK Biobank participants were calculated based on the GWAS significant SNPs (with P < 5E − 08) identified from our meta-GWAS analyses, using PRSice (Euesden et al. 2015). In the UK Biobank genotype data, we identified the 78 (for PC1-SA) and 55 (for PC2-SA) SNPs. After clumping based on LD (r2 < 0.1), the final scores were obtained using the 16 and the 14 independent SNPs, respectively, for PC1-SA and PC2-SA. Associations for the polygenic scores for PC1-SA were tested with the respective PC scores and with the General Learning Ability and the Form Perception aptitude, using multiple linear regression models. For the regression models, the outcome variable was either PC-SA score or ability/aptitude value, and the predictor variable was the polygenic score, both of which were standardized (i.e., z-scored) so that the coefficient for the polygenic score can be interpreted as the number of standard deviations the outcome increases for every standard deviation increase in the polygenic score, holding all the other covariates constant. All the analyses were adjusted for age, sex, and/or MRI site. The association tests for the Form Perception aptitude variable were additionally adjusted for General Learning Ability.
Results
Participants
Here we report findings obtained in 23 784 participants assessed across 19 cohorts from the CHARGE Consortium and the UK Biobank (Supplementary Table E1 in Extended Data) and replicated in a subsequent release of the UK Biobank participants (n = 6234) as well as in silico (Nieuwboer et al. 2016) using region-based summary statistics provided by the ENIGMA Consortium (n = 19 512).
Principal Component Analysis of Cortical Phenotypes
To identify components of shared variance of the surface area and of the thickness of the cerebral cortex, we have carried out principal component analyses of regional values of surface area and of thickness (34 regions segmented by FreeSurfer or FSL FIRST [in 2 cohorts]) in each cohort. For surface area and thickness, respectively, the first (PC1) and second (PC2) components were loaded by similar sets of cortical regions across all 13 cohorts available for loading calculations (Supplementary Figure E1 [surface area], Supplementary Figure E2 [cortical thickness] and Supplementary Table E2 in Extended Data). Figure 1 illustrates the median loadings for each of the 34 cortical regions in PC1 (Fig. 1A) and PC2 (Fig. 1B) for surface area. Note that PC2 includes only a handful of cortical regions in the medial aspect of the occipital lobe, including the pericalcarine (primary visual) cortex.
Figure 1.
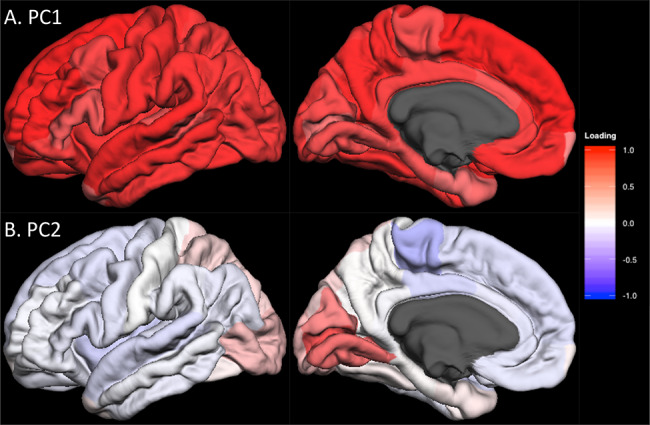
Lateral and medial views of median unrotated principal component (PC) loadings for the surface area of the 34 cortical regions in CHARGE consortium cohorts. Lateral (left column) and medial (right column) views of the median PC loadings are shown for PC1 (A, top) and PC2 (B, bottom). The red-to-blue color indicates the positive-to-negative loading values (i.e., correlation between PC scores and raw data) as indicated by the color bar. The regional median PC loading values are shown in Supplementary Table E2.
Genome-Wide Association Studies and their Meta-analysis
We then executed a GWAS in each of the 19 cohorts and, subsequently, meta-analyzed these cohort-based results for each of the four phenotypes, namely, PC1 and PC2 of surface area and PC1 and PC2 of thickness.
For surface area, the first (global) component was associated with 42 independent SNPs in 18 independent loci (Supplementary Table E3 in Extended Data). The strongest signal was found in a broad region of chromosome 17, which contained a number of genes, including those associated previously with intracranial volume (Hibar et al. 2015; Adams et al. 2016) and/or general cognitive function (Trampush et al. 2017) (Fig. 2 and Supplementary Table E4A in Extended Data). The second (visual) component was associated with 57 SNPs in 17 independent loci (Supplementary Table E3 in Extended Data). The strongest signal was found at a locus on chromosome 14 containing DAAM1 (Fig. 3 and Supplementary Table E4B in Extended Data). We replicated 695/807 (global PC1) and 952/1155 (visual PC2) GWAS-significant SNPs (Supplementary Tables E4A and E4B in Extended Data).
Figure 2.
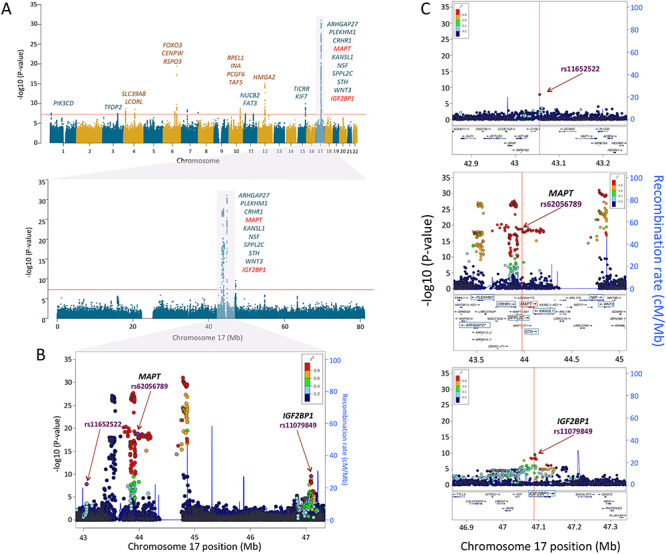
Genetic loci of global variation of cortical surface area. (A) Manhattan plots of loci associated with PC1 of cortical surface area across the genome (upper panel) and in chromosome 17 (lower panel). The vertical axes represent the –log10 (P-values), and the horizontal axes represent the chromosome number (upper) or the hg19 position on chromosome 17 (lower). All the labeled genes indicate the protein-coding genes mapped to the GWAS-significant SNPs with P < 5E − 08 (indicated by the red-horizontal lines). Among these genes on chromosome 17, the red-colored genes MAPT and IGF2BP1 have higher gene expression levels in the entire cerebral cortex during the prenatal period than the postnatal period (Fig. 4). Regional association plots of the 42.9–47.3 Mb region of chromosome 17 containing the GWAS-significant SNPs (panel B) and the 3 independent genetic loci (panel C) identified by FUMA (Watanabe et al. 2017). The horizontal axis indicates the genomic position on the human chromosome 17 (hg19). The left-vertical axis indicates the –log10 (P-values) obtained from the GWAS meta-analysis and the right-vertical axis, the estimated recombination rate from the HapMap samples. The purple-colored index SNP indicates the top SNP (rs11652522, P = 1.6E − 08) within the region (top panel), the top SNP within MAPT (rs62056789, P = 1.6E − 19: middle panel), or IGF2BP1 (rs11079849 P = 2.8E − 10: bottom panel). The red-to-blue colors indicate the degree of linkage disequilibrium (LD) between each SNP and the index SNP. The LD was measured by the pairwise squared allelic correlation r2 estimated in the 1000 Genomes European reference panels (Nov 2014 EUR). The plot was created using LocusZoom (http://locuszoom.org/).
Figure 3.
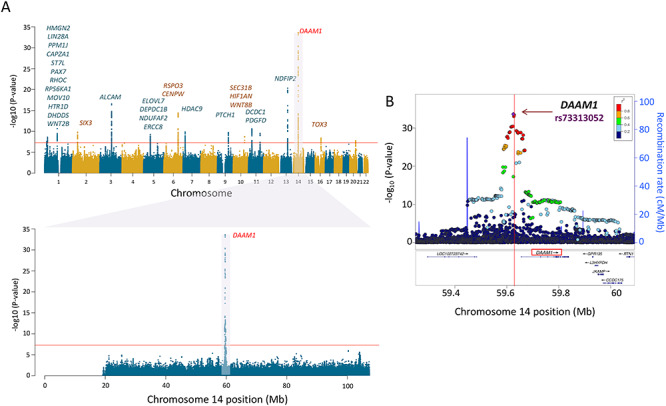
Genetic locus of regional variation of cortical surface area (SA). (A) Manhattan plot of loci associated with PC2 of surface area. The labeled gene on chromosome 14 is DAAM1, the protein-coding gene mapped to the GWAS-significant SNPs. DAAM1 shows high levels of expression in the entire cerebral cortex during the prenatal period (see Fig. 4). (B) Regional association plot for genomic locus containing DAAM1. The horizontal axis represents the genomic position on the human chromosome 14 (hg19). The left-vertical axis indicates the –log10 (P-values) obtained from the GWAS meta-analysis and the right-vertical axis, the estimated recombination rate from the HapMap samples. The red-to-blue colors indicate the degree of linkage disequilibrium (LD) between each SNP and the top SNP (rs73313052, P = 2.4E − 34). The LD was based on the pairwise squared allelic correlation r2 estimated in the 1000 Genomes European reference panels (Nov 2014 EUR). The plot was created using LocusZoom (http://locuszoom.org/).
For cortical thickness, meta-GWAS identified no locus associated with PC1 and one independent SNP in 1 locus on chromosome 15 associated with PC2 (Supplementary Figure E4).
Genomic Landscape and Genome Biology of the Main Genetic Loci
Next, we focus on the two genetic loci containing SNPs with the strongest associations with the cortex-wide (global PC1) and regional (visual PC2) variations in the cortical surface area on chromosomes 17 and 14, respectively (Figs 2 and 3).
The locus on chromosome 17, associated with the global component of surface area (PC1), spans 4.5 Mb and contains 10 protein-coding genes (Fig. 2A). Given that surface area is determined mainly by the growth of the human cerebral cortex (most pronounced before birth), we have examined spatiotemporal pattern of their expression using the BrainSpan dataset (Supplementary Table E5 in Extended Data). As shown in Figure 4A, only 2 of the 10 protein-coding genes within this locus show high levels of expression in the entire cerebral cortex during the prenatal period: insulin-like growth factor 2 MRNA-binding protein 1 (IGF2BP1) and microtubule-associated protein Tau (MAPT). Other genes either show low expression during this period (e.g., ARHGAP27) or appear to be expressed only in subcortical structures (e.g., WNT3). Next, we have examined several publically available databases to ascertain whether top SNPs in/near the two genes could influence their expression. IGF2BP1 “functions by binding to the mRNAs of certain genes, including insulin-like growth factor 2, beta-actin and beta-transducin repeat-containing protein, and regulating their translation” (Stelzer et al. 2016). The top SNP (rs11079849) lies in an enhancer/promoter region of IGF2BP1 in fetal brain and alters regulatory motifs of several transcription factors (Ward and Kellis 2012). The most differential binding affinity occurs for specificity protein 2 transcription factor (SP2), a cell cycle regulator in neural stem and progenitor cells (Liang et al. 2013). This top SNP is also associated with differential methylation at 16 CpGs located within 1Mbps of the SNP (chr17: 46 908 359–47 102 000) (Gaunt et al. 2016). The minor allele (T, associated with higher values of PC1) is associated with lower DNA methylation in the promotor region of IGF2BP1. MAPT “promotes microtubule assembly and stability, and might be involved in the establishment and maintenance of neuronal polarity” (Stelzer et al. 2016). The top SNP (rs62056789) is found in the promotor/enhancer regions in fetal brain (Ward and Kellis 2012) and, via motif change, is in a position to influence binding affinity of Forkhead box (Fox) transcription factors, such as FOXF1 and FOXF2, that play important roles in regulating expression of genes involved in cell growth, proliferation, differentiation, and longevity (Tuteja and Kaestner 2007a, 2007b). In addition to FOX-family transcription factors, the top SNP’s LD-proxy (rs625056790, r2 = 1) alters motifs for POU3F2, a POU-homeodomain transcription factor shown to have a critical role in neuronal differentiation in mouse brain (Nakai et al. 1995). The top SNP is also associated with differential methylation at 35 CpG sites located within 1Mbp of the SNP (chr17: 43 099 144–44 439 469) (Gaunt et al. 2016). The minor allele (C, associated with lower values of PC1) is associated with lower DNA methylation of CpGs in the promotor region and higher methylation of CpGs in the enhancer regions of MAPT. Finally, we evaluated the spatiotemporal pattern of coexpression of the two genes in the cerebral cortex during the prenatal period, followed by the assessment of gene enrichment using Gene Ontology. Using the BrainSpan dataset (Supplementary Table E5 in Extended Data; prenatal period, cerebral cortex), we carried out a coexpression analysis for each of the two genes and retained the top 1% genes whose expression varied in time (8–37 postconception weeks) and space (11 cortical regions) in the same directions as the seed gene (IGF2BP1 or MAPT) for Gene Ontology analysis using the PANTHER overrepresentation test tool (Mi et al. 2019). The IGF2BP1 coexpression network (555 genes) is enriched for a number of biological processes related to cell division (e.g., mitotic G2/M transition checkpoint, DNA replication, negative regulation of DNA endoreduplication), gene transcription (e.g., regulation of transcription by RNA polymerase II), and translation (regulation of gene silencing by miRNA); see Supplementary Table E6 in Extended Data. The MAPT coexpression network (424 genes) is enriched for a number of biological processes related to neurite development (e.g., regulation of neuron projection development, regulation of axon extension, axon development), axonal transport (e.g., axonal transport, organelle transport along microtubule), and neurotransmission (synaptic vesicle exocytosis, glutamate secretion); see Supplementary Table E7 in Extended Data.
Figure 4.
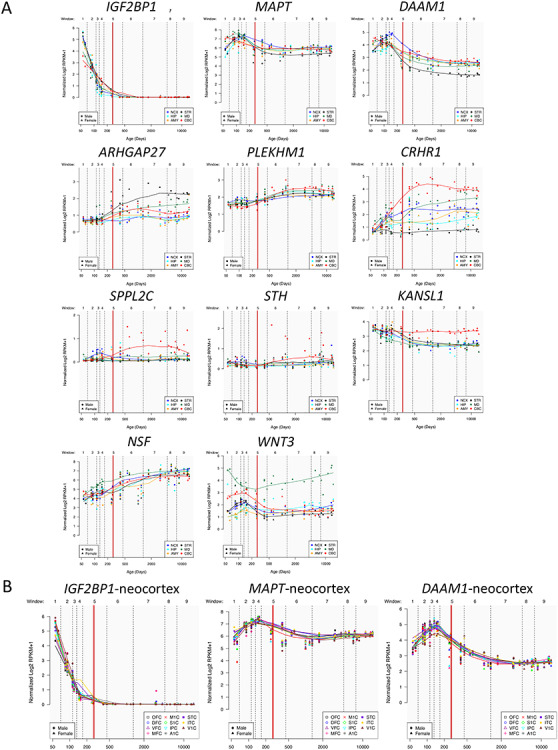
Spatial and temporal expression of genes mapped to GWAS-significant SNPs on chromosomes 17 (PC1) or 14 (PC2). (A) Gene expression levels for the 10 (chromosome 17) and the 1 (chromosome 14) protein-coding genes measured by RMKM (reads per kilobase per million) as a function of human brain developmental time. All the genes except for DAAM1 are on chromosome 17. The time is split into 9 windows based on postconception days (indicated by vertical dashed lines): 52–69 (window 1), 70–111 (window 2), 112–132 (window 3), 133–167 (window 4), 168–447 (window5), 448–1299 (window 6), 1300–4648 (window 7), 4649–7570 (window 8), and 7571–14 876 (window 9). The boundary between pre- and postnatal periods is indicated by a red vertical line. Each colored point represents the transformed expression level of each gene across 16 anatomical brain regions and ages. Brain structure includes 11 neocortical areas (NCX, blue) and 5 subcortical regions: hippocampus (HIP, cyan), amygdala (AMY, orange), striatum (STR, black), mediodorsal nucleus of thalamus (MD, dark green), and cerebellar cortex (CBC, red). (B) Expression levels of the three “developmental” genes IGF2BP1, MPAT, and DAAM1 that have higher expression (RPKM) in the 11 neocortical areas during prenatal (vs. postnatal) periods. The included neocortical regions are orbitofrontal cortex (OFC, black), dorsolateral prefrontal cortex (DFC, blue), ventrolateral prefrontal cortex (VFC, purple), medial frontal cortex (MFC, magenta), primary motor cortex (M1C, red), primary sensory cortex (S1C, green), inferior parietal cortex (IPC, cyan), primary auditory cortex (A1C, gray), superior temporal cortex (STC, violet), inferior temporal cortex (ITC, yellow), and primary visual cortex (V1C, brown).
The locus on chromosome 14, associated with the regional—visual cortex—component of surface area (PC2), contains a single gene, namely, DAAM1 (Fig. 3). This gene is a key component of the planar-cell-polarity signaling pathway (Tissir and Goffinet 2010; Beane et al. 2012); it acts as a bridging factor between disheveled, Rho-family GTPases and Rho-associated kinases (Habas et al. 2001), a molecular complex involved in organizing actin cytoskeleton (Yang and Mlodzik 2015). As shown in Figure 4B, DAAM1 is expressed in the human cerebral cortex between ~80 and ~150 postconception days; after birth, its expression is very low. Note that, in monkeys, neurogenesis of the primary visual cortex begins around embryonic day 40 (E40) and ends at E100 (165-day gestation) (Rakic 1988). There are multiple lines of evidence supporting the notion that the dates of neurogenesis in macaque can be translated to those in human (Zhu et al. 2018). Based on models for matching neurodevelopment across species, a neurogenesis event in the cerebral cortex of the rhesus macaque at postconception day 50 translates to postconception day 55 in the human (http://translatingtime.org/translate (Workman et al. 2013).
Next, we have examined several publicly available databases to ascertain whether top SNPs in/near DAAM1 could influence its expression. Based on HaploReg (Ward and Kellis 2012) analysis, the top SNP (rs73313052) is found in an enhancer region. According to the mQTL Database (Gaunt et al. 2016), the DAAM1 top SNP did not have any differentially methylated CpGs within 1Mbp; but SNPs in strong linkage disequilibrium (LD) with the top SNP (r2 > 0.6) are associated with differential methylation at two CpG sites, cg18819791 and cg22995959. The minor allele of the top SNP (A, associated with lower values of PC2) is associated (via its proxy) with higher DNA methylation at these two CpG sites, which may influence DAAM1 expression; one can predict lower expression values in the carriers of the rs73313052_A minor allele. Finally, as above, we examined coexpression of DAAM1 across all cortical regions and prenatal time points using the same BrainSpan dataset. As expression of DAAM1 increases, so does expression of genes enriched in pathways involving neuron migration and cytoskeleton organization, among others (Supplementary Table E8 in Extended Data). To ascertain the pattern of DAAM1 coexpression specific to the primary visual cortex (V1), we have identified genes coexpressed highly (top 1%) in V1 but not in any other cortical region (i.e., not present among top 1% in any of the other eight regions). This analysis yielded striking enrichment for mitochondria-related genes coexpressed strongly in the same direction as DAAM1 in V1 but not in the other cortical regions (Supplementary Table E9 in Extended Data). This observation turned our attention to the well-known parcellation of the visual cortex to cytochrome oxidase–rich subregions and so-called blobs (V1) and stripes (V2/V3) (Livingstone and Hubel 1982). We then examined colocalization of DAAM1 and a mitochondrial marker ATP5A in the developing (22nd postconception week) visual cortex (Supplementary Figure E5); its colocalization is consistent with the coexpression analyses described above.
Genetic Correlations with Relevant Complex Traits
We evaluated similarities in the genetic architecture of the global PC1 and the visual PC2 of surface area with that of several traits studied by GWAS in a large number of individuals (n > 20 000). We focused on traits related to general growth (birth weight, adult height), brain function (global cognition, educational attainment), and several brain disorders (attention deficit hyperactivity disorder, schizophrenia, bipolar disorder, major depressive disorder, and late-onset Alzheimer’s disease; see Supplementary Table E10 for the GWAS sources). As shown in Figure 5 (see also Supplementary Table E11 in Extended Data), the SNPs associated with the global PC1 overlapped with SNPs associated with the general growth and brain function but not with any psychiatric disorders. This was not the case for the regional growth of the cerebral cortex (PC2); none of the assessed traits showed genetic correlations (after correction for multiple comparisons). This pattern is consistent with the enrichment of biological processes implicating cell proliferation (e.g., centrosome duplication, chromosome separation) and early brain development (e.g., neural tube closure, brain morphogenesis) in a gene set coexpressed (during prenatal development) with genes associated with PC1 (Supplementary Table E12A in Extended Data); note, nonetheless, that a number of biological processes are equally enriched for the PC1- and PC2-based coexpression datasets (e.g., mitotic nuclear division, neurogenesis; Supplementary Table E12B). Note also that SNP-based heritabilities for surface area PC1 and PC2 were enriched in genomic regions with higher accessibility in the germinal zone (vs. cortical plate), as identified by de la Torre-Ubieta and colleagues (de la Torre-Ubieta et al. 2018) (Supplementary Table E13).
Figure 5.
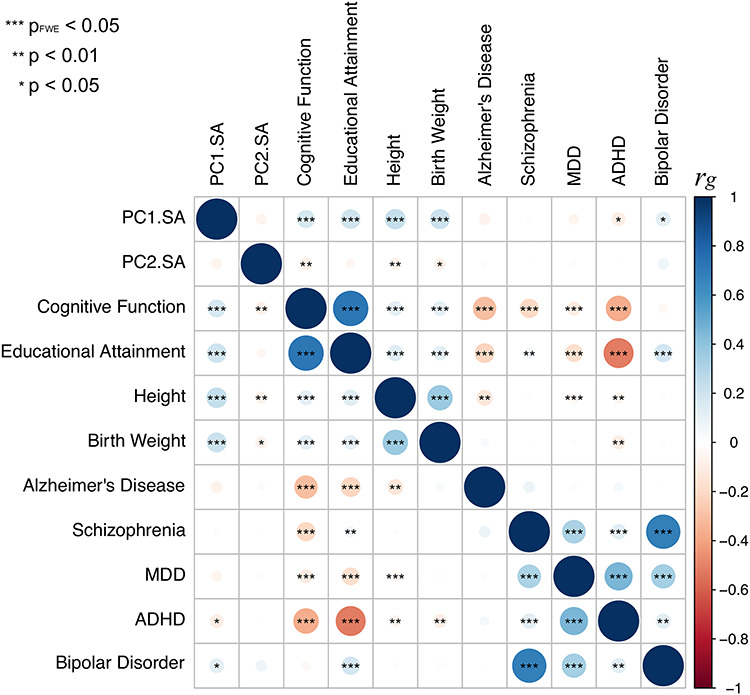
Genetic correlations between PC1 and PC2 (surface area) and a number of complex traits related to general growth, brain function, and several brain disorders. See Supplementary Table E10 for the GWAS sources.
Associations with Occupational Aptitudes
As a proof of concept, we have tested the possibility that the global and regional components, and/or their molecular architecture, predict complex behavior captured by the individual’s occupation. We took advantage of the DOT (National Research Council 1980) that contains 28 801 titles of which 12 099 are so-called “base” titles (corresponding to occupations). Each base title is associated with scores capturing, among other things, aptitudes defined as “the capacities or abilities required of a worker to facilitate the learning of job tasks” (National Research Council 1980, p. 29); DOT contains scores for a total of 11 aptitudes, including General Learning Ability and Form Perception, where lower values indicating greater ability/aptitude (range: 1–5). We chose to test the relationship between the global component (PC1) and the General Learning Ability based on the observed genetic correlation between PC1 and educational attainment (see above). We predicted that individuals with larger PC1 would be more likely to hold occupations associated with a greater General Learning Ability, defined as “The ability to ‘catch on’ or understand instructions and underlying principles; the ability to reason and make judgments. This is closely related to doing well in school” (US Department of Labor 1991, p. 9–3). We chose to test the relationship between the regional (visual) component (PC2) and Form Perception based on comparative studies of the primate visual cortex. These studies suggest that a larger primary visual cortex (containing more neurons) can process visual information with a greater granularity (de Sousa and Proulx 2014). Thus, a model proposed by de Sousa and Proulx states that “the size of visual field represented by a neuron is inversely related to the size of the cortical region” (de Sousa and Proulx 2014). We predicted that individuals with a larger PC2 would be more likely to hold occupations associated with a greater Form Perception aptitude, defined as follows: “The ability to perceive pertinent details in objects or in pictorial or graphic material. Ability to make visual comparisons and discrimination and see slight differences in shapes and shadings of figures and widths and lengths of lines” (US Department of Labor 1991, p. 9–17). Given that greater aptitude for Form Perception is found in occupations requiring also greater General Learning Ability, General Learning Ability can be either a precision or a confounding variable. Hence, in order to increase statistical power or prevent spurious inference, we adjusted for this variable, in addition to sex and age, when testing the association between Form Perception aptitude and PC2. To test these predictions, we used data on occupations of the UK Biobank participants with available MRI data (n = 21 609) and linked the (UK) Standard Occupation Codes (Office for National Statistics 2000) with the DOT codes (National Research Council 1980). To increase homogeneity of this sample, we included only participants classified as “White” in the UK Biobank database (a combination of self-reported ethnicity and genetic principal components; n = 18 890). Both MRI and occupation data were available in 12 829 of these individuals. In this sample, PC1 was associated with General Learning Ability (beta = −0.08; t = −8.4, P = 3.6E − 17; adjusted for sex and age). PC2 was associated with Form Perception aptitude (beta = −0.0059; t = −2.5, P = 0.014; adjusted for sex, age and General Learning Ability, which correlates with Form Perception aptitude [r = 0.38, P < 0.001]). In both cases, higher values of the principal components (i.e., higher global and regional surface area) predicted higher values of the respective occupational aptitudes. We then tested associations between PC1-related MAPT (rs62056789) and PC2-related DAAM1 (rs2164950, an LD-proxy of [the imputed] rs73313052, r2 = 1) and, respectively, General Learning Ability and Form Perception aptitude in all UK Biobank (“White”) individuals with both occupation and genetic data (n = 222 562). For MAPT (rs62056789), we observed an association between General Learning Ability and this polymorphism (P = 8.3E − 10, 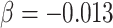 , t = −6.1; adjusted for sex and age). For DAAM1 (rs2164950), we observed no association between Form Perception aptitude and this polymorphism (P = 0.24,
, t = −6.1; adjusted for sex and age). For DAAM1 (rs2164950), we observed no association between Form Perception aptitude and this polymorphism (P = 0.24, 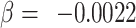 , t = −1.2; adjusted for sex, age and General Learning Ability).
, t = −1.2; adjusted for sex, age and General Learning Ability).
Associations with Polygenic Scores for Surface Area PC1 and PC2
As a proof of concept, we derived polygenic scores based on the GWAS-significant SNPs identified from surface area PC1 and PC2 meta-GWAS and tested their associations with the respective phenotypes in UK Biobank “White” participants (n = 18 036). As expected, the polygenic scores were strongly correlated with the PC1 scores (P = 4.8E − 55, 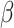 = 0.095, t = 15.7; adjusted for age, sex and MRI site) and PC2 scores (P = 8.5E − 107,
= 0.095, t = 15.7; adjusted for age, sex and MRI site) and PC2 scores (P = 8.5E − 107, 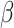 = 0.16, t = 22.1; adjusted for age, sex, MRI site). Finally, in all “White” UK Biobank participants with both the genetic and occupational data (n = 249 690), we tested for the relationship between polygenic scores and the two occupational aptitudes. The polygenic score for PC1 was strongly associated with General Learning Ability in the expected direction, namely, higher polygenic score predicted higher ability (P = 8.4E − 14,
= 0.16, t = 22.1; adjusted for age, sex, MRI site). Finally, in all “White” UK Biobank participants with both the genetic and occupational data (n = 249 690), we tested for the relationship between polygenic scores and the two occupational aptitudes. The polygenic score for PC1 was strongly associated with General Learning Ability in the expected direction, namely, higher polygenic score predicted higher ability (P = 8.4E − 14, 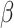 = −0.015, t = −7.5; adjusted for age and sex). We observed no association between Form Perception and the polygenic score for PC2 (P = 0.77,
= −0.015, t = −7.5; adjusted for age and sex). We observed no association between Form Perception and the polygenic score for PC2 (P = 0.77, 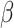 = −0.0005, t = −0.3; adjusted for age, sex and General Learning Ability).
= −0.0005, t = −0.3; adjusted for age, sex and General Learning Ability).
Discussion
Here we discovered a nonoverlapping set of 99 “independent” SNPs contributing to the global and regional tangential growth of the human cerebral cortex. On the other hand, our meta-GWAS of cortical thickness, carried out in the same individuals, yielded only one locus. This negative finding (similar to those reported by the CHARGE (Hofer 2019) and ENIGMA (Grasby 2019) Consortia) may reflect substantial dynamics of cortical thickness during puberty (Walhovd et al. 2017) and aging (Vinke et al. 2018) and the contribution of different genetic variants throughout the lifespan.
The robustness of our findings with regard to surface area is remarkable. It is likely that this phenotype—measured in adults of different ages—provides a high-fidelity proxy of prenatal and early postnatal brain development. As reviewed previously, surface area of the human cerebral cortex reaches its peak around 2 years of age (Gilmore et al. 2018); it remains relatively stable throughout adulthood (less than 4% decrease between 40 and 80 years of age in our UK Biobank sample). The developmental origin of this phenotype is reflected in the results of our meta-GWAS in several respects. First, many genes associated with variations in surface area are highly expressed during prenatal development; expression of DAAM1 is a prime example in that its expression peaks during midgestation and decreases to negligible levels by birth. Second, genes coexpressed with the three genes studied here in more detail (i.e., MAPT, IGF2BP1 and DAAM1) are enriched for biological processes associated with cell growth (e.g., regulation of cell cycle). Third, genetic variants associated with both the global PC1 and visual PC2 are enriched in chromatin regions with higher accessibility in the germinal zone (vs. cortical plate) in the fetal (human) brains.
The global (PC1) component yielded a set of consistent results. First, the main genetic locus (chromosome 17) associated with this component has been observed previously in our meta-GWAS of intracranial volume (Adams et al. 2016); intracranial volume correlates strongly with surface area. Second, genetic variations associated with this component overlap with a number of traits related to both the overall growth of the body, namely, birth weight and adult height, but also with traits associated with cognitive abilities, namely, educational attainment and cognitive function. Third, using occupational aptitudes, we have confirmed the latter by showing a strong association between the global PC1 and General Learning Ability, a trait expected to relate strongly to educational attainment. It is also of note that General Learning Ability was associated with genetic variations in MAPT, one of the genes in the chromosome 17 locus. Mutations in this gene occur more frequently in patients with frontotemporal dementia (Ciani et al. 2019; Strang et al. 2019).
The visual (PC2) component provided an unexpected set of new insights. To start with, the grouping of the primary and secondary visual cortices by their shared variance in surface area suggests that a set of common processes guiding their development may be, to a certain extent, distinct from those involved in the global cortical growth. Experimental work in nonhuman primates points strongly to the eye as a source of this distinction: removing the eyes during fetal development decreases the surface area of the visual cortex (Bourgeois and Rakic 1996; Dehay et al. 1996). This effect is consistent with a tight relationship between the volume of the lateral geniculate nucleus (LGN) and the surface area of the primary visual cortex in the human (adult) brain, as assessed postmortem (Andrews et al. 1997). In both cases, it is likely that waves of spontaneous activity in the retina (Shatz and Stryker 1988; Feller et al. 1996), transmitted to the developing cortex via LGN, play a key role in mediating these effects. At a molecular level, our meta-GWAS of the visual PC2 revealed a robust association in a genetic locus containing DAAM1. DAAM1 is a key component in the planar-cell-polarity signaling pathway (Tissir and Goffinet 2010; Beane et al. 2012); it acts as a bridging factor between disheveled, Rho-family GTPases and Rho-associated kinases (Habas et al. 2001), a molecular complex involved in organizing actin cytoskeleton (Yang and Mlodzik 2015). We showed that DAAM1 is highly expressed between 12th and 22nd postconception weeks in the human cerebral cortex and that genes coexpressed with DAAM1 in the primary visual cortex are enriched in mitochondria-related pathways. We formulated a working model by which DAAM1 regulates tangential expansion of the visual cortex by interacting with LGN inputs, likely at the level of cortical subplate, during midgestation.
A larger visual cortex is likely to possess more interhemispheric connections; DAAM1 polymorphism is associated with a structure-predicted functional connectivity of the human visual cortex (Mollink et al. 2019). Comparative studies in nonhuman primates also suggest that a larger primary visual cortex contains a larger number of neurons and, in turn, a more fine-grained representation of the visual field (de Sousa and Proulx 2014). It has been shown, in both human and nonhuman primates, that species/individuals with a larger visual cortex are less prone to size illusions (e.g., Ebbinghaus illusion), possibly due to less of an overlap between visual representations of the central object and the surrounding context (Schwarzkopf et al. 2011; de Sousa and Proulx 2014). Our findings of a significant—albeit weak—relationship between the visual PC2 and Form Perception aptitude are consistent with a more granular visual representation in individuals with a larger visual cortex.
Overall, these findings illustrate how specification of cortical areas, and their relative growth, might be guided by an interaction between fetal environment and generic developmental mechanisms, such as those constituting planar-cell-polarity signaling pathway. They also illustrate that interindividual variations in global and regional development of the human cerebral cortex (and its molecular architecture) can cascade—albeit in a very limited manner—to behaviors as complex as the choice of one’s occupation.
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Numbers 23509 and 43688.
Conflict of Interest: None declared.
ENIGMA Consortium Authors
Katrina L. Grasby (Psychiatric Genetics, QIMR Berghofer Medical Research Institute, Brisbane, Australia), Neda Jahanshad (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Jodie N. Painter (Psychiatric Genetics, QIMR Berghofer Medical Research Institute, Brisbane, Australia), Lucía Colodro-Conde (Psychiatric Genetics, QIMR Berghofer Medical Research Institute, Brisbane, Australia), Janita Bralten (Department of Human Genetics, Radboud University medical center, Nijmegen, The Netherlands), Derrek P. Hibar (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Penelope A. Lind (Psychiatric Genetics, QIMR Berghofer Medical Research Institute, Brisbane, Australia), Fabrizio Pizzagalli (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Christopher R.K. Ching (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Mary Agnes B. McMahon (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Natalia Shatokhina (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Leo Zsembik (Department of Genetics and UNC Neuroscience Center, University of North Carolina at Chapel HIll, Chapel Hill, USA), Ingrid Agartz (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Saud Alhusaini (Department of Molecular and Cellular Therapeutics, Royal College of Surgeons in Ireland, Dublin, Ireland), Marcio A.A. Almeida (Department of Human Genetics and South Texas Diabetes and Obesity Institute, Rio Grande Valley School of Medicine, University of Texas, Brownsville, USA), Dag Alnæs (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Inge K. Amlien (Centre for Lifespan Changes in Brain and Cognition, Department of Psychology, University of Oslo, Oslo, Norway), Micael Andersson (Department of Integrative Medical Biology, Umeå University, Umeå, Sweden), Tyler Ard (Laboratory of Neuro Imaging, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, University of Southern California, Los Angeles, USA), Nicola J. Armstrong (Mathematics and Statistics, Murdoch University, Murdoch, Australia), Allison Ashley-Koch (Duke Molecular Physiology Institute, Duke University Medical Center, Durham, USA), Manon Bernard (The Hospital for Sick Children, University of Toronto, Toronto, Canada), Rachel M. Brouwer (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands), Elizabeth E.L. Buimer (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands), Robin Bülow (Institute for Radiology and Neuroradiology, University Medicine, Ernst-Moritz-Arndt University, Greifswald, Germany), Christian Bürger (Department of Psychiatry, University of Münster, Münster, Germany), Dara M. Cannon (Centre for Neuroimaging and Cognitive Genomics, National University of Ireland Galway, Galway, Ireland), Mallasr Chakravarty (Douglas Mental Health University Institute, McGill University, Montreal, Canada), Qiang Chen (Lieber Institute for Brain Development, Baltimore, USA), Joshua W. Cheung (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Baptiste Couvy-Duchesne (Institute for Molecular Bioscience, The University of Queensland, Brisbane, Australia), Anders M. Dale (Department of Neurosciences, University of California, San Diego, La Jolla), Shareefa Dalvie (Department of Psychiatry and Mental Health, University of Cape Town, Cape Town, South Africa), Tânia K. de Araujo (Department of Medical Genetics, School of Medical Sciences, University of Campinas—UNICAMP, Campinas, Brazil), Greig I. de Zubicaray (Faculty of Health, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, Australia), Sonja M.C. de Zwarte (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands), Anouk den Braber (Department of Biological Psychology, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Nhat Trung Doan (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Katharina Dohm (Department of Psychiatry, University of Münster, Münster, Germany), Stefan Ehrlich (Division of Psychological and Social Medicine and Developmental Neurosciences, Technische Universität Dresden, Dresden, Germany), Hannah-Ruth Engelbrecht (Division of Human Genetics, Institute of Infectious Disease and Molecular Medicine, University of Cape Town, Cape Town, South Africa), Susanne Erk (Division of Mind and Brain Research, Department of Psychiatry and Psychotherapy, Campus Charité Mitte, Charité—Universitätsmedizin Berlin, Berlin, Germany), Chun Chieh Fan (Department of Cognitive Science, University of California San Diego, San Diego, USA), Iryna O. Fedko (Department of Biological Psychology, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Sonya F. Foley (Cardiff University Brain Research Imaging Centre, Cardiff University, Cardiff, UK), Judith M. Ford (San Francisco Veterans Administration Medical Center, San Francisco, USA), Masaki Fukunaga (Division of Cerebral Integration, National Institute for Physiological Sciences, Okazaki, Japan), Melanie E. Garrett (Duke Molecular Physiology Institute, Duke University Medical Center, Durham, USA), Tian Ge (Psychiatric and Neurodevelopmental Genetics Unit, Center for Genomic Medicine, Massachusetts General Hospital, Boston, USA), Sudheer Giddaluru (NORMENT—K.G. Jebsen Centre for Psychosis Research, Department of Clinical Science, NORMENT University of Bergen, Bergeb, Norway), Aaron L. Goldman (Lieber Institute for Brain Development, Baltimore, USA), Nynke A. Groenewold (Department of Psychiatry and Mental Health, University of Cape Town, Cape Town, South Africa), Dominik Grotegerd (Department of Psychiatry, University of Münster, Münster, Germany), Tiril P. Gurholt (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Boris A. Gutman (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Narelle K. Hansell (Queensland Brain Institute, University of Queensland, St Lucia, Australia), Mathew A. Harris (Centre for Clinical Brain Sciences and Edinburgh Imaging, University of Edinburgh, Edinburgh, UK), Marc B. Harrison (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Courtney C. Haswell (Duke UNC Brain Imaging and Analysis Center, Duke University Medical Center, Durham, USA), Michael Hauser (Duke Molecular Physiology Institute, Duke University Medical Center, Durham, USA), Dirk J. Heslenfeld (Department of Cognitive and Clinical Neuropsychology, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), David Hoehn (Max Planck Institute of Psychiatry, Munich, Germany), Laurena Holleran (Centre for Neuroimaging and Cognitive Genomics, National University of Ireland Galway, Galway, Ireland), Martine Hoogman (Department of Human Genetics, Radboud university medical center, Nijmegen, The Netherlands), Jouke-Jan Hottenga (Department of Biological Psychology, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Masashi Ikeda (Department of Psychiatry, Fujita Health University School of Medicine, Toyoake, Japan), Deborah Janowitz (Department of Psychiatry and Psychotherapy, University Medicine Greifswald, Greifswald, Germany), Iris E. Jansen (Department of Complex Trait Genetics, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Tianye Jia (Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China), Christiane Jockwitz (Institute of Neuroscience and Medicine (INM-1), Research Centre Jülich, Jülich, Germany), Ryota Kanai (Department of Neuroinformatics, Araya, Inc., Inc., Tokyo), Sherif Karama (Department of Psychiatry, McGill University, Montreal, Canada), Dalia Kasperaviciute (Department of Clinical and Experimental Epilepsy, University College London, London, UK), Tobias Kaufmann (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Sinead Kelly (Public Psychiatry Division, Massachusetts Mental Health Center, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, USA), Masataka Kikuchi (Department of Genome Informatics, Graduate School of Medicine, Osaka University, Suita, Japan), Marieke Klein (Department of Human Genetics, Radboud university medical center, Nijmegen, The Netherlands), Michael Knapp (Department of Medical Biometry, Informatics and Epidemiology, University Hospital Bonn, Bonn, Germany), Annchen R. Knodt (Department of Psychology and Neuroscience, Duke University, Durham, USA), Bernd Krämer (Section for Experimental Psychopathology and Neuroimaging, Department of General Psychiatry, Heidelberg University Hospital, Heidelberg, Germany), Thomas M. Lancaster (Cardiff University Brain Research Imaging Centre, Cardiff University, Cardiff, UK), Phil H. Lee (Psychiatric and Neurodevelopmental Genetics Unit, Center for Genomic Medicine, Massachusetts General Hospital, Boston, USA), Tristram A. Lett (Division of Mind and Brain Research, Department of Psychiatry and Psychotherapy, Campus Charité Mitte, Charité—Universitätsmedizin Berlin, Berlin, Germany), Lindsay B. Lewis (McGill Centre for Integrative Neuroscience, McGill University, Montreal, Canada), Iscia Lopes-Cendes (Department of Medical Genetics, School of Medical Sciences, University of Campinas—UNICAMP, Campinas, Brazil), Michelle Luciano (Department of Psychology, University of Edinburgh, Edinburgh, UK), Fabio Macciardi (Department of Psychiatry and Human Behavior, School of Medicine, University of California, Irvine, Irvine, USA), Andre F. Marquand (Department of Cognitive Neuroscience, Radboud university medical center, Nijmegen, The Netherlands), Samuel R. Mathias (Department of Psychiatry, Yale University School of Medicine, New Haven, USA), Tracy R. Melzer (Department of Medicine, University of Otago, Christchurch, Christchurch, New Zealand), Yuri Milaneschi (Psychiatry, Amsterdam UMC Vrije Universiteit, Psychiatry, The Netherlands), Nazanin Mirza-Schreiber (Max Planck Institute of Psychiatry, Munich, Germany), Jose C.V. Moreira (, BRAINN—Brazilian Institute of Neuroscience and Neurotechnology, Campinas, Brasil), Thomas W. Mühleisen (Institute of Neuroscience and Medicine (INM-1), Research Centre Jülich, Jülich, Germany), Bertram Müller-Myhsok (Max Planck Institute of Psychiatry, Munich, Germany), Pablo Najt (Centre for Neuroimaging and Cognitive Genomics, National University of Ireland Galway, Galway, Ireland), Soichiro Nakahara (Department of Psychiatry and Human Behavior, School of Medicine University of California, University of California, Irvine, Irvine, USA), Kwangsik Nho (Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, USA), Loes M. Olde Loohuis (Center for Neurobehavioral Genetics, University of California Los Angeles, Los Angeles, USA), Dimitri Papadopoulos Orfanos (NeuroSpin, CEA, Université Paris-Saclay, Gif sur Yvette, France), John F. Pearson (Biostatistics and Computational Biology Unit, University of Otago, Christchurch, Christchurch, New Zealand), Toni L. Pitcher (Department of Medicine, University of Otago, Christchurch, Christchurch, New Zealand), Benno Pütz (Max Planck Institute of Psychiatry, Munich, Germany), Anjanibhargavi Ragothaman (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Faisal M. Rashid (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Ronny Redlich (Department of Psychiatry, University of Münster, Münster, Germany), Céline S. Reinbold (Department of Biomedicine, University of Basel, Basel, Switzerland), Jonathan Repple (Department of Psychiatry, University of Münster, Münster, Germany), Geneviève Richard (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Brandalyn C. Riedel (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Shannon L. Risacher (Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, USA), Cristiane S. Rocha (Department of Medical Genetics, School of Medical Sciences, University of Campinas—UNICAMP, Campinas, Brazil), Nina Roth Mota (Department of Human Genetics, Radboud university medical center, Nijmegen, The Netherlands), Lauren Salminen (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Arvin Saremi (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Andrew J. Saykin (Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, USA), Fenja Schlag (Language and Genetics Department, Max Planck Institute for Psycholinguistics, Nijmegen, The Netherlands), Lianne Schmaal (Orygen, The National Centre of Excellence for Youth Mental Health, Melbourne, Australia), Peter R. Schofield (Neuroscience Research Australia, Sydney, Australia), Rodrigo Secolin (Department of Medical Genetics, School of Medical Sciences, University of Campinas—UNICAMP, Campinas, Brazil), Chin Yang Shapland (Language and Genetics Department, Max Planck Institute for Psycholinguistics, Nijmegen, The Netherlands), Li Shen (Department of Biostatistics, Epidemiology and Informatics, University of Pennsylvania, Philadelphia, USA), Jean Shin (The Hospital for Sick Children, University of Toronto, Toronto, Canada), Elena Shumskaya (Department of Human Genetics, Radboud university medical center, Nijmegen, The Netherlands), Ida E. Sønderby (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Emma Sprooten (Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, The Netherlands), Lachlan T. Strike (Queensland Brain Institute, University of Queensland, St Lucia, Australia), Katherine E. Tansey (MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, UK), Alexander Teumer (Institute for Community Medicine, University Medicine Greifswald, Greifswald, Germany), Anbupalam Thalamuthu (Centre for Healthy Brain Ageing, University of New South Wales, Sydney, Australia), Sophia I. Thomopoulos (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Diana Tordesillas-Gutiérrez (Neuroimaging Unit, Valdecilla Biomedical Research Institute IDIVAL, Santander, Spain), Jessica A. Turner (Department of Psychology, Georgia State University, Atlanta, USA), Anne Uhlmann (Department of Psychiatry and Mental Health, University of Cape Town, Cape Town, South Africa), Costanza Ludovica Vallerga (Institute for Molecular Bioscience, The University of Queensland, Brisbane, Australia), Dennis van der Meer (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Marjolein M.J. van Donkelaar (Department of Human Genetics, Radboud university medical center, Nijmegen, The Netherlands), Liza van Eijk (School of Psychology, University of Queensland, Brisbane, Australia), Theo G.M. van Erp (Department of Psychiatry and Human Behavior, School of Medicine University of California, University of California, Irvine, Irvine, USA), Neeltje E.M. van Haren (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands), Daan van Rooij (Department of Cognitive Neuroscience, Radboud university medical center, Nijmegen, The Netherlands), Marie-José van Tol (Cognitive Neuroscience Center, Department of Neuroscience, Cardiff University, Groningen, The Netherlands), Jan H. Veldink (Department of Neurology, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands), Ellen Verhoef (Language and Genetics Department, Max Planck Institute for Psycholinguistics, Nijmegen, The Netherlands), Esther Walton (Department of Psychology, Georgia State University, Atlanta, USA), Yunpeng Wang (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Joanna M. Wardlaw (Centre for Clinical Brain Sciences and Edinburgh Imaging, University of Edinburgh, Edinburgh, UK), Wei Wen (Centre for Healthy Brain Ageing, University of New South Wales, Sydney, Australia), Lars T. Westlye (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Christopher D. Whelan (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Stephanie H. Witt (Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany), Katharina Wittfeld (German Center for Neurodegenerative Diseases Rostock/Greifswald, Greifswald, Germany), Christiane Wolf (Department of Psychiatry, Psychosomatics and Psychotherapy, University of Würzburg, Würzburg, Germany), Thomas Wolfers (Department of Human Genetics, Radboud university medical center, Nijmegen, The Netherlands), Clarissa L. Yasuda (Department of Neurology, FCM, University of Campinas—UNICAMP, Campinas, Brasil), Dario Zaremba (Department of Psychiatry, University of Münster, Münster, Germany), Zuo Zhang (Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK), Alyssa H. Zhu (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Marcel P. Zwiers (Department of Cognitive Neuroscience, Radboud university medical center, Nijmegen, The Netherlands), Eric Artiges (INSERM Unit 1000—Neuroimaging and Psychiatry, Paris-Saclay University, Gif sur Yvette, France), Amelia A. Assareh (Centre for Healthy Brain Ageing, University of New South Wales, Sydney, Australia), Rosa Ayesa-Arriola (Department of Psychiatry, University Hospital Marqués de Valdecilla, School of Medicine, University of Cantabria–IDIVAL, Santander, Spain), Aysenil Belger (Duke UNC Brain Imaging and Analysis Center, Duke University Medical Center, Durham, USA), Christine L. Brandt (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Gregory G. Brown (Department of Psychiatry, University of California San Diego, San Diego, USA), Sven Cichon (Department of Biomedicine, University of Basel, Basel, Switzerland), Joanne E. Curran (Department of Human Genetics and South Texas Diabetes and Obesity Institute, Rio Grande Valley School of Medicine, University of Texas, Brownsville, USA), Gareth E. Davies (Avera Institute for Human Genetics, Sioux Falls, USA), Franziska Degenhardt (Institute of Human Genetics, School of Medicine and University Hospital Bonn, University of Bonn, Bonn, Germany), Bruno Dietsche (Department of Psychiatry and Psychotherapy, Philipps-University Marburg, Marburg, Germany), Srdjan Djurovic (Department of Medical Genetics, Oslo University Hospital, Oslo, Norway), Colin P. Doherty (Department of Neurology, St James’s Hospital, Dublin, Ireland), Ryan Espiritu (Information Sciences Institute, University of Southern California, Los Angeles, USA), Daniel Garijo (Information Sciences Institute, University of Southern California, Los Angeles, USA), Yolanda Gil (Information Sciences Institute, University of Southern California, Los Angeles, USA), Penny A. Gowland (Sir Peter Mansfield Imaging Centre, University of Nottingham, Nottingham, UK), Robert C. Green (Brigham and Women’s Hospital, Boston, USA), Alexander N. Häusler (Center for Economics and Neuroscience, University of Bonn, Bonn, Germany), Walter Heindel (Department of Clinical Radiology, University of Münster, Münster, Germany), Beng-Choon Ho (Department of Psychiatry, University of Iowa College of Medicine, Iowa City, USA), Wolfgang U. Hoffmann (Institute for Community Medicine, University Medicine Greifswald, Greifswald, Germany), Florian Holsboer (HMNC Holding GmbH, Munich, Germany), Georg Homuth (Department of Functional Genomics, Interfaculty Institute for Genetics and Functional Genomics, University Medicine Greifswald, Greifswald, Germany), Norbert Hosten (Institute of Diagnostic Radiology and Neuroradiology, Greifswald, Germany), Clifford R. Jack Jr (Dept of Radiology, Mayo Clinic, Rochester, USA), MiHyun Jang (Information Sciences Institute, University of Southern California, Los Angeles, USA), Andreas Jansen (Department of Psychiatry and Psychotherapy, Philipps-University Marburg, Marburg, Germany), Knut Kolskår (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Sanne Koops (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands), Axel Krug (Department of Psychiatry and Psychotherapy, Philipps-University Marburg, Marburg, Germany), Kelvin O. Lim (Department of Psychiatry, University of Minnesota, Minneapolis, USA), Jurjen J. Luykx (Department of Translational Neuroscience, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands), Daniel H. Mathalon (Department of Psychiatry and Weill Institute for Neurosciences, University of California San Francisco, San Francisco, USA), Karen A. Mather (Centre for Healthy Brain Ageing, University of New South Wales, Sydney, Australia), Venkata S. Mattay (Lieber Institute for Brain Development, Baltimore, USA), Sarah Matthews (MRC Integrative Epidemiology Unit, Department of Population Health Sciences, Bristol Medical School, Bristol, UK), Jaqueline Mayoral Van Son (Department of Psychiatry, University Hospital Marqués de Valdecilla, School of Medicine, University of Cantabria–IDIVAL, Santander, Spain), Sarah C. McEwen (Department of Psychiatry, University of California San Diego, La Jolla, USA), Ingrid Melle (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Derek W. Morris (Centre for Neuroimaging and Cognitive Genomics, National University of Ireland Galway, Galway, Ireland), Bryon A. Mueller (Department of Psychiatry, University of Minnesota, Minneapolis, USA), Matthias Nauck (Institute of Clinical Chemistry and Laboratory Medicine, University Medicine Greifswald, Greifswald, Germany), Jan E. Nordvik (Sunnaas Rehabilitation Hospital HT, Nesodden, Norway), Markus M. Nöthen (Institute of Human Genetics, School of Medicine and University Hospital Bonn, University of Bonn, Bonn, Germany), Daniel S. O’Leary (Department of Psychiatry, University of Iowa College of Medicine, Iowa City, USA), Nils Opel (Department of Psychiatry, University of Münster, Münster, Germany), Marie—Laure Paillère Martinot (INSERM Unit 1000—Neuroimaging and Psychiatry, Paris-Saclay University, Gif sur Yvette, France), G. Bruce Pike (Radiology and Clinical Neurosciences, Hotchkiss Brain Institute, University of Calgary, Calgary, Canada), Adrian Preda (School of Medicine, University of California Irvine, Irvine, USA), Erin B. Quinlan (Social, Genetic and Developmental Psychiatry Centre, King’s College London, London, UK), Varun Ratnakar (Information Sciences Institute, University of Southern California, Los Angeles, USA), Simone Reppermund (Centre for Healthy Brain Ageing, University of New South Wales, Sydney, Australia), Vidar M. Steen (NORMENT—K.G. Jebsen Centre for Psychosis Research, Department of Clinical Science, NORMENT University of Bergen, Bergen, Norway), Fábio R. Torres (Department of Medical Genetics, School of Medical Sciences, University of Campinas—UNICAMP, Campinas, Brazil), Dick J. Veltman (Psychiatry, Amsterdam UMC Vrije Universiteit, Psychiatry, The Netherlands), James T. Voyvodic (Duke UNC Brain Imaging and Analysis Center, Duke University Medical Center, Durham, USA), Robert Whelan (School of Psychology, Trinity College Dublin, Dublin, Ireland), Tonya White (Department of Child and Adolescent Psychiatry/Psychology, Erasmus Medical Center-Sophia Children’s Hospital, Rotterdam, The Netherlands), Hidenaga Yamamori (Department of Psychiatry, Osaka University Graduate School of Medicine, Suita, Japan), Marina K.M. Alvim (Department of Neurology, FCM, University of Campinas—UNICAMP, Campinas, Brasil), David Ames (Academic Unit for Psychiatry of Old Age, University of Melbourne, Melbourne, Australia), Tim J. Anderson (Department of Medicine, University of Otago, Christchurch, Christchurch, New Zealand), Ole A. Andreassen (NORMENT—K.G. Jebsen Centre for Psychosis Research, Division of Mental Health and Addiction, NORMENT Oslo University Hospital, Oslo, Norway), Alejandro Arias-Vasquez (Department of Psychiatry, Radboud university medical center, Nijmegen, The Netherlands), Mark E. Bastin (Centre for Clinical Brain Sciences and Edinburgh Imaging, University of Edinburgh, Edinburgh, UK), Bernhard T. Baune (Department of Psychiatry, The University of Melbourne, Melbourne, Australia.), John Blangero (Department of Human Genetics and South Texas Diabetes and Obesity Institute, Rio Grande Valley School of Medicine, University of Texas, Brownsville, USA), Dorret I. Boomsma (Department of Biological Psychology, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Henry Brodaty (Centre for Healthy Brain Ageing, University of New South Wales, Sydney, Australia), Han G. Brunner (Department of Human Genetics, Radboud university medical center, Nijmegen, The Netherlands), Randy L. Buckner (Department of Psychology and Center for Brain Science, Harvard University, Boston, USA), Jan K. Buitelaar (Department of Cognitive Neuroscience, Radboud university medical center, Nijmegen, The Netherlands), Juan R. Bustillo (Department of Psychiatry, University of New Mexico, Albuquerque, USA), Wiepke Cahn (Department of Psychiatry, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands), Vince Calhoun (Department of Electrical and Computer Engineering, The University of New Mexico, Albuquerque, USA), Xavier Caseras (MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, UK), Svenja Caspers (Institute for Anatomy I Medical Faculty, Heinrich-Heine University, Düsseldorf, Germany), Gianpiero L. Cavalleri (Molecular and Cellular Therapeutics, The Royal College of Surgeons In Ireland, Dublin, Ireland), Fernando Cendes (Department of Neurology, FCM, University of Campinas—UNICAMP, Campinas, Brasil), Aiden Corvin (Department of Psychiatry, School of Medicine, Trinity College Dublin, Dublin, Ireland), Benedicto Crespo-Facorro (Department of Psychiatry, University Hospital Marqués de Valdecilla, School of Medicine, University of Cantabria–IDIVAL, Santander, Spain), John C. Dalrymple-Alford (Department of Psychology, University of Canterbury, Christchurch, New Zealand), Udo Dannlowski (Department of Psychiatry, University of Münster, Münster, Germany), Eco J.C. de Geus (Department of Biological Psychology, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Ian J. Deary (Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, Edinburgh, UK), Norman Delanty (FutureNeuro Research Centre, Royal College of Surgeons in Ireland, Dublin, Ireland), Chantal Depondt (Department of Neurology, Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium), Sylvane Desrivières (Social, Genetic and Developmental Psychiatry Centre, King’s College London, London, UK), Gary Donohoe (Centre for Neuroimaging and Cognitive Genomics, National University of Ireland Galway, Galway, Ireland), Thomas Espeseth (Department of Psychology, University of Oslo, Oslo, Norway), Guillén Fernández (Department of Cognitive Neuroscience, Radboud university medical center, Nijmegen, The Netherlands), Simon E. Fisher (Language and Genetics Department, Max Planck Institute for Psycholinguistics, Nijmegen, The Netherlands), Herta Flor (Department of Cognitive and Clinical Neuroscience, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany), Andreas J. Forstner (Institute of Human Genetics, School of Medicine and University Hospital Bonn, University of Bonn, Bonn, Germany), Clyde Francks (Language and Genetics Department, Max Planck Institute for Psycholinguistics, Nijmegen, The Netherlands), Barbara Franke (Department of Human Genetics, Radboud university medical center, Nijmegen, The Netherlands), David C. Glahn (Department of Psychiatry, Yale University School of Medicine, New Haven, USA), Randy L. Gollub (Department of Psychiatry, Massachusetts General Hospital, Boston, USA), Hans J. Grabe (German Center for Neurodegenerative Diseases Rostock/Greifswald, Greifswald, Germany), Oliver Gruber (Section for Experimental Psychopathology and Neuroimaging, Department of General Psychiatry, Heidelberg University Hospital, Heidelberg, Germany), Asta K. Håberg (Department of Neuroscience, Norwegian University of Science and Technology, Trondheim, Norway), Ahmad R. Hariri (Department of Psychology and Neuroscience, Duke University, Durham, USA), Catharina A. Hartman (Department of Psychiatry, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands), Ryota Hashimoto (Molecular Research Center for Children’s Mental Development, United Graduate School of Child Development, Osaka University, Suita, Japan), Andreas Heinz (Division of Mind and Brain Research, Department of Psychiatry and Psychotherapy, Campus Charité Mitte, Charité—Universitätsmedizin Berlin, Berlin, Germany), Manon H.J. Hillegers (Department of Child and Adolescent Psychiatry/Psychology, Erasmus Medical Center-Sophia Children’s Hospital, Rotterdam, The Netherlands), Pieter J. Hoekstra (Department of Psychiatry, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands), Avram J. Holmes (Department of Psychology, Yale University, New Haven, USA), L. Elliot Hong (Department of Psychiatry, Maryland Psychiatry Research Center, University of Maryland School of Medicine, Baltimore, USA), William D. Hopkins (Neuroscience Institute, Georgia State University, Atlanta, USA), Hilleke E. Hulshoff Pol (Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands), Terry L. Jernigan (Center for Human Development, University of California San Diego, La Jolla, USA), Erik G. Jönsson (Centre for Psychiatric Research, Department of Clinical Neuroscience, Karolinska Institutet, Stockholm, Sweden), René S. Kahn (Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, USA), Martin A. Kennedy (Department of Pathology and Biomedical Science, University of Otago, Christchurch, Christchurch, New Zealand), Tilo T.J. Kircher (Department of Psychiatry and Psychotherapy, Philipps-University Marburg, Marburg, Germany), Peter Kochunov (Department of Psychiatry, Maryland Psychiatry Research Center, University of Maryland School of Medicine, Baltimore, USA), John B.J. Kwok (Neurogenetics and Epigenetics, Brain and Mind Centre, The University of Sydney, Sydney, Australia), Stephanie Le Hellard (NORMENT—K.G. Jebsen Centre for Psychosis Research, Department of Clinical Science, NORMENT University of Bergen, Bergen, Norway), Nicholas G. Martin (Genetic Epidemiology, QIMR Berghofer Medical Research Institute, Brisbane, Australia), Jean—Luc Martinot (INSERM Unit 1000—Neuroimaging and Psychiatry, Paris-Saclay University, Gif sur Yvette, France), Colm McDonald (Centre for Neuroimaging and Cognitive Genomics, National University of Ireland Galway, Galway, Ireland), Katie L. McMahon (Herston Imaging Research Facility, School of Clinical Sciences, Queensland University of Technology, Brisbane, Australia), Andreas Meyer-Lindenberg (Department of Psychiatry and Psychotherapy, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany), Rajendra A. Morey (Duke UNC Brain Imaging and Analysis Center, Duke University Medical Center, Durham, USA), Lars Nyberg (Department of Integrative Medical Biology, Umeå University, Umeå, Sweden), Jaap Oosterlaan (Emma Children’s Hospital, Academic Medical Center, Amsterdam, The Netherlands), Roel A. Ophoff (Center for Neurobehavioral Genetics, University of California Los Angeles, Los Angeles, USA), Tomas Paus (Holland Bloorview Kids Rehabilitation Hospital, Bloorview Research Institute, University of Toronto, Toronto, ON, Canada), Zdenka Pausova (The Hospital for Sick Children, University of Toronto, Toronto, Canada), Brenda W.J.H. Penninx (Psychiatry, Amsterdam UMC Vrije Universiteit, Psychiatry, The Netherlands), Tinca J.C. Polderman (Complex Trait Genetics, Center for Neurogenomics and Cognitive Research, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Danielle Posthuma (Department of Biological Psychology, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Marcella Rietschel (Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany), Joshua L. Roffman (Department of Psychiatry, Massachusetts General Hospital, Boston, USA), Laura M. Rowland (Department of Psychiatry, Maryland Psychiatry Research Center, University of Maryland School of Medicine, Baltimore, USA), Perminder S. Sachdev (Centre for Healthy Brain Ageing, University of New South Wales, Sydney, Australia), Philipp G. Sämann (Max Planck Institute of Psychiatry, Munich, Germany), Gunter Schumann (Social, Genetic and Developmental Psychiatry Centre, King’s College London, London, UK), Kang Sim (General Psychiatry, Institute of Mental Health, Singapore, Singapore), Sanjay M. Sisodiya (Department of Clinical and Experimental Epilepsy, University College London, London, UK), Jordan W. Smoller (Psychiatric and Neurodevelopmental Genetics Unit, Center for Genomic Medicine, Massachusetts General Hospital, Boston, USA), Iris E. Sommer (Department of Medical and Biological Psychology, University of Bergen, Bergen, Norway), Beate St Pourcain (MRC Integrative Epidemiology Unit, Department of Population Health Sciences, Bristol Medical School, Bristol, UK), Dan J. Stein (Department of Psychiatry and Mental Health, University of Cape Town, Cape Town, South Africa), Arthur W. Toga (Laboratory of Neuro Imaging, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, University of Southern California, Los Angeles, USA), Julian N. Trollor (Department of Developmental Disability Neuropsychiatry, University of New South Wales, Sydney, Australia), Nic J.A. Van der Wee (Department of Psychiatry, Leiden University Medical Center, Leiden, The Netherlands), Dennis van’t Ent (Department of Biological Psychology, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands), Henry Völzke (Institute for Community Medicine, University Medicine Greifswald, Greifswald, Germany), Henrik Walter (Division of Mind and Brain Research, Department of Psychiatry and Psychotherapy, Campus Charité Mitte, Charité—Universitätsmedizin Berlin, Berlin, Germany), Bernd Weber (Institute of Experimental Epileptology and Cognition Research, University Hospital Bonn, Bonn, Germany), Daniel R. Weinberger (Lieber Institute for Brain Development, Baltimore, USA), Margaret J. Wright (Queensland Brain Institute, University of Queensland, St Lucia, Australia), Juan Zhou (Center for Cognitive Neuroscience, Neuroscience and behavioral disorders program, Duke-National University of Singapore Medical School, Singapore, Singapore), Jason L. Stein (Department of Genetics and UNC Neuroscience Center, University of North Carolina at Chapel HIll, Chapel Hill, USA), Paul M. Thompson (Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, USA), Sarah E. Medland (Psychiatric Genetics, QIMR Berghofer Medical Research Institute, Brisbane, Australia)
Supplementary Material
Contributor Information
Jean Shin, The Hospital for Sick Children, University of Toronto, Toronto, M5G 0A4 ON, M5G 0A4, Canada; Holland Bloorview Kids Rehabilitation Hospital, Bloorview Research Institute, University of Toronto, Toronto, M4G 1R8 ON, Canada.
Shaojie Ma, Department of Genetics, Yale University School of Medicine, New Haven, 06510 CT, USA; Department of Neuroscience, Yale University School of Medicine, New Haven, 06510 CT, USA.
Edith Hofer, Clinical Division of Neurogeriatrics, Department of Neurology, Medical University of Graz, 8036 Graz, Austria; Institute for Medical Informatics, Statistics and Documentation, Medical University of Graz, 8036 Graz, Austria.
Yash Patel, Holland Bloorview Kids Rehabilitation Hospital, Bloorview Research Institute, University of Toronto, Toronto, M4G 1R8 ON, Canada.
Daniel E Vosberg, Holland Bloorview Kids Rehabilitation Hospital, Bloorview Research Institute, University of Toronto, Toronto, M4G 1R8 ON, Canada.
Steven Tilley, Holland Bloorview Kids Rehabilitation Hospital, Bloorview Research Institute, University of Toronto, Toronto, M4G 1R8 ON, Canada.
Gennady V Roshchupkin, Department of Radiology and Nuclear Medicine, Erasmus University Medical Center, 3015 Rotterdam, The Netherlands; Department of Medical Informatics, Erasmus MC, 3015 Rotterdam, The Netherlands; Department of Epidemiology, Erasmus University Medical Center, 3015 Rotterdam, The Netherlands.
André M M Sousa, Department of Neuroscience, Kavli Institute for Neuroscience, Yale School of Medicine, New Haven, 06510 CT, USA.
Xueqiu Jian, Institute of Molecular Medicine and Human Genetics Center, University of Texas Health Science Center at Houston, 77030 Houston, 77030 TX, USA.
Rebecca Gottesman, Johns Hopkins School of Medicine, Baltimore, 21205 MD, USA.
Thomas H Mosley, University of Mississippi Medical Center, Jackson, 39216 MS, USA.
Myriam Fornage, Institute of Molecular Medicine and Human Genetics Center, University of Texas Health Science Center at Houston, 77030 Houston, 77030 TX, USA.
Yasaman Saba, Gottfried Schatz Research Center for Cell Signaling, Metabolism and Aging, Medical University of Graz, 8036 Graz, Austria.
Lukas Pirpamer, Clinical Division of Neurogeriatrics, Department of Neurology, Medical University of Graz, 8036 Graz, Austria.
Reinhold Schmidt, Clinical Division of Neurogeriatrics, Department of Neurology, Medical University of Graz, 8036 Graz, Austria.
Helena Schmidt, Gottfried Schatz Research Center for Cell Signaling, Metabolism and Aging, Medical University of Graz, 8036 Graz, Austria.
Amaia Carrion-Castillo, Language and Genetics Department, Max Planck Institute for Psycholinguistics, 6525 Nijmegen, The Netherlands.
Fabrice Crivello, Groupe d’Imagerie Neurofonctionnelle, Institut des Maladies Neurodégénératives, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique, et Université de Bordeaux, F-33000 Bordeaux, France.
Bernard Mazoyer, Groupe d’Imagerie Neurofonctionnelle, Institut des Maladies Neurodégénératives, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique, et Université de Bordeaux, F-33000 Bordeaux, France.
Joshua C Bis, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, 98101 WA, USA.
Shuo Li, Department of Biostatistics, Boston University School of Public Health, Boston, 02118, MA, USA.
Qiong Yang, Department of Biostatistics, Boston University School of Public Health, Boston, 02118, MA, USA.
Michelle Luciano, Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, EH8 9YL Edinburgh, UK; Department of Psychology, University of Edinburgh, EH8 9JZ Edinburgh, UK.
Sherif Karama, Montreal Neurological Institute, McGill University, H3A 2B4 Montreal, QC, Canada.
Lindsay Lewis, Montreal Neurological Institute, McGill University, H3A 2B4 Montreal, QC, Canada.
Mark E Bastin, Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, EH8 9YL Edinburgh, UK; Centre for Clinical Brain Sciences, University of Edinburgh, EH8 9YL Edinburgh, UK.
Mathew A Harris, Centre for Clinical Brain Sciences, University of Edinburgh, EH8 9YL Edinburgh, UK; Division of Psychiatry, University of Edinburgh, EH8 9JZ Edinburgh, UK.
Joanna M Wardlaw, Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, EH8 9YL Edinburgh, UK; UK Dementia Research Institute, University of Edinburgh, EH8 9JZ Edinburgh, UK.
Ian E Deary, Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, EH8 9YL Edinburgh, UK; Department of Psychology, University of Edinburgh, EH8 9JZ Edinburgh, UK.
Markus Scholz, Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, 04109 Leipzig, Germany; LIFE Research Center for Civilization Diseases, 04103 Leipzig, Germany.
Markus Loeffler, Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, 04109 Leipzig, Germany; LIFE Research Center for Civilization Diseases, 04103 Leipzig, Germany.
A Veronica Witte, Department of Neurology, Max Planck Institute for Human Cognitive and Brain Sciences, 04103 Leipzig, Germany; Faculty of Medicine, CRC 1052 Obesity Mechanisms, University of Leipzig, 04109 Leipzig, Germany; Day Clinic for Cognitive Neurology, University Hospital Leipzig, 04103 Leipzig, Germany.
Frauke Beyer, Department of Neurology, Max Planck Institute for Human Cognitive and Brain Sciences, 04103 Leipzig, Germany; Faculty of Medicine, CRC 1052 Obesity Mechanisms, University of Leipzig, 04109 Leipzig, Germany.
Arno Villringer, Department of Neurology, Max Planck Institute for Human Cognitive and Brain Sciences, 04103 Leipzig, Germany; Faculty of Medicine, CRC 1052 Obesity Mechanisms, University of Leipzig, 04109 Leipzig, Germany; Day Clinic for Cognitive Neurology, University Hospital Leipzig, 04103 Leipzig, Germany.
Nicola J Armstrong, Mathematics and Statistics, Murdoch University, 6150 Perth, WA, Australia.
Karen A Mather, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, 2052 Sydney, NSW, Australia; Neuroscience Research Australia, 2031 Sydney, NSW, Australia.
David Ames, National Ageing Research Institute, Royal Melbourne Hospital, 3052 Melbourne, VIC, Australia; Academic Unit for Psychiatry of Old Age, St. Vincent's Health, The University of Melbourne, 3010 Melbourne, VIC, Australia.
Jiyang Jiang, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, 2052 Sydney, NSW, Australia.
John B Kwok, Brain and Mind Centre, The University of Sydney, 2050 Sydney, NSW, Australia; School of Medical Sciences, University of New South Wales, 2052 Sydney, NSW, Australia.
Peter R Schofield, Neuroscience Research Australia, 2031 Sydney, NSW, Australia; School of Medical Sciences, University of New South Wales, 2052 Sydney, NSW, Australia.
Anbupalam Thalamuthu, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, 2052 Sydney, NSW, Australia.
Julian N Trollor, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, 2052 Sydney, NSW, Australia; Department of Developmental Disability Neuropsychiatry, School of Psychiatry, University of New South Wales, 2031 Sydney, NSW, Australia.
Margaret J Wright, Queensland Brain Institute, The University of Queensland, 4072 St Lucia, QLD, Australia; Centre for Advanced Imaging, The University of Queensland, 4072 St Lucia, QLD, Australia.
Henry Brodaty, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, 2052 Sydney, NSW, Australia; Dementia Centre for Research Collaboration, University of New South Wales, 2052 Sydney, NSW, Australia.
Wei Wen, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, 2052 Sydney, NSW, Australia.
Perminder S Sachdev, Centre for Healthy Brain Ageing, School of Psychiatry, University of New South Wales, 2052 Sydney, NSW, Australia; Neuropsychiatric Institute, Prince of Wales Hospital, 2031 Sydney, NSW, Australia.
Natalie Terzikhan, Department of Epidemiology, Erasmus University Medical Center, 3015 Rotterdam, The Netherlands.
Tavia E Evans, Department of Radiology and Nuclear Medicine, Erasmus University Medical Center, 3015 Rotterdam, The Netherlands; Department of Epidemiology, Erasmus University Medical Center, 3015 Rotterdam, The Netherlands.
Hieab H H H Adams, Department of Radiology and Nuclear Medicine, Erasmus University Medical Center, 3015 Rotterdam, The Netherlands; Department of Epidemiology, Erasmus University Medical Center, 3015 Rotterdam, The Netherlands.
M Arfan Ikram, Department of Radiology and Nuclear Medicine, Erasmus University Medical Center, 3015 Rotterdam, The Netherlands; Department of Epidemiology, Erasmus University Medical Center, 3015 Rotterdam, The Netherlands; Department of Neurology, Erasmus MC University Medical Centre, 3015 Rotterdam, The Netherlands.
Stefan Frenzel, Department of Psychiatry and Psychotherapy, University Medicine Greifswald, 17489 Greifswald, Germany.
Sandra van der Auwera-Palitschka, Department of Psychiatry and Psychotherapy, University Medicine Greifswald, 17489 Greifswald, Germany; 44German Center for Neurodegenerative Diseases (DZNE), Site Rostock/Greifswald, Greifswald, 37075, Germany.
Katharina Wittfeld, Department of Psychiatry and Psychotherapy, University Medicine Greifswald, 17489 Greifswald, Germany; 44German Center for Neurodegenerative Diseases (DZNE), Site Rostock/Greifswald, Greifswald, 37075, Germany.
Robin Bülow, Institute for Diagnostic Radiology and Neuroradiology, University Medicine Greifswald, 17489 Greifswald, Germany.
Hans Jörgen Grabe, Department of Psychiatry and Psychotherapy, University Medicine Greifswald, 17489 Greifswald, Germany; 44German Center for Neurodegenerative Diseases (DZNE), Site Rostock/Greifswald, Greifswald, 37075, Germany.
Christophe Tzourio, Inserm, Bordeaux Population Health Research Center, University of Bordeaux, Team VINTAGE, UMR 1219, F-33000 Bordeaux, France; Department of Neurology, CHU de Bordeaux, F-33000 Bordeaux, France.
Aniket Mishra, Inserm, Bordeaux Population Health Research Center, University of Bordeaux, Team VINTAGE, UMR 1219, F-33000 Bordeaux, France.
Sophie Maingault, Institut des Maladies Neurodégénratives, UMR 5293, CEA, CNRS, University of Bordeaux, Ubordeaux, F-33000 Bordeaux, France.
Stephanie Debette, Inserm, Bordeaux Population Health Research Center, University of Bordeaux, Team VINTAGE, UMR 1219, F-33000 Bordeaux, France; Department of Neurology, CHU de Bordeaux, F-33000 Bordeaux, France; Department of Neurology, Boston University School of Medicine, Boston, 02118 MA, USA.
Nathan A Gillespie, Virginia Institute for Psychiatric and Behavioural Genetics, Virginia Commonwealth University, Richmond, 23284 VA, USA.
Carol E Franz, Department of Psychiatry, University of California, San Diego, 92093 CA, USA; Center for Behavior Genetics of Aging, University of California, San Diego, 92093 CA, USA.
William S Kremen, Department of Psychiatry, University of California, San Diego, 92093 CA, USA; Center for Behavior Genetics of Aging, University of California, San Diego, 92093 CA, USA; VA San Diego Center of Excellence for Stress and Mental Health, San Diego, 92161 CA, USA.
Linda Ding, Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, 90033 CA, USA.
Neda Jahanshad, Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, University of Southern California, Los Angeles, 90033 CA, USA.
Nenad Sestan, Department of Genetics, Yale University School of Medicine, New Haven, 06510 CT, USA; Department of Neuroscience, Yale University School of Medicine, New Haven, 06510 CT, USA.
Zdenka Pausova, The Hospital for Sick Children, University of Toronto, Toronto, M5G 0A4 ON, M5G 0A4, Canada; Department of Physiology, University of Toronto, Toronto, M5S 1A8 ON, Canada; Department of Nutritional Sciences, University of Toronto, Toronto, M5S 1A8 ON, Canada.
Sudha Seshadri, Department of Neurology, Boston University School of Medicine, Boston, 02118 MA, USA; 55Department of Epidemiology and Biostatistics, Glenn Biggs Institute for Alzheimer’s and Neurodegenerative Diseases, UT Health San Antonio, San Antonio, 78229 TX, USA.
Tomas Paus, Holland Bloorview Kids Rehabilitation Hospital, Bloorview Research Institute, University of Toronto, Toronto, M4G 1R8 ON, Canada; Department of Psychology, University of Toronto, Toronto, M5S 3G3 ON, Canada; Department of Psychiatry, University of Toronto, Toronto, M5T 1R8 ON, Canada.
References
- Adams HH, Hibar DP, Chouraki V, Stein JL, Nyquist PA, Renteria ME, Trompet S, Arias-Vasquez A, Seshadri S, Desrivieres S, et al. 2016. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 19:1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TJ, Halpern SD, Purves D. 1997. Correlated size variations in human visual cortex, lateral geniculate nucleus, and optic tract. J Neurosci. 17:2859–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane WS, Tseng AS, Morokuma J, Lemire JM, Levin M. 2012. Inhibition of planar cell polarity extends neural growth during regeneration, homeostasis, and development. Stem Cells Dev. 21:2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois JP, Rakic P. 1996. Synaptogenesis in the occipital cortex of macaque monkey devoid of retinal input from early embryonic stages. Eur J Neurosci. 8:942–950. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JR, Patterson N, Robinson EB, et al. 2015a. An atlas of genetic correlations across human diseases and traits. Nat Genet. 47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C, Patterson N, Daly MJ, Price AL, Neale BM. 2015b. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Fiecas M, Gutierrez ED, Panizzon MS, Eyler LT, Vuoksimaa E, Thompson WK, Fennema-Notestine C, Hagler DJ Jr, Jernigan TL, et al. 2013. Genetic topography of brain morphology. Proc Natl Acad Sci U S A. 110:17089–17094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Gutierrez ED, Thompson W, Panizzon MS, Jernigan TL, Eyler LT, Fennema-Notestine C, Jak AJ, Neale MC, Franz CE, et al. 2012. Hierarchical genetic organization of human cortical surface area. Science. 335:1634–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani M, Benussi L, Bonvicini C, Ghidoni R. 2019. Genome wide association study and next generation sequencing: a glimmer of light toward new possible horizons in frontotemporal dementia research. Front Neurosci. 13:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosca T, Emmel A. 2010. Revising the standard occupational classification system for 2010. Mon Labor Rev. 133:32–41. [Google Scholar]
- de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, Geschwind DH. 2018. The dynamic landscape of open chromatin during human cortical neurogenesis. Cell. 172:289–304. e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa AA, Proulx MJ. 2014. What can volumes reveal about human brain evolution? A framework for bridging behavioral, histometric, and volumetric perspectives. Front Neuroanat. 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Giroud P, Berland M, Killackey H, Kennedy H. 1996. Contribution of thalamic input to the specification of cytoarchitectonic cortical fields in the primate: effects of bilateral enucleation in the fetal monkey on the boundaries, dimensions, and gyrification of striate and extrastriate cortex. J Comp Neurol. 367:70–89. [DOI] [PubMed] [Google Scholar]
- Emmel A, Cosca T. 2010. The 2010 standard occupational classification (soc): a classification system gets an update. OCCUP Outlook Q. 54:13–19. [Google Scholar]
- Euesden J, Lewis CM, O'Reilly PF. 2015. Prsice: polygenic risk score software. Bioinformatics. 31:1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller MB, Wellis DP, Stellwagen D, Werblin FS, Shatz CJ. 1996. Requirement for cholinergic synaptic transmission in the propagation of spontaneous retinal waves. Science. 272:1182–1187. [DOI] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K. 2015. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 47:1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. 2000. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 97:11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, Zheng J, Duggirala A, McArdle WL, Ho K, et al. 2016. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore JH, Knickmeyer RC, Gao W. 2018. Imaging structural and functional brain development in early childhood. Nat Rev Neurosci. 19:123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby KL, Jahanshad N, Painter JN, Colodro-Conde L, Bralten J, Hibar DP, Lind PA, Pizzagalli F, Ching CRK, McMahon MAB, et al. 2019. The genetic architecture of the human cerebral cortex. bioRxiv. 399402; doi: 10.1101/399402. [DOI] [Google Scholar]
- Habas R, Kato Y, He X. 2001. Wnt/frizzled activation of rho regulates vertebrate gastrulation and requires a novel formin homology protein daam1. Cell. 107:843–854. [DOI] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M, et al. 2015. Common genetic variants influence human subcortical brain structures. Nature. 520:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer E, Roshchupkin GV, Adams HHH, Knol MJ, Lin H, Li S, Zare H, Ahmad S, Armstrong NJ, Satizabal CL, et al. 2019. Genetic determinants of cortical structure (thickness, surface area and volumes) among disease free adults in the CHARGE Consortium. bioRxiv. doi: 10.1101/409649. [DOI] [Google Scholar]
- International Labour Office . 2012. International Standard Classification of Occupations 2008 (ISCO-08): structure, group definitions and correspondence tables. Geneva: International Labour Office. [Google Scholar]
- United States Department of Labor & Bureau of Labor Statistics . 2012. Crosswalk between the 2008 International Standard Classification of Occupations to the 2010 SOC. https://www.bls.gov/soc/ISCO_SOC_Crosswalk_process.pdf (03 October 2019, date last accessed).
- Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, et al. 2018. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 362:eaat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Xiao G, Yin H, Hippenmeyer S, Horowitz JM, Ghashghaei HT. 2013. Neural development is dependent on the function of specificity protein 2 in cell cycle progression. Development. 140:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone MS, Hubel DH. 1982. Thalamic inputs to cytochrome oxidase-rich regions in monkey visual cortex. Proc Natl Acad Sci U S A. 79:6098–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. 2019. Panther version 14: more genomes, a new panther go-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47:D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollink J, Smith SM, Elliott LT, Kleinnijenhuis M, Hiemstra M, Alfaro-Almagro F, Marchini J, van Cappellen van Walsum AM, Jbabdi S, et al. 2019. The spatial correspondence and genetic influence of interhemispheric connectivity with white matter microstructure. Nat Neurosci. 22:809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, et al. 1995. The pou domain transcription factor brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 9:3109–3121. [DOI] [PubMed] [Google Scholar]
- National Research Council . 1980. Work, jobs, and occupations: a critical review of the dictionary of occupational titles. Washington (DC): The National Academic Press. [Google Scholar]
- Nieuwboer HA, Pool R, Dolan CV, Boomsma DI, Nivard MG. 2016. GWIS: genome-wide inferred statistics for functions of multiple phenotypes. Am J Hum Genet. 99:917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Office for National Statistics . 2000. Standard occupational classification. Basingstoke (UK): Palgrave Macmillan. [Google Scholar]
- Office for National Statistics . 2010a. Mapping Standard Occupational Classification 2010 (SOC2010) unit group with size of organisation to ISCO08 unit group.
- Office for National Statistics . 2010b. Mapping Standard Occupational Classification 2010 (SOC2010) unit group with size of organisation to ISCO08 unit group. https://www.ons.gov.uk/file?uri=/methodology/classificationsandstandards/standardoccupationalclassificationsoc/soc2010/ug201002soc2010toisco08v2_tcm77-283163.xls (03 October 2019, date last accessed). [Google Scholar]
- Office for National Statistics . 2012. Relationship between: Standard Occupational Classification 2010 (SOC2010) and Standard Occupational Classification 2000 (SOC2000). Available at: https://www.ons.gov.uk/file?uri=/methodology/classificationsandstandards/standardoccupationalclassificationsoc/soc2010/relationship-between-soc2010-and-soc2000.zip (03 October 2019, date last accessed).
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M. 2011. A bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 56:907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JC, Boerwinkle E, et al. 2009. Cohorts for heart and aging research in genomic epidemiology (charge) consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. 1988. Specification of cerebral cortical areas. Science. 241:170–176. [DOI] [PubMed] [Google Scholar]
- Schwarzkopf DS, Song C, Rees G. 2011. The surface area of human v1 predicts the subjective experience of object size. Nat Neurosci. 14:28–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selemon LD, Ceritoglu C, Ratnanather JT, Wang L, Harms MP, Aldridge K, Begovic A, Csernansky JG, Miller MI, Rakic P. 2013. Distinct abnormalities of the primate prefrontal cortex caused by ionizing radiation in early or midgestation. J Comp Neurol. 521:1040–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatz CJ, Stryker MP. 1988. Prenatal tetrodotoxin infusion blocks segregation of retinogeniculate afferents. Science. 242:87–89. [DOI] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann O, et al. 2012. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 44:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et al. 2016. The genecards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 54:1.30.31–1.30.33. [DOI] [PubMed] [Google Scholar]
- Strang KH, Golde TE, Giasson BI. 2019. Mapt mutations, tauopathy, and mechanisms of neurodegeneration. Lab Invest. 99:912–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. 2015. Uk biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. 2010. Planar cell polarity signaling in neural development. Curr Opin Neurobiol. 20:572–577. [DOI] [PubMed] [Google Scholar]
- Trampush JW, Yang ML, Yu J, Knowles E, Davies G, Liewald DC, Starr JM, Djurovic S, Melle I, Sundet K, et al. 2017. Gwas meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the cogent consortium. Mol Psychiatry. 22:336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G, Kaestner KH. 2007a. Forkhead transcription factors II. Cell. 131:192. [DOI] [PubMed] [Google Scholar]
- Tuteja G, Kaestner KH. 2007b. Snapshot: Forkhead transcription factors I. Cell. 130:1160. [DOI] [PubMed] [Google Scholar]
- US Department of Labor, United States. Employment, & Training Administration . 1991. The revised handbook for analyzing jobs. Indianapolis (US): Jist Works. [Google Scholar]
- Vinke EJ, de Groot M, Venkatraghavan V, Klein S, Niessen WJ, Ikram MA, Vernooij MW. 2018. Trajectories of imaging markers in brain aging: the Rotterdam study. Neurobiol Aging. 71:32–40. [DOI] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Giedd J, Dale AM, Brown TT. 2017. Through thick and thin: a need to reconcile contradictory results on trajectories in human cortical development. Cereb Cortex. 27:1472–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. 2012. Haploreg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D. 2017. Functional mapping and annotation of genetic associations with fuma. Nat Commun. 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. 2010. Metal: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Magi R, Ferreira T, Fall T, Graff M, Justice AE, et al. 2014. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 9:1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman AD, Charvet CJ, Clancy B, Darlington RB, Finlay BL. 2013. Modeling transformations of neurodevelopmental sequences across mammalian species. J Neurosci. 33:7368–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Mlodzik M. 2015. Wnt-frizzled/planar cell polarity signaling: cellular orientation by facing the wind (wnt). Annu Rev Cell Dev Biol. 31:623–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, Esteller-Cucala P, Juan D, Ferrandez-Peral L, Gulden FO, et al. 2018. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science. 362:eaat8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


