Abstract
Sterol response element binding protein (SREBP) is a master regulator of cellular lipogenesis. One key step in the regulation of SREBP activity is its sequential cleavage and trans-location by several different proteinases including SREBP cleavage activating protein (SCAP). We have previously reported that Brahma related gene 1 (BRG1) directly interacts with SREBP1c and SREBP2 to activate pro-lipogenic transcription in hepatocytes. We report here that BRG1 deficiency resulted in reduced processing and nuclear accumulation of SREBP in the murine livers in two different models of non-alcoholic steatohepatitis (NASH). Exposure of hepatocytes to lipopolysaccharide (LPS) and palmitate (PA) promoted SREBP accumulation in the nucleus whereas BRG1 knockdown or inhibition blocked SREBP maturation. Further analysis revealed that BRG1 played an essential role in the regulation of SCAP expression. Mechanistically, BRG1 interacted with Sp1 and directly bound to the SCAP promoter to activate SCAP transcription. Forced expression of exogenous SCAP partially rescued the deficiency in the expression of SREBP target genes in BRG1-null hepatocytes. In conclusion, our data uncover a novel mechanism by which BRG1 contributes to SREBP-dependent lipid metabolism.
Keywords: transcriptional regulation, hepatocyte, lipid metabolism, transcription factor, chromatin remodeling protein, steatosis
Introduction
Lipid is a major biological macromolecule that plays diverse roles in orchestrating differentiation, proliferation, migration, communication, survival, and death of mammalian cells (Glatz, 2011; Patil et al., 2019; Nechipurenko, 2020; Xie et al., 2020). On the other hand, disorders of lipid metabolism, typically characterized excessive lipid synthesis and defective lipid catabolism, contribute to a host of human diseases including coronary heart disease, obesity, hypertension, and non-alcoholic steatohepatitis/NASH (Cifkova, 2014; Islam et al., 2019; Magni, 2019; Pei et al., 2020). Liver is a major hub of lipid metabolism (Nguyen et al., 2008). The hepatocytes rely on thousands of proteins, which include transmembrane receptors, transcriptional regulators, and transporters, to coordinate lipid metabolism tailoring to various cellular events. This normally well-programmed process can be hijacked by both intrinsic and extrinsic pathogenic stimuli to skew lipid metabolism and to promote the development of diseases (Neuschwander-Tetri, 2010; Mashek et al., 2015).
Sterol response element binding protein (SREBP), initially identified and characterized by the Brown and Goldstein laboratory, represents a family of transcription factors considered master regulators of lipid metabolism (Shimano and Sato, 2017). Three members have been identified for this family, SREBP1a, SREBP1c, and SREBP2. It is generally agreed that SREBP1a/1c are primarily responsible for the synthesis of fatty acids whereas SREBP2 selectively regulates cholesterol synthesis although functional redundancies among the three SREBP isoforms have been observed (Horton et al., 2002b). All three SREBP proteins can be detected to exhibit a unanimous expression pattern but SREBP1c and SREBP2 are the predominantly expressed isoforms in most tissues in vivo (Shimomura et al., 1997). The relevance of SREBPs in lipid homeostasis and in the pathogenesis of lipid disorder-associated human diseases has been mostly supported by transgenic animal models. Deletion of SREBP1c in mice, for instance, results in approximately 50% reduction in hepatic fatty acid production accompanying an across-the-board down-regulation of enzymes involved in lipogenesis (Liang et al., 2002). On the contrary, mice harboring over-expression of either SREBP1c or SREBP2 develop fatty liver (steatosis) spontaneously although SREBP1c over-expression preferentially leads to an increase in plasma triglyceride levels whereas SREBP2 over-expression causes hypercholesterolemia (Horton et al., 2002a).
Innate SREBP proteins, once translated, dwell in the ER as trans-membrane factors (Goldstein et al., 2006). Processing and consequently nuclear translocation of SREBP entails the participation of three accessory proteins, SREBP cleavage activating protein (SCAP), site 1 protease (S1P), and site 2 protease (S2P). Upon stimulation by various factors, SCAP escorts SREBP to the Golgi apparatus where they are clipped, sequentially, by S1P and S2P (Brown and Goldstein, 1997). The liberated/mature SREBP, designated as nSREBP, moves into the nucleus functioning as a pro-lipogenic transcription factor to regulate lipid homeostasis and disorder (Hampton, 2002). Consistent with this model, liver specific deletion of SCAP results in defective processing of all three nSREBPs and a drastic reduction in hepatic lipid production (Horton et al., 2002a). In addition, genetic polymorphism of the SCAP gene has been found to be associated with NASH and hypertension in humans (Sun et al., 2013; Yang et al., 2017). However, the epigenetic regulation of SCAP transcription during NASH pathogenesis remains poorly defined.
Brahma related gene 1 (BRG1) utilizes the energy derived from ATP hydrolysis to mobilize nucleosomes along the chromatin and modulate chromatin accessibility. We have previously shown that BRG1 is an important moderator of liver injury linking specific transcriptional events to the alteration of liver function (Fan et al., 2019; Li et al., 2019a, c, d, e; Dong et al., 2020; Shao et al., 2020). Importantly, liver specific deletion of BRG1 attenuates steatosis in several different animal models (Kong et al., 2018; Li et al., 2018; Liu et al., 2019a; Fan et al., 2020). Here we describe a novel mechanism whereby BRG1 contributes to hepatic liver metabolism by regulating SCAP-mediated SREBP processing.
Materials and Methods
Animals
All animal experiments were reviewed and approved by the intramural Ethics Committee on Humane Treatment of Experimental Animals. Smarca4f/f mice (Dong et al., 2020) were crossed to Alb-Cre mice (Kong et al., 2019b) to generate hepatocyte conditional BRG1 knockout (LKO) mice. To induce NASH, 8 week-old male mice were fed a high fat diet (D12492, Research Diets) for 16 consecutive weeks (Fan et al., 2019). Alternatively, 8 week-old male mice were fed a methionine- and choline-deficient (MCD) diet (A02082002B, Research Diets) for four consecutive weeks as previously described (Kong et al., 2019a).
Cell Culture, Plasmids, Transient Transfection, and Reporter Assay
Primary mouse hepatocytes were isolated and maintained as previously described (Li et al., 2019a). HepG2 cells were maintained in DMEM supplemented with 10% FBS. The human SCAP promoter-luciferase constructs (Nakajima et al., 1999), FLAG-tagged BRG1, Myc-tagged SCAP (Chen et al., 2016), have been described previously. Small interfering RNAs targeting BRG1 are: #1, AACATGCACCAGATGCACAAG and #2, GCCCATGGAGTCCATGCAT. Transient transfection was performed with Lipofectamine 2000. Cells were harvested 48 h after transfection. Luciferase activities were assayed 24–48 h after transfection using a luciferase reporter assay system (Promega) as previously described (Yang et al., 2019a, b).
Protein Extraction and Western Blot
Whole cell lysates were obtained by re-suspending cell pellets in RIPA buffer (50 mM Tris pH7.4, 150 mM NaCl, and 1% Triton X-100) with freshly added protease inhibitor (Roche) as previously described (Lv et al., 2020; Mao et al., 2020; Yang et al., 2020a, b). Nuclear proteins were prepared with the NE-PER Kit (Pierce) following manufacturer’s recommendation. Western blot analyses were performed with anti-BRG1 (Santa Cruz, sc-10768), anti-SREBP1 (Proteintech, 14088-1), anti-SREBP2 (Proteintech, 19811-1), anti-SCAP (Cell Signaling Tech, 13102), and anti-β-actin (Sigma, A2228) antibodies. For densitometrical quantification, densities of target proteins were normalized to those of b-actin as previously described (Sun et al., 2020; Wu T. et al., 2020). Data are expressed as relative protein levels compared to the control group which is arbitrarily set as one.
RNA Isolation and Real-Time PCR
RNA was extracted with the RNeasy RNA isolation kit (Qiagen). Reverse transcriptase reactions were performed using a SuperScript First-strand Synthesis System (Invitrogen) as previously described (Liu et al., 2019b; Zhao et al., 2019). Real-time PCR reactions were performed on an ABI Prism 7500 system with the following primers: human SCAP, 5′-TCACGTTGCAGCCGTCTTCCTT-3′ and 5′-CAGGATGCCA ATCCAGACAACG-3′; human BRG1, 5′-TCATGTTGGCG AGCTATTTCC-3′ and 5′-GGTTCCGAAGTCTCAACGATG-3′; human FASN, 5′-CTTCCGAGATTCCATCCTACGC-3′ and 5′-TGGCAGTCAGGCTCACAAACG-3′; human LDLR, 5′-GAC GTGGCGTGAACATCTG-3′ and 5′-CTGGCAGGCAATGCTT TGG-3′; mouse Scap, 5′-CCGAGGATGACCCTGACTGA-3′ and 5′-AGAGCAGCCCATGGTTGTAGA-3′; mouse S1p, 5′-CT ACTATGGAGGAATGCCGACAG-3′ and 5′-CTCCGTTCTGT GGCAAATAGGG-3′; mouse S2p, 5′-ACGGCGGAAAGCAA GGATGCTT-3′ and 5′-GTGCCAAAGTCTGCATCAGCGT-3′; mouse Fasn, 5′-TTTAAAGGGAGGGAGGGAGA-3′ and 5′-GG CAGGATAGGGAAACACTGA-3′; mouse Ldlr, 5′-TGTGAAT TTGGTGGCTGAAAAC-3′ and 5′-AATAGGGAAGAAGATG GACAGGAAC-3′. Ct values of target genes were normalized to the Ct values of housekeekping control gene (18s, 5′-CGCGGTTCTATTTTGTTGGT-3′ and 5′-TCGTCTTCGAAACTCCGACT-3′ for both human and mouse genes) using the ΔΔCt method and expressed as relative mRNA expression levels compared to the control group which is arbitrarily set as one.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described before (Li et al., 2019b, f, 2020a, b, c; Shao et al., 2019; Weng et al., 2019; Chen et al., 2020a; Wu X. et al., 2020; Hong et al., 2021). In brief, chromatin in control and treated cells were cross-linked with 1% formaldehyde. Cells were incubated in lysis buffer (150 mM NaCl, 25 mM Tris pH 7.5, 1% Triton X-100, 0.1% SDS, and 0.5% deoxycholate) supplemented with protease inhibitor tablet and PMSF. DNA was fragmented into ∼200 bp pieces using a Branson 250 sonicator. Aliquots of lysates containing 200 μg of protein were used for each immunoprecipitation reaction with anti-BRG1 (Santa Cruz, sc-10768), anti-Sp1 (Abcam, ab227383), or pre-immune IgG. Precipitated genomic DNA was amplified by real-time PCR with the following primers: SCAP proximal promoter, 5′-ATACTTCCCTCCGGTGTCCAC-3′ and 5′-ACCTCTCACCTCCACCTTTAC-3′; SCAP distal promoter, 5′-AAATGCGAGGACATGTACAATAC-3′ and 5′-ATTTAAAAGCTAAGTTGAC-3′. A total of 10% of the starting material is also included as the input. Data are then normalized to the input and expressed as % recovery relative the input as previously described (Chen et al., 2020b, c). All experiments were performed in triplicate wells and repeated three times.
Statistical Analysis
Sample sizes reflected the minimal number needed for statistical significance based on power analysis and prior experience. Two-tailed Student’s t-test was performed using an SPSS package. Unless otherwise specified, p values smaller than 0.05 were considered statistically significant.
Results
BRG1 Regulates SREBP Maturation in the Liver
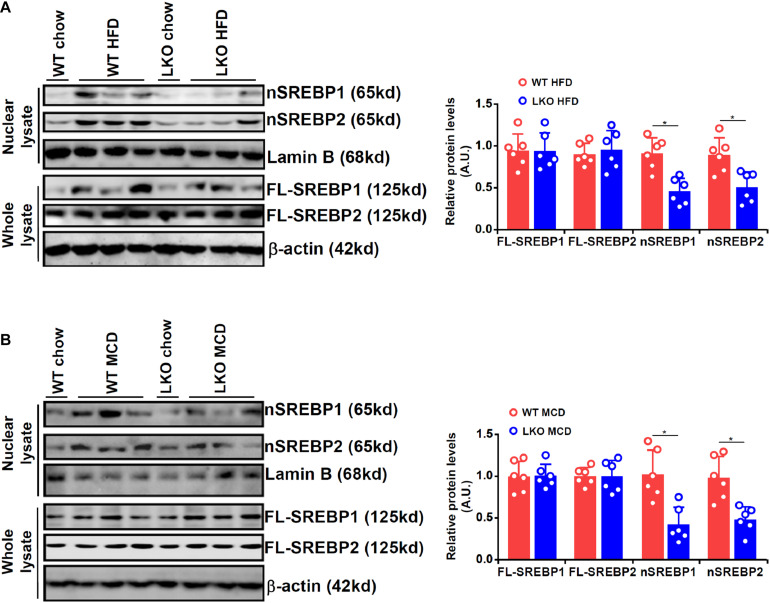
We first evaluated the effect of BRG1 deficiency on SREBP maturation in the liver in two different models of steatosis. To this end, we generated liver conditional BRG1 knockout (LKO) mice by crossing the Smarca4-flox mice with the Alb-Cre mice. In the first model in which the mice were fed a high-fat diet (HFD) for 16 weeks, it was observed that nuclear SREBP1/2 levels were appreciably increased in the livers of the HFD-fed mice compared to the control diet-fed (chow) mice; up-regulation of nuclear SREBP1/2 by HFD feeding was more modest in the LKO livers than in the WT livers (Figure 1A). In the second model in which the mice were fed a methionine-and-choline deficient (MCD) diet for 4 weeks, a similar observation was made that BRG1 deficiency dampened the accumulation of SREBP1/2 in the nucleus.
FIGURE 1.
BRG1 regulates SREBP maturation in the liver. (A) WT and BRG1 LKO mice were fed a high-fat diet (HFD) for 16 weeks. SREBP levels were examined in whole liver lysates and liver nuclear lysates by Western blotting. (B) WT and BRG1 LKO mice were fed a methionine-and-choline deficient diet (MCD) for 4 weeks. SREBP levels were examined in whole liver lysates and liver nuclear lysates by Western blotting. N = 6 mice for each group. Data represent averages of three independent experiments and error bars represent SEM. *p < 0.05, two-tailed Student’s t-test.
BRG1 Is Essential for SREBP Maturation in Hepatocytes
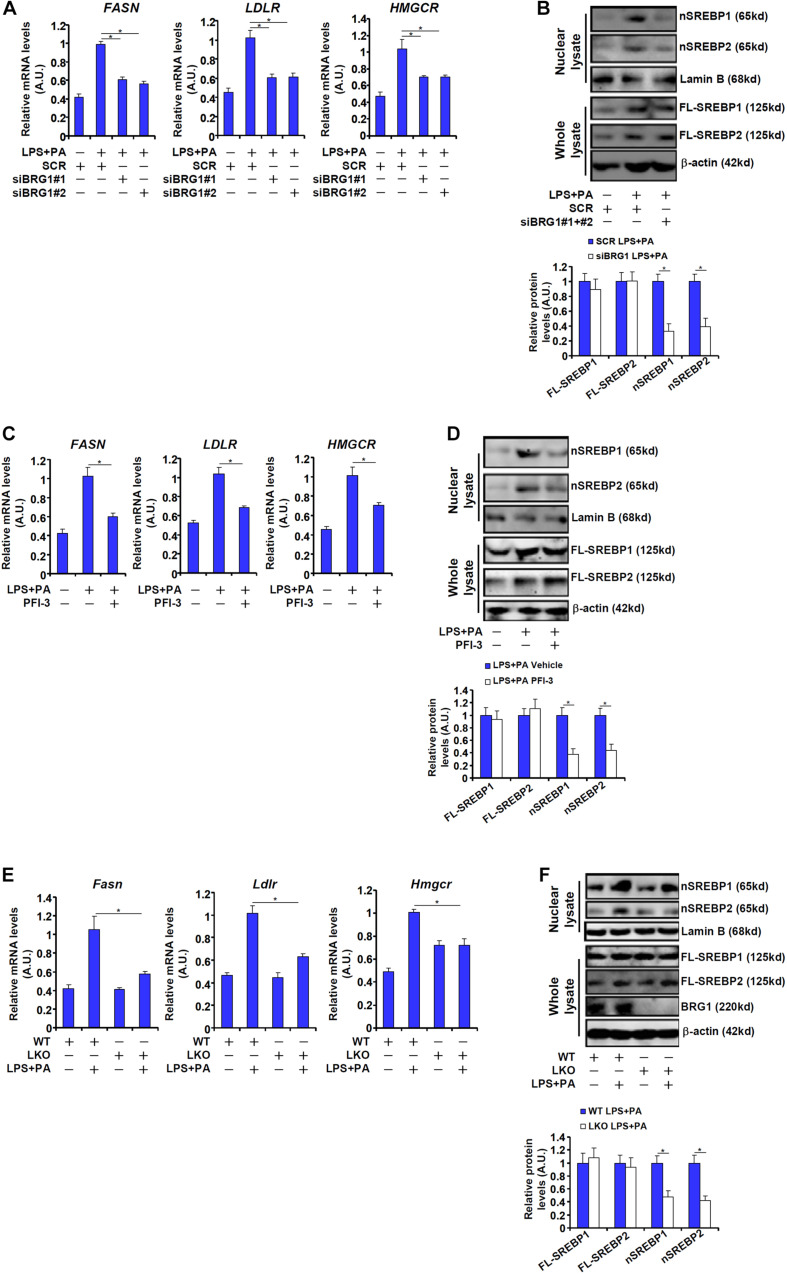
We next verified the observation that BRG1 might contribute to SREBP maturation in cultured hepatocytes. The combined treatment of LPS plus free fatty acids (palmitate, PA), which has been reported to induce SREBP target gene expression in macrophages (Li et al., 2013), induced the expression of FASN, a prototypical SREBP1 target gene, and the expression of LDLR, prototypical SREBP2 target gene, in HepG2 cells (Figure 2A). Knockdown of BRG1 by siRNA attenuated the induction of both FASN and LDLR, indicative of repressed SREBP activity (Figure 2A). Consistent with these changes, we found that BRG1 knockdown reduced the levels of nSREBP1/2 without significantly altering full-length SREBP1/2 in LPS + PA-treated HepG2 cells (Figure 2B). Similarly, treatment with the small-molecule BRG1 inhibitor PFI-3 resulted in a down-regulation of SREBP target genes and partially blocked SREBP processing in HepG2 cells (Figures 2C,D). Finally, when primary hepatocytes were isolated from wild type and BRG1 LKO mice and exposed to LPS + PA treatment, the induction of SREBP target genes and SREBP maturation were attenuated in the LKO cells compared to the WT cells (Figures 2E,F).
FIGURE 2.
BRG1 is essential for SREBP maturation in hepatocytes. (A,B) HepG2 cells were transfected with siRNA targeting BRG1 or scrambled siRNA (SCR) followed by treatment with LPS (1 μg/ml) + PA (0.2 mM). SREBP target genes were examined by qPCR. SREBP levels were examined in whole cell lysates and nuclear lysates by Western blotting. (C,D) HepG2 cells were treated with LPS (1 μg/ml) + PA (0.2 mM) in the presence or absence of PFI-3 (3 μM). SREBP target genes were examined by qPCR. SREBP levels were examined in whole cell lysates and nuclear lysates by Western blotting. (E,F) Primary hepatocytes isolated from WT and BRG1 LKO mice were treated with LPS (1 μg/ml) + PA (0.2 mM). SREBP levels were examined in whole cell lysates and nuclear lysates by Western blotting. Data represent averages of three independent experiments and error bars represent SEM. *p < 0.05, two-tailed Student’s t-test.
BRG1 Regulates SCAP Expression
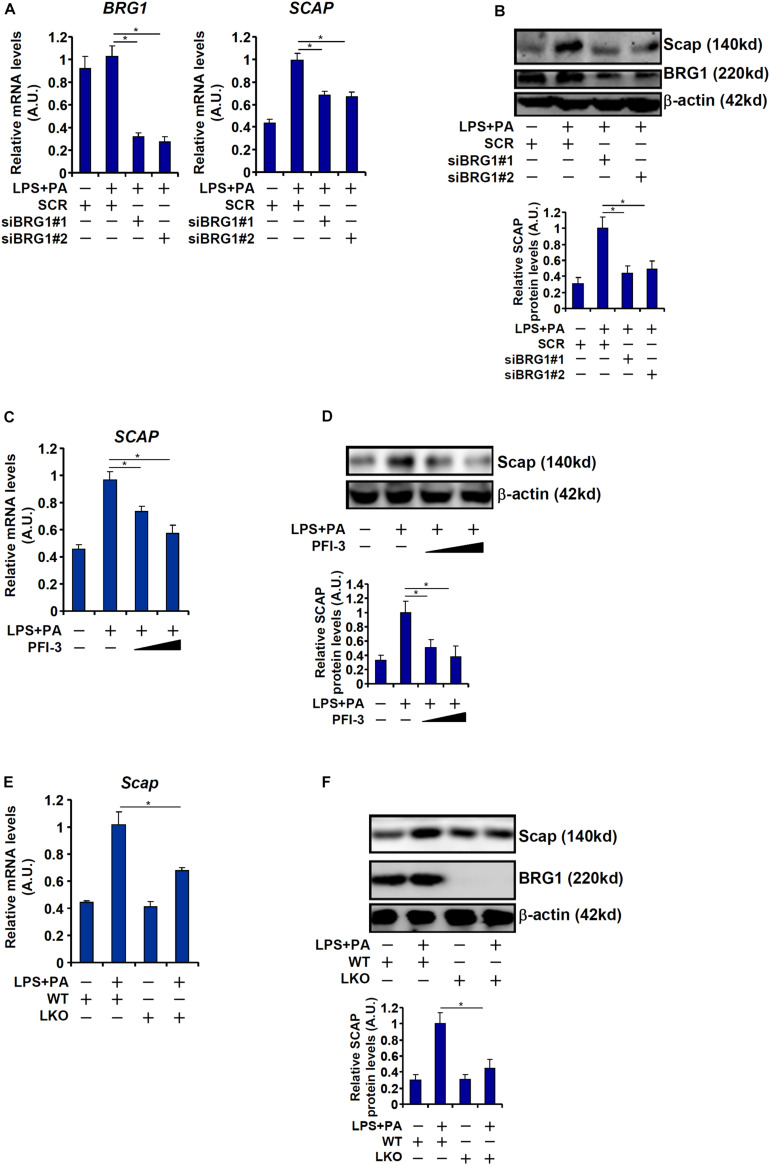
Because BRG1 deficiency was associated with impaired SREBP maturation in hepatocytes, we hypothesized that BRG1 might play a role regulating the expression levels of SREBP processing enzyme(s). We therefore compared the expression of the three SREBP processing enzymes, SCAP, S1P, and S2P, in WT and LKO livers. As shown in Figures 3A,B, all three SREBP processing proteins were up-regulated in HFD-fed livers compared to the chow-fed livers; BRG1 deficiency, however, dampened the up-regulation of SCAP expression without altering either S1P or S2P expression. Similarly, MCD diet feeding induced the mRNA (Figure 3C) and protein (Figure 3D) levels of SCAP, S1P, and S2P in the murine livers; the induction of SCAP expression by MCD diet feeding was more modest in the LKO livers than the WT livers whereas the induction of S1P/S2P expression was comparable between the LKO livers and WT livers.
FIGURE 3.
BRG1 regulates SCAP expression in vivo. (A,B) WT and BRG1 LKO mice were fed a high-fat diet (HFD) for 16 weeks. SCAP expression was examined by qPCR and Western blotting. N = 5 mice for the chow groups and N = 6 mice for the HFD groups. (C,D) WT and BRG1 LKO mice were fed a methionine-and-choline deficient diet (MCD) for 8 weeks. SCAP expression was examined by qPCR and Western blotting. N = 5 mice for the chow groups and N = 6 mice for the MCD groups. Data represent averages of three independent experiments and error bars represent SEM. *p < 0.05, two-tailed Student’s t-test.
Next, we evaluated the effect of BRG1 depletion or inhibition on SCAP expression in cultured hepatocytes. Exposure of HepG2 cells to LPS + PA treatment robustly augmented SCAP expression at both mRNA (Figure 4A) and protein (Figure 4B) levels. BRG1 knockdown by two independent siRNAs attenuated the induction of SCAP expression by LPS + PA treatment. Alternatively, the addition of a small-molecule BRG1 inhibitor (PFI-3) dose-dependently ameliorated the induction of SCAP expression by LPS + PA treatment (Figures 4C,D). Finally, when primary hepatocytes were isolated from the WT and the LKO mice and exposed to LPS + PA stimulation, the induction of SCAP molecules was not as strong in the LKO cells as in the WT cells (Figures 4E,F). Consistent with the changes in SCAP expression, it was also observed that there were fewer lipid droplets in the LKO hepatocytes than in the WT hepatocytes exposed to LPS + PA treatment (Supplementary Figure 1).
FIGURE 4.
BRG1 regulates SCAP expression in vitro. (A,B) HepG2 cells were transfected with siRNA targeting BRG1 or scrambled siRNA (SCR) followed by treatment with LPS (1 μg/ml) + PA (0.2 mM). SREBP target genes were examined by qPCR. SCAP expression was examined by qPCR and Western blotting. (C,D) HepG2 cells were treated with LPS (1 μg/ml) + PA (0.2 mM) in the presence or absence of PFI-3 (3 μM). SCAP expression was examined by qPCR and Western blotting. (E,F) Primary hepatocytes isolated from WT and BRG1 LKO mice were treated with LPS (1 μg/ml) + PA (0.2 mM). SCAP expression was examined by qPCR and Western blotting. Data represent averages of three independent experiments and error bars represent SEM. *p < 0.05, two-tailed Student’s t-test.
BRG1 Interacts With Sp1 to Activate SCAP Transcription in Hepatocytes
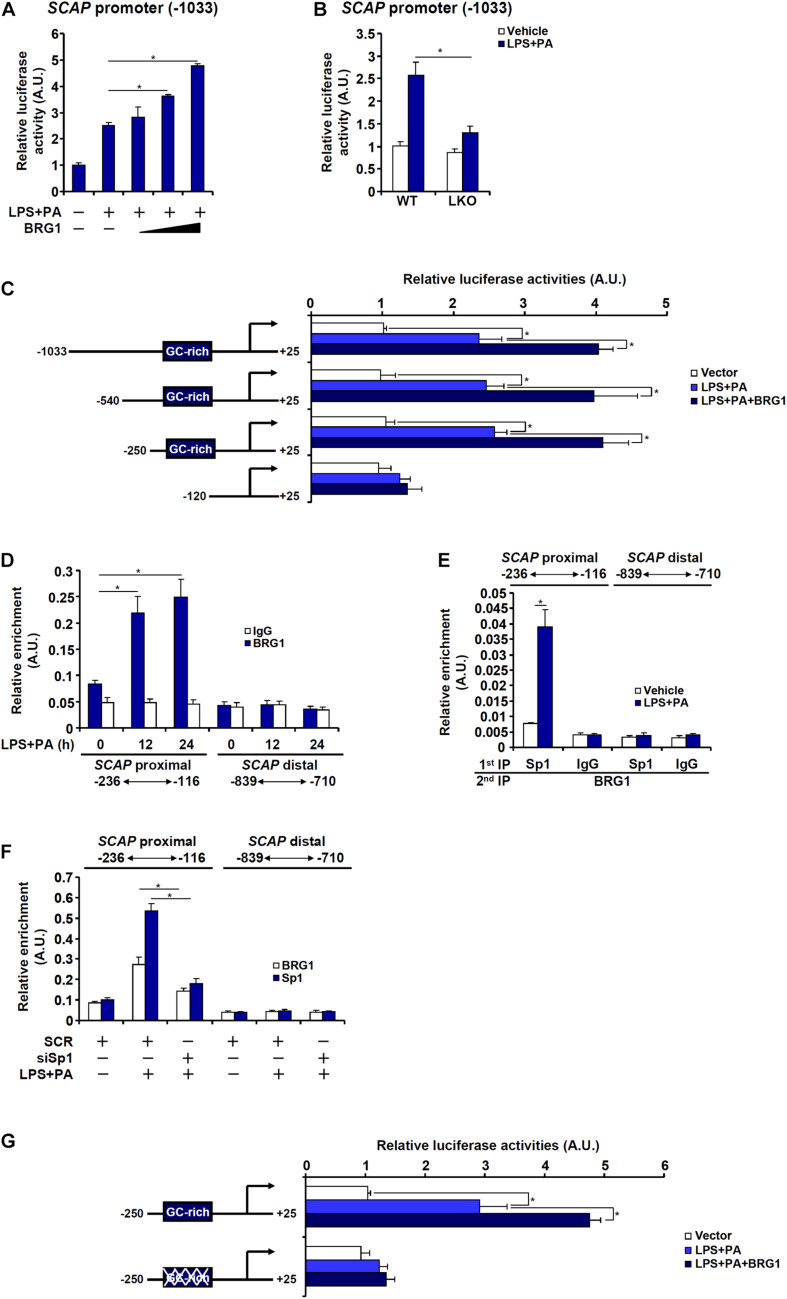
Next, it was determined whether regulation of SCAP expression by BRG1 in hepatocytes occurred at the transcriptional level. An SCAP promoter-luciferase fusion construct (−1,033) was transfected into HepG2 cells. Treatment with LPS + PA stimulated the SCAP promoter activity by more than twofold and BRG1 over-expression further augmented the SCAP promoter activity in a dose-dependent manner (Figure 5A). When the same SCAP promoter-luciferase construct was transfected into primary hepatocytes isolated from WT and BRG1 LKO mice, treatment with LPS + PA stimulated the SCAP promoter activity much more potently in the WT hepatocytes than in the LKO hepatocytes (Figure 5B). Serial deletions were then introduced to the full-length SCAP promoter-luciferase construct and the mutated constructs were tested for the responsiveness to the stimulation of LPS + PA treatment plus BRG1 over-expression. As shown in Figure 5C, the −540 construct and the −250 construct, but not the −120 construct, responded to the stimulation of LPS + PA treatment plus BRG1 over-expression comparably as the −1,033 construct. ChIP assay confirmed that LPS + PA treatment induced BRG1 recruitment to the SCAP proximal promoter between −250 and −120; as a negative control, BRG1 occupancy on the SCAP distal promoter was not detected with or without LPS + PA treatment (Figure 5D). Further analysis revealed a GC-rich region that could potentially occupied by the transcription factor Sp1 between −250 and −120 of the SCAP promoter. Furthermore, Re-ChIP assay showed that an Sp1-BRG1 complex was detectable on the SCAP promoter only when the cells were stimulated by LPS + PA (Figure 5E). ChIP assay confirmed that Sp1 knockdown abrogated the binding of both Sp1 and BRG1 to the proximal SCAP promoter (Figure 5F). Adding further support to the model that Sp1 recruits BRG1 to activate SCAP transcription was the observation that the SCAP promoter construct with Sp1 site mutated could not be induced by BRG1 over-expression (Figure 5G).
FIGURE 5.
BRG1 interacts with Sp1 to activate SCAP transcription in hepatocytes. (A) An SCAP promoter-luciferase construct (–1033/+25) was transfected into HepG2 cells with or without BRG1 followed by treatment with LPS (1 μg/ml) + PA (0.2 mM). Luciferase activities were normalized by protein concentration and GFP fluorescence. (B) An SCAP promoter-luciferase construct (–1033/+25) was transfected into primary hepatocytes isolated from WT and BRG1 LKO mice followed by treatment with LPS (1 μg/ml) + PA (0.2 mM). Luciferase activities were normalized by protein concentration and GFP fluorescence. (C) SCAP promoter-luciferase constructs of various lengths were transfected into HepG2 cells with or without BRG1 followed by treatment with LPS (1 μg/ml) + PA (0.2 mM). Luciferase activities were normalized by protein concentration and GFP fluorescence. (D) HepG2 cells were treated with or without LPS (1 μg/ml) + PA (0.2 mM) and harvested at indicated time points. ChIP assays were performed with anti-BRG1 or IgG. (E) HepG2 cells were treated with or without LPS (1 μg/ml) + PA (0.2 mM) for 24 h. Re-ChIP assays were performed with indicated antibodies. (F) HepG2 cells were transfected with siRNA targeting Sp1 or scrambled siRNA (SCR) followed by treatment with LPS (1 μg/ml) + PA (0.2 mM). ChIP assays were performed with anti-Sp1 or anti-BRG1. (G) Wild type or Sp1 site mutant SCAP promoter-luciferase construct was transfected into HepG2 cells with or without BRG1 followed by treatment with LPS (1 μg/ml) + PA (0.2 mM). Luciferase activities were normalized by protein concentration and GFP fluorescence. Data represent averages of three independent experiments and error bars represent SEM. *p < 0.05, two-tailed Student’s t-test.
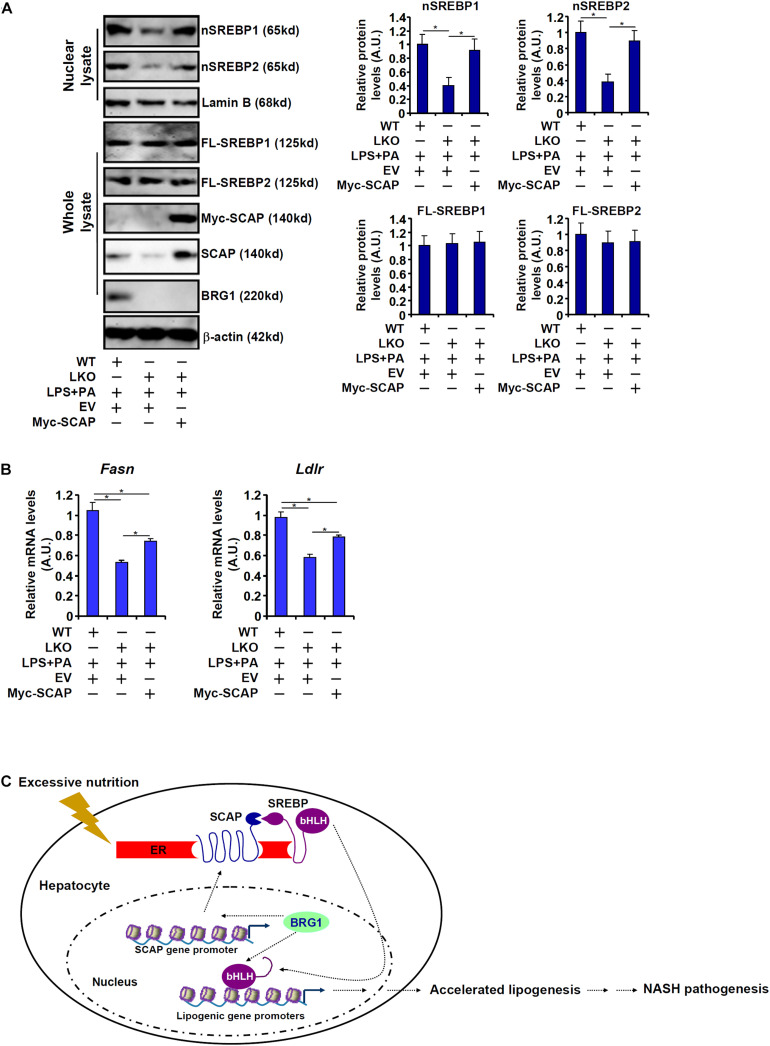
SCAP Re-introduction Partially Rescues the Expression of SREBP Target Genes in BRG1 Deficient Hepatocytes
Finally, exogenous SCAP was introduced into BRG1-deficient hepatocytes to address the question as to whether it could rescue the expression of SREBP target genes. As shown in Figure 6A, over-expression of Myc-tagged SCAP largely restored SREBP processing in the LKO hepatocytes despite the absence of BRG1. Consistent with this observation, SCAP over-expression also partially normalized the levels of SREBP target genes: there was a 39% increase in Fasn expression and a 36% increase in Ldlr expression in the SCAP over-expressed LKO cells compared to the mock over-expressed LKO cells (Figure 6B). Accordingly, forced expression of exogenous SCAP partially restored accumulation of lipid droplet in the LKO hepatocytes (Supplementary Figure 2).
FIGURE 6.
SCAP re-introduction partially rescues the expression of SREBP target genes in BRG1 deficient hepatocytes. (A,B) A Myc-tagged SCAP expression construct was transfected into primary hepatocytes isolated from BRG1 LKO mice followed by treatment with LPS (1 μg/ml) + PA (0.2 mM). Gene expression levels were examined by qPCR and Western blotting. Data represent averages of three independent experiments and error bars represent SEM. *p < 0.05, two-tailed Student’s t-test. (C) A schematic model. BRG1 may contribute to pro-lipogenic transcription in hepatocytes via at least two independent mechanisms. On the one hand, BRG1 stimulates SCAP expression to promote SREBP maturation. On the other hand, BRG1 interacts with mature SREBPs in the nucleus to directly activate pro-lipogenic gene transcription. Consequently, accelerated lipogenesis in hepatocytes leads to steatosis.
Discussion
The trans-location and subsequent cleavage of nascent SREBPs catapults these master regulators of lipid metabolism into the nucleus to orchestrate pro-lipogenic transcription. This process is mediated by the well-conserved SCAP-S1P-S2P axis. We have previously shown that the chromatin BRG1 contributes to hepatic lipid metabolism by functioning as a co-activator for SREBP1/2 (Li et al., 2018; Fan et al., 2020). Here we detail a novel mechanism in which BRG1 modulates SREBP maturation via activating SCAP transcription (Figure 6C). Our data add to a growing body of evidence that cements the crucial role BRG1 plays in regulating cellular metabolism. For instance, Miao et al. (2009) have reported that BRG1 is essential for the synthesis of bile acids, a key function of hepatocyte, by interacting with the nuclear receptor FXR. Imbalzano and colleagues have demonstrated that BRG1 activates the transcription of several enzymes involved in fatty acid synthesis, including ATP citrate lyase (ACLY) and acetyl CoA carboxylase (ACC), to drive breast cancer proliferation (Wu et al., 2016; Nickerson et al., 2017). Metabolomic analysis has revealed that BRG1 deficiency in cardiomyocytes results in skewed fatty acid utilization, glycolysis, and glycogen synthesis (Banerjee et al., 2015). Genomewide transcriptomic studies suggest that BRG1 preferentially, at least in epithelial cells, binds to the SREBP target promoters to remodel chromatin structure (Barutcu et al., 2016). It is interesting to note that BRG1 deficiency or inhibition did not alter basal levels of SCAP in the livers (Figure 3) and in cultured hepatocytes (Figure 4), suggesting that BRG1 is dispensable for the maintenance of lipid homeostasis under physiological conditions. This observation is consistent with our previous findings showing that basal lipid profiles (triglycerides and cholesterol) are comparable between the WT mice and the BRG1 LKO mice. Rather, stimuli-induced SCAP expression appeared to rely on BRG1 suggesting that BRG1 is a pathogenic factor-driven regulator of lipid metabolism disorders. These data present Brg1 as an attractive target for intervention: since the absence of Brg1 presumably will not interfere with normal tissue/organ function, the predicament of selective drug delivery could be effectively circumvented.
Another noteworthy finding is that although SCAP re-introduction in the BRG1 LKO cells largely restored SREBP processing (Figure 6A), SREBP target gene expression was only partially, but not completely, corrected (Figure 6B). These data allude to a model in which SCAP-mediated SREBP maturation and nuclear translocation serves as a necessary but not adequate step for pro-lipogenic transcription. Indeed, several epigenetic factors have been shown to interact with BRG1 to regulate SREBP1 activity. The histone H4K16 acetyltransferase MOF (Liu et al., 2019a) and the H3K9 demethylase KDM3A can both bind to SREBP target promoters to regulate SREBP-dependent transcription (Fan et al., 2020). An intriguing question that remains to be answered is whether the functional relevance of MOF and/or KDM3A can be extended to the regulation of SCAP transcription and thus SREBP processing. There are a few indications wherein the SCAP promoter appears to be influenced by epigenetic modifications. Carraway et al. (2020) have shown that histone H3K9 acetylation and DNA methylation can differentially regulate SCAP expression in leukemia cells although the involvement of specific histone/DNA modifying enzymes is not clear at this point. It would be of great interest to further examine the epigenetic mechanism whereby SCAP transcription is regulated to provide more flexibility in targeting the SREBP pathway.
There are several lingering issues that deserve further attention in future studies. First, the broader biological/pathobiological significance of our finding, i.e., whether BRG1 can contribute to SREBP maturation in other pathophysiological conditions such as insulin stimulation or fast-feed cycle remains unclear. Of note, it has been previously shown that insulin-induced transcription of SREBP target genes in pre-adipocyte correlates with increased recruitment of BRG1 to the SREBP target promoters although it was not determined whether BRG1 could directly promote insulin-induced SREBP nuclear translocation (Lee et al., 2007). Second, we focused our study on the liver (hepatocytes), one of the three major peripheral tissues targeted by insulin. Because SREBP maturation plays a role in the metabolism of both adipose tissue (adipocytes) (Kim and Spiegelman, 1996) and skeletal muscle (myocytes) (Guillet-Deniau et al., 2002), it is tempting to speculate that BRG1 could be involved in the regulation of metabolic homeostasis in these tissues as well by virtue of contributing to SREBP maturation. Third, it is noteworthy that despite the decreased presence of nuclear SREBP in the LKO hepatocytes, overall SREBP levels remained undisturbed. A likely explanation could be that the SREBP expression, at the transcriptional level, might be subjected to regulation by BRG1. It has long been documented that both SREBP1 (Amemiya-Kudo et al., 2000) and SREBP2 (Sato et al., 1996) can bind to their respective promoters and activate the transcription in cis. Because BRG1 can interact with both SREBP1 (Li et al., 2018) and SREBP2 (Fan et al., 2020), it is possible that reduced FL-SREBP processing in the LKO cells may be offset by a proportional reduction in its expression such that overall FL-SREBP levels remain marginally affected.
In summary, we present evidence to demonstrate that BRG1 can contribute to cellular lipid metabolism by, in addition to acting as an SREBP co-factor, promoting SCAP-dependent SREBP maturation. These data not only place BRG1 in the center of metabolic programming but provide renewed rationale for targeting BRG1 in the intervention of human diseases related to lipid metabolic disorders.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the Nanjing Medical University Ethics Committee on Humane Treatment of Experimental Animals.
Author Contributions
ZF and YX conceived the project. MK, YZ, and JS designed the experiments, performed experiments, collected data, and analyzed data. YX wrote the manuscript. ZF handled the funding. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported in part by grants from the National Natural Science Foundation of China (81700554 and 81800520), the Nanjing Municipal Administration of Health and Human Services (YKK17061), the Fundamental Research Funds for Central Universities (021414380323), and the Nanjing Medical Science and Technique Development Foundation (QRX17004).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.622866/full#supplementary-material
References
- Amemiya-Kudo M., Shimano H., Yoshikawa T., Yahagi N., Hasty A. H., Okazaki H., et al. (2000). Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J. Biol. Chem. 275 31078–31085. [DOI] [PubMed] [Google Scholar]
- Banerjee R., Bultman S. J., Holley D., Hillhouse C., Bain J. R., Newgard C. B., et al. (2015). Non-targeted metabolomics of Brg1/Brm double-mutant cardiomyocytes reveals a novel role for SWI/SNF complexes in metabolic homeostasis. Metabolomics 11 1287–1301. 10.1007/s11306-015-0786-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu A. R., Lajoie B. R., Fritz A. J., Mccord R. P., Nickerson J. A., Van Wijnen A. J., et al. (2016). SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 26 1188–1201. 10.1101/gr.201624.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. (1997). The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89 331–340. 10.1016/s0092-8674(00)80213-5 [DOI] [PubMed] [Google Scholar]
- Carraway H. E., Malkaram S. A., Cen Y., Shatnawi A., Fan J., Ali H. E. A., et al. (2020). Activation of SIRT6 by DNA hypomethylating agents and clinical consequences on combination therapy in leukemia. Sci. Rep. 10:10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Fan Z., Sun L., Chen J., Feng Y., Fan X., et al. (2020a). Epigenetic activation of the small GTPase TCL contributes to colorectal cancer cell migration and invasion. Oncogenesis 9:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Yuan Y., Sun L., Chen J., Yang M., Yin Y., et al. (2020b). MKL1 mediates TGF-β induced RhoJ transcription to promote breast cancer cell migration and invasion. Front. Cell Dev. Biol. 8:832. 10.3389/fcell.2020.00832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Zhao Q., Xu T., Yu L., Zhuo L., Yang Y., et al. (2020c). BRG1 activates PR65A transcription to regulate NO bioavailability in vascular endothelial cell. Front. Cell Dev. Biol. 8:774. 10.3389/fcell.2020.00774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Li S., Yu H., Liu X., Huang L., Wang Q., et al. (2016). ER adaptor SCAP translocates and recruits IRF3 to perinuclear microsome induced by cytosolic microbial DNAs. PLoS Pathog. 12:e1005462. 10.1371/journal.ppat.1005462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifkova R. (2014). Antiplatelet and lipid-lowering drugs in hypertension. Eur. Cardiol. 9 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Kong M., Zhu Y., Shao Y., Wu D., Lu J., et al. (2020). Activation of TWIST transcription by chromatin remodeling protein BRG1 contributes to liver fibrosis in mice. Front. Cell Dev. Biol. 8:340. 10.3389/fcell.2020.00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Kong M., Li M., Hong W., Fan X., Xu Y. (2020). Brahma related Gene 1 (Brg1) regulates cellular cholesterol synthesis by acting as a co-factor for SREBP2. Front. Cell Dev. Biol. 8:259. 10.3389/fcell.2020.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Li N., Xu Z., Wu J., Fan X., Xu Y. (2019). An interaction between MKL1, BRG1, and C/EBPbeta mediates palmitate induced CRP transcription in hepatocytes. Biochim. Biophys. Acta Gene Regul. Mech. 1862:194412. 10.1016/j.bbagrm.2019.194412 [DOI] [PubMed] [Google Scholar]
- Glatz J. F. (2011). Challenges in fatty acid and lipid physiology. Front. Physiol. 2:45. 10.3389/fphys.2011.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J. L., Debose-Boyd R. A., Brown M. S. (2006). Protein sensors for membrane sterols. Cell 124 35–46. 10.1016/j.cell.2005.12.022 [DOI] [PubMed] [Google Scholar]
- Guillet-Deniau I., Mieulet V., Le Lay S., Achouri Y., Carre D., Girard J., et al. (2002). Sterol regulatory element binding protein-1c expression and action in rat muscles: insulin-like effects on the control of glycolytic and lipogenic enzymes and UCP3 gene expression. Diabetes 51 1722–1728. 10.2337/diabetes.51.6.1722 [DOI] [PubMed] [Google Scholar]
- Hampton R. Y. (2002). Proteolysis and sterol regulation. Annu. Rev. Cell Dev. Biol. 18 345–378. 10.1146/annurev.cellbio.18.032002.131219 [DOI] [PubMed] [Google Scholar]
- Hong W., Kong M., Qi M., Bai H., Fan Z., Zhang Z., et al. (2021). BRG1 mediates nephronectin activation in hepatocytes to promote T lymphocyte infiltration in ConA-induced hepatitis. Front. Cell Dev. Biol. 8:587502. 10.3389/fcell.2020.587502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. D., Goldstein J. L., Brown M. S. (2002a). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109 1125–1131. 10.1172/jci0215593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. D., Goldstein J. L., Brown M. S. (2002b). SREBPs: transcriptional mediators of lipid homeostasis. Cold Spring Harb. Symp. Quant. Biol. 67 491–498. 10.1101/sqb.2002.67.491 [DOI] [PubMed] [Google Scholar]
- Islam M. A., Amin M. N., Siddiqui S. A., Hossain M. P., Sultana F., Kabir M. R. (2019). Trans fatty acids and lipid profile: a serious risk factor to cardiovascular disease, cancer and diabetes. Diabetes Metab. Syndr. 13 1643–1647. 10.1016/j.dsx.2019.03.033 [DOI] [PubMed] [Google Scholar]
- Kim J. B., Spiegelman B. M. (1996). ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 10 1096–1107. 10.1101/gad.10.9.1096 [DOI] [PubMed] [Google Scholar]
- Kong M., Chen X., Lv F., Ren H., Fan Z., Qin H., et al. (2019a). Serum response factor (SRF) promotes ROS generation and hepatic stellate cell activation by epigenetically stimulating NCF1/2 transcription. Redox Biol. 26:101302. 10.1016/j.redox.2019.101302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M., Chen X., Xu H., Wenping, Fang M., Xu Y. (2018). Hepatocyte-specific deletion of Brg1 alleviates methionine-and-choline-deficient diet (MCD) induced non-alcoholic steatohepatitis in mice. Biochem. Biophys. Res. Commun. 503 344–351. 10.1016/j.bbrc.2018.06.027 [DOI] [PubMed] [Google Scholar]
- Kong M., Hong W., Shao Y., Lv F., Fan Z., Li P., et al. (2019b). Ablation of serum response factor in hepatic stellate cells attenuates liver fibrosis. J. Mol. Med. 97 1521–1533. 10.1007/s00109-019-01831-8 [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Sohn D. H., Han D., Lee H. W., Seong R. H., Kim J. B. (2007). Chromatin remodeling complex interacts with ADD1/SREBP1c to mediate insulin-dependent regulation of gene expression. Mol. Cell. Biol. 27 438–452. 10.1128/mcb.00490-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. C., Varghese Z., Moorhead J. F., Lee C. T., Chen J. B., Ruan X. Z. (2013). Cross-talk between TLR4-MyD88-NF-kappaB and SCAP-SREBP2 pathways mediates macrophage foam cell formation. Am. J. Physiol. Heart Circ. Physiol. 304 H874–H884. [DOI] [PubMed] [Google Scholar]
- Li N., Kong M., Zeng S., Hao C., Li M., Li L., et al. (2019a). Brahma related gene 1 (Brg1) contributes to liver regeneration by epigenetically activating the Wnt/beta-catenin pathway in mice. FASEB J. 33 327–338. 10.1096/fj.201800197r [DOI] [PubMed] [Google Scholar]
- Li N., Li M., Hong W., Shao J., Xu H., Shimano H., et al. (2018). Brg1 regulates pro-lipogenic transcription by modulating SREBP activity in hepatocytes. Biochim. Biophys. Acta Mol. Basis Dis. 1864 2881–2889. 10.1016/j.bbadis.2018.05.022 [DOI] [PubMed] [Google Scholar]
- Li N., Liu S., Zhang Y., Yu L., Hu Y., Wu T., et al. (2020a). Transcriptional activation of matricellular protein Spondin2 (SPON2) by BRG1 in vascular endothelial cells promotes macrophage chemotaxis. Front. Cell Dev. Biol. 8:794. 10.3389/fcell.2020.00794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen B., Dong W., Kong M., Fan Z., Yu L., et al. (2019b). MKL1 promotes endothelial-to-mesenchymal transition and liver fibrosis by activating TWIST1 transcription. Cell Death Dis. 10:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen B., Dong W., Kong M., Shao Y., Fan Z., et al. (2019c). The chromatin remodeler Brg1 integrates ROS production and endothelial-mesenchymal transition to promote liver fibrosis in mice. Front. Cell Dev. Biol. 7:245. 10.3389/fcell.2019.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Kong X., Zhang Y., Yu L., Guo J., Xu Y. (2020b). Dual roles of chromatin remodeling protein BRG1 in angiotensin II-induced endothelial-mesenchymal transition. Cell Death Dis. 11:549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Li P., Lu Y., Sun D., Zhang X., Xu Y. (2019d). A non-autonomous role of MKL1 in the activation of hepatic stellate cells. Biochim. Biophys. Acta Gene Regul. Mech. 1862 609–618. 10.1016/j.bbagrm.2019.03.001 [DOI] [PubMed] [Google Scholar]
- Li Z., Lv F., Dai C., Wang Q., Jiang C., Fang M., et al. (2019e). Activation of galectin-3 (LGALS3) transcription by injurious stimuli in the liver is commonly mediated by BRG1. Front. Cell Dev. Biol. 7:310. 10.3389/fcell.2019.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Xia J., Fang M., Xu Y. (2019f). Epigenetic regulation of lung cancer cell proliferation and migration by the chromatin remodeling protein BRG1. Oncogenesis 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Yu L., Xiao B., Li T., Kong X., et al. (2020c). BRG1 stimulates endothelial derived alarmin MRP8 to promote macrophage infiltration in an animal model of cardiac hypertrophy. Front. Cell Dev. Biol. 8:569. 10.3389/fcell.2020.00569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Yang J., Horton J. D., Hammer R. E., Goldstein J. L., Brown M. S. (2002). Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 277 9520–9528. 10.1074/jbc.m111421200 [DOI] [PubMed] [Google Scholar]
- Liu L., Hong W., Li M., Ren H., Wang J., Xu H., et al. (2019a). A cross talk between BRG1 and males absent on the first contributes to reactive oxygen species production in a mouse model of nonalcoholic steatohepatitis. Antioxid. Redox Signal. 30 1539–1552. 10.1089/ars.2016.6822 [DOI] [PubMed] [Google Scholar]
- Liu L., Mao L., Wu X., Wu T., Liu W., Yang Y., et al. (2019b). BRG1 regulates endothelial-derived IL-33 to promote ischemia-reperfusion induced renal injury and fibrosis in mice. Biochim. Biophys. Acta Mol. Basis Dis. 1865 2551–2561. 10.1016/j.bbadis.2019.06.015 [DOI] [PubMed] [Google Scholar]
- Lv F., Li N., Kong M., Wu J., Fan Z., Miao D., et al. (2020). CDKN2a/p16 antagonizes hepatic stellate cell activation and liver fibrosis by modulating ROS levels. Front. Cell Dev. Biol. 8:176. 10.3389/fcell.2020.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni P. (2019). Bicuspid aortic valve, atherosclerosis and changes of lipid metabolism: are there pathological molecular links? J. Mol. Cell. Cardiol. 129 231–235. 10.1016/j.yjmcc.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Mao L., Liu L., Zhang T., Qin H., Wu X., Xu Y. (2020). Histone deacetylase 11 contributes to renal fibrosis by repressing KLF15 transcription. Front. Cell Dev. Biol. 8:235. 10.3389/fcell.2020.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashek D. G., Khan S. A., Sathyanarayan A., Ploeger J. M., Franklin M. P. (2015). Hepatic lipid droplet biology: getting to the root of fatty liver. Hepatology 62 964–967. 10.1002/hep.27839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Fang S., Lee J., Comstock C., Knudsen K. E., Kemper J. K. (2009). Functional specificities of Brm and Brg-1 Swi/Snf ATPases in the feedback regulation of hepatic bile acid biosynthesis. Mol. Cell. Biol. 29 6170–6181. 10.1128/mcb.00825-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T., Hamakubo T., Kodama T., Inazawa J., Emi M. (1999). Genomic structure and chromosomal mapping of the human sterol regulatory element binding protein (SREBP) cleavage-activating protein (SCAP) gene. J. Hum. Genet. 44 402–407. 10.1007/s100380050187 [DOI] [PubMed] [Google Scholar]
- Nechipurenko I. V. (2020). The enigmatic role of lipids in cilia signaling. Front. Cell Dev. Biol. 8:777. 10.3389/fcell.2020.00777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuschwander-Tetri B. A. (2010). Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology 52 774–788. 10.1002/hep.23719 [DOI] [PubMed] [Google Scholar]
- Nguyen P., Leray V., Diez M., Serisier S., Le Bloc’h J., Siliart B., et al. (2008). Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. 92 272–283. [DOI] [PubMed] [Google Scholar]
- Nickerson J. A., Wu Q., Imbalzano A. N. (2017). Mammalian SWI/SNF enzymes and the epigenetics of tumor cell metabolic reprogramming. Front. Oncol. 7:49. 10.3389/fonc.2017.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil M., Henderson J., Luong H., Annamalai D., Sreejit G., Krishnamurthy P. (2019). The art of intercellular wireless communications: exosomes in heart disease and therapy. Front. Cell Dev. Biol. 7:315. 10.3389/fcell.2019.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei K., Gui T., Kan D., Feng H., Jin Y., Yang Y., et al. (2020). An overview of lipid metabolism and nonalcoholic fatty liver disease. Biomed Res. Int. 2020:4020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato R., Inoue J., Kawabe Y., Kodama T., Takano T., Maeda M. (1996). Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 271 26461–26464. 10.1074/jbc.271.43.26461 [DOI] [PubMed] [Google Scholar]
- Shao J., Weng X., Zhuo L., Yu L., Li Z., Shen K., et al. (2019). Angiotensin II induced CSF1 transcription is mediated by a crosstalk between different epigenetic factors in vascular endothelial cells. Biochim. Biophys. Acta Gene Regul. Mech. 1862 1–11. 10.1016/j.bbagrm.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Shao J., Xu Y., Fang M. (2020). BRG1 deficiency in endothelial cells alleviates thioacetamide induced liver fibrosis in mice. Biochem. Biophys. Res. Commun. 521 212–219. 10.1016/j.bbrc.2019.10.109 [DOI] [PubMed] [Google Scholar]
- Shimano H., Sato R. (2017). SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat. Rev. Endocrinol. 13 710–730. 10.1038/nrendo.2017.91 [DOI] [PubMed] [Google Scholar]
- Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. (1997). Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99 838–845. 10.1172/jci119247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Chen B., Wu J., Jiang C., Fan Z., Feng Y., et al. (2020). Epigenetic regulation of a disintegrin and metalloproteinase (ADAM) promotes colorectal cancer cell migration and invasion. Front. Cell Dev. Biol. 8:581692. 10.3389/fcell.2020.581692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Wang M., Song H., Wu T., Wei H., He S., et al. (2013). SCAP gene polymorphisms decrease the risk of nonalcoholic fatty liver disease in females with metabolic syndrome. J. Genet. 92 565–570. 10.1007/s12041-013-0280-9 [DOI] [PubMed] [Google Scholar]
- Weng X., Zhang Y., Li Z., Yu L., Xu F., Fang M., et al. (2019). Class II transactivator (CIITA) mediates IFN-gamma induced eNOS repression by enlisting SUV39H1. Biochim. Biophys. Acta Gene Regul. Mech. 1862 163–172. 10.1016/j.bbagrm.2019.01.005 [DOI] [PubMed] [Google Scholar]
- Wu Q., Madany P., Dobson J. R., Schnabl J. M., Sharma S., Smith T. C., et al. (2016). The BRG1 chromatin remodeling enzyme links cancer cell metabolism and proliferation. Oncotarget 7 38270–38281. 10.18632/oncotarget.9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Wang H., Xin X., Yang J., Hou Y., Fang M., et al. (2020). An MRTF-A-Sp1-PDE5 axis mediates angiotensin-II-induced cardiomyocyte hypertrophy. Front. Cell Dev. Biol. 8:839. 10.3389/fcell.2020.00839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Dong W., Zhang T., Ren H., Wang J., Shang L., et al. (2020). Epiregulin (EREG) and myocardin related transcription factor A (MRTF-A) form a feedforward loop to drive hepatic stellate cell activation. Front. Cell Dev. Biol. 8:591246. 10.3389/fcell.2020.591246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Li J., Kang R., Tang D. (2020). Interplay between lipid metabolism and autophagy. Front. Cell Dev. Biol. 8:431. 10.3389/fcell.2020.00431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Li Z., Guo J., Xu Y. (2020a). Deacetylation of MRTF-A by SIRT1 defies senescence induced down-regulation of collagen type I in fibroblast cells. Biochim. Biophys. Acta Mol. Basis Dis. 1866:165723. 10.1016/j.bbadis.2020.165723 [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu L., Fang M., Bai H., Xu Y. (2019a). The chromatin remodeling protein BRM regulates the transcription of tight junction proteins: implication in breast cancer metastasis. Biochim. Biophys. Acta Gene Regul. Mech. 1862 547–556. 10.1016/j.bbagrm.2019.03.002 [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu L., Li M., Cheng X., Fang M., Zeng Q., et al. (2019b). The chromatin remodeling protein BRG1 links ELOVL3 trans-activation to prostate cancer metastasis. Biochim. Biophys. Acta Gene Regul. Mech. 1862 834–845. 10.1016/j.bbagrm.2019.05.005 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yang G., Yu L., Lin L., Liu L., Fang M., et al. (2020b). An interplay between MRTF-A and the histone acetyltransferase TIP60 mediates hypoxia-reoxygenation induced iNOS transcription in macrophages. Front. Cell Dev. Biol. 8:484. 10.3389/fcell.2020.00484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. D., Song J. Y., Wang S., Liu F. H., Zhang Y. N., Shang X. R., et al. (2017). Genetic variations in sterol regulatory element binding protein cleavage-activating protein (SCAP) are associated with blood pressure in overweight/obese Chinese children. PLoS One 12:e0177973. 10.1371/journal.pone.0177973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Yang J., Chen H., Li J., Que L., Zhu G., et al. (2019). Peli1 induction impairs cardiac microvascular endothelium through Hsp90 dissociation from IRE1alpha. Biochim. Biophys. Acta Mol. Basis Dis. 1865 2606–2617. 10.1016/j.bbadis.2019.06.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.