Abstract
Background
Although lung adenocarcinoma (LADC) with sensitizing mutations of the epidermal growth factor receptor (EGFR) is highly sensitive to EGFR tyrosine kinase inhibitors (EGFR-TKIs), in most cases disease progression inevitably occurs. Our aim was to investigate the predictive and prognostic significance of adjusted tumoral EGFR variant allele frequency (EGFR-aVAF) in the above setting.
Methods
Eighty-nine Caucasian advanced-stage LADC patients with known exon-specific EGFR mutations undergoing EGFR-TKI treatment were included. The correlations of EGFR-aVAF with clinicopathological variables including progression-free and overall survival (PFS and OS, respectively) were retrospectively analyzed.
Results
Of 89 EGFR-mutant LADC patients, 46 (51.7%) had exon 19 deletion, while 41 (46.1%) and 2 (2.2%) patients had exon 21- and exon 18-point mutations, respectively. Tumoral EGFR-aVAF was significantly higher in patients harboring EGFR exon 19 mutations than in those with exon 21-mutant tumors (P<0.001). Notably, patients with EGFR exon 19 mutant tumors demonstrated significantly improved PFS (P=0.003) and OS (P=0.02) compared to patients with exon 21 mutations. Irrespective of specific exon mutations, a statistically significant positive linear correlation was found between EGFR-aVAF of tumoral tissue and PFS (r=0.319; P=0.002). High (≥70%) EGFR-aVAF was an independent predictor of longer PFS [vs. low (<70%) EGFR-aVAF; median PFSs were 52 vs. 26 weeks, respectively; P<0.001]. Additionally, patients with high EGFR-aVAF also had significantly improved OS than those with low EGFR-aVAF (P=0.011).
Conclusions
Our study suggests that high (≥70%) EGFR-aVAF of tumoral tissue predicts benefit from EGFR-TKI treatment in advanced LADC and, moreover, that exon 19 EGFR mutation is associated with high EGFR-aVAF and improved survival outcomes.
Keywords: Epidermal growth factor receptor mutation (EGFR mutation), lung adenocarcinoma (LADC), variant allele frequency (VAF)
Introduction
Lung cancer is the most frequently diagnosed malignancy worldwide (11.6% of the total cases) and the leading cause of cancer-related mortality (18.4% of the total cancer deaths) (1). Histologically, non-small cell lung cancer (NSCLC) is the predominant lung cancer subtype and more than 40% of all NSCLCs diagnosed are lung adenocarcinomas (LADCs) (1). However, not all LADCs are the same, and inter-tumoral heterogeneity exists both in terms of pathological and molecular features (2).
Epidermal growth factor receptor (EGFR) mutations are the second most common oncogenic driver events in LADC, accounting for approximately 15% of all LADCs in Caucasian patients and about 40% to 50% in Asian patients (3,4). EGFR is a member of the ErbB family of tyrosine kinase receptors that is expressed in some normal epithelial, mesenchymal, and neurogenic tissue with cytoplasmic kinase activity transducing important growth factor signaling (5,6). However, in malignant tumors including LADC, EGFR is often constantly stimulated due to the sustained production in the tumor microenvironment of EGFR ligands, or as a result of a mutation in EGFR itself that locks the receptor in a state of continuous activation (7,8). About 90% of activating EGFR mutations are short in-frame deletions in exon 19 or point mutations in exon 21 often referred to as “classical” EGFR mutations (9,10). Exon 18 mutations are rare and relatively homogenous (compared to other rare mutations such as EGFR exon 20 insertions) as they represent about 4% of all EGFR mutations (9,10). Importantly, in LADC, these EGFR-sensitizing mutations confer sensitivity both to first-, second- and third-generation EGFR tyrosine kinase inhibitors (EGFR-TKIs) such as gefitinib, erlotinib, dacomitinib, afatinib and osimertinib in patients with advanced-stage disease (11-13).
Over the past decade, the application of EGFR-TKIs have led to a new era in the treatment of LADC. Accordingly, EGFR-TKIs improve both the progression-free survival (PFS) [10.8 vs. 5.4 months in the chemotherapy (CHT) group; P<0.001] and overall survival (OS) (30.5 vs. 23.6 months in the CHT group; P=0.31) in patients who were selected on the basis of EGFR-sensitizing mutations (14). Still, the objective response rate to EGFR-TKIs in patients carrying EGFR-sensitizing mutations is only 70% to 80%, and while some patients show clear survival benefit to TKIs others failed to respond properly (15,16). Therefore, in order to assess the effectiveness of current treatment options, it is crucial to understand the intrinsic and extrinsic factors that influence the responsiveness to TKIs in these patients.
Sensitivity to EGFR-TKIs is associated with female sex, never-smoking status and Asian ethnicity, however, such clinical factors are in fact predictors of EGFR mutations rather than true treatment-related prognosticators for TKI efficacy (14,15,17,18). Nevertheless, different EGFR mutation subtypes and molecular characteristics can also determine different predictive and prognostic features (15). In addition, differences in the proportion of tumor cells (TCs) harboring EGFR mutations might also contribute to therapy response, since only a fraction of cancer cells in an individual patient carry heterozygous activating mutations, whereas other cancer cells carry wild-type EGFR (19-22). Accordingly, previous studies on Asian patients suggest that higher relative EGFR mutational abundance might predict benefit from EGFR-TKI treatment (19,23,24).
Presently, the biological and clinical relevance of adjusted tumoral EGFR variant allele frequency (EGFR-aVAF) in terms of prognosis and clinical response to EGFR-TKIs is still mostly unclear. Therefore, in order to improve patient selection and to better understand the influence of EGFR-aVAF in this setting with regards to therapeutic approaches, our aim was to assess the relationship between EGFR-aVAF and response to EGFR-TKIs in a homogenous patient cohort of Caucasian LADC patients.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-814).
Methods
Ethics statement
The present study was directed in accordance with the guidelines of the Helsinki Declaration (revised in 2013) of the World Medical Association. The study was approved by the national level ethics committee (Hungarian Scientific and Research Ethics Committee of the Medical Research Council, ETT-TUKEB, 7214-1/2016/EKU). The need for individual informed consent for this retrospective study was waived. After clinical information was collected, patient identifiers were removed, and subsequently, patients cannot be identified either directly or indirectly.
Study population
Based on our inclusion/exclusion criteria, 89 pathologically confirmed advanced-stage LADC patients were included in this multi-center, retrospective study, who received EGFR-TKI therapy mainly in the following two Hungarian medical centers between 2008 and 2020: Torokbalint County Institute of Pulmonology, Torokbalint; and Department of Pulmonology of the Semmelweis University, Budapest, Hungary. Of note, all ten participating medical centers are enlisted in Table S1. All tumor tissues were tested for EGFR mutations required for anti-EGFR therapy and all samples were retrieved from treatment-naïve patients. Based on our inclusion criteria, cytologically or histologically verified unresectable stage IIIb or stage IV patients were included who received either gefitinib or erlotinib as first- or second-line treatment. According to the therapy guidelines of the host institutes, only patients with Eastern Cooperative Oncology Group (ECOG) performance status (PS) 0–1 were included. With regards to our exclusion criteria, patients with concomitant mutations in two or more exons, or patients harboring resistance mutations such as T790M substitution in exon 20 were excluded. Additionally, all cases where the exact percentage of neoplastic cells was not available were also subsequently excluded. Patients treated with afatinib were excluded due to the relatively small number of cases. Similarly, patients in whom TKI therapy was suspended by the reason of drug-related toxicities like hepato- and cardiotoxicities, or patients treated with EGFR-TKIs as third-line therapy were also excluded. Finally, patients who received EGFR-TKI therapy for a period less than 4 weeks or the cause of death was not related to lung cancer progression were also excluded. Clinicopathological data regarding gender, age at lung cancer diagnosis, smoking history, type of EGFR exon mutation, EGFR-aVAF, treatment and survival data for the included patients were retrospectively collected from medical records and/or records from the National Health Insurance Office or Central Statistical Office.
Treatment
Diagnostic and therapeutic approaches were conducted in accordance with the individual institutional guidelines and with the current National Comprehensive Cancer Network (NCCN) guidelines with no differences across the host institutes (25). All included patients received either gefitinib or erlotinib as first- or second-line systemic therapy on a daily basis (250 and 150 mg, respectively) until disease progression. In case of patients who received first-line CHT before the initiation of TKI therapy, patients were treated with platinum-based standard of care CHT regimens. According to the national treatment financing scheme, all EGFR-TKI-treated patients had to return to the hospital every month for chest X-ray and clinical check-up, and the clinical response to treatment was classified based on follow-up CT scans every 3 months by using the response evaluation criteria in solid tumors (RECIST 1.0) (26).
EGFR mutation analysis
Tissue samples were acquired during diagnostic procedures including wedge resection surgery and bronchoscopic- or transthoracic needle biopsy. The diagnosis of LADC of each case was confirmed on a freshly prepared hematoxylin and eosin stained slide. As this estimate is critical for the study, the exact proportion of neoplastic cells was reassessed by two independent expert histopathologists. All mutational analyses were performed at the 1st Department of Pathology and Experimental Cancer Research of the Semmelweis University. Genomic DNA was extracted using the High Pure PCR Template Preparation Kit (Roche, Basel, Switzerland) in accordance with the manufacturer’s instructions, and all samples underwent testing for mutation in EGFR codon (in exon 18, 19, 21) using the Therascreen EGFR Pyro Kit (Qiagen, Germany) on a PyroMarkTM Q24 (Qiagen) pyrosequencing instrument. The percentage of mutated nucleic acid was calculated with the equipment software (Qiagen PyroMarkTM), relating the peak of mutated base to that of the wild-type base, which was considered 100%. The obtained VAF for each patient was then normalized to the proportion of neoplastic cells in each specimen using the following formula:
| [1] |
Where VAF represents the percentage of the EGFR variant alleles determined by the pyrosequencing assay and TC% is the estimated percentage of neoplastic cells.
Statistical analyses
All statistical analyses were performed using the SPSS Statistics 23.0 package (SPSS Inc., Chicago, IL, USA). Data distribution was verified by the Kolmogorov-Smirnov normality test. EGFR-aVAF of tumoral tissue as a continuous variable was analyzed with regards to dichotomized clinicopathological variables by Mann-Whitney U test and Kruskal–Wallis test. The co-primary endpoints were PFS and OS. PFS was defined as the time from commencement of gefitinib or erlotinib treatment to disease progression according to the aforementioned RECIST 1.0 criteria. OS was defined as the interval between the initiation of medication and death related to progressive disease. Clinical follow-up was closed on the 1st of April, 2020. Survival curves were estimated by Kaplan-Meier plots and the differences between different groups were compared using the log-rank test. The association between EGFR-aVAF as continuous variable and PFS and OS was also evaluated by using the Spearman’s correlation coefficient. The value of linear correlation coefficient (r) varies from –1 to 1 both values inclusive. No linear correlation (r=0), weak positive correlation (0< r ≤0.3), moderate positive linear correlation (0.3< r ≤0.7), strong positive linear correlation (0.7< r ≤1) (27). The independent prognostic value of the clinicopathological variables was studied with Cox proportional hazard regression model, which was adjusted for EGFR-aVAF and age (as continuous variables), gender (male versus female), EGFR exon mutation (exon 19 versus exon 21), therapeutic agents (gefitinib versus erlotinib) and treatment line (first- versus second-line). All reported P values are two-sided, and a level of 0.05 or less was considered statistically significant.
Results
Patient characteristics and EGFR-aVAF
After applying the exclusion criteria, 89 LADC patients with known EGFR gene mutations were enrolled in the study whose clinicopathological characteristics are summarized in Table 1. All patients had advanced-stage disease and Caucasian background. Median age of all cases was 67 (range, 34–92) years and patients were predominantly female (71.9%). A total of 46 (51.7%) patients had exon 19 deletion, while 41 (46.1%) and 2 (2.2%) patients had exon 21- and exon 18-point mutations, respectively. Median age was 61, 66 and 70 years in exon mutation subgroups 18, 19 and 21, respectively (with no significant differences in age distribution, P=0.332; data not shown). As for therapeutic approaches, 58 (65.2%) patients received gefitinib, while 31 (34.8%) patients were treated with erlotinib.
Table 1. Patient characteristics and adjusted tumoral EGFR-VAF in human LADC.
| Characteristics | Number of patients (%) | Mean EGFR-aVAF, % | P valuea |
|---|---|---|---|
| All patients | 89 (100.0) | ||
| Age (years) | 0.93b | ||
| <65 | 36 (40.4) | 63.53 | |
| ≥65 | 53 (59.6) | 64.6 | |
| Gender | 0.809b | ||
| Male | 25 (28.1) | 64.12 | |
| Female | 64 (71.9) | 64.19 | |
| Smoking history | 0.467c | ||
| Never smoker | 48 (51.7) | 64.46 | |
| Ex-smoker | 10 (11.2) | 73.3 | |
| Current smoker | 14 (15.7) | 58.5 | |
| No data | 19 (21.3) | – | |
| Therapeutic agent | 0.428b | ||
| Gefitinib | 58 (65.2) | 61.64 | |
| Erlotinib | 31 (34.8) | 68.9 | |
| Treatment line | 0.882b | ||
| First-line | 46 (51.7) | 63.35 | |
| Second-line | 43 (48.3) | 65.05 | |
| EGFR exon mutation | |||
| Exon 18 | 2 (2.2) | – | <0.001b |
| Exon 19 | 46 (51.7) | 75.04 | |
| Exon 21 | 41 (46.1) | 51.44 |
a, P values refer to mean EGFR-aVAF between patient subgroups; b, Mann-Whitney U test; c, Kruskal-Wallis test; d, not included in the statistical calculation. EGFR, epidermal growth factor receptor; EGFR-aVAF, adjusted EGFR variant allele frequency; LADC, lung adenocarcinoma.
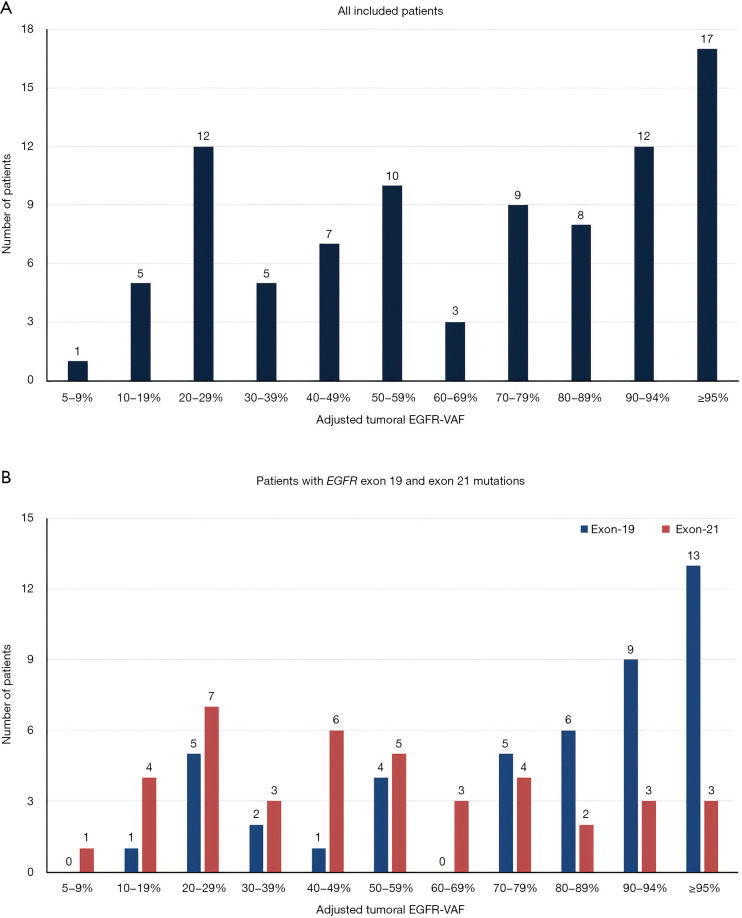
In order to study the clinical relevance of mutational percentage of tumoral tissue, we performed comparative statistical analyses of EGFR-aVAF and clinicopathological variables. Out of all 89 cases, 72 cases showed EGFR-aVAF between 5% and 94% and 17 patients exhibited EGFR-aVAF ≥95% (Figure 1A). In case of six patients the EGFR-aVAF of tumoral tissue was <20%. Interestingly, the adjusted VAF was significantly higher in patients harboring EGFR exon 19 mutations than those with exon 21 mutant tumors (P<0.001; Table 1, Figure 1B). There were no statistically significant differences in the mean EGFR-aVAF with respect to age (P=0.93), gender (P=0.809), or smoking history (P=0.467).
Figure 1.
EGFR-aVAF of tumoral tissue in LADC patients. (A) Bar chart illustrating the distribution of all included LADC patients (n=89), according to tumoral EGFR-aVAF irrespective of specific exon mutations. (B) Distribution of LADC patients diagnosed with EGFR exon 19 and exon 21 mutations (n=46 and n=41, respectively). EGFR, epidermal growth factor receptor; EGFR-aVAF, adjusted EGFR variant allele frequency; LADC, lung adenocarcinoma.
EGFR exon 19 mutation associates with superior survival outcomes
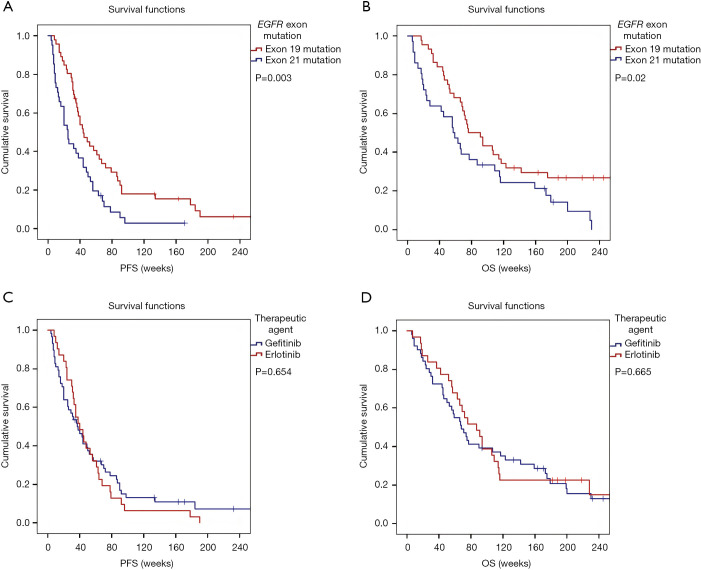
The median PFS and OS of the full cohort was 38 and 72 weeks, respectively. At the time of the closing date of the clinical follow-up, all patients with EGFR exon 18 mutations, 42 patients with exon 19 mutations and 39 patients with exon 21 mutations had experienced disease progression after EGFR-TKI therapy. Due to the small number of patients in EGFR exon 18-mutated subgroup, statistical analyses were performed solely by comparing the median PFS and OS of exon subgroups 19 and 21. Accordingly, as shown in Figure 2A, LADC patients with tumors harboring EGFR exon 19 mutations had significantly improved median PFS than those with exon 21 mutations (median PFSs were 44 vs. 25 weeks, respectively; P=0.003). In line with the PFS data, EGFR exon 19 mutations were significantly associated with longer OS as well (vs. exon 21 mutation, median OSs were 76 vs. 57 weeks, respectively; P=0.02; Figure 2B). With regards to the administered therapeutic agents, no significant differences have been observed neither in PFS (P=0.654; Figure 2C) nor in OS (P=0.665; Figure 2D) in patients treated with gefitinib vs. erlotinib. Of note, the treatment line of EGFR-TKI did not influence the survival outcomes neither (Figure S1A,B). As for smoking history, there was no significant difference in PFS between never-smoker versus ever-smoker patients (P=0.099; Figure S1C). Interestingly, however, Kaplan-Meyer curves demonstrated significantly longer median OS in never-smoker patients (vs. ever-smokers, median OSs were 106 vs. 52 weeks, respectively, P=0.007; Figure S1D).
Figure 2.
Kaplan-Meier plots for PFS and OS in patients with LADC according to specific EGFR exon mutations and therapeutic approaches. (A) LADC patients with tumors harboring EGFR exon 21 mutations had significantly shorter median PFS than those with exon 19 mutations (median PFSs were 25 vs. 44 weeks, respectively; P=0.003, log-rank test). (B) EGFR exon 21 mutation was also associated with significantly shorter OS in these patients (vs. EGFR exon 19 mutations, median OSs were 57 vs. 76 weeks, respectively; P=0.02, log-rank test). (C) No significant differences in PFS have been observed in patients treated with gefitinib vs. erlotinib (median PFSs were 37 vs. 40 weeks, respectively; P=0.654, log-rank test). (D) Similarly, the OS also did not differ significantly between the patients treated with gefitinib vs. erlotinib (median OSs were 68 vs. 87 weeks, respectively; P=0.665, log-rank test). PFS, progression-free survival; OS, overall survival; LADC, lung adenocarcinoma; EGFR, epidermal growth factor receptor.
EGFR-aVAF has clinical utility in predicting survival outcomes in LADC patients treated with EGFR-TKIs
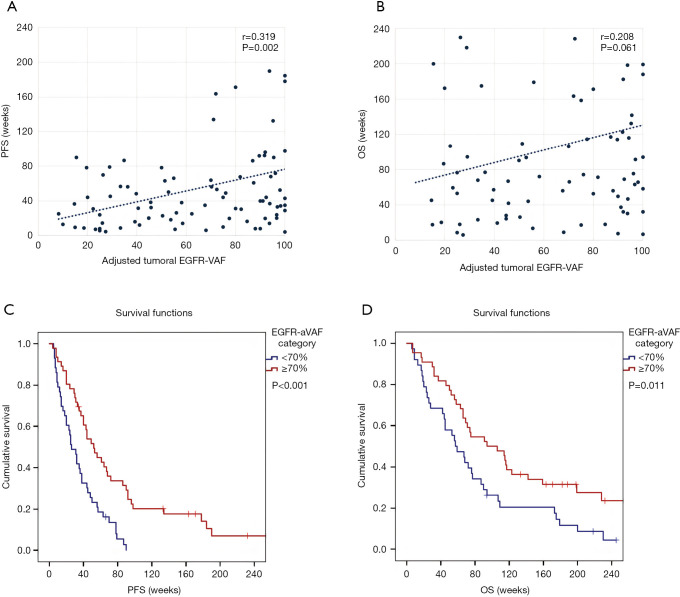
Next, we evaluated the survival outcomes of TKI-treated EGFR-mutant LADC patients with regards to adjusted tumoral variant allele frequencies. Importantly, a statistically significant positive linear correlation was found between EGFR-aVAF and PFS (r=0.319; P=0.002, Spearman’s correlation; Figure 3A). In contrast, no significant correlation was found between EGFR-VAF and OS, although the correlation coefficient was found to be clinically notable (r=0.208; P=0.061, Spearman’s correlation; Figure 3B). In order to rule out the potential confounding effects of Spearman’s correlation and to evaluate the survival outcomes with Kaplan-Meier methods, patients were categorized by the median EGFR-aVAF (70%) of tumoral tissue. Therefore, we grouped patients into low (<70%) and high (≥70%) EGFR-aVAF categories and found that patients with high adjusted tumoral EGFR-VAF had significantly longer PFS than those in the low EGFR-aVAF group (median PFSs were 52 vs. 26 weeks, respectively; P<0.001, Figure 3C). Additionally, patients with high EGFR-aVAF also had significantly improved OS (vs. those with low EGFR-aVAF; median OSs were 94 vs. 57 weeks, respectively; P=0.011, Figure 3D).
Figure 3.
Scatter plots and Kaplan-Meier estimates for PFS and OS in LADC patients according to EGFR-aVAF. (A) Scatter plot showing significant positive linear correlation between tumoral EGFR-aVAF and PFS (r=0.319; P=0.002, Spearman’s correlation) (each dot represents a single patient, and the dashed line shows the linear trendline). (B) Statistically non-significant, although clinically notable correlation was found between EGFR-VAF and OS (r=0.208; P=0.061, Spearman’s correlation). (C) Patients with tumoral EGFR-aVAF ≥70% had significantly longer PFS than those in the EGFR-aVAF low (<70%) group (median PFSs were 52 vs. 26 weeks, respectively; P<0.001, log-rank test). (D) Similarly, the median OS was also significantly increased in patients with high (≥70%) EGFR-aVAF [vs. those with low (<70%) EGFR-aVAF, median OSs were 94 vs. 57 weeks, respectively; P=0.011, log-rank test]. PFS, progression-free survival; OS, overall survival; LADC, lung adenocarcinoma; EGFR, epidermal growth factor receptor; EGFR-aVAF, adjusted EGFR variant allele frequency.
In order to assess if the predictive value of tumoral EGFR-aVAF was independent from other clinicopathological factors, we performed a multivariate Cox regression analysis (Table 2). The model was adjusted for clinicopathological variables such as EGFR-aVAF, age, gender, EGFR exon mutation, therapeutic agents and treatment line. Importantly, we found that EGFR-aVAF of tumoral tissue remained a significant prognostic factor for PFS [continuous variable, hazard ratio (HR): –0.009, 95% confidence interval (CI): 0.982–0.999; P=0.042; Table 2]. Besides, Cox regression analysis revealed that the specific exon mutations (nominal variable, HR: 0.284, 95% CI: 1.017–1.735; P=0.037) also influence the PFS independently.
Table 2. Multivariate Cox regression model for clinicopathological variables influencing the PFS.
| Clinicopathological parameters | PFS |
|---|---|
| EGFR-aVAF (continuous) | |
| HR | –0.009 |
| 95% CI | (0.982–0.999) |
| P | 0.042 |
| EGFR exon mutation (exon 19 vs. exon 21) | |
| HR | 0.284 |
| 95% CI | (1.017–1.735) |
| P | 0.037 |
| Age (continuous) | |
| HR | –0.021 |
| 95% CI | (0.958–1.001) |
| P | 0.06 |
| Gender (male vs. female) | |
| HR | 0.460 |
| 95% CI | (0.913–2.747) |
| P | 0.102 |
| Therapeutic agent (gefitinib vs. erlotinib) | |
| HR | –0.032 |
| 95% CI | (0.595–1.579) |
| P | 0.899 |
| Treatment line (first- vs. second-line) | |
| HR | –0.013 |
| 95% CI | (0.607–1.603) |
| P | 0.957 |
PFS, progression-free survival; EGFR, epidermal growth factor receptor; EGFR-aVAF, adjusted EGFR variant allele frequency; HR, hazard ratio; CI, confidence interval.
Discussion
In the era of precision and individualized cancer therapy precise definition of tumor type including comprehensive histological classification, and description of clinically relevant molecular pathological characteristics is crucial (28,29). Targeting EGFR is a promising strategy for treating LADC patients, since numerous studies over the past decade have shown that the TKI inhibitors gefitinib and erlotinib are effective for advanced-stage NSCLCs harboring EGFR sensitizing mutations (30,31). Still, the efficacy of TKIs is not consistent for every patient and not all patients with EGFR-activating mutation show similar response rates and PFSs (18). Hence, there is an urgent need for identifying valid predictive and prognostic factors that enable clinicians to effectively select the patients who may benefit more from EGFR-TKI treatment. Early in 2011, Zhou et al. reported that the relative EGFR mutational abundance might predict the therapy response to gefitinib in advanced-stage Asian NSCLC patients, yet the predictive value and clinicopathological significance of EGFR-aVAF is still controversial, especially in Caucasian patients (19). Therefore, the aim of this study was to assess the clinicopathological significance of EGFR-aVAF and to evaluate its predictive and prognostic relevance in a homogenous cohort of Hungarian LADC patients treated with EGFR-TKIs.
First, we analyzed the association of major clinicopathological characteristics and tumoral EGFR-aVAF. Our results revealed that a considerable proportion of LADCs contain a heterogeneous population of both EGFR mutated and non-mutated cancer cells since the majority of all included cases showed an EGFR-aVAF between 5% and 94% and only 17 patients exhibited EGFR-aVAF ≥95%. This finding is in line with previously published data also suggesting that only a certain percentage of TCs carry heterozygous activating mutations in NSCLC patients, while other TCs carry wild-type EGFR (21,22). Accordingly, this might explain the controversial response rates seen in EGFR-TKI-treated patients. In the current study, 2.2% of patients carried exon 18 EGFR mutations, therefore the incidence rate is similar to other studies. However, due to the small number of patients harboring exon 18 mutations, subgroup specific statistical calculations were performed without these patients (15). Importantly, we found that the aVAF of the tumoral tissue was significantly higher in patients harboring EGFR exon 19 mutations than those with exon 21 mutated tumors. This ratio is in line with a previously published Asian study, however, to the best of our knowledge, ours is the first detailed evaluation of tumoral EGFR-aVAF with regards to specific EGFR exon mutations in Caucasian patients (23).
Next, in order to assess the clinical relevance of this heterogeneity in EGFR-aVAF between the patients harboring exon 19 vs. exon 21 mutations, we investigated the prognostic and predictive relevance of the aforementioned EGFR exon alterations. As expected, patients harboring EGFR exon 19 mutations indeed had significantly longer PFS than those with EGFR exon 21 mutations. These findings are in line with previously published data also suggesting a significant advantage in PFS for patients carrying exon 19 deletions in comparison with those carrying EGFR exon 21 mutations (32-35). In addition, based on a recent study on 55 metastatic NSCLC patients, exon 19-mutated patients tend to have better survival outcomes than patients with exon 18 point-mutations as well (15). To date, the mechanism underlying the different sensitivities to EGFR-TKI treatment between exon 19 and exon 21 mutated tumors remains to be elucidated (34). Based on our results a possible explanation might be that EGFR-aVAF of tumoral tissue is significantly higher in EGFR exon 19 mutated patients compared to patients harboring exon 21 mutations and thus EGFR-TKIs might be more effective in these patients. Meanwhile, others suggest that the better survival outcomes with EGFR exon 19 than exon 21 mutations might be due to differential inhibition of downstream signals, since EGFR-TKIs inhibit the phosphorylation of EGFR, Akt, and Erk to a greater degree in exon 19 deletion cells than in exon 21 mutated cells (36). Furthermore, an additional explanation might be that exon 19 deletions and 21 mutations present different intrinsic sensitivities to the EGFR-TKIs (34,37). Importantly, different mutations in the same exon might also indicate different predictive roles since non-L747 to E749 (LRE) deletions has a worse response to TKIs than LRE deletions but we had no data on the type of deletions in exon 19 (38). Altogether, the biology that lies behind the responsiveness to EGFR-TKIs with regards to EGFR mutational subtypes is yet to be elucidated, however, our findings might provide background for future studies. In line with the PFS data, EGFR exon 19 mutations were also associated with improved OS compared to exon 21 mutations. As for treatment-related data, no significant differences were observed in PFS or OS regarding treatment line and therapeutic agents, which is in line with the findings of others (39-42).
Finally, we investigated the predictive and prognostic relevance of tumoral EGFR-aVAF and a statistically significant moderate positive linear correlation was found between EGFR-aVAF and PFS. Notably, we also found that high (≥70%) tumoral EGFR-aVAF was associated both with improved median PFS and OS, with a clinically relevant difference between low and high subgroups of 26 and 37 weeks, respectively. It should be noted, however, that the patients were divided into low and high EGFR-aVAF subgroups based on the median value in our dataset, therefore, until further validation, caution is needed when using it as a cut-off value in future studies. Our results are of high clinical importance because previous studies have only focused on whether the mutation is positive, and only a few investigated the predictive role of the relative EGFR mutational abundance (19,23,24). Yet, to our knowledge, our study is the first investigating the predictive and prognostic relevance of the exact value of EGFR-aVAF in Caucasian patients and, moreover, the first suggesting a clinically relevant threshold for predicting treatment response in these patients. In support of this, multivariate Cox regression analysis also revealed that EGFR-aVAF at diagnosis influenced PFS independently from age, gender, therapeutic agent, treatment line, and type of EGFR exon mutation. These results might partly explain why the efficacy of TKIs is not consistent for every patient harboring a certain type of EGFR mutation. Accordingly, quantitative diagnosis methods of EGFR-aVAF may help to select patients who are most or least likely to benefit from EGFR-TKIs. Importantly, however, current clinical treatment protocols with regards to EFGR-TKI are still primarily based on the absence or presence of activating EGFR mutations (25). Accordingly, until future validation, the clinicians should choose the most appropriate treatment for their patients regardless of EGFR-aVAF status. Nevertheless, changes in EGFR-aVAF might also occur during cancer progression and therapy. For instance, a recent study suggests that cancer genome in colorectal cancer patients adapts dynamically to pulsatile drug schedules and the abundance of resistance mutations could increase after long-time targeted therapies (43). Therefore, dynamic monitoring of EGFR-aVAF during therapy is also warranted.
There are several limitations in our study. Despite the fact that our cohort was homogenous the final number of patients harboring EGFR mutations was relatively small due to our strict inclusion/exclusion criteria. Nevertheless, our cohort provided the opportunity to draw some conclusions that evidently need to be validated in additional studies. Another limitation of our study is its retrospective nature with given limitations in interpreting the results. Thus, some of our results need to be confirmed in a prospective setting. Loss of heterozygosity and EGFR amplification occurs frequently in LADC patients harboring EGFR activating mutations and could serve as an indicator for better response from EGFR-TKI treatment (44-46). Accordingly, both of the aforementioned genetic alterations might also correlate with higher aVAF values, yet we did not investigate the presence of these alterations since they are not part of the routine mutational analyses in Hungary. Finally, all included patients were treated with first-generation EGFR-TKI erlotinib and gefitinib, yet these inhibitors are being slowly replaced by second- and third-generation EGFR-TKIs in the clinical practice. All in all, taken into account all the aforementioned potential study limitations, caution is needed when interpreting the results of the present study and further analyses are warranted to clarify the exact predictive role of EGFR-aVAF in EGFR-TKI-treated LADC patients.
Conclusions
To conclude, our study suggests that EGFR-aVAF of tumoral tissue predicts the extent of benefit from EGFR-TKI treatment. Moreover, in regards with exon specific mutations, the average EGFR-aVAF is higher among patients with exon 19 deletions thus confirming the longer PFS and OS of these patients. Our results might as well explain why the duration of response of some EGFR mutant patients was not as long as expected when no resistance related abnormality was found. Altogether, by shedding light on the predictive and prognostic relevance of EGFR-aVAF, our results might help to improve patient selection and treatment in advanced-stage LADC patients harboring EGFR-sensitizing mutations.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors are grateful to Laszlo Gobel for his technical help in the development of the clinical database. The authors would also like to thank the following clinicians for providing information concerning the clinicopathological data of included patients: Zsuzsanna Szalai, Gabriella Meszegeto and Istvan Albert.
Funding: BD, JM and CB acknowledge funding from the Hungarian National Research, Development and Innovation Office (KNN121510, KH130356, BD; NAP2-2017-1.2.1- NKP-0002, K129065, JM; NVKP_16-1-2016-0004, CB). BD and VL were supported by the Austrian Science Fund (FWF I3522, VL; FWF I3977 and I4677, BD). VL is a recipient of the Bolyai Research Scholarship of the Hungarian Academy of Sciences and the UNKP-19-4 New National Excellence Program of the Ministry for Innovation and Technology. ZM was supported by the UNKP-20-3 New National Excellence Program of the Ministry for Innovation and Technology. CB was supported by the Higher Education Institutional Excellence Programme of the Ministry of Human Capacities in Hungary within the framework of the Molecular Biology Thematic Programme of the Semmelweis University.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study was directed in accordance with the guidelines of the Helsinki Declaration (revised in 2013) of the World Medical Association. The study was approved by the national level ethics committee (Hungarian Scientific and Research Ethics Committee of the Medical Research Council, ETT-TUKEB, 7214-1/2016/EKU). The need for individual informed consent for this retrospective study was waived. After clinical information was collected, patient identifiers were removed, and subsequently, patients cannot be identified either directly or indirectly.
Footnotes
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-814
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-814
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-814). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Revannasiddaiah S, Thakur P, Bhardwaj B, et al. Pulmonary adenocarcinoma: implications of the recent advances in molecular biology, treatment and the IASLC/ATS/ERS classification. J Thorac Dis 2014;6:S502-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 2015;5:2892-911. [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison PT, Vyse S, Huang PH. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin Cancer Biol 2020;61:167-79. 10.1016/j.semcancer.2019.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bethune G, Bethune D, Ridgway N, et al. Epidermal growth factor receptor (EGFR) in lung cancer: an overview and update. J Thorac Dis 2010;2:48-51. [PMC free article] [PubMed] [Google Scholar]
- 6.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. 10.1146/annurev-pathol-011110-130206 [DOI] [PubMed] [Google Scholar]
- 7.Inamura K, Ninomiya H, Ishikawa Y, et al. Is the epidermal growth factor receptor status in lung cancers reflected in clinicopathologic features? Arch Pathol Lab Med 2010;134:66-72. 10.5858/2008-0586-RAR1.1 [DOI] [PubMed] [Google Scholar]
- 8.Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol 2018;12:3-20. 10.1002/1878-0261.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 2004;64:8919-23. 10.1158/0008-5472.CAN-04-2818 [DOI] [PubMed] [Google Scholar]
- 10.Beau-Faller M, Prim N, Ruppert AM, et al. Rare EGFR exon 18 and exon 20 mutations in non-small-cell lung cancer on 10 117 patients: a multicentre observational study by the French ERMETIC-IFCT network. Ann Oncol 2014;25:126-31. 10.1093/annonc/mdt418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. 10.1056/NEJMoa044238 [DOI] [PubMed] [Google Scholar]
- 12.Yu HA, Riely GJ. Second-generation epidermal growth factor receptor tyrosine kinase inhibitors in lung cancers. J Natl Compr Canc Netw 2013;11:161-9. 10.6004/jnccn.2013.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews Wright NM, Goss GD. Third-generation epidermal growth factor receptor tyrosine kinase inhibitors for the treatment of non-small cell lung cancer. Transl Lung Cancer Res 2019;8:S247-64. 10.21037/tlcr.2019.06.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 15.Rossi S, D'Argento E, Basso M, et al. Different EGFR gene mutations in exon 18, 19 and 21 as prognostic and predictive markers in NSCLC: a single institution analysis. Mol Diagn Ther 2016;20:55-63. 10.1007/s40291-015-0176-x [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, Okamoto I, Kashii T, et al. Multicentre prospective phase II trial of gefitinib for advanced non-small cell lung cancer with epidermal growth factor receptor mutations: results of the West Japan Thoracic Oncology Group trial (WJTOG0403). Br J Cancer 2008;98:907-14. 10.1038/sj.bjc.6604249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SY, Myung JK, Kim HR, et al. Factors that predict clinical benefit of EGFR TKI therapy in patients with EGFR wild-type lung adenocarcinoma. Tuberc Respir Dis (Seoul) 2019;82:62-70. 10.4046/trd.2018.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Chang A. Molecular predictors of EGFR-TKI sensitivity in advanced non-small cell lung cancer. Int J Med Sci 2008;5:209-17. 10.7150/ijms.5.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Q, Zhang XC, Chen ZH, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol 2011;29:3316-21. 10.1200/JCO.2010.33.3757 [DOI] [PubMed] [Google Scholar]
- 20.Zhu Y, Guo Z, Liu Y, et al. A novel ARMS-based assay for the quantification of EGFR mutations in patients with lung adenocarcinoma. Oncol Lett 2018;15:2905-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi K, Okami J, Kodama K, et al. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci 2008;99:929-35. 10.1111/j.1349-7006.2008.00782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang SX, Yamashita K, Yamamoto M, et al. EGFR genetic heterogeneity of nonsmall cell lung cancers contributing to acquired gefitinib resistance. Int J Cancer 2008;123:2480-6. 10.1002/ijc.23868 [DOI] [PubMed] [Google Scholar]
- 23.Li X, Cai W, Yang G, et al. Comprehensive analysis of EGFR-mutant abundance and its effect on efficacy of EGFR TKIs in advanced NSCLC with EGFR mutations. J Thorac Oncol 2017;12:1388-97. 10.1016/j.jtho.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Zhang M, Tang W, et al. Mutation abundance affects the therapeutic efficacy of EGFR-TKI in patients with advanced lung adenocarcinoma: a retrospective analysis. Cancer Biol Ther 2018;19:687-94. 10.1080/15384047.2018.1450115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ettinger DS, Wood DE, Aggarwal C, et al. NCCN guidelines insights: non-small cell lung cancer, version 1.2020. J Natl Compr Canc Netw 2019;17:1464-72. 10.6004/jnccn.2019.0059 [DOI] [PubMed] [Google Scholar]
- 26.Schwartz LH, Litiere S, de Vries E, et al. RECIST 1.1-update and clarification: from the RECIST committee. Eur J Cancer 2016;62:132-7. 10.1016/j.ejca.2016.03.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratner B. The correlation coefficient: Its values range between +1/−1, or do they? J Target Meas Anal Mark 2009;17:139-42. 10.1057/jt.2009.5 [DOI] [Google Scholar]
- 28.Lohinai Z, Hoda MA, Fabian K, et al. Distinct epidemiology and clinical consequence of classic versus rare EGFR mutations in lung adenocarcinoma. J Thorac Oncol 2015;10:738-46. 10.1097/JTO.0000000000000492 [DOI] [PubMed] [Google Scholar]
- 29.Raparia K, Villa C, DeCamp MM, et al. Molecular profiling in non-small cell lung cancer: a step toward personalized medicine. Arch Pathol Lab Med 2013;137:481-91. 10.5858/arpa.2012-0287-RA [DOI] [PubMed] [Google Scholar]
- 30.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer). Lancet 2005;366:1527-37. 10.1016/S0140-6736(05)67625-8 [DOI] [PubMed] [Google Scholar]
- 31.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005;353:123-32. 10.1056/NEJMoa050753 [DOI] [PubMed] [Google Scholar]
- 32.Jackman DM, Yeap BY, Sequist LV, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res 2006;12:3908-14. 10.1158/1078-0432.CCR-06-0462 [DOI] [PubMed] [Google Scholar]
- 33.Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res 2006;12:839-44. 10.1158/1078-0432.CCR-05-1846 [DOI] [PubMed] [Google Scholar]
- 34.Hong W, Wu Q, Zhang J, et al. Prognostic value of EGFR 19-del and 21-L858R mutations in patients with non-small cell lung cancer. Oncol Lett 2019;18:3887-95. 10.3892/ol.2019.10715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang H, Zhu M, Li Y, et al. Association between EGFR exon 19 or exon 21 mutations and survival rates after first-line EGFR-TKI treatment in patients with non-small cell lung cancer. Mol Clin Oncol 2019;11:301-8. 10.3892/mco.2019.1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu JQ, Zhong WZ, Zhang GC, et al. Better survival with EGFR exon 19 than exon 21 mutations in gefitinib-treated non-small cell lung cancer patients is due to differential inhibition of downstream signals. Cancer Lett 2008;265:307-17. 10.1016/j.canlet.2008.02.064 [DOI] [PubMed] [Google Scholar]
- 37.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 38.Chung KP, Wu SG, Wu JY, et al. Clinical outcomes in non-small cell lung cancers harboring different exon 19 deletions in EGFR. Clin Cancer Res 2012;18:3470-7. 10.1158/1078-0432.CCR-11-2353 [DOI] [PubMed] [Google Scholar]
- 39.Zhang W, Wei Y, Yu D, et al. Gefitinib provides similar effectiveness and improved safety than erlotinib for advanced non-small cell lung cancer: a meta-analysis. Medicine (Baltimore) 2018;97:e0460. 10.1097/MD.0000000000010460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chai CS, Liam CK, Pang YK, et al. Gefitinib versus erlotinib as first-line treatment in EGFR mutant advanced lung adenocarcinoma. Eur Respir J 2016;48:OA3341. [Google Scholar]
- 41.Wu JY, Yu CJ, Yang CH, et al. First- or second-line therapy with gefitinib produces equal survival in non-small cell lung cancer. Am J Respir Crit Care Med 2008;178:847-53. 10.1164/rccm.200803-389OC [DOI] [PubMed] [Google Scholar]
- 42.Massuti B, Morán T, Porta R, et al. Multicenter prospective trial of customized erlotinib for advanced non-small cell lung cancer (NSCLC) patients (p) with epidermal growth factor receptor (EGFR) mutations: Final results of the Spanish Lung Cancer Group (SLCG) trial. J Clin Oncol 2009;27:8023. 10.1200/jco.2009.27.15_suppl.8023 [DOI] [Google Scholar]
- 43.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:795-801. 10.1038/nm.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma ES, Wong CL, Siu D, et al. Amplification, mutation and loss of heterozygosity of the EGFR gene in metastatic lung cancer. Int J Cancer 2007;120:1828-31; author reply 1832-3. 10.1002/ijc.22506 [DOI] [PubMed] [Google Scholar]
- 45.Shan L, Wang Z, Guo L, et al. Concurrence of EGFR amplification and sensitizing mutations indicate a better survival benefit from EGFR-TKI therapy in lung adenocarcinoma patients. Lung Cancer 2015;89:337-42. 10.1016/j.lungcan.2015.06.008 [DOI] [PubMed] [Google Scholar]
- 46.Hayes DN, McLeod HL. EGFR regulation by microRNA in lung cancer: a rose by any other name ... is an increasingly complicated rose. Ann Oncol 2008;19:1036-7. 10.1093/annonc/mdn357 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as