Abstract
Background
N6-methyladenosine (m6A) is found to be associated with promoting tumorigenesis in different types of cancers, however, the function of m6A-related genes in testicular germ cell tumors (TGCT) development remains to be illuminated. This study aimed to investigated the prognostic value of m6A RNA methylation regulators in TGCT.
Methods
We collected TGCT patients’ information about clinicopathologic parameters and twenty-two m6A regulatory genes expression from The Cancer Genome Atlas (TCGA) database and Genotype-Tissue Expression (GTEx). We analyzed the differentially expressed m6A RNA methylation regulators between tumor tissues and normal tissues, as well as the correlation of m6A RNA methylation regulators. By using Cox univariate analysis, last absolute shrinkage and selection operator (LASSO) Cox regression algorithm and Cox multivariate proportional hazards regression analysis, a risk score was constructed based on a TCGA training cohort, and further verified in the TCGA testing cohort. Then, univariate and multivariate Cox regression analyses were used to evaluate the relationship between risk score and progression-free survival (PFS) in TGCT. Finally, the six-gene risk score was further verified by two gene expression profiles (GSE3218 and GSE10783) as an independent external validation cohort.
Results
Distinct expression patterns of m6A regulatory genes were identified between TGCT tissues and normal tissues in TCGA and GTEx datasets. To predict prognosis of TGCT patients, a risk score was calculated based on six selected m6A RNA methylation regulators (YTHDF1, RBM15, IGF2BP1, ZC3H13, METTL3, and FMR1). Additionally, we found significant differences between the high-risk and low-risk groups in serum marker study levels and histologic subtype. Univariate and multivariate analysis indicated that high risk score was associated with unfavorable PFS. Ultimately, the risk score was further verified by two gene expression profiles (GSE3218 and GSE10783).
Conclusions
Based on six selected m6A RNA methylation regulators, we developed a m6A methylation related risk score that can independently predict the prognosis of TGCT patients, and verify the prediction efficiency in TCGA and GEO datasets. Patients in high-risk group were associated with serum tumor marker study levels beyond the normal limits, non-seminoma, and unfavorable survival time. However, further prospective experiments should be carried out to verify our results.
Keywords: Testicular germ cell tumors (TGCT), N6-methyladenosine (m6A), m6A RNA methylation, risk score, prognosis
Introduction
As the most common malignancy in men between 15 and 44 years of age, testicular germ cell tumors (TGCT) have been increasing over the past few decades in most populations (1,2). TGCTs can be classified by histologic subtype into three groups: seminoma, non-seminoma, and spermatocytic tumor (3). Non-seminomas and seminomas together represent 98–99% of all TGCTs, and the peak incidence of them are at approximately ages 25 years and 35 years, respectively (3). Spermatocytic tumor are very rare at all ages (median age 54 years) (4,5). The patients’ life expectancy at age 30 was estimated as 45.2 years of age (6). Although Testicular cancer has become a model for a curable neoplasm and over 90% of TGCT patients will be cured of their disease by undergoing surgery, radiotherapy and chemotherapy, young men diagnosed with testicular cancer presented an excess mortality during the first 2 years post diagnosis (6,7).
Therefore, there is an urgent need to clarify the possible mechanisms underlying the development of TGCT, discover novel biomarkers for early screening, and develop therapeutic targets.
RNA methylation modifications compose over 60 percent of all RNA modifications, and the most common type of RNA methylation modification is N6-methyladenosine (m6A) RNA methylation (8). m6A RNA methylation regulators are widely distributed in various types of RNA, such as mRNA, transport RNA (tRNA) and ribosomal RNA (rRNA) (9). Recent studies reveal that the prevalence of m6A sites in numerous transcripts and their unique and conserved distribution mostly centered near the stop codons or enriched at 3’ untranslated regions (3'UTRs) in the transcriptomes of humans and mice (10,11). RNA m6A modification has been shown to play vital roles in regulating mRNA splicing, translation, stability and RNA-protein interaction (9,12,13). It is well known that RNA modification is mediated by a methyltransferase complex. The factors in m6A pathways include “writers” (METTL3, METTL14, WTAP, VIRMA, RBM15/15B, and ZC3H13) and “erasers” (FTO and ALKBH5) that respectively install and reverse the methylation, and RNA-binding proteins “readers” (YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, HNRNPC, HNRNPA2B1, FMR1, IGF2BP1, IGF2BP2, IGF2BP3, and PRRC2A) that recognize mRNA m6A sites (14-16).
Accumulating evidences have suggested the fact that m6A modification could be associated with promoting tumorigenesis in different types of cancers. Previous study has suggested that METTL14, a major RNA N6-adenosine methyltransferase, suppresses the metastatic potential of hepatocellular carcinoma by modulating m6A-dependent primary microRNA processing (17). Additionally, it has been revealed that METTL3 might be essential in lung cancer cells by directly promoting translation independently of methyltransferase activity and downstream m6A reader proteins (18). Over-expressed FTO is detected in acute myeloid leukemia (AML) and can inhibit the AML cell differentiation (19). The key role of m6A RNA methylation regulators are also noted in different cancers, such as renal cell carcinoma, bladder cancer, and glioblastoma (20-22). Recently, a study reported that m6A is detectable in RNA of TGCT cell lines and tissues by quantification of m6A in RNA. They further found that METTL3, ALKBH5, YTHDC1, YTHDF1, YTHDF2 and HNRNPC might be the main writers, erasers, and readers of the m6A modification in TGCT by RT-qPCR (23). However, the function of m6A RNA methylation modification in TGCT development and prognosis remains to be illuminated. In the present study, we used data from TCGA and GTEx databases to analyze the expression of m6A RNA methylation regulators in TGCT and their relationship with the clinicopathological features of TGCT. Then, we selected six m6A methylation regulators to construct a risk score and analyzed the prognostic role of the risk score in TGCT. Ultimately, we validated the prediction accuracy of the prognostic model in GEO dataset. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tau-20-963).
Methods
Patient samples
All of these normalized clinical data and RNA-seq data of patients were obtained from TCGA (http://cancergenome.nih.gov/) and GTEx (Genotype-Tissue Expression). Finally, we identified the gene expression profiles and of 134 TGCT cases and 165 cases of normal control for further analysis. Ultimately, we extracted complete clinical data which included survival time from 121 of 134 TGCT patients (Table 1).
Table 1. Clinical information of included TCGA cohort.
| Clinical parameters | Variables | TCGA cohort |
|---|---|---|
| Survival status | Progress | 35 |
| Progress-free | 99 | |
| Age(years) | <35 | 90 |
| ≥35 | 44 | |
| Race | White | 119 |
| Black | 6 | |
| Asian | 4 | |
| NR | 5 | |
| Pathology stage | I | 93 |
| II | 17 | |
| III | 15 | |
| NR | 9 | |
| Type of tumor | Seminoma | 72 |
| Non-seminoma | 62 | |
| Stage based on SM | S0 | 43 |
| S1 | 38 | |
| S2 | 34 | |
| S3 | 5 | |
| SX | 12 | |
| NR | 2 | |
| Lymphovascular invasion | Yes | 55 |
| No | 75 | |
| NR | 4 |
TCGA, The Cancer Genome Atlas; NR, not recorded; SM, serum markers.
Selection of m6A RNA methylation regulators and correlation analysis
According to the previous studies (24), 22 m6A RNA methylation regulators, including METTL3, METTL14, METTL16, WTAP, RBM15, RBM15, VIRMA, ZC3H13, YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, HNRNPC, HNRNPA2B1, FTO, ALKBH5, FMR1, IGF2BP1, IGF2BP2, IGF2BP3, and PRRC2A, were used for our analysis. By using Wilcoxon’s Test, we assessed the different expression of m6A RNA methylation regulators between TGCT patients and controls. Then, the correlation analysis among m6A RNA methylation regulators was carried out by Pearson correlation analysis.
Consensus clustering analysis
To determine whether the expression levels of m6A RNA methylation regulators were associated with prognosis, the TCGA TGCT cohort was clustered into different groups by consensus expression of m6A RNA methylation regulators with “Consensus Cluster Plus” in R. The Kaplan-Meier method and log-rank test were carried out to evaluate the progression-free survival (PFS) difference between different clusters. Chi-square test was used to compare the distribution of age, race, gender, stage, histologic subtype, serum marker study levels, lymph node status and TNM stage between different clusters.
Prognostic signatures generation and prediction
We carried out Cox univariate analysis to identify possible prognostic m6A RNA methylation regulators in the entire TCGA TGCT cohort. For further analysis, 79 of 121 patients were selected as a training cohort. Then, we excluded some genes highly correlated with one another using last absolute shrinkage and selection operator (LASSO) Cox regression algorithm for avoiding overfitting the model. The ten m6A RNA methylation regulators were selected for further Cox multivariate proportional hazards regression analysis. Finally, six genes (YTHDF1, RBM15, IGF2BP1, ZC3H13, METTL3, and FMR1) were identified to construct a risk score. The risk score was calculated with the following formula:
| [1] |
where n, coef(i), and x(i) represent the number of genes, the coefficient, and the relative expression value of each gene selected by multivariate analysis, respectively. Based on the median risk score of all samples, patients were divided into high-risk group and low-risk group. Kaplan-Meier curve and log-rank test were carried out to evaluate the relationship between PFS and risk score. Receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) were completed to evaluate the prognostic ability of risk score by using the package of “survivalROC” in R.
Validation of the prognostic signature
Thirty-two of 121 patients in TCGA cohort were selected as a testing cohort. Based on the same cut-off risk score, the patients in the testing cohort were classified into high-risk group and low-risk group. Kaplan-Meier curve and log-rank test were carried out to evaluate the relationship between PFS and risk score. ROC and AUC were completed to evaluate the prognostic ability of risk score by using the package of “survivalROC” in R.
Ultimately, we validated prediction accuracy of the prognostic model in the entire TCGA cohort. Kaplan-Meier curve and ROC were carried out. The chi-square test was performed to evaluate the association between the risk score and clinicopathological parameters in the entire TCGA cohort. To investigate whether risk score was an independent prognostic factor, univariate and multivariate Cox regression analyses were used in the entire TCGA cohort. Prognostic value of the risk score stratified by clinicopathological parameters was further assessed.
External validations of the six-gene risk score using GEO database
The six-gene risk score was further verified by two gene expression profiles (GSE3218 and GSE10783) extracted from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The “sva” package was used to remove batch effects and other needless variables. Overall, 108 samples from GSE3218 and GSE10783 were used as an independent external validation cohort. Kaplan-Meier curve and log-rank test were carried out to evaluate the relationship between overall survival (OS) and risk score. ROC and AUC were completed to evaluate the prognostic ability of risk score.
Gene set enrichment analysis (GSEA)
GSEA is a systematic method used to figure out whether the hallmark gene sets predicted statistically significant differences between two different groups (25). Here, GSEA was performed to analyze the significant survival differences between two groups divided by risk scores in the entire TCGA cohort. It had been considered to be significantly enriched that normalized P value less than 0.05 and false discovery rate (FDR) less than 0.25.
Statistical analysis
All statistical data and figures were analyzed by using R 3.6.0. By using Wilcoxon’s Test, we assessed the different expression of m6A RNA methylation regulators between TGCT patients and controls. The correlation analysis among m6A RNA methylation regulators was carried out by Pearson correlation analysis. The chi-square test was performed to evaluate the association between the risk score and clinicopathological parameters. To investigate whether risk score was an independent prognostic factor, univariate and multivariate Cox regression analyses were used in the entire TCGA cohort. All statistical results with P<0.05 were considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Relative expression of m6A regulatory genes in TGCT
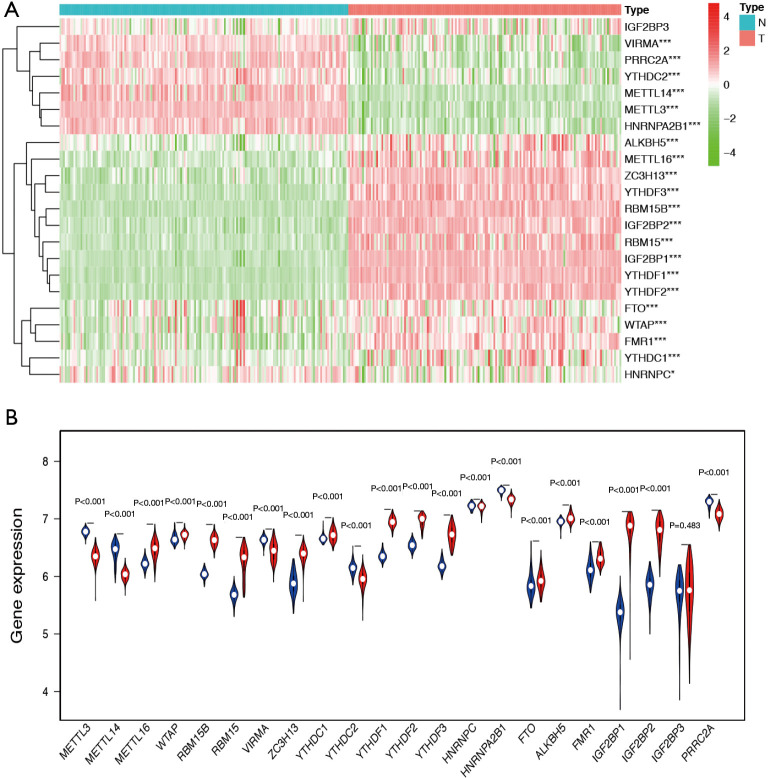
We analysed the mRNA expression levels of the 22 m6A regulatory genes in TCGA and GTEx databases to identify the distinct expression patterns between TGCT tissues and normal tissues. Figure 1A is the heat map which indicated the expression levels of m6A regulatory genes. Compared with normal tissues, METTL16, WTAP, RBM15, RBM15B, ZC3H13, YTHDC1, YTHDF1, YTHDF2, YTHDF3, FTO, ALKBH5, FMR1, IGF2BP1, and IGF2BP2 were up-regulated, while METTL3, METTL14, VIRMA, and YTHDC2, HNRNPA2B1, and PRRC2A were down-regulated in TGCT tissue samples (Figure 1B).
Figure 1.
The differential expression of m6A RNA methylation regulators between TGCT samples and normal tissues. (A) Hierarchical clustering of TGCT and normal tissues expressing the 22 m6A regulatory genes in TCGA and GTEx databases. (B) The expression of 22 m6A regulatory genes between TGCT and normal tissues. The red represents tumor group and blue represents normal tissue group. TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression. *, P<0.05 ; ***, P<0.001.
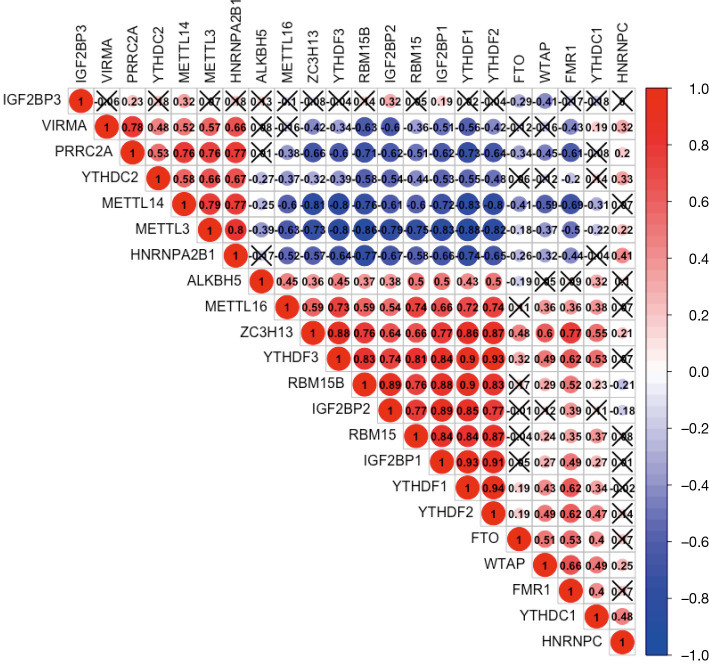
We furthor investigated the correlation between each two types of m6A regulatory genes in TGCT using “corrplot” package in R software (Figure 2). The expressions of ZC3H13, RBM15, RBM15B, YTHDF1, YTHDF2, YTHDF3, IGF2BP1, and IGF2BP2 had remarkably high positive correlations with each other (Pearson’s r>0.6). In addition, the results suggested that METTL3 was negatively associated with ZC3H13, RBM15, RBM15B, YTHDF1, YTHDF2, YTHDF3, IGF2BP1, and IGF2BP2 (Pearson’s r<−0.7). Hence, we indicated that there were close and complicated correlations between the nine m6A regulatory genes.
Figure 2.
Correlation matrix of 22 m6A regulatory genes in the TCGA and GTEx databases. An X represents P>0.001, which means there was no statistically significant correlation between two m6A regulatory genes. TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression.
Consensus clustering of m6A RNA methylation regulators distinguished two clusters of TGCT with different prognosis
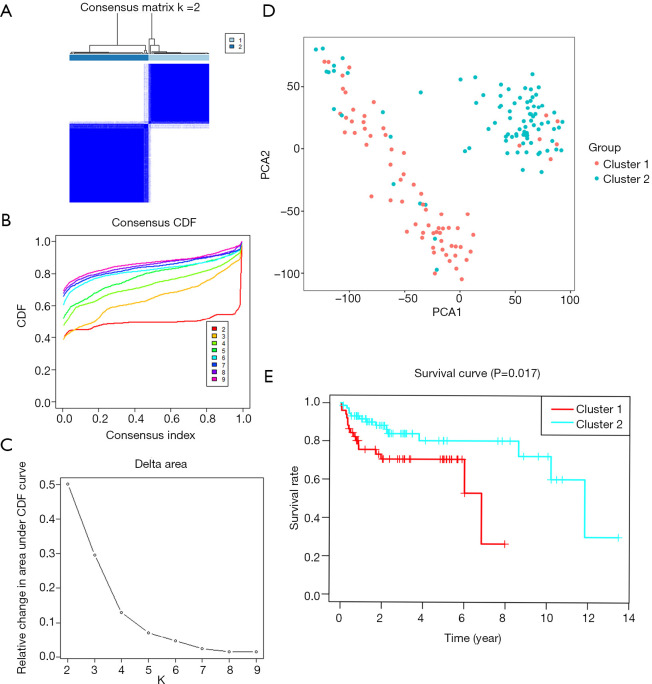
Based on the expression similarity of m6A RNA methylation regulators, k=2 was demonstrated to be the most appropriated selection to divide the TGCT patient cohort into two clusters, namely cluster 1 and cluster 2 (Figure 3A,B,C). Moreover, we performed principal component analysis (PCA) for comparison of the transcriptional profile between cluster1 and cluster2. The result suggested that there was a significant distinction between the two subgroups (Figure 3D). A significant shorter PFS was observed in TGCT patients in the cluster 1 (5-year PFS =69%) than those in the cluster 2 (5-year PFS =79%) (P=0.017) (Figure 3E). Then the associations between the clustering and clinicopathological features were evaluated. Significant difference was found between the cluster 1 and cluster 2 for the serum marker study levels and histologic subtype (P<0.001), while no significant difference was observed for other parameters such as age, race, stage, and TNM stage (Figure 4). Additionally, the levels of AFP (P<0.001) and hCG (P<0.05) increased in cluster 1, compared with cluster 2. However, there was no significant difference between the level of LDH in cluster 1 and that in cluster 2 (Figure S1).
Figure 3.
Overall survival of TGCT patients in the two different clusters. (A) The TCGA TGCT cohort was divided into two distinct clusters when k=2. (B) Consensus clustering cumulative distribution function (CDF) for k=2 to 10. (C) Relative change in area under CDF curve for k=2 to 10. (D) Principal component analysis of the total RNA expression profile. TGCT in cluster 1 and 2 are marked in red and blue, respectively. (E) Kaplan-Meier PFS curve for TGCT patients in cluster 1 and 2. TCGA, The Cancer Genome Atlas; PFS, progression-free survival; TGCT, testicular germ cell tumors.
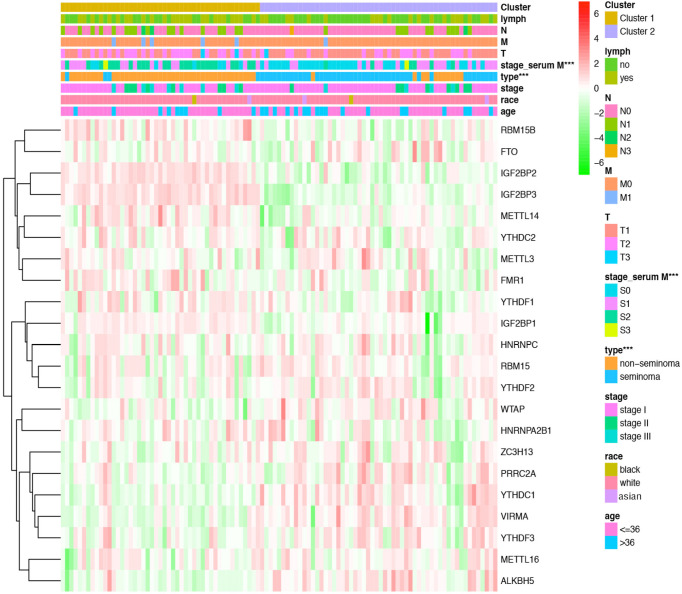
Figure 4.
The distribution of clinicopathological variables between different clusters. Significant difference was found for the serum marker study levels and histologic subtype between cluster 1 and cluster 2. ***, P<0.001.
Prognostic signature of m6A RNA methylation regulators in TGCT
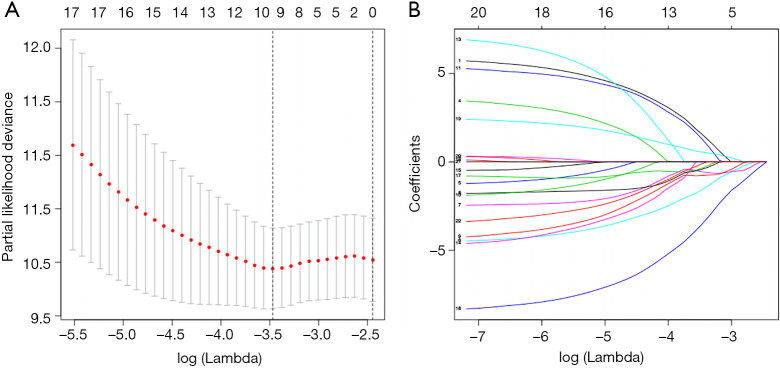
In the entire TCGA TGCT cohort, we used univariate Cox regression to investigate the effect of m6A RNA methylation regulators on TGCT prognosis (Table 2). Then, LASSO multivariate Cox regression was performed to avoid overfitting the model in the training cohort. Hence, ten candidate genes were selected (Figure 5A,B). Finally, the risk score of TGCT was constructed by selecting YTHDF1, RBM15, IGF2BP1, ZC3H13, METTL3 via multivariate Cox regression analysis (Table S1).
Table 2. Cox univariate analysis of m6A RNA methylation regulators to investigate the effect of m6A RNA methylation regulators on TGCT prognosis.
| Gene | HR (95% CI) | P value |
|---|---|---|
| FMR1 | 0.049092 (0.002285–1.054691) | 0.054112 |
| VIRMA | 0.105596 (0.010457–1.066322) | 0.056711 |
| YTHDC1 | 0.032825 (0.000721–1.493478) | 0.079424 |
| ZC3H13 | 0.065814 (0.003068–1.41185) | 0.081952 |
| PRRC2A | 0.021448 (0.000226–2.037345) | 0.098195 |
| RBM15 | 0.366743 (0.100793–1.334426) | 0.127967 |
| ALKBH5 | 0.030539 (1.334426–3.21472) | 0.141982 |
| YTHDF2 | 0.083202 (0.002673–2.590295) | 0.156363 |
| HNRNPA2B1 | 0.01239 (0.000016–9.590852) | 0.195735 |
| METTL16 | 0.268298 (0.028964–2.48528) | 0.246702 |
| IGF2BP3 | 1.616413 (0.71076–3.676055) | 0.251994 |
| IGF2BP1 | 2.155541 (0.477137–9.737995) | 0.318165 |
| IGF2BP2 | 2.368073 (0.430614–13.02274) | 0.321583 |
| HNRNPC | 0.058167 (0.000207–16.31963) | 0.322646 |
| METTL3 | 3.217734 (0.122173–84.74715) | 0.483761 |
| YTHDC2 | 0.447566 (0.04586–4.367978) | 0.489175 |
| YTHDF3 | 0.437247 (0.033203–5.75815) | 0.529372 |
| FTO | 0.537583 (0.069118–4.181175) | 0.55315 |
| WTAP | 0.220542 (0.001001–48.58904) | 0.582888 |
| RBM15B | 2.072659 (0.072891–58.9362) | 0.669586 |
| YTHDF1 | 2.083396 (0.047455–91.46694) | 0.703659 |
| METTL14 | 0.58829 (0.012586–27.49706) | 0.786803 |
TGCT, testicular germ cell tumors; HR, hazard ratio.
Figure 5.
Multivariate Cox regression via LASSO is presented, and ten candidate m6A RNA methylation regulators were selected. (A) Cross-validation for tuning parameter screening in the LASSO regression model. (B) LASSO coefficient profiles of the common genes. LASSO, last absolute shrinkage and selection operator.
The risk score for each patient was calculated with the following formula:
| Risk score =4.95105371407088*METTL3+(−4.61228230602799)*RBM15 (−4.29950269353698)*ZC3H13+5.38635751376823*YTHDF1+(−5.32912179904041)*FMR1+2.6414130443825*IGF2BP1 |
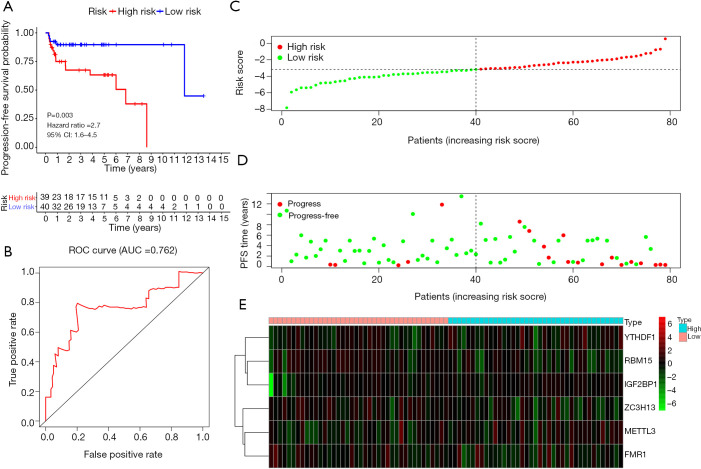
A total of 39 and 40 TGCT patients were categorized into the high-risk group and low-risk group, respectively. Kaplan-Meier survival analysis showed that the high-risk group had a better PFS than low-risk group in TGCT (Figure 6A, P=0.003). The time-dependent ROC curve analysis was performed to assess prediction efficiency of risk score in TGCT patients. AUC for was 0.762, which demonstrated the good performance of the risk score for prognosis prediction in TGCT (Figure 6B). The risk score distribution of the patients on in the training cohort was displayed in Figure 6C. The PFS and risk scores of patients with TGCT in the training cohort were shown in Figure 6D. Figure 6E indicated that the heatmap of the six key genes expression profiles in the training cohort.
Figure 6.
Validation of the prognostic signature in the TCGA TGCT training cohort (A) Kaplan-Meier plot represents that patients in the high-risk group had significantly shorter PFS than those in the low-risk group. (B) Time-dependent ROC curve analysis for survival prediction by the risk score in the training test based on the TCGA dataset. (C) The risk score distribution of patients in the training cohort. (D) The distributions of risk scores and PFS status. The red and green dots indicated the progress and progress free respectively. (E) The heatmap of the six key genes expression profiles in the training cohort. TCGA, The Cancer Genome Atlas; TGCT, testicular germ cell tumors; ROC, receiver operating characteristic; PFS, progression-free survival.
Validation of the prognostic signature in the TCGA testing cohort
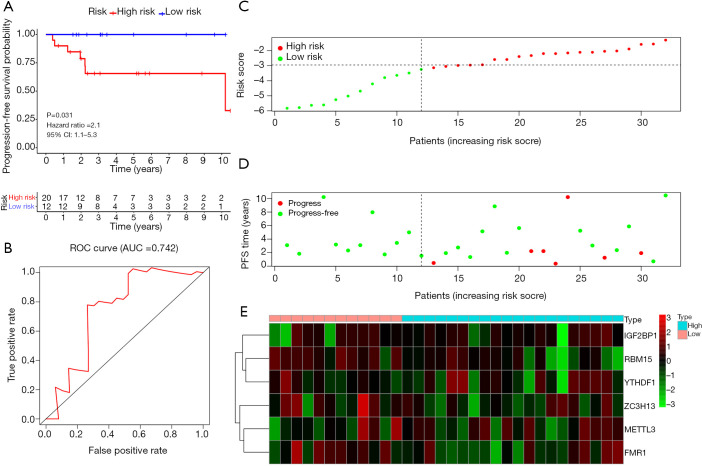
A total of 32 cases were included in the TGCT testing cohort. Based on the same cut-off value of the risk score, 20 patients were categorized into high-risk group and the remaining 12 cases were grouped into low-risk group. Kaplan-Meier survival analysis showed that the low-risk group had a better PFS than high-risk group in TGCT (Figure 7A, P=0.031). The time-dependent ROC curve analysis was performed to assess prediction efficiency of risk score in TGCT patients. The AUC for was 0.742, which demonstrated the good performance of the risk score for prognosis prediction in TGCT (Figure 7B). The risk score distribution of the patients on in the testing cohort was displayed in Figure 7C. The PFS and risk scores of patients with TGCT in the testing cohort were shown in Figure 7D. Figure 7E indicated that the heatmap of the six key genes expression profiles in the testing cohort.
Figure 7.
Validation of the prognostic signature in an independent TGCT cohort. (A) Kaplan-Meier survival curve showing PFS outcomes according to relative high-risk and low-risk patients. (B) Time-dependent ROC curve analysis for survival prediction by the risk score in the testing cohort based on the TCGA dataset. (C) The risk score distribution of patients in the testing cohort. (D) The distributions of risk scores and PFS status. The red and green dots indicated the progress and progress free respectively. (E) The heatmap of the six key genes expression profiles in the testing cohort. TGCT, testicular germ cell tumors; TCGA, The Cancer Genome Atlas; PFS, progression-free survival; ROC, receiver operating characteristic.
Identification of the independent prognostic factors in the entire TCGA cohort
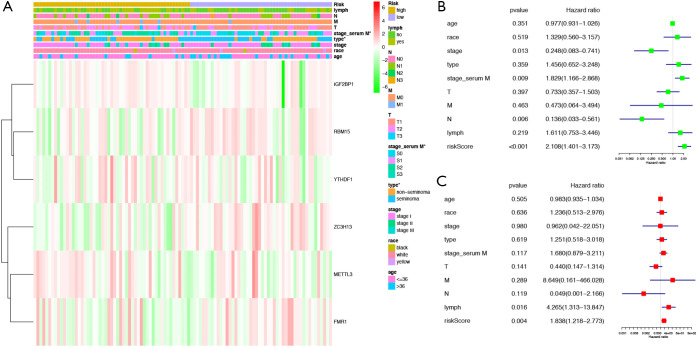
A total of 111 cases were included in the entire TGCT cohort. Based on the same cut-off value of the risk score, patients were categorized into high-risk group and low-risk group. The heat map indicated that the differential expression of the six selected m6A RNA methylation regulators and clinicopathological variables between high-risk and low-risk groups. We found significant differences between the two groups in serum marker study levels and histologic subtype (P<0.05) (Figure 8A). We used Cox univariate and multivariate analyses to determine whether the risk signature was an independent predictor. Univariate analyses showed that stage (P=0.013), serum marker study levels (P=0.009), N stage (P=0.006), and risk score (P<0.001) were significantly linked with PFS (Figure 8B). Multivariate analyses showed that lymph node status (P=0.016) and risk score (P<0.004) were significantly linked with PFS (Figure 8C). These results suggest that the risk signature is a risk factor for TGCT patients and can independently predict the prognosis of TGCT patients.
Figure 8.
Effects of the risk score and clinicopathological variables on the prognosis of TGCT patients (A) The heat map shows the expression of six m6A RNA methylation regulators and the distribution of clinicopathological variables between the high- and low-risk groups. (B) Cox univariate analyses of clinicopathological variables (including the risk score) and overall survival. (C) Cox multivariate analyses of clinicopathological variables (including the risk score) and PFS. TGCT, testicular germ cell tumors; PFS, progression-free survival. *, P<0.05.
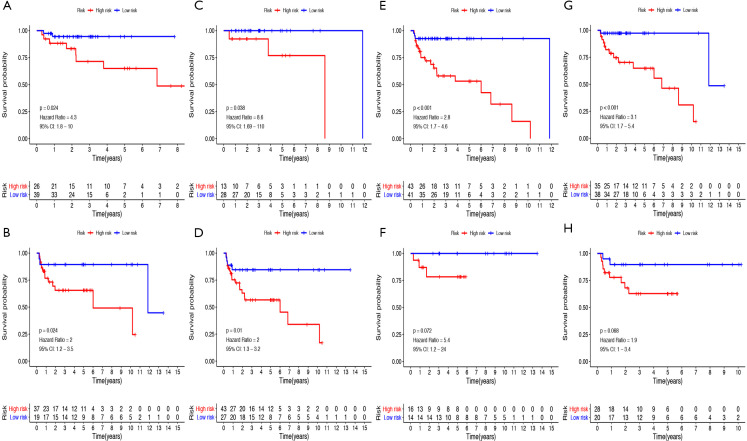
The patients were also divided into groups according to clinicopathological variables to investigate the prognostic value of risk score. As shown in Figure 9A,B,C,D,E,F,G,H, the PFS rate was significantly lower in the high-risk group compared to that in the low-risk group for the cases with seminoma (P=0.0024), or cases with non-seminoma (P=0.0024), or cases at serum marker study levels within normal limits (P<0.038), or patients at serum tumor marker study levels beyond the normal limits (P=0.01), or patients at the stage I (P<0.001) or those with no lymphovascular invasion (P<0.001). However, no significant difference was found for PFS between high- and low-risk groups for cases with stage II-III (P=0.072), or patients with lymphovascular invasion (P=0.068).
Figure 9.
Prognostic value of the risk score in TGCT patients classified into specific cohorts. Kaplan-Meier survival curve of PFS for patients with (A) seminoma, (B) non-seminoma, (C) serum marker study levels within normal limits, (D) serum marker study levels beyond the normal limits, (E) stage I, (F) stage II-III, (G) no lymphovascular invasion, (H) lymphovascular invasion. TGCT, testicular germ cell tumors; PFS, progression-free survival.
GSEA identifies a signaling pathway related with risk score
In consideration of risk score as an independent prognostic factor for PFS of TGCT patients, we were eager to explore how risk score was involved in TGCT pathogenesis. We carried out GSEA between tissues with different risk scores.
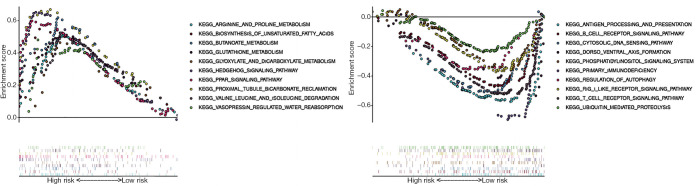
Based on the NOM p-val (normalized P value <0.05), FDR q-val (FDR<0.25) and normalized enrichment score (NES), we selected the most significantly enriched biological pathways. The results indicated that high risk scores were associated with some essential signaling pathways including arginine and proline metabolism pathway, biosynthesis of unsaturated fatty acids, butanoate metabolism, glutathione metabolism, glyoxylate and dicarboxylate metabolism, hedgehog signaling pathway, PPAR signaling pathway, proximal tubule bicarbonate reclamation, valine leucine and isoleucine degradation, vasopressin regulated water reabsorption (Figure 10A). The results indicated that low risk scores were associated with signaling pathways including antigen processing and presentative, B cell receptor signaling pathway, cytosolic DNA sensing, dorsoventral axis formation, phosphatidylinositol signaling system, primary immunodeficiency, regulation of autophagy, rig like receptor signaling pathway, T cell receptor signaling pathway, ubiquitin mediated proteolysis (Figure 10B). The above results give a clue of the underlying mechanism in the pathogenesis of TGCT.
Figure 10.
Enrichment plots from gene set enrichment analysis (GSEA). (A) The ten most significantly enriched signaling pathways in the high-risk score subgroup. (B) The ten most significantly enriched signaling pathways in the low risk score subgroup. ES, enrichment score; NES, normalized ES; NOM p-val, normalized P value.
External validations of the six-gene risk score using GEO database
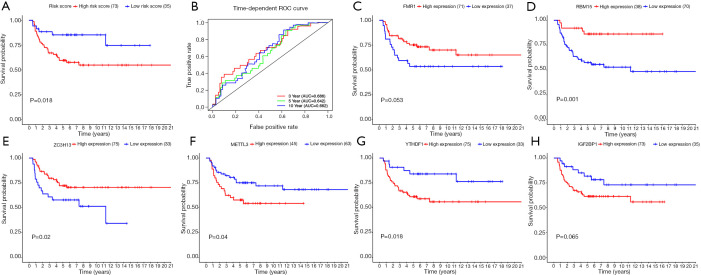
Besides calculating risk scores of patients in TCGA dataset as an internal validation, we further calculated risk scores of patients in the GSE3218 and GSE10783 datasets as the independent external validations based on the same formula. Similarly, patients were arrayed according to the cut-off risk score in the training cohorts. As expected, TGCT patients were divided into two groups based on risk scores with a significantly different OS (P=0.018; Figure 11A). The ROC analysis of 3/5/10-year OS was performed for evaluating predictive efficiency of the risk score. The 3-year AUC for risk score was 0.686. The 5-year AUC for risk score was 0.642. The 10-year AUC for risk score was 0.662 (Figure 11B). High FMR1 expression group had longer OS than the low expression group but no difference in OS (P=0.053; Figure 11C). Compared with low RBM15 or ZC3H13 expression group, the high expression group had better OS (P<0.05; Figure 11D,E). Kaplan-Meier curves analysis demonstrated that the patients with high METTL3, YTHDF1, and IGF2BP1 expression had shorter OS than those with low expression, however, the difference in OS between high IGF2BP1 expression and low IGF2BP1 expression was not statistically significant (P=0.065) (Figure 11F,G,H).
Figure 11.
The prognostic value of six-gene risk score of TGCT patients based on the GSE3218 and GSE10783 datasets. (A) Kaplan-Meier survival curve showing OS outcomes according to relative high-risk and low-risk patients. (B) ROC analysis of 3/5/10-year OS for risk score showed the predictive efficiency of the risk score. Kaplan-Meier curves of OS in different expression levels of (C) FMR1, (D) RBM15, (E) ZC3H13, (F) METTL3, (G) YTHDF1, and (H) IGF2BP1. TGCT, testicular germ cell tumors; OS, overall survival; ROC, receiver operating characteristic.
Discussion
TGCT, as the common malignancy in young men, have been increasing over the past few years. Over 80% of TGCT patients are cured of their disease by surgery, radiotherapy and chemotherapy (26,27). However, it is worth noticing that almost one quarter of patients show resistance to standard chemotherapy, which suggested that there are some molecular mechanisms influencing the prognosis of TGCT. The expression levels of m6A regulators are demonstrated to be comparable between TGCT and normal tissues (23). In the present study, we found that most m6A RNA methylation regulators were abnormally expressed in TGCT. In addition, based on the expression of the ten m6A RNA methylation regulators, the TGCT training cohort in TCGA could be divided into different clusters with significant differences for PFS, serum marker study levels and histologic subtype. Moreover, based on TCGA dataset, a robust risk score was constructed based on YTHDF1, RBM15, IGF2BP1, ZC3H13, METTL3, and FMR1. The results showed that the risk score had good performance for predicting the clinical outcome of TGCT. More importantly, this six-gene risk score was further successfully validated as an independent prognostic marker in the TCGA texting cohort and an external independent TGCT cohort (GSE3218 and GSE10783), indicating that this prognostic model is highly robust for prognosis prediction.
Univariate analyses showed that stage, serum marker study levels, N stage, and risk score were significantly associated with PFS. Multivariate analyses showed that lymph node status and risk score were significantly linked with PFS. When patients were also grouped according to clinicopathological variables, the risk signature could also distinguish the PFS outcomes of the cases with seminoma, or cases with non-seminoma, or cases at serum marker study levels within normal limits, or patients at serum tumor marker study levels beyond the normal limits, or patients at the stage I or those with no lymphovascular invasion. Our results indicated that the high-risk group suffered a more unfavorable clinical outcome compared to the low-risk group in TGCT.
Our study showed that the expression levels of FMR1, RBM15, and ZC3H13 were positively correlated with clinical outcome of TGCT, suggesting that these three m6A regulatory genes might be suppressor genes in TGCT. Recent studies reported that the oncogenic role of FXR1 in melanoma and prostate cancer (28,29). Another research found that downregulating FMRP in testicular embryonal carcinoma cells could enhance miR-383-mediated suppression of cell proliferation, which is contrary to our results (30). This opposite result might be due to the small sample size. Similarly, down-regulated RBM15 was proved to inhibit the growth, proliferation, and induce apoptosis in chronic myelogenous leukemia cells (31). In different types of tumors, it is possible that different roles of m6A regulators can be discovered. Additionally, Zhu et al. reported that ZC3H13 could act as a tumor suppressor in colorectal cancer by regulating proliferation and invasion (32), indicating that ZC3H13 may play similar roles in TGCT. On contrast, the expression of the other gene METTL3, YTHDF1, and IGF2BP1 were negatively associated with the prognosis in TGCT, indicating that these three genes might act as tumor promoter genes for TGCT tumorigenesis. At present, the role of METTL3 is not very clear. Although METTL3 can exhibit a contrary role in same cancer, the main role of METTL3 is to facilitating cancer initiation and progression (33-35). YTHDF1 was usually reported to be an oncogene and was related with poor outcomes in colorectal cancer and ovarian cancer (36,37). IGF2BP1, as a RNA binding protein, contains tandem common RNA-binding domains, including K homology domains, arginine/glycine-rich domains, and RNA recognition motifs (38). In various types of cancers, IGF2BP1 acts as a post-transcriptional regulator modulating the expression of mRNA targets thus controlling cancer cell proliferation, and is associated with an unfavorable prognosis (39).
In order to explore how m6A RNA methylation regulators were involved in TGCT pathogenesis, we carried out GSEA between tissues with different risk scores and found that high-risk group was associated with some signaling pathways including biosynthesis of unsaturated fatty acids, butanoate metabolism, glutathione metabolism, glyoxylate and dicarboxylate metabolism, PPAR signaling pathway, valine leucine and isoleucine degradation. Increased glutathione levels are detected in various types of cancers, making tumor tissues more resistant to chemotherapy (40,41). In fact, polyunsaturated acids are one of the main targets of free radical damage (42). To prevent lipoperoxidative damage, Testicular germ cells are endowed with enzymatic and non-enzymatic scavenger systems (40). Among the enzymatic systems, glutathione plays a main role in antioxidant defense (43). Oxidative stress is involved in cancer development and progression, which suggests that antioxidant defense may provide protection from cancer (44). Interestingly, in the process of testicular neoplastic growth, spermatogonial cells evade the toxic effects of endogenous oxidants due to loss of CCDC6 (45). By inhibiting Nrf2-mediated antioxidant response, m6A RNA methylation may be involved in oxidative stress in DEHP-related testicular injury (46). Hence, we speculate that m6A RNA methylation may play vital roles in the prognosis of TGCT by regulate oxidative stress and antioxidant response.
Some limitations of this study are noteworthy. First, we only analyzed the mRNA expression of m6A RNA methylation regulators. As we all know, it is not a perfect method to predict protein expression based on mRNA expression (47). Furthermore, the current study is based on bioinformatic analysis. Further experiments and clinical researches are required.
Conclusions
In summary, our results demonstrated that 21 out of 22 m6A RNA methylation regulators are dysregulated between TGCT tissues and normal tissues. Based on six selected m6A methylation regulators, we developed a m6A methylation related risk score that can independently predict the prognosis of TGCT patients, and verify the prediction efficiency in TCGA and GEO datasets. Patients in high-risk group were associated with serum tumor marker study levels beyond the normal limits, non-seminoma, and unfavorable survival time. However, further prospective experiments should be carried out to verify our results.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This article was funded by the National Natural Science Foundation of China (grant number: 81871151; 81801438) and Medical key talent of Jiangsu Province: ZDRCA2016009.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-963
Peer Review File: Available at http://dx.doi.org/10.21037/tau-20-963
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-963). The authors have no conflicts of interest to declare.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Ghazarian AA, Trabert B, Devesa SS, et al. Recent trends in the incidence of testicular germ cell tumors in the United States. Andrology 2015;3:13-8. 10.1111/andr.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajpert-De Meyts E, McGlynn KA, Okamoto K, et al. Testicular germ cell tumours. Lancet 2016;387:1762-74. 10.1016/S0140-6736(15)00991-5 [DOI] [PubMed] [Google Scholar]
- 4.Chia VM, Quraishi SM, Devesa SS, et al. International trends in the incidence of testicular cancer, 1973-2002. Cancer Epidemiol Biomarkers Prev 2010;19:1151-9. 10.1158/1055-9965.EPI-10-0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carriere P, Baade P, Fritschi L. Population based incidence and age distribution of spermatocytic seminoma. J Urol 2007;178:125-8. 10.1016/j.juro.2007.03.024 [DOI] [PubMed] [Google Scholar]
- 6.Capocaccia R, Gatta G, Dal Maso L. Life expectancy of colon, breast, and testicular cancer patients: an analysis of US-SEER population-based data. Ann Oncol 2015;26:1263-8. 10.1093/annonc/mdv131 [DOI] [PubMed] [Google Scholar]
- 7.Einhorn LH. Curing metastatic testicular cancer. Proc Natl Acad Sci U S A 2002;99:4592-5. 10.1073/pnas.072067999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Li J, Feng G, et al. MicroRNA-145 Modulates N(6)-Methyladenosine Levels by Targeting the 3'-Untranslated mRNA Region of the N(6)-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem 2017;292:3614-23. 10.1074/jbc.M116.749689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen T, Hao YJ, Zhang Y, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015;16:289-301. 10.1016/j.stem.2015.01.016 [DOI] [PubMed] [Google Scholar]
- 10.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012;485:201-6. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 11.Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 2012;149:1635-46. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 2014;15:313-26. 10.1038/nrm3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu N, Dai Q, Zheng G, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015;518:560-4. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li A, Chen YS, Ping XL, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res 2017;27:444-7. 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018;28:616-24. 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholler E, Weichmann F, Treiber T, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. Rna 2018;24:499-512. 10.1261/rna.064063.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 2017;65:529-43. 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- 18.Lin S, Choe J, Du P, et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 2016;62:335-45. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Weng H, Su R, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 2017;31:127-41. 10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Wang J, Hong B, et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma - a retrospective study using TCGA database. Aging (Albany NY) 2019;11:1633-47. 10.18632/aging.101856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han J, Wang JZ, Yang X, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer 2019;18:110. 10.1186/s12943-019-1036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Zhao BS, Zhou A, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017;31:591-606.e6. 10.1016/j.ccell.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nettersheim D, Berger D, Jostes S, et al. N6-Methyladenosine detected in RNA of testicular germ cell tumors is controlled by METTL3, ALKBH5, YTHDC1/F1/F2, and HNRNPC as writers, erasers, and readers. Andrology 2019;7:498-506. [DOI] [PubMed] [Google Scholar]
- 24.Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother 2019;112:108613. 10.1016/j.biopha.2019.108613 [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldman DR, Patil S, Trinos MJ, et al. Progression-free and overall survival in patients with relapsed/refractory germ cell tumors treated with single-agent chemotherapy: endpoints for clinical trial design. Cancer 2012;118:981-6. 10.1002/cncr.26375 [DOI] [PubMed] [Google Scholar]
- 27.Porcu P, Bhatia S, Sharma M, et al. Results of treatment after relapse from high-dose chemotherapy in germ cell tumors. J Clin Oncol 2000;18:1181-6. 10.1200/JCO.2000.18.6.1181 [DOI] [PubMed] [Google Scholar]
- 28.Zalfa F, Panasiti V, Carotti S, et al. The fragile X mental retardation protein regulates tumor invasiveness-related pathways in melanoma cells. Cell Death Dis 2017;8:e3169. 10.1038/cddis.2017.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao H, Gao R, Yu C, et al. The RNA-binding protein FXR1 modulates prostate cancer progression by regulating FBXO4. Funct Integr Genomics 2019;19:487-96. 10.1007/s10142-019-00661-8 [DOI] [PubMed] [Google Scholar]
- 30.Tian H, Cao YX, Zhang XS, et al. The targeting and functions of miRNA-383 are mediated by FMRP during spermatogenesis. Cell Death Dis 2013;4:e617. 10.1038/cddis.2013.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Wang S, Zhang Y, et al. Biological effects of decreasing RBM15 on chronic myelogenous leukemia cells. Leuk Lymphoma 2012;53:2237-44. 10.3109/10428194.2012.684350 [DOI] [PubMed] [Google Scholar]
- 32.Zhu D, Zhou J, Zhao J, et al. ZC3H13 suppresses colorectal cancer proliferation and invasion via inactivating Ras-ERK signaling. J Cell Physiol 2019;234:8899-907. 10.1002/jcp.27551 [DOI] [PubMed] [Google Scholar]
- 33.Cui Q, Shi H, Ye P, et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 2017;18:2622-34. 10.1016/j.celrep.2017.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visvanathan A, Patil V, Arora A, et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2018;37:522-33. 10.1038/onc.2017.351 [DOI] [PubMed] [Google Scholar]
- 35.Vu LP, Pickering BF, Cheng Y, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 2017;23:1369-76. 10.1038/nm.4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishizawa Y, Konno M, Asai A, et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget 2018;9:7476-86. 10.18632/oncotarget.23554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu T, Wei Q, Jin J, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res 2020;48:3816-31. 10.1093/nar/gkaa048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wachter K, Kohn M, Stohr N, et al. Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Biol Chem 2013;394:1077-90. 10.1515/hsz-2013-0111 [DOI] [PubMed] [Google Scholar]
- 39.Muller S, Glass M, Singh AK, et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res 2019;47:375-90. 10.1093/nar/gky1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calvert P, Yao KS, Hamilton TC, et al. Clinical studies of reversal of drug resistance based on glutathione. Chem Biol Interact 1998;111-112:213-24. 10.1016/S0009-2797(98)00008-8 [DOI] [PubMed] [Google Scholar]
- 41.Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci 2006;43:143-81. 10.1080/10408360500523878 [DOI] [PubMed] [Google Scholar]
- 42.Lenzi A, Gandini L, Picardo M, et al. Lipoperoxidation damage of spermatozoa polyunsaturated fatty acids (PUFA): scavenger mechanisms and possible scavenger therapies. Front Biosci 2000;5:E1-E15. [DOI] [PubMed] [Google Scholar]
- 43.Meister A. Glutathione metabolism. Methods Enzymol 1995;251:3-7. 10.1016/0076-6879(95)51106-7 [DOI] [PubMed] [Google Scholar]
- 44.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer 2003;3:276-85. 10.1038/nrc1046 [DOI] [PubMed] [Google Scholar]
- 45.Staibano S, Ilardi G, Leone V, et al. Critical role of CCDC6 in the neoplastic growth of testicular germ cell tumors. BMC Cancer 2013;13:433. 10.1186/1471-2407-13-433 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Zhao TX, Wang JK, Shen LJ, et al. Increased m6A RNA modification is related to the inhibition of the Nrf2-mediated antioxidant response in di-(2-ethylhexyl) phthalate-induced prepubertal testicular injury. Environ Pollut 2020;259:113911. 10.1016/j.envpol.2020.113911 [DOI] [PubMed] [Google Scholar]
- 47.Guo Y, Xiao P, Lei S, et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim Biophys Sin (Shanghai) 2008;40:426-36. 10.1111/j.1745-7270.2008.00418.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as