Abstract
Background
The etiopathogenesis of benign prostatic hyperplasia (BPH) is extremely complicated which involving epithelial-mesenchymal transition (EMT) of epithelial cells and growth of stromal cells. Long non-coding RNAs (lncRNAs) belong to a group of noncoding RNAs which has been widely studied in other diseases but rarely in BPH. Here, we intend to investigate the roles of a lncRNA DIO3 opposite strand (DIO3OS) in BPH progression.
Methods
BPH-1 cells were used to study EMT and WPMY-1 cells were applied to study proliferation induced by TGF-β1, resveratrol, DIO3OS and miRNAs.
Results
DIO3OS was over-expressed in BPH tissues and could be upregulated by Transforming growth factor beta 1 (TGF-β1) and downregulated by resveratrol. Smad2/Smad3/Smad4 complex could bind to the DIO3OS promotor region and thereby enhanced its transcription which was responsible for the regulation of TGF-β1 and resveratrol on DIO3OS expression. TGF-β1 promoted BPH-1 cells EMT and WPMY-1 cells proliferation via DIO3OS and this effect could be blocked by resveratrol. MiR-656-3p and miR-485-5p were targets of DIO3OS and DIO3OS promoted BPH-1 cells EMT and WPMY-1 cells proliferation via miR-656-3p and miR-485-5p. Connective tissue growth factor (CTGF) and zinc finger e-box binding homeobox 1 (ZEB1) were confirmed to be targets of both miR-656-3p and miR-485-5p and could be modulated by TGF-β1, resveratrol, DIO3OS, miR-656-3p and miR-485-5p.
Conclusions
DIO3OS is highly expressed in BPH tissues and regulated by TGF-β1 as well as resveratrol in a Smads dependent manner. DIO3OS facilitates BPH-1 cells EMT and WPMY-1 cells proliferation by upregulating CTGF and ZEB1 via miR-656-3p and miR-485-5p.
Keywords: Benign prostatic hyperplasia (BPH), long non-coding RNA (lncRNAs), DIO3 opposite strand (DIO3OS), epithelial-mesenchymal transition (EMT), miRNAs
Introduction
Benign prostatic hyperplasia (BPH) is a high incidence disorder in elderly men the pathogenesis of which is multifactorial and largely unknown (1). Epithelial-mesenchymal transition (EMT), a process whereby epithelial cells losing epithelial characteristics and acquiring a mesenchymal phenotype, has been confirmed to participate in the pathogenesis of BPH (2-4). Moreover, the aberrant growth of prostate stromal cells has also been discovered to be a contributing factor in the processes of BPH (5-7).
Transforming growth factor beta 1 (TGF-β1) has been demonstrated excessively secreted by stromal cells into the stromal microenvironment during BPH, affecting the biological processes of epithelial cells and stromal cells (8,9). Resveratrol (trans-3,5,4-trihydroxystilbene) is a natural polyphenol and is widely exist in mulberries, grapes, peanuts, polygonum cuspidatum and other plants (10). Studies have identified resveratrol as an effective agent in treating various illnesses, including BPH (11,12). In addition, many researches indicated resveratrol could reverse the biological effects induced by TGF-β1 via repressing phosphorylation of Smad2 and Smad3 (13,14).
Long noncoding RNAs (lncRNAs) belong to a group of noncoding RNAs which have over 200 nucleotides in length (15). They participate in progressions of a wide range of diseases through regulating several biological proceedings, including cell differentiation, EMT, apoptosis and proliferation (16). Nevertheless, there are extremely few studies on lncRNAs in BPH. A recent study identified a highly expressed lncRNA DNM3OS in prostate stromal tissues of BPH and uncovered the roles of DNM3OS in promoting prostate stromal cells transformation (17). In that study, they also found another lncRNA DIO3 opposite strand (DIO3OS) significantly upregulated in prostate stromal tissues of BPH. Nevertheless, the functions and mechanisms of DIO3OS in BPH remained unclear. Hence, we intended to investigate DIO3OS in BPH in the present study.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1169).
Methods
Patients and specimens
A total of 16 patients with BPH who underwent transurethral resection of the prostate were recruited at the at Shanghai Ninth People’s Hospital. Thirteen normal prostate tissues were acquired from bladder cancer patients who underwent radical cystoprostatectomy. Written informed consents were signed by all the participants. Current research was approved by the Ethical Committee of Shanghai Ninth People’s Hospital (No. 2017207) and strictly conformed to the 2013 Declaration of Helsinki.
Cell culture, treatment, and transfection
Benign prostatic hyperplasia epithelial-1 (BPH-1) cells were purchased from the German Collection of Microorganisms and Cell cultures (Leibniz Institute DSMZ, Germany). SV40 large-T antigen-immortalized stromal cell line WPMY-1 was obtained from the Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China). RPMI 1640 medium (Gibico, USA) containing 10% fetal bovine serum was applied to maintain BPH-1 cells and DMEM medium (Gibico) containing 5% fetal bovine serum was used to culture WPMY-1 cells. Cells were treated with 5 ng/mL TGF-β1 (RD Biosciences, USA) or 20µM resveratrol (MedChemExpress, USA) for 72 h (12,18,19). Cell transfection was carried out by using Lipofectamine 3000 (Invitrogen, USA) reagent. A pcDNA 3.1-DIO3OS overexpression plasmids, DIO3OS shRNA, miRNA mimics and inhibitors were designed by Genepharma (Shanghai, China).
RNA extraction and real-time quantitative PCR (qRT-PCR)
Total RNAs were extracted from tissues and cells by employing TRIzol (Invitrogen) following the manufacturer’s instructions. To obtain nuclear and cytosolic RNAs, a PARIS Kit was used to separate subcellular fractions of cells before extracting RNAs. RNAs were reversely transcribed into cDNAs with a PrimeScript RT reagent kit (Takara, Japan). qPCR was carried out using SYBR real-time PCR kits (Takara). mRNA expression was calculated based on an internal control GAPDH. Expression of miRNAs were detected by a mirVanaTM qRT-PCR microRNA Detection Kit (Ambion, USA) and normalized to U6 expression.
Western blot assay
RIPA buffer (Beyotime, China) added protease inhibitors was employed to extract total proteins from cells. Concentration of total proteins was determined by a BCA kit (Thermo Scientific, USA). Equivalent proteins were subjected to SDS-polyacrylamide gels and electotransferred onto PVDF membranes (Millipore, USA). The membranes were the incubated with primary antibodies and secondary antibodies in turn. Protein bands were visualized at a Molecular Imager ChemiDoc XRS System (Bio-Rad, USA). The following primary antibodies were used: Smad2 (#5339, Cell Signaling Technology, USA); Phospho-Smad2 (Ser465/Ser467) (#18338, Cell Signaling Technology, USA); Smad3 (#9523, Cell Signaling Technology, USA); Phospho-Smad3 (Ser423/425) (#9520, Cell Signaling Technology, USA); α-Smooth Muscle Actin (α-SMA) (#19245, Cell Signaling Technology); N-cadherin (#13116, Cell Signaling Technology); E-cadherin (ab1416, Abcam); CCND1 (ab16663, Abcam); p21 (ab188224, Abcam); Connective tissue growth factor (CTGF) (ab6992, Abcam); Zinc Finger E-Box Binding Homeobox 1 (ZEB1) (ab181451, Abcam); GAPDH (ab9485, Abcam).
Chromatin immunoprecipitation (ChIP)
ChIP assays were conducted with a Simple Chip Enzymatic Chromatin IP kit (Cell Signaling Technology) according to the manufacturer’s instructions. Antibodies including Smad2 (#5339, Cell Signaling Technology), Smad3 (#9523, Cell Signaling Technology) and Smad4 (#46535, Cell Signaling Technology) were used to immunoprecipitated the DNA-protein cross-links with IgG as a negative control. Chromatin DNA in the immunoprecipitates were purified and subjected to qRT-PCR analysis.
Luciferase reporter assay
The promotor region of DIO3OS containing wild type (wt) or mutant type (mut) of predicted Smad2/Smad3/Smad4 complex recognition site was constructed into a PGL3-Basic luciferase reporter (Promega, USA). The luciferase reporters were co-transfected into cells together with expression plasmids of Smads (Smad2/Smad3/Smad4). a Dual-Luciferase reporter assay system (Promega) was employed to test luciferase activities after 48 h.
The wild type or mutant type fragment of DIO3OS, CTGF 3’UTR or ZEB1 3’UTR containing predicted miR-656-3p or miR-485-5p binding site was constructed into a pMIR-REPORT vector (Thermo Scientific). The luciferase reporters were co-transfected into cells together with miRNAs mimics. Luciferase reporter assay system (Promega) was employed to test luciferase activities after 48 h.
EMT evaluation
EMT of BPH-1 cells was evaluated by observing elongated fibroblast-like morphology with scattered distribution using a phase contrast microscope. The expressions of EMT biomarkers including α-SMA, E-cadherin and N-cadherin were analyzed through qRT-PCR and Western blot assays as described above.
Cell proliferation assay
Proliferation of WPMY-1 cells were estimated by CCK-8 assay. Cells (5×103) were cultivated in a 96-well plate for 24 h before reagents treatment and transfection. CCK-8 reagents (MedChemExpress) were added into each well 24 h, 48 h and 72 h later. A microplate reader (Bio-Rad) was then applied to measure the absorbance at 450 nm. The expressions of cell cycle related genes including CCND1 and p21 were tested through qRT-PCR and Western blot assays as described above.
Fluorescence in situ hybridization (FISH)
Subcellular localization of DIO3OS was identified via a FISH assay using a DIO3OS specific probe and Fluorescent in Situ Hybridization Kit (RiboBio, China). 4’,6-diamidino-2-phenylindole (DAPI) was used for nuclear labeling. Images were acquired with a fluorescence microscope (OLYMPUS, Japan).
RNA pull-down
RNA pull-down assay was used to find miRNAs which could bind to DIO3OS using a biotin-labeled DIO3OS probe (RiboBio) and streptavidin magnetic beads (Life Technologies, USA). RNAs were extracted with TRIzol from the complexes and subjected to qRT-PCR analysis to detect expressions of miRNAs.
Statistical analysis
Research data from at least three independent experiments were shown as the means ± SD. Differences among different groups were analyzed by Student’s t-test or one-way ANOVA with SPSS 17.0 software (IBM, USA). P<0.05 was considered statistically significant.
Results
DIO3OS is highly expressed in BPH tissues and regulated by TGF-β1 and resveratrol
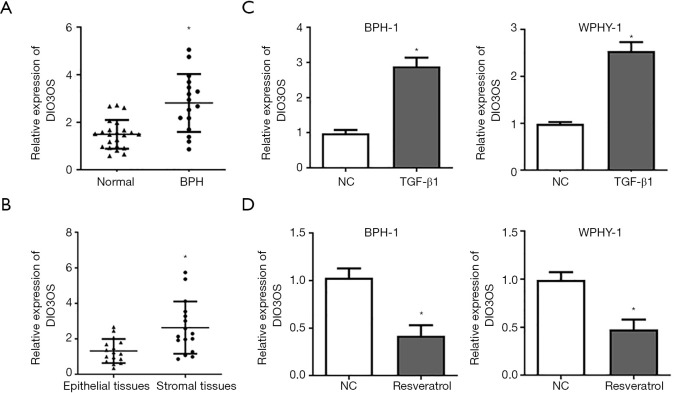
In order to probe the role of DIO3OS in BPH, the expressions of DIO3OS in epithelial tissues and stromal tissues were detected by RT-qPCR, which showed that DIO3OS expressions were prominently upregulated in BPH tissues compared with normal prostate tissues (Figure 1A). DIO3OS level was also found to be higher in stromal tissues than that of epithelial tissues of BPH (Figure 1B). We also demonstrated that DIO3OS could be significantly upregulated by TGF-β1 and downregulated by resveratrol in both BPH-1 cells and WPMY-1 cells (Figure 1C,D).
Figure 1.
DIO3OS is highly expressed in BPH tissues and regulated by TGF-β1 and resveratrol. (A) Comparison of DIO3OS expressions in BPH tissues and normal prostate tissues. (B) Comparison of DIO3OS expressions in stromal tissues and epithelial tissues of BPH. (C,D) DIO3OS expressions in BPH-1 cells and WPMY-1 cells were detected with RT-qPCR after TGF-β1 or resveratrol treatment. *, P<0.05.
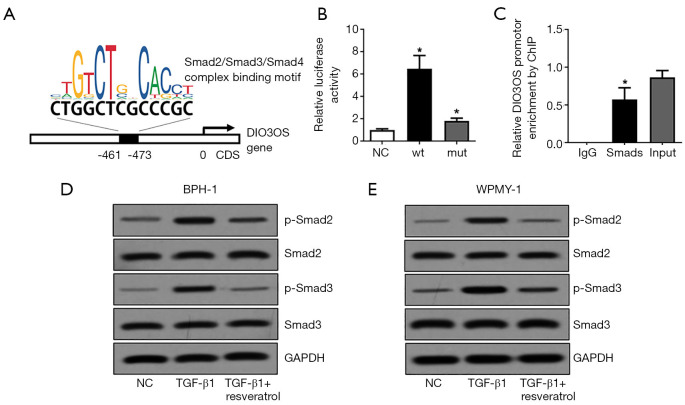
TGF-β1 and resveratrol mediates DIO3OS expression in a Smads dependent manner
To explore the mechanism of TGF-β1 and resveratrol in mediating DIO3OS expression, we searched the JASPAR database and noticed a potential Smad2/Smad3/Smad4 complex binding site in DIO3OS promotor region (Figure 2A). Then, this site was validated by a luciferase reporter assay in which the luciferase activity was significantly enhanced by Smads (Figure 2B). Moreover, ChIP assay confirmed that the DIO3OS promotor region could interact with Smad2/Smad3/Smad4 complex (Figure 2C). More important, TGF-β1 could stimulate phosphorylation of Smad2 and Smad3 and resveratrol blocked TGF-β1 signaling via repressing phosphorylation of Smad2 and Smad3 in both BPH-1 cells and WPMY-1 cells (Figure 2D,E). Consider with the previous results, we concluded that TGF-β1 and resveratrol mediated DIO3OS expression in a Smads dependent manner.
Figure 2.
TGF-β1 and resveratrol mediates DIO3OS expression in a Smads dependent manner. (A) Potential Smad2/Smad3/Smad4 complex binding site in DIO3OS promotor region. (B) Validation of the predicted binding site by a luciferase reporter assay. (C) Validation of the interaction between DIO3OS promotor region and Smad2/Smad3/Smad4 complex by ChIP assay. (D,E) Phosphorylation of Smad2 and Smad3 in both BPH-1 cells and WPMY-1 cells mediated by TGF-β1 and resveratrol were analyzed by Western blot assay. *, P<0.05.
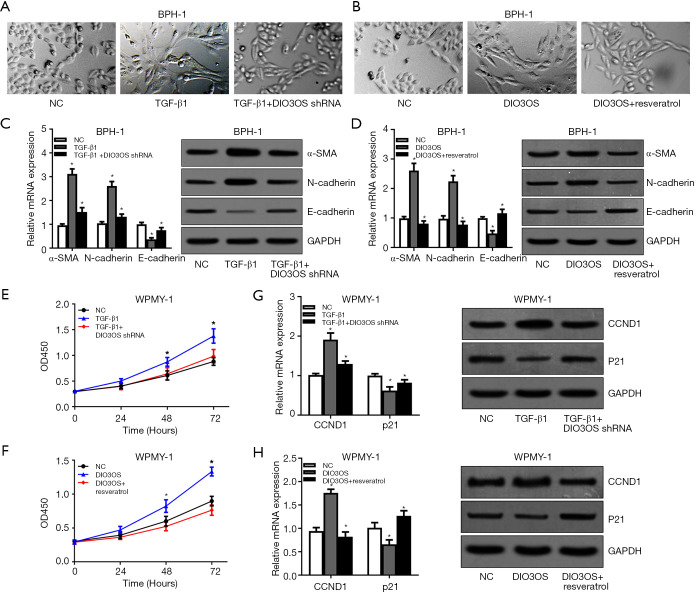
TGF-β1 promotes BPH-1 cells EMT and WPMY-1 cells proliferation via DIO3OS which can be blocked by resveratrol
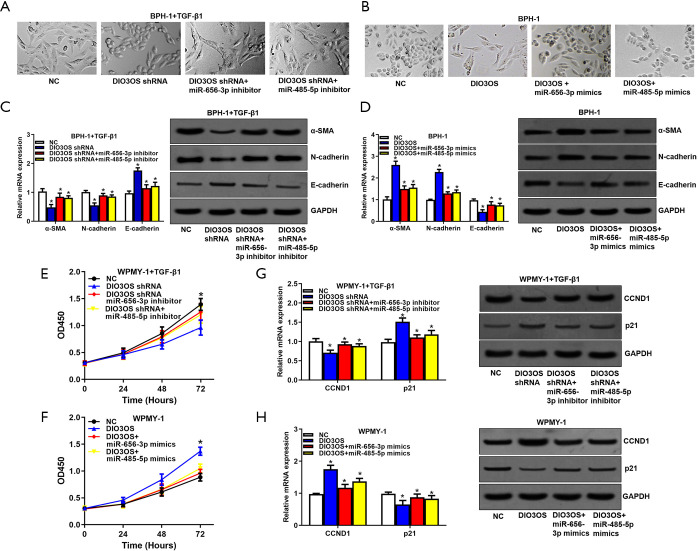
Next, to investigate the biological functions of DIO3OS, we transfected DIO3OS shRNAs into BPH-1 cells and WPMY-1 cells stimulated with TGF-β1, and transfected DIO3OS overexpression plasmids into BPH-1 cells and WPMY-1 cells. Then, we observed that TGF-β1 could obviously induce EMT of BPH-1 cells and this effect could be attenuated by DIO3OS silencing (Figure 3A). At the same time, overexpression of DIO3OS enhanced the EMT of BPH-1 cells which could be repressed by resveratrol (Figure 3B). Accordingly, the mRNA and protein levels of EMT biomarkers including α-SMA, N-cadherin and E-cadherin were modulated by TGF-β1 and DIO3OS silencing, as well as by DIO3OS overexpression and resveratrol (Figure 3C,D). Similarly, TGF-β1 could promote proliferation of WPMY-1 cells and this effect could be reversed by DIO3OS silencing (Figure 3E), and overexpression of DIO3OS facilitated proliferation of WPMY-1 cells which could be inhibited by resveratrol (Figure 3F). As expected, the mRNA and protein expressions of cell cycle related genes including CCND1 and p21 were modulated by TGF-β1 and DIO3OS silencing, as well as by DIO3OS overexpression and resveratrol (Figure 3G,H). All these data indicated that TGF-β1 promoted BPH-1 cells EMT and WPMY-1 cells proliferation via DIO3OS and this effect could be blocked by resveratrol.
Figure 3.
TGF-β1 promotes BPH-1 cells EMT and WPMY-1 cells proliferation via DIO3OS which can be blocked by resveratrol. (A,B) EMT of BPH-1 cells caused by TGF-β1, DIO3OS and resveratrol were evaluated by observing elongated fibroblast-like morphology with scattered distribution. (C,D) The mRNA and protein levels of EMT biomarkers including α-SMA, E-cadherin and N-cadherin were analyzed by qRT-PCR and Western blot assays. (E,F) Proliferation of WPMY-1 cells modulated by TGF-β1, DIO3OS and resveratrol were determined by CCK8 assays. (G,H) The mRNA and protein expressions of cell cycle related genes including CCND1 and p21 were tested with qRT-PCR and Western blot assays. *, P<0.05.
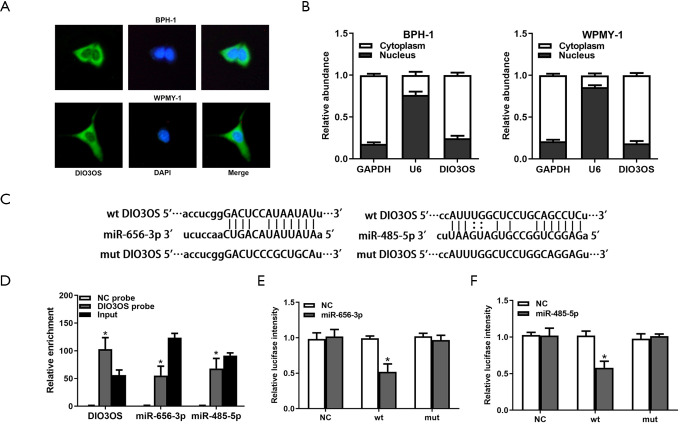
MiR-656-3p and miR-485-5p are targets of DIO3OS
Through FISH assays and qRT-PCR assays, we identified DIO3OS were abundant in cytoplasm of BPH-1 and WPMY-1 cells (Figure 4A,B), which indicating that DIO3OS might potentially interact with miRNAs. By consulting starBase database, we predicted miR-656-3p and miR-485-5p might be targets of DIO3OS (Figure 4C). Thereafter, RNA pull-down assay was employed to examine whether DIO3OS could bind with miR-656-3p and miR-485-5p. MiR-656-3p and miR-485-5p were pulled-down by DIO3OS probe but not by NC probe (Figure 4D). Additionally, luciferase reporter assay verified the specific binding sites of DIO3OS with miR-656-3p and miR-485-5p (Figure 4E). To sum up, miR-656-3p and miR-485-5p were targets of DIO3OS.
Figure 4.
MiR-656-3p and miR-485-5p are targets of DIO3OS. (A,B) Subcellular localization of DIO3OS was determined by FISH assays and qRT-PCR assays. (C) Predicted miR-656-3p and miR-485-5p binding sequences in DIO3OS sequence. (D) Combination of DIO3OS with miR-656-3p and miR-485-5p was verified using RNA pull-down assay. (E) Predicted binding sequences of DIO3OS with miR-656-3p and miR-485-5p were verified by luciferase reporter assay. wt, wild type; mut, mutant type; *, P<0.05.
DIO3OS promotes BPH-1 cells EMT and WPMY-1 cells proliferation via miR-656-3p and miR-485-5p
In BPH-1 cells stimulated with TGF-β1, miR-656-3p and miR-485-5p inhibitors was able to reverse the EMT inhibition caused by DIO3OS shRNA (Figure 5A), while miR-656-3p and miR-485-5p mimics could alleviate EMT caused by overexpression of DIO3OS (Figure 5B). The EMT biomarkers showed corresponding expression changes (Figure 5C,D). Analogously, In WPMY-1 cells treated with TGF-β1, miR-656-3p and miR-485-5p inhibitors attenuated the proliferation suppression induced by DIO3OS shRNA (Figure 5E), while miR-656-3p and miR-485-5p mimics abated the proliferation acceleration caused by overexpression of DIO3OS (Figure 5F). Also, the proliferation biomarkers showed corresponding expression changes (Figure 5G,H). All these results revealed that DIO3OS promoted BPH-1 cells EMT and WPMY-1 cells proliferation via miR-656-3p and miR-485-5p.
Figure 5.
DIO3OS promotes BPH-1 cells EMT and WPMY-1 cells proliferation via miR-656-3p and miR-485-5p. (A,B) EMT of BPH-1 cells regulated by DIO3OS together with miR-656-3p and miR-485-5p were evaluated by observing elongated fibroblast-like morphology with scattered distribution. (C,D) The mRNA and protein expressions of EMT biomarkers including α-SMA, N-cadherin and E-cadherin were detected by qRT-PCR and Western blot assays. (E,F) Proliferation of WPMY-1 cells modulated by DIO3OS together with miR-656-3p and miR-485-5p were determined by CCK8 assays. (G,H) The mRNA and protein expressions of cell cycle related genes including CCND1 and p21 were tested using qRT-PCR and Western blot assays. *, P<0.05.
CTGF and ZEB1 are targets of miR-656-3p and miR-485-5p
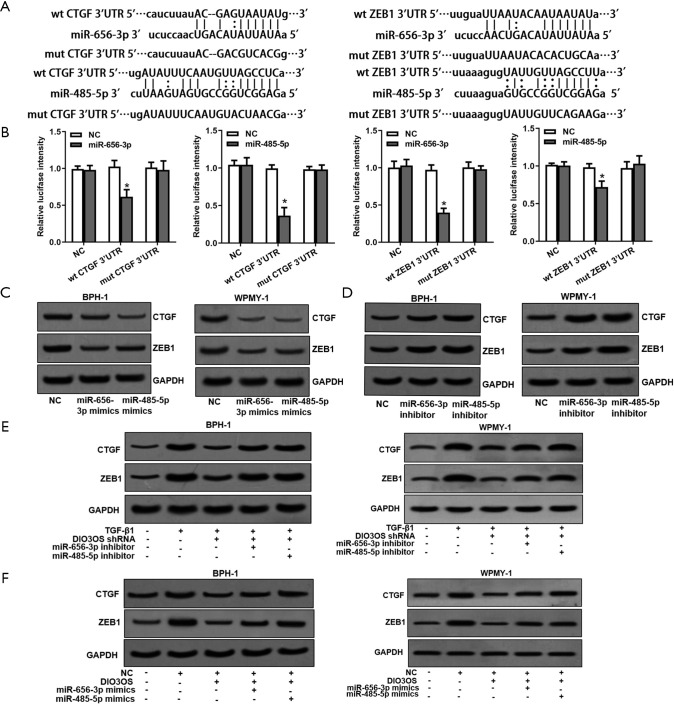
By searching microRNA.org database, we found CTGF and ZEB1 were possible targets of both miR-656-3p and miR-485-5p (Figure 6A). These targets were subsequently validated by employing luciferase reporter assay (Figure 6B). Western blot assays displayed that protein levels of CTGF and ZEB1 were affected by both miR-656-3p and miR-485-5p (Figure 6C,D). As a result, we demonstrated that TGF-β1 could upregulate CTGF and ZEB1 expression via DIO3OS/miR-656-3p and miR-485-5p axis (Figure 6E), and DIO3OS was able to increase CTGF and ZEB1 expression which could be eliminate by resveratrol, miR-656-3p and miR-485-5p axis in BPH cells (Figure 6F).
Figure 6.
CTGF and ZEB1 are targets of miR-656-3p and miR-485-5p. (A) Potential miR-656-3p and miR-485-5p binding sequences in 3’UTR of CTGF and ZEB1 mRNA. (B) Predicted miR-656-3p and miR-485-5p binding sites in 3’UTR of CTGF and ZEB1 mRNA were verified by luciferase reporter assay. (C,D) Regulation of CTGF and ZEB1 protein levels by TGF-β1, resveratrol, DIO3OS, miR-656-3p and miR-485-5p was determined by Western blot assay. (E,F) Regulation of CTGF and ZEB1 protein levels by TGF-β1, resveratrol, DIO3OS, miR-656-3p and miR-485-5p was tested by Western blot assay. wt, wild type; mut, mutant type; *, P<0.05.
Discussion
Emerging evidences indicate that the changes in lncRNAs are associated with the development of multiple diseases (16). Nevertheless, there are extremely few studies on lncRNAs in BPH. Herein, we identified a lncRNA DIO3OS which was significantly upregulated in BPH. In inflammatory bowel disease, the expression of DIO3OS is evidently decreased in tissues and plasmas of patients compared with in healthy individuals (20). DIO3OS probably also plays a role in Zearalenone mediated toxic effects on reproduction and development (21). In pancreatic cancer, DIO3OS facilitates PC cells growth and invasion by modulating the expression of ALDOA via miR-122 (22).
In current study, we displayed that DIO3OS could be upregulated by TGF-β1 and downregulated by resveratrol in both BPH-1 and WPMY-1 cells. Smad2/Smad3/Smad4 complex could bind to the DIO3OS promotor region and thereby enhanced its transcription. Hence, TGF-β1 and resveratrol mediated DIO3OS expression in a Smads dependent manner. Then, we showed that TGF-β1 promoted BPH-1 cells EMT and WPMY-1 cells proliferation via DIO3OS and this effect could be blocked by resveratrol.
A common way for lncRNAs to perform its functions is to influence the activity of miRNAs by a competing endogenous RNAs (ceRNA) manner (23,24). Our data showed that miR-656-3p and miR-485-5p were targets of DIO3OS. Previous studies uncovered that miR-656-3p and miR-485-5p inhibited proliferation, migration and EMT of some cancer cells (25-28). Then, our data implied DIO3OS promoted BPH-1 cells EMT and WPMY-1 cells proliferation via miR-656-3p and miR-485-5p.
Furthermore, our data revealed CTGF and ZEB1 were targets of both miR-656-3p and miR-485-5p and could be restrained by miR-656-3p and miR-485-5p. CTGF is an extracellular matrix-associated protein and a member of the CCN proteins family which functions in many cellular proceedings, including cell migration, proliferation, adhesion, and fibrosis (29). ZEB1 is the basic element of transcription factor network that mediates EMT and has been confirmed to be a vital molecule in the control of proliferation, differentiation, and metastasis (30). Both CTGF and ZEB1 are critical downstream molecules that response to TGF-β signals and involved in cellular and biological processes of cell proliferation and EMT (31,32). We consider that these factors including TGF-β1, resveratrol, DIO3OS, miR-656-3p and miR-485-5p finally realize their biological effects through modulating CTGF and ZEB1.
Conclusions
All in all, in our study, we verified that DIO3OS is highly expressed in BPH tissues and regulated by TGF-β1 and resveratrol in a Smads dependent manner. DIO3OS promotes BPH-1 cells EMT and WPMY-1 cells proliferation by upregulating CTGF and ZEB1 via miR-656-3p and miR-485-5p.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by a foundation from Shanghai Jiao Tong University School of Medicine Multi-center Clinical Research Project (DLY201809) and and Huangpu Distric Industrial Support Fund (XK2020011).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Written informed consents were signed by all the participants. Current research was approved by the Ethical Committee of Shanghai Ninth People’s Hospital (No. 2017207) and strictly conformed to the 2013 Declaration of Helsinki.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1169
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1169). The authors have no conflicts of interest to declare.
References
- 1.De Nunzio C, Presicce F, Tubaro A, Inflammatory mediators in the development and progression of benign prostatic hyperplasia. Nat Rev Urol 2016;13:613-26. 10.1038/nrurol.2016.168 [DOI] [PubMed] [Google Scholar]
- 2.Zhang B, Chen X, Xie C, et al. Leptin promotes epithelial-mesenchymal transition in benign prostatic hyperplasia through downregulation of BAMBI. Exp Cell Res 2020;387:111754. 10.1016/j.yexcr.2019.111754 [DOI] [PubMed] [Google Scholar]
- 3.Alonso-Magdalena P, Brössner C, Reiner A, et al. A role for epithelial-mesenchymal transition in the etiology of benign prostatic hyperplasia. Proc Natl Acad Sci U.S.A. 2009;106:2859-63. 10.1073/pnas.0812666106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi X, Peng Y, Du X, et al. Estradiol promotes epithelial-to-mesenchymal transition in human benign prostatic epithelial cells. Prostate 2017;77:1424-37. 10.1002/pros.23404 [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Wang Y, Gratzke C, et al. Ghrelin Aggravates Prostate Enlargement in Rats with Testosterone-Induced Benign Prostatic Hyperplasia, Stromal Cell Proliferation, and Smooth Muscle Contraction in Human Prostate Tissues. Oxid Med Cell Longev 2019;2019:4748312. 10.1155/2019/4748312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Q, Gratzke C, Wang R, et al. A NAV2729-sensitive mechanism promotes adrenergic smooth muscle contraction and growth of stromal cells in the human prostate. J Biol Chem 2019;294:12231-49. 10.1074/jbc.RA119.007958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Li B, Ciotkowska A, et al. Onvansertib, a polo-like kinase 1 inhibitor, inhibits prostate stromal cell growth and prostate smooth muscle contraction, which is additive to inhibition by α-blockers. Eur J Pharmacol 2020;873:172985. 10.1016/j.ejphar.2020.172985 [DOI] [PubMed] [Google Scholar]
- 8.Afdal A, Darwin E, Yanwirasti Y, et al. The Expression of Transforming Growth Factor Beta-1 and Interleukin-6 on Human Prostate: Prostate Hyperplasia and Prostate Cancer. Open Access Maced J Med Sci 2019;7:1905-10. 10.3889/oamjms.2019.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu S, Yu W, Lv TJ, et al. Evidence of TGF-β1 mediated epithelial-mesenchymal transition in immortalized benign prostatic hyperplasia cells. Mol Membr Biol 2014;31:103-10. 10.3109/09687688.2014.894211 [DOI] [PubMed] [Google Scholar]
- 10.Breuss JM, Atanasov AG, Uhrin P. Resveratrol and Its Effects on the Vascular System. Int J Mol Sci 2019;20:1523. 10.3390/ijms20071523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia N, Daiber A, Förstermann U, et al. Antioxidant effects of resveratrol in the cardiovascular system. Br J Pharmacol 2017;174:1633-46. 10.1111/bph.13492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Hu WL, Lu MX, et al. Resveratrol induces apoptosis of benign prostatic hyperplasia epithelial cell line (BPH-1) through p38 MAPK-FOXO3a pathway. BMC Complement Altern Med 2019;19:233. 10.1186/s12906-019-2648-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Zhou QM, Lu YY, et al. Resveratrol Inhibits the Migration and Metastasis of MDA-MB-231 Human Breast Cancer by Reversing TGF-β1-Induced Epithelial-Mesenchymal Transition. Molecules 2019;24:1131. 10.3390/molecules24061131 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Song Y, Chen Y, Li Y, et al. Resveratrol Suppresses Epithelial-Mesenchymal Transition in GBM by Regulating Smad-Dependent Signaling. Biomed Res Int 2019;2019:1321973. 10.1155/2019/1321973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsagakis I, Douka K, Birds I, et al. Long non-coding RNAs in development and disease: conservation to mechanisms. J Pathol 2020;250:480-95. 10.1002/path.5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, Zhang M, Ou Z, et al. Long noncoding RNA DNM3OS promotes prostate stromal cells transformation via the miR-29a/29b/COL3A1 and miR-361/TGFβ1 axes. Aging (Albany NY) 2019;11:9442-60. 10.18632/aging.102395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slabáková E, Pernicová Z, Slavíčková E, et al. TGF-β1-induced EMT of non-transformed prostate hyperplasia cells is characterized by early induction of SNAI2/Slug. Prostate 2011;71:1332-43. 10.1002/pros.21350 [DOI] [PubMed] [Google Scholar]
- 19.Starsíchová A, Lincová E, Pernicová Z, et al. TGF-beta1 suppresses IL-6-induced STAT3 activation through regulation of Jak2 expression in prostate epithelial cells. Cell Signal 2010;22:1734-44. 10.1016/j.cellsig.2010.06.014 [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Hou Y, Chen W, et al. KIF9 AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease. Mol Med Rep 2018;17:2195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie H, Wang H, Li X, et al. ZEA exerts toxic effects on reproduction and development by mediating DIO3OS in mouse endometrial stromal cells. J Biochem Mol Toxicol 2019;33:e22310. 10.1002/jbt.22310 [DOI] [PubMed] [Google Scholar]
- 22.Cui K, Jin S, Du Y, et al. Long noncoding RNA DIO3OS interacts with miR-122 to promote proliferation and invasion of pancreatic cancer cells through upregulating ALDOA. Cancer Cell Int 2019;19:202. 10.1186/s12935-019-0922-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol 2019;13:46-60. 10.1002/1878-0261.12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Huo C, Lin X, et al. Computational Identification of Cross-Talking ceRNAs. Adv Exp Med Biol 2018;1094:97-108. 10.1007/978-981-13-0719-5_10 [DOI] [PubMed] [Google Scholar]
- 25.Chen T, Qin S, Gu Y, et al. Long non-coding RNA NORAD promotes the occurrence and development of non-small cell lung cancer by adsorbing MiR-656-3p. Mol Genet Genomic Med 2019;7:e757. 10.1002/mgg3.757 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Wang R, Zheng B, Liu H, et al. Long non-coding RNA PCAT1 drives clear cell renal cell carcinoma by upregulating YAP via sponging miR-656 and miR-539. Cell Cycle 2020;19:1122-31. 10.1080/15384101.2020.1748949 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Lin XJ, He CL, Sun T, et al. hsa-miR-485-5p reverses epithelial to mesenchymal transition and promotes cisplatin-induced cell death by targeting PAK1 in oral tongue squamous cell carcinoma. Int J Mol Med 2017;40:83-9. 10.3892/ijmm.2017.2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Zhou X, Zeng F, et al. miR-485-5p inhibits the progression of breast cancer cells by negatively regulating MUC1. Breast Cancer 2020;27:765-75. 10.1007/s12282-020-01075-2 [DOI] [PubMed] [Google Scholar]
- 29.Ohkawara B, Kobayakawa A, Kanbara S, et al. CTGF/CCN2 facilitates LRP4-mediated formation of the embryonic neuromuscular junction. EMBO Rep 2020;21:e48462. 10.15252/embr.201948462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drápela S, Bouchal J, Jolly MK, et al. ZEB1: A Critical Regulator of Cell Plasticity, DNA Damage Response, and Therapy Resistance. Front Mol Biosci 2020;7:36. 10.3389/fmolb.2020.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Cai QY, Liu J, et al. Ursolic acid suppresses the invasive potential of colorectal cancer cells by regulating the TGF-β1/ZEB1/miR-200c signaling pathway. Oncol Lett 2019;18:3274-82. 10.3892/ol.2019.10604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valle-Tenney R, Rebolledo DL, Lipson KE, et al. Role of hypoxia in skeletal muscle fibrosis: Synergism between hypoxia and TGF-β signaling upregulates CCN2/CTGF expression specifically in muscle fibers. Matrix Biol 2020;87:48-65. 10.1016/j.matbio.2019.09.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as