Abstract
Background
Renal cell carcinoma (RCC) is a common urologic malignancy. Although the relationship between clear cell RCC (ccRCC) and obesity has been well-established by several large-scale retrospective studies, the molecular mechanisms and genetic characteristics behind this correlation remains unclear. In the current study, several bioinformatics tools were used to identify the key genes in ccRCC related to obesity.
Methods
Microarray data comparing ccRCC with normal renal tissues in patients with and without obesity were downloaded from the GEO database for screening of differentially expressed genes (DEGs). The DEGs were verified with expression level and survival analysis using several online bioinformatics tools.
Results
In the current study, the differential expression of five genes correlated with both ccRCC and obesity; IGHA1 and IGKC as oncogenes, and MAOA, MUC20 and TRPM3 as tumor suppressor genes. These genes were verified by comparing the relationship between the expression levels and survival outcomes from open-source data in The Cancer Genome Atlas (TCGA) dataset.
Conclusions
In conclusion, the five genes differentially expressed in ccRCC and obesity are related to disease progression and prognosis, and therefore could provide prognostic value for patients with ccRCC.
Keywords: Clear cell renal cell carcinoma (ccRCC), obesity, prognosis, biomarker
Introduction
Renal cell carcinoma (RCC) is a common urologic malignancy with several histopathological subtypes, among which clear cell RCC (ccRCC) remains the most common. According to the latest global cancer statistics in 2018, there were approximately 400,000 cases of newly diagnosed RCC and 175,000 RCC-related deaths annually (1). Although an increasing number of RCC cases were diagnosed at their early stages due to the improvements in imaging techniques, and managed by nephron sparing surgery, nearly 30% of RCCs were diagnosed at the locally advanced stage or with distant metastasis (2). Currently, the first-line therapy for metastatic RCC (mRCC) is either molecular-targeted drugs, such as sorafenib, axitinib, sunitinib, pazopanib and everolimus, or immune checkpoint inhibitors such as nivolumab (3). However, due to the strong side effects of these drugs as well as the individualized sensitivity of patients to these drugs, the overall survival (OS) and quality-of-life of patients with mRCC has not improved much over the past decade. Thus, the main focus for improving OS should be on early screening, diagnosis and intervention.
The etiology of RCC is still not clear; however, the relationship between ccRCC and obesity has been well-established by several large-scale retrospective studies (4-7). Nevertheless, the mechanism of obesity leading to high incidence of ccRCC has not been fully studied. Several obesity prognostic biomarkers, including leptin, leptin receptor, adiponectin, visfatin and apelin have been studied. However, the significance of these prognostic biomarkers is controversial. Circulating leptin had a negative correlation with RCC risk in a study by Spyridopoulos et al. but was reported to be associated with a high risk of RCC occurrence by Liao et al. (8,9). The prognostic value of circulating adiponectin may also need further study, since the present studies have come to contradictory conclusions regarding the relationship between circulating adiponectin and RCC prognosis (10,11). Therefore, it is essential to identify the key genes that correlate RCC and obesity so that the potential mechanism behind the obesity/RCC relationship can be further studied.
Currently, with the gradual development and popularization of high-throughput sequencing and gene chips in oncological study, we can identify key genes related to carcinogenesis and prognosis based on open-source platforms and bioinformatics. In this study, we used bioinformatics methods to identify and validate the genes significantly related to both obesity and ccRCC.
We present the following article in accordance with the STREGA reporting checklist (available at http://dx.doi.org/10.21037/tau-20-891).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and the Harmonized Tripartite Guideline for Good Clinical Practice from the International Conference on Harmonization. The study was approved by the research medical ethics committee of Shanghai Renji Hospital {Ethics board approval number: Renji Ethics [2017]091}.
Gene expression profile data
GSE46699, the gene expression data and related clinical data of ccRCC, was obtained from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). GSE46699 contains 60 patients with ccRCC divided into two groups; 12 obese patients compared with13 non-obese patients, and 16 smoking patients compared with 19 non-smoking patients. The tissue samples included 60 tumor and 56 normal tissues. GSE46699 was used to screen for genes differentially expressed in both ccRCC and obesity. Hub genes were verified with sequencing data and clinical information from The Cancer Genome Atlas (TCGA) data portal (https://portal.gdc.cancer.gov/).
Data standardization and screening of differentially expressed genes (DEG)
Raw biological data in GSE46699 downloaded from GEO dataset was first preprocessed and standardized. The data platform for GSE46699 was GPL19109 (HG-U133_Plus_2) Affymetrix Human Genome U133Plus 2.0 Array. The Robust Multi-array Average method was used to eliminate noise and guarantee integrity.
DEGs between ccRCC vs. normal kidney tissues and between obese vs. non-obese cases were detected using the “limma” (linear models for microarray data) R package. The screening criteria for DEGs was based on fold change and t-test. The criteria for statistical significance included either absolute value of |log2FC| (fold change) >1.00 and t-test P value <0.05. DEGs with log FC <0 were defined as downregulated genes, while DEGs with log FC >0 were defined as upregulated genes.
KEGG and GO enrichment analyses of DEGs
To explore the functional annotations of the DEGs including the associated biological processes, molecular functions, and cellular components, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses were conducted. Based on the GO database, GO analysis obtains all functions involving a gene and then uses Fisher’s exact test and multiple comparison tests to calculate the significance level (P value) and false discovery rate (FDR) of each function, so as to determine the significant functions embodied by the DEGs (12). KEGG is a database that systematically analyzes the relationships between genes (and their coding products), gene functions, and genomic information (13). Fisher’s exact test and chi-square test were used in KEGG pathway analysis to identify significant pathways. A P value <0.05 was set as the cutoff criteria for GO and KEGG analysis. Hierarchical clustering was also conducted.
Validation and survival analysis of DEGs
The expression of DEGs in ccRCC was validated using the Gene Expression Profiling Interactive Analysis (GEPIA) web server, an online tool that provides gene expression profiling of cancer and normal tissues, as well as interactive analyses based on TCGA and Genotype-Tissue Expression (GTEx) data portals (14). The survival analysis of the DEGs was also conducted through GEPIA, with the OS as the primary end point. The expression of the DEGs in different stages of ccRCC were also validated based on the TCGA data in GEPIA.
Statistical analysis
R 4.0.0 was used for the analysis of DEGs. χ2 test was used to analyze the statistical significance of GO and KEGG pathway analysis. Differences with P<0.05 were considered statistically significant.
Results
Screening of DEGs in ccRCC
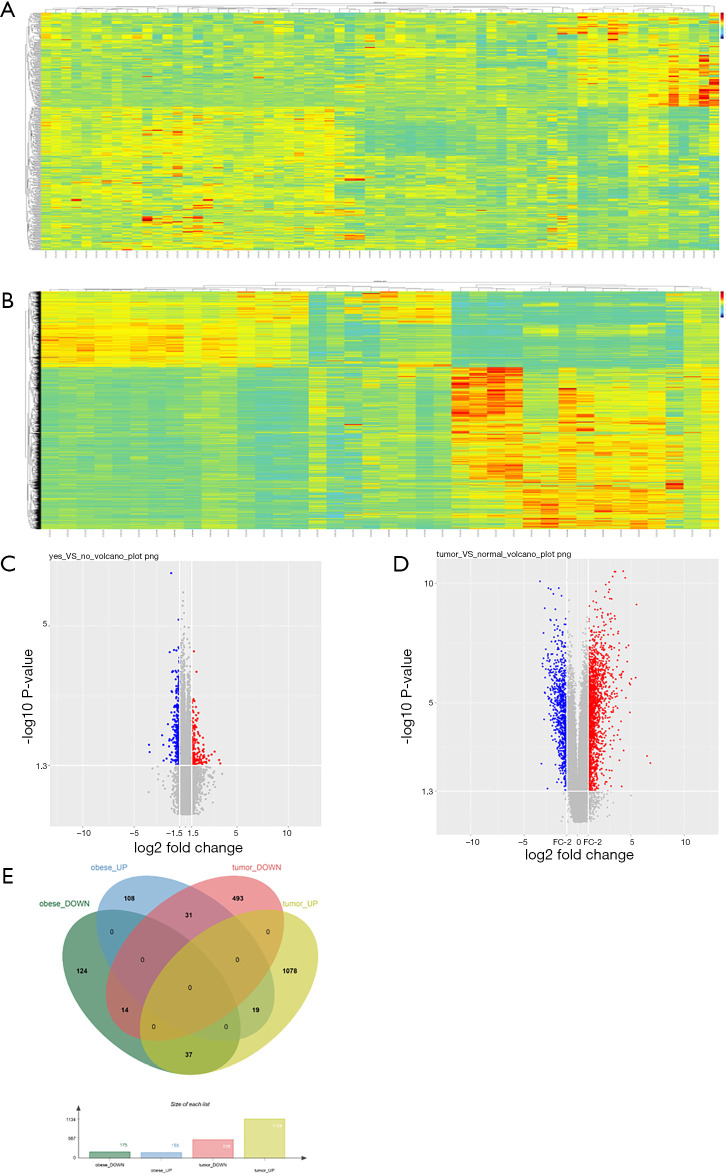
After searching the GEO datasets with the key words ‘Obesity’ and ‘Clear Cell Renal Cell Carcinoma’ (ccRCC), the GSE46699 dataset which includes smoking- and obesity-related molecular alterations in ccRCC was used for further analysis. Gene expression was compared between tumor samples from obese vs. non-obese patients to identify tumor genes that correlate with obesity. Additionally, we compared gene expression in tumor vs. para-cancer tissue samples from patients with both ccRCC and obesity to identify genes that correlate with ccRCC. The DEGs obtained from these two analyses are shown as heat maps and volcano plots in Figure 1. Twelve genes were differentially expressed in both analyses, including 8 upregulated and 4 downregulated genes.
Figure 1.
Differentially expressed genes (DEGs) and commonly DEGs in the GSE46699 dataset. Heatmaps of differential gene expression in (A) tumor samples from obese vs. non-obese patients and (B) tissue samples from patients with vs. without ccRCC. Volcano plots of gene expression in (C) tumor samples from obese vs. non-obese patients with ccRCC and (D) tissue samples from patients with vs. without ccRCC. (E) Venn diagram of commonly DEGs in the GSE46699 dataset. ccRCC, clear cell renal cell carcinoma.
Functional annotation and KEGG pathway enrichment of DEGs
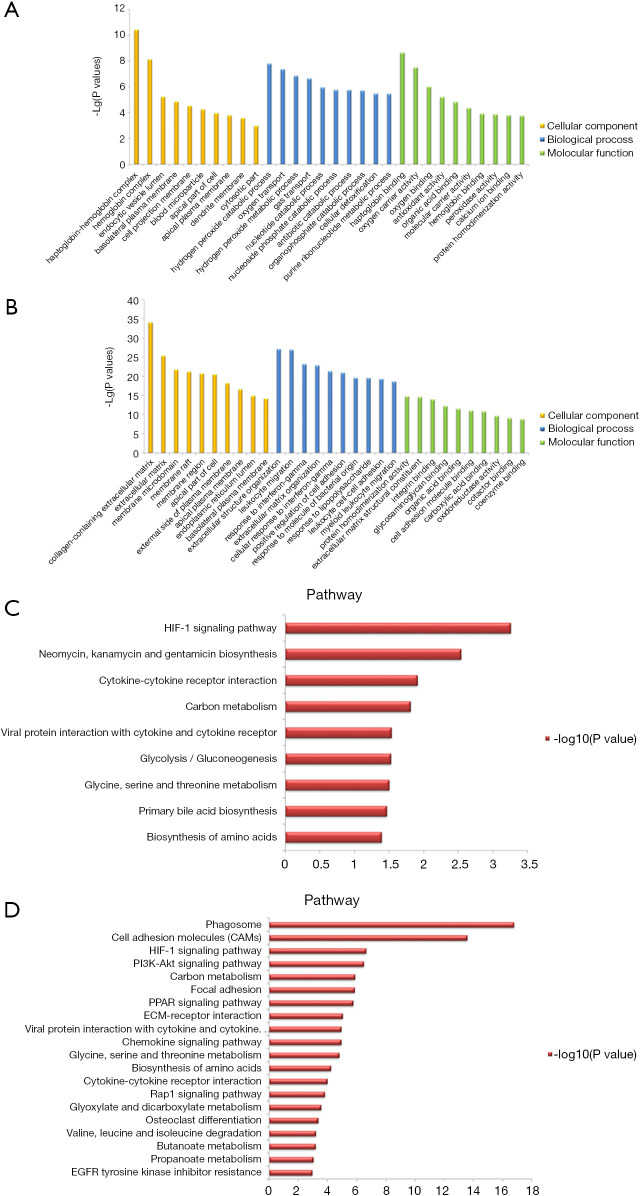
The GO analysis and KEGG pathway enrichment were performed on the two groups of DEGs to further explore potential biological processes in ccRCC (Figure 2). GO analysis indicated that the DEGs play roles in the hydrogen peroxide catabolic process, extracellular structure organism construction, and leukocyte migration (Figure 2A,B). Based on KEGG pathway enrichment analysis, the most significant pathway was “Phagosome”; other significant pathways included “HIF-1 signaling pathway”, and “Cell adhesion molecules” (CAMs) (Figure 2C,D). Hierarchical clustering showed that the DEGs can distinguish RCC tissue from normal tissue (Figure 3).
Figure 2.
GO analysis and KEGG pathway enrichment. GO analysis of (A) obesity-related DEGs and (B) ccRCC-related DEGs. KEGG pathway enrichment of (C) obesity-related and (D) ccRCC-related DEGs. GO, Gene Ontology; DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes; ccRCC, clear cell renal cell carcinoma.
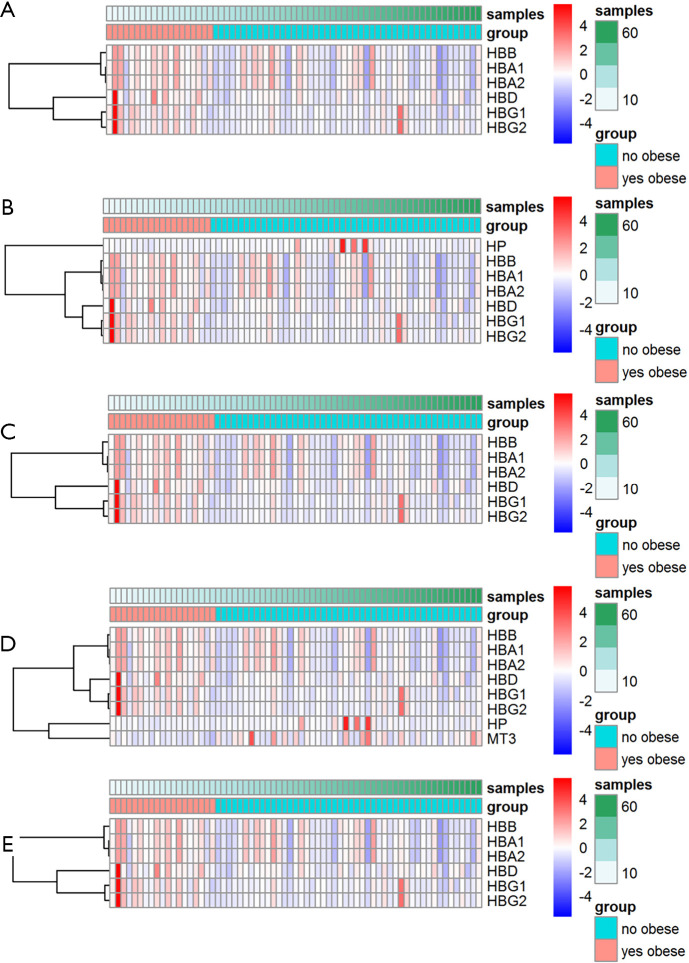
Figure 3.
Hierarchical clustering analysis of ccRCC-related DEGs. Hierarchical clustering of ccRCC-related DEG expression in relation to (A) haptoglobin binding, (B) haptoglobin-hemoglobin complex, (C) hemoglobin complex, (D) hydrogen peroxide catabolic process, and (E) oxygen transport. ccRCC, clear cell renal cell carcinoma; DEGs, differentially expressed genes.
Validation of DEGs
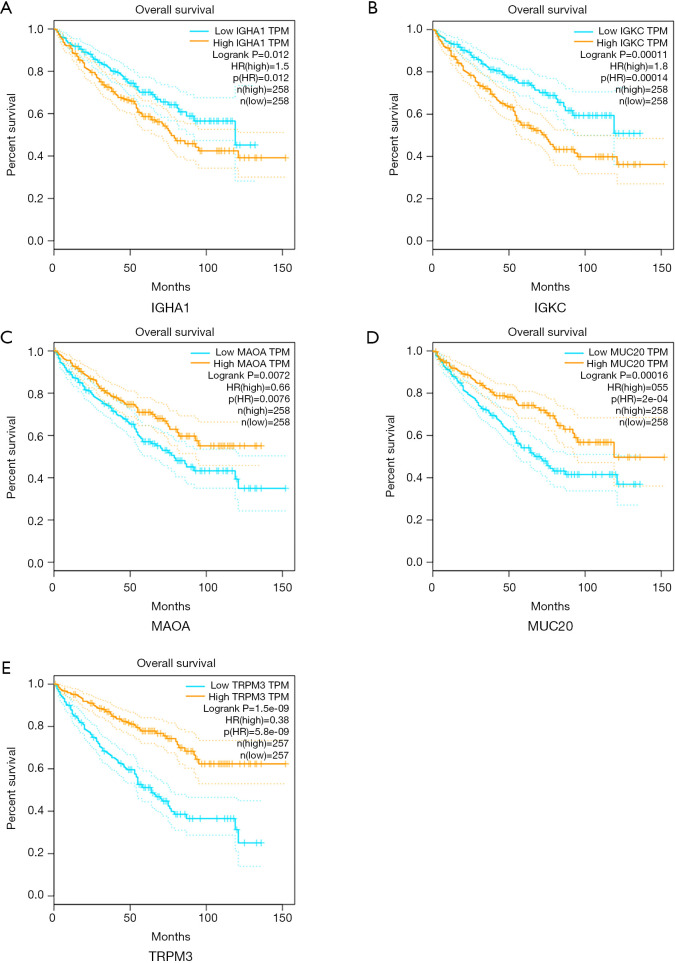
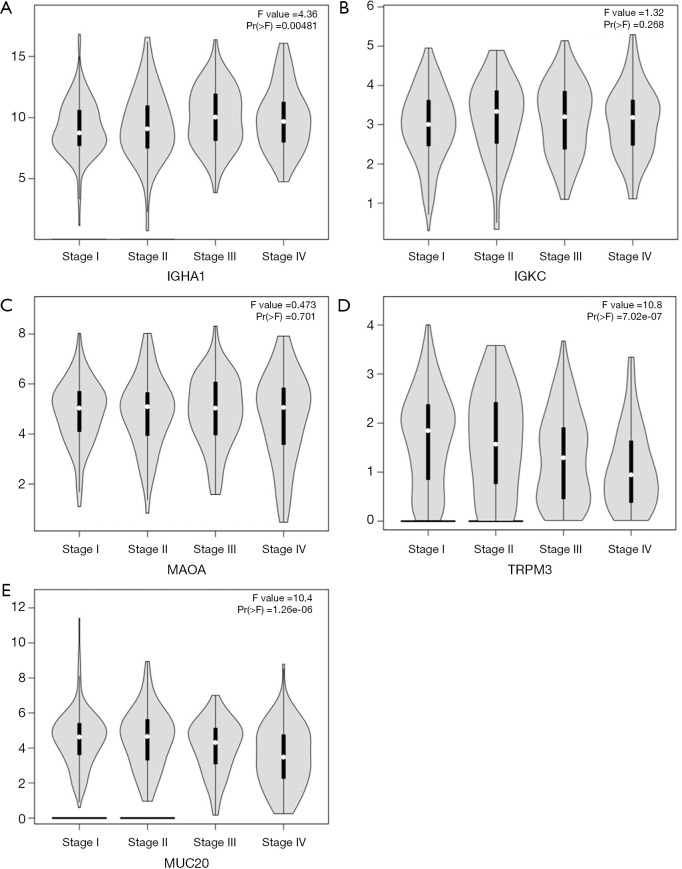
The 12 DEGs were further validated using the patients’ data from GEPIA, an online, interactive, gene expression profiling analysis tool. The expression levels of the 12 DEGs were all significantly different in ccRCC tissues compared with normal para-cancerous tissues (Figure 4). Furthermore, among the 12 DEGs, there were 5 genes that statistically correlated with OS; IGHA1 and IGKC were negatively correlated with OS, and MAOA, MUC20 and TRPM3 were positively correlated with OS (Figure 5). The expression level of these five DEGs at different stages of ccRCC showed similar trends in that the expression of both IGHA1 and IGKC were elevated in the later stages (stage III–IV) of ccRCC during which the survival rate drops dramatically, while MAOA, MUC20 and TRPM3 expression decreased in the later stages of ccRCC (Figure 6).
Figure 4.
Expression levels of the DEGs in normal para-cancerous kidney and ccRCC tumor-associated tissues in GEPIA database. Gene expression levels of (A) CCL18, (B) COL8A1, (C) HS3ST2, (D) IGHA1, (E) IGKC, (F) IGSF6, (G) MS4A4A and (H) S100A8 were significantly upregulated in ccRCC compared with normal tissues. Gene expression levels of (I) MAOA, (J) MUC20, (K) PP7080 and (L) TRPM3 were significantly downregulated in ccRCC compared with normal tissues. GEPIA, Gene Expression Profiling Interactive Analysis. DEGs, differentially expressed genes; ccRCC, clear cell renal cell carcinoma. *, P<0.05.
Figure 5.
Overall survival analysis of key genes in ccRCC (based on TCGA data in GEPIA). Expression levels of (A) IGHA1, (B) IGKC, (C) MAOA, (D) MUC20 and (E) TRPM3 were significantly correlated, either negatively or positively, with the overall survival of patients with ccRCC. GEPIA, Gene Expression Profiling Interactive Analysis; ccRCC, clear cell renal cell carcinoma; TCGA, The Cancer Genome Atlas.
Figure 6.
Gene expression levels of key genes at progressive stages of ccRCC (based on TCGA data in GEPIA) Expression levels of (A) IGHA1, (B) IGKC, (C) MAOA, (D) TRPM3 and (E) MUC20 were significantly correlated, either positively or negatively, to specific stages of ccRCC. GEPIA, Gene Expression Profiling Interactive Analysis.
Discussion
Currently, mRCC is detected in nearly 30% of newly diagnosed RCC cases. Even though the application of molecular-targeted drugs and immune checkpoint inhibitors has improved the survival of patients with mRCC, the outcome of these interventions is far from ideal.
Obesity is well recognized as a risk factor for many diseases, including type II diabetes mellitus, cardiovascular disease, hypertension, etc. (15). Epidemiological studies have also revealed the relationship between obesity and various types of human malignancies, including RCC (7,16). Approximately 3.6% of all newly diagnosed cancer cases have been connected with an increased body mass index (17), and obesity likely increases cancer mortality since a previous study showed that weight reduction reduces the risk (18). The mechanisms by which obesity causes carcinogenesis and tumor progression remain unknown. One possible reason is that adipose tissue, now viewed as one of the largest endocrine organs in human body, is believed to cause a chronic state of inflammation and to have mitogenic, proangiogenic and antiapoptotic effects (19,20). After reviewing previous studies, we realized that the mechanism and the genes involved have not been clearly defined. There is still controversy regarding the biological function of obesity-related genes in the progression and prognosis of ccRCC.
In the current study, bioinformatics methods were used to identify five genes whose expression correlate, either positively or negatively, with both ccRCC and obesity, i.e., IGHA1, IGKC, MAOA, MUC20 and TRPM3. IGHA1 (immunoglobulin heavy constant alpha 1) encodes the N-terminal sequence of the constant region the IgA1 isoform of immunoglobulin IgA. In previous studies, IGHA1 was reported to be a prognostic biomarker for better outcomes in patients with triple-negative breast cancer (21,22). In another study, it was reported to also have a relationship with altered adipose tissue homeostasis in prostate cancer and could predict high-risk cases (23). IGKC (immunoglobulin κ constant) is a polymorphic gene which is characterized by the segregation of three Km alleles—Km1, Km2, and Km3—on chromosome 2p12 (24). IGKC has been recognized as an effective prognostic biomarker for several human malignancies (25), and most notably for breast cancer (26,27). MAOA (monoamine oxidase A) is a mitochondrial enzyme, which degrades monoamine neurotransmitters and dietary amines, and produces H2O2 (28). Its biological roles in prostate cancer have been studied, including its participation in epithelial-mesenchymal transition, tumor hypoxia, and oxidative stress, which are related to high-grade Gleason score prostate cancer as well as progression and metastasis (29-31). Additionally, MAOA is involved in mediating neuroendocrine differentiation of prostate cancer cells, a feature associated with hormone-refractory prostate cancer (32). Mucin-20, encoded by MUC20, is a member of the mucin family, a group of heavily glycosylated proteins expressed by a variety of epithelial cells. Previous literature reported that the mucin family plays a critical role in a variety of malignancies, and family members have also been viewed as potential diagnostic markers and therapeutic targets. Chen et al. reported that Mucin-20 promotes aggressive and metastatic behavior of epithelial ovarian cancer by activating the integrin beta 1 signaling pathway (33). A similar result was found in colorectal cancer by Xiao et al. and in pancreatic duct carcinoma by Chen et al. (34,35). The biological function of TRPM3 (transient receptor potential melastatin 3) in ccRCC has been previously studied by Hall et al. who showed that an increase in TRPM3 expression in ccRCC correlated with Ca2+ influx, activation of CAMKK2, AMPK, and ULK1, and phagophore formation, which ultimately leads to tumor progression (36). However, in our present study, TRPM3 appears to be a tumor suppressor gene.
The present study identified five key genes involved in ccRCC progression that are also related to obesity, i.e., IGHA1 and IGKC as oncogenes and MAOA, MUC20 and TRPM3 as tumor suppressor genes. This study has the following limitations. First, the data used were downloaded from a publicly available database, and we did not validate the findings with in vivo or in vitro experiments. Second, the study was a prospective study, and we did not explore the underling mechanisms of the genes linking obesity and ccRCC. Thus, a multicenter prospective study will be conducted to evaluate the possible applications of these key genes to predict disease outcomes, and further study will focus on the related mechanisms of these genes.
In conclusion, with the help of several bioinformatics tools, the current study identified five obesity-associated genes that correlate with the progression and prognosis of ccRCC and could provide prognostic value in the treatment of ccRCC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the Three-Year-Action-Project (No. 16CR3062B) of Shanghai Shen Kang Hospital Development Center, the Medical-Engineering cross fund of Shanghai Jiao Tong University (No. YG2017QN46), and the National Natural Science Foundation of China (No. 81972369).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and the Harmonized Tripartite Guideline for Good Clinical Practice from the International Conference on Harmonization. The study was approved by the research medical ethics committee of Shanghai Renji Hospital {Ethics board approval number: Renji Ethics [2017]091}.
Footnotes
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-891
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-891). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018:68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Cairns P. Renal cell carcinoma. Cancer Biomark 2010;9:461-73. 10.3233/CBM-2011-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bex A, Albiges L, Ljungberg B, et al. Updated European Association of Urology Guidelines Regarding Adjuvant Therapy for Renal Cell Carcinoma. Eur Urol 2017;71:719-22. 10.1016/j.eururo.2016.11.034 [DOI] [PubMed] [Google Scholar]
- 4.Hidayat K, Du X, Shi BM. Body fatness at a young age and risks of eight types of cancer: systematic review and meta-analysis of observational studies. Obes Rev 2018;19:1385-94. 10.1111/obr.12705 [DOI] [PubMed] [Google Scholar]
- 5.Landberg A, Falt A, Montgomery S, et al. Overweight and obesity during adolescence increases the risk of renal cell carcinoma. Int J Cancer 2019;145:1232-7. 10.1002/ijc.32147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson M, Carreras-Torres R, Scelo G, et al. The influence of obesity-related factors in the etiology of renal cell carcinoma-A mendelian randomization study. PLoS Med 2019;16:e1002724. 10.1371/journal.pmed.1002724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capitanio U, Bensalah K, Bex A, et al. Epidemiology of Renal Cell Carcinoma. Eur Urol 2019;75:74-84. 10.1016/j.eururo.2018.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spyridopoulos TN, Petridou ET, Dessypris N, et al. Inverse association of leptin levels with renal cell carcinoma: results from a case-control study. Hormones (Athens) 2009;8:39-46. 10.14310/horm.2002.1220 [DOI] [PubMed] [Google Scholar]
- 9.Liao LM, Schwartz K, Pollak M, et al. Serum leptin and adiponectin levels and risk of renal cell carcinoma. Obesity (Silver Spring) 2013;21:1478-85. 10.1002/oby.20138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao LM, Weinstein SJ, Pollak M, et al. Prediagnostic circulating adipokine concentrations and risk of renal cell carcinoma in male smokers. Carcinogenesis 2013;34:109-12. 10.1093/carcin/bgs322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajandram R, Perumal K, Yap NY. Prognostic biomarkers in renal cell carcinoma: is there a relationship with obesity? Transl Androl Urol 2019;8:S138-S146. 10.21037/tau.2018.11.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heymsfield SB, Wadden TA. Mechanisms, Pathophysiology, and Management of Obesity. New Eng J Med 2017;376:254-66. 10.1056/NEJMra1514009 [DOI] [PubMed] [Google Scholar]
- 16.Lauby-Secretan B, Scoccianti C, Loomis D, et al. Body Fatness and Cancer--Viewpoint of the IARC Working Group. New Eng J Med 2016;375:794-8. 10.1056/NEJMsr1606602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold M, Pandeya N, Byrnes G, et al. Global burden of cancer attributable to high body-mass index in 2012: a population-based study. Lancet Oncol 2015;16:36-46. 10.1016/S1470-2045(14)71123-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodwin PJ, Stambolic V. Impact of the obesity epidemic on cancer. Ann Rev Med 2015;66:281-96. 10.1146/annurev-med-051613-012328 [DOI] [PubMed] [Google Scholar]
- 19.Iyengar NM, Gucalp A, Dannenberg AJ, et al. Obesity and Cancer Mechanisms: Tumor Microenvironment and Inflammation. J Clin Oncol 2016;34:4270-6. 10.1200/JCO.2016.67.4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng T, Lyon CJ, Bergin S, et al. Obesity, Inflammation, and Cancer. Annu Rev Pathol 2016;11:421-49. 10.1146/annurev-pathol-012615-044359 [DOI] [PubMed] [Google Scholar]
- 21.Hsu HM, Chu CM, Chang YJ, et al. Six novel immunoglobulin genes as biomarkers for better prognosis in triple-negative breast cancer by gene co-expression network analysis. Sci Rep 2019;9:4484. 10.1038/s41598-019-40826-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Li M, Hua Q, et al. Identification of an eight-lncRNA prognostic model for breast cancer using WGCNA network analysis and a Coxproportional hazards model based on L1-penalized estimation. Int J Mol Med 2019;44:1333-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangiola S, Stuchbery R, Macintyre G, et al. Periprostatic fat tissue transcriptome reveals a signature diagnostic for high-risk prostate cancer. Endocr Relat Cancer 2018;25:569-81. 10.1530/ERC-18-0058 [DOI] [PubMed] [Google Scholar]
- 24.Pandey JP, Kistner-Griffin E, Black L, et al. IGKC and FcγR genotypes and humoral immunity to HER2 in breast cancer. Immunobiology 2014;219:113-7. 10.1016/j.imbio.2013.08.005 [DOI] [PubMed] [Google Scholar]
- 25.Schmidt M, Hellwig B, Hammad S, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clin Cancer Res 2012;18:2695-703. 10.1158/1078-0432.CCR-11-2210 [DOI] [PubMed] [Google Scholar]
- 26.Denkert C, von Minckwitz G, Brase JC, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol 2015;33:983-91. 10.1200/JCO.2014.58.1967 [DOI] [PubMed] [Google Scholar]
- 27.Schmidt M, Micke P, Hengstler JG. IGKC and prognosis in breast cancer. Clin Cancer Res 2013;19:304. 10.1158/1078-0432.CCR-12-2818 [DOI] [PubMed] [Google Scholar]
- 28.Shih JC, Chen K, Ridd MJ. Monoamine oxidase: from genes to behavior. Annu Rev Neurosci 1999;22:197-217. 10.1146/annurev.neuro.22.1.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.True L, Coleman I, Hawley S, et al. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci USA 2006;103:10991-6. 10.1073/pnas.0603678103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peehl DM, Coram M, Khine H, et al. The significance of monoamine oxidase-A expression in high grade prostate cancer. J Urol 2008;180:2206-11. 10.1016/j.juro.2008.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu JB, Yin L, Shi C, et al. MAOA-Dependent Activation of Shh-IL6-RANKL Signaling Network Promotes Prostate Cancer Metastasis by Engaging Tumor-Stromal Cell Interactions. Cancer Cell 2017;31:368-82. 10.1016/j.ccell.2017.02.003 [DOI] [PubMed] [Google Scholar]
- 32.Lin YC, Chang YT, Campbell M, et al. MAOA-a novel decision maker of apoptosis and autophagy in hormone refractory neuroendocrine prostate cancer cells. Sci Rep 2017;7:46338. 10.1038/srep46338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen CH, Shyu MK, Wang SW, et al. MUC20 promotes aggressive phenotypes of epithelial ovarian cancer cells via activation of the integrin beta1 pathway. Gynecol Oncol 2016;140:131-7. 10.1016/j.ygyno.2015.11.025 [DOI] [PubMed] [Google Scholar]
- 34.Xiao X, Wang L, Wei P, et al. Role of MUC20 overexpression as a predictor of recurrence and poor outcome in colorectal cancer. J Transl Med 2013;11:151. 10.1186/1479-5876-11-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen ST, Kuo TC, Liao YY, et al. Silencing of MUC20 suppresses the malignant character of pancreatic ductal adenocarcinoma cells through inhibition of the HGF/MET pathway. Oncogene 2018;37:6041-53. 10.1038/s41388-018-0403-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall DP, Cost NG, Hegde S, et al. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell 2014;26:738-53. 10.1016/j.ccell.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as