Abstract
Background
To develop machine learning classifiers at admission for predicting which patients with coronavirus disease 2019 (COVID-19) who will progress to critical illness.
Methods
A total of 158 patients with laboratory-confirmed COVID-19 admitted to three designated hospitals between December 31, 2019 and March 31, 2020 were retrospectively collected. 27 clinical and laboratory variables of COVID-19 patients were collected from the medical records. A total of 201 quantitative CT features of COVID-19 pneumonia were extracted by using an artificial intelligence software. The critically ill cases were defined according to the COVID-19 guidelines. The least absolute shrinkage and selection operator (LASSO) logistic regression was used to select the predictors of critical illness from clinical and radiological features, respectively. Accordingly, we developed clinical and radiological models using the following machine learning classifiers, including naive bayes (NB), linear regression (LR), random forest (RF), extreme gradient boosting (XGBoost), adaptive boosting (AdaBoost), K-nearest neighbor (KNN), kernel support vector machine (k-SVM), and back propagation neural networks (BPNN). The combined model incorporating the selected clinical and radiological factors was also developed using the eight above-mentioned classifiers. The predictive efficiency of the models is validated using a 5-fold cross-validation method. The performance of the models was compared by the area under the receiver operating characteristic curve (AUC).
Results
The mean age of all patients was 58.9±13.9 years and 89 (56.3%) were males. 35 (22.2%) patients deteriorated to critical illness. After LASSO analysis, four clinical features including lymphocyte percentage, lactic dehydrogenase, neutrophil count, and D-dimer and four quantitative CT features were selected. The XGBoost-based clinical model yielded the highest AUC of 0.960 [95% confidence interval (CI): 0.913–1.000)]. The XGBoost-based radiological model achieved an AUC of 0.890 (95% CI: 0.757–1.000). However, the predictive efficacy of XGBoost-based combined model was very close to that of the XGBoost-based clinical model, with an AUC of 0.955 (95% CI: 0.906–1.000).
Conclusions
A XGBoost-based based clinical model on admission might be used as an effective tool to identify patients at high risk of critical illness.
Keywords: COVID-19, critical illness, chest CT, machine learning, prediction
Introduction
The emergence and rapid spread of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as a potentially fatal disease is a major and urgent threat to global health. As of July 24, 2020, there are more than 15.64 million confirmed cases by World Health Organization (WHO) with 636,384 deaths. The clinical spectrum of COVID-19 pneumonia ranges from mild to critically ill. Most patients of COVID-19 had mild acute respiratory infection symptoms, such as fever, dry cough, and fatigue, but some could rapidly develop fatal complications, including acute respiratory distress syndrome (ARDS) or respiratory failure, multiple organ dysfunction or failure, septic shock or even death (1). Until now, no specific treatments were recommended for COVID-19 except for meticulous supportive care (2); thus, early identification of patients with a high-risk of progression to critical illness may facilitate the provision of proper supportive treatment in advance and reduce mortality.
Some attempts have been made to develop forewarning models by taking into account possible prognostic biomarkers to predict poor outcomes in patients with COVID-19. Ji et al. established a clinical nomogram to predict progression risk in COVID-19 (3). Liu et al.identified patients at elevated risk of severe illness according to quantitative computed tomography (CT) features of pneumonia lesions in the early days (4). Liang et al. developed a clinical score consisting of 10 clinical variables at hospital admission for predicting which patients with COVID-19 will develop critical illness (5). Yan et al. developed a clinical model based on lactic dehydrogenase (LDH), lymphocyte and high-sensitivity C-reactive protein (hs-CRP) that can predict the mortality rates of COVID-19 patients >10 days in advance with >90% accuracy (6). Dong et al. developed a scoring system based on D-dimer, lymphocyte, and erythrocyte sedimentation rate to predict the severity of patients with COVID-19 (7). Wang et al. constructed clinical-laboratory model to predict in-hospital mortality of COVID-19 patients (8). However, the role of quantitative CT features has not been fully investigated and the majority of these studies follow the standard scientific methods, such as Cox regression and binary logistic regression analysis. While undeniably successful, these standard methods might have inherent limitations.
Machine learning is broadly defined as a body of computational methods/models that use patterns in data to improve performance or make accurate predictions (9). It provides a powerful set of tools to unravel the relationship between the variables and outcomes, particularly when data are nonlinear and complex (10). It is best applied when there are lots of variables and overfitting can be a problem for traditional statistical methods (10). The profusion of data requires machine learning to improve and accelerate the management of COVID-19 (11). Recent studies have identified the ability of machine learning and artificial intelligence (AI) using CT findings or radiomic/deep learning features extracted from CT images to detect, triage, and assess the severity and prognosis of COVID-19 patients (12-23). The machine learning models might serve to augment human diagnostic performance and show great potentials for assisting decision-making in the management of COVID-19 patients by assessing disease severity and predicting clinical outcomes.
Considering the machine learning method is purely data-driven, it is essential to compare multiple models for optimal prediction of a specific task (24). Therefore, the primary aims of this study are to compare the performance of multiple machine learning models based on clinical, laboratory, and radiological data for predicting critical illness in patients with COVID-19 pneumonia. Early detection of patients who are likely to develop critical illness is of great importance in the clinical settings, which may help clinicians to better choose treatment strategy and improve the use of limited resources.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-2580).
Methods
Data sources
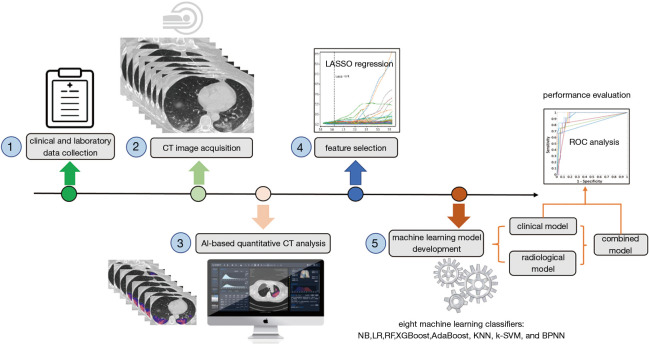
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the institutional review Board of the First Affiliated Hospital of Guangzhou Medical University (approval number: 202056); the need for informed consent was waived due to the retrospective nature of the study. The reporting follows the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) checklist (25). We included laboratory-confirmed hospitalized cases with COVID-19 admitted to three designated hospitals (Huangpi District Hospital of Traditional Chinese Medicine, Hankou Hospital of Wuhan, and The First Affiliated Hospital of Guangzhou Medical University) for COVID-19 treatment between December 31, 2019 and March 31, 2020. COVID-19 cases were confirmed by real-time reverse transcription-polymerase chain reaction (RT-PCR) assay of nasal and pharyngeal swab specimens (at least two samples were taken, at least 24 hours apart) for COVID-19 according to the protocol established by the WHO. Patients aged <18 years or patients with no available clinical/CT records or patients were critically ill on admission were excluded. Finally, 158 patients with COVID-19 were included, 123 (77.8%) were non-critical and 35 (22.2%) were critical cases. On admission, clinical data including age, sex, and comorbidities of patients were collected. The laboratory parameters, mainly including routine blood tests, coagulation profile, liver and renal function, and myocardial enzyme were examined at admission. The data in source documents were confirmed independently by two researchers. Figure 1 illustrates the workflow of this study.
Figure 1.
The framework of predicting progression to critical illness in COVID-19 patients. The workflow mainly consists of five steps: (1) clinical and laboratory data collection; (2) chest CT image acquisition; (3) AI-based quantitative CT analysis; (4) feature selection; and (5) development of clinical, radiological, and combined models using eight machine learning classifiers. The performance of models was evaluated by receiver operating characteristic curve analysis.
CT image acquisition
All patients underwent chest CT scans by a 64-slice CT scanner (Siemens Definition AS + 128, Forchheim, Germany). Each patient was scanned from the lung apex to the diaphragm during a breath-hold at the end full inspiration and at end normal-expiration. To reduce breathing artifacts, patients were instructed on breath-holding. No contrast agent was administered. CT acquisition was executed as follows: tube voltage, 120 Kilovolt (kV); tube current, auto milliampere second (mAs); pitch, 1.2; Rotation time, 0.5 s; the field of view (FOV), 330 mm ×330 mm.Lung images were reconstructed at a slice thickness of 1.0–1.25 mm using the I50 medium sharp algorithm. Lung window level and window width were set as −530–430 Hounsfield units (HU) and 1,400–1,600 HU, respectively.
Quantitative CT analysis
The quantitative analysis of lung infected by COVID-19 was performed by a care.ai Intelligent Multi-disciplinary Imaging Diagnosis Platform Intelligent Evaluation System of Chest CT for COVID-19 (YT-CT-Lung, YITU Healthcare Technology Co., Ltd., China). This system used a multi-scale convolutional neural network with adaptive thresholding and morphological operations for the segmentation of lungs and pneumonia lesions (26,27). By thresholding on CT values in the pneumonia lesions, three quantitative features were generated, including ground-glass opacities (GGO) with value ranges of −1,000–−500 HU, semi-consolidation with value ranges of −500–−250 HU and consolidation with density ranges of -250–60 HU (4).A quantitative analysis of pneumonia lesions, GGO, consolidation, and whole lungs was performed based on the segmentation results. All images were independently reviewed and assessed by two radiologists (with 10 and 20 years of experience in thoracic imaging) and discrepancies were resolved by consensus. A total of 201 quantitative CT features were extracted, which were listed below: (I) Volumes of pneumonia lesion, GGO, and consolidation in both lungs, left lung, right lung, and five lobes (n=24). (II) Volumes and percentages of pneumonia lesion, GGO, and consolidation in 18 lung segments (n=36). (III) Percentages of pneumonia volume, GGO volume, and consolidation volume in both lungs, left lung, right lung, and each lobe (n=24). (IV) CT values (mean, standard deviation, median, maximum, interquartile range) of pneumonia lesions, GGO, and consolidation in both lungs, left lung, and right lung (n=45); Hellinger distance, intersection over union (IOU), volume, CT values (mean, standard deviation, median, maximum, interquartile range) of total lung, volumes and percentages of whole lung with density of −1,000 to −700 HU, −700 to −600 HU, −600 to −500 HU, −500 to −300 HU, −300 to −200 HU, −200 to 60 HU, and 60 to 1,000 HU (n=22); herein, Hellinger distance is used to measure the similarity of two distributions. The closer the value is to 0, the higher the similarity. IOU is also called an overlap ratio, which is the ratio of the intersection and union of two distributions. Ideally, they are completely overlapping, that is, the ratio is 1.0. (V) Hellinger distance, IOU, volume, CT values (mean, standard deviation, median, maximum, interquartile range) of left lung, volumes and percentages of left lung with density of −1,000 to −700 HU, −700 to −600 HU, −600 to −500 HU, −500 to −300 HU, −300 to −200 HU, −200 to 60 HU, and 60 to 1,000 HU (n=22). (6)Hellinger distance, IOU, volume, CT values (mean, standard deviation, median, maximum, interquartile range) of the right lung, volumes and percentages of right lung with density of −1,000 to −700 HU, −700 to −600 HU, −600 to −500 HU, −500 to −300 HU, −300 to −200 HU, −200 to 60 HU, and 60 to 1,000 HU (n=22). (7) Each of the five lung lobes was scored with the following formula: 3× the volume ratio of consolidation to total lung + 2× the volume ratio of GGO to total lung (n=5). Accordingly, the total lung score was computed by summarizing the scores of five lobes (n=1).
Definition of endpoint
We defined the severity of COVID-19 according to the newest COVID-19 guidelines released by the National Health Commission of China (28) and the guidelines of the American Thoracic Society for community-acquired pneumonia (29). We defined critical illness as a composite of admission to intensive care unit (ICU), respiratory failure requiring mechanical ventilation, shock during hospitalization, or death.
Feature selection and machine learning model development
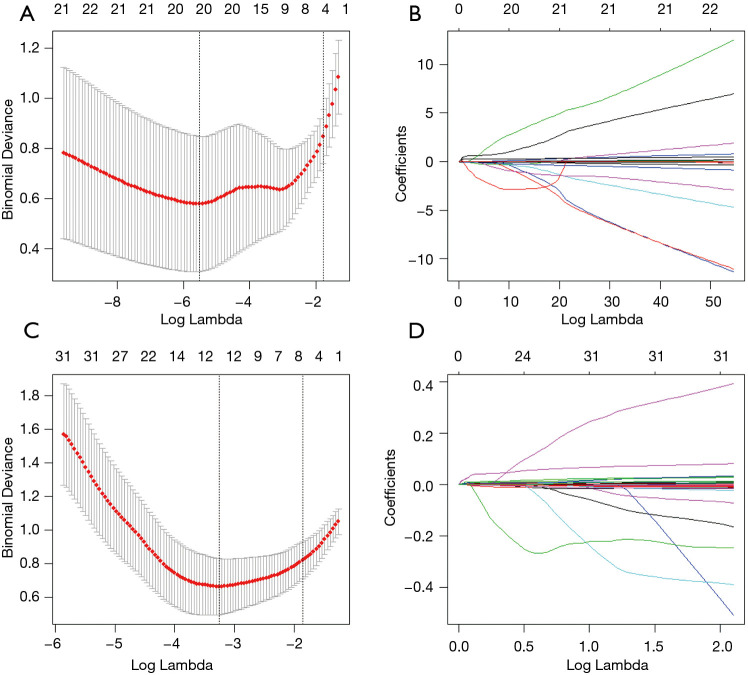
COVID-19 patients in the training dataset were included for feature selection and machine learning based model development. Imputation for missing variables was considered if missing values were less than 20%. Five laboratory variables (C-reactive protein, myohemoglobin, creatine kinase, erythrocyte sedimentation rate, and brain natriuretic peptide) with missing values >50% were excluded. Finally, a total of 27 clinical data and 201 quantitative CT features were entered into the selection process, respectively. We used mean value to impute numeric features. The least absolute shrinkage and selection operator (LASSO) logistic regression algorithm was used to select the most significant predictors from among all the candidate variables. It can minimize the potential collinearity of variables measured from the same patient and over-fitting of variables. The penalty parameter lambda was selected in the LASSO regression by 5-fold cross-validation based on the error within one standard error range of the minimum.
We firstly constructed the clinical and radiological models based on the corresponding clinical and radiological features selected by LASSO and then built the combined model based on the combination of the selected clinical and radiological features. Eight machine learning classifiers were used to develop those models for predicting critical illness, including Naive Bayes (NB), Linear Regression (LR), Random Forest (RF), Extreme Gradient Boosting (XGBoost), Adaptive Boosting (AdaBoost), K-Nearest Neighbor (KNN), Kernel Support Vector Machine (k-SVM), and Back Propagation Neural Networks (BPNN). The predictive value of the models is validated by 5-fold cross-validation. Classification performance of the machine learning models was measured using the area under the curve (AUC), F1 score, accuracy, positive predictive value (PPV), negative predictive value (NPV), sensitivity, and specificity. Machine learning models were implemented in open source Python 3X and Project Jupyter version 1.2.3 (Anaconda, Inc, https://jupyter.org/about).
Statistical analysis
Categorical variables were expressed as counts and percentages, while continuous variables are shown as mean and standard deviation (SD) or median and interquartile range. All the statistical analyses were performed using R software, version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria). The packages were used as follows: “glmnet” for LASSO logistic regression, “xgboost” for XGBoost, “adabag” for AdaBoost, “naivebayes” for NB, “mlr” for LR, “class” for KNN, “randomForest” for RF, “e1071” for SVM, and “nnet” for BPNN. Differences of clinical and laboratory characteristics between the non-critical and critical COVID-19 cases were compared using the Chi-square test or Fisher’s exact test or Mann-Whitney U test, if appropriate. The comparison of different models used the Delong test. A P<0.05 was considered significant.
Results
Clinical characteristics of patients
Among the 158 patients with COVID-19, 123 (77.8%) were non-critical cases, and 35 (22.2%) were critical cases including 12 deaths during hospitalization. The relatively high critically ill rate seen in our study was related to the fact that the First Affiliated Hospital of Guangzhou Medical University only admitted severe/critical cases transferred from other designated hospitals of Guangzhou (10 critical cases were included). The mean age of all patients was 58.9±13.9 years (range, 25–95 years), 89 of 158 patients (56.3%) were male. Fever (72.8%) was the most common symptom, followed by dry cough (67.7%), shortness of breath (48.7%) and fatigue (41.8%). 67 patients (42.4%) had at least one underlying comorbidity, with hypertension (25.3%) being the most common, followed by diabetes (13.3%) and heart diseases (8.9%). Baseline clinical and laboratory characteristics of non-critically ill and critically ill patients are shown in Table 1.
Table 1. The baseline characteristics and laboratory findings at admission.
| Non-critical (n=123) | Critical (n=35) | P value | |
|---|---|---|---|
| Age (years) | 58.2±14.4 | 61.5±11.7 | 0.213 |
| Sex | |||
| Male | 64 (52.0) | 25 (71.4) | 0.041 |
| Female | 59 (48.0) | 10 (28.6) | |
| Comorbidities | |||
| COPD | 3 (2.4) | 2 (5.7) | 0.307 |
| Heart disease | 10 (8.1) | 4 (11.4) | 0.513 |
| Hypertension | 23 (18.7) | 17 (48.6) | <0.001 |
| Diabetes | 9 (7.3) | 12 (34.3) | <0.001 |
| Malignancy | 1 (0.8) | 1 (2.9) | 0.395 |
| Cerebropathy | 3 (2.4) | 0 | 1.000 |
| Others | 23 (18.7) | 7 (20.0) | 0.863 |
| No. of comorbidities | |||
| 0 | 80 (65.0) | 11 (31.4) | <0.001 |
| 1 | 25 (20.3) | 11 (31.4) | 0.167 |
| 2 | 13 (10.6) | 9 (25.7) | 0.049 |
| 3 | 3 (2.4) | 2 (5.7) | 0.307 |
| 4 | 2 (1.6) | 2 (5.7) | 0.213 |
| WBC (×109/L) | 5.4±2.1 | 9.2±5.3 | <0.001 |
| Neutrophil (×109/L) | 4.4±6.2 | 8.2±5.2 | 0.002 |
| Neutrophil (%) | 67.0±15.4 | 86.6±6.9 | <0.001 |
| Lymphocyte (×109/L) | 1.3±1.9 | 0.6±0.3 | 0.031 |
| Lymphocyte (%) | 22.8±11.3 | 8.0±4.7 | <0.001 |
| Eosinophil (×109/L) | 0.1±0.3 | 0.01±0.02 | 0.143 |
| Eosinophil (%) | 2.3±10.2 | 0.1±0.2 | 0.227 |
| Monocyte (×109/L) | 0.4±0.2 | 0.5±0.3 | 0.186 |
| Monocyte (%) | 7.7±3.3 | 5.1±2.9 | 0.002 |
| Hemoglobin (g/L) | 126.4±19.4 | 131.9±16.0 | 0.138 |
| Platelet (g/L) | 216.4±76.7 | 172.5±56.4 | 0.001 |
| Fibrinogen (g/L) | 3.8±2.0 | 5.4±1.8 | 0.002 |
| D-dimer (μg/mL) | 20.7±102.3 | 963.9±2,241.8 | 0.011 |
| hs-CRP (mg/L) | 19.1±19.9 | 34.8±4.3 | <0.001 |
| ALT (U/L) | 26.1±17.0 | 43.0±28.2 | 0.004 |
| AST (U/L) | 25.9±14.2 | 50.3±34.6 | 0.001 |
| TBIL (μmol/L) | 12.5±7.7 | 13.4±6.4 | 0.554 |
| DBIL (μmol/L) | 6.1±19.3 | 6.0±4.2 | 0.983 |
| ALP (U/L) | 67.0±30.6 | 73.2±36.7 | 0.436 |
| LDH (U/L) | 231.8±105.9 | 458.4±161.4 | <0.001 |
| Procalcitonin (ng/mL) | 0.14±0.13 | 0.52±0.84 | 0.032 |
| Creatinine (μmol/L) | 69.7±17.8 | 91.0±46.8 | 0.020 |
| Urea nitrogen (mmol/L) | 4.6±1.6 | 7.6±4.3 | 0.001 |
Data were mean ± standard deviation (SD) or number (percentage). P values were calculated by t test, Mann-Whitney U test, χ2 test or Fisher’s exact test, as appropriate. Abbreviations: COPD, chronic obstructive pulmonary disease; CT, computed tomography; WBC, white blood cells; hs-CRP, high-sensitivity C-reactive protein; ALT, alanine transaminase; AST, aspartate aminotransferase; TBIL, total bilirubin; DBIL, direct bilirubin; ALP, alkaline phosphatase; LDH, lactate dehydrogenase.
Predictors of developing critical illness in COVID-19 patients
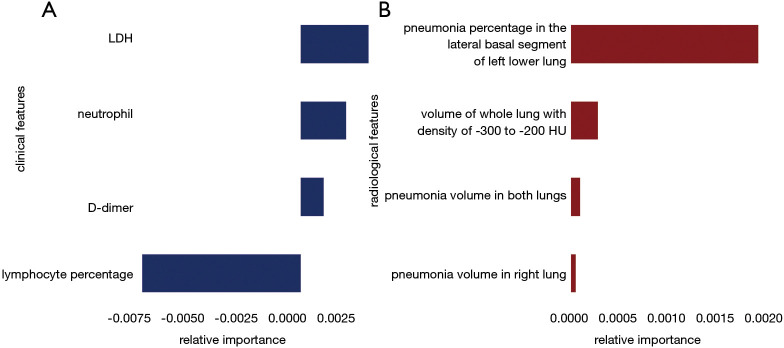
A total of 27 clinical and laboratory variables measured at hospital admission (Table 1) were included in the LASSO regression. After LASSO regression selection (Figure 2), four variables remained significant predictors of critical illness, which were ranked as lymphocyte percentage, LDH, neutrophil count, and D-dimer according to the absolute value of regression coefficient (Figure 3A). Of the 201 quantitative CT features, the vast majority of them were redundant and only four features were selected (Figure 2), which were ranked as pneumonia percentage in the lateral basal segment of left lower lung, volume of whole lung with density of −300 to −200 HU, pneumonia volume in both lungs, and pneumonia volume in right lung according to the absolute value of regression coefficient (Figure 3B). Figure 4 illustrates the CT findings and clinical parameters in two representative cases of non-critical and critical COVID-19 patients.
Figure 2.
Feature selection using the LASSO binary logistic regression model. (A) Tuning parameter (lambda) selection in the LASSO regression used 5-fold cross-validation via 1 standard error criteria, four laboratory features with non-zero coefficient were selected. (B) LASSO coefficient profiles of the 27 clinical features. (C) Tuning parameter (lambda) selection in the LASSO regression used 5-fold cross-validation via 1 standard error criteria, four quantitative CT features with non-zero coefficient were selected. (D) LASSO coefficient profiles of the 201 radiological features.
Figure 3.
Relative importance of the selected clinical (A) and radiological (B) features according to the LASSO regression coefficient.
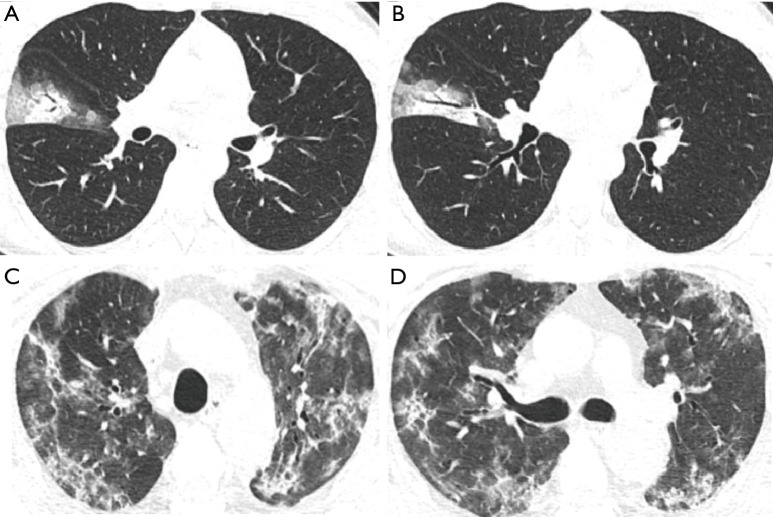
Figure 4.
Two representative cases of non-critical and critical COVID-19 patients. The non-critical case was a 25-year-old female presented with fever for one day. Her initial chest CT images show GGO and consolidation with crazy paving and air bronchogram sign in the lateral segment of right middle lobe of lung (A,B). The laboratory tests show WBC of 4.3×109/L, neutrophil of 2.7×109/L, lymphocyte count of 1.1×109/L, lymphocyte percentage of 26.1%, d-dimer of 263 µg/mL, and LDH of 47.6 U/L. The critical case was a 58-year-old male who had fever for 10 days and shortness of breath for 3 days. The admission thin-section chest CT images demonstrate extensive GGO and consolidation with crazy paving and bronchial wall thickening in both lungs (C,D). The laboratory findings show WBC of 10.2×109/L, neutrophil of 9.6×109/L, lymphocyte count of 0.2×109/L, lymphocyte percentage of 2.2%, d-dimer of 1,807 µg/mL, and LDH of 811.7 U/L.
Performance of the clinical model, radiological model, and combined model
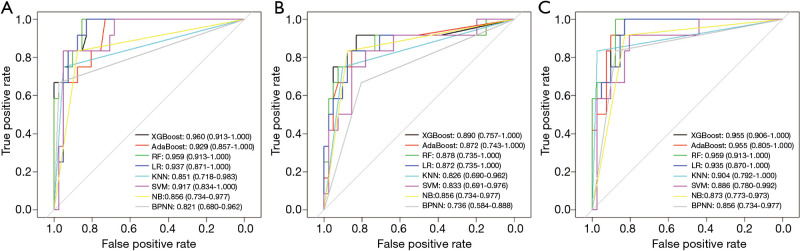
Machine learning models were formulated according to the above risk factors associated with critical illness, and validated by internal bootstrap validation. Tables 2-4 and Figure 5A,B,C show the predictive performance of eight classifiers in the clinical, radiological, and combined models, respectively. In the validation phase of the clinical model (Table 2 and Figure 5A), the AUCs of eight machine learning classifiers ranged from 0.821 to 0.960. The AUCs of XGBoost, AdaBoost, RF, LR, and SVM exceeded 0.900. The SVM showed the highest discriminatory powers of AUC of 0.960 (95% CI: 0.913–1.000), with sensitivity of 100.0% (95% CI: 83.3–100.0%), specificity of 87.8% (95% CI: 75.6–100.0%), accuracy of 90.6% (95% CI: 81.1–98.1%), F1 score of 82.8% (95% CI: 65.9–100.0%), PPV of 70.6% (54.5–100.0%), and NPV of 100.0% (95.1–100.0%). In the validation phase of radiological model (Table 3 and Figure 5B), the AUCs of all classifiers exceed 0.800 except BNPP. The XGBoost-based model achieved an AUC of 0.890 (95% CI: 0.757–1.000), sensitivity of 91.7% (95% CI: 66.7–100.0%), specificity of 90.2% (95% CI: 75.6–100.0%), accuracy of 90.6% (95% CI: 77.4–96.2%), F1 score of 80.3% (95% CI: 57.1–100.0%), PPV of 71.4% (95% CI: 50.0–100.0%), and NPV of 97.2% (95% CI: 90.7–100.0%). In the validation phase of combined model (Table 4 and Figure 5C), the AUCs of eight classifiers ranged from 0.856 to 0.959. The XGBoost-based combined model performed similarly with the XGBoost-based clinical model, with an AUC of 0.955 (95% CI: 0.906–1.000), sensitivity of 100.0% (91.7–100.0%), specificity of 87.8% (75.6–97.6%), accuracy of 90.6% (81.1–98.1%), F1 score of 82.8% (95% CI: 68.4–96.0%), PPV of 70.6% (54.5–92.3%), and NPV of 100.0% (97.1–100.0%). The clinical model outperformed the radiological model in predicting the risk of developing critical illness in patients with COVID-19, however, with no significant difference (P=0.330). Adding the quantitative CT features to the clinical model achieved no significant improvement (P=0.763).
Table 2. Comparison of clinical model based on eight machine learning classifiers in predicting critical illness among patients with COVID-19.
| Classifiers | Measured metrics | ||||||
|---|---|---|---|---|---|---|---|
| AUC (95% CI) | Accuracy% (95% CI) | F1 score (95% CI) | PPV% (95% CI) | NPV% (95% CI) | Specificity% (95% CI) | Sensitivity% (95% CI) | |
| XGBoost | 0.960 (0.913–1.000) | 90.6 (81.1–98.1) | 82.8 (65.9–100.0) | 70.6 (54.5–100.0) | 100.0 (95.1–100.0) | 87.8 (75.6–100.0) | 100.0 (83.3–100.0) |
| AdaBoost | 0.929 (0.857–1.000) | 84.9 (71.7–98.1) | 75.0 (53.3–100.0) | 60.0 (44.4–100.0) | 100.0 (91.1–100.0) | 80.5 (63.4–100.0) | 100.0 (66.7–100.0) |
| RF | 0.959 (0.913–1.000) | 90.6 (81.1–98.1) | 82.8 (68.4–100.0) | 70.6 (54.5–100.0) | 100.0 (97.1–100.0) | 87.8 (75.6–100.0) | 100.0 (91.7–100.0) |
| LR | 0.937 (0.871–1.000) | 90.6 (81.1–98.1) | 82.8 (68.4–96.0) | 70.6 (54.5–92.3) | 100.0 (97.1–100.0) | 87.8 (75.6–97.6) | 100.0 (91.7–100.0) |
| KNN | 0.851 (0.718–0.983) | 90.6 (83.0–98.1) | 78.9 (55.6–100.0) | 83.3 (62.5–100.0) | 92.9 (86.7–100.0) | 95.1 (87.8–100.0) | 75.0 (50.0–100.0) |
| SVM | 0.917 (0.834–1.000) | 92.5 (73.6–98.1) | 86.5 (57.1–100.0) | 81.8 (46.2–100.0) | 97.4 (92.7–100.0) | 95.1 (65.9–100.0) | 91.7 (75.0–100.0) |
| NB | 0.856 (0.734–0.977) | 86.8 (77.4–94.3) | 74.1 (53.8–94.7) | 66.7 (50.0–90.0) | 94.9 (87.8–100.0) | 87.8 (75.6–97.6) | 83.3 (58.3–100.0) |
| BPNN | 0.821 (0.680–0.962) | 90.6 (83.0–96.2) | 76.6 (52.0–95.7) | 90.0 (69.2–100.0) | 90.9 (84.8–97.6) | 97.6 (92.7–100.0) | 66.7 (41.7–91.7) |
The confusion matrix in our study was given as a 2×2 contingency table that reported the number of true positives, false positives, false negatives, and true negatives. Sensitivity = true positives/(true positives + false negatives) ×100%. Specificity = True negatives/(true negatives + false positives) ×100%. Accuracy = (true positives + true negatives)/n ×100%. The F1 score is equivalent to harmonic mean of the precision and recall, where the best value is 1.0 and the worst value is 0.0. The formula for F1 score is: F1 =2 * (precision * recall)/(precision + recall), precision = true positives/(true positives + false positives), recall = true positives/(true positives + false negatives). PPV was the probability that the disease was present when the test was positive (expressed as a percentage). NPV was the probability that the disease was not present when the test was negative (expressed as a percentage). The ROC curve was created by plotting the true positive rate (sensitivity) against the false positive rate (1-sensitivity). By varying the predicted probability threshold, we calculated AUC values. We calculated 95% CIs with the bootstrap (100 iterations) method. AUC, area under the curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; NB, Naive Bayes; LR, Linear Regression; RF, Random Forest; XGBoost, Extreme Gradient Boosting; AdaBoost, Adaptive Boosting; KNN, K-Nearest Neighbor; k-SVM, Kernel Support Vector Machine; BPNN, Back Propagation Neural Networks.
Table 3. Comparison of radiological model based on eight machine learning classifiers in predicting critical illness among patients with COVID-19.
| Classifiers | Measured metrics | ||||||
|---|---|---|---|---|---|---|---|
| AUC (95% CI) | Accuracy% (95% CI) | F1 score (95% CI) | PPV% (95% CI) | NPV% (95% CI) | Specificity% (95% CI) | Sensitivity% (95% CI) | |
| XGBoost | 0.890 (0.757–1.000) | 90.6 (77.4–96.2) | 80.3 (57.1–100.0) | 71.4 (50.0–100.0) | 97.2 (90.7–100.0) | 90.2 (75.6–100.0) | 91.7 (66.7–100.0) |
| AdaBoost | 0.872 (0.743–1.000) | 86.8 (71.7–96.2) | 77.2 (53.3–95.7) | 66.7 (44.4–91.7) | 96.5 (89.5–100.0) | 87.8 (65.9–97.6) | 91.7 (66.7–100.0) |
| RF | 0.878 (0.735–1.000) | 88.7 (75.5–96.2) | 76.9 (55.6–100.0) | 71.4 (47.6–100.0) | 95.3 (89.7–100.0) | 90.2 (70.7–100.0) | 83.3 (66.7–100.0) |
| LR | 0.872 (0.735–1.000) | 86.8 (73.6–96.2) | 78.6 (54.3–96.0) | 68.8 (45.8–92.3) | 96.6 (89.7–100.0) | 87.8 (68.3–97.6) | 91.7 (66.7–100.0) |
| KNN | 0.826 (0.690–0.962) | 86.8 (77.4–94.3) | 72.4 (50.0–95.2) | 70.0 (50.0–90.9) | 92.5 (86.0–100.0) | 90.2 (80.5–97.6) | 75.0 (50.0–100.0) |
| SVM | 0.833 (0.691–0.976) | 83.0 (67.9–92.5) | 68.3 (47.5–94.7) | 57.9 (40.0–90.0) | 94.9 (88.2–100.0) | 82.9 (61.0–97.6) | 83.3 (58.3–100.0) |
| NB | 0.856 (0.734–0.977) | 86.8 (77.4–96.2) | 74.1 (53.8–94.7) | 66.7 (50.0–90.0) | 94.9 (88.1–100.0) | 87.8 (78.0–97.6) | 83.3 (58.3–100.0) |
| BPNN | 0.736 (0.584–0.888) | 77.4 (66.0–88.7) | 57.1 (37.0–81.1) | 50.0 (33.3–72.7) | 89.5 (81.8–97.1) | 80.5 (68.3–92.7) | 66.7 (41.7–91.7) |
The confusion matrix in our study was given as a 2×2 contingency table that reported the number of true positives, false positives, false negatives, and true negatives. Sensitivity = true positives/(true positives + false negatives) ×100%. Specificity = True negatives/(true negatives + false positives) ×100%. Accuracy = (true positives + true negatives)/n ×100%. The F1 score is equivalent to harmonic mean of the precision and recall, where the best value is 1.0 and the worst value is 0.0. The formula for F1 score is: F1 =2 * (precision * recall)/(precision + recall), precision = true positives/(true positives + false positives), recall = true positives/(true positives + false negatives). PPV was the probability that the disease was present when the test was positive (expressed as a percentage). NPV was the probability that the disease was not present when the test was negative (expressed as a percentage). The ROC curve was created by plotting the true positive rate (sensitivity) against the false positive rate (1-sensitivity). By varying the predicted probability threshold, we calculated AUC values. We calculated 95% CIs with the bootstrap (100 iterations) method. AUC, area under the curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; NB, Naive Bayes; LR, Linear Regression; RF, Random Forest; XGBoost, Extreme Gradient Boosting; AdaBoost, Adaptive Boosting; KNN, K-Nearest Neighbor; k-SVM, Kernel Support Vector Machine; BPNN, Back Propagation Neural Networks.
Table 4. Comparison of combined model based on eight machine learning classifiers in predicting critical illness among patients with COVID-19.
| Classifiers | Measured metrics | ||||||
|---|---|---|---|---|---|---|---|
| AUC (95% CI) | Accuracy% (95% CI) | F1 score (95% CI) | PPV% (95% CI) | NPV% (95% CI) | Specificity% (95% CI) | Sensitivity% (95% CI) | |
| XGBoost | 0.955 (0.906–1.000) | 90.6 (81.1–98.1) | 82.8 (68.4–96.0) | 70.6 (54.5–92.3) | 100.0 (97.1–100.0) | 87.8 (75.6–97.6) | 100.0 (91.7–100.0) |
| AdaBoost | 0.955 (0.905–1.000) | 92.5 (83.0–98.1) | 85.7 (70.4–96.0) | 75.0 (57.1–92.3) | 100.0 (97.6–100.0) | 90.2 (78.0–97.6) | 100.0 (91.7–100.0) |
| RF | 0.959 (0.913–1.000) | 90.6 (83.0–98.1) | 82.8 (70.4–100.0) | 70.6 (57.1–100.0) | 100.0 (97.6–100.0) | 87.8 (78.0–100.0) | 100.0 (91.7–100.0) |
| LR | 0.935 (0.870–1.000) | 88.7 (79.2–96.2) | 80.0 (66.5–95.7) | 66.7 (52.2–91.7) | 100.0 (97.1–100.0) | 85.4 (73.2–97.6) | 100.0 (91.7–100.0) |
| KNN | 0.904 (0.792–1.000) | 94.3 (86.8–100.0) | 87.0 (65.6–100.0) | 90.9 (75.0–100.0) | 95.2 (88.9–100.0) | 97.6 (92.7–100.0) | 83.3 (58.3–100.0) |
| SVM | 0.886 (0.780–0.992) | 84.9 (73.6–94.3) | 74.3 (54.1–100.0) | 62.5 (45.5–100.0) | 97.1 (90.7–100.0) | 82.9 (68.3–100.0) | 91.7 (66.7–100.0) |
| NB | 0.873 (0.773–0.973) | 84.9 (75.5–94.3) | 73.3 (58.1–88.9) | 61.1 (47.4–80.0) | 97.2 (91.4–100.0) | 82.9 (70.7–92.7) | 91.7 (75.0–100.0) |
| BPNN | 0.856 (0.734–0.977) | 86.8 (77.4–94.3) | 74.1 (53.8–94.7) | 66.7 (50.0–90.0) | 94.9 (88.1–100.0) | 87.8 (78.0–97.6) | 83.3 (58.3–100.0) |
The confusion matrix in our study was given as a 2×2 contingency table that reported the number of true positives, false positives, false negatives, and true negatives. Sensitivity = true positives/(true positives + false negatives) ×100%. Specificity = True negatives/(true negatives + false positives) ×100%. Accuracy = (true positives + true negatives)/n ×100%. The F1 score is equivalent to harmonic mean of the precision and recall, where the best value is 1.0 and the worst value is 0.0. The formula for F1 score is: F1 =2 * (precision * recall)/(precision + recall), precision = true positives/(true positives + false positives), recall = true positives/(true positives + false negatives). PPV was the probability that the disease was present when the test was positive (expressed as a percentage). NPV was the probability that the disease was not present when the test was negative (expressed as a percentage). The ROC curve was created by plotting the true positive rate (sensitivity) against the false positive rate (1-sensitivity). By varying the predicted probability threshold, we calculated AUC values. We calculated 95% CIs with the bootstrap (100 iterations) method. AUC, area under the curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; NB, Naive Bayes; LR, Linear Regression; RF, Random Forest; XGBoost, Extreme Gradient Boosting; AdaBoost, Adaptive Boosting; KNN, K-Nearest Neighbor; k-SVM, Kernel Support Vector Machine; BPNN, Back Propagation Neural Networks.
Figure 5.
Receiver operating characteristic curve analyses of eight machine learning classifiers in predicting critical illness among COVID-19 patients. (A) clinical model; (B) radiological model; and (C) combined model.
Discussion
In this study, we developed and validated multiple machine learning models to predict the risk of developing critical illness among patients hospitalized for COVID-19 pneumonia. The results demonstrated that the clinical model including decreased lymphocyte percentage, increased LDH, neutrophil count, and D-dimer could achieve the highest performance in predicting critical illness in COVID-19 patients, with an AUC of 0.960 (95% CI: 0.913–1.000) and accuracy of 90.6% (95% CI: 81.1–98.1%).
Currently, predicted risk factors associated with a fatal outcome have been often identified from clinical and laboratory parameters. Although the COVID-19 more likely infected older males with pre-existing comorbidities, they were not good predictors of developing critical illness. Previous studies have determined many risk factors related to disease severity or poor prognosis using traditional statistical methods or LASSO regression (3-8). In fact, the identification of predictors depends on available features, feature selection method used and sample size of studies. Our findings showed that lymphocyte percentage, LDH, neutrophil count, and D-dimer were four significant predictors of severity of COVID-19. Lymphocytopenia was a prominent feature of patients with COVID-19 because targeted invasion by viral particles damages the cytoplasmic component of the lymphocyte and causes its destruction, which may reflect the severity of COVID-19 (2). In this study, lymphocyte percentage seems to play the most crucial role in prediction of critical illness of COVID-19. For critically ill patients with COVID-19, the rise in LDH level indicates an increase of the activity and extent of lung injury (30). Neutrophilia is one of the biomarkers of acute infection. Neutrophils are recruited early to sites of infection where they kill pathogens (bacteria, fungi, and viruses) by oxidative burst and phagocytosis (31). Some literature supported the hypothesis that a little known yet powerful function of neutrophils—the ability to form neutrophil extracellular traps—may contribute to organ damage and death in COVID-19 (32). Neutrophil count, either individually or paired in a ratio with lymphocytes, also predicts disease severity in COVID-19 patients (33-35). Elevation of D-dimer indicated a hypercoagulable state in patient with COVID-19, which was an independent predictor of requiring critical care support or in-hospital mortality (36). Our SVM-based clinical model selected the above four biomarkers that predict the critical illness of individual patients in advance with accuracy of more than 90%.
Chest CT plays an indispensable role in the detection, diagnosis, and follow-up of COVID-19 pneumonia (37). Visual CT findings such as GGO, consolidation, crazy paving, and bronchial wall thickening are key clues to COVID-19. However, chest CT images are usually visually interpreted by radiologists in the clinical setting, which is somewhat subjective with large variability that unable to quantitatively assess the disease severity and is also time-consuming and labor-intensive. Recently, many studies used AI algorithms integrate chest CT findings with or without other variables, such as clinical symptoms, exposure history, and laboratory testing to rapidly diagnose COVID-19 (15-18,38-54). Also, other studies have used quantitative CT features derived from artificial intelligence to quantify pneumonia lesions and the risk of poor outcomes in patients with COVID-19 (4,19-22,55-61). In particular, Yin et al. concluded that quantitative CT features were superior to that of a semiquantitative visual CT score in the assessment of the severity of COVID-19 (60). Liu et al. found that quantitative CT features on day 0 and day 4 could predict the progression to severe illness in COVID-19 patients, which outperformed the acute physiology and chronic health evaluation II score, neutrophil-to-lymphocyte ratio, and D-dimer (4). Yu et al. observed that larger consolidation lesions in the upper lung on admission CT would increase the risk of poor prognosis in COVID-19 patients (61). In this study, although the XGBoost-based radiological model achieved a good accuracy in predicting the risk of developing critical illness in patients with COVID-19, it was hard to provide additional improvement to the XGBoost-based clinical model, maybe due to the high enough performance of the clinical model.
This study also has some potential limitations. Firstly, the retrospective nature of this study with a relatively small sample size. Secondly, the data for machine learning training and validation were all from China, which could limit the generalizability of the models in other areas of the world. Therefore, other validations of the proposed models outside China would be helpful. Thirdly, our AI system has not evaluated the radiological features (such as crazy paving, lymphadenopathy, bronchial wall thickening, and pleural effusion) extracting by radiologists (38,62,63), which may help to improve the model performance. However, the CT findings are mainly used to diagnose COVID-19 not to predict the outcome of COVID-19. Finally, future external validation is needed to identify the generalizability of our machine learning models. Although the external validation was not performed due to insufficient data for machine learning, the testing results of our clinical model might be good because it was built by four simple and strong predictors that proven in previous studies.
In conclusion, in this study, we identified the SVM-based clinical model with lymphocyte percentage, LDH, neutrophil count, and D-dimer as the optimal tool to estimate the risk of developing critical illness among patients with COVID-19. Early detection of patients who are likely to develop critical illness is of great importance in the clinical settings, which may help select patients at risk of rapid deterioration who should require high-level monitoring. If a patient’s predicted risk for critical illness is low, regular monitoring may be enough, whereas high-risk patients might need aggressive treatment or ICU care. However, large-scale prospective studies in the future are warranted to validate the effectiveness of our proposed machine learning models.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the institutional review Board of the First Affiliated Hospital of Guangzhou Medical University (approval number: 202056); the need for informed consent was waived due to the retrospective nature of the study.
Footnotes
Reporting Checklist: The authors have completed the STROBE reporting checklist, available at http://dx.doi.org/10.21037/jtd-20-2580
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-2580
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-2580). All authors have no conflicts of interest to declare.
References
- 1.Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 2020;323:1239-42. 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- 2.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507-13. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji D, Zhang D, Xu J, et al. Prediction for Progression Risk in Patients with COVID-19 Pneumonia: the CALL Score. Clin Infect Dis 2020;71:1393-9. 10.1093/cid/ciaa414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu F, Zhang Q, Huang C, et al. CT quantification of pneumonia lesions in early days predicts progression to severe illness in a cohort of COVID-19 patients. Theranostics 2020;10:5613-22. 10.7150/thno.45985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang W, Liang H, Ou L, et al. Development and Validation of a Clinical Risk Score to Predict the Occurrence of Critical Illness in Hospitalized Patients With COVID-19. JAMA Intern Med 2020;180:1081-9. 10.1001/jamainternmed.2020.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan L, Zhang H, Goncalves J, et al. An interpretable mortality prediction model for COVID-19 patients. Nat Mach Intell 2020;2:283-8. 10.1038/s42256-020-0180-7 [DOI] [Google Scholar]
- 7.Dong Y, Zhou H, Li M, et al. A novel simple scoring model for predicting severity of patients with SARS-CoV-2 infection. Transbound Emerg Dis 2020;67:2823-9. 10.1111/tbed.13651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Zuo P, Liu Y, et al. Clinical and laboratory predictors of in-hospital mortality in patients with COVID-19: a cohort study in Wuhan, China. Clin Infect Dis 2020;71:2079-88. 10.1093/cid/ciaa538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol 2019;20:e262-e273. 10.1016/S1470-2045(19)30149-4 [DOI] [PubMed] [Google Scholar]
- 10.Nicholls M. Machine Learning-state of the art. Eur Heart J 2019;40:3668-9. 10.1093/eurheartj/ehz801 [DOI] [PubMed] [Google Scholar]
- 11.Peiffer-Smadja N, Maatoug R, Lescure F, et al. Machine Learning for COVID-19 needs global collaboration and data-sharing. Nat Mach Intell 2020;2:293-4. 10.1038/s42256-020-0181-6 [DOI] [Google Scholar]
- 12.Lalmuanawma S, Hussain J, Chhakchhuak L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: A review. Chaos Solitons Fractals 2020;139:110059. 10.1016/j.chaos.2020.110059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Q, Shuo W, Liang L, et al. Radiomics Analysis of Computed Tomography helps predict poor prognostic outcome in COVID-19. Theranostics 2020;10:7231-44. 10.7150/thno.46428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Q, Du SY, Gao S, et al. A model based on CT radiomic features for predicting RT-PCR becoming negative in coronavirus disease 2019 (COVID-19) patients. BMC Med Imaging 2020;20:118. 10.1186/s12880-020-00521-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attallah O, Ragab DA, Sharkas M. MULTI-DEEP: A novel CAD system for coronavirus (COVID-19) diagnosis from CT images using multiple convolution neural networks. PeerJ 2020;8:e10086. 10.7717/peerj.10086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin C, Chen W, Cao Y, et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat Commun 2020;11:5088. 10.1038/s41467-020-18685-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmon SA, Sanford TH, Xu S, et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat Commun 2020;11:4080. 10.1038/s41467-020-17971-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song J, Wang H, Liu Y, et al. End-to-end automatic differentiation of the coronavirus disease 2019 (COVID-19) from viral pneumonia based on chest CT. Eur J Nucl Med Mol Imaging 2020;47:2516-24. 10.1007/s00259-020-04929-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haimovich AD, Ravindra NG, Stoytchev S, et al. Development and Validation of the Quick COVID-19 Severity Index: A Prognostic Tool for Early Clinical Decompensation. Ann Emerg Med 2020;76:442-53. 10.1016/j.annemergmed.2020.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai W, Liu T, Xue X, et al. CT Quantification and Machine-learning Models for Assessment of Disease Severity and Prognosis of COVID-19 Patients. Acad Radiol 2020;27:1665-78. 10.1016/j.acra.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Zhang Q, Tan Y, et al. Prediction of COVID-19 Severity from Chest CT and Laboratory Measurements: Evaluation of a Machine Learning Approach. JMIR Med Inform 2020;8:e21604. 10.2196/21604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue H, Yu Q, Liu C, et al. Machine learning-based CT radiomics method for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: a multicenter study. Ann Transl Med 2020;8:859. 10.21037/atm-20-3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma X, Ng M, Xu S, et al. Development and validation of prognosis model of mortality risk in patients with COVID-19. Epidemiol Infect 2020;148:e168. 10.1017/S0950268820001727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo DP, Zorn KM, Clark AM, et al. Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction. Mol Pharm 2018;15:4361-70. 10.1021/acs.molpharmaceut.8b00546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. PLoS Med 2007;4:e296. 10.1371/journal.pmed.0040296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention: Springer; 2015. p. 234-41. [Google Scholar]
- 27.Wang S, Zhou M, Liu Z, Liu Z, Gu D, Zang Y, et al. Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Med Image Anal 2017;40:172-83. 10.1016/j.media.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guidelines for the diagnosis and treatment of novel coronavirus (2019-nCoV) infection (trial version 7) (in Chinese). National Health Commission of the People's Republic of China. March 04, 2020; doi: 10.7661/j.cjim.20200202.064. [DOI]
- 29.Metlay JP, Waterer GW, Long AC, et al. Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 2019;200:e45-e67. 10.1164/rccm.201908-1581ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishaba T, Tamaki H, Shimaoka Y, et al. Staging of acute exacerbation in patients with idiopathic pulmonary fibrosis. Lung 2014;192:141-9. 10.1007/s00408-013-9530-0 [DOI] [PubMed] [Google Scholar]
- 31.Schönrich G, Raftery MJ. Neutrophil Extracellular Traps Go Viral. Front. Immunol. 2016;7:366. 10.3389/fimmu.2016.00366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barnes BJ, Adrover JM, Baxter-Stoltzfus A, et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J Exp Med 2020;217:e20200652. 10.1084/jem.20200652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Yang L, Gui S, et al. Association of clinical and radiographic findings with the outcomes of 93 patients with COVID-19 in Wuhan, China. Theranostics 2020;10:6113-21. 10.7150/thno.46569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Liu Y, Xiang P, et al. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J Transl Med 2020;18:206. 10.1186/s12967-020-02374-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan X, Li F, Wang X, et al. Neutrophil to lymphocyte ratio as prognostic and predictive factor in patients with coronavirus disease 2019: A retrospective cross-sectional study. J Med Virol 2020;92:2573-81. 10.1002/jmv.26061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Yan X, Fan Q, et al. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J Thromb Haemost. 2020;18:1324-9. 10.1111/jth.14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Yu H, Zhang S. The indispensable role of chest CT in the detection of coronavirus disease 2019 (COVID-19). Eur J Nucl Med Mol Imaging 2020;47:1638-9. 10.1007/s00259-020-04795-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abbasian Ardakani A, Acharya UR, Habibollahi S, et al. COVIDiag: a clinical CAD system to diagnose COVID-19 pneumonia based on CT findings. Eur Radiol 2021;31:121-30. 10.1007/s00330-020-07087-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang H, Xia L, Yan F, et al. Diagnosis of Coronavirus Disease 2019 (COVID-19) With Structured Latent Multi-View Representation Learning. IEEE Trans Med Imaging 2020;39:2606-14. 10.1109/TMI.2020.2992546 [DOI] [PubMed] [Google Scholar]
- 40.Mei X, Lee HC, Diao KY, et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med 2020;26:1224-8. 10.1038/s41591-020-0931-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren HW, Wu Y, Dong JH, et al. Analysis of clinical features and imaging signs of COVID-19 with the assistance of artificial intelligence. Eur Rev Med Pharmacol Sci 2020;24:8210-8. [DOI] [PubMed] [Google Scholar]
- 42.Zhang K, Liu X, Shen J, et al. Clinically Applicable AI System for Accurate Diagnosis, Quantitative Measurements, and Prognosis of COVID-19 Pneumonia Using Computed Tomography. Cell 2020;181:1423-1433.e11. 10.1016/j.cell.2020.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ardakani AA, Kanafi AR, Acharya UR, et al. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput Biol Med 2020;121:103795. 10.1016/j.compbiomed.2020.103795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ko H, Chung H, Kang WS, et al. COVID-19 Pneumonia Diagnosis Using a Simple 2D Deep Learning Framework With a Single Chest CT Image: Model Development and Validation. J Med Internet Res 2020;22:e19569. 10.2196/19569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Zhong Z, Li Y, et al. From community-acquired pneumonia to COVID-19: a deep learning-based method for quantitative analysis of COVID-19 on thick-section CT scans. Eur Radiol 2020;30:6828-37. 10.1007/s00330-020-07042-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Lure FYM, Miao H, et al. Using artificial intelligence to assist radiologists in distinguishing COVID-19 from other pulmonary infections. J Xray Sci Technol, 2020. [Epub ahead of print]. doi: 10.3233/XST-200735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou L, Li Z, Zhou J, et al. A Rapid, Accurate and Machine-Agnostic Segmentation and Quantification Method for CT-Based COVID-19 Diagnosis. IEEE Trans Med Imaging 2020;39:2638-52. 10.1109/TMI.2020.3001810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pham TD. A comprehensive study on classification of COVID-19 on computed tomography with pretrained convolutional neural networks. Sci Rep 2020;10:16942. 10.1038/s41598-020-74164-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X, Hui H, Niu M, et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: A multicentre study. Eur J Radiol 2020;128:109041. 10.1016/j.ejrad.2020.109041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang B, Jin S, Yan Q, et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system. Appl Soft Comput 2021;98:106897. 10.1016/j.asoc.2020.106897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan T, Wong PK, Ren H, et al. Automatic distinction between COVID-19 and common pneumonia using multi-scale convolutional neural network on chest CT scans. Chaos Solitons Fractals 2020;140:110153. 10.1016/j.chaos.2020.110153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu X, Jiang X, Ma C, et al. A Deep Learning System to Screen Novel Coronavirus Disease 2019 Pneumonia. Engineering (Beijing) 2020;6:1122-9. 10.1016/j.eng.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Javor D, Kaplan H, Kaplan A, et al. Deep learning analysis provides accurate COVID-19 diagnosis on chest computed tomography. Eur J Radiol 2020;133:109402. 10.1016/j.ejrad.2020.109402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang SH, Govindaraj VV, Górriz JM, et al. Covid-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inf Fusion 2021;67:208-29. 10.1016/j.inffus.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura-Sandoval Y, Arévalo-Molina ME, Cristancho-Rojas CN, et al. Validation of Chest Computed Tomography Artificial Intelligence to Determine the Requirement for Mechanical Ventilation and Risk of Mortality in Hospitalized Coronavirus Disease-19 Patients in a Tertiary Care Center In Mexico City. Rev Invest Clin 2020. [Epub ahead of print]. doi: 10.24875/RIC.20000451 [DOI] [PubMed] [Google Scholar]
- 56.Salvatore C, Roberta F, Angela L, et al. Clinical and laboratory data, radiological structured report findings and quantitative evaluation of lung involvement on baseline chest CT in COVID-19 patients to predict prognosis. Radiol Med 2021;126:29-39. 10.1007/s11547-020-01293-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao LS, Li P, Sun F, et al. Development and Validation of a Deep Learning-Based Model Using Computed Tomography Imaging for Predicting Disease Severity of Coronavirus Disease 2019. Front Bioeng Biotechnol 2020;8:898. 10.3389/fbioe.2020.00898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lessmann N, Sánchez CI, Beenen L, et al. Automated Assessment of CO-RADS and Chest CT Severity Scores in Patients with Suspected COVID-19 Using Artificial Intelligence. Radiology 2021;298:E18-E28. 10.1148/radiol.2020202439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanza E, Muglia R, Bolengo I, et al. Quantitative chest CT analysis in COVID-19 to predict the need for oxygenation support and intubation. Eur Radiol 2020;30:6770-8. 10.1007/s00330-020-07013-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin X, Min X, Nan Y, et al. Assessment of the Severity of Coronavirus Disease: Quantitative Computed Tomography Parameters versus Semiquantitative Visual Score. Korean J Radiol 2020;21:998-1006. 10.3348/kjr.2020.0423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu Q, Wang Y, Huang S, et al. Multicenter cohort study demonstrates more consolidation in upper lungs on initial CT increases the risk of adverse clinical outcome in COVID-19 patients. Theranostics 2020;10:5641-8. 10.7150/thno.46465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prokop M, van Everdingen W, van Rees Vellinga T, et al. CO-RADS: A Categorical CT Assessment Scheme for Patients Suspected of Having COVID-19-Definition and Evaluation. Radiology 2020;296:E97-E104. 10.1148/radiol.2020201473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Dong C, Hu Y, et al. Temporal Changes of CT Findings in 90 Patients with COVID-19 Pneumonia: A Longitudinal Study. Radiology 2020;296:E55-E64. 10.1148/radiol.2020200843 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as