Abstract
Background
Lung cancer is a chronic, progressive and malignant disease associated with ever-growing incidence and mortality. Targeted therapy plays an important role in the clinical treatment of lung cancer. Besides, neuron-specific enolase (NSE), an intracellular enzyme, is highly correlated with the targeted treatment outcome in patients with non-small cell lung cancer (NSCLC). The present study aimed to explore the correlation of NSE with the detection of gene mutations.
Methods
It is a case-control study. From June 2017 to October 2019, the newly diagnosed patients with lung adenocarcinoma were enrolled from the First Affiliated Hospital of Anhui Medical University. Next-generation sequencing (NGS) was conducted in these patients. Kruskal-Wallis test was used to calculate the difference in NSE levels between mutant and non-mutant group and the differences were compared between blood and tissue samples.
Results
Compared with patients with no gene mutation (15.4±7.8 mmol/L), the NSE levels in patients with gene mutations were remarkably increased in blood sample group (22.2±12.9 mmol/L) (P<0.05). Besides, the linear regression model was applied for analysis which further emphasized the close relationship between them. The area under the ROC curve (AUC) of NSE was 0.7300 [95% confidence interval (CI): 0.6059–0.8541] and optimal threshold was 18.5650 U/mL with a sensitivity of 87.50% and a specificity of 52.08%. In addition, NSE levels increased in blood sample group, suggesting that the occurrence of polygenic mutation with dismal prognosis, but no correlation was detected in tissue sample group.
Conclusions
This study elucidates the functional role of NSE, and findings in this study notably increase the gene detection efficiency for lung adenocarcinoma.
Keywords: Adenocarcinoma of lung, neuron-specific enolase (NSE), genetic testing, next-generation sequencing (NGS)
Introduction
According to the American Cancer Society and the Cancer Statistics Center, lung cancer is a chronic, progressive and malignant disease, which causes an annual of over 130,000 deaths worldwide (expected to be 135,720 by 2020), and an annual of 200,000 newly-diagnosed cases (expected to be 228,820 by 2020) (1). Lung cancer is also the leading cause of cancer-related death in 93 countries, among which, the highest incidence rates are observed in Micronesia/Polynesia, and Eastern Asia, including China (2). It is estimated that, 85% lung cancer cases have non-small cell lung cancer (NSCLC), which can be further classified as adenocarcinoma (65%) and squamous cell carcinoma (30%) (3).
Among the existing treatments, targeted therapy is the least harmful and most tolerable one for patients who have lost the opportunities of surgery (4). Currently, samples for genetic detection are mainly collected from the blood, pleural effusion, urine or tumor tissues of patients, among which, tumor tissues achieve the highest specificity and sensitivity in genetic detection (5-7). However, invasive procedures are not applicable to patients with metastatic disease or compromised health status (6). Because of drug resistance, patients have to undergo multiple genetic tests. In addition, tissue samples are not always available, so the choice of targeted drug often depends on blood sample detection (8). To reduce drug resistance, the alternative use of two-targeted drugs has been proposed, but the effect remains unclear so far (9). Thus, it is crucial to early and effectively identify patients with genetic mutations to improve patient survival and reduce the financial burden on patients.
Neuron-specific enolase (NSE) is an intracellular enzyme generally detected in medullary-thyroid carcinoma and in small cell lung cancer (SLCL) (10). NSE plays an important role in the diagnosis of SCLC patients. Enormous studies have indicated the role of NSE in predicting response to chemotherapy or chemo-radiotherapy in SCLC patients (11,12). Therefore, NSE is considered as an independent predictor of survival compared with other prognostic factors. The NSE level is stable, and it has little association with age, sex, smoking status and cumulative smoking dose (13). NSE can be detected in various circulating cells, such as erythrocytes, plasma cells, and platelets (13,14). However, the association of NSE with the detection of gene mutations in lung cancer has been rarely investigated for the time being. Therefore, this study aimed to investigate the correlation between serum NSE levels and the detection of gene mutations in patients with lung adenocarcinoma. We present the following article in accordance with the STREGA reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-1633).
Methods
Patients
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by our institutional ethics board of The First Affiliated Hospital of Anhui Medical University (No. 2020-09-14) and individual consent for this retrospective analysis was waived. This retrospective, case-control study was carried out between June 2017 to October 2019, 120 lung adenocarcinoma patients admitted at the Department of Geriatric Respiratory and Critical Care in the First Affiliated Hospital of Anhui Medical University were enrolled into the present study.
All patients participating in this study met the following criteria: (I) all patients underwent lung biopsy, pleural fluid cytology or lymph node biopsy to obtain a definite pathological type, and they were diagnosed with lung adenocarcinoma. (II) Sufficient blood or tissues were collected from patients to complete next-generation sequencing (NGS). (III) All patients were not treated with chemotherapy, radiotherapy, radiofrequency ablation, or surgery. (IV) All patients had no histories of tumor, endocrine and metabolic disorders.
Analysis of gene mutations
In this study, the targeted sequencing approach based on Illumina platform was conducted to detect the common gene mutations in tissue biopsies or plasma samples from 120 lung adenocarcinoma patients. The specific procedure was as follows.
Sample pretreatment: the sample was collected into a sterile centrifuge tube, and the precipitate was collected after centrifugation. DNA was isolated and purified using the Blood/Tissue Genomic DNA Isolation Kit [Tiangen Biotech (Beijing) Co., Ltd.].
Library preparation: In brief, 1 µg genomic DNA was sheared into fragments, then, the processes of end-repairing, A-tailing, and ligation with indexed adapters were conducted in turn, and finally the size was selected. After polymerase chain reaction (PCR) amplification, all libraries were purified and enriched.
Quantitative PCR (Q-PCR) for quantitation: the gradient dilution of libraries was performed on ice. Preparation of the PCR system: the Novo SYBR qPCR Mix Novo 50 ROX reference dye and Novo Quantification Primer were melt and centrifuged at once for preparation. Preparation of the reaction system: a total volume of 20 µL was prepared for the reaction system. Two parallel experiments were conducted for each sample. 16 µL sample was distributed to the eight-row fluorescent quantitative PCR tube, which were then blended and centrifuged.
Genome sequencing: genome sequencing was performed using the HiSeq X-TEN System (Illumina, Novogene Bioinformatics Technology Co., Ltd., Novogene, Beijing, China).
We provided patients with 23-gene panel test (ALK, BRAF, EGFR, AKT1, HER2/ERBB2, FGFR1, FGFR2, GNA11, GNAQ, HRAS, KIT, KRAS, MET, NRAS, DDR2, PDGFRA, PIK3CA, MAP2K1, PTEN, RET, ROS1, SMO, TSC1) and 10-gene panel test (ALK, BRAF, EGFR, HER2/ERBB2, KRAS, MET, PIK3CA, RET, ROS1, TP53). What type of test was decided by the patients based on the cost of test differing from 23 to 10 gene panel. Thirty-nine patients received the 23-gene panel and 81 patients received the 10-gene panel. Those without any mutation were classified as non-mutant group, and single or multiple gene mutations were classified as mutant group. The genes that tested positive included EGFR, ALK, TP53, ROS1, HER2/ERBB2, MET, BRAF, RET, KRAS, NRAS, PTEN, PDGFRA, KIT, FGFR2, TSC1 and PIK3CA.
Detection of NSE
Detection of the serum NSE levels: blood samples were taken when patients were first admitted to hospital before any treatment and the NSE levels were determined immediately according to the double-antibody sandwich magnetic particle chemiluminescent method. The matched kit, Roche (e601, Basel, Switzerland) automatic electrochemical luminescent immunoassay analyzer, was also used.
Statistical analysis
The Empower(R) (www.empowerstats.com, X&Y solutions, Inc, Boston, MA, USA) and R (http://www.R-project.org)softwares were employed for all statistical analyses. Continuous variables were presented as mean and standard deviation and categorical data were shown as number of cases and percentage. The t-test was used for those normal distribution and Kruskal-Wallis test was used for non-normal variables. Pearson’s χ2 test and Fisher’s exact test were used to analyze the differences in categorical variables. The association of serum NSE level with gene mutation was evaluated using the multiple logistic regression models. Kruskal-Wallis test was carried out to revise the correlation between the NSE level and the gene mutation types. The receiver operating characteristic (ROC) curve was plotted to assess the value of NSE in predicting gene mutation in blood sample group, and the relationships of different NSE contents with gene mutation were evaluated by the linear regression model. A difference of P<0.05 indicated statistical significance. Delete the cases of missing data.
Results
Objects of study
Three hundred and forty-seven lung cancer patients who underwent NGS were enrolled in this study. Fifty-five patients were excluded from the study because they had undergone surgery, targeted therapy, or chemotherapy. One hundred and twenty-three cases were excluded due to the pathological squamous cell carcinoma, small cell or unclassified and the pleural effusion for NGS. Sixty-nine others were excluded because of incomplete data. The age of the enrolled 120 patients ranged from 31 to 87 years, among them, 40.00% were females. The blood and tissue samples were collected from all the enrolled patients for genetic detection, including 86 mutant cases (mutant group) and 34 non-mutant cases (non-mutant group). In the blood sample group, the percentage of lung cancer patients with distant metastasis was significantly higher in mutant group than in non-mutant group (P=0.01).
Relationship of NSE with gene mutation
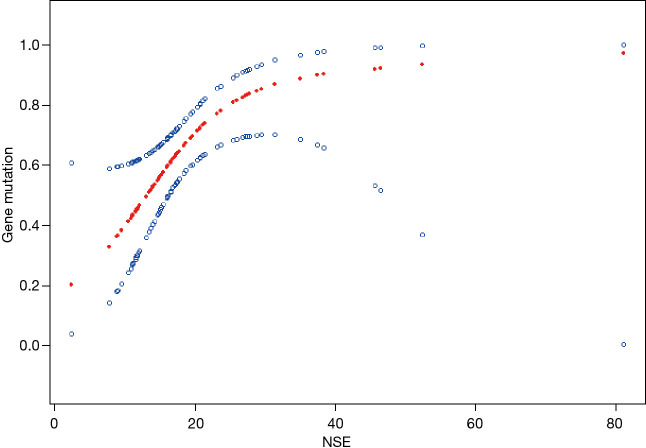
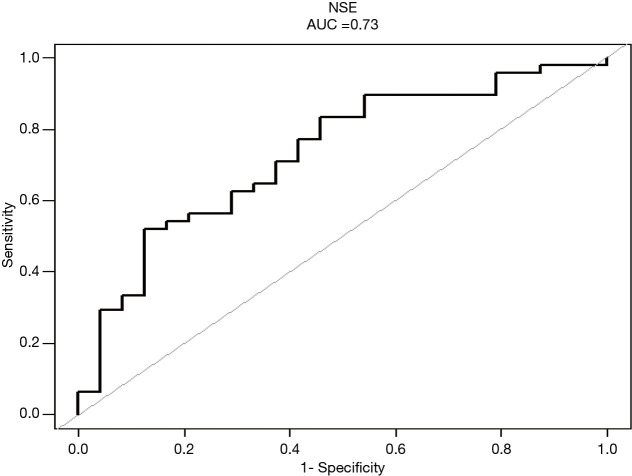
In the blood samples, the NSE levels were higher in mutant group than in non-mutant group (P=0.002), but in tissue samples, the difference was not obvious (P=0.819) (Table 1). Results of multivariate analysis revealed significant correlation of NSE with gene mutation (OR =7.6; 95% CI, 2.0–28.9; P=0.003) (Table 2). Furthermore, variables were adjusted for age, gender, hemoglobin, platelet, and leukocyte levels and smoking history, with no significant changes (OR =8.5; 95% CI, 1.9–38.7; P=0.006). In addition, the linear regression model further highlighted the tight relationship between NSE and gene mutation in blood sample group (Figure 1). A higher NSE value indicated a higher probability of mutation detection. Results of ROC curve analysis are shown in Figure 2. The area under the curve (AUC) of NSE was 0.7300 [95% confidence interval (CI): 0.6059–0.8541) and optimal threshold was 18.5650 U/mL with a sensitivity of 87.50% and a specificity of 52.08%.
Table 1. Patient characteristics.
| Gene mutation | Blood sample | Tissue sample | |||||
|---|---|---|---|---|---|---|---|
| No | Yes | P value | No | Yes | P value | ||
| No. | 24 | 48 | 10 | 38 | |||
| Age | 65.5±8.7 | 62.6±12.4 | 0.318 | 60.7±18.8 | 65.7±11.0 | 0.281 | |
| NSE (U/mL) | 15.4±7.8 | 22.2±12.9 | 0.002 | 16.7±4.4 | 20.2±12.0 | 0.819 | |
| Gender | 0.864 | 0.488 | |||||
| Female | 9 (37.5%) | 19 (39.6%) | 3 (30.0%) | 17 (44.7%) | |||
| Male | 15 (62.5%) | 29 (60.4%) | 7 (70.0%) | 21 (55.3%) | |||
| Metastasis | 0.010 | 1.000 | |||||
| No | 9 (39.1%) | 6 (12.5%) | 3 (30.0%) | 12 (31.6%) | |||
| Yes | 14 (60.9%) | 42 (87.5%) | 7 (70.0%) | 26 (68.4%) | |||
| Leukocyte (10^9/L) | 6.4±1.7 | 6.8±2.4 | 0.444 | 6.9±2.7 | 6.7±2.6 | 0.761 | |
| Hemoglobin (10^9/L) | 124.0±14.7 | 120.9±17.8 | 0.470 | 124.1±9.2 | 123.8±15.2 | 0.951 | |
| Platelet (10^9/L) | 226.3±62.3 | 233.0±72.4 | 0.701 | 253.1±103.3 | 232.5±78.8 | 0.494 | |
| Smoking history | 0.099 | 0.487 | |||||
| No | 14 (58.3%) | 37 (77.1%) | 5 (50.0%) | 24 (63.2%) | |||
| Yes | 10 (41.7%) | 11 (22.9%) | 5 (50.0%) | 14 (36.8%) | |||
NSE, neuron-specific enolase.
Table 2. Multivariate regression for the relationship between NSE and gene mutation in blood samples.
| Variable | Non-adjusted | Adjust I | Adjust II | |||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |||
| NSE (U/mL) | ||||||||
| <18.5 | 1.0 | 1.0 | 1.0 | |||||
| ≥18.5 | 7.6 (2.0, 28.9) | 0.003 | 7.7 (2.0, 29.8) | 0.003 | 8.5 (1.9, 38.7) | 0.006 | ||
Model I adjusted for age and gender. Model II adjusted for age, gender, hemoglobin, platelet, leukocyte levels and smoking history. CI, confidence interval; OR, odds ratio; NSE, neuron-specific enolase.
Figure 1.
NSE and positive rate of gene mutations dose-response relationship in blood sample. Adjusted for age, gender, hemoglobin, platelet, and leukocyte levels. NSE, neuron-specific enolase.
Figure 2.
ROC curve of the relationship between NSE and positive rate of gene mutations in blood sample group. AUC of NSE was 0.7300 (95% CI: 0.6059–0.8541). AUC, area under the curve; ROC, receiver operating characteristic; NSE, neuron-specific enolase.
Specific differences in NSE levels between the blood and tissue sample groups
The single gene mutation in blood samples was significantly correlated with NSE (P=0.045) (Table 3). Among them, the NSE levels in MET, KRAS, ROS1 and TP53 mutations increased, while NSE level showed weak correlations with EGFR, TSC1, FGFR2 and NRAS mutations. Correspondingly, EGFR, MET, ERBB2 and KRAS mutations showed relatively high correlations with NSE levels in tissue samples, while ALK and TP53 mutations showed weak correlation. EGFR mutation was not associated with NSE level in either tissue or blood sample group. In polygenic mutations, the association of NSE level with gene mutation was still stronger in blood sample group (P=0.032). In terms of polygenic mutations containing EGFR, TP53, ERBB2 and MET, the NSE level increased in blood sample group (P<0.05). But no such relationship was detected in tissue sample group. More analysis results were run than can be included in the article. The interested reader can find them in a supplementary appendix online.
Table 3. NSE levels of single-gene and multi-gene mutations.
| Gene mutation | Blood sample | Tissue sample | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-mutation group NSE levels (U/mL) | Mutation group NSE levels (U/mL) | P values | Non-mutation group NSE levels (U/mL) | Mutation group NSE levels (U/mL) | P values | ||||
| Single gene mutation | |||||||||
| EGFR | 15.4±7.8 | 14 | 18.6±7.5 | 0.223 | 16.7±4.4 | 12 | 22.7±16.6 | 0.281 | |
| ERBB2 | 2 | 13.0±1.4 | 0.672 | 2 | 34.6±20.3 | 0.013 | |||
| MET | 1 | 35.0± NA | 1 | 35.4± NA | |||||
| TP53 | 1 | 27.2± NA | 2 | 16.4±0.7 | 0.935 | ||||
| ALK | 2 | 26.1±16.1 | 0.095 | 2 | 20.3±9.8 | 0.389 | |||
| BRAF | |||||||||
| NRAS | 1 | 11.1± NA | |||||||
| PIK3CA | |||||||||
| PTEN | |||||||||
| PDGFRA | |||||||||
| KRAS | 4 | 38.2±13.2 | 0.003 | 2 | 28.4±19.0 | 0.066 | |||
| KIT | |||||||||
| ROS1 | 1 | 27.1± NA | |||||||
| TSC1 | 1 | 9.6± NA | |||||||
| FGFR2 | 1 | 2.4± NA | |||||||
| RET | |||||||||
| Total | 28 | 21.6±11.8 | 0.045 | 21 | 24.2±14.9 | 0.135 | |||
| Polygenic mutations contain | |||||||||
| EGFR | 11 | 21.3±5.4 | 0.029 | 11 | 14.9±3.2 | 0.298 | |||
| ERBB2 | 3 | 37.7±37.6 | 0.010 | 2 | 15.1±0.0 | 0.629 | |||
| TP53 | 11 | 26.7±18.8 | 0.002 | 7 | 15.9±3.0 | 0.675 | |||
| MET | 3 | 25.9±5.9 | 0.021 | 3 | 16.8±3.3 | 0.982 | |||
| ALK | 3 | 18.3±2.7 | 0.226 | 4 | 13.9±3.9 | 0.295 | |||
| BRAF | 1 | 19.1± NA | 1 | 15.5± NA | |||||
| NRAS | |||||||||
| PIK3CA | 2 | 22.9±5.4 | 0.2 | 3 | 15.2±3.0 | 0.585 | |||
| PTEN | 2 | 23.7±4.2 | 0.155 | ||||||
| PDGFRA | 1 | 19.6± NA | |||||||
| KRAS | 2 | 20.5±1.3 | 0.373 | 2 | 18.0±0.9 | 0.693 | |||
| KIT | 2 | 14.9±6.0 | 0.925 | ||||||
| ROS1 | 2 | 16.4±0.5 | 0.923 | ||||||
| TSC1 | 2 | 23.7±4.2 | 0.155 | 1 | 12.5± NA | ||||
| FGFR2 | |||||||||
| RET | 2 | 15.2±0.4 | 0.967 | 1 | 15.5± NA | ||||
| Total | 20 | 23.0±14.6 | 0.032 | 17 | 15.3±2.8 | 0.314 | |||
Discussion
To some extent, lung cancer is a genetic disease with various histological subtypes. Only a minority of mutant genes will promote or drive the development of lung cancer, regardless of the complex gene mutations (15). With the deepening of research, an increasing number of genes have been proved to be related to the development of lung cancer, and they are incorporated into the treatment guidelines (16).
However, it is always difficult to obtain sufficient tissue samples for gene mutation analysis in clinical studies, especially for some patients with end-stage disease. Serum carcinoembryonic antigen (CEA) level was reported to be an independent predictor for plasma EGFR mutation, which was associated with the effect of EGFR-tyrosine kinase inhibitor (TKI) treatment (8,17). These studies have prompted EGFR detection to some extent, and EGFR mutation may be sensitively detected in patient plasma with a higher CEA level when tumor tissues is not available. This is also the significance of our study.
We also examined the relationship between other tumor markers [CEA and cytokeratin-19 fragment (CYFRA21-1)] and the positive rate of gene detection in blood samples. However, our data cannot indicate that CEA levels are associated with positive rate of gene detection (Appendix). There is a certain correlation for CYFRA21-1 in all blood samples, but the P value in the 10-gene panel is 0.069. Therefore, we don’t know for sure whether there is a positive correlation.
Serum NSE level reflects cell proliferation and mortality, and the elevated levels may indicate distant metastasis (18). NSE has been identified to predict mutations in some genes, but no significant correlation is detected so far (19,20). Some studies have suggested that, NSE is associated with EGFR mutations in adenocarcinoma cases, but no consensus is reached for the time being (21). The existing studies focus on individual gene mutation, and no comprehensive analysis is conducted. Therefore, the diagnostic value of NSE for lung adenocarcinoma gene mutation was evaluated in this study.
Our data demonstrated that, for blood sample group, the NSE levels were significantly higher in patients with genetic mutation than those without mutation. Meanwhile, more patients from whom the blood samples were collected for genetic detection were enrolled in the study. This phenomenon has also accelerated the studies of using blood sample for gene mutation detection (22,23). A series of large clinical trials have adopted blood samples for genetic detection, such as B-F1RST (NCT02848651) and BFAST (NCT03178552). In the 2019 EUROPEAN SOCIETY FOR MEDICAL ONCOLOGY (ESMO) congress(https://www.esmo.org/), the BFAST (NCT03178552) study on ALK mutations treated with alectinib shows that, blood test may be used to replace the invasive biopsy for more lung cancer patients. Cells exfoliate from the primary tumor and enter the circulatory system, which marks the beginning of malignant tumor metastasis. Typically, these cells are called the circulating tumor cells (CTCs). CTCs have been proven to be independently correlated with distant metastases in NSCLC and the elevated serum NSE level (24). This may explain the increased positive rates for genetic mutations detected in blood samples with the increase in NSE levels. The tissue samples contain a large number of tumor cells, so the detection rate of gene mutation is high, which may partly explain the weak correlation between NSE levels and the positive rate of gene mutation detection in the tissue samples.
Suh et al. also reported that, patients with elevated serum NSE levels had shorter overall survival (OS) and progression-free survival (PFS), and the poor prognosis of first-line EGFR-TKI treatment in NSCLC patients with EGFR mutation was associated with the elevated NSE levels (25). In our study, NSE level increased in single gene mutation group, in which MET, KRAS, ROS1 and TP53 showed obvious correlations, but NSE level was weakly correlated with conventional single-gene mutations, such as ALK and EGFR. This might be due to the insufficient sample size. A large percentage of lung cancer is not caused by a single oncogene mutation, which accounts for the cause of drug resistance and poor prognosis (26,27). In our study, multi-gene mutations predict higher NSE levels and earlier metastasis in the blood.
Patients with EGFR, ALK, or ROS1 mutation combined with TP53 mutation have shorter survival (26). Notably, MET amplification and PIK3CA mutations have been reported as the resistance mechanism in patients with EGFR mutation (27). Our results showed that, in the diagnosis of lung adenocarcinoma, the NSE levels markedly elevated in patients with polygenic mutations combined TP53 mutation compared with patients with non-gene mutation. Among the 11 patients with polygenic mutations containing EGFR, 9 were associated with TP53 among which 1 was EGFR/TP53/MET mutation, and the remaining one was associated with ALK, while the others were associated with KRAS and TSC1. We knew quite early on that papillary thyroid carcinoma cells have high expression of NSE and mutant forms of p53 protein (28). Both increased NSE levels and TP53 mutation could more easily be found in adenocarcinoma and squamous cell lines of lung cancer (29). To the best of our knowledge, no studies have identified the predictive role of NSE in lung adenocarcinoma with TP53 mutation. The prevalence of MET mutation detection was low, but the NSE levels increased significantly in both single-gene and polygenic mutations with MET. Li et al. reported that MET DNA expression in blood was independently correlated with NSE levels in patients with lung cancer (19). There were also research data showing that NSE was not associated with c-MET expression in lung cancer patients (30). This is contrary to our results. KRAS mutations have also been reported as the acquired resistance to TKIs (31), indicating that EGFR mutation with poor prognosis is more detectable in the blood with a high NSE level. However, there was no significant correlation between ALK mutation and the NSE level.
In this study, some limitations should be noted. Firstly, the sample size was small, especially for the tissue samples without gene mutation, which might be responsible for the higher success rate of genetic detection in tissue samples. Secondly, the relationships between NSE levels with the mutation subtypes had not been fully elucidated yet. Thirdly, the long-term effect of NSE on gene mutation was not determined.
Conclusions
In conclusion, NSE content effectively improves the diagnostic value of gene mutation detected using the blood samples for lung adenocarcinoma. This phenomenon is particularly significant in the polygenic mutations. Our findings may provide new insights into the prognostic value of targeted therapy and the use of blood samples for gene detection.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was funded by the Colleges and Universities in Anhui Province Natural Science Research Projects (No. KJ2018A0208).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by our institutional ethics board of The First Affiliated Hospital of Anhui Medical University (No. 2020-09-14) and individual consent for this retrospective analysis was waived.
Footnotes
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-1633
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-1633
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1633). Dr. FZX reports grants from Colleges and Universities in Anhui Province Natural Science Research Projects (No. KJ2018A0208), during the conduct of the study; Dr. YBZ reports grants from Colleges and Universities in Anhui Province Natural Science Research Projects (No. KJ2018A0208), during the conduct of the study.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Friedlaender A, Banna G, Malapelle U, et al. Next Generation Sequencing and Genetic Alterations in Squamous Cell Lung Carcinoma: Where Are We Today? Front Oncol 2019;9:166. 10.3389/fonc.2019.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heist RS, Engelman JA. SnapShot: non-small cell lung cancer. Cancer Cell 2012;21:448.e2. 10.1016/j.ccr.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 5.Deng Q, Qiu Y, Jia J, et al. Genetic alteration profile of EGFR-mutant resected IIB-IIIA stage NSCLC and correlation to clinical outcomes. Transl Lung Cancer Res 2019;8:838-46. 10.21037/tlcr.2019.10.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franovic A, Raymond VM, Erlander MG, et al. Urine test for EGFR analysis in patients with non-small cell lung cancer. J Thorac Dis 2017;9:S1323-31. 10.21037/jtd.2017.06.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei S, Lieberman D, Morrissette JJ, et al. Using "residual" FNA rinse and body fluid specimens for next-generation sequencing: An institutional experience. Cancer Cytopathol 2016;124:324-9. 10.1002/cncy.21666 [DOI] [PubMed] [Google Scholar]
- 8.Gao Y, Song P, Li H, et al. Elevated serum CEA levels are associated with the explosive progression of lung adenocarcinoma harboring EGFR mutations. BMC Cancer 2017;17:484. 10.1186/s12885-017-3474-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solomon B, Kok P, Livingstone A, et al. P2.03-044 OSCILLATE - Phase 2 Trial of nating Osimertinibwith Gefitinib in Patients with EGFR-T790M Mutation Positive Advanced NSCLC. J Thorac Oncol 2017. 10.1016/j.jtho.2017.09.1295 [DOI] [Google Scholar]
- 10.Sung HJ, Cho JY. Biomarkers for the lung cancer diagnosis and their advances in proteomics. BMB Rep 2008;41:615-25. 10.5483/BMBRep.2008.41.9.615 [DOI] [PubMed] [Google Scholar]
- 11.Holdenrieder S, von Pawel J, Dankelmann E, et al. Nucleosomes, ProGRP, NSE, CYFRA 21-1, and CEA in monitoring first-line chemotherapy of small cell lung cancer. Clin Cancer Res 2008;14:7813-21. 10.1158/1078-0432.CCR-08-0678 [DOI] [PubMed] [Google Scholar]
- 12.Choi N, Baumann M, Flentjie M, et al. Predictive factors in radiotherapy for non-small cell lung cancer: present status. Lung Cancer 2001;31:43-56. 10.1016/S0169-5002(00)00156-2 [DOI] [PubMed] [Google Scholar]
- 13.Schneider J. Tumor markers in detection of lung cancer. Adv Clin Chem 2006;42:1-41. 10.1016/S0065-2423(06)42001-1 [DOI] [PubMed] [Google Scholar]
- 14.Qi W, Li X, Kang J. Advances in the study of serum tumor markers of lung cancer. J Cancer Res Ther 2014;10 Suppl:C95-C101. 10.4103/0973-1482.145801 [DOI] [PubMed] [Google Scholar]
- 15.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol 2018;13:323-58. 10.1016/j.jtho.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 17.Que D, Xiao H, Zhao B, et al. EGFR mutation status in plasma and tumor tissues in non-small cell lung cancer serves as a predictor of response to EGFR-TKI treatment. Cancer Biol Ther 2016;17:320-7. 10.1080/15384047.2016.1139238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrigno D, Buccheri G, Giordano C. Neuron-specific enolase is an effective tumour marker in non-small cell lung cancer (NSCLC). Lung Cancer 2003;41:311-20. 10.1016/S0169-5002(03)00232-0 [DOI] [PubMed] [Google Scholar]
- 19.Li D, Li F, Wu Y, et al. Quantification of serum MET in non-small-cell lung cancer and its clinical significance. Clin Biochem 2015;48:110-4. 10.1016/j.clinbiochem.2014.11.021 [DOI] [PubMed] [Google Scholar]
- 20.Tan Q, Huang Q, Ma G, et al. Relationship between serum tumor markers and Anaplastic Lymphoma Kinase mutations in stage IV lung adenocarcinoma in Hubei province, Central China. J Clin Lab Anal 2020;34:e23027. 10.1002/jcla.23027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang R, Wang X, Li K. Predictive and prognostic value of preoperative serum tumor markers is EGFR mutation-specific in resectable non-small-cell lung cancer. Oncotarget 2016;7:26823-36. 10.18632/oncotarget.8662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. 10.1038/s41591-018-0134-3 [DOI] [PubMed] [Google Scholar]
- 23.Mohan S, Foy V, Ayub M, et al. Profiling of Circulating Free DNA Using Targeted and Genome-wide Sequencing in Patients with SCLC. J Thorac Oncol 2020;15:216-30. 10.1016/j.jtho.2019.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Ma K, Yang Z, et al. Systematic Correlation Analyses of Circulating Tumor Cells with Clinical Variables and Tumor Markers in Lung Cancer Patients. J Cancer 2017;8:3099-104. 10.7150/jca.18032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suh KJ, Keam B, Kim M, et al. Serum Neuron-Specific Enolase Levels Predict the Efficacy of First-Line Epidermal Growth Factor Receptor (EGFR) Tyrosine Kinase Inhibitors in Patients With Non-Small Cell Lung Cancer Harboring EGFR Mutations. Clin Lung Cancer 2016;17:245-52.e1. 10.1016/j.cllc.2015.11.012 [DOI] [PubMed] [Google Scholar]
- 26.Aisner DL, Sholl LM, Berry LD, et al. The Impact of Smoking and TP53 Mutations in Lung Adenocarcinoma Patients with Targetable Mutations-The Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res 2018;24:1038-47. 10.1158/1078-0432.CCR-17-2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reis H, Metzenmacher M, Goetz M, et al. MET Expression in Advanced Non-Small-Cell Lung Cancer: Effect on Clinical Outcomes of Chemotherapy, Targeted Therapy, and Immunotherapy. Clin Lung Cancer 2018;19:e441-63. 10.1016/j.cllc.2018.03.010 [DOI] [PubMed] [Google Scholar]
- 28.Fabien N, Fusco A, Santoro M, et al. Description of a human papillary thyroid carcinoma cell line. Morphologic study and expression of tumoral markers. Cancer 1994;73:2206-12. [DOI] [PubMed] [Google Scholar]
- 29.Gasperi-Campani A, Roncuzzi L, Ricotti L, et al. Molecular and biological features of two new human squamous and adenocarcinoma of the lung cell lines. Cancer Genet Cytogenet 1998;107:11-20. 10.1016/S0165-4608(98)00076-4 [DOI] [PubMed] [Google Scholar]
- 30.Miao L, Lu Y, Xu Y, et al. PD-L1 and c-MET expression and survival in patients with small cell lung cancer. Oncotarget 2017;8:53978-88. 10.18632/oncotarget.9765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A 2012;109:E2127-33. 10.1073/pnas.1203530109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as