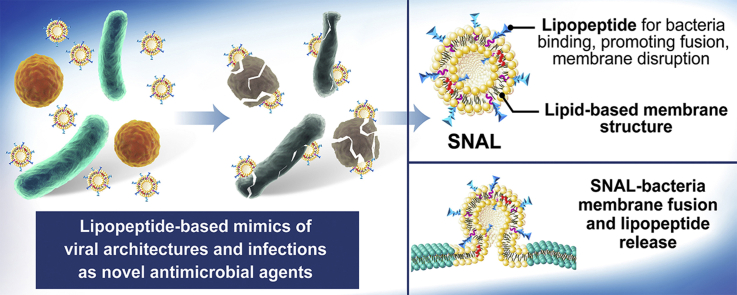
Abstract
Enveloped viruses such as SARS-CoV-2 frequently have a highly infectious nature and are considered effective natural delivery systems exhibiting high efficiency and specificity. Since simultaneously enhancing the activity and selectivity of lipopeptides is a seemingly unsolvable problem for conventional chemistry and pharmaceutical approaches, we present a biomimetic strategy to construct lipopeptide-based mimics of viral architectures and infections to enhance their antimicrobial efficacy while avoiding side effects. Herein, a surface-nanoengineered antimicrobial liposome (SNAL) is developed with the morphological features of enveloped viruses, including a moderate size range, lipid-based membrane structure, and highly lipopeptide-enriched bilayer surface. The SNAL possesses virus-like infection to bacterial cells, which can mediate high-efficiency and high-selectivity bacteria binding, rapidly attack and invade bacteria via plasma membrane fusion pathway, and induce a local “burst” release of lipopeptide to produce irreversible damage of cell membrane. Remarkably, viral mimics are effective against multiple pathogens with low minimum inhibitory concentrations (1.6–6.3 μg mL−1), high bactericidal efficiency of >99% within 2 h, >10-fold enhanced selectivity over free lipopeptide, 99.8% reduction in skin MRSA load after a single treatment, and negligible toxicity. This bioinspired design has significant potential to enhance the therapeutic efficacy of lipopeptides and may create new opportunities for designing next-generation antimicrobials.
Keywords: Virus-inspired mimics, Antimicrobial lipopeptides, Liposomes, Virus-like infections, Activity and selectivity
Graphical abstract
Highlights
-
•
Virus-inspired surface-nanoengineered antimicrobial liposomes (SNALs) are developed.
-
•
SNALs are lipopeptide-based mimics with virus-like architectures and infectivity.
-
•
The loaded lipopeptides contribute to bacteria binding and membrane disruption.
-
•
SNALs simultaneously enhance the antimicrobial activity and selectivity of lipopeptides.
-
•
SNALs represent promising antimicrobials to combat antibiotic-resistance.
1. Introduction
The fight against antibiotic resistance is as old as the antibiotic era itself. With each new antibiotic progressing into a clinical setting, resistance generally develops within several months to a decade but can appear as rapidly as several months [1]. Recently, antimicrobial lipopeptides, consisting of linear or cyclic peptide chains and lipophilic hydrocarbon tails, have been widely considered to be promising treatment options to combat infections. Their major advantage is that they exhibit direct membrane-disruptive effects against cells in a phospholipid-dependent manner [2,3]. Since they do not act on specific targets such as enzymes or DNA, the development of resistance to lipopeptides may be slow and limited [1,3]. Therefore, recent inroads have contributed to remarkable advances in understanding the unique therapeutic potential of lipopeptides and their mimics, as studied by many research groups [[4], [5], [6], [7], [8], [9], [10], [11], [12]].
Despite remarkable progress, the unsatisfactory activity and high toxicity of lipopeptides remain responsible for the decline in U.S. Food and Drug Administration (FDA)–approved antibiotics in recent years [1,[6], [7], [8]]. For cationic lipopeptides, their amphiphilic nature enables the compounds to electrostatically interact with the negatively charged bacterial membrane, while their lipophilic lipid tails of appropriate lengths are generally critical for lipopeptide insertion, membrane destabilization, and microbial lysis [3,7]. The strategy of chemical modification can increase the selectivity of amphiphilic lipopeptides to some extent; however, simultaneously achieving high antimicrobial activity and selectivity is still very difficult. Although the increase in the hydrophilic oligopeptide moieties can generally promote lipopeptides to recognize bacteria rather than mammalian cells, it may also weaken the bacterial cell membrane penetration capacity of lipopeptides. Correspondingly, although increasing the length or number of lipid moieties can improve antibacterial activity, excessive hydrophobic content will result in undesired hemolysis and the deterioration of selectivity owing to a significant decrease in the charging effect [1,7]. Therefore, the structural optimization of cationic lipopeptides has been encountering a significant bottleneck that must be addressed using alternative approaches.
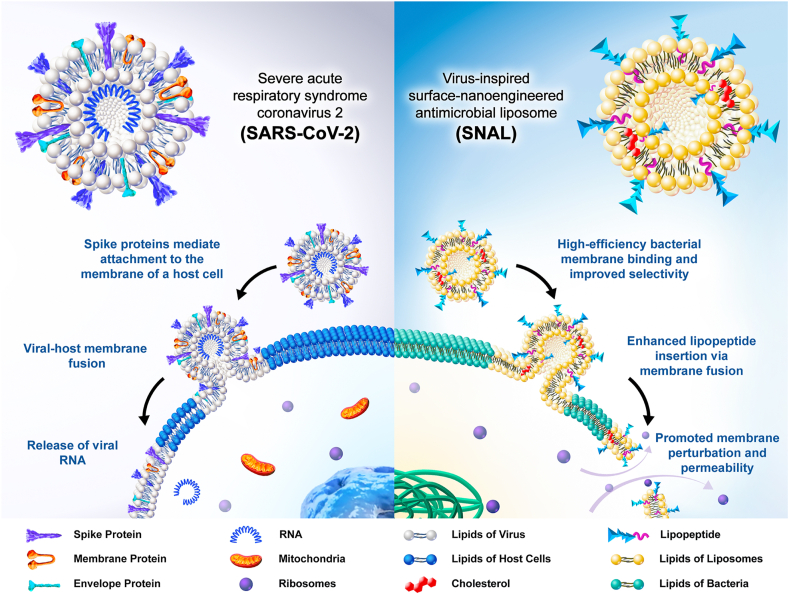
Viruses, some of the highly evolved nanoparticulate organisms in nature, are generally considered effective natural delivery systems that exhibit high efficiency and specificity. Over the past several decades, the potential valuable application of viruses, particularly enveloped viruses (viruses with a surrounding outer lipid bilayer membrane), has inspired scientists to construct morphological and structural mimics that mirror the distinctive architecture and infection of viruses [4,5,[13], [14], [15], [16], [17]]. In the 20th and 21st centuries, enveloped viruses, including severe acute respiratory syndrome (SARS), Ebola, human immunodeficiency virus (HIV), Middle East respiratory syndrome (MERS), influenza, etc., have caused many devastating effects. Among these, SARS coronavirus 2 (SARS-CoV-2), the virus behind coronavirus disease 2019 (COVID-19), have spread throughout the world, causing over 2.3 million deaths and an astonishing 106.7 million-plus confirmed cases by February 8, 2021 [18,19]. The high infectivity and specific binding ability of SARS-CoV-2 depend on the spike protein anchored in the outer surface of the virus, which is responsible for recognizing, binding to host cell membranes and mediating virus-cell membrane fusion (Scheme 1) [14,[20], [21], [22]]. With natural affinity to bacteria cells, we expected that antimicrobial cationic lipopeptides could have potential to exert spike protein-like effects. Therefore, we hypothesized that fabricating surface-functionalized lipid-based nanovectors with enriched targeting moieties of lipopeptides on the bilayer surface may be a promising method of improving the performance of antimicrobial lipopeptides via the virus-like infection pathway.
Scheme 1.
Schematic illustrations for the structure of SARS-CoV-2 and its mechanism of entry into host cells via plasma membrane fusion pathway (left), and the structure of SNAL and its proposed mechanism of antimicrobial action (right).
To prove our concept, we designed a virus-inspired surface-nanoengineered antimicrobial liposome (SNAL) with lipopeptides intercalated within the phospholipid bilayers to endow lipopeptide formulations with virus-like high infectivity and specificity. Since conventional vehicles (e.g., micelles, microspheres, and nanocapsules) frequently encapsulate lipopeptides in their interior and shield their targeting oligopeptide moiety from binding bacterial cell membranes [23,24], SNALs were considered to be a more promising system to simultaneously achieve high activity and selectivity (the structure of conventional formulations and their mechanism of action are shown in Fig. S1). As a proof-of-concept, the structural characterization of the SNAL was conducted to confirm the morphology, liposomal stability, drug loading capacity, and intercalation of lipopeptides within the liposome membrane. Meanwhile, the antimicrobial and hemolytic activities are considered the gold standard for assessing liposomal lipopeptides, followed by drug delivery behavior and antibacterial mechanism studies. The virus-inspired lipopeptide nanovectors are expected to provide the following benefits as depicted in Scheme 1: (i) While amphipathic lipopeptides are encapsulated by anchoring the hydrocarbon tail of lipopeptides onto the lipid bilayer interior via hydrophobic interaction, the nonspecific binding between lipopeptides and neutral membranes of mammalian cells can be reduced, causing lower hemolysis and higher selectivity; (ii) the targeting oligopeptide moieties of cationic lipopeptides clustered on the SNAL surface may mimic the bioactivity of envelope-anchored components (e.g., cell-penetrating peptides and spike proteins) to mediate high-efficiency bacterial cell membrane binding; (iii) the liposomes with concentrated membrane-active peptides on the lipid bilayer could promote their fusion with target cell membranes and subsequently induce a local “burst” release of a high dose of the lipopeptides into the bacterial membrane; (iv) relatively high local lipopeptide concentrations in the lipid bilayer can promote membrane perturbations and permeability, resulting in the irreversible damage of cell membrane, large leakage of cytoplasmic contents, and ultimate bacterial cell death. Briefly, this bioinspired strategy focuses on the development of excellent viral mimics from structural bionics to functional implementation to address the contradictions in simultaneously achieving the high antimicrobial activity and selectivity of lipopeptides. The findings of this research may provide a promising strategy and rationale for the development, optimization, and reuse of some failed, abandoned, or yet-to-be-pursued clinical candidate lipopeptides. Furthermore, it is to be noted that although the entry pathway of a virus was imitated to improve the properties of antimicrobial lipopeptides, SNALs tend to exhibit antimicrobial effects rather than infect mammalian cells because their therapeutic actions are dependent on the loaded drugs and no gene is delivered by these systems to achieve viral DNA replication.
2. Materials and methods
2.1. Materials and reagents
Hydrogenated l-α-phosphatidylcholine (Hydro Egg PC), cholesterol, (N-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-phytosphingosine (C6-NBD) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (DOPE-RhB) were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL). Mueller-Hinton broth (MHB) and Mueller-Hinton agar (MHA) were purchased from Oxoid (Basingstoke, England). Melittin was obtained from Aladdin (Shanghai, China). Methicillin-resistant S. aureus (MRSA, ATCC 33591), vancomycin-resistant E. faecalis (VRE, ATCC 700802), Pseudomonas aeruginosa (P. aeruginosa, ATCC 27853), and Klebsiella pneumoniae (K. pneumoniae, ATCC 13383) were obtained from ATCC (USA) and reconstituted according to the suggested protocols. Triton X-100 was purchased from Sigma-Aldrich (Shanghai, China). Dulbecco's Modified Eagle Medium (DMEM), Fetal Bovine Serum (FBS), and 1% penicillin/streptomycin were purchased from Gibco Life Technologies (Grand Island, NY, USA). All other chemicals and solvents were of analytical grade and used without further purification.
2.2. Determining the minimum inhibitory concentration
The antimicrobial activities of various lipopeptides and liposomal lipopeptides were tested by the microdilution method using MRSA, VRE, P. aeruginosa, and K. pneumoniae. Briefly, bacteria were grown on a shaker at 37 °C to mid-logarithmic phase and diluted to 2 × 105 CFU mL−1 using an MHB medium. Subsequently, samples were serially diluted 2-fold in PBS buffer to obtain a range of 0.2–400 μg mL−1. Aliquots of 50 μL of bacterial suspensions were added to 50 μL of serial dilutions of medium containing lipopeptides or liposomal lipopeptides. After incubation for 18–20 h at 37 °C, the absorbance of each well at 630 nm was measured using an ELx8000 microplate reader (Biotek Instruments, USA). The minimum inhibitory concentration (MIC, the minimum drug concentration that completely inhibited bacterial growth) values were determined as the lowest sample concentration that completely inhibited the growth of bacteria throughout the incubation period.
2.3. Killing kinetics assay
MRSA, VRE, P. aeruginosa, and K. pneumoniae isolates were grown to mid-logarithmic phase and diluted in MHB to approximately 2 × 107 CFU mL−1 as the working suspension. Bacteria were treated with lipopeptide or its liposomal formulation at concentrations of 1 × MIC and 4 × MIC of the lipopeptide. Bacterial suspensions without any treatment were used as negative controls. The suspensions were incubated with constant shaking at 100 rpm at 37 °C. At various incubation times (0 min, 15 min, 30 min, 1 h, 2 h, 4 h, 6 h, and 10 h), the bacterial suspension was sampled and 10-fold serially diluted to various concentrations. The diluted bacterial solution (10 μL) was spotted on an MHA plate and incubated at 37 °C overnight. Subsequently, the visible colonies on the plates were counted to obtain the total viable cell numbers.
2.4. Preparation and characterization of liposomal lipopeptides
To determine the effect of the preparation method on lipopeptide encapsulation, we prepared liposomal lipopeptides using three various methods: thin-film hydration, reverse-phase evaporation, and ethanol injection. Different liposomal formulations were prepared using 80 mg of Hydro Egg PC, cholesterol, and lipopeptide at a weight ratio of 7 : 3: 4.3. For the thin-film hydration, the Hydro Egg PC, cholesterol, and lipopeptide were dissolved in 500 μL of a solvent mixture of methanol and chloroform (v/v = 1/1) and subsequently dried using nitrogen gas for 10 min to remove all solvents. The thin film of the lipid mixture was resolubilized in 2 mL of deionized water. Subsequently, the lipid suspensions were vortexed for 1 min and sonicated for 3 min to obtain the final liposomes. For the reverse-phase evaporation, known quantities of the Hydro Egg PC and cholesterol were dissolved in 4 mL of chloroform as the oil phase, while lipopeptides were dissolved in 1 mL of deionized water in the aqueous phase. The aqueous phase was added dropwise into the stirred oil phase and continuously stirred for 10 min at 65 °C. Subsequently, the organic solvents were removed under reduced pressure with a rotary evaporator. Liposomes were obtained by adding 2 mL of PBS and sonication for 3 min. For the ethanol injection, Hydro Egg PC and cholesterol were dissolved in 1 mL of ethanol along with lipopeptides. Subsequently, the mixture was added dropwise into 6 mL of deionized water under stirring at 1000 rpm for 30 min at 37 °C to produce liposomal lipopeptides. The bare liposomes were prepared using the ethanol injection method as mentioned above without the addition of lipopeptides.
2.5. FRET assay
The interaction of lipopeptides within the liposome lipid bilayer was investigated using Förster resonance energy transfer (FRET), and two lipid-derived dyes, C6-NBD (excitation/emission = 460/530 nm) and DOPE-RhB (excitation/emission = 560/583 nm) were employed as FRET pairs [25]. Briefly, 0.17 wt% of C6-NBD and 1.74 wt% of DOPE-RhB were dissolved in ethanol along with Hydro Egg PC, cholesterol, and lipopeptides. Thereafter, dye-labeled bare liposomes and liposomal lipopeptides were prepared using the ethanol injection method as mentioned above. Subsequently, excess dyes were removed by centrifugation at 1000 g for 15 min. The fluorescence spectrum of each sample was measured between 500 and 650 nm using an excitation wavelength of 460 nm using a luminescence spectrometer (PerkinElmer LS55, USA). Fluorescence recovery of the donor (C6-NBD) at the lower emission peak (530 nm) was used to indicate the interaction between lipopeptide and the bilayer of the liposome.
2.6. SNAL-bacterium fusion assay
The SNAL formulation was further examined for its fusion capability with P. aeruginosa bacteria using a previously reported method [26]. DOPE-RhB at a concentration of 1.74 wt% was mixed with Hydro Egg PC, cholesterol, and lipopeptides to prepare the fluorescently labeled bare liposomes and SNAL, followed by centrifugation to remove free DOPE-RhB. Subsequently, 0.75 mL of DOPE-RhB-labeled liposome suspension at various concentrations was mixed with an equal volume of 2 × 109 CFU mL−1 P. aeruginosa. After incubation for 10 min, cells were harvested by centrifugation at 5000×g for 5 min, washed with deionized water, and fixed at room temperature in 2% (v/v) glutaraldehyde in PBS for 15 min. After incubation, the samples were washed and incubated in 1 mL 10 μg mL−1 DAPI solution at room temperature for 10 min. After washing, resuspension, the samples were transferred to sterilized poly-d-lysine-coated Lab-Tek II chambered coverglass for 2 h. The cells were washed in deionized water, air-dried, and mounted using a Vectashield H1000 mounting medium. Subsequently, images were acquired using a DeltaVision OMX 3D-SIM Super-Resolution system controlled by AcquireSR software (GE Healthcare).
2.7. In vivo murine model of MRSA skin infections
All procedures involving mice were approved by the Institutional Animal Care and Use Committee of Sun Yat-Sen University (SYSU-IACUC-2019-000203), and all animal care was performed according to the Guide for Care and Use of Laboratory Animals. Male Institute of Cancer Research (ICR) mice (6 weeks old, 20–30 g) obtained from the Laboratory Animal Center of Sun Yat-Sen University were used in the murine abscess model of MRSA infection. The dorsal skin was depilated by shaving and applying a depilatory cream 1 day before the experiment, and then randomly divided into the control, melittin-treated, lipopeptide-treated, and SNAL groups. The bacteria were washed thrice with PBS and diluted to 1 × 108 CFU mL−1. Subsequently, 100 μL of MRSA suspension was injected subcutaneously into the dorsum of each mouse. Thirty minutes after inoculation, 100 μL of PBS, melittin (5 mg mL−1), lipopeptide (5 mg mL−1), and SNAL (5 mg mL−1) were injected into to the bacterial injection site. These mice were euthanized after 48 h of infection, and the infected skins were collected. For histological evaluation, the skin samples fixed in 4% paraformaldehyde were paraffin-embedded, sectioned, stained with hematoxylin and eosin dye (H&E), then analyzed under light microscopy. To evaluate bacterial clearance, we homogenized the collected infected skin tissues in 1 mL of sterile PBS, 10-fold serially diluted; subsequently, aliquots were spread onto sterile MHA plates. After overnight incubation at 37 °C, colonies on the plates were counted and photographed digitally. The levels of interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α) in the skin homogenate solutions were determined using commercial ELISA kits (Dakewe, Shenzhen, China).
2.8. In vivo toxicology assessments
The biocompatibility of the SNALs was further investigated in both local effects and systemic toxicity evaluations. First, the local toxicity assessment of lipopeptides and SNALs was conducted by subcutaneously injecting 100 μL of lipopeptides (5 mg mL−1) and SNALs (5 mg mL−1) into the backs of shaved ICR male mice (20–30 g). 48 h after injection, the skin of the injection site was harvested and fixed for histological analysis using H&E staining. Meanwhile, blood serum samples of the mice at 48 h were collected for analysis of alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), creatinine (Cr), sodium, and potassium ions levels. For the systemic toxicity assessment, male ICR mice (20–30 g) were randomly divided into four groups and injected intravenously through the tail vein with PBS, melittin, lipopeptides, and SNALs at a dose of 6 mg kg−1. After 7 days of treatment, major organs (heart, liver, spleen, lung and kidney) were harvested and fixed for H&E histological analysis. All tissue slices for H&E staining were evaluated by pathologists in the Department of Pathology, Shantou University Medical College, blind to the experimental condition.
2.9. Statistical analysis
All experiments were conducted with at least three different samples per group, and all data points were shown as mean ± SD. Statistical evaluations were performed using the software SPSS (version 17.0, SPSS Inc., Chicago, IL, USA). Statistical analysis among groups was conducted using one-way analysis of variance (ANOVA) followed by a least significant difference (LSD) multiple comparison test, where differences were considered statistically significant with a probability of p < 0.05.
Further detailed materials and methods (including synthesis and characterization of lysine/arginine-based lipopeptides, characterization of SNAL, as well as hemolysis, cytotoxicity, membrane permeability and bacterial morphology assay) can be found in the supporting information.
3. Result and discussion
3.1. Synthesis and screening of candidate lipopeptide
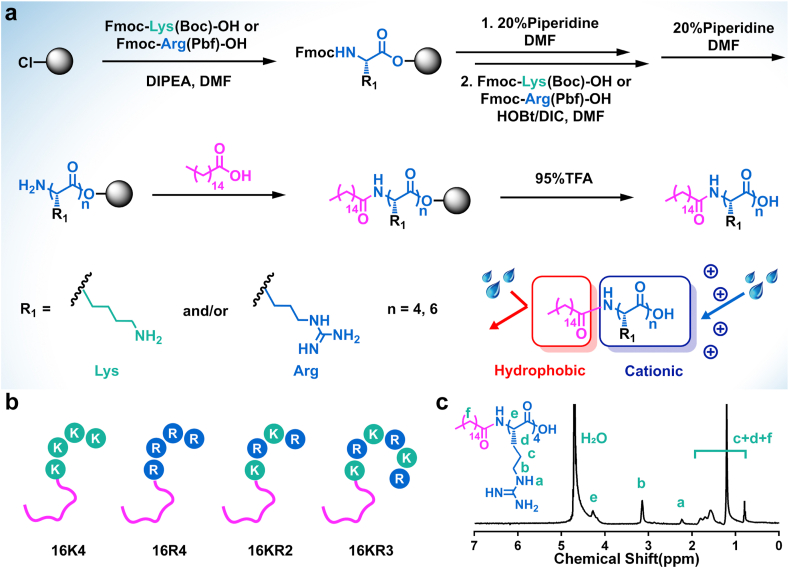
Cationic lipopeptides is a unique and diverse group of amphipathic molecules with antibacterial activity. Their common structural feature is the presence of lipophilic hydrocarbon tail and basic amino acid residues (i.e., Lys, Arg) providing a net positive charge [7]. Our previous study exhibited that conjugating palmitic acid (C16) molecule to the N terminus of cationic oligopeptides could endow them with potent and broad-spectrum antimicrobial activity [27,28]. Therefore, a series of lysine (K)/arginine (R)-based cationic lipopeptides were coupled with a palmitic acid (C16) molecule to obtain model lipopeptides, including C16-KKKK (16K4), C16-RRRR (16R4), C16-KRKR (16KR2), and C16-KRKRKR (16KR3), employing standard Fmoc-based solid-phase peptide chemistry. The synthesis routes and structural representations of these model molecules are shown in Fig. 1a and b. HPLC, ESI-MS, and 1H NMR were used to confirm the successful synthesis of the lipopeptides with their purity, molecular weights, and corresponding 1H NMR spectra, respectively (Figs. 1c,S2,S3 and Table S1). Since hospital-acquired infections are mainly caused by the pathogens acronymically referred to as ESKAPE, four of which including Gram-positive MRSA and VRE as well as Gram-negative P. aeruginosa and K. pneumoniae were chosen as representative strains to demonstrate the antimicrobial activity of various agents. MIC values were determined by standard broth micro-dilution method, and lower MIC values indicate more potent antimicrobial activity. According to the in vitro antibacterial test results, the MIC values of all model lipopeptides ranged from 6.3 to 25 μg mL−1 (Table 1). These lipopeptides have been shown to have a potential for antibacterial application because of their broad-spectrum activity against typical multidrug-resistant bacteria (i.e., MRSA and VRE) and clinically important microbes (i.e., P. aeruginosa and K. pneumoniae). While lysine-based lipopeptides 16K4 exhibited slightly better activity against Gram-negative organisms, Gram-positive bacteria were observed to be more sensitive to arginine-rich lipopeptides. In this study, 16R4 exhibited the highest antimicrobial activity with MICs in the range of 6.3–12.5 μg mL−1 and the lowest geometric mean (Gm) value of MICs (9.4 μg mL−1). Many envelope-anchored components, such as the transactivator of transcription (TAT) protein of HIV, have been reported to be frequently rich in arginine, while their guanidinium groups have a unique advantage in facilitating cellular uptake of the vesicles [29]. Therefore, the effective bacterial membrane insertion and corresponding enhanced membrane perturbation may also be responsible for the high activity of 16R4. Since hemolysis is one of the most common side effects of many antimicrobial peptides [30,31], the hemolytic activities of various lipopeptides were evaluated, and the selectivity for bacterial cells over mammalian cells was determined by the ratio of HC25 (the concentration of a polymer that causes 25% hemolysis of red blood cells) to the Gm value of MICs (HC25/Gm) (Table 1). As shown in Table 1, 16R4 displayed the best antimicrobial activity in this study. Meanwhile, considered that the selectivity of 16R4 (HC25/Gm = 6.7) is comparable to that of many typical antimicrobial peptides and synthetic analogs, such as magainins, protegrin-1, and pelgipeptin D [32,33], 16R4 was identified as a representative lipopeptide and candidate for further preparation method studies of the virus-inspired SNAL.
Fig. 1.
Synthesis and characterization of various amphipathic lipopeptides. (a) General scheme for the synthesis of various model lipopeptides. (b) Structural representations of various model lipopeptides. (c) Representative 1H NMR spectrum of 16R4 in D2O.
Table 1.
Antimicrobial and hemolytic activity of lipopeptides.
| Diagram | Lipopeptide | Sequence | MIC [μg·mL−1] |
Gm [μg·mL−1] | HC25 [μg·mL−1] | Selectivity (HC25/Gm) | |||
|---|---|---|---|---|---|---|---|---|---|
| Gram-positive |
Gram-negative |
||||||||
| MRSA | VRE | P. aeruginosa | K. pneumoniae | ||||||
 |
16K4 | C16-KKKK | 25 | 25 | 25 | 12.5 | 21.8 | 1000 | 46.0 |
 |
16R4 | C16-RRRR | 6.3 | 6.3 | 12.5 | 12.5 | 9.4 | 62.5 | 6.7 |
 |
16KR2 | C16-KRKR | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 | 15.6 | 1.3 |
 |
16KR3 | C16-KRKRKR | 12.5 | 12.5 | 6.3 | 25 | 14.1 | 250 | 17.8 |
3.2. Preparation and characterization of the virus-inspired SNAL
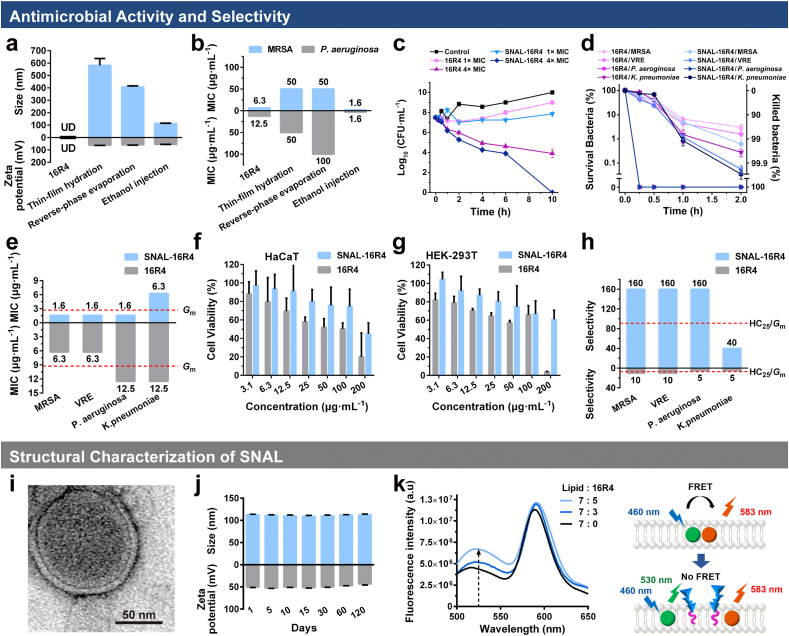
While amphipathic lipopeptides can be entrapped in either the lipophilic phase of the bilayer or the interior water phase of liposomes, various methods, including thin-film hydration, reverse-phase evaporation, and ethanol injection, have been used to prepare optimized virus-like liposomal lipopeptides. Most enveloped viruses have been reported to be spherical and approximately 100 nm in diameter [34], and small liposomes with a size of ~100 nm are prone to fuse with biological membranes owing to their high surface tension [35]. In this study, the liposomal 16R4 prepared using the ethanol injection method had an average diameter of 112.7 ± 2.4 nm, which was much smaller than that of other formulation (Fig. 2a). While bare liposome exhibited negative zeta potential values of −10.3 ± 0.4 mV and the zeta penitential of free 16R4 was undetectable, the positive zeta potential of various liposomal formulations of 16R4 indicated that amphiphilic lipopeptide can be spontaneously enriched on liposome surface to a certain extent.
Fig. 2.
Preparation and characterization of the SNAL. (a) Hydrodynamic size, surface zeta potential and (b) in vitro antimicrobial activity of liposomes prepared by various methods; time-kill curves of 16R4 and SNAL-16R4 against MRSA at different concentrations (c) and various bacteria at 4 × MIC (d); (e) in vitro antimicrobial activity of 16R4 and SNAL-16R4 against various bacteria; Cytocompatibility of 16R4 and SNAL-16R4 toward (f) HaCaT and (g) HEK-293T; (h) selectivity indexes of 16R4 and SNAL-16R4 for various bacteria; (i) representative TEM images of SNAL-16R4; (j) hydrodynamic size and surface zeta potential change of SNAL-16R4 within 3 months of storage; (k) fluorescence spectra of bare liposome (Lipid: 16R4 = 7 : 0) and SNAL-16R4 (Lipid: 16R4 = 7 : 3 or 7 : 5) labeled with a pair of FRET fluorescent dyes. The data represent the mean ± standard deviation (SD) of three individual experiments. UD: undetectable.
To further identify the best process for preparing antibacterial liposomes, we evaluated in vitro antibacterial activities of various liposomal 16R4 against MRSA and P. aeruginosa. Fig. 2b shows that the MIC values of liposomal 16R4 prepared using thin-film hydration and the reverse-phase evaporation method ranged from 50 to 100 μg mL−1 and exhibited approximately 4–8-fold lower antibacterial activity than free 16R4 (MIC = 6.3–12.5 μg mL−1). In contrast, liposomal 16R4 prepared using the ethanol injection method demonstrated improved antimicrobial activity and the best overall antibacterial activity against MRSA, VRE, P. aeruginosa, and K. pneumoniae with MICs of 1.6, 1.6, 1.6, and 6.3 μg mL−1, respectively, while its Gm value of the MIC was 2.7 μg mL−1 (Fig. 2e). Furthermore, the optimum liposomal 16R4 possessed much lower MICs than those of most antibacterial polymers or well-known highly effective antimicrobial peptides such as melittin, magainin, and LL-37 [33,36]. Time-kill studies further demonstrated the advantage of our liposome formulation in enhancing the killing efficiency of lipopeptides. As Fig. 2c and d shows, free 16R4 and liposomal 16R4 eradicated bacteria in a dose- and time-dependent manner, and killed Gram-negative bacteria much faster than they did Gram-positive bacteria. More than 99% of the bacteria, including MRSA, VRE, P. aeruginosa, and K. pneumoniae, could be killed by liposomal 16R4 after treatment for 2 h (Fig. 2d). At a concentration of 4 × MIC, the optimum liposomal 16R4 demonstrated remarkably enhanced antimicrobial efficiency, achieved more than a 99.9% killing of MRSA within 4 h and 100% killing of P. aeruginosa in 15 min. Our group previously demonstrated a nanoparticle formation of star-shaped polypeptides with surface radial peripheral side chains based on lysine [37,38] or arginine [30] to fight bacterial infections. Studies demonstrated that these nanoparticles had virus-like highly functionalized surfaces, providing an increasing trend for electrostatic binding with anionic bacterial membranes; thus, they exhibited excellent broad-spectrum antimicrobial activities and high selectivity. While the MIC values of our designed arginine-based star-shaped polypeptides 4R6G6 against MRSA and P. aeruginosa were 3.1 and 12.5 μg mL−1, respectively [30], the enhanced antimicrobial activity of our optimum liposomal arginine-based 16R4 may be attributed to the high-efficiency bacterial membrane binding mediated by its virus-like highly functionalized surface and the existence of potential fusion process between liposome with bacteria cells, which induced a local “burst” release of a lethal dose of membrane-active agents to produce irreversible damage to bacterial membrane.
Furthermore, since intercalating lipopeptide within the liposome membrane is beneficial for protecting the hydrocarbon tail of cationic lipopeptides from nonspecific interactions with mammalian cells, hemolysis and cytotoxicity assays were conducted to investigate the cytocompatibility of liposomal 16R4 prepared using the ethanol injection method. We found that the hemolysis of 16R4 was alleviated after liposome encapsulation and the HC25 of liposomal 16R4 remarkably increased from 62.5 to 250 μg mL−1. Similarly, the results in Fig. 2f,g and Fig. S4 showed that the liposomal formulation of 16R4 clearly reduced the toxicity of 16R4 toward normal epidermal keratinocyte cell line (HaCaT), human embryonic kidney 293 cell line (HEK-293T), and human hepatocyte cell line (L02). By comparing the HC25 values with the MICs of the tested microbes (HC25/MIC), the selectivity indexes of the liposomal 16R4 for MRSA, VRE, P. aeruginosa, and K. pneumoniae were observed to be greater than 160, 160, 160, and 40, respectively (Fig. 2h). The optimum liposomal 16R4 demonstrated superior selectivity toward microbes over mammalian cells (HC25/Gm = 91.4), which was 13.7 times higher than that of free 16R4 (HC25/Gm = 6.7). Antimicrobials with selectivity indexes higher than 10 have been reported to have the potential for both external and systemic applications [30]. In practice, it is important to maintain a drug dose or concentration within the therapeutic window that produces a therapeutic response without causing significant adverse effect in patients. The improved selectivity indexes of liposomal 16R4 indicate broader therapeutic window. The Gm value of MICs and HC25 of liposomal 16R4 are 2.7 μg mL−1 and 250 μg mL−1, respectively, such wide therapeutic window may have broad implications in the treatment of various diseases derived from bacterial infections. Furthermore, the excellent broad-spectrum antimicrobial activity and high selectivity of the optimum 16R4-loaded liposome (SNAL-16R4) suggested that its structure was most probably consistent with the virus-inspired SNAL, as we assumed, which achieved the transformation from structural bionics to functional implementation endowing the systems with virus-like infectivity.
To further characterize the structure of SNAL-16R4, we performed experiments to confirm the morphology, drug loading capacity, liposomal stability, and intercalation of the lipopeptides within the liposome membrane. Transmission electron microscopy (TEM) imaging (Figs. 2i and S5) revealed that SNAL-16R4 existed as homogeneous spheres with a typical lipid bilayer. 16R4 was loaded in SNAL-16R4 with a drug loading capacity of 28.0% and encapsulation efficacy of 93.5%. Since the encapsulation efficacy of SNAL-16R4 was high, the as-prepared SNAL-16R4 was utilized for subsequent investigations without additional purification. We monitored the structural change of SNAL-16R4 using dynamic light scattering (DLS) measurements and observed that SNAL-16R4 had a homogeneous distribution and remained stable with the diameter of approximately 100 nm during 3 months of storage (Figs. 2j and S6). Meanwhile, SNAL-16R4 maintained a high surface zeta potential of approximately +50 mV during the storage period, which could have an increasing trend for electrostatic binding with anionic bacterial membranes. The high zeta potential and liposomal stability of SNAL-16R4 suggested that 16R4 was stably encapsulated and enriched on the liposome surface, providing strong electrostatic repulsion between cationic nanoparticles that prevent aggregation. To further confirm the successful intercalation of 16R4 within the liposome lipid bilayer, we performed FRET analysis as previously reported [25]. In this analysis, the lipid bilayer of bare liposomes and SNAL-16R4 were doped with two different dyes (C6-NBD and DOPE-RhB) that constituted a FRET pair (Fig. 2k). We observed that the bare liposome lipid bilayer exhibited a noticeable FRET effect with a 460 nm excitation, as strong fluorescence was observed at approximately 583 nm. With the introduction of lipopeptide 16R4, a recovery of fluorescence occurred at the lower emission wavelength of approximately 530 nm. The results suggested that 16R4 was inserted into the liposome lipid bilayer; thus, it weakened the FRET effect of the dyes in the original liposome membrane. In general, we concluded that SNAL-16R4 adopted a structure similar to enveloped viruses, such as SARS-CoV-2, HIV, and influenza (Scheme 1 and Fig. S7), because the novel liposomal formulation possessed the structural features of a virus, including a particle size approximately 100 nm, a membranous structure composed of lipids, and a highly functionalized surface by biologically active peptides.
3.3. Antimicrobial mechanism of the virus-inspired SNAL
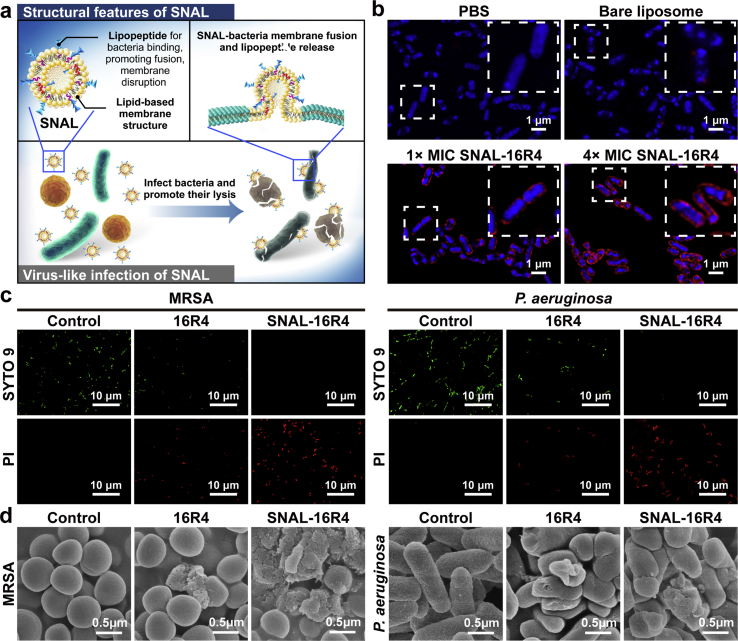
To elucidate the mechanism underlying the antibacterial activity of the virus-like SNAL (Fig. 3a), we evaluated the membrane fusion, permeabilization, and morphological damage effect of SNAL-16R4 on bacteria. First, the lipid bilayer of both SNAL-16R4 and bare liposomes was labeled with a red fluorophore DOPE-RhB, nucleoids of bacteria were stained blue using DAPI. As shown in Fig. 3b, while untreated bacteria only exhibited uniform DAPI staining, a strong signal of red fluorescence surrounding the bacterial nucleoids after 10 min of SNAL-16R4 treatment suggested the occurrence of a fast liposome-bacteria fusion process. In addition, when bacteria were treated with a higher concentration of SNAL-16R4, the red fluorescence increased in a dose-dependent manner, indicating a larger quantity of SNAL-16R4 fused with bacterial membrane. The results may explain the dose-dependent in vitro antibacterial effect of SNAL-16R4. In the bare liposome treatment group, sporadic red dots near the area of bacterial nucleoid were visible. The results suggested that liposomes without membrane-active peptides on the lipid bilayer may mediate poor liposome internalization through a mechanism distinct from SNAL-16R4. Subsequently, the membrane permeabilizing activity of 16R4 and SNAL-16R4 was studied using an SYTO 9 green-fluorescent nucleic acid stain and propidium iodide (PI) red-fluorescent nucleic acid stain, and imaged using confocal laser scanning microscopy (CLSM). SYTO 9 is a membrane-permeable dye that stains both intact cells and membrane-damaged bacterial cells. Conversely, membrane-impermeable dye PI can only permeate those bacterial cells whose cell membrane structures have been compromised. Furthermore, PI has higher affinity to bind DNA than SYTO 9. When bacterial cytoplasmic membrane is damaged and both stains are exposed to the same DNA, PI will replace SYTO 9 resulting in red fluorescent signal and a reduction in the SYTO 9 fluorescence [37,39,40]. As Fig. 3c shows, the control bacteria exhibited green-colored cells, indicating that the cell membranes of most bacteria were intact. In contrast, both 16R4 and SNAL-16R4 were observed to damage bacteria using a membrane-disruption mechanism generating red fluorescence. The CLSM images indicated that SNAL-16R4 induced more significant membrane permeabilization than free 16R4. Similarly, field emission scanning electron microscopy (FE-SEM) images showed that SNAL-16R4 could result in more distinct damage to the integrity of the bacterial cell walls and membranes than free 16R4 (Fig. 3d). Dramatic morphological changes, including collapse, deformation, lysis, and leakage of cell content, were visually observed in MRSA and P. aeruginosa cells after treatment using SNAL-16R4. In this study, lipopeptide 16R4 played a significant role in promoting both the binding and entry of liposomes as a membrane component similar to the spike protein of SARS-CoV-2, and eradicating bacterial infections as an effective membrane-disruptive agent. As attaining a certain threshold concentration in the membrane is indispensable for antimicrobial peptides and their analogs to disrupt the bacterial membrane bilayer [41], the enhanced membrane disruption may be caused by the fast fusion process of the SNAL in delivering a lethal dose of lipopeptide to the bacterial membranes. SNAL-16R4 not only has virus-like components and nanostructures, but also possesses virus-like infections as was expected.
Fig. 3.
Antimicrobial mechanism of action of SNAL-16R4. (a) Schematic of SNAL-16R4 fusing with a bacterial membrane for antibacterial. (b) Fluorescence study of the fusion interaction between SNAL-16R4 and P. aeruginosa after 10-min incubation, while an enlarged image of boxed regions is shown at the top right corner. (c) CLSM images of MRSA and P. aeruginosa treated with 16R4 or SNAL-16R4 at 8 × MIC for 2 h. (d) Morphological changes of the MRSA and P. aeruginosa with 16R4 or SNAL-16R4 treatment at 8 × MIC for 4 h.
3.4. In vivo antimicrobial efficacy of the virus-inspired SNAL
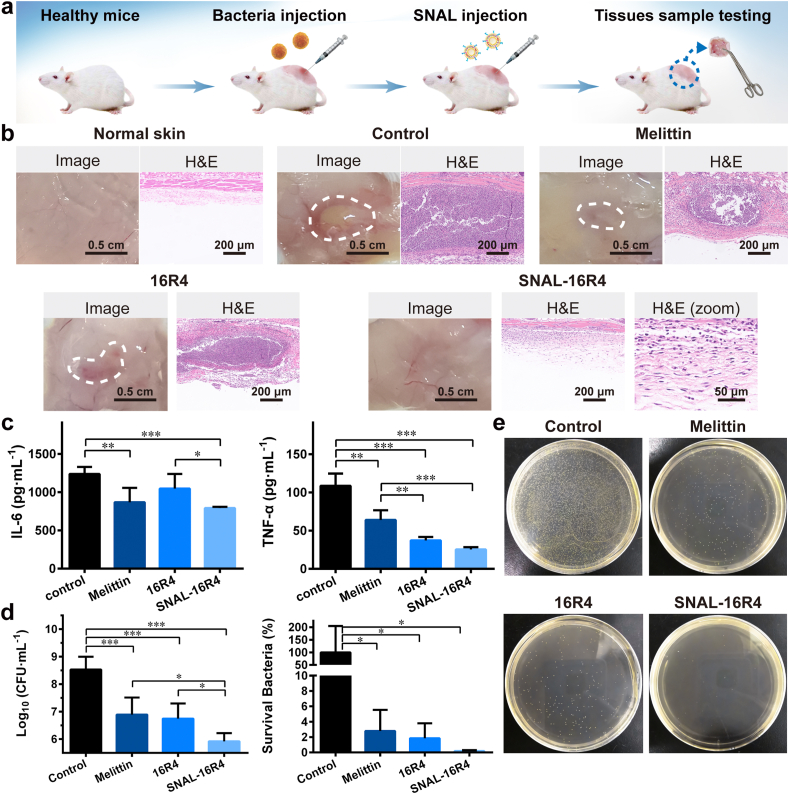
We further investigated the in vivo anti-bacterial efficacy of the virus-inspired SNAL-16R4 using a murine abscess model of MRSA infection following the reported method (Fig. 4a) [37,38]. Melittin, a potent natural antimicrobial peptide, was used as a positive control. As Fig. 4b shows, the representative photograph of control group mice showed noticeable skin abscess in the subcutaneous tissues, while the observed abscess or erythema in both melittin-treated and 16R4-treated mice was lower, and the subcutaneous tissues in SNAL-16R4-treated mice had no apparent skin lesions. A H&E histological examination revealed a massive infiltration of acute and chronic inflammatory cells, primarily neutrophils, in the subcutaneous connective tissue of control mice, accompanied by local abscesses. Inflammatory cell infiltration was observed to markedly reduce but remained observable after melittin and 16R4 treatments. H&E staining analysis indicated that the skin morphology of MRSA-infected mice after SNAL-16R4 treatment was nearly identical to that of normal healthy mice. Interestingly, compared with the control group mice, mice treated with SNAL-16R4 had significantly reduced MRSA-induced proinflammatory cytokines including IL-6 (p < 0.001) and TNF-α (p < 0.001), while SNAL-16R4 exhibited an improved therapeutic effect than free 16R4 (p < 0.05 for IL-6) (Fig. 4c). Suppressing the proinflammatory response is expected to reduce the inflammatory reaction responsible for perpetuating tissue injury.
Fig. 4.
In vivo antibacterial efficacy study of SNAL-16R4. (a) Schematic of the infection, treatment, and evaluation of the therapeutic efficacy used in mice with MRSA-induced cutaneous abscesses. (b) Representative infected skin photographs and H&E-stained sections of the infected mice skins after various treatments. (c) Levels of proinflammatory (IL-6 and TNF-α) cytokines measured using ELISA in the infected skin with various treatments. (d) Microbial burden and bacterial survival of each group after 48 h of MRSA injection in a murine model of skin. (e) Photographs of MRSA colonies in the agar plates from the homogenate of infected skin after being appropriately diluted. Mean ± SD, n = 4. *p < 0.05, **p < 0.01, ***p < 0.001.
To further evaluate the in vivo therapeutic efficacy of SNAL-16R4 against MRSA, we quantified the microbial burden in the infected tissues (Fig. 4d and e). Compared with the control group, SNAL-16R4 exhibited a potent in vivo bactericidal activity with approximately 405 times reduction in colony forming units (CFU), which was comparable and even superior to the performance of melittin. In this study, a single dose of SNAL-16R4 achieved a 99.8% reduction in the bacterial load in the skin abscess. Mimicking the viral architecture to construct SNAL-16R4 was observed to be particularly beneficial for enhancing antimicrobial activity; thus, the number of MRSA bacteria remaining in the dorsal skin of the SNAL-16R4-treated mice was 6.7 times lower than that in the 16R4-treated mice (p < 0.05).
3.5. In vivo safety of the virus-inspired SNAL
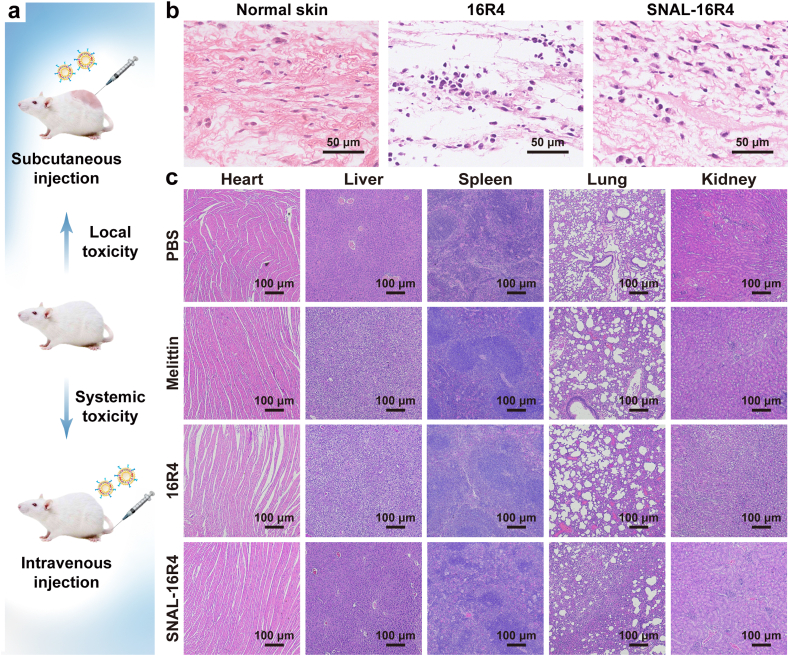
We evaluated the local and systemic toxicity of SNAL-16R4 in uninfected mice (Fig. 5a). Local irritation was analyzed using H&E staining images of the skin tissue at the injection site 48 h post-injection. As Fig. 5b shows, the skins injected with 16R4 were characterized by the loss of normal skin architecture and lymphocyte infiltration. Conversely, the subcutaneous injection of SNAL-16R4 exhibited good skin compatibility and no apparent skin irritation in mice. To study the metabolism and biocompatibility of drugs after subcutaneous SNAL-16R4 administration, we monitored the acute toxicity that SNAL-16R4 might cause to major metabolic organs and the balance of electrolytes at 48 h post-injection by analyzing the serum levels of ALT, AST, BUN, Cr, sodium, and potassium ions. As shown in Table S2, the levels of the functional parameters of the liver (ALT and AST) and kidney (BUN and Cr) as well as serum ion concentration in the blood remained unchanged. These preliminary results suggested that SNAL-16R4 could be metabolized and would not induce acute toxicity. Furthermore, we examined the major organs of mice 7 days after intravenous injection of various agents. The total blood volume in a mouse was estimated as ~6% of body weight; thus, a dose of 6 mg kg−1 was applied because it was 16–64 × MIC of SNAL-16R4 and greater than the median lethal doses (LD50) of many antibiotics, such as gramicidin (1.5 mg kg−1) and polymyxin B (5.4 mg kg−1) [37]. Additionally, melittin was selected as the control group because it is a typical antimicrobial peptide that has limited clinical use owing to its high systemic toxicity [30]. Fig. 5c shows significant pathological damage of melittin-treated mice and 16R4-treated mice, including liver cell degeneration and lung congestion. However, 7 days of SNAL-16R4 treatment did not cause animal weight loss and significant histological changes in different organs, indicating that the viral-like SNAL successfully reduced the systemic side effects of the lipopeptide.
Fig. 5.
Local and systemic toxicity evaluation of SNAL-16R4. (a) Schematic experimental protocol for in vivo safety evaluation of SNAL-16R4; (b) H&E-stained skin sections from mice injected subcutaneously with PBS, 16R4, or SNAL-16R4 for 48 h; (c) H&E-stained sections of major organs from the mice sacrificed on Day 7 after tail vein injection of PBS, melittin, 16R4, or SNAL-16R4.
4. Conclusion
We propose a biomimetic virus-like strategy to enhance the antibacterial efficacy of lipopeptides while avoiding severe side effects. Our designed SNAL-16R4 can mediate high-efficiency and high-selectivity bacteria cell membrane binding, rapidly attack and invade bacteria via plasma membrane fusion pathway, and induce a local “burst” release of lipopeptide to produce irreversible damage of cell membrane, achieving the transformation from structural bionics to functional implementation. Therefore, the virus-like infectivity of SNAL significantly improves the in vitro and in vivo antimicrobial activity, bactericidal efficiency, biocompatibility, and selectivity of loading lipopeptide 16R4. Overall, the results of this study show that SNAL-16R4 has significant potential as an effective and safe therapeutic agent for the treatment of bacterial infections.
CRediT authorship contribution statement
Yin Shi: Investigation, Methodology, Formal analysis, Data curation, Writing – original draft. Xiaoqian Feng: Methodology, Formal analysis. Liming Lin: Methodology, Formal analysis. Jing Wang: Investigation, Data curation. Jiaying Chi: Methodology, Formal analysis. Biyuan Wu: Investigation, Formal analysis. Guilin Zhou: Investigation, Data curation. Feiyuan Yu: Methodology, Data curation. Qian Xu: Investigation, Formal analysis. Daojun Liu: Conceptualization, Validation. Guilan Quan: Investigation, Validation. Chao Lu: Supervision, Conceptualization, Methodology, Visualization, Writing – review & editing. Xin Pan: Supervision, Resources, Validation. Jianfeng Cai: Supervision, Conceptualization, Writing – review & editing. Chuanbin Wu: Supervision, Resources, Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 81803467, 81773660), and the Research and Development Plan for Key Areas in Guangdong Province (No. 2019B020204002).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.02.038.
Contributor Information
Chao Lu, Email: chaolu@jnu.edu.cn.
Xin Pan, Email: panxin2@mail.sysu.edu.cn.
Jianfeng Cai, Email: jianfengcai@usf.edu.
Chuanbin Wu, Email: chuanbin_wu@126.com.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Cochrane S.A., Vederas J.C. Lipopeptides from bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med. Res. Rev. 2016;36:4–31. doi: 10.1002/med.21321. [DOI] [PubMed] [Google Scholar]
- 2.de Breij A., Riool M., Cordfunke R.A., Malanovic N., de Boer L., Koning R.I., Ravensbergen E., Franken M., van der Heijde T., Boekema B.K., Kwakman P.H.S., Kamp N., El Ghalbzouri A., Lohner K., Zaat S.A.J., Drijfhout J.W., Nibbering P.H. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aan4044. [DOI] [PubMed] [Google Scholar]
- 3.Mandal S.M., Barbosa A.E.A.D., Franco O.L. Lipopeptides in microbial infection control: scope and reality for industry. Biotechnol. Adv. 2013;31:338–345. doi: 10.1016/j.biotechadv.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z.J., Zhang X., Xu X.H., Li Y.K., Li Y.C., Zhong D., He Y.Y., Gu Z.W. Virus-inspired mimics based on dendritic lipopeptides for efficient tumor-specific infection and systemic drug delivery. Adv. Funct. Mater. 2015;25:5250–5260. [Google Scholar]
- 5.Jiang Q., Nie Y., Chen X.B., He Y., Yue D., Gu Z.W. pH-triggered pinpointed cascading charge-conversion and redox-controlled gene release design: modularized fabrication for nonviral gene transfection. Adv. Funct. Mater. 2017;27:1701571. [Google Scholar]
- 6.Blaskovich M.A.T., Hansford K.A., Gong Y.J., Butler M.S., Muldoon C., Huang J.X., Ramu S., Silva A.B., Cheng M., Kavanagh A.M., Ziora Z., Premraj R., Lindahl F., Bradford T.A., Lee J.C., Karoli T., Pelingon R., Edwards D.J., Amado M., Elliott A.G., Phetsang W., Daud N.H., Deecke J.E., Sidjabat H.E., Ramaologa S., Zuegg J., Betley J.R., Beevers A.P.G., Smith R.A.G., Roberts J.A., Paterson D.L., Cooper M.A. Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat. Commun. 2018;9:22. doi: 10.1038/s41467-017-02123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makovitzki A., Avrahami D., Shai Y. Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15997–16002. doi: 10.1073/pnas.0606129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koopmans T., Wood T.M., t Hart P., Kleijn L.H.J., Hendrickx A.P.A., Willems R.J.L., Breukink E., Martin N.I. Semisynthetic lipopeptides derived from nisin display antibacterial activity and lipid II binding on par with that of the parent compound. J. Am. Chem. Soc. 2015;137:9382–9389. doi: 10.1021/jacs.5b04501. [DOI] [PubMed] [Google Scholar]
- 9.Shen W., Zhang Y., Wan P.Q., An L., Zhang P., Xiao C.S., Chen X.S. Antineoplastic drug-free anticancer strategy enabled by host-defense-peptides-mimicking synthetic polypeptides. Adv. Mater. 2020;32:2001108. doi: 10.1002/adma.202001108. [DOI] [PubMed] [Google Scholar]
- 10.Perriman A.W., Williams D.S., Jackson A.J., Grillo I., Koomullil J.M., Ghasparian A., Robinson J.A., Mann S. Synthetic viruslike particles and hybrid constructs based on lipopeptide self-assembly. Small. 2010;6:1191–1196. doi: 10.1002/smll.200901186. [DOI] [PubMed] [Google Scholar]
- 11.Mowery B.P., Lindner A.H., Weisblum B., Stahl S.S., Gellman S.H. Structure-activity relationships among random nylon-3 copolymers that mimic antibacterial host-defense peptides. J. Am. Chem. Soc. 2009;131:9735–9745. doi: 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]
- 12.Chen W.Y., Chang H.Y., Lu J.K., Huang Y.C., Harroun S.G., Tseng Y.T., Li Y.J., Huang C.C., Chang H.T. Self-assembly of antimicrobial peptides on gold nanodots: against multidrug-resistant bacteria and wound-healing application. Adv. Funct. Mater. 2015;25:7189–7199. [Google Scholar]
- 13.Rohovie M.J., Nagasawa M., Swartz J.R. Virus-like particles: next-generation nanoparticles for targeted therapeutic delivery. Bioeng. Transl. Med. 2017;2:43–57. doi: 10.1002/btm2.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Lin J.Y., Wang P.Y., Luo Q., Lin H.R., Zhang Y., Hou Z.Q., Liu J.F., Liu X.L. Tumor microenvironment responsive shape reversal self-targeting virus-inspired nanodrug for imaging-guided near-infrared-II photothermal chemotherapy. ACS Nano. 2019;13:12912–12928. doi: 10.1021/acsnano.9b05425. [DOI] [PubMed] [Google Scholar]
- 15.Wu H., Zhong D., Zhang Z.J., Li Y.C., Zhang X., Li Y.K., Zhang Z.Z., Xu X.H., Yang J., Gu Z.W. Bioinspired artificial tobacco mosaic virus with combined oncolytic properties to completely destroy multidrug-resistant cancer. Adv. Mater. 2020;32:1904958. doi: 10.1002/adma.201904958. [DOI] [PubMed] [Google Scholar]
- 16.Boato F., Thomas R.M., Ghasparian A., Freund-Renard A., Moehle K., Robinson J.A. Synthetic virus-like particles from self-assembling coiled-coil lipopeptides and their use in antigen display to the immune system. Angew. Chem. Int. Ed. 2007;46:9015–9018. doi: 10.1002/anie.200702805. [DOI] [PubMed] [Google Scholar]
- 17.Sun S.Y., Wang R.M., Huang Y.D., Xu J.L., Yao K., Liu W.S., Cao Y.M., Qian K. Design of hierarchical beads for efficient label-free cell capture. Small. 2019;15:1902441. doi: 10.1002/smll.201902441. [DOI] [PubMed] [Google Scholar]
- 18.Ye Y.Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- 19.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., Baysal E., Mangold M., Henze L., Lauster R., Mall M.A., Beyer K., Rohmel J., Voigt S., Schmitz J., Miltenyi S., Demuth I., Muller M.A., Hocke A., Witzenrath M., Suttorp N., Kern F., Reimer U., Wenschuh H., Drosten C., Corman V.M., Giesecke-Thiel C., Sander L.E., Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 20.Wrapp D., Wang N.S., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia S., Liu M.Q., Wang C., Xu W., Lan Q.S., Feng S.L., Qi F.F., Bao L.L., Du L.Y., Liu S.W., Qin C., Sun F., Shi Z.L., Zhu Y., Jiang S.B., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su H., Su H., Zhou F., Huang Z., Ma X., Natarajan K., Zhang M., Huang Y. Molecular insights into small molecule drug discovery for SARS-CoV-2. Angew. Chem. Int. Ed. 2021;60:2–16. doi: 10.1002/anie.202008835. [DOI] [PubMed] [Google Scholar]
- 23.Thapa R.K., Diep D.B., Tonnesen H.H. Topical antimicrobial peptide formulations for wound healing: current developments and future prospects. Acta Biomater. 2020;103:52–67. doi: 10.1016/j.actbio.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 24.Nordstrom R., Malmsten M. Delivery systems for antimicrobial peptides. Adv. Colloid Interface Sci. 2017;242:17–34. doi: 10.1016/j.cis.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Dehaini D., Wei X.L., Fang R.H., Masson S., Angsantikul P., Luk B.T., Zhang Y., Ying M., Jiang Y., Kroll A.V., Gao W.W., Zhang L.F. Erythrocyte-platelet hybrid membrane coating for enhanced nanoparticle functionalization. Adv. Mater. 2017;29:1606209. doi: 10.1002/adma.201606209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thamphiwatana S., Gao W.W., Obonyo M., Zhang L.F. In vivo treatment of Helicobacter pylori infection with liposomal linolenic acid reduces colonization and ameliorates inflammation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17600–17605. doi: 10.1073/pnas.1418230111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y.Q., Smith C., Wu H.F., Teng P., Shi Y., Padhee S., Jones T., Nguyen A., Cao C.H., Yin H., Cai J.F. Short antimicrobial lipo-a/g-AA hybrid peptides. Chembiochem. 2014;15:2275–2280. doi: 10.1002/cbic.201402264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gide M., Nimmagadda A., Su M., Wang M.H., Teng P., Li C.P., Gao R.X., Xu H., Li Q., Cai J.F. Nano-sized lipidated dendrimers as potent and broad-spectrum antibacterial agents. Macromol. Rapid Commun. 2018;39:1800622. doi: 10.1002/marc.201800622. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt N., Mishra A., Lai G.H., Wong G.C.L. Arginine-rich cell-penetrating peptides. FEBS Lett. 2010;584:1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 30.Wang J., Lu C., Shi Y., Feng X.Q., Wu B.Y., Zhou G.L., Quan G.L., Pan X., Cai J.F., Wu C.B. Structural superiority of guanidinium-rich, four-armed copolypeptides: role of multiple peptide-membrane interactions in enhancing bacterial membrane perturbation and permeability. ACS Appl. Mater. Interfaces. 2020;12:18363–18374. doi: 10.1021/acsami.0c02752. [DOI] [PubMed] [Google Scholar]
- 31.Lei R.Y., Hou J.C., Chen Q.X., Yuan W.R., Cheng B.L., Sun Y.Q., Jin Y., Ge L.J., Ben-Sasson S.A., Chen J., Wang H.X., Lu W.Y., Fang X.M. Self-assembling myristoylated human alpha-defensin 5 as a next-generation nanobiotics potentiates therapeutic efficacy in bacterial infection. ACS Nano. 2018;12:5284–5296. doi: 10.1021/acsnano.7b09109. [DOI] [PubMed] [Google Scholar]
- 32.Chin W., Zhong G.S., Pu Q.Q., Yang C., Lou W.Y., De Sessions P.F., Periaswamy B., Lee A., Liang Z.C., Ding X., Gao S.J., Chu C.W., Bianco S., Bao C., Tong Y.W., Fan W.M., Wu M., Hedrick J.L., Yang Y.Y. A macromolecular approach to eradicate multidrug resistant bacterial infections while mitigating drug resistance onset. Nat. Commun. 2018;9:917. doi: 10.1038/s41467-018-03325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li P., Zhou C., Rayatpisheh S., Ye K., Poon Y.F., Hammond P.T., Duan H.W., Chan-Park M.B. Cationic peptidopolysaccharides show excellent broad-spectrum antimicrobial activities and high selectivity. Adv. Mater. 2012;24:4130–4137. doi: 10.1002/adma.201104186. [DOI] [PubMed] [Google Scholar]
- 34.Prussin A.J., Schwake D.O., Lin K., Gallagher D.L., Buttling L., Marr L.C. Survival of the enveloped virus phi6 in droplets as a function of relative humidity, absolute humidity, and temperature. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.00551-18. e00551-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang C.M., Chen C.H., Pornpattananangkul D., Zhang L., Chan M., Hsieh M.F., Zhang L.F. Eradication of drug resistant Staphylococcus aureus by liposomal oleic acids. Biomaterials. 2011;32:214–221. doi: 10.1016/j.biomaterials.2010.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li P., Poon Y.F., Li W.F., Zhu H.Y., Yeap S.H., Cao Y., Qi X.B., Zhou C.C., Lamrani M., Beuerman R.W., Kang E.T., Mu Y.G., Li C.M., Chang M.W., Leong S.S.J., Chan-Park M.B. A polycationic antimicrobial and biocompatible hydrogel with microbe membrane suctioning ability. Nat. Mater. 2011;10:149–156. doi: 10.1038/nmat2915. [DOI] [PubMed] [Google Scholar]
- 37.Lu C., Quan G.L., Su M., Nimmagadda A., Chen W.D., Pan M., Teng P., Yu F.Y., Liu X., Jiang L., Du W.Y., Hu W., Yao F., Pan X., Wu C.B., Liu D.J., Cai J.F. Molecular architecture and charging effects enhance the in vitro and in vivo performance of multi-arm antimicrobial agents based on star-shaped poly(L-lysine) Adv. Ther. 2019;2:1900147. [Google Scholar]
- 38.Zheng M.C., Pan M., Zhang W.C., Lin H.C., Wu S.L., Lu C., Tang S.J., Liu D.J., Cai J.F. Poly(α-L-lysine)-based nanomaterials for versatile biomedical applications: current advances and perspectives. Bioact. Mater. 2021;6:1878–1909. doi: 10.1016/j.bioactmat.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahman A., Jui M.S., Bam M., Cha Y.J., Luat E., Alabresm A., Nagarkatti M., Decho A., Tang C.B. Facial amphiphilicity-induced polymer nanostructures for antimicrobial applications. ACS Appl. Mater. Interfaces. 2020;12:21221–21230. doi: 10.1021/acsami.9b19712. [DOI] [PubMed] [Google Scholar]
- 40.Wang Peng, Yuan Yonghui, Xu Ke, Zhong Hongshan, Yang Yinghui, Jin Shiyu, Yang Ke, Qi Xun. Biological applications of copper-containing materials. Bioact. Mater. 2021;6:916–927. doi: 10.1016/j.bioactmat.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melo M.N., Ferre R., Castanho M.A.R.B. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009;7:245–250. doi: 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.