Abstract
Dioecy, the presence of separate sexes on distinct individuals, has evolved repeatedly in multiple plant lineages. However, the specific mechanisms by which sex systems evolve and their commonalities among plant species remain poorly understood. With both XY and ZW sex systems, the family Salicaceae provides a system to uncover the evolutionary forces driving sex chromosome turnovers. In this study, we performed a genome-wide association study to characterize sex determination in two Populus species, P. euphratica and P. alba. Our results reveal an XY system of sex determination on chromosome 14 of P. euphratica, and a ZW system on chromosome 19 of P. alba. We further assembled the corresponding sex-determination regions, and found that their sex chromosome turnovers may be driven by the repeated translocations of a Helitron-like transposon. During the translocation, this factor may have captured partial or intact sequences that are orthologous to a type-A cytokinin response regulator gene. Based on results from this and other recently published studies, we hypothesize that this gene may act as a master regulator of sex determination for the entire family. We propose a general model to explain how the XY and ZW sex systems in this family can be determined by the same RR gene. Our study provides new insights into the diversification of incipient sex chromosomes in flowering plants by showing how transposition and rearrangement of a single gene can control sex in both XY and ZW systems.
Keywords: dioecy, sex determination, sex chromosome turnover, genome, Populus
Introduction
The origin and evolution of dioecy (separate sexes) has long been one of the most fascinating topics for biologists (Henry et al. 2018; Feng et al. 2020). In flowering plants, dioecy occurs in only ∼6% of all species and has independently evolved thousands of times from hermaphroditic ancestors (Renner and Ricklefs 1995; Renner 2014). Many of these species have sex determined by a pair of sex chromosomes that differ in morphology and/or sequence, in the form of male heterogamety (XY system) or female heterogamety (ZW system) (Ming et al. 2011; Charlesworth 2016). Theory predicts that sex chromosomes evolve from ancestral autosomes via successive mutations in two linked genes with complementary dominance (Charlesworth and Charlesworth 1978; Charlesworth 1991). Subsequently, the suppression of recombination between these two sex-determining genes progressively spreads along Y or W chromosomes, and permits the accumulation of repetitive elements and duplication or translocation of genomic fragments, which in turn leads to the formation of a sex-specific region and finally degeneration of the sex chromosome (Bergero and Charlesworth 2009; Bachtrog 2013; Charlesworth 2013). Characterizing the genomic architecture of sex in dioecious species is critical for understanding the origin of sex chromosomes, especially in their early stage of evolution.
Over the past decade, impressive progress has been made in unraveling the genetic basis of sex determination in several dioecious plants and the evolutionary history of their sex chromosomes, including papaya (Wang et al. 2012), persimmon (Akagi et al. 2014), asparagus (Harkess et al. 2017), strawberry (Tennessen et al. 2018), date palm (Torres et al. 2018), and kiwifruit (Akagi et al. 2018; Akagi et al. 2019). Consistent with the independent origins of sex chromosomes, the sex-determining genes identified in these species differ from each another, although most of them function in similar hormone response pathways (Feng et al. 2020). In addition, recent studies suggest rapid turnover of sex-determination regions (SDRs) in some plant lineages (Charlesworth 2015; Moore et al. 2016; Henry et al. 2018). For example, the sex chromosome turnover in strawberries is driven by repeated translocation of a female-specific sequence (Tennessen et al. 2018); a heterogamety change from female to male was also proposed in section Otites of the genus Silene (Balounova et al. 2019). The combined evidence from these studies demonstrates the high variation of plant sex-determination mechanisms, and so understanding the factors that drive the convergent evolution of sex chromosomes in plants remains elusive (Zhang et al. 2014).
The family Salicaceae provides an excellent system to study the drivers of sex chromosome evolution. This family includes two sister genera, Populus and Salix, which are comprised exclusively of dioecious species (Peto 1938; Zhang et al. 2018; Li et al. 2020). Previous studies in multiple Salix species have consistently mapped the SDRs to chromosome 15, and proposed a ZW system in which females are the heterogametic sex (Hou et al. 2015; Pucholt et al. 2015; Chen et al. 2016; Pucholt et al. 2017; Zhou et al. 2018; Zhou, Macaya-Sanz, Carlson, et al. 2020). However, an XY system was recently identified on chromosome 7 in S. nigra (Sanderson et al., unpublished data). In comparison, the SDR has been mapped to multiple locations in different Populus species, indicating a dynamic evolutionary history of the sex chromosomes. The SDR has been mapped to the proximal telomeric end of chromosome 19 in P. trichocarpa and P. nigra (sections Tacamahaca and Aigeiros) (Gaudet et al. 2007; Yin et al. 2008; Geraldes et al. 2015; Zhou, Macaya-Sanz, Schmutz, et al. 2020), and to a pericentromeric region of chromosome 19 in P. tremula, P. tremuloides, and P. alba (section Populus) (Pakull et al. 2009; Paolucci et al. 2010; Kersten et al. 2014; Pakull et al. 2015). Most Populus species display an XY sex-determination system, but there is some evidence that P. alba has a ZW system (Paolucci et al. 2010). Thus far, the only SDRs that has been assembled in Populus are those of P. trichocarpa, P. deltoides, and P. tremula, and these appear to be much smaller than those observed in Salix (Geraldes et al. 2015; Müller et al. 2020; Zhou, Macaya-Sanz, Schmutz, et al. 2020; Xue et al., unpublished data). Our recent study on the W chromosome of S. purpurea showed intriguing palindromic structures, in which four copies of the gene encoding a type A cytokinin response regulator (RR) were identified (Zhou, Macaya-Sanz, Carlson, et al. 2020). Interestingly, the ortholog of this gene has also been reported to be associated with sex in Populus, where inverted repeats of truncated portions of the gene on the Y chromosome are likely responsible for silencing the intact gene in males (Geraldes et al. 2015; Brautigam et al. 2017; Melnikova et al. 2019; Müller et al. 2020). This gene is therefore an excellent candidate for a common sex-determination mechanism in the Salicaceae. However, it is still unclear whether this candidate gene is present in all of the SDRs in the family. Here, we identify the sex-determination systems of two additional Populus species, P. euphratica and P. alba, which are from sects. Turanga and Populus, respectively (Wang et al. 2020). We report their complete SDR assemblies and propose a general model to illustrate the potentially shared mechanism of sex determination in this family.
Results
Genome Assembly
We have previously reported the assembly of the genomes of a male P. euphratica (Zhang et al. 2020) and a male P. alba var. pyramidalis (a variety of P. alba) (Ma et al. 2019). Here, we further sequenced and de novo assembled female genomes for both species using Oxford Nanopore reads. The assembly for the female P. euphratica consists of 1,229 contigs with an N50 of 1.7 Mb and a total size of ∼529.0 Mb, whereas the female P. alba var. pyramidalis assembly has 357 contigs with an N50 of 3.08 Mb, covering a total of ∼358.5 Mb (supplementary table S1, Supplementary Material online). Both assemblies showed extensive synteny with their respective male reference genomes, and therefore, based on their syntenic relationships, the assembled contigs were anchored onto 19 pseudochromosomes (supplementary figs. S1 and S2, Supplementary Material online). The chromosome identities were then assigned by comparison to P. trichocarpa (Tuskan et al. 2006).
XY Sex Determination on Chromosome 14 in P. euphratica
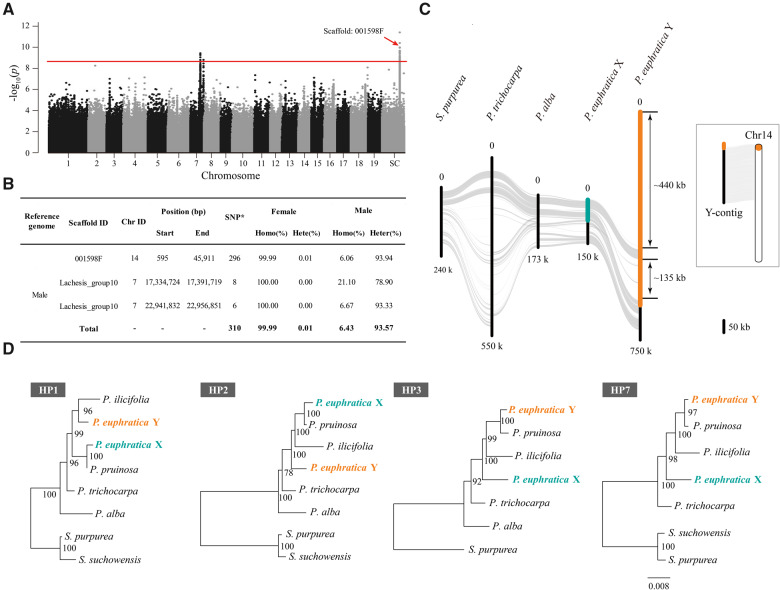
In order to characterize the sex-determination system of P. euphratica, we resequenced the genomes of 30 male and 30 female individuals (supplementary table S2, Supplementary Material online) and performed a genome-wide association study (GWAS). Using the male assembly as the reference genome, a total of 24,651,023 high-quality single-nucleotide polymorphisms (SNPs) were identified. After Bonferroni correction, we recovered 310 SNPs significantly associated with sex (α < 0.05; figs. 1A and supplementary fig. S3A and table S3, Supplementary Material online). In-depth analysis found that almost all genotypes (99.99%) of these sex-associated loci were homozygous in females, whereas 93.57% of the genotypes were heterozygous in males (fig. 1B). A similar pattern was observed when the sex association analysis was performed by using the female assembly as the reference genome (supplementary figs. S3B and S4 and tables S4 and S5, Supplementary Material online). These results consistently indicate that an XY system is involved in sex determination of P. euphratica.
Fig. 1.
XY sex determination on chromosome 14 in Populus euphratica. (A) Manhattan plot of P. euphratica based on the results of genome-wide association study (GWAS) with the male genome as reference. The y axis represents the strength of association (−log10(P value)) for each SNP sorted by chromosomes and scaffolds (SC; x axis). The red line indicates the significance after Bonferroni multiple testing correction (α < 0.05). Note that the scaffold “001598F” is located on chromosome 14 based on its syntenic relationship with the proximal end of chromosome 14 of P. trichocarpa. (B) Summary of male P. euphratica genome regions containing SNPs significantly associated with sex. “SNP*,” significantly associated SNPs; “Homo,” Homozygous; “Hete,” Heterozygous. (C) Synteny relationships between our assembled Y-contig and X chromosome of P. euphratica, as well as the corresponding region of chromosome 14 for P. alba, P. trichocarpa, and Salix purpurea. The highlighted part represents the sex-determination region (SDR), yellow for Y-SDR and green for X-SDR. Schematic diagram showing the corresponding position of the SDR on chromosome 14 of P. euphratica. (D) Phylogenetic relationships of the homologous pairs (HP) shared between Y- and X-SDR of P. euphratica and their orthologous genes in other Salicaceae species. Detailed information about these genes is listed in supplementary table S7, Supplementary Material online and additional phylogenetic trees are shown in supplementary figure S7, Supplementary Material online. Note that only the orthologous genes located on the corresponding region of chromosome 14 were used for phylogenetic analysis.
The vast majority of the sex-associated markers occurred on a scaffold from chromosome 14 (the unanchored scaffold “001598F” in male genome was located onto chromosome 14 based on its syntenic relationship with P. trichocarpa genome), whereas a few other SNPs were present at chromosomes 7, 9, 12, and 19 (fig. 1A and B and supplementary fig. S4 and table S5, Supplementary Material online). We then attempted to use ultralong nanopore reads generated from a male individual (supplementary table S6, Supplementary Material online) to further reconstruct a new assembly with X and Y haplotypes as separate contigs. This led to the identification of a large contig (∼14 Mb) that was highly similar to the sex-associated regions and specifically contained Y-linked alleles (supplementary fig. S5, Supplementary Material online). The Y-linked region was further determined by examining the relative depth of coverage when aligning male versus female resequencing reads against the reference (supplementary fig. S6, Supplementary Material online). Based on the syntenic relationship, the SDR of P. euphratica can be mapped to the proximal end of chromosome 14 and the Y-linked region is ∼658 kb in length, corresponding to ∼84 kb on the X chromosome (fig. 1C). We found that two segments spanning 440 and 135 kb, respectively, were specific to the Y-linked region (fig. 1C), suggesting the occurrence of significant chromosome divergence between the X and Y haplotypes, which can be maintained by suppressed recombination.
We predicted a total of 37 protein-coding genes in the Y-linked region, many of which have high similarity with genes on other autosomes and are considered as translocated genes (supplementary table S7, Supplementary Material online). Among these, we found that nine of the Y-specific genes were annotated as members of the LONELY GUY (LOG) family, which encodes cytokinin-activating enzymes that play a dominant role in the maintenance of the shoot apical meristem and in the establishment of determinate floral meristems (Kuroha et al. 2009; Tokunaga et al. 2012; Han and Jiao 2015). Ten genes were identified in both X and Y haplotypes. A phylogenetic analysis of these genes showed that the X and Y alleles began to diverge after their split with P. trichocarpa and P. alba, but before the diversification of section Turanga (fig. 1D and supplementary fig. S7, Supplementary Material online), suggesting that the SDR of P. euphratica appears to be established relatively recently. Although we cannot separate the X and Y haplotypes in P. ilicifolia and P. pruinosa, the unstable phylogenetic pattern suggests a shared SDR among the species of section Turanga. The average synonymous substitution rate (dS) of the ten pairs of X–Y homologs of P. euphratica was 0.035 ± 0.0066 SE (supplementary fig. S8 and table S7, Supplementary Material online). Assuming the divergence time between Salix and Populus was 60 Ma (Tuskan et al. 2006) and the average dS was 0.146 ± 0.0022 SE between S. purpurea and P. trichocarpa (Zhou, Macaya-Sanz, Carlson, et al. 2020), the estimated age of the X–Y divergence of P. euphratica was ∼14.4 Ma. This is further evidence that the SDR of P. euphratica was established before the diversification of section Turanga (Chen et al. 2020).
ZW Sex Determination on Chromosome 19 in P. alba
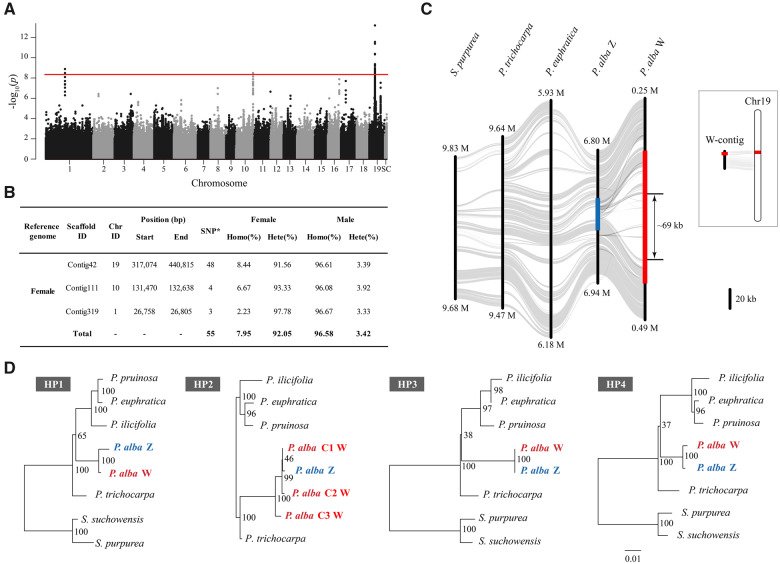
We used a similar GWAS strategy for 30 male and 30 female resequenced individuals to characterize the sex-determination system of P. alba (supplementary table S8, Supplementary Material online). When the male and female assembly was used as a reference genome, respectively, 173 and 55 SNPs that were significantly associated with sex were identified (fig. 2A and B and supplementary figs. S9 and S10 and tables S9–11, Supplementary Material online). Most of the sex-associated SNPs are heterozygous in females and homozygous in males (fig. 2B and supplementary table S10, Supplementary Material online), confirming the ZW sex-determination system in P. alba, which was also suggested based on genetic mapping in a previous study (Paolucci et al. 2010), and confirmed by local genome assemblies more recently (Müller et al. 2020).
Fig. 2.
ZW sex determination on chromosome 19 in Populus alba. (A) Manhattan plot of P. alba based on the results of GWAS with female genome as reference. The y axis represents the strength of association (−log10(P value)) for each SNP sorted by chromosomes and scaffolds (SC; x axis). The red line indicates the significance after Bonferroni multiple testing correction (α < 0.05). (B) Summary of female P. alba genome regions containing SNPs significantly associated with sex. “SNP*,” significantly associated SNPs; “Homo,” Homozygous; “Hete,” Heterozygous. (C) Synteny relationships between our assembled W-contig and Z chromosome of P. alba, as well as the corresponding region of chromosome 19 for P. euphratica, P. trichocarpa, and Salix purpurea. The highlighted part represents SDR, red for W-SDR and blue for Z-SDR. Schematic diagram showing the corresponding position of the SDR on chromosome 19 of P. alba. (D) Phylogenetic relationships of the homologous pairs (HP) shared between W- and Z-SDR of P. alba and their orthologous genes in other Salicaceae species. Detailed information about these genes is listed in supplementary table S12, Supplementary Material online. Note that, there are three copies for “HP2” on the W-SDR of P. alba, and only the orthologous genes located on the corresponding region of chromosome 19 were used for phylogenetic analysis.
We found that these sex-associated SNPs are mainly located on a nonterminal region of chromosome 19 (fig. 2A and B and supplementary fig. S9 and table S10, Supplementary Material online). Next, we examined the female-specific depth profile, combined with the support of ultralong nanopore reads (supplementary table S6, Supplementary Material online), to delineate the W haplotype of P. alba to a region of ∼140 kb on chromosome 19, with a corresponding Z haplotype that is only 33 kb in length (fig. 2C and supplementary figs. S11 and S12, Supplementary Material online). Compared with the Z haplotype and corresponding autosomal regions of the other Salicaceae species, a specific insertion of 69 kb was observed in the W haplotype, indicating a recent origin of the SDR in P. alba.
Sequence annotation predicted 18 protein-coding genes in the W haplotype, six of which were also found in the Z haplotype (supplementary table S12, Supplementary Material online). One of these genes is a homolog of SOMBRERO (SMB), a NAC-domain transcription factor which has a similar function to the VND/NST transcription factors that regulate secondary cell wall thickening in woody tissues and maturing anthers of Arabidopsis (Mitsuda et al. 2005; Bennett et al. 2010). This gene was expanded from one member in the Z haplotype to three copies in the W haplotype (“HP2” in fig. 2D). Because the estimated dS between Z and W alleles varied greatly among the four gene pairs (from 0.013 to 0.236, supplementary fig. S8 and table S12, Supplementary Material online), we cannot accurately assess the age of the Z–W divergence. However, the close relationship of these alleles between the W and Z haplotype suggests that recombination suppression occurred very recently in P. alba (fig. 2D). There are 12 genes specific to the W haplotype (supplementary table S12, Supplementary Material online), including DM2H (DANGEROUS MIX2H), which encodes a nucleotide-binding domain and leucine-rich repeat immune receptor protein (Chae et al. 2014); CCR2 (Cinnamoyl CoA reductase), which is involved in lignin biosynthesis and plant development (Thévenin et al. 2011); and STRS1 (STRESS RESPONSE SUPPRESSOR1), a gene encoding a DEAD-box RNA helicase, which is involved in epigenetic gene silencing related to stress responses (Khan et al. 2014). More interestingly, we identified three copies of the gene encoding a type A cytokinin response regulator (RR) in the W-specific region (fig. 3A), the ortholog of which has also been identified to be associated with sex determination in poplar and willow (Geraldes et al. 2015; Brautigam et al. 2017; Melnikova et al. 2019; Müller et al. 2020; Zhou, Macaya-Sanz, Carlson, et al. 2020). Few sequence differences were found among these three copies, and combined with the fact that the ortholog of the RR gene is located at the distal end of chromosome 19 in P. trichocarpa and P. euphratica (fig. 3), we conclude that the RR gene was translocated from the end of chromosome 19 to the W haplotype of P. alba and then underwent at least two rounds of recent duplication.
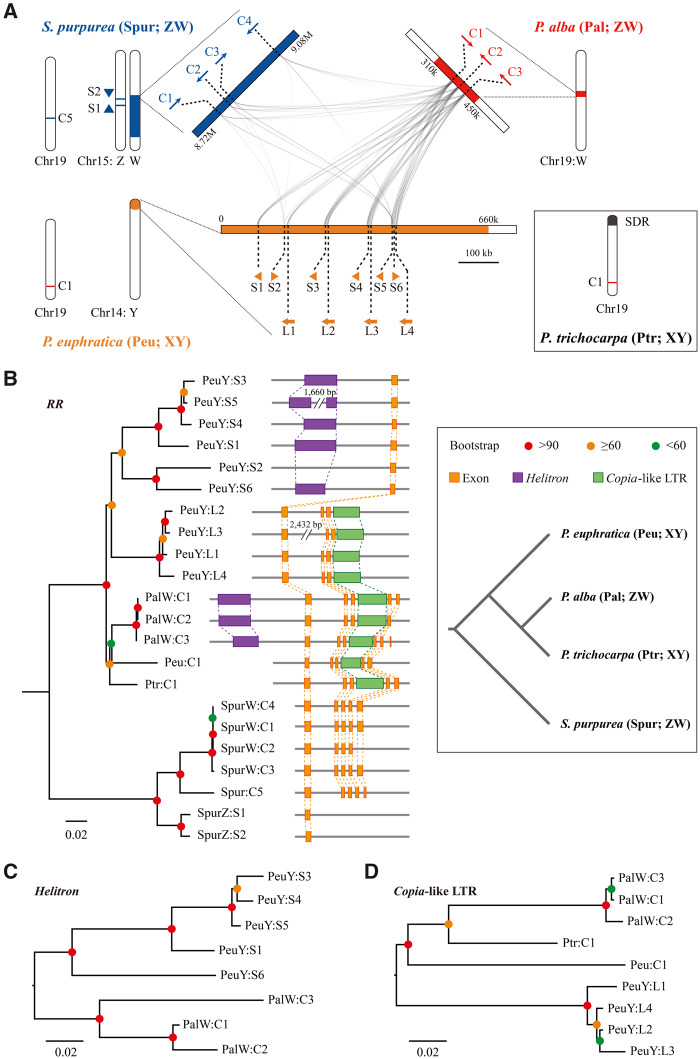
Fig. 3.
Evidence for SDR turnover in Salicaceae. (A) Synteny relationships among the Y-SDR of Populus euphratica (yellow) and the W-SDRs of P. alba (red) and Salix purpurea (blue), showing the copies of RR intact gene (“C”) and partial duplicates (S, small duplicate; L, large duplicate) on their SDRs. For each species, corresponding positions for other RR gene copies or partial duplicates on the autosome are also shown. (B) Phylogenetic relationship of the RR sequences (including intact genes and partial duplicates) identified in the four species. The tree was rooted by a paralogous gene “RR16” (ID: Potri.019G058900). The gene structures and relative positions of Helitron and Copia-like LTR are also shown. The phylogeny of the four species was also showed in the box. Phylogenetic relationships of the Helitron (C) and Copia-like LTR (D) around the RR sequences. All the sequences were named according to figure 3A. Peu, P. euphratica; Pal, P. alba; Ptr, P. trichocarpa; Spur, S. purpurea.
Evidence for SDR Turnover in Salicaceae
We have shown that P. euphratica and P. alba have different sex-determination systems, and that the SDRs are different from those reported in P. trichocarpa and S. purpurea, indicating extraordinarily high diversity of sex determination in the Salicaceae. In order to examine whether the SDRs originated independently in each lineage, or evolved into the current SDRs separately after a common ancient origin, we performed syntenic analysis on these SDRs in P. euphratica and P. alba, and the corresponding autosomal regions in P. trichocarpa and S. purpurea. We found that although the pseudoautosomal regions of these sex chromosomes are highly collinear with their corresponding autosomal regions in other species, the sequences in the sex-specific regions are not alignable (figs. 1C and 2C). In contrast, although there was little collinearity among these SDRs, a homologous sequence with multiple duplicates was identified between the Y haplotype of P. euphratica and the W haplotype of P. alba (fig. 3A). Interestingly, the locations of the duplicates overlapped with the three predicted RR genes in P. alba. In the corresponding regions of the Y haplotype of P. euphratica, we identified ten partial duplicates of the RR gene including four covering the first three exons (large duplicate) and six covering only the first exon (small duplicate) of the RR gene (fig. 3). No premature stop codons were detected within these duplicates, and the nonsynonymous to synonymous substitution rate ratios (dN/dS) between the large duplicates and intact RR gene of P. euphratica were <1 (supplementary fig. S13, Supplementary Material online), suggesting that they have been undergoing purifying selection and might be functional. However, it was observed that the dN/dS between the small duplicates and intact RR gene were close to 1, which is consistent with the characteristic of pseudogenes evolving without selection pressure (supplementary fig. S13, Supplementary Material online). Remarkably, small RNA-Seq revealed that these partial duplicates could generate small-interfering RNAs (siRNAs) that may affect the expression of the intact RR gene (supplementary fig. S14, Supplementary Material online), which was further supported by no expression of the RR gene in male flower bud of P. euphratica (supplementary fig. S15, Supplementary Material online). In addition, phylogenetic analysis of these duplicates showed that the three RR genes in P. alba clustered together and are closely related to the intact orthologs of P. euphratica and P. trichocarpa, whereas the partial duplicates from P. euphratica divided into two main clades, one with only large duplicates and a second clade with only small duplicates (fig. 3B).
As the RR duplicates were found in the SDRs of all of the current and previously studied species, we believe that they may play important roles in sex determination of the Salicaceae species. These results also lead to the hypothesis that these species shared an ancient origin of sex chromosomes, followed by frequent turnover events due to translocation of the RR duplicates. This is further supported by the distant relationship between the partial and intact RR duplicates (fig. 3B), which indicate that the partial duplicates originated before the divergence of these poplar species and were repeatedly inserted into the SDRs of P. euphratica. Further support is provided by a broader phylogenetic analysis, which shows that the partial duplicates of P. trichocarpa, P. deltoides, and P. euphratica occur together in a separate clade from the full-length genes. However, the partial duplicates from P. tremula occur together with the full-length genes from P. tremula and P. alba (both from section Populus), suggesting a more recent, independent origin (supplementary fig. S16, Supplementary Material online).
We did not detect any structurally intact long-terminal repeat retrotransposons (LTR-RTs) around these RR duplicates, which made it impossible to estimate their insertion time. However, around the RR duplicates in P. euphratica, we identified a Helitron-like transposable element upstream of each small duplicate except the second one (“PeuY:S2”), and a Copia-like LTR fragment in the downstream region of each large duplicate (fig. 3B). These two repetitive elements were also identified in all three RR duplicates of P. alba, and are located upstream and in the third intron of the RR gene, respectively, similar to that in P. euphratica. The phylogenetic trees of the two elements and the RR duplicates exhibited a similar topological relationship, suggesting that they may be transposed together as a unit (fig. 3C and D). The extremely high similarity of these sequences indicates that they were recently transposed into the SDRs of P. euphratica and P. alba, respectively, consistent with the observation that their sex chromosomes have not been severely degenerated. In addition, we found that the Helitron-like element was not present in the upstream region of the intact RR genes on chromosome 19 of P. euphratica and P. trichocarpa (fig. 3B), which led us to speculate that this element may be the main driving force for gene replication during the evolution of the SDRs in P. euphratica and P. alba. However, we failed to detect the same pattern in S. purpurea, in which multiple Copia LTR-RTs were predicted instead of the Helitron elements (Zhou, Macaya-Sanz, Carlson, et al. 2020). This implies that poplar and willow may have different SDR turnover mechanisms, which requires further evidence from more species to confirm.
Discussion
It is notoriously difficult to assemble the complete sequence of SDRs or sex chromosomes, which usually have a high repeat density and many translocated segments from autosomes (Bachtrog 2013; Charlesworth 2013). In our study, the sex-associated loci were initially mapped onto multiple different chromosomes (figs. 1 and 2), although they consistently revealed an XY sex-determination system in P. euphratica and a ZW system in P. alba. These results may be caused by the lack and/or misassembly of SDRs in the reference genome, especially when the genome from a homozygous (XX or ZZ) individual was used as reference, the reads from Y- or W-specific regions of hemizygous (XY or ZW) individuals may be misaligned to homologous sequences on autosomes and lead to false associations. Similar phenomena were also observed in the sex association analysis of P. trichocarpa, P. balsamifera, and S. purpurea, which may lead to an inaccurate localization of SDRs in assemblies (Geraldes et al. 2015; Zhou, Macaya-Sanz, Carlson, et al. 2020). The high sequence similarity between these sex-associated regions and the SDRs we finally established strongly supports this possibility (supplementary figs. S5 and S11, Supplementary Material online). Therefore, our research emphasizes the importance and necessity for precise assembly of SDRs using multiple complementary methods, including the ultralong read sequencing, haplotype-phased assembly, and the sex-specific depth of read mapping.
Our results further indicate that the SDRs of poplar species are generally shorter in length and contain relatively fewer genes than the SDR recently reported in S. purpurea (Zhou, Macaya-Sanz, Carlson, et al. 2020), though the size of this SDR may be inflated due to overlap with the centromere (Zhou et al. 2018). Comparative analysis between X/Z and Y/W haplotypes suggests that the SDRs of P. euphratica and P. alba were established relatively recently, which is a common feature of the sex chromosomes of the Salicaceae species studied so far (Geraldes et al. 2015; Pucholt et al. 2017; Zhou et al. 2018; Zhou, Macaya-Sanz, Carlson, et al. 2020). Along with this, our results also suggest that the Y and W chromosomes have expanded in content, a pattern that is common in young sex chromosomes of plants (Hobza et al. 2015; Hobza et al. 2017). Moreover, our results simultaneously showed that the Salicaceae exhibit an extremely fast rate of sex-chromosome turnover. In previous studies, SDRs have been reported only on chromosome 15 with female heterogamety (ZW) in willow except S. nigra (Hou et al. 2015; Pucholt et al. 2015; Chen et al. 2016; Pucholt et al. 2017; Zhou et al. 2018; Zhou, Macaya-Sanz, Carlson, et al. 2020; Sanderson et al., unpublished data), and on chromosome 19 of poplar with most species showing male heterogamety (XY) (Gaudet et al. 2007; Yin et al. 2008; Geraldes et al. 2015). However, our study identified an XY system with the SDR on chromosome 14 of P. euphratica for the first time, and confirmed a ZW system with SDR on chromosome 19 of P. alba. These results highlight the complexity and diversity of sex determination in this family. Comparative analysis showed that translocation of genes from autosomes to the SDR and gene replication frequently occurred both on the Y chromosomes of P. euphratica and on the W chromosomes of P. alba, indicating that these two events are likely to be important contributors during SDR turnover. The regulatory mechanisms and functions of these genes in sex determination and sexual dimorphism in these two species need further investigation.
Among all genes on SDRs, the cytokinin response regulator is the most likely candidate for controlling sex determination in the Salicaceae, not only because the orthologs of this gene have been found to be sex-associated in most of the reported species in the family but also because it is the only homologous sequence found in the sex chromosomes of P. euphratica, P. alba, P. trichocarpa, P. deltoides, P. tremula, and S. purpurea (fig. 3 and supplementary fig. S16, Supplementary Material online), the Salicaceae species with SDR precisely assembled (Müller et al. 2020; Zhou, Macaya-Sanz, Carlson, et al. 2020; Zhou, Macaya-Sanz, Schmutz, et al. 2020; Xue et al., unpublished data). Recent progress has revealed that the genes involved in cytokinin signaling play important roles in the regulation of unisexual flower development in plants (Kieber and Schaller 2018; Wybouw and De Rybel 2019; Feng et al. 2020). Specifically, a Y-specific type-C cytokinin response regulator (Shy Girl, SyGI) was recently identified as a suppressor of carpel development and therefore is a strong candidate of sex determination in kiwifruit (Akagi et al. 2018). Similar to the pattern of the RR genes found in the Salicaceae species, in kiwifruit SyGI was duplicated from an autosome and subsequently gained a new function on its Y chromosome. However, the type-A RR genes we identified here are not orthologous to the SyGI gene, so we speculate that they may have different functions in the cytokinin signaling pathway. Based on our results, it is reasonable to suspect that the RR genes are more likely to function as a dominant promoter of female function (fig. 4), as they exist on the W chromosomes of both P. alba and S. purpurea in intact duplicates and exhibit female-specific expression in flower buds of P. euphratica and P. alba (supplementary fig. S15, Supplementary Material online). In contrast, the RR gene fragments on the Y chromosome of P. euphratica exist as two partial duplicates with different sizes. This may serve as a female suppressor by encoding an siRNA that targets the intact RR gene at the distal end of chromosome 19, possibly through RNA-directed DNA methylation (Brautigam et al. 2017; Müller et al. 2020). A recent functional study in P. tremula has provided direct evidence for this model using CRISPR-Cas9 to knock out the full-length RR gene and convert individuals from female to male (Müller et al. 2020). Therefore, our findings show that Salicaceae species potentially share a common mechanism of sex determination, in which the specific duplication of the RR orthologs on SDRs may have played an important role in the acquisition of separate sexes in these species.
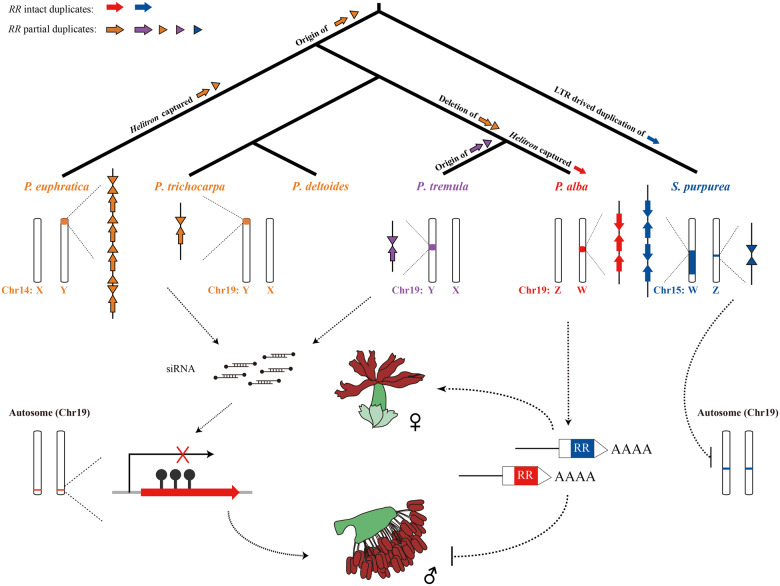
Fig. 4.
Hypothetical model for sex system turnovers in Salicaceae. The W chromosomes of Populus alba and Salix purpurea both carry several intact RR genes and are likely to serve as a dominant promoter of female function. On the Y chromosomes of P. euphratica, P. trichocarpa, P. deltoides, and P. tremula, partial duplicates of the RR gene likely act as suppressors of female development by encoding an siRNA that targets the intact RR gene through RNA-directed DNA methylation. The SDR information for S. purpurea (Zhou, Macaya-Sanz, Carlson, et al. 2020), P. trichocarpa (Zhou, Macaya-Sanz, Schmutz, et al. 2020), P. deltoides, and P. tremula (Müller et al. 2020) comes from recently published studies.
More interestingly, we identified Helitron-like repetitive elements upstream of the RR duplicate in both SDRs of P. euphratica and P. alba, regardless of whether the RR duplicate is intact or partial (fig. 3). As a major class of DNA transposons, Helitrons are hypothesized to transpose by a rolling circle replication mechanism, and have been found to frequently capture genes or gene fragments and move them around the genome, which is believed to be important in the evolution of host genomes (Morgante et al. 2005; Kapitonov and Jurka 2007). Our results suggest that the RR fragments and intact gene sequences appear to have been captured by Helitrons in P. euphratica and P. alba, and subsequently replicated in their SDRs (figs. 3 and 4). Furthermore, our phylogenetic analysis revealed two independent origins of the RR partial duplicates in the genus Populus (supplementary fig. S16, Supplementary Material online). The clustering of the RR partial duplicates of P. euphratica, P. pruinosa, P. ilicifolia, P. trichocarpa, and P. deltoides separately from the intact RR genes in the phylogeny indicates that these partial duplicates were present before the diversification of poplar species. On the other hand, the clustering of partial RR duplicates from P. tremula and P. tremuloides with full-length genes from P. tremula and P. alba suggests an independent origin in these species (supplementary fig. S16, Supplementary Material online). Moreover, our phylogenetic analysis also indicated that the intact RR gene was captured very recently in P. alba, at least after its split with P. tremula (supplementary fig. S16, Supplementary Material online). Based on the fossil record, this would suggest an origin within the past 20 My (Cronk 2005). We found that the partial duplicate of the RR gene is lacking in P. alba, which may be another key event in addition to the duplication of the intact RR gene, in the transition of the sex-determination system from XY to ZW (fig. 4). In addition, the high nucleotide identity among intact RR genes of S. purpurea reflects another possible SDR turnover event in willow, which might be driven by the replication of a Copia LTR (Zhou, Macaya-Sanz, Carlson, et al. 2020), rather than by a Helitron as we found in poplar. Moreover, we also identified an inverted repeat of the first exon of the RR gene and an intact copy on the chromosomes 15Z and 19 of S. purpurea, respectively (fig. 3). This suggests a model whereby the inverted repeat is suppressing the RR gene of chromosome 19 in males, but the SDR on the W chromosome may be dominant to this effect in females, possibly due to increased dosage or another mechanism (fig. 4). These observations further indicate that the sex-determination system of S. purpurea may have been changed from XY to ZW relatively recently, since the suppressing mechanism from the RR partial duplication is still retained. Therefore, our results suggest that the high activity of these repetitive elements is the most likely cause of the recently established SDRs in these species, and further indicate that at least four turnover events have occurred in the evolution of sex chromosomes of the Salicaceae species (fig. 4).
In conclusion, here, we present an XY system of sex determination with the SDR on the proximal end of chromosome 14 in P. euphratica, and a ZW system with the SDR on a nonterminal region of chromosome 19 in P. alba. Both SDRs appear to have evolved relatively recently and are characterized by frequent translocations from autosomes and gene replication events. Our comparative analysis also demonstrated an extremely fast rate of sex chromosome turnover among Salicaceae species, which may be driven by Helitron transposons in poplar and by Copia LTRs in willow. Most importantly, we propose a model showing that poplar and willow have a common underlying mechanism of sex determination, which controls the XY and ZW systems simultaneously through a type-A RR gene. In the future, it will be necessary to conduct transgenic function experiments and comparative analysis from more species in this family to further support our model.
Materials and Methods
Genome Sequencing
We have previously reported the reference genome of a male P. euphratica (Zhang et al. 2020) and a male P. alba (Ma et al. 2019). In this study, we further collected the fresh leaves of a female P. euphratica and a female P. alba for genome sequencing and assembly. Genomic DNA was extracted using the QIAGEN Genomic DNA extraction kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. To generate Oxford Nanopore long reads, ∼15 μg of genomic DNA was size-selected using the BluePippin system (Sage Science), and processed according to the protocol of Ligation Sequencing Kit (SQK-LSK109). The final library was sequenced on a PromethION sequencer (Oxford Nanopore Technologies, UK) with a running time of 48 h. The Oxford Nanopore proprietary base-caller, Albacore v2.1.3, was used to perform base calling of the raw signal data and convert the FAST5 files into FASTQ files.
In addition, paired-end libraries with insert size of ∼300 bp were constructed using NEB Next Ultra DNA Library Prep Kit (NEB), with the standard protocol provided by Illumina (San Diego, CA). The library was sequenced on an Illumina HiSeq X Ten platform (Illumina, San Diego, CA). These sequencing data were used for correction of errors inherent to long read data for genome assembly.
Genome Assembly
For genome assembly, we first removed the Nanopore long reads shorter than 1 kb and the low-quality reads with a mean quality ≤7. The long reads underwent self-correction using the module “NextCorrect” and then assembled into contigs using “NextGraph” implemented in Nextdenovo v2.2.0 (https://github.com/Nextomics/NextDenovo) with default parameters. Subsequently, the filtered Nanopore reads were mapped to the initial assembly using the program Minimap2 v2.17-r941 (Li 2018) and NextPolish v1.0 (https://github.com/Nextomics/NextPolish) was used with three iterations to polish the genome. In addition, we further aligned the Illumina reads to the genome using BWA-MEM v0.7.15 (Li and Durbin 2009) and corrected base-calling by an additional three rounds of NextPolish runs with default parameters. Finally, the corrected genome was aligned to their respective male reference genome using the LAST program (Kielbasa et al. 2011) and the syntenic relationships were used to anchor the assembled contigs onto 19 chromosomes. The chromosome identities were then assigned based on comparison with P. trichocarpa.
Population Sample Collection, Resequencing, and Mapping
Silica gel-dried leaves of P. euphratica and P. alba were collected from wild populations in western China. For each species, the sex of 30 male and 30 female individuals was identified from flowering catkins. Genomic DNA of each sample was extracted using the Qiagen DNeasy Plant Minikit (Qiagen, Hilden, Germany). Paired-end libraries were prepared using the NEBNext Ultra DNA Library Prep Kit (NEB) and sequenced on an Illumina HiSeq X Ten platform, according to the manufacturer’s instructions.
The generated raw reads were first subjected to quality control and low-quality reads were removed if they met either of the following criteria (Ma et al. 2018): 1) ≥10% unidentified nucleotides (N); 2) a phred quality ≤7 for >65% of read length; and 3) reads overlapping >10 bp with the adapter sequence, allowing <2-bp mismatch. Reads shorter than 45 bp after trimming were also discarded. The obtained high-quality cleaned reads were subsequently mapped to the male and female reference genomes of each species, respectively, using BWA-MEM v0.7.15 with default parameters (Li and Durbin 2009). The alignment results and marked duplicate reads were sorted using SAMtools v0.1.19 (Li et al. 2009). Finally, Genome Analysis Toolkit (GATK) (DePristo et al. 2011) was used to process base quality recalibrations to enhance alignments in regions around putative indels with two steps: 1) “RealignerTargetCreator” was applied to identify regions where realignment was needed and 2) “IndelRealigner” was used to realign these regions.
SNP Calling, Filtering, and GWAS
To prevent biases in SNP calling accuracy due to the difference of samples size between groups, single-sample SNP and genotype calling were first implemented using GATK (Yang et al. 2018) with “HaplotypeCaller,” and then multisample SNPs were identified after merging the results of each individual by “GenotypeGVCFs.” A series of filtering steps were performed to reduce false positives (Yang et al. 2018), including removal of 1) indels with a quality scores <30, 2) SNPs with more than two alleles, 3) SNPs at or within 5 bp from any indels, 4) SNPs with a genotyping quality scores (GQ) <10, and 5) SNPs with extremely low (less than one-third average depth) or extremely high (>3-fold average depth) coverage. The identified SNPs were used for subsequent GWAS analysis. A standard case/control model between allele frequencies and sex phenotype was performed using Plink v1.9 (Purcell et al. 2007). For each species, associations at α < 0.05 after Bonferroni correction for multiple testing were reported as the significantly sex-associated SNPs. These sex-associated SNPs that occurred within 10 kb on the same chromosome were merged into the same interval.
Construction of P. euphratica Y Contig and P. alba W Contig
To construct the Y contig of P. euphratica and the W contig of P. alba, we further generated ultralong sequences from a male (XY) P. euphratica and a female (ZW) P. alba, using an optimized DNA extraction followed by modified library preparation based on the Nanopore PromethION sequencer (Jain et al. 2018; Gong et al. 2019). For P. euphratica, we did not find contigs that clearly contained Y-linked sequences in its male genome, which may be due to assembly errors, so we used multiple methods to determine its Y contig. At first, we attempted to find the male-specific k-mers from the high-quality resequencing reads of both male and female samples. Briefly, all 32-bp k-mers starting with the “AG” dinucleotide were extracted from all resequencing reads, and the number of occurrences of each specific subsequence in female and male individuals was counted, respectively. The use of the “AG” dinucleotide is to reduce the number of k-mer sequences and effectively speed up the analysis. The k-mer counts were then compared between male and female, and the male-specific k-mers (female count was 0) were obtained. Next, we extracted the ultralong nanopore reads containing at least one of the identified male-specific k-mers, and assembled these ultralong reads using the software Canu v1.7 (Koren et al. 2017), resulting in a “male-specific contig” that was 450 kb in length. Simultaneously, we also de novo assembled all of the ultralong nanopore reads into a draft male genome using Nextdenovo v2.2.0. By comparing the “male-specific contig” with the obtained male genome, we identified a candidate Y contig that contained a large number of male-specific alleles and exhibited a widespread synteny and continuity with the “male-specific contig.” To further refine the SDR along this candidate Y contig, we remapped the resequencing data to the draft genome by BWA-MEM v0.7.15 (Li and Durbin 2009), and extracted the average depth of coverage using a nonoverlapping sliding window (1 kb in length) by SAMtools v0.1.19 (Li et al. 2009). Finally, we compared the relative depth of coverage between male and female individuals, and found that the region between 0 and 658 kb of this contig showed male-specific depth and was therefore considered to be the SDR on the Y chromosome of P. euphratica.
For P. alba, we first performed a whole-genome alignment between its male and female genome using the program LAST (Kielbasa et al. 2011). Fortunately, we found that the sex-associated region in the female genome contained a large insert compared with the corresponding region in the male genome. We used the same method as above to count the relative depth of coverage between male and female individuals of P. alba, and found that the region between 310 and 450 kb of this contig exhibited female-specific depth. Therefore, this region was directly considered to be the SDR on the W chromosome of P. alba, and the assembly accuracy of this region was also confirmed by our ultralong nanopore reads.
Annotation and Comparison of the Y and W Contigs
Transposable elements in our assembled Y and W contigs were identified and classified using the software RepeatMasker (Tarailo-Graovac and Chen 2009). Gene annotation was conducted by combining the results of de novo prediction from the program Augustus v.3.2.1 (Stanke et al. 2006) with the parameter “–species=arabidopsis,” and homology-based prediction using the protein sequences of A. thaliana, P. trichocarpa, and S. purpurea downloaded from Phytozome 12 (https://phytozome.jgi.doe.gov/), as well as transcriptome data of P. euphratica and P. alba generated from our previous studies using the software of EVidenceModeler (1.1.1 version) (Haas et al. 2008; Ma et al. 2019; Hu et al. 2020; Zhang et al. 2020). The predicted genes were searched against predicted proteins from P. trichocarpa, S. suchowensis, and A. thaliana to find the closest homologous annotation.
To construct the phylogenetic relationships among the allelic genes on the X/Y or Z/W contigs, we further identified their orthologous genes in P. pruinosa (Yang et al. 2017), P. ilicifolia (Chen et al. 2020), and S. suchowensis (Dai et al. 2014) genomes by combining reciprocal BLAST results and their syntenic relationships. The sequences were aligned using ClustalW with default parameters provided in MEGA5 (Tamura et al. 2011) and the resulting alignments were adjusted manually. A maximum likelihood tree was built using MEGA5 with default parameters. The nonsynonymous (dN) and synonymous (dS) substitution rate and their ratio (dN/dS) were estimated using the yn00 function in PAML (Yang 2007).
Expression Analysis of Intact and Partial RR Duplicates in Flower Buds
The male and female flower buds of P. euphratica and P. alba were collected in Lanzhou (Gansu) and Urumqi (Xinjiang) of China, respectively, on July 20, 2020. The total RNA was extracted and purified using Plant Total RNA purification kit with DNase I (Aidlab), and following analysis of qualification and quantification was performed with Nano Drop (NanoDrop, Madison) and Agilent 2100 bioanalyzer (Agilent, Santa Clara). To determine the expression patterns of the intact RR gene, RNA (2 μg) was reverse-transcribed using a Tiangen Fast Quant RT Kit (Tiangen), and quantitative Real-Time PCR (qRT–PCR) was performed with BIO-RAD CFX96TM Real-Time System using the forward primer 5′-AATGACCGGTTATGAGCTGT and the reverse primer 5′-TTACTGATTAATTCGTGTAG. The qRT–PCR program consisted of an initial temperature of 95 °C for 3 min, followed by 39 cycles of 95 °C for 10 s, 60 °C for 30 s. Melt curve was constructed by increasing the temperature from 65 to 95 °C with increment of 0.5 °C. The gene expression level was calculated by the 2-ΔCT method (Livak and Schmittgen 2001). To detect the expression of RR partial duplications in P. euphratica, the small RNA regions corresponding to the 18–30 nt bands in the marker lane (14–30 ssRNA Ladder Marker, TAKARA) were excised and recovered. The small RNA libraries were then produced using standardized procedures including adapter ligation, cDNA synthesis, and size selection. The prepared library was sequenced using the DNBSEQ platform (BGI, Shenzhen, China) and the obtained clean reads were mapped to the male genome of P. euphratica using Bowtie v.1.2.2 (Langmead and Salzberg 2012). Only sequences with a perfect match were used for further analysis. The Integrative Genomics Viewer (IGV) (Robinson et al. 2011) was used for visualization of the small RNA-seq mappings.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (31561123001, 31922061, 41871044, 31500502), NSF Dimensions of Biodiversity Program (1542509 to S.P.D. and 1542599 to M.O.), National Key Research and Development Program of China (2016YFD0600101), and Fundamental Research Funds for the Central Universities (SCU2019D013).
Data Availability
The whole-genome sequencing raw data, small RNA-seq data, genome assembly of female P. euphratica and P. alba, and the sequences and annotation information of P. euphratica Y-contig and P. alba W-contig have been deposited in the Genome Warehouse in BIG Data Center (BIG Data Center Members, 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number PRJCA002485 that is publicly accessible at https://bigd.big.ac.cn/bioproject.
References
- Akagi T, Henry IM, Ohtani H, Morimoto T, Beppu K, Kataoka I, Tao R.. 2018. A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell 30(4):780–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Henry IM, Tao R, Comai L.. 2014. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346(6209):646–650. [DOI] [PubMed] [Google Scholar]
- Akagi T, Pilkington SM, Varkonyi-Gasic E, Henry IM, Sugano SS, Sonoda M, Firl A, McNeilage MA, Douglas MJ, Wang T, et al. 2019. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat Plants. 5(8):801–809. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 14(2):113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balounova V, Gogela R, Cegan R, Cangren P, Zluvova J, Safar J, Kovacova V, Bergero R, Hobza R, Vyskot B, et al. 2019. Evolution of sex determination and heterogamety changes in section Otites of the genus. Sci Rep. 9(1):1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B.. 2010. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22(3):640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D.. 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 24(2):94–102. [DOI] [PubMed] [Google Scholar]
- Brautigam K, Soolanayakanahally R, Champigny M, Mansfield S, Douglas C, Campbell MM, Cronk Q.. 2017. Sexual epigenetics: gender-specific methylation of a gene in the sex determining region of Populus balsamifera. Sci Rep. 7(1):45388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Bomblies K, Kim ST, Karelina D, Zaidem M, Ossowski S, Martin-Pizarro C, Laitinen RA, Rowan BA, Tenenboim H, et al. 2014. Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159(6):1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 1978. A model for the evolution of dioecy and gynodioecy. Am Nat. 112(988):975–997. [Google Scholar]
- Charlesworth B. 1991. The evolution of sex chromosomes. Science 251(4997):1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2013. Plant sex chromosome evolution. J Exp Bot. 64(2):405–420. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2015. Plant contributions to our understanding of sex chromosome evolution. New Phytol. 208(1):52–65. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2016. Plant sex chromosomes. Annu Rev Plant Biol. 67(1):397–420. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang T, Fang L, Li X, Yin T.. 2016. Confirmation of single-locus sex determination and female heterogamety in willow based on linkage analysis. PLoS One 11(2):e0147671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ai F, Zhang J, Ma X, Yang W, Wang W, Su Y, Wang M, Yang Y, Mao K, et al. 2020. Survival in the Tropics despite isolation, inbreeding and asexual reproduction: insights from the genome of the world’s southernmost poplar (Populus ilicifolia). Plant J. 103(1):430–442. [DOI] [PubMed] [Google Scholar]
- Cronk QCB. 2005. Plant eco-devo: the potential of poplar as a model organism. New Phytol. 166(1):39–48. [DOI] [PubMed] [Google Scholar]
- Dai X, Hu Q, Cai Q, Feng K, Ye N, Tuskan GA, Milne R, Chen Y, Wan Z, Wang Z, et al. 2014. The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 24(10):1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Sanderson BJ, Keefover-Ring K, Liu J, Ma T, Yin T, Smart LB, DiFazio SP, Olson MS.. 2020. Pathways to sex determination in plants: how many roads lead to Rome? Curr Opin Plant Biol. 54:61–68. [DOI] [PubMed] [Google Scholar]
- Gaudet M, Jorge V, Paolucci I, Beritognolo I, Mugnozza GS, Sabatti M.. 2007. Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genet Genomes. 4(1):25–36. [Google Scholar]
- Geraldes A, Hefer CA, Capron A, Kolosova N, Martinez-Nuñez F, Soolanayakanahally RY, Stanton B, Guy RD, Mansfield SD, Douglas CJ, et al. 2015. Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol Ecol. 24(13):3243–3256. [DOI] [PubMed] [Google Scholar]
- Gong L, Wong CH, Idol J, Ngan CY, Wei CL.. 2019. Ultra-long read sequencing for whole genomic DNA analysis. J Vis Exp. 145:e58954. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR.. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Jiao Y.. 2015. APETALA1 establishes determinate floral meristem through regulating cytokinins homeostasis in Arabidopsis. Plant Signal Behav. 10(11):e989039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Zhou J, Xu C, Bowers JE, Van der Hulst R, Ayyampalayam S, Mercati F, Riccardi P, McKain MR, Kakrana A, et al. 2017. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun. 8(1):1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Akagi T, Tao R, Comai L.. 2018. One hundred ways to invent the sexes: theoretical and observed paths to dioecy in plants. Annu Rev Plant Biol. 69(1):553–575. [DOI] [PubMed] [Google Scholar]
- Hobza R, Cegan R, Jesionek W, Kejnovsky E, Vyskot B, Kubat Z.. 2017. Impact of repetitive elements on the Y chromosome formation in plants. Genes. 8(11):302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobza R, Kubat Z, Cegan R, Jesionek W, Vyskot B, Kejnovsky E.. 2015. Impact of repetitive DNA on sex chromosome evolution in plants. Chromosome Res. 23(3):561–570. [DOI] [PubMed] [Google Scholar]
- Hou J, Ye N, Zhang D, Chen Y, Fang L, Dai X, Yin T.. 2015. Different autosomes evolved into sex chromosomes in the sister genera of. Sci Rep. 5(1):9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Yang W, Zheng Z, Niu Z, Yang Y, Wan D, Liu J, Ma T.. 2020. Analysis of alternative splicing and alternative polyadenylation in Populus alba var. pyramidalis by single-molecular long-read sequencing. Front Genet. 11:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, Tyson JR, Beggs AD, Dilthey AT, Fiddes IT, et al. 2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 36(4):338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J.. 2007. Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet. 23(10):521–529. [DOI] [PubMed] [Google Scholar]
- Kersten B, Pakull B, Groppe K, Lueneburg J, Fladung M.. 2014. The sex-linked region in Populus tremuloides Turesson 141 corresponds to a pericentromeric region of about two million base pairs on P. trichocarpa chromosome 19. Plant Biol J. 16(2):411–418. [DOI] [PubMed] [Google Scholar]
- Khan A, Garbelli A, Grossi S, Florentin A, Batelli G, Acuna T, Zolla G, Kaye Y, Paul LK, Zhu JK, et al. 2014. The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing. Plant J. 79(1):28–43. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE.. 2018. Cytokinin signaling in plant development. Development 145(4):dev.149344. [DOI] [PubMed] [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith MC.. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21(3):487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM.. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H.. 2009. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21(10):3152–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang D, Zhang L, Kang M, Lu Z, Zhu R, Mao X, Xi Z, Ma T.. 2020. Intergeneric relationships within the family Salicaceae s.l. based on plastid phylogenomics. Int J Mol Sci. 20:3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25(4):402–408. [DOI] [PubMed] [Google Scholar]
- Ma J, Wan D, Duan B, Bai X, Bai Q, Chen N, Ma T.. 2019. Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol J. 17(2):451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Wang K, Hu Q, Xi Z, Wan D, Wang Q, Feng J, Jiang D, Ahani H, Abbott RJ, et al. 2018. Ancient polymorphisms and divergence hitchhiking contribute to genomic islands of divergence within a poplar species complex. Proc Natl Acad Sci U S A. 115(2):E236–E243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova NV, Kudryavtseva AV, Borkhert EV, Pushkova EN, Fedorova MS, Snezhkina AV, Krasnov GS, Dmitriev AA.. 2019. Sex-specific polymorphism of MET1 and ARR17 genes in Populus × sibirica. Biochimie 162:26–32. [DOI] [PubMed] [Google Scholar]
- Ming R, Bendahmane A, Renner SS.. 2011. Sex chromosomes in land plants. Annu Rev Plant Biol. 62(1):485–514. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M.. 2005. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17(11):2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A.. 2005. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 37(9):997–1002. [DOI] [PubMed] [Google Scholar]
- Moore RC, Harkess AE, Weingartner LA.. 2016. How to be a seXY plant model: a holistic view of sex-chromosome research. Am J Bot. 103(8):1379–1382. [DOI] [PubMed] [Google Scholar]
- Müller NA, Kersten B, Leite Montalvão AP, Mähler N, Bernhardsson C, Bräutigam K, Carracedo Lorenzo Z, Hoenicka H, Kumar V, Mader M, et al. 2020. A single gene underlies the dynamic evolution of poplar sex determination. Nat Plants. 6(6):630–637. [DOI] [PubMed] [Google Scholar]
- Pakull B, Groppe K, Meyer M, Markussen T, Fladung M.. 2009. Genetic linkage mapping in aspen (Populus tremula L. and Populus tremuloides Michx.). Tree Genet Genomes. 5(3):505–515. [Google Scholar]
- Pakull B, Kersten B, Lüneburg J, Fladung M.. 2015. A simple PCR-based marker to determine sex in aspen. Plant Biol J. 17(1):256–261. [DOI] [PubMed] [Google Scholar]
- Paolucci I, Gaudet M, Jorge V, Beritognolo I, Terzoli S, Kuzminsky E, Muleo R, Scarascia Mugnozza G, Sabatti M.. 2010. Genetic linkage maps of Populus alba L. and comparative mapping analysis of sex determination across Populus species. Tree Genet Genomes. 6(6):863–875. [Google Scholar]
- Peto F. 1938. Cytology of poplar species and natural hybrids. Can J Res. 16c(11):445–455. [Google Scholar]
- Pucholt P, Rönnberg-Wästljung AC, Berlin S.. 2015. Single locus sex determination and female heterogamety in the basket willow (Salix viminalis L.). Heredity 114(6):575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucholt P, Wright AE, Conze LL, Mank JE, Berlin S.. 2017. Recent sex chromosome divergence despite ancient dioecy in the willow Salix viminalis. Mol Biol Evol. 34(8):1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS, Ricklefs RE.. 1995. Dioecy and its correlates in the flowering plants. Am J Bot. 82(5):596–606. [Google Scholar]
- Renner SS. 2014. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am J Bot. 101(10):1588–1596. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP.. 2011. Integrative Genomics Viewer. Nat Biotechnol. 29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson BJ, Feng G, Hu N, Grady J, Carlson CH, Smart LB, Keefover-Ring K, Yin T, Ma T, Liu J, et al. Unpublished data. Sex determination through X-Y heterogamety in Salix nigra. bioRxiv doi: 10.1101/2020.03.23.000919. [DOI] [PMC free article] [PubMed]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B.. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N.. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 25(1):4–10. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Wei N, Straub SCK, Govindarajulu R, Liston A, Ashman TL.. 2018. Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. PLoS Biol. 16(8):e2006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenin J, Pollet B, Letarnec B, Saulnier L, Gissot L, Maia-Grondard A, Lapierre C, Jouanin L.. 2011. The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol Plant. 4(1):70–82. [DOI] [PubMed] [Google Scholar]
- Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, Sakakibara H.. 2012. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 69(2):355–365. [DOI] [PubMed] [Google Scholar]
- Torres MF, Mathew LS, Ahmed I, Al-Azwani IK, Krueger R, Rivera-Nuñez D, Mohamoud YA, Clark AG, Suhre K, Malek JA.. 2018. Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat Commun. 9(1):3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313(5793):1596–1604. [DOI] [PubMed] [Google Scholar]
- Wang J, Na JK, Yu Q, Gschwend AR, Han J, Zeng F, Aryal R, VanBuren R, Murray JE, Zhang W, et al. 2012. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci U S A. 109(34):13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhang L, Zhang Z, Li M, Wang D, Zhang X, Xi Z, Keefover-Ring K, Smart LB, DiFazio SP, et al. 2020. Phylogenomics of the genus Populus reveals extensive interspecific gene flow and balancing selection. New Phytol. 225(3):1370–1382. [DOI] [PubMed] [Google Scholar]
- Wybouw B, De Rybel B.. 2019. Cytokinin – a developing story. Trends Plant Sci. 24(2):177–185. [DOI] [PubMed] [Google Scholar]
- Xue L, Wu H, Chen Y, Li X, Hou J, Lu J, Wei S, Dai X, Olson MS, Liu J, et al. Unpublished data. Two antagonistic effect genes mediate separation of sexes in a fully dioecious plant. bioRxiv doi: 10.1101/2020.03.15.993022. [DOI]
- Yang W, Wang K, Zhang J, Ma J, Liu J, Ma T.. 2017. The draft genome sequence of a desert tree Populus pruinosa. Gigascience 6(9):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ma T, Wang Z, Lu Z, Li Y, Fu C, Chen X, Zhao M, Olson MS, Liu J.. 2018. Genomic effects of population collapse in a critically endangered ironwood tree Ostrya rehderiana. Nat Commun. 9(1):5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yin T, Difazio SP, Gunter LE, Zhang X, Sewell MM, Woolbright SA, Allan GJ, Kelleher CT, Douglas CJ, Wang M, et al. 2008. Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res. 18(3):422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Boualem A, Bendahmane A, Ming R.. 2014. Genomics of sex determination. Curr Opin Plant Biol. 18:110–116. [DOI] [PubMed] [Google Scholar]
- Zhang L, Xi Z, Wang M, Guo X, Ma T.. 2018. Plastome phylogeny and lineage diversification of Salicaceae with focus on poplars and willows. Ecol Evol. 8(16):7817–7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen Y, Zhang J, Ma X, Li Y, Li M, Wang D, Kang M, Wu H, Yang Y, et al. 2020. Improved genome assembly provides new insights into genome evolution in a desert poplar (Populus euphratica). Mol Ecol Resour. 20(3):781–794. [DOI] [PubMed] [Google Scholar]
- Zhou R, Macaya-Sanz D, Carlson CH, Schmutz J, Jenkins JW, Kudrna D, Sharma A, Sandor L, Shu S, Barry K, et al. 2020. A willow sex chromosome reveals convergent evolution of complex palindromic repeats. Genome Biol. 21(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Macaya-Sanz D, Rodgers-Melnick E, Carlson CH, Gouker FE, Evans LM, Schmutz J, Jenkins JW, Yan J, Tuskan GA, et al. 2018. Characterization of a large sex determination region in Salix purpurea L. (Salicaceae). Mol Genet Genomics. 293(6):1437–1452. [DOI] [PubMed] [Google Scholar]
- Zhou R, Macaya-Sanz D, Schmutz J, Jenkins JW, Tuskan GA, DiFazio SP.. 2020. Sequencing and analysis of the sex determination region of Populus trichocarpa. Genes (Basel) 11(8):843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome sequencing raw data, small RNA-seq data, genome assembly of female P. euphratica and P. alba, and the sequences and annotation information of P. euphratica Y-contig and P. alba W-contig have been deposited in the Genome Warehouse in BIG Data Center (BIG Data Center Members, 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number PRJCA002485 that is publicly accessible at https://bigd.big.ac.cn/bioproject.