Abstract
How animals, particularly livestock, adapt to various climates and environments over short evolutionary time is of fundamental biological interest. Further, understanding the genetic mechanisms of adaptation in indigenous livestock populations is important for designing appropriate breeding programs to cope with the impacts of changing climate. Here, we conducted a comprehensive genomic analysis of diversity, interspecies introgression, and climate-mediated selective signatures in a global sample of sheep and their wild relatives. By examining 600K and 50K genome-wide single nucleotide polymorphism data from 3,447 samples representing 111 domestic sheep populations and 403 samples from all their seven wild relatives (argali, Asiatic mouflon, European mouflon, urial, snow sheep, bighorn, and thinhorn sheep), coupled with 88 whole-genome sequences, we detected clear signals of common introgression from wild relatives into sympatric domestic populations, thereby increasing their genomic diversities. The introgressions provided beneficial genetic variants in native populations, which were significantly associated with local climatic adaptation. We observed common introgression signals of alleles in olfactory-related genes (e.g., ADCY3 and TRPV1) and the PADI gene family including in particular PADI2, which is associated with antibacterial innate immunity. Further analyses of whole-genome sequences showed that the introgressed alleles in a specific region of PADI2 (chr2: 248,302,667–248,306,614) correlate with resistance to pneumonia. We conclude that wild introgression enhanced climatic adaptation and resistance to pneumonia in sheep. This has enabled them to adapt to varying climatic and environmental conditions after domestication.
Keywords: climate adaptation, genome-wide SNPs, whole-genome sequences, introgression, ovine, pneumonia
Introduction
In the face of climate change, sufficient genetic variations are crucial for a population’s ability to adapt to climatic stress and thereby maintain biodiversity (Han et al. 2014; Kristensen et al. 2018). Although standing variation and new mutations are important resources for the adaptive process, genomic introgression between divergent evolutionary lineages may also produce a variety of adaptive benefits in a wide spectrum of organisms (Arnold and Kunte 2017). Recently, interspecies genetic introgression has been the focus of much empirical and theoretical interest (Larson and Burger 2013; Harrison and Larson 2014). In particular, the contribution of wild-relative introgression to indigenous populations has been studied at the genome-wide level in livestock based on modern and ancient genomic data (Bosse et al. 2014; Wu et al. 2018; Hu et al. 2019; Frantz et al. 2020). Nevertheless, wild introgression associated with climatic adaptation has been rarely investigated in livestock, which can provide important information in molecular breeding for improved adaptation.
The genus Ovis contains domestic sheep (O. aries) and seven extant wild relatives (argali O. ammon, Asiatic mouflon O. orientalis, European mouflon O. musimon, urial O. vignei, bighorn sheep O. canadensis, thinhorn sheep O. dalli, and snow sheep O. nivicola; Rezaei et al. 2010). After domestication from Asian mouflon (O. orientalis) in the Near East ∼8,000–10,000 years ago (Ryder 1984; Zeder 2008, 2015; Stiner et al. 2014), sheep have acquired a worldwide range while adapting to a wide range of environments (e.g., climate and pathogen) and production systems and developing into as many as 1,400 breeds (Scherf 2000; Lv et al. 2014). For example, some breeds (e.g., Bashibai and Awassi) showed a certain degree of resistance to pneumonia (Valle Zárate et al. 2006; Du 2018), which is a common infectious disease in domestic sheep worldwide, causing huge losses to the global sheep industry. In contrast to domestic breeds, wild sheep species inhabit specific ranges that overlap with that of domestic sheep. On rare occasions, hybridization between wild (argali, urial, Asiatic mouflon, European mouflon, and snow sheep) and domestic sheep has been documented to produce viable and fertile interspecific hybrids in captivity as well as in the wild (Woronzow et al. 1972; Bunch and Foote 1977; Arnold 2004; Schröder et al. 2016; Alberto et al. 2018). With the availability of the reference genome sequence of O. aries (Oar_v4.0), the wild and domestic sheep offer the opportunity to investigate the contribution of genomic introgression from congeneric wild species to rapid local climatic adaptation in domestic populations.
Here, we generated a large data set of genome-wide high-density (600K) single nucleotide polymorphisms (SNPs) and whole-genome sequences of wild and domestic sheep. These include European, Near East, and Central Asian populations from regions underrepresented in earlier work but being essential to assess levels of wild-relative introgression. We combined these data with publicly available genotypes to build a collection of 3,938 samples from 129 domestic populations and all of the 7 wild sheep species (fig. 1A, supplementary fig. S1 and tables S1 and S2, Supplementary Material online). We further collected data for 117 climatic variables over 40 years to serve as a proxy for the environment to which populations or species have adapted. We applied selection tests to detect genomic introgression and climatic-induced selective signatures based on the synthesis of molecular and climatic data. Our main aims are 2-fold: 1) to detect the genomic traces left by centuries of climatic selective pressures and genomic introgression from wild relatives in domestic sheep; and 2) to evaluate the role of wild introgression in shaping the adaptive genome landscape of native populations.
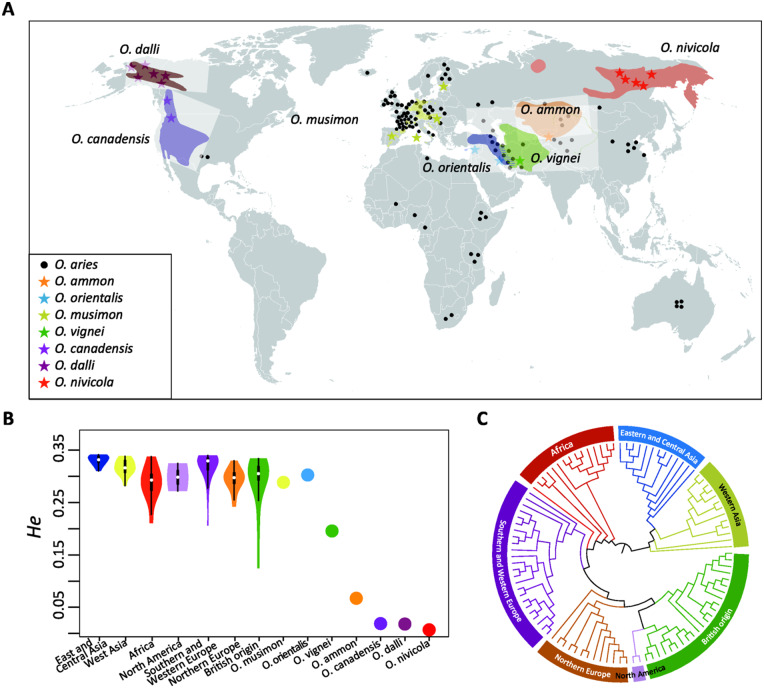
Fig. 1.
Geographic origin, genetic diversity, and genetic differentiation of domestic sheep populations. Population names and their abbreviations are detailed in supplementary table S1, Supplementary Material online. Colored regions represent the distribution range areas of wild sheep. (A) Geographic origin of wild and domestic sheep studied. (B) Distribution of expected heterozygosity (He) for the seven wild sheep species and domestic sheep from different geographic regions. The white dot indicates the median value. (C) NJ tree of domestic populations based on the Reynolds’ distances (Reynolds et al. 1983).
Results and Discussion
Genomic Diversity and Differentiation
Among domestic populations, we observed the highest level of within-population genomic diversity (e.g., proportion of polymorphic SNPs [Pn], observed [Ho] and expected heterozygosity [He]) in Eastern and Central Asian populations, whereas the lowest level of genomic variability was observed in African populations (fig. 1B, supplementary table S4, Supplementary Material online). Our findings agreed with previous findings using a lower density of SNPs (Kijas et al. 2012).
The principal component analysis (PCA), ADMIXTURE with K = 7 and neighbor-joining (NJ) tree revealed a clear and consistent geographic pattern of domestic sheep coupled with the influence of elite breeds, such as Merino and Leicester (fig. 1, supplementary figs. S2–S4, Supplementary Material online). We observed seven major regional genetic groups, including populations from Eastern and Central Asia, Western Asia, Africa, North America, Southern and Western Europe, Northern Europe, and British origin. Both Asiatic and European mouflon possess similar levels of genomic diversity, whereas lower values were observed in the other wild sheep species (fig. 1B, supplementary table S4, Supplementary Material online). This may partly reflect their divergence from domestic sheep for which the Illumina SNP array has been designed (Miller et al. 2012, supplementary table S5, Supplementary Material online). Nevertheless, the two BeadChip were designed from a range of domestic breeds and wild sheep which harbor high levels of polymorphism, and, thus, the ascertainment bias in the comparison of heterozygosity between species or breeds introduced by the design of these chips would be small (Kijas et al. 2012). As allele frequency-dependent diversity estimates are sensitive to ascertainment bias, we have removed the SNPs in high linkage disequilibrium (LD) to counter the effect of the bias and, thus, generated meaningful comparisons between species (Lopez Herraez et al. 2009; Kijas et al. 2012).
Interspecies Common Introgression
We searched for potential gene flow between wild relatives and their sympatric domestic populations using different approaches. In the TreeMix analysis involving different combinations of wild (argali, Asiatic mouflon, European mouflon, and snow sheep) and domestic sheep, the strict drift models assuming 1, 4, 7, and 7 migration events could explain 99.8%, 99.7%, 96.4%, and 95.6% of the variance in relatedness among populations, respectively. It revealed signals of genomic introgressions from argali to Bashibai (fig. 2B), Asiatic mouflon to Grey Shirazi, European mouflon to Shetland and Corse, and snow sheep to Kulundin and Tuva populations (supplementary fig. S5B, Supplementary Material online).
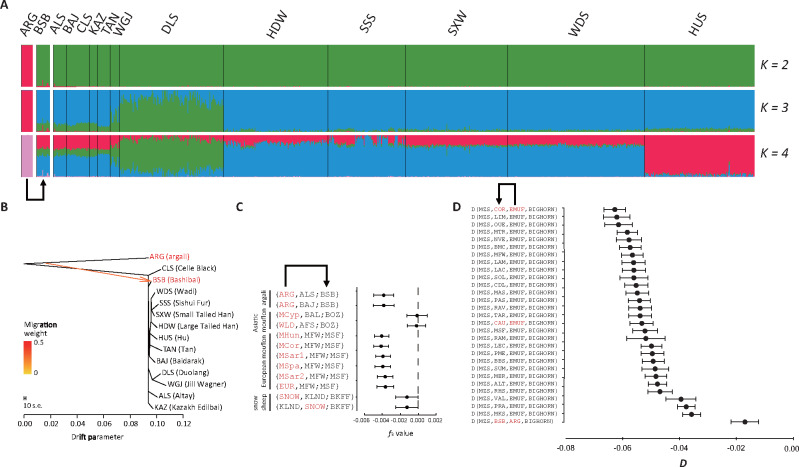
Fig. 2.
Gene flow signals inferred by the analyses of ADMIXTURE, TreeMix, f3, and D statistics. (A) Admixture analysis of all the domestic sheep. (B) Phylogenetic network analysis by TreeMix v.1.12 showing the inferred gene flow between argali and East and Central Asian populations. The branch length is proportional to the drift of each population. ARG (argali) was used as the outgroup to root the tree. The scale bar shows ten units of standard error (SE) of the entries in the sample covariance matrix. The migration weight represents the fraction of ancestry derived from the migration edge. (C) Three-way admixture analysis between wild and domestic populations. The triples show the most significant f3 statistics for various species, with sympatric domestic sheep as the target population and wild sheep species as one of the source population. Asiatic mouflon (MCyp, WLD); Baluchi (BAL); Bozakh (BOZ); Afshari (AFS); European mouflon (MHun, MCor, MSar1, MSpa, MSar2, EUR); Chinese fine wool Merino (MFW); Chinese superfine Merino (MSF); snow sheep (SNOW); Kulundin (KLND); Baikal fine-fleeced (BKFF). (D) D-statistics with the form D (W, X; Y, Z) for wild and domestic populations. D-statistics were implemented to detect admixture from species of the tribe of Y (ARG, argali; EMUF, European mouflon) to either W (MZS, Menz sheep) or X (sympatric domestic populations). Negative D statistic values indicate gene flow from Y to X. COR(Corse), LIM(Limousine), OUE (Ouessant), MTR (Manech Tête Rouge), NVE (Noire du Velay), BMC (Blanche du Massif Central), MFW (Chinese fine wool Merino), LAM (Meat Lacaune), LAC (Dairy Lacaune), SOL (Solognote), CDL (Causse du Lot), MAS (Mérinos d’Arles), PAS (Préalpes du Sud), RAV (Rava), TAR (Tarasconnaise), CAU (Caucasian), MSF (Chinese superfine Merino), RAM (Mérinos de Rambouillet), LEC (Leccese), PME (Poll Merino), BBS (Brown Mountain), SUM (Sumavska), MER (Merino), ALT (Altamurana), RHS (Rhoen sheep), VAL (Valachian), PRA (Pramenka), MKS (Carpathian Mountain), BSB (Bashibai).
The f3 tests indicate significant evidence admixture (f3 < 0) from argali in Bashibai, from Asiatic mouflon in Bozakh, from European mouflon in Chinese Superfine Merino, and from snow sheep in Baikal fine-fleeced (fig. 2C). D-statistics showed significant values (D = −0.0628 to −0.017; |Z| > 3) for introgression of argali ancestry in Bashibai, and for European mouflon ancestry in 28 Southwestern European populations (fig. 2D). Additionally, the ADMIXTURE analysis showed the genetic components of wild species in several sympatric domestic populations such as the genetic components of argali in Bashibai and Altay sheep, Asiatic mouflon in Grey Shirazi and Bozakh, European mouflon in Corse and Shetland, snow sheep in Buubei and Tuva (fig. 2A;supplementary fig. S5A, Supplementary Material online). It is noteworthy that rams of these domestic populations show similar large and curled horns and coat colors as those of wild sheep (fig. 3).
Fig. 3.
Images of introgression donors (A) argali, (B) Asiatic mouflon, (C) European mouflon, (D) snow sheep, and introgressed domestic sheep (E) Bashibai, (F) Grey Shiraz, (G) Corse, (H) Shetland, and (I) Kulundin. Photo credits are showed in supplementary table S8, Supplementary Material online.
In particular, we detected the introgression signal of European mouflon in Shetland sheep, which probably occurred before their arrival at the Shetland isles via the Viking invasions (Ryder 1981). Shetland sheep is one of the smallest British breeds and it retains many of the characteristics of wild sheep such as spiral horn, mouflon markings, and natural hardiness (fig. 3H, Chessa et al. 2009). Today, they are considered a primitive or “unimproved” breed (Noddle and Ryder 1974). Similar morphological features have been observed between primitive domesticates and their wild ancestors of other taxa, such as Capra (Horwitz and Bar-Gal 2006), Bos (Upadhyay et al. 2017), Equus (Kvist et al. 2019; Chodkiewicz 2020), and Canis (Jordana et al. 1999).
We next sought to estimate the average proportion of the domestic sheep genome that may have a wild sheep origin using PCAdmix with PP (posterior probabilities) ≥0.95. We found the average proportions of their genomes from the wild species were 0.29% (0.05–2.12%), 5.09% (3.68–6.99%), 0.97% (0.59–1.3%), and 0.076% (0.05–0.09%) for Eastern and Central Asian, Western Asian, Southwestern European, and Russian populations, respectively (supplementary table S9, Supplementary Material online). The range in proportion of introgressed genomes identified here is similar to that previously reported in the introgression from argali to Tibetan sheep (5.23–5.79%; Hu et al. 2019), European mouflon to European domestic sheep (1.0–4.1%; Barbato et al. 2017), cattle to yak (∼1.3%; Medugorac et al. 2017), and banteng to domestic cattle (∼2.93%; Chen et al. 2018).
Nevertheless, we could not differentiate that after centuries the wild introgression left in domestic sheep was introduced by intercrossing in captivity or in the wild in the past. We noted that it is difficult to discriminate the signals of introgression and common ancestral polymorphisms at the whole-genome scale (Lowery et al. 2013; Barbato et al. 2017; Hu et al. 2019), particularly between the phylogenetically close species such as between Asiatic mouflon/European mouflon and domestic sheep. However, a very small amount of common signals (argali: 0–4.57%, snow sheep: 0–3.55%, Asiatic mouflon: 0–9.5%, and European mouflon: 0–21.59%) were detected in the introgression tests between the same wild species and different domestic populations, which might preclude the possibility of incomplete lineage sorting (ILS; Huerta-Sánchez et al. 2014).
In the CIWI (consistently introgressed windows of interest) analysis, we identified SNP alleles in a total of 1,499, 1,410, 3,493, and 801 genes in domestic sheep that originate from argali, Asiatic mouflon, European mouflon or snow sheep, respectively, using the 95th percentile. Our analyses identified 283 shared genes with alleles originating from the four wild relatives (fig. 4D). In particular, introgression signal in RXFP2, a prime candidate gene for horn type and size in ruminants (Johnston et al. 2011; Allais-Bonnet et al. 2013), was also detected from cattle to yak (Medugorac et al. 2017). The findings suggested convergent genetic evolution in these genes linked to interspecies introgression, which might be due to similar adaptive functions of the genes in the species.
Fig. 4.
Introgression signals at the genomic level and their impact on genomic diversity. (A) Phylogenetic relationships among the Ovis species. The signals of gene flow observed in this study were showed. (B) Graphical representation of the inferred local ancestry for Bashibai sheep (BSB) by PCAdmix. The color scheme indicates the assignment of each block to one of the two reference populations. Genomic regions not analyzed by the software due to the absence of SNPs are indicated as white gaps. The top panel is the inset expanding the genomic region on chromosome 2, where genes related to the citrullination function are located. The second panel is the within-analysis concordance scores (A-scores) calculated along the chromosome to represent the concordance of ancestry assignment among the 15 Bashibai individuals. The A-scores relative to the four reference populations are represented by the colored segments. The CIWI (consistently introgressed windows of interest) score is calculated from the A-scores and is represented by the black solid line. The third panel shows the PCAdmix results for one individual (number 14) of Bashibai, identified from the comparisons with four different domestic reference populations (SSS, HDW, HUS and WDS). SSS, Sishui Fur sheep; HDW, Large Tailed Han sheep; HUS, Hu sheep; WDS, Wadi sheep. The bottom panel is the 15 diploid individuals of Bashibai, and they are represented by the 15 numbered lines. Each line represents a diploid individual of Bashibai. (C) Graphical representation of the inferred local ancestry for Tuva sheep (TUVA) from the PCAdmix analysis. The top panel is the inset expanding the genomic region within chromosome 3. The second panel is within-analysis concordance scores (A-scores). They are calculated along the chromosome to represent the concordance of ancestry assignment among the 16 focal individuals. The third panel is the PCAdmix results for one individual (number 6) of Tuva, identified from the comparison with four different domestic reference populations (BAJ, CAU, KUI, and ROM). BAJ, Baidarak; CAU, Caucasian; KUI, Kuibyshev; ROM, Romanov. The bottom panel is the 16 focal diploid individuals from Tuva. Each line represents a diploid individual of Tuva and extends for the total length of chromosome 3. (D) Overlapped genes identified with introgression signals from argali, Asiatic mouflon, European mouflon, and snow sheep to domestic sheep. (E) Expected heterozygosity (He), observed (Ho), and the proportion of polymorphic SNPs (Pn) of introgressed and nonintrogressed populations domestic populations (*P < 0.05).
For the 283 genes, we identified significant (P < 0.05) gene ontology (GO) terms associated with sensory perception of chemical stimulus (GO:0007606, P = 0.0332), lipid phosphorylation (GO:0046834, P = 0.0033), and neurological system process (GO:0050877, P = 0.042) (supplementary table S10, Supplementary Material online). In particular, the GO term (GO:0007606) contains olfactory-related genes ADCY3 (fig. 4C) and TRPV1, suggesting that genomic introgression from wild relatives contributed to the sensory perception of chemicals (e.g., pheromones for reproduction success) in domestic populations.
We observed more shared functional genes (1,119 = 283 + 836; fig. 4D) with high CIWI scores when only the introgression signals from argali, Asiatic mouflon, and European mouflon to sympatric domestic populations were considered (fig. 4B, supplementary table S11, Supplementary Material online). Of the 1,119 genes, we revealed significantly enriched GO terms associated with immune function including protein citrullination (GO:0018101, P = 0.001), citrulline biosynthetic process (GO:0019240, P = 0.0049), and citrulline metabolic process (GO:0000052, P = 0.0176) (supplementary table S11, Supplementary Material online). We detected common introgression signals in the PADI (peptidyl arginine deiminases) gene family (e.g., PADI1, PADI2, PADI3, PADI4, and PADI6, fig. 4B), which are involved in antibacterial innate immunity mediated by neutrophil extracellular traps (Li et al. 2010). The genes PADI2 and PADI4 can citrullinate the important host defense peptide LL-37, resulting in impaired bacterial destruction (Kilsgård et al. 2012).
Common Introgression in PADI2 Inferred from Whole-Genome Sequences
To further test for introgression in the target genomic region (chr2: 248,302,667–248,306,614), we calculated D-statistics by three forms [[[MEN, BSB], ARGS], BIGS], [[[MEN, GSS], AMUF], BIGS], and [[[MEN, CAUC], EMUF], BIGS] using high-coverage whole-genome sequences. The D-statistics showed that the ABBA pattern (donors to recipients) is significantly higher than the BABA pattern (donors to the reference population of nonintrogressed animals) among the three forms in the target region (fig. 5A, supplementary fig. S9A, Supplementary Material online). We concatenated the introgressed tracks around PADI2 (chr2: 248.26–248.58 Mb). We identified the total length of 140 kb (without interval), 200 kb (interval < 80 kb), and 300 kb (interval < 40 kb) in the three forms above, respectively.
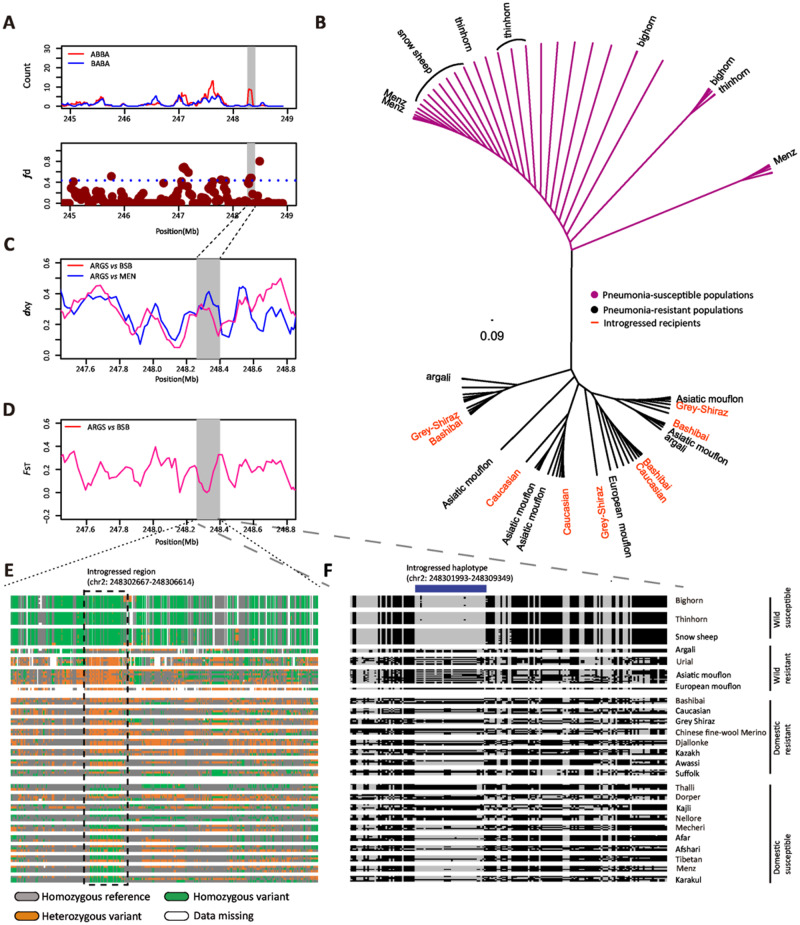
Fig. 5.
Introgression in a specific region of PADI2 (chr2: 248,302,667–248,306,614). (A) Counts of observed site patterns (ABBA and BABA sites) and genome-wide distribution of fd values calculated for 100-kb windows with a 20-kb step across the genome for Bashibai (BSB) sheep. Each dot represents a 100-kb window and the dashed line indicates the significance threshold (P < 0.05). (B) NJ tree of 38 individuals from pneumonia-resistant and 50 individuals from pneumonia-susceptible populations based on p-distances using the SNPs in the introgressed genomic region (chr2: 248,302,667–248,306,614). (C) Mean pairwise sequence divergence (dxy) of the introgressed genomic region on chromosome 2 between argali (ARGS) and either Bashibai (BSB) or Menz (MEN) sheep. Bashibai (BSB) represents pneumonia-resistant sheep with signatures of argali introgression. (D) Population differentiation (FST) around the introgressed genomic region on chromosome 2 between argali (ARGS) and Bashibai (BSB). (E) The pattern of SNP genotypes in PADI2 among the pneumonia-susceptible (bighorn, thinhorn, snow, Thalli, Dorper, Kajli, Nellore, Mecheri, Afar, Afshari, Tibetan, Menz, Karakul Sheep' has been changed to 'bighorn sheep, thinhorn sheep, snow sheep, Thalli sheep, Dorper sheep, Kajli sheep, Nellore sheep, Mecheri sheep, Afar sheep, Afshari sheep, Tibetan sheep, Menz sheep, Karakul sheep) and pneumonia-resistant (argali, urial, Asiatic mouflon, European mouflon, Bashibai, Chinese fine-wool Merino, Djallonké, Kazakh, Awassi, Suffolk) populations of wild and domestic sheep. (F) Haplotype pattern in the region (chr2: 248,301,993–248,309,349, 39 SNPs) of PADI2 gene. The introgressed haplotype is at high frequency in pneumonia-resistant populations of wild (e.g., argali, Asiatic mouflon, European mouflon, and urial) and domestic (e.g., Bashibai) sheep, whereas at low frequency in pneumonia-susceptible populations of wild (e.g., bighorn, thinhorn, and snow sheep) and domestic (e.g., Menz and Karakul) sheep. Each column is a polymorphic genomic SNP, and each row is a phased haplotype. The reference and alternative allele are indicated by black and gray, respectively.
To further locate the introgressed regions in the genomes of Bashibai, Grey Shiraz, and Caucasian sheep, we computed the f-statistic (fd) value for each 100-kb window (with a 20-kb step) across the genomes (Martin et al. 2015). The fd values in the detected tracks were significantly higher than the threshold values (P < 0.05). Based on the cutoff P < 0.05, we concatenated the potentially introgressed regions with interval <100 kb and detected 2,067, 2,176, and 1,434 significantly introgressed genomic tracts for Bashibai, Grey Shiraz, and Caucasian sheep, respectively. The genomic tracts have a total length of 307,560, 350,480, and 239,460 kb and cover ∼12.56%, 14.31%, and 9.78% of the whole genomes, respectively.
In order to rule out the possibility of ILS for the putatively introgressed tract, we calculated the probability of ILS using a generation time of 3 years (Zhao et al. 2017), a recombination rate of 1.0 × 10−8 (Kijas et al. 2012) and a divergence time of 2.36 Ma between argali and domestic sheep (Yang et al. 2017), and 0.01–0.021 Ma as the split time between mouflon (including Asiatic mouflon and European mouflon) and domestic sheep (0.021 Ma between O. musimon and O. aries; Yang et al. 2017). This analysis produced the expected length of shared ancestral tracts of L(argali) = 63.56 bp and L(mouflon) = 7,142.86–15,000 bp. The probability of a length of 140, 200, and 300 kb (i.e., the observed introgressed regions from argali, Asiatic mouflon, and European mouflon) was 0, 2.01 × 10−11–2.32 × 10−5, and 0–4.33 × 10−8, respectively. All the estimated probabilities were much lower than 0.05. Thus, the identified introgressed genomic regions in Bashibai, Grey Shiraz, and Caucasian sheep were unlikely due to ILS.
For the domestic breeds and wild species, we collected the information of pneumonia resistance and pneumonia susceptibility from the records or description in previous publications (supplementary table S12, Supplementary Material online). Only the breeds and species which have been reported to be pneumonia resistant and pneumonia susceptible or show clear evidence of high/low mortality from pneumonia were included in the following analyses. In total, 16 domestic breeds and 7 wild species fit into the categories (supplementary table S12, Supplementary Material online). We computed the mean pairwise sequence divergence (dxy) between wild sheep and either the pneumonia-resistant (Bashibai, Grey Shiraz, and Caucasian sheep) or pneumonia-susceptible populations (Menz sheep). The results showed that the dxy value in the focused regions was significantly reduced in Bashibai, Grey Shiraz, and Caucasian sheep but highly elevated in Menz sheep (fig. 5C, supplementary fig. S9B, Supplementary Material online), suggesting genetic introgression in the target region. The FST values (argali vs. Bashibai, Asiatic mouflon vs. Grey Shiraz, European mouflon vs. Caucasian) were lower in the common introgressed genomic region than in the rest of the genome (fig. 5D, supplementary fig. S9C, Supplementary Material online). These results support the common introgression of alleles in PADI2 from wild relatives to domestic populations.
Resistance to Pneumonia by Genetic Introgression
The NJ tree based on the p-distance of the common introgressed genomic region displayed a clear divergence pattern between the pneumonia-susceptible and the pneumonia-resistant populations of wild and domestic sheep (fig. 5B), rather than the phylogenetic relationships inferred from chromosome-wise SNPs (supplementary fig. S10, Supplementary Material online). In addition, we compared the genotypes and haplotypes of the target genomic region for pneumonia-susceptible and pneumonia-resistant populations based on well-documented records of pneumonia incidence (supplementary table S12, Supplementary Material online). Particularly, we found that the genotype and haplotype patterns of Bashibai, Grey Shiraz, and Caucasian sheep were highly similar to that of argali, Asiatic mouflon, and European mouflon, respectively, but were strikingly different from that of Menz sheep. Noteworthy, the genotype and haplotype pattern of Menz sheep were similar to that of highly susceptible wild species of bighorn, thinhorn, and snow sheep (figs. 5E and F).
In the estimation of Tajima’s D, the introgressed region in PADI2 showed positive values in the resistant populations but negative values in the susceptible populations. Also, the estimates showed a higher |Tajima’s D susceptible-populations − Tajima’s D resistant-populations| value (fig. 6A), which suggested a signature of positive selection in the putatively introgressed region within the gene. VolcanoFinder detected a strong signal (maximum log likelihood ratio = 14.63) beside PADI2 in pneumonia-resistant and pneumonia-susceptible populations, indicating an introgressed sweep (fig. 6B). Furthermore, the genome scan across chromosome 2 by the cross-population composite likelihood ratio (XP-CLR) approach showed significant signatures of positive selection in PADI2 between pneumonia-susceptible and pneumonia-resistant populations of domestic sheep (fig. 6C, supplementary table S13, Supplementary Material online). We also detected the largest LD block (chr2: 248,292,698–248,306,614, 13 kb) in PADI2, overlapping largely with the introgressed region inferred (fig. 6D). These results suggested that the introgression of PADI2 from wild relatives could have enhanced the resistance to pneumonia in domestic sheep.
Fig. 6.
Selective sweep analysis across chromosome 2 and LD pattern in the PADI2 gene. (A) PADI2 gene with signal of positive selection in pneumonia-resistant population. Tajima’s D values are plotted using a 1-kb sliding window. The gray shadow is the introgressed regions (chr2: 248,301,993–248,309,349). (B) Plot of the maximum likelihood test statistic based on pneumonia-resistant and pneumonia-susceptible populations shows an introgressed sweep. (C) XP-CLR genome scan by comparing pneumonia-resistant (Bashibai, Caucasian, Grey Shiraz, Chinese fine-wool Merino, Djallonke, Kazakh, Awassi, and Suffolk sheep) with pneumonia-susceptible populations (Thalli, Dorper, Kajli, Nellore, Mecheri, Afar, Afshari, Tibetan, Menz, and Karakul sheep) of domestic sheep. The top 1% cutoff level (XP-CLR score = 6.568220) was indicate by a red horizontal line. Significant selective signatures in PADI2 are marked in green. (D) LD plot of SNPs located in PADI2. LD values (D′) between two loci are detailed in boxes (D′ = 0 − 1). D′ = 1 indicates perfect disequilibrium.
For the susceptible wild species of bighorn, thinhorn, and snow sheep, infection by relevant pathogens following contact with domestic sheep will cause serious mortality events (Foreyt and Jessup 1982). As a result, no hybrid animal between them and domestic sheep was observed in the wild, and, thus, no introgression could have occurred between domestic sheep and the three susceptible wild species. Nevertheless, hybrid animals between domestic sheep and the species of argali, Asian mouflon, and European mouflon have been found in the wild (Woronzow et al. 1972; Arnold 2004; Schröder et al. 2016). These observations further supported the introgressed alleles of PADI2 from the wild relatives such as argali, Asian mouflon, and European mouflon could have contributed to pneumonia resistance in domestic sheep.
Overall, our study suggested PADI2 as a candidate gene for future gene editing and molecular breeding for pneumonia-resistant animals. Nevertheless, we cautioned that ovine pneumonia can be caused by multiple factors such as lung worms, maedi-visna, bacterial bronchopneumonia, enzootic pneumonia, and fungal infection (Tibbo et al. 2001). Here we only have the general species/breed information of resistance or susceptibility to pneumonia for the wild and domestic sheep, whereas no clinical data were recorded for the specific individuals sequenced. Thus, further investigations on the molecular mechanisms underlying pneumonia resistance are needed.
Climate-Driven Selective Signatures
Using the Samβada approach, we selected the top 20,000 significant univariate models (∼0.01% of a total of 182,230,776 univariate models [1,557,528 genotypes × 117 environmental parameters]) based on the Wald Score. We detected 2,265 SNPs associated with climatic variables (supplementary table S17, Supplementary Material online). Using the latent factor mixed model (LFMM) approach, we identified 3,437 SNPs with |z| scores ≥10 (supplementary table S18, Supplementary Material online), suggesting that they were significantly associated with climatic variables in the 70 autochthonous breeds (supplementary table S14, Supplementary Material online). Overall, 393 SNPs were detected by both the approaches, and were annotated to be located near or within 317 functional genes associated with climate-mediated selection (fig. 7A;supplementary table S19, Supplementary Material online). This number of shared genes between Samβada (n = 808) and LFMM method (n = 1,101) was significantly greater than that expected by chance (the total number of genes in sheep n = 17,666, the observed number of overlapping genes n = 317, the number of overlapping genes expected by chance n = 49; P < 0.05) (fig. 7C).
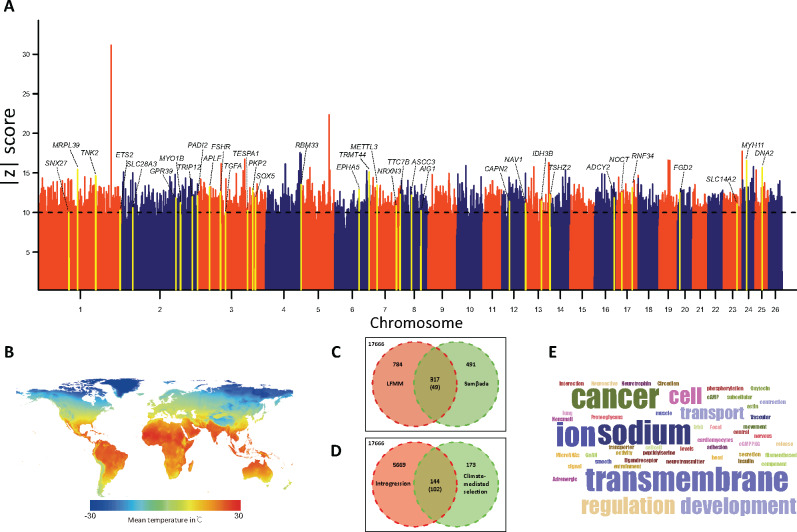
Fig. 7.
The overlapped genes of genetic introgression and climate-mediated selection signals. (A) Genome-wide distribution of significance values [−log10 (P)] for the correlation between SNPs and the environmental variables in the LFMM test. Arrows indicate the overlapped genes between climate-mediated selection and introgression signals. (B) The geographic pattern of mean temperature in °C used in the SNP-environment association analysis. (C) Overlapped genes identified from the LFMM and Samβada approaches; the number in parenthesis shows the number of overlapping genes expected by chance. (D) Overlapped genes identified from introgression and climate-mediated selection; the number in parenthesis shows the number of overlapping genes expected by chance. (E) Word cloud of GO terms and KEGG pathways with significant (P < 0.05) enrichments for 144 overlapped genes between genetic introgression and climate-mediated selection.
A close examination of the function of the 317 annotated genes found: 1) 85 genes (e.g., METTL3, RNF34, ETS2, and CAPN2) associated with tumor growth and suppression, cell cycle progression, apoptosis, and cellular transformation; 2) 64 genes (e.g., NAV1, NRXN3, and SOX5) involved in the regulation of adrenergic neurons and receptors and semaphorins; 3) 46 genes (e.g., ASCC3, TNK2, and TESPA1) affecting metabolism toward glycolysis and gluconeogenesis, GTPase activity and lipid metabolism, and endocrine hormones; 4) 47 genes (e.g., PADI2, RBM33, AIG1, and FGD2) responsible for autoimmune regulation and immunodeficiency; and 5) 54 genes (e.g., GPR39, PKP2, and NOCT) accounting for tissue and organ morphology, and organismal abnormalities.
Functions of the climate-associated genes (e.g., METTL3 and NAV1) implied a strong impact of solar radiation (including UV radiation), temperature, and climate-related spread of pathogens on livestock (Bertolini et al. 2018; Flori et al. 2019). Of genes with evidence for putative selection, several (e.g., TTC7B, SLC28A3, IDH3B, and SLC14A2) were involved in the regulation of the cardiovascular system and the development of urinary and respiratory tract, which could be due to the difference in altitude and precipitation of their distribution regions (Flori et al. 2019). These three genes were also associated with climatic changes in a recent study in cattle (Flori et al. 2019).
Also, we detected 25 reproduction-related genes (e.g., FSHR, TRIP12, and TSHZ2) involved in oocyte and follicular development, premature ovarian failure or control of ovulation, some of which (e.g., FSHR and TSHZ2) were previously found to be associated with climatic conditions in sheep and goat (Lv et al. 2014; Bertolini et al. 2018; Osei-Amponsah et al. 2019). In particular, we found the gene FSHR related to seasonal breeding in sheep and other mammals (Rosa and Bryant 2003; Hobbs et al. 2012). These observations are reasonable because reproduction in mammals is closely associated with their response to light cycles and temperature (Chemineau et al. 2010; Lv et al. 2014).
GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the climate-driven selective genes detected 12 and 11 significantly (P < 0.05) enriched GO categories and pathways, respectively (supplementary table S20, Supplementary Material online). Some of them were reported previously to be associated with climate-mediated adaptation in mammals, such as calcium ion binding (GO:0005509, P = 0.00864) (Morenikeji et al. 2020), ATP binding (GO:0005524, P = 0.01503) (Mintoo et al. 2019), and neurotrophin signaling pathway (oas04722, P = 0.001984) (Mitre et al. 2017). Overall, the significant GO categories and pathways highlighted the main functional categories of the selective genes associated with climate-driven selective pressures, such as 1) nervous system development and function, 2) energy metabolism and endocrine regulation, 3) immune and inflammatory response, 4) organ development and morphology, and 5) cancer and cell development and proliferation. We noted that the climatic data covered a period of only 40 years. This might not reflect the exact climatic conditions since several hundred or several thousand years ago when the native breeds have inhabited in their distribution regions. Thus, future investigations with the climatic data over a longer period would show more informative results.
Influence of Wild Introgression on the Genomes of Domestic Populations
We examined genomic diversity in the 48 domestic populations which showed potential introgression from wild relatives. We found that 15 introgressed populations have significantly (the Kruskal–Wallis test, P < 0.05) higher level of genomic diversity than the 33 nonintrogressed populations in terms of the indices of Ho, He, and Pn (introgressed populations: mean Ho = 0.3387, mean He = 0.3267, and mean Pn = 0.946; nonintrogressed populations: mean Ho = 0.3221, mean He = 0.31, and mean Pn = 0.9036; fig. 4E). This finding demonstrated that introgression from wild relatives has increased the genomic diversity in domestic populations. Similarly, genomic introgressions from their progenitors have increased the genetic diversity in managed populations of honey bees (Harpur et al. 2012).
Further, we evaluated the contribution of introgression from wild sheep to climatic adaptation of domestic populations. Of the 5,813 genes with introgressed alleles from wild relatives, we found 144 genes associated with climate-mediated selection (the observed number of genes associated with climate-mediated selection n = 317; fig. 7D). The observed number of overlapping selective signals is significantly higher than the overlap expected by chance between the methods (the total number of genes in sheep n = 17,666, the observed number of overlapping genes n = 144; the number of overlapping genes expected by chance n = 102; P < 0.05; fig. 7D). A close examination of the 144 shared genes detected in the wild introgression and climatic association analyses showed that the overlapped genes have functions closely related to local environmental and climatic adaptation such as central nervous system development (EPHA5; Olivieri and Miescher 1999), angiogenesis (NRP2; Sulpice et al. 2008), oxytocin signaling pathway (RYR3; Matsuo et al. 2009), and lung development (ADCY2; Hardin et al. 2012).
In particular, we found a set of genes associated with immune response, response to virus and xenophagy and bacterial invasion. The two immune-related genes PADI1 and PADI2, which showed common introgression from various wild relatives in domestic populations, were also identified to be under climate-mediated selective pressure. Thus, the two genes might play an important role in immune response to local pathogens via adaptive introgression. Therefore, our results provided strong evidence that genomic introgression from congeneric wild sheep species has played an important role in shaping the climate-induced adaptive genome landscape of domestic populations.
Conclusions
We performed a comprehensive genomic investigation on the world’s wild and domestic sheep. We unveiled frequent wild introgression, which have increased the genomic diversity of domestic sheep. A proportion of allelic variability in functional genes received from wild relatives appears to provide locally adaptive phenotypes such as sensory perception of chemical stimulus and odorants and in particular innate immunity to pneumonia in domestic sheep. Coupled with the evidence of climatic selective signatures, our results revealed that frequent common introgression from wild relatives into sympatric domestic sheep has shaped the adaptive genomic landscape of indigenous populations and contributed to their rapid adaptation to local climates. These findings add to our understanding the evolutionary mechanisms of climate-mediated adaptation for local livestock populations. The newly generated genome-wide data are valuable resources for the future genetic improvement of sheep to develop new tolerant populations in the face of climate change.
Materials and Methods
Ovine Infinium HD SNP BeadChip Data
The Ovine Infinium HD SNP BeadChip data consisted of SNP genotypes generated in this study and those already available, totaling 3,850 individuals from 111 domestic sheep populations (3,447 individuals, hereafter referred to as “data set I”) and 7 wild sheep species (403 individuals, including 15 argali, 16 Asiatic mouflon, 4 urial, 98 European mouflon, 80 snow sheep, 135 bighorn, and 55 thinhorn sheep). The data generated here included genotypes of 888 individuals from 63 domestic sheep populations genotyped using the Ovine Infinium HD SNP BeadChip. DNA samples of the genotyped individuals were provided by the contributors (supplementary tables S1 and S2, Supplementary Material online).
The already available data include genotypes of 2,684 individuals (2,490 domestic sheep, 2 Asiatic mouflon, 2 European mouflon, 135 bighorn sheep, and 55 thinhorn sheep) with Ovine Infinium HD SNP BeadChip and 278 individuals (69 domestic sheep, 14 Asiatic mouflon, 4 urial, 96 European mouflon, 15 argali, and 80 snow sheep; supplementary tables S1 and S2, Supplementary Material online) with the OvineSNP50 BeadChip. Summary information, including breed names, abbreviations, geographic origins, sampling sizes, and contributors is detailed in figure 1A, supplementary figure S1 and tables S1 and S2, Supplementary Material online. Approximate coordinates of the geographic origins for the samples were provided by the contributors, or were assigned as the centroid of their known range area.
In domestic sheep, we selected 70 autochthonous populations genotyped with the Ovine Infinium HD SNP BeadChip (data set II) in the analyses of climatic selective pressures. To assess genomic introgression from wild relatives to sympatric domestic populations, 13 populations (n = 896) from Eastern and Central Asia, 13 populations (n = 176) from Western Asia, 34 populations (n = 608) from Southwestern Europe, and 9 populations (n = 284) from Russia selected from data set I were merged with argali (n = 15), Asiatic mouflon (n = 18), European mouflon (n = 98), and snow sheep (n = 80), respectively (hereafter named as data set III [832 individuals and 41,358 common SNPs], IV [194 individuals and 41,057 common SNPs], V[695 individuals and 39,443 common SNPs], and VI [298 individuals and 41,737 common SNPs]).
Climatic Data from 1961 to 2001
A total of 117 climatic and elevation parameters for the geographic origins of 70 worldwide autochthonous breeds (supplementary table S14, Supplementary Material online) were extracted from the global climate data set (http://www.cru.uea.ac.uk/data, last accessed September 24, 2020) of the Climatic Research Unit, Norwich (New et al. 2002). Data collected over a period of 40 years from 1961 to 2001 included yearly and monthly trends of the following variables: mean diurnal temperature range in °C (DTR), number of days with ground frost (FRS), the coefficient of variation of monthly precipitation in percent (PRCV), precipitation in mm/month (PR), number of days with >0.1 mm rain per month (RDO), relative humidity in percentage (REH), percent of maximum possible sunshine (percent of day length, SUN), mean temperature in °C (TMP), and 10-m wind speed in m/s (WND) (fig. 7B;supplementary fig. S11, Supplementary Material online). High-resolution global distributions of elevation and the environmental variables (yearly mean values), and the Spearman’s rho correlation coefficient among the elevation and climatic parameters are shown (supplementary fig. S12 and table S15, Supplementary Material online).
SNP Data Quality Control
We first merged the Ovine Infinium HD BeadChip data sets and updated the SNP physical positions based on the assembly Oar v.4.0 (http://www.ncbi.nlm.nih.gov/ assembly/GCF_ 000298735.2, last accessed September 24, 2020) using the program PLINK v1.09 (Purcell et al. 2007). Owing to differences in the number of SNPs and samples in the data sets, we then implemented quality control for the data sets using various criteria in PLINK v.1.09 software (supplementary table S3, Supplementary Material online). After filtering, we identified 495,965 SNPs and 3,217 individuals from data set I for genomic diversity and selective sweep analyses, 53,279 SNPs and 3,217 individuals from data set I for population structure analysis, 519,176 SNPs and 1,156 individuals of the 70 autochthonous breeds from data set II for climatic association analyses, 36,395 SNPs and 832 individuals from data set III, 36,508 SNPs and 194 individuals from data set IV, 36,148 SNPs and 695 individuals from data set V, and 29,562 SNPs and 298 individuals from the data set VI for genomic introgression analysis (supplementary table S3, Supplementary Material online).
Whole-Genome Sequences and SNP Calling
Chromosome 2 in 88 whole-genome sequences from our other studies was included in the analyses, comprising 54 domestic and 34 wild sheep (Deng et al. 2020; Li et al. 2020; Chen ZH, Xu YX, and Li MH , unpublished data). The domestic sheep samples represent eight pneumonia-resistant (24 individuals) and ten pneumonia-susceptible populations (30 individuals) of different geographic origins from Asia (including the Middle East), Africa, and Europe. The wild sheep samples represent four pneumonia-resistant (two argali, four urial, seven Asiatic mouflon, and one European mouflon) and three pneumonia-susceptible species (six bighorn, six thinhorn, and eight snow sheep). The record of resistance or susceptibility to pneumonia for the wild species and domestic populations was from well-documented literature (supplementary table S12, Supplementary Material online). All the samples were sequenced on the Illumina Hiseq X Ten sequencer to a depth of 20–30× with an average depth of 20.49×. We filtered the raw reads using three criteria: 1) reads with unidentified nucleotides (N) more than 10%, 2) reads having adapters, and 3) reads with a phred quality score <5 were excluded. Clean reads were used for further analysis.
Using the Burrows–Wheeler Aligner (BWA) v.0.7.17 MEM module (Li and Durbin 2009) with the parameter “bwa –k 32 –M -R,” we mapped the clean reads onto the sheep reference genome Oar v.4.0. Duplicate reads were excluded using Picard MarkDuplicates and were sorted using Picard SortSam. We only kept reads that were properly mapped with mapping quality more than 20. Base quality score recalibrate (BQSR) with ApplyBQSR module of the Genome Analysis Toolkit (GATK) v.4.0 (McKenna et al. 2010) was applied to eliminate potential sequence error. After mapping, SNP calling was carried out by GATK best practice workflow of joint genotyping strategy. Two steps were carried out: 1) We called variants for each sample using Haplotypecaller module with parameter “--genotyping-mode DISCOVERY --min-base-quality-score 20 --output-mode EMIT_ALL_ SITES --emit-ref-confidence GVCF”; and 2) we performed joint genotyping by combining all the GVCFs using GenotypeGVCFs module and CombineGVCFs module together. Variant sites with QUAL <30.0, QD <2.0, FS >60.0, MQ <40.0, HaplotypeScore >13.0, MappingQualityRankSum <12.5, and ReadPosRankSum <8.0 were discarded using VariantFiltration module of GATK. We called SNP separately for each species and then merged them using BCFtools v1.9 (Li 2011). PLINK v1.09 was used for further filtering of SNPs in the populations. We excluded SNPs that met any of the following criteria: 1) proportion of missing genotypes >10% and 2) SNPs with minor allele frequency <0.05. In total, we identified 119,884 SNPs on chromosome 2 for further analysis.
Genetic Diversity and Population Genetic Structure
We evaluated the genomic diversity for the populations of wild and domestic sheep based on seven metrics, including the proportion of polymorphic SNPs (Pn), observed (Ho) and expected heterozygosity (He), inbreeding coefficient (F) using PLINK v1.09, and allelic richness (Ar) and private allele richness (pAr) using ADZE v1.0 (Szpiech et al. 2008). We investigated the levels of LD between pairs of autosomal SNPs with the r2 estimate (the parameters “--r2 --ld-window 99999 --ld-window-r2 0”) using PLINK v1.09. Recent effective population size (Ne) for each population was then calculated based on the LD estimates (Kijas et al. 2012).
To examine population genetic structure, we first implemented PCA based on the SmartPCA program from the EIGENSOFT v.6.0beta (Patterson et al. 2006). Additionally, we constructed an NJ tree based on pairwise Reynolds’ genetic distances (Reynolds et al. 1983) using PHYLIP v.3.695 (Felsenstein 1993). The NJ tree was visualized with FigTree v.1.4.3 (Rambaut 2014), and the robustness of a specific tree topology was tested by 1,000 bootstraps. We explored the genetic components of the populations using the maximum-likelihood clustering program ADMIXTURE v1.23 with K = 2–10 (Alexander et al. 2009). We then performed additional ADMIXTURE analyses for the domestic populations from different geographic regions selected in the genomic introgression analyses (see below) using the sympatric wild species as the outgroup (i.e., argali for 13 Eastern and Central Asia populations, Asiatic mouflon for 13 Western Asian populations, European mouflon for 34 Southwestern European populations, and snow sheep for 20 Russian populations). We processed the results with Clumpp v1.1.2 (Jakobsson and Rosenberg 2007) and visualized them with Distruct v1.1 (Rosenberg 2003).
Genomic Introgression
To determine the potential genomic introgressions from wild relatives into sympatric domestic breeds, such as argali to Eastern and Central Asian breeds, Asiatic mouflon to Western Asian breeds, European mouflon to Southwestern European breeds, and snow sheep to Russian breeds (fig. 4A), we implemented three different statistical analyses in the selected populations of wild and domestic sheep (supplementary table S9, Supplementary Material online), including TreeMix (Pickrell and Pritchard 2012), f3-statistics (Patterson et al. 2012), and D-statistics (Patterson et al. 2012). In the TreeMix analysis, we constructed the maximum likelihood trees using blocks of 1,000 SNPs and the wild species of argali, Asiatic mouflon, European mouflon, and snow sheep as the roots. We set the number of tested migration events (M) from 1 to 20 and quantified the model by the covariance for each migration event. Additionally, we computed f3-statistics using the qp3Pop program with the default parameters in the AdmixTools package (Patterson et al. 2012). We used the f3-statistics based on the following scenarios: f3(C; argali, B), f3(C; Asiatic mouflon, B), f3(C; European mouflon, B), and f3(C; snow sheep, B), where B and C represent domestic breeds from the same geographic regions such as Eastern and Central Asia, Western Asia, Southwestern Europe, and Russia. We considered f3-statistics <0 as statistically significant, indicating historical events of admixture (Reich et al. 2009). Also, we estimated D-statistics using the program AdmixTools (Patterson et al. 2012) for the following scenarios: D (MZS, X, argali, BIGHORN), D (MZS, X, Asiatic mouflon, BIGHORN), D (MZS, X, European mouflon, BIGHORN), and D (MZS, X, snow sheep, BIGHORN), where MZS, X, and BIGHORN represent Menz sheep (the nonintrogressed reference population), domestic populations from the four geographic regions above, and the outgroup of bighorn sheep, respectively. |Z| scores >3 indicated statistically significant (P < 0.001) deviations from D = 0, suggesting gene flow occurred between the donor wild species and tested domestic population X.
Further, we characterized the introgressed genomic regions on the basis of local-ancestry assignment and CIWIs (Barbato et al. 2017). We first phased the genotypes using the program fastPHASE v1.239 with the default parameters (Scheet and Stephens 2006). For the focal domestic populations, we identified the genomic segments of the wild ancestry based on the reference of a wild and four domestic populations using the program PCAdmix v1.0 (Brisbin et al. 2012). We further used a sliding window to identify the introgressed genomic segments following the method developed by Barbato et al. (2017). We filtered the results of highly concordant introgression signals along chromosomes and assigned a concordance score to each sliding window. We took the 95th percentile of the genome-wide concordance score distribution as the CIWIs for each autosome.
Based on the results of TreeMix, Admixture, f3-statistics, D-statistics, and CIWI analyses, breeds showing introgression signals in both population-level and genomic-level analyses were selected as “introgressed populations,” whereas others showing no signals of introgression in either population-level or genomic-level analysis were considered as “nonintrogressed populations.” To examine the contribution of wild introgression to genomic diversity in domestic populations, we compared the genomic diversity estimates (e.g., He, Ho, and Pn) between the two groups of domestic populations (introgressed vs. nonintrogressed) using the Kruskal–Wallis test in the software SPSS v24.0 (IBM Corp. 2016).
Whole-Genome Sequence Analyses and ILS
Furthermore, we focused on the introgressed region chr2: 245–249 Mb (especially the focal gray-shaded region: 248.28–248.4 Mb, 248.28–248.58 Mb, and 248.26–248.5 Mb in argali introgression, Asiatic mouflon introgression, and European mouflon introgression, respectively) that harbors the PADI2 gene using high-depth whole-genome sequences. We validated the introgressed signals detected above using ABBA, BABA, D-statistics, and f-statistic (fd) value (Martin et al. 2015). We used the D-statistics [[[MEN, BSB], ARGS], BIGS], [[[MEN, GSS], AMUF], BIGS], and [[[MEN, CAUC], EMUF], BIGS], in which BIGS (bighorn sheep) is the outgroup; ARGS (argali), AMUF (Asiatic mouflon), and EMUF (European mouflon) represent the donor species; MEN (Menz sheep) and populations of BSB, GSS, and CAUC (Bashibai, Grey Shiraz, and Caucasian sheep) represent the domestic sheep susceptible and resistant to pneumonia, respectively. To further locate the introgressed genomic regions, we computed the f-statistic (fd) value for each 100-kb sliding window with 20-kb step across the whole genome of argali versus Bashibai sheep, Asiatic mouflon versus Grey Shiraz sheep, and European mouflon versus Caucasian sheep, respectively, with Menz sheep as the nonintrogressed reference population and bighorn sheep as the outgroup. We phased the genotypes of chromosome 2 using Shapeit v2.12 (Delaneau et al. 2014).
Further, we calculated the mean pairwise sequence divergence (dxy) between argali and Menz/Bashibai, between Asiatic mouflon and Menz/Grey Shiraz, as well as between European mouflon and Menz/Caucasian. Also, the FST value of the common introgressed genomic region was calculated for the pairwise comparisons of argali vs. Bashibai, Asiatic mouflon vs. Grey Shiraz, and European mouflon vs. Caucasian. The introgressed tract could be due to ILS. To verify that the target genomic region was derived from introgression rather than common ancestry, we calculated the probability of ILS for the three tracks, which were inferred to be introgressed from argali to Bashibai, Asiatic mouflon to Grey Shiraz, and European mouflon to Caucasian sheep. The expected length of a shared ancestral sequence is L = 1/(r × t), in which r is the recombination rate per generation per base pair (bp), and t is the branch length between argali/mouflon and domestic sheep since divergence. The probability of a length of at least m is 1 − GammaCDF (m, shape = 2, r = 1/L), in which GammaCDF is the Gamma distribution function (Huerta-Sánchez et al. 2014; Hu et al. 2019).
In addition, we constructed the NJ tree based on the p-distance for the target genomic region among the pneumonia-susceptible and pneumonia-resistant populations using PLINK v1.09 (Purcell et al. 2007). To view the specific genotype pattern of the common introgressed gene PADI2, we extracted the genotypes of 435 SNPs in the PADI2 gene (chr2: 248,285,826–248,352,997) from the high-depth whole-genome sequences and compared the pattern of the genotypes between the pneumonia-susceptible (i.e., 3 pneumonia-susceptible wild species, 20 individuals; 10 pneumonia-susceptible domestic breeds, 30 individuals) and pneumonia-resistant samples (i.e., 4 pneumonia-resistant wild species, 14 individuals; 8 pneumonia-resistant domestic breeds, 24 individuals).
Signatures for Local Climatic Adaptation
We performed two different algorithms to detect signatures of local adaptation across the 70 worldwide autochthonous breeds (supplementary table S14, Supplementary Material online). First, we used an individual-based spatial analysis method (Stucki et al. 2017) in the Samβada software (https://lasig.epfl.ch/sambada, last accessed September 24, 2020). The algorithm measured explanatory variables incorporating population structure into the models to decrease the occurrences of spurious genotype–environment associations. The Samβada calculated multiple univariate logistic regression analyses, whereby individuals were coded as either present or absent (i.e., binary information: 1 or 0) for a given SNP allele and the association between all possible pairs of allele and environmental variable was measured across the sampling sites. We considered the significant models based on the Wald tests with the Bonferroni correction at the level of P < 0.01.
We further calculated the correlations between SNPs and climatic variables using the LFMM (http://membres-timc.imag.fr/Olivier.Francois/lfmm/index.htm, last accessed September 24, 2020) program, which is based on population genetics, ecological modeling, and statistical learning techniques (Frichot et al. 2013). The LFMM can efficiently control for random effects that represent population history and isolation-by-distance patterns and lower the risk of false-positive associations in landscape genomics (Frichot et al. 2013). We summarized all the climatic variables by using the first axis from a PCA (supplementary table S16, Supplementary Material online). We identified K = 4 latent factors on the basis of population structure analyses using the programs SmartPCA and STRUCTURE v2.3.4 (supplementary figs. S13 and S14, Supplementary Material online). We computed LFMM parameters (|z| scores) for all the variants based on the MCMC (Markov chain Monte Carlo) algorithm with a burn-in of 5,000 and 10,000 iterations. The threshold for identifying candidate genes in LFMM analyses was set to |z| scores ≥10, indicating significant SNP effects at the level of P < 10−22 after Bonferroni correction for a type I error α = 10−16 and L ≈ 1 × 106 loci (Frichot et al. 2013; Lv et al. 2014).
Haplotype, Selective Signature, and LD Analysis within PADI2
To examine the haplotypes in PADI2, we first excluded SNPs with proportion of missing genotypes >2% and minor allele frequency <0.01 in the 88 individuals with whole-genome sequences. Shapeit v2.12 (Delaneau et al. 2014) was then used to infer the haplotypes. Furthermore, genome scan across chromosome 2 was performed to identify potential regions under positive selection between pneumonia-resistant (Bashibai, Caucasian, Grey Shiraz, Chinese fine-wool Merino, Djallonke, Kazakh, Awassi, and Suffolk) and pneumonia-susceptible (Thalli, Dorper, Kajli, Nellore, Mecheri, Afar, Afshari, Tibetan, Menz, and Karakul) breeds of domestic sheep using XP-CLR v.1.0 (Chen et al. 2010). The SNPs with <10% missing data were allowed (--max-missing 0.9) in the analysis. XP-CLR scores were calculated using the grid points spaced by 2,000 bp with a maximum of 200 SNPs in a window of 0.5 cM and the down-weighting contributions of highly correlated variants (r2 > 0.95) with the parameters -w1 0.005 200 2000 2 -p0 0.95. The top 1% genomic regions with the highest XP-CLR scores were considered to be the selective signatures. In addition, the sliding-window approach (window size = 1 kb) was used to quantify the Tajima’s D of pneumonia-resistant and pneumonia-susceptible populations using vcftools v0.1.14 (Danecek et al. 2011). Also, we implemented genome-wide scans of adaptive introgression using VolcanoFinder v1.0 (Setter et al. 2020). We first generated the allele frequency file and the unnormalized site frequency spectrum using SweepFinder2 (DeGiorgio et al. 2016). The two files were then used as input in the program VolcanoFinder with the options of ./VolcanoFinder –ig 1000 and the Model 1. Haploview software (Barrett et al. 2005) was then used to examine the LD pattern in PADI2.
Gene Annotation, GO, and Pathway Analysis
We annotated genes located within 20 kb upstream and downstream of these selective regions or SNPs using the O. aries assembly Oar_v.4.0 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000298735.2, last accessed September 24, 2020). We implemented GO enrichment and KEGG pathway analyses for the annotated genes using the sheep genome background in the DAVID (database for annotation, visualization, and integrated discovery) Bioinformatics Resources v.6.8 (Huang et al. 2009). The threshold was set to P < 0.05 and at least two genes from the input gene list in the enriched category were considered as the enriched GO terms and KEGG pathways. We created word clouds for the significance GO terms and KEGG pathways on the Wordcloud generator (https://www.jasondavies.com/wordcloud/, last accessed September 24, 2020).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was financially supported by the grants from the National Key Research and Development Program-Key Projects of International Innovation Cooperation between Governments (2017YFE0117900), the National Natural Science Foundation of China (Nos. 31661143014, 31825024, 31972527, 31660651, and 31760661), the External Cooperation Program of Chinese Academy of Sciences (152111KYSB20150010), the Second Tibetan Plateau Scientific Expedition and Research Program (STEP) (No. 2019QZKK0501), and the Taishan Scholars Program of Shandong Province (No. ts201511085). The genotypes were in part produced under financial support of the Russian Ministry of Higher Education and Science. The Chinese government’s contribution to CAAS-ILRI Joint Laboratory on Livestock and Forage Genetic Resources in Beijing and to ICARDA is appreciated. We thank Kathiravan Periasamy (Animal Production and Health Laboratory, Joint FAO/IAEA Division, International Atomic EnergyAgency, Vienna, Austria), Amadou Traore (Institut de l’Environnement et de Recherches Agricoles [INERA], Ouagadougou, Burkina Faso), Masroor Ellahi Babar (Virtual University, Lahore, Pakistan), Pradeepa Silva (University of Peradeniya, Peradeniya, Sri Lanka), Seyed Abbas Rafat (University of Tabriz, College of Agriculture, Tabriz, Iran), Thiruvenkadan A.K. and Saravanan Ramasamy (Tamil Nadu Veterinary and Animal Sciences University, Chennai, India), Innokenty Ammosov, Ming-Jun Liu and Wen-Rong Li for providing the samples of unpublished data, and Guang-Jian Liu (Novogene Bioinformatics Institute) for his kind help in SNP calling.
Author Contributions
M.H.L. designed the study. M.H.L., Y.-H.C., M.S., L.G., F.-H.L., X.-L.X., X.-H.W., H.Y., C.-B.L., P.Z., P.-C.W., Y.-S.Z., J.-Q.Y., W.-H.P., E.H., D.P.B., M.B., A.E., M.N., H.S.-D., M.D.-Q., A.V.D., T.E.D., N.A.Z., G.B., O.S., E.C., C.W., G.E., J.M.M., A.A., J.-L.H., O.H., J.M.M., Z.S., D.C., J.K., M.W.B., J.A.L., and J.K. contributed samples and SNP genotypes or provided help during the sample collection. Y.-H.C., S.-S.X., and Z.-H.C.performed the genome data analyses. M.-H.L., Y.-H.C., S.-S.X., and Z.-H.C. wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Data Availability
The BeadChip SNP genotypes and whole-genome sequences reported in this study are available upon request for research purpose.
References
- Alberto FJ, Boyer F, Orozco-terWengel P, Streeter I, Servin B, De Villemereuil P, Benjelloun B, Librado P, Biscarini F, Colli L, et al. 2018. Convergent genomic signatures of domestication in sheep and goats. Nat Commun. 9:813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allais-Bonnet A, Grohs C, Medugorac I, Krebs S, Djari A, Graf A, Fritz S, Seichter D, Baur A, Russ I, et al. 2013. Novel insights into the bovine polled phenotype and horn ontogenesis in Bovidae. PLoS One 8(5):e63512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML. 2004. Natural hybridization and the evolution of domesticated, pest and disease organisms. Mol Ecol. 13(5):997–1007. [DOI] [PubMed] [Google Scholar]
- Arnold ML, Kunte K.. 2017. Adaptive genetic exchange: a tangled history of admixture and evolutionary innovation. Trends Ecol Evol. 32(8):601–611. [DOI] [PubMed] [Google Scholar]
- Barbato M, Hailer F, Orozco-terWengel P, Kijas J, Mereu P, Cabras P, Mazza R, Pirastru M, Bruford MW.. 2017. Genomic signatures of adaptive introgression from European mouflon into domestic sheep. Sci Rep. 7(1):7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ.. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. [DOI] [PubMed] [Google Scholar]
- Bertolini F, Servin B, Talenti A, Rochat E, Kim ES, Oget C, Palhière I, Crisà A, Catillo G, Steri R, The AdaptMap consortium, et al. 2018. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet Sel Evol. 50(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse M, Megens HJ, Frantz LA, Madsen O, Larson G, Paudel Y, Duijvesteijn N, Harlizius B, Hagemeijer Y, Crooijmans RP, et al. 2014. Genomic analysis reveals selection for Asian genes in European pigs following human-mediated introgression. Nat Commun. 5:4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisbin A, Bryc K, Byrnes J, Zakharia F, Omberg L, Degenhardt J, Reynolds A, Ostrer H, Mezey JG, Bustamante CD.. 2012. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol. 84(4):343–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch T, Foote W.. 1977. Evolution of the 2n = 54 karyotype of Domestic sheep (Ovis aries). Genet Sel Evol. 9(4):509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemineau PP, Malpaux B, Brillard JP, Fostier A.. 2010. Photoperiodic treatments and reproduction in farm animals. Bull Acad Vet Fr. 163(1):19–26. [Google Scholar]
- Chen H, Patterson N, Reich D.. 2010. Population differentiation as a test for selective sweeps. Genome Res. 20(3):393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Cai Y, Chen Q, Li R, Wang K, Huang Y, Hu S, Huang S, Zhang H, Zheng Z, et al. 2018. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat Commun. 9:2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessa B, Pereira F, Arnaud F, Amorim A, Goyache F, Mainland I, Kao RR, Pemberton JM, Beraldi D, Stear MJ, et al. 2009. Revealing the history of sheep domestication using retrovirus integrations. Science 324(5926):532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodkiewicz A. 2020. Advantages and disadvantages of Polish primitive horse grazing on valuable nature areas – a review. Glob Ecol Conserv. 21:e00879. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011; 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGiorgio M, Huber CD, Hubisz MJ, Hellmann I, Nielsen R.. 2016. SweepFinder2: increased sensitivity, robustness and flexibility. Bioinformatics 32(12):1895–1897. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, McVean GA, Donnelly P, Lunter G, Marchini JL, Myers S, Gupta-Hinch A, Iqbal Z, Mathieson I; The 1000 Genomes Project Consortium, et al. 2014. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 5:3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Xie XL, Wang DF, Zhao C, Lv FH, Li X, Yang J, Yu JL, Shen M, Gao L, et al. Forthcoming 2020. Paternal origins and migratory episodes of domestic sheep. Curr Biol. doi: 10.1016/j.cub.2020.07.077. [DOI] [PubMed] [Google Scholar]
- Du ZH. 2018. Screening of differentially expressed genes against Mycoplasma ovipneumoniae [master’s thesis] based on RNA-Seq.
- Felsenstein J. 1993. PHYLIP (phylogeny inference package). Version 3.5c. Seattle (WA): Department of Genetics, University of Washington.
- Flori L, Moazami‐Goudarzi K, Alary V, Araba A, Boujenane I, Boushaba N, Casabianca F, Casu S, Ciampolini R, Coeur D’Acier A, et al. 2019. A genomic map of climate adaptation in Mediterranean cattle breeds. Mol Ecol. 28(5):1009–1029. [DOI] [PubMed] [Google Scholar]
- Foreyt WJ, Jessup DA.. 1982. Fatal pneumonia of bighorn sheep following association with domestic sheep. J Wildl Dis. 18(2):163–168. [DOI] [PubMed] [Google Scholar]
- Frantz LAF, Bradley DG, Larson G, Orlando L.. 2020. Animal domestication in the era of ancient genomics. Nat Rev Genet. 21(8):449–460. [DOI] [PubMed] [Google Scholar]
- Frichot E, Schoville SD, Bouchard G, Francois O.. 2013. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol Biol Evol. 30(7):1687–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Smyth RL, Young BE, Brooks TM, de Lozada AS, Bubb P, Butchart SHM, Larsen FW, Hamilton H, Hansen MC, et al. 2014. A biodiversity indicators dashboard: addressing challenges to monitoring progress towards the Aichi biodiversity targets using disaggregated global data. PLoS One 9(11):e112046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin M, Zielinski J, Wan ES, Hersh CP, Castaldi PJ, Schwinder E, Hawrylkiewicz I, Sliwinski P, Cho MH, Silverman EK.. 2012. CHRNA3/5, IREB2, and ADCY2 are associated with severe chronic obstructive pulmonary disease in Poland. Am J Respir Cell Mol Biol. 47(2):203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpur BA, Minaei S, Kent CF, Zayed A.. 2012. Management increases genetic diversity of honey bees via admixture. Mol Ecol. 21(18):4414–4421. [DOI] [PubMed] [Google Scholar]
- Harrison RG, Larson EL.. 2014. Hybridization, introgression, and the nature of species boundaries. J Hered. 105(S1):795–809. [DOI] [PubMed] [Google Scholar]
- Hobbs RJ, Howard J, Wildt DE, Comizzoli P.. 2012. Absence of seasonal changes in FSHR gene expression in the cat cumulusoocyte complex in vivo and in vitro. Reproduction 144(1):111–122. [DOI] [PubMed] [Google Scholar]
- Horwitz LK, Bar-Gal GK.. 2006. The origin and genetic status of insular caprines in the Eastern Mediterranean: a case study of free-ranging goats (Capra aegagrus cretica) on Crete. Hum Evol. 21(2):123–138. [Google Scholar]
- Hu XJ, Yang J, Xie XL, Lv FH, Cao YH, Li WR, Liu MJ, Wang YT, Li JQ, Liu YG, et al. 2019. The genome landscape of Tibetan sheep reveals adaptive introgression from argali and the history of early human settlements on the Qinghai-Tibetan Plateau. Mol Biol Evol. 36(2):283–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- Huerta-Sánchez E, Jin X, Bianba Z, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, Ni P, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBM Corp. 2016. IBM SPSS Statistics for Windows. Version 24.0. Armonk (NY): IBM Corp.
- Jakobsson M, Rosenberg NA.. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14):1801–1806. [DOI] [PubMed] [Google Scholar]
- Johnston SE, McEwan JC, Pickering NK, Kijas JW, Beraldi D, Pilkington JG, Pemberton JM, Slate J.. 2011. Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol Ecol. 20(12):2555–2566. [DOI] [PubMed] [Google Scholar]
- Jordana J, Manteca X, Ribo O.. 1999. Comparative analysis of morphological and behavioral characters in the domestic dog and their importance in the reconstruction of phylogenetic relationships in canids. Genet Mol Biol. 22(1):49–57. [Google Scholar]
- Kijas JW, Lenstra JA, Hayes B, Boitard S, Neto LRP, San Cristobal M, Servin B, McCulloch R, Whan V, Gietzen K, other members of the International Sheep Genomics Consortium, et al. 2012. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10(2):e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilsgård O, Andersson P, Malmsten M, Nordin SL, Linge HM, Eliasson M, Sörenson E, Erjefält JS, Bylund J, Olin AI, et al. 2012. Peptidylarginine deiminases present in the airways during tobacco smoking and inflammation can citrullinate the host defense peptide LL-37, resulting in altered activities. Am J Respir Cell Mol Biol. 46(2):240–248. [DOI] [PubMed] [Google Scholar]
- Kristensen TN, Ketola T, Kronholm I.. 2018. Adaptation to environmental stress at different timescales. Ann NY Acad Sci. Special Issue: 1–18. [DOI] [PubMed] [Google Scholar]
- Kvist L, Niskanen M, Mannermaa K, Wutke S, Aspi J.. 2019. Genetic variability and history of a native Finnish horse breed. Genet Sel Evol. 51(1):35–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Burger J.. 2013. A population genetics view of animal domestication. Trends Genet. 29(4):197–205. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y.. 2010. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 207(9):1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang J, Shen M, Xie XL, Liu GJ, Xu YX, Lv FH, Yang H, Yang YL, Liu CB, et al. 2020. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat Commun. 11:2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Herraez D, Bauchet M, Tang K, Theunert C, Pugach I, Li J, Nandineni MR, Gross A, Scholz M, Stoneking M.. 2009. Genetic variation and recent positive selection in worldwide human populations: evidence from nearly 1 million SNPs. PLoS One 4(11):e7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery RK, Uribe G, Jimenez EB, Weiss MA, Herrera KJ, Regueiro M, Herrera RJ.. 2013. Neanderthal and Denisova genetic affinities with contemporary humans: introgression versus common ancestral polymorphisms. Gene 530(1):83–94. [DOI] [PubMed] [Google Scholar]
- Lv FH, Agha S, Kantanen J, Colli L, Stucki S, Kijas JW, Joost S, Li MH, Ajmone Marsan P.. 2014. Adaptations to climate-mediated selective pressures in sheep. Mol Biol Evol. 31(12):3324–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD.. 2015. Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol Biol Evol. 32(1):244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N, Tanda K, Nakanishi K, Yamasaki N, Toyama K, Takao K, Takeshima H, Miyakawa T.. 2009. Comprehensive behavioral phenotyping of ryanodine receptor type 3 (RyR3) knockout mice: decreased social contact duration in two social interaction tests. Front Behav Neurosci. 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medugorac I, Graf A, Grohs C, Rothammer S, Zagdsuren Y, Gladyr E, Zinovieva N, Barbieri J, Seichter D, Russ I, et al. 2017. Whole-genome analysis of introgressive hybridization and characterization of the bovine legacy of Mongolian yaks. Nat Genet. 49(3):470–475. [DOI] [PubMed] [Google Scholar]
- Miller JM, Kijas JW, Heaton MP, McEwan JC, Coltman DW.. 2012. Consistent divergence times and allele sharing measured from cross-species application of SNP chips developed for three domestic species. Mol Ecol Resour. 12(6):1145–1150. [DOI] [PubMed] [Google Scholar]
- Mintoo AA, Zhang H, Chen C, Moniruzzaman M, Deng T, Anam M, Emdadul Huque QM, Guang X, Wang P, Zhong Z, et al. 2019. Draft genome of the river water buffalo. Ecol Evol. 9(6):3378–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitre M, Mariga A, Chao MV.. 2017. Neurotrophin signalling: novel insights into mechanisms and pathophysiology. Clin Sci. 131(1):13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morenikeji OB, Ajayi OO, Peters SO, Mujibi FD, De Donato M, Thomas BN, Imumorin IG.. 2020. RNA-seq profiling of skin in temperate and tropical cattle. J Anim Sci Technol. 62(2):141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New M, Lister D, Hulme M, Makin I.. 2002. A high-resolution data set of surface climate over global land areas. Clim Res. 21:1–25. [Google Scholar]
- Noddle BA, Ryder ML.. 1974. Primitive sheep in the Aran Islands. J Archaeol Sci. 1(1):109–112. [Google Scholar]
- Olivieri G, Miescher GC.. 1999. Immunohistochemical localization of EphA5 in the adult human central nervous system. J Histochem Cytochem. 47(7):855–861. [DOI] [PubMed] [Google Scholar]
- Osei-Amponsah R, Chauhan SS, Leury BJ, Cheng L, Cullen B, Clarke IJ, Dunshea FR.. 2019. Genetic selection for thermotolerance in ruminants. Animals (Basel) 9(11):948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 192(3):1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D.. 2006. Population structure and eigenanalysis. PLoS Genet. 2(12):e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2014. FigTree, ver. 1.4. 3. Edinburgh: Program distributed by the author. Available from: http://tree.bio.ed.ac.uk/software/figtree. Accessed September 24, 2020.
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L.. 2009. Reconstructing Indian population history. Nature 461(7263):489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC.. 1983. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105(3):767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei HR, Naderi S, Chintauan-Marquier IC, Taberlet P, Virk AT, Naghash HR, Rioux D, Kaboli M, Pompanon F.. 2010. Evolution and taxonomy of the wild species of the genus Ovis (Mammalia, Artiodactyla, Bovidae). Mol Phylogenet Evol. 54(2):315–326. [DOI] [PubMed] [Google Scholar]
- Rosa HJD, Bryant MJ.. 2003. Seasonality of reproduction in sheep. Small Rumin Res. 48(3):155–171. [Google Scholar]
- Rosenberg NA. 2003. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 4(1):137–138. [Google Scholar]
- Ryder ML. 1981. A survey of European primitive breeds of sheep. Genet Sel Evol. 13(4):381–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder ML. 1984. Evolution of domesticated animals. New York: Longman. [Google Scholar]
- Scheet P, Stephens M.. 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 78(4):629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf BD. 2000. World watch list for domestic animal diversity (No. Ed. 3). Rome: Food and Agriculture Organization of the United Nations.
- Schröder O, Lieckfeldt D, Lutz W, Rudloff C, Frölich K, Ludwig A.. 2016. Limited hybridization between domestic sheep and the European mouflon in Western Germany. Eur J Wildl Res. 62(3):307–314. [Google Scholar]
- Setter D, Mousset S, Cheng X, Nielsen R, DeGiorgio M, Hermisson J.. 2020. VolcanoFinder: genomic scans for adaptive introgression. PLoS Genet. 16(6):e1008867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiner MC, Buitenhuis H, Duru G, Kuhn SL, Mentzer SM, Munro ND, Pollath N, Quade J, Tsartsidou G, Ozbasaran M.. 2014. A forager-herder trade-off, from broad-spectrum hunting to sheep management at Aşıklı Höyük, Turkey. Proc Natl Acad Sci U S A. 111(23):8404–8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki S, Orozco-terWengel P, Forester BR, Duruz S, Colli L, Masembe C, Negrini R, Landguth E, Jones MRNEXTGEN ConsortiumBruford MW, et al. 2017. High performance computation of landscape genomic models including local indicators of spatial association. Mol Ecol Resour. 17(5):1072–1089. [DOI] [PubMed] [Google Scholar]
- Sulpice E, Plouet J, Berge M, Allanic D, Tobelem G, Merkulova-Rainon T.. 2008. Neuropilin-1 and neuropilin-2 act as coreceptors, potentiating proangiogenic activity. Blood 111(4):2036–2045. [DOI] [PubMed] [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA.. 2008. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24(21):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbo M, Woldemeskel M, Gopilo A.. 2001. An outbreak of respiratory disease complex in sheep in central Ethiopia. Trop Anim Health Prod. 33(5):355–365. [DOI] [PubMed] [Google Scholar]
- Upadhyay MR, Chen W, Lenstra JA, Goderie CR, MacHugh DE, Park SD, Magee DA, Matassino D, Ciani F, Megens HJ, et al. ; European Cattle Genetic Diversity Consortium. 2017. Genetic origin, admixture and population history of aurochs (Bos primigenius) and primitive European cattle. Heredity 118(2):169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle Zárate A, Musavaya K, Schäfer C.. 2006. Gene flow in animal genetic resources: a study on status, impact and trends. Stuttgart: commissioned by Federal Ministry for Economic Cooperation and Development. . [Google Scholar]
- Woronzow NN, Korobizgna KW, Nadler CF, Hofman R, Esalojnitskow TN, Gorelow JK.. 1972. Chromossomi dikich baranow i proisschojdjenije domaschnich owjez. Lriroda 3:74–81. [Google Scholar]
- Wu DD, Ding XD, Wang S, Wójcik JM, Zhang Y, Tokarska M, Li Y, Wang MS, Faruque O, Nielsen R, et al. 2018. Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat Ecol Evol. 2(7):1139–1145. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang Y, Zhao Y, Zhang X, Li R, Chen L, Zhang G, Jiang Y, Qiu Q, Wang W, et al. 2017. Draft genome of the Marco Polo Sheep (Ovis ammon polii). GigaScience 6(12):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder MA. 2008. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proc Natl Acad Sci U S A. 105(33):11597–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder MA. 2015. Core questions in domestication research. Proc Natl Acad Sci U S A. 112(11):3191–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YX, Yang J, Lv FH, Hu XJ, Xie XL, Zhang M, Li WR, Liu MJ, Wang YT, Li JQ, et al. 2017. Genomic reconstruction of the history of native sheep reveals the peopling patterns of nomads and the expansion of early pastoralism in East Asia. Mol Biol Evol. 34(9):2380–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The BeadChip SNP genotypes and whole-genome sequences reported in this study are available upon request for research purpose.