Abstract
Visible pigmentation phenotypes can be used to explore the regulation of gene expression and the evolution of coat color patterns in animals. Here, we performed whole-genome and RNA sequencing and applied genome-wide association study, comparative population genomics and biological experiments to show that the 2,809-bp-long LINE-1 insertion in the ASIP (agouti signaling protein) gene is the causative mutation for the white coat phenotype in swamp buffalo (Bubalus bubalis). This LINE-1 insertion (3′ truncated and containing only 5′ UTR) functions as a strong proximal promoter that leads to a 10-fold increase in the transcription of ASIP in white buffalo skin. The 165 bp of 5′ UTR transcribed from the LINE-1 is spliced into the first coding exon of ASIP, resulting in a chimeric transcript. The increased expression of ASIP prevents melanocyte maturation, leading to the absence of pigment in white buffalo skin and hairs. Phylogenetic analyses indicate that the white buffalo-specific ASIP allele originated from a recent genetic transposition event in swamp buffalo. Interestingly, as a similar LINE-1 insertion has been identified in the cattle ASIP gene, we discuss the convergent mechanism of coat color evolution in the Bovini tribe.
Keywords: white coat color, water buffalo, ASIP gene, LINE-1, transposon, convergent evolution
Introduction
Animal pigmentation is one of the most visible and variable traits shaped by natural and/or artificial selection. As a visible phenotypic marker, pigmentation has played an important role in our understanding of inheritance, development, and evolutionary theory (Hoekstra 2006; Mort et al. 2015; Cuthill et al. 2017). In mammals, basic coat coloration is determined by the ratio of two pigments—eumelanin and pheomelanin. There are almost 200 color genes identified in mice (Mort et al. 2015). These genes act during developmental and cellular processes, including melanocyte development, melanogenesis, pigment transport, and transfer (Cieslak et al. 2011). Coat color, an important form of camouflage in the wild ancestors of domestic animals, is likely under strong purifying selection. The relaxation of natural selective constraints and human mediated positive selection for different coat color phenotypes in domestic animals are believed to be the primary driving mechanisms leading to their significantly enriched allelic variation in coat-color-associated genes (Norris and Whan 2008; Fang et al. 2009; Cieslak et al. 2011; Henkel et al. 2019; Bruders et al. 2020). Domestic animal genetic resources provide an excellent opportunity to study the causative mutations and regulatory mechanisms responsible for coat color diversity (Henkel et al. 2019; Bruders et al. 2020).
The domestic Asian water buffalo (Bubalus bubalis) is an important animal resource, with a current global population of approximately 202 million supplying draught power, milk and/or meat in at least 67 countries on 5 continents (http://www.fao.org/faostat/). Two types have been recognized—river and swamp (Zhang et al. 2020). River buffaloes are native to the Indian subcontinent and have spread west as far as to the Balkans, Greece, Egypt, and Italy within recorded historical times, whereas swamp buffaloes are found throughout south-east Asia, from Assam in India and Bangladesh in the west to the Yangtze valley of China in the east. Swamp buffalo is usually dark gray to black with white chevrons (one or two white stripes on the throat) and socks, and relatively straight, long, and pale-colored horns. White swamp buffaloes (a common variant) have white hairs over the entire body, overlying pink skin, but their eyes are dark, the same as those of black buffaloes (supplementary fig. S1, Supplementary Material online). In some Asian countries, such as China and Indonesia, white buffaloes are particularly valued because the white coat phenotype is preferred for cultural (ceremonial slaughter at funerals) or religious reasons. Although a dominant gene was shown to determine the white coat phenotype nearly 60 years ago (Rife and Buranamanas 1959; Rife 1962), its molecular basis remains unknown. Nevertheless, two independent loss-of-function mutations in microphthalmia-associated transcription factor gene (MITF) are responsible for white-spotted coat color, a well-known phenotype in swamp buffalo from Indonesia (Yusnizar et al. 2015) while a premature stop codon mutation in tyrosinase gene (TYR) for oculocutaneous albinism in river buffalo (Damé et al. 2012).
In this study, we performed whole-genome and RNA sequencing using both next-generation sequencing and long-read sequencing strategies, and applied genome-wide association study (GWAS), population genomics and biological experiments to explore the genetic mechanism underlying the white coat phenotype of swamp buffaloes. We demonstrated that a transposable element (TE) insertion, functioning as the active promoter of the ASIP (agouti signaling protein) gene, is the causal mutation for the white coat phenotype.
Results
Mapping of the White Coat Phenotype in Swamp Buffaloes
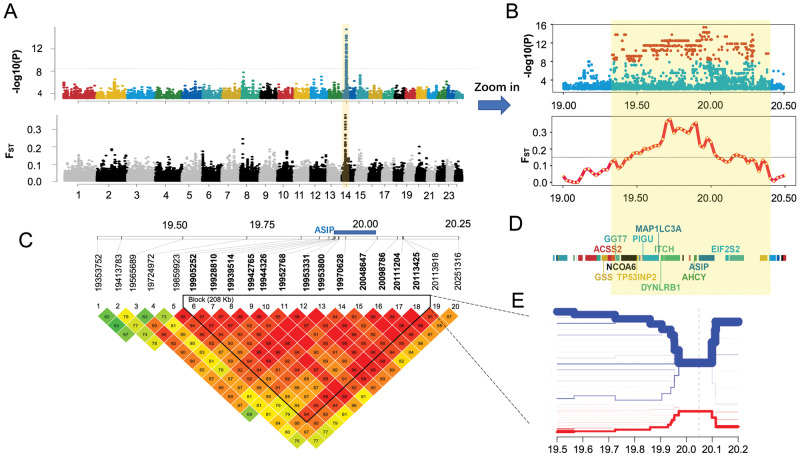
Whole-genome sequencing (WGS) was conducted for 22 white and 41 black swamp buffaloes that were randomly sampled from five populations (supplementary table S1A, Supplementary Material online). In total, 2,003 Gb of sequence data were generated, resulting in averages over the 63 animals for depth (11.95×) and coverage (98.27%) of the river buffalo reference genome (UOA_WB_1, GCF_003121395.1; Low et al. 2019) (supplementary table S2, Supplementary Material online). After quality control, 10,999,832 SNPs remained for GWAS analysis of the white coat phenotype. We applied Fisher’s exact test with the dominant gene effect model in the PLINK software v1.07 (Purcell et al. 2007) and identified the most significant peak with the highest level of significance located on buffalo chromosome (BBU) 14 associated with the white coat phenotype (fig. 1A). This region contained 407 genome-wide statistically significant SNPs (Fisher’s exact test, Bonferroni-corrected P-value <0.05, −log10P = 8.34; fig. 1B;supplementary table S3, Supplementary Material online), spanning 1.07 Mb (BBU14:19332562 − 20392733). It harbors five pseudogenes and 23 genes (16 protein-coding genes and 7 RNA genes), including the well-known color gene ASIP (fig. 1E;supplementary table S4, Supplementary Material online).
Fig. 1.
Whole-genome sequencing identifies that a 1.07-Mb-long region including ASIP on buffalo chromosome 14 (BBU14) is associated with the white coat phenotype. (A) Manhattan plot showing a single genomic region on BBU14 significantly associated with the white coat phenotype. (B) Zoomed association signals of GWAS and genetic differentiation (FST) in the significant region. (C) Validation of the strong association of the 1.07-Mb-long region including ASIP with the white coat phenotype, by genotyping 20 variants in an independent sample and LD analysis. The location of ASIP is indicated by a blue bar. (D) Genes in the significant region. (E) Bifurcation diagram showing the distinct long-range haplotypes in white (red) and black (blue) buffaloes. Moving from the core SNP (vertical dot line) in two directions, the diagram divides if two alleles are present at the SNP. The x-axis is the chromosome location. The thickness of the lines represents the counts of long-range haplotype in the sample.
Meanwhile, population genomic analyses were done to detect any signatures of selection underlying the white coat phenotype. A genomic scan for population genetic differentiation measured by FST was conducted using a sliding window of 50 kb length with an increment of 25 kb. The results showed that the top 20 windows of FST values were mainly located in a region on BBU14 (BBU14:19575001 − 20000000), overlapping with the GWAS signals (fig. 1A and B; supplementary table S5, Supplementary Material online). This finding suggested that this region carried a signature of positive selection in white buffaloes. However, the measure of polymorphism (nucleotide diversity [Pi]), showed similar patterns in the GWAS signal region in white and black buffaloes (data not shown), which could largely be explained by the dominant inheritance of white coat color, so that the majority of the white buffaloes were heterozygotes.
Validation of the GWAS Signals Using Independent Samples
The above analyses provided the first line of evidence that the peak on BBU14 likely represented a candidate locus responsible for white coat color in swamp buffaloes. To validate the GWAS signals, we performed an association analysis using a panel of 20 significant variants, but based on a larger collection of samples, including 80 white and 122 black buffaloes that were sampled from Thailand, Bangladesh, and China (supplementary tables S1A and S6, Supplementary Material online). These variants were generally distributed evenly in the target genomic region with relatively more variants falling in the location of ASIP, the strong candidate gene. As expected, the larger sample size resulted in more significant associations with −log10P-values of 18.7–50.2 (Fisher’s exact test) (supplementary tables S7 and S8, Supplementary Material online). In particular, four adjacent SNPs located in the first intron and upstream of ASIP (BBU14:19970628, BBU14:20048647, BBU14:20098786, and BBU14:20111204) showed the strongest signals (supplementary table S8, Supplementary Material online). In addition, linkage disequilibrium (LD) analysis indicated that 13 SNPs in this region were tightly linked, forming an LD block that harbored ASIP (fig. 1C). As shown in a haplotype bifurcation diagram (see Materials and Methods), the white buffalo-specific core haplotype in this region showed long-range homozygosity, which was distinct from that of black buffaloes (fig. 1D). Taken together, these evidences suggested that the genomic region harboring ASIP was strongly associated with the white coat phenotype.
Given its critical role in mammalian pigmentation (Cieslak et al. 2011), we considered ASIP as a candidate gene and explored for any functional mutations within this gene that may be responsible for white coat phenotype. A total of 51 SNPs in ASIP were significantly associated with white coat phenotype (supplementary tables S3 and S9, Supplementary Material online) in the GWAS analysis. However, none was predicted to have any functional effects on the agouti protein (supplementary table S9, Supplementary Material online). Furthermore, as a validation, we used Sanger sequencing to detect the variants of ASIP in three DNA pools (one for white buffaloes and two for black buffaloes; see Materials and Methods). We identified 11 SNPs, of which nine were shared with the WGS results whereas the other two showed no association with the white coat phenotype because they occurred in only one of the two black buffalo DNA pools (supplementary table S10, Supplementary Material online). The translation of ASIP was validated to ensure that there was no missense mutation that altered the agouti protein structure and led to the white coat.
We then sought to explore if large structural variants were associated with coat color in the region of the GWAS signals. Three software tools, mrFAST v2.6.1.0 (Alkan et al. 2009), CNVnator v0.3.3 (Abyzov et al. 2011), and BreakDancer v1.1.2 (Chen et al. 2009), were used (see Materials and Methods), but based on our short-read WGS data, no structural variant private to white buffaloes was detected (supplementary tables S11−S14, Supplementary Material online).
Upregulated Expression of ASIP in White Buffalo Skin
To track down the potential causative gene(s), we compared transcription profiles of the 23 genes annotated in the significant GWAS region. Skin biopsies of six animals (three each of white and black swamp buffaloes) were used for whole transcriptome sequencing (RNA-seq) (supplementary table S15, Supplementary Material online). Based on the RNA-seq data, ASIP showed a 10.3-fold increase in transcription in the skin of white buffaloes (transcripts per million, TPM: 28.04 ± 6.34) as compared with black buffaloes (TPM: 2.96 ± 0.73) (fig. 2A;supplementary table S16, Supplementary Material online). This striking difference was further verified by real-time quantitative PCR (relative expression: 1.25 ± 0.75 for black buffaloes vs 12.86 ± 4.88 for white buffaloes; Student’s t-test P < 0.001; fig. 2A;supplementary tables S17−S18, Supplementary Material online). Furthermore, we characterized the tissue-specific expression profile using our newly generated RNA-seq data and published data from 55 tissue and cell types of river buffaloes (supplementary table S19, Supplementary Material online). Interestingly, among the genes in the significant GWAS region, only ASIP showed tissue-specific expression in skin tissue (supplementary fig. S2, Supplementary Material online). These findings showed that the white coat phenotype in swamp buffaloes might be the result of a cis-regulatory variant that elevated ASIP expression in the skin.
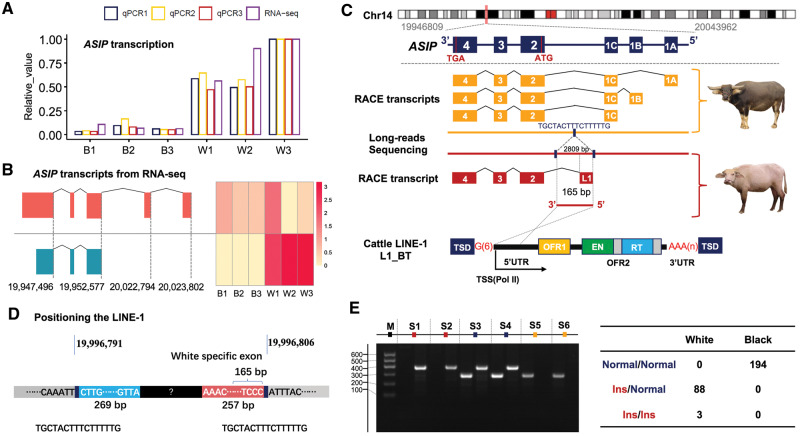
Fig. 2.
Identification of the LINE-1 insertion in ASIP of white buffaloes. (A) Significantly higher expression of ASIP in white buffalo skin (W1, W2, and W3) than in black buffalo skin (B1, B2, and B3) revealed by RNA-seq and qPCR. Three experimental replicates of qPCR are shown separately (qPCR1, qPCR2, and qPCR3). (B) Distinct transcripts assembled from RNA-seq data of skin samples of white (blue) and black (red) buffaloes. (C) Full-length transcripts of ASIP generated using the RACE-PCR and characterization of the LINE-1 insertion in the white buffalo ASIP. The structure of a full-length LINE-1 element from cattle (L1-BT; Girardot et al. 2006) is shown as reference. (D) Schematic representation showing the chromosomal position of the LINE-1 insertion determined by soft-clipped reads analysis and the partial sequences of the LINE-1 insertion obtained by de novode novo assembling of the soft-clipped reads. (E) Genotyping the LINE-1 insertion using the allele-specific PCR and its perfect association with white coat phenotype. PCR products of wild allele (Normal; 296 bp) and mutant allele (Ins; 387 bp) are separated for six samples (S1−S6) using agarose gel electrophoresis.
Identification of the White Buffalo-Specific ASIP Transcript
To address whether ASIP transcripts were different between white and black buffaloes, we initially visualized RNA-seq reads in IGV (Integrative Genomics Viewer; Robinson et al. 2017). This revealed distinct patterns of overlapping reads that were mapped to ASIP exons (supplementary fig. S3, Supplementary Material online). In black buffaloes, as expected, reads were aligned to both noncoding exons and coding exons with similar read counts (depth) across all three coding exons. In white buffaloes, however, reads were mainly aligned to the coding exons. In particular, read counts on exon 2 (the first coding exon) decreased gradually from the 5′ end to the 3′ end, implicating a distinct transcript. Transcript assembly and quantification based on the RNA-seq data using the Stringtie software v2.0 (Pertea et al. 2015) showed two abundantly expressed transcripts, one in black buffaloes and another in white buffaloes (fig. 2B).
To isolate full-length transcripts and characterize their transcription initiation sites, 5′ and 3′ Rapid Amplification of cDNA Ends (RACE) PCR experiments for skin samples of one white buffalo and one black buffalo were done (supplementary table S17, Supplementary Material online). The RACE-PCR products were subject to conventional cloning, followed by Sanger sequencing. Sequences were determined for multiple clones of white (16 clones in 3′ end and 17 in 5′ end) and black (14 in 3′ end and 14 in 5′ end) buffaloes. Comparison of clones from black buffalo showed six alternative transcripts that shared the same coding exons and 3′ UTR but differed in 5′ UTR (fig. 2C;supplementary fig. S4, Supplementary Material online). White buffalo, however, had only one transcript. Interestingly, whereas sharing the same coding exons and 3′ UTR with black buffalo, the white buffalo-specific transcript contained an unknown 165-bp sequence at 5′ UTR that could not be aligned to the buffalo reference genome (fig. 2C;supplementary fig. S4, Supplementary Material online).
To further characterize this white buffalo-specific transcript, a BlastN search assigned this unknown 165-bp fragment to a bovine LINE-1 transposon element (L1-BT, GenBank accession number DQ000238) with a sequence identity of 98%. This suggested that the presence of a LINE-1 insertion upstream of ASIP led to a chimeric transcript in white buffalo.
Genomic Position of the White Buffalo-Specific LINE-1 Insertion in ASIP
To position the LINE-1 insertion, we analyzed the soft-clipped reads that mapped upstream of ASIP (BBU14:19952567 − 20083962) in our WGS data. In contrast to aligned reads, soft-clipped reads were partially mapped to the buffalo reference genome and contained unmapped sequences, suggestive of structural variants. To improve the efficiency of comparative analysis, we pooled data of 10 randomly selected samples from each of the two coat color phenotypes, and compared the counts of soft-clipped reads on each genomic position between white and black buffaloes (supplementary file 1, Supplementary Material online). The position at BBU14:19996806 showed the top signal (the difference of 38 in the counts of soft-clipped reads) that was further verified in the IGV (supplementary fig. S5, Supplementary Material online). The soft-clipped reads mapped to this position were divided into two categories: left soft-clipped reads (truncated at BBU14:19996806) and right soft-clipped reads (truncated at BBU14:19996791). The 16-bp fragment (TGCTACTTTCTTTTTG) between these two reads showed much higher read depth than its flanking regions in white buffaloes (supplementary fig. S5, Supplementary Material online), indicating the presence of an insertion variant. Then, we de novo assembled all the soft-clipped reads containing the 16-bp fragment, yielding two contigs: one of 269 bp on the left connecting to the upstream flanking sequence at position BBU14:19996791 and another of 257 bp on the right joining the downstream flanking sequence at position BBU14:19996806 (fig. 2D;supplementary fig. S5, Supplementary Material online). The 165-bp 5′ UTR of the white buffalo-specific transcript was perfectly aligned to the contig on the right. Thus, we positioned the white buffalo ASIP LINE-1 insertion at BBU14:19996791 − 19996806 and obtained its head- and tail-end DNA sequences. This LINE-1 insertion was 44.2 kb away from the first coding exon (exon 2, located at BBU14:19952408 − 19952577) of ASIP.
Sequence of the Complete LINE-1 Insertion in White Buffalo ASIP
To determine the complete sequence of the LINE-1 insertion, we sequenced one white buffalo and one black buffalo using Nanopore long-read sequencing technology. We generated 80.64 Gb of filtered data, including 7,833,594 and 9,759,939 reads with mean lengths of 5,103 and 4,166 bp in white buffalo and black buffalo, respectively. The reads aligned to the genomic region of ASIP (BBU14:19946809 − 20113374) were extracted for de novo assembling. Using the Canu assembler v1.8 (Koren et al. 2017), 33 contigs were assembled for the white buffalo with a mean length of 12,024 bp, of which the longest contig was 67,524 bp (supplementary file 2, Supplementary Material online). By aligning the two partial LINE-1 fragments (269 and 257 bp) assembled from short-reads to this longest contig, we resolved the complete structure of the LINE-1 insertion. It was 2,809 bp in length flanked by the 16-bp direct repeat (TGCTACTTTCTTTTTG) that was characterized as the target site duplication (TSD) of the LINE-1 element (fig. 2C). In black buffalo, however, there was only one copy of this 16-bp sequence and the LINE-1 fragment was not detected.
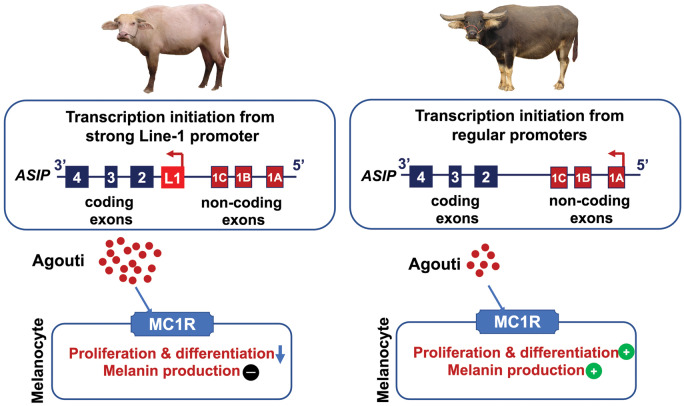
This LINE-1 was 3′ truncated and contained only 5′ UTR of a full-length LINE element. It was located upstream of the first coding exon and in the same orientation as that of the ASIP transcription. Therefore, the promoter of LINE-1 could act as a strong alternative promoter to drive ASIP expression in white buffaloes. The 165 bp of 5′ UTR transcribed from the LINE-1 was spliced into the first coding exon, creating the chimeric ASIP transcript in white buffaloes (fig. 2C).
To validate the association of the presence of this LINE-1 element with the white coat phenotype, we developed a genotyping assay and examined 91 white and 194 black buffaloes (fig. 2E;supplementary table S1B, Supplementary Material online). The result showed that the LINE-1 was perfectly associated with the white coat phenotype. All black buffaloes were wild-type homozygotes. White buffaloes were either heterozygous or homozygous for the LINE-1 insertion, confirming that the white coat phenotype is inherited as a dominant Mendelian trait.
Effect of Increased ASIP Expression on Melanocyte Development
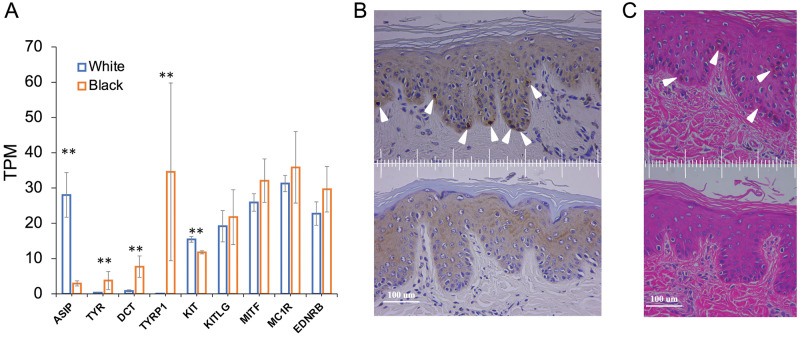
To explore the regulatory mechanism of the white coat phenotype, we investigated the gene expression profiles based on skin RNA-seq data generated in the current study. Transcriptome analysis revealed a total of 344 differentially expressed genes (DEGs), of which 148 DEGs were downregulated whereas 196 DEGs were upregulated in white buffalo skin (false discovery rate [FDR] <0.01 and fold change ≥2 as the thresholds). A functional annotation showed that the downregulated DEGS were enriched in melanocyte biology-related Gene Ontology (GO) terms (e.g., melanin metabolic process, melanin biosynthetic process) and KEGG pathways (e.g., tyrosine metabolism, melanogenesis), indicating that the melanocyte function might be diminished in white buffalo skin (supplementary table S20, Supplementary Material online). We found that 5 out of the 11 skin-color-associated genes (TYR, DCT/TYRP2, TYRP1, PMEL, and OCA2) showed significantly lower or no expression (P < 0.01) in white buffaloes, five (KITLG, MITF, MC1R, EDNRB, and SOX10) displayed no difference (P > 0.05) between white and black buffaloes whereas only KIT had a slightly higher expression (P < 0.05) in white buffalo skin (fig. 3A;supplementary table S16, Supplementary Material online). We then focused on the tyrosinase-related family genes TYRP2 and TYRP1 that are consecutively expressed in melanocytes during their migration in the dermis and maturation, as markers of early and late differentiation, respectively (Steel et al. 1992; Botchkareva et al. 2003; Manceau et al. 2011). Although TYRP2 was expressed in both white and black buffaloes, TYRP1 was expressed only in black buffaloes (fig. 3A;supplementary table S16, Supplementary Material online), indicating that the melanocyte was fully differentiated in black buffaloes but not in white buffaloes. This was further supported by the immunohistochemical staining of skin samples. The fully differentiated (Trp1+) melanocytes were observed at the dermal–epidermal junction in black buffaloes, whereas no Trp1+ signal was present in white buffaloes (fig. 3B). Melanin pigment was present near the melanocytes in black buffaloes but not in white buffaloes (fig. 3C). Collectively, these results indicated that the overexpression of ASIP prevented melanocyte maturation, leading to the absence of pigment in white buffalo skin and hairs (fig. 4).
Fig. 3.
Functional consequences of ASIP over-expression in white buffalo skin. (A) Transcription levels of ASIP, TRY, DCT (TYRP2), TYRP1, KIT, KITLG, MITF, MC1R, and EDNRB. Error bars indicate standard deviation (SD). “**”Significance level (P < 0.01) of a Wald test using the DESeq2 package. (B) Immunohistochemical analysis of skin samples of black (top) and white (bottom) buffaloes. Arrowheads indicate melanocytes with tyrosinase-related protein 1 (TYRP1) expression (Trp+). (C) Hematoxylin−Eosin (HE) staining of skin samples of black (top) and white (bottom) buffaloes. Arrowheads indicate melanin pigment near the melanocytes.
Fig. 4.
Model of molecular mechanism for swamp buffalo white coat phenotype. The gene is represented 3′−5′ from left to right to be consistent with that shown in figure 2.
Interestingly, the upregulated DEGs in white buffalo skin were also overrepresented in the growth-related GO terms, such as the development of skeletal system, tissue, organ, and connective tissue (supplementary table S20, Supplementary Material online), implicating possible pleiotropic effect of ASIP overexpression on physiology and metabolism in white buffaloes.
Origin of the LINE-1 Insertion in White Buffalo ASIP
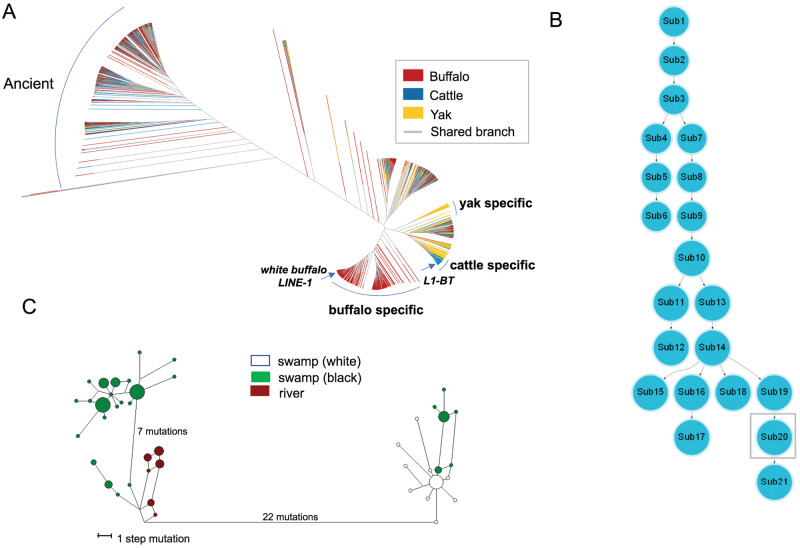
Mammalian genomes host hundreds of thousands of LINE-1 elements that have accumulated since the origin of mammals (Boissinot and Sookdeo 2016). Although the great majority of LINE-1s are inactive, some retain the ability to retrotranspose (Sassaman et al. 1997; Richardson et al. 2015). To investigate if the LINE-1 insertion in white buffalo ASIP occurred recently in the buffalo species or was shared with the other related bovine species, we explored its evolutionary origin based on the sequence similarity at two levels—among and within species. First, full-length LINE-1 elements were identified from the reference genomes of water buffalo and two related species (taurine cattle and yak), using the RepeatMasker software v4.07 (http://www.repeatmasker.org) and compared with the bovine L1-BT transposon element (GenBank accession number DQ000238) as the reference library. A total of 6,986 full-length LINE-1 copies, including 2,516 from river buffalo, 1,571 from swamp buffalo, 1,617 from taurine cattle, and 1,282 from yak genomes (supplementary tables S21−S24, Supplementary Material online), were obtained and used to construct an approximately maximum-likelihood tree. A primary analysis showed that the LINE-1 copies were closely related between swamp and river buffaloes (supplementary fig. S6, Supplementary Material online). To improve the visualization of the phylogeny of LINE-1s from different species, we used the same color for swamp and river buffaloes in the tree (fig. 5A). This tree held two major clades: 1) One on the left consisted of water buffalo LINE-1 copies mixed with those of taurine cattle and yak. They all had comparably long branch lengths, representing ancient and inactive LINE-1 copies; and 2) On the right another clade was divided into several subclades of those with mixed LINE-1 copies of all three species, but having relatively short branch lengths as well as three species-specific subclades. These species-specific LINE-1 copies displayed the shortest branch lengths and tended to cluster tightly to each other within species, suggesting their recent evolutionary origins. The LINE-1 copy in white buffalo ASIP clustered with the water buffalo-specific subclade, indicating it to be a young copy derived from the water buffalo-specific LINE-1 copies.
Fig. 5.
Evolutionary analyses of LINE-1 elements in the genome and ASIP haplotypes. (A) An approximate maximum-likelihood tree of 6,986 full-length LINE-1 elements in buffalo, cattle, and yak reference genomes. The buffalo includes both swamp and river buffaloes. The clade on the left are LINE-1 copies that are mixed among species and have long branch lengths, indicating that they are ancient and amplified before the split of these species. Species-specific clades are found on the right, in which the buffalo-specific clade contains both swamp and river buffaloes. One arrow points to the LINE-1 clustering with that of white buffalo ASIP and another to the LINE-1 of cattle ASIP (L1-BT). The shared branch is shown in gray color. (B) A minimum spanning tree showing the evolutionary relationship among LINE-1 subfamilies in the water buffalo genomes. The ASIP LINE-1 insertion of white buffaloes belongs to the sub20 subfamily (in the gray square), a relatively young subfamily. (C) Median-joining network of water buffalo ASIP haplotypes. Colors: Green, swamp buffalo; brown, river buffalo; and white, white swamp buffalo.
Next, we characterized water buffalo-specific LINE-1 copies and the evolutionary origin of the ASIP LINE-1. Using the 2,809-bp-long ASIP LINE-1 as a probe, we identified 1,500 and 1,267 LINE-1 elements in the river and swamp buffalo reference genomes, respectively, of which 1,009 and 766 were retained after filtering based on their sequence identities (>80%; supplementary tables S25−S26, Supplementary Material online). A minimum spanning (MS) tree analysis categorized these LINE-1 elements into 21 distinct subfamilies, each containing from 51 to 139 copies (P-values for subfamily partition ranging from 8E-215 to 7E-124) (fig. 5B;supplementary table S27, Supplementary Material online). The LINE-1 copy in white buffalo ASIP belonged to a young subfamily (sub20 in fig. 5B).
Finally, we did a population genetics analysis to characterize the relationship of haplotypes in the genomic region of 10-kb flanking the insertion point of LINE-1 upstream of ASIP (BBU14:19991854 − 20001504) in both white and black buffaloes. We identified 42 haplotypes in 73 buffaloes (63 swamp buffaloes from our WGS data and 10 river buffaloes from published data; Whitacre et al. 2017). A median-joining network defined three haplogroups—one for river buffaloes and two for swamp buffaloes, namely SW1 and SW2. All white buffaloes were in the SW2 group (fig. 5C). This finding was also supported by the maximum likelihood evolutionary tree and sequence alignment (supplementary fig. S7, Supplementary Material online). These results indicated that the haplotype carrying the LINE-1 insertion was closely related to a haplogroup of the nonwhite swamp buffaloes, in line with the scenario that white buffaloes originated from a recent genetic transposition event within the swamp buffalo rather than due to introgression from the river buffalo or another species.
Discussion
In this study, we combined evidence from GWAS, RNA-seq, long-read sequencing, and histological data to demonstrate that a LINE-1 insertion functioning as the active promoter of ASIP is the causal mutation for the white coat phenotype in swamp buffaloes. To our knowledge, this is the first morphological trait in water buffalo (Online Mendelian Inheritance in Animals [OMIA] 000213 − 89462 at https://omia.org/OMIA000213/89462/) to have its molecular mechanism uncovered.
White coat color, a common phenotypic variant in mammals, may be due to albinism or leucism. The former results from a disruption of pigment synthesis, usually caused by mutations of TYR, whereas the latter is caused by the absence of mature melanocytes in skin (Cieslak et al. 2011; Damé et al. 2012). Mutations in genes involved in melanocyte development, such as KIT (e.g., Haase et al. 2007) and MITF (e.g., Karlsson et al. 2007), can lead to leucism. ASIP encodes the agouti signaling protein, which has been well characterized as having an important role in melanin synthesis. It acts as an antagonist to the alpha-MSH (melanocyte-stimulating hormone) for the melanocortin-1 receptor (MC1R), leading to an increased pheomelanin synthesis in melanocytes (Furumura et al. 1998; Schiaffino 2010). The elevated expression of ASIP increases pheomelanin production whereas its decreased expression or loss of function mutations tends to result in the exclusive production of eumelanin and thus dark pigmentation (nonagouti phenotype) in rodents (Kingsley et al. 2009; Hubbard et al. 2010; Tanave et al. 2019). Recent studies also demonstrate that ASIP is involved in melanocyte development. It not only inhibits forward differentiation of melanoblasts (unpigmented melanocyte precursors) (Sviderskaya et al. 2001), but also induces rapid dedifferentiation of cultured melanocytes to the morphology of melanoblasts (Hida et al. 2009; Le Pape et al. 2009). ASIP is involved in the formation of a stripe pattern and dorso-ventral patterning in mammals (Girardot et al. 2006; Manceau et al. 2011; Mallarino et al. 2016), birds (Haupaix et al. 2018; Inaba et al. 2019; Robic et al. 2019), and teleost fishes (Ceinos et al. 2015; Kratochwil et al. 2018; Cal et al. 2019). The cis-regulatory variation in ASIP has also been shown to facilitate the adaptive winter camouflage polymorphism in snowshoe hares (Jones et al. 2018). In this study, we illustrate that a regulatory mutation leading to the 10-fold increase of ASIP expression prevents melanocyte differentiation and thus results in the white coat phenotype in swamp buffaloes.
TEs are a key source of genomic structural variations (SVs) in both eukaryotic and prokaryotic genomes. Recent evidence indicates that, in humans and model organisms, TEs play important roles in gene regulation, by contributing promoters and transcription factor binding sites and by affecting chromatin structures to change the expression of nearby genes (Merenciano et al. 2016; Burns 2017; De Cecco et al. 2019; Jang et al. 2019; Diehl et al. 2020). For example, viable yellow agouti (Avy) mice carry an intracisternal A-particle (IAP) retrotransposon inserted into the ASIP locus and the cryptic promoter within the IAP 5′ long-terminal repeat acts to drive the ectopic expression of ASIP, resulting in altered coat color, obesity, and an increased incidence of tumors (Duhl et al. 1994; Michaud et al. 1994; Klebig et al. 1995). However, the regulatory function and evolution of TEs have not been well characterized in agricultural animals (Girardot et al. 2006; Dreger and Schmutz 2011).
LINE-1 is the most abundant type of retrotransposon in mammalian genomes (Richardson et al. 2015), and mounting evidence indicates that, in humans, the insertion of a LINE-1 element can affect the expression of neighboring genes, causing phenotypic variation and diseases (Burns 2017; De Cecco et al. 2019; Jang et al. 2019). One potential mechanism by which LINE-1 affects gene expression is by introducing regulatory elements or promoters (Faulkner et al. 2009; Elbarbary et al. 2016). A full-length LINE-1 is typically 6 − 8 kb and contains a promoter within its 5′ UTR, two open reading frames (ORF1 and ORF2), a short 3′ UTR and a poly(A) tail (Moran et al. 1996). The 5′ UTR of LINE-1 has bidirectional promoter activity—a sense promoter that drives the transcription of the ORF-1 and ORF-2 proteins required for retrotransposition and an antisense promoter that affects the transcription of its upstream genomic region (Speek 2001; Nigumann et al. 2002; Beck et al. 2011). The transcribed 5′ LINE-1 antisense sequences are usually spliced to the exons of neighboring genes to form chimeric transcripts (Speek 2001). Recent studies show that LINE-1 antisense promoter-driven transcriptions are common in humans (Faulkner et al. 2009; Criscione et al. 2016). In this study, we identify a 2,809-bp-long LINE-1 insertion upstream of the first coding exon of ASIP, which acts as an active proximal promoter (∼44 kb away from the first coding exon) to initiate the transcription of ASIP in white buffaloes. In contrast, the wild type allele of ASIP initiates the transcription from a distal promoter (∼72 kb away from the first coding exon) in black buffaloes. The promoter activity of LINE-1 could be enhanced by the upstream flanking sequence (Lavie et al. 2004), inducing an increased expression of ASIP. However, a different mechanism is found in white sheep, where Norris and Whan (2008) identified a tandem duplication encompassing ASIP and two neighboring genes, AHCY and ITCH, which enhanced the expression of ASIP activated by a duplicated copy of the nearby ITCH promoter.
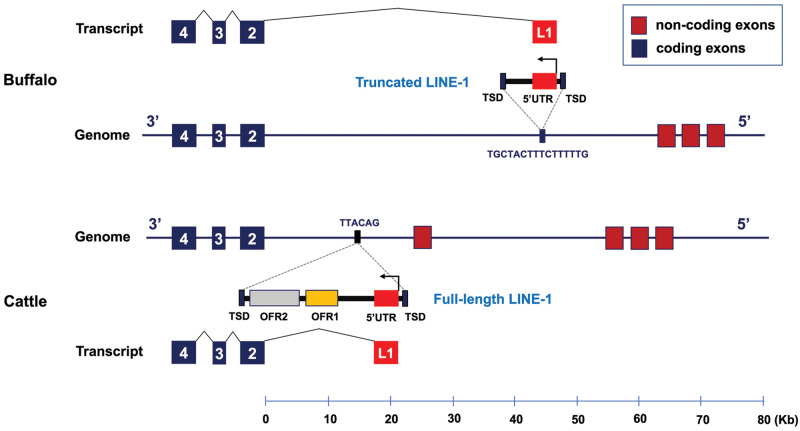
In nature, although convergence in phenotype is common, convergence at the molecular level is rather rare (Zou and Zhang 2015). However, in cattle as in white buffaloes, a LINE-1 element (L1-BT) located between the noncoding and coding exons of ASIP is associated with the brindle coat color in Normande cattle (Girardot et al. 2006). Sequence comparisons indicate independent origins of the LINE-1 elements in white buffaloes and cattle. First, the two LINE-1 insertions are located in different genomic positions relative to ASIP, that is, 44 and 15 kb from the first coding exon in white buffaloes and in cattle, respectively (fig. 6). Second, they belong to species-specific LINE-1 subclades (fig. 5A). Third, the two LINE-1 insertions have distinct TSDs to facilitate independent transposition events. Although the DNA structures of the LINE-1 elements are different in the two species, that is, full-length LINE-1 (8.4 kb) in cattle and 3′ truncated LINE-1 (2.8 kb) in white buffaloes, they share significant functional similarities (fig. 6). First, both LINE-1 elements have the same orientation as ASIP and transcribe a conserved sequence (∼160 bp, 98% identity) from the LINE-1 that is spliced to the coding exons forming a chimeric transcript. Second, consistent with that in white buffaloes, the LINE-1 insertion also led to the overexpression of ASIP in cattle (Girardot et al. 2006; Albrecht et al. 2012). Therefore, the two independent LINE-1 insertions in ASIP lead to similar functional impacts, and our study presents a compelling case for a convergent mechanism affecting coat color evolution in the Bovini tribe (Martin and Orgogozo 2013; Cuthill et al. 2017).
Fig. 6.
Two independent LINE-1 insertions occurred in ASIP of water buffalo (Bubalus bubalis) and cattle (Bos taurus). The genomes, transcripts, and LINE-1 elements are represented 3′−5′ from left to right to be consistent with that in figure 2. The LINE-1 of cattle ASIP is based on Girardot et al. (2006).
In the human genome, more than 99% of LINE-1 copies are unable to move due to 5′ truncation, rearrangement or mutation (Goodier and Kazazian 2008; Beck et al. 2011,Hancks and Kazazian 2016), with only a few remaining capable of retrotransposition (Brouha et al. 2003; Beck et al. 2011). Frequent 5′ truncation is explained by an integration mechanism of LINE-1 retrotransposon—target primed reverse transcription (TPRT) (Luan et al. 1993). During TPRT, the LINE-1 endonuclease nicks genomic DNA, freeing a 3′ hydroxyl that serves as a primer for polymerizing the cDNA copy onto the host DNA. This process is frequently aborted, resulting in 5′ truncated LINE-1 copies. However, the LINE-1 copy of white buffalo ASIP is 3′ truncated and contains only 5′ UTR, which might be generated by an unconventional integration mechanism. This 3′ truncation could be a special consequence of TPRT coupled with a reverse transcription/integration reaction to create an inversion in LINE-1 retrotransposition, a mechanism called “twin priming” by Ostertag and Kazazian (2001). This result also highlights the important role that LINEs play in the evolution of many species.
Materials and Methods
Study Samples
Three sets of swamp buffaloes were sampled from China, Thailand, and Bangladesh (supplementary table S1B, Supplementary Material online): 63 from China that were used for WGS and GWAS analysis, 202 from China, Thailand, and Bangladesh that were used for an association study to validate the GWAS signals and 285 that were used to verify the candidate causative mutation (LINE-1 insertion), which combined those used for WGS and the validation experiment, plus samples used only for genotyping the LINE-1 insertion.
Genomic DNA was extracted from blood or ear tissue using the phenol/chloroform method. The integrity and yield of genomic DNA were assessed and verified using agarose gel electrophoresis and a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA), respectively.
WGS Data Generation and Variant Detection
Paired-end short insert (350 bp) libraries were constructed from genomic DNA and sequenced using the Illumina HiSeq X Ten system (Illumina, San Diego, CA). Read pairs were aligned to the river buffalo reference genome (UOA_WB_1) using the BWA-MEM algorithm (http://bio-bwa.sourceforge.net/bwa.shtml) with the default parameters. PCR duplicates were removed with the MarkDuplicates module in the Picard Tools package v2.9.0 (http://broadinstitute.github.io/picard/). Realignment around indels was done using the GATK module IndelRealigner v3.8 (https://software.broadinstitute.org/gatk/). After variant calling by the GATK module UnifiedGenotyper, variant filtering was done using the parameters “QUAL < 30, QualByDepth (QD) < 2.0, RMS Mapping Quality (MQ) < 40.0, Mapping Quality Rank Sum Test (MQRankSum) < −12.5, Read Pos Rank Sum Test (ReadPosRankSum) < −8.0, Haplotype Score > 13.0.”
GWAS and Population Genomics Analyses
The VCFtools software v0.1.16 (Danecek et al. 2011) was used to convert the variant data file from VCF format to Plink format. For quality control filtering, we removed SNPs with call rates <90% or with minor allele frequencies <0.05 or departure from Hardy−Weinberg equilibrium <10−6 and discarded individuals with >10% missing genotypes. GWAS analysis was done using Fisher’s exact test and the dominance gene effect model in the Plink software v1.07 (Purcell et al. 2007) with the parameters “−model −modeldom −Fisher.” The GWAS results were visualized using the qqman R package v0.1.4 (https://CRAN.R-project.org/package=qqman).
Genetic differentiation (FST) between the populations was calculated using a sliding window approach (window size of 50 kb with step size of 25 kb) using the VCFtools. We reviewed the plots of average FST calculated using 50% overlapping windows of variable sizes (1, 10, 30, 50, and 70 kb) and found that the genome-wide pattern was reasonably smooth for the larger window sizes (30, 50, and 70 kb) but relatively noisy for small window sizes (1 and 10 kb). Therefore, we showed the results of 50-kb window size from this analysis.
Variant Genotyping Using the KASP Method
For validation of the GWAS signals, we genotyped a panel of 19 SNPs and one indel that showed significant associations with the white color phenotype in the target genomic region. Following the KASP assay guidelines (https://biosearch-cdn.azureedge.net/assetsv6/KASP-genotyping-chemistry-User-guide.pdf), the wild-type and mutant-type allele-specific upstream PCR primers and a common downstream primer were designed for each variant (supplementary table S6, Supplementary Material online).
The microfluidic-based IMAP platform (CapitalBio Technology, Beijing, China) was used for PCR amplification. The final reaction was done in a total volume of 1 μl, which contained 0.3 μl DNA template (50 ng/μl), 0.14 μl primer mix (12 µM each of the two allele-specific forward primers and 30 µM reverse primer), 0.5 μl 2× universal KASP Master Mix (LGC, United Kingdom) and 0.06 μl ddH2O. PCR thermocycling was done as follows: initiation at 95 °C for 15 min; 10 cycles of denaturation at 95 °C for 20 s and touchdown annealing from 61 °C (−0.6 °C/cycle) for 60 s; followed by 26 cycles of denaturation at 95 °C for 20 s and annealing at 55 °C for 60 s and finished by an extension at 37 °C for 60 s. An end-point fluorescent read of the PCR products was done using the LuxScan-10K/D instrument (CapitalBio Technology, Beijing, China).
LD Analysis
LD (pairwise r2 statistic) was calculated and visualized using the Haploview software v4.1 (Barrett et al. 2005). The R package rehh v3.0.1 (Gautier et al. 2017) was used to draw a haplotype bifurcation diagram (Sabeti et al. 2002) that visualizes the breakdown of LD at increasing distances from the focal core allele. The haplotypes used for drawing the bifurcation diagram were phased using the Beagle software v4.1 (Browning and Browning 2007).
Variant Annotation in the 1.07-Mb Genomic Region of GWAS Signals
The Annovar program v2018Apr16 (Wang et al. 2010) was applied to annotate each variant, using an annotation file of GTF format prepared for the river buffalo reference genome (UOA_WB_1).
Amplification and Sanger Sequencing of ASIP Exons and Flanking Regions
Primers (supplementary table S10, Supplementary Material online) were designed based on the river buffalo reference genome (UOA_WB_1) to amplify and sequence the coding exons and flanking regions (2,000 bp upstream of the first coding exon and 1,000 bp downstream of the last coding exon) of ASIP. Three DNA pools (Guizhou [white], Guizhou [black] and Yanjin [black] buffalo breeds) were prepared as templates for PCR amplification. Each pool represented six individuals with genomic DNA equally mixed. PCR products were used for Sanger sequencing.
SV Detection in the 1.07-Mb Genomic Region
SVs were detected using three software tools—the mrFAST v2.6.1.0 (Alkan et al. 2009), the BreakDancer v1.1.2 (Chen et al. 2009), and the CNVnator v0.3.3 (Abyzov et al. 2011). For the mrFAST analysis, paired-end sequencing reads were first mapped to the river buffalo reference genome (UOA_WB_1) with the parameters “--search -- pe-e 5,” followed by calculating read depth to detect segmental duplications and deletions. SVs were detected using the BreakDancer with the default parameters “-q 35-c 3-s 7-b 100-t-d” and using the CNVnator with bin size set to 500 bp (-tree -his 500 -stat 500 -partition500 -call 500). The statistic Vst was used to test the difference in copy numbers at each SV between white buffaloes and black buffaloes: Vst = (Vt−Vs)/Vt, where Vt is the overall variance of copy number and Vs the average variance within populations.
RNA Extraction and qPCR
Total RNA was prepared from ear skin samples using the TRIzol reagent (Thermo Scientific, 15596026) in accordance with the manufacturer’s recommendation. RNA purity, concentration, and integrity were assessed using the LabChip GX Touch Nucleic Acid Analyzer (PerkinElmer, Waltham, MA). Reverse transcription was done using the PrimeScript RT Reagent Kit (TAKARA Bio, Mountain View, CA). Real-time quantitative PCR was done on the LightCycler 480 Instrument II (Roche Diagnostics, Mannheim, Germany) using the SYBR Green I Master (Roche) kit. The designed primers are listed in supplementary table S17, Supplementary Material online. The 18S rRNA was used as the internal reference gene.
RNA-Seq and Data Analysis
Sequencing libraries were constructed using the NEBNext UltraTM RNA Library Prep Kit (NEB, Ipswich, MA) following the manufacture’s recommendations. The library preparations were sequenced using the Illumina HiSeq X Ten system. After quality control, the paired-end reads were mapped to the river buffalo reference genome (UOA_WB_1) using the HISAT2 software v2.6.1.0 (Kim et al. 2019). Transcripts were assembled and quantified with the StringTie software v2.1.1 (Pertea et al. 2015). The Cuffcompare tool of the Cufflink suite v2.2.1 (Trapnell et al. 2012) was used to compare the alternative transcripts among individuals. The results were visualized using the pheatmap R package v1.0.12 with Ward’s hierarchical clustering method (https://CRAN.R-project.org/package=pheatmap).
The differential expression analysis was performed using the DESeq2 package v1.4.5 (Love et al. 2014). The significance of the difference in gene expression was determined using a Wald test in the DESeq2 package. The results with an FDR ≤0. 05 were considered noteworthy. A functional annotation of DEGs was conducted through GO enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using the open access WebGestalt tool (http://www.webgestalt.org, Liao et al. 2019). The gene expression level was quantified using TPM with the StringTie software v2.1.1 (Pertea et al. 2015). It normalizes sequencing depth and gene length. To analyze gene expression profiles across different buffalo tissues for the 23 genes in the genomic region of the GWAS signals, RNA-seq data of six skin samples generated in this study were combined with published data of 248 samples from NCBI (PRJEB4351 [30 tissues of a male and a female Mediterranean buffalo calves from Italy] and PRJEB25226 [218 of the 220 tissue and cell samples, except ERX2403664 and ERX2403645 with no runs of data, of six Mediterranean water buffaloes from Italy and four river buffaloes of Pandharpuri and Bhadawari breeds from India]; Williams et al. 2017; Li et al. 2019; Low et al. 2019; Young et al. 2019). The gene expression values (TPM) were log2 transformed and then visualized using the pheatmap R package v1.0.12 with Ward’s hierarchical clustering method.
5′ and 3′ RACE
The 5′ and 3′ RACE PCR experiments were done using the SMARTer RACE 5′/3′ kit (TAKARA Bio) according to the manufacturer’s instructions. The Primer Premier 5.0 software (http://www.premierbiosoft.com) was used to design specific primers (supplementary table S17, Supplementary Material online). The PCR product was cloned into the pClone007 vector using the pClone007 vector kit (Beijing TsingKe Biotech Co., Ltd, China) and multiple individual clones were sequenced.
Genotyping of the LINE-1 Insertion
Two pairs of primers were designed based on the assembled white buffalo-specific ASIP sequence (supplementary table S17, Supplementary Material online). The PCR was set up in a final volume of 25 µl containing 18 µl dd H2O, 5 pmol of each primer, 200 ng of genomic DNA, 2.5 µl 10× PCR buffer (TaKaRa, Dalian, China), 200 µM dNTP mixture (TaKaRa), and 1 U Taq polymerase (TaKaRa). PCR conditions were: 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 60 °C for 1 min, and 72 °C for 1 min, and the final extension for 7 min at 72 °C.
Immunohistochemistry and HE Staining
The skin samples were embedded in paraffin and then sectioned for Hematoxylin−Eosin (HE) staining and immunohistochemical staining. The rabbit polyclonal antibody anti-TRP1 (ab83774, Abcam, Cambridge, MA) was used for immunohistochemical staining.
Nanopore Long-Read Sequencing
DNA samples from one each of white and one black buffaloes were used for Nanopore long read sequencing in accordance with the standard protocol provided by Oxford Nanopore Technologies (ONT, Oxford, United Kingdom).
The FAST5 files containing signal data generated by the Nanopore sequencer were converted into the FASTQ format using the Albacore software in the MinKNOW package (ONT). Clean reads were obtained by removing the adaptor sequences, low-quality sequence reads, and short reads (length <500 bp). The Minimap2 software v2.17 (Li 2018) was used to map clean reads to BBU14 of the river buffalo reference genome (UOA_WB_1). Reads mapped to the target region were extracted using the Samtools software v1.10 (Li et al. 2009). Regional de novo assembly was done using the Canu software v1.8 (Koren et al. 2017) with the parameters “correctedErrorRate 0.144; CorOutCoverage 40.”
Phylogenetic Analyses of LINE-1 Repeats
LINE-1 repeat elements were extracted from the reference genomes of river buffalo (UOA_WB_1), swamp buffalo (GWHAAJZ00000000, https://bigd.big.ac.cn/search/? dbId=gwh&q=GWHAAJZ00000000; Luo et al. 2020), taurine cattle (ARS-UCD1.2, GCA_002263795.2; Rosen et al. 2020), and yak (BosGru3.0, GCA_005887515.2), using the RepeatMasker software v4.07 (http://www.repeatmasker.org) with the slow search option, based on the Repbase repeat database v9.04 (http://www.girinst.org/). The ParseRM_GetNesting.pl script was used to filter out the nested LINE-1 elements from the RepeatMasker output. To extract the full-length LINE-1, the resulting non-nested LINE-1s were aligned to the full-length bovine LINE-1 L1-BT transposon element sequence (DQ000238), followed by a filtering to remove elements with length <7,000 bp, truncation at 5′ UTR <200 bp, and truncation at 3′ UTR <300 bp. Finally, to ensure that the LINE-1 elements were highly homologous, a clustering-based approach was used to keep the LINE-1s with a sequence identity of >80%, implemented in the CD-HIT software v4.6.8 (Fu et al. 2012) with the parameter “-T 0 -c 0.8 -M 0 -n 5 -p 0.”
The Mafft software v7.407 (Katoh et al. 2019) was used for multiple sequence alignment of the qualified full-length LINE-1s with the parameter “mafft --quiet --thread 24 --retree 1.” An approximately maximum-likelihood phylogeny was constructed based on the output (aligned.fa) from the Mafft alignment using the FastTree software v2.2.11 (Price et al. 2009) with the default settings “Nucleotide distances: Jukes–Cantor Joins; balanced Support: SH-like 1000; Search: Normal + NNI + SPR (2 rounds range 10) + ML-NNI opt-each = 1; TopHits: 1.00*sqrtN close = default refresh = 0.80; ML Model: Jukes–Cantor, CAT approximation with 20 rate categories” and visualized using the iTOL online website (https://itol.embl.de/).
For evolutionary analysis within the water buffalo species, the LINE-1 elements homologous with the 2,809-bp-long white buffalo ASIP LINE-1 insertion were identified using the RepeatMasker software. Sequence homology analysis was done using the cross_match software v1.09 (http://www.phrap.org/phredphrapconsed.html) with the parameters “-gap_init -25 -gap_ext -5 -minscore 10 -minmatch 6 -alignments -bandwidth 50 -word_raw.” The MS trees of LINE-1s were constructed using the COSEG software v0.2.2 (http://www.repeatmasker.org/) to define the subfamilies.
Haplotype Network of the LINE-1 Insertion Region
SNPs in a genomic region of 10-kb flanking the LINE-1 insertion point upstream of ASIP (BBU14:19991854 − 20001504) were used for haplotype analysis. In addition to 63 swamp buffaloes that were whole-genome sequenced in this study, we used 10 river buffaloes for comparative purpose (NCBI Sequence Read Archive SRR4477876−SRR4477880, SRR4477882−SRR4477884, SRR4477888, and SRR4477890; Whitacre et al. 2017). Haplotypes were phased using the Beagle software v4.1 (Browning and Browning 2007). A median-joining network and a maximum likelihood (ML) evolutionary tree based on Tamura–Nei model were constructed using the Network software v5.0.1.1 (Bandelt et al. 1999) and the MEGA7 software (Kumar et al. 2016), respectively. To construct the ML tree, a discrete Gamma distribution (5 categories) was used to model evolutionary rate differences among sites.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Scientific Foundation of China (Grant no. 31561143010) and the China Agricultural Research System (Grant no. CARS-36). Linzhao Fang was funded through Health Data Research UK (HDR-UK) (Grant no. HDR-9004) and the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Actions (MSCA) (Grant No. 801215). We appreciate the Chinese Government’s contribution to the Chinese Academy of Agricultural Sciences (CAAS)-International Livestock Research Institute (ILRI) Joint Laboratory on Livestock and Forage Genetic Resources in Beijing (2018-GJHZ-01) and this article contributes to the Consortium of International Agricultural Research Centers (CGIAR) Research Program on Livestock. We thank Ian J. Jackson (University of Edinburgh) for his insightful comments on the manuscript. We also gratefully acknowledge the critical review of our manuscript by three anonymous reviewers.
Data Availability
Whole-genome sequencing data generated in this study have been submitted to the NCBI Sequence Read Archive (SRA) as BioProject ID PRJNA633919.
References
- Abyzov A, Urban AE, Snyder M, Gerstein M.. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 21(6):974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht E, Komolka K, Kuzinski J, Maak S.. 2012. Agouti revisited: transcript quantification of the ASIP gene in bovine tissues related to protein expression and localization. PLoS One. 7(4):e35282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. 2009. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 41(10):1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A.. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16(1):37–48. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ.. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. [DOI] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV.. 2011. LINE-1 elements in structural variation and disease. Annu Rev Genom Hum Genet. 12(1):187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Sookdeo A.. 2016. The evolution of LINE-1 in vertebrates. Genome Biol Evol. 8(12):3485–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkareva NV, Botchkarev VA, Gilchrest BA.. 2003. Fate of melanocytes during development of the hair follicle pigmentary unit. J Investig Dermatol Symp Proc. 8(1):76–79. [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH. Jr.. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 100(9):5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL.. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81(5):1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruders R, Van Hollebeke H, Osborne EJ, Kronenberg Z, Maclary E, Yandell M, Shapiro MD.. 2020. A copy number variant is associated with a spectrum of pigmentation patterns in the rock pigeon (Columba livia). PLoS Genet. 16(5):e1008274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH. 2017. Transposable elements in cancer. Nat Rev Cancer 17(7):415–424. [DOI] [PubMed] [Google Scholar]
- Cal L, Suarez-Bregua P, Comesana P, Owen J, Braasch I, Kelsh R, Cerda-Reverter JM, Rotllant J.. 2019. Countershading in zebrafish results from an Asip1 controlled dorsoventral gradient of pigment cell differentiation. Sci Rep. 9(1):3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceinos RM, Guillot R, Kelsh RN, Cerda-Reverter JM, Rotllant J.. 2015. Pigment patterns in adult fish result from superimposition of two largely independent pigmentation mechanisms. Pigment Cell Melanoma Res. 28(2):196–209. [DOI] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6(9):677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslak M, Reissmann M, Hofreiter M, Ludwig A.. 2011. Colours of domestication. Biol Rev Camb Philos Soc. 86(4):885–899. [DOI] [PubMed] [Google Scholar]
- Criscione SW, Theodosakis N, Micevic G, Cornish TC, Burns KH, Neretti N, Rodic N.. 2016. Genome-wide characterization of human L1 antisense promoter-driven transcripts. BMC Genomics 17(1):463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthill IC, Allen WL, Arbuckle K, Caspers B, Chaplin G, Hauber ME, Hill GE, Jablonski NG, Jiggins CD, Kelber A, et al. 2017. The biology of color. Science 357(6350):eaan0221. [DOI] [PubMed] [Google Scholar]
- Damé MCF, , Xavier GM, , Oliveira-Filho JP, , Borges AS, , Oliveira HN, , Riet-Correa F, , Schild AL. 2012. A nonsense mutation in the tyrosinase gene causes albinism in water buffalo. BMC Genet. 13:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, Caligiana A, Brocculi G, Adney EM, Boeke JD, et al. 2019. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566(7742):73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl AG, Ouyang N, Boyle AP.. 2020. Transposable elements contribute to cell and species-specific chromatin looping and gene regulation in mammalian genomes. Nat Commun. 11(1):1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger DL, Schmutz SM.. 2011. A SINE insertion causes the black-and-tan and saddle tan phenotypes in domestic dogs. J Hered. 102(Suppl 1):S11–18. [DOI] [PubMed] [Google Scholar]
- Duhl DMJ, Vrieling H, Miller KA, Wolff GL, Barsh GS.. 1994. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 8(1):59–65. [DOI] [PubMed] [Google Scholar]
- Elbarbary RA, Lucas BA, Maquat LE.. 2016. Retrotransposons as regulators of gene expression. Science 351(6274):aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Larson G, Ribeiro HS, Li N, Andersson L.. 2009. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5(1):e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 41(5):563–571. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumura M, Sakai C, Potterf SB, Vieira WD, Barsh GS, Hearing VJ.. 1998. Characterization of genes modulated during pheomelanogenesis using differential display. Proc Natl Acad Sci U S A. 95(13):7374–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Klassmann A, Vitalis R.. 2017. rehh 2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol Ecol Resour. 17(1):78–90. [DOI] [PubMed] [Google Scholar]
- Girardot M, Guibert S, Laforet MP, Gallard Y, Larroque H, Oulmouden A.. 2006. The insertion of a full-length Bos taurus LINE element is responsible for a transcriptional deregulation of the Normande Agouti gene. Pigment Cell Res. 19(4):346–355. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH. Jr.. 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135(1):23–35. [DOI] [PubMed] [Google Scholar]
- Haase B, Brooks SA, Schlumbaum A, Azor PJ, Bailey E, Alaeddine F, Mevissen M, Burger D, Poncet PA, Rieder S, et al. 2007. Allelic heterogeneity at the equine KIT locus in dominant white (W) horses. PLoS Genet. 3(11):e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr. 2016. Roles for retrotransposon insertions in human disease. Mob DNA 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupaix N, Curantz C, Bailleul R, Beck S, Robic A, Manceau M.. 2018. The periodic coloration in birds forms through a prepattern of somite origin. Science 361(6408):eaar4777. [DOI] [PubMed] [Google Scholar]
- Henkel J, Saif R, Jagannathan V, Schmocker C, Zeindler F, Bangerter E, Herren U, Posantzis D, Bulut Z, Ammann P, et al. 2019. Selection signatures in goats reveal copy number variants underlying breed-defining coat color phenotypes. PLoS Genet. 15(12):e1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida T, Wakamatsu K, Sviderskaya EV, Donkin AJ, Montoliu L, Lynn Lamoreux M, Yu B, Millhauser GL, Ito S, Barsh GS, et al. 2009. Agouti protein, mahogunin, and attractin in pheomelanogenesis and melanoblast-like alteration of melanocytes: a cAMP-independent pathway. Pigment Cell Melanoma Res. 22(5):623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE. 2006. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 97(3):222–234. [DOI] [PubMed] [Google Scholar]
- Hubbard JK, Uy JAC, Hauber ME, Hoekstra HE, Safran RJ.. 2010. Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet. 26(5):231–239. [DOI] [PubMed] [Google Scholar]
- Inaba M, Jiang TX, Liang YC, Tsai S, Lai YC, Widelitz RB, Chuong CM.. 2019. Instructive role of melanocytes during pigment pattern formation of the avian skin. Proc Natl Acad Sci USA. 116(14):6884–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, Zhang D, Li D, Xing X, Kim S, et al. 2019. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet. 51(4):611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Mills LS, Alves PC, Callahan CM, Alves JM, Lafferty DJR, Jiggins FM, Jensen JD, Melo-Ferreira J, Good JM.. 2018. Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science 360(6395):1355–1358. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas IIE, et al. 2007. Efficient mapping of Mendelian traits in dogs through genome-wide association. Nat Genet. 39(11):1321–1328. [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD.. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37(8):907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley EP, Manceau M, Wiley CD, Hoekstra HE.. 2009. Melanism in peromyscus is caused by independent mutations in agouti. PLoS ONE 4(7):e6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebig ML, Wilkinson JE, Geisler JG, Woychik RP.. 1995. Ectopic expression of the agouti gene in transgenic mice causes obesity, features of type II diabetes, and yellow fur. Proc Natl Acad Sci U S A. 92(11):4728–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM.. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil CF, Liang Y, Gerwin J, Woltering JM, Urban S, Henning F, Machado-Schiaffino G, Hulsey CD, Meyer A.. 2018. Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science 362(6413):457–460. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie L, Maldener E, Brouha B, Meese EU, Mayer J.. 2004. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. 14(11):2253–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pape E, Passeron T, Giubellino A, Valencia JC, Wolber R, Hearing VJ.. 2009. Microarray analysis sheds light on the dedifferentiating role of agouti signal protein in murine melanocytes via the Mc1r. PNAS. 106(6):1802–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Bickhart DM, Ramunno L, Iamartino D, Williams JL, Liu GE.. 2019. Comparative sequence alignment reveals river buffalo genomic structural differences compared with cattle. Genomics 111(3):418–425. [DOI] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B.. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47(W1):W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WY, Tearle R, Bickhart DM, Rosen BD, Kingan SB, Swale T, Thibaud-Nissen F, Murphy TD, Young R, Lefevre L, et al. 2019. Chromosome-level assembly of the water buffalo genome surpasses human and goat genomes in sequence contiguity. Nat Commun. 10:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH.. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 72(4):595–605. [DOI] [PubMed] [Google Scholar]
- Luo X, Zhou Y, Zhang B, Zhang Y, Wang X, Feng T, Li Z, Cui K, Zhang Z, Luo C, et al. 2020. Understanding divergent domestication traits from the whole-genome sequencing of swamp-and river-buffalo populations. Natl Sci Rev. 7(3):686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallarino R, Henegar C, Mirasierra M, Manceau M, Schradin C, Vallejo M, Beronja S, Barsh GS, Hoekstra HE.. 2016. Developmental mechanisms of stripe patterns in rodents. Nature 539(7630):518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Mallarino R, Hoekstra HE.. 2011. The developmental role of Agouti in color pattern evolution. Science 331(6020):1062–1065. [DOI] [PubMed] [Google Scholar]
- Martin A, Orgogozo V.. 2013. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67(5):1235–1250. [DOI] [PubMed] [Google Scholar]
- Merenciano M, Ullastres A, de Cara MA, Barron MG, Gonzalez J.. 2016. Multiple independent retroelement insertions in the promoter of a stress response gene have variable molecular and functional effects in Drosophila. PLoS Genet. 12(8):e1006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud EJ, Van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP.. 1994. Differential expression of a new dominant agouti allele (AIAPY) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 8(12):1463–1472. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87(5):917–927. [DOI] [PubMed] [Google Scholar]
- Mort RL, Jackson IJ, Patton EE.. 2015. The melanocyte lineage in development and disease. Development 142(4):620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Matlik K, Speek M.. 2002. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79(5):628–634. [DOI] [PubMed] [Google Scholar]
- Norris BJ, Whan VA.. 2008. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res. 18(8):1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr. 2001. Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 11(12):2059–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26(7):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV.. 2015. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol Spectr. 3(2):MDNA3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rife DC. 1962. Color and horn variations in water buffalo: the inheritance of coat color, eye color and shape of horns. J Hered. 53(5):239–246. [DOI] [PubMed] [Google Scholar]
- Rife DC, Buranamanas P.. 1959. Inheritance of white coat color in the water buffalo of Thailand. J Hered. 50(6):269–272. [Google Scholar]
- Robic A, Morisson M, Leroux S, Gourichon D, Vignal A, Thebault N, Fillon V, Minvielle F, Bed’Hom B, Zerjal T, et al. 2019. Two new structural mutations in the 5’ region of the ASIP gene cause diluted feather color phenotypes in Japanese quail. Genet Sel Evol. 51(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP.. 2017. Variant review with the integrative genomics viewer (IGV). Cancer Res. 77(21):e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E, Rowan TN, Low WY, Zimin A, Couldrey C, et al. 2020. De novo assembly of the cattle reference genome with single-molecule sequencing. GigaScience 9(3):giaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419(6909):832–837. [DOI] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH.. 1997. Many human L1 elements are capable of retrotransposition. Nat Genet. 16(1):37–43. [DOI] [PubMed] [Google Scholar]
- Schiaffino MV. 2010. Signaling pathways in melanosome biogenesis and pathology. Int J Biochem Cell Biol. 42(7):1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 21(6):1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel KP, Davidson DR, Jackson IJ.. 1992. TRP-2/DT, a new early melanoblast marker, shows that steel growth factor (c-kit ligand) is a survival factor. Development 115(4):1111–1119. [DOI] [PubMed] [Google Scholar]
- Sviderskaya EV, Hill SP, Balachandar D, Barsh GS, Bennett DC.. 2001. Agouti signaling protein and other factors modulating differentiation and proliferation of immortal melanoblasts. Dev Dyn. 221(4):373–379. [DOI] [PubMed] [Google Scholar]
- Tanave A, Imai Y, Koide T.. 2019. Nested retrotransposition in the East Asian mouse genome causes the classical nonagouti mutation. Commun Boil. 2:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitacre LK, Hoff JL, Schnabel RD, Albarella S, Ciotola F, Peretti V, Strozzi F, Ferrandi C, Ramunno L, Sonstegard TS, et al. 2017. Elucidating the genetic basis of an oligogenic birth defect using whole genome sequence data in a non-model organism. Sci Rep. 7(1):39719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JL, Iamartino D, Pruitt KD, Sonstegard T, Smith TPL, Low WY, Biagini T, Bomba L, Capomaccio S, Castiglioni B, et al. 2017. Genome assembly and transcriptome resource for river buffalo, Bubalus bubalis (2n = 50). Gigascience 6(10): gix088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R, Lefevre L, Bush SJ, Joshi A, Singh SH, Jadhav SK, Dhanikachalam V, Lisowski ZM, Iamartino D, Summers KM, et al. 2019. A gene expression atlas of the domestic water buffalo (Bubalus bubalis). Front Genet. 10(6):668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusnizar Y, , Wilbe M, , Herlino AO, , Sumantri C, Rachman , Noor R, , Boediono A, , Andersson L, , Andersson G. 2015. Microphthalmia‐associated transcription factor mutations are associated with white‐spotted coat color in swamp buffalo. Anim Genet. 46(6):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Colli L, Barker JSF.. 2020. Asian water buffalo: domestication, history and genetics. Anim Genet. 51(2):177–191. [DOI] [PubMed] [Google Scholar]
- Zou Z, Zhang J.. 2015. Are convergent and parallel amino acid substitutions in protein evolution more prevalent than neutral expectations? Mol Biol Evol. 32(8):2085–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome sequencing data generated in this study have been submitted to the NCBI Sequence Read Archive (SRA) as BioProject ID PRJNA633919.