Abstract
Context
Accurate methods for early gestational diabetes mellitus (GDM) (during the first trimester of pregnancy) prediction in Chinese and other populations are lacking.
Objectives
This work aimed to establish effective models to predict early GDM.
Methods
Pregnancy data for 73 variables during the first trimester were extracted from the electronic medical record system. Based on a machine learning (ML)-driven feature selection method, 17 variables were selected for early GDM prediction. To facilitate clinical application, 7 variables were selected from the 17-variable panel. Advanced ML approaches were then employed using the 7-variable data set and the 73-variable data set to build models predicting early GDM for different situations, respectively.
Results
A total of 16 819 and 14 992 cases were included in the training and testing sets, respectively. Using 73 variables, the deep neural network model achieved high discriminative power, with area under the curve (AUC) values of 0.80. The 7-variable logistic regression (LR) model also achieved effective discriminate power (AUC = 0.77). Low body mass index (BMI) (≤ 17) was related to an increased risk of GDM, compared to a BMI in the range of 17 to 18 (minimum risk interval) (11.8% vs 8.7%, P = .09). Total 3,3,5′-triiodothyronine (T3) and total thyroxin (T4) were superior to free T3 and free T4 in predicting GDM. Lipoprotein(a) was demonstrated a promising predictive value (AUC = 0.66).
Conclusions
We employed ML models that achieved high accuracy in predicting GDM in early pregnancy. A clinically cost-effective 7-variable LR model was simultaneously developed. The relationship of GDM with thyroxine and BMI was investigated in the Chinese population.
Keywords: GDM, early prediction, machine learning models, early pregnancy, BMI, thyroxine
Gestational diabetes mellitus (GDM) is a common complication during pregnancy (1) that affects up to 15% of pregnant women worldwide (2). Hyperglycemia is not, by itself, life-threatening for pregnant women, but can be harmful to the fetus, leading to complications, including stillbirth, premature delivery, macrosomia, fetal hyperinsulinemia, and clinical neonatal hypoglycemia (1). The American Diabetes Association (ADA) and the International Association of Diabetes and Pregnancy Study Groups (IADPSG) recommend diagnosing GDM via a 2-hour, 75-g oral glucose tolerance test (OGTT) at 24 to 28 weeks of pregnancy (3, 4). There is accumulating evidence indicating that the exposure of embryos or fetuses to a hyperglycemic environment in the uterus can lead to chronic health problems later in life (5), including obesity, diabetes, and cardiovascular diseases (6-8). Theoretically, GDM patients could have hyperglycemia for a long or short period of time before the GDM diagnosis, so the fetus will be more or less exposed to an intrauterine hyperglycemic environment in the second trimester (from 13 weeks of pregnancy to the day of the OGTT). Previous studies confirmed that fetal growth can already be abnormal preceding the diagnosis of GDM, including smaller fetuses at 24 weeks of gestation (9) and increased abdominal circumference growth rates compared with the non-GDM group (10). Our previous study indicated that insulin therapy after GDM diagnosis cannot fully protect offspring from diet-related metabolic disorders in adulthood (11). Therefore, a hysteretic diagnosis of GDM at 24 to 28 weeks of gestation might be too late for intervention and cannot completely reverse the adverse effects (including changes in epigenetics and abnormal fetal growth that occurred before 24 weeks of gestation) of the intrauterine hyperglycemia exposure on the offspring. It is thus essential to establish a prediction model to identify the high-risk group of GDM in the first trimester and provide an opportunity for interventions prior to diagnosis in the third trimester.
In GDM prediction, prior research has sought to find a threshold value of fasting plasma glucose (FPG) in the first trimester through large sample studies (12). Although elevating diagnostic criteria from an FPG greater than or equal to 5.1 mM to an FPG greater than or equal to 6.1 mM can obtain nearly 100% specificity, the corresponding low sensitivity (1%) greatly limits the feasibility (12). In recent years, some novel biomarkers have been reported as potential GDM predictors, including angiopoietin-like protein 8, plasma fatty acid-binding protein 4, and various adipokines (13-15), but the low availability of these biomarkers in clinical practice limits their application. The exploration of prediction models based on multiple common risk factors, such as advanced maternal age, body mass index (BMI), and family history of diabetes, provides a new perspective in solving the problem (16). Recently, artificial intelligence technology, particularly supervised machine learning (ML) methods, has been reported to demonstrate a powerful self-learning ability with improved GDM prediction accuracy (17). However, GDM predictions are often made during the second trimester (20th week of gestation), creating a limited time frame for doctors to intervene (17). Therefore, in this study, we generated ML algorithms to predict GDM in the first trimester of pregnancy.
Materials and Methods
Data source
The training data set included patients that were recruited from the 2017 obstetrical electronic medical record data from the International Peace Maternal and Child Health Hospital, Shanghai Jiao Tong University School of Medicine. Women with pre-GDM (FPG ≥ 7.0 mM or glycated hemoglobin [HbA1c] ≥ 6.5%) were excluded. Samples that had a missing observation of greater than 20% were excluded from the data set. Candidate variables including sociodemographic characteristics, clinical variables, and laboratory indexes in the first trimester were collected. Following this, the 2018 obstetrical electronic medical record data were collected and curated, which served as the testing group to evaluate the prediction models. The details of the research subject selection are presented in Supplementary Fig. S1 (18). The GDM diagnostic criteria followed the IADPSG guidelines (FPG ≥ 5.1 mM, 1-h ≥ 10 mM, and/or 2-h ≥ 8.5 mM). This study was approved by the medical ethical committee of International Peace Maternity and Child Health Hospital, School of Medicine, Shanghai Jiao Tong University (No. GKLW2019-05).
Variable selection
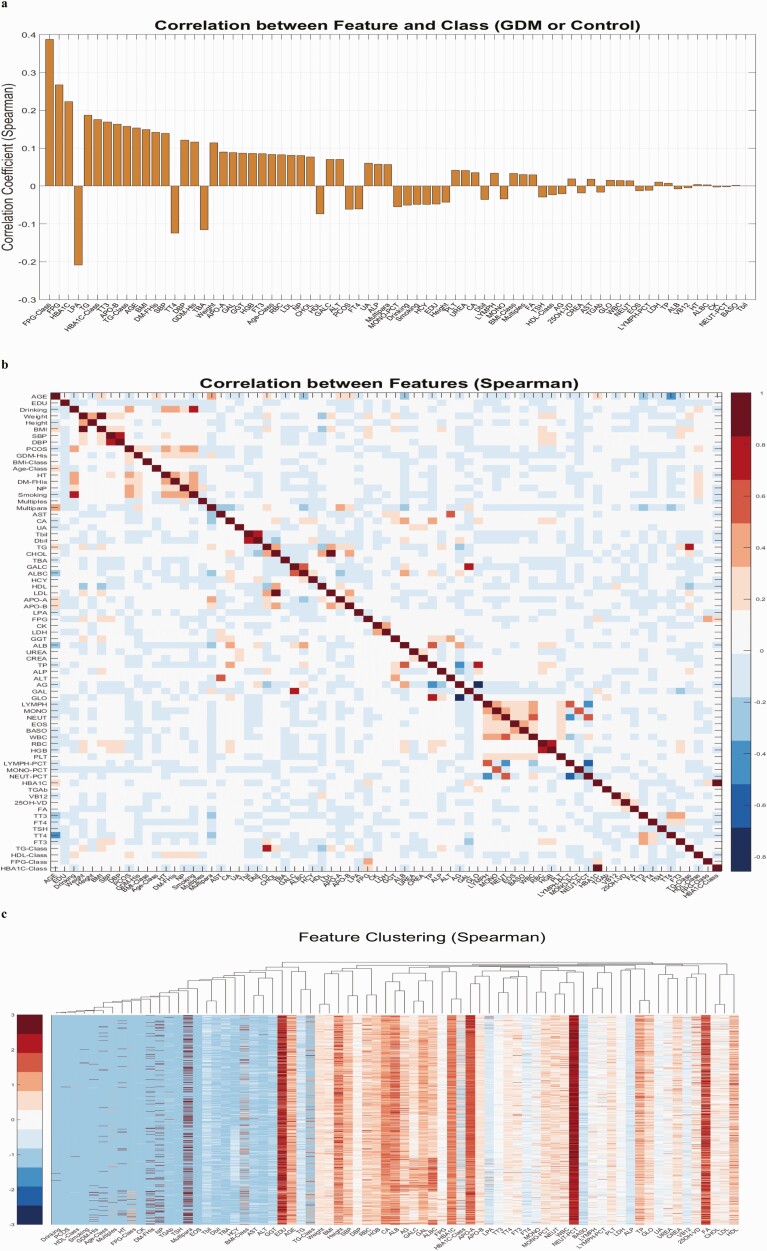
To ensure better model discrimination and create an efficient approach for clinical practice with fewer redundant variables, variable selection was conducted to select a panel of biomarkers with the most discriminative power for our outcome. All of the variables were sorted based on their absolute Spearman correlation coefficients and Pearson correlation coefficients with respect to the GDM and control groups, as demonstrated in Fig. 1A and Supplementary Fig. S2A (18). Fig. 1A shows that the indicators related to glucose and lipid metabolism have the strongest correlation with GDM, including FPG, HbA1c, lipoprotein(a), triglyceride (TG), and apolipoprotein-B. Total 3,3,5′-triiodothyronine (TT3) and GDM are also significantly correlated. Initial analyses using Spearman or Pearson correlation showed that several variables were highly correlated to each other and formed small clusters, as shown in Fig. 1B and 1C and Supplementary Fig. S2B and S2C (18). Correlation coefficient values are presented in Supplementary Fig. S5 (18). This indicated that a representative small cluster of variables may provide enough discriminative power for a simplified model. The rationale for conducting correlation coefficient analyses before applying the model-free sequential forward variable selection is that in situations when many features exhibit at least a weak correlation (eg, |corr| > 0.05) with the outcome vector and when the features belong to multiple clusters, the sequential forward feature selection method tends to select representative features from each orthogonal cluster, spanning a more diverse feature space while excluding redundant information.
Figure 1.
Variable selection results. A, Spearman correlation coefficients between each variable and the gestational diabetes mellitus (GDM)/non-GDM label vector, over all the samples. The bar plots from left to right represent absolute values from high to low. B, Spearman correlation coefficients between all the variables over vectors of all the samples. Detailed correlation coefficient values can be found in Supplementary Table S5 (18). C, Variable-way hierarchical clustering results using distance metrics based on Spearman correlation coefficients.
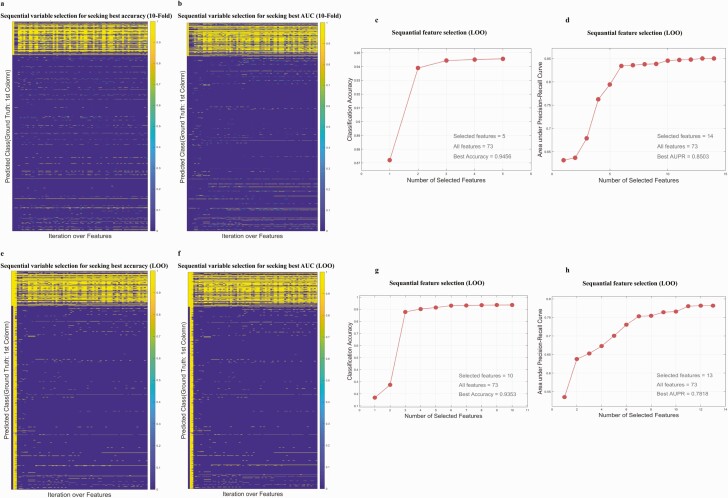
We applied a variable selection strategy that was previously successfully used in gene selection (19, 20). In short, variable selection was completed using a cross-validation (CV) framework of 10-fold 100-repeat CV and leave-one-out (LOO) CV. The details of the CV method are shown in the Supplementary text (18). The variables were sorted using absolute correlation coefficients (both Pearson and Spearman correlation were tested and Spearman was chosen) with respect to the GDM and control group, and an iterative approach of variable inclusion was used to assess the predictive power of each individual variable, using the average prediction accuracy or the area under the receiver operating characteristic (ROC) curve (AUC) as the indicator of model improvement. Fig. 2A, 2B, 2E, and 2F demonstrate the selected variable in each iteration. Fig. 2C, 2D, 2G, and 2H show the incremental trajectory of accuracy or AUC when including a contributing variable that remained in the selected variable pool in the 10-fold and LOO CV, respectively.
Figure 2.
A and B, Ten-fold cross-validation (CV)-based detailed prediction outcomes of each variable selection iteration. The yellow and blue elements represent predicted gestational diabetes mellitus (GDM) cases and predicted non-GDM cases, respectively. A, Seeking optimal accuracy. B, Seeking optimal area under the curve (AUC). C and D, Variable selection trajectory guided by classification accuracy and AUC, respectively, under a 10-fold CV framework. E and F, Leave-one-out CV-based detailed prediction outcomes of each variable selection iteration. The yellow and blue elements represent predicted GDM cases and predicted non-GDM cases, respectively. E, Seeking optimal accuracy. F, Seeking optimal AUC. G and H, Variable selection trajectory guided by classification accuracy and AUC, respectively, under a leave-one-out CV framework.
Prediction methods
Using the selected variable panel, 4 ML methods were tested: logistic regression (LR) (21), k-nearest neighbor (KNN) (22), support vector machine (SVM) (23, 24), and deep neural network (DNN) (25, 26). For the DNN classifier, a sequential model with 2 densely connected hidden layers and a single continuous output layer was devised (more details are shown in the Supplementary text [18]). The LR classifier involved a linear combination of variables using a sigmoid function. The SVM can identify classes by creating a hyperplane of decision within a higher feature space in a nonlinear fashion (27). For the SVM classifier, a radial basis function (Gaussian) support vector model was used after considering the linear kernel, polynomial kernel, and radial basis function kernels, for which the default parameters were set as per the LIBSVM package (23). For the KNN classifier, the hyperparameter k = 20 was chosen after testing k = 1, 5, 10, 15, 20, 50, and 100, so that the KNN’s majority voting was adopted as the prediction value.
Model evaluation
The discrimination of the models was assessed using the ROC curves and the AUC. The Hosmer-Lemeshow (HL) test was used to evaluate the calibration of each model. Finally, decision curve analysis (DCA) was introduced to evaluate the clinical application of each of the models. DCA is a useful method for evaluating the clinical net benefit of prediction models by comparing it to scenarios where all or none of the patients are treated.
Results
Sample size
In total, 16 819 cases were included in the training data set, and 15 371 cases were included in the testing data set. Sociodemographic characteristics are presented in Tables 1–3. The incidence of GDM between the training data set and the testing data set had no statistical difference (16.0% vs 14.4%, P = .07). The difference of multipara rates between the training data set and the testing data set showed statistically significance (P = .004). However, the difference in multipara rates between the 2 cohorts is very small (32.9% and 31.4%). A plausible explanation for this is that the large sample size magnifies the small difference between the 2 cohorts. Generally, the sociodemographic characteristics of the 2 groups are very similar. Good consistency in the data between the training data set and the testing data set is very important, because (i) this is in line with the real clinical setting (cohort data from the same hospital in adjacent years should be similar) and (ii) if the sociodemographic characteristics of the training data set and the testing data set are too different, this will jeopardize the calibration of the model.
Table 1.
Sociodemographic characteristics of the training group and testing group
| Characteristic | 2017 training group n = 16 819 | 2018 testing group n = 15 371 | P |
|---|---|---|---|
| Age, y, median (IQR) | 31 (28-34) | 31 (28-34) | .68 |
| BMI before pregnancy (kg/m2), median (IQR) | 20.8 (19.3-22.6) | 20.5 (19.5-22.5) | .51 |
| Smoking | 95 (0.6) | 74 (0.5) | .30 |
| Educational background | |||
| Primary school degree | 15 (0.1) | 9 (0.1) | .70 |
| Junior high school degree | 388 (2.3) | 360 (2.3) | |
| High school degree | 889 (5.3) | 789 (5.1) | |
| University degree and above | 15 527 (92.3) | 14 213 (92.5) | |
| Family history of diabetes in a first-degree relative | 1202 (7.1) | 1046 (6.8) | .23 |
| GDM | 2696 (16.0) | 2216 (14.4) | .07 |
| Personal history of GDM | 176 (1.0) | 138 (0.9) | .18 |
| Natural pregnancy | 14 504 (86.2) | 13 258 (86.3) | .96 |
| Multiple pregnancy | 489 (2.9) | 466 (3.0) | .51 |
| Multipara | 5539 (32.9) | 4833 (31.4) | .004 |
Abbreviations: BMI, body mass index; GDM, gestational diabetes mellitus; IQR, interquartile range.
Table 2.
Sociodemographic characteristics of gestational diabetes mellitus (GDM) and non-GDM cases
| Characteristic | 2017 training group | P | 2018 testing group | P | ||
|---|---|---|---|---|---|---|
| GDM cases | Controls | GDM cases | Controls | |||
| n = 2696 | n = 14 123 | n = 2216 | n = 13 155 | |||
| n (%) | n (%) | n (%) | n (%) | |||
| Age, y, median (IQR) | 32 (29-36) | 30 (28-34) | < .001 | 33 (30-36) | 30 (28-33) | < .001 |
| < 38 | 2340 (86.8) | 13240 (93.7) | < .00 | 1933 (87.2) | 12324 (93.7) | < .00 |
| ≥ 38 | 356 (13.2) | 883 (6.3) | 1 | 283 (12.8) | 831 (6.3) | 1 |
| Weight, kg, before pregnancy, median (IQR) | 56.0 (52.0-62.0) | 54.5 (50.0-59.0) | < .001 | 58.0 (52.0-64.0) | 55.0 (50.0-59.0) | < .001 |
| Height, cm, median (IQR) | 161.0 (158.0-165.0) | 162.0 (159.0-165.0) | < .001 | 161.0 (158.0-165.0) | 162.0 (160.0-165.0) | < .001 |
| BMI before pregnancy (kg/m2), median (IQR) | 21.6 (20.1-23.6) | 20.7 (19.3-22.3) | < .001 | 22.1 (20.1-24.4) | 20.8 (19.5-22.1) | < .001 |
| ≤ 23 | 1620 (60.1) | 9926 (70.2) | < .00 | 1386 (62.5) | 11 064 (84.1) | < .00 |
| > 23 | 1076 (39.9) | 4197 (29.7) | 1 | 830 (37.5) | 2091 (15.9) | 1 |
| Drinking | 7 (0.3) | 39 (0.3) | 1.00 | 35 (1.6) | 218 (1.7) | 0.79 |
| Smoking | 14 (0.5) | 81 (0.6) | .89 | 13 (0.6) | 61 (0.5) | 0.44 |
| Educational background | ||||||
| Primary school degree | 5 (0.2) | 10 (0.1) | < .001 | 3 (0.1) | 6 (0.05) | < .001 |
| Junior high school degree | 102 (3.8) | 286 (2) | 65 (2.9) | 295 (2.2) | ||
| High school degree | 162 (6) | 727 (5.1) | 142 (6.4) | 647 (4.9) | ||
| University degree and above | 2427 (90.0) | 13 100 (92.8) | 2006 (90.5) | 12 207 (92.8) | ||
| Family history of diabetes in a first-degree relative | 439 (16.3) | 763 (5.4) | < .001 | 341 (15.4) | 705 (5.4) | < .001 |
Abbreviations: BMI, body mass index; GDM, gestational diabetes mellitus; IQR, interquartile range.
Table 3.
Clinical features of gestational diabetes mellitus (GDM) and non-GDM cases in the first trimester
| Characteristic | 2017 training group | P | 2018 testing group | P | ||
|---|---|---|---|---|---|---|
| GDM cases | Controls | GDM cases | Controls | |||
| n = 2696 | n = 14 123 | n = 2216 | n = 13 155 | |||
| n (%) | n (%) | n (%) | n (%) | |||
| SBP, mm Hg, median (IQR) | 114 (10-122) | 110 (102-117) | < .001 | 115 (106-124) | 110 (102-117) | < .001 |
| DBP, mm Hg, median (IQR) | 71 (65-77) | 68 (62-73) | < .001 | 71 (64-79) | 68 (62-74) | < .001 |
| PCOS | 13 (0.5) | 30 (0.2) | .02 | 30 (1.4) | 65 (0.5) | < .001 |
| Personal history of GDM | 132 (4.9) | 44 (0.3) | < .001 | 94 (4.2) | 44 (0.3) | < .001 |
| Natural pregnancy | 2180 (80.9) | 12 324 (87.3) | < .001 | 1796 (81.0) | 11 462 (87.1) | < .001 |
| Multiple pregnancy | 110 (4.1) | 379 (2.7) | < .001 | 80 (3.6) | 386 (2.9) | < .001 |
| Multipara | 1053 (39.1) | 4486 (31.8) | < .001 | 825 (37.2) | 4008 (30.5) | < .001 |
Abbreviations: DBP, diastolic blood pressure; GDM, gestational diabetes mellitus; IQR, interquartile range; PCOS, polycystic ovary syndrome; SBP, systolic blood pressure.
Variable setting
The 73 alternative variables, including sociodemographic characteristics, clinical variables, and laboratory indexes in the first trimester, are provided in Tables 1 and 2 and Supplementary Table S1 (18). Six variables, namely, age, BMI, FPG, HbA1c, high-density lipoprotein (HDL), and TG, were set as categorical variables apart from continuous variables. Previously, the ADA developed screening standards for women at high risk for gestational diabetes (28), which included BMI greater than 25 (> 23 if Asian American) and one or more of the following risk factors: HDL less than 35 mg/dL (0.9 mM); TG greater than 250 mg/dL (2.8 mM); and HbA1c greater than 5.7%. Therefore, in this study, the BMI, HDL, TG, and HbA1c binary classification threshold standards were adopted per ADA recommendations. The testing data set was used to perform the optimal scaling regression analysis between age and gestational diabetes. With the increase of age, the risk of GDM gradually increases, but the increase is not linear; after age 38 years, the risk of GDM increases faster with age (Supplementary Fig. A [18]). Therefore, we set the categorical age cutoff at age 38 years. The IADPSG uses 5.1 mM as the diagnostic criterion for early pregnancy gestational diabetes, but this has not been adopted in China because of high false-positive rates. However, pregnant women with fasting blood glucose exceeding 5.1 mM in early pregnancy will receive nutrition and exercise intervention, and thus, the FPG classification standard was set at 5.1 mM herein (12). The criteria by category are discussed in detail in the Supplementary text (18).
Variable selection
To use as much as possible of the data, we considered 2-by-2 combinations of (10-fold CV or LOO CV) and (accuracy or AUC) to select feature sets. Specifically, when using the 10-fold CV to seek the optimal accuracy for predicting GDM (accuracy = sensitivity + specificity − 1) in the training data set, 5 variables were selected and the accuracy was 0.9456. When using the LOO CV to seek the optimal accuracy, 9 variables were selected and the accuracy was 0.9356. When using the 10-fold CV to seek the optimal AUC, 14 variables were selected and the AUC was 0.8503. For the combination of the LOO CV and optimal AUC, 13 variables were selected and the AUC was 0.8503. Details are shown in Table 4. We merged all of the selected variables to obtain a 17-feature panel, namely, age, agea, FPG, FPGa, HbA1c, HbA1ca, lipoprotein(a), apolipoprotein A, apolipoprotein B, TG, TT3, total thyroxin (TT4), multiple pregnancy, multipara, smoking, family history of diabetes in a first-degree relative, and GDM history (categorical variables are denoted by a). BMI was not selected by our variable selection model. The statistics of these 17 variables in the GDM group and the control group are shown in Table 5. Compared with the control group, the GDM group is older and has higher FPG, HbA1c, apolipoprotein A, apolipoprotein B, TG, multiple pregnancy rate, multipara rate, and TT3 and lower TT4 (P < .001). The incidence of previous history of GDM and family history of diabetes in the GDM group was significantly higher than that in the control group. The obvious difference in these variables between the 2 groups indicates that these variables have strong predictive potentials. There was no significant difference in smoking rate between the GDM group and the control group (0.5% vs 0.6% in the 2017 cohort, P = .89; 0.6% vs 0.5% in the 2018 cohort, P = .44). Interestingly, smoking was still being screened out by ML as a potential GDM predictor. This agrees with a recent study that indicates smoking is an independent risk factor for GDM (29). Based on prior clinical experience and a close examination of each variable, the selected variables were further narrowed to 7 variables, practically useful for clinical implementation. To validate the selected 7 features are of high discriminatory power, we performed a simulation test comparing the selected 7 features and 7 randomly selected features. We first enumerated all of the 7-feature combinations out of the 17 features using the nchoosek function in MATLAB. Specifically, the command “nchoosek ([1:17], 7)” generated 19 448 combinations. Then, to randomly select combinations, we sequentially drew every tenth combination to obtain 1945 combinations (as detailed in Supplementary Table S6 [18]). Based on each randomly generated feature set, we performed SVM, KNN, and LR prediction using the same parameters we used for the selected 7-feature–based predictions. As demonstrated in Supplementary Fig. S3 (18), the average AUCs based on randomly selected features are significantly lower than the AUC computed based on the selected 7-feature–based predictions, and in the best LR prediction model, the AUC based on the selected 7 features is higher than the maximum AUC of all randomly drawn feature combinations (0.77 vs 0.70, P < .001).
Table 4.
Selecting variables by k-nearest neighbor
| 10-fold (accuracy) | LOO (accuracy) | 10-fold (AUC) | LOO (AUC) | |
|---|---|---|---|---|
| Selected variables | FPGa | FPG | FPB | FPG |
| Lipoprotein(a) | FPGa | FPGa | HbA1c | |
| Total 3,5,3′-triiodothyronine | Lipoprotein(a) | Lipoprotein(a) | Family history of diabetes in a first-degree relative | |
| Agea | Total 3,5,3′-triiodothyronine | Total 3,5,3′-triiodothyronine | Triglyceride | |
| Multiple pregnancy | Age | Triglyceride | Age | |
| Total thyroxin | Age | Total 3,5,3′-triiodothyronine | ||
| ApoA | HbA1ca | Lipoprotein(a) | ||
| Multipara | Total thyroxin | Agea | ||
| Multiple pregnancy | Agea | Total thyroxin | ||
| ApoB | Multipara | |||
| Multipara | ApoA | |||
| Previous GDM | Multiple pregnancy | |||
| Multiple pregnancy | Previous GDM | |||
| Smoking |
Using the 10-fold method, 5 variables were selected to obtain optimal accuracy; using the LOO method, 9 variables were selected to obtain optimal accuracy; using the 10-fold method, 14 variables were selected to obtain the optimal ROC area; using the LOO method, 13 variables were selected to obtain the optimal ROC area.
Abbreviations: ApoA, apolipoprotein A; ApoB, apolipoprotein B; AUC, area under the curve; FPG, fasting plasma glucose; GDM, gestational diabetes mellitus; HBA1c, glycated hemoglobin; LOO, leave-one-out; ROC, receiver operating characteristic.
a Categorical variable: age younger than 38 years: 0, age 38 years or older: 1; FPG less than 5.1 mmol/L: 0, FPG 5.1 or greater and less than 7.0 mmol/L: 1; HbA1c 5.7 or less: 0, HbA1c greater than 5.7 and less than 6.5: 1.
Table 5.
Selected 17 variables in the training group and testing group
| Characteristic | 2017 training group | P | 2018 testing group | P | ||
|---|---|---|---|---|---|---|
| GDM cases | Controls | GDM cases | Control | |||
| n = 2696 n (%) | n = 14 123 n (%) | n = 2216 n (%) | n = 13 155 n (%) | |||
| Age, y, median (IQR) | 32 (29-36) | 30 (28-34) | < .001 | 33 (30-36) | 30 (28-33) | < .001 |
| Agea, ≥ 38 y | 356 (13.2) | 883 (6.3) | < .001 | 283 (12.8) | 831 (6.3) | < .001 |
| Smoking | 14 (0.5) | 81 (0.6) | .89 | 13 (0.6) | 61 (0.5) | .44 |
| Family history of diabetes in a first-degree relative | 439 (16.3) | 763 (5.4) | < .001 | 341 (15.4) | 705 (5.4) | < .001 |
| Personal history of GDM | 132 (4.9) | 44 (0.3) | < .001 | 94 (4.2) | 44 (0.3) | < .001 |
| Multiple pregnancy | 110 (4.1) | 379 (2.7) | < .001 | 80 (3.6) | 386 (2.9) | < .001 |
| Multipara | 1053 (39.1) | 4486 (31.8) | < .001 | 825 (37.2) | 4008 (30.5) | < .001 |
| ApoA | 2.01 (1.93-2.08) | 1.98 (1.94-2.02) | < .001 | 2.15 (2.01-2.29) | 2.14 (2.01-2.27) | .17 |
| ApoB | 0.89 (0.84-0.94) | 0.85 (0.84-0.88) | < .001 | 0.79 (0.70-0.91) | 0.74 (0.65-0.85) | < .001 |
| Triglyceride | 1.47 (1.15-1.89) | 1.22 (0.97-1.52) | < .001 | 1.49 (1.16-1.93) | 1.24 (0.98-1.57) | < .001 |
| Lipoprotein(a) | 157.8 (101.5-185.9) | 191.2 (173.3-210.9) | < .001 | 103.0 (46.0-216.3) | 123.0 (57.0-232.0) | < .001 |
| FPG, mM | 4.77 (4.49-5.13) | 4.50 (4.30-4.70) | < .001 | 4.78 (4.50-5.14) | 4.54 (4.33-4.73) | < .001 |
| FPGa, ≥ 5.1 and < 7.0 mM, n (%) | 766 (28.4) | 494 (3.5) | < .001 | 614 (27.7) | 400 (3.0) | < .001 |
| HbA1c, % | 5.3 (5.1-5.5) | 5.1 (5.0-5.3) | < .001 | 5.4 (5.2-5.6) | 5.2 (5.1-5.4) | < .001 |
| HbA1ca, > 5.7 and < 6.5, n (%) | 179 (6.6) | 71 (0.5) | < .001 | 241 (10.9) | 131 (1.0) | < .001 |
| Total thyroxin, pM | 114.2 (106.6-119.0) | 116.0 (112.6-120.1) | < .001 | 115.7 (99.4-132.9) | 118.9 (102.8-134.2) | < .001 |
| Total 3,3,5′-triiodothyronine, nM | 2.10 (2.00-2.23) | 2.02 (1.97-2.08) | < .001 | 2.10 (1.90-2.40) | 2.00 (1.80-2.30) | < .001 |
Abbreviations: ApoA, apolipoprotein A; ApoB, apolipoprotein B; FPG, fasting plasma glucose; GDM, gestational diabetes mellitus; HBA1c, glycated hemoglobin.
a Categorical variable: age younger than 38 years: 0, age 38 years or older: 1; FPG less than 5.1 mmol/L: 0, FPG 5.1 or greater and less than 7.0 mmol/L: 1; HbA1c 5.7 or less: 0, HbA1c greater than 5.7 and less than 6.5: 1.
Development of prediction models
Eight prediction models were developed: KNN, SVM, LR, and DNN models were developed for both 7-variable and all-variable sets. The adjusted odds ratios (ORs) and coefficients from the LR model with 7 variables are shown in Table 6.
Table 6.
Multivariate analysis for the 7-variable logistic regression model
| β | Adjusted odds ratio (95% CI) | P | |
|---|---|---|---|
| Intercept | −14.2334 | – | < .001 |
| Age | .0681 | 1.070 (1.058-1.083) | < .001 |
| Previous GDM | 2.6181 | 13.710 (9.532-19.718) | < .001 |
| Family history of diabetes in a first-degree relative | 1.1062 | 3.023 (2.610-3.501) | < .001 |
| Multiple pregnancy | .4349 | 1.545 (1.208-1.976) | .001 |
| FPGa | 2.8165 | 16.718 (14.125-19.788) | < .001 |
| HBA1c | 1.6925 | 5.433 (4.472-6.600) | < .001 |
| Triglyceride | .5005 | 1.650 (1.528-1.781) | < .001 |
Abbreviations: FPG, fasting plasma glucose; GDM, gestational diabetes mellitus; HBA1c, glycated hemoglobin.
a Categorical variable: FPG less than 5.1 mM: 0, FPG 5.1 mM or greater: 1.
Discrimination of different models
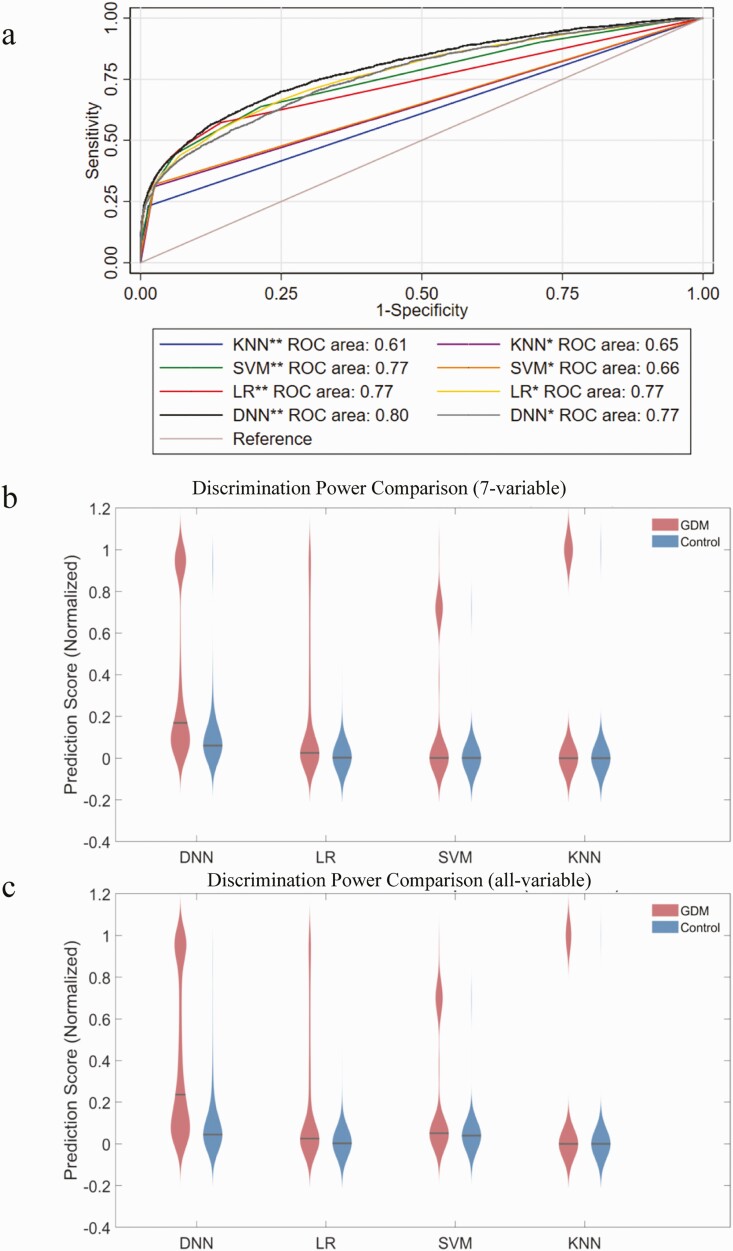
The AUCs of different models are provided in Fig. 3A and Table 7. The all-variable DNN, SVM, KNN, and LR models had AUCs and 95% CIs of 0.80 (95% CI, 0.79-0.81), 0.77 (95% CI, 0.76-0.78), 0.61 (95% CI, 0.59-0.62), and 0.77 (95% CI, 0.76-0.78), respectively. The 7-variable DNN, SVM, KNN, and LR models had AUCs and 95% CIs of 0.77 (95% CI, 0.76-0.78), 0.66 (95% CI, 0.65-0.67), 0.65 (95% CI, 0.63-0.66), and 0.77 (95% CI, 0.76-0.78), respectively. The discrimination effect of each model is shown visually using violin plots (Fig. 3B and 3C). The all-variable DNN demonstrated the best discrimination ability, and the traditional LR models produced higher AUCs than the KNN and SVM models. The optimal sensitivity and specificity of each model in certain threshold probability value ranges are given in Table 7. The accuracy of previous prediction models has also been summarized; existing models do not exceed 0.70 and our model achieved the highest discrimination (Supplementary Table S2 [18]).
Figure 3.
Discriminative power comparison between different prediction models. A, Receiver operating characteristic (ROC) curves of different prediction models based on the 7-variable panel (*) and all-variable panel (**). B and C, Violin plot comparisons of predicted score distribution using different prediction models with the 7-variable panel and the all-variable panel.
Table 7.
Sensitivity and specificity of different models
| Prediction model | AUC (95% CI) | Optimum threshold probability | Sensitivity, % | Specificity, % | Youden index |
|---|---|---|---|---|---|
| LRa | 0.77 (0.76-0.78) | 0.13 | 59 | 82 | 0.41 |
| LRb | 0.77 (0.76-0.78) | 0.02 | 58 | 86 | 0.44 |
| KNNa | 0.65 (0.63-0.66) | – | 31 | 98 | 0.29 |
| KNNb | 0.61 (0.59-0.62) | – | 23 | 99 | 0.22 |
| SVMa | 0.66 (0.65-0.67) | 0.14 | 32 | 98 | 0.30 |
| SVMb | 0.77 (0.76-0.78) | 0.15 | 32 | 98 | 0.30 |
| DNNa | 0.77 (0.76-0.78) | 0.10 | 70 | 69 | 0.39 |
| DNNb | 0.80 (0.79-0.81) | 0.15 | 63 | 82 | 0.45 |
Abbreviations: AUC, area under the curve; DNN, deep neural network; KNN, k-nearest neighbor; LR, logistic regression; SVM, support vector machine.
a Seven-variable model.
b All-variable model.
Calibration of different models
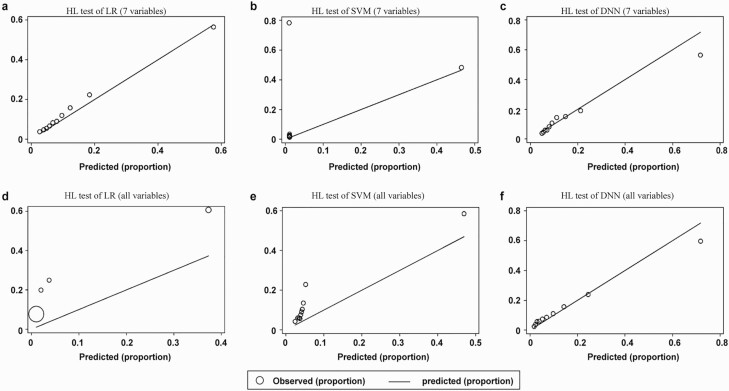
The HL test was used to test the calibration of the LR, SVM, and DNN models (Fig. 4). The HL test was not applied to the KNN models because the model did not provide individual risk probabilities. The P values of 6 different models were less than .001 in the HL test. The 7-variable models (Fig. 4A-4C) showed superior HL test performance compared to the all-variable models (Fig. 4D-4F). The 7-variable LR model provided the most accurate calibration among all the prediction models.
Figure 4.
Calibration of different models. The P values of all prediction models in Hosmer-Lemeshow (HL) tests are less than .001. The 7-variable models, A to C, show superior HL test performance compared to D to F, the all-variable models. This is because if the model incorporates all of the variables without selection, it will inevitably overfit, which will significantly affect the model calibration.
Clinical use
The DCA results of the models are presented in Supplementary Fig. S4 (18). Compared to treating all patients or none of the patients, our prediction models provide a net benefit.
Discussion
Our paper explores prediction models based on a large sample of the Chinese population using clinical data before 12 weeks of gestation, 2 months earlier than previous state-of-the-art ML models. We used ML variable selection methods to screen for risk factors for early development of GDM. Of the 73 extracted variables, 17 variables were selected for our models, which included sociodemographic data (age, agea, smoking, and family history of diabetes in a first-degree relative), clinical characteristics (multiple pregnancy, multipara, and previous GDM history), glucose metabolism (FPG, FPGa, HbA1c, and HbA1ca), lipid metabolism (lipoprotein[a], apolipoprotein A, apolipoprotein B, TG), and thyroid function (TT3, TT4). Of these 17 variables, 7 were selected based on intravariable correlation and clinical importance for our parsimonious model: age, family history of diabetes in a first-degree relative, multiple pregnancy, previous GDM history, FPGa, HbA1c, and TG. Details of how the 7 variables were selected are discussed in the Supplementary text (18). As shown in Fig. 3, our all-variable DNN model achieved the highest accuracy in predicting GDM in early pregnancy, followed by SVM and KNN. Our parsimonious models using 7 variables performed similarly and with increased efficiency. The DCA of the different models also showed similar results (Supplementary text; Supplementary Fig. S4 [18]).
Model comparisons
The advantage of DNN is its ability to capture subtle nonlinear relationships between variables and outcomes. However, DNNs have a risk of overfitting, and because DNN is a black box to end users, the individual weighted contribution of each variable can be difficult to explain (30). On the other hand, LR highlights a clear contribution of each variable, making it useful for real-time clinical implementation. Our method of including only the important variables in each model resulted in a negligible running time difference between prediction models.
The HL test was adopted to evaluate the calibration of prediction models (31). As KNN only results in a binary outcome rather than individual predicted probabilities, the HL test and DCA curve were not used to evaluate these models. The P values of all the models for the HL test were less than .001, which implied that the model calibrations were not optimal. This shows that although the models were to be able to distinguish high-risk status of GDM in early pregnancy, the specific risk probabilities provided by these models can be further improved (32). However, the 7-variable LR model revealed slightly better calibration than the DNN model. This may be due to the poor correlation between threshold probability and risk probability in DNN and SVM, indicating the HL test is not optimal to measure calibration for complex ML models. Furthermore, compared to existing LR prediction models (Supplementary Table S2 [18]), our 7-variable LR and DNN models demonstrated very promising results in predicting GDM in early pregnancy.
There have been limited studies predicting GDM using ML algorithms. A retrospective electronic medical record study with 580 000 pregnancies in Israel reported an AUC of 0.85 using all variables and an AUC of 0.80 using only 9 variables (17). However, the clinical data collected from studies in Israel were obtained at 20 weeks of pregnancy, unlike our prediction using variables extracted only from the first trimester. This allowed them to use variables that are useful only during the second and third trimesters to predict GDM, such as human placental growth hormone, human chorionic somatomammotropin, progesterone, and placental growth hormone (33).
Risk factor evaluation
The selected variables were found to be consistent with previous clinical studies. Advanced age, previous GDM history, family history of diabetes, and blood glucose are well-known risk factors of GDM (34). Women with twin pregnancies have an increased risk of GDM, and higher rates of adverse pregnancy outcomes occur in GDM twin pregnancies (35). HbA1c reflects the average blood glucose levels over the last 1 to 2 months (36, 37). Previous studies hypothesized that the link between higher parity and insulin resistance could be explained by the decreasing β-cell reserve in consecutive pregnancies (38, 39). However, prediction models showed that parity plays a more complicated role, with multipara without previous GDM history reducing the risk of future GDM (OR = 0.5, P = .05), and multipara with previous GDM history increasing the risk of future GDM (OR = 1.6, P = .55) (40, 41). We therefore believe that parity, when used with other selected variables, is conditionally correlated to GDM, and that its predictive power can be increased through such a combination.
Lipoprotein(a) was one of the 17 predictors and demonstrated high prediction power (AUC = 0.66, 95% CI, 0.65-0.68). Previous studies indicated that high levels of TGs and apolipoproteins are risk factors for GDM (42, 43). However, lipoprotein(a) transports oxidized phospholipids that have proinflammatory activity, so the possible association of higher lipoprotein(a) levels with GDM remains controversial (44, 45). For our model, the predicted effect of lipoprotein was better than that of apolipoproteins (Supplementary Table S3 [18]). The reasons for this are not known.
Despite obesity being a well-known risk factor for GDM, our variable selection model did not choose BMI, instead highlighting biochemical indicators that reflect the level of lipid metabolism, such as TG. There are several explanations for this. First, compared to Europeans, Asians have more subcutaneous fat and higher s-leptin levels in early pregnancy, despite having lower BMI (46). Second, the relationship between BMI and GDM is complex, with high BMI individuals having an insulin resistance mechanism and low BMI individuals having a defective insulin secretion mechanism in GDM (47, 48). Our study showed that both an increased BMI and a very low BMI (≤ 17) (n = 432) are related to an increased risk of GDM (Supplementary Fig. S5 [18]), compared to a BMI in the range of 17 to 18 (minimum risk interval) (n = 915), but this association was not statistically significant (11.8% vs 8.7%, P = .09). Existing studies have not shown that extremely low BMI could increase the risk of GDM (17), but it has been found that BMI had J-shaped associations with overall mortality and diabetes mortality (48), supporting our findings.
A large portion of the selected variables were of a biochemical nature (Supplementary Table S1 [18]). For example, TT3 and TT4 were selected as predictors of GDM, strongly suggesting the existence of a close relationship between thyroid function and GDM. In our training group, the GDM group had higher levels of TT3 (median, 2.1 nM vs 2.02 nM, P < .001) and free 3,5,3′-triiodothyronine (FT3) (median, 4.80 pM vs 4.60 pM, P < .001) and lower levels of TT4 (median, 114.2 nM vs 116.0 nM, P < .001) and free thyroxin (FT4) (median, 13.6 pM vs 14.0 pM, P < .001) compared to the non-GDM group. This result was consistent with previous studies (49, 50). Current research findings remain divided with respect to the question whether high T3 or low T4 in early pregnancy is a risk factor for GDM, as this may be affected by variations between populations (49-51). A study from a US cohort showed that FT4 was not associated with GDM, but high FT4-FT3 conversion efficiency (increased FT3/FT4 ratio) increased the risk of GDM (51). Several studies noted that FT3 levels were positively associated with insulin secretion and hyperinsulinemia (52). A study of the Chinese population suggested that increasing FT4 levels functioned as a protective mechanism against GDM, in that higher FT4 levels were associated with a lower incidence of GDM (P < .001) (49). Most prior research has focused on the relationship between FT3 and FT4 and GDM, because FT3 and FT4 have much higher biological activity than TT3 and TT4 and can directly reflect thyroid function (51). Interestingly, when we included thyroxine in the prediction model, the TT3 and TT4 levels had better predictive power than FT3 and FT4 (Supplementary Table S4 [18]). This suggests that the relationship between thyroxine and GDM is conditionally dependent on factors such as TT3 and TT4. However, further research on the relationships among TT4, FT4, and the risk of GDM in the Chinese population is needed.
Limitations
The limitations of this study include the limited sample size, the fact that all the data were collected from a single center, and a lack of external verification. The prediction model is based on retrospective electronic medical data that many have inherent biases. However, electronic medical records are easily available clinical data resources, and predicting GDM based on electronic medical records is often the most feasible option. The diversity of laboratory testing between different hospitals caused by different laboratory instruments may also influence the effects of prediction and extrapolation. However, these shortcomings do not change the fact that the proposed variable selection and ML-based methodology itself are worthy of attention in early GDM prediction. In future work, we plan to collect multicenter clinical data to verify the extrapolation of these prediction models.
Conclusions
This study established state-of-the-art prediction models in early pregnancy for the early intervention of GDM in Chinese women. Using an ML-based variable selection approach, 17 important GDM predictive variables were selected. These 17 indicators are worthy of in-depth study in the GDM field; in particular, lipoprotein(a) may be closely related to GDM. A 7-variable LR model was developed for more practical clinical applications. Further research is required to clarify the relationship among TT4, FT4, and GDM and between excessively low BMI and GDM in the Chinese population.
Acknowledgments
We thank all those who helped to collect the data and the graduate students who took part in the statistical analysis.
Financial Support: This work was supported by the National Key Research and Development Program of China (grant Nos. 2018YFC1002804 and 2016YFC1000203), the National Natural Science Foundation of China (grant Nos. 81671412 and 81661128010), Program of Shanghai Academic Research Leader (grant No. 20XD1424100), the Outstanding Youth Medical Talents of Shanghai Rising Stars of Medical Talent Youth Development Program, Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (grant No. 2019-12M-5-064), the Foundation of Shanghai Municipal Commission of Health and Family Planning (grant No. 20144Y0110), the Natural Science Foundation of Shanghai (grant Nos. 20511101900 and 20ZR1427200), the Shanghai Shenkang Hospital Development Center, the Clinical Technology Innovation Project (grant Nos. SHDC12019107), and the Clinical Skills Improvement Foundation of Shanghai Jiaotong University School of Medicine (grant No. JQ201717).
Author Contributions: Yan-Ting Wu contributed to the research idea and was the project coordinator. Chen-Jie Zhang contributed to the manuscript drafting. Ben Willem Mol and Andrew Kawai contributed to manuscript modification. Cheng Li contributed to figure production and modification. Lei Chen contributed to the data export from the hospital electronic medical record system. Yu Wang, Jian-Zhong Sheng, and Jian-Xia Fan provided useful suggestions. Yi Shi contributed to the ML algorithm and statistical analysis. He-Feng Huang provided project guidance and financial support. All authors read and approved the final manuscript.
Glossary
Abbreviations
- ADA
American Diabetes Association
- AUC
area under the curve
- BMI
body mass index
- CV
cross-validation
- DCA
decision curve analysis
- DNN
deep neural network
- FPG
fasting plasma glucose
- FT3
free 3,5,3′-triiodothyronine
- FT4
free thyroxin
- GDM
gestational diabetes mellitus
- HbA1c
glycated hemoglobin
- HDL
high-density lipoprotein
- HL
Hosmer-Lemeshow
- IADPSG
International Association of Diabetes and Pregnancy Study Groups
- KNN
k-nearest neighbor
- LDL
low-density lipoprotein
- LOO
leave-one-out
- LR
logistic regression
- ML
machine learning
- OGTT
oral glucose tolerance test
- ROC
receiver operating characteristic
- SVM
support vector machine
- TG
triglyceride
- TT3
total 3,3,5′-triiodothyronine
- TT4
total thyroxin
Additional Information
Disclosures: The authors have nothing to disclose.
Data Availability
Some or all data sets generated during and/or analyzed during the present study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Gabbe SG. Gestational diabetes mellitus. N Engl J Med. 1986;315(16):1025-1026. [DOI] [PubMed] [Google Scholar]
- 2. Chiefari E, Arcidiacono B, Foti D, Brunetti A. Gestational diabetes mellitus: an updated overview. J Endocrinol Invest. 2017;40(9):899-909. [DOI] [PubMed] [Google Scholar]
- 3. American Diabetes Association. Classification and diagnosis of diabetes: Standards of Medical Care in Diabetes—2018. Diabetes Care. 2018;41(Suppl 1):S13-S27. [DOI] [PubMed] [Google Scholar]
- 4. Weinert LS. International Association of Diabetes and Pregnancy Study Groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy: comment to the International Association of Diabetes and Pregnancy Study Groups Consensus Panel. Diabetes Care. 2010;33(7):e97; author reply e98. [DOI] [PubMed] [Google Scholar]
- 5. Ding GL, Wang FF, Shu J, et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61(5):1133-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dabelea D, Hanson RL, Lindsay RS, et al. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes. 2000;49(12):2208-2211. [DOI] [PubMed] [Google Scholar]
- 7. Dabelea D, Mayer-Davis EJ, Lamichhane AP, et al. Association of intrauterine exposure to maternal diabetes and obesity with type 2 diabetes in youth: the SEARCH Case-Control Study. Diabetes Care. 2008;31(7):1422-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tam WH, Ma RCW, Ozaki R, et al. In utero exposure to maternal hyperglycemia increases childhood cardiometabolic risk in offspring. Diabetes Care. 2017;40(5):679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sletner L, Jenum AK, Yajnik CS, et al. Fetal growth trajectories in pregnancies of European and South Asian mothers with and without gestational diabetes, a population-based cohort study. PLoS One. 2017;12(3):e0172946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sovio U, Murphy HR, Smith GC. Accelerated fetal growth prior to diagnosis of gestational diabetes mellitus: a prospective cohort study of nulliparous women. Diabetes Care. 2016;39(6):982-987. [DOI] [PubMed] [Google Scholar]
- 11. Zhu H, Chen B, Cheng Y, et al. Insulin therapy for gestational diabetes mellitus does not fully protect offspring from diet-induced metabolic disorders. Diabetes. 2019;68(4):696-708. [DOI] [PubMed] [Google Scholar]
- 12. Zhu WW, Yang HX, Wei YM, et al. Evaluation of the value of fasting plasma glucose in the first prenatal visit to diagnose gestational diabetes mellitus in china. Diabetes Care. 2013;36(3):586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leong I. Diabetes: ANGPTL8 as an early predictor of gestational diabetes mellitus. Nat Rev Endocrinol. 2018;14(2):64. [DOI] [PubMed] [Google Scholar]
- 14. Ning H, Tao H, Weng Z, Zhao X. Plasma fatty acid-binding protein 4 (FABP4) as a novel biomarker to predict gestational diabetes mellitus. Acta Diabetol. 2016;53(6):891-898. [DOI] [PubMed] [Google Scholar]
- 15. Bao W, Baecker A, Song Y, Kiely M, Liu S, Zhang C. Adipokine levels during the first or early second trimester of pregnancy and subsequent risk of gestational diabetes mellitus: a systematic review. Metabolism. 2015;64(6):756-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naylor CD, Sermer M, Chen E, Farine D. Selective screening for gestational diabetes mellitus. Toronto Trihospital Gestational Diabetes Project Investigators. N Engl J Med. 1997;337(22):1591-1596. [DOI] [PubMed] [Google Scholar]
- 17. Artzi NS, Shilo S, Hadar E, et al. Prediction of gestational diabetes based on nationwide electronic health records. Nat Med. 2020;26(1):71-76. [DOI] [PubMed] [Google Scholar]
- 18. Huang HF. Data from: supplementary materials for JCEM. OSF. Deposited July 18, 2020. 10.17605/OSF.IO/H4E78 [DOI] [Google Scholar]
- 19. Shi Y, Cai Z, Xu L, Ren W, Goebel R, Lin G. A model-free greedy gene selection for microarray sample class prediction. 2006 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (IEEE CIBCB), Toronto, Ontario, Canada, September 28-29. 2006:406-417. [Google Scholar]
- 20. Xiong M, Fang X, Zhao J. Biomarker identification by feature wrappers. Genome Res. 2001;11(11):1878-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cramer JS. The Origins of Logistic Regression (Technical Report). 119. Tinbergen Institute; 2002:167-178. https://papers.tinbergen.nl/02119.pdf [Google Scholar]
- 22. Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc. 2002;97(457):77-87. [Google Scholar]
- 23. Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2(27):1-27. [Google Scholar]
- 24. Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46:389-422. [Google Scholar]
- 25. Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504-507. [DOI] [PubMed] [Google Scholar]
- 26. Deng L, Yu D. Deep learning: methods and applications. Found Trends Signal Process. 2014;7(3-4):197-387. [Google Scholar]
- 27. Nanayakkara S, Fogarty S, Tremeer M, et al. Characterising risk of in-hospital mortality following cardiac arrest using machine learning: a retrospective international registry study. PLoS Med. 2018;15(11):e1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buschur E, Stetson B, Barbour L. Diabetes in pregnancy. In: Feingold KR, Anawalt B, Boyce A, et al, eds. Endotext, MDText.com Inc; 2018:1- 14. https://www.ncbi.nlm.nih.gov/books/NBK279010/ [Google Scholar]
- 29. Bar-Zeev Y, Haile ZT, Chertok IA. Association between prenatal smoking and gestational diabetes mellitus. Obstet Gynecol. 2020;135(1):91-99. [DOI] [PubMed] [Google Scholar]
- 30. Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet. 2019;393(10181):1577-1579. [DOI] [PubMed] [Google Scholar]
- 31. Bertolini G, D’Amico R, Nardi D, Tinazzi A, Apolone G. One model, several results: the paradox of the Hosmer-Lemeshow goodness-of-fit test for the logistic regression model. J Epidemiol Biostat. 2000;5(4):251-253. [PubMed] [Google Scholar]
- 32. Alba AC, Agoritsas T, Walsh M, et al. Discrimination and calibration of clinical prediction models: users’ guides to the medical literature. JAMA. 2017;318(14):1377-1384. [DOI] [PubMed] [Google Scholar]
- 33. Barbour LA, McCurdy CE, Hernandez TL, Kirwan JP, Catalano PM, Friedman JE. Cellular mechanisms for insulin resistance in normal pregnancy and gestational diabetes. Diabetes Care. 2007;30(Suppl 2):S112-S119. [DOI] [PubMed] [Google Scholar]
- 34. Zhu Y, Zhang C. Prevalence of gestational diabetes and risk of progression to type 2 diabetes: a global perspective. Curr Diab Rep. 2016;16(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ooi S, Wong VW. Twin pregnancy with gestational diabetes mellitus: a double whammy? Diabetes Care. 2018;41(2):e15-e16. [DOI] [PubMed] [Google Scholar]
- 36. Rahbar S, Blumenfeld O, Ranney HM. Studies of an unusual hemoglobin in patients with diabetes mellitus. Biochem Biophys Res Commun. 1969;36(5):838-843. [DOI] [PubMed] [Google Scholar]
- 37. Koenig RJ, Peterson CM, Jones RL, Saudek C, Lehrman M, Cerami A. Correlation of glucose regulation and hemoglobin AIc in diabetes mellitus. N Engl J Med. 1976;295(8):417-420. [DOI] [PubMed] [Google Scholar]
- 38. Peters RK, Kjos SL, Xiang A, Buchanan TA. Long-term diabetogenic effect of single pregnancy in women with previous gestational diabetes mellitus. Lancet. 1996;347(8996):227-230. [DOI] [PubMed] [Google Scholar]
- 39. Schwartz N, Nachum Z, Green MS. The prevalence of gestational diabetes mellitus recurrence—effect of ethnicity and parity: a metaanalysis. Am J Obstet Gynecol. 2015;213(3):310-317. [DOI] [PubMed] [Google Scholar]
- 40. Sweeting AN, Appelblom H, Ross GP, et al. First trimester prediction of gestational diabetes mellitus: a clinical model based on maternal demographic parameters. Diabetes Res Clin Pract. 2017;127:44-50. [DOI] [PubMed] [Google Scholar]
- 41. van Leeuwen M, Opmeer BC, Zweers EJ, et al. Estimating the risk of gestational diabetes mellitus: a clinical prediction model based on patient characteristics and medical history. BJOG. 2010;117(1):69-75. [DOI] [PubMed] [Google Scholar]
- 42. Ryckman KK, Spracklen CN, Smith CJ, Robinson JG, Saftlas AF. Maternal lipid levels during pregnancy and gestational diabetes: a systematic review and meta-analysis. BJOG. 2015;122(5):643-651. [DOI] [PubMed] [Google Scholar]
- 43. Ravnsborg T, Andersen LL, Trabjerg ND, Rasmussen LM, Jensen DM, Overgaard M. First-trimester multimarker prediction of gestational diabetes mellitus using targeted mass spectrometry. Diabetologia. 2016;59(5):970-979. [DOI] [PubMed] [Google Scholar]
- 44. Todoric J, Handisurya A, Leitner K, Harreiter J, Hoermann G, Kautzky-Willer A. Lipoprotein(a) is not related to markers of insulin resistance in pregnancy. Cardiovasc Diabetol. 2013;12:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aydemir B, Behice Serinkan Cinemre F, Cinemre H, et al. Paraoxonase 1 (PON1) Q192R and L55M polymorphisms, lipid profile, lipid peroxidation and lipoprotein-a levels in Turkish patients with pregnancy-related disorders. Gynecol Endocrinol. 2019;35(5):417-421. [DOI] [PubMed] [Google Scholar]
- 46. Sommer C, Jenum AK, Waage CW, Mørkrid K, Sletner L, Birkeland KI. Ethnic differences in BMI, subcutaneous fat, and serum leptin levels during and after pregnancy and risk of gestational diabetes. Eur J Endocrinol. 2015;172(6): 649-656. [DOI] [PubMed] [Google Scholar]
- 47. Kautzky-Willer A, Prager R, Waldhausl W, et al. Pronounced insulin resistance and inadequate β-cell secretion characterize lean gestational diabetes during and after pregnancy. Diabetes Care. 1997;20(11):1717-1723. [DOI] [PubMed] [Google Scholar]
- 48. Bhaskaran K, Dos-Santos-Silva I, Leon DA, Douglas IJ, Smeeth L. Association of BMI with overall and cause-specific mortality: a population-based cohort study of 3·6 million adults in the UK. Lancet Diabetes Endocrinol. 2018;6(12):944-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang S, Shi FT, Leung PC, Huang HF, Fan J. Low thyroid hormone in early pregnancy is associated with an increased risk of gestational diabetes mellitus. J Clin Endocrinol Metab. 2016;101(11):4237-4243. [DOI] [PubMed] [Google Scholar]
- 50. Agarwal MM, Dhatt GS, Punnose J, Bishawi B, Zayed R. Thyroid function abnormalities and antithyroid antibody prevalence in pregnant women at high risk for gestational diabetes mellitus. Gynecol Endocrinol. 2006;22(5):261-266. [DOI] [PubMed] [Google Scholar]
- 51. Rawal S, Tsai MY, Hinkle SN, et al. A longitudinal study of thyroid markers across pregnancy and the risk of gestational diabetes. J Clin Endocrinol Metab. 2018;103(7):2447-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bakker SJ, ter Maaten JC, Popp-Snijders C, Heine RJ, Gans RO. Triiodothyronine: a link between the insulin resistance syndrome and blood pressure? J Hypertens. 1999;17(12 Pt 1):1725-1730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data sets generated during and/or analyzed during the present study are not publicly available but are available from the corresponding author on reasonable request.