Abstract
The mitochondrial genetic code is much more varied than the standard genetic code. The invertebrate mitochondrial code, for instance, comprises six initiation codons, including five alternative start codons. However, only two initiation codons are known in the echinoderm and flatworm mitochondrial code, the canonical ATG and alternative GTG. Here, we analyzed 23 Asteroidea mitogenomes, including ten newly sequenced species and unambiguously identified at least two other start codons, ATT and ATC, both of which also initiate translation of mitochondrial genes in other invertebrates. These findings underscore the diversity of the genetic code and expand upon the suite of initiation codons among echinoderms to avoid erroneous annotations. Our analyses have also uncovered the remarkable conservation of gene order among asteroids, echinoids, and holothuroids, with only an interchange between two gene positions in asteroids over ∼500 Ma of echinoderm evolution.
Keywords: mitochondrial phylogeny, Asteroidea, mitochondrial gene evolution, mitochondrial gene order, Echinodermata, translation table
In 1968, Crick proposed that although the overall genetic code is likely to be conserved through deep evolutionary time, translation initiation codons may differ among taxa (Crick 1968). Deviations from the standard code were first discovered in the human mitochondrion (Barrell et al. 1979), with a multitude of code variations that have since been uncovered (Lobanov et al. 2010). A key difference between the standard and alternative codes is in the initiation codon, for which the canonical ATG is now known to be just one of numerous possibilities. For example, up to 47 possible start codons have been found in the model bacterium Escherichia coli (Hecht et al. 2017).
The mitochondrial genes of echinoderms are translated according to a variant genetic code (translation table 9) distinct from the standard code. Based on a 3,849-bp fragment of mitochondrial DNA from Asterina pectinifera (Echinodermata: Asteroidea), Himeno et al. (1987) uncovered a number of variations in the genetic code, such as AGA and AGG coding for serine instead of arginine, and hypothesized that ATT and ATA could be alternative initiation codons for the NADH dehydrogenase subunit 3 (ND3) and NADH dehydrogenase subunit 5 (ND5) genes, respectively (Asakawa et al. 1995). Apart from the standard ATG, the only accepted alternative mitochondrial initiation codon for echinoderms is GTG (Jacobs et al. 1988; Cantatore et al. 1989). The aforementioned studies have suggested ATA, ATC, and ATT as possible start codons, although there have been reservations (Cantatore et al. 1989). To date, the validity of initiation codons in the echinoderm mitochondrial code other than ATG and GTG remains uncertain. Conversely, the mitochondrial genetic code of other invertebrates (translation table 5) comprises five alternative initiation codons—the two present in the echinoderm mitochondrial code, along with ATC, ATT, and TTG. Interestingly, ATT is the most frequently annotated start codon for ND3 among invertebrates in an investigation of over 900 mitogenomes, although ATG remains the dominant initiation codon for all protein-coding genes (PCGs) in translation tables 5 and 9 (Donath et al. 2019). The same study also found that multiple other initiation codons have been annotated for echinoderms, including all alternative start codons in translation table 5 plus TTT, TAT, GAT, and CTG.
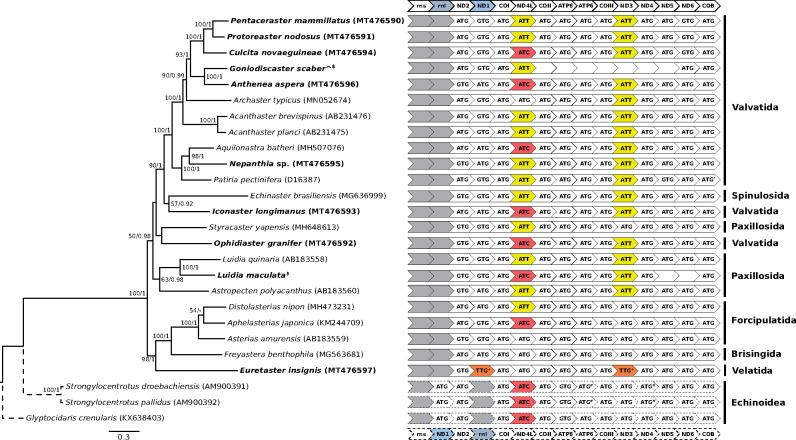
In this study, we assembled eight new complete and three incomplete mitogenomes across ten species of asteroid sea stars for analysis with 16 published mitogenomes. For two genes, ND3 and NADH dehydrogenase subunit 4L (ND4L), we found unambiguous evidence for several species initiating with either ATT (Asteroidea only; ND3, n = 16; and ND4L, n = 12) or ATC (ND4L only; Asteroidea, n = 7; and Echinoidea, n = 3) (Fig. 1), but no taxa had mitochondrial initiation codon ATA (Materials and Methods and see Cantatore et al. 1989). We furthermore found preliminary evidence in Euretaster insignis for TTG being a fourth alternative initiation codon through reannotation of the NADH dehydrogenase subunit 1 (ND1) to be in frame with the other mitogenomes. TTG was also the initiation codon for ND3 (as annotated by MITOS2; Bernt et al. 2013b). Indeed, inspection of our ND3 alignment revealed that E. insignis had one less codon at the 5′ end than other echinoderm species. The codon in ND3 that preceded TTG, AAT, is not a known mitochondrial initiation codon in echinoderms or even invertebrates more generally, although it is a possible start codon in Escherichia coli (Hecht et al. 2017). Nevertheless, TTG is an established alternative initiation codon in invertebrate mitochondria (Okimoto et al. 1990), so it possibly also initiates translation of ND1 and ND3 in E. insignis and other echinoderms (see Donath et al. 2019). More mitogenomes from its close relatives (order Velatida) would clarify this relationship with other asteroids and the components of the mitochondrial genetic code.
Fig. 1.
Maximum likelihood phylogeny of Asteroidea mitochondrial genomes (solid lines) with Echinoidea as outgroup (dotted lines) (left) and mitochondrial initiation codons arranged in gene order (right). Values on nodes represent maximum likelihood bootstrap (≥50)/posterior probability (≥0.90) values. Samples sequenced in this study are in bold. ^Two Goniodiscaster scaber samples pooled into a single terminal. */#Annotated as ATT and ATA respectively in GenBank but corrected to ATG (see main text for details). +Possible novel initiation codon but inconclusive (see main text for details). $See Data availability.
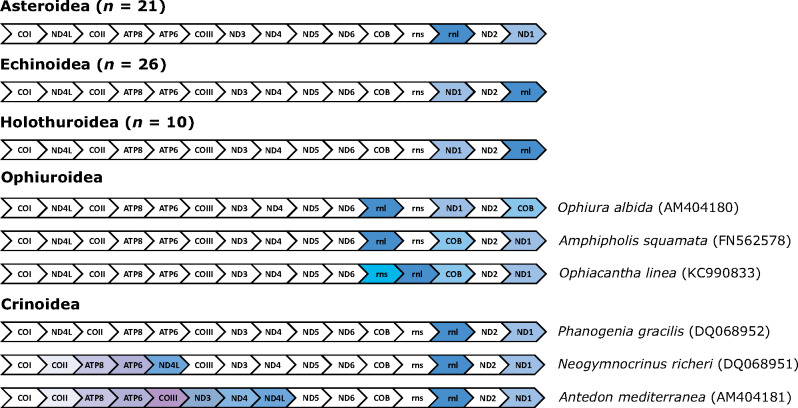
Considering the deep divergence between Asteroidea and Echinoidea in the Cambrian (O’Hara et al. 2014), the conservation of mitochondrial gene order within each of these taxa is remarkable (Fig. 1). Examination of 21 asteroid, 26 echinoid, and ten holothuroid mitogenomes revealed that there has only been an interchange in ND1 and 16S rRNA gene positions, and both echinoids and holothuroids share the same gene order (Fig. 2). These echinoderm taxa have a relatively stable order of mitochondrial genes despite diverging from one another as far back as ∼500 Ma (O’Hara et al. 2014). In contrast, clades that have diversified over similar geological time (e.g., Ophiuroidea [n = 3] and Crinoidea [n = 3] [Perseke et al. 2008], as well as Corallimorpharia [Lin et al. 2014; Quattrini et al. 2020]) display much greater variability in gene arrangements (Fig. 2).
Fig. 2.
Mitochondrial gene order among the five living classes of Echinodermata.
In particular, the remaining echinoderm classes, Ophiuroidea and Crinoidea, exhibit larger variations in gene order compared with their relatives in Echinodermata. Lee et al. (2019) examined 15 species of ophiuroid mitogenomes and found limited gene order conservation compared with asteroids, echinoids, and holothuroids, although the block of genes between cytochrome c oxidase subunit I (COI) and NADH dehydrogenase subunit 6 (ND6) were ordered identically among the four classes (see Scouras et al. 2004). Based on 17 mitogenomes, Galaska et al. (2019) found similar patterns and estimated that rearrangements of rRNA genes occurred ∼175–205 Ma for two ophiuroid species (Ophiacantha linea and Amphipholis squamata; see also O’Hara et al. 2017). Mitochondrial gene order in Crinoidea is most varied, with some crinoids (e.g., Antedon mediterranea and Neogymnocrinus richeri) no longer showing the conserved block of genes present in the other four echinoderm classes. In general, nucleotide substitution rate is positively linked to gene order variation (Shao et al. 2003; Xu et al. 2006; Bernt et al. 2013a; see also Yokobori et al. 2004, 2005). Therefore, the more rapid mitochondrial nucleotide substitution in ophiuroids and crinoids (Scouras et al. 2004) could be driving the differences seen among echinoderm classes.
Although targeted sequencing of fast-evolving mitochondrial markers remains useful for phylogenetic inference (e.g., mitochondrial control region, Bronstein et al. 2018), complete mitochondrial genes and genomes are now readily obtainable via whole-genome sequencing (e.g., Galaska et al. 2019; this study), and even as by-products of restriction site-associated DNA (Terraneo et al. 2018a, 2018b) or hybrid-capture (Allio et al. 2020; Quek et al. 2020) sequencing. Mitogenome sequences are frequently used in downstream applications such as population genomics and phylogenomics (Quek et al. 2019; Barrett et al. 2020; Inoue et al. 2020; Poliseno et al. 2020). Apart from homologous gene sequences isolated from mitogenomes, gene order can also be useful for elucidating phylogenetic relationships, particularly for ancient divergences as mitochondrial DNA sequences are likely to be substitution saturated due to the rapid evolutionary rates (Boore and Brown 1998; Oxusoff et al. 2018). For example, Fritzsch et al. (2006) reconstructed the phylogeny of protostomes based on mitochondrial gene arrangements and recovered the monophyly of arthropods, annelids, platyhelminths, and nematodes. However, such methods are more applicable for deep relationships across a broad range of taxa (Boore and Brown 1998; Lavrov and Lang 2005; Fritzsch et al. 2006), or among species with a number of gene rearrangements (Chen et al. 2018; but see Tyagi et al. 2020).
The mitogenome phylogeny reconstructed here (Fig. 1) is consistent with the phylotranscriptomic analysis of Linchangco et al. (2017) in showing that Velatida, Brisingida, and Forcipulatida share a close relationship and are sister to Valvatida, Spinulosida, and Paxillosida. The paraphyly of Valvatida has been reported in previous studies (Janies et al. 2011; Mah and Blake 2012) and also noted by Linchangco et al. (2017). The latter recovered Paxillosida as a clade, though we note that Ophidiasteridae (represented here by Ophidiaster granifer) was not analyzed. Although Ophidiasteridae is conventionally placed within Valvatida, Mah and Blake (2012) have reported that Ophidiasteridae is more closely related to Paxillosida. Nevertheless, we note that the sister relationship between Styracaster yapensis (Paxillosida) and Ophidiaster granifer as well as the general polyphyly of Paxillosida is not well supported by both reconstructions (Fig. 1). Resolution of these relationships and revision of the Asteroidea classification are warranted but require broader species and gene sampling.
Overall, this study has highlighted not just the diversity of the genetic code and the conserved gene order of many echinoderm mitogenomes but also improved gene annotations arising from the accurate identification of initiation codons.
Materials and Methods
New mitogenomes sequenced in this study consisted of 11 taxa, of which two were previously sequenced together with Archaster typicus in Quek et al. (2019) and the remaining nine samples were from Ip et al. (2019) and the cryogenic collection of the Lee Kong Chian Natural History Museum (for sample information, see http://dx.doi.org/10.5281/zenodo.3834170). DNA extraction, library preparation, sequencing, read quality trimming, and assembly followed by mitogenome contig identification and annotation largely followed Quek et al. (2019), but sequencing was performed on a HiSeq 4000 (150×150 bp). Raw reads were trimmed using Trimmomatic v0.38 (Bolger et al. 2014) and assembled with SPAdes v3.12.0 (Bankevich et al. 2012) under default settings. A BlastN (e-value = 10e−6) was conducted against two Acanthaster mitogenomes (Yasuda et al. 2006) to identify mitochondrial contigs, which were then annotated using MITOS2 (Bernt et al. 2013b).
Data obtained were combined with published, annotated mitochondrial sequences for an additional 16 species from GenBank. From the 27 mitogenomes, 13 PCGs and both rRNAs were extracted and aligned. Annotations of some PCGs by MITOS2 (Bernt et al. 2013b) had discrepancies in lengths when aligned to reference mitogenomes downloaded from GenBank, so all PCGs were aligned and annotations adjusted following visual inspection to ensure that they were in frame and accurately annotated.
Cytochrome b, ATPase subunit 6, and NADH dehydrogenase subunit 4 in three published mitochondrial genomes were originally annotated with alternative initiation codons ATA (Strongylocentrotus droebachiensis [AM900391] and S. pallidus [AM900392]) or ATT (Patiria pectinifera [D16387]), but inspection of gene alignments allowed the start codon to be corrected as the standard ATG codon (Fig. 1). Sequences from three samples could not be assembled into complete mitochondrial genomes (Fig. 1), so only full-length genes were extracted for phylogenetic analysis.
Sequence matrix preparation and phylogenetic analyses were conducted as described in Quek et al. (2019) with phylogenetic reconstruction performed based on maximum likelihood implemented in RAxML v8.2.11 (Stamatakis 2014) and Bayesian inference carried out in MrBayes v3.2.6 (Ronquist et al. 2012).
Acknowledgments
This work was supported by the National Research Foundation, Prime Minister’s Office, Singapore under its Marine Science R&D Programme (MSRDP-P03). We would like to thank Iffah Binte Iesa and Foo Maosheng for assistance with the museum loans, as well as David J.W. Lane for confirming species identifications. Computational work was partially performed on resources of the National Supercomputing Centre, Singapore (https://www.nscc.sg).
Author Contributions
Z.B.R.Q. designed the research and conducted the analyses; Z.B.R.Q., J.J.M.C., Y.C.A.I., and Y.K.S.C. procured the samples and performed the lab work; and Z.B.R.Q., J.J.M.C., Y.C.A.I., and D.H. wrote the paper.
Data Availability
All sequences and mitogenomes assembled have been deposited in GenBank (accession numbers MT444489–MT444514 and MT476590–MT476597, respectively). Raw sequencing reads are available in the NCBI Sequence Read Archive under BioProject PRJNA660967 (SRR12622297–SRR12622308). Sample information, gene alignments, and the concatenated matrix used for phylogenetic analyses are available at Zenodo (http://dx.doi.org/10.5281/zenodo.3834170).
References
- Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F.. 2020. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 20(4):892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa S, Himeno H, Miura K, Watanabe K.. 1995. Nucleotide sequence and gene organisation of the starfish Asterina pectinifera mitochondrial genome. Genetics 140(3):1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell BG, Bankier AT, Drouin J.. 1979. A different genetic code in human mitochondria. Nature 282(5735):189–194. [DOI] [PubMed] [Google Scholar]
- Barrett NJ, Hogan RI, Allcock AL, Molodtsova T, Hopkins K, Wheeler AJ, Yesson C.. 2020. Phylogenetics and mitogenome organisation in black corals (Anthozoa: Hexacorallia: Antipatharia): an order-wide survey inferred from complete mitochondrial genomes. Front Mar Sci. 7:440. [Google Scholar]
- Bernt M, Bleidorn C, Braband A, Dambach J, Donath A, Fritzsch G, Golombek A, Hadrys H, Jühling F, Meusemann K, et al. 2013. a. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol. 69(2):352–364. [DOI] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. b. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore L, Brown WM.. 1998. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 8(6):668–674. [DOI] [PubMed] [Google Scholar]
- Bronstein O, Kroh A, Haring E.. 2018. Mind the gap! The mitochondrial control region and its power as a phylogenetic marker in echinoids. BMC Evol Biol. 18(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantatore P, Roberti M, Rainaldi G, Gadaleta MN, Saccone C.. 1989. The complete nucleotide sequence, gene organization, and genetic code of the mitochondrial genome of Paracentrotus lividus. J Biol Chem. 264(19):10965–10975. [PubMed] [Google Scholar]
- Chen L, Chen P, Xue X, Hua H, Li Y, Zhang F, Wei S.. 2018. Extensive gene rearrangements in the mitochondrial genomes of two egg parasitoids, Trichogramma japonicum and Trichogramma ostriniae (Hymenoptera: Chalcidoidea: Trichogrammatidae). Sci Rep. 8(1):7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FHC. 1968. The origin of the genetic code. J Mol Biol. 38(3):367–379. [DOI] [PubMed] [Google Scholar]
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M.. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch G, Schlegel M, Stadler PF.. 2006. Alignments of mitochondrial genome arrangements: applications to metazoan phylogeny. J Theor Biol. 240(4):511–520. [DOI] [PubMed] [Google Scholar]
- Galaska MP, Li Y, Kocot KM, Mahon AR, Halanych KM.. 2019. Conservation of mitochondrial genome arrangements in brittle stars (Echinodermata, Ophiuroidea). Mol Phylogenet Evol. 130:115–120. [DOI] [PubMed] [Google Scholar]
- Hecht A, Glasgow J, Jaschke PR, Bawazer LA, Munson MS, Cochran JR, Endy D, Salit M.. 2017. Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res. 45(7):3615–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H, Masaki H, Kwai T, Ohta T, Kumagai I, Miura K, Wantanabe K.. 1987. Unusual genetic codes and a novel gene structure for tRNA in starfish mitochondrial DNA. Gene 56(2–3):219–230. [DOI] [PubMed] [Google Scholar]
- Inoue J, Hisata K, Yasuda N, Satoh N.. 2020. An investigation into the genetic history of Japanese populations of three starfish, Acanthaster planci, Linckia laevigata, and Asterias amurensis, based on complete mitochondrial DNA sequences. G3 (Bethesda) 10(7):2519–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip YCA, Tay YC, Gan SX, Ang HP, Tun K, Chou LM, Huang D, Meier R.. 2019. From marine park to future genomic observatory? Enhancing marine biodiversity assessments using a biocode approach. Biodivers Data J. 7:e46833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HT, Elliot DJ, Math VB, Farquharson A.. 1988. Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J Mol Biol. 202(2):185–217. [DOI] [PubMed] [Google Scholar]
- Janies DA, Voight JR, Daly M.. 2011. Echinoderm phylogeny including Xyloplax, a progenetic asteroid. Syst Biol. 60(4):420–438. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Lang BF.. 2005. Poriferan mtDNA and animal phylogeny based on mitochondrial gene arrangements. Syst Biol. 54(4):651–659. [DOI] [PubMed] [Google Scholar]
- Lee T, Bae YJ, Shin S.. 2019. Mitochondrial gene rearrangement and phylogenetic relationships in the Amphilepidida and Ophiacanthida (Echinodermata, Ophiuroidea). Mar Biol Res. 15(1):26–35. [Google Scholar]
- Lin MF, Kitahara MV, Luo H, Tracey D, Geller J, Fukami H, Miller DJ, Chen CA.. 2014. Mitochondrial genome rearrangements in the Scleractinia/Corallimorpharia complex: implications for coral phylogeny. Genome Biol Evol. 6(5):1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linchangco GV, Foltz DW, Reid R, Williams J, Nodzak C, Kerr AM, Miller AK, Hunter R, Wilson NG, Nielsen WJ, et al. 2017. The phylogeny of extant starfish (Asteroidea: 241 Echinodermata) including Xyloplax, based on comparative transcriptomics. Mol Phylogenet Evol. 115:161–170. [DOI] [PubMed] [Google Scholar]
- Lobanov AV, Turanov AA, Hatfield DL, Gladyshev VN.. 2010. Dual functions of codons in the genetic code. Crit Rev Biochem Mol. 45(4):257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah CL, Blake DB.. 2012. Global diversity and phylogeny of the Asteroidea (Echinodermata). PLoS One 7(4):e35644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara TD, Hugall AF, Thuy B, Moussalli A.. 2014. Phylogenomic resolution of the class Ophiuroidea unlocks a global microfossil record. Curr Biol. 24(16):1874–1879. [DOI] [PubMed] [Google Scholar]
- O’Hara TD, Hugall AF, Thuy B, Stöhr S, Martynov AV.. 2017. Restructuring higher taxonomy using broad-scale phylogenomics: the living Ophiuroidea. Mol Phylgenet Evol. 107:415–430. [DOI] [PubMed] [Google Scholar]
- Okimoto R, Macfarlane JL, Wolstenholme DR.. 1990. Evidence for the frequent use of TTG as the translation initiation codon of mitochondrial protein genes in the nematodes, Ascaris suum and Caenorhabditis elegans. Nucleic Acids Res. 18(20):6113–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxusoff L, Préa P, Perez Y.. 2018. A complete logical approach to resolve the evolution and dynamics of mitochondrial genome in bilaterians. PLoS One 13(3):e0194334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perseke M, Fritzsch G, Ramsch K, Bernt M, Merkle D, Middendorf M, Bernhard D, Stadler PF, Schlegel M.. 2008. Evolution of mitochondrial gene orders in echinoderms. Mol Phylogenet Evol. 47(2):855–864. [DOI] [PubMed] [Google Scholar]
- Poliseno A, Santos MEA, Hiroki K, Macdonald B, Quattrini AM, McFadden CS, Reimer JD. Forthcoming 2020. Evolutionary implications of analyses of complete mitochondrial genomes across order Zoantharia (Cnidaria: Hexacorallia). J Zool Syst Evol Res. [Google Scholar]
- Quattrini AM, Rodríguez E, Faircloth BC, Cowman PF, Brugler MR, Farfan GA, Hellberg ME, Kitahara MV, Morrison CL, Paz-García DA, et al. Forthcoming 2020. Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat Ecol Evol. [DOI] [PubMed] [Google Scholar]
- Quek RZB, Jain SS, Neo ML, Rouse GW, Huang D.. 2020. Transcriptome‐based target‐enrichment baits for stony corals (Cnidaria: Anthozoa: Scleractinia). Mol Ecol Resour. 20(3):807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek ZBR, Chang JJM, Ip YCA, Huang D.. 2019. Complete mitochondrial genome of the sea star Archaster typicus (Asteroidea: Archasteridae). Mitochondrial DNA B 4(2):3130–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scouras A, Beckenbach K, Arndt A, Smith MJ.. 2004. Complete mitochondrial genome DNA sequence for two ophiuroids and a holothuroid: the utility of protein gene sequence and gene maps in the analyses of deep deuterostome phylogeny. Mol Phylogent Evol. 31(1):50–65. [DOI] [PubMed] [Google Scholar]
- Shao R, Dowton M, Murrell A, Barker SC.. 2003. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol Biol Evol. 20(10):1612–1619. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terraneo TI, Arrigoni R, Benzoni F, Forsman ZH, Berumen ML.. 2018. a. Using ezRAD to reconstruct the complete mitochondrial genome of Porites fontanesii (Cnidaria: Scleractinia). Mitochondrial DNA B 3(1):173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terraneo TI, Arrigoni R, Benzoni F, Forsman ZH, Berumen ML.. 2018. b. The complete mitochondrial genome of Porites harrisoni (Cnidaria: Scleractinia) obtained using next-generation sequencing. Mitochondrial DNA B 3(1):286–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi K, Chakraborty R, Cameron SL, Sweet AD, Chandra K, Kumar V.. 2020. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta). Sci Rep. 10(1):695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jameson D, Tang B, Higgs PG.. 2006. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J Mol Evol. 63(3):375–392. [DOI] [PubMed] [Google Scholar]
- Yasuda N, Hamaguchi M, Sasaki M, Nagai S, Saba M, Nadaoka K.. 2006. Complete mitochondrial genome sequences for Crown-of-thorns starfish Acanthaster planci and Acanthaster brevispinus. BMC Genomics. 7(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S, Fukuda N, Nakamura M, Aoyama T, Oshima T.. 2004. Long-term conservation of six duplicated structural genes in cephalopod mitochondrial genomes. Mol Biol Evol. 21(11):2034–2046. [DOI] [PubMed] [Google Scholar]
- Yokobori S, Oshima T, Wada H.. 2005. Complete nucleotide sequence of the mitochondrial genome of Doliolum nationalis with implications for evolution of urochordates. Mol Phylogenet Evol. 34(2):273–283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences and mitogenomes assembled have been deposited in GenBank (accession numbers MT444489–MT444514 and MT476590–MT476597, respectively). Raw sequencing reads are available in the NCBI Sequence Read Archive under BioProject PRJNA660967 (SRR12622297–SRR12622308). Sample information, gene alignments, and the concatenated matrix used for phylogenetic analyses are available at Zenodo (http://dx.doi.org/10.5281/zenodo.3834170).