Abstract
The mitoribosome, as known from studies in model organisms, deviates considerably from its ancestor, the bacterial ribosome. Deviations include substantial reduction of the mitochondrial ribosomal RNA (mt-rRNA) structure and acquisition of numerous mitochondrion-specific (M) mitoribosomal proteins (mtRPs). A broadly accepted view assumes that M-mtRPs compensate for structural destabilization of mt-rRNA resulting from its evolutionary remodeling. Since most experimental information on mitoribosome makeup comes from eukaryotes having derived mitochondrial genomes and mt-rRNAs, we tested this assumption by investigating the mitochondrial translation machinery of jakobids, a lineage of unicellular protists with the most bacteria-like mitochondrial genomes. We report here proteomics analyses of the Andalucia godoyi small mitoribosomal subunit and in silico transcriptomic and comparative genome analyses of four additional jakobids. Jakobids have mt-rRNA structures that minimally differ from their bacterial counterparts. Yet, with at least 31 small subunit and 44 large subunit mtRPs, the mitoriboproteome of Andalucia is essentially as complex as that in animals or fungi. Furthermore, the relatively high conservation of jakobid sequences has helped to clarify the identity of several mtRPs, previously considered to be lineage-specific, as divergent homologs of conserved M-mtRPs, notably mS22 and mL61. The coexistence of bacteria-like mt-rRNAs and a complex mitoriboproteome refutes the view that M-mtRPs were ancestrally recruited to stabilize deviations of mt-rRNA structural elements. We postulate instead that the numerous M-mtRPs acquired in the last eukaryotic common ancestor allowed mt-rRNAs to pursue a broad range of evolutionary trajectories across lineages: from dramatic reduction to acquisition of novel elements to structural conservatism.
Keywords: jakobids, ribosome, proteomics, LECA, ribosomal protein, mitochondria
Introduction
During the pregenomic era, the most convincing evidence for the endosymbiotic origin of mitochondria was the organelle’s translation system, as it shares numerous features, from rRNA structure to protein constituents, with those of bacteria (Gray and Doolittle 1982). The most detailed knowledge about the bacterial ribosome came initially from crystal structure determination in Thermus thermophilus (Yusupov et al. 2001) and Escherichia coli (Schuwirth et al. 2005) and concurrent seminal works from the groups of Steitz, Yonath, and Ramakrishnan (Rodnina and Wintermeyer 2010; Ramakrishnan 2014). Typically, the bacterial ribosome contains three ribosomal RNA (rRNA) species, the small subunit (SSU) or 16S rRNA, the large subunit (LSU) or 23S rRNA, and the 5S rRNA (also associated with the LSU), in addition to 54 ribosomal proteins (RPs) of which 21 are associated with the SSU and 33 with the LSU. Only minor deviations in bacterial RP content have been reported (Yutin et al. 2012; Grosjean et al. 2014; Brown et al. 2015). For example, the SSU of T. thermophilus lacks the otherwise broadly distributed S21 RP and instead contains THX (Tsiboli et al. 1994; Wimberly et al. 2000). In turn, a putative new RP was recently postulated in Bacteroidetes (Sberro et al. 2019).
Although bacterial ribosomes are close to invariable, their descendants—present-day mitochondrial ribosomes (mitoribosomes)—display an extraordinarily large array of conspicuous differences, both in RNA structure and in protein composition. Mitochondrial ribosomal RNAs (mt-rRNAs) have diverged in various organisms in sequence and undergone both loss of ancestral secondary structure elements and, to a lesser extent, gain of structural extensions. Deviations from bacterial RNA structures range from moderate, as in plants and several protist lineages (see Gray et al. 2004; Valach, Burger, et al. 2014) to extreme, for example, in bilaterian animals, kinetoplastids, diplonemids, and apicomplexans (de la Cruz, Lake, et al. 1985; de la Cruz, Simpson, et al. 1985; Klimov and Knowles 2011; Feagin et al. 2012; Valach, Moreira, et al. 2014). For example, whereas bacteria-like 5S mt-rRNAs are present in various protists (Valach, Burger, et al. 2014) and plants (Waltz et al. 2020), this rRNA species has been lost in other lineages like fungi and metazoans. With few exceptions, this loss has been functionally compensated in different ways such as by a repurposed tRNA in mammals, expansions of LSU mt-rRNA in yeast, and new mitoribosomal proteins (mtRPs) in trypanosomes (Amunts et al. 2014; Brown et al. 2014; Greber et al. 2014; Ramrath et al. 2018; Tobiasson and Amunts 2020).
The difference in protein components between bacterial and mitochondrial ribosomes is even more conspicuous. According to comparative phylogenomic analyses (Smits et al. 2007; Desmond et al. 2011), the mitoribosome of the last eukaryotic common ancestor (LECA) has maintained all bacterial RPs except S20. Mitochondrial RPs of bacterial origin (B-mtRPs) are on average almost twice as large as their bacterial counterparts, due both to additional conserved protein domains such as the RNA recognition motif (Smits et al. 2007) and to nonglobular extensions (Melnikov et al. 2018). More importantly, LECA’s mitoribosome has apparently recruited numerous novel proteins—here referred to as mitochondrion-specific mitoribosomal proteins (M-mtRPs)—whose most recent count has been established at 18 (Gray et al. 2020). Subsequent divergence into present-day eukaryotic clades was accompanied by the differential loss of LECA mtRPs and gain of new lineage-specific proteins, leading to a striking diversity of mitoribosome architecture across eukaryotes (Waltz and Giegé 2020).
The genes that specify mitoribosomal components reside in both the nuclear and mitochondrial genome. In all eukaryotes examined, mt-rRNA genes have remained in mitochondrial DNA (mtDNA). In contrast, the 53 B-mtRPs have migrated to the nucleus to various extents (100% in mammals but only ∼50% in jakobids [Burger et al. 2013]). The unicellular protist clade jakobids (supergroup Discoba), and in particular Andalucia godoyi, possess the most bacteria-like mitochondrial genomes known, not only with regard to the number of genes retained, but also other features such as bacteria-like LSU, SSU, and 5S mt-rRNAs, RNA polymerase, and putative Shine–Dalgarno motifs (Lang et al. 1997; Burger et al. 2013; Valach, Burger, et al. 2014). Indeed, our recent in silico mitoproteome analysis of Andalucia characterizes this species as “the eukaryote whose mitochondrion likely resembles the LECA mitochondrion more closely than does the mitochondrion in any other eukaryote studied to date” (Gray et al. 2020). Therefore, Andalucia is the system of choice for testing hypotheses regarding the transition from LECA’s mitoribosome to contemporary mitochondrial translation machineries.
Our previous in silico analyses predicted that the Andalucia mitoribosome includes as many as 70 mtRPs. Of these, 28 were predicted to be associated with the SSU and 42 with the LSU, of which 12 and 16, respectively, are mtDNA-encoded (Gray et al. 2020). However, since these predictions were based on sequence similarity (and across large evolutionary distances), a protein in Andalucia that is a homolog of a yeast or human mtRP might not necessarily be associated with the same mitoribosomal subunit, or may not even be a constituent of the mitoribosome. The in silico assignment for proteins with dual function is particularly tricky, as in the case of yeast mS47, which appears to be a catalytically active 3-hydroxyisobutyryl-CoA hydrolase, HIBCH (Ehd3), involved in amino acid degradation (Hiltunen et al. 2003; Desai et al. 2017).
Therefore, we set out to examine experimentally the mitochondrial translation machinery of A. godoyi. In addition, we identified in silico mtRPs from four other jakobids—Jakoba bahamiensis, J. libera, Reclinomonas americana, and Seculamonas ecuadoriensis—as well as broadly across eukaryotes. This work addressed several questions: 1) Are the mtRP candidates predicted for Andalucia conserved across jakobids? 2) Are these proteins indeed associated with the Andalucia mitoribosome? 3) Does the ancestral structure of Andalucia mt-rRNAs correlate with the absence of mtRPs that have been proposed to compensate for and stabilize deviant RNA structure elements in model organisms?
In the discussion that follows, we will use the now widely accepted new unifying nomenclature of RPs that reflects the phylogenetic distribution and subcellular localization of these proteins (Ban et al. 2014). For instance, uL1m designates the mitoribosomal protein L1 of the LSU that occurs universally in all domains of life, whereas bS1m designates the mitoribosomal protein S1 of the SSU that occurs also in bacteria, with RPs specific to mitoribosomes assigned the prefix “m” (e.g., mS22 and mL61) (Amunts et al. 2014; Brown et al. 2014).
Results
Secondary Structures of Jakobid Mitochondrial rRNAs
The mitochondrial LSU and SSU rRNAs from jakobids are much more similar to their bacterial homologs than those from other eukaryotes (Burger et al. 2013). Among examined jakobids, Andalucia has the most bacteria-like mt-rRNAs. Specifically, sequence identity of Andalucia SSU mt-rRNA with the E. coli 16S rRNA is upwards of 70% (supplementary fig. S1, Supplementary Material online). Comparison with the bacterial 16S rRNA secondary structure model indicates loss of only two terminal helices in Andalucia SSU mt-rRNA, as well as minor apical truncations in three helices. Nucleotide substitutions occur mostly in the central and 3′ major domains, and are compensated by covariational changes of the base-pairing partners (fig. 1A; for sequence–structure alignments of jakobid SSU mt-rRNAs, see supplementary fig. S2, Supplementary Material online).
Fig. 1.
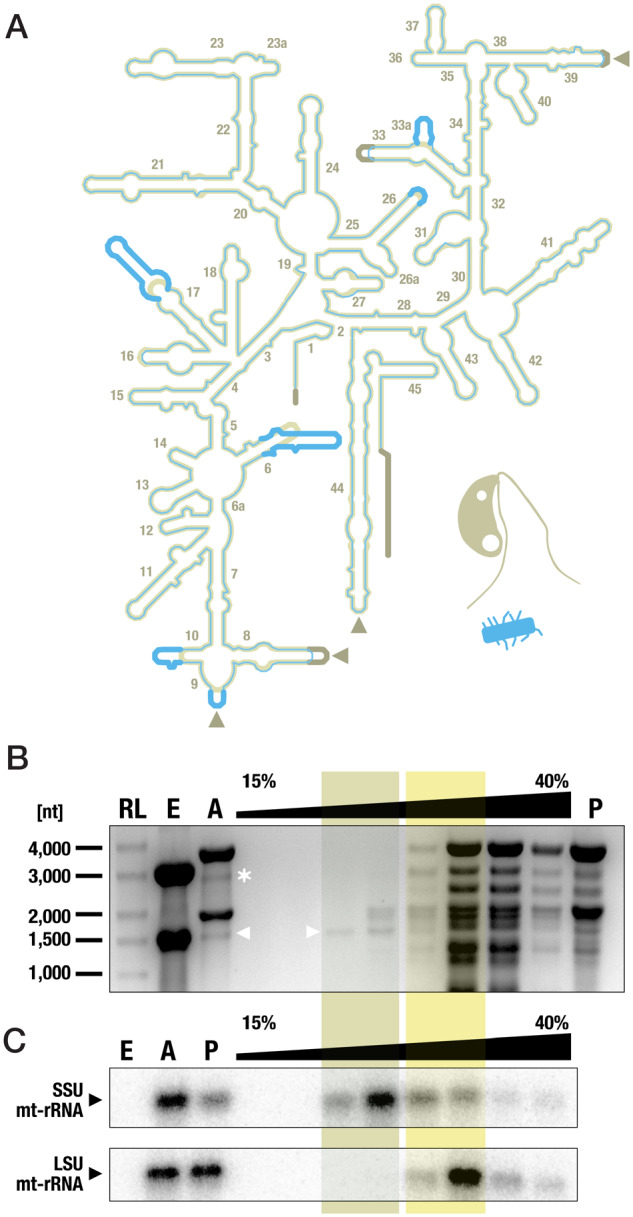
Enrichment of the bacteria-like SSU mt-rRNA of A. godoyi. (A) Comparison of the secondary structure models of Andalucia SSU mt-rRNA (beige; NC_021124.1 [66176..67656]) and E. coli SSU (16S) rRNA (blue; GenBank NC_000913.3 [4166659..4168200]). Thick lines highlight the structural differences between the two rRNAs. Arrowheads indicate where expansions occur in SSU mt-rRNA of other jakobids (see also supplementary fig. S2, Supplementary Material online). (B) Analysis of RNA content after sucrose velocity gradient fractionation of Andalucia cell lysates. Arrowhead and asterisk indicate the band of SSU and LSU mt-rRNAs, respectively. Lanes: RL, RNA ladder; E, RNA from Enterobacter cells (prey); A, RNA from Andalucia cells; P, pooled fractions from 15–40% sucrose gradient. (C) Northern blot hybridization analysis of mt-rRNA distribution (sample A23) using SSU mt-rRNA-specific (ag03) and LSU mt-rRNA-specific (ag04) probes. The enrichment peaks of mtSSU and mtLSU are highlighted in beige and yellow, respectively. For further details, see supplementary figure S3, Supplementary Material online.
Compared with jakobid SSU mt-rRNA, the secondary structure model of LSU mt-rRNA deviates more noticeably from the E. coli 23S rRNA model (for a secondary structure diagram of R. americana LSU mt-rRNA, see [Petrov et al. 2019]; the only difference in Andalucia is a slightly reduced H63). In Andalucia (and the other jakobids), several helices have been lost and others truncated in Domain III (i.e., H54 to H59). Furthermore, helices H63 and H68 are apically truncated, and H98 is absent. Most sequence variation occurs in Domain I (helices H4–H10) and Domain V (H76–H78), yet with little or no impact on the inferred secondary structure.
The termini of jakobid SSU and LSU mt-rRNA sequences available in public sequence repositories were annotated based on sequence comparison with 16S and 23S rRNAs from E. coli (Burger et al. 2013). Here, we determined the termini experimentally by mapping previously generated RNA-Seq reads from A. godoyi and J. bahamiensis (Valach, Burger, et al. 2014) to the gene sequences. We found that, compared with the in silico-predicted termini, the correct 5′ end of SSU mt-rRNA is located further upstream (3 and 5 nt in A. godoyi and J. bahamiensis, respectively) whereas the 3′ end is further downstream (21 and 3 nt in A. godoyi and J. bahamiensis, respectively) (fig. 1A and supplementary fig. S2, Supplementary Material online). For the LSU mt-rRNA from Andalucia, the correct 5′ and 3′ termini are 11 nt downstream and 11 nt upstream, respectively, of the in silico-assigned positions (the corresponding data from J. bahamiensis were inconclusive). The newly established ends of Andalucia LSU mt-rRNA can be folded into a 10-bp long expansion of the helix H1 that brings together the terminal regions of the rRNA; the sequences from the other jakobids have the same base-pairing capability. Interestingly, this terminal helix in jakobid LSU mt-rRNA resembles the terminal expansion segment (H0-ES) of the yeast LSU mt-rRNA, which forms a membrane-facing protuberance together with its associated proteins mL44, mL57, and mL58 (Amunts et al. 2014). The jakobid H0-ES could serve as a binding platform for the jakobid mL44 counterpart (see also supplementary material).
Enrichment of the Small Subunit of the Andalucia Mitochondrial Ribosome
To examine the protein makeup of a jakobid mitoribosome in the absence of protein-tagging techniques or antibodies, we established a protocol to purify A. godoyi mitoribosomes from a whole cell lysate by ultracentrifugation (see Materials and Methods section). This undertaking was a challenging task, because the protist grows slowly (∼72 h generation time) and only to low titers (∼2 × 106/ml). Andalucia requires live bacteria as a food source, which have to be added repeatedly in limited amounts to reduce contamination in downstream analyses. Another hurdle was endogenous ribonuclease activity in the cell lysate, which was countered by low magnesium ion concentrations during mitoribosome isolation, a measure that increases the risk of destabilizing ribonucleoprotein complexes.
The Andalucia mitoribosomes were purified by sucrose gradient centrifugation, and their enrichment was monitored by semiquantitative RT-PCR and Northern hybridization using SSU and LSU mt-rRNA-specific oligonucleotide probes. Mitoribosomal SSU (mtSSU) separated well from cytosolic SSU (fig. 1B and C, supplementary fig. S3A, B, E, and F, Supplementary Material online), whereas mitoribosomal LSU (mtLSU) extensively overlapped with both the cytosolic SSU and LSU (fig. 1C and supplementary fig. S3C, Supplementary Material online); contamination with bacterial ribosomes was negligible (supplementary fig. S3D, Supplementary Material online). None of the fractions combined enrichment of both mtSSU and mtLSU, indicating that suppression of RNases by low magnesium ion concentration came at the price of dissociating the mitoribosomal subunits. Therefore, proteomics validation of Andalucia mtRPs was performed exclusively on mtSSU, whereas in silico approaches were applied to the complete mitoribosome.
Composition of the Small Subunit of the Andalucia Mitochondrial Ribosome
The proteins of the Andalucia mtSSU preparation (supplementary fig. S3H, Supplementary Material online) were extracted and analyzed by tandem mass spectrometry (MS/MS). In parallel, we examined the protein composition of whole cell lysates to gauge the relative enrichment of individual proteins in mtSSU samples (supplementary fig. S3H, Supplementary Material online).
According to the composition of the Andalucia mitoribosome inferred in silico by sequence similarity (Gray et al. 2020), SSU and LSU contain 28 and 40 proteins, respectively. By MS/MS analysis, we detected all of the expected SSU mtRPs with the exception of two (mS25 and mS38). In contrast, whole cell-lysate controls contained only about 30% of the SSU mtRPs, testifying to a significant enrichment of mtSSU by our purification protocol (table 1 and supplementary tables S1–S3, Supplementary Material online).
Table 1.
MS Identifications of 31 Assigned mtSSU RPs from A. godoyi.
| Andalucia IDa | Assignmentb | Length [aa] | Predicted size [kDa] | MTPc | No. of Unique Tryptic Peptidesd |
||
|---|---|---|---|---|---|---|---|
| Enriched mtSSU | Whole-cell lysate | Mitochondria-enriched lysate | |||||
| Rps1 | bS1m | 186 | 21.52 | n.a. | 8 | ||
| Rps2 | uS2m | 214 | 24.72 | n.a. | 11 | 1 | 2 |
| Rps3 | uS3m | 206 | 23.27 | n.a. | 11 | ||
| Rps4 | uS4m | 208 | 24.27 | n.a. | 9 | 1 | |
| ANDGO_05657 | uS5m | 187 | 20.08 | + | 9 | 1 | 3 |
| ANDGO_04075 | bS6m | 193 | 21.05 | − | 7 | 3 | |
| Rps7 | uS7m | 150 | 17.48 | n.a. | 6 | ||
| Rps8 | uS8m | 131 | 14.60 | n.a. | 10 | 1 | 3 |
| ANDGO_06340 | uS9m | 289 | 31.01 | + | 11 | 1 | 5 |
| Rps10 | uS10m | 109 | 12.88 | n.a. | 7 | 3 | |
| Rps11 | uS11m | 127 | 13.62 | n.a. | 6 | ||
| Rps12 | uS12m | 128 | 14.14 | n.a. | 7 | ||
| Rps13 | uS13m | 123 | 14.31 | n.a. | 8 | 5 | |
| Rps14 | uS14m | 100 | 11.62 | n.a. | 3 | 1 | |
| ANDGO_03225 | uS15m | 127 | 13.78 | + | 4 | 3 | |
| ANDGO_00845 | bS16m | 114 | 12.40 | + | 6 | 1 | |
| ANDGO_08788 | uS17m | 106 | 11.87 | + | 4 | ||
| ANDGO_06513 | bS18m | 146 | 16.32 | + | 7 | ||
| Rps19 | uS19m | 78 | 8.97 | n.a. | 6 | 1 | 2 |
| ANDGO_04082 | bS21m | 76 | 9.08 | − | 4 | 1 | |
| ANDGO_06241* | mS22 | 249 | 26.66 | + | 10 | 2 | 3 |
| ANDGO_07482 | mS23 | 258 | 29.20 | − | 6 | 1 | 1 |
| ANDGO_08032 | mS26 | 168 | 19.49 | + | 10 | 2 | |
| ANDGO_05980 | mS29 | 353 | 38.78 | + | 22 | 3 | 9 |
| ANDGO_04851* | mS31 | 266 | 29.06 | − | 14 | ||
| ANDGO_02870 | mS33 | 89 | 9.66 | + | 5 | 1 | |
| ANDGO_08763 | mS35 | 137 | 14.39 | + | 6 | 1 | 3 |
| ANDGO_02863 | mS37 | 80 | 9.03 | + | 4e | ||
| ANDGO_00506 | mS38 | 91 | 10.51 | + | 0f | ||
| ANDGO_06438* | mS42 | 207 | 23.52 | + | 12 | 5 | 14 |
| ANDGO_00091* | mS47 | 406 | 44.47 | + | 20 | 4 | 11 |
Identifier of A. godoyi proteins in the genome-inferred proteome. Asterisk, newly assigned components compared with (Gray et al. 2020).
Assignment according to the new nomenclature (Ban et al. 2014).
Prediction of an MTP (Gray et al. 2020). +, predicted; −, not predicted; n.a., not applicable (mtDNA-encoded protein).
Number of unique peptides according to Mascot analyses (see Materials and Methods section). Blank, protein not detected.
Protein only detected in the sample A23.
Protein is not detectable by the experimental approach (see Results section).
Retrospectively, it is not surprising that mS25 (ANDGO_07531) and mS38 (ANDGO_00506) (Gray et al. 2020) were not retrieved by MS/MS of mtSSU-enriched samples. For ANDGO_07531, we now have evidence that despite sequence similarity this protein is not a homolog of mS25, but rather of mL61, and thus absent from the mtSSU. The mS25 and mL61 proteins have been confounded because of their shared L51_S25_CI-B8 domain (Pfam ID PF05047), which is also present in additional mtRPs and other non-RPs. However, as we dissect in detail below, the two proteins are distinguishable by phylogenetic analysis, which places mS25 and mL61 into separate clades and clearly affiliates ANDGO_07531 with experimentally confirmed mL61 sequences from model organisms.
The absence of the second protein, mS38, from MS/MS data most probably reflects an experimental issue, that is, the protease digestion procedure applied. The Andalucia protein is likely processed in the same way as the mammalian and yeast homologs (Greber et al. 2015; Desai et al. 2017), yielding only a 29-residue long mature protein that represents the highly conserved C-terminal portion of the precursor protein. Due to the high lysine and arginine content of the mature Andalucia mS38 protein, trypsin digestion will generate peptides too short for conventional MS/MS detection.
Newly Identified Small Subunit Mitoribosomal Proteins of Andalucia
To identify potentially unrecognized components of the Andalucia mtSSU, we examined which of the MS/MS-detected proteins have an enrichment profile (whole cell vs. mtSSU preparations) that resembles that of the trusted mtRPs. For details about the procedure applied, see the flowchart in supplementary fig. S4, Supplementary Material online. We found 104 proteins with SSU mtRP-like distribution (supplementary fig. S5, Supplementary Material online). From these 104 proteins, we filtered out 63, which according to the in silico functional annotation were evidently copurified fortuitously, for example, components of the cytoskeleton or endoplasmatic reticulum. In this step, we also removed proteins predicted to contain trans-membrane and membrane-interacting domains, since in all systems so far examined, anchoring of the ribosome to the mitochondrial inner membrane is achieved by the LSU and not the SSU (Ott and Herrmann 2010; Pfeffer et al. 2015; Englmeier et al. 2017). Among the remaining 41 proteins, 25 were conserved SSU mtRPs, leaving 16 candidates of unrecognized SSU mtRPs, which we then ranked by their propensity of residing in mitochondria and conservation across jakobids (supplementary table S4, Supplementary Material online). The six highest ranking candidates included proteins annotated as a hypothetical protein, various dehydrogenases, a hydrolase, and a superoxide dismutase. These six proteins are referred to in the following as the top SSU mtRP candidates.
All candidates were examined by highly sensitive profile Hidden Markov Model (HMM) searches and phylogenetic analyses, and working hypotheses were tested by inspecting mitoribosomal structures from model organisms. These in-depth studies allowed us to identify four mitoribosomal proteins (mS22, mS31, mS42, and mS47), which had remained unrecognized by our previous sequence similarity-based analyses (Gray et al. 2020) (table 1).
Identification of Andalucia mS22
The top SSU mtRP candidate ANDGO_06241 is prominently enriched in Andalucia mtSSU preparations and, lacking obvious sequence similarity with functionally defined proteins, was initially annotated as a mitochondrion-targeted hypothetical protein. HMM searches with profiles from the Pfam database (El-Gebali et al. 2019) returned a weak match to the MRP-S22 domain (PF10245; E∼ 8.7e−03), which is characteristic for animal and trypanosome mS22. However, ANDGO_06241 matches also, and with a somewhat stronger signal (E∼ 1.5e−07), the MRP-L20 protein domain (PF12824), which is typical for fungal mL58 (previously designated MRP-L20). In ANDGO_06241, the MRP-S22 domain is located C-terminally and the MRP-L20 domain N-terminally. Homologs with the precisely same domain arrangement are present in the inferred proteomes of all other jakobids examined, suggesting that this domain arrangement plays a vital role (supplementary table S1, Supplementary Material online). Inspection of the experimentally validated mS22 from Trypanosoma and homologs from other kinetoplastids revealed that these mtRPs are also composed of a C-terminal MRP-S22 and an N-terminal mL58-like domain, which supports that ANDGO_0624 is an mS22 homolog.
Identification of Andalucia mS47 with Dual Function
ANDGO_00091 is another top SSU mtRP candidate (supplementary table S4, Supplementary Material online). However, Blast searches of this sequence against NCBI’s nonredundant protein sequences (nr) retrieved the mitochondrial matrix enzymes HIBCHs with convincingly strong support (E∼ 3e−142, query coverage ∼90%). ANDGO_00091 contains all residues necessary for a catalytically active HIBCH, a mitochondrial matrix enzyme that is known to participate in the degradation of certain amino acids, as well as in the metabolism of beta-alanine and propanoate. Therefore, we did not consider it as an mtRP in our initial in silico proteome analysis (Gray et al. 2020). Incidentally, in the mitoribosomal 3D structure model from yeast, a putatively enzymatically active HIBCH is associated peripherally with mtSSU and has been designated mS47 (Desai et al. 2017) (supplementary fig. S6, Supplementary Material online). Sequences of mS47 homologs from Arabidopsis (Waltz et al. 2019) and Trypanosoma (Ramrath et al. 2018) also resemble HIBCHs, but lack the enzyme’s substrate-binding site. We therefore conclude that ANDGO_00091 represents mS47 and that it is catalytically active like its yeast counterpart.
Identification of Andalucia mS42/mS43
A third top SSU mtRP candidate from Andalucia is ANDGO_06438 (supplementary table S4, Supplementary Material online), which had been initially annotated as mitochondrial iron/manganese superoxide dismutase based on high-scoring matches (E∼ 4e−121, query coverage ∼96%) in Blast searches against GenBank nr database. The presence of the function-critical metal ion-binding site in the ANDGO_06438 sequence suggests that this protein is catalytically active, which at first sight made it an unlikely mitoribosome component. However, there are precedents. Trypanosoma and yeast mitoribosomes contain the paralogous mS42 and mS43, which are characterized by a divergent, noncatalytic superoxide dismutase-fold. The paralog pair associates as a heterodimer that structurally resembles the dimeric mitochondrial matrix enzyme superoxide dismutase (Desai et al. 2017; Ramrath et al. 2018) (supplementary table S1 and fig. S6, Supplementary Material online). A recent study revealed that the Neurospora mitoribosome includes an mS42 homodimer instead of an mS42/mS43 heterodimer (Itoh et al. 2020). Sequence comparison of fungal mS42 and mS43 indicates that the Neurospora protein is actually more closely related to the yeast mS43 than mS42; still, similarly to its yeast counterparts, the Neurospora mtRP also lacks the catalytic Fe ion-binding site and is therefore considered an enzyme paralog that lost its activity upon recruitment to the mitoribosome.
The sequence resemblance to Fe/Mn superoxide dismutase strongly suggests that ANDGO_06438 is an mS42/mS43 homolog. In contrast to its counterparts from fungi and Trypanosoma, the Andalucia mS42/mS43 has apparently retained its catalytic activity as a radical-scavenging enzyme. Evolutionary relationships between mS42 and mS43 from various organisms are difficult to resolve, which could indicate either that superoxide dismutases or superoxide dismutase-like proteins have been recruited to the mitoribosome at multiple occasions, or that the original mtRPs were replaced at different times in different lineages by paralogs. For simplicity, we refer to the Andalucia protein as mS42 (table 1 and fig. 2).
Fig. 2.

Distribution of currently known mtSSU and mtLSU RPs in jakobids and other eukaryotes. Presence or absence of mtRP homologs is indicated in black and light gray, respectively. Dark gray highlights tentative assignments as an mtRP, requiring further validation (e.g., mitoribosome isolation). Circles, updated assignments compared with previous publications (Desmond et al. 2011; Gray et al. 2020; Hammond et al. 2020; Tobiasson and Amunts 2020; Waltz and Giegé 2020). All RPs with the prefix “b” or “u” (bS/bL, uS/uL) are present in bacteria. Notes to boxes and circles labeled a–k: a, mS22 and Saccharomyces/Neurospora/Arabidopsis mS45 were recognized as homologs, and Tetrahymena and Euglena also contain an A3T2-domain-only mS22 (see Results section). b, mS22, mS37, mL59, and mL60 were not retrieved from Acanthamoeba, but homologs are present in some amoebozoans (for mS22, see supplementary fig. S8B, Supplementary Material online). c, mS27 and mS44 have been recognized as homologs (Itoh et al. 2020). d, mS31 and mS46 have been recognized as homologs (see Waltz et al. 2020; supplementary fig. S7, Supplementary Material online). e, mS42/mS43 is a homodimer, assuming the role of distinct paralogous mS42 and mS43, in Neurospora (Itoh et al. 2020), most likely in jakobids (see the Results section), and possibly also in Acanthamoeba. f, mL59 and mL64 have been recognized as homologs (Waltz and Giegé 2020). g, mL74 of Trypanosoma and mL108 of Neurospora represent mL61 (see the Results). h, presence of mL42 and mL63 in LECA is uncertain because due to an extremely short and weak sequence signature of the proposed conserved domains between animal and trypanosome counterparts, it is currently impossible to retrieve homologs in other lineages by sequence similarity searches and verify that these RPs are indeed orthologs, that is, not a result of convergent evolution. i, presence of mL44 in LECA is uncertain because the mL44 homolog in jakobids has not been confirmed experimentally as an mtRP. k, mtRPs missing from mitoribosome cryo-EM 3D reconstructions. The following mtRP assignments have become obsolete: mS45, mS46, mL74, and mL108 (this work), mS44 (Itoh et al. 2020), and mL64 (Waltz and Giegé 2020) (see also supplementary material).
Identification of Andalucia mS31
Akin to ANDGO_06241 (Andalucia mS22, see above), ANDGO_04851 was initially annotated as a hypothetical protein. While lacking a recognizable conserved protein domain, a closer inspection revealed that the C-terminal moiety of ANDGO_04851 contains a region with moderate similarity to the ∼40 amino acid-long core of mS31 from ciliates and metazoans. Across eukaryotes, the core mS31 region can be considerably degenerate, which makes it sometimes difficult to retrieve homologs even from closely related species (see also supplementary fig. S7, Supplementary Material online). This may explain why we did not identify homologs in three out of the other four jakobids (supplementary table S4, Supplementary Material online). Nevertheless, the enrichment of ANDGO_04851 in MS/MS experiments supports the proposed functional assignment (supplementary table S2 and fig. S5, Supplementary Material online), even if the protein does not carry a recognizable mitochondrial targeting peptide (MTP), as in three other undisputed instances (table 1 and supplementary table S4, Supplementary Material online).
Tentative Clade-Specific Components of Jakobid mtSSU
The three remaining top SSU mtRP candidates from Andalucia are ANDGO_08608 with high sequence similarity to a 3-hydroxybutyryl-CoA dehydrogenase (Blast E∼ 3e−170, query coverage ∼94%), an enzyme involved in the mitochondrial isoleucine metabolism; ANDGO_06552, which resembles mitochondrial glutamate dehydrogenases (E∼ 0, query coverage ∼93%); and ANDGO_06037, which retrieves malate dehydrogenases (E∼ 0, query coverage ∼96%) (supplementary table S4, Supplementary Material online). All three Andalucia proteins have sequence features of functional enzymes, with homologs also present in the inferred proteomes from the other jakobids examined here. We speculate that these three proteins are jakobid-specific recruits of mitochondrial metabolic matrix proteins to the mitoribosome and have retained their enzymatic activity. It would be most interesting to validate our inferences by structural analyses of the jakobid mitoribosome.
Broadly Distributed mS34 and mS41 Are Missing from the Andalucia mtSSU
Proteome and 3D-structure analyses confirmed that mS34 is an integral constituent of mitoribosomes in mammals, Arabidopsis and Trypanosoma, but not in Saccharomyces or Neurospora. Andalucia also appears to lack the mS34 gene as do Jakoba spp., whereas unambiguous homologs are present in the R. americana and S. ecuadoriensis nuclear genomes (supplementary table S1, Supplementary Material online). Previously it seemed that the presence of mS34 was correlated with the absence or truncation of the mt-rRNA structure elements h6, h8–h10, and h12 in human and Trypanosoma SSU mt-rRNA (PDB 5aj3-Aj and 6hiw-Cj, respectively; Greber et al. 2015; Ramrath et al. 2018), and vice versa, that the presence of these rRNA elements is associated with the absence of mS34 in yeast. However, recent data from the Arabidopsis and Tetrahymena mitoribosomes show that the retention of these rRNA elements does not preclude mS34 being part of the mitoribosome (see PDB 6xyw-By and 6z1p-Bz, respectively; Tobiasson and Amunts 2020; Waltz et al. 2020) (supplementary fig. S6, Supplementary Material online). In these species with mt-rRNA structure more similar to that of bacteria and jakobids, mS34 binds in between h6 and h8–h10, a region that most likely corresponds to the ancestral binding site of the protein.
Another protein apparently absent from Andalucia and the other examined jakobids is mS41. This mtRP has been experimentally confirmed in yeast (Desai et al. 2017), trypanosome (Ramrath et al. 2018), ciliate (Tobiasson and Amunts 2020), and Arabidopsis (Waltz et al. 2020), but is apparently not part of the metazoan mitoribosomes (supplementary fig. S6, Supplementary Material online). Otherwise, this mtRP appears widespread throughout eukaryotes as seen in sequence similarity searches across protist genomes (fig. 2). In the structural model of the yeast and Arabidopsis mitoribosomes, mS41 interacts almost exclusively with the SSU mt-rRNA helices h6, h15, and h17, but the corresponding region is completely protein-free in bacterial ribosomes. The same situation most likely applies to jakobid SSU mt-rRNAs, given their particularly bacteria-like structures.
Previously Unrecognized or Incorrectly Annotated Mitoribosomal Proteins in Other Eukaryotes
In the course of this study, we realized that certain mtRP classes are difficult to distinguish because of shared sequence domain signatures. One easily confounded pair is mS22/“mS45” & mL58 and the other is mS25 & mL61, the distinction between which we attempted to resolve by eukaryote-wide comparative analyses, as detailed in the following two sections.
Fungal “mS45” Is a Derived mS22 Homolog
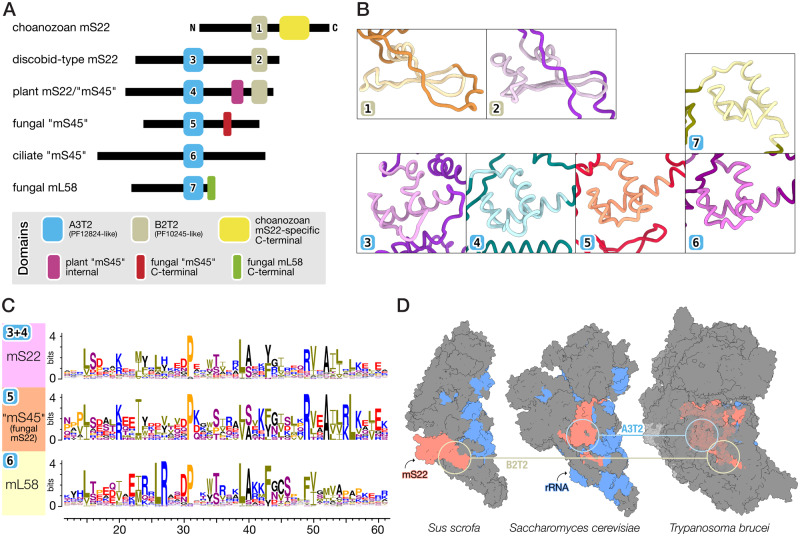
The identification of Andalucia’s mS22 was complicated by the fact that homologs from animals include a single distinctive protein domain MRP-S22, whereas those from trypanosomes include an additional domain otherwise typical for fungal mL58. To understand the domain composition of mS22 across eukaryotes, we created specific profile HMMs for each of these domains, first based on metazoan, trypanosome, and jakobid homologs, and then iteratively including newly identified additional homologs from other eukaryotes. The profile HMM so generated and covering the MRP-S22 (PF10245) domain will be referred to as B2T2 (∼40 residues), alluding to the secondary structure of the corresponding protein region that consists of two beta strands and two turns. Similarly, the profile HMM including the mL58 domain (MRP-L40 or PF12824) will be referred to as A3T2 (∼50 residues) because it contains three alpha helices and two turns (fig. 3A and B, supplementary fig. S8A, Supplementary Material online). With these profile HMMs in hand, we performed HMM searches to identify in eukaryotic proteomes all sequences carrying B2T2, and in independent searches those containing A3T2 (for the collection of inferred eukaryotic proteomes examined here, see Materials and Methods section). Both sets of retrieved sequences were then inspected for additional protein domains.
Fig. 3.
In-depth domain analyses identify fungal “mS45” as a derived mS22. (A) Domain arrangement of choanozoan mS22 (in metazoans+choanoflagellates; note the distinct >100 amino acid-long C-terminal extension), discobid-type mS22 (in discobids, ichthyosporeans, amoebozoans, stramenopiles, cryptophytes, haptophytes, apusozoans, rhodophytes, and chytrid fungi), plant mS22/“mS45” (green algae and land plants), fungal “mS45” (i.e., derived mS22), ciliate “mS45,” and fungal mL58. Cartoon depictions are approximately to scale (relative to median lengths across screened species). For details, see also supplementary figure S8B and C, Supplementary Material online. (B) Snapshots of B2T2 domain structures from pig (S. scrofa) mS22 (1; PDB 5aj3) and T. brucei mS22 (2; PDB 6hiw), as well as of A3T2 domain structures from Trypanosoma mS22 (3; PDB 6hiw), A. thaliana mS22 (4; PDB 6xyw), yeast (S. cerevisiae) “mS45” (5; PDB 5mrc), ciliate (T. thermophila) “mS45” (6; PDB 6z1p), and yeast mL58 (7; PDB 3j6b). For full structural models, see supplementary figure S8A, Supplementary Material online. (C) Sequence logo representations of A3T2 domains typical for mS22 across eukaryotes, fungal “mS45,” and fungal mL58 demonstrate substantial similarity between “mS45” and mS22 (For further details, see supplementary figure S8C and D, Supplementary Material online.). (D) Comparison of the relative positions of the A3T2 and B2T2 domains of mS22 homologs in the mtSSU structural models from pig, yeast, and Trypanosoma. Surfaces of mS22 and SSU mt-rRNA are colored orange and blue, respectively. In the Trypanosoma mtSSU model, mS47, mS48, and mS61 surfaces are displayed semitransparent to better show the location of mS22.
Sequences retrieved with the B2T2 profile fall into two distinct groups (fig. 3A). The first contains proteins carrying exclusively a B2T2 domain, found in choanozoans, that is, metazoans and their closest protozoan relatives, the choanoflagellates (supplementary fig. S8B, Supplementary Material online). The second group contains proteins that carry not only B2T2, but also a second domain, A3T2. The corresponding sequences come from essentially all nonchoanozoan eukaryotic groups including jakobids (supplementary fig. S8B, Supplementary Material online).
Sequences retrieved with the profile A3T2 were highly diverse. Phylogenetic analysis based on this domain showed moderate resolution due to the limited length of the protein region, but still revealed an informative pattern. Among the four clades is one that combines the above-identified two-domain mS22 RPs, together with several proteins lacking the B2T2 domain. The latter proteins are mostly from fungi (but also from alveolates), annotated as homologs of the yeast mS45, which has been considered to be a clade-specific mtRP (fig. 3C and supplementary fig. S8C and D, Supplementary Material online).
Our finding that the fungal and alveolate mS45—which lack the mS22-typical B2T2 domain—affiliate in this tree with the two-domain mS22 suggests that “mS45” is not a distinct mtRP but rather a variant mS22 that has lost its B2T2 domain. The view that mS45 is an mS22 homolog is corroborated by the fact that in the 3D-structure model of yeast mtSSU, this protein interacts with a similar set of conserved mtRPs and occupies a similar location as do animal and trypanosome mS22, in particular the A3T2 domain of the latter (Greber et al. 2015; Desai et al. 2017; Ramrath et al. 2018) (fig. 3D; see also supplementary material). Based on the phylogenetic tree (supplementary fig. S8C, Supplementary Material online), we conjecture that alveolates and stramenopiles as well have lost the B2T2 domain from their mS22 homologs making these mtRPs similar to the fungal “mS45.”
Shared Ancestry of mS25 and mL61, the Latter Encompassing Clade-Specific “mL74” and “mL108”
As mentioned above, mS25 and mL61 share considerable sequence similarity, which led us to the initial misannotation of ANDGO_07531 (Gray et al. 2020). Distinction of the two mtRP classes is impeded by their shared L51_S25_CI-B8 domain (Pfam ID PF05047). In addition, this domain occurs also in mL42 and mL53, and further in nonmitoribosomal proteins such as respiratory chain Complex I subunit NDUFA2 (aka “CI-B8”).
We constructed eukaryote-wide phylogenetic trees with the L51_S25_CI-B8 domains, using NDUFA2 homologs as outgroups. The mtRPs clearly separated into three clades, each represented by experimentally confirmed homologs of mL43, mL53, and mS25 & mL61, respectively. Within the mS25 & mL61 clade, metazoan mS25 on the one hand and fungal mL61 on the other coherently cluster (fig. 4A and supplementary fig. S9A, Supplementary Material online). Although the exact relationship of metazoan mS25 and fungal mL61 to the various protist members within this clade was inconclusive and varied with the applied phylogenetic methodology (supplementary fig. S9B, Supplementary Material online), HMM profile comparisons indicated that the protist sequences were overall more similar to mL61 than to mS25 (supplementary fig. S9C and D, Supplementary Material online).
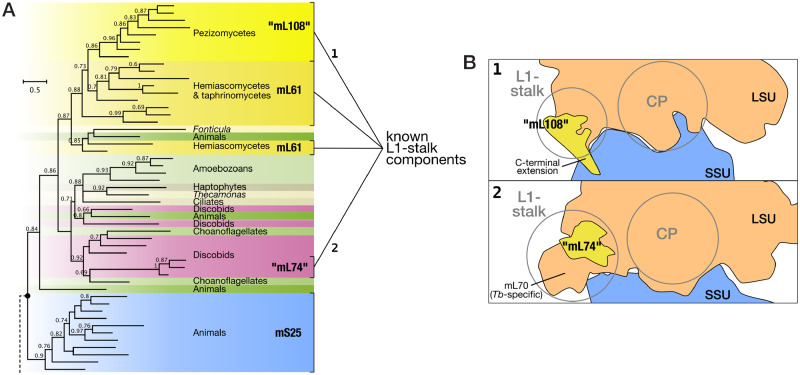
Fig. 4.
In-depth domain analyses indicate that animal mS25, kinetoplastid “mL74,” and pezizomycete “mL108” are derived from mL61. (A) Section of the phylogenetic tree of “L51_S25_CI-B8” domains across eukaryotes that focuses on mS25 and mL61-like sequences (full tree in supplementary fig. S9A, Supplementary Material online). A clade containing all known mS25 representatives (blue) branches separately from the fungal mL61 homologs (yellow shades), as well as mL61-like sequences from other eukaryotes, whose phylogenetic affiliations are indicated by different background colors (support values below 60 are not shown). Homologs of pezizomycete mL61 (aka “mL108”) and kinetoplastid mL61 (aka “mL74”) emerge from within the larger mL61/mL61-like diversity. Dark green background highlights mL61-like sequences from animals, whose genomes also code for a regular, metazoan-type mS25. (B) The yeast mL61, “mL108,” and “mL74” are all known components of the mitoribosomal L1-stalk (see Results section). Although mL61 has not been observed by cryo-EM, locations of the “mL108” (1; Itoh et al. 2020) and “mL74” (2; PDB 6hix) have been determined. As shown in the top views at the mitoribosome subunit interface, the two proteins clearly occupy a similar place in their respective mtLSU, corroborating their homology. For further details, see supplementary figure S9, Supplementary Material online.
Interestingly, deeply-diverging metazoans such as placozoa, sponges, and sea anemones possess two versions of L51_S25_CI-B8 domain-containing proteins, one of which adheres to animal mS25, the other to sequences from nonmetazoan species within the mS25 & mL61 clade (fig. 4A and supplementary fig. S9E, Supplementary Material online). This result strongly suggests that mS25 and mL61 are paralogs that emerged by gene duplication early in metazoan evolution, and that only mS25 was retained in the lineage leading to bilaterian animals.
In addition, the phylogenetic tree sheds light on the descendants of two mtRPs that were previously considered clade-specific. More specifically, mL74 of kinetoplastids (Trypanosoma and relatives; Ramrath et al. 2018) and the recently identified mL108 of pezizomycetes (Neurospora and relatives; Itoh et al. 2020) associate with the mS25 & mL61 clade to the exclusion of other L51_S25_CI-B8 domain-containing protein groups. In particular, the phylogeny shows that “mL108” is the pezizomycete homolog of the yeast mL61, albeit containing a clade-specific ∼40 amino acid-long C-terminal extension (supplementary fig. S9F, Supplementary Material online). The close relationship between mL61, “mL108,” and “mL74” is further corroborated by mitoribosomal 3D-structure models in which Trypanosoma mL74 (Ramrath et al. 2018) and Neurospora mL108 (Itoh et al. 2020) interact with the same partners (uL1m and helices H76–78; also called the L1 stalk) as does yeast mL61 (Kaur and Stuart 2011; Zeng et al. 2018) (fig. 4B).
Discussion
The Andalucia Mitoribosome Combines Highly Bacteria-like and Typical Mitochondrial Features
Jakobid mitochondria are renowned for their extraordinarily bacteria-like rRNAs (Burger et al. 2013; Valach, Burger, et al. 2014) (fig. 1A), but as we have shown earlier (Gray et al. 2020) and now here, this notion applies much less to mitoribosomal proteins. For example, we did not detect more than the conventional number of mtRPs of bacterial provenance (20 SSU and 33 LSU proteins), that is, a mitochondrial counterpart of bS20 appears to be missing in jakobids just as it is in all other eukaryotes. Similarly conventional is the number of M-mtRPs, with at least 11 and 12 such proteins in mtSSU and mtLSU, respectively (fig. 2). Extrapolating from the set of conserved mtRPs that we detected in Andalucia, its mitoribosome has an overall RNA-to-protein ratio of 1:1, which is half the ratio in bacterial ribosomes (2:1) (supplementary table S1, Supplementary Material online). Due to the additional M-mtRPs, the jakobid mitoribosome is estimated to be ∼20% larger than its bacterial counterpart and thus comparable in size to the yeast mitoribosome.
Still, the jakobid mitoribosomal proteome has several minimally derived features setting it apart from other eukaryotes. For instance, in sequence, B-mtRPs of jakobids more closely resemble their bacterial homologs than do their counterparts in other eukaryotes and therefore are placed in proximity to bacterial outgroups in phylogenetic trees (Derelle and Lang 2012; Derelle et al. 2015). Even M-mtRPs from jakobids appear to be less derived compared with those from other eukaryotes. Examples are the aforementioned homologs of mS42 and mS47 that in jakobids have apparently retained their metabolic function. In this aspect, jakobids represent an evolutionary transition state, where proteins co-opted to the mitoribosome still exert their ancestral role, while in more rapidly evolving lineages gene duplication occurred followed by subfunctionalization of paralogs and elimination of functional redundancy.
A Glimpse at the Evolutionary Trajectories of Mitoribosomal Proteins
Eukaryote-wide comparative analyses conducted in the course of this study led us to revise the functional assignment of several mtRPs from animals, fungi, plants, and trypanosomes. The reassignment was facilitated by three assets. One is the availability of near-atomic resolution structures of mitoribosomes, which allowed us to corroborate postulated homologs based on topological properties and interaction partners (Amunts et al. 2014; Greber et al. 2015; Desai et al. 2017; Ramrath et al. 2018; Itoh et al. 2020; Waltz et al. 2020). The second asset is the availability of experimental mitoribosomal and mitoproteome data from several eukaryotic groups, especially protists (Smith et al. 2007; Gawryluk et al. 2014; Rugen et al. 2019; Waltz et al. 2019; Hammond et al. 2020). Third, the moderate divergence of jakobid proteins allowed us to build highly specific profile HMMs for sensitive homology assignment and to reconstruct phylogenetic trees stabilized by short branches.
For example, we provide evidence that mL74 and mL108, previously considered clade-specific to the kinetoplastids and pezizomycete fungi, respectively, are in fact derived versions of the otherwise widely distributed mL61 (fig. 4). In addition, we disentangled the assignment of several mitoribosomal proteins that are easily confused when relying on sequence information and conserved-protein content alone. The first pair of easily confounded mtRPs is mL61 & mS25. Phylogenetic analyses indicate that the ancestral form is actually mL61, from which mS25 arose by gene duplication in the metazoan lineage. In this instance, evolution took an additional step: in bilaterian animals, the orthologous mL61 was lost, leaving behind the paralogous mS25 (fig. 4 and supplementary fig. S8, Supplementary Material online). This is a typical case of hidden paralogy, a well-known problem in phylogenetic reconstruction leading to erroneous phylogenetic inferences.
The second confounding pair is mS22/mS45 & mL58. According to our in silico analyses, mS22, which is present in most eukaryotes including jakobids (fig. 3 and supplementary fig. S8, Supplementary Material online), contains two conserved protein domains (B2T2 and A3T2, see Results). The corresponding orthologs from metazoans and choanoflagellates have lost the N-terminal domain, and thus only possess B2T2, whereas those in fungi have retained just the A3T2 domain and have therefore not been previously recognized as mS22, but incorrectly assigned as a novel mtRP, mS45. Further complicating matters, fungal mitoribosomes contain the LSU mtRP mL58, which is characterized by a single A3T2 domain similarly to “mS45,” that is, the actual fungal mS22. The most parsimonious scenario is that the fungal mL58 originated from mS22 by gene duplication (likely via another paralog, Rrg9; supplementary fig. S8C and D, Supplementary Material online) and subsequently lost the B2T2 protein domain.
The mtRP Complement of Andalucia Implies an Extended Set of Mitoribosomal Proteins in LECA
The phylogenetic distribution of paralogs not only allows us to formulate hypotheses about the timing of gene duplications, subfunctionalization, and losses of one of the two paralogs, but most importantly, about the ancestral gene at the origin of these events. For two paralogous pairs of mtRP-encoding genes, we provide reasonable ancestry evidence, notably, we posit that mS22 gave rise to mL58 and mL61 to mS25.
A related longstanding question pertains to the set of ancestral genes present in LECA. The array of postulated mitoribosomal components in LECA was first established over a decade ago (Smits et al. 2007). Through the present and earlier studies by us (Gray et al. 2020) and work by others (Desmond et al. 2011; Waltz and Giegé 2020), this list has now expanded from initially 30 SSU and 38 LSU mt-RPs to 33 and 46 (possibly 49), respectively (see fig. 2). Thus, LECA’s mitoribosome, with about 50% more RPs than its bacterial predecessor, possessed a much more complex mitochondrial translation machinery than generally assumed.
Was Recruitment of Mitochondrion-Specific Ribosomal Proteins Driven by mt-rRNA Defects?
The observation that in mammals and fungi streamlined mt-rRNAs are associated with high mitoriboproteome complexity led to the common assumption that M-mtRPs structurally replace evolutionarily deleted rRNA elements. High-resolution analyses of mitoribosomal architecture showed that this notion fully applies only to certain mitoribosomes (e.g., from Trypanosoma; Ramrath et al. 2018), whereas in others (e.g., from pig; Greber et al. 2015), the volume of certain missing rRNA segments has remained void. A variation of the RNA-replacement-by-protein view is that evolutionary changes to mt-rRNA structure resulted in its structural instability, which led to the incorporation of stabilizing M-mtRPs (Petrov et al. 2019).
As we show here, the hypothesis of “structural patching” is at odds with our findings in Andalucia and other jakobids, in which a remarkably bacteria-like rRNA architecture coexists with numerous M-mtRPs. Although many M-mtRPs or even B-mtRPs have evolved to compensate for absence of certain rRNA elements (Hosseini et al. 2018), this does not appear to have been their ancestral role. For instance, the mtSSU proteins mS42/mS43 and mS47 nowadays compensate for rRNA helix truncations in Trypanosoma (Ramrath et al. 2018). However, as illustrated here by Andalucia, these M-mtRPs were recruited early on into a mitoribosome with virtually no rRNA structural deviations, which implies that their initial incorporation did not serve to counter rRNA lability. Similarly, five M-mtRPs that compensate for helix truncations and losses of certain rRNA helices in mammalian mtSSU (Hosseini et al. 2018) are conserved in other lineages that do contain full-length versions of these helices with the corresponding proteins binding close by. This strongly suggests that the compensatory role of these proteins could arise because they already resided in the corresponding location (supplementary table S5, Supplementary Material online). These and other examples (see supplementary material) illustrate that deviation of mt-rRNAs and acquisition of most, if not all ancestral M-mtRPs were independent evolutionary events and presumably the result of very different evolutionary forces.
Although we argue about the “purpose” of protein acquisitions to the ancestral mitoribosome, rRNA-stabilizing M-mtRPs may well have been recruited in lineages such as plants and fungi, whose mitoribosomes have undergone a relatively recent constructive phase. In the Arabidopsis mitoribosome, for example, numerous extended mt-rRNA segments are stabilized by dedicated proteins of the pentatricopeptide repeat (PPR) family (Waltz et al. 2020), which has undergone a recent expansion in land plants (Gutmann et al. 2020). Likewise, M-mtRPs interacting with rRNA expansions in the Saccharomyces mitoribosome are recent acquisitions in the fungal lineage, many having nucleic acid-interaction domains (Amunts et al. 2014) (The case of mL44 is discussed in more detail in supplementary material.). It will be interesting to retrace the order of evolutionary events once more information is available about mitoribosomes of early-diverging plants and fungi.
What Then Drives the Recruitment of Organelle-Specific Proteins?
The mitoribosome is not the only mitochondrial complex that is considerably inflated compared with its bacterial ancestor. The same phenomenon applies to mitochondrial bioenergetic complexes where numerous accessory proteins were added to the ancient bacterial core (Acestor et al. 2011; Gawryluk et al. 2012; Hirst 2013; Jett and Leary 2018). It is widely assumed that the newly acquired proteins serve mitochondrion-specific functions, and thus are a manifestation of adaptive evolutionary forces (Greber and Ban [2016] for mitoribosomes and Hirst [2011] for respiratory Complex I). Up to now, special biological functions in mitochondrial translation have only been documented for three mtRPs. The mtRP mL45 (and, in yeast, its evolutionarily unrelated counterpart Mba1) mediates the binding of the mitoribosome to the inner mitochondrial membrane (Greber et al. 2014; Englmeier et al. 2017), which is necessary for the cotranslational insertion of the many highly hydrophobic mitochondrion-encoded proteins synthesized on the mitoribosome (Ott and Herrmann 2010). The other two examples are lineage-specific mtRPs of the PPR family: mS39, which helps to usher leader-less mt-mRNAs to the mammalian mitoribosome (Kummer et al. 2018), and mS83, which forms the mt-mRNA channel in the Arabidopsis mitoribosome (Waltz et al. 2020). Actually, the majority of eukaryote-wide conserved M-mtRPs are of lesser importance in mitoribosome biogenesis, since they are incorporated at intermediate or late stages of assembly, and two particularly broadly conserved RPs, mL53 and mL61, are even nonessential in yeast (Zeng et al. 2018).
As outlined earlier (van der Sluis et al. 2015), the fact that a given protein plays a specific functional role does not imply that the initial recruitment was driven by adaptation to mitochondrion-specific demands. Instead, the expansion of mitochondrial protein content is more likely due to neutral (constructive) evolution, which invokes neutrally fixed complexity and only gradual increasing dependency from the newly recruited components (Stoltzfus 1999; Gray et al. 2010; Lukeš et al. 2011).
It is noteworthy that the evolutionary trajectory of complexes and pathways in the much younger “sister” organelle, the plastid, is quite similar, but the number of accessory proteins is apparently less. For instance the plastoribosome from land plants comprises only six such accessory proteins (Graf et al. 2017) and the plastid NAD(P)H dehydrogenase complex only 10 (compared with ∼30 in the mitochondrial Complex I) (Peng et al. 2011;), indicating that protein acquisition is intimately linked to the establishment of organelles in the eukaryotic cell. However, available plastid proteome data are limited in number and taxonomic breadth, with conspicuous lack of information from slowly evolving plastids that retain more (cyano)bacterial features (similar to mitochondria in jakobids), precluding us from drawing a picture of the evolutionary trajectory of the plastid as detailed as the one we currently have for mitochondria.
Current Limitations and Outlook
As a cautionary note, we feel it is important to emphasize the difficulty in recognizing homologs of mitoribosomal proteins, especially in rapidly evolving systems. Extreme cases are the mitoribosomes of Tetrahymena and Trypanosoma, which appeared to have lost nearly half of the mtRPs predicted to have been present in LECA based on sequence information alone. However, many of the seemingly lost mtRPs have in fact remained unrecognized due to extreme sequence divergence (this study; Ramrath et al. 2018; Tobiasson and Amunts 2020). Although proteins with low sequence conservation are best recognized by HMM searches, this approach fails when moderately derived and taxonomically broad homologous sequences are in short supply for profile-HMM construction as most aptly illustrated by the case of mS31 (see supplementary material).
A second issue is that our view of mitoribosome composition and architecture is shaped by the biochemical procedures by which ribonucleoprotein complexes are currently isolated. In contrast to bacterial or cytosolic ribosomes, which are distinct subcellular entities, mitoribosomes are tightly bound to the mitochondrial membrane (Pfeffer et al. 2015; Englmeier et al. 2017), explaining why the isolation of free mitoribosomes has been a challenge in whichever system it has been attempted. Procedures for isolating mitoribosomes rely on detergents, potentially disrupting critical protein-protein or protein-RNA interactions. In particular, if a protein of poor sequence conservation and short length (e.g., mL42 or mL63) is absent from the 3D-structure inference, it was not necessarily lost during evolution, but potentially during ribosome isolation. Noteworthy examples are components of the L1 stalk, mL61 and uL1m, which have not been observed in cryo-EM-based structures either due to fragility or high mobility of the L1 stalk (Amunts et al. 2014; Desai et al. 2017). Similarly, the absence of this structural element in a recent model of the Tetrahymena mitoribosome (Tobiasson and Amunts 2020) is unlikely a consequence of genuinely missing L1 stalk components because candidate uL1m and mL61 (identified here) were previously detected by proteomics of the organism’s purified mitochondria (Smith et al. 2007) (see also supplementary fig. S9A and table S1, Supplementary Material online). Conversely, copurification of extra proteins with the mitoribosome does not provide unequivocal evidence that such proteins participate in translation (see Kehrein et al. 2015). The mitoribosome may just serve as a foothold for certain proteins such as the matrix enzyme HIBCH in yeast and Andalucia.
The last and most important point is that it is currently not feasible to prepare a minimal functional mitoribosome capable of in vitro translation, so it is difficult to argue that isolated mitoribosomes are pure and intact. Therefore, key for future 3D and proteomics studies will be the availability of an in vitro translation assay, which will pinpoint, in conjunction with gene-knockout or gene-editing, the components that are necessary and sufficient for translation. Once a functional test is available, our view of the mitoribosome as we see it today may undergo further unexpected transformation.
Materials and Methods
Strains and Culture Conditions
Andalucia godoyi (strain PRA-185) was kindly provided by Alastair Simpson (Dalhousie University, Halifax, Canada). Andalucia godoyi starter cultures were grown at 18–20 °C for 2 weeks in 25 cm2 horizontal plastic culture flasks containing 15 ml of WCL medium (http://megasun.bch.umontreal.ca/People/lang/FMGP/methods/wcl.html, last accessed June 11, 2020), feeding on live Klebsiella (Enterobacter) aerogenes (ATCC 13048). To scale up, the starter culture was first transferred into a 225 cm2 horizontal plastic culture flask containing 100 ml fresh medium, grown for a further two weeks, and then transferred into a 700 cm2 sterile glass container with 400 ml additional medium. The culture was regularly inspected under a microscope and new live bacteria were added once most had been consumed by the protist. As soon as the late exponential phase was reached at ∼2 × 106 cells/ml (usually after two weeks), Andalucia cells were harvested by centrifugation at 8,000 × g for 20 min at 4 °C. The supernatant was discarded and the pellet resuspended in WCL. The suspension was transferred into 1.5-ml tubes and centrifuged at 8,000 × g for 3 min. This WCL wash was repeated twice. The pelleted cells were either directly frozen in liquid nitrogen or resuspended in 1% DMSO and stored at −70 °C until further use.
RNA Extraction, Reverse Transcription, PCR, and Northern Blot Hybridization
RNA was extracted with a “home-made” Trizol substitute (Rodríguez-Ezpeleta et al. 2009). Residual DNA in RNA preparations was removed by digestion with TURBO DNase (Invitrogen) followed by extraction with the Trizol substitute. Reverse transcription was performed with AMV reverse transcriptase (Roche) using target-specific primers or with SuperScript IV Reverse Transcriptase (Thermo) using random hexamers. DNA was amplified with Q5 High-Fidelity DNA Polymerase (New England BioLabs). Radio-labeling of oligonucleotide probes employed T4 polynucleotide kinase (Thermo). Northern blotting and hybridization were performed essentially as described previously (Valach, Moreira, et al. 2014), except that agarose gel electrophoresis was performed in the TT buffer system (1× TT: 30 mM tricine, 30 mM triethanolamine) with 0.4 M formaldehyde in the gel to improve the separation of long, highly structured RNAs (Mansour and Pestov 2013). Each RNA sample was mixed with an equal volume of the loading buffer (50% formamide, 30 mM tricine, 30 mM triethanolamine, 0.5 mM EDTA, 0.02% bromophenol blue), denatured (70 °C, 5 min), chilled on ice, and supplemented with formaldehyde to a final concentration of 0.4 M. Electrophoretic separation of the samples was carried out at 6 V/cm in a 1% or 2% agarose gel in 1× TT buffer. Gels were stained with ethidium bromide and RNA was visualized under UV light. Oligonucleotides (Integrated DNA Technologies) used as primers and probes are listed in supplementary table S6, Supplementary Material online.
Mapping of mt-rRNA Ends
Transcriptome sequencing (RNA-Seq) data from a previous study (Valach, Burger, et al. 2014) were used to determine the 5′ and 3′ termini of mature mt-rRNAs of A. godoyi and J. bahamiensis. Briefly, total RNA including small RNAs was extracted from whole cells. RNA-Seq libraries were constructed using the TruSeq Small RNA Sample Prep kit (Illumina) without size-fractionation and sequenced on Illumina HiSeq2500 (A. godoyi) and MiSeq (J. bahamiensis) platforms at the Genome Quebec Innovation Center (Montreal, Canada). After quality trimming and adaptor clipping with cutadapt v1.2.1 (Martin 2011), reads were mapped onto mitochondrial genome sequences (GenBank Acc. Nos KC353352 and KC353354, respectively) by bowtie2 in the local mode (Langmead and Salzberg 2012). The bowtie2-output SAM file was then parsed to locate the mt-rRNA termini at single nucleotide resolution. The same procedure allowed us earlier to precisely map 5S mt-rRNA termini (Valach, Burger, et al. 2014).
Enrichment of Mitochondria
Mitochondrial preparations were obtained by the nitrogen cavitation approach described earlier (Valach et al. 2018). Briefly, cells were grown until late exponential phase as detailed above, harvested by centrifugation (8,000 × g, 4 °C, 20 min), washed twice in WCL medium, once in ice-cold SoTE buffer (0.6 M sorbitol, 10 mM Tris-HCl pH7.5, 5 mM EDTA pH8.0), and then resuspended in the same buffer. Cells were lysed in a nitrogen cavitation chamber (Parr Instrument Company) under 90-bar nitrogen pressure for 20 min. The lysate was centrifuged (1,000 × g, 4 °C, 20 min) and the mitochondria-containing supernatant was transferred to a new tube. After an additional centrifugation (20,000 × g, 4 °C, 20 min), the mitochondria-enriched pellet was washed once in ice-cold SoTE buffer (20,000 × g, 4 °C, 10 min). The mitochondrial material was either directly used or frozen in liquid nitrogen and stored at −70 °C. Mitochondrial enrichment was assessed by measuring the relative proportion of mitochondrial to cytosolic rRNAs using Northern blotting.
Sucrose Gradient, Protein Extraction, and Sample Preparation for Mass Spectrometry
Andalucia godoyi mtSSU was enriched by sucrose gradient centrifugation. Andalucia cells harvested from a 4–6 l culture were resuspended in one volume homogenization buffer (5 mM Tris-HCl pH7.5, 1 mM MgCl2, 1 mM DTT, 0.25 mM EDTA; the buffer composition was optimized to minimize mt-rRNA degradation). Cells were then lysed with two volumes homogenization buffer supplemented with 2% Triton X-100, 1× EDTA-free protease inhibitor (Roche), and 4 U/μl SUPERase·In (Invitrogen). The lysate was incubated on ice for 5 min followed by centrifugation (18,000 × g, 10 min, 4 °C). From the resulting supernatant of ∼250 µl, ∼1/10 (∼100 µg proteins) was set aside to be later used as the “input reference,” while the remaining 9/10 portion was loaded on top of a 5-ml 15–40% sucrose gradient (containing the homogenization buffer and 0.05% Triton X-100) and centrifuged in an AH-650 swinging-bucket rotor (∼250,000 × g, 3 h, 4 °C); alternatively, we used a 10-ml 10–40% sucrose gradient in an TH-641 swinging-bucket rotor (∼250,000 × g, 5 h, 4 °C). After centrifugation, gradient fractions of 250 μl were collected from the top, snap-frozen in liquid nitrogen, and stored at −70 °C until further use. To determine the migration of the mtSSU, RNA was extracted from every second gradient fraction, and the mt-rRNA content was measured by Northern blotting and RT-PCR as described above.
Fractions enriched in the mtSSU were concentrated to a volume of 50–65 µl on a 10-kDa MWCO Amicon Ultra-0.5 (Millipore Sigma) centrifugal filter (14,000 × g, 30–120 min, 4 °C) following the manufacturer’s instructions. The concentrate was mixed with a 20-fold larger volume of UT buffer (6 M urea, 100 mM Tris pH8.5) and concentrated to ∼120 µl using a 3-kDa MWCO filter device, essentially following a previously published procedure (Wiśniewski et al. 2009) to remove detergents and sucrose. A small aliquot (∼1/5) was analyzed by Tricine SDS–PAGE (Schägger 2006). After electrophoresis, protein bands were visualized by staining with SYPRO Ruby fluorescent dye (Invitrogen). Similarly, the aliquot of the sucrose-gradient input reference was diluted 200 times in UT buffer and then concentrated to ∼250 µl by passage through a 3-kDa MWCO filter (Amicon Ultra-15) to remove Triton X-100 prior to MS/MS analysis. Samples enriched in mitochondrial material (see above) were heat-denaturated in the presence of 2% SDS and concentrated by electrophoresis in a SDS–PAGE stacking gel. Subsequently, the protein-containing zone was cut out and the proteins were fixed by methanol and acetic acid ( see Valach et al. 2018). Replicates were prepared for each sample, the mtSSU-enriched material (samples A23 and A26), whole-cell lysate reference (A31 and A32), and mitochondria-enriched fraction (A10 and A11) (supplementary tables S2, S4, and S6, Supplementary Material online).
Protein Identification, Quantification, and Ranking
Detergent-free protein samples (∼2 to 4 µg each) were submitted to proteomics technology platforms at the Institut de Recherches Cliniques de Montréal (IRCM) and the Institut de Recherche en Immunologie et en Cancérologie (IRIC) in Montreal, Canada, for in-solution trypsin digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis using an Orbitrap Fusion (Thermo) instrument. Peptide searches in the raw MS/MS data set were performed by the platforms using Mascot (Proteome Discoverer, Matrix Science), whereas we used MaxQuant v1.6.1.0 (Cox and Mann 2008; Tyanova et al. 2016) (supplementary table S7, Supplementary Material online).
In both Mascot and MaxQuant analyses, we searched for peptide-spectrum matches (PSMs) in a custom database of A. godoyi proteins inferred from mitochondrial and nuclear genome sequences (Burger et al. 2013; Gray et al. 2020). Trypsin digestion parameters were set allowing up to two missed cleavage sites per protein. As a fixed modification, we specified carbamidomethylation of cysteine; as variable modifications (up to four per peptide), we specified methionine oxidation, asparagine and glutamine deamidation, serine and threonine phosphorylation, and conversion of glutamine and glutamate at peptide N-termini to pyrrolidone-carboxylic acid. Minimum and maximum peptide lengths were set to 7 and 50 amino acid residues, respectively (up to 6,000 Da). False discovery rates for PSMs and protein identification probability were determined by the target-decoy approach and set to 1%.
Proteins were quantified essentially as in an earlier publication (Valach et al. 2018) by calculating PAI (Protein Abundance Index) (Rappsilber et al. 2002) and iBAQ (intensity-Based Absolute Quantification) values (Schwanhäusser et al. 2011). Briefly, spectral (MS/MS) counts and identified peptide intensities (for PAI and iBAQ, respectively) of each protein were normalized by the theoretical number of peptides to which the protein could give rise using the MS-Digest tool from the ProteinProspector v5.23.2 tool suite (available from: http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi? form=msdigest, last accessed April 15, 2020). We used the following parameters: trypsin digest; no missed cleavage; carbamidomethyl at Cys residues as fixed modification; no variable modification; minimal length of 7 amino acids; and peptide mass range from 720 to 3,000 Da. The selected mass range thus covered >95% of all identified peptides.
The enrichment of a protein in mtSSU samples (A23, A26) was calculated as the ratio of the protein quantity in this sample versus its average abundance in the reference samples (A31, A32). Threshold values for proteins to be considered candidates for new SSU mtRPs were determined based on the abundance and enrichment of the identified mtSSU RPs (supplementary table S2 and fig. S5, Supplementary Material online). This was done separately for the results of Mascot and MaxQuant, as well as the particular quantification scheme (PAI, iBAQ), that is, with four different metrics. Proteins were analyzed further (see below) if in both mtSSU-enriched samples they displayed an SSU mtRP-like distribution for at least one metric (supplementary fig. S4, Supplementary Material online).
Protein identification in the whole-cell lysate replicates was consistent, with a correlation factor ∼0.89 for both quantitative measures (iBAQ and PAI) and both search tools (Mascot and MaxQuant). The composition of mtSSU-enriched samples across different preparations was more variable (correlation ∼0.63).
Analyses of Protein and RNA Sequences and Structure Modeling
The Andalucia-specific SSU mtRP candidates identified by quantitative analyses of MS/MS data had been mostly annotated as hypothetical proteins (Gray et al. 2020). For these candidates, we analyzed their protein-domain composition with SMART (Letunic and Bork 2018), Pfam (Finn et al. 2016), and NCBI CDD (Marchler-Bauer et al. 2015). Potential transmembrane helices (THM) were predicted using TMHMM2.0 (webservice at http://www.cbs.dtu.dk/services/TMHMM/, last accessed January 28, 2020) and PolyPhobius (http://phobius.sbc.su.se/poly.html, last accessed January 28, 2020) employing default parameters. Mitochondrial protein targeting was predicted previously (Gray et al. 2020).
We searched by local Blast (Altschul et al. 1997) for homologs of Andalucia SSU and LSU mtRPs in the genome-inferred proteomes of four additional jakobid species (J. bahamiensis, J. libera, R. americana, and S. ecuadoriensis). The corresponding proteome data sets were constructed as described previously for A. godoyi (Gray et al. 2020). Procedures described for jakobids in the following were also applied to reference proteome data sets from a wide collection of eukaryotes downloaded from NCBI RefSeq or from UniProtKB (supplementary table S8, Supplementary Material online). For comparative purposes, we identified homologs of SSU and LSU mtRPs in the five jakobid species by local Blast searches (Altschul et al. 1997) in custom databases of predicted jakobid proteins with a collection of previously characterized RPs as queries. When Blast failed to retrieve a homolog, the proteomes were queried with HMMER3 (Eddy 2011) using in-house generated profile HMMs. For building these profiles, as well as for constructing phylogenetic trees, multiple sequence alignments were generated by Clustal Omega (Sievers et al. 2011) or MAFFT v7.388 (Katoh and Standley 2013). Trees were computed by FastTree v2.1.11 (Price et al. 2010) or IQ-TREE v1.6.10 (Nguyen et al. 2015). Sequence logos were generated using WebLogo 3 (Crooks et al. 2004). HHsuite v3.0.3 (Remmert et al. 2012) was then used to generate profile HMMs from the alignments and to compare pairs of profile HMMs. Alignment annotations and peptide-to-protein mapping were done in Geneious Prime 2020.1.1 (Biomatters).
Multiple nucleotide-sequence alignments of SSU and LSU mt-rRNAs from seven jakobid species (A. godoyi, R. americana, S. ecuadoriensis, J. bahamiensis, J. libera, Histiona aroides, Ophirina amphinema; available from GenBank) and E. coli 16S and 23S rRNAs were also generated using MAFFT and were manually adjusted based on secondary structure conservation. Secondary structures were modeled following known structural elements of the E. coli rRNAs (Yusupov et al. 2001) (see also the Comparative RNA website at http://www.rna.ccbb.utexas.edu/, last accessed February 29, 2020). Folding of divergent and additional elements in the jakobid sequences was predicted by thermodynamics- and covariation-based tools implemented in the Vienna RNA package 2.0 (Lorenz et al. 2011). Models were drawn using XRNA v1.1.12 (http://rna.ucsc.edu/rnacenter/xrna/, last accessed February 29, 2020) and Affinity Designer v1.8 (Serif Europe).
3D models of the E. coli ribosome (Noeske et al. 2015) and mitochondrial ribosomes from Sus scrofa (Greber et al. 2015), Saccharomyces cerevisiae (Amunts et al. 2014; Desai et al. 2017), Trypanosoma brucei (Ramrath et al. 2018), Arabidopsis thaliana (Waltz et al. 2020), and Tetrahymena thermophila (Tobiasson and Amunts 2020) were visually inspected in the UCSF Chimera (Pettersen et al. 2004) and UCSF ChimeraX 0.91/1.0 (Goddard et al. 2018) environments.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Mohamed Aoulad Aissa (Université de Montréal, Montreal, Canada) for excellent technical assistance during protist cultivation and help with the initial experiments. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) (grants RGPIN-2014-05286 and RGPIN-2019-04024 to G.B.; and RGPIN-2017-05411 to B.F.L.) and from the Fonds de Recherche du Quebec—Nature et Technologies (FRQNT) (grant 2018-PR-206806 to B.F.L., G.B.). M.W.G. acknowledges past support of research in this area by CIHR (grant MOP-4124).
Author Contributions
G.B., M.V. conceptualization; M.V., M.S., M.W.G. data curation; M.V., J.A.G.A., M.W.G., G.B. investigation; M.V. formal analysis; J.A.G.A., M.V. methodology; G.B. project administration, supervision; G.B., B.F.L., M.S. resources; G.B., B.F.L. funding acquisition; M.V. visualization; M.V., G.B. writing—original draft; M.V., G.B., M.W.G., B.F.L., J.A.G.A., M.S. writing—review and editing.
Data Availability
New data reported in this article are available in the online supplementary material.
References
- Acestor N, Zíková A, Dalley RA, Anupama A, Panigrahi AK, Stuart KD.. 2011. Trypanosoma brucei mitochondrial respiratome: composition and organization in procyclic form. Mol Cell Proteomics 10(9):M110.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Brown A, Bai X-C, Llácer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SHW, Ramakrishnan V.. 2014. Structure of the yeast mitochondrial large ribosomal subunit. Science 343(6178):1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JHD, Dinman JD, Dragon F, Ellis SR, Lafontaine DLJ, Lindahl L, Liljas A, Lipton JM, et al. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Amunts A, Bai X-C, Sugimoto Y, Edwards PC, Murshudov G, Scheres SHW, Ramakrishnan V.. 2014. Structure of the large ribosomal subunit from human mitochondria. Science 346(6210):718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF.. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523(7559):208–211. [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF.. 2013. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 5(2):418–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26(12):1367–1372. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 14(6):1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz VF, Lake JA, Simpson AM, Simpson L.. 1985. A minimal ribosomal RNA: sequence and secondary structure of the 9S kinetoplast ribosomal RNA from Leishmania tarentolae. Proc Natl Acad Sci U S A. 82(5):1401–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz VF, Simpson AM, Lake JA, Simpson L.. 1985. Primary sequence and partial secondary structure of the 12S kinetoplast (mitochondrial) ribosomal RNA from Leishmania tarentolae: conservation of peptidyl-transferase structural elements. Nucleic Acids Res. 13(7):2337–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, Lang BF.. 2012. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol. 29(4):1277–1289. [DOI] [PubMed] [Google Scholar]
- Derelle R, Torruella G, Klimeš V, Brinkmann H, Kim E, Vlček Č, Lang BF, Eliáš M.. 2015. Bacterial proteins pinpoint a single eukaryotic root. Proc Natl Acad Sci U S A. 112(7):E693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Brown A, Amunts A, Ramakrishnan V.. 2017. The structure of the yeast mitochondrial ribosome. Science 355(6324):528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond E, Brochier-Armanet C, Forterre P, Gribaldo S.. 2011. On the last common ancestor and early evolution of eukaryotes: reconstructing the history of mitochondrial ribosomes. Res Microbiol. 162(1):53–70. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47(D1):D427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englmeier R, Pfeffer S, Förster F.. 2017. Structure of the human mitochondrial ribosome studied in situ by cryoelectron tomography. Structure 25(10):1574–1581.e2. [DOI] [PubMed] [Google Scholar]
- Feagin JE, Harrell MI, Lee JC, Coe KJ, Sands BH, Cannone JJ, Tami G, Schnare MN, Gutell RR.. 2012. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum. PLoS One 7(6):e38320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawryluk RMR, Chisholm KA, Pinto DM, Gray MW.. 2012. Composition of the mitochondrial electron transport chain in Acanthamoeba castellanii: structural and evolutionary insights. Biochim Biophys Acta 1817(11):2027–2037. [DOI] [PubMed] [Google Scholar]
- Gawryluk RMR, Chisholm KA, Pinto DM, Gray MW.. 2014. Compositional complexity of the mitochondrial proteome of a unicellular eukaryote (Acanthamoeba castellanii, supergroup Amoebozoa) rivals that of animals, fungi, and plants. J Proteomics 109:400–416. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE.. 2018. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27(1):14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf M, Arenz S, Huter P, Dönhöfer A, Novácek J, Wilson DN.. 2017. Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 45(5):2887–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Derelle R, Klimeš V, Leger MM, Sarrasin M, Vlček Č, Roger AJ, Eliáš M, Lang BF.. 2020. The draft nuclear genome sequence and predicted mitochondrial proteome of Andalucia godoyi, a protist with the most gene-rich and bacteria-like mitochondrial genome. BMC Biol. 18(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Doolittle WF.. 1982. Has the endosymbiont hypothesis been proven? Microbiol Rev. 46(1):1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G.. 2004. Mitochondria of protists. Annu Rev Genet. 38(1):477–524. [DOI] [PubMed] [Google Scholar]
- Gray MW, Lukeš J, Archibald JM, Keeling PJ, Doolittle WF.. 2010. Irremediable complexity? Science 330(6006):920–921. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Ban N.. 2016. Structure and function of the mitochondrial ribosome. Annu Rev Biochem. 85(1):103–132. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D, Ban N.. 2015. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348(6232):303–308. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N.. 2014. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505(7484):515–519. [DOI] [PubMed] [Google Scholar]
- Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barré A, Yoshizawa S, Fourmy D, de Crécy-Lagard V, et al. 2014. Predicting the minimal translation apparatus: lessons from the reductive evolution of Mollicutes. PLoS Genet. 10(5):e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann B, Royan S, Schallenberg-Rüdinger M, Lenz H, Castleden IR, McDowell R, Vacher MA, Tonti-Filippini J, Bond CS, Knoop V, et al. 2020. The expansion and diversification of pentatricopeptide repeat RNA-editing factors in plants. Mol Plant 13(2):215–230. [DOI] [PubMed] [Google Scholar]
- Hammond MJ, Nenarokova A, Butenko A, Zoltner M, Dobáková EL, Field MC, Lukeš J.. 2020. A uniquely complex mitochondrial proteome from Euglena gracilis. Ávila-Arcos MC, editor. Mol Biol Evol . 37:2173–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen JK, Mursula AM, Rottensteiner H, Wierenga RK, Kastaniotis AJ, Gurvitz A.. 2003. The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 27(1):35–64. [DOI] [PubMed] [Google Scholar]
- Hirst J. 2011. Why does mitochondrial complex I have so many subunits? Biochem J. 437(2):e1–3. [DOI] [PubMed] [Google Scholar]
- Hirst J. 2013. Mitochondrial complex I. Annu Rev Biochem. 82(1):551–575. [DOI] [PubMed] [Google Scholar]
- Hosseini M, Roy P, Sissler M, Zirbel CL, Westhof E, Leontis N.. 2018. How to fold and protect mitochondrial ribosomal RNA with fewer guanines. Nucleic Acids Res. 46(20):10946–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Naschberger A, Mortezaei N, Herrmann J, Amunts A. 2020. Analysis of translating mitoribosome reveals functional characteristics of protein synthesis in mitochondria of fungi. Nat Commun 11, 5187. 10.1038/s41467-020-18830-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jett KA, Leary SC.. 2018. Building the CuA site of cytochrome c oxidase: a complicated, redox-dependent process driven by a surprisingly large complement of accessory proteins. J Biol Chem. 293(13):4644–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Stuart RA.. 2011. Truncation of the Mrp20 protein reveals new ribosome-assembly subcomplex in mitochondria. EMBO Rep. 12(9):950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrein K, Schilling R, Möller-Hergt BV, Wurm CA, Jakobs S, Lamkemeyer T, Langer T, Ott M.. 2015. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 10(6):843–853. [DOI] [PubMed] [Google Scholar]
- Klimov PB, Knowles LL.. 2011. Repeated parallel evolution of minimal rRNAs revealed from detailed comparative analysis. J Hered. 102(3):283–293. [DOI] [PubMed] [Google Scholar]
- Kummer E, Leibundgut M, Rackham O, Lee RG, Boehringer D, Filipovska A, Ban N.. 2018. Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 560(7717):263–267. [DOI] [PubMed] [Google Scholar]
- Lang BF, Burger G, O'Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW.. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387(6632):493–497. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46(D1):D493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL.. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol. 6(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš J, Archibald JM, Keeling PJ, Doolittle WF, Gray MW.. 2011. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life 63(7):528–537. [DOI] [PubMed] [Google Scholar]
- Mansour FH, Pestov DG.. 2013. Separation of long RNA by agarose-formaldehyde gel electrophoresis. Anal Biochem. 441(1):18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43(D1):D222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. [Google Scholar]
- Melnikov S, Manakongtreecheep K, Söll D.. 2018. Revising the structural diversity of ribosomal proteins across the three domains of life. Mol Biol Evol. 35(7):1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, Haeseler von A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Wasserman MR, Terry DS, Altman RB, Blanchard SC, Cate JHD.. 2015. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 22(4):336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Herrmann JM.. 2010. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta 1803(6):767–775. [DOI] [PubMed] [Google Scholar]
- Peng L, Yamamoto H, Shikanai T.. 2011. Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim Biophys Acta 1807(8):945–953. [DOI] [PubMed] [Google Scholar]
- Petrov AS, Wood EC, Bernier CR, Norris AM, Brown A, Amunts A.. 2019. Structural patching fosters divergence of mitochondrial ribosomes. Mol Biol Evol. 36(2):207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE.. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Woellhaf MW, Herrmann JM, Förster F.. 2015. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat Commun. 6(1):110–118. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. Poon AFY, editor. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. 2014. The ribosome emerges from a black box. Cell 159(5):979–984. [DOI] [PubMed] [Google Scholar]
- Ramrath DJ, Niemann M, Leibundgut M, Bieri P, Prange C, Horn EK, Leitner A, Boehringer D, Schneider A, Ban N.. 2018. Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes. Science 362(6413):eaau7735. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M.. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12(8):1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, Söding J.. 2012. HHblits: lightning-fast iterative protein sequence searching by HMM–HMM alignment. Nat Methods 9(2):173–175. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W.. 2010. The ribosome goes Nobel. Trends Biochem Sci. 35(1):1–5. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Teijeiro S, Forget L, Burger G, Lang BF.. 2009. Construction of cDNA libraries: focus on protists and fungi. Methods Mol Biol. 533:33–47. [DOI] [PubMed] [Google Scholar]
- Rugen N, Straube H, Franken LE, Braun H-P, Eubel H.. 2019. Complexome profiling reveals association of PPR proteins with ribosomes in the mitochondria of plants. Mol Cell Proteomics 18(7):1345–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sberro H, Fremin BJ, Zlitni S, Edfors F, Greenfield N, Snyder MP, Pavlopoulos GA, Kyrpides NC, Bhatt AS.. 2019. Large-scale analyses of human microbiomes reveal thousands of small, novel genes. Cell 178(5):1245–1259.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. 2006. Tricine-SDS–PAGE. Nat Protoc. 1(1):16–22. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD.. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310(5749):827–834. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M.. 2011. Global quantification of mammalian gene expression control. Nature 473(7347):337–342. [DOI] [PubMed] [Google Scholar]
- Sievers FWilm ADineen DGibson TJKarplus KLi WLopez RMcWilliam HRemmert MSöding J, et al. . 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DGS, Gawryluk RMR, Spencer DF, Pearlman RE, Siu KWM, Gray MW.. 2007. Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila: direct analysis by tandem mass spectrometry. J Mol Biol. 374(3):837–863. [DOI] [PubMed] [Google Scholar]
- Smits P, Smeitink JAM, van den Heuvel LP, Huynen MA, Ettema TJG.. 2007. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 35(14):4686–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A. 1999. On the possibility of constructive neutral evolution. J Mol Evol. 49(2):169–181. [DOI] [PubMed] [Google Scholar]
- Tobiasson V, Amunts A.. 2020. Ciliate mitoribosome illuminates evolutionary steps of mitochondrial translation. eLife 9:e59264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiboli P, Herfurth E, Choli T.. 1994. Purification and characterization of the 30s ribosomal proteins from the bacterium Thermus thermophilus. Eur J Biochem. 226(1):169–177. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J.. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 11(12):2301–2319. [DOI] [PubMed] [Google Scholar]
- Valach M, Burger G, Gray MW, Lang BF.. 2014. Widespread occurrence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res. 42(22):13764–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valach M, Léveillé-Kunst A, Gray MW, Burger G.. 2018. Respiratory chain complex I of unparalleled divergence in diplonemids. J Biol Chem. 293(41):16043–16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valach M, Moreira S, Kiethega GN, Burger G.. 2014. Trans-splicing and RNA editing of LSU rRNA in Diplonema mitochondria. Nucleic Acids Res. 42(4):2660–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis EO, Bauerschmitt H, Becker T, Mielke T, Frauenfeld J, Berninghausen O, Neupert W, Herrmann JM, Beckmann R.. 2015. Parallel structural evolution of mitochondrial ribosomes and OXPHOS complexes. Genome Biol Evol. 7(5):1235–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltz F, Giegé P.. 2020. Striking diversity of mitochondria-specific translation processes across eukaryotes. Trends Biochem Sci. 45(2):149–162. [DOI] [PubMed] [Google Scholar]
- Waltz F, Nguyen T-T, Arrivé M, Bochler A, Chicher J, Hammann P, Kuhn L, Quadrado M, Mireau H, Hashem Y, et al. 2019. Small is big in Arabidopsis mitochondrial ribosome. Nat Plants 5(1):106–117. [DOI] [PubMed] [Google Scholar]
- Waltz F, Soufari H, Bochler A, Giegé P, Hashem Y.. 2020. Cryo-EM structure of the RNA-rich plant mitochondrial ribosome. Nat Plants 20:745–747. [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V.. 2000. Structure of the 30S ribosomal subunit. Nature 407(6802):327–339. [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M.. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6(5):359–362. [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF.. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292(5518):883–896. [DOI] [PubMed] [Google Scholar]
- Yutin N, Puigbò P, Koonin EV, Wolf YI.. 2012. Phylogenomics of prokaryotic ribosomal proteins. PLoS One 7(5):e36972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R, Smith E, Barrientos A.. 2018. Yeast mitoribosome large subunit assembly proceeds by hierarchical incorporation of protein clusters and modules on the inner membrane. Cell Metab. 27(3):645–656.e647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New data reported in this article are available in the online supplementary material.