Abstract
The emergence of bacterial resistance to antibiotics has led to the search for alternate antimicrobial treatment strategies. Engineered nanoparticles (NPs) for efficient penetration into a living system have become more common in the world of health and hygiene. The use of microbial enzymes/proteins as a potential reducing agent for synthesizing NPs has increased rapidly in comparison to physical and chemical methods. It is a fast, environmentally safe, and cost-effective approach. Among the biogenic sources, fungi and bacteria are preferred not only for their ability to produce a higher titer of reductase enzyme to convert the ionic forms into their nano forms, but also for their convenience in cultivating and regulating the size and morphology of the synthesized NPs, which can effectively reduce the cost for large-scale manufacturing. Effective penetration through exopolysaccharides of a biofilm matrix enables the NPs to inhibit the bacterial growth. Biofilm is the consortia of sessile groups of microbial cells that are able to adhere to biotic and abiotic surfaces with the help extracellular polymeric substances and glycocalyx. These biofilms cause various chronic diseases and lead to biofouling on medical devices and implants. The NPs penetrate the biofilm and affect the quorum-sensing gene cascades and thereby hamper the cell-to-cell communication mechanism, which inhibits biofilm synthesis. This review focuses on the microbial nano-techniques that were used to produce various metallic and non-metallic nanoparticles and their “signal jamming effects” to inhibit biofilm formation. Detailed analysis and discussion is given to their interactions with various types of signal molecules and the genes responsible for the development of biofilm.
Keywords: micronanotechnique, nanoparticles, antibiofilm, quorum-sensing, quorum quencher
Introduction
Most chronic infections in humans are found to be caused by biofilm. Biofilm is the syntrophic association of microbial cells that remain adhered to biotic or abiotic surfaces with self-synthesized hydrated polymeric substances (Costerton et al., 1999). The development of biofilm occurs via the adherence of planktonic bacterial cells to a surface such as medical devices and prosthetics. It also leads to the development of valve endocarditis, chronic otitis media, cystic fibrosis, and wound-associated infections (Donlan, 2001; Santos et al., 2011; Abidi et al., 2013).
The ability of the bacterial cells to adapt to and monitor various environmental conditions depends on the mechanism of cell-to-cell communication, which is density-dependent and based on contact-associated exchange of chemical substances (Lobedanz and Søgaard-Andersen, 2003; Phelan et al., 2012), chemical signaling (Eberhard et al., 1981), and signaling associated with electrical impulse (Nielsen et al., 2010; Shrestha et al., 2013).
Such a density-dependent communication system in bacteria attributed by small chemical substance (auto inducers) is termed “quorum sensing” (QS). This process was first observed within Vibrio fischeri (Nealson et al., 1970) and the term QS was given by Fuqua et al. (1994). The QS machinery comprise of acyl homoserine lactones (AHLs) which are the group of auto inducing peptides that play an important role in causing bacterial pathogenesis. Various virulence factors are produced through QS which include exotoxin A, lection, pyocyanin, and elastase in Pseudomonas aeruginosa, whereas protein A, enterotoxin, lipases, hemolysins, and fibronectin were reported in Staphylococcus aureus (Yarwood et al., 2004; Carnes et al., 2010). The developed virulence factors help the bacterial cells to evade the host immune system pathogenicity. The formation of biofilm is a multistep process that involves the transcription of various genes with respect to the planktonic form of microbial cells of the same organism (Donlan, 2002). The conversion of the planktonic to its sessile forms results in the enhancement of various chemical substances, resulting in genetic changes within the cells. Thus the sessile micro colonies develop a thick extracellular polymeric substance (EPS) comprising of exopolysaccharides, proteins, extracellular DNA (e DNA), and other polymeric substances that act as a physical barrier around the bacterial cells. This condition results in the maturation of biofilm via the process of quorum sensing (QS; Lahiri et al., 2019). The development of biofilm occurs through the mechanism of irreversible attachment of the bacterial cells upon the surface followed by the production of QS molecules, transportation of substances within the biofilm, metabolism of substrate by various sessile micro colonies, development of EPS, and finally the metastasis of the sessile colonies (Lahiri et al., 2019). Although use of antibiotics is the first choice to combat bacterial infection, the rapid increase in antibiotic resistance due to uncontrolled use of antibiotics has become a major health concern (Laxminarayan et al., 2013; Chioro et al., 2015; Zhao et al., 2017; Zhong and Zhao, 2018; Ma et al., 2019; Sarkar et al., 2020). The development of most antibiotics is based on targeting the protein synthesis machinery that in turn results in the destruction of the pathogenic cells (Nikaido, 2009) – precisely the planktonic cells. However, the biofilm-causing sessile cells often remain unaffected and lead to survival of the pathogen.
The traditional approach that was involved in treating biofilm was the combinatorial use of various antibiotics that exhibits various mechanisms of killing. But with the development of antibiotic resistance, conventional drugs are failing to inhibit the formation of biofilm. The development of EPS around the micro colonies prevents the penetration or zero diffusion of antibiotics within the biofilm. The development of EPS around the micro colonies prevents the penetration causing zero or reduced diffusion of antibiotics within the biofilm. Moreover, alterations in the microenvironment within biofilm matrices result in the development of the concentration gradient of metabolites causing reduced or almost no growth of bacteria. It has also been observed that the fluctuation in the microenvironment results in the alteration of nutrient supply, generation of oxidative stress, low availability of water, starvation, and change in temperature which in turn results in the development of adaptive stress responses within the bacterial cells (Singh et al., 2017). It has also been observed that the fluctuation in the microenvironment results in the alteration of nutrient supply, oxidative stress, low availability of water, starvation, and change in temperature which results in the development of adaptive stress responses within the bacterial cells. This is followed by the transformation of the bacterial cells to a highly protected spore-like state, known as persisters, which is also a potent cause of developing resistances towards antibiotics (Stewart, 2002). This issue warrants the exploration of new drugs or drug-like compounds to combat the biofilm.
Recently, the use of various nanoparticles (NPs) has become popular to treat bacterial infections as an alternative to antibiotics. Since NPs follow a totally different mechanism of action to target the bacteria and do not need to penetrate the bacterial cell, new pathways emerged (Wang et al., 2017a).
Microbiologically-synthesized nanoparticles are found to be more advantageous as compared to chemically-synthesized counterparts, as the former does not need very stringent conditions like a pure starting material. The requirement of optimal conditions and clement temperatures (20–30°C) make microbiologically-synthesized NPs more commercially viable (Vaseghi et al., 2018). Moreover, the presence of a biological capping agent on some micro biogenic nanoparticles, as a protective covering against oxidation, agglomeration, and aggregation, offer a higher stability (Durán and Seabra, 2012). Hence, microbiologically-synthesized NPs are often considered to be a better option for antibacterial therapies (Capeness et al., 2019).
Hence, the aim of the present review is to describe the efficiencies of various microbiologically-synthesized nanoparticles as potent antibiofilm agents with special reference to their ability to inhibit quorum sensing through affecting their regulator gene cascade.
Synthesis of Microbial Nanoparticles
Recently, technological advancements in the field of nanoparticles (NPs) have revolutionized their applicability in healthcare sectors due to the adjustable physico-chemical properties, which include thermal and electrical conductivities, absorption of light, melting point, and enhancement of catalytic activity by altering the surface-to-volume ratio. The field of nanotechnology encompasses the process of synthesizing nano-dimensional particles possessing various shape‐ and size-dependent properties (Rafique et al., 2017). Various types of NPs, including silver nanoparticles (AgNPs), show wide applications in the healthcare domain that include hyperthermia of tumors (Iravani, 2014), delivery of drugs, medical imagining, chemical sensors, catalysis, wireless electronic logic, computer transistors, memory chips, and its antimicrobial efficacy (Das et al., 2014).
Conventionally, NPs are synthesized via various physical, chemical, and mechanical processes such as ultra-sonication, radiolysis, microwave, spray pyrolysis, electro spinning, sol-gel method, chemical reduction, and inert condensation methods. But the urgent need for a less time consuming, low cost, high yield, non-toxic, and environment-friendly process has shifted the focus towards greener approaches (Khandel and Kumar-Shahi, 2016; Fang et al., 2019).
Biogenic sources like bacteria fungi and various parts of plants play an effective role in stabilizing the NPs (Durán et al., 2005). The green synthesis of NPs utilizes microbial cells like fungi, yeast, and bacteria as the process can be controlled by manipulating the culture conditions, like nutrient, pH, pressure, and temperature. The microbial system possesses an intrinsic mechanism of synthesizing NPs from metallic salts (Li et al., 2011).
Studies have shown that bacterial cells play an important role in the conversion of heavy metals to metallic NPs. The existence of various types of interactive pathways present within the bacterial cells is responsible for the synthesis of the metallic NPs. Another advantage of implementing bacterial cells is their ability to produce sustainable nanoparticles at a large scale (Fariq et al., 2017). It has also been observed that fungi play a predominant role in the synthesis of NPs via both extracellular and intracellular enzymes that are present within cells (Fariq et al., 2017). Enzymes like nicotinamide adenine dinucleotide (NADH)-dependent reductase was responsible for the synthesis of metallic NPs (Guilger-Casagrande and Lima, 2019). Nitrate reductase enzyme and anthraquinones from Fusarium oxysporum was responsible for the reduction of silver ions. Another study revealed that extracellular NADH-dependent nitrate reductase from the same fungi and quinolones were used to synthesize AgNPs (Anil Kumar et al., 2007). NADH-dependent oxidoreductase from fungi is also responsible for the synthesis of AuNPs (Kitching et al., 2015). Studies also showed that α-NADH-dependent reductase and nitrate reductase was used for the synthesis of NPs. Due to the presence of larger amounts of biomass in fungi, the yield of NPs is usually higher compared to bacterial cells. Although bacteria are more commonly used in synthesizing metallic NPs, the fungi could be more advantageous due to the presence of mycelia that provide greater surface area for interactions. The amount of enzyme produced by the fungi is higher compared to that of bacteria; thus the rate of conversion of the metallic salts to metallic NPs is faster. The fungal cell wall also played an important role in the mechanism of absorption and the reduction of metal ions for the formation of NPs (Khandel and Shahi, 2018).
The components of fungal cells, like the cell wall, cell membrane, protein, enzymes, and other intracellular components, play a vital role in the synthesis of the nanoparticle. Various parameters like temperature, pH, biomass, and other physical factors participate in regulating the synthesis of metallic NPs like AgNPs. These nanoparticles have various properties which have proven to be useful for human welfare, largely with antimicrobial (antibacterial, antifungal, and antiviral) activities. In fact, the mechanism of biosynthesis of AgNPs by fungi or fungal-based materials does not require any toxic agents during the NP recovery and purification process (Wei et al., 2009). But, like other nanoparticles, mycogenic AgNPs have some disadvantages. The biosafety of the use of AgNPs and their biocompatibility need to be tested prior to application, especially in the field of healthcare. The main obstacle for industrial production of mycogenic metallic NPs lie in the fact that most of the fungal species known for nanoparticle production have been reported to be pathogenic to human and plant. On the contrary, Trichoderma reesei, being a nonpathogenic fungus, is now well accepted as an industrially-adapted strain for the production of AgNPs (Dorcheh and Vahabi, 2016). Some other limitations that are associated the fungi-mediated NPs synthesis are higher cost of production and longer time of biosynthesis (Jeevanandam et al., 2016). The advantage of using bacterial species for the synthesis of NPs is to its fast growth and easier mechanism of manipulating genetic expressions (Lovley and Woodward, 1996). The use of bacterial species for the purpose of synthesizing metallic NPs is due to its ability to survive at higher concentrations of metallic ions (Haefeli et al., 1984).
Mechanism of Synthesis of Microorganism-Assisted Nanoparticles
Processes of both intracellular and extracellular synthesis of nanoparticles (NP) by microorganisms from metals, metal oxides, or metalloids have been well documented in literature (Patil and Chandrasekaran, 2020). The extracellular process involves reduction of metal ions for NPs synthesis by microbial enzymes and proteins, bacterial or fungal cell wall components, or organic molecules present in the culture medium, whereas the intracellular process involves initial electrostatic attraction of metal ions by carboxyl groups of the microbial cell wall, resulting in passage of metal ions through the cells and reduction by intracellular proteins and cofactors to produce NPs (Siddiqi et al., 2018). Biochemical mechanisms involving microorganism-mediated nanoparticle synthesis can be seen as a part of microbial resistance mechanisms for cellular detoxification. This involves alterations in the solubility of inorganic and toxic ions by enzymatic reduction and/or precipitation in the form of nanostructures. Both extracellular and intracellular bio-catalytic synthesis mechanisms have been proposed, which mainly involves oxidoreductase enzymes (e.g., NADH-dependent nitrate reductase, NADPH-dependent sulfite reductase flavoprotein subunit α, and cysteine desulfhydrase) and cellular transporters (Grasso et al., 2019). Nano-dimension materials are biosynthesized within the microorganisms by binding target ions from the surroundings and converting these toxic metal ions into the corresponding element metal through cellular enzymes. Based on the location of synthesis of nanoparticles, it can be classified into intracellular or extracellular. The intracellular method involves transporting ions into the microbial cell to form nanoparticles in the presence of enzymes. The extracellular mode involves trapping the metal ions on the cell surface and reducing ions in the presence of enzymes (Li et al., 2011).
Microbial Enzymes in Bio Reduction of Metal, Metalloid, and Non-Metal Ions to Nanoparticles
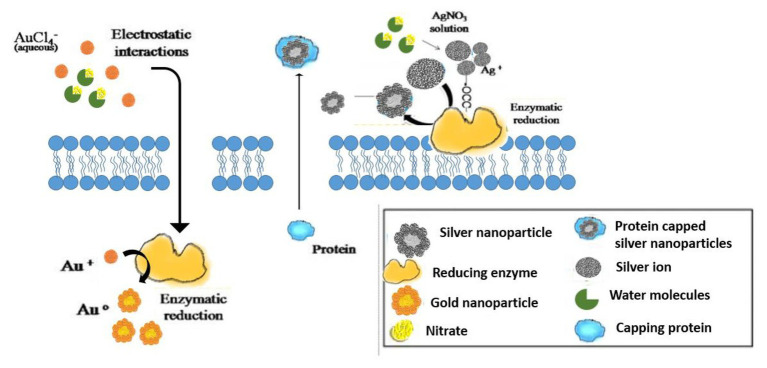
Microbial conversion of metal and metalloids to respective nanoparticles can be accomplished by the extracellular enzymes produced by different bacteria and fungi. Extracellular enzymes, such as nitrate reductase, can help in electron transfer from certain donors (e.g., hydroxyl groups) to Ag+ and thus helps in conversion to metallic AgNPs. It has been observed that the functional groups, such as  ,
,  ,
,  , or
, or 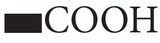 , of microbial proteins help in stabilization of the NPs by providing binding sites to the metal ions followed by its reduction into NPs on the cell wall or in the periplasmic space. In some cases, proteins act as the main reducing or capping agents during the formation and stabilization of NPs. Intracellular enzymes like cytochrome oxidases are also found to help in the reduction of metal ions to NPs via electron transfer between cytoplasm components (e.g., NADH/NADPH), vitamins, and organic acids. Intracellular reductase can initiate the biosynthesis and stabilization of NPs in three possible ways: periplasmic reductase can directly reduce M+ to M, bio reduction at the cytoplasm or periplasm produces M from M+, or bioconversion of M2+ to M+ in cytoplasm and M formation can occur (Klaus et al., 1999; Mishra et al., 2017; Lv et al., 2018; Siddiqi et al., 2018).
, of microbial proteins help in stabilization of the NPs by providing binding sites to the metal ions followed by its reduction into NPs on the cell wall or in the periplasmic space. In some cases, proteins act as the main reducing or capping agents during the formation and stabilization of NPs. Intracellular enzymes like cytochrome oxidases are also found to help in the reduction of metal ions to NPs via electron transfer between cytoplasm components (e.g., NADH/NADPH), vitamins, and organic acids. Intracellular reductase can initiate the biosynthesis and stabilization of NPs in three possible ways: periplasmic reductase can directly reduce M+ to M, bio reduction at the cytoplasm or periplasm produces M from M+, or bioconversion of M2+ to M+ in cytoplasm and M formation can occur (Klaus et al., 1999; Mishra et al., 2017; Lv et al., 2018; Siddiqi et al., 2018).
Metalloids such as Te2+ and Se2+ are harmful to both health and the environment and also involve toxic chemical reductants during their degradation (Presentato et al., 2018). Bio-inspired reductants can be an option for their efficient degradation and decontamination as they produce minimum or no toxic products in the entire degradation process. One such example is the aerobic disintegration of SeO32− by Actinomycete rhodococcus into Se-NPs. The reduction of SeO32− to Se-NPs is based on the LaMer mechanism where Se-nucleation seeds were formed, which assemble to form the nanoparticles that precipitated as nano-crystals from the suspension due to higher free energy and lower stability in solutions (Jana, 2015). In another study, Se-NPs were produced both intra-cellularly and extra-cellularly by Enterobacter cloacae Z0206 via the enzyme fumarate reductase possessing selenite reducing factor. Se-NPs were also produced by microorganisms such as Citrobacter freundii Y9 (anaerobic synthesis) and Pseudomonas putida (aerobic synthesis). In the latter case, it is found that thiol-containing amino acids (such as cysteine) help in the chelation of SeO32− which in turn forms seleno di-glutathione. This again can act as the substrate of glutathione reductase, producing an unstable intermediate Se0. Spherical nanoparticles of Se and Te are also formed by microbial species such as Ochrobactrum sp. MPV1 and Stenotrophomonas maltophilia SeITE02. The detoxification of tellurite to black Te-NPs can be achieved with the help of NADH-dependent reductase (Song et al., 2017; Wang et al., 2017b; Xu et al., 2018).
Intracellular magnetosomes in some bacteria like Magnetospirillum magneticum help in the encapsulation of Fe2O3-NPs in its dissolved form with the help of some multicellular proteins (e.g., ferritin or iron reductase enzymes). Bacterial magnetosomes are organelles of magnetotactic bacteria for geomagnetic navigation and comprises of magnetic nano crystals of magnetic minerals magnetite (Fe3O4) or greigite (Fe3S4) that remain surrounded by biological membranes made up of proteins, glycolipids, and phospholipids. The synthesis of magnetosomes is dependent on various environmental conditions, cellular stress, and cell proliferation cycles. The development of the magnetosomes involves transportation of iron outside the bacterial cell membranes via the vesicles that are formed, alignment of magnetosomes in a chain, development of crystals, and maturation of the crystals that are being formed (Kuzajewska et al., 2020). The membrane of the magnetosome differs from the plasmalemma on the basis of its composition and provides an appropriate environment for the purpose of biomineralization. This development of magnetosome is a highly controlled process that is regulated by unique protein sets encoded by the magnetosome island (Barber-Zucker and Zarivach, 2017). Supersaturating concentrations of iron also caused nucleation of magnetite at the interface of magnetosome membranes. It has been observed that the formation of vesicles occurs prior to the biomineralization event. Thus, pumping of supersaturating amounts of iron into the vesicles could be performed easier via the MamB and MamM proteins, such as in the case of Magnetospirillum magneticum. Interactions between the ions of the crystal and the surface proteins help in achieving a better nucleation process. It was also observed that the morphology of magnetite nanoparticles is dependent on the solution chemistry and physical conditions such as super saturation state, iron supply direction, concentration of activator and inhibitor ions or molecules, pH, redox potential, and temperature (Faivre and Schüler, 2008).
These metal oxide nanoparticles use FeCl3 as a common precursor. For example, metal oxide nanoparticles of CuO and SnO2 were previously made using microorganisms Morganella morganii and Erwinia herbicola, respectively, utilizing enzymes such as NADH involving redox reactions. The metabolites secreted by the bacteria in the culture broth can induce the reduction and stabilize the newly-formed metal NPs (Srivastava and Mukhopadhyay, 2014; Obayemi et al., 2015).
Nanoparticles of transition metal chalgonides have been synthesized by various researchers. For example, CdS-NPs can be formed by Moorella thermoacetica extra-cellularly by the addition of Cd(NO3)2 in bacterial culture media which helps in the photosynthetic reduction of CO2 to acetic acid. CdS-NPs can be produced both extra-cellularly and intra-cellularly by Desulfovibrio caledoniensis. It is a three-step process that involves ATP sulfurylase-mediated activation of the anaerobic reduction of sulfate being present within bacteria as well as ferredoxin or NADH-mediated reduction of the resultant adenosine-phosphosulfate (APS) complex to sulfite followed by assimilatory or dissimilatory sulfite reductase which reduced sulfite to sulfide. PbS nano crystals can also be biosynthesized by controlling the concentration of poly-etheneglycol in the Clostridiaceae sp. where SO42− is first reduced to S2− by the sulfate-reducing bacteria, and then S2− gradually combined with Pb+2 to precipitate as PbS-NPs (Qi et al., 2016; Yue et al., 2016). Studies have also shown that grapheme-associated highly dispersed Pd-Ag bimetallic NPs can be synthesized using Shewanella oneidensis MR-1(Han et al., 2019). The conversion of graphene oxides to grapheme nano sheets can be achieved through the use of crude polysaccharides obtained from Pleurotus flabellatus (Dasgupta et al., 2017; Figure 1).
Figure 1.
Microbial enzymes in bio reduction of metal, metalloid, and nonmetal ions to nanoparticles.
Microbial Exopolysaccharides for Synthesis of Nanoparticles
Exopolysaccharides (EPSs) from bacterial cells are produced extra-cellularly and play vital roles in surface adherence and cell-cell communication. EPSs possess the ability of reducing metal ions to produce nanoparticles and also help in the stabilization of the NPs by acting as a capping agent. Therefore, the EPSs are used as an alternate choice for microbiological production of numerous metal nanoparticles. Bacterial EPSs mainly comprise of carbohydrates such as D-glucose, L-fucose, D-mannose, D-galactose, N-acetyl-D-glucosamine, N-acetyl D-galactosamine, and non-carbohydrate components which are responsible for the anionic nature to the EPSs. These organic groups tend to increase the lipophilicity of the EPSs and directly influence their interaction with cations such as metal ions. Metal ions in contact with EPS are chelated and then reduced and stabilized by various functional groups via electrostatic bonds. For example, oxidation of  groups to form
groups to form 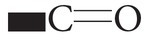 groups and oxidation of
groups and oxidation of  groups to form
groups to form 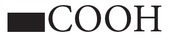 groups play an important role during metal nanoparticles synthesis. The polymeric structure of EPSs create a network by
groups play an important role during metal nanoparticles synthesis. The polymeric structure of EPSs create a network by  bonding in which nanoparticles stabilize with subsequent preventions of their agglomeration and precipitation (Escárcega-González et al., 2018). Various types of functional groups that are associated with EPS of both Gram positive and Gram negative acts as reductive and stabilizing agents for the purpose of synthesizing NPs with the capping and chelating processes (Emam and Ahmed, 2016). This helps in the regulation of size, particle dispersion, and shape of the NPs (Kanmani and Lim, 2013). The ability of the NPs to develop broader applicability is due to the muco-adhesion properties that help in the development of non-specific protein receptor recognition by the NPs (Kanmani and Lim, 2013). In previous research it was revealed that structurally known EPS from succinoglycanbacteria were used for the synthesis of AgNPs. It is a polymeric substance produced by Sinorhizobium meliloti that helped in the reduction of the metal by inducing oxidation of the aldehyde group to a carboxyl group with the help of nucleophilic insertion (Kwon et al., 2009). Curdlan is another type of EPS comprising of (1, 3)-β-D-glucan repeated units that are joined by β-(1, 3)-glycosidic bonds and are produced predominantly by Alcaligenes faecalis, Rhizobium sp., and Agrobacterium sp. These are used for the purpose of synthesizing and stabilizing NPs (Zhang and Edgar, 2014). Curdlan, a water insoluble polymeric substance, can be carboxylated or oxidized to curdlan derivatives for the purpose of synthesizing NPs. Leung et al. (2010) synthesized AgNPs with the help of carboxymethylated-curdlan. The persistence of the negatively-charged hydroxyl group and carboxyl groups resulted in better reduction of the silver ions. Dextran is another predominant component of EPS that helps in the synthesis of grapheme NPs (Hu et al., 2016). Chemically, dextran is a multifaceted branched glucan comprising of glucose residues that remain interlinked with α-(1,6) glycosidic linkages and are predominantly produced by certain groups of lactic acid bacteria like Leuconostoc mesenteroides and Streptococcus mutans. Bankura et al. (2012) synthesized size-controlled AgNPs using the aqueous solution of dextran which acted as both reductant and stabilizer.
bonding in which nanoparticles stabilize with subsequent preventions of their agglomeration and precipitation (Escárcega-González et al., 2018). Various types of functional groups that are associated with EPS of both Gram positive and Gram negative acts as reductive and stabilizing agents for the purpose of synthesizing NPs with the capping and chelating processes (Emam and Ahmed, 2016). This helps in the regulation of size, particle dispersion, and shape of the NPs (Kanmani and Lim, 2013). The ability of the NPs to develop broader applicability is due to the muco-adhesion properties that help in the development of non-specific protein receptor recognition by the NPs (Kanmani and Lim, 2013). In previous research it was revealed that structurally known EPS from succinoglycanbacteria were used for the synthesis of AgNPs. It is a polymeric substance produced by Sinorhizobium meliloti that helped in the reduction of the metal by inducing oxidation of the aldehyde group to a carboxyl group with the help of nucleophilic insertion (Kwon et al., 2009). Curdlan is another type of EPS comprising of (1, 3)-β-D-glucan repeated units that are joined by β-(1, 3)-glycosidic bonds and are produced predominantly by Alcaligenes faecalis, Rhizobium sp., and Agrobacterium sp. These are used for the purpose of synthesizing and stabilizing NPs (Zhang and Edgar, 2014). Curdlan, a water insoluble polymeric substance, can be carboxylated or oxidized to curdlan derivatives for the purpose of synthesizing NPs. Leung et al. (2010) synthesized AgNPs with the help of carboxymethylated-curdlan. The persistence of the negatively-charged hydroxyl group and carboxyl groups resulted in better reduction of the silver ions. Dextran is another predominant component of EPS that helps in the synthesis of grapheme NPs (Hu et al., 2016). Chemically, dextran is a multifaceted branched glucan comprising of glucose residues that remain interlinked with α-(1,6) glycosidic linkages and are predominantly produced by certain groups of lactic acid bacteria like Leuconostoc mesenteroides and Streptococcus mutans. Bankura et al. (2012) synthesized size-controlled AgNPs using the aqueous solution of dextran which acted as both reductant and stabilizer.
Au NPs can also be green synthesized using Cupriavidus metallidurans and Delftia acidovorans Au nuggets can be obtained from the bacterial biofilms (Johnston et al., 2013). Experimental observations revealed that nano particulate Au can inhibit biofilm formation (Reith et al., 2010). Au NPs bring about inhibition of biofilm by altering the surface chemistry and hydrophobicity as well as interacting with lipids and proteins that are present on the bacterial cell membrane (Ikuma et al., 2015). This alters the penetration of the NPs within the biofilm. The ability of NPs to penetrate the biofilm depend on the charge existing on the surface, size of the NPs, and the chemistry and concentration of NPs (Ikuma et al., 2015). The penetration of the NPs is followed by its interaction with the structural components of the biofilm, causing the disintegration of the biofilm (Qayyum and Khan, 2016; Pinto et al., 2019). It has been further observed that modified groups of the Au NPs can increase its inhibition potential either on single or multiple types of cells present within the biofilm. Various types of forces, like Van der Waals, hydrogen-bonds, electrostatic, and hydrophobic interactions, can effectively inhibit the biofilm (Yu et al., 2018).
Microbial Bio Surfactants for Synthesis of Nanoparticles
Bio surfactants are microbial surface-active amphiphilic molecules produced mainly by bacteria, fungi, and yeasts. Their hydrophilic moiety consists of carbohydrates, cyclic peptides, amino acids, carboxylic acids, or phosphates and the hydrophobic moiety comprises mainly of long-chain fatty acid or hydroxyl fatty acid. These are divided to two types viz. low-molecular-weight surface active agents (LMW) such as glycolipids, lipopeptides, and phospholipids and high molecular-weight polymers (HMW) mainly referred to as bio emulsifiers such as emulsan (Pati et al., 2020). They can also be classified as: (i) hydroxylated and cross-linked fatty acids (mycolic acids); (ii) glycolipids (rhamnolipids); or (iii) lipopolysaccharides. Bio surfactants can act as excellent capping agents during the synthesis of metallic nanoparticles via biogenic processes (Płaza et al., 2014). Their mechanism of action involves adsorption onto metallic nanoparticles, surface-stabilizing the nanoparticles, and preventing subsequent aggregation, thereby helping in stabilization process (Kiran et al., 2011; Gahlawat and Choudhury, 2019). Biosurfactants are the groups of amphipathic molecules possessing both hydrophobic and hydrophilic moieties creating partitions at the interface between fluid phases possessing various degrees of hydrogen bonding and polarity (Rodrigues et al., 2006). The micro emulsions, i.e., the water-soluble droplets that are present within, act as a micro-reactor. The increase in the concentration of the surfactants results in a decrease in the size of droplets, thereby reducing the particle size. The presence of water plays an important role in regulating the size and the morphology of the NPs. The size of the particle and mono dispersity is dependent on the molar ratio (R) of the water (Han et al., 2008).
Microbial Synthesis of Nanoparticles Through Bio Mineralization
Some microorganisms possess the unique property of mobilizing/immobilizing the metal salts by reducing them into metal ions which precipitate within or outside the microbial cells. They, with the help of efflux pumps, accomplish the complexation and inactivation of metals, with their subsequent precipitation by changing the oxidation state of the metals via redox reactions.
For example, gold (I)-thiosulfate enters Acidithiobacillus thiooxidans cells metabolically and are broken down to Au(I) and thiosulfate (S2O32−) ions. Thiosulfate acts as an energy source while Au(I) reduces itself to elemental gold intracellularly. These elemental gold precipitates inside the bacterial cells to form NPs during the late stationary growth phase and are released from the cells later on. Finally, the gold particles in the bulk solution are grown into micrometer-scale wire and octahedral gold (Lengke and Southam, 2005).
In another study, it was proposed that the sulfate-reducing bacteria bring about the reduction of the gold (I)-thiosulfate complex through three possible pathways: iron sulfide formation, localized reducing effects, or a metabolic pathway. The first process involves the adsorption of gold (I)-thiosulfate on the surfaces of iron sulfide freshly formed by sulfate-reducing bacteria producing elemental gold. The second process involves localized reducing conditions causing the gold (I)-thiosulfate complex reduce to hydrogen sulfide (HS−) by sulfate-reducing bacteria which was released through the outer membrane pores resulting into precipitation of elemental gold. The third process involved decomplexation of gold (I)-thiosulfate complex to Au(I) and thiosulfate ions which are later released from the cells (Lengke and Southam, 2005; Lengke et al., 2006).
Microbial Synthesis of Magnetic Nanoparticles
The bacterial magnetic nanoparticle (BMP) formed from magnetotactic bacteria (MTB), known as magnetosomes, which are a type of magnetic nanoparticle, has a lot of possibilities in nano biotechnology (Vargas et al., 2018). These are intracellular magnetic particles comprising of oxides and sulfides of iron within the bacterial cell that act as a bacterial compass needle helping bacterium to migrate along oxygen gradients in aquatic environments, under the influence of the Earth’s geomagnetic field. BMPs have the ability to disperse in aquatic medium and are usually carried by phospholipid vesicles.
Bio mineralization of BMPs occurs via multiple steps; the first step involves a GTPase-mediated invagination of the cytoplasmic membrane followed by assembly into a linear chain along the cytoskeletal filaments. The second step involves trans membrane iron-transporter-mediated accumulation of ferrous ions inside the vesicles. The final step involves magnetite crystal nucleation by triggering BMP proteins which includes the accumulation of supersaturating iron concentrations, and partial reduction and dehydration of ferrihydrite to magnetite (Arakaki et al., 2008).
In another study, magnetite was synthesized by Shewanella oneidensis involving both passive and active methods. Active utilization of ferrihydrite as a source of electron acceptor to form Fe2+in a high pH environment followed by localized conversion of Fe2+ and Fe3+ near negatively charged cell walls leads to super saturation, causing magnetite to precipitate (Li et al., 2011). The membrane controls the particle size, crystallization, and particle morphology in the particle size, crystallization, and particle morphology. The phospholipids bilayer can entrap the bacterial nanoparticles and contains about 20–40 species of membrane proteins (Grünberg et al., 2004).
The BMPs used in the nano biotechnology and nano medicine are mainly extracted from species of Magnetospirillum gryphiswaldense MSR-1 and Magnetospirillum magneticum AMB-1, although there are other species of MTB that can be cultivated (Chen et al., 2016).
Microbial Synthesis of Stable Quantum Dot Nanoparticles
Recently, fluorescent or quantum dots (QDs) nanoparticles have been widely used in several biological, biomedical, optical, and optoelectronic applications such as biosensors, photovoltaics, optoelectronics, transistors, oil exploration, biomedicine, imaging, and solar cells due to its unique size-dependent properties. Their increased utility is due to their biocompatibility and lesser toxic by-product generation during its synthesis, indicating a pathway to greener technology. So far, synthesis of CdS, CdSe, and CdTe QDs involved the use of harmful chemicals such as bidentate thiols [e.g., dithiothreitol (DTT), mercaptosuccinic acid (MSA), mercaptopropionic acid (MPA)], and ligands with different functional groups (amino, hydroxyl, and carboxylic acid, among others). Researchers have isolated bacteria that are cadmium‐ and tellurite-resistant Antarctic bacteria Pseudomonas (eight isolates), Psychrobacter (three isolates) and Shewanella (one isolate) capable of synthesizing CdS and CdTe QDs when exposed to toxic oxidizing heavy metals like Cd and Te with a time-dependent change in fluorescence emission color (Plaza et al., 2016). The CdSe nanoparticles are one of the examples that exhibit fluorescent tags. Cui et al. (2009) reported intracellular synthesis of CdSe nanoparticles in S. cerevisiae using genetic engineering techniques. The genes involved in glutathione biosynthesis, namely GSH1, GSH2, and GLR1, become silent in yeast and after interaction with inorganic ions resulted in a significant reduction in fluorescence, which in turn was proportional to the amount of CdSe nanoparticle synthesis. It was found that Na2SeO3 was reduced to selenocysteine (Cys-Se)2, a complex of selenium-containing cysteine, which after the addition of CdCl2 generated CdSe nanoparticles. In another work by Bruna et al. (2019), CdS QDs by polyextremophile halophilic bacteria Halobacillus sp. DS2 were synthesized with increased tolerance to NaCl (Bruna et al., 2019). A tunable ternary CdSAg QD involving cation exchange were synthesized by Órdenes-Aenishanslins et al. (2020). Nanoparticles were also produced within bacterial cells extra-cellularly via exposure to cysteine and CdCl2 in a reaction. This reaction was dependent on S2− generation mediated by cysteine desulfhydrase enzymes and utilized cellular biomolecules to stabilize the nanoparticle.
Microbial Organic Particles for Nanoparticle Synthesis
Bacterial cellulose (BC) are used to form nano fibers and to instill a bactericidal property in the nano fibers. A combination of bactericidal chitin (Ch) with bacterial cellulose (BC) nano fibers was developed to form a nanocomposite of BC-Ch in a process that is considered a green approach. Acetobacter aceti was also fed with Ch79d to biosynthesize bio-BC-Ch79d nanocomposites to produce 50–100-nm-wide nanofibrils (Butchosa et al., 2013).
A number of microbial components are found to be involved in the formation of nanoparticles (Table 1), which are found to act as potent antibiofilm agents through inhibition of the quorum-sensing process.
Table 1.
Contribution of microbes in the formation of nanoparticles.
| Classification | Microorganism | Microbial component | Raw material | Element used | Nanoparticle size (nm) | Morphology | Synthesis location | References |
|---|---|---|---|---|---|---|---|---|
| Bacteria | Lactobacillus rhamnosus | EPS | Ag | 10 | Spherical, triangular, rod, and hexagonal | Extracellular | Kanmani and Lim, 2013 | |
| Bacillus licheniformis Dahb1 | EPS | ZnO | 100 | Hexagonal | Extracellular | Abinaya et al., 2018 | ||
| Brevibacterium casei MSA19 | Biosurfactant | Ag | Extracellular | Kiran et al., 2010 | ||||
| Bacillus amyloliquifaciens KSU-109 | Biosurfactant | Surfactin | CdS | Extracellular | Singh et al., 2011 | |||
| Pseudomonas aeruginosa BS-161R. | Biosurfactant | Rhamnolipid | Ag | Extracellular | Kumar et al., 2010 | |||
| Bacillus cereus | Ag | 20–40 | Spherical | Extracellular | Sunkar and Nachiyar, 2012 | |||
| Kocuria flava | Cu | 5–30 | Spherical | Extracellular | Kaur et al., 2015 | |||
| Pseudomonas aeruginosa JP-11 | CdS | 20–40 | Spherical | Extracellular | Raj et al., 2016 | |||
| Shewanella loihica PV-4 | Pd and Pt | 2–7 | Spherical | Intracellular | Ahmed et al., 2018 | |||
| Ochrobactrum sp. MPV | Te | Spherical, rod | Intracellular | Zonaro et al., 2017 | ||||
| Bacillus cereus | Ag | 20–40 | Spherical | Extracellular | Sunkar and Nachiyar, 2012 | |||
| Escherichia coli | Ag | 8 | Spherical | Extracellular | Mahanty et al., 2013 | |||
| Geobacillus sp. | Au | 5–50 | Quasi hexagonal | Extracellular | Correa-Llantén et al., 2013 | |||
| Bacillus subtilis | Ag | 20–25 | Spherical | Extracellular | Srinath et al., 2018 | |||
| Nocardiopsis sp. MBRC-1 | Ag | 45 | Spherical | Extracellular | Manivasagan et al., 2013 | |||
| Pseudomonas fluorescens | Au | 50–70 | Spherical | Extracellular | Rajasree and Suman, 2012 | |||
| Serratianematodiphila | Ag | 10–31 | Spherical crystalline | Extracellular | Malarkodi et al., 2013 | |||
| Shewanellaoneidensis | U | 150 | Extracellular | Satyanarayana Reddy et al., 2010 | ||||
| Magnetospirillummagneticum | Fe3O4 | 10–60 | Cuboidal, rectangular and spherical NPs | Intracellular | Obayemi et al., 2015 | |||
| Shewanellaoneidensis MR-1 | CdSe | 3.3 | Intracellular | Tian et al., 2017 | ||||
| Escherichia coli | CdTe | 62 | Extracellular | Kominkova et al., 2017 | ||||
| Clostridiaceae sp | PbS | Cubic NPs | 50 | Extracellular | Yue et al., 2016 | |||
| Erwinia herbicola | SnO2 | 15–45 | Spherical andtetragonal NPs | Extracellular | Srivastava and Mukhopadhyay, 2014 | |||
| Serratia marcescens | Bi | <150 | Irregular | Intracellular | Nazari et al., 2012 | |||
| Actinomycetes | Streptacidiphilus durhamensis | Ag | 8–48 | Spherical | Buszewski et al., 2018 | |||
| Streptomyces griseoruber | Au | 5–50 | Spherical, hexagonaland triangular | Ranjitha and Rai, 2017 | ||||
| Streptomyces xinghaiensis OF1 | Ag | 5–20 | Spherical | Wypij et al., 2018 | ||||
| Rhodococcus sp. NCIM2891 | Ag | 10–15 | Spherical | Otari et al., 2014 | ||||
| Fungi | Penicillium diversum | Ag | 10–15 | Spherical | Ganachari et al., 2012 | |||
| Fusarium oxysporum JT1 | Au | 22 | - | Thakker et al., 2013 | ||||
| Trichoderma harzianum | CdS | 3–8 | Spherical | Bhadwal et al., 2014 | ||||
| Aspergillus terreus | ZnO | 28–63 | Spherical | Baskar et al., 2015 | ||||
| Colletotrichum sp. | Al2O3 | 30–50 | Spherical | Suryavanshi et al., 2017 | ||||
| Yeast | Rhodosporidium diobovatum | PbS | 2–5 | Spherical | Seshadri et al., 2011 | |||
| Saccharomyces cerevisiae | Ag, Au Nanoplates | 2–20 | Spherical, hexagonal and triangularnanoplates | Korbekandi et al., 2016; Yang et al., 2017 | ||||
| Pichia kudriavzevii | ZnO | 10–60 | Hexagonal wurtzitestructure | Moghaddam et al., 2017 | ||||
| Rhodotorula glutinis | Ag | 15 | Spherical | Cunha et al., 2018 | ||||
| Virus | Tobacco mosaic virus (TMV) | Pd, Au | 3–4, 5 | Carbon nanotubes, spherical | Kobayashi et al., 2012; Fan et al., 2013 | |||
| M13 virus | TiO2 | 20–40 | Mesoporous nanowires | Chen et al., 2013 | ||||
| Hepatitis E virus | Nanoconjugates | 27–34 | Icosahedral | Chen et al., 2018 | ||||
| Potato virus X | Nanocarriers | 13 | Helical | Le et al., 2017 | ||||
| Algae | Sargassum muticum | ZnO | 0–57 | Hexagonal | Sanaeimehr et al., 2018 | |||
| Gelidium amansii | Ag | 27–54 | Spherical | Pugazhendhi et al., 2018 | ||||
| Laminaria japonica | Ag | 31 | Spherical to Oval | Kim et al., 2016 | ||||
| Cystoseira baccata | Au | 8 | Spherical | González-Ballesteros et al., 2017 | ||||
| Chlorella vulgaris | Pd | 5–20 | Spherical | Garole et al., 2019 | ||||
| Spirogyra varians | Ag | 35 | Quasi spherical | Salari et al., 2016 | ||||
| Chlorella vulgaris | Au | 2–10 | Spherical | Annamalai and Nallamuthu, 2015 |
Mechanism of Quorum Sensing
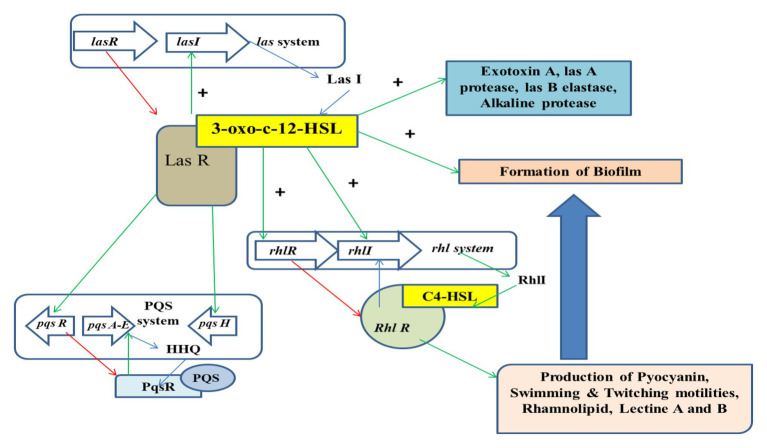
The process of QS is the mechanism of interaction between the bacterial cells via the production of extracellular chemicals known as Auto inducers (AIs). This mechanism helps in the process of synchronizing the bacterial cells and their various expressions to respond to the changes of the environment. This process is observed in both Gram-positive and Gram-negative bacterial cells. Studies have shown that Gram-negative bacterial cells possess three major groups of Ais, whereas the Gram positive bacterial cells communicate with the help of auto inducing peptides (AIPs; Raffa et al., 2005). The mechanism of QS can be inhibited by the process of quorum quenching (QQ; Dong et al., 2002). Various mechanisms are involved in the process of QQ that comprise of competitive inhibition and cleavage of the QS signals. This predominantly brings about the inhibition of QS. The chemical that inhibits the mechanism is referred to as Quorum sensing inhibitor (QSI), whereas the enzymes involved in such inhibition are QQ enzymes. It has often been noticed that QQ enzymes can inhibit QS by targeting AHLs. Several classes of enzymes are capable of degrading AHL signal, such as acylases and oxido reductases that are predominantly of a bacterial origin. It has been also observed that the half-life of AHL is dependent on pH and temperature (Yates et al., 2002; Delalande et al., 2005). There are two novel AHL enzymes, namely N-acyl homoserine lactonase (AiiA) and esterase (Est), that can be isolated from Altererythrobacter sp. S1-5 (Wang et al., 2019). The former enzyme can hydrolyze and inactivate a variety of acyl homoserine lactones (AHLs). Quorum sensor molecules involved in bacterial quorum sensing (QS) were extracted and purified from Bacillus sp. 240B1. After its covalent immobilization onto magnetic nanoparticles (MNPs), the quorum quenching ability of r-AiiA-MNP nano biocatalyst was evaluated and was found to be effective in inhibiting QS (Beladiya et al., 2015). The las system comprises of the lasR regulatory gene that codes for the Las R protein, while lasI synthase is regulated by lasI gene which is associated with the synthesis of signaling molecule of the AHL family, i.e., 3-oxo-C12-HSL. The LasR/3-oxo-C12-HSL is responsible for activating the virulence gene. The rhl system comprises of rhlR and rhlI genes. These further results in the activation of the las system that is responsible for the production of pyocyanin, rhamnolipids, and swarming motilities. The rhl system is regulated by the las system. PQS intermediates between the two other systems. The PqsA_E acts as a precursor for 2-heptyl-4-quinolones (HHQ) and regulates the conversion of HHQ to 2-heptyl-3-hydroxy-4-quinolone (PQS; Figure 2; Carette et al., 2020).
Figure 2.
Regulation of the mechanism of QS.
Mechanism of Quorum Sensing in Gram-Negative Bacteria
The Gram-negative bacterial cells communicate via signaling molecules, termed as auto inducers (AI), such as acetyl homoserine lactone (AHL) and other chemical molecules whose synthesis is dependent on S-adenosylmethionine (SAM; Walker et al., 2011). SAM acts as an amino acid subtract required for the synthesis of acyl homoserine lactones (Whitehead et al., 2001). A study showed that E. coli comprising of plasmid associated lux I requires the presence of SAM for the synthesis of N-(3-oxooctanoyl)-L-homoserine lactone (Hanzelka and Greenberg, 1996).
The AI produced by the bacterial cells can easily diffuse through the outer layer of the cell membrane. Enhancement of AIs is observed during high cell density (HCD) and thus regulates the transcriptional factors for the genes that are associated with the process of QS. Various types of signaling molecules are associated with the Gram-negative bacterial cells like 3-hydroxy-palmitic acid from Ralstonia solanacearum and 2-heptyl-3-hydroxy-4-quinolone from P. aeruginosa (Flavier et al., 1997). The latter is an opportunistic nosocomial disease-causing organism remaining associated with infections like cystic fibrosis (CF), lung infection, and various types of dermal and burn wound infections (Ammons et al., 2009). These Gram-negative bacteria comprise of three major QS circuits. One such circuit is the lasI encoding protein LasI that is associated with the production of auto inducer as well as the gene LasR that in turn encodes for the transcriptional activator LasR. Another profound QS circuit comprises of gene rhlI, associated with the synthesis of autoinducer-N-(butonyl)-L-Homoserine lactone and rhlR that is responsible for the transcriptional activator RhlR (Pearson et al., 1994, 1995). The third QS circuit which is also observed in P. aeruginosa is related to alkyl quinolones, especially 2-heptyl-3-hydroxy-4-quinolones that are predominantly regulated by pqsABCDEH and PqsR regulator genes (Pesci et al., 1999).
Mechanism of Inhibition of Quorum Sensing by Micro Biogenic NPs
Information pertaining to the inhibition in the mechanism of QS by NPs are very limited as only a few studies have been performed; this therefore remains a somewhat underexplored field despite being interesting. NPs act as potent inhibitors of QS by affecting the mechanism of cell-cell communication or inhibition of the signals associated with the mechanism of QS, thereby hindering the synthesis of various types of the signaling molecules and preventing the formation of molecule-receptor complex. This in turn stops the signal transduction cascade (Sadekuzzaman et al., 2015) Silver nanoparticles (AgNPs) have been used as QQ agents due to their strong antimicrobial activity (Castellano et al., 2007; Chen and Schluesener, 2008).
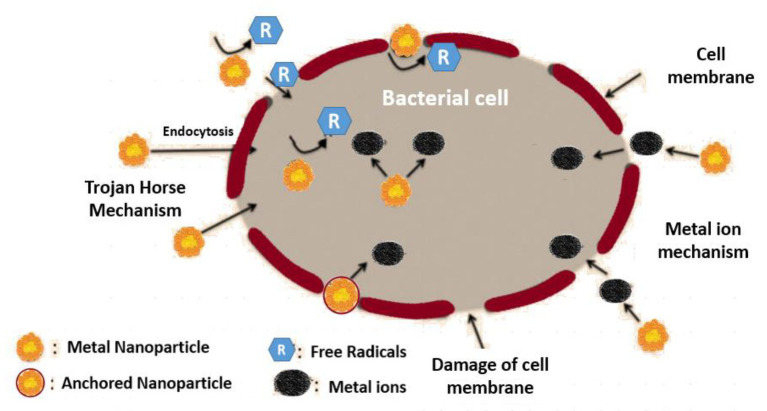
The scientific communities have shown their interest in the use of AgNPs due to their broad range of antimicrobial activities (Kim et al., 2007; Lara et al., 2011; Brandt et al., 2012) and the convenience during application for their physico-chemical properties and surface area to volume ratio (Kim et al., 2007; Figure 3). Various other types of NPs, like AuNPs, TiO2, SiO2, and ZnO, from microbial sources possess the efficacy of inhibiting the QS cascade and thus inhibit the formation of the biofilm (Shah et al., 2008; Samanta et al., 2017; Al-Shabib et al., 2018).
Figure 3.
Mechanism of NPs to bring about damage to the bacterial cells.
NP-Associated Inhibition of Quorum Sensing Cascade
The growth of the sessile communities of bacteria within the biofilm can only be checked by quorum sensing (QS), a bacterial cellular communication system that is a non-cytotoxic process (Høiby et al., 2010). The success in the treatment of any kind of chronic infections is devising an efficient mechanism of delivering the drug molecules up to the target cells. NPs have played a pivotal role in the mechanism of inhibiting the process of QS, and thus could inhibit biofilm formation.
The nano materials possess dimensions at the scale of nanometers possessing chemical and physical properties different from that of the bulk materials (Wang et al., 2017a). NPs are proven to be the most accepted drug delivering vehicle, possessing the ability to inhibit the growth of the microbial cells and thus can fight against pathogenic organisms. NPs show various mechanisms pertaining to the inhibition of biofilm and microbial growth. Various studies were conducted to predict the probable inhibition mechanism of microbial growth by the NPs. It was observed that ZnO NP has the ability of inhibiting the NorA efflux pump present in S. aureus (Banoee et al., 2010). It has been further observed ZnO NP, along with the antibiotic ciprofloxacin, could enhance the zone of inhibition by 22–27% of E. coli and S. aureus, respectively (Banoee et al., 2010). Studies also showed that iron oxide NPs coated with polyacrylic acid help in inhibiting Mycobacterium smegmatis by enhancing the efflux inhibition (Padwal et al., 2015).
Inhibition of Quorum Sensing by Micro Biogenic Silver Nanoparticles
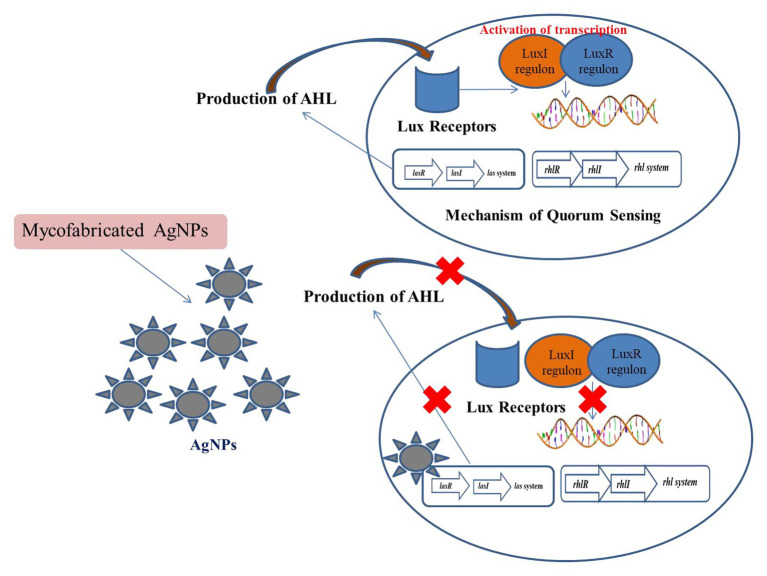
The antimicrobial potential of AgNPs has made it an important therapeutic agent. But advancements in the era of antibiotics has resulted in the minimization of the use of silver (Castellano et al., 2007; Li et al., 2009). The presence of wide spectrum antimicrobial activities by AgNPs due to its high surface area to volume ratio has been a key factor in its success (Kim et al., 2007; Lara et al., 2011; Brandt et al., 2012). Various studies have been conducted that showed that silver composites and silver have significant roles as antimicrobial agents (Panáček et al., 2006; Naik et al., 2013). AgNPs also act as potent bactericidal agents against S. aureus, P. aeruginosa, and E. coli (Khurana et al., 2016). Literature has shown that AgNPs can be used as potent anti-QS agents, thus hindering the formation of the biofilm and production of violacein by C. violaceium (Jagtap and Priolkar, 2013). It has been further observed that green-synthesized NPs played a key role in controlling infections associated with microbes. Studies have revealed that Ag NPs have the potentiality of blocking the synthesis of signaling molecules by inhibiting LasI/Rhl I synthase. Ag NPs had the potential of inhibiting the QS of P. aeruginosa (Ali et al., 2017). In-silico studies comprising of molecular docking revealed that Ag NPs have the potency of locking the active sites of various proteins comprising of LasI or RhlI synthase along with their surrounding residues being present. Ag NPs effectively block the active sites and thus efficiently inhibit the mechanism of QS. Ag NPs possess the ability of inhibiting the QS-genes by blocking the transcriptional regulatory proteins which inactivates the LasR or RhlR system. Ag NPs also possess the ability to act effectively as anti-QS agents by inhibiting signaling molecules like LasI and RhlI. Studies have also shown that micro-fabricated forms of Ag NPs synthesized from Rhizopus arrhizus metabolites inhibit the QS mechanism of P. aeruginosa (Singh et al., 2015b). It was observed that micro fabricated Ag NPs were able to bring about a marked reduction in the production of signaling molecules at a concentration of 0-25 μg/ml. It was further observed that these micro-fabricated NPs were able to bring about a reduction of 79–84% of lasA and lasB genes’ expression and down regulated these genes. The expression of the targeted QS genes, like lasA, lasB, lasI, lasR, rhlI, rhlR, rhlA, phzA1, and fabH2, is activated by the AHL-LasR complex. The down regulation of the QS genes was achieved by the myco-fabricated Ag NPs. Its mechanism against various virulence factors such as LasB ealastase, LasA protease, rhamnolipid, and pyocyanin occurred via phzA1, rhlA, and lasAB operons. The production of AHL is enhanced by rhlR via and the signal cell receptor RhlR. Similarly, RhlR is also associated with the activation of fabH2 and rhlAB operons. Thus myco-fabricated Ag NPs help in the down regulation of rhlR and thereby decrease the production of RhlR (Singh et al., 2015a; Figure 4).
Figure 4.
Mechanism of inhibition of QS by myco-fabricated Ag NPs.
Inhibition of Quorum Sensing Using Micro Biogenic Gold Nanoparticles
In recent times, the use of Au NPs have attracted various researchers due to their catalytic properties that are largely used in the field of diagnostics and biologics (Mesbahi, 2010; Jain et al., 2012; Giasuddin et al., 2013). The wide applicability of Au NPs is due to its simple mechanism of synthesis, convenience to use, and relatively low toxicity in comparison to the other types of nano materials in use (Capek, 2014). Au NPS showed efficient antimicrobial activity against various types of microbes like methicillin-resistant S. aureus (MRSA), Salmonella typhi, and Bacillus Calmette-Gu’erin (Zhao et al., 2010; Lima et al., 2013; Bindhu and Umadevi, 2014). Studies also revealed that acyl homoserine lactone lactonase protein associated with An NP help in inhibiting QS of Proteus sp. (Vinoj et al., 2015). These NPs were also able to degrade N-hexanoyl-L-homoserine lactone with the help of N-acyl homoserine lactonase that was present on the surface of the NPs. The enzyme also helps in degrading the moiety associated with acyl homoserine lactone and brings about the conformational changes of the signaling molecule, thus preventing the binding with LuxR transcriptional regulator resulting in the inhibition of QS (Kaufmann et al., 2005; Bai et al., 2008). They were also able to inhibit the metabolic activities and production of EPS, preventing the formation of biofilm and changing the hydrophobicity of the bacterial cells (Samanta et al., 2017). The Au NP produced from the mycelium of Laccaria fraternal was responsible for stabilizing the NPs which in turn plays an important role in the reduction of pyocyanin production from P. aeruginosa (Samanta et al., 2017).
Inhibition of Quorum Sensing by Micro Biogenic Titanium Dioxide NPs
Titanium dioxide NPs possess the ability of inhibiting QS and also have a wide range of activities that includes photo catalysis of organic dyes, usage within the photochromic appliances, gas sensors, dye sensitized solar cells, and antimicrobial activities (Banerjee, 2011). It has been observed that TiO2NPs in the presence of UV rays produce super-oxides that help in inhibiting the growth of MRSA (Shah et al., 2008). These NPs possess the ability to oxidize the organic substances that are present within the bacterial cells and thus kill the cells (Fujishima et al., 2000; Cho et al., 2005; Shiraishi and Hirai, 2008). Experimental observations indicated that AgCl-TiO2 NPs was an effective anti-quorum sensing compound against C. violaceum (Naik and Kowshik, 2014). It has also been found that the silver of Ag NPs can prevent the synthesis of violacein which can precisely block the mechanism of QS. Moreover, the inhibition of QS was observed in the absence of oxo-octanoyl homoserine lactone by AgCl-TiO2 NPs.
Inhibition of Quorum Sensing by Micro Biogenic Silicon Oxide NPs
The mechanism of QS was also inhibited significantly using SiO2 NPs which are found naturally in the form of quartz that is present as a major element within minerals, rocks, and sands. These NPs are widely used in biomedicines due to their small particle size and biocompatibility (Malvindi et al., 2012). These NPs are predominantly used in healthcare, chemical, cosmetics, composites, energy microelectronics, aerospace, pharmaceutical, and textile industries (Weiss et al., 2006; Santos et al., 2015). The ROS released by these NPs can also cause damage to DNA, resulting in the death of the cell. Hence, these are regarded as useful substances with antimicrobial activities (Tang and Cheng, 2013). NPs coated with various types of organic components were used to inhibit QS and treat resistant microorganisms by removing signaling molecules from the external environment. Research showed that NPs coated with β-cyclodextrin help in the inhibition of AHL‐ dependent QS of V. fischeri (Miller, 2015). The study showed that the presence of β-cyclodextrin associated with Si-NP helps in taking up the AHL-molecule from the environment and reduction in bio luminescence. It has been further observed that these NPs were also able to down regulate LuxA and LuxR genes.
Inhibition of Quorum Sensing by Micro Biogenic ZnO Nps
ZnO NPs are used largely in the field of dentistry (Tomczak et al., 2009). These NPs possess the ability of utilizing proteins from the environment, thus bringing about inhibition of metabolism and cytotoxicity and hindering various cellular processes (Horie et al., 2009). They exhibit a potent antibacterial property by inhibiting bacterial growth and the adherence property of the cells, thus also preventing the formation of biofilm (Yamamoto, 2001; Brayner et al., 2006). Various research works have been performed that demonstrated the strength of ZnO NPs to act as a potent anti-QS agent. The inhibition of the QS mechanism greatly influenced the biofilm formation in a P. aeruginosa strain isolated from Cystic fibrosis (CF; García-Lara et al., 2015). These NPs possess the ability of down regulating the QS genes within the Gram-negative bacterial cells. Researchers showed that ZnO NPs were able to inhibit QS in P. aeruginosa by down regulating lasR. lasI, rhl I, and rhl R (Saleh et al., 2019). Another study showed that ZnO NPs were able to reduce the swimming and swarming motility within P. aeruginosa and also showed its efficacy against the pqs and las system of QS (Khan et al., 2020). ZnO NPs resulted in the efflux of the zinc cation efflux pump of czc operon and various other regulators of transcription such as type III repressor ptrA and porin gene opdT followed by the repression in the production of pyocyanin-associated phz operon. The ZnO NPs also possess the ability of enhancing membrane hydrophobicity in P. aeruginosa (Lee et al., 2014).
Conclusion
The bacterial cells possess the ability to sense their surrounding population by analyzing the production of auto inducers, known as quorum sensing. This mechanism also helps the bacterial cells to communicate with one another, resulting in the development of biofilm. The mode of action of QS inhibition is mainly via inhibition of signal generation, blockage of signal receptors, and disruption of QS signals. Currently, research is focused on finding ways to interrupt the process of bacterial QS using compounds of microbial origin. Microbial biofilm are increasingly causing medical-implant-associated infections. Thus, novel treatment strategies are urgently required for device-associated biofilms infections. The use of nano materials has emerged as a promising approach in preventing biofilm formation by destroying the exopolysaccharides (EPS) of the biofilm matrix and killing the bacteria. Key factors responsible for using nanomaterials for biofilm treatment are their low cytotoxicity and novel mechanisms of action. Nanoparticles’ toxicity strongly depends on their physicochemical properties such as shape, size, surface chemistry, structure, agglomeration state, and cell types in contact with the nano materials (Tran et al., 2020).
The few disadvantages involving microbial nanoparticle synthesis are the tedious purification steps and poor understanding of the mechanisms. Additionally, controlling the shape, size, and mono dispersity in the solution phase is a matter of concern. An important challenge is scaling up the production level processing for industrial applications. This includes addressing a few important issues such as selection of ideal bacteria depending on growth rate, enzyme activities, and biochemical pathways, selection of the biocatalyst state (bacterial enzymes) either of whole cells, crude enzymes, or purified enzymes that could increase the rate of reaction, and optimal conditions for cell growth and enzyme activity. Optimization is also needed for higher biomass synthesis, optimal reaction conditions for better removal of unwanted residual nutrients and metabolites, better extraction and purification processes (freeze-thawing, heating processes, and osmotic shock) of the nanoparticles, and better stabilization of the produced NPs without aggregation (Iravani, 2014).
Microbiologically-synthesized nanoparticles have emerged as a new agent that can be used to either down regulate the operon associated with quorum sensing proteins, or to enhance quorum-quenching activity to prevent biofilm formation. Although QS inhibition shows good potential for treatment of infections, further development and research are necessary to fully understand the mechanisms of action and suitability for clinical applications. Nanomaterial-based treatment methods are expected to continue developing more sophisticated or more complex mechanisms of destroying the EPS and killing the bacteria, although the need for future developments to prevent recurrence after biofilm treatment is still needed.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by Fundamental Research Grant Scheme (304/PPSK/615059) awarded by Ministry of Higher Education, Malaysia, and Odisha Higher Education Program for Excellence and Equity (OHEPEE), Govt of Odisha.
References
- Abidi S. H., Sherwani S. K., Siddiqui T. R., Bashir A., Kazmi S. U. (2013). Drug resistance profile and biofilm forming potential of Pseudomonas aeruginosa isolated from contact lenses in Karachi-Pakistan. BMC Ophthalmol. 13:57. 10.1186/1471-2415-13-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abinaya M., Vaseeharan B., Divya M., Sharmili A., Govindarajan M., Alharbi N. S., et al. (2018). Bacterial exopolysaccharide (EPS)-coated ZnO nanoparticles showed high antibiofilm activity and larvicidal toxicity against malaria and Zika virus vectors. J. Trace Elem. Med. Biol. 45, 93–103. 10.1016/j.jtemb.2017.10.002 [DOI] [PubMed] [Google Scholar]
- Ahmed E., Kalathil S., Shi L., Alharbi O., Wang P. (2018). Synthesis of ultra-small platinum, palladium and gold nanoparticles by Shewanella loihica PV-4 electrochemically active biofilms and their enhanced catalytic activities. J. Saudi Chem. Soc. 22, 919–929. 10.1016/j.jscs.2018.02.002 [DOI] [Google Scholar]
- Ali S. G., Ansari M. A., Sajid Jamal Q. M., Khan H. M., Jalal M., Ahmad H., et al. (2017). Antiquorum sensing activity of silver nanoparticles in P. aeruginosa: an in silico study. Silico Pharmacol. 5:12. 10.1007/s40203-017-0031-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shabib N. A., Husain F. M., Hassan I., Khan M. S., Ahmed F., Qais F. A., et al. (2018). Biofabrication of zinc oxide nanoparticle from Ochradenus baccatus leaves: broad-spectrum antibiofilm activity, protein binding studies and in vivo toxicity and stress studies. J. Nanomater. 2018:8612158. 10.1155/2018/8612158 [DOI] [Google Scholar]
- Ammons M. C. B., Ward L. S., Fisher S. T., Wolcott R. D., James G. A. (2009). In vitro susceptibility of established biofilms composed of a clinical wound isolate of Pseudomonas aeruginosa treated with lactoferrin and xylitol. Int. J. Antimicrob. Agents 33, 230–236. 10.1016/j.ijantimicag.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anil Kumar S., Abyaneh M. K., Gosavi S. W., Kulkarni S. K., Pasricha R., Ahmad A., et al. (2007). Nitrate reductase-mediated synthesis of silver nanoparticles from AgNO3. Biotechnol. Lett. 29, 439–445. 10.1007/s10529-006-9256-7 [DOI] [PubMed] [Google Scholar]
- Annamalai J., Nallamuthu T. (2015). Characterization of biosynthesized gold nanoparticles from aqueous extract of Chlorella vulgaris and their anti-pathogenic properties. Appl. Nanosci. 5, 603–607. 10.1007/s13204-014-0353-y [DOI] [Google Scholar]
- Arakaki A., Nakazawa H., Nemoto M., Mori T., Matsunaga T. (2008). Formation of magnetite by bacteria and its application. J. R. Soc. Interface 5, 977–999. 10.1098/rsif.2008.0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F., Han Y., Chen J., Zhang X. H. (2008). Disruption of quorum sensing in Vibrio harveyi by the AiiA protein of Bacillus thuringiensis. Aquaculture 274, 36–40. 10.1016/j.aquaculture.2007.11.024 [DOI] [Google Scholar]
- Banerjee A. N. (2011). The design, fabrication photocatalytic utility of nanostructured semiconductors: focus on TiO2-based nanostructures. Nanotechnol. Sci. Appl. 4:35. 10.2147/NSA.S9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankura K. P., Maity D., Mollick M. M. R., Mondal D., Bhowmick B., Bain M. K., et al. (2012). Synthesis, characterization and antimicrobial activity of dextran stabilized silver nanoparticles in aqueous medium. Carbohydr. Polym. 89, 1159–1165. 10.1016/j.carbpol.2012.03.089 [DOI] [PubMed] [Google Scholar]
- Banoee M., Seif S., Nazari Z. E., Jafari-Fesharaki P., Shahverdi H. R., Moballegh A., et al. (2010). ZnO nanoparticles enhanced antibacterial activity of ciprofloxacin against Staphylococcus aureus and Escherichia coli. J. Biomed. Mater. Res. B Appl. Biomater. 93, 5572–5561. 10.1002/jbm.b.31615 [DOI] [PubMed] [Google Scholar]
- Barber-Zucker S., Zarivach R. (2017). A look into the biochemistry of magnetosome biosynthesis in magnetotactic bacteria. ACS Chem. Biol. 12, 13–22. 10.1021/acschembio.6b01000 [DOI] [PubMed] [Google Scholar]
- Baskar G., Chandhuru J., Sheraz Fahad K., Praveen A. S., Chamundeeswari M., Muthukumar T. (2015). Anticancer activity of fungal l-asparaginase conjugated with zinc oxide nanoparticles. J. Mater. Sci. Mater. Med. 26:43. 10.1007/s10856-015-5380-z [DOI] [PubMed] [Google Scholar]
- Beladiya C., Tripathy R. K., Bajaj P., Aggarwal G., Pande A. H. (2015). Expression, purification and immobilization of recombinant AiiA enzyme onto magnetic nanoparticles. Protein Expr. Purif. 113, 56–62. 10.1016/j.pep.2015.04.014 [DOI] [PubMed] [Google Scholar]
- Bhadwal A. S., Tripathi R. M., Gupta R. K., Kumar N., Singh R. P., Shrivastav A. (2014). Biogenic synthesis and photocatalytic activity of CdS nanoparticles. RSC Adv. 4, 9484–9490. 10.1039/c3ra46221h [DOI] [Google Scholar]
- Bindhu M. R., Umadevi M. (2014). Antibacterial activities of green synthesized gold nanoparticles. Mater. Lett. 120, 122–125. 10.1016/j.matlet.2014.01.108 [DOI] [Google Scholar]
- Brandt O., Mildner M., Egger A. E., Groessl M., Rix U., Posch M., et al. (2012). Nanoscalic silver possesses broad-spectrum antimicrobial activities and exhibits fewer toxicological side effects than silver sulfadiazine. Nanomed. Nanotechnol. Biol. Med. 8, 478–488. 10.1016/j.nano.2011.07.005 [DOI] [PubMed] [Google Scholar]
- Brayner R., Ferrari-Iliou R., Brivois N., Djediat S., Benedetti M. F., Fiévet F. (2006). Toxicological impact studies based on Escherichia coli bacteria in ultrafine ZnO nanoparticles colloidal medium. Nano Lett. 6, 866–870. 10.1021/nl052326h [DOI] [PubMed] [Google Scholar]
- Bruna N., Collao B., Tello A., Caravantes P., Díaz-Silva N., Monrás J. P., et al. (2019). Synthesis of salt-stable fluorescent nanoparticles (quantum dots) by polyextremophile halophilic bacteria. Sci. Rep. 9:1953. 10.1038/s41598-018-38330-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszewski B., Railean-Plugaru V., Pomastowski P., Rafińska K., Szultka-Mlynska M., Golinska P., et al. (2018). Antimicrobial activity of biosilver nanoparticles produced by a novel Streptacidiphilusdurhamensis strain. J. Microbiol. Immunol. Infect. 51, 45–54. 10.1016/j.jmii.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Butchosa N., Brown C., Larsson P. T., Berglund L. A., Bulone V., Zhou Q. (2013). Nanocomposites of bacterial cellulose nanofibers and chitin nanocrystals: fabrication, characterization and bactericidal activity. Green Chem. 15, 3404–3413. 10.1039/c3gc41700j [DOI] [Google Scholar]
- Capek I. (2014). Preparation and functionalization of gold nanoparticles. J. Surf. Sci. Technol. 29, 1–18. 10.18311/jsst/2013/1859 [DOI] [Google Scholar]
- Capeness M. J., Echavarri-Bravo V., Horsfall L. E. (2019). Production of biogenic nanoparticles for the reduction of 4-Nitrophenol and oxidative laccase-like reactions. Front. Microbiol. 10:997. 10.3389/fmicb.2019.00997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J., Nachtergael A., Duez P., El Jaziri M., Rasamiravaka T. (2020). Natural compounds inhibiting Pseudomonas aeruginosa biofilm formation by targeting quorum sensing circuitry, in bacterial biofilms. London, UK: Intechopen. [Google Scholar]
- Carnes E. C., Lopez D. M., Donegan N. P., Cheung A., Gresham H., Timmins G. S., et al. (2010). Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat. Chem. Biol. 6, 41–45. 10.1038/nchembio.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano J. J., Shafii S. M., Ko F., Donate G., Wright T. E., Mannari R. J., et al. (2007). Comparative evaluation of silver-containing antimicrobial dressings and drugs. Int. Wound J. 4, 114–122. 10.1111/j.1742-481X.2007.00316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. Y., Dang X., Klug M. T., Qi J., Dorval Courchesne N. M., Burpo F. J., et al. (2013). Versatile three-dimensional virus-based template for dye-sensitized solar cells with improved electron transport and light harvesting. ACS Nano 7, 6563–6574. 10.1021/nn4014164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Schluesener H. J. (2008). Nanosilver: a nanoproduct in medical application. Toxicol. Lett. 176, 1–12. 10.1016/j.toxlet.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Chen C. C., Stark M., Baikoghli M., Cheng R. H. (2018). Surface functionalization of hepatitis E virus nanoparticles using chemical conjugation methods. J. Vis. Exp. 2018:e57020. 10.3791/57020, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Wang P., Li L. (2016). Applications of bacterial magnetic nanoparticles in nanobiotechnology. J. Nanosci. Nanotechnol. 16, 2164–2171. 10.1166/jnn.2016.10954 [DOI] [PubMed] [Google Scholar]
- Chioro A., Coll-Seck A. M., Høie B., Moeloek N., Motsoaledi A., Rajatanavin R., et al. (2015). Antimicrobial resistance: a priority for global health action. Bull. World Health Organ. 93:439. 10.2471/BLT.15.158998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K. H., Park J. E., Osaka T., Park S. G. (2005). The study of antimicrobial activity and preservative effects of nanosilver ingredient. Electrochim. Acta 51, 956–960. 10.1016/j.electacta.2005.04.071 [DOI] [Google Scholar]
- Correa-Llantén D. N., Muñoz-Ibacache S. A., Castro M. E., Muñoz P. A., Blamey J. M. (2013). Gold nanoparticles synthesized by Geobacillus sp. strain ID17 a thermophilic bacterium isolated from Deception Island, Antarctica. Microb. Cell Factories 12:75. 10.1186/1475-2859-12-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Cui R., Liu H. H., Xie H. Y., Zhang Z. L., Yang Y. R., Pang D. W., et al. (2009). Living yeast cells as a controllable biosynthesizer for fluorescent quantum dots. Adv. Funct. Mater. 19, 2359–2364. 10.1002/adfm.200801492 [DOI] [Google Scholar]
- Cunha F. A., da Cunha M. C. S. O., da Frota S. M., Mallmann E. J. J., Freire T. M., Costa L. S., et al. (2018). Biogenic synthesis of multifunctional silver nanoparticles from Rhodotorula glutinis and Rhodotorula mucilaginosa: antifungal, catalytic and cytotoxicity activities. World J. Microbiol. Biotechnol. 34:127. 10.1007/s11274-018-2514-8 [DOI] [PubMed] [Google Scholar]
- Das V. L., Thomas R., Varghese R. T., Soniya E. V., Mathew J., Radhakrishnan E. K. (2014). Extracellular synthesis of silver nanoparticles by the Bacillus strain CS 11 isolated from industrialized area. 3 Biotech 4, 121–126. 10.1007/s13205-013-0130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Sarkar J., Ghosh M., Bhattacharya A., Mukherjee A., Chattopadhyay D., et al. (2017). Green conversion of graphene oxide to graphene nanosheets and its biosafety study. PLoS One 12:e0171607. 10.1371/journal.pone.0171607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delalande L., Faure D., Raffoux A., Uroz S., D’Angelo-Picard C., Elasri M., et al. (2005). N-hexanoyl-L-homoserine lactone, a mediator of bacterial quorum-sensing regulation, exhibits plant-dependent stability and may be inactivated by germinating Lotus corniculatus seedlings. FEMS Microbiol. Ecol. 52, 13–20. 10.1016/j.femsec.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Dong Y. H., Gusti A. R., Zhang Q., Xu J. L., Zhang L. H. (2002). Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68, 1754–1759. 10.1128/AEM.68.4.1754-1759.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M. (2001). Biofilms and device-associated infections. Emerg. Infect. Dis. 7, 277–281. 10.3201/eid0702.010226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. 10.3201/eid0809.020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorcheh S., Vahabi K. (2016). “Biosynthesis of nanoparticles by fungi: large-scale production” in Fungal Metabolites. eds. Mérillon J., Ramawat K.(Switzerland: Springer; ), 395–414. [Google Scholar]
- Durán N., Marcato P. D., Alves O. L., De Souza G. I. H., Esposito E. (2005). Mechanistic aspects of biosynthesis of silver nanoparticles by several Fusarium oxysporum strains. J. Nanobiotechnology 3:8. 10.1186/1477-3155-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán N., Seabra A. B. (2012). Metallic oxide nanoparticles: state of the art in biogenic syntheses and their mechanisms. Appl. Microbiol. Biotechnol. 95, 275–288. 10.1007/s00253-012-4118-9 [DOI] [PubMed] [Google Scholar]
- Eberhard A., Burlingame A. L., Eberhard C., Kenyon G. L., Nealson K. H., Oppenheimer N. J. (1981). Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20, 2444–2449. 10.1021/bi00512a013 [DOI] [PubMed] [Google Scholar]
- Emam H. E., Ahmed H. B. (2016). Polysaccharides templates for assembly of nanosilver. Carbohydr. Polym. 135, 300–307. 10.1016/j.carbpol.2015.08.095 [DOI] [PubMed] [Google Scholar]
- Escárcega-González C. E., Garza-Cervantes J. A., Vázquez-Rodríguez A., Morones-Ramírez J. R. (2018). Bacterial exopolysaccharides as reducing and/or stabilizing agents during synthesis of metal nanoparticles with biomedical applications. Int. J. Polym. Sci. 2018:7045852. 10.1155/2018/7045852 [DOI] [Google Scholar]
- Faivre D., Schüler D. (2008). Magnetotactic bacteria and magnetosomes. Chem. Rev. 108, 4875–4898. 10.1021/cr078258w [DOI] [PubMed] [Google Scholar]
- Fan X. Z., Pomerantseva E., Gnerlich M., Brown A., Gerasopoulos K., McCarthy M., et al. (2013). Tobacco mosaic virus: a biological building block for micro/nano/bio systems. J. Vac. Sci. Technol. A 31:050815. 10.1116/1.4816584 [DOI] [Google Scholar]
- Fang X., Wang Y., Wang Z., Jiang Z., Dong M. (2019). Microorganism assisted synthesized nanoparticles for catalytic applications. Energies 12:190. 10.3390/en12010190 [DOI] [Google Scholar]
- Fariq A., Khan T., Yasmin A. (2017). Microbial synthesis of nanoparticles and their potential applications in biomedicine. J. Appl. Biomed. 15, 241–248. 10.1016/j.jab.2017.03.004 [DOI] [Google Scholar]
- Flavier A. B., Clough S. J., Schell M. A., Denny T. P. (1997). Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26, 251–259. 10.1046/j.1365-2958.1997.5661945.x [DOI] [PubMed] [Google Scholar]
- Fujishima A., Rao T. N., Tryk D. A. (2000). Titanium dioxide photocatalysis. J Photochem Photobiol C: Photochem Rev 1, 1–21. 10.1016/S1389-5567(00)00002-2 [DOI] [Google Scholar]
- Fuqua W. C., Winans S. C., Greenberg E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density‐ responsive transcriptional regulators. J. Bacteriol. 176, 269–275. 10.1128/JB.176.2.269-275.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlawat G., Choudhury A. R. (2019). A review on the biosynthesis of metal and metal salt nanoparticles by microbes. RSC Adv. 9:12944. 10.1039/C8RA10483B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganachari S. V., Bhat R., Deshpande R., Venkataraman A. (2012). Extracellular biosynthesis of silver nanoparticles using fungi Penicillium diversum and their antimicrobial activity studies. Bionanoscience 2, 316–321. 10.1007/s12668-012-0046-5 [DOI] [Google Scholar]
- García-Lara B., Saucedo-Mora M. A., Roldán-Sánchez J. A., Pérez-Eretza B., Ramasamy M., Lee J., et al. (2015). Inhibition of quorum-sensing-dependent virulence factors and biofilm formation of clinical and environmental Pseudomonas aeruginosa strains by ZnO nanoparticles. Lett. Appl. Microbiol. 61, 299–305. 10.1111/lam.12456 [DOI] [PubMed] [Google Scholar]
- Garole V. J., Choudhary B. C., Tetgure S. R., Garole D. J., Borse A. U. (2019). Palladium nanocatalyst: green synthesis, characterization and catalytic application. Int. J. Environ. Sci. Technol. 16, 7885–7892. 10.1007/s13762-018-2173-1 [DOI] [Google Scholar]
- Giasuddin A., Jhuma K., Haq A. M. (2013). Use of gold nanoparticles in diagnostics, surgery and medicine: a review. Bangladesh J. Med. Biochem. 5, 56–60. 10.3329/bjmb.v5i2.13346 [DOI] [Google Scholar]
- González-Ballesteros N., Prado-López S., Rodríguez-González J. B., Lastra M., Rodríguez-Argüelles M. C. (2017). Green synthesis of gold nanoparticles using brown algae Cystoseira baccata: its activity in colon cancer cells. Colloids Surf. B: Biointerfaces 153, 190–198. 10.1016/j.colsurfb.2017.02.020 [DOI] [PubMed] [Google Scholar]
- Grasso G., Zane D., Dragone R. (2019). Microbial nanotechnology: challenges and prospects for green biocatalytic synthesis of nanoscale materials for sensoristic and biomedical applications. Nanomaterials 10:11. 10.3390/nano10010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg K., Müller E. -C., Otto A., Reszka R., Linder D., Kube M., et al. (2004). Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70, 1040–1050. 10.1128/AEM.70.2.1040-1050.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilger-Casagrande M., de Lima R. (2019). Synthesis of silver nanoparticles mediated by fungi: a review. Front. Bioeng. Biotechnol. 7:287. 10.3389/fbioe.2019.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haefeli C., Franklin C., Hardy K. (1984). Plasmid-determined silver resistance in Pseudomonas stutzeri isolated from a silver mine. J. Bacteriol. 158, 389–392. 10.1128/JB.158.1.389-392.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R., Song X., Wang Q., Qi Y., Deng G., Zhang A., et al. (2019). Microbial synthesis of graphene-supported highly-dispersed Pd-Ag bimetallic nanoparticles and its catalytic activity. J. Chem. Technol. Biotechnol. 94, 3375–3383. 10.1002/jctb.6150 [DOI] [Google Scholar]
- Han D., Yang H., Zhu C., Wang F. (2008). Controlled synthesis of CuO nanoparticles using TritonX-100-based water-in-oil reverse micelles. Powder Technol. 185, 286–290. 10.1016/j.powtec.2007.10.018 [DOI] [Google Scholar]
- Hanzelka B. L., Greenberg E. P. (1996). Quorum sensing in Vibrio fischeri: evidence that S-adenosylmethionine is the amino acid substrate for autoinducer synthesis. J. Bacteriol. 178, 5291–5294. 10.1128/JB.178.17.5291-5294.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby N., Ciofu O., Bjarnsholt T. (2010). Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 21, 595–599. 10.2217/fmb.10.125 [DOI] [PubMed] [Google Scholar]
- Horie M., Nishio K., Fujita K., Endoh S., Miyauchi A., Saito Y., et al. (2009). Protein adsorption of ultrafine metal oxide and its influence on cytotoxicity toward cultured cells. Chem. Res. Toxicol. 22, 543–553. 10.1021/tx800289z [DOI] [PubMed] [Google Scholar]
- Hu Y., He L., Ding J., Sun D., Chen L., Chen X. (2016). One-pot synthesis of dextran decorated reduced graphene oxide nanoparticles for targeted photo-chemotherapy. Carbohydr. Polym. 144, 223–229. 10.1016/j.carbpol.2016.02.062 [DOI] [PubMed] [Google Scholar]
- Ikuma K., Decho A. W., Lau B. L. T. (2015). When nanoparticles meet biofilms-interactions guiding the environmental fate and accumulation of nanoparticles. Front. Microbiol. 6:591. 10.3389/fmicb.2015.00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iravani S. (2014). Bacteria in nanoparticle synthesis: current status and future prospects. Int. Sch. Res. Not. 44, 235–239. 10.1155/2014/359316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap S., Priolkar K. R. (2013). Evaluation of ZnO nanoparticles and study of ZnO-TiO2 composites for lead free humidity sensors. Sensors Actuators B Chem. 183, 411–418. 10.1016/j.snb.2013.04.010 [DOI] [Google Scholar]
- Jain S., Hirst D. G., O’Sullivan J. M. (2012). Gold nanoparticles as novel agents for cancer therapy. Br. J. Radiol. 85, 103–113. 10.1259/bjr/59448833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S. (2015). Advances in nanoscale alloys and intermetallics: low temperature solution chemistry synthesis and application in catalysis. Dalton Trans. 44, 18692–18717. 10.1039/C5DT03699b [DOI] [PubMed] [Google Scholar]
- Jeevanandam J., Chan Y. S., Danquah M. K. (2016). Biosynthesis of metal and metal oxide nanoparticles. Chem. Bio. Eng. Rev. 3, 55–67. 10.1002/cben.201500018 [DOI] [Google Scholar]
- Johnston C. W., Wyatt M. A., Li X., Ibrahim A., Shuster J., Southam G., et al. (2013). Gold biomineralization by a metallophore from a gold-associated microbe. Nat. Chem. Biol. 9, 241–243. 10.1038/nchembio.1179 [DOI] [PubMed] [Google Scholar]
- Kanmani P., Lim S. T. (2013). Synthesis and structural characterization of silver nanoparticles using bacterial exopolysaccharide and its antimicrobial activity against food and multidrug resistant pathogens. Process Biochem. 48, 1099–1109. 10.1016/j.procbio.2013.05.011 [DOI] [Google Scholar]
- Kaufmann G. F., Sartorio R., Lee S. H., Rogers C. J., Meijler M. M., Moss J. A., et al. (2005). Revisiting quorum sensing: discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc. Natl. Acad. Sci. U. S. A. 102, 309–314. 10.1073/pnas.0408639102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H., Dolma K., Kaur N., Malhotra A., Kumar N., Dixit P., et al. (2015). Marine microbe as nano-factories for copper biomineralization. Biotechnol. Bioprocess Eng. 20, 51–57. 10.1007/s12257-014-0432-7 [DOI] [Google Scholar]
- Khan M. F., Husain F. M., Zia Q., Ahmad E., Jamal A., Alaidarous M., et al. (2020). Anti-quorum sensing and anti-biofilm activity of zinc oxide nanospikes. ACS Omega 5, 32203–32215. 10.1021/acsomega.0c03634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandel P., Kumar-Shahi S. (2016). Microbes mediated synthesis of metal nanoparticles: current status and future prospects. Int. J. Nanomater. Bios. 6, 1–24. [Google Scholar]
- Khandel P., Shahi S. K. (2018). Mycogenic nanoparticles and their bio-prospective applications: current status and future challenges. J. Nanostruct. Chem. 8, 369–391. 10.1007/s40097-018-0285-2 [DOI] [Google Scholar]
- Khurana C., Sharma P., Pandey O. P., Chudasama B. (2016). Synergistic effect of metal nanoparticles on the antimicrobial activities of antibiotics against biorecycling microbes. J. Mater. Sci. Technol. 32, 524–532. 10.1016/j.jmst.2016.02.004 [DOI] [Google Scholar]
- Kim J. S., Kuk E., Yu K. N., Kim J. H., Park S. J., Lee H. J., et al. (2007). Antimicrobial effects of silver nanoparticles. Nanomedicine nanotechnology. Biol. Med. 3, 95–101. 10.1016/j.nano.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Kim D. Y., Saratale R. G., Shinde S., Syed A., Ameen F., Ghodake G. (2016). Green synthesis of silver nanoparticles using Laminaria japonica extract: characterization and seedling growth assessment. J. Clean. Prod. 172, 2910–2918. 10.1016/j.jclepro.2017.11.123 [DOI] [Google Scholar]
- Kiran G. S., Sabu A., Selvin J. (2010). Synthesis of silver nanoparticles by glycolipid biosurfactant produced from marine Brevibacterium casei MSA19. J. Biotechnol. 148, 221–225. 10.1016/j.jbiotec.2010.06.012 [DOI] [PubMed] [Google Scholar]
- Kiran G. S., Selvin J., Manilal A., Sujith S. (2011). Biosurfactants as green stabilizers for the biological synthesis of nanoparticles. Crit. Rev. Biotechnol. 31, 354–364. 10.3109/07388551.2010.539971 [DOI] [PubMed] [Google Scholar]
- Kitching M., Ramani M., Marsili E. (2015). Fungal biosynthesis of gold nanoparticles: mechanism and scale up. Microb. Biotechnol. 8, 904–917. 10.1111/1751-7915.12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus T., Joerger R., Olsson E., Granqvist C. G. (1999). Silver-based crystalline nanoparticles, microbially fabricated. Proc. Natl. Acad. Sci. U. S. A. 96, 13611–13614. 10.1073/pnas.96.24.13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Tomita S., Sawada K., Shiba K., Yanagi H., Yamashita I., et al. (2012). Chiral meta-molecules consisting of gold nanoparticles and genetically engineered tobacco mosaic virus. Opt. Express 20, 24856–24863. 10.1364/OE.20.024856 [DOI] [PubMed] [Google Scholar]
- Kominkova M., Milosavljevic V., Vitek P., Polanska H., Cihalova K., Dostalova S., et al. (2017). Comparative study on toxicity of extracellularly biosynthesized and laboratory synthesized CdTe quantum dots. J. Biotechnol. 241, 193–200. 10.1016/j.jbiotec.2016.10.024 [DOI] [PubMed] [Google Scholar]
- Korbekandi H., Mohseni S., Jouneghani R. M., Pourhossein M., Iravani S. (2016). Biosynthesis of silver nanoparticles using Saccharomyces cerevisia. Artif. Cells Nanomed. Biotechnol. 44, 235–239. 10.3109/21691401.2014.937870 [DOI] [PubMed] [Google Scholar]
- Kumar C. G., Mamidyala S. K., Das B., Sridhar B., Sarala Devi G., Karuna M. S. L. (2010). Synthesis of biosurfactant-based silver nanoparticles with purified rhamnolipids isolated from Pseudomonas aeruginosa BS-161R. J. Microbiol. Biotechnol. 20, 1061–1068. 10.4014/jmb.1001.01018 [DOI] [PubMed] [Google Scholar]
- Kuzajewska D., Wszołek A., Żwierełło W., Kirczuk L., Maruszewska A. (2020). Magnetotactic bacteria and magnetosomes as smart drug delivery systems: anew weapon on the battlefield with cancer? Biology 9:102. 10.3390/biology9050102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C., Park B.-h., Kim H.-w., Jung S.-h. (2009). Green synthesis of silver nanoparticles by sinorhizobial octasaccharide isolated from Sinorhizobium meliloti. Bull. Kor. Chem. Soc. 30, 1651–1654. 10.5012/bkcs.2009.30.7.1651 [DOI] [Google Scholar]
- Lahiri D., Dash S., Dutta R., Nag M. (2019). Elucidating the effect of anti-biofilm activity of bioactive compounds extracted from plants. J. Biosci. 44:52. 10.1007/s12038-019-9868-4 [DOI] [PubMed] [Google Scholar]
- Lara H. H., Garza-Treviño E. N., Ixtepan-Turrent L., Singh D. K. (2011). Silver nanoparticles are broad-spectrum bactericidal and virucidal compounds. J. Nanobiotechnology 9:30. 10.1186/1477-3155-9-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan R., Duse A., Wattal C., Zaidi A. K. M., Wertheim H. F. L., Sumpradit N., et al. (2013). Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098. 10.1016/S1473-3099(13)70318-9 [DOI] [PubMed] [Google Scholar]
- Le D. H. T., Lee K. L., Shukla S., Commandeur U., Steinmetz N. F. (2017). Potato virus X, a filamentous plant viral nanoparticle for doxorubicin delivery in cancer therapy. Nanoscale 9, 2348–2357. 10.1039/C6NR09099K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. -H., Kim Y. -G., Cho M. H., Lee J. (2014). ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol. Res. 169, 888–896. 10.1016/j.micres.2014.05.005 [DOI] [PubMed] [Google Scholar]
- Lengke M. F., Fleet M. E., Southam G. (2006). Synthesis of plantinum nanoparticles by reaction of filamentous cyanobacteria with plantinum(IV)-chloride complex. Langmuir. 22, 7318–7323. 10.1021/la060873s [DOI] [PubMed] [Google Scholar]
- Lengke M. F., Southam G. (2005). The effect of thiosulfate-oxidizing bacteria on the stability of the gold-thiosulfate complex. Geochim. Cosmochim. Acta 69, 3759–3772. 10.1016/j.gca.2005.03.012 [DOI] [Google Scholar]
- Leung T. C. -Y., Wong C. K., Xie Y. (2010). Green synthesis of silver nanoparticles using biopolymers, carboxymethylated-curdlan and fucoidan. Mater. Chem. Phys. 121, 402–405. 10.1016/j.matchemphys.2010.02.026 [DOI] [Google Scholar]
- Li X., Chen S., Hu W., Shi S., Shen W., Zhang X., et al. (2009). In situ synthesis of CdS nanoparticles on bacterial cellulose nanofibers. Carbohydr. Polym. 76, 509–512. 10.1016/j.carbpol.2008.11.014 [DOI] [Google Scholar]
- Li X., Xu H., Chen Z. S., Chen G. (2011). Biosynthesis of nanoparticles by microorganisms and their applications. J. Nanomater. 2011:270974. 10.1155/2011/270974 [DOI] [Google Scholar]
- Lima E., Guerra R., Lara V., Guzmán A. (2013). Gold nanoparticles as efficient antimicrobial agents for Escherichia coli and Salmonella typhi. Chem. Cent. J. 7:11. 10.1186/1752-153X-7-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobedanz S., Søgaard-Andersen L. (2003). Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 17, 2151–2161. 10.1101/gad.274203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Woodward J. C. (1996). Mechanisms for chelator stimulation of microbial Fe(III)-oxide reduction. Chem. Geol. 132, 19–24. 10.1016/S0009-2541(96)00037-X [DOI] [Google Scholar]
- Lv Q., Zhang B., Xing X., Zhao Y., Cai R., Wang W., et al. (2018). Biosynthesis of copper nanoparticles using Shewanella loihica PV-4 with antibacterial activity: novel approach and mechanisms investigation. J. Hazard. Mater. 347, 141–149. 10.1016/j.jhazmat.2017.12.070 [DOI] [PubMed] [Google Scholar]
- Ma Y., Lan G., Li C., Cambaza E. M., Liu D., Ye X., et al. (2019). Stress tolerance of Staphylococcus aureus with different antibiotic resistance profiles. Microb. Pathog. 133:103549. 10.1016/j.micpath.2019.103549 [DOI] [PubMed] [Google Scholar]
- Mahanty A., Bosu R., Panda P., Netam S. P., Sarkar B. (2013). Microwave assisted rapid combinatorial synthesis of silver nanoparticles using E. coli culture supernatant. Int J Pharm. Bio. Sci 4, 1030–1103. [Google Scholar]
- Malarkodi C., Rajeshkumar S., Paulkumar K., Vanaja M., Jobitha G. D. G., Annadurai G. (2013). Bactericidal activity of bio mediated silver nanoparticles synthesized by Serratia nematodiphila. Drug Invent. Today 5, 119–125. 10.1016/j.dit.2013.05.005 [DOI] [Google Scholar]
- Malvindi M. A., Brunetti V., Vecchio G., Galeone A., Cingolani R., Pompa P. P. (2012). SiO2 nanoparticles biocompatibility and their potential for gene delivery and silencing. Nanoscale 4, 486–495. 10.1039/C1NR11269D [DOI] [PubMed] [Google Scholar]
- Manivasagan P., Venkatesan J., Senthilkumar K., Sivakumar K., Kim S. K. (2013). Biosynthesis, antimicrobial and cytotoxic effect of silver nanoparticles using a novel Nocardiopsis sp. MBRC-1. Biomed. Res. Int. 2013:287638. 10.1155/2013/287638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbahi A. (2010). A review on gold nanoparticles radiosensitization effect in radiation therapy of cancer. Rep. Pract. Oncol. Radiother. 15, 176–180. 10.1016/j.rpor.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. P. (2015). Bacterial communication and its role as a target for nanoparticle-based antimicrobial therapy (Doctoral dissertation).
- Mishra S., Singh B. R., Naqvi A. H., Singh H. B. (2017). Potential of biosynthesized silver nanoparticles using Stenotrophomonas sp. BHU-S7 (MTCC 5978) for management of soil-borne and foliar phytopathogens. Sci. Rep. 7:45154. 10.1038/srep45154, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam A. B., Moniri M., Azizi S., Rahim R. A., Ariff A. B., Saad W. Z., et al. (2017). Biosynthesis of ZnO nanoparticles by a new Pichia kudriavzevii yeast strain and evaluation of their antimicrobial and antioxidant activities. Molecules 22:872. 10.3390/molecules22060872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik K., Chatterjee A., Prakash H., Kowshik M. (2013). Mesoporous TiO2 nanoparticles containing Ag ion with excellent antimicrobial activity at remarkable low silver concentrations. J. Biomed. Nanotechnol. 9, 664–673. 10.1166/jbn.2013.1567 [DOI] [PubMed] [Google Scholar]
- Naik K., Kowshik M. (2014). Anti-quorum sensing activity of AgCl-TiO2 nanoparticles with potential use as active food packaging material. J. Appl. Microbiol. 117, 972–983. 10.1111/jam.12589 [DOI] [PubMed] [Google Scholar]
- Nazari P., Faramarzi M. A., Sepehrizadeh Z., Mofid M. R., Bazaz R. D., Shahverdi A. R. (2012). Biosynthesis of bismuth nanoparticles using Serratia marcescens isolated from the Caspian Sea and their characterisation. IET Nanobiotechnol. 6, 58–62. 10.1049/iet-nbt.2010.0043 [DOI] [PubMed] [Google Scholar]
- Nealson K. H., Platt T., Hastings J. W. (1970). Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322. 10.1128/JB.104.1.313-322.1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen L. P., Risgaard-Petersen N., Fossing H., Christensen P. B., Sayama M. (2010). Electric currents couple spatially separated biogeochemical processes in marine sediment. Nature 463, 1071–1074. 10.1038/nature08790 [DOI] [PubMed] [Google Scholar]
- Nikaido H. (2009). Multidrug resistance in bacteria. Annu. Rev. Biochem. 78, 119–146. 10.1146/annurev.biochem.78.082907.145923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayemi J. D., Dozie-Nwachukwu S., Danyuo Y., Odusanya O. S., Anuku N., Malatesta K., et al. (2015). Biosynthesis and the conjugation of magnetite nanoparticles with luteinizing hormone releasing hormone (LHRH). Mater. Sci. Eng. C 46, 482–496. 10.1016/j.msec.2014.10.081 [DOI] [PubMed] [Google Scholar]
- Órdenes-Aenishanslins N., Anziani-Ostuni G., Monrás J. P., Tello A., Bravo D., Toro-Ascuy D., et al. (2020). Bacterial synthesis of ternary cdsag quantum dots through cation exchange: tuning the composition and properties of biological nanoparticles for bioimaging and photovoltaic applications. Microorganisms 8:631. 10.3390/microorganisms8050631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otari S. V., Patil R. M., Nadaf N. H., Ghosh S. J., Pawar S. H. (2014). Green synthesis of silver nanoparticles by microorganism using organic pollutant: its antimicrobial and catalytic application. Environ. Sci. Pollut. Res. 21, 1503–1513. 10.1007/s11356-013-1764-0 [DOI] [PubMed] [Google Scholar]
- Padwal P., Bandyopadhyaya R., Mehra S. (2015). Biocompatible citric acid-coated iron oxide nanoparticles to enhance the activity of first-line anti-TB drugs in Mycobacterium smegmatis. J. Chem. Technol. Biotechnol. 90, 1773–1781. 10.1002/jctb.4766 [DOI] [Google Scholar]
- Panáček A., Kvítek L., Prucek R., Kolář M., Večeřová R., Pizúrová N., et al. (2006). Silver colloid nanoparticles: synthesis, characterization their antibacterial activity. J. Phys. Chem. B 110, 16248–16253. 10.1021/jp063826h [DOI] [PubMed] [Google Scholar]
- Pati S., Chatterji A., Dash B. P., Nelson B. R., Sarkar T., Shahimi S., et al. (2020). Structural characterization and antioxidant potential of chitosan by γ-irradiation from the carapace of horseshoe crab. Polymers 12:2361. 10.3390/polym12102361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S., Chandrasekaran R. (2020). Biogenic nanoparticles: a comprehensive perspective in synthesis, characterization, application and its challenges. J. Genet. Eng. Biotechnol. 18:67. 10.1186/s43141-020-00081-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. P., Gray K. M., Passador L., Tucker K. D., Eberhard A., Iglewski B. H., et al. (1994). Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U. S. A. 91, 197–201. 10.1073/pnas.91.1.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. P., Passador L., Iglewski B. H., Greenberg E. P. (1995). A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92, 1490–1494. 10.1073/pnas.92.5.1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci E. C., Milbank J. B. J., Pearson J. P., Mcknight S., Kende A. S., Greenberg E. P., et al. (1999). Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96, 11229–11234. 10.1073/pnas.96.20.11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan V. V., Liu W. T., Pogliano K., Dorrestein P. C. (2012). Microbial metabolic exchangeg-the chemotype-to-phenotype link. Nat. Chem. Biol. 8, 26–35. 10.1038/nchembio.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto R. M., Lopes-de-Campos D., Martins M. C. L., Van Dijck P., Nunes C., Reis S. (2019). Impact of nanosystems in Staphylococcus aureus biofilms treatment. FEMS Microbiol. Rev. 43, 622–641. 10.1093/femsre/fuz021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Płaza G. A., Chojniak J., Banat I. M. (2014). Biosurfactant mediated biosynthesis of selected metallic nanoparticles. Int. J. Mol. Sci. 15, 13720–13737. 10.3390/ijms150813720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza D. O., Gallardo C., Straub Y. D., Bravo D., Pérez-Donoso J. M. (2016). Biological synthesis of fluorescent nanoparticles by cadmium and tellurite resistant Antarctic bacteria: exploring novel natural nanofactories. Microb. Cell Factories 15:76. 10.1186/s12934-016-0477-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presentato A., Piacenza E., Anikovskiy M., Cappelletti M., Zannoni D., Turner R. J. (2018). Biosynthesis of selenium-nanoparticles and -nanorods as a product of selenite bioconversion by the aerobic bacterium Rhodococcus aetherivorans BCP1. New Biotechnol. 41, 1–8. 10.1016/j.nbt.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Pugazhendhi A., Prabakar D., Jacob J. M., Karuppusamy I., Saratale R. G. (2018). Synthesis and characterization of silver nanoparticles using Gelidium amansii and its antimicrobial property against various pathogenic bacteria. Microb. Pathog. 114, 41–45. 10.1016/j.micpath.2017.11.013 [DOI] [PubMed] [Google Scholar]
- Qayyum S., Khan A. U. (2016). Nanoparticles vs. biofilms: a battle against another paradigm of antibiotic resistance. MedChemComm 7, 1479–1498. 10.1039/C6MD00124F [DOI] [Google Scholar]
- Qi P., Zhang D., Zeng Y., Wan Y. (2016). Biosynthesis of CdS nanoparticles: a fluorescent sensor for sulfate-reducing bacteria detection. Talanta 147, 142–146. 10.1016/j.talanta.2015.09.046 [DOI] [PubMed] [Google Scholar]
- Raffa R. B., Iannuzzo J. R., Levine D. R., Saeid K. K., Schwartz R. C., Sucic N. T., et al. (2005). Bacterial communication (“quorum sensing”) via ligands and receptors: a novel pharmacologic target for the design of antibiotic drugs. J. Pharmacol. Exp. Ther. 312, 417–423. 10.1124/jpet.104.075150 [DOI] [PubMed] [Google Scholar]
- Rafique M., Sadaf I., Rafique M. S., Tahir M. B. (2017). A review on green synthesis of silver nanoparticles and their applications. Artif. Cells Nanomed. Biotechnol. 45, 1272–1291. 10.1080/21691401.2016.1241792 [DOI] [PubMed] [Google Scholar]
- Raj R., Dalei K., Chakraborty J., Das S. (2016). Extracellular polymeric substances of a marine bacterium mediated synthesis of CdS nanoparticles for removal of cadmium from aqueous solution. J. Colloid Interface Sci. 462, 166–175. 10.1016/j.jcis.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Rajasree S. R., Suman T. Y. (2012). Extracellular biosynthesis of gold nanoparticles using a gram negative bacterium Pseudomonas fluorescens. Asian Pac. J. Trop. Dis. 2, 796–799. 10.1016/S2222-1808(12)60267-9 [DOI] [Google Scholar]
- Ranjitha V. R., Rai V. R. (2017). Actinomycetes mediated synthesis of gold nanoparticles from the culture supernatant of Streptomyces griseoruber with special reference to catalytic activity. 3 Biotech 7:299. 10.1007/s13205-017-0930-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith F., Fairbrother L., Nolze G., Wilhelmi O., Clode P. L., Gregg A., et al. (2010). Nanoparticle factories: biofilms hold the key to gold dispersion and nugget formation. Geology 63, 1227–1230. 10.1130/G31052.1 [DOI] [Google Scholar]
- Rodrigues L., Banat I. M., Teixeira J., Oliveira R. (2006). Biosurfactants: potential applications in medicine. J. Antimicrob. Chemother. 57, 609–618. 10.1093/jac/dkl024 [DOI] [PubMed] [Google Scholar]
- Sadekuzzaman M., Yang S., Mizan M. F. R., Ha S. D. (2015). Current and recent advanced strategies for combating biofilms. Compr. Rev. Food Sci. Food Saf. 14, 491–509. 10.1111/1541-4337.12144 [DOI] [Google Scholar]
- Salari Z., Danafar F., Dabaghi S., Ataei S. A. (2016). Sustainable synthesis of silver nanoparticles using macroalgae Spirogyra varians and analysis of their antibacterial activity. J. Saudi Chem. Soc. 20, 459–464. 10.1016/j.jscs.2014.10.004 [DOI] [Google Scholar]
- Saleh M. M., Sadeq R. A., Latif H. K. A., Abbas H. A., Askoura M. (2019). Zinc oxide nanoparticles inhibits quorum sensing and virulence in Pseudomonas aeruginosa. Afr. Health Sci. 19, 2043–2055. 10.4314/ahs.v19i2.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S., Singh B. R., Adholeya A. (2017). Intracellular synthesis of gold nanoparticles using an Ectomycorrhizal strain EM-1083 of Laccaria fraterna and its nanoanti-quorum sensing potential against Pseudomonas aeruginosa. Indian J. Microbiol. 57, 448–460. 10.1007/s12088-017-0662-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaeimehr Z., Javadi I., Namvar F. (2018). Antiangiogenic and antiapoptotic effects of green-synthesized zinc oxide nanoparticles using Sargassum muticum algae extraction. Cancer Nanotechnol. 9:3. 10.1186/s12645-018-0037-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos C. S. C., Gabriel B., Blanchy M., Menes O., García D., Blanco M., et al. (2015). Industrial applications of nanoparticles – a prospective overview. Mater. Today Proc. 2, 456–465. 10.1016/j.matpr.2015.04.056 [DOI] [Google Scholar]
- Santos A. P. A., Watanabe E., de Andrade D. (2011). Biofilm on artificial pacemaker: fiction or reality? Arq. Bras. Cardiol. 97, 113–120. 10.1590/S0066-782X2011001400018 [DOI] [PubMed] [Google Scholar]
- Sarkar T., Salauddin M., Chakraborty R. (2020). In-depth pharmacological and nutritional properties of bael (Aegle marmelos): a critical review. J. Agric. Food Res. 2:100081. 10.1016/j.jafr.2020.100081 [DOI] [Google Scholar]
- Satyanarayana Reddy A., Chen C. Y., Chen C. C., Jean J. S., Chen H. R., Tseng M. J., et al. (2010). Biological synthesis of gold and silver nanoparticles mediated by the bacteria Bacillus subtilis. J. Nanosci. Nanotechnol. 10, 6567–6574. 10.1166/jnn.2010.2519 [DOI] [PubMed] [Google Scholar]
- Seshadri S., Saranya K., Kowshik M. (2011). Green synthesis of lead sulfide nanoparticles by the lead resistant marine yeast, Rhodosporidium diobovatum. Biotechnol. Prog. 27, 1464–1469. 10.1002/btpr.651 [DOI] [PubMed] [Google Scholar]
- Shah M. S. A. S., Nag M., Kalagara T., Singh S., Manorama S. V. (2008). Silver on PEG-PU-TiO2 polymer nanocomposite films: An excellent system for antibacterial applications. Chem. Mater. 20, 2455–2460. 10.1021/cm7033867 [DOI] [Google Scholar]
- Shiraishi Y., Hirai T. (2008). Selective organic transformations on titanium oxide-based photocatalysts. J Photochem Photobiol C: Photochem Rev 9, 157–170. 10.1016/j.jphotochemrev.2008.05.001 [DOI] [Google Scholar]
- Shrestha P. M., Rotaru A. E., Summers Z. M., Shrestha M., Liu F., Lovley D. R. (2013). Transcriptomic and genetic analysis of direct interspecies electron transfer. Appl. Environ. Microbiol. 79, 2397–2404. 10.1128/AEM.03837-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi K. S., Husen A., Rao R. A. K. (2018). A review on biosynthesis of silver nanoparticles and their biocidal properties. J. Nanobiotechnology 16:14. 10.1186/s12951-018-0334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B. R., Dwivedi S., Al-Khedhairy A. A., Musarrat J. (2011). Synthesis of stable cadmium sulfide nanoparticles using surfactin produced by Bacillus amyloliquifaciens strain KSU-109. Colloids Surf. B: Biointerfaces 85, 207–213. 10.1016/j.colsurfb.2011.02.030 [DOI] [PubMed] [Google Scholar]
- Singh R., Shedbalkar U. U., Wadhwani S. A., Chopade B. A. (2015b). Bacteriagenic silver nanoparticles: synthesis, mechanism applications. Appl. Microbiol. Biotechnol. 99, 4579–4593. 10.1007/s00253-015-6622-1 [DOI] [PubMed] [Google Scholar]
- Singh S., Singh S. K., Chowdhury I., Singh R. (2017). Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol. J. 11, 53–62. 10.2174/1874285801711010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B. R., Singh B. N., Singh A., Khan W., Naqvi A. H., Singh H. B. (2015a). Mycofabricated biosilver nanoparticles interrupt Pseudomonas aeruginosa quorum sensing systems. Sci. Rep. 5:13719. 10.1038/srep13719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Li X., Cheng Y., Xiao X., Lu Z., Wang Y., et al. (2017). Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction. Sci. Rep. 7:3239. 10.1038/s41598-017-03558-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinath B. S., Namratha K., Byrappa K. (2018). Eco-friendly synthesis of gold nanoparticles by Bacillus subtilis and their environmental applications. Adv. Sci. Lett. 24, 5942–5946. 10.1166/asl.2018.12224 [DOI] [Google Scholar]
- Srivastava N., Mukhopadhyay M. (2014). Biosynthesis of SnO2 nanoparticles using bacterium Erwinia herbicola and their photocatalytic activity for degradation of dyes. Ind. Eng. Chem. Res. 53, 13971–13979. 10.1021/ie5020052 [DOI] [Google Scholar]
- Stewart P. S. (2002). Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292, 107–113. 10.1078/1438-4221-00196 [DOI] [PubMed] [Google Scholar]
- Sunkar S., Nachiyar C. V. (2012). Biogenesis of antibacterial silver nanoparticles using the endophytic bacterium Bacillus cereus isolated from Garcinia xanthochymus. Asian Pac. J. Trop. Biomed. 2, 953–959. 10.1016/S2221-1691(13)60006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi P., Pandit R., Gade A., Derita M., Zachino S., Rai M. (2017). Colletotrichum sp.‐ mediated synthesis of sulphur and aluminium oxide nanoparticles and its in vitro activity against selected food-borne pathogens. LWT 81, 188–194. 10.1016/j.lwt.2017.03.038 [DOI] [Google Scholar]
- Tang L., Cheng J. (2013). Nonporous silica nanoparticles for nanomedicine application. Nano Today 8, 290–312. 10.1016/j.nantod.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker J. N., Dalwadi P., Dhandhukia P. C. (2013). Biosynthesis of gold nanoparticles using Fusarium oxysporum f. sp. cubense JT1, a plant pathogenic fungus. ISRN Biotechnol. 2013:515091. 10.5402/2013/515091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L. J., Li W. W., Zhu T. T., Chen J. J., Wang W. K., An P. F., et al. (2017). Directed biofabrication of nanoparticles through regulating extracellular electron transfer. J. Am. Chem. Soc. 139, 12149–12152. 10.1021/jacs.7b07460 [DOI] [PubMed] [Google Scholar]
- Tomczak M. M., Gupta M. K., Drummy L. F., Rozenzhak S. M., Naik R. R. (2009). Morphological control and assembly of zinc oxide using a biotemplate. Acta Biomater. 5, 876–882. 10.1016/j.actbio.2008.11.011 [DOI] [PubMed] [Google Scholar]
- Tran H. M., Tran H., Booth M. A., Fox K. E., Nguyen T. H., Tran N., et al. (2020). Nanomaterials for treating bacterial biofilms on implantable medical devices. Nano 10, 1–19. 10.3390/nano10112253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas G., Cypriano J., Correa T., Leão P., Bazylinski D. A., Abreu F. (2018). Applications of magnetotactic bacteria, magnetosomes and magnetosome crystals in biotechnology and nanotechnology: mini-review. Molecules 23, 1–25. 10.3390/molecules23102438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseghi Z., Nematollahzadeh A., Tavakoli O. (2018). Green methods for the synthesis of metal nanoparticles using biogenic reducing agents: a review. Rev. Chem. Eng. 34, 529–559. 10.1515/revce-2017-0005 [DOI] [Google Scholar]
- Vinoj G., Pati R., Sonawane A., Vaseeharan B. (2015). In vitro cytotoxic effects of gold nanoparticles coated with functional acyl homoserine lactone lactonase protein from Bacillus licheniformis and their antibiofilm activity against proteus species. Antimicrob. Agents Chemother. 59, 763–771. 10.1128/AAC.03047-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. K., Jacobs R. L., Watts J. L., Rottiers V., Jiang K., Finnegan D. M., et al. (2011). A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell 147, 840–852. 10.1016/j.cell.2011.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. -N., Guan Q. -T., Pain A., Kaksonen A. H., Hong P. -Y. (2019). Discovering, characterizing and applying acyl homoserine lactone-quenching enzymes to mitigate microbe-associated problems under saline conditions. Front. Microbiol. 10:823. 10.3389/fmicb.2019.03139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hu C., Shao L. (2017a). The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomedicine 12, 1227–1249. 10.2147/IJN.S121956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang D., Pan X., Lee D. J., Al-Misned F. A., Mortuza M. G., et al. (2017b). Aerobic and anaerobic biosynthesis of nano-selenium for remediation of mercury contaminated soil. Chemosphere 170, 266–273. 10.1016/j.chemosphere.2016.12.020 [DOI] [PubMed] [Google Scholar]
- Wei D., Sun W., Qian W., Ye Y., Ma X. (2009). The synthesis of chitosan-based silver nanoparticles and their antibacterial activity. Carbohydr. Res. 344, 2375–2382. 10.1016/j.carres.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Weiss J., Takhistov P., McClements D. J. (2006). Functional materials in food nanotechnology. J. Food Sci. 71, 107–116. 10.1111/j.1750-3841.2006.00195.x [DOI] [Google Scholar]
- Whitehead N. A., Barnard A. M. L., Slater H., Simpson N. J. L., Salmond G. P. C. (2001). Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25, 365–404. 10.1111/j.1574-6976.2001.tb00583.x, [DOI] [PubMed] [Google Scholar]
- Wypij M., Czarnecka J., Świecimska M., Dahm H., Rai M., Golinska P. (2018). Synthesis, characterization and evaluation of antimicrobial and cytotoxic activities of biogenic silver nanoparticles synthesized from Streptomyces xinghaiensis OF1 strain. World J. Microbiol. Biotechnol. 34:23. 10.1007/s11274-017-2406-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Qiao L., Guo Y., Ma L., Cheng Y. (2018). Preparation, characteristics and antioxidant activity of polysaccharides and proteins-capped selenium nanoparticles synthesized by Lactobacillus casei ATCC 393. Carbohydr. Polym. 195, 576–585. 10.1016/j.carbpol.2018.04.110 [DOI] [PubMed] [Google Scholar]
- Yamamoto O. (2001). Influence of particle size on the antibacterial activity of zinc oxide. Int. J. Inorg. Mater. 3, 643–646. 10.1016/S1466-6049(01)00197-0 [DOI] [Google Scholar]
- Yang Z., Li Z., Lu X., He F., Zhu X., Ma Y., et al. (2017). Controllable biosynthesis and properties of gold nanoplates using yeast extract. Nano-Micro Lett. 9:5. 10.1007/s40820-016-0102-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood J. M., Bartels D. J., Volper E. M., Greenberg E. P. (2004). Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850. 10.1128/JB.186.6.1838-1850.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates E. A., Philipp B., Buckley C., Atkinson S., Chhabra S. R., Sockett R. E., et al. (2002). N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature- acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70, 5635–5646. 10.1128/IAI.70.10.5635-5646.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Liu J., Yin Y., Shen M. (2018). Interactions between engineered nanoparticles and dissolved organic matter: a review on mechanisms and environmental effects. J. Environ. Sci. 63, 198–217. 10.1016/j.jes.2017.06.021 [DOI] [PubMed] [Google Scholar]
- Yue L., Wang J., Zhang Y., Qi S., Xin B. (2016). Controllable biosynthesis of high-purity lead-sulfide (PbS) nanocrystals by regulating the concentration of polyethylene glycol in microbial system. Bioprocess Biosyst. Eng. 39, 1839–1846. 10.1007/s00449-016-1658-x [DOI] [PubMed] [Google Scholar]
- Zhang R., Edgar K. J. (2014). Properties, chemistry applications of the bioactive polysaccharide curdlan. Biomacromolecules 15, 1079–1096. 10.1021/bm500038g [DOI] [PubMed] [Google Scholar]
- Zhao Y., Tian Y., Cui Y., Liu W., Ma W., Jiang X. (2010). Small molecule-capped gold nanoparticles as potent antibacterial agents that target gram-negative bacteria. J. Am. Chem. Soc. 132, 12349–12356. 10.1021/ja1028843 [DOI] [PubMed] [Google Scholar]
- Zhao X., Zhao F., Wang J., Zhong N. (2017). Biofilm formation and control strategies of foodborne pathogens: food safety perspectives. RSC Adv. 7, 36670–36683. 10.1039/C7RA02497E [DOI] [Google Scholar]
- Zhong J., Zhao X. (2018). Isothermal amplification technologies for the detection of foodborne pathogens. Food Anal. Methods 11, 1543–1560. 10.1007/s12161-018-1177-2 [DOI] [Google Scholar]
- Zonaro E., Piacenza E., Presentato A., Monti F., Dell’Anna R., Lampis S., et al. (2017). Ochrobactrum sp. MPV1 from a dump of roasted pyrites can be exploited as bacterial catalyst for the biogenesis of selenium and tellurium nanoparticles. Microb. Cell Factories 16:215. 10.1186/s12934-017-0826-2 [DOI] [PMC free article] [PubMed] [Google Scholar]