Abstract
Attention deficit hyperactivity disorder (ADHD) is a common and highly heritable neurodevelopmental disorder with poorly understood pathophysiology and genetic mechanisms. A balanced chromosomal translocation interrupts CTNND2 in several members of a family with profound attentional deficit and myopia, and disruption of the gene was found in a separate unrelated individual with ADHD and myopia. CTNND2 encodes a brain-specific member of the adherens junction complex essential for postsynaptic and dendritic development, a site of potential pathophysiology in attentional disorders. Therefore, we propose that the severe and highly penetrant nature of the ADHD phenotype in affected individuals identifies CTNND2 as a potential gateway to ADHD pathophysiology similar to the DISC1 translocation in psychosis or AUTS2 in autism.
Keywords: attention deficit hyperactivity disorder (ADHD), translocation mapping, whole genome sequencing, Nanopore sequencing, familial ADHD, refractive error of the eye, myopia, neurodevelopment, gene identification, protein-truncating variants (PTVs)
Introduction
Attention deficit hyperactivity disorder (ADHD [MIM: 143465]) is a common and highly heritable disorder. Family, twin, and adoption studies confirm a strong genetic influence on ADHD vulnerability, and quantitative genetic studies indicate that ADHD genetic risk is continuously distributed throughout the population.1 Despite heritability estimates of 60%–90% for categorically defined ADHD,2 the genetic factors involved have been elusive. Most genetic and environmental risk factors are likely to have only a small effect on causal pathways, although rare genetic variants3,4 might have a major influence in some individuals. Furthermore, family environment adversity factors (high degree of psychosocial stress, maternal mental disorder, paternal criminality, low socioeconomic status, and foster care) are associated with increased rates of ADHD.5,6 The heterogeneity of presentation and socioenvironmental phenocopies has made it difficult to isolate the specific genetic factors involved.
Genetic variation in catenin delta 2 (CTNND2 [MIM: 604275]) has been associated with human neurodevelopmental phenotypes, including autism7 and intellectual disability within the del(5) (p15) cri-du-chat syndrome [MIM: 123450].8 CTNND2 encodes delta-catenin, an adhesive junction-associated protein of the armadillo/beta-catenin superfamily that is expressed within proliferating neuronal progenitor cells of the neuroepithelium and in the dendritic compartment of postmitotic neurons.9 In humans, CTNND2 is located at 5p15.2 and encodes four alternatively spliced isoforms, three of which use a downstream start codon compared to the full-length 21-exon variant. All are predominantly confined to the brain, with variation in their regional expression patterns. The 6.2 kb mRNA isoform has the most abundant whole brain expression overall, while the full length 7.0 kb isoform is detected primarily in cortical regions and the 4.2 kb isoform is present in subcortical regions and only just detectable in the cerebral cortex.9 Data from the Genome Aggregation Database (gnomAD) indicate that CTNND2 has a high probability of loss-of-function (LOF) intolerance with a LOEUF (loss-of-function observed/expected upper bound fraction) of 0.098, indicating that only 9.8% of the expected LOF variants were observed and, therefore, CTNND2 is likely under selection against LOF variants.10
Involvement of CTNND2 in the phenotypes of ADHD and myopia is suggested by genetic studies of inherited structural variation in ADHD probands, leading to identification of an intronic deletion in CTNND2 within a derived gene set of ADHD-enriched copy number variants (CNVs)4 and several genome-wide studies implicating polymorphisms within CTNND2 in myopia susceptibility in East Asian populations.11, 12, 13, 14, 15 However, a definitive role for CTNND2 in pathogenesis of these conditions has yet to be established.
Here, we describe four individuals with protein-truncating CTNND2 disruptions, three of whom are members of a multigenerational family with marked attention deficit disorder, and all of whom present with ADHD and myopia. A 5;6 chromosome translocation was present in all familial individuals with ADHD tested and was absent in tested unaffected family members. The chromosome 6 breakpoint directly impacted no coding genes, while the chromosome 5 breakpoint disrupted intron 2 of CTNND2. Identification of an unrelated individual with a CTNND2 disruption and attention deficit provided independent confirmation and strongly indicates that haploinsufficiency of CTNND2 is a high-penetrance cause of a human ADHD phenotype.
Subjects and methods
Recruitment, consent, and sample collection
Participants were recruited from one of the following: Boston Children’s Hospital, Harvard Medical School; University Hospitals of Cleveland and Case Western Reserve University; or the University of Nebraska Medical Center. All individual study protocols were approved by local institutional review boards (IRBs; 2009P002139, 08-10-15, and 20200792). Procedures were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national). Informed consent from all participants was obtained for publication of the data. Medical and psychiatric histories were taken as well as detailed phenotyping by clinical geneticists with expertise in dysmorphology. All families were offered genetic counseling.
Whole exome sequencing and variant screening
For the index family (family 1), array-based comparative genomic hybridization analysis of whole-blood extracted DNA was performed in duplicate using two platforms: the Agilent 244K microarray chip and the Illumina Infinium Omni5Exome-4 kit. Whole genome sequencing (WGS) was performed on short-read and long-read platforms using the 10x Genomics Chromium System (for all three affected individuals tested) and the Oxford Nanopore PromethION Sequencing Platform (for only the individual identified as II.2 in Figure 1). Locus validation of the breakpoint and fused sequences on the derivative chromosomes was carried out by Sanger sequencing.
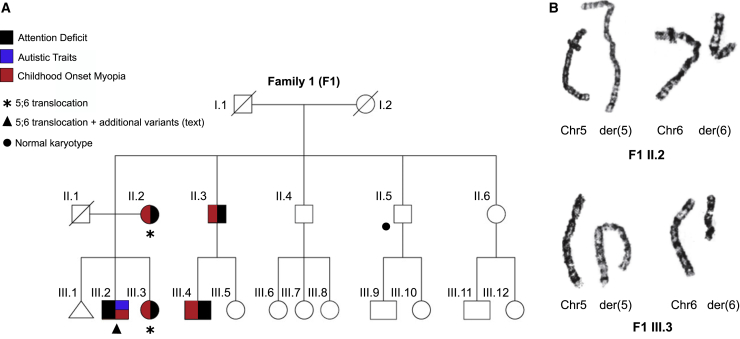
Figure 1.
A family with highly penetrant attention deficit and childhood myopia
(A) Pedigree and clinical manifestations of the individuals from family 1 with ADHD and myopia. Black, attention deficit; blue, autistic traits; red, childhood onset myopia. Karyotypes of tested individuals are indicated. For others, DNA was not available. Asterisk, translocation; filled triangle, translocation plus additional genetic findings, text; filled circle, normal karyotype.
(B) Karyotypes of individual F1 II.2 and F1 III.3 demonstrating a translocation between chromosomes 5 and 6.
For whole genome phasing and structural variant calling, Long Ranger’s Whole Genome Mode was used to analyze FASTQ files generated from a 10x Genomics Chromium-prepared library using Illumina’s bcl2fastq software. Paired sequences obtained from each sample were mapped to human genome reference GRCh37/hg19 using Long Ranger 2.1.6 with default parameters. Variant calling of SNPs and indels was carried out with the general purpose FreeBayes caller incorporated into Long Ranger. The variants were filtered by requiring a depth of coverage of at least 10 and a quality score of 30. Opal 4.0 from Fabric Genomics was used for variant annotation and analysis using the following filters: coverage ≥ 10 reads, quality score ≥ 40, Omicia Score ≥ 0.7, and minor allele frequency (MAF) in Exome Aggregation Consortium (ExAC), Exome Variant Server (EVS) and 1000 Genomes Project < 5%. The Omicia score is a proprietary score that assesses whether a variant is likely to be deleterious. It is a meta-classifier that combines scores from the following variant scoring algorithms: SIFT,16 PolyPhen,17 MutationTaster,18 and PhyloP.19 All flagged variants were automatically annotated with pathogenicity scores from the VAAST Variant Prioritizer.20
For individual I.2 from family 2, whole exome sequencing, copy-number variant (CNV) analysis, and qPCR were performed based on potential involvement of delta-catenin. DNA was barcoded and exonic regions and flanking splice junctions of the genome were captured using a custom-modified captureset based on the IDT xGen Exome Research Panel v1.0. Massively parallel sequencing was done on an Illumina system with 100 bp or greater paired-end reads. Logarithmically transformed coverage ratios were scaled to ensure regions with no deletions had ratios of 1.0 and regions with a heterozygous deletion would be expected to have a ratio of 0.5. Putative deletions or duplications were confirmed by qPCR.21,22
Results
Identification of a family with ADHD and myopia segregating with a balanced translocation
Five of 20 individuals (25%) in a three-generation family of European descent had ADHD clustered in two arms of the family (75% offspring in those families) in a pattern suggestive of Mendelian autosomal dominant inheritance (family 1; Figure 1A). Three affected individuals were assessed: a 9-year-old boy (F1 III.2), a 10-year-old girl (F1 III.3), and their 46-year-old mother (F1 II.2). All were found to be non-dysmorphic. Phenotypes with high penetrance were ADHD and early-onset myopia, which were shared common features in all affected individuals. A typical optical strength was −6.75 diopters (F1 III.3). ADHD diagnoses were made by two independent child psychiatrists at Children’s Hospital Boston using DSM-IV diagnostic criteria23 current at that time and confirmed to meet newer DSM-V criteria for ADHD.
F1 II.2 had a partial college education, while F1 III.2 and F1 III.3 were in grade school and receiving treatment for ADHD. F1 III.3 was unable to tolerate psychostimulants due to side effects and received primarily behavioral interventions. F1 III.2 responded substantially to amphetamine and dextroamphetamine mixed salts and clonidine, which are conventional first- and second-line medications for ADHD.24
Notably, the male subject alone (F1 III.2) had short stature (2nd growth percentile) and autistic traits but did not meet criteria for autism spectrum disorder. This additional phenotype was not observed in any of the other 19 individuals in the pedigree.
We identified a t(5;6)(p15.1;q14.1) chromosome translocation [t(5;6)(6qter→6q14::5p15.1→5qter;6pter→6q14.1::5p15.1→5pter] on karyotype analysis of three assessed affected individuals (Figure 1B). The karyotype in F1 II.5 (unaffected) was negative and therefore imputed negative in F1 III.9 and F1 III.10 (unaffected offspring). A 244K Array comparative genomic hybridization (CGH) indicated that the translocation was balanced with respect to coding genes, while the denser Illumina SNP array confirmed these findings in F1 II.2 (Figure 2A). Individual F1 III.2, who displayed additional autism-like traits and short stature, had a novel 1.1 Mb microduplication on chromosome 6 (Figure 2B) that was not found in the other familial translocation carriers encompassing ethylmalonyl-CoA decarboxylase 1 (ECHDC1 [MIM: 612136]), ring finger protein 146 (RNF146 [MIM: 612137]), R-spondin 3 (RSPO3 [MIM: 610574]), and centromeric protein W, CENPW [MIM: 611264]). This structural variant was not present in the DECIPHER database25 or Database of Genomic Variants.26 F1 III.2 additionally harbored truncating variants in developmental genes identified via WGS. These were lysine methyltransferase 2B (KMT2B [MIM: 606834]), NC_000019.10 :g.35718139G>T (NM_014727.2:c.121G>T; NP_055542.1:p.Glu41Ter) and short stature homeobox (SHOX [MIM: 312865]), NC_000023.11:g.630946A>T (NM_000451.3:c.49A>T; NP_000442.1(LRG_710p1):p. Lys17Ter). These variants were not present in the gnomAD, ClinVar or Human Gene Mutation Database (HGMD) databases and were absent in other family members with ADHD. These additional genetic variants unique to F1 III.2 are possible contributors to F1 III.2′s ancillary phenotypic features, consistent with a multiple-hit model.27
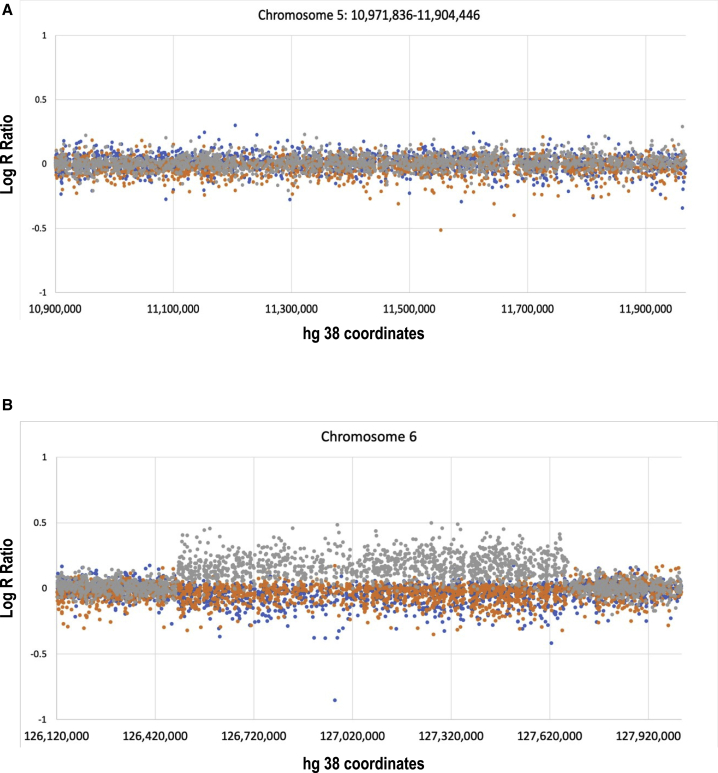
Figure 2.
Analysis of copy number variation in the chromosome 5;6 translocation
(A) Illumina Chip data demonstrating balanced sequence of chromosome 5 in the region that contains CTNND2 (chr5:109,000,000–119,000,000) in three F1 translocation carriers (II.2 in brown, III.2 in gray, and III.3 in blue).
(B) 1.2 Mb duplication (chr6:126121880–127921880) in the more severely-affected III.2 (gray), which was not present in other translocation carriers (II.2 in brown and III.3 in blue). This region encompasses ECHDC1, RNF146, RSPO3, and CENPW genes.
Identification of CTNND2 disruption at the translocation breakpoint
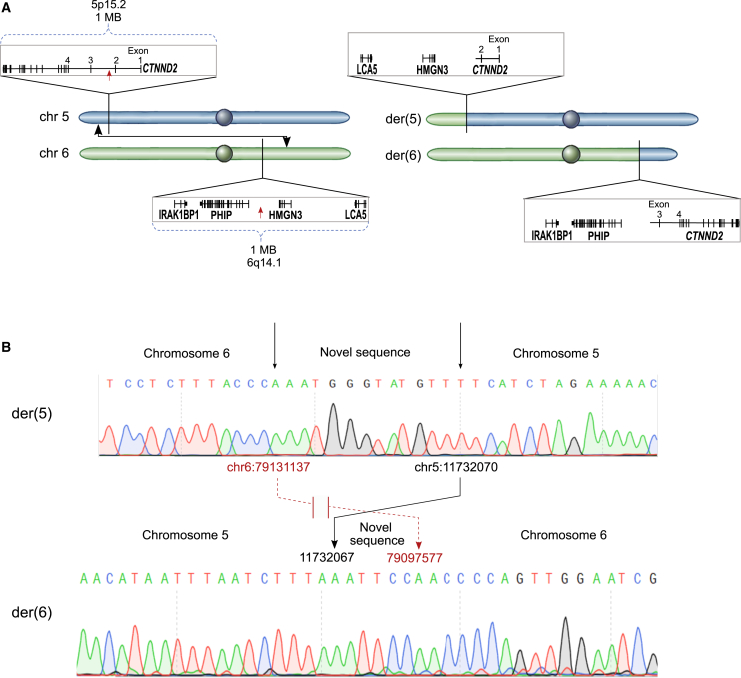
Both short- and long-read WGS localized the chromosome 5 breakpoint to intron 2 of CTNND2 (GenBank: NM_001332.4) on 5p15.2, while the chromosome 6 breakpoint did not disrupt any coding genes, being approximately 20 kb removed from the nearest gene on chromosome 6 (Figure 3A). Direct sequencing of the derivative chromosome breakpoint regions confirmed the novel sequences and indicated that neither of the derivative chromosomes harbored novel promoter-gene or gene-gene fusions (Figure 3B). A 12 bp insertion bridged chromosomes 5 and 6 on the derivative chromosome 5, as did a 5 bp insertion on the derivative chromosome 6. The final rearrangement was determined to be Seq[GRCh38] t(5;6)(p15.1;q14.1) g.[chr5:qter_11732070::TTGTATGGGTAA::chr6:79131137_qter] g.[chr6:pter_79,097,577::CCTTAA::chr5:11,732,067_pter].
Figure 3.
Chromosome 5;6 translocation
(A) Graphic representation of the results obtained from the quantitative analysis of derivative chromosomes by short-read and long-read sequencing, which demonstrated chromosome breakpoints on Chr5 and Chr6 and flanking genes (left). Final positions after rearrangements are shown on the right.
(B) Breakpoint sequences were confirmed by Sanger sequencing of PCR products of the regions flanking the breakpoints in blood cells from F1 II.2. Connecting arrows from Der 5 to Der 6 indicate hg38 coordinates at breakpoint and final reattachment locations on both chromosomes, with novel sequences inserted at junctions. There is a 3 bp loss in CTNND2 intron 2 between derivative chromosome 5 and derivative chromosome 6. A loss of approximately 30 kb on chromosome 6 is in a non-genic region.
No enhancers were bioinformatically identified within 20 kb of either breakpoint, which was determined using the Genehancer track on the University of California, Santa Cruz (UCSC) genome browser. Neither breakpoint boundary disrupted a topologically associating domain (TAD) boundary in ENCODE data from IMR90 lung cells and human embryonic stem cells (hESCs),28 further confirmed using the UCSC genome browser Hi-C and Micro-C functions.
The translocation separated exon 1 and exon 2 of CTNND2 from the other 19 exons, consistent with isolated haploinsufficiency of CTNND2 (Figure 3A).
Identification of an individual with ADHD and myopia with an exon 2 deletion of CTNND2
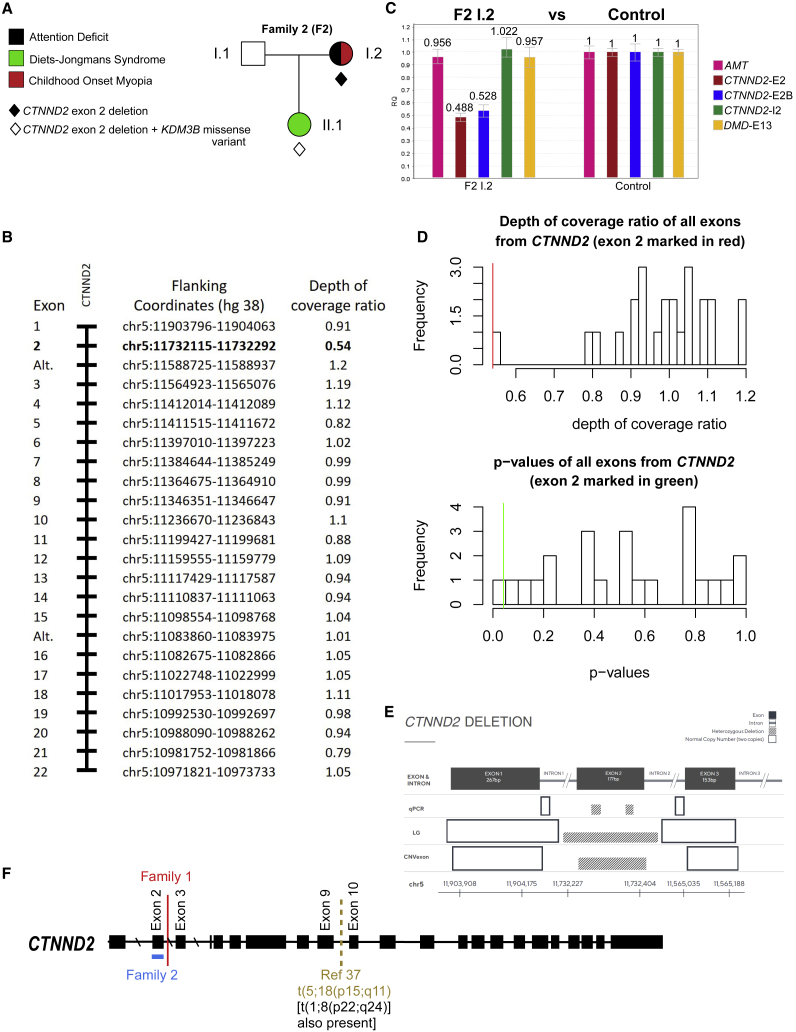
We searched for individuals with CTNND2 variants of unknown significance (VUSs) in a larger cohort of unrelated probands with developmental conditions, excluding missense mutations for clarity, focusing on early LOF variants. Subjects were ascertained from the case group that had undergone genetic testing for autism or developmental delay at genetic testing companies that specifically test for CTNND2 variants (Fulgent Genetics, Prevention Genetics, and Blueprint Genetics). In 24 instances, a positive result for a variant had been reported, one of which was a truncating VUS. This variant, a deletion of exon 2 of CTNND2 (Figures 4A–4D), was present in family 2 (Figure 4A). The family was comprised of a parent-child trio wherein the female child (II.1, referred to as F2 II.1) had developmental delay and the mother (I.2, referred to as F2 I.2) carried a diagnosis of ADHD. All members of the trio underwent exome sequencing and qPCR, while, in addition, F2 II.1 had low-coverage sequencing (8–10 × ). For qPCR on F2 I.2, a resampling approach was used to estimate the probability of obtaining a log transformed coverage ratio value as low as CTNND2 exon 2 (0.54) from the set of 24 CTNND2 exons’ values. For each CTNND2 exon, we sampled one exon at a time (with replacement) from the log transformed coverage ratio values of all 24 exons 1 million times at random and calculated the probability (p value) of obtaining exons in which a given coverage ratio value was lower than or equal to the coverage ratio value of the exon being tested (Figure 4D). All three modalities confirmed an exon 2 deletion of CTNND2 in F2 II.1, while exome and qPCR confirmed the exon 2 deletion in F2 I.2 (Figure 4E).
Figure 4.
Genetic analysis of family 2 and summary of genetic findings
(A–E) The presence of a CTNND2 exon 2 deletion was established by low-coverage whole genome sequencing, exome sequencing, and qPCR as described in Subjects and methods.
(A) Pedigree and phenotypes of family 2.
(B) F2 I.2: relative read count for all exome-sequenced CTNND2 exons. Alt. refers to alternate exons not found in the longest transcript.
(C) F2 I.2: qPCR of exon 2 of CTNND2 and estimated gene dosage ratios. For F2 I.2, the autosomal AMT and X-linked DMD control targets were within the normal expected range, while two different targets within CTNND2 exon 2 were indicative of a heterozygous deletion (CTNND2-E2 and CTNND2-E2B). Pink, biallelic exon 6 of AMT gene; brown and blue, two probes for CTNND2 exon 2; green, CTNND2 intron 2; blue, DMD gene exon 13. Error bars are ± 1 SD from 3 replicates.
(D) Analysis of depth of coverage ratios for F2 I.2 CTNND2 exons. Top panel: frequency distribution histogram of log-transformed coverage ratio values of all exons from CTNND2. Bars represent frequency of observations within bins of depth of coverage values on the horizontal axis. The red vertical line marks the position of the depth of coverage ratio value of CTNND2 exon 2 within that specific bin. Bottom panel: frequency distribution of probability values (p values) from all CTNND2 exons from the resampling analysis. The horizontal axis represents bins of p values of the CTNND2 exons getting a depth of coverage ratio less than or equal to the depth of coverage ratio of a particular exon from CTNND2. The green vertical line marks the p value of CTNND2 exon 2 within its specific bin. Exon 2 had a coverage ratio value of 0.54, consistent with a hemizygous deletion (p = 0.0415) and 3 standard deviations (SDs) below the expected value for this exon, while all other exons are within ± 1 SD.
(E) Summary schematic of qPCR and next-generation sequencing (NGS) approaches showing loss of CTNND2 exon 2. Results from three methods are shown: quantitative PCR (qPCR), low-pass whole genome sequencing (LG), and exon-by-exon copy number variant calling (CNVexon). Concordance across all methods for a heterozygous deletion is demonstrated. The critical region confirmed by all three methods includes exon 2 of CTNND2.
(F) Summary of CTNND2 genetic findings in individuals with ADHD, supporting a haploinsufficiency mechanism. Red, translocation breakpoint in family 1; blue line, deletion in family 2; dashed brown line, translocation breakpoint from Hofmeister et al.37
F2 I.2, a 31-year-old individual, completed an adult ADHD self-report scale and was evaluated by a board-certified adult psychiatrist, with both results consistent with ADHD. The subject carries an independent community-based medical diagnosis of ADHD and undergoes treatment with atomoxetine.29 She was in special education classes as a child for reading and comprehension difficulties, the latter of which has since resolved. She is high-functioning and currently completing an advanced life science degree (Master’s degree in psychology after previously obtaining a Bachelor’s degree). This subject has myopia and has worn glasses since the age of three. The inheritance of this participant’s deletion mutation could not be established.
F2 II.1 is a 23-month-old female who carries the same heterozygous maternally inherited exon 2 deletion in CTNND2. She also harbored a de novo heterozygous NC_000005.10:g.138393234T>C (NM_016604.3: c.2693T>C; NP_057688.2 p.Phe898Ser) change in lysine demethylase 3B (KDM3B [MIM: 609373]), predicted to be benign by PolyPhen-2,17 deleterious by SIFT16 and Provean,30 and disease causing by MutationTaster.18 This variant was not found in the gnomAD,10 ClinVar, or HGMD databases. F2 II.1 displays developmental delay, autism, moderate hearing loss, short stature, and dysmorphic features consistent with Diets-Jongmans syndrome [MIM: 618846], which is caused by heterozygous KDM3B mutations31 and is consistent with the heterozygous c.2693T>C, p.Phe898Ser change in KDM3B being the causative mutation for her dysmorphia and intellectual disability. The severity of this latter condition, however, masks milder neurodevelopmental phenotypes, such as ADHD. F2 II.1 was too young to undergo detailed vision testing and was removed from further analyses (Table 1).
Table 1.
Phenotypic characteristics of subjects in this study
| F1 II.2 | F1 III.2 | F1 III.3 | F2 I.2 | F2 II.1 | |
|---|---|---|---|---|---|
| ADHD | + | + | + | + | NAa |
| Myopia/refractive error | + | + | + | + | NAb |
| Autism or autistic features | – | +c | – | – | + |
| Growth deficits (small stature) | – | +d | – | – | +d |
| Dysmorphic features | – | – | – | – | + |
| Moderate hearing loss | – | – | – | – | + |
| CTNND2 disruption | translocation | translocation | translocation | exon 2 deletion | exon 2 deletion |
| Other genetic findings | – | 6q22.32 microduplicationeKMT2B c.121G>T (p.Glu41Ter) variante,f SHOX c.49A>T (p.Lys17Ter) variante,f | – | – |
KDM3B c.2693T>C (p.Phe898Ser) variantg,h |
| Additional OMIM diagnoses | – | short stature, idiopathic, familial (MIM: 300582) | – | – | Diets-Jongmans syndrome (MIM: 618846) |
Abbreviations are as follows: ADHD, attention deficit hyperactivity disorder; OMIM, Online Mendelian Inheritance in Man.
F2 II.1 is still too young to determine her development.
F2 II.1 is still too young to accurately determine her vision.
F1 II.2 had an original diagnosis of DSM-IV pervasive developmental disorder, not otherwise specified.
F1 III.2′s height is approximately 2nd percentile. F2 II.1’s height is approximately 6th percentile and weight is 2nd percentile.
Not found in other familial translocation carriers.
Deleterious by SIFT and MutationTaster.
De novo.
Deleterious by SIFT, Provean, and MutationTaster.
Discussion
Here, we describe unrelated individuals from two families with remarkably consistent penetrant ADHD, early onset myopia, and disruptions in CTNND2. Although ADHD is one of the most heritable childhood neuropsychiatric disorders, a range of genome-wide association study (GWAS)-implicated genes involved in various cell and neuronal functions (cell division, cell adhesion, neuronal migration, and neuronal plasticity) have mostly failed to pass genome-wide levels of significance, with a handful of recent exceptions.32 ADHD displayed by individuals with CTNND2 lesions ranged in severity from a relatively mild presentation in F2 I.2 to a severe presentation in the familial translocation carriers. The specificity of the genetic defects—the balanced translocation in the index family and exon 2 deletion in F2 I.2—strongly supports that CTNND2 disruptions are responsible for the ADHD and myopia phenotypes displayed by these individuals.
Delta-catenin is an armadillo family member with a coiled-coil domain, an armadillo domain important in binding cadherin and beta-catenin,33 a polyproline tract able to bind src receptor kinases,34 and a PDZ domain at the C terminus that is able to bind erbin.35 Both the translocation and exon deletion we describe disrupt the coiled-coiled domain in the N-terminal section of the protein, leaving other domains intact. As the coiled-coiled domain is unique to the single longest isoform, but not to three shorter isoforms, and there are intragenic promoter elements that are unaffected by the translocation or exonic deletion,36 we wished to determine if truncating lesions elsewhere in the gene are associated with ADHD. We found that an intronic deletion within CTNND2 intron 4 has previously been reported within an ADHD cohort,4 but it would not be expected to affect coding exons. Furthermore, we identified published individuals in an unrelated family with a rearrangement affecting CTNND2 consisting of two separate balanced translocations.37 In this family with reading disabilities, CTNND2 was disrupted by a translocation breakpoint in intron 9 as well as a second uncharacterized gene at another breakpoint, zinc finger and SCAN domain containing 30 (ZSCAN30), with both genes fused together in opposite orientations. In the index family of the current study and in F2 I.2, the genetic lesions are more specific relative to the genetic complexity of the Hofmeister et al.37 family, where it might be challenging to assign the relative contributions of each of multiple genetic rearrangements to the observed phenotypes. However, affected individuals in the Hofmeister et al.37 study were reported to have attention deficit consistent with our interpretation that CTNND2 disruption is consistent with highly penetrant ADHD. Finally, subjective attentional deficits were reported in a large family with a p.Glu1044Lys CTNND2 substitution, which altered dendrite morphology in vitro;38 however, no formal assessment, testing, or follow-up for ADHD was done.
All confirmed translocation carriers in this study and the Hofmeister et al.37 study developed severe early onset myopia from early toddler and school years, as did F2 I.2 in this study. Individuals in the index family with ADHD (individuals II.3 and III.4) who did not submit to genetic analysis also had childhood onset myopia, which was not present in non-ADHD family members. Thus, in the index family, ADHD and severe childhood myopia segregated together (Figure 1). Notably, Ctnnd2 is implicated in eye development in mice39 and is consistent with an independent association of CTNND2 SNPs with myopia in selected human populations.11, 12, 13, 14 Like ADHD, myopia is a common complex disorder, and CTNND2 lies 5 Mb telomeric to the autosomal dominant high myopia locus MYP19 (MIM: 613969).15 The autosomal dominant ADHD4 locus (MIM: 608906) is centromeric to CTNND2 on 5p13. Figure 4F summarizes the CTNND2 disruptions from this study and Hofmeister et al.37
A substantial amount of in vivo and in vitro data support an important role for delta-catenin in behavior and neural development, including proliferation of neural progenitors, neurogenesis, dendritic arborization, and synaptic function. Unlike other members of the p120ctn protein family, delta-catenin is unique in that it has restricted expression to central neurons40, 41, 42, 43, 44 and is widely expressed in different regions of the brain,44,45 both during early development and in the mature brain.46,47 Delta-catenin is first expressed within proliferating neuronal progenitor cells of the developing neuroepithelium, becomes downregulated during neuronal migration, and is later re-expressed in the dendritic compartment of postmitotic neurons.9 In the mouse, delta-catenin expression increases through fetal life and peaks at postnatal day 7, with lower levels thereafter, and shifts from embryonic expression at the cell periphery to nascent apical dendrites in postnatal life and adulthood.9,46,48 Delta-catenin is likely to have neuron type-specific expression patterns and functional roles.46 Analysis of the primary visual cortical region in adult mice indicates that the expression of members of the p120ctn family is varied within neuronal subtypes,49,50 with delta-catenin enriched in inhibitory somatosensory neurons, growth cones,34 dendritic segments, and postsynaptic terminals in central neurons.51,52
A mouse model with a truncated form of delta-catenin that includes the first 461 amino acids of delta-catenin fused to GFP at its C terminus (delta-catenin N-term mice) has been described.53 Mutant mice have severe deficits in the hidden version of the water maze (H-WM), a sensitive measure of spatial learning and hippocampal function,54 failing to acquire the H-WM task even after 14 days of training. Normal performance on the visible water maze task suggests that the deficits observed are probably spatial in nature. Mutant animals have normal levels of exploratory activity and anxiety. In implicit motor-skill learning on an accelerating rotarod, mutant mice displayed an overall deficit in motor coordination. Psychostimulants improved rotation behavior, motor coordination, and motor skill learning.55 On a Pavlovian fear-conditioning task, mutant animals froze significantly less than wild-type animals 24 h later, while auditory conditioning assessed 48 h later was also impaired in delta-catenin mutants, confirming that δ-cat−/− animals have learning deficits that extend beyond spatial-learning impairments.
Delta-catenin is a key regulator of dendrite arborization in pyramidal neurons. Loss of delta-catenin in hippocampal neurons by short hairpin RNA (shRNA)-mediated knockdown during the stages coinciding with dendrite development reduces dendritic arbors,51 similar to what is observed in neurons from delta-catenin N-term mice.51 In vivo two photon imaging studies in cortical neurons of wild-type and delta-catenin N-term mice indicate no alterations in the dendritic arbor at 5 weeks of age56 but, thereafter, delta-catenin N-term mice have progressive retraction of the dendritic arbor, leading to functional deficits in cortical responsiveness.56 Overexpression of delta-catenin enhances the branching number of dendrites in primary hippocampal neurons.57,58
Taken together, these data suggest that delta-catenin likely regulates multiple aspects of dendrite morphogenesis, including developmental dendrite arborization and dendrite stability in the adult. This is enabled by its interaction with erbin, which binds to delta-catenin at the PDZ domain and promotes its appropriate subcellular localization.35 The C-terminal region of delta-catenin is the site of interaction with cortactin, actin and 14-3-3ε/ζ34,48 and is a critical determinant of its ability to regulate dendrite morphogenesis.59 Delta-catenin also interacts with cyclin-dependent kinase 5 (CDK5) and with regulators of the Rho family of small GTPases,34,57,60, 61, 62 well-known regulators of the actin cytoskeleton that have profound influence on dendritic morphogenesis.63, 64, 65 The increase in density of synapses observed with overexpression of delta-catenin in mature primary hippocampal neurons is inhibited by the use of a dominant negative Rac construct or a dominant negative Cdc42,60 indicating a G protein-mediated role in delta-catenin’s effects on regulating the actin cytoskeleton. Abolishing delta-catenin phosphorylation at Thr454 by Akt57 inhibits delta-catenin’s effect on dendrite-like branching in NIH 3T3 cells,57 while loss of CDK5-mediated phosphorylation of delta-catenin at residues Ser300 and Ser357 enhances the density of dendritic protrusions and enhances the association between delta-catenin and N-cadherin, GRIP, and PSD95, resulting in an increase in the surface AMPA receptor (AMPAR) and ratio of AMPA/N-methyl-d-aspartate (NMDA) compared to the wild-type delta-catenin.66,67
Thus, delta-catenin plays a critical role in dendritic architecture via several effectors in convergent downstream pathways that assemble actin filaments. Other actin modeling proteins within this pathway have been associated with ADHD-like behaviors in relevant mouse models.68, 69, 70, 71, 72, 73 Our finding that CTNND2 is a high penetrance ADHD gene in human subjects provides additional evidence that dendritic aberrations are a potential pathophysiologic cause of ADHD.
Dendritic lesions are linked to a wide range of neurodevelopmental phenotypes.74, 75, 76 Functional CTNND2 variants are likely to act cumulatively with other risk genetic variants when present to produce a more complex or extended phenotype, consistent with multiple-hit developmental models27 that suggest that phenotypes with two deleterious mutations are distinct and/or more severe than cases carrying only the co-occurring mutation. Thus, de novo CTNND2 variants have been associated with distinct or more severe phenotypes7,8 albeit a putative Mendelian phenotype attributed to a CTNND2 missense variant38 was recently revised to assign ultimate causation to a different gene.77
In summary, the complexity and heterogeneity of the ADHD phenotype has hindered the search for underlying genetic causes. Consistent with evidence characterizing ADHD subjects as carrying a higher burden of inherited rare CNVs in neurodevelopmental genes or a higher burden of rare protein-truncating single nucleotide variants,4 we describe a rare family carrying a penetrant translocation that, by interrupting CTNND2, causes an ADHD/myopia phenotype that is also found in unrelated individuals with disruption of the gene. The severe and highly penetrant nature of the ADHD phenotype in the subjects in this study identifies CTNND2 as a potential gateway to ADHD pathophysiology similar to the DISC1 translocation in psychosis.78
Data and code availability
The datasets and variants generated during this study are available at GenBank (Whole genome sequencing; SRA accession PRJNA64834) and ClinVar (Gene variants; Accessions SUB7836623, SUB7824655).
Acknowledgments
We thank Dr. Neil Molyneaux and Dr. Alexander Miron (Case Western Reserve University) for bioinformatics assistance. We thank Mirandy Teguh (Fulgent Genetics) for custom qPCR work and Jessa Humanski (Fulgent Genetics) for coordination. This publication was made possible by the Clinical and Translational Science Collaborative of Cleveland, 4UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research (A.A) and the US National Institutes of Health (R01MH113106; A.W.-B.).
Declaration of interests
S.P.S. and E.N. are employees and shareholders of Fulgent Genetics. J.P. is a founder of Global Gene Corp and a member of its scientific advisory board.
Web resources
OMIM, https://www.omim.org
The International Genome Sample Resource, http://www.internationalgenome.org/
NHLBI Exome Sequencing Project Exome Variant Server, https://evs.gs.washington.edu/EVS/
The Human Gene Mutation Database, http://www.hgmd.cf.ac.uk/ac/index.php
NCBI ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
ENCODE: Encyclopedia of DNA Elements, https://www.encodeproject.org
University of California Santa Cruz Genomics Institute Genome Browser, https://genome.ucsc.edu/
NCBI dbSNP, https://www.ncbi.nlm.nih.gov/snp/
Fabric Genomics, https://fabricgenomics.com/
References
- 1.Faraone S.V., Perlis R.H., Doyle A.E., Smoller J.W., Goralnick J.J., Holmgren M.A., Sklar P. Molecular genetics of attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Larsson H., Chang Z., D’Onofrio B.M., Lichtenstein P. The heritability of clinically diagnosed attention deficit hyperactivity disorder across the lifespan. Psychol. Med. 2014;44:2223–2229. doi: 10.1017/S0033291713002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochukova E.G., Huang N., Keogh J., Henning E., Purmann C., Blaszczyk K., Saeed S., Hamilton-Shield J., Clayton-Smith J., O’Rahilly S., et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elia J., Gai X., Xie H.M., Perin J.C., Geiger E., Glessner J.T., D’arcy M., deBerardinis R., Frackelton E., Kim C., et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevens S.E., Sonuga-Barke E.J., Kreppner J.M., Beckett C., Castle J., Colvert E., Groothues C., Hawkins A., Rutter M. Inattention/overactivity following early severe institutional deprivation: presentation and associations in early adolescence. J. Abnorm. Child Psychol. 2008;36:385–398. doi: 10.1007/s10802-007-9185-5. [DOI] [PubMed] [Google Scholar]
- 6.Biederman J., Milberger S., Faraone S.V., Kiely K., Guite J., Mick E., Ablon S., Warburton R., Reed E. Family-environment risk factors for attention-deficit hyperactivity disorder. A test of Rutter’s indicators of adversity. Arch. Gen. Psychiatry. 1995;52:464–470. doi: 10.1001/archpsyc.1995.03950180050007. [DOI] [PubMed] [Google Scholar]
- 7.Turner T.N., Sharma K., Oh E.C., Liu Y.P., Collins R.L., Sosa M.X., Auer D.R., Brand H., Sanders S.J., Moreno-De-Luca D., et al. Loss of δ-catenin function in severe autism. Nature. 2015;520:51–56. doi: 10.1038/nature14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medina M., Marinescu R.C., Overhauser J., Kosik K.S. Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics. 2000;63:157–164. doi: 10.1006/geno.1999.6090. [DOI] [PubMed] [Google Scholar]
- 9.Ho C., Zhou J., Medina M., Goto T., Jacobson M., Bhide P.G., Kosik K.S. delta-catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J. Comp. Neurol. 2000;420:261–276. [PubMed] [Google Scholar]
- 10.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y.J., Goh L., Khor C.C., Fan Q., Yu M., Han S., Sim X., Ong R.T., Wong T.Y., Vithana E.N., et al. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118:368–375. doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu B., Jiang D., Wang P., Gao Y., Sun W., Xiao X., Li S., Jia X., Guo X., Zhang Q. Replication study supports CTNND2 as a susceptibility gene for high myopia. Invest. Ophthalmol. Vis. Sci. 2011;52:8258–8261. doi: 10.1167/iovs.11-7914. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., Su S., Yang M., Hu N., Yao Y., Zhu R., Zhou J., Liang C., Guan H. Association of ZNF644, GRM6, and CTNND2 genes with high myopia in the Han Chinese population: Jiangsu Eye Study. Eye (Lond.) 2016;30:1017–1022. doi: 10.1038/eye.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Z., Zhou J., Chen X., Zhou X., Sun X., Chu R. Polymorphisms in the CTNND2 gene and 11q24.1 genomic region are associated with pathological myopia in a Chinese population. Ophthalmologica. 2012;228:123–129. doi: 10.1159/000338188. [DOI] [PubMed] [Google Scholar]
- 15.Li J., Zhang Q. Insight into the molecular genetics of myopia. Mol. Vis. 2017;23:1048–1080. [PMC free article] [PubMed] [Google Scholar]
- 16.Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 17.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7 doi: 10.1002/0471142905.hg0720s76. Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 19.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flygare S., Hernandez E.J., Phan L., Moore B., Li M., Fejes A., Hu H., Eilbeck K., Huff C., Jorde L., et al. The VAAST Variant Prioritizer (VVP): ultrafast, easy to use whole genome variant prioritization tool. BMC Bioinformatics. 2018;19:57. doi: 10.1186/s12859-018-2056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schenkel L.C., Kerkhof J., Stuart A., Reilly J., Eng B., Woodside C., Levstik A., Howlett C.J., Rupar A.C., Knoll J.H.M., et al. Clinical Next-Generation Sequencing Pipeline Outperforms a Combined Approach Using Sanger Sequencing and Multiplex Ligation-Dependent Probe Amplification in Targeted Gene Panel Analysis. J. Mol. Diagn. 2016;18:657–667. doi: 10.1016/j.jmoldx.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Kerkhof J., Schenkel L.C., Reilly J., McRobbie S., Aref-Eshghi E., Stuart A., Rupar C.A., Adams P., Hegele R.A., Lin H., et al. Clinical Validation of Copy Number Variant Detection from Targeted Next-Generation Sequencing Panels. J. Mol. Diagn. 2017;19:905–920. doi: 10.1016/j.jmoldx.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 23.American Psychiatric Association . American Psychiatric Association; 2000. Diagnostic and statistical manual of mental disorders: DSM-IV-TR. [Google Scholar]
- 24.Caye A., Swanson J.M., Coghill D., Rohde L.A. Treatment strategies for ADHD: an evidence-based guide to select optimal treatment. Mol. Psychiatry. 2019;24:390–408. doi: 10.1038/s41380-018-0116-3. [DOI] [PubMed] [Google Scholar]
- 25.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girirajan S., Rosenfeld J.A., Cooper G.M., Antonacci F., Siswara P., Itsara A., Vives L., Walsh T., McCarthy S.E., Baker C., et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K., et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spencer T., Biederman J., Wilens T., Prince J., Hatch M., Jones J., Harding M., Faraone S.V., Seidman L. Effectiveness and tolerability of tomoxetine in adults with attention deficit hyperactivity disorder. Am. J. Psychiatry. 1998;155:693–695. doi: 10.1176/ajp.155.5.693. [DOI] [PubMed] [Google Scholar]
- 30.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diets I.J., van der Donk R., Baltrunaite K., Waanders E., Reijnders M.R.F., Dingemans A.J.M., Pfundt R., Vulto-van Silfhout A.T., Wiel L., Gilissen C., et al. De Novo and Inherited Pathogenic Variants in KDM3B Cause Intellectual Disability, Short Stature, and Facial Dysmorphism. Am. J. Hum. Genet. 2019;104:758–766. doi: 10.1016/j.ajhg.2019.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demontis D., Walters R.K., Martin J., Mattheisen M., Als T.D., Agerbo E., Baldursson G., Belliveau R., Bybjerg-Grauholm J., Bækvad-Hansen M., et al. ADHD Working Group of the Psychiatric Genomics Consortium (PGC) Early Lifecourse & Genetic Epidemiology (EAGLE) Consortium. 23andMe Research Team Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Q., Paredes M., Medina M., Zhou J., Cavallo R., Peifer M., Orecchio L., Kosik K.S. delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J. Cell Biol. 1999;144:519–532. doi: 10.1083/jcb.144.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez M.C., Ochiishi T., Majewski M., Kosik K.S. Dual regulation of neuronal morphogenesis by a delta-catenin-cortactin complex and Rho. J. Cell Biol. 2003;162:99–111. doi: 10.1083/jcb.200211025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laura R.P., Witt A.S., Held H.A., Gerstner R., Deshayes K., Koehler M.F., Kosik K.S., Sidhu S.S., Lasky L.A. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J. Biol. Chem. 2002;277:12906–12914. doi: 10.1074/jbc.M200818200. [DOI] [PubMed] [Google Scholar]
- 36.Dreos R., Ambrosini G., Périer R.C., Bucher P. The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015;43:D92–D96. doi: 10.1093/nar/gku1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofmeister W., Nilsson D., Topa A., Anderlid B.M., Darki F., Matsson H., Tapia Páez I., Klingberg T., Samuelsson L., Wirta V., et al. CTNND2-a candidate gene for reading problems and mild intellectual disability. J. Med. Genet. 2015;52:111–122. doi: 10.1136/jmedgenet-2014-102757. [DOI] [PubMed] [Google Scholar]
- 38.van Rootselaar A.F., Groffen A.J., de Vries B., Callenbach P.M.C., Santen G.W.E., Koelewijn S., Vijfhuizen L.S., Buijink A., Tijssen M.A.J., van den Maagdenberg A.M.J.M. δ-Catenin (CTNND2) missense mutation in familial cortical myoclonic tremor and epilepsy. Neurology. 2017;89:2341–2350. doi: 10.1212/WNL.0000000000004709. [DOI] [PubMed] [Google Scholar]
- 39.Duparc R.H., Boutemmine D., Champagne M.P., Tétreault N., Bernier G. Pax6 is required for delta-catenin/neurojugin expression during retinal, cerebellar and cortical development in mice. Dev. Biol. 2006;300:647–655. doi: 10.1016/j.ydbio.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 40.Keirsebilck A., Bonné S., Staes K., van Hengel J., Nollet F., Reynolds A., van Roy F. Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics. 1998;50:129–146. doi: 10.1006/geno.1998.5325. [DOI] [PubMed] [Google Scholar]
- 41.Sirotkin H., O’Donnell H., DasGupta R., Halford S., St Jore B., Puech A., Parimoo S., Morrow B., Skoultchi A., Weissman S.M., et al. Identification of a new human catenin gene family member (ARVCF) from the region deleted in velo-cardio-facial syndrome. Genomics. 1997;41:75–83. doi: 10.1006/geno.1997.4627. [DOI] [PubMed] [Google Scholar]
- 42.Paulson A.F., Mooney E., Fang X., Ji H., McCrea P.D. Xarvcf, Xenopus member of the p120 catenin subfamily associating with cadherin juxtamembrane region. J. Biol. Chem. 2000;275:30124–30131. doi: 10.1074/jbc.M003048200. [DOI] [PubMed] [Google Scholar]
- 43.Hatzfeld M., Nachtsheim C. Cloning and characterization of a new armadillo family member, p0071, associated with the junctional plaque: evidence for a subfamily of closely related proteins. J. Cell Sci. 1996;109:2767–2778. doi: 10.1242/jcs.109.11.2767. [DOI] [PubMed] [Google Scholar]
- 44.Paffenholz R., Franke W.W. Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation. 1997;61:293–304. doi: 10.1046/j.1432-0436.1997.6150293.x. [DOI] [PubMed] [Google Scholar]
- 45.Kawamura Y., Fan Q.W., Hayashi H., Michikawa M., Yanagisawa K., Komano H. Expression of the mRNA for two isoforms of neural plakophilin-related arm-repeat protein/delta-catenin in rodent neurons and glial cells. Neurosci. Lett. 1999;277:185–188. doi: 10.1016/s0304-3940(99)00875-7. [DOI] [PubMed] [Google Scholar]
- 46.Földy C., Darmanis S., Aoto J., Malenka R.C., Quake S.R., Südhof T.C. Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl. Acad. Sci. USA. 2016;113:E5222–E5231. doi: 10.1073/pnas.1610155113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munton R.P., Tweedie-Cullen R., Livingstone-Zatchej M., Weinandy F., Waidelich M., Longo D., Gehrig P., Potthast F., Rutishauser D., Gerrits B., et al. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol. Cell. Proteomics. 2007;6:283–293. doi: 10.1074/mcp.M600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Toyo-oka K., Wachi T., Hunt R.F., Baraban S.C., Taya S., Ramshaw H., Kaibuchi K., Schwarz Q.P., Lopez A.F., Wynshaw-Boris A. 14-3-3ε and ζ regulate neurogenesis and differentiation of neuronal progenitor cells in the developing brain. J. Neurosci. 2014;34:12168–12181. doi: 10.1523/JNEUROSCI.2513-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tasic B., Menon V., Nguyen T.N., Kim T.K., Jarsky T., Yao Z., Levi B., Gray L.T., Sorensen S.A., Dolbeare T., et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Ebbert A., Riley Z.L., Royall J.J., Aiona K., et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arikkath J., Israely I., Tao Y., Mei L., Liu X., Reichardt L.F. Erbin controls dendritic morphogenesis by regulating localization of delta-catenin. J. Neurosci. 2008;28:7047–7056. doi: 10.1523/JNEUROSCI.0451-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan L., Seong E., Beuscher J.L., Arikkath J. δ-Catenin Regulates Spine Architecture via Cadherin and PDZ-dependent Interactions. J. Biol. Chem. 2015;290:10947–10957. doi: 10.1074/jbc.M114.632679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Israely I., Costa R.M., Xie C.W., Silva A.J., Kosik K.S., Liu X. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr. Biol. 2004;14:1657–1663. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- 54.Cho Y.H., Friedman E., Silva A.J. Ibotenate lesions of the hippocampus impair spatial learning but not contextual fear conditioning in mice. Behav. Brain Res. 1999;98:77–87. doi: 10.1016/s0166-4328(98)00054-0. [DOI] [PubMed] [Google Scholar]
- 55.Bourdy R., Sánchez-Catalán M.J., Kaufling J., Balcita-Pedicino J.J., Freund-Mercier M.J., Veinante P., Sesack S.R., Georges F., Barrot M. Control of the nigrostriatal dopamine neuron activity and motor function by the tail of the ventral tegmental area. Neuropsychopharmacology. 2014;39:2788–2798. doi: 10.1038/npp.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matter C., Pribadi M., Liu X., Trachtenberg J.T. Delta-catenin is required for the maintenance of neural structure and function in mature cortex in vivo. Neuron. 2009;64:320–327. doi: 10.1016/j.neuron.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim H., Han J.R., Park J., Oh M., James S.E., Chang S., Lu Q., Lee K.Y., Ki H., Song W.J., Kim K. Delta-catenin-induced dendritic morphogenesis. An essential role of p190RhoGEF interaction through Akt1-mediated phosphorylation. J. Biol. Chem. 2008;283:977–987. doi: 10.1074/jbc.M707158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edbauer D., Cheng D., Batterton M.N., Wang C.F., Duong D.M., Yaffe M.B., Peng J., Sheng M. Identification and characterization of neuronal mitogen-activated protein kinase substrates using a specific phosphomotif antibody. Mol. Cell. Proteomics. 2009;8:681–695. doi: 10.1074/mcp.M800233-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee H.B., He Y., Yang G.S., Oh J.A., Ha J.S., Song O.H., Lee D.J., Jung S.C., Kim K.K., Kim K., Kim H. Determination of C-Terminal δ-Catenin Responsible for Inducing Dendritic Morphogenesis. J. Nanosci. Nanotechnol. 2015;15:5589–5592. doi: 10.1166/jnn.2015.10460. [DOI] [PubMed] [Google Scholar]
- 60.Abu-Elneel K., Ochiishi T., Medina M., Remedi M., Gastaldi L., Caceres A., Kosik K.S. A delta-catenin signaling pathway leading to dendritic protrusions. J. Biol. Chem. 2008;283:32781–32791. doi: 10.1074/jbc.M804688200. [DOI] [PubMed] [Google Scholar]
- 61.Gu D., Sater A.K., Ji H., Cho K., Clark M., Stratton S.A., Barton M.C., Lu Q., McCrea P.D. Xenopus delta-catenin is essential in early embryogenesis and is functionally linked to cadherins and small GTPases. J. Cell Sci. 2009;122:4049–4061. doi: 10.1242/jcs.031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghose S., Min Y., Lin P.C. δ-Catenin activates Rho GTPase, promotes lymphangiogenesis and growth of tumor metastases. PLoS ONE. 2015;10:e0116338. doi: 10.1371/journal.pone.0116338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newey S.E., Velamoor V., Govek E.E., Van Aelst L. Rho GTPases, dendritic structure, and mental retardation. J. Neurobiol. 2005;64:58–74. doi: 10.1002/neu.20153. [DOI] [PubMed] [Google Scholar]
- 64.Threadgill R., Bobb K., Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 65.Carlisle H.J., Kennedy M.B. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28:182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 66.Poore C.P., Sundaram J.R., Pareek T.K., Fu A., Amin N., Mohamed N.E., Zheng Y.L., Goh A.X., Lai M.K., Ip N.Y., et al. Cdk5-mediated phosphorylation of delta-catenin regulates its localization and GluR2-mediated synaptic activity. J. Neurosci. 2010;30:8457–8467. doi: 10.1523/JNEUROSCI.6062-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ochiishi T., Futai K., Okamoto K., Kameyama K., Kosik K.S. Regulation of AMPA receptor trafficking by delta-catenin. Mol. Cell. Neurosci. 2008;39:499–507. doi: 10.1016/j.mcn.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 68.Zimmermann A.M., Jene T., Wolf M., Görlich A., Gurniak C.B., Sassoè-Pognetto M., Witke W., Friauf E., Rust M.B. Attention-Deficit/Hyperactivity Disorder-like Phenotype in a Mouse Model with Impaired Actin Dynamics. Biol. Psychiatry. 2015;78:95–106. doi: 10.1016/j.biopsych.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 69.Arcos-Burgos M., Jain M., Acosta M.T., Shively S., Stanescu H., Wallis D., Domené S., Vélez J.I., Karkera J.D., Balog J., et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol. Psychiatry. 2010;15:1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- 70.Lionel A.C., Crosbie J., Barbosa N., Goodale T., Thiruvahindrapuram B., Rickaby J., Gazzellone M., Carson A.R., Howe J.L., Wang Z., et al. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci. Transl. Med. 2011;3:95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- 71.O’Sullivan M.L., de Wit J., Savas J.N., Comoletti D., Otto-Hitt S., Yates J.R., 3rd, Ghosh A. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Won H., Mah W., Kim E., Kim J.W., Hahm E.K., Kim M.H., Cho S., Kim J., Jang H., Cho S.C., et al. GIT1 is associated with ADHD in humans and ADHD-like behaviors in mice. Nat. Med. 2011;17:566–572. doi: 10.1038/nm.2330. [DOI] [PubMed] [Google Scholar]
- 73.Rivero O., Selten M.M., Sich S., Popp S., Bacmeister L., Amendola E., Negwer M., Schubert D., Proft F., Kiser D., et al. Cadherin-13, a risk gene for ADHD and comorbid disorders, impacts GABAergic function in hippocampus and cognition. Transl. Psychiatry. 2015;5:e655. doi: 10.1038/tp.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaufmann W.E., Moser H.W. Dendritic anomalies in disorders associated with mental retardation. Cereb. Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 75.Phillips M., Pozzo-Miller L. Dendritic spine dysgenesis in autism related disorders. Neurosci. Lett. 2015;601:30–40. doi: 10.1016/j.neulet.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dierssen M., Ramakers G.J. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes Brain Behav. 2006;5(Suppl 2):48–60. doi: 10.1111/j.1601-183X.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 77.Florian R.T., Kraft F., Leitão E., Kaya S., Klebe S., Magnin E., van Rootselaar A.F., Buratti J., Kühnel T., Schröder C., et al. FAME consortium Unstable TTTTA/TTTCA expansions in MARCH6 are associated with Familial Adult Myoclonic Epilepsy type 3. Nat. Commun. 2019;10:4919. doi: 10.1038/s41467-019-12763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets and variants generated during this study are available at GenBank (Whole genome sequencing; SRA accession PRJNA64834) and ClinVar (Gene variants; Accessions SUB7836623, SUB7824655).