ABSTRACT
Background
Whole grain wheat (WGW) products are advocated as a healthy choice when compared with refined wheat (RW). One proposed mechanism for these health benefits is via the microbiota, because WGW contains multiple fibers. WGW consumption has been proposed to ameliorate nonalcoholic fatty liver disease, in which microbiota might play a role.
Objectives
We investigated the effect of WGW compared with RW intervention on the fecal microbiota composition and functionality, and correlated intervention-induced changes in bacteria with changes in liver health parameters in adults with overweight or obesity.
Methods
We used data of a 12-wk double-blind, randomized, controlled, parallel trial to examine the effects of a WGW (98 g/d) or RW (98 g/d) intervention on the secondary outcomes fecal microbiota composition, predicted microbiota functionality, and stool consistency in 37 women and men (aged 45–70 y, BMI 25–35 kg/m2). The changes in microbiota composition, measured using 16S ribosomal RNA gene sequencing, after a 12-wk intervention were analyzed with nonparametric tests, and correlated with changes in liver fat and circulating concentrations of liver enzymes including alanine transaminase, aspartate transaminase, γ-glutamyltransferase, and serum amyloid A.
Results
The WGW intervention increased the mean (± SD) relative abundances of Ruminococcaceae_UCG-014 (baseline: 2.2 ± 4.6%, differential change over time (Δ) 0.51 ± 4.2%), Ruminiclostridium_9 (baseline: 0.065 ± 0.11%, Δ 0.054 ± 0.14%), and Ruminococcaceae_NK4A214_group (baseline: 0.37 ± 0.56%, Δ 0.17 ± 0.83%), and also the predicted pathway acetyl-CoA fermentation to butyrate II (baseline: 0.23 ± 0.062%, Δ 0.035 ± 0.059%), compared with the RW intervention (P values <0.05). A change in Ruminococcaceae_NK4A214_group was positively correlated with the change in liver fat, in both the WGW (ρ = 0.54; P = 0.026) and RW (ρ = 0.67; P = 0.024) groups.
Conclusions
In middle-aged overweight and obese adults, a 12-wk WGW intervention increased the relative abundance of a number of bacterial taxa from the family Ruminococcaceae and increased predicted fermentation pathways when compared with an RW intervention. Potential protective health effects of replacement of RW by WGW on metabolic organs, such as the liver, via modulation of the microbiota, deserve further investigation.
This trial was registered at clinicaltrials.gov as NCT02385149.
Keywords: fatty liver, human, gut microbiota, overweight/obesity, refined wheat, whole grain wheat, fermentation
See corresponding commentary on page 459.
Introduction
The consumption of whole grains (WGs) is associated with lower risk of diseases such as type 2 diabetes, cardiovascular diseases, obesity, and certain types of cancer in observational studies (1–3). In addition, consumption of WGs rather than refined grain has been proposed to prevent nonalcoholic fatty liver disease (NAFLD) (4). The presence of numerous bioactive compounds makes WGs, including whole grain wheat (WGW), nutritionally superior to refined wheat (RW) products (5). In refined products, both the bran and germ in the wheat kernel, which are rich sources of dietary fiber, polyphenols, B-complex vitamins, betaine, choline, and minerals, have been removed (5, 6). However, multiple previous WG interventions in human subjects have reported conflicting results in improvements in health parameters such as insulin sensitivity and cholesterol concentrations (7–10).
The potential health benefits of WGW have also been ascribed to fiber. WGW is a source of both fermentable and nonfermentable dietary fibers, such as (hemi)cellulose including arabinoxylan and β-glucan, lignin, and the oligosaccharides raffinose, stachyose, and fructan (11). Dietary fibers are not hydrolyzed or absorbed in the small intestine, and therefore can directly interact with the colonic gut microbiota, resulting in the production of metabolites that are relevant to health (12, 13). Diet affects the microbiota, and consequently dietary changes (for instance, via interventions) can possibly modulate the intestinal microbiota composition and functionality (13). Cereal fibers and other components in WGW such as iron (14) have shown an ability to modulate the gut microbiota composition in vitro and in vivo (15, 16). Moreover, multiple human trials demonstrated an increase in specific bacteria associated with host health benefits after WGW interventions, including enhanced abundance of Bifidobacterium (17, 18) and Lactobacillus (18, 19). Two trials reported an increase in butyrate-producing bacteria upon WGW or WG consumption (19, 20). However, other human trials did not find effects of WGs on the microbiota composition (21–23). The effects of WGW and WGs on both the composition and functions of the gut microbiota in humans are therefore not yet well understood. Some previous trials investigated only a selected subset of bacteria (17, 18, 24) and could have failed to capture the full effects of WGW and RW on the gut microbiota. It remains to be elucidated if the gut microbiota mediates the health benefits of WGW in humans.
The fermentable dietary fibers can be broken down by the gut microbiota (11, 25), producing SCFAs, mainly acetate, butyrate, and propionate. SCFAs can reach the liver and the peripheral circulation, where they can affect organ function and metabolism (26, 27). In vitro studies and animal experiments have shown that SCFAs increase fat oxidation in liver tissue (28, 29), suggesting a link between microbial-produced SCFAs and liver fat. Modulation of the gut microbiota is thought to play a role in the development of NAFLD and nonalcoholic steatohepatitis (30–33). In the review of Ross et al. (4) the effect of WGs on the microbiota was proposed as a potential mechanism in the prevention of NAFLD. The presence of a perturbed metabolic health status, such as being middle-aged and being overweight or obese, is often a prerequisite for the development of NAFLD (4).
Recently, we performed the Graandioos study (7, 34), and observed that a 12-wk RW intervention increased intrahepatic triglycerides (IHTGs) and decreased microbiota diversity in middle-aged adults with overweight and obesity, whereas a WGW intervention prevented an increase in liver fat (7). We have previously reported the effect of WGW and RW on the abundances of a preselected subset of bacteria (7). In the current article we describe the effects of this randomized, controlled, double-blind, parallel trial with a 12-wk WGW intervention compared with an RW intervention on the complete fecal microbiota composition with additional analyses, as well as the effects of the interventions on predicted microbial functionality. Moreover, we investigated the relation between intervention-induced changes in bacterial composition and changes in various liver health parameters, such as IHTGs and liver enzymes, to examine the potential role of microbiota in the preventive effect of WGW in hepatic fat accumulation.
Methods
Subjects
Fifty middle-aged Dutch men and postmenopausal women (aged 45–70 y, BMI 25–35 kg/m2) with mildly elevated plasma total cholesterol (>5 mmol/L) participated. Subjects using cholesterol-lowering medication or subjects that used antibiotics <1 mo prior to day 1 of the study were excluded (for details see reference 7). Subjects had no history of medical/surgical events that could affect the study outcomes. During the screening visit, subjects filled in an FFQ to determine habitual dietary intake. WG consumption was quantified by research dietitians using The Dutch Food Composition Database (NEVO) table 2010. This trial was approved by the Medical Ethical Committee of Wageningen University and registered at clinicaltrials.gov under NCT02385149. All participants gave written consent before participation.
Study design and procedures
This study was a randomized, controlled, double-blind, parallel trial (Supplemental Figure 1). Recruitment and study logistics are described in reference 7. Before the start of the intervention, a 4-wk run-in period with uncolored RW products was included to reduce WGW intake variation at baseline. Afterwards, subjects were randomly assigned to a 12-wk intervention with either WGW products or colored RW products. Age, gender, BMI, and cholesterol concentrations were stratified among the intervention groups. The randomization was conducted using block randomization (Microsoft Excel) by a researcher who was not involved in the study. Participants were asked to continue their dietary pattern and dietary habits during the intervention period. They were not allowed to lose or gain weight during the intervention. Before and after the 12-wk intervention, feces were collected. At test days, liver fat accumulation and metabolic health parameters were measured. Participants recorded stool consistency weekly using the Bristol Stool Chart, which describes seven types of stools ranging from 1: hard/lumpy to 7: watery without solid pieces. The day prior to the test day, participants consumed a standardized low-fat evening meal, refrained from alcohol or strenuous exercise, and were not allowed to eat or drink anything except water after 20:00 to ensure a fasting state.
Intervention products
Participants received either WGW or RW products to replace their habitual intake of grain products. Four slices of bread (in total 100 g/d), and one serving of ready-to-eat-cereals (33.4 g/d) were consumed daily. In total, this added up to 98 g of RW or WGW flour per day. RW products were colored with roasted wheat malt and caramelized sugar to match the appearance of WGW products. On the macronutrient and energy level, the RW and WGW products were similar, except for fiber content. The WGW products contained 17.6 g fiber/100 g, and the RW products 7.2 g fiber/100 g. The nutritional composition (macronutrients, vitamins, and minerals) of the intervention products is provided in Supplemental Table 1. During the run-in and intervention periods, consumption of additional WG food products was not allowed, including products from other grain sources (i.e., brown rice), in both the WGW and RW groups. All participants received a list of WG products to avoid during the intervention. Subjects were allowed to complement their daily diet with additional refined grain products.
Clinical chemistry and IHTG accumulation
Lipid content in the liver was quantified with proton magnetic resonance spectroscopy using a 3-T whole-body MRI scanner (7). Plasma alanine transaminase (ALT), aspartate transaminase (AST), γ-glutamyltransferase (GGT), C-reactive protein (CRP), and serum amyloid A (SAA) were included as liver health markers, which were analyzed as described previously (7). β-Hydroxybutyrate was also included as marker because it can be synthesized in the liver via the metabolism of butyrate. As a biomarker for WGW intake, plasma alkylresorcinol was analyzed as described previously (7).
Fecal samples and microbiota profiling
Feces were collected at home ≤3 d before the test day using collection pots and equipment and were stored at −20°C for ≤3 d. Samples were transferred to −80°C. Fecal material was mechanically homogenized, and genomic DNA was isolated with the use of an AGOWA mag Mini kit (AGOWA, Berlin, Germany) according to the manufacturer's instructions. The V4 hypervariable region of the 16S ribosomal RNA (rRNA) gene was PCR amplified using F515/R806 primers. PCR products were purified, followed by paired-end sequencing on an Illumina MiSeq platform, as described previously (7). Twenty-two technical replicate samples were included in the dataset, representing 2 aliquots taken from the same fecal sample at the same time from which all consecutive steps were performed separately to isolate DNA. Raw sequencing data were first demultiplexed by trimming barcodes and primer sequences. Afterwards, the data were processed and amplicon sequence variants (ASVs) were picked with NG-Tax using default settings (35, 36). Chimeras were detected and filtered when the forward and reverse read of that ASV was identical to two different ASVs, and the abundance of the matched ASVs were ≥2 times the abundance of that specific ASV. The SILVA reference database version 128 was used to assign taxonomy, with a confidence >80% for genus-level classification.
Microbiota composition analyses
R version 3.5.1 (R Foundation) was used for all analyses (37). Raw counts were transformed to relative abundance. Technical replicates were compared by calculating pairwise Pearson correlation using the genus-level relative abundance. Afterwards, bacterial abundance in technical replicates was averaged. α-Diversity was calculated using Faith phylogenetic diversity based on branch length connecting taxa in those samples and the root node of the phylogenetic tree. Pairwise weighted UniFrac (38) distance-based principal coordinate analysis was used to visualize overall microbial community variation (39). Permutational multivariate analysis of variance (PERMANOVA) (40) was used to test for significant differences in overall community composition between groups.
Prediction of microbiota functionality and markers for SCFA production
Microbial functions were predicted based on the 16S sequences using the PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) algorithm (41). The default workflow was followed (41). Compositional count data of predicted pathways and Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs were transformed to relative abundance. More details are described in Supplemental Methods. KEGG orthologs were selected that could be used as markers for SCFA production (see Supplemental Methods).
Statistical analysis
The nonparametric Wilcoxon test was applied for paired comparisons of bacteria and predicted pathway relative abundances within intervention group over time, due to the nonnormal distribution of the data. The unpaired Mann–Whitney U test was used to determine the differences between groups at baseline and the differential changes over time (Δ) between intervention groups. Differences in baseline characteristics and FFQ data were assessed using independent t tests, or Mann–Whitney U tests, depending on the variable distribution. Bristol Stool Chart scores were analyzed using linear mixed effects modeling with interventions, time, and the interaction as fixed effects, and subject ID as a random effect (R package lme4). Correlations between (changes of) relative abundances of bacteria at the genus level and (changes of) liver health parameters between paired samples were assessed by Spearman rank correlation tests. Analyses were performed without and with adjustment for multiple comparisons using false discovery rate (FDR). Nonadjusted P values < 0.05 were considered significant, because modest changes were expected due to the explorative character of this study.
Results
Subject characteristics and stool consistency
Participants with missing fecal samples before or after the intervention (n = 5) or use of antibiotics during the intervention (n = 8) were removed from the analysis, resulting in 16 participants in the RW group, and 21 participants in the WGW group. No significant differences in baseline characteristics were found between groups, except for β-hydroxybutyrate (P = 0.043), which was higher in the RW group (Table 1). The mean (± SD) habitual WG intake before the start of the intervention was 60.1 ± 59.2 g/d in the RW group, and 54.4 ± 38.0 g/d in the WGW group (P = 0.73; Supplemental Table 2). No significant differences in the habitual intake of WGs, total carbohydrates, or dietary fibers were found between groups. Habitual medication use did not change during intervention, and analgesic use during the whole study period was ≤15 d in total. Stool consistency showed a nonspecific trend during the run-in and intervention periods (Supplemental Figure 2), without a significant effect of time (P = 0.58) or intervention (P = 0.64) on the consistency score. The main findings in this subset of the study population reflected those in the total study population, published previously (7), namely RW significantly increased IHTGs compared with the WGW group (P = 0.033), and WGW consumption decreased SAA (P = 0.042) and increased β-hydroxybutyrate (P = 0.003), compared with the RW group (Supplemental Table 3).
TABLE 1.
Baseline characteristics of middle-aged overweight and obese subjects in the RW or WGW group in a subset of the total study population1
| Variables | RW group | WGW group | P value |
|---|---|---|---|
| Gender,2 n males (%) | 9 (60) | 12 (60) | 0.96 |
| Age, y | 60 ± 6.0 | 60 ± 5.4 | 0.97 |
| Body weight, kg | 84 ± 7.0 | 86 ± 9.4 | 0.36 |
| BMI, kg/m2 | 27 ± 2.2 | 28 ± 2.0 | 0.14 |
| IHTGs,3 % | 3.6 ± 2.5 | 5.5 ± 6.4 | 0.83 |
| Total cholesterol, mmol/L | 6.0 ± 0.76 | 6.2 ± 0.71 | 0.38 |
| CRP,3 μg/mL | 2.9 ± 3.1 | 4.8 ± 7.3 | 0.95 |
| SAA,3 μg/mL | 2.1 ± 1.9 | 7.2 ± 16 | 0.76 |
| GGT, U/L | 19 ± 13 | 21 ± 13 | 0.61 |
| AST, U/L | 19 ± 5.6 | 19 ± 4.7 | 0.60 |
| ALT, U/L | 31 ± 9.4 | 36 ± 12 | 0.18 |
| β-Hydroxybutyrate, mmol/L | 0.30 ± 0.20 | 0.20 ± 0.20 | 0.043 |
Values are presented as group means ± SD, n = 16 (RW) or n = 21 (WGW). ALT, alanine transaminase; AST, aspartate transaminase; CRP, C-reactive protein; GGT, γ-glutamyltransferase; IHTG, intrahepatic triglyceride; RW, refined wheat; SAA, serum amyloid A; WGW, whole grain wheat.
Presented as the number and percentage of males.
Nonparametric distribution, evaluated with the Mann–Whitney U test.
Microbiota composition and predicted pathways at baseline
The microbiota composition and predicted pathway datasets were of high quality (Supplemental Figure 3), and all details are provided in the Supplemental Results. In total, 1450 unique ASVs were identified in the microbiota dataset within 181 unique genera, and 356 pathways were predicted to be active in the dataset. Substantial interindividual variation in microbiota composition at the genus level was observed at baseline (Supplemental Figure 4), without clear differences between intervention groups for the average composition. Without correcting for multiple testing, 7 bacterial taxa and 11 predicted pathways were significantly different between groups at baseline (Figure 1, Supplemental Table 4). After FDR correction, none of the bacterial taxa or pathways were significantly different at baseline.
FIGURE 1.
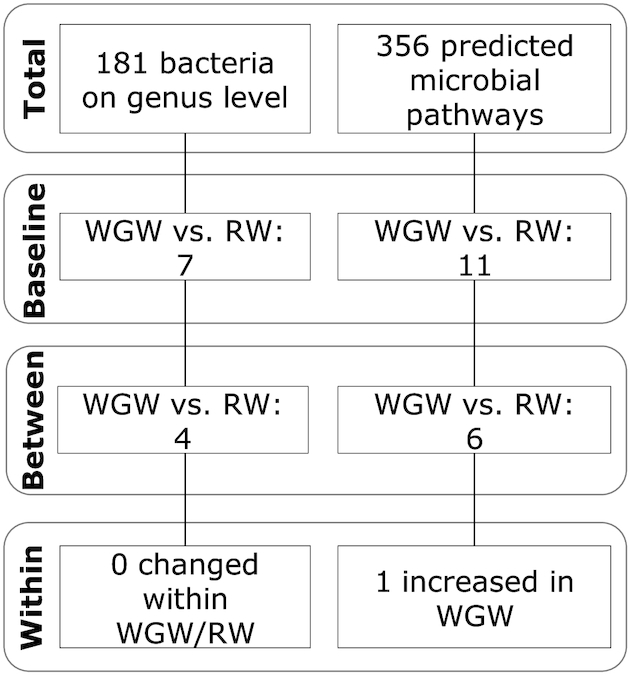
Significantly different fecal bacterial taxa and bacterial pathways at baseline and after 12-wk of an RW or WGW intervention in middle-aged overweight and obese adults. The flow diagram shows the number of bacterial taxa at the genus level and the predicted pathways for which the change in relative abundance was significantly different among interventions. RW, refined wheat; WGW, whole grain wheat.
The gut microbiota diversity
No significant clustering effect was found between baseline and postintervention within and between groups based on overall microbiota profiles (PERMANOVA P = 0.63, Figure 2A). However, the overall microbiota community of most individuals showed a shift over time. The change in microbial diversity (phylogenetic diversity, PD) (Figure 2B) was not significantly different between the RW and WGW interventions (P = 0.21). Both RW and WGW interventions did not change microbial diversity (RW: baseline 11.5 ± 1.38 PD, Δ −0.67 ± 1.66 PD, P = 0.19; WGW: baseline 11.0 ± 1.50 PD, Δ −0.09 ± 1.10 PD, P = 0.97). On the group level, no differences were found in the overall microbiota community composition. Overall, the interventions changed neither microbiota phylogenetic diversity (α-diversity) nor the overall community composition (β-diversity).
FIGURE 2.
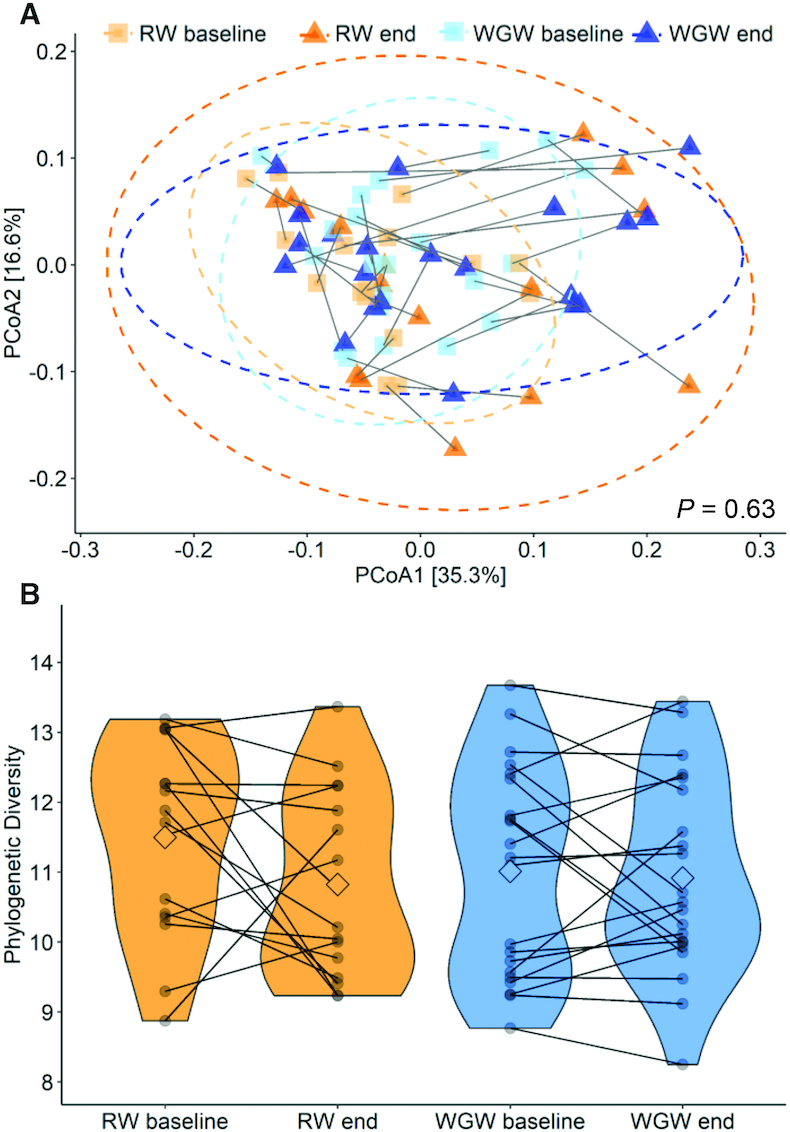
The effect of 12 wk of an RW or WGW intervention on the fecal microbiota diversity in middle-aged overweight and obese adults. (A) PCoA plot using weighted UniFrac dissimilarity to visualize the overall microbiota community variation. Color and shape highlight the intervention groups before or after the intervention. The lines connect the within-person samples over time; 95% CIs are plotted. (B) Microbial diversity as assessed using Faith phylogenetic diversity at baseline and after the 12-wk intervention (end). Individual paired samples are connected by a line. The width of the colored shapes indicates the sample density, the squared shape inside indicates the group mean. PCoA, principal coordinate analysis; RW, refined wheat; WGW, whole grain wheat.
The effect of RW and WGW interventions on the microbiota composition
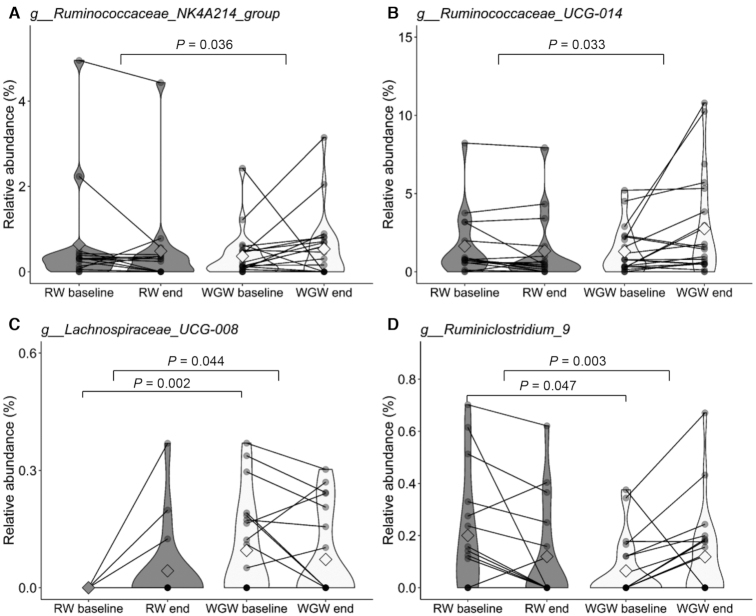
Intervention effects on microbiota abundances were found at the genus level between and within the WGW and RW groups (Table 2). Between groups, 4 bacterial taxa were significantly different over time (Figure 3), namely Ruminiclostridium_9 (P = 0.003),Ruminococcaceae_NK4A214_group (P = 0.036),Lachnospiraceae_UCG-008 (P = 0.044), and Ruminococcaceae_UCG-014 (P = 0.033), without significant differences within intervention groups. Not all of the aforementioned bacterial taxa were present in all individuals; in Supplemental Table 5, the number of subjects in which the bacterial taxa were detected is shown. Overall, the WGW intervention increased 3 bacterial taxa (within the Ruminococcaceae family) and decreased 1 bacterial taxon (within the Lachnospiraceae family), whereas the RW intervention mostly decreased these bacteria.
TABLE 2.
Fecal bacterial taxa abundances at the genus level at baseline and change after 12 wk of intervention that were found to be significantly different within and/or between the WGW and RW group in middle-aged overweight and obese adults1
| RW group | WGW group | Within RW | Within WGW | Group comparison | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | Baseline relative abundance, % | Δ Relative abundance after 12 wk, % | Baseline relative abundance, % | Δ Relative abundance after 12 wk, % | P value | FDR P value | P value | FDR P value | P value | FDR P value |
| f_Ruminococcaceae; g_Ruminiclostridium_9 | 0.20 ± 0.23 | −0.081 ± 0.17 | 0.065 ± 0.11 | 0.054 ± 0.14 | 0.07 | NS2 | 0.07 | NS | 0.003 | NS |
| f_Ruminococcaceae; g_Ruminococcaceae_NK4A214_group | 0.64 ± 1.3 | −0.15 ± 0.49 | 0.37 ± 0.56 | 0.17 ± 0.83 | 0.12 | NS | 0.11 | NS | 0.036 | NS |
| f_Lachnospiraceae; g_Lachnospiraceae_UCG-008 | 0.00 ± 0.00 | 0.043 ± 0.10 | 0.10 ± 0.12 | −0.023 ± 0.083 | 0.18 | NS | 0.19 | NS | 0.044 | NS |
| f_Ruminococcaceae; g_Ruminococcaceae_UCG-014 | 1.6 ± 2.1 | −0.31 ± 0.78 | 2.2 ± 4.6 | 0.51 ± 4.2 | 0.12 | NS | 0.13 | NS | 0.033 | NS |
| f_Lachnospiraceae; g_Dorea | 1.7 ± 1.1 | −0.72 ± 0.91 | 1.6 ± 0.81 | −0.018 ± 0.99 | 0.005 | NS | 0.61 | NS | 0.055 | NS |
| f_Tannerellaceae; g_Parabacteroides | 0.37 ± 0.30 | 0.60 ± 1.42 | 0.45 ± 0.41 | 0.35 ± 0.97 | 0.007 | NS | 0.18 | NS | 0.32 | NS |
| f_Lachnospiraceae; g_[Eubacterium]_ventriosum_group | 0.17 ± 0.23 | −0.14 ± 0.24 | 0.14 ± 0.14 | −0.050 ± 0.15 | 0.044 | NS | 0.71 | NS | 0.18 | NS |
| f_Peptostreptococcaceae_g_unknown | 1.2 ± 2.8 | −0.64 ± 1.5 | 0.56 ± 0.71 | −0.23 ± 0.69 | 0.024 | NS | 0.18 | NS | 0.57 | NS |
| f_Ruminococcaceae; g_Ruminococcus_2 | 1.4 ± 1.3 | −0.24 ± 1.5 | 2.1 ± 1.8 | −1.1 ± 2.1 | 0.11 | NS | 0.024 | NS | 0.44 | NS |
| f_Ruminococcaceae; g_Subdoligranulum | 6.8 ± 4.7 | −1.7 ± 4.6 | 6.1 ± 5.3 | −1.8 ± 3.7 | 0.10 | NS | 0.024 | NS | 0.89 | NS |
| f_Burkholderiaceae; g_Sutterella | 0.27 ± 0.44 | 0.085 ± 0.45 | 0.35 ± 0.92 | 0.30 ± 0.49 | 0.31 | NS | 0.010 | NS | 0.46 | NS |
| f_Ruminococcaceae; g_Ruminococcaceae_UCG-005 | 1.1 ± 1.1 | 0.38 ± 1.5 | 0.91 ± 1.1 | 0.21 ± 1.0 | 0.75 | NS | 0.042 | NS | 0.47 | NS |
Values are presented as group means ± SD, n = 16 (RW) or n = 21 (WGW). FDR, false discovery rate; RW, refined wheat; WGW, whole grain wheat; Δ, differential change over time.
2NS, nonsignificant: P value > 0.05.
FIGURE 3.
Fecal bacterial taxa at the genus level at baseline and after the 12-wk RW or WGW intervention (end) that were found to be significantly different between the groups in middle-aged overweight and obese adults. The relative abundances of (A) Ruminococcaceae_NK4A214_group; (B) Ruminococcaceae_UCG-014; (C) Lachnospiraceae_UCG-008; and (D) Ruminiclostridium_9 are shown. Data are presented as group mean (the squared shape), n = 16 (RW) or n = 21 (WGW), and the width of the colored shapes indicates the sample density. Individual paired samples are connected by a line. RW, refined wheat; WGW, whole grain wheat.
Correlations between the microbiota and liver health parameters
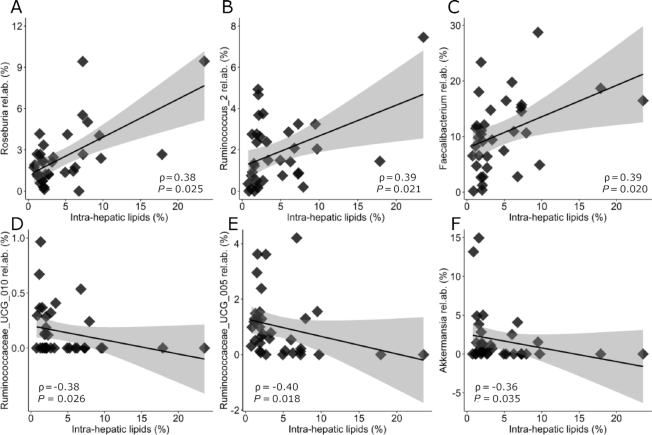
The relative abundances of all bacteria at baseline were then correlated with baseline IHTGs. Significant positive correlations were found between IHTG and Roseburia (Figure 4, ρ = 0.38, P = 0.025), Ruminococcus_2 (ρ = 0.39, P = 0.021), and Faecalibacterium (ρ = 0.39, P = 0.020). Significant negative correlations were found between IHTGs and Ruminococcaceae_UCG-010 (ρ = −0.38, P = 0.026), Ruminococcaceae_UCG-005 (ρ = −0.40, P = 0.018), and Akkermansia (ρ = −0.36, P = 0.035). After removal of 2 outliers (18% and 26% IHTG), the correlation between IHTGs and Ruminococcus_2 was still significant (P = 0.050), whereas for the other bacteria there was a trend toward significance (P values between 0.05 and 0.10). After FDR correction, none of these correlations were significant.
FIGURE 4.
Fecal bacterial taxa at the genus level that were found to be significantly correlated with IHTGs at baseline in middle-aged overweight and obese adults. The relative abundances of (A) Roseburia; (B) Ruminococcus_2; (C) Faecalibacterium; (D) Ruminococcaceae_UCG-010; (E) Ruminococcaceae_UCG-005; and (F) Akkermansia are shown, fitted with a linear regression model with a 95% CI. Noncorrected P values are shown. Data of n = 35 participants are shown, who had both microbiota and IHTG data at baseline available. IHTG, intrahepatic triglyceride.
Correlations between changes in liver health markers including IHTGs, ALT, AST, GGT, SAA, CRP, and β-hydroxybutyrate with changes in bacterial abundances after 12 wk of intervention were tested (Supplemental Figure 5). Change in IHTGs was positively correlated with change in Ruminococcaceae_NK4A214_group abundance in both the WGW (ρ = 0.54, P = 0.026) and RW (ρ = 0.67, P = 0.024) groups (Table 3). Moreover, change in ALT was significantly correlated with changes of 2 bacterial taxa in the RW and 2 in the WGW intervention. Change in AST was correlated with changes of 3 bacterial taxa in the RW group, 1 in the WGW group, and 1 (Lachnoclostridium) in both groups. Changes in GGT or SAA were significantly correlated with changes in abundance of 1 bacterial taxon in the RW group and 1 in the WGW group, or 3 bacterial taxa in the WGW group, respectively. Change in CRP was correlated with changes of 3 bacterial taxa in the RW group. Furthermore, change in β-hydroxybutyrate was positively correlated with 3 bacterial taxa in the RW group and 1 (Streptococcus) in both groups. After FDR correction, none of these correlations were significant.
TABLE 3.
The significant correlations between changes in fecal bacterial abundances at the genus level, and changes in liver health parameters after 12 wk of an RW or WGW intervention in middle-aged overweight and obese adults1
| RW | WGW | |||||
|---|---|---|---|---|---|---|
| Taxa (Δ 12 wk) | Spearman ρ | P value | FDR P value | Spearman ρ | P value | FDR P value |
| IHTG (Δ 12 wk) | ||||||
| g_Ruminococcaceae_NK4A214_group | 0.67 | 0.024 | NS2 | 0.54 | 0.026 | NS |
| ALT (Δ 12 wk) | ||||||
| g_Bacteroides | 0.30 | NS | NS | −0.44 | 0.048 | NS |
| g_Bifidobacterium | 0.77 | 0.001 | NS | −0.14 | NS | NS |
| g_Faecalibacterium | −0.11 | NS | NS | −0.46 | 0.037 | NS |
| g_Lachnoclostridium | 0.60 | 0.014 | NS | −0.20 | NS | NS |
| AST (Δ 12 wk) | ||||||
| g_[Eubacterium]_coprostanoligenes_group | 0.53 | 0.034 | NS | 0.065 | NS | NS |
| g_Butyricicoccus | 0.51 | 0.045 | NS | −0.22 | NS | NS |
| g_Lachnoclostridium | 0.51 | 0.045 | NS | −0.48 | 0.027 | NS |
| g_Lachnospiraceae_NK4A136_group | 0.53 | 0.036 | NS | 0.35 | NS | NS |
| g_Ruminiclostridium_6 | 0.10 | NS | NS | 0.46 | 0.036 | NS |
| CRP (Δ 12 wk) | ||||||
| g_Butyricicoccus | −0.58 | 0.025 | NS | −0.052 | NS | NS |
| g_Lachnospiraceae_NK4A136_group | −0.68 | 0.007 | NS | 0.36 | NS | NS |
| g_Parabacteroides | 0.54 | 0.041 | NS | 0.25 | NS | NS |
| g_Uncultured | −0.59 | 0.023 | NS | −0.16 | NS | NS |
| GGT (Δ 12 wk) | ||||||
| g_[Eubacterium]_coprostanoligenes_group | −0.21 | NS | NS | 0.50 | 0.021 | NS |
| g_Erysipelotrichaceae_UCG-003 | 0.55 | 0.028 | NS | −0.071 | NS | NS |
| β-Hydroxybutyrate (Δ 12 wk) | ||||||
| g_Erysipelotrichaceae_UCG-003 | −0.027 | NS | NS | 0.67 | 0.002 | NS |
| g_Lachnospiraceae_ND3007_group | −0.10 | NS | NS | 0.48 | 0.036 | NS |
| g_Ruminococcus_2 | 0.13 | NS | NS | 0.46 | 0.049 | NS |
| g_Streptococcus | 0.61 | 0.011 | NS | 0.48 | 0.040 | NS |
| SAA (Δ 12 wk) | ||||||
| g_Anaerostipes | −0.23 | NS | NS | 0.52 | 0.018 | NS |
| g_Blautia | −0.29 | NS | NS | 0.53 | 0.015 | NS |
| g_Butyricicoccus | −0.46 | NS | NS | 0.46 | 0.036 | NS |
Values represent the Spearman ρ correlation coefficients, n = 16 (RW) or n = 21 (WGW). ALT, alanine transaminase; AST, aspartate transaminase; CRP, C-reactive protein; FDR, false discovery rate; GGT, γ-glutamyltransferase; IHTG, intrahepatic triglyceride; RW, refined wheat; SAA, serum amyloid A; WGW, whole grain wheat; Δ, differential change over time.
2NS, nonsignificant: P value > 0.05.
Changes in predicted microbial pathways
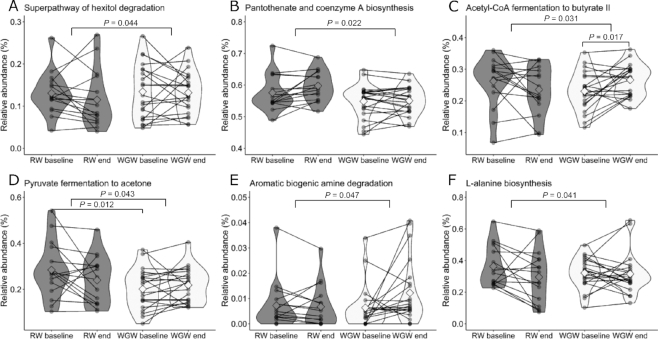
The intervention effects on predicted microbial pathways were investigated. Between the WGW and the RW interventions, 6 predicted pathways were significantly different over time (Table 4), namely hexitol degradation, pantothenate and coenzyme A biosynthesis, acetyl-CoA fermentation to butyrate II, pyruvate fermentation to acetone, aromatic biogenic amine degradation, and l-alanine biosynthesis. Subjects in both groups displayed a variation in response over time for the significantly different predicted pathways (Figure 5). Within the WGW group, acetyl-CoA fermentation to butyrate II was significantly increased (P = 0.017). Within the RW group, a second pathway related to fermentation was predicted to be decreased, namely pyruvate fermentation to acetate and lactate II (P = 0.049; Table 4).
TABLE 4.
Predicted fecal microbial pathway relative abundance at baseline and change after 12 wk of intervention that were found to be significantly different within and/or between the WGW and RW groups in middle-aged overweight and obese adults1
| RW group | WGW group | Within RW | Within WGW | Group comparison | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathway name | Pathway number | Baseline relative abundance, % | Δ Relative abundance after 12 wk, % | Baseline relative abundance, % | Δ Relative abundance after 12 wk, % | P value | FDR P value | P value | FDR P value | P value | FDR P value |
| Superpathway of hexitol degradation | HEXITOLDEGSUPER.PWY | 0.13 ± 0.049 | −0.015 ± 0.056 | 0.13 ± 0.065 | −0.00 ± 0.056 | 0.70 | NS2 | 0.97 | NS | 0.044 | NS |
| Pantothenate and coenzyme A biosynthesis | PANTOSYN.PWY | 0.58 ± 0.057 | 0.022 ± 0.039 | 0.55 ± 0.054 | 0.00 ± 0.034 | 0.11 | NS | 0.76 | NS | 0.022 | NS |
| Acetyl-CoA fermentation to butyrate II | PWY.5676 | 0.26 ± 0.079 | −0.026 ± 0.081 | 0.23 ± 0.062 | 0.035 ± 0.059 | 0.12 | NS | 0.017 | NS | 0.031 | NS |
| Pyruvate fermentation to acetone | PWY.6588 | 0.28 ± 0.12 | −0.044 ± 0.12 | 0.20 ± 0.087 | 0.017 ± 0.068 | 0.32 | NS | 0.18 | NS | 0.043 | NS |
| Aromatic biogenic amine degradation | PWY.7431 | 0.010 ± 0.010 | 0.00 ± 0.010 | 0.010 ± 0.010 | 0.0059 ± 0.012 | 0.30 | NS | 0.56 | NS | 0.047 | NS |
| l-Alanine biosynthesis | PWY0.1061 | 0.36 ± 0.13 | −0.067 ± 0.16 | 0.32 ± 0.10 | −0.010 ± 0.12 | 0.30 | NS | 0.55 | NS | 0.041 | NS |
| l-Arginine degradation II | AST.PWY | 0.0048 ± 0.010 | 0.00 ± 0.011 | 0.010 ± 0.023 | −0.00 ± 0.012 | 0.048 | NS | 0.23 | NS | 0.15 | NS |
| l-Lysine biosynthesis I | DAPLYSIESYN.PWY | 0.77 ± 0.10 | −0.065 ± 0.14 | 0.75 ± 0.079 | −0.030 ± 0.079 | 0.041 | NS | 0.15 | NS | 0.82 | NS |
| (Kdo)2-lipid A biosynthesis | KDO.NAGLIPASYN.PWY | 0.024 ± 0.036 | −0.010 ± 0.045 | 0.024 ± 0.045 | 0.010 ± 0.027 | 0.043 | NS | 0.28 | NS | 0.50 | NS |
| Pyruvate fermentation to acetate and lactate II | PWY.5100 | 0.94 ± 0.14 | −0.049 ± 0.086 | 0.91 ± 0.084 | −0.020 ± 0.071 | 0.049 | NS | 0.79 | NS | 0.13 | NS |
| PreQ0 biosynthesis | PWY.6703 | 0.32 ± 0.063 | 0.057 ± 0.077 | 0.31 ± 0.054 | 0.021 ± 0.044 | 0.046 | NS | 0.82 | NS | 0.95 | NS |
| Vitamin K-2 biosynthesis | PWY.5845 | 0.023 ± 0.029 | 0.00 ± 0.038 | 0.025 ± 0.039 | 0.0022 ± 0.025 | 0.19 | NS | 0.037 | NS | 0.52 | NS |
| Superpathway of ubiquinol-8 biosynthesis | UBISYN.PWY | 0.025 ± 0.023 | 0.00 ± 0.022 | 0.028 ± 0.028 | 0.0013 ± 0.021 | 0.61 | NS | 0.024 | NS | 1.0 | NS |
Values are presented as group means ± SD, n = 16 (RW) or n = 21 (WGW). FDR, false discovery rate; PWY, pathway; RW, refined wheat; WGW, whole grain wheat; Δ, differential change over time.
2NS, nonsignificant: P value >0.05.
FIGURE 5.
Predicted fecal microbial pathway relative abundance at baseline and after 12 wk of RW or WGW intervention that were found to be significantly different between the groups in middle-aged overweight and obese adults. The relative abundances of the predicted pathways (A) superpathway of hexitol degradation; (B) pantothenate and coenzyme A biosynthesis; (C) acetyl-CoA fermentation to butyrate II; (D) pyruvate fermentation to acetone; (E) aromatic biogenic amine degradation; and (F) l-alanine biosynthesis are shown. Data are presented as group mean (the squared shape), n = 16 (RW) or n = 21 (WGW), and the width of the colored shapes indicates the sample density. Individual paired samples are connected by a line. RW, refined wheat; WGW, whole grain wheat.
Because changes in predicted fermentation pathways were found, we examined the effect of the interventions on selected genes involved in SCFA production (Supplemental Table 6). No significant differences in changes of these predicted genes were observed over time between groups, but butyrate kinase was significantly increased within the WGW group (P = 0.038), and phosphate acetyltransferase was significantly decreased within the RW group (P = 0.021). Overall, WGW intervention showed a trend toward increased predicted fermentation pathways by the microbiota, whereas RW intervention showed opposite effects.
Discussion
We investigated the effects of 12-wk consumption of RW or WGW products on the gut microbiota composition and predicted microbiota functional pathways in men and women with overweight or obesity. We found significant differences between the WGW and RW intervention for a number of bacterial taxa from carbohydrate-degrading families and on predicted fermentation pathways, with a trend toward increased fermentation to butyrate within the WGW group, and a trend toward decreased fermentation within the RW group (a graphical overview is provided in Supplemental Figure 6). Although WGW consumption is already quite high in The Netherlands, we provided an intervention with 98 g/d WGW, which was higher than the mean habitual WG intake at baseline of participants in both the RW and WGW groups, of 60.1 and 54.4 g/d WG, respectively.
Even though the difference in fiber between the WGW and RW intervention was 10 g/d, we observed only subtle differences between intervention groups over time with respect to microbiota composition and functionality, and no effect on stool consistency. Microbial diversity within samples as calculated previously by the Shannon Index was decreased after RW intervention in Schutte et al. (7), but we did not find a significant difference in microbial diversity changes between groups in a subset of study participants with the phylogenetic diversity index, which takes into account phylogenetic relatedness between bacteria. Twelve weeks of a WGW intervention increased relative abundances of Ruminiclostridium_9, Ruminococcaceae_NK4A214_group, and Ruminococcaceae_UCG-014, and decreased Lachnospiraceae_UCG-008, whereas a 12-wk RW intervention decreased abundance, without significant effects within the groups. Ruminococcaceae genera have previously been shown to increase after a resistant starch or nonstarch-polysaccharide diet high in wheat bran mainly comprised of hemicellulose (42,43). Members of the Ruminococcaceae can degrade cellulose and hemicellulose fibers (44–46), also present in WGW, whereas members of the Lachnospiraceae are known to ferment a wide variety of fibers (44, 46). In line with our results, Vuholm et al. (47) found an increase in an unassigned genus of the family Ruminococcaceae after 6 wk of WGW consumption, in healthy overweight females.
We found the change in relative abundance of Ruminococcaceae_NK4A214_group and the intervention-induced change in liver fat correlated in both groups. Several studies showed that Ruminococcaceae was lower in the feces of NAFLD patients compared with controls (48–50). However, in this study we found a positive correlation between a change in Ruminococcaceae_NK4A214_group and a change in liver fat in both intervention groups, although the RW intervention increased liver fat and decreased abundance of Ruminococcaceae_NK4A214_group indicating that the changes relative to each other did not have a strong correlation. At baseline, however, we found a positive correlation between liver fat and Ruminococcaceae_UCG-010, and Ruminococcaceae_UCG-005. We could not identify any human trials involving these specific bacteria in relation to liver fat. As in many previous dietary trials, the microbiota response to diet was subject-dependent. The microbiota at the start of intervention of any individual can determine the magnitude of response upon dietary changes (43). Korem et al. (51) found that the gut microbiota composition was person-specific and generally resilient to bread interventions, but their intervention lasted only 1 wk whereas our study encompassed a 12-wk intervention. In contrast to previous findings (17, 18), WGW consumption did not increase Bifidobacterium and Lactobacillus in our study. This discrepancy could be partly explained by differences in the fermentable fiber composition and fractions in the intervention products. For instance, fructans can have prebiotic activity by inducing specific changes in the composition and/or activity of the microbiota such as Bifidobacterium and Lactobacillus (52). It has previously been described that the concentration of fructans differs in WGW (53).
Ruminococcaceae and Lachnospiraceae, together with Bacteroidetes, encompass ∼85% of the total butyrate-producing potential of the gut microbiota (54). Changes in bacterial taxa within the Ruminococcaceae might point toward an effect on carbohydrate breakdown, which could fit the predicted changes in SCFA fermentation pathways in feces. WGW increased the predicted relative abundance of fermentation to butyrate, whereas RW lowered the fermentation to butyrate as well as fermentation to acetone. The latter pathway can lead to formation of products such as acetone, butanol, ethanol, acetate, and butyrate. Moreover, a decreased fermentation to acetate and lactate was found within the RW group. Lactate-utilizing bacteria can use acetate and lactate for the production of butyrate (55), indicating that a lowered capacity to produce acetate and lactate potentially leads to a reduced butyrate production. Despite the run-in period, the RW group showed a higher relative abundance for the predicted pyruvate fermentation to acetone pathway and for Ruminiclostridium_9 compared with the WGW group at baseline. Both were decreased after RW and increased after WGW. Similarly, a higher abundance of Lachnospiraceae_UCG-008 was observed in the RW group at baseline, which increased after RW and decreased after WGW. Therefore, the differential effects on this pathway and these bacteria between RW or WGW intervention could have been partly caused by regression to the mean. In this study, the effects on microbiota and predicted pathways lost statistical significance after correction for multiple comparisons, which shows that a 12-wk WGW and RW intervention induced subtle changes in gut microbiota. This is not surprising considering the relatively modest modulation of diet, only altering wheat products.
In line with our finding that WGW increased 2 predicted fermentation pathways, Vanegas et al. (20) reported increased concentrations of acetate and total SCFAs in feces after 6 wk of a WG, predominantly wheat, intervention compared with refined grain in middle-aged adults. Moreover, Vuholm et al. (47) found that a 6-wk RW intervention decreased fecal butyrate concentrations when compared with a WGW intervention in adults with overweight. Additionally, a study in rats showed that a 6-wk WGW intervention increased total SCFAs in colonic content and butyrate in cecum content compared with RW intervention (56). Resistant starch and nonstarch polysaccharide inside the whole-grain matrix were associated with increased production of metabolites (57). Overall, incorporating feasible doses of WGW in the diet favorably affects the gut microbiota phenotype as indicated by the increased predicted potential to produce butyrate. These findings can be explained by the decreased fiber content in the RW intervention products (12, 13), although only a fraction of the 17.6 g/d fiber in the WGW intervention is fermentable. The fermentability of fibers in WGW is relatively low, for instance, when compared with fibers in WG rye (58, 59). Eriksen et al. (60) showed that WG rye resulted in increased Bifidobacterium, known to be stimulated by some fermentable fibers, in men with the metabolic syndrome, whereas WG wheat did not.
The reduced potential to produce SCFAs after RW intervention can have implications for health. For instance, SCFAs can decrease intestinal inflammation, as demonstrated by human and animal in vivo studies (61). In addition to local health effects, SCFAs might also influence liver fat through stimulation of hepatic fat oxidation via activation of AMP-activated protein kinase (28, 29). As published previously, in this trial, the 12-wk RW intervention significantly increased liver fat, whereas liver fat did not change in the WGW group (7). Therefore, we hypothesized that RW might increase liver fat content indirectly via decreased cereal fiber fermentation and SCFA production. We found that butyrate-producing (62) Roseburia positively correlated with liver fat at baseline. In line with our data, the study of Raman et al. (48) showed a significant overrepresentation of Roseburia in the microbiota of NAFLD patients. In contrast, Roseburia was decreased in nonobese NAFLD patients compared with healthy controls, and this depletion was linked to increased plasma ALT in 126 nonobese subjects (63). Several of our correlations of other bacteria with liver fat at baseline were in line with previous findings on bacterial composition in NAFLD patients compared with controls (64, 65). This was the case for Ruminococcus_2 (64) and Akkermansia (65), but for Faecalibacterium opposite effects were observed (50). Although a previous study showed that ALT and GGT were correlated with changes in fecal bacteria over time in a prospective, cross-sectional study (63), we could not confirm these findings in our correlation analysis. Although correlations do not provide information on causality, these outcomes might be of interest for future studies into the relation between WGW, RW, and liver health.
A limitation of our study is the analysis of microbiota functional metagenomes by PICRUSt2 based on the 16S rRNA sequencing data, instead of using more direct methods, such as metagenomics. Although the quality control indicated that closely related reference genomes were available for the bacteria present in this dataset, indicative for reliable pathway predictions, PICRUSt is subject to inherit biases. Another restraint of our study is that the SCFA concentrations in feces were not measured. Therefore, the predicted pathway findings as well as the potential role of SCFAs for liver health could not be validated in this study. Another limitation of our study is the small sample size of 37 subjects. Because the study was powered to detect changes in plasma cholesterol concentrations and participants had to be excluded due to missing fecal samples or antibiotic use, we might have missed the effects of either RW or WGW on the microbiota or predicted microbiota functionality.
The strengths of our study include the long study duration of 16 wk in total, and the good compliance with the diet based upon alkylresorcinol data (7). Therefore, our findings could show more long-term changes rather than acute effects. Comparisons with outcomes of existing literature about WGW, RW, and microbiota composition revealed few similarities with our trial (17, 18, 19, 24, 47). Some previous trials could have missed the subtle effects of WGW and RW on the microbiota because only a few selected bacterial taxa were targeted in these studies via, for example, qPCR (17, 18, 24), whereas the fecal microbiota is a highly complex community with multiple species present. The use of 16S rRNA sequencing provided important insights into the effect of WGW and RW on microbiota as a whole, as well as effects on predicted microbial community functionality.
In conclusion, we demonstrated that a 12-wk 98 g/d WGW intervention increased relative abundances of a number of bacterial taxa that are involved in carbohydrate degradation and SCFA production and predicted fermentation pathways, whereas an RW intervention decreased abundance of these bacteria or predicted fermentation capacity, pointing toward a less healthy gut microbiota phenotype. The difference in fiber intake during the WGW intervention (17.6 g/d fiber) compared with the RW intervention (7.2 g/d fiber) likely resulted in differences in predicted bacterial fermentation capacity. This may be one of the mechanisms underlying the significant increases of liver fat observed with RW intervention. Potential health effects of replacement of RW by WGW via modulation of the microbiota, and consequent protective effects on metabolic organs such as the liver, deserve further investigation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the participants involved in the study. The MyNewGut project (financed by the European Commission in the 7th Framework Program FP7, grant agreement 613979) is acknowledged for sponsoring the microbiota analyses.
The authors’ responsibilities were as follows––DE, SW, and LAA: designed the study; SS, DE, and LAA: responsible for the execution of the trial and data collection; MPHvT: analyzed the data and discussed with LAA and GJEJH; MPHvT: wrote the first draft, which was improved by LAA; GJEJH, LAA, SW, and FPMH: critically reviewed and improved the manuscript; and all authors: read and approved the final manuscript.
Notes
This research was supported by the public-private partnership “Combining innovation with tradition: improving resilience with essential nutrients and whole wheat bread,” financed by Topsector Agri & Food (TKI-AF 12083) and TNO roadmap Nutrition & Health and co-funded by Cereal Partners Worldwide, the Dutch Bakery Center, and GoodMills Innovation GmbH.
Author disclosures: The authors report no conflicts of interest.
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Supplemental Methods, Supplemental Results, Supplemental Figures 1–6, and Supplemental Tables 1–6 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviation used: ALT, alanine transaminase; AST, aspartate transaminase; ASV, amplicon sequence variant; CRP, C-reactive protein; FDR, false discovery rate; GGT, γ-glutamyltransferase; IHTG, intrahepatic triglyceride; KEGG, Kyoto Encyclopedia of Genes and Genomes; NAFLD, nonalcoholic fatty liver disease; PD, phylogenetic diversity; PERMANOVA, permutational multivariate analysis of variance; PICRUSt, phylogenetic investigation of communities by reconstruction of unobserved states; rRNA, ribosomal ribonucleic acid; RW, refined wheat; SAA, serum amyloid A; WG, whole grain; WGW, whole grain wheat; Δ, differential change over time.
Contributor Information
Mara P H van Trijp, Nutrition, Metabolism & Genomics Group, Division of Human Nutrition and Health, Wageningen University, Wageningen, The Netherlands.
Sophie Schutte, Nutrition, Metabolism & Genomics Group, Division of Human Nutrition and Health, Wageningen University, Wageningen, The Netherlands.
Diederik Esser, Nutrition, Metabolism & Genomics Group, Division of Human Nutrition and Health, Wageningen University, Wageningen, The Netherlands.
Suzan Wopereis, TNO, Netherlands Organization for Applied Scientific Research, Zeist, The Netherlands.
Femke P M Hoevenaars, TNO, Netherlands Organization for Applied Scientific Research, Zeist, The Netherlands.
Guido J E J Hooiveld, Nutrition, Metabolism & Genomics Group, Division of Human Nutrition and Health, Wageningen University, Wageningen, The Netherlands.
Lydia A Afman, Nutrition, Metabolism & Genomics Group, Division of Human Nutrition and Health, Wageningen University, Wageningen, The Netherlands.
References
- 1. de Munter JS, Hu FB, Spiegelman D, Franz M, van Dam RM. Whole grain, bran, and germ intake and risk of type 2 diabetes: a prospective cohort study and systematic review. PLoS Med. 2007;4(8):e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Truswell A. Cereal grains and coronary heart disease. Eur J Clin Nutr. 2002;56(1):1. [DOI] [PubMed] [Google Scholar]
- 3. Sahyoun NR, Jacques PF, Zhang XL, Juan W, McKeown NM. Whole-grain intake is inversely associated with the metabolic syndrome and mortality in older adults. Am J Clin Nutr. 2006;83(1):124–31. [DOI] [PubMed] [Google Scholar]
- 4. Ross AB, Godin J-P, Minehira K, Kirwan JP. Increasing whole grain intake as part of prevention and treatment of nonalcoholic fatty liver disease. Int J Endocrinol. 2013;2013:585876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartłomiej S, Justyna R-K, Ewa N. Bioactive compounds in cereal grains—occurrence, structure, technological significance and nutritional benefits—a review. Food Sci Technol Int. 2012;18(6):559–68. [DOI] [PubMed] [Google Scholar]
- 6. McKevith B. Nutritional aspects of cereals. Nutr Bull. 2004;29(2):111–42. [Google Scholar]
- 7. Schutte S, Esser D, Hoevenaars FP, Hooiveld GJ, Priebe MG, Vonk RJ, Wopereis S, Afman LA. A 12-wk whole-grain wheat intervention protects against hepatic fat: the Graandioos study, a randomized trial in overweight subjects. Am J Clin Nutr. 2018;108(6):1264–74. [DOI] [PubMed] [Google Scholar]
- 8. Kirwan JP, Malin SK, Scelsi AR, Kullman EL, Navaneethan SD, Pagadala MR, Haus JM, Filion J, Godin JP, Kochhar S et al. A whole-grain diet reduces cardiovascular risk factors in overweight and obese adults: a randomized controlled trial. J Nutr. 2016;146(11):2244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tighe P, Duthie G, Vaughan N, Brittenden J, Simpson WG, Duthie S, Mutch W, Wahle K, Horgan G, Thies F. Effect of increased consumption of whole-grain foods on blood pressure and other cardiovascular risk markers in healthy middle-aged persons: a randomized controlled trial. Am J Clin Nutr. 2010;92(4):733–40. [DOI] [PubMed] [Google Scholar]
- 10. Hollænder PL, Ross AB, Kristensen M. Whole-grain and blood lipid changes in apparently healthy adults: a systematic review and meta-analysis of randomized controlled studies. Am J Clin Nutr. 2015;102(3):556–72. [DOI] [PubMed] [Google Scholar]
- 11. Fardet A. New hypotheses for the health-protective mechanisms of whole-grain cereals: what is beyond fibre? Nutr Res Rev. 2010;23(1):65–134. [DOI] [PubMed] [Google Scholar]
- 12. Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23(6):705–15. [DOI] [PubMed] [Google Scholar]
- 13. Holscher HD. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut microbes. 2017;8(2):172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kortman GA, Dutilh BE, Maathuis AJ, Engelke UF, Boekhorst J, Keegan KP, Nielsen FG, Betley J, Weir JC, Kingsbury Z. Microbial metabolism shifts towards an adverse profile with supplementary iron in the TIM-2 in vitro model of the human colon. Front Microbiol. 2016;6:1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fehlbaum S, Prudence K, Kieboom J, Heerikhuisen M, van den Broek T, Schuren F, Steinert R, Raederstorff D. In vitro fermentation of selected prebiotics and their effects on the composition and activity of the adult gut microbiota. IJMS. 2018;19(10):3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jefferson A, Adolphus K. The effects of intact cereal grain fibers, including wheat bran on the gut microbiota composition of healthy adults: a systematic review. Front Nutr. 2019;6(33). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Christensen EG, Licht TR, Kristensen M, Bahl MI. Bifidogenic effect of whole-grain wheat during a 12-week energy-restricted dietary intervention in postmenopausal women. Eur J Clin Nutr. 2013;67(12):1316–21. [DOI] [PubMed] [Google Scholar]
- 18. Costabile A, Klinder A, Fava F, Napolitano A, Fogliano V, Leonard C, Gibson GR, Tuohy KM. Whole-grain wheat breakfast cereal has a prebiotic effect on the human gut microbiota: a double-blind, placebo-controlled, crossover study. Br J Nutr. 2008;99(1):110–20. [DOI] [PubMed] [Google Scholar]
- 19. Vitaglione P, Mennella I, Ferracane R, Rivellese AA, Giacco R, Ercolini D, Gibbons SM, La Storia A, Gilbert JA, Jonnalagadda S et al. Whole-grain wheat consumption reduces inflammation in a randomized controlled trial on overweight and obese subjects with unhealthy dietary and lifestyle behaviors: role of polyphenols bound to cereal dietary fiber. Am J Clin Nutr. 2015;101(2):251–61. [DOI] [PubMed] [Google Scholar]
- 20. Vanegas SM, Meydani M, Barnett JB, Goldin B, Kane A, Rasmussen H, Brown C, Vangay P, Knights D, Jonnalagadda S. Substituting whole grains for refined grains in a 6-wk randomized trial has a modest effect on gut microbiota and immune and inflammatory markers of healthy adults. Am J Clin Nutr. 2017;105(3):635–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lappi J, Salojärvi J, Kolehmainen M, Mykkänen H, Poutanen K, de Vos WM, Salonen A. Intake of whole-grain and fiber-rich rye bread versus refined wheat bread does not differentiate intestinal microbiota composition in Finnish adults with metabolic syndrome. J Nutr. 2013;143(5):648–55. [DOI] [PubMed] [Google Scholar]
- 22. Ampatzoglou A, Atwal KK, Maidens CM, Williams CL, Ross AB, Thielecke F, Jonnalagadda SS, Kennedy OB, Yaqoob P. Increased whole grain consumption does not affect blood biochemistry, body composition, or gut microbiology in healthy, low-habitual whole grain consumers. J Nutr. 2015;145(2):215–21. [DOI] [PubMed] [Google Scholar]
- 23. Cooper DN, Kable ME, Marco ML, De Leon A, Rust B, Baker JE, Horn W, Burnett D, Keim NL. The effects of moderate whole grain consumption on fasting glucose and lipids, gastrointestinal symptoms, and microbiota. Nutrients. 2017;9(2):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bird AR, Vuaran MS, King RA, Noakes M, Keogh J, Morell MK, Topping DL. Wholegrain foods made from a novel high-amylose barley variety (Himalaya 292) improve indices of bowel health in human subjects. Br J Nutr. 2008;99(5):1032–40. [DOI] [PubMed] [Google Scholar]
- 25. Slavin J. Why whole grains are protective: biological mechanisms. Proc Nutr Soc. 2003;62(1):129–34. [DOI] [PubMed] [Google Scholar]
- 26. Boets E, Gomand SV, Deroover L, Preston T, Vermeulen K, Preter V, Hamer HM, den Mooter G, Vuyst L, Courtin CM. Systemic availability and metabolism of colonic‐derived short‐chain fatty acids in healthy subjects: a stable isotope study. J Physiol. 2017;595(2):541–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11(10):577. [DOI] [PubMed] [Google Scholar]
- 28. den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, Oosterveer MH, Jonker JW, Groen AK, Reijngoud D-J. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes. 2015;64(7):2398–408. [DOI] [PubMed] [Google Scholar]
- 29. Mollica MP, Raso GM, Cavaliere G, Trinchese G, De Filippo C, Aceto S, Prisco M, Pirozzi C, Di Guida F, Lama A. Butyrate regulates liver mitochondrial function, efficiency, and dynamic, in insulin resistant obese mice. Diabetes. 2017;66(5):1405–18. [DOI] [PubMed] [Google Scholar]
- 30. Kobyliak N, Abenavoli L, Mykhalchyshyn G, Kononenko L, Boccuto L, Kyriienko D, Dynnyk O. A multi-strain probiotic reduces the fatty liver index, cytokines and aminotransferase levels in NAFLD patients: evidence from a randomized clinical trial. J Gastrointestin Liver Dis. 2018;27(1):41–9. [DOI] [PubMed] [Google Scholar]
- 31. Malaguarnera M, Vacante M, Antic T, Giordano M, Chisari G, Acquaviva R, Mastrojeni S, Malaguarnera G, Mistretta A, Li Volti G et al. Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Dig Dis Sci. 2012;57(2):545–53. [DOI] [PubMed] [Google Scholar]
- 32. Manzhalii E, Virchenko O, Falalyeyeva T, Beregova T, Stremmel W. Treatment efficacy of a probiotic preparation for non-alcoholic steatohepatitis: a pilot trial. J Dig Dis. 2017;18(12):698–703. [DOI] [PubMed] [Google Scholar]
- 33. Aller R, De Luis DA, Izaola O, Conde R, Gonzalez Sagrado M, Primo D, De La Fuente B, Gonzalez J. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: a double blind randomized clinical trial. Eur Rev Med Pharmacol Sci. 2011;15(9):1090–5. [PubMed] [Google Scholar]
- 34. Hoevenaars FPM, Esser D, Schutte S, Priebe MG, Vonk RJ, van den Brink WJ, van der Kamp JW, Stroeve JHM, Afman LA, Wopereis S. Whole grain wheat consumption affects postprandial inflammatory response in a randomized controlled trial in overweight and obese adults with mild hypercholesterolemia in the Graandioos study. J Nutr. 2019;149(12):2133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramiro-Garcia J, Hermes GD, Giatsis C, Sipkema D, Zoetendal EG, Schaap PJ, Smidt H. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Res. 2018;5:1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Poncheewin W, Hermes GDA, van Dam JCJ, Koehorst JJ, Smidt H, Schaap PJ. NG-Tax 2.0: a semantic framework for high-throughput amplicon analysis. Front Genet. 2019;10:1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Team RC . R: A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2007. [Google Scholar]
- 38. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oksanen J, Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P, O'Hara R, Simpson G, Solymos P. vegan: Community Ecology Package. R package version 2.5-2. R Foundation; 2018. [Google Scholar]
- 41. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, Date P, Farquharson F, Johnstone AM, Lobley GE et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB. Bergey's manual of systematic bacteriology. Vol. 3. The Firmicutes: Springer Science & Business Media; 2011. [Google Scholar]
- 45. Robert C, Bernalier-Donadille A. The cellulolytic microflora of the human colon: evidence of microcrystalline cellulose-degrading bacteria in methane-excreting subjects. FEMS Microbiol Ecol. 2003;46(1):81–9. [DOI] [PubMed] [Google Scholar]
- 46. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3(4):289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vuholm S, Nielsen DS, Iversen KN, Suhr J, Westermann P, Krych L, Andersen JR, Kristensen M. Whole-grain rye and wheat affect some markers of gut health without altering the fecal microbiota in healthy overweight adults: a 6-week randomized trial. J Nutr. 2017;147(11):2067–75. [DOI] [PubMed] [Google Scholar]
- 48. Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11(7):868–75.e3. [DOI] [PubMed] [Google Scholar]
- 49. Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, Hu Y, Li J, Liu Y. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5(1):8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Iino C, Endo T, Mikami K, Hasegawa T, Kimura M, Sawada N, Nakaji S, Fukuda S. Significant decrease in Faecalibacterium among gut microbiota in nonalcoholic fatty liver disease: a large BMI- and sex-matched population study. Hepatol Int. 2019;13(6):748–56. [DOI] [PubMed] [Google Scholar]
- 51. Korem T, Zeevi D, Zmora N, Weissbrod O, Bar N, Lotan-Pompan M, Avnit-Sagi T, Kosower N, Malka G, Rein M. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab. 2017;25(6):1243. [DOI] [PubMed] [Google Scholar]
- 52. Vandeputte D, Falony G, Vieira-Silva S, Wang J, Sailer M, Theis S, Verbeke K, Raes J. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut. 2017;66(11):1968–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Markowiak P, Śliżewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients. 2017;9(9):1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vital M, Karch A, Pieper DH. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems. 2017;2(6):e00130–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Muñoz-Tamayo R, Laroche B, Walter E, Doré J, Duncan SH, Flint HJ, Leclerc M. Kinetic modelling of lactate utilization and butyrate production by key human colonic bacterial species. FEMS Microbiol Ecol. 2011;76(3):615–24. [DOI] [PubMed] [Google Scholar]
- 56. Han F, Wang Y, Han Y, Zhao J, Han F, Song G, Jiang P, Miao H. Effects of whole-grain rice and wheat on composition of gut microbiota and short-chain fatty acids in rats. J Agric Food Chem. 2018;66(25):6326–35. [DOI] [PubMed] [Google Scholar]
- 57. Rose DJ. Impact of whole grains on the gut microbiota: the next frontier for oats? Br J Nutr. 2014;112(S2):S44–S9. [DOI] [PubMed] [Google Scholar]
- 58. Andersson AA, Andersson R, Piironen V, Lampi A-M, Nyström L, Boros D, Fraś A, Gebruers K, Courtin CM, Delcour JA. Contents of dietary fibre components and their relation to associated bioactive components in whole grain wheat samples from the HEALTHGRAIN diversity screen. Food Chem. 2013;136(3-4):1243–8. [DOI] [PubMed] [Google Scholar]
- 59. Andersson R, Fransson G, Tietjen M, Aman P. Content and molecular-weight distribution of dietary fiber components in whole-grain rye flour and bread. J Agric Food Chem. 2009;57(5):2004–8. [DOI] [PubMed] [Google Scholar]
- 60. Eriksen AK, Brunius C, Mazidi M, Hellström PM, Risérus U, Iversen KN, Fristedt R, Sun L, Huang Y, Nørskov NP et al. Effects of whole-grain wheat, rye, and lignan supplementation on cardiometabolic risk factors in men with metabolic syndrome: a randomized crossover trial. Am J Clin Nutr. 2020;111(4):864–76. [DOI] [PubMed] [Google Scholar]
- 61. Bach Knudsen KE, Lærke HN, Hedemann MS, Nielsen TS, Ingerslev AK, Gundelund Nielsen DS, Theil PK, Purup S, Hald S, Schioldan AG et al. Impact of diet-modulated butyrate production on intestinal barrier function and inflammation. Nutrients. 2018;10(10):1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294(1):1–8. [DOI] [PubMed] [Google Scholar]
- 63. Wang B, Jiang X, Cao M, Ge J, Bao Q, Tang L, Chen Y, Li L. Altered fecal microbiota correlates with liver biochemistry in nonobese patients with non-alcoholic fatty liver disease. Sci Rep. 2016;6:32002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D, Furlanello C, Zandonà A, Paci P, Capuani G et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65(2):451–64. [DOI] [PubMed] [Google Scholar]
- 65. Nistal E, Sáenz de Miera LE, Ballesteros Pomar M, Sánchez-Campos S, García-Mediavilla MV, Álvarez-Cuenllas B, Linares P, Olcoz JL, Arias-Loste MT, García-Lobo JM et al. An altered fecal microbiota profile in patients with non-alcoholic fatty liver disease (NAFLD) associated with obesity. Rev Esp Enferm Dig. 2019;111(4):275–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.