Abstract
Uveal melanoma is the most common eye cancer in adults and is clinically and genetically distinct from skin cutaneous melanoma. In a subset of cases, the oncogenic driver is an activating mutation in CYSLTR2, the gene encoding the G protein–coupled receptor cysteinyl-leukotriene receptor 2 (CysLTR2). The mutant CYSLTR2 encodes for the CysLTR2–L129Q receptor, with the substitution of Leu to Gln at position 129 (3.43). The ability of CysLTR2–L129Q to cause malignant transformation has been hypothesized to result from constitutive activity, but how the receptor could escape desensitization is unknown. Here, we characterize the functional properties of CysLTR2–L129Q. We show that CysLTR2–L129Q is a constitutively active mutant that strongly drives Gq/11 signaling pathways. However, CysLTR2–L129Q only poorly recruits β-arrestin. Using a modified Slack–Hall operational model, we quantified the constitutive activity for both pathways and conclude that CysLTR2–L129Q displays profound signaling bias for Gq/11 signaling pathways while escaping β-arrestin–mediated downregulation. CYSLTR2 is the first known example of a G protein–coupled receptor driver oncogene that encodes a highly biased constitutively active mutant receptor. These results provide new insights into the mechanism of CysLTR2–L129Q oncoprotein signaling and suggest CYSLTR2 as a promising potential therapeutic target in uveal melanoma.
Keywords: G protein–coupled receptor, G protein, β-arrestins, uveal melanoma, signaling, CysLTR2, biased signaling, constitutive activity
Abbreviations: BRET, bioluminescent resonance energy transfer; BRET2, bioluminescent resonance energy transfer 2; BSA, bovine serum albumin; CA, constitutive activity; CAM, constitutively active mutant; CI, confidence interval; CysLTR2, cysteinyl-leukotriene receptor 2; DPBS, Dulbecco’s PBS; EPAC, exchange protein activated by cAMP; FBS, fetal bovine serum; GPCR, G protein–coupled receptor; HTRF, homogeneous time-resolved fluorescence immunoassay; IP1, d-myo-inositol-1-phosphate; LiCl, lithium chloride; LTD4, leukotriene D4; MV, missense variant; PLCβ, phospholipase C-β; RLuc3, Renilla Luciferase; UVM, uveal melanoma; V2(A)6, hexa-alanine variant
The superfamily of G protein–coupled receptors (GPCRs) is the largest gene family encoding cell-signaling transmembrane proteins, and approximately one-quarter of ∼400 nonolfactory GPCRs are therapeutic drug targets. Large-scale genomic analysis has revealed that one in five individuals carries a missense variant (MV) in a clinically relevant GPCR gene. The rate of de novo germline MVs in a GPCR gene is one in every 300 newborns, and one in 7 MVs is observed at a potential functionally relevant site (1). In addition, GPCR genes are commonly mutated in cancer. Somatic mutations are found in 20% of tumor samples, but the lack of specific “hotspot” variants makes it difficult to identify and validate individual receptors as driver oncogenes (2).
We recently reported the discovery of a recurrent “hotspot” somatic missense mutation of the GPCR gene CYSLTR2. The mutant CYSLTR2 encodes cysteinyl-leukotriene receptor 2 (CysLTR2)–L129Q that carries a single amino acid substitution at a highly conserved residue in helix 3 (Ballesteros–Weinstein generic position 3.43) (3) and serves as a driver oncogene in patients with uveal melanoma (UVM) (2). UVM is the most common intraocular malignancy and is associated with a high rate of metastasis with short survival time for patients (4). UVM shows a characteristic pattern of mutually exclusive activating mutations in the CysLTR2–Gq/11–PLCβ4 (phospholipase C-β4) pathway in almost all tumors (2, 5, 6). The same CysLTR2–L129Q mutation has also been identified as an oncogenic driver mutation in several other melanocytic tumors (7). CysLTR2 is a significantly mutated GPCR not only in UVM but also in gastrointestinal adenocarcinoma (8).
Here, we report that the UVM oncogene product CysLTR2–L129Q has a unique gain-of-function phenotype and report its precise signaling mechanism. We show that CysLTR2–L129Q is a constitutively active mutant (CAM) receptor that strongly couples to Gq/11 cellular signaling pathways. However, the receptor CAM only very weakly recruits β-arrestins and thereby avoids cellular downregulation mechanisms. We quantified the signaling bias of the mutant receptor by comparing the constitutive activity (CA) for the WT and L129Q mutant using values calculated from the modified Slack–Hall operational model. Finally, we showed that the receptor bias of the L129Q CAM toward Gq and away from β-arrestins is due to the unusual C-terminal sequence of the receptor.
Results and discussion
CysLTR2–L129Q signals through Gq/11-PLCβ pathways
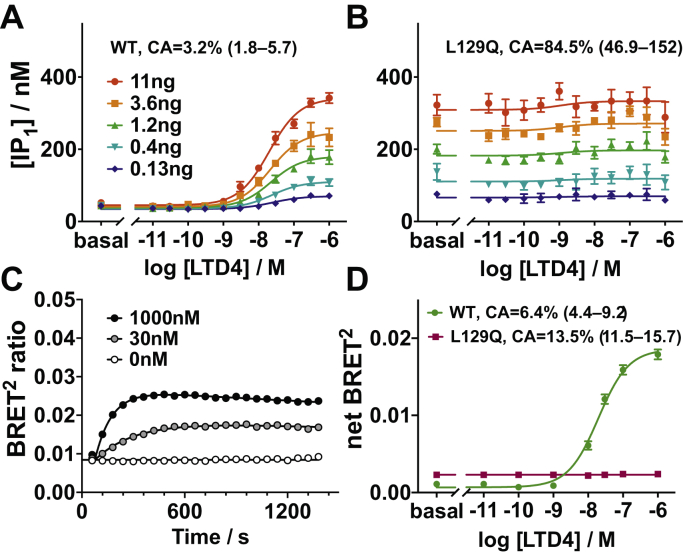
To characterize the functional phenotype of CysLTR2–L129Q, we first determined the agonist-dependent signaling for CysLTR2–L129Q and CysLTR2 WT. CysLTR2 predominantly couples to Gq/11 when treated with the agonist leukotriene D4 (LTD4) (9). Phospholipase C-β (PLCβ) is the classical effector of Gq/11 and results in receptor-stimulated phosphoinositide hydrolysis that is conveniently monitored as an accumulation of d-myo-inositol-1-phosphate (IP1) in the presence of lithium chloride (LiCl) (10). We first obtained a time course of basal and LTD4-dependent IP1 accumulation in HEK293T cells transiently transfected with plasmids for CysLTR2 WT, CysLTR2–L129Q, and mock controls. LTD4-stimulated CysLTR2 WT showed increasing IP1 accumulation over the first 100 min before reaching a plateau (Fig. S1A), whereas the unstimulated CysLTR2 WT samples were indistinguishable from mock-transfected controls with and without LTD4 treatment. The samples transfected with the same amount of DNA encoding for the CysLTR2–L129Q mutant showed ligand-independent IP1 accumulation of comparable magnitude as LTD4-treated CysLTR2 WT. After 100 min, the basal IP1 accumulation of CysLTR2–L129Q kept increasing, whereas the ligand-dependent signaling of the WT receptor reached a plateau (Fig. S1B).
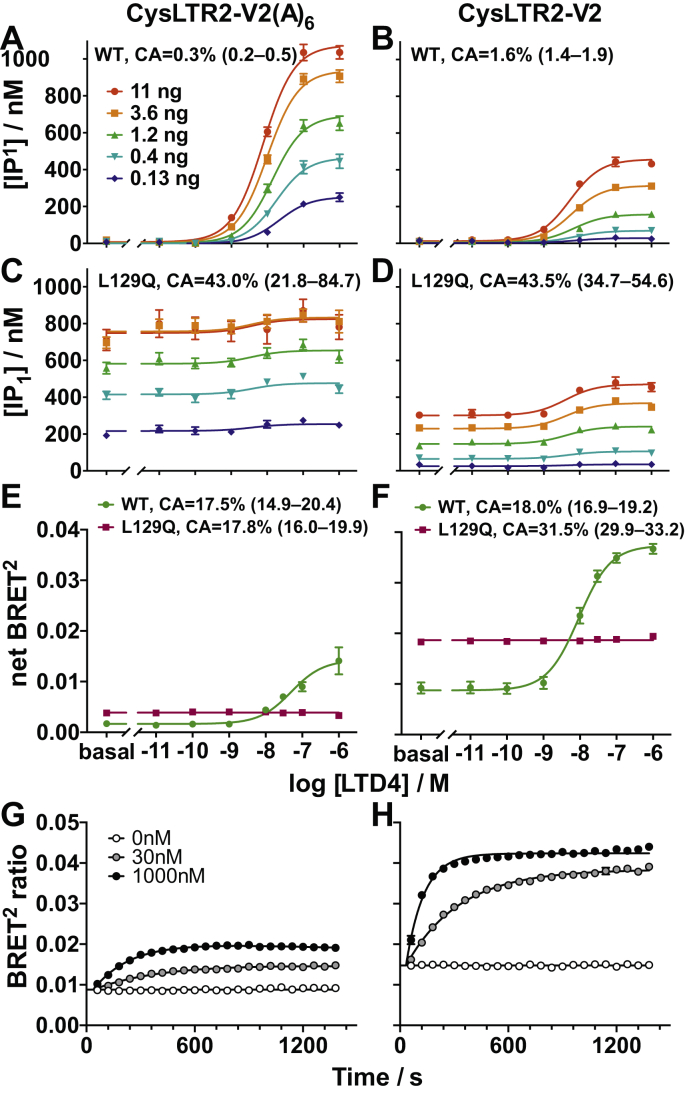
We generated fusion constructs of CysLTR2 WT and CysLTR2–L129Q, with a version of GFP (GFP10) at the C-terminus. These fusion constructs enable quantification of basal and agonist-dependent Gq/11 cellular signaling and β-arrestin recruitment activity under comparable conditions. The agonist LTD4 induces a dose-dependent increase in IP1 accumulation for CysLTR2 WT (Fig. 1A) that scales as expected with the receptor density controlled by the gene dosage as described by a modified Slack–Hall operational model (Fig. 2, Table S1B). In contrast, the CysLTR2–L129Q mutant shows little or no response to treatment with LTD4, but the ligand-independent basal IP1 accumulation dramatically increases with increasing gene dosage (Fig. 1B). Therefore, the mutation is meeting the essential criteria for a CAM because the hallmark of CAM receptors is agonist-independent signaling that scales with receptor density. The results also show that under the experimental conditions, the Gq signaling pathway is not saturated, which suggests that the high basal receptor activation of Gq for CysLTR2–L129Q is not due to a high amplification of the Gq signaling pathway.
Figure 1.
Oncoprotein CysLTR2–L129Q is a Gq-biased CAM that only weakly recruits β-arrestin2. (A and B) Gq second messenger IP1 accumulation assay. A, Agonist LTD4 leads to dose-dependent IP1 accumulation in HEK293T cells transfected with different amounts (0.13–11 ng DNA, blue to red) of CysLTR2-GFP10 WT. B, The corresponding experiment with CysLTR2–L129Q shows dramatic agonist-independent basal IP1 accumulation that scales with the amount of CysLTR2–L129Q–encoding DNA. Only a very small agonist-dependent response to LTD4 can be detected. The results show that CysLTR2–L129Q is a CAM with 85% constitutive activity relative to fully agonist-stimulated WT receptor. Data points are the mean ± SEM of the accumulated IP1 concentration and from one experiment with four replicates each. The set of curves are fits to the Slack–Hall operational model (Table S1B, Fig. 2). C and D, β-arrestin2-recruitment BRET2 assays. C, Time course of LTD4-stimulated β-arrestin2 recruitment for three LTD4 concentrations (0 nM, 30 nM, and 1000 nM). β-arrestin2 recruitment exhibits a biphasic time course, increasing for about 10 min after LTD4 addition before slowly decreasing. The data points are the mean ± SEM from three independent experiments with eight replicates each. Curves are double exponential fits (Table S1C). D, The LTD4 dose-dependent β-arrestin2 recruitment to CysLTR2 WT (dark green; EC50 is 30 nM (95% CI: 25–36 nM)). In comparison, CysLTR2–L129Q (maroon) shows higher basal β-arrestin2 recruitment, corresponding to 13.5% constitutive activity, and no response to agonist. The data points are the mean ± SEM from three independent experiments with three replicates each. CAM, constitutively active mutant; IP1, d-myo-inositol-1-phosphate; LTD4, leukotriene D4; BRET2, bioluminescence resonance energy transfer 2; CysLTR2, cysteinyl-leukotriene receptor 2.
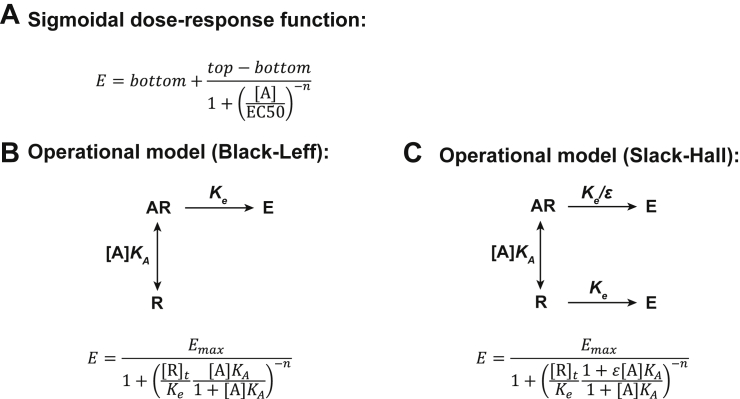
Figure 2.
Modeling the pharmacology of constitutively active receptors. The different models and corresponding equations proposed to describe the pharmacology of the constitutively active CysLTR2-L129Q receptor are described here. A, the sigmoidal dose-response model, B, the Black–Leff operational model (11), and (C) the Slack–Hall operational model (12, 13). CysLTR2, cysteinyl-leukotriene receptor 2.
Characterizing the β-arrestin recruitment to CysLTR2–L129Q
Signals from active GPCRs are normally terminated by β-arrestin–dependent mechanisms, including desensitization, sequestration, and downregulation. We next asked the question, how is CysLTR2–L129Q capable of sustained strong signaling at a level comparable with the fully agonist-stimulated WT receptor? CysLTR2 has been shown to bind β-arrestin2 in response to several agonists (14). However, little is known about the β-arrestin–dependent desensitization, trafficking, and downregulation of CysLTR2 and CysLTR2–L129Q.
We designed a bioluminescence resonance energy transfer 2 (BRET2) experiment to quantify the basal and agonist-dependent binding of β-arrestins to CysLTR2 variants. The BRET2 experiments are performed on HEK293T cells expressing CysLTR2–GFP10 fusion construct in combination with β-arrestins fused to an engineered variant of Renilla luciferase (RLuc3), β-arrestin–RLuc3 (15). We performed a time-course experiment to characterize the agonist-dependent β-arrestin recruitment. The BRET2 ratio shows an agonist concentration–dependent increase for approximately 10 min after the addition of the agonist LTD4, before starting to decrease again slowly (Fig. 1C). The initial increase in the slope increases with higher concentrations of the agonist. The shapes of the time courses were similar when comparing samples expressing β-arrestin1–RLuc3 and β-arrestin2–RLuc3, but the peak increase seen for β-arrestin2–RLuc3 was almost twice that of β-arrestin1–RLuc3 (Fig. S2). Such a biphasic BRET2 β-arrestin recruitment time course is typical for GPCRs with “class A” β-arrestin–recruitment phenotype that have transient, weak interactions with β-arrestins. These class-A receptors rapidly recycle after internalization (16).
The LTD4 dose-dependent increase of the BRET2 ratio for samples transfected with CysLTR2–GFP10 WT and β-arrestin–RLuc3 substantiate the finding from the time-course assay that the agonist-dependent increase of BRET2 is larger for β-arrestin2–RLuc3 as compared with β-arrestin1–RLuc3 (Fig. S3A). Although the agonist-dependent increase was different, the midpoints of the sigmoidal fits of the agonist dose-dependent data for both β-arrestins were identical (Fig. S3B). From these findings, we decided to proceed with the subsequent experiments using only β-arrestin2, which gave the larger BRET2 ratios.
CysLTR2–L129Q poorly recruits β-arrestins
To characterize the effect of the L129Q mutation on β-arrestin recruitment, we included a set of samples expressing CysLTR2–L129Q in the BRET2 experiments. The results from the LTD4 dose-response experiment show a basal, ligand-independent net BRET2 ratio of 0.0028 ± 0.00007, with no ligand dose dependence (Fig. 1D). In comparison, the basal net BRET2 ratio for the WT receptor is 0.0007 ± 0.0002 that increases to 0.0186 ± 0.0004 at saturating LTD4 concentrations (Table S1A).
CysLTR2–L129Q is a Gq-biased CAM that escapes β-arrestin–mediated downregulation
We next quantified the CA for both Gq/11 signaling and β-arrestin recruitment using the modified Slack–Hall operational model to enable the calculation of receptor bias between Gq/11 and β-arrestin pathways (12) for L129Q relative to WT. The term receptor bias was introduced to describe the pathway preference of the basal signaling activity of a receptor (12), in contrast to the term agonist bias that describes ligand-dependent pathway preferences of a receptor (17).
Figure 2 introduces two operational models: the Black–Leff and Slack–Hall models. The key insight of Zhou et al. is that the Slack–Hall model can be used to quantify the inherent agonist-independent pathway bias of the constitutive signaling of a receptor referred to as receptor bias (12, 13). The Slack–Hall model is an expansion of the classical Black–Leff operational model, which underlies methods to calculate functional selectivity or agonist bias (17). In the Slack–Hall model, both the free receptor [R] and agonist-bound receptor [AR] can produce a stimulus, . The parameter ε describes the efficacy of an agonist (A), to produce a stimulus. The Slack–Hall model splits the τ parameter of the Black-Leff model into a product of two parameters, χ and ε. The basal response is determined by χ and is defined as the ratio of [R]t, the total receptor concentration, and Ke, the receptor concentration producing half-maximal effect in the absence of an agonist. In contrast, the τ parameter in the Black-Leff model is the ratio of [R]t and a different Ke, which is defined as the receptor concentration producing half-maximal effect in the presence of a saturating agonist concentration. The ε parameter measures the intrinsic efficacy of the ligand. We slightly modified the original form of this equation to account for fitting problems for χ, the basal response parameter, in cases where the CA is very low. Taking the log τ parameter from the Black-Leff model, we implicitly calculate log χ from log τ–log ε (refer to Experimental procedures).
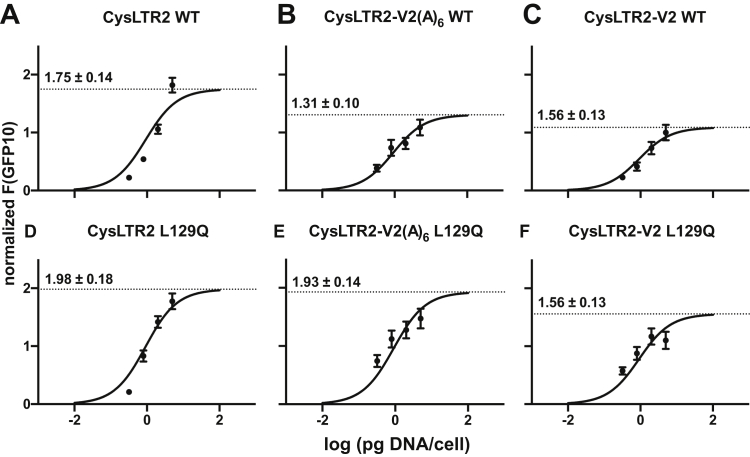
The parameter χ determines the value of the basal response. The challenge is that χ is proportional to the receptor density, which requires standardization for the comparison of receptor mutants with potential impact on receptor expression levels. In our experiments, we control the receptor density by the gene dosage and measure the fluorescence from the GFP10 fusion to calibrate the relative expression levels (Fig. 3). The GFP10 readings were normalized and fit to a sigmoidal model, where GFP10 fluorescence, designated as F(GFP10), is the response as a function of DNA dosage.
Figure 3.
Gene dosage-dependent receptor expression. Normalized GFP10 fluorescence (F(GFP10)) of HEK293T cells transfected with varying amounts of receptor-encoding plasmid are fit to a sigmoidal model, where F(GFP10) is the output as a function of DNA dosage. 40,000 cells were transfected with 12.5, 32, 80, and 200 ng of receptor-encoding plasmid, to yield 0.3125, 0.8, 2, and 5 pg/cell of DNA. The log EC50 value (log EC50 = −0.038 ± 0.072) is shared for all CysLTR2 WT (A, normal; B, V2(A)6 tail; C, V2 tail) and L129Q (D, normal; E, V2(A)6 tail; F, V2 tail) variants. The bottom parameter is set to zero, whereas the top parameter is left free to capture the different expression levels of each variant and is indicated on the graphs. These sigmoidal fits are used to interpolate F(GFP10) for subsequent plots (Fig. 4) because the F(GFP10) is not strong enough to reliably measure the total receptor concentrations, especially at lower DNA/cell levels used in the Gq second messenger IP1 assays. IP1, d-myo-inositol-1-phosphate; V2(A)6, hexa-alanine variant; CysLTR2, cysteinyl-leukotriene receptor 2.
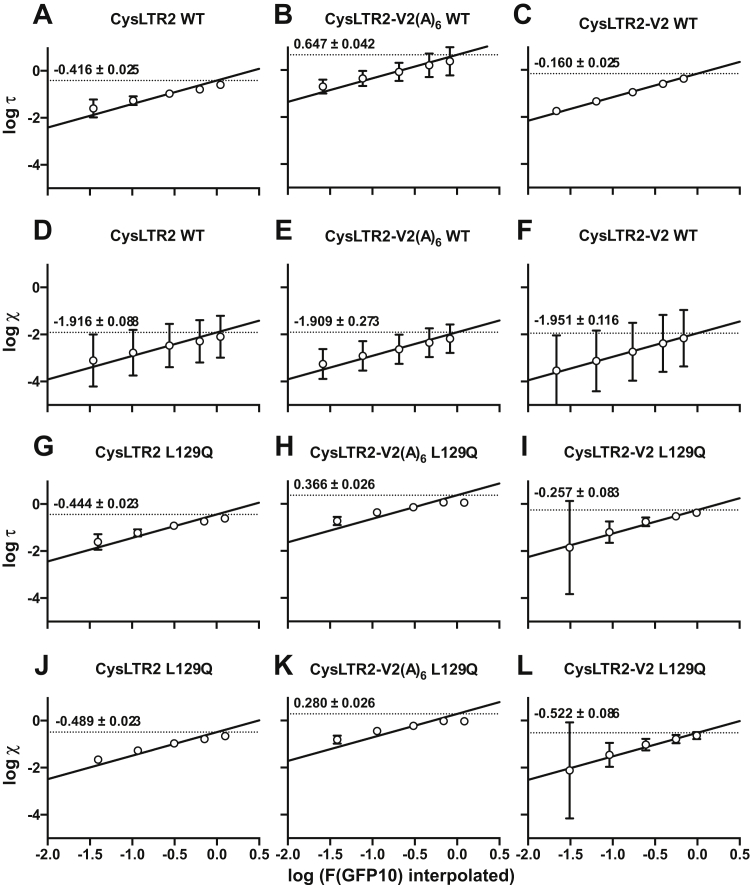
We used the sigmoidal fits of the F(GFP10) to adjust for lower DNA/cell levels used in the Gq second messenger IP1 assays, and thus the interpolated F(GFP10) values were plotted against log τ values and log χ values from fitting the data to the modified Slack–Hall model (Fig. 4, Table S1B). Assuming the F(GFP10) is proportional to the total receptor concentration by some scaling constant, c, and rearranging gives the following:
Figure 4.
Basal and agonist-dependent receptor activity for Gq pathway as a function of receptor expression. The F(GFP10) readings for HEK293T cells transfected with varying amounts of receptor-encoding plasmid from the Gq second messenger IP1 assays (Figs. 1, A–B, 6, A–D) were interpolated from the sigmoidal fits in Figure 3. 7000 cells were transfected with 0.1, 0.4, 1.2, 3.6, and 11 ng of receptor-encoding plasmid, to yield 0.019, 0.057, 0.17, 0.51, and 1.57 pg/cell of DNA. These were plotted against log τ values (WT, A–C; L129Q, G–I) and log χ values (WT, D–F; L129Q, J–L) obtained from fitting the data to the modified Slack–Hall model (Table S1B). Assuming the F(GFP10) is proportional to the total receptor concentration by some scaling constant, c, and rearranging gives . Plotting log χ against log (F(GFP10)) and fitting to a line with a slope of 1 gives y-intercept of log c – log Ke. We can similarly plot log τ against log (F(GFP10)) to get a y-intercept of log c – log Ke + log ε. The y0 values are indicated on graphs and allow for an accurate quantification of log Ke and log ε, of different receptor constructs at standard density. IP1, D-myo-inositol-1-phosphate.
Thus, plotting log χ against log (F(GFP10)) and fitting to a line with a slope of 1 gives y-intercept of log c – log Ke. We can similarly plot log τ against log (F(GFP10)) to get a y-intercept of log c – log Ke + log ε. These allow for an accurate quantification of log ε and of differences of log Ke for different receptor constructs at a standard density.
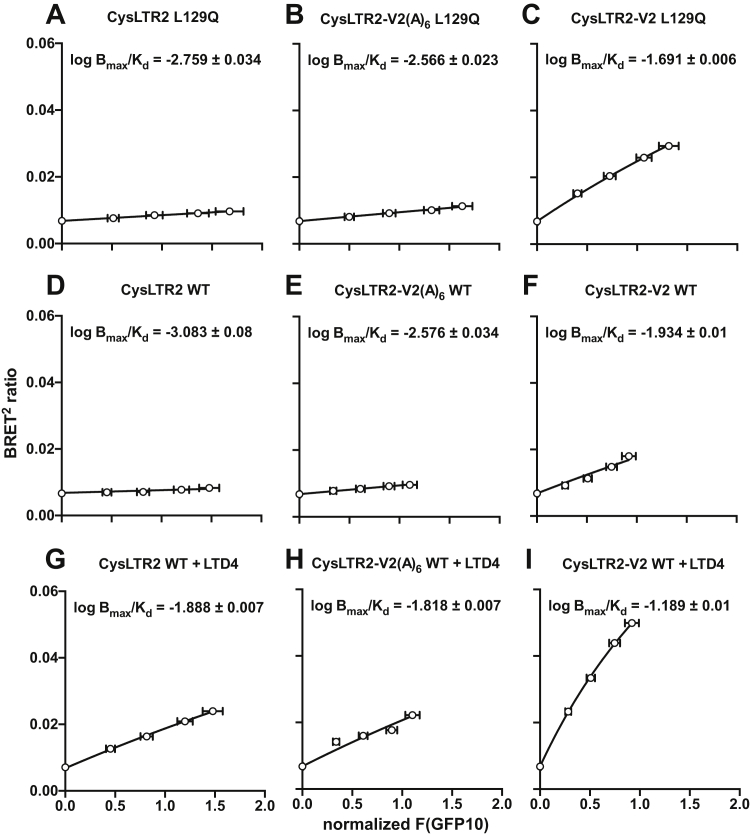
We noticed that in the absence of a ligand, the Slack–Hall model reduces to the mathematical form of a one-site saturation-binding function (18). We plotted the BRET2 ratios against normalized F(GFP10) readings for each CysLTR2 variant and fit the data to a one-site saturation-binding isotherm accordingly (Fig. 5).
Figure 5.
Basal and agonist-dependent receptor activity for β-Arrestin2 pathway as a function of receptor expression. Saturation-binding BRET2 experiment with CysLTR2–L129Q (A–C), unstimulated WT (D–F) and WT stimulated with 1000-nM LTD4 (G–I). The data are fit to a one-site saturation binding function. Tight independent estimates of Kd and Bmax are not required because at low concentrations, only the ratio Bmax:Kd determines the concentration-dependent binding, which can be estimated from the initial slope. The initial slopes are well defined by samples even at low expression levels of receptors and avoid the need for very high receptor concentrations to reach saturation. All data are fit with a shared log Bmax, and log Bmax/Kd are indicated on the graphs. This enables direct comparison of the slopes for unstimulated CysLTR2 WT and CysLTR2–L129Q to subsequently calculate the constitutive activity values in Figures 1D and 6, E–F. BRET2, bioluminescence resonance energy transfer 2; LTD4, leukotriene D4; CysLTR2, cysteinyl-leukotriene receptor 2.
Tight independent estimates of Kd and Bmax are not required because at low concentrations, only the ratio Bmax:Kd determines the concentration-dependent binding, which can be estimated from the initial slope. The initial slopes are well defined by samples even at low expression levels of receptors and avoid the need for very high receptor concentrations to reach saturation. The initial slope of a saturation binding experiment as a function of the total receptor concentration is Bmax/Kd. The initial slope of the Slack–Hall model as a function of the total receptor concentration is Emax/Ke. The equivalence of Bmax/Kd and Emax/Ke enables subsequent calculations of Δlog χ, the differences of log χ for the WT and mutant receptors that determine changes in the CA, and the double difference ΔΔlog χ that determines the receptor bias. Those differences eliminate the constant Bmax and Emax terms and focus on changes of the bias-relevant terms Kd and Ke.
Using the values obtained from the fits in Figures 4 and 5, we are finally able to compare CAs for the different receptor constructs at a standard density. The CAs are normalized relative to the fully agonist-stimulated WT receptor. The ligand-independent CA of CysLTR2–L129Q corresponds to 84.5% (95% confidence interval [CI]: 46.9%–152%) of the maximally LTD4-stimulated CysLTR2 WT at a comparable total receptor concentration. The CA of the WT receptor is 3.2% (95% CI: 1.8%–5.7%). Therefore, the L129Q mutation results in a 26-fold increase of the CA in the Gq pathway. The ligand-dependent increase in IP1 signaling of CysLTR2–L129Q is statistically insignificant as reflected by the efficacy parameter ε of 1.11 (95% CI: 1.03–1.19) close to unity (Table S1B). Overall, the results suggest that CysLTR2–L129Q displays a gain-of-function phenotype for ligand-independent basal signaling and a loss-of-function phenotype for agonist-dependent signaling in the Gq/11 signaling pathway. Compared with the agonist-dependent β-arrestin recruitment of the WT receptor (set to 100%), the CA of the WT receptor is 6.4% and that of the L129Q mutant is 13.5%. Therefore, the effect of the L129Q mutation on the CA in the β-arrestin pathway is a 2-fold increase, which is much smaller than the 26-fold increase in the Gq pathway. Therefore, the L129Q mutation introduces a strong bias of the constitutive signaling (“receptor bias”) toward Gq and away from β-arrestins.
It is challenging to quantify receptor bias. Although ligand bias or biased agonism has been studied for many GPCR–agonist pairs with well-developed mathematical approaches, the concept of receptor bias is still a relatively underexplored area. To our knowledge, this is the first time a rigorous mathematical method has been developed for the detailed analysis of receptor bias. The method described here is generalizable and can be used to study mutations that might cause receptor bias in any GPCR.
Enhanced recruitment of β-arrestin has only a small effect on basal Gq activation in CysLTR2–L129Q
We speculated that the receptor bias away from β-arrestins may be due to a lack of phosphorylation sites in the C-terminal tail of the receptor. The CysLTR2 sequence SVWLRKE has been predicted to be a partial phosphorylation code characteristic of a “class A” β-arrestin–recruitment phenotype (19), with transient and weak interactions with β-arrestins, consistent with our findings of a biphasic β-arrestin–recruitment time course. We hypothesized that adding a sequence of the vasopressin V2 receptor, which carries a strong phosphorylation code, would enhance the recruitment of β-arrestins and switch the receptor to a “class B” β-arrestin–recruitment phenotype. Class B recruitment phenotype corresponds to tight and more stable binding of β-arrestins, which leads to the GPCR-β–arrestin complex being sustained for a longer time (16). We used a hexa-alanine variant (V2(A)6) as a negative control in which we replaced six Ser and Thr residues with Ala residues (20). To explore the strong bias of CysLTR2–L129Q toward Gq, we must investigate the relationship between β-arrestin and Gq binding to CysLTR2. To this end, we enhanced the β-arrestin recruitment to CysLTR2 through the addition of a strong phosphorylation code and observed the corresponding effects on Gq binding and activation. By enhancing the β-arrestin binding to CysLTR2–L129Q, is it possible to observe a shift in the bias?
The results from IP1 and BRET2 assays of the V2 and V2(A)6 tail variants for CysLTR2 WT and L129Q are shown in Figure 6. The V2 tail reduces the agonist-dependent signaling at comparable gene dosage for WT as compared with the V2(A)6 tail variant (Fig. 6, A–B). Similarly, the basal signaling of the L129Q mutant is reduced for V2 as compared with V2(A)6 (Fig. 6, C–D). Once more, Figure 3, Figure 4, Figure 5 were used to compare the CAs for the different receptor constructs at a standard density. The CA values of the L129Q mutants are comparable with 43.5% for the -V2 tail and 43.0% for the V2(A)6 tail variant. Surprisingly, the V2 tail restores some of the agonist-dependent signaling of L129Q as indicated by an efficacy parameter ε of 1.84 (95% CI: 1.70–1.99) different from unity (Table S1B). The V2 tail enhances both basal and agonist-dependent increase of BRET2 for CysLTR2 WT (Fig. 6, E–F). Interestingly, the enhancement of basal, agonist-independent β-arrestin recruitment to CysLTR2–L129Q was much more pronounced for the V2 tail variant than with the V2(A)6 control, which showed only weak β-arrestin recruitment similar to that of CysLTR2–L129Q without the added sequences. We also determined the agonist-dependent β-arrestin–recruitment time course for CysLTR2 WT that showed a biphasic time course for the V2(A)6 control and a monophasic increase for the V2 construct, which is consistent with a “class B” β-arrestin–recruitment phenotype and with tightly bound β-arrestin (Fig. 6, G–H). The CA significantly increases from 18.0% for WT to 31.5% for L129Q in the -V2 tail variant, whereas the V2(A)6 variants have comparable CA values or 17.5% (WT) and 17.8% (L129Q). Therefore, the V2 tail reduces the receptor bias of L129Q away from β-arrestins. We conclude that the receptor bias of the L129Q CAM toward Gq and away from β-arrestins is due to the C-terminal sequence of the receptor.
Figure 6.
Recruitment of β-arrestin has only small effect on basal Gq activation in CysLTR2–L129Q. We added the C-terminal 27 residues of the vasopressin V2 receptor to full-length CysLTR2 to promote high-affinity interactions with β-arrestins (construct CysLTR2-V2). Construct CysLTR2–V2(A)6 is the corresponding phosphorylation-resistant control. A–D, Gq second messenger IP1 accumulation assay. E–H, β-arrestin2–recruitment BRET2 assay. A and B, Agonist-stimulated Gq signaling is reduced by the addition of the V2 sequence as compared with the addition of V2(A)6 sequences at comparable gene dosages. C and D, Similarly, basal Gq signaling of L129Q is reduced by half in CysLTR2-V2 as compared with -V2(A)6. Data points are the mean ± SEM from three independent experiments with six replicates each. Sets of curves are fits to the Slack–Hall operational model (Table S1B). E and F, Agonist-stimulated β-arrestin recruitment is enhanced by the V2 sequence and reduced by the V2(A)6 sequence. Basal, agonist-independent β-arrestin recruitment of L129Q is 5-fold stronger for CysLTR2-V2 than for -V2(A)6. Data points are the mean ± SEM from three independent experiments with three replicates each. G and H, Time course of LTD4-stimulated β-arrestin2 recruitment for three LTD4 concentrations (0 nM, white circles; 30 nM, gray circles; 1000 nM, black circles). While the CysLTR2-V2(A)6 WT exhibited a biphasic time course, CysLTR2-V2 WT reached a plateau after about 8 min. Data are fit to double-exponential curves (Table S1C), and points are the mean ± SEM from three independent experiments with eight replicates. LTD4, leukotriene D4; V2(A)6, hexa-alanine variant; CysLTR2, cysteinyl-leukotriene receptor 2.
Why does the enhanced β-arrestin recruitment not interfere more strongly with the Gq-activation–dependent IP1 accumulation? The binding of β-arrestins with an activated receptor includes two interaction modes, one mode that preactivates β-arrestin, with interactions only with the phosphorylated tail of the receptor and a second fully activated mode, with simultaneous interactions with the core and the phosphorylated tail of the receptor blocking G protein signaling (21). The recent structure of a GPCR–G protein–β-arrestin megacomplex demonstrates how one receptor can simultaneously interact with a G protein, bound to the transmembrane core, and with β-arrestin, bound to the tail, without blocking G protein signaling (22). In contrast to the G protein, the structural determinants of the β-arrestin interactions with the receptor core are largely unknown because only the complex with rhodopsin has been solved so far (21, 23).
Based upon the available structural and biochemical data, we conclude that with CysLTR2–L129Q, the β-arrestin is unable to compete strongly with the G protein for the core binding site. Even by adding the V2 tail sequence to the mutant, where the β-arrestin is forced to interact strongly with the phosphorylated tail of the receptor, the β-arrestin is unable to simultaneously interact with the core to enter the fully activated mode. This is why a marked decrease in Gq activity is not observed. It is possible that this weak competition with the G protein is due to a receptor core conformation that is incompatible with β-arrestin binding but suitable, and even favorable, for G protein binding and activation. We speculate that the strong receptor bias of CysLTR2–L129Q toward Gq signaling is due to a selective stabilization of an intermediate state that is partially activated, perhaps facilitated by the L129Q mutation. In the intermediate state, G protein binding is promoted by the open pocket at the core. Catalytic activation of the nucleotide exchange in the G protein only transiently requires a fully active receptor state, whereas the stable interaction of β-arrestins with the receptor core requires a fully active state, consistent with our findings. We plan to investigate further the mechanisms behind the receptor bias in CysLTR2–L129Q.
In conclusion, we characterized a CYSLTR2 oncogene in UVM and established that CysLTR2–L129Q drives Gq/11 signaling activity in malignant UVM and serves as a driver oncogene. We established that the CysLTR2–L129Q CAM is highly biased toward Gq/11 cellular signaling pathways and fails to recruit significantly β-arrestins. The lack of a strong phosphorylation code in the cytoplasmic tail contributes to the signaling bias of the extremely high CA of the mutant receptor. The biased constitutive signaling pattern of CysLTR2–L129Q explains why it can persistently activate Gq and avoid β-arrestin–dependent cellular downregulation mechanisms.
Experimental procedures
We generated a CysLTR2–GFP10 fusion construct that enables quantification of basal and agonist-dependent Gq/11 cellular signaling and β-arrestin recruitment activity, as well as total receptor expression, under comparable conditions. Experiments were conducted on HEK293T cells transfected with the plasmids encoding the fusion constructs. The downstream Gq/11 activity was quantified using the CisBio IP-One homogeneous time-resolved fluorescence immunoassay (HTRF). The β-arrestin–recruitment activity was quantified using a BRET2 assay.
Materials
LTD4 was from Cayman Chemical (Ann Arbor, MI). YM-254890 was from Wako Pure Chemical Industries (Richmond, VA). BRET substrate methoxy e-Coelenterazine was from NanoLight Technology (Pinetop, AZ). The IP-One HTRF kit was from CisBio (Codolet, France). Bovine serum albumin (BSA) fraction V, fatty acid-free, was from Roche (Basel, Switzerland). Poly-d-lysine and LiCl were from Sigma-Aldrich (St Louis, MO). HEK293T cells were from the American Type Culture Collection (Manassas, VA). Dulbecco’s modified Eagle’s medium (DMEM) GlutaMAX, FluoroBrite DMEM, Dulbecco’s PBS (DPBS) without calcium and magnesium, and HEPES buffer were from Thermo Fisher Scientific (Waltham, MA). Penicillin/Streptomycin (10,000 U/ml), L-glutamine, and Lipofectamine 2000 were from Thermo Fisher Scientific (Waltham, MA). Fetal bovine serum (FBS) was from Gemini Bio-Products (West Sacramento, CA). Black and white low-volume 384-well microplates, and black CELLSTAR 96-well microplates (polystyrene wells, flat bottom) were from Greiner (Monroe, NC). NEBuilder Hifi DNA Assembler, Dpn1, T4 DNA Ligase, Q5 Hot Start High-Fidelity DNA Polymerase, and dNTPs were from New England BioLabs (Ipswich, MA). QuikChange Lightning Site-Directed Mutagenesis Kit was from Agilent Technologies (Santa Clara, CA) and TagMaster Site-Directed Mutagenesis Kit was from GM Biosciences Inc (Frederick, MD). Oligonucleotides which are listed in Table S2 were purchased at the standard desalting grade from Integrated DNA Technologies (Coralville, IA). QIAGEN Plasmid Maxi Kits and QIAprep Spin Miniprep Kit were from QIAGEN (Germantown, MD).
Molecular biology
CysLTR2–1D4 expression constructs
The synthetic vector encodes human CysLTR2 cDNA in pcDNA3.1(+) fused to an N-terminal FLAG tag (DYKDDDDK) and a C-terminal 1D4 epitope tag (TETSQVAPA) (2). The FLAG tag was then deleted by site-directed mutagenesis using the TagMaster Site-Directed Mutagenesis Kit according to the manufacturer’s instructions to generate CysLTR2-1D4. TagMaster primers that were used to generate the 1D4 constructs are listed in Table S2A and were purchased from Integrated DNA Technologies. All constructs were confirmed by sequencing (Genewiz, South Plainfield, NJ).
CysLTR2–GFP10 fusion protein construct
Primer design
The NEBuilder HiFi DNA Assembly Tool was used to assemble the BRET2 acceptor constructs CysLTR2–GFP10. These were assembled from three parts: pcDNA3.1(+) backbone from construct HA-CLIP-CLR (24), CysLTR2, and full-length C-terminal 1D4 epitope tag from FLAG-CysLTR2-1D4 mentioned above, and GFP10 from YB124_CXCR4–GFP10 (15). The primers (Table S2B) were designed using the NEBuilder Assembly Tool on the NEB website with a specific sequence to prime to the gene of interest for template priming (3’ end), as well as an overlap sequence to aid in assembly (5’ end).
PCR amplification of fragments
The fragments introduced above were PCR-amplified using Q5 Hot Start High-Fidelity DNA Polymerase and fresh dNTPs purchased from NEB. Briefly, the PCRs were performed in 25-μl total volume containing: 1× Q5 reaction buffer, 0.2-mM dNTPs, 0.5 μM of the forward primer, 0.5 μM of the reverse primer, 1-ng template DNA, and 1 unit of Q5 Hot Start High-Fidelity DNA Polymerase. The PCR thermocycle was as follows: initial denaturation at 98 °C for 30 s, followed by 25 cycles of denaturation (98 °C, 10s), annealing (varied from 50–70 °C, 30 s), and elongation (72 °C, 3 min), and ending with a final elongation (72 °C, 2 min). The recommended annealing temperature calculated on the NEBuilder Assembly Tool was used for each primer pair. After PCR, 1 unit of DpnI was added and the mixture was incubated at 37 °C for 30 min to digest any remaining template DNA. This was cleaned up, and any enzymes were removed using a DNA Clean and Concentrator (Zymo Research). The concentrations of all PCR-amplified fragments were determined using a NanoDrop.
Isothermal assembly
The NEBuilder Hifi DNA Assembler includes three enzymes: the exonuclease to create 3’ overhangs to aid annealing of neighboring fragments sharing a complimentary overlap region, the polymerase to fill the gaps of each annealed fragment, and the DNA ligase to seal nicks in the assembled DNA. The assembly reaction was performed in 20-μl total volume, with 50 ng of the vector, 100 ng of insert(s), and 10 μl of the NEBuilder HiFi DNA Assembly Master Mix. This was incubated at 50 °C for 60 min, and 2 μl of the assembled product was used to transform NEB 5-alpha competent Escherichia coli cells. Cells were spread on LB-Amp plates, and colonies were picked and confirmed by sequencing.
After the assembled product was confirmed by sequencing, the second NotI site that is flanked by two XhoI sites, which had been part of the pcDNA3.1(+) backbone in HA-CLR-CLIP, was removed. This was simply performed by digesting at the XhoI sites and self-ligating the vector using T4 DNA Ligase. We then sequenced the NotI-removed CysLTR2–GFP10 in its entirety to check for any erroneous modifications or linkages.
The β-arrestin2-RLuc3 BRET2 donor was constructed previously by fusing the coding sequence of RLuc3 to the C-terminus of β-arrestin2 (15).
CysLTR2-V2 and CysLTR2-V2(A)6 constructs
The cDNA encoding the 27 amino acids from the C-terminal tail of the vasopressin V2 receptor, GRTPPSLGPQDESCTTASSSLAKDTSS, was fused to the end of the full-length CysLTR2 receptor in the CysLTR2–GFP10 construct introduced above. As a negative control, we also fused a hexa-Ala variant (20) of the 27 amino acids with the phosphorylation sites (Ser and Thr) replaced with Ala (GRTPPSLGPQDESCTTAAAALAKDAAA, Ala substitutions underlined) to the full-length CysLTR2 receptor. For both of these amino acid sequences, we used the GeneOptimizer algorithm in GeneArt (Thermo Fisher Scientific, Waltham, MA) to get a DNA sequence optimized for humans and avoiding major restriction sites. The NEBuilder HiFi DNA Assembly Tool was used to assemble these constructs from two parts: the single-stranded oligonucleotide of the V2 receptor tail sequence and the linearized CysLTR2–GFP10 construct cut after the full-length receptor and before the GFP10. The primers were designed using the NEBuilder Assembly Tool on the NEB website, and their sequences are shown in Table S2C. The PCR amplification of the CysLTR2–GFP10 fragment and the isothermal assembly of these two parts were performed as outlined above. Note that the NEBuilder HiFi DNA Assembly method allows the assembly of a single-stranded oligonucleotide to a double-stranded DNA strand. All constructs were prepared from the QIAGEN Plasmid Maxi Kits and confirmed by Sanger sequencing using standard BGH reverse-sequencing primers.
Signaling assays
IP1 accumulation assay
HEK293T cells were maintained in DMEM GlutaMAX supplemented with 10% FBS (passage numbers 5–14) at 37 °C under 5% CO2. Cells were transiently transfected directly ‘in-plate’ in low-volume 384-well plates using Lipofectamine 2000 according to manufacturer’s instructions with some modifications. The total DNA amount was kept constant at 11 ng per well using empty vector pcDNA3.1(+). All transfection reagents mixes were performed in DMEM GlutaMAX, unless specifically noted as being performed in FluoroBrite DMEM (DMEM without phenol red, described later). Briefly, the appropriate amount of plasmid DNA was mixed with DMEM (no FBS). In a separate mixture, the total Lipofectamine 2000 (2.5 μl per μg DNA) was mixed in DMEM (no FBS) and incubated for 5 min. The appropriate amount of Lipofectamine 2000/DMEM mixture was mixed with the DNA/DMEM and incubated for 20 min. Cells were then trypsinized, resuspended in DMEM supplemented with 20% FBS, and counted. Cells were mixed with the DNA/Lipofectamine 2000/DMEM mixture and directly plated onto 0.01% poly-d-lysine–coated, white, clear-bottom, tissue culture–treated low-volume 384-well plates at a density of 7000 cells per well in 7 μl. For the DNA titration assay with the CysLTR2-V2 tail variants, cells were similarly transfected in FluoroBrite DMEM and plated in 0.01% poly-d-lysine–coated, black, clear-bottom, tissue culture–treated low volume 384-well plates. All assays were conducted 24 h after the transfection. All plasmids were prepared from QIAGEN Plasmid Maxi Kits, resulting in a high-quality and high-concentration plasmid solution (about 1000–4000 ng/μl) unless otherwise specified. For specific experiments, the plasmids were prepared from the QIAprep Spin Miniprep Kit resulting in 10-fold lower concentrations.
The CisBio IP-One HTRF immunoassay quantifies IP1, a degradation product of the second messenger d-myo-inositol-1,4,5-trisphosphate, to measure the activation of PLCβ by Gq-coupled GPCRs (25). The IP1 assay is a competitive HTRF assay where the d2-labeled IP1 analogue acts as the fluorescence acceptor and the terbium cryptate–labeled anti-IP1 monoclonal antibody (mAb) acts as the fluorescence donor. The terbium cryptate is a long-lifetime fluorescence donor that can be excited by UV light. LiCl is added during the stimulation period of the assay to block further degradation of IP1 by the enzyme inositol monophosphatase. It has been suggested that PLCβ-dependent IP1 accumulation also includes contributions from the d-myo-inositol-1,4-bisphosphate formed by direct hydrolysis of phosphatidylinositol-4-phosphate, instead of d-myo-inositol-1,4-bisphosphate formed by the dephosphorylation of d-myo-inositol-1,4,5-trisphosphate formed by hydrolysis of phosphatidylinositol-4,5-bisphophate (10). Because this novel pathway does not depend on phosphatidylinositol-4,5-bisphophate, which is predominantly found at the plasma membrane, the phosphatidylinositol-4-phosphate–dependent IP1 accumulation could have a substantial contribution to PLCβ signaling from the endosomal compartment.
Agonist dose-response of CysLTR2 DNA titration assay
HEK293T cells were transiently transfected with a serial dilution of 11, 3.6, 1.2, 0.4, and 0.1 ng of WT and mutants of CysLTR2-GFP10 per well as described above. For the DNA titration assay of CysLTR2 WT and L129Q, either WT or L129Q assays were performed in individual plates with mock-transfected cells as controls. For the DNA titration assay of CysLTR2-V2 variants, each experiment was performed in three individual plates, with three sets of two technical replicates each, with WT and mock-transfected cells as controls. Then, 24 h after transfection, the assay plate was placed on an aluminum heating block maintained at 37 °C, and cells were treated with 7 μl/well of various concentrations of LTD4 (final concentrations from 1 μM to 10 pM) diluted in a prewarmed stimulation assay buffer provided by the manufacturer (10-mM HEPES, 1-mM CaCl2, 0.5-mM MgCl2, 4.2-mM KCl, 146-mM NaCl, 5.5-mM glucose, 50-mM LiCl, pH 7.4) supplemented with 0.2% (w/v) BSA and 50-mM LiCl to prevent IP1 degradation. The plate was incubated at 37 °C for 2 h. After incubation, cells were lysed by addition of 3 μl/well of d2-labeled IP1 analogue and 3 μl/well of terbium cryptate–labeled anti-IP1 mAb diluted in the lysis and detection buffers. The plates were incubated overnight, in the dark, at RT. Time-resolved fluorescence signals were read on the BioTek Synergy NEO plate reader (for cells transfected in DMEM GlutaMAX and assayed in white microplates) or the BioTek Synergy NEO2-TRF Hybrid multi-mode reader (for cells transfected in FluoroBrite DMEM and assayed in black microplates) (BioTek Instruments, Winooski, VT) in the Rockefeller University’s High-Throughput and Spectroscopy Resource Center. The plate is first subjected to flash lamp excitation at 320 nm (BioTek Synergy NEO) or to laser excitation (BioTek Synergy NEO2-TRF) at 337 nm, and then the fluorescence is measured at wavelengths centered at 620 nm and 665 nm simultaneously.
Time-course assay
We obtained a time-course of basal and LTD4-dependent IP1 accumulation in HEK293T cells transiently transfected with plasmids for CysLTR2 WT, CysLTR2–L129Q, and mock-transfected control. HEK293T cells were transiently transfected with 11 ng of CysLTR2-1D4 (WT and L129Q) per well. Then, 24 h after transfection, the assay plate was placed on an aluminum heating block as described above, and the cells were treated every 20 min over 180 min with 3.5 μl/well of LiCl diluted in a prewarmed stimulation assay buffer at a final concentration of 50 mM. Cells were incubated at 37 °C. At 1 h after the start point, 3.5 μl/well of LTD4 diluted in prewarmed DMEM GlutaMAX at a final concentration of 100 nM (agonist stimulated) or 3.5 μl/well of DMEM GlutaMAX alone (basal) was added in appropriate wells and incubated for 2 h. The reaction was then stopped by successively adding 3 μl/well of the IP1 analogue and the anti-IP1 mAb in a reverse chronological order.
BRET2 assays
General procedure
We then generated a fusion construct of CysLTR2 with GFP10 that can be used in BRET2 assays in combination with β-arrestin fused to an engineered variant of Renilla luciferase, β-arrestin2-RLuc3 (15). HEK293T cells were transiently cotransfected with β-arrestin2-RLuc3 and CysLTR2-GFP10 WT or the L129Q mutant directly ‘in-plate’ in 96-well plates using Lipofectamine 2000 as described above with slight modifications to account for the larger well volume. The total DNA amount was kept constant at 205 ng per well using an empty vector pcDNA3.1(+). Briefly, a master mix of the β-arrestin2-RLuc3 was made in FluoroBrite DMEM (DMEM without phenol red and suitable for fluorescence experiments) and the CysLTR2–GFP10 DNA were added to these after appropriate distribution. In a separate mixture, the total Lipofectamine 2000 was mixed in FluoroBrite DMEM and incubated for 5 min. The appropriate amount of Lipofectamine 2000/FluoroBrite DMEM mixture was mixed with the DNA/FluoroBrite DMEM and incubated for 20 min. Cells were then trypsinized, resuspended in FluoroBrite DMEM, 20% FBS, 30-mM HEPES, and 8-mM glutamine, and counted. Cells were mixed with the DNA/Lipofectamine 2000/FluoroBrite DMEM mixture and directly plated onto 0.01% poly-d-lysine–coated, black, clear-bottom, tissue culture–treated 96-well plates at a density of 40,000 cells per well in 100-μl FluoroBrite DMEM 10% FBS, 15-mM HEPES, 4-mM glutamine. All assays were conducted 24 h after the transfection. All plasmids were prepared from QIAGEN Plasmid Maxi Kits.
Saturation-binding assays
HEK293T cells were transiently transfected with 5 ng of β-arrestin2-RLuc3 and 0, 12.8, 32, 80, or 200 ng of CysLTR2-GFP10 WT/-L129Q, CysLTR2-V2 WT/-L129Q, or CysLTR2-V2(A)6 WT/-L129Q per well. Then, 24 h after transfection, media were aspirated carefully from all wells. Then, 30 μl of the prewarmed BRET buffer (DMEM FluoroBrite, 15-mM HEPES, 0.1% (w/v) BSA, 4-mM glutamine) was added to each well. Half of the WT wells were stimulated with LTD4, so 10 μl of LTD4 in the BRET buffer (final concentration 1 μM) was added to these. Then, 10 μl of the BRET buffer was added to all other wells. Cells were incubated for 10 min at RT. After the incubation, BRET2 measurements were taken on the BioTek Synergy NEO2 microplate reader using filter set 109 (center wavelength/band width) of 410/80 nm (donor) and 515/30 nm (acceptor). First, the GFP fluorescence was read using the monochromator (ex: 395 nm, em: 510 nm ± 20 nm from the bottom, autogain) to quantify total expression levels. After this procedure, the cell-permeable substrate methoxy e-Coelenterazine (Me-O-e-CTZ/Prolume Purple) was added to each well at a final concentration of 5 μM, and the luminescence at the two wavelengths was read simultaneously.
Time-course assays
For the time-course assay, HEK293T cells were transiently transfected with 5 ng of β-arrestin2-RLuc3 and 80 ng of CysLTR2–GFP10 WT, CysLTR2–V2 WT or CysLTR2–V2(A)6 WT per well. Then, 24 h after transfection, media were aspirated and 30 μl of the prewarmed BRET buffer was added to each well and the GFP fluorescence was read. Methoxy e-Coelenterazine was added to three columns at 5-μM final concentration followed by addition of 0 nM, 30 nM, and 1000 nM of LTD4 to appropriate wells in the three columns. The plate was quickly placed into the microplate reader so that there was as little lag time between the addition of the ligand and BRET2 readings as possible. The three columns take about 60 s to read, and this was repeated 24 times such that a BRET2 reading was recorded every 60 s for about 24 min.
Agonist dose-response assay
HEK293T cells were transiently transfected with 5 ng of β-arrestin2-RLuc3 and 80 ng of CysLTR2-GFP10 WT/-L129Q, CysLTR2-V2 WT/-L129Q, or CysLTR2-V2(A)6 WT/-L129Q per well. Then, 24 h after transfection, media were aspirated and 30 μl of prewarmed BRET buffer was added to each well. Various concentrations of LTD4 (final concentrations from 1 μM to 10 pM) were added to appropriate wells and incubated for 10 min at RT. After the incubation, GFP signals were measured, the substrate was added, and the BRET2 signals were obtained.
Quantification and statistical analysis
Data reduction, standard calibration, and transformation of HTRF data from IP1 assays
The raw signals from the IP1 assay were transformed into a fluorescence ratio (665 nm/620 nm), and IP1 concentrations were interpolated from a standard curve prepared using the supplied IP1 calibrator. The IP1 standard curve was fit to a sigmoidal curve using the equation,
| (1) |
The Bottom and Top parameters are the minimum and maximum fluorescence ratios obtained from the standard curve, respectively, and the IC50 and concentration of IP1 in nM (x) are calculated as logarithmic values.
The fluorescence ratios obtained from individual experiments were then converted into the corresponding IP1 concentration (nM, linear) using the following equation and the standard curve:
| (2) |
The IC50 and IP1 concentrations are now calculated as linear values and not logarithmic values. In some cases, these concentrations were further analyzed to obtain normalized IP1 values relative to the unstimulated mock-transfected cells (set to 0%) and to the fully stimulated WT receptor (set to 100%).
Modeling agonist dose-response and receptor density from IP1 assays
IP1 concentrations or normalized IP1 data were fitted to specific models summarized in Figure 2 and introduced below.
Sigmoidal dose response
For the LTD4 dose response of HEK293T cells transfected with varying amounts of CysLTR2 encoding plasmid DNA, the IP1 concentrations in nM were plotted against the logarithmic concentration of LTD4. These data were first fit to a three-parameter sigmoidal dose-response function described below:
| (3) |
The Bottom and Top parameters describe the lower and upper asymptotic values, respectively. The logarithmic form of the log EC50 parameter ensures positive solution for the EC50. Moreover, the logarithmic fitting parameters account for the fact that the solutions for EC50 should be log-normally distributed. We use an alternative form for dose-response experiments with competitors or inverse agonists, where the log IC50 parameter replaces log EC50 and ensures positive fitting solutions for the half maximal inhibitory concentration IC50.
| (4) |
To account for data sets that show no significant dose response, we also fit each data set with a horizontal line function as an alternative hypothesis. We chose the best model, either sigmoidal curve or horizontal line, by the Akaike Information Criterion (Table S1A). Note that the horizontal line fitting function in the GraphPad Prism 8 software has the form because the software requires the use of the independent variable x, which is multiplied by zero to negate its influence. Effectively, this fit is a horizontal line plotting the mean IP1 concentrations for all LTD4 doses.
Slack–Hall operational model
Figure 2 introduces two operational models: the Black–Leff and Slack–Hall models. The key insight of Zhou et al. is that the Slack–Hall model can be used to quantify the agonist-independent, inherent pathway bias of the constitutive signaling of a receptor referred to as receptor bias (12, 13). The Slack–Hall model is an expansion of the classical Black–Leff operational model, which underlies methods to calculate functional selectivity or agonist bias (17). In the Slack–Hall model, both the free receptor [R] and agonist-bound receptor [AR] can produce a stimulus, . The parameter ε describes the efficacy of an agonist (A) to produce a stimulus. The Slack–Hall model splits the τ parameter of the Black–Leff model into a product of two parameters, χ and ε. The basal response is determined by χ and is defined as the ratio of [R]t, the total receptor concentration, and Ke, the receptor concentration producing half-maximal effect in the absence of an agonist. In contrast, the τ parameter in the Black–Leff model is the ratio of [R]t and a different Ke, which is defined as the receptor concentration producing half-maximal effect in the presence of a saturating agonist concentration. The ε parameter measures the intrinsic efficacy of the ligand. We slightly modified the original form of this equation to account for fitting problems for χ, the basal response parameter, in cases where the CA is very low. Taking the log τ parameter from the Black–Leff model, we implicitly calculate log χ from log τ–log ε. The final equation of our modified Slack–Hall model is as follows:
| (5) |
Table S1B shows the parameters for the Slack–Hall operational model fitted to the experiments shown in Fig. 1, A–B, 6, A–D. In these fits, x is the log of the agonist concentration and y is the response to the agonist. Emax, maximal IP1 concentration for the system, was first fit individually for each condition, and then the highest Emax value from this was used as a shared, fixed value for all final fits. The parameter log KA is the logarithm of the agonist-receptor association constant, KA. Note that KA is the inverse of the dissociation constant Ka. The fitting parameters log ε and log KA were shared for all conditions, whereas log τ was left free to give an independent value for each condition. The optimal value and error for log χ was separately calculated from the difference log τ–log ε.
Modeling the time course of IP1 accumulation using IP1 assays
For the IP1 accumulation time course (Fig. S1, A–B), the corrected IP1 concentrations, in nM, were plotted against time in minutes. These data are then fitted to a one-phase decay model using the following equation:
| (6) |
Here y0 is the IP1 concentration at time zero, while Plateau is the IP1 concentration at infinite time, k is the rate constant of the decay, x is the time of incubation, and y is the IP1 concentration.
Data reduction for BRET2 assays
Raw BRET2 ratios were determined by calculating the ratio of the light intensity emitted by the GFP10 (515 nm) over the light intensity emitted by the RLuc3 (410 nm). The BRET2 signals minus the basal BRET2 signals (β-arrestin-RLuc3 only signals) give the net BRET2 values.
Two-phase decay model for time course
For all time-course assays, the BRET2 ratios (y) were plotted against time (x), in seconds, to assess the time dependence of the LTD4-stimulated β-arrestin recruitment (Figs. 1C and 6, G–H). These data were fitted to a two-phase decay model using the following equation:
| (7) |
This model is the sum of two decay processes, one fast and the other slow. The fast process describes the rise of the curve, whereas the slow process determines the subsequent decay. We use logarithmic fitting parameters, log kfast and log kslow, to constrain the fitting space to positive values of the rate constants, kfast and kslow, which are rate constants for the two decay processes. The Plateau parameter scales the peak height, and y0 is the β-arrestin recruitment at time zero. kfast describes the initial recruitment of β-arrestin, which is dependent on the concentration of the active receptor and is represented as the initial increase in the signal. kslow describes the disassembly of the receptor–β-arrestin complex and is represented by the decay of the signal over time. For this fit, x0 and log kslow are shared for all three curves (0-nM, 30-nM, and 1000-nM LTD4). The y0 is determined by first fitting the 0-nM LTD4 curve to a horizontal line and then using this constant as the fixed y0 value for the fits of the 30-nM and 1000-nM LTD4 data where log kfast is varied independently for each condition. The data for the fits are provided in Table S1C.
Sigmoidal dose-response and normalization
For the agonist dose-response assays, the BRET2 ratios were plotted against logarithmic concentrations of LTD4 (Figs. 1D and 6, E–F). The data were fit to a sigmoidal curve (Equation 3), with an alternative model as a horizontal line, , as described above. Table S1A summarizes the fitting parameters.
CA and receptor bias for Gq and β-arrestins
We quantified the CA using a modified Slack–Hall operational model to enable the calculation of the receptor bias between Gq/11 and β-arrestin pathways for L129Q relative to WT and to characterize the effect of the V2 tail variants. The term receptor bias was introduced to describe the pathway preference of the basal signaling activity of a receptor (12), in contrast to the agonist bias that describes ligand-dependent pathway preferences of a receptor (17). The parameter χ determines the value of the basal response. The challenge is that χ is proportional to the receptor density, which requires standardization for the comparison of receptor mutants with potential impact on receptor expression levels. In our experiments, we control the receptor density by the gene dosage and measure the fluorescence from the GFP10 fusion to calibrate the relative expression levels (Fig. 3). The GFP10 readings were first normalized by dividing the sample F(GFP10) by the basal F(GFP10) (β-arrestin2-RLuc3 only), and then further divided this by the normalized F(GFP10) of the exchange protein activated by cAMP (EPAC) BRET2 biosensor (RLuc3-EPAC-GFP10 (15) developed from the guanine nucleotide EPAC that acts as the positive control) to give the following:
| (8) |
The inner averages are for technical replicates per experiment, and the outer averages are for all experiments. These data are fit to a sigmoidal model, where F(GFP10) is the response as a function of DNA dosage. The log EC50 parameter is shared in the global fit, the bottom parameter is set to zero, whereas the top parameter is left unconstrained to capture the different expression levels of each variant. We use these sigmoidal fits to interpolate F(GFP10) for lower DNA/cell levels used in the Gq second messenger IP1 assays, and thus the interpolated F(GFP10) were plotted against log τ values and log χ values from fitting the data to the modified Slack–Hall model (Fig. 4, Table S1B). Assuming the F(GFP10) is proportional to the total receptor concentration by some scaling constant, c, and rearranging gives the following:
| (9) |
Thus, plotting log χ against log (F(GFP10)) and fitting to a line with a slope of 1 gives y-intercept of log c – log Ke. We can similarly plot log τ against log (F(GFP10)) to get a y-intercept of log c – log Ke + log ε. These allow for an accurate quantification of log ε; and of differences of log Ke for different receptor constructs at a standard density.
We noticed that in the absence of a ligand, the Slack–Hall model reduces to the mathematical form of a one-site saturation-binding function (18). We then plotted the BRET2 ratios against normalized F(GFP10) readings of each CysLTR2 variant (Fig. 5). These data are fitted to a one-site saturation-binding isotherm using the following equation:
| (10) |
The logarithmic fitting parameters, log Bmax and log Kd, ensure positive fitting solutions for Bmax and Kd. Bmax is the maximal increase of the BRET2 ratio due to β-arrestin binding. log Bmax is a shared value for the global fits of all variants. Kd is the equilibrium dissociation of the F(GFP10), which gives half-maximal β-arrestin binding. The log Kd varied independently for each curve. The parameter background is constrained to 0.0068, the BRET2 ratio for the β-arrestin2-RLuc3 sample without receptor.
Tight independent estimates of Kd and Bmax are not required because at low concentrations, only the ratio Bmax:Kd determines the concentration-dependent binding, which can be estimated from the initial slope. The initial slopes are well defined by samples even at low expression levels of receptors and avoid the need for very high receptor concentrations to reach saturation. The initial slope of a saturation binding experiment as a function of the total receptor concentration is Bmax/Kd. The initial slope of the Slack–Hall model as function of the total receptor concentration is Emax/Ke. The equivalence of Bmax/Kd and Emax/Ke enables subsequent calculations of Δlog χ, the differences of log χ for the WT and mutant receptors that determine changes in the CA, and the double difference ΔΔlog χ that determines the receptor bias. Those differences eliminate the constant Bmax and Emax terms and focus on changes of the bias-relevant terms Kd and Ke.
Using Figures 4 and 5, we are able to compare the CAs for the different receptor constructs at a standard density. We further normalize the CAs relative to the fully agonist-stimulated WT receptor. These values are reported above the corresponding graphs in Figures 1, A–B and D and 6, A–F.
Data availability
All data needed to evaluate the conclusions in the article are presented in the article or the Supporting Information. Additional requests should be made to the corresponding author.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Michel Bouvier for providing the EPAC BRET2 reporter, Dr Fraser Glickman at the Rockefeller University's High-Throughput and Spectroscopy Resource Center for training, discussions, and assistance with the experiments.
Author contributions
E. C., M. H., J. M. M., T. D. H., A. R. M., P. C., Y. C., T. P. S., and T. H. designed the research; E. C., M. H., J. M. M., T. D. H., A. R. M., M. A. K. performed the research; E. C., M. H., J. M. M., T. D. H., A. R. M., P. C., Y. C., T. P. S., and T. H. analyzed and interpreted the data; E. C., M. H., J. M. M., T. P. S., and T. H. wrote the manuscript.
Funding and additional information
E. C. was funded by the François Wallace Monahan Fellowship and the Tom Haines Fellowship in Membrane Biology. M. H. and J. M. M. were supported by the Tri-Institutional Training Program in Chemical Biology and M. H. by T32 GM115327. We also acknowledge grant support from the Robertson Foundation, the Crowley Family Fund, and the Danica Foundation to T. P. S. and T. H. The work was further supported by Memorial Sloan Kettering Cancer Center (MSKCC) support grant/core grant (P30 CA008748) and grants from the National Cancer Institute (NCI) (K08CA140946 Y. C.; R01CA193837, Y. C.; P50CA092629, Y. C.; P50CA140146, P. C.; K08CA151660, P. C.; DP2 CA174499, P. C.), U.S. Department of Defense (US DOD) (W81XWH-10-1-0197, P. C.), Prostate Cancer Foundation (Y. C.), Starr Cancer Consortium (Y. C. and P. C.), Geoffrey Beene Cancer Research Center (Y. C. and P. C.), Gerstner Family Foundation (Y. C.), Bressler Scholars Fund (Y. C.), and Cycle for Survival (Y. C.).
Edited by Henrik Dohlman
Footnotes
Present address for Amanda R. Moore: Department of Discovery Oncology, Genentech Inc, South San Francisco, California, USA.
This article contains supporting information.
Contributor Information
Thomas P. Sakmar, Email: sakmar@rockefeller.edu.
Thomas Huber, Email: hubert@rockefeller.edu.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Hauser A.S., Chavali S., Masuho I., Jahn L.J., Martemyanov K.A., Gloriam D.E., Babu M.M. Pharmacogenomics of GPCR drug targets. Cell. 2018;172:41–54.e19. doi: 10.1016/j.cell.2017.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore A.R., Ceraudo E., Sher J.J., Guan Y., Shoushtari A.N., Chang M.T., Zhang J.Q., Walczak E.G., Kazmi M.A., Taylor B.S., Huber T., Chi P., Sakmar T.P., Chen Y. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandy-Szekeres G., Munk C., Tsonkov T.M., Mordalski S., Harpsoe K., Hauser A.S., Bojarski A.J., Gloriam D.E. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chua V., Mattei J., Han A., Johnston L., LiPira K., Selig S.M., Carvajal R.D., Aplin A.E., Patel S.P. The Latest on Uveal Melanoma Research and Clinical Trials: Updates from the Cure Ocular Melanoma (CURE OM) Science Meeting (2019) Clin. Cancer Res. 2020 doi: 10.1158/1078-0432.CCR-20-2536. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore A.R., Ran L., Guan Y., Sher J.J., Hitchman T.D., Zhang J.Q., Hwang C., Walzak E.G., Shoushtari A.N., Monette S., Murali R., Wiesner T., Griewank K.G., Chi P., Chen Y. GNA11 Q209L Mouse model reveals RasGRP3 as an essential signaling Node in uveal melanoma. Cell Rep. 2018;22:2455–2468. doi: 10.1016/j.celrep.2018.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hitchman T.D., Bayshtok G., Ceraudo E., Moore A.R., Lee C., Jia R., Wang N., Pachai M.R., Shoushtari A.N., Francis J.H., Guan Y., Chen J., Chang M.T., Taylor B.S., Sakmar T.P. Combined Inhibition of Gαq and MEK Enhances Therapeutic Efficacy in Uveal Melanoma. Clin. Cancer Res. 2020 doi: 10.1158/1078-0432.CCR-20-2860. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kusters-Vandevelde H.V.N., Germans M.R., Rabbie R., Rashid M., Ten Broek R., Blokx W.A.M., Prinsen C.F.M., Adams D.J., Ter Laan M. Whole-exome sequencing of a meningeal melanocytic tumour reveals activating CYSLTR2 and EIF1AX hotspot mutations and similarities to uveal melanoma. Brain Tumor Pathol. 2018;35:127–130. doi: 10.1007/s10014-018-0308-1. [DOI] [PubMed] [Google Scholar]
- 8.Wu V., Yeerna H., Nohata N., Chiou J., Harismendy O., Raimondi F., Inoue A., Russell R.B., Tamayo P., Gutkind J.S. Illuminating the Onco-GPCRome: novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 2019;294:11062–11086. doi: 10.1074/jbc.REV119.005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heise C.E., O'Dowd B.F., Figueroa D.J., Sawyer N., Nguyen T., Im D.S., Stocco R., Bellefeuille J.N., Abramovitz M., Cheng R., Williams D.L., Jr., Zeng Z., Liu Q., Ma L., Clements M.K. Characterization of the human cysteinyl leukotriene 2 receptor. J. Biol. Chem. 2000;275:30531–30536. doi: 10.1074/jbc.M003490200. [DOI] [PubMed] [Google Scholar]
- 10.de Rubio R.G., Ransom R.F., Malik S., Yule D.I., Anantharam A., Smrcka A.V. Phosphatidylinositol 4-phosphate is a major source of GPCR-stimulated phosphoinositide production. Sci. Signal. 2018;11:eaan1210. doi: 10.1126/scisignal.aan1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black J.W., Leff P. Operational models of pharmacological agonism. Proc. R. Soc. Lond. B Biol. Sci. 1983;220:141–162. doi: 10.1098/rspb.1983.0093. [DOI] [PubMed] [Google Scholar]
- 12.Zhou B., Hall D.A., Giraldo J. Can adding constitutive receptor activity Redefine biased signaling quantification? Trends Pharmacol. Sci. 2019;40:156–160. doi: 10.1016/j.tips.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Slack R.J., Hall D.A. Development of operational models of receptor activation including constitutive receptor activity and their use to determine the efficacy of the chemokine CCL17 at the CC chemokine receptor CCR4. Br. J. Pharmacol. 2012;166:1774–1792. doi: 10.1111/j.1476-5381.2012.01901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan D., Stocco R., Sawyer N., Nesheim M.E., Abramovitz M., Funk C.D. Differential signaling of cysteinyl leukotrienes and a novel cysteinyl leukotriene receptor 2 (CysLT(2)) agonist, N-methyl-leukotriene C(4), in calcium reporter and beta arrestin assays. Mol. Pharmacol. 2011;79:270–278. doi: 10.1124/mol.110.069054. [DOI] [PubMed] [Google Scholar]
- 15.Berchiche Y.A., Sakmar T.P. CXC chemokine receptor 3 alternative Splice variants selectively activate different signaling pathways. Mol. Pharmacol. 2016;90:483–495. doi: 10.1124/mol.116.105502. [DOI] [PubMed] [Google Scholar]
- 16.DeWire S.M., Ahn S., Lefkowitz R.J., Shenoy S.K. β-Arrestins and cell signaling. Annu. Rev. Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 17.Kenakin T., Watson C., Muniz-Medina V., Christopoulos A., Novick S. A simple method for quantifying functional selectivity and agonist bias. ACS Chem. Neurosci. 2012;3:193–203. doi: 10.1021/cn200111m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulme E.C., Trevethick M.A. Ligand binding assays at equilibrium: validation and interpretation. Br. J. Pharmacol. 2010;161:1219–1237. doi: 10.1111/j.1476-5381.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X.E., He Y., de Waal P.W., Gao X., Kang Y., Van Eps N., Yin Y., Pal K., Goswami D., White T.A., Barty A., Latorraca N.R., Chapman H.N., Hubbell W.L., Dror R.O. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell. 2017;170:457–469.e413. doi: 10.1016/j.cell.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oakley R.H., Laporte S.A., Holt J.A., Barak L.S., Caron M.G. Association of beta-arrestin with G protein-coupled receptors during clathrin-mediated endocytosis dictates the profile of receptor resensitization. J. Biol. Chem. 1999;274:32248–32257. doi: 10.1074/jbc.274.45.32248. [DOI] [PubMed] [Google Scholar]
- 21.Kang Y., Zhou X.E., Gao X., He Y., Liu W., Ishchenko A., Barty A., White T.A., Yefanov O., Han G.W., Xu Q., de Waal P.W., Ke J., Tan M.H., Zhang C. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen A.H., Thomsen A.R.B., Cahill T.J., 3rd, Huang R., Huang L.Y., Marcink T., Clarke O.B., Heissel S., Masoudi A., Ben-Hail D., Samaan F., Dandey V.P., Tan Y.Z., Hong C., Mahoney J.P. Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex. Nat. Struct. Mol. Biol. 2019;26:1123–1131. doi: 10.1038/s41594-019-0330-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flock T., Hauser A.S., Lund N., Gloriam D.E., Balaji S., Babu M.M. Selectivity determinants of GPCR-G-protein binding. Nature. 2017;545:317–322. doi: 10.1038/nature22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenzen E., Dodig-Crnkovic T., Kotliar I.B., Pin E., Ceraudo E., Vaughan R.D., Uhlen M., Huber T., Schwenk J.M., Sakmar T.P. Multiplexed analysis of the secretin-like GPCR-RAMP interactome. Sci. Adv. 2019;5:eaaw2778. doi: 10.1126/sciadv.aaw2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trinquet E., Fink M., Bazin H., Grillet F., Maurin F., Bourrier E., Ansanay H., Leroy C., Michaud A., Durroux T., Maurel D., Malhaire F., Goudet C., Pin J.P., Naval M. D-myo-inositol 1-phosphate as a surrogate of D-myo-inositol 1,4,5-tris phosphate to monitor G protein-coupled receptor activation. Anal Biochem. 2006;358:126–135. doi: 10.1016/j.ab.2006.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the article are presented in the article or the Supporting Information. Additional requests should be made to the corresponding author.