Abstract
Automated tracking of wound‐healing progress using images from smartphones can be useful and convenient for the patient to perform at home. To evaluate the feasibility, 119 images were taken with an iPhone smartphone during the treatment of a chronic wound at one patient's home. An image analysis algorithm was developed to quantitatively classify wound content as an index of wound healing. The core of the algorithm involves transforming the colour image into hue‐saturation‐value colour space, after which a threshold can be reliably applied to produce segmentation using the Black‐Yellow‐Red wound model. Morphological transforms are used to refine the classification. This method was found to be accurate and robust with respect to lighting conditions for smartphone‐captured photos. The wound composition percentage showed a different trend from the wound area measurements, suggesting its role as a complementary metric. Overall, smartphone photography and automated image analysis is a promising cost‐effective way of monitoring patients. While the current setup limits our capability of measuring wound area, future smartphones equipped with depth‐sensing technology will enable accurate volumetric evaluation in addition to composition analysis.
Keywords: biomedical image analysis, image segmentation, image thresholding, photography, wound healing
1. INTRODUCTION
Information on the wound‐healing process is clinically important. For example, it is recommended to monitor the rate of wound healing in diabetic foot ulcers to determine whether the treatment was optimal.1 With the recent rapid development and wide adoption of smartphone digital photography technology, we explore the feasibility of tracking wound changes with smartphones at home. Our vision is that, eventually, this image collection process can be performed by the patient him/herself. The benefit of using a portable camera for telemedicine has been discussed long before the current generation of modern smartphones.2, 3, 4 In these studies, the potential of cell phones in tracking wound‐healing progress was recognised. They especially highlighted camera phones' capability of storing and transmitting useful clinical data, that is, wound images. The saving on transportation and clinician time was reported to be significant and well‐received.
However, most of these studies were performed at a time when camera phones took photos of much worse quality than even today's most affordable smartphones. Moreover, the old telemedicine setup still required clinicians to manually interpret individual images. In comparison, today's smartphones are powered by high‐quality imaging systems, fast Internet connections, and powerful processors capable of performing simple image analysis to produce useful quantitative wound‐healing statistics. This may reduce the burden on clinicians to manually review the images if the images are taken in a manner that allows automated wound area contouring and segmentation. Our goal here is to develop and evaluate a workflow to extract clinically relevant information from a smartphone wound image, with the hope that, in the future, patients can take these pictures themselves at home to save hospital visits and clinician time. This means that, for image acquisition, no additional device or calibration should be required, and the analysis methods should be agnostic to the capturing device and robust under different lighting conditions. Then, the image will be processed by an algorithm to report clinically useful information.
We recognised that two different types of output information are potentially useful: wound dimension and wound composition. Most wound image analysis research has been focused on the photogrammetric measurement of physical wound dimensions. This usually relies on placing a calibrated marker or grid next to the wound,5, 6, 7, 8, 9 from which the pixel dimensions can be approximately correlated to physical dimensions. However, such a setup can become cumbersome and confusing if one would like to ask the patient to acquire the image him/herself. Moreover, each type of grid or marking needs to be individually segmented and calibrated for image analysis, making the analysis ill‐suited for a non‐supervised workflow. Alternatively, it is also possible to directly obtain approximate physical dimensions from 2D images if the intrinsic and extrinsic camera matrices are calibrated and assuming the wound is planar, a reasonable assumption for small wounds. For instance, a calibrated, specially constructed camera system is described in Reference 10, and other commercial (usually expensive) devices are also available. While this may be a feasible solution at a hospital clinic or research facility, it does not fit our ultimate goal of making the data collection process accessible for everyone at home without special equipment. We would like each image to be acquired freehand from different poses and different cameras. Moreover, each camera has a distinct intrinsic calibration, which can only be determined through a non‐trivial camera calibration process.
On the other hand, wound composition (mixture of slough, necrosis, granulation, epithelium, etc.) can be highly variable during healing processes like granulation and reepithelisation. Therefore, quantification and reporting of such processes by calculating the percentage of each type of tissue can provide useful information.11 To report wound composition, semantic segmentation of the wound must be performed. Because physical dimensions are not required, this can be performed on physically uncalibrated images. One of the earliest attempts at this was performed by Arnqvist et al9 who used a semi‐automatic approach on red‐green‐blue (RGB) images, where an operator would manually determine the best among 16 classifiers for classification. While the semi‐automatic approach was a novel method then, later research focused more on automatic methods because of the development in fields of digital photography, machine learning, and computer vision. The method developed by Mekkes et al11 is based on colour coordinates on the RGB cube scale. They realised the importance of tissue colour inside the wound and proposed applying the Black‐Yellow‐Red wound colour model to the analysis. They semi‐automatically generated a mapping matrix for each possible RGB combination for classification. Hansen et al12 provided a related application for determining the severity of wound after injury using colour information.
With increased computational power and higher bit depth acquired in digital colour images, more modern methods have been developed. More researchers started considering transforming the image colour space instead of using the colour cube classifier approach. Hoppe et al13, 14 was one of the first to use the hue‐saturation‐intensity (HSI) colour model to quantify the overall colour into five different grades of slough content. Their method involved calibration of the camera colour intensity. Their treated images include mostly red granulation and yellow slough. With recent advancements in photography, especially much more robust white balancing algorithms today, the calibration step is likely no longer necessary. More recently, Mukherjee et al15 described a method where the saturation component of the HSI colour space is considered using fuzzy divergence‐based thresholding and machine learning methods to classify the percentage composition of different wound tissue. Such machine learning‐based methods can often provide better results but are harder to implement on mobile devices where software frameworks are not as developed, and computational power is limited. We would like to take an approach similar to Mukherjee's but develop a method that is (a) easily implementable on all of today's smartphone platforms, (b) does not involve calibration, and (c) is robust enough to work well under different lighting conditions. As a secondary goal, we would like the algorithm to work well on large chronic wounds (because of the population of patient we see at our clinic; see example images in Figure 1) as most literature mentioned above treats much smaller wounds.
Figure 1.
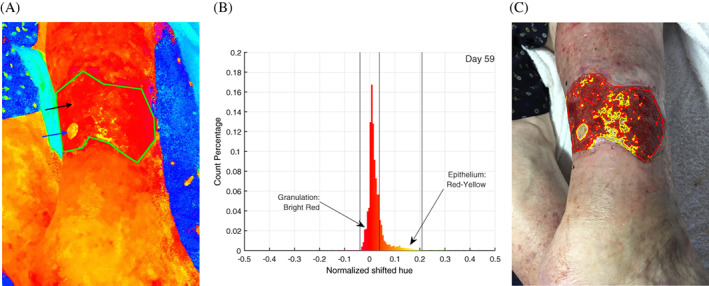
An example of the image analysis pipeline of image, taken on day 59 of treatment. A, Image converted to hue‐saturation‐value (HSV) space, with the hue space visualised with maximum saturation. The top black arrow points at the large red area, which corresponds to granulation tissue. The bottom blue arrow points at a yellow patch, which is new grown epithelial tissue. B, The distribution of hue values inside the green contour is counted and plotted in the histogram shown. C, Thresholds are applied on the histogram result followed by morphological transforms and visualisation operation to produce the wound classification shown. The vertical lines indicate thresholds found to be the best for red granulation at and for yellow at
Recently, given the consent of a patient seen at the William Osler Health System (Toronto, Canada) Palliative Medicine unit, we collected colour photos to track the healing progress of a chronic wound on the patient's leg. Photos were taken at the patient's home using the rear camera on an Apple iPhone 7 Plus smartphone. The camera has a 12‐megapixel (4032 × 3024 pixels) sensor and f/1.8 aperture. Over the interval of the study, we saw visually significant healing of this wound. One aspect of the progress is the shrinking physical dimension. Therefore physical dimensions (ie, longest length and widest width) were recorded with a ruler (because this information cannot be easily determined from an uncalibrated camera system as discussed above). The composition of the wound over the treatment period has been highly variable and is the focus of our analysis that is described below.
2. METHODS
A total of 119 wound images were collected for a chronic wound on the patient's leg during a 90‐day treatment period. Patient consent was obtained in compliance with the Declaration of Helsinki. Wound boundaries were manually contoured on each image. The use of the manual approach is because currently reliable non‐supervised wound segmentation methods available5, 6, 16, 17, 18 are based on machine learning approaches, such as using support vector machine and convolutional neural networks. These methods are well‐understood but can only be trained and implemented when large wound image datasets are available. Therefore, wound contouring algorithms were deemed to not be the focus of the current study.
The wound area was extracted from the contour using a polygon area mask defined by the contour points. This area is defined as the region of interest (ROI). A colour space transformation from the RGB space to the hue‐saturation‐value (HSV) space was performed on the image. The HSV space is commonly used to perform segmentation of different objects5, 13, 14, 15 because of its much higher contrast between semantically different objects or tissues. The hue space value is, by convention, represented by a non‐dimensional value from 0 to 360 and wraps around. The values 0 and 360 represent the colour red, while green is at 120 and blue is at 240. The algorithm for conversion between RGB colour space and HSV colour space is:
| (1) |
Because most of our colour of interest in the Black‐Yellow‐Red model11 is centred around the red region, we took a different period in the hue space: H ∈ (−180, 180). As a result, 0 and 120 still represent red and green, respectively, but blue is shifted one negative period to (240 − 360) = − 120. Most importantly, all pixels with red colour would now be next to each other for histogram analysis, allowing much more convenient binning and intuitive visualisation. Finally, the range (−180, 180) is normalised to (−0.5, 0.5) for interoperability between different software platforms, which may define a different range for hue space (one of the most common alternatives being 0‐255 to fit within an 8‐bit image depth). The entire operation can be expressed mathematically as:
| (2) |
Notably, this is a different shifting approach from the one taken by Hoppe et al.13 Their approach, as described in the Table 1 in their study, can be described as H shifted = (H − 210) mod 360. Their goal was also to solve the same issue of a discontinuous red value. The advantage of Hoppe's method is that all numbers are kept positive, thus making it sometimes easier for software implementation, but the disadvantage is that the absolute location of the hue locations is shifted (eg, red is no longer at 0 or 360).
All pixels in the hue channel inside the ROI were counted on a histogram. The best thresholds were experimentally determined to classify the pixels inside the wound as “slough,” “epithelium,” and “no label.” Assuming each type of area would be rather contiguous, morphological operations were performed to improve the accuracy of classification. First, an image erosion structuring element of appropriate size was applied so that small misclassified regions would be eliminated. Then, a dilation filter using the same structuring element is applied to recover the correct size of classified areas. For visualisation, a small dilation structuring element was applied to each area, followed by a subtraction of the area. This series of operation allows drawing out the outer contour of the classified area. Areas of different classifications were delineated by different contour colours.
Finally, the percentage of each type of pixel classification inside the wound was calculated to be interpreted as the percent of the area inside the wound being the labelled type of tissue:
| (3) |
The entire pipeline of the analysis was implemented in the Python programming language using the OpenCV computer vision library. A sample analysis procedure is illustrated in Figure 1. Compared with some of the similar existing methods, our method is friendly for different programming platforms and libraries (eg, no machine learning library required), requiring only basic operations commonly found in image manipulation libraries. Moreover, we do not perform any physical or colour calibration procedure.
In addition to the image analysis data, an upper‐bound estimate of the wound area is provided by the measured widest length and longest length for comparison:
| (4) |
Statistical analysis was performed in Python (version 3.6.2, Anaconda Software Distribution) and MATLAB (version 9.0 R2016a, The MathWorks, Inc.).
3. RESULTS
The algorithm was run on all 119 images collected. Representative images on days 15, 41, and 87 and their analyses are shown in Figure 2. The wound was on the patient's right leg. Multiple images were required to capture the full extent of the large wound. Before treatment (on day 0), the entire chronic wound area was covered by slough. As treatment progressed, we saw significant granulation in the wound area. Around day 30, all the original slough had been replaced by granulation tissue, with signs of reepithelialisation. Afterwards, we observed a significant epithelisation process. It was also during this period where significant shrinking of the overall wound boundary was observed. (Figure 3) Therefore, the wound‐healing process can be distinguished into roughly two distinct phases: the first phase consisting primarily of granulation before day 30 and the second phase after day 30 when granulation process has saturated inside the wound, and reepithelialisation starts.
Figure 2.
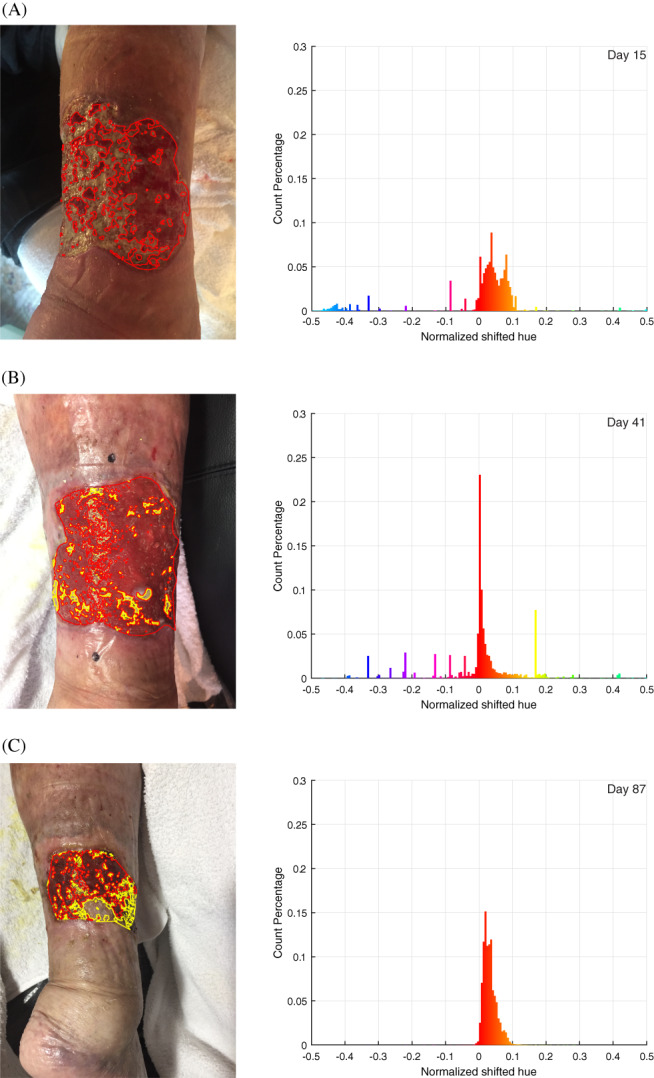
Representative analysis performed on (A) day 15, (B) day 41, and (C) day 87. The three images presented are chosen to represent different stages in the treatment process and varying image lighting conditions. The left column shows the images taken with each classified area contoured (red: granulation, yellow: reepithelialised tissue). The right column shows the hue value histogram inside the wound area. Note the peak becomes significantly higher and narrower from day 15 to 41 because of the much‐increased percentage of granulation tissue, and the peak becomes slightly wider on day 87 because of the growing amount of epithelial tissue. The general trend is also visualised in Figure 5
Figure 3.
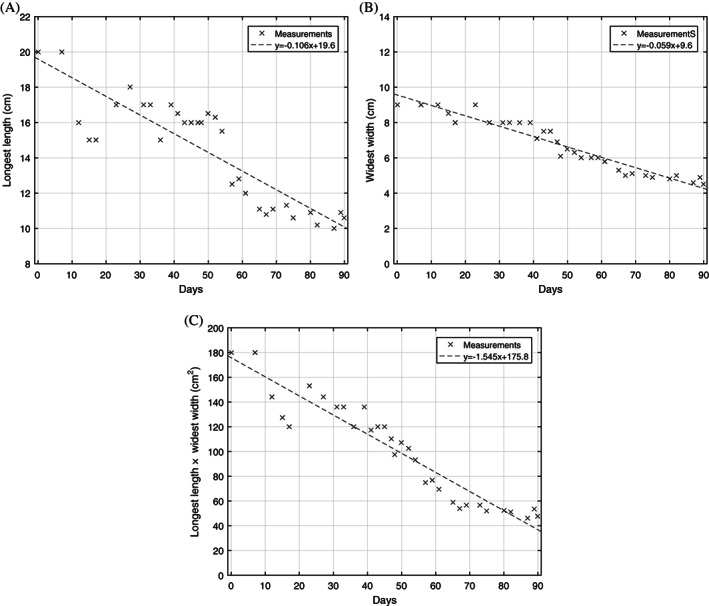
Wound size as measured by (A) longest length, (B) widest width, and (C) product of longest length and widest width, an estimate of the total wound area. Least squares linear regression is shown for each statistic: (A) R 2 = 0.774, (B) R 2 = 0.930, and (C) R 2 = 0.903
To validate the effectiveness of using the HSV colour space, we can observe the trend of hue histogram progression through the treatment process and correlate the trend with our expectation from visual inspection. As illustrated in Figure 2, the initial phase of granulation is characterised by the narrowing of the histogram peak and the increase in magnitude of the peak. The narrowing of the peak represents the decreasing amount of yellow slough, which has a higher hue number than red granulation tissue, while the increase of the peak corresponds to the increase in the number of pixels classified as red granulation tissue. In the second phase of healing, where granulation saturates and epithelium starts to form, the histogram gradually shifts into the yellow region with a decreased magnitude of peak. This represents granulation tissue turning into epithelial tissue. This overall trend of the hue distribution progression is visualised in Figure 5, where all individual histograms from the collected images on different dates are stacked on the vertical axis, and the height of the histogram is represented by the colour intensity. When the segmentation was visualised and checked on individual images manually, the result appears accurate and is robust with respect to the different lighting condition, which can be seen in the three images chosen in Figure 2.
Figure 5.
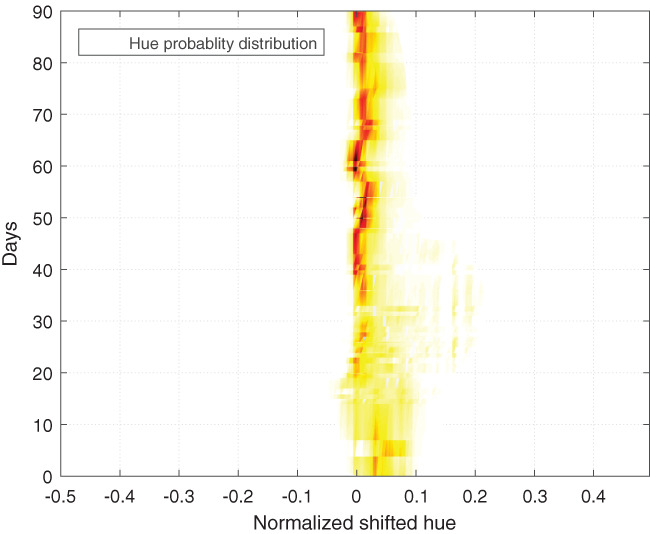
Trend of wound healing from all images visualised as tissue type distribution progression over the 90‐day tracking period. Darker colour represents higher probability density. The overall density is normalised to 1 for each analysed image. Then, the distribution for each day is averaged between images. Values between days with observations are interpolated. From bottom to top, first, we obverse a wide distribution with a peak on the right side of 0 (which represents yellow pixels). As treatment starts, this peak shifts left, and distribution becomes narrower and peaks much higher at 0 (which represents red pixels). When the reepithelisation process starts, the distribution and peak shift slightly right into yellow
The trend of measured wound size is shown in Figure 3. There is a strong linear decreasing trend (wound area vs days into treatment, R 2 = 0.903). In comparison, Figure 4 shows rapid initial granulation and epithelium growth and saturation of the growth after a few days and does not have a clear linear trend. Therefore, it is clear that the two different wound progression monitoring approaches quantifies two different aspects of the healing process. This suggests that, despite wound area being the more commonly cited metric, wound composition as calculated from our setup can provide additional insight into the healing process. Such a segmentation algorithm is especially suited for the current patient case because of the large, non‐planar wound area (thus hard to accurately measure) and the highly heterogeneous and variable wound composition (which makes the wound composition analysis an ideal way of tracking the progression).
Figure 4.
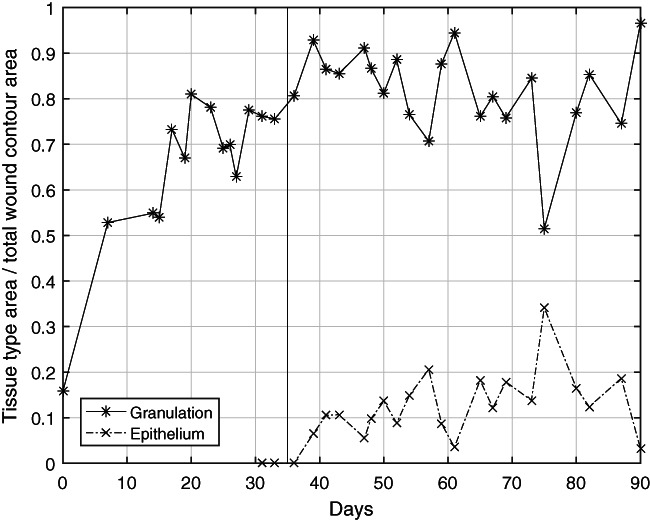
Trend of wound healing as represented by the granulation and epithelial tissue. Measured statistic across multiple images on the same day is averaged. In the beginning, there was a rapid and steady trend of slough being replaced by granulation tissue as evident on the plot as the rise in granulation tissue percentage. Accounting for epithelial tissue started on approximately day 30 when significant epithelium growth started. With an initial rise of epithelium percentage around days 35 to 60, the level was steady afterwards. This plot provides different and complementary information to the wound size information in Figure 3. A limitation of the quantification of epithelium tissue inside the wound is that it does not adequately illustrate the reepithelialisation process outside the wound because it is no longer inside the wound area
4. DISCUSSION
We have demonstrated the feasibility to track wound‐healing process using smartphone photography, where all images were acquired at the patient's home without special preparation. The focus was to develop and evaluate an algorithm for the quantification of different wound contents as an index to track the healing progress.
Our approach of transforming the image into the HSV colour space has been shown to be robust with respect to varying lighting conditions and can be used to capture both granulation and epithelial growth for this case study. Intuitively, our approach is similar to how humans distinguish different tissue types by colour. The robustness of the algorithm comes from the fact that, theoretically, the hue value should be minimally affected by the overall luminance of the image. In practice, one needs to make sure that the ROI in the photo is neither over‐exposed or under‐exposed. Fortunately, with the advancement in smartphone technology, nowadays, the image quality taken with an average smartphone is sufficient for such analysis. In the future, we would also like to verify the algorithm on subjects with different skin tones and textures. An adaptive thresholding method might be required for such cases, such as the one described in Reference 19.
In addition, our current approach consisted of an erosion and then dilation morphological transformation process that is designed to eliminate small pixel areas that are misclassified. A common source of this misclassification is because of varying lighting conditions, such as specular reflections. The morphological transforms were found to be very effective at removing the effect of such artefacts on the overall analysis.
We note that the current study is limited because of the limited one‐patient‐camera dataset available to us, and the next step would be to validate the algorithm design on multiple cases under different conditions. In addition, with a large amount of tagged image data at different stages of the wound collected, instead of correlating images to physical dimensions, one can construct a classifier correlating images to The National Pressure Ulcer Advisory Panel wound stages or other potentially more meaningful clinical metrics.
A smartphone‐based algorithmic approach to home wound care has significant advantages over traditional telemedicine. With the current feasibility validation, we propose building a centralised patient record service where chronic wound patients can take daily images of the wound and upload to the database. The wound analysis can be performed on the smartphone itself to provide real‐time feedback to the patient if desired. Meanwhile, the database is set up to be associated with existing patient records, track monitor wound‐healing progress, and automatically alerts the physician when intervention is necessary. Home care nurses can also monitor patients remotely through the statistics and image records instead of having to physically visit the patient. Such a technology can potentially play an important role in a more efficient long‐term care system.
An important limitation of the current setup is that the wound area needs to be manually measured using a ruler, which can be cumbersome and inaccurate. This is because, as discussed above, the wound area cannot be extracted from a 2D image taken in an uncalibrated setup. Moreover, large wound areas over a non‐planar geometry, such as the current case of large wound on leg, cannot be captured in one single image. Transparent soft grids, such as the Visitrak film,7 for tracing are likely the most accurate solution currently but require skin contact and are a specialised equipment that the patient needs to acquire.
A promising technology to solve this limitation is 3D cameras. These cameras provide not only colour but also physical distance information of the captured scene. This allows the characterisation of both the colours and physical dimension of the wound. With 3D reconstruction technology, the wound can be of any arbitrary large size and 3D geometry in space. Because the algorithm reconstructs the wound fully in 3D, the measured physical dimensions can be much more accurate than existing methods. Depth information can also make wound contouring much easier, eliminating the need for a complex wound area detection algorithm. The 3D camera technology has been demonstrated to be very useful in wound contouring and wound size characterisation.20, 21 Moreover, the technology can also accurately provide information about the depth of wound (in addition to area), which was previously impossible. Finally, such a technique does not require contact with the wound bed.
Traditionally, the 3D camera technology has been limited by its resolution, portability, and cost. In recent years, these issues have largely been solved with new products that are of High‐definition (HD, 1280×720) resolution, small form factor, and can be purchased for around $200.22 In addition, stereo rear camera and depth camera setups have started to appear on mainstream smartphones. This is partly because of the recent surge of interest in smartphone‐based augmented reality and 3D facial recognition. For example, the Apple iPhone X has a depth camera using structured illumination technology for 3D facial recognition. Google Project Tango is another experimental project of incorporating depth‐sensing technology into smartphones, which will enable “3D modeling on the go.”23 Once adopted, we can potentially have the unprecedented capability to accurately measure both wound volume and wound composition with a single photo shot from smartphones. Therefore, we would like to suggest this as a future direction in wound characterisation efforts.
CONFLICTS OF INTEREST
The authors have no conflict of interest to declare.
ACKNOWLEDGEMENTS
The authors would like to thank members of the Guided Therapeutics (GTx) Lab at University Health Network (UHN) for their support. The authors received no specific funding for this work.
Shi RB, Qiu J, Maida V. Towards algorithm‐enabled home wound monitoring with smartphone photography: A hue‐saturation‐value colour space thresholding technique for wound content tracking. Int Wound J. 2019;16:211–218. 10.1111/iwj.13011
REFERENCES
- 1. Steed DL, Attinger C, Colaizzi T, et al. Guidelines for the treatment of diabetic ulcers. Wound Repair Regen. 2006;14(6):680‐692. 10.1111/j.1524-475X.2006.00176.x. [DOI] [PubMed] [Google Scholar]
- 2. Hofmann‐Wellenhof R, Salmhofer W, Binder B, Okcu A, Kerl H, Soyer HP. Feasibility and acceptance of telemedicine for wound care in patients with chronic leg ulcers. J Telemed Telecare. 2006;12(1_suppl):15‐17. 10.1258/135763306777978407. [DOI] [PubMed] [Google Scholar]
- 3. Braun RP, Vecchietti JL, Thomas L, et al. Telemedical wound care using a new generation of mobile telephones. Arch Dermatol. 2005;141(2):254‐258. 10.1001/archderm.141.2.254. [DOI] [PubMed] [Google Scholar]
- 4. Terry M, Halstead LS, O'Hare P, et al. Feasibility study of home care wound management using telemedicine. Adv Skin Wound Care. 2009;22(8):358‐364. 10.1097/01.ASW.0000358638.38161.6b. [DOI] [PubMed] [Google Scholar]
- 5. Perez AA, Gonzaga A, Alves JM. Segmentation and analysis of leg ulcers color images. Proceedings International Workshop on Medical Imaging and Augmented Reality; Shatin, Hong Kong, China. 2001:262‐266. 10.1109/MIAR.2001.930300. [DOI] [Google Scholar]
- 6. Wang C, Yan X, Smith M, et al. A unified framework for automatic wound segmentation and analysis with deep convolutional neural networks. 2015 37th Annu Int Conf IEEE Eng Med Biol Soc; Milan. 2015:2415‐2418. 10.1109/EMBC.2015.7318881. [DOI] [PubMed] [Google Scholar]
- 7. Chang AC, Dearman B, Greenwood JE. A comparison of wound area measurement techniques: visitrak versus photography. Eplasty. 2011;11:e18. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3080766&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 8. Moustris GP, Hiridis SC, Deliparaschos KM, Konstantinidis KM. Evolution of autnomous and semi‐autnomous robotic surgical systems: a review of the literature. Int J Med Robot. 2011;7(April):375‐392. 10.1002/rcs. [DOI] [PubMed] [Google Scholar]
- 9. Arnqvist J, Hellgren J, Vincent J. Semiautomatic classification of secondary healing ulcers in multispectral images. [1988 Proceedings] 9th International Conference on Pattern Recognition. IEEE Comput. Soc. Press; Rome, Italy. 459‐461. 10.1109/ICPR.1988.28266. [DOI] [Google Scholar]
- 10. Krouskop TA, Baker R, Wilson MS. A noncontact wound measurement system. J Rehabil Res Dev. 2002;39(3):337‐345. http://www.rehab.research.va.gov/jour/02/39/3/krouskop.htm. [PubMed] [Google Scholar]
- 11. Mekkes JR, Westerhof W. Image processing in the study of wound healing. Clin Dermatol. 1995;13(4):401‐407. 10.1016/0738-081X(95)00071-M. [DOI] [PubMed] [Google Scholar]
- 12. Hansen GL, Sparrow EM, Kokate JY, Leland KJ, Iaizzo PA. Wound status evaluation using color image processing. IEEE Trans Med Imaging. 1997;16(1):78‐86. 10.1109/42.552057. [DOI] [PubMed] [Google Scholar]
- 13. Hoppe A, Wertheim D, Melhuish J, Morris H, Harding KG, Williams RJ. Computer assisted assessment of wound appearance using digital imaging. Annu Reports Res React Institute, Kyoto Univ. 2001;3:2595–2597. doi: 10.1109/IEMBS.2001.1017312 [DOI] [Google Scholar]
- 14. Oduncu H, Hoppe A, Clark M, Williams RJ, Harding KG. Analysis of skin wound images using digital color image processing: a preliminary communication. Int J Low Extrem Wounds. 2004;3(3):151‐156. 10.1177/1534734604268842. [DOI] [PubMed] [Google Scholar]
- 15. Mukherjee R, Manohar DD, Das DK, Achar A, Mitra A, Chakraborty C. Automated tissue classification framework for reproducible chronic wound assessment. Biomed Res Int. 2014;2014:1‐9. 10.1155/2014/851582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kolesnik M, Fexa A. How robust is the SVM wound segmentation? Proc 7th Nord Signal Process Symp NORSIG 2006; 2007; Rejkjavik. (Section 4): :50‐53. 10.1109/NORSIG.2006.275274. [DOI] [Google Scholar]
- 17. Lu H, Li B, Zhu J, et al. Wound intensity correction and segmentation with convolutional neural networks. Concurr Comput Pract Exp. 2017;29(6):1‐10. 10.1002/cpe.3927. [DOI] [Google Scholar]
- 18. Kolesnik M, Fexa A. Multi‐dimensional color histograms for segmentation of wounds in images. In: Kamel M, Campilho A, eds. Image Analysis and Recognition. Berlin, Heidelberg: Springer Berlin Heidelberg; 2005:1014‐1022. [Google Scholar]
- 19. Yang G, Li H, Zhang L, Cao Y. Research on a skin color detection algorithm based on self‐adaptive skin color model. Int Conf Commun Intell Inf Secur. Vol 2010; Nanning, Guangxi Province, China. 2010:266‐270. 10.1109/ICCIIS.2010.67. [DOI] [Google Scholar]
- 20. Gaur A, Sunkara R, Noel A, Raj J, Celik T. Efficient wound measurements using RGB and depth images. Int J Biomed Eng Technol. 2015;18(4):333‐358. 10.1504/IJBET.2015.071009. [DOI] [Google Scholar]
- 21. Plassmann P, Jones BF, Ring EFJ. A structured light system for measuring wounds. Photogramm Rec. 1995;15(86):197‐204. 10.1111/0031-868X.00025. [DOI] [Google Scholar]
- 22. Carfagni M, Furferi R, Governi L, Servi M, Uccheddu F, Volpe Y. On the performance of the intel SR300 depth camera: metrological and critical characterization. IEEE Sens J. 2017;17(14):4508‐4519. 10.1109/JSEN.2017.2703829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schops T, Sattler T, Hane C, Pollefeys M. 3D modeling on the go: interactive 3D reconstruction of large‐scale scenes on mobile devices. Proc – 2015 Int Conf 3D Vision, 3DV 2015; Lyon. 2015:291‐299. 10.1109/3DV.2015.40. [DOI] [Google Scholar]


