ABSTRACT
Background
Gut microbiota composition as influenced by long-term diet may be associated with the risk of adult chronic diseases. Thus, establishing the relation of long-term diet, particularly starting from early life, with adult microbiota composition would be an important research advance.
Objective
We aimed to investigate the association of long-term intake of energy, carbohydrate, fiber, protein, and fat from infancy to late adolescence with microbiota composition in adulthood.
Methods
Within the prospective DOrtmund Nutritional and Anthropometric Longitudinally Designed (DONALD) Study, we sampled stool 1 or 2 times within 1 y from 128 adults (median age: 29 y). Microbiota composition was profiled by 16S ribosomal RNA sequencing. Annual dietary records from age 1 to 18 y were retrieved. We estimated trajectories of energy, energy-adjusted carbohydrate, fiber, protein, and fat intake with multilevel models, producing predicted intake at age 1 y and rates of change in intake. A multivariate, zero-inflated, logistic-normal model was used to model the association between intake trajectories and the composition of 158 genera in single-sampled individuals. Associations found in this model were confirmed in double-sampled individuals using a zero-inflated Beta regression model.
Results
Adjusting for covariates and temporal differences in microbiota composition, long-term carbohydrate intake was associated with 3 genera. Specifically, carbohydrate intake at age 1 y was negatively associated with Phascolarctobacterium [coefficient = −4.31; false discovery rate (FDR)–adjusted P = 0.006] and positively associated with Dialister (coefficient = 3.06; FDR-adjusted P = 0.003), and the rate of change in carbohydrate intake was positively associated with Desulfovibrio (coefficient = 13.16; FDR-adjusted P = 0.00039). Energy and other macronutrients were not associated with any genus.
Conclusions
This work links long-term carbohydrate intake to microbiota composition. Considering the associations of high carbohydrate intake and microbiota composition with some diseases, these findings could inform the development of gut microbiota–targeted dietary recommendations for disease prevention.
Keywords: long-term diet, carbohydrate intake, 3-day weighed dietary records, gut microbiota composition, Phascolarctobacterium, Dialister, Desulfovibrio, DONALD study
Introduction
Habitual or long-term diet crucially influences the development of several chronic diseases in adults (1). Moreover, the role of the gut microbiota in the development of some of these chronic diseases is becoming increasingly evident (2). The fact that gut microbiota modulates the impact of diet on health (3) suggests that changes in gut microbiota composition may be one of the mechanisms underlying the relation between long-term diet and chronic diseases. Indeed, long-term diet exerts a profound and sustained impact on the gut microbiota (3). It also induces the growth and proliferation of specific gut bacteria in order to achieve a state of gut eubiosis (4). Thus, it is necessary to continue to explore the link between long-term diet and microbiota composition. The relation between long-term diet and the highly interacting members of the gut microbiota is exceptionally complex. Therefore, pinpointing the nature of the relation of long-term diet with individual bacterial abundances could provide additional and important insights. This might reveal tractable interactions between diet and gut bacteria, both in terms of bacteria-specific metabolism and identifying the specific bacteria that might potentially respond best to tailored dietary alterations.
Evidently, the nutrient composition of diets primarily drives their relations with gut microbiota composition (5, 6). While food and dietary pattern approaches acknowledge the synergistic effect of nutrients and foods with a matrix, exploring singular nutrients, especially in longitudinal studies, will enhance our understanding of diet–microbiota relations (4). Indeed, a large and growing body of epidemiological evidence has revealed the significant impact of macronutrient intake on microbiota composition (5–9). The 3 macronutrients—carbohydrates (including fibers), proteins, and fat—may reach the gut microbiota after escaping primary digestion due to intake exceeding the rate of digestion, inherent structural complexities, and inability of human enzymes to metabolize them (10). Since macronutrients differently influence microbiota composition and different microbes have different metabolic capabilities (11), additional information on the independent associations of macronutrients with microbiota composition is required.
While the relation of long-term macronutrient intake in adulthood with adult microbiota composition is well documented (12–14), there is insufficient evidence on the impact of long-term diet, particularly starting early in life on adult microbiota composition. Such an investigation would be vital since the association of diet with gut microbiota composition appears to be life-stage dependent (5). Exploring long-term diet over the life stages requires repeated dietary assessments. A variable measured repeatedly over time can be comprehensively modeled as individual growth trajectories, producing potentially interesting dimensions of initial status or starting point and rates of change (15). Evaluating the relation between individuals’ initial status and rates of change over time in macronutrient intake and microbiota composition would improve our understanding of the impact of macronutrient intake on later microbiota composition. Although the core gut microbiota appear to be temporally stable, large variation in the less-abundant microbes, even in a short period, has been reported (16). In adults, evidence of this temporal variation ranges from moderate (17) to considerable (18). Unsurprisingly, temporal variations in gut microbiota composition correlate with dietary changes (19, 20). Aside from 1 study (14), previous studies linking long-term macronutrient intake with microbiota composition in adults have understandably been restricted to gut microbiota composition profiling at a single time point (12, 13). Repeated measurements of microbiota composition will additionally control for intraindividual differences, thereby strengthening long-term diet–gut microbiota associations.
The sufficiently large number of dietary assessments of the participants of the DOrtmund Nutritional and Anthropometric Longitudinally Designed (DONALD) Study across the life course and tracking of these individuals into adulthood (21) present an opportunity to explore long-term diet–gut microbiota associations. To this end, we analyzed annual 3-d weighed dietary records from infancy to late adolescence in the DONALD study in order to capture long-term intake of energy and 4 major macronutrients—carbohydrate, fiber, protein, and fat—as individual growth trajectories, and examine their prospective associations with microbiota composition in adulthood, taking into account covariates and temporal differences in microbiota composition.
Methods
Study population
Commencing in 1985, the DONALD study is an ongoing, open prospective cohort study of individuals living in the German town of Dortmund and surrounding cities. The DONALD study uses a convenient sampling scheme. Aside from the higher educational attainment and higher socioeconomic status of DONALD study participants’ parents, there are no major deviations from the reference German population (21). The main aim of this study is to evaluate the relations between dietary intake, metabolism, and growth from infancy to adulthood. Participants’ examinations included annually repeated anthropometric measurements and 3-d weighed dietary records. Early-life factors such as birth weight and delivery mode (cesarean compared with vaginal delivery) and socioeconomic status of study participants’ parents were extracted from maternal delivery records and parental interviews. The DONALD study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the University of Bonn (approval number 098/06). Written informed consent was obtained from the parents or legal guardians of the participants in childhood and later on from the participants themselves. Details of the recruitment and follow-up in the DONALD study are presented elsewhere (21).
Study design
This current microbiota composition–focused study is nested within the DONALD study. All study participants who were adults, aged ≥18 y, were eligible. Three hundred stool-sampling kits and questionnaires on history of gastrointestinal conditions and intake of antibiotics and probiotics within 6 mo were sent out by mail in 2 phases within 1 y. The participants were randomly selected. For the individuals who provided stool samples, archived self-reported dietary data and other covariates were retrieved. The study population's flow chart is shown in Supplemental Figure 1.
Assessment of dietary intake
In the DONALD study, dietary intake was assessed annually using 3-d weighed dietary records on 3 consecutive days. Dietary records were coded and linked to the continuously updated in-house food-composition database, LEBTAB, in order to calculate the intake of energy and nutrients (21). Individual means of daily energy (kcal/d) and carbohydrate, fiber, protein, and fat (g/d) intakes were calculated. Since we planned to model trajectories of intake and nonlinear trajectory modeling typically requires >2 assessment points per individual (15, 22), an additional inclusion criterion in the current study was the availability of ≥3 repeated dietary records between age 1 and 18 y.
Stool sampling, DNA extraction, and 16S ribosomal RNA sequencing of stool samples
Stool sampling and DNA extraction
Participants provided stool samples 1 or 2 times within 1 y. Samples were collected in participants’ homes into tubes containing RNAlater(Qiagen). The stability of gut microbiota composition for 7 d at room temperature in RNAlater is documented (23). Within 24 h of collection, samples were sent to the German Center for Neurodegenerative Diseases Biobank. On arrival at the biobank, samples were homogenized, placed into aliquots, and stored at −80°C. Bacterial genomic DNA was extracted from 0.25 g of stool sample using the repeat bead-beating plus column protocol as described by Yu and Morrison (24), in combination with the QIAamp Fast DNA Stool Mini Kit and protocol (Qiagen) with slight modifications. Details of the DNA extraction can be found in the Supplemental Methods. The DNA samples were stored at −80°C until 16S ribosomal RNA (rRNA) sequencing.
16S rRNA sequencing of stool samples
Following DNA extraction, the V3-V4 regions of the 16S rRNA gene were amplified through 30 cycles of PCR reactions on the template DNA using the primer pair 5′-TCGT CGGC AGCG TCAG ATGT GTAT AAGA GACA GCCT ACGG GNGG CWGC AG-3′ and 5′-GTCT CGTG GGCT CGGA GATG TGTA TAAG AGAC AGGA CTAC HVGG GTAT CTAA TCC-3′, according to the 16S Metagenomic Sequencing preparation protocol for Illumina MiSeq and as previously described by Fouhy et al. (25). Indexed products were visualized using gel electrophoresis and cleaned with AMPure XP magnetic-based beads prior to DNA quantification using the Qubit (BioSciences), along with the Qubit High Sensitivity DNA kit (Life Technologies). Samples were pooled at equimolar concentrations, and high-throughput sequencing was completed on the Illumina MiSeq platform in the Teagasc sequencing facility using a 2 × 300-cycle kit, following standard Illumina sequencing protocols.
Assessment of other covariates
Demographic information such as sex, early-life data such as birth weight and length, and delivery mode was obtained from maternal delivery records, while breastfeeding duration and maternal and socioeconomic variables such as maternal education and occupation were collected by parental interviews. For the current study, we considered maternal variables such as maternal BMI from the first visit. Anthropometric measurements were conducted annually at the study center and BMI trajectories from age 4 to 18 y were computed, as previously reported (26). Physical activity was assessed using standardized questionnaires and the mean of physical activity [metabolic equivalent of task hours (MET-h)/week] between age 4 and 18 y was computed. Lifestyle factors from age 18 y, such as alcohol consumption and smoking, as well as the history of gastrointestinal conditions and intake of antibiotics and probiotics around stool sampling were obtained by questionnaires.
Statistical analyses
Basic characteristics
Basic characteristics of the study sample are presented as medians with 25th and 75th percentiles or as counts (with percentages) as appropriate. Comparisons of basic characteristics between individuals who provided 1 stool sample (single-sampled individuals) and 2 stool samples (double-sampled individuals) were explored using the Mann–Whitney U and the chi-square tests for continuous and categorical variables, respectively. These analyses were conducted using SAS 9.4 (SAS®; SAS Institute).
Modeling of trajectories of intake of energy, carbohydrate, fiber, protein, and fat
Energy-adjusted macronutrient intakes were computed using the residual method with the energy intake as the independent variable and the absolute macronutrient intake as dependent variables. To model actual macronutrient intake, the residuals from the regression models were standardized to their age-specific predicted macronutrient value at the mean energy intake (27).
Energy and macronutrient intake trajectories were modeled using multilevel models with level-1 (age) and level-2 (individual) random effects, such that each individual had his/her own trajectories (random intercepts and slopes or rates of change over time). All multilevel models were fitted with PROC MIXED in SAS 9.4. The “between/within” method was used to compute the denominator degrees of freedom for tests of fixed effects and SE variance-covariance structure. To model initial intake, we centered age at the minimum age of 1 y. For the trajectory of intake for each variable—energy, carbohydrate, fiber, protein, and fat—we considered 2 age models: linear and quadratic. We examined the optimal age models by comparing model fits using the corrected Akaike's Information Criterion. For the optimal age models, we included sex if P < 0.05. The current intercepts were the predicted intake at age 1 y, and the linear and quadratic slope(s) (rates of change) were the amount of increase/decrease in intake for each unit of increase in age and age-squared, respectively. These dietary trajectory variables, intake at age 1 y and rates of change in intake, were used for the subsequent analyses.
16S rRNA sequencing data preprocessing
The resulting 300-bp paired-end reads from the MiSeq analysis were assembled using FLASH (fast length adjustment of short reads to improve genome assemblies). QIIME (Quantitative Insights into Microbial Ecology; version 1.8.0) was used for quality filtering of paired-end reads, which is based on a quality score of >25 and the removal of mismatching barcodes. Prior to the analysis as part of the quality-control process, sequences that produced <40,000 reads were manually removed. Open reference operational taxonomic units were formed from the sequences at 97% similarity. Finally, using the R package Phyloseq, operational taxonomic units were assigned to 341 genera and their relative abundances were estimated.
Multivariable linear regression
For the regression analyses of the association of trajectories of energy and macronutrient intake with gut bacterial taxa, we considered 158 adequately abundant taxa using the criteria of relative abundance ≥0.2% in at least 10% of the samples by Wu et al. (12). Of these 158 taxa, Bacteroides, Blautia, and Lachnoclostridium were present in all participants. These 158 genera and their variability are presented in Supplemental Table 1. We adopted a 2-step statistical analysis in order to fit models that are appropriate for the number of assessments of the dependent variables (taxa) and to follow the recommendation of the need for multiple statistical methods to identify associations between diet and microbial abundances (28).
In the first step, we examined associations of intake at age 1 y and rates of change in intake with the relative abundance of the 158 genera in the single-sampled individuals. We regressed the relative abundance of the 158 genera simultaneously on these dietary intake variables by using the multivariate zero-inflated logistic-normal (MZILN) model (29). The MZILN handles compositional structure and high dimensionality of microbiota data. It is appropriate for modeling microbial taxa data as dependent variables. Its ability to account for the complex correlation among simultaneously modeled taxa is an advantage over analyzing taxa one by one with multiple testing correction. The model automatically takes the last taxon as the reference. Thus, the last genus out of the 158 genera, Succiniclasticum, was used as the first reference genus. To ensure valid results, we randomly selected 2 new reference genera, Acinetobacter and Prevotellaceae UCG-001, and reran our analysis. The dietary variable–genus associations that were observed across 2 or 3 reference taxa were considered consistent and hence selected. The average of the coefficients was reported. For this analysis, model 1 represents associations of each intake trajectory (intake at age 1 y and their rates of change) with each genus. Model 2 (full multivariable-adjusted) comprises all dietary intake variables, sex, an indicator variable for whether an individual has a sibling in the study sample (reference category) or not, birth weight and length, maternal BMI, maternal gestational weight, maternal education and occupation, cesarean (compared with vaginal) delivery, first-born (compared with other) birth order, smoking household, alcohol consumption, smoking, physical activity, BMI trajectory, and age at fecal sampling. All dietary variables and continuous covariates were standardized by subtracting their means and dividing them over their standard deviations, and all categorical variables were dichotomized. In sensitivity analysis, we fitted the same models excluding participants who reported gastrointestinal disease or took antibiotics or probiotics within 6 mo and compared the results of these models.
In the second step, associations with P values <0.05 in the above full multivariable-adjusted model were validated in the double-sampled individuals. For this analysis, we used a zero-inflated Beta regression model with random effects (ZIBR) model (30). Like the MZILN, the ZIBR also handles the compositional and zero-inflated nature of the microbiota data. Importantly, the ZIBR includes random effects that take the correlation of the repeated sampling of microbiota in the same individual into account. The model produces a joint test of association (P) for its logistic and Beta components. The estimates of these components were interpreted as effects of a dietary variable on the presence of a genus and on the level of relative abundance given the presence of the genus, respectively. For this analysis, we modeled all dietary variables and all covariates that were found to be different (P < 0.05) between single-sampled and double-sampled individuals. Each genus was analyzed one by one. When we tested the association of a dietary variable with >1 genus, we corrected for multiple testing by selecting an associated genus at a false discovery rate (FDR)–adjusted P < 0.05. For the whole dataset, missing covariate values were multiply imputed to give 10 imputed datasets and covariate values for each individual were computed as averages across imputations. All multivariable regression analyses were performed in R (version 3.6.2).
Results
Description of study population
Table 1 presents the characteristics of the 128 individuals in the current study. The median age of the participants at the time of stool sampling was 29 y. Fifty-one individuals provided 1 sample (single-sampled) and 77 individuals provided 2 samples (double-sampled) at an average of 280 d apart. There was a median of 16 dietary records per individual. One-third of the study sample completed all 18 annual dietary intake records. More than half (60%) were females. Furthermore, the birth weight was ∼3.5 kg. The majority of the mothers (90%) were highly educated. They had ∼45 MET-h/wk of physical activity. Approximately 8% followed the overweight trajectory. In adulthood, most of the participants smoked and consumed alcohol. Except for history of gastrointestinal conditions and use of antibiotics or probiotics within 6 mo of sampling, we found no differences in the median or proportion of other characteristics between the single-sampled and double-sampled individuals. The median energy intake and energy-adjusted carbohydrate, fiber, protein, and fat intakes at age 1 year and 18 y were 769 and 2056 kcal/d, 100 and 281 g/d, 9 and 19 g/d, 25 and 73 g/d, and 31 and 79 g/d, respectively (Supplemental Table 2). As shown in Supplemental Figure 2A–E, intake generally increased over time, with a few dietary variables appearing to increase nonlinearly. Furthermore, interindividual variation in intake appeared smaller at age 1 y as compared with age 18 y.
TABLE 1.
Basic characteristics of the study population1
| n | Values | Single-sampled (n = 51) | Double-sampled (n = 77) | P | |
|---|---|---|---|---|---|
| Dietary intake | |||||
| Annual dietary records | 128 | 16 (11, 18)2 | 16 (10, 18) | 16 (12, 18) | 0.79 |
| Complete annual dietary records, n (%) | 128 | 39 (30.47) | 15 (29.41) | 24 (31.17) | 0.83 |
| Demographic variables | |||||
| Female, n (%) | 128 | 82 (64.06) | 28 (54.90) | 54 (70.13) | 0.09 |
| Age, y | 128 | 29.3 (23.7, 32.8) | 29.85 (23.96, 32.99) | 27.99 (23.43, 32.18) | 0.32 |
| Early-life, socioeconomic, and maternal variables | |||||
| Birth weight, g | 115 | 3460 (3150, 3730) | 3425 (3080, 3630) | 3480 (3185, 3745) | 0.35 |
| Birth length, cm | 115 | 52 (50, 53) | 52 (50, 53) | 52 (51, 53.5) | 0.68 |
| Cesarean delivery, n (%) | 25 | 16 (64) | 8 (61.54) | 8 (66.67) | 0.79 |
| First-born child, n (%) | 115 | 63 (58.26) | 26 (55.32) | 41 (60.29) | 0.79 |
| Siblings, n (%) | 128 | 26 (20.31) | 8 (15.69) | 18 (23.38) | 0.29 |
| Breastfeeding duration, wk | 115 | 26 (8, 41) | 28 (8, 42) | 26 (7, 41) | 0.94 |
| Maternal BMI, kg/m2 | 112 | 22.61 (20.4, 26.34) | 22.26 (20.34, 26.34) | 23.12 (20.5, 26.34) | 0.92 |
| Maternal gestational weight gain, kg | 115 | 13 (10, 16) | 13 (10, 16) | 13 (10.5, 15) | 0.67 |
| Maternal education, high, n (%) | 92 | 83 (90.21) | 34 (91.89) | 49 (89) | 0.96 |
| Maternal employment, employed, n (%) | 92 | 26 (28.26) | 8 (21.62) | 18 (32.73) | 0.42 |
| Smoking household, n (%) | 93 | 28 (30.11) | 13 (36.12) | 15 (26.31) | 0.53 |
| Overweight trajectory, n (%) | 115 | 9 (7.83) | 4 (8.51) | 5 (7.35) | 0.85 |
| Physical activity, MET-h/wk | 120 | 44.98 (28.94, 60.85) | 53.14 (30.6, 71.69) | 43.10 (28.73, 55.89) | 0.14 |
| Lifestyle factors in adulthood, n (%) | |||||
| Current smokers | 90 | 78 (86.67) | 30 (81.08) | 48 (90.57) | 0.35 |
| Current alcohol consumers | 96 | 80 (83.33) | 19 (92.31) | 61 (77.19) | 0.06 |
| GIT condition | 103 | 19 (18.45) | 1 (3.85) | 18 (23.38) | 0.04 |
| Antibiotics use | 103 | 29 (28.16) | 2 (7.69) | 27 (35.06) | <0.001 |
| Probiotics intake | 103 | 35 (33.98) | 2 (7.69) | 33 (42.86) | <0.001 |
n = 128. Age in double-sampled subsets represents the mean age of the 2 sampling occasions; P values were obtained by the Mann–Whitney U test and chi-square tests for continuous and categorical variables, respectively; n of covariates are <128 due to missing data; missing covariate values were imputed in the regression analysis. GIT, gastrointestinal; MET-h, metabolic equivalent of task hours.
Median; 25th, 75th percentile in parentheses (all such values).
Modeling intake of energy and macronutrient trajectories
In the entire study sample (n = 128), energy and all macronutrients except for fiber showed quadratic trajectories with age—that is, the quadratic age models performed better than the linear age models according to the corrected Akaike's Information Criterion. Since we observed P < 0.05 for sex in the energy, carbohydrate, and fat models, it was included as a covariate in these models. Thus, energy, carbohydrate, and fat were fitted as sex-conditioned quadratic age models, protein as an unconditional quadratic age model, and fiber as an unconditional linear age model. Consequently, each of energy, carbohydrate, protein, and fat had 3 dietary intake trajectory variables: intake at age 1 y and linear and quadratic rates of change; and fiber had 2 dietary intake variables: intake at age 1 y and linear rate of change.
Multivariable linear regression
Figure 1 shows the coefficient estimates of associations between each set of dietary intake trajectories and genus-level relative abundance of 158 genera with P < 0.05 in the single-sampled individuals (n = 51), accounting for inter-taxa correlation. Energy intake at age 1 y was associated with 23 genera (negatively with 16 and positively with 7), its linear rate of change was not associated with any genera, and its quadratic rate of change was associated with 7 genera (negatively with 3 and positively with 4). Carbohydrate intake at age 1 y was associated with 11 genera (negatively with 7 and positively with 4), its linear rate of change was associated with 8 genera (negatively with 5 and positively with 3), and its quadratic rate of change was associated with 5 genera (negatively with 4 and positively with 1). Fiber intake at age 1 y was associated with 13 genera (negatively with 7 and positively with 6) and its rate of change was associated with 12 genera (negatively with 8 and positively with 4). Protein intake at age 1 y was associated with 5 genera (negatively with 1 and positively with 4). The linear rate of change in protein intake was associated with 20 genera (negatively with 10 and positively with 10), and the quadratic rate of change was associated with 6 genera (negatively with 4 and positively with 2). Finally, fat intake at age 1 y was positively associated with 7 genera, its linear rate of change was associated with 9 genera (negatively with 5 and positively with 4), and its quadratic rate of change was not found to be associated with any genus.
FIGURE 1.
Heatmap showing associations between each set of dietary intake trajectory variables and the relative abundance of bacterial genera at P < 0.05 in single-sampled individuals (n = 51). Associations for each set of dietary intake trajectory variables with the 158 taxa at P < 0.05. Values represent beta coefficients. Coefficients were obtained by using a multivariate zero-inflated logistic-normal. Positive and negative associations are shown in shades of green and red, respectively (the darker the color, the higher the absolute value of the coefficients).
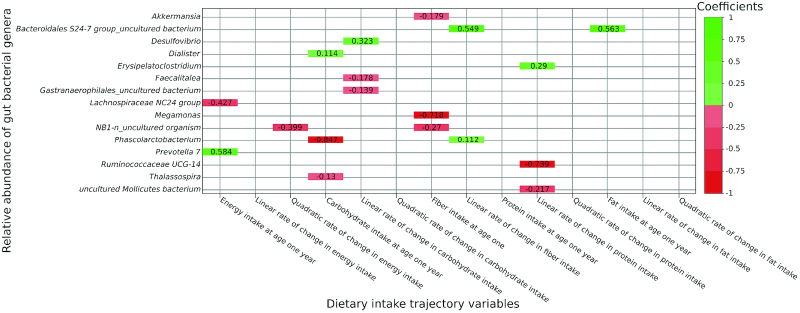
Figure 2 shows the coefficient estimates for the associations with P < 0.05 in the full multivariable-adjusted models, following adjustment for covariates. These include the association of energy intake at age 1 y with 2 genera, negative with Lachnospiraceae NC2004 group and positive with Prevotella 7, as well as the negative association of quadratic rate of change in energy intake with NB1-n_uncultured organism. Furthermore, carbohydrate intake at age 1 y was negatively associated with Phascolarctobacterium and Thalassospira and positively associated with Dialister. Further, the linear rate of change in carbohydrate intake was negatively associated with Gastranaerophilales_uncultured bacterium and Faecalitalea and positively associated with Desulfovibrio. In addition, fiber intake at age 1 y was negatively associated with Akkermansia, NB1-n_uncultured organism, and Megamonas and the linear rate of change in fiber intake was positively associated with Phascolarctobacterium and Bacteroidales S24–7 group_uncultured bacterium. Furthermore, the linear rate of change in protein intake was negatively associated with Ruminococcaceae UCG-014 and uncultured Mollicutes bacterium and positively associated with Erysipelatoclostridium. Finally, fat intake at age 1 y was positively associated with Bacteroidales S24–7 group_uncultured bacterium. These associations were not found to be altered either in direction or substantially in magnitude after excluding 3 participants who reported gastrointestinal disease and/or taking antibiotics or probiotics within 6 mo of fecal sampling, suggesting that these results were robust.
FIGURE 2.
Heatmap showing full multivariable-adjusted associations of all dietary intake trajectory variables and relative abundance of bacterial genera at P < 0.05 in single-sampled individuals (n = 51). Associations at P < 0.05. Values represent B-coefficients. Coefficients were obtained by using a multivariate zero-inflated logistic-normal. Positive and negative associations are shown in shades of green and red, respectively (the darker the color, the higher the absolute value of the coefficients)
Table 2 presents the coefficients, P values, and FDR-adjusted P values of the above 18 dietary intake–genera associations that were validated in the double-sampled individuals (n = 77) using the ZIBR model. This model included all dietary intake variables and history of gastrointestinal disease and/or taking antibiotics or probiotics within 6 mo since these were the only covariates that were found to be different between the single- and double-sampled subsets (P < 0.05). Overall, 3 genera—Phascolarctobacterium, Dialister, and Desulfovibrio—were associated with a dietary intake variable at an FDR-adjusted P < 0.05. Given the presence of these bacteria, the relative abundance of Phascolarctobacterium decreased (coefficient = −4.31, FDR-adjusted P = 0.006) and Dialister increased (coefficient = 3.06, FDR-adjusted P = 0.003) as the carbohydrate intake at age 1 y increased. Additionally, the relative abundance of Desulfovibrio increased (coefficient = 13.16, FDR-adjusted P = 0.00039) as the linear rate of change in carbohydrate intake increased.
TABLE 2.
Association of long-term energy and macronutrient intake with relative abundance of bacterial genera in double-sampled individuals1
| Logistic regression | B-regression | |||||
|---|---|---|---|---|---|---|
| Coefficient | P | Coefficient | P | Joint P | FDR-adjusted P | |
| Energy intake at age 1 y | ||||||
| Lachnospiraceae NC2004 group | −1.07 | 0.63 | −0.17 | 0.58 | 0.77 | 0.77 |
| Prevotella 7 | −0.58 | 0.36 | −0.37 | 0.30 | 0.38 | 0.76 |
| Quadratic rate of change in energy intake | ||||||
| NB1-n_uncultured organism | −0.52 | 0.62 | −2.05 | 0.05 | 0.13 | |
| Carbohydrate intake at age 1 y | ||||||
| Phascolarctobacterium | 14.91 | 0.04 | −4.31 | 0.01 | 0.004 | 0.006 |
| Thalassospira | 2.24 | 0.70 | -0.78 | 0.56 | 0.78 | 0.78 |
| Dialister | −1534.76 | 0.001 | 3.06 | 0.09 | 0.001 | 0.003 |
| Linear rate of change in carbohydrate intake | ||||||
| Gastranaerophilales_uncultured bacterium | −17.08 | 0.09 | 2.08 | 0.64 | 0.21 | 0.21 |
| Faecalitalea | −0.95 | 0.95 | 8.43 | 0.11 | 0.08 | 0.12 |
| Desulfovibrio | 8.88 | 0.55 | 13.16 | 0.000028 | 0.00013 | 0.00039 |
| Fiber intake at age 1 y | ||||||
| Akkermansia | −0.43 | 0.20 | −0.08 | 0.45 | 0.34 | 0.78 |
| NB1-n_uncultured organism | −0.21 | 0.46 | −0.04 | 0.86 | 0.75 | 0.78 |
| Megamonas | 0.11 | 0.62 | 0.06 | 0.61 | 0.78 | 0.78 |
| Linear rate of change in fiber intake | ||||||
| Phascolarctobacterium | −0.40 | 0.56 | −0.12 | 0.48 | 0.66 | 0.66 |
| Bacteroidales S24–7 group_uncultured bacterium | −0.50 | 0.19 | 0.14 | 0.34 | 0.27 | 0.54 |
| Linear rate of change in protein intake | ||||||
| Erysipelatoclostridium | 0.89 | 0.91 | −0.76 | 0.56 | 0.03 | 0.10 |
| Ruminococcaceae UCG-014 | −24.81 | 1.00 | 0.58 | 0.70 | 0.93 | 1.00 |
| Uncultured Mollicutes bacterium | −3.25 | 0.38 | −0.56 | 0.71 | 1.00 | 1.00 |
| Fat intake at age 1 y | ||||||
| Bacteroidales S24 group_uncultured bacterium | 0.71 | 0.87 | −2.38 | 0.15 | 0.35 | |
1 n = 77. Coefficients were obtained by using zero-inflated Beta regression model with random-effects model. A few coefficients of the logistic model may be biased due to the small proportion of individuals without these genera. FDR, false discovery rate.
Discussion
The aim of this investigation among 128 DONALD study participants was to examine the relation of long-term intakes of energy, carbohydrate, fiber, protein, and fat from infancy to late adolescence with gut microbiota composition in adulthood. We estimated energy and macronutrients intake from annual 3-d dietary records from age 1 to 18 y and modeled long-term intake as intake trajectories (intake at age 1 y and rates of change in intake representing the temporal sequence of intake). The microbiota composition was profiled by 16S rRNA sequencing and relative abundance was estimated. Independent of several covariates and inter- and intraindividual differences in microbiota composition, we observed that 1) carbohydrate intake at age 1 y was negatively associated with the relative abundance of Phascolarctobacterium and positively associated with the relative abundance of Dialister and 2) the linear rate of change in carbohydrate intake was positively associated with the relative abundance of Desulfovibrio.
To our knowledge, no study has yet investigated the association of long-term diet from infancy to late adolescence with adult microbiota composition. The high quality of the longitudinal dietary data from the DONALD study makes this unique investigation possible. Consistent with our findings is a prospective study in 22 children, aged 4 to 8 y, that showed that a dietary pattern highly loaded with carbohydrate-rich grains and starchy foods was inversely associated with Phascolarctobacterium (31). Considering that this study profiled microbiota in childhood (31), it is possible that the relation that we observed in our study commenced early on in life. Additionally, cross-sectional studies in 43 children, aged 7 to 9 y (32), and 561 adults, aged 30–70 y (33), observed that carbohydrate intake was positively associated with Dialister. These findings and ours suggest that the impact on Dialister may be a consistent feature of carbohydrate intake regardless of age. Moreover, our result for Desulfovibrio is in line with studies in adults showing the positive association of long-term intake of carbohydrates and simple sugars with an enterotype that includes Desulfovibrio (12) and an association between carbohydrate intake and Desulfovibrio (14).
A possible explanation for the association of carbohydrate intake at age 1 y with the abundance of Phascolarctobacterium and Dialister in adulthood is that high carbohydrate intake in infancy influences the assembly, maturation, and maintenance of gut microbiota composition via alteration of local and systemic tissue structure and functions such as the innate and adaptive immune system (34). This critical timing of high intake may set the foundation for the abundance of these bacteria over the life course. This is supported by the fact that the postweaning diet is decisive for gut microbiota development, succession, and consolidation that persist into adulthood (35). In addition, there is evidence of early-life nutritional programming of adult health status through variation in the quantity of nutrient intake during the first year of life (36). Our study thus suggests that the gut microbiota composition may be one of the adult health parameters that is programmed by early-life nutrition, specifically carbohydrate intake. Remarkably, Phascolarctobacterium (family: Acidaminococcaceae; class: Negativicutes; phylum: Firmicutes) and Dialister (family: Veillonellaceae; class: Negativicutes; phylum: Firmicutes) utilize carbohydrate-derived succinate as a carbon source to generate propionate (37). Furthermore, Phascolarctobacterium, Dialister, and other bacteria exhibit lottery-like mutual exclusion, such that 1 bacterium occupies the entirety of the community's abundance quota and the other members are excluded (38). Hence, the intake of a high-carbohydrate substrate might have resulted in a substantial increase in the abundance of Dialister, depleting the succinate nutrient pool, and subsequently inhibiting the growth of Phascolarctobacterium. This may suggest a complete reliance of Phascolarctobacterium on succinate and a low ability to shift to using other carbohydrate substrates. Additionally, it is possible that the metabolism of carbohydrate by Dialister creates a metabolic environment that directly eradicates Phascolarctobacterium. Interestingly, contrasting associations of Phascolarctobacterium and Dialister with some conditions such as successful weight loss (39) and insulin sensitivity (40) have been reported. This suggests that the current findings of Phascolarctobacterium and Dialister with carbohydrate intake are unlikely to be spurious. Moderate carbohydrate intake at this age in infancy could influence the relative abundance of these bacteria in adulthood. Desulfovibrio (family: Desulfovibrionaceae; class: Deltaproteobacteria; phylum: Proteobacteria), on the other hand, is the most abundant sulfate-reducing bacterium in humans (41). Importantly, the ability of Desulfovibrio to utilize many carbohydrate substrates such as fructose and glucose is also documented (42). Thus, an explanation for our finding of the positive association of the linear rate of change in carbohydrate intake with Desulfovibrio is that the persistently high carbohydrate intake afforded Desulfovibrio a continuous supply of substrates and subsequent increase in abundance.
Our data only show 3 temporally stable, Phascolarctobacterium, Dialister, and Desulfovibrio associations with long-term carbohydrate intake. These 3 bacterial genera may be promising biomarkers of carbohydrate intake. Our results are in agreement with the notion that carbohydrates, among all macronutrients, are the preferred energy source for the gut microbiota. High carbohydrate intake driving the depletion of Phascolarctobacterium and the increase in Dialister and Desulfovibrio suggests that carbohydrate intake, in particular, drives changes in microbiota composition, as reported in a recent review (43). This finding is also in support of a large study that showed that the intake of carbohydrate is the largest dietary predictor of gut bacterial diversity (44). The association of carbohydrate intake with these 3 genera may have been influenced by the variation of the composition of these genera in the study sample. However, since many genera with higher variance, which includes the core gut microbiota, were not associated with either carbohydrate intake or other dietary intake variables, it appears that our findings are unlikely to be influenced by variation in the composition of the bacteria. In addition, the fact that we did not find associations of diet with any of our core gut microbiota (Bacteroides, Blautia, and Lachnoclostridium) after covariate adjustment is in line with a study showing that the core microbiota are resilient to temporary extrinsic factors that include diet (45). In addition, because Bacteroides and Blautia are among the core genera reported in a multicenter large-scale study (46) and observed in our study suggests that microbiota composition in the current study is well estimated. Some associations reported by others (12–14) that could not be confirmed by this study perhaps indicate the versatility of long-term diet–microbiota associations.
Given that the associations of carbohydrate intake with Phascolarctobacterium, Dialister, and Desulfovibrio were independent of fiber intake and we did not observe an association of fiber intake with these genera, it seems plausible that the association of carbohydrate intake with these bacteria is specific to simple carbohydrates and starch. These results by no means imply that the intake of fiber or its subtypes does not influence microbiota composition; our analyses only indicated that long-term fiber intake was not associated with microbiota composition in adult participants in the current study. High simple carbohydrate and starch intakes are associated with unfavorable metabolic profiles, inflammatory bowel diseases, obesity, type 2 diabetes, cardiovascular disease, and cancer (43). Although it is common that a gut bacterium associated with health in one setting could be associated with disease in another context, Phascolarctobacterium seems to be generally health promoting while Dialister and Desulfovibrio are more commonly associated with disease conditions. Specifically, a high abundance of Phascolarctobacterium was associated with gastrointestinal health (47), high insulin sensitivity (39), and a reduction in systemic inflammation (48). Further, a high abundance of Dialister was associated with impaired glucose tolerance (49), low insulin sensitivity (40), type 2 diabetes (50), and gastric cancer (51). Desulfovibrio abundance was increased in individuals with inflammatory bowel diseases (52, 53) and type 2 diabetes (50, 54). Therefore, how the abundance of these bacteria might influence the association between high carbohydrate intake and the aforementioned conditions warrants further research. Maternal BMI and participants’ smoking status were related to most bacteria taxa in our multivariate analysis. This is in line with the association of maternal obesity status and the offspring's microbiota composition (55) and the association between smoking and microbiota composition (56). Dietary guidelines and recommendations that limit the amount of foods high in sugar and added-sugar consumption and that encourage a balanced diet as well as age-appropriate guidance exist in most countries. Therefore, it is imperative that parents and guardians help and encourage their children, starting from infancy, to adhere to these recommendations.
Some important strengths of this study are its prospective design, the large number (an average of 16) of repeated dietary assessments, the double sampling of microbiota composition in most of the study sample, and the deployment of 2 complementary statistical methods. These strengths helped us reach reasonable conclusions. In addition, to our knowledge, this is the first study to report on the relation of long-term carbohydrate intake from infancy to late adolescence with adult microbiota composition. Furthermore, we included a number of potential confounders, which was not considered in previous studies. We also controlled for inter-taxa correlations and adjusted for multiple testing. Nevertheless, our study comes with some potential limitations. First, it is an observational study, so no causality can be inferred. Second, our sample size, despite being larger (12, 14) and comparable (13) to previous studies exploring long-term diet–microbiota relations, is remarkably smaller than the 1070–1730 individuals recommended for evaluating the determinants of gut microbiota composition (46). Therefore, this study might have missed some associations. Like every self-reported dietary assessment, misreporting is likely to exist in this study, particularly with parental proxies. Nonetheless, there have been successful applications of these self-reported dietary data to address many research questions. Further, our food-composition table may have influenced the estimation of intake of energy and macronutrients. Indeed, a re-evaluation of our findings with accurate biomarkers of intake would be desirable in the future. Moreover, this study involves a taxonomy-based microbiota profiling of the 16S rRNA gene and the drawbacks of taxonomy-based classification are well acknowledged. Our taxonomic resolution up to genus level precludes inferences to the species level. Associations of intake with species-level compositions and functions will be addressed in follow-up work. Participant factors such as genetics, early-life health conditions, and other covariates across the life course could be important unmeasured confounders. Finally, our study sample is relatively homogeneous with respect to ethnicity, socioeconomic status, and geographical location; thus, it is not certain to what extent our results can be generalized beyond this group of individuals. Larger and more heterogeneous studies are required to validate our findings. Although the sample for this microbiota-based study within the DONALD study was randomly selected, the participation rate of <50% suggests that we cannot rule out bias due to nonparticipation. Additionally, other studies should explore the underlying mechanisms and functional relevance of our findings.
The current study provides novel evidence regarding the association of long-term carbohydrate intake from infancy to late adolescence with the composition of Phascolarctobacterium, Dialister, and Desulfovibrio in adulthood. This study gives helpful insights and fosters further research into the relation between long-term dietary intake at young ages and microbiota composition later in life. Considering the associations of high carbohydrate intake and these bacteria with some diseases, our findings could help inform gut microbiota–targeted dietary recommendations that would be of critical importance for health and disease prevention.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the support of the staff at the DONALD study center for data collection, staff of the German Center for Neurodegenerative Diseases Biobank for processing of stool samples, and staff of the Teagasc sequencing facility for the 16S rRNA sequencing. We also thank Stefan Benda for his support in formatting the figures.
The authors’ responsibilities were as follows—KO, JC, and UN: conception and the design of the study; ANO, KB, KL, and CS: fecal DNA extraction, 16S rRNA gene sequencing, and bioinformatics; KO: data analysis and interpretation and drafting of the manuscript; MS: supported data analysis; UA: supported review of study population and data collection; KB, MS, UA, GC, JC, and UN: contributed to data interpretation; and all authors: critically revised the manuscript and read and approved the final manuscript. The authors reported no conflicts of interest.
Notes
The current study was financially supported by the Metabolic HEALTH through nutrition, microbiota and tryptophan bioMARKers (HEALTHMARK) project in the frame of the EU Joint Programming Initiative “A Healthy Diet for a Healthy Life” through the Federal Ministry of Education and Research, Germany (grant number: 01EA1705A), and Science Foundation Ireland, Ireland (grant number: 16/ERA-HDHL/3362). This study was also supported by the “Diet-Body-Brain” Competence Cluster in Nutrition Research funded by the Federal Ministry of Education and Research (grant number: FKZ: 01EA1809A). The DONALD study was supported by the Ministry of Science and Research of North Rhine Westphalia, Germany.
Supplemental Methods, Supplemental Figures 1 and 2, and Supplemental Tables 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: DONALD, DOrtmund Nutritional and Anthropometric Longitudinally Designed; FDR, false discovery rate; MET-h, metabolic equivalent task hours; MZILN, multivariate zero-inflated logistic-normal; rRNA, ribosomal RNA; ZIBR, zero-inflated Beta regression model with random effects.
Contributor Information
Kolade Oluwagbemigun, Unit of Nutritional Epidemiology, Department of Nutrition and Food Sciences, University of Bonn, Bonn, Germany.
Aoife N O'Donovan, APC Microbiome Ireland, University College Cork, Cork, Ireland; Teagasc Moorepark Food Research Centre, Fermoy, Ireland; School of Microbiology, University College Cork, Cork, Ireland.
Kirsten Berding, APC Microbiome Ireland, University College Cork, Cork, Ireland.
Katriona Lyons, APC Microbiome Ireland, University College Cork, Cork, Ireland; Teagasc Moorepark Food Research Centre, Fermoy, Ireland.
Ute Alexy, Unit of Nutritional Epidemiology, Department of Nutrition and Food Sciences, University of Bonn, Bonn, Germany.
Matthias Schmid, Department of Medical Biometry, Informatics, and Epidemiology, University Hospital Bonn, University of Bonn, Bonn, Germany.
Gerard Clarke, APC Microbiome Ireland, University College Cork, Cork, Ireland; INFANT Research Centre, University College Cork, Cork, Ireland; Department of Psychiatry and Neurobehavioral Science, University College Cork, Cork, Ireland.
Catherine Stanton, APC Microbiome Ireland, University College Cork, Cork, Ireland; Teagasc Moorepark Food Research Centre, Fermoy, Ireland.
John Cryan, APC Microbiome Ireland, University College Cork, Cork, Ireland.
Ute Nöthlings, Unit of Nutritional Epidemiology, Department of Nutrition and Food Sciences, University of Bonn, Bonn, Germany.
Data Availability
The data described in the manuscript, code book, and analytic code will not be made available because of data protection regulations.
References
- 1. Shim J-S, Oh K, Kim HC. Dietary assessment methods in epidemiologic studies. Epidemiol Health. 2014;36:e2014009–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. 2019;216(1):20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leeming ER, Johnson AJ, Spector TD, Le Roy CI. Effect of diet on the gut microbiota: rethinking intervention duration. Nutrients. 2019;11(12):2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16(1):35–56. [DOI] [PubMed] [Google Scholar]
- 6. Klingbeil E, de La Serre CB. Microbiota modulation by eating patterns and diet composition: impact on food intake. Am J Physiol Regul Integr Comp Physiol. 2018;315(6):R1254–60. [DOI] [PubMed] [Google Scholar]
- 7. Sheflin AM, Melby CL, Carbonero F, Weir TL. Linking dietary patterns with gut microbial composition and function. Gut Microbes. 2017;8(2):113–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang Q, Liang Q, Balakrishnan B, Belobrajdic DP, Feng QJ, Zhang W. Role of dietary nutrients in the modulation of gut microbiota: a narrative review. Nutrients. 2020;12(2):381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. 2018;362(6416):776–80. [DOI] [PubMed] [Google Scholar]
- 10. Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. 2019;7(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gong L, Wen T, Wang J. Role of the microbiome in mediating health effects of dietary components. J Agric Food Chem. 2020;68(46):12820–35. [DOI] [PubMed] [Google Scholar]
- 12. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight Ret al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang Z-Z, Chen G, Hong Q, Huang S, Smith HM, Shah RD, Scholz M, Ferguson JF. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Front Genetics. 2019;10:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fragiadakis GK, Wastyk HC, Robinson JL, Sonnenburg ED, Sonnenburg JL, Gardner CD. Long-term dietary intervention reveals resilience of the gut microbiota despite changes in diet and wei. ght. Am J Clin Nutr. 2020;111(6):1127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duncan TE, Duncan SC. An introduction to latent growth curve modeling. Behav Ther. 2004;35(2):333–63. [Google Scholar]
- 16. Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RLet al. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brüssow H. How stable is the human gut microbiota? And why this question matters. Environ Microbiol. 2016;18(9):2779–83. [DOI] [PubMed] [Google Scholar]
- 18. Rajilić-Stojanović M, Heilig H, Tims S, Zoetendal EG, de Vos WM. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol. 2013;15(4):1146–59. [DOI] [PubMed] [Google Scholar]
- 19. Davenport ER, Mizrahi-Man O, Michelini K, Barreiro LB, Ober C, Gilad Y. Seasonal variation in human gut microbiome composition. PLoS One. 2014;9(3):e90731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang J, Guo Z, Lim AA, Zheng Y, Koh EY, Ho D, Qiao J, Huo D, Hou Q, Huang Wet al. Mongolians core gut microbiota and its correlation with seasonal dietary changes. Sci Rep. 2015;4:5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kroke A, Manz F, Kersting M, Remer T, Sichert-Hellert W, Alexy U, Lentze MJ. The DONALD study: history, current status and future perspectives. Eur J Nutr. 2004;43(1):45–54. [DOI] [PubMed] [Google Scholar]
- 22. Curran PJ, Obeidat K, Losardo D. Twelve frequently asked questions about growth curve modeling. J Cogn Dev. 2010;11(2):121–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flores R, Shi J, Yu G, Ma B, Ravel J, Goedert JJ, Sinha R. Collection media and delayed freezing effects on microbial composition of human stool. Microbiome. 2015;3(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques. 2004;36(5):808–12. [DOI] [PubMed] [Google Scholar]
- 25. Fouhy F, Deane J, Rea MC, O'Sullivan O, Ross RP, O'Callaghan G, Plant BJ, Stanton C. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One. 2015;10(3):e0119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oluwagbemigun K, Buyken AE, Alexy U, Schmid M, Herder C, Nothlings U. Developmental trajectories of body mass index from childhood into late adolescence and subsequent late adolescence-young adulthood cardiometabolic risk markers. Cardiovasc Diabetol. 2019;18(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Willett W. Nutritional epidemiology. 3rd ed. New York: Oxford University Press; 2013. [Google Scholar]
- 28. Zhang X, Nieuwdorp M, Groen AK, Zwinderman AH. Statistical evaluation of diet-microbe associations. BMC Microbiol. 2019;19(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Lee K, Karagas MR, Madan JC, Hoen AG, O'Malley AJ, Li H. Conditional regression based on a multivariate zero-inflated logistic-normal model for microbiome relative abundance data. Stat Biosci. 2018;10(3):587–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen EZ, Li H. A two-part mixed-effects model for analyzing longitudinal microbiome compositional data. Bioinformatics. 2016;32(17):2611–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berding K, Holscher HD, Arthur AE, Donovan SM. Fecal microbiome composition and stability in 4- to 8-year old children is associated with dietary patterns and nutrient intake. J Nutr Biochem. 2018;56:165–74. [DOI] [PubMed] [Google Scholar]
- 32. Nakayama J, Yamamoto A, Palermo-Conde LA, Higashi K, Sonomoto K, Tan J, Lee Y. Impact of westernized diet on gut microbiota in children on Leyte Island. Front Microbiol. 2017;8:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nuli R, Cai J, Kadeer A, Zhang Y, Mohemaiti P. Integrative analysis toward different glucose tolerance-related gut microbiota and diet. Front Endocrin. 2019;10:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tanaka M, Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergology Int. 2017;66(4):515–22. [DOI] [PubMed] [Google Scholar]
- 35. Laforest-Lapointe I, Arrieta M-C. Patterns of early-life gut microbial colonization during human immune development: an ecological perspective. Front Immunol. 2017;8:788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langley-Evans SC, Muhlhausler B. Early life nutritional programming of adult health status. : Vaiserman Aeditor. Early life origins of ageing and longevity. Cham (Switzerland): Springer International Publishing; 2019. p.87–120. [Google Scholar]
- 37. Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–45. [DOI] [PubMed] [Google Scholar]
- 38. Verster AJ, Borenstein E.. Competitive lottery-based assembly of selected clades in the human gut microbiome. Microbiome. 2018; 6(1):186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muniz Pedrogo DA, Jensen MD, Van Dyke CT, Murray JA, Woods JA, Chen J, Kashyap PC, Nehra V. Gut microbial carbohydrate metabolism hinders weight loss in overweight adults undergoing lifestyle intervention with a volumetric diet. Mayo Clin Proc. 2018;93(8):1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Naderpoor N, Mousa A, Gomez-Arango LF, Barrett HL, Dekker Nitert M, de Courten B. Faecal microbiota are related to insulin sensitivity and secretion in overweight or obese adults. J Clin Med. 2019;8(4):452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scanlan PD, Shanahan F, Marchesi JR. Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol Ecol. 2009;69(2):213–21. [DOI] [PubMed] [Google Scholar]
- 42. Labes A, Schönheit P. Sugar utilization in the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324: starch degradation to acetate and CO2 via a modified Embden-Meyerhof pathway and acetyl-CoA synthetase (ADP-forming). Arch Microbiol. 2001;176(5):329–38. [DOI] [PubMed] [Google Scholar]
- 43. Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva Set al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Salonen A, Salojärvi J, Lahti L, de Vos WM. The adult intestinal core microbiota is determined by analysis depth and health status. Clin Microbiol Infect. 2012;18:16–20. [DOI] [PubMed] [Google Scholar]
- 45. Seo YS, Lee H-B, Kim Y, Park H-Y. Dietary carbohydrate constituents related to gut dysbiosis and health. Microorganisms. 2020;8(3):427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte Det al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–4. [DOI] [PubMed] [Google Scholar]
- 47. Wu F, Guo X, Zhang J, Zhang M, Ou Z, Peng Y. Phascolarctobacterium faeciumabundant colonization in human gastrointestinal tract. Exp Ther Med. 2017;14(4):3122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Citronberg JS, Curtis KR, White E, Newcomb PA, Newton K, Atkinson C, Song X, Lampe JW, Hullar MAJ. Association of gut microbial communities with plasma lipopolysaccharide-binding protein (LBP) in premenopausal women. ISME J. 2018;12(7):1631–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ciubotaru I, Green SJ, Kukreja S, Barengolts E. Significant differences in fecal microbiota are associated with various stages of glucose tolerance in African American male veterans. Transl Res. 2015;166(5):401–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sanchez-Alcoholado L, Castellano-Castillo D, Jordán-Martínez L, Moreno-Indias I, Cardila-Cruz P, Elena D, Muñoz-Garcia AJ, Queipo-Ortuño MI, Jimenez-Navarro M. Role of gut microbiota on cardio-metabolic parameters and immunity in coronary artery disease patients with and without type-2 diabetes mellitus. Front Microbiol. 2017;8:1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJYet al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67(6):1024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Loubinoux J, Bronowicki JP, Pereira IA, Mougenel JL, Faou AE. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol Ecol. 2002;40(2):107–12. [DOI] [PubMed] [Google Scholar]
- 53. Rowan F, Docherty NG, Murphy M, Murphy B, Calvin Coffey J, O'Connell PR. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum. 2010;53(11):1530–6. [DOI] [PubMed] [Google Scholar]
- 54. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen Det al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. [DOI] [PubMed] [Google Scholar]
- 55. Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS One. 2014;9(11):e113026–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang C, Shi G. Smoking and microbiome in oral, airway, gut and some systemic diseases. J Transl Med. 2019;17(1):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data described in the manuscript, code book, and analytic code will not be made available because of data protection regulations.