Abstract
Oxidation of 5-methylcytosine (5mC) in DNA by the ten-eleven translocation (TET) family of enzymes is indispensable for gene regulation in mammals. More recently, evidence has emerged to support a biological function for TET-mediated m5C oxidation in messenger RNA. Here, we describe a previously uncharacterized role of TET-mediated m5C oxidation in transfer RNA (tRNA). We found that the TET-mediated oxidation product 5-hydroxylmethylcytosine (hm5C) is specifically enriched in tRNA inside cells and that the oxidation activity of TET2 on m5C in tRNAs can be readily observed in vitro. We further observed that hm5C levels in tRNA were significantly decreased in Tet2 KO mouse embryonic stem cells (mESCs) in comparison with wild-type mESCs. Reciprocally, induced expression of the catalytic domain of TET2 led to an obvious increase in hm5C and a decrease in m5C in tRNAs relative to uninduced cells. Strikingly, we also show that TET2-mediated m5C oxidation in tRNA promotes translation in vitro. These results suggest TET2 may influence translation through impacting tRNA methylation and reveal an unexpected role for TET enzymes in regulating multiple nodes of the central dogma.
Keywords: 5-methylcytocine, transfer RNA, translational regulation, demethylation, RNA modification
Abbreviations: BSA, bovine serum albumin; FBS, fetal bovine serum; LC-MS/MS, liquid chromatography and tandem mass spectrometry; mESCs, mouse embryonic stem cell; m5C, 5-methylcytosine; NSUN2, NOP2/Sun domain protein 2; PBST, 1× phosphate-buffered saline, 0.1% Tween; SSC, saline-sodium citrate; TET, ten-eleven translocation; TET2-CD, TET2 catalytic domain; tRNA, transfer RNA; TRDMT1, tRNA aspartic acid MTase1; 5hmc, 5-hydroxymethylcytosine; 5fc, 5-formylcytosine; 5caC, 5-carboxylcytosine
Along with the post-translational modifications of histone proteins, the direct, reversible methylation of cytosines in CG dinucleotides in DNA (called CpG sites) is one of several layers of regulatory information that determines chromatin state (1, 2, 3). The ten-eleven translocation (TET) family of 5-methylcytosine dioxygenases catalyzes the successive oxidation of 5-methylcytosine (abbreviated as “5mC” in DNA) to 5-hydroxymethylcytosine (5hmC), to 5-formylcytosine (5fC), and lastly to 5-carboxylcytosine (5caC), providing an additional layer of epigenetic regulation to the mammalian genome (4, 5, 6). Biochemical assays suggest that one member of the TET family, TET2, works on both DNA and RNA as well (7), and recent findings have begun to reveal the biological function of TET-mediated oxidation in RNA. One study suggested that TET-mediated oxidation in mRNA promotes global protein synthesis in Drosophila (8). Several earlier studies have also shown that TET-mediated mRNA oxidation decreases stability. These effects may result from ADAR1-mediated repression of the target genes (9). Conversely, 5-methylcytosine in RNA (abbreviated as m5C) can also promote mRNA stability through specific m5C reader proteins (10, 11). m5C oxidation has been postulated to disrupt the binding of these m5C-specific readers and thereby tune stability.
While the abundance and occupancy of m5C sites in mRNA remain under investigation, m5C is highly abundant in tRNA. The majority of known functional roles of m5C in RNA species are also from the studies of m5C sites in tRNAs. In tRNA, m5C sites occur most often at the junction of the variable loop and the T stem-loop. The modification of three cytosines spanning positions 47 to 5-0 has been suggested to stabilize the tRNA structure (12, 13). Cytosine 38 in the anticodon loop of the tRNA is another frequently modified site. m5C38 in mouse tRNAAsp has been shown to stimulate amino acid charging of the tRNA and to facilitate translation of poly-Asp–containing proteins (14). In addition, m5C38 can protect tRNAs from stress-induced endonuclease-mediated fragmentation (15, 16) and help to maintain correct translational read-out of near-cognate codons (17). m5C also exists at C34 in tRNALeu(CAA) and mitochondrial (mt) tRNAMet (18, 19) and mt-tRNAMet in mammals (20, 21, 22). Lastly, m5C installation has been shown to be important for the final steps of tRNAThr and tRNACys biogenesis (23). In addition to its well-documented functions in tRNA, m5C also exists in rRNA and is important for translational fidelity (24, 25, 26). The m5C sites in RNA are installed by several methyltransferases, including tRNA aspartic acid MTase1 (TRDMT1), Dnmt2, NOP2/Sun domain protein 2 (NSUN2), Nsun3 (20, 21), and Nsun6 (27). The biological significance of m5C in RNA is further emphasized by genetic studies. For instance, knockout of NSUN2 in mice causes male infertility and reduced growth (28), while mutations in human NSUN2 are involved in intellectual disability (29). Lastly, DNMT2 deficiency has also been shown to affect polypeptide synthesis in humans (16).
Interestingly, cytoplasmic and mitochondrial tRNAs have been shown to carry f5C, an oxidation product of m5C. The alpha-ketoglutaric acid–dependent dioxygenase ALKBH1 has been shown to be involved in the biogenesis of f5C at the first position of the anticodon (position 34 of canonical tRNAs) in mitochondrial tRNAMet (20, 30). f5C of mt-tRNAMet is important for the decoding of AUA methionine codons during mitochondrial translation (31). Additionally, ALKBH1 had also been shown to catalyze the formation of 5-hydroxymethyl-2-O-methylcytidine (hm5Cm) and 5-formyl-2-O-methylcytidine (f5Cm) at the same position in cytoplasmic tRNALeu (32). ALKBH1-catalyzed oxidation reactions are important for translation and mitochondrial function. However, m5C oxidation in tRNALeu and mitochondrial tRNAiMet is specifically carried out by ALKBH1, not the TET enzymes (30). Whether TET-mediated m5C oxidation occurs on tRNA species as well as their biological functions are not fully understood. All these previous studies have highlighted the importance of the reversible regulation of m5C in tRNA. Here, we investigated whether TET2 can catalyze tRNA m5C oxidation and the potential biological function from this oxidation reaction.
Results
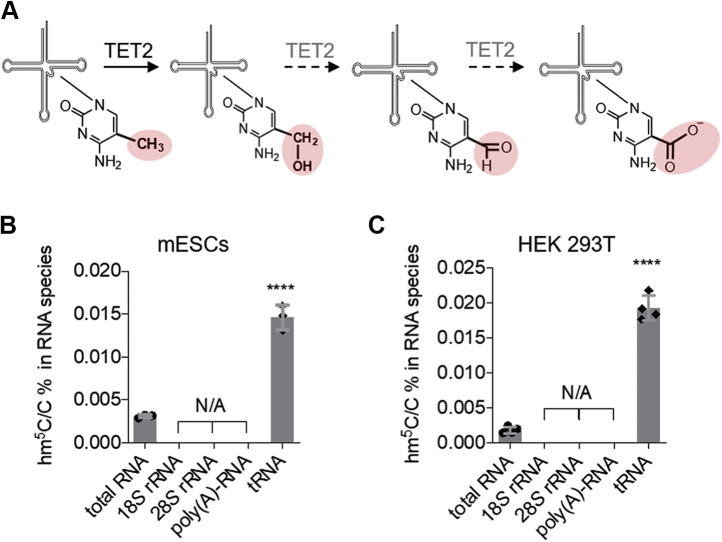
To study whether TET enzymes catalyze oxidation on m5C in other RNA species, we first quantified the levels of hm5C (the first m5C oxidation product, Fig. 1A) in several major RNA species. We extracted total RNA, 18S rRNA, 28S rRNA, polyadenylated RNA (poly(A)-RNA), and tRNA from mouse embryonic stem cells (mESCs) and HEK 293T cells (Fig. S1, A–D). The extracted RNAs were then degraded to single nucleosides before being analyzed via triple quadrupole liquid chromatography and tandem mass spectrometry (LC-MS/MS). The LC-MS/MS results show that among these different RNA species, hm5C is more significantly enriched in tRNAs in both mESCs and HEK293T cells (Fig. 1, B–C, Fig. S1E, and Fig. S2).
Figure 1.
hm5C is enriched in tRNA. A, schematic of TET2-mediated m5C oxidation products. B, LC-MS/MS quantification of hm5C in total RNA, 18S rRNA, 28S rRNA, poly(A)-RNA, and tRNA extracted from mESCs. C, LC-MS/MS quantification of hm5C in total RNA, 18S rRNA, 28S rRNA, poly(A)-RNA, and tRNA extracted from mESCs HEK293T cells. p values were determined using two-tailed Student's t test for unpaired samples. Error bars represent mean ± s.d., n = 4 (four biological replicates × two technical replicates) ∗∗∗∗ p < 0.001. LC-MS/MS, liquid chromatography and tandem mass spectrometry; mESCs, mouse embryonic stem cells.
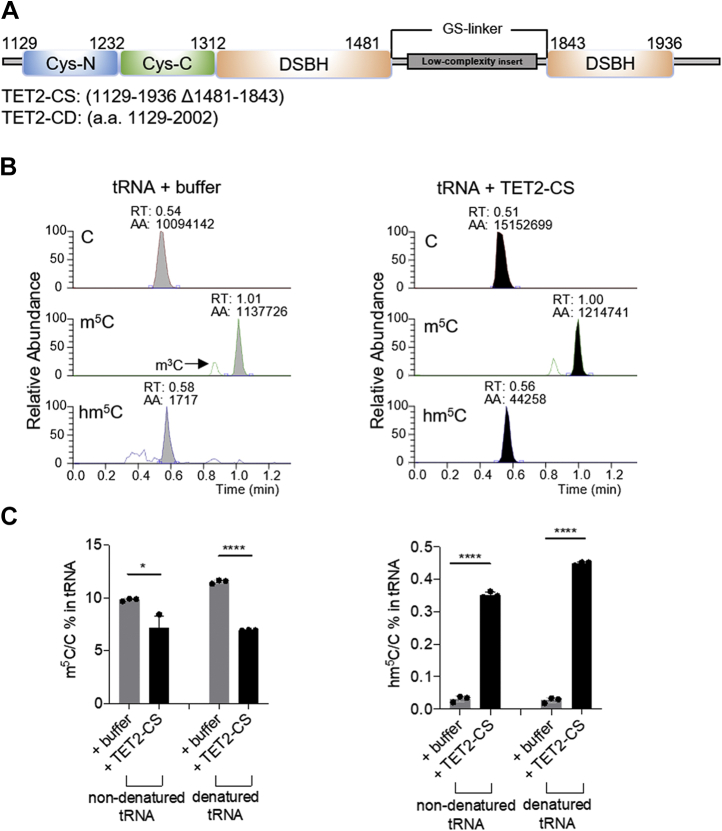
In light of recent discoveries that TET2 may be responsible for modifying RNA in cells (33), we focused on the role of TET2 in accounting for the observed enrichment of hm5C in tRNA. To investigate if m5C can be oxidized by TET2, we conducted in vitro oxidation assays. The activity of a purified TET2 truncation variant (TET2-CS, previously crystallized (34), Fig. 2A) was first confirmed on a 5mC-containing ssDNA oligo (Fig. S3, A–C). We then incubated tRNAs purified from HEK293T cells either with TET2-CS or in buffer alone. To investigate whether tRNA structure has an impact in TET-mediated oxidation, we used both native and denatured tRNAs in this biochemical oxidation reaction. Analysis of the oxidation products via LC-MS/MS clearly shows that TET2-CS is capable of oxidizing m5C to hm5C in tRNA in vitro (Fig. 2, B–C), while the further oxidation products, f5C and ca5C, were not detected (Fig. S3, D–E). However, in the presence of EDTA (an iron chelator), TET2-CS cannot lead to decreased m5C and increased hm5C in tRNA (Fig. S4).
Figure 2.
TET2-CS–mediated in vitro oxidation reaction. A, constructs of TET2-CS and TET2-CD. B, LC-MS/MS tracks of C, m5C, and hm5C from in vitro oxidation reaction of TET2-CS with purified tRNAs. tRNAs purified from HEK293T cells were incubated with either TET2-CS or buffer in the presence of alpha-ketoglutarate. C, LC-MS/MS quantification of C, m5C, and hm5C from in vitro oxidation reaction of TET2-CS with purified tRNAs. p values were determined using two-tailed Student's t test for unpaired samples. Error bars represent mean ± s.d., n = 3 (three biological replicates × two technical replicates). ∗ p < 0.05, ∗∗∗∗ p < 0.001. AA, atomic absorption; Cys-C, cysteine (Cys)-rich domain C terminal; Cys-N, cysteine (Cys)-rich domain N terminal; DSBH, double-stranded β-helix; LC-MS/MS, liquid chromatography and tandem mass spectrometry; N/A, not detectable; RT, retention time.
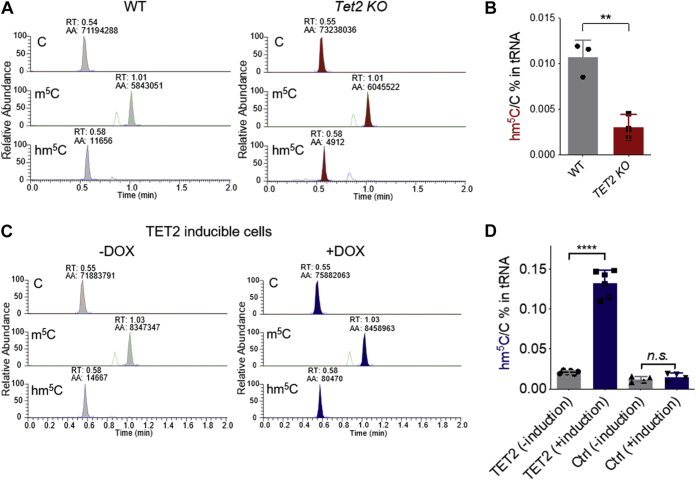
Given that we observed that the enrichment of hm5C in tRNA and that TET2 is capable of generating this modification in vitro, we sought to study if TET2-mediated oxidation generates tRNA hm5C in cells. To this end, we quantified m5C and hm5C levels in tRNAs extracted from wild-type and Tet2 KO mESCs (Fig. 3). The results show that Tet2 KO leads to a significant decrease of hm5C in tRNA. We also observed a noticeable, but not significant, increase of m5C in tRNA in Tet2 KO mESCs in comparison with the wild-type mESCs (Fig. 3, A–B and Fig. S5A). Although the expression of TET2 in mESCs leads to the starkly obvious increases of hm5C in tRNAs, we observed a low level of hm5C in tRNA. These results suggest that TET2 does not demethylate all m5C sites in tRNAs. It is possible that TET2 works with other demethylase enzymes such as ALKBH1 (30) to work on individual subsets of tRNAs.
Figure 3.
TET2-mediated m5C oxidation inside cells. A, LC-MS/MS tracks of C, m5C, and hm5C from purified tRNAs extracted from wild-type and Tet2 KO mESCs. B, LC-MS/MS quantification of hm5C in tRNA extracted from wild-type and Tet2 KO mESCs. C, LC-MS/MS tracks of C, m5C, and hm5C from purified tRNAs extracted from TET2-inducible expression HEK293T cells before and after doxycycline induction. D, LC-MS/MS quantification of hm5C in tRNA extracted from TET2 inducible expression HEK293T cells and control HEK293T cells before and after doxycycline induction. p values were determined using two-tailed Student's t test for unpaired samples. Error bars represent mean ± s.d., n = 3 (three biological replicates × two technical replicates) ∗∗ p < 0.01, ∗∗∗∗ p < 0.001, n.s. means p > 0.05. AA, atomic absorption; KO, knock-out; LC-MS/MS, liquid chromatography and tandem mass spectrometry; mESCs, mouse embryonic stem cells; RT, retention time.
To further confirm that TET2 is responsible for tRNA m5C oxidation in cells, we first constructed a doxycycline-inducible expression system for the TET2 catalytic domain (TET2-CD) in HEK293T cell line (35). Using this expression system, TET2-CD expression can be titrated to near-physiological expression levels, thereby avoiding any potential artifacts caused by overexpression (Fig. 2A and Fig. S5, B–C). We then validated that this increase in TET2-CD expression resulted in an increase in TET2-CD activity. To do this, we performed dot-blot assays using anti-5mC and anti-5hmC antibodies to further validate our LC-MS/MS findings (Fig. S5D). In addition, we quantified and compared 5mC and 5hmC levels in DNA in our cells before and after doxycycline treatment using LC-MS/MS. The results show that induced expression of TET2-CD leads to decreased levels of 5mC and increased 5hmC levels in DNA (Fig. S5, E–F). These results were consistent with the results from the dot-blot assay and together collectively suggested that our doxycycline-inducible system can successfully induce the expression of functional TET2-CD. After we validated the inducible cell line, we next quantified m5C and hm5C levels in tRNA before and after doxycycline treatment. The LC-MS/MS results show that induced expression of the catalytic domain of TET2 leads to significantly increased hm5C levels in tRNA (Fig. 3, C–D) with no obvious change of m5C in tRNA (Fig. S5G). We reasoned that it is due to the higher level of m5C in comparison with hm5C in tRNA; also TET2 is not the only tRNA m5C demethylase. In contrast, the parental HEK293T cell line (which we used to construct the inducible cell line) did not show any change of both m5C and hm5C levels before and after doxycycline treatment (Fig. 3D and Fig. S5, G–H). Together with the results from Tet2 KO mESCs, these results revealed that TET2-mediated m5C oxidation occurs on tRNA inside cells.
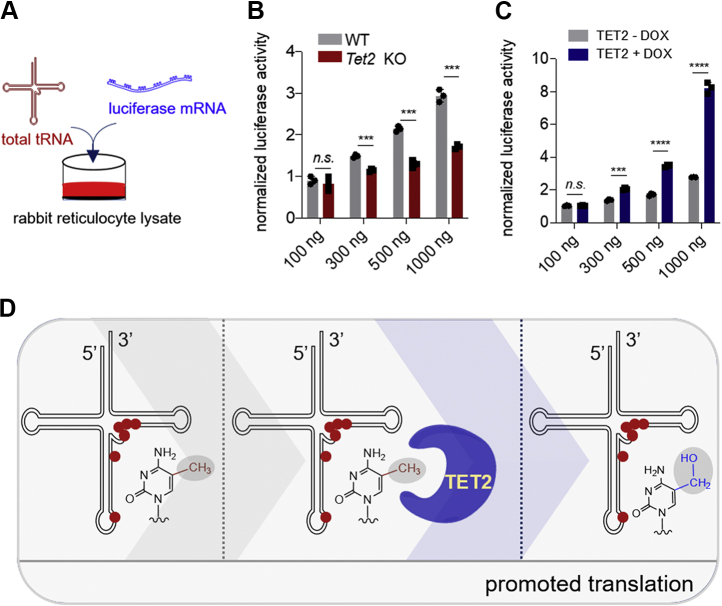
After observing tRNA as a target for TET-mediated oxidation, we investigated the possible biological consequences of TET2-mediated oxidation on m5C in tRNA. Given that tRNA modifications can tune translational efficiency (36), it is possible that TET-mediated oxidation of tRNAs could affect translation. To investigate this hypothesis, we utilized a rabbit reticulocyte-based in vitro translation system to measure the production of active luciferase protein from a fixed amount of luciferase mRNA (Fig. 4A). To probe how TET2-mediated oxidation of tRNAs affects translation, we first extracted tRNAs from either wild-type or Tet2 KO mESCs. We then spiked-in increasing amounts (100, 300, 500, and 1000 ng) of purified tRNAs into separate translation reactions along with the luciferase mRNA (uncapped in vitro-transcribed RNA containing a 30-base poly(A) tail from Promega) and measured luciferase activity. The results show that tRNAs originating from wild-type mESC cells lead to significant increase of luciferase activity. In contrast, spiking in tRNAs extracted from the Tet2 KO cells did not lead to increased luciferase signals (Fig. 4B). We also performed the in vitro translation assays using tRNAs extracted from TET2-inducible expression cells before and after administration of doxycycline. The results show that tRNAs originating from cells after doxycycline addition lead to an obvious increase of luciferase signal whereas the tRNAs from untreated cells elicit a relatively lower luciferase signal (Fig. 4C). In this in vitro translation reaction, we can rule out the possibility that TET2 proteins bind to the luciferase mRNA to promote translation since the reaction system only contains supplemented tRNAs, luciferase mRNA, and reticulocytes. Furthermore, we studied whether the expression of TET2 leads to increased translation inside cells. To this end, we quantified protein synthesis in TET2-induced and uninduced conditions using puromycin incorporation followed by Western blot analysis. The results showed that the expression of TET2 did not lead to an obvious impact on overall translation (Fig. S6A). It would be insightful to investigate the impact of TET2-mediated tRNA oxidation on specific transcripts in future studies.
Figure 4.
TET2-mediated tRNA oxidation significantly promotes translation. A, illustration of the in vitro translation assay. B, quantification of luciferase signals from the in vitro translation system supplemented with 100 to 1000 ng of tRNAs extracted from wild-type and Tet2 KO mESCs. C, quantification of luciferase signals from the in vitro translation system supplemented with 100 to 1000 ng of tRNAs extracted from HEK293T cells with inducible TET2-CD before and after doxycycline induction. Error bars represent mean ± s.d., n = 3 (three biological replicates × two technical replicates). D, illustration of TET2-mediated m5C oxidation promotes translation. The previously confirmed m5C sites (C34, C38, C47-C50) in tRNA are highlighted as red balls. KO, knock-out; mESCs, mouse embryonic stem cells; TET2-CD, TET2 catalytic domain.
We also sought to identify the specific tRNA targets of TET2. Since NSUN2 is one of the most studied tRNA m5C methyltransferase enzymes, we examined whether TET2 works on the m5C-containing tRNA targets of NSUN2 (16). As shown in updated Figure S6B, we quantified the levels of m5C and hm5C levels in four tRNAs extracted from Tet2 KO and wild-type mESCs. The results showed that Tet2 KO leads to a significant increase of hm5C and a noticed decrease of m5C level in tRNAGly (Fig. S6B). The effects of TET2-mediated oxidation are not obvious on the other three tRNAs including tRNAAsp, tRNAVal, and tRNALeu (Fig. S6B). These results collectively suggested that tRNAGly is a possible target of TET2.
Discussion
The presence of an appreciable level of hm5C in cellular RNA and the involvement of the TET family of enzymes in producing this modification support the hypothesis that the function of TET enzymes is not restricted to epigenetic regulation at the DNA level. Our results suggest that TET2 can oxidize m5C and generate hm5C on tRNAs in mammalian cells. We show that hm5C is particularly abundant in tRNAs in comparison with other major RNA species including rRNA and poly(A)-RNAs. This suggests that tRNA is possibly a major RNA target of TET2.
Our finding that TET2 can oxidize m5C on tRNA inside cells raises questions regarding the localization and timing of this activity. Given that TET2 is known to localize to the nucleus (37), it is likely that the oxidation of tRNAs is performed in that compartment as a step in pre-tRNA maturation following transcription and m5C installation by NSUN2/DNMT2 (16). These nascent pre-tRNAs would likely present accessible m5C sites, some of which may become occluded once fully folded and mature. While pre-tRNAs are the most likely primary tRNA substrate of the TET2-CD in our system, this is not the only possibility. Mature tRNAs are known to be transported into the nucleus via a retrograde importation mechanism (38, 39, 40); thus, it is also possible that mature tRNAs are a substrate of the TET2-CD.
As our results suggest, the decrease of m5C levels in tRNA upon expression of the TET2-CD is reproducible yet not significant while the increase of hm5C in tRNA is significant, (Fig. 3, C–D and Fig. S7G). This implies that only a subset of tRNAs and possibly only specific but not all m5C sites in tRNAs are the targets of the TET2-CD in our system, a hypothesis which is supported by the previous findings of ALKBH1-mediated tRNA m5C oxidation (30). Furthermore, whether TET1 and TET3 can oxidize m5C in tRNAs is unknown. Given that TET3 resides in both the cell nucleus and the cytoplasm (41), cytoplasmic TET3 might have optimal accessibility to mature tRNAs. It is possible that tRNA m5C sites are under special and temporal regulation. Along the tRNA maturation process, it is possible that several demethylase enzymes can work on different tRNA m5C sites in both the nucleus and the cytoplasm to facilitate proper tRNA biogenesis and regulation. In addition, it is known that reprogramming of m5C34 in tRNA facilitates translation of specific ribosomal proteins upon oxidative stress to sustain life (42). The dynamic regulation of the individual sites of m5C in tRNAs by distinct enzymes may represent another layer of translational control, especially when faced with different types of environmental stresses. This work provides a starting point for future studies to determine which tRNAs are targeted by TET2, what effects these oxidized tRNAs exert on translation, and how target and nontarget tRNAs are discriminated by TET2.
Lastly, while we were able to see a clear enrichment of hm5C in tRNAs, further oxidation products (namely f5C and ca5C) were not detectable in our analyses. Given that the role of these modifications in DNA is currently a matter of research (43), their apparent absence in tRNA in our system raises questions about TET2's activity on RNA. We have previously described two important characteristics of TET activity (7): (1, 7) DNA is generally the preferred substrate as opposed to RNA for TET oxidation, and (2) the first oxidation step (m5C to hm5C) is the most efficient and preferred step, while f5C and ca5C are less efficient products in both DNA and RNA. These two points may explain why the further oxidation products are absent in tRNA; the disfavored substrate type compounded with the disfavored products result in an extremely low efficiency of f5C and ca5C. Moreover, the lack of further oxidative products could be due to a number of additional biological factors, ranging from the more structured nature of tRNA than that of DNA to possible steric hindrance from other tRNA-modifying enzymes that install the plethora of modifications present on mature tRNA, or possibly from other tRNA modifications themselves.
Taken together, this study reveals that tRNAs are a target for TET-mediated oxidation of m5C to hm5C and that this conversion may play a role in supporting mRNA translation (Fig. 4D). These findings provide a starting point for future investigations into the effects of TET2-mediated oxidation of m5C in tRNAs, the mechanism by which this oxidation modulates translation, and, more generally, expanding our view of the regulatory controls exerted by TET family enzymes across both epigenetics and epitranscriptomics.
Experimental procedures
Mammalian cell culture and plasmid transfection
HEK293T cells were cultured with Dulbecco's Modified Eagle Medium (GIBCO) media supplemented with 10% fetal bovine serum (FBS) (GIBCO), 1% Pen/Strep (GIBCO). mESCs were cultured with Dulbecco's Modified Eagle Medium (GIBCO) media supplemented with 10% FBS (GIBCO), 1% pen/strep (GIBCO), 1× non-essential amino acid (GIBCO), 1× L-glutamine (GIBCO), 50 μM 2-mercaptoethanol (Bio-Rad), and 1000 μ/ml mouse leukemia inhibitory factor (LIF) (Gemini). All the cells were cultured in a humidified cell culture incubator with 5% CO2 at 37 °C. For mESCs culture, MEF feeder cells (Sigma) were used. For passaging, cells were washed with PBS once, and then 0.25% trypsin was added and incubated at 37 °C for 3 min. After medium was added to inactivate trypsin digestion, cells were split for different purposes. Tet2 KO mESC cell line was derived as previously described (44).
Doxycycline-induced TET2-CD expression cell line construction
To generate stable cell lines capable of doxycycline-inducible TET2 expression, we used high-efficiency, low-background (HILO) recombination-mediated cassette exchange (RMCE) technology (35). This started with a plasmid containing a RIPE cassette that consists of a tetracycline-inducible EGFP gene with a puromycin resistance marker. The human TET2 catalytic domain (residues 1129–2002) was cloned in place of EGFP to yield a TET2-RIPE construct. These constructs could be transfected into HILO acceptor cells, which are variants of HEK293T cells that contain a matching recombination locus encoding blasticidin resistance. The acceptor cells were maintained in media containing DMEM, high-glucose+GlutaMAX (Thermo), 10% FBS (Sigma), 1 mM sodium pyruvate (Thermo), 1× pen/strep (Thermo), and 2.5 μg/ml of blasticidin S (Thermo). For transfection, cells were plated into antibiotic-free medium to ∼70% confluency. Cells were then cotransfected with 990 ng of TET2-RIPE plasmid plus 10 ng of Cre recombinase plasmid, with the aim to insert the TET2-RIPE cassette into the acceptor locus via Cre-mediated recombination, thus swapping blasticidin for puromycin resistance. The next day, cells were split 1:2 and, after about 6 h, puromycin was added to a concentration of 2 μg/ml. Every 24 h, medium was changed, and the puromycin concentration was increased to 4 μg/ml for more stringent selection. Once colonies formed and began to expand, puromycin was reduced back to 2 μg/ml for maintenance.
PCR was used to confirm locus-specific recombination in the TET2 stable cell lines. Cells were harvested, and genomic DNA was extracted. Primers that flanked the region where recombination takes place were paired with primers complementary to sequences in the acceptor or TET2 cell line, but not both. GAPDH was used as a control.
PCR primers
EF: 5ʹ-CCAGCTTGGCACTTGATGT-3ʹ; BR: 5ʹ-TAGCCC TCCCACACATAACC-3ʹ; PR: 5ʹ-TCGTAGAAGGGGAGGT TGC-3ʹ; 4F: 5ʹ-CCAAAAGAGAGCTGCACGCTAC-3ʹ; BF: 5ʹ-GCAACGGCTACAATCAACAG-3ʹ; WR: 5ʹ-GGGCCACAAC TCCTCATAAA-3ʹ; hGAPDH_F: 5ʹ-CCTGACCTGCCGTCT AGAAA-3ʹ; hGAPDH_R: 5ʹ-CCCTGTTGCTGTAGCCAAAT-3ʹ.
For induction with doxycycline, cells were split 1 day prior into 6-well plates so that they would be 60 to 70% confluent on the day of induction. Doxycycline was dissolved in PBS and then added in equal volumes to the wells to achieve a final concentration of 100 ng/ml. Cells were harvested by trypsinization at 24, 48, or 72 h, and genomic DNA was extracted for LC-MS/MS analysis or dot blotting for 5mC and 5hmC modifications. Genomic DNA Purification Kit (Invitrogen) was used to extract the genome DNA following the manufacturer's instruction. RNA isolation was carried out as described below for cells induced 48 h.
RNA isolation
Total RNA extraction
TRIzol (Invitrogen) was used to isolate total RNA following the manufacturer's instructions.
Poly-A RNA extraction
mRNA was extracted from the total RNA by using Dynabeads mRNA Purification Kit (Ambion) following the manufacturer's instructions. The rRNA was further removed by RiboMinus Eukaryote Kit (Invitrogen). The mRNA purity and integrity were tested by Bioanalyzer with RNA nano Chips (Agilent Technologies).
Small-RNA extraction
Small RNAs were extracted from total RNA using an RNA Clean and Concentrator -25 Kit (Zymo Research) following the manufacturer's protocol for small RNAs.
tRNA extraction
Purified tRNAs were obtained by first migrating 1 to 2 μg of small RNAs through a 6% urea-TBE polyacrylamide denaturing gel (Invitrogen). Nucleic acids were stained using SYBR Gold nucleic acid gel stain (Invitrogen). Migrated tRNAs were recovered from the gel using the ZR small-RNA PAGE Recovery Kit (Zymo Research).
tRNA isolation
The procedure was adapted from a previous report (45). Briefly, RNA species smaller than 200 nt were extracted from the total RNA using RNA Clean & Concentrator (RCC) Kits (Zymo Research). The tRNA fraction was further extracted from the small RNAs by using 15% TBE-Urea gel. For specific tRNA isolation, streptavidin-conjugated M-280 magnetic Dynabeads (Invitrogen) were used. Twenty microliters of beads were washed once with buffer A (10 mM Tris, pH 7.5, 2 M NaCl, 2 mM EDTA) and then resuspended in 20 μl of buffer A. Biotinylated DNA probes (200 μM), complementing with the sequences of specific tRNA, in 10 μl of water were mixed with the same volume of beads in buffer A. After incubation at room temperature for 30 min with gentle mixing, the probe-coated beads were washed four times with buffer B (5 mM Tris, pH 7.5, 1 M NaCl, 1 mM EDTA) and resuspended with 6 × saline-sodium citrate (SSC) solution. After mixing the total tRNA and probe-coated beads in 6 × SSC solution, they were incubated at 75 °C for 10 min. Then, the mixture was rotated at room temperature for 3 h to allow the annealing. The probe-coated beads were washed three times with 3 × SSC solution and twice with 1 × SSC. Finally, the tRNA was eluted three times with RNase-free water.
Liquid chromatography and tandem mass spectrometry
The RNA and DNA samples were digested using Nucleoside Digestion Mix (NEB) at 37 °C for 2 h to get single nucleosides. Ten microliters of the sample or standard was injected into an HPLC-QQQ-MS/MS system. C18 column in reverse-phase ultra-performance liquid chromatography system was used to separate nucleosides with online mass spectrometry detection by the Altis (Thermo Fisher Scientific) QQQ triple-quadruple LC mass spectrometer in positive electrospray ionization mode with Buffer A (0.1% formic acid solution) and buffer B (30% acetonitrile in 0.1% formic acid solution). The nucleosides were quantified using retention time and the nucleoside-to-base ion mass transitions of 272.1 → 156.0 (5caC), 256.1 → 140.0 (5fC), 258.1 → 124.1 (5hmC), 242.1 → 126.1 (5mC), 228.1 → 112.1 (dC), 288.1 → 156.0 (ca5C), 272.1 → 140.0 (f5C), 274.1 → 124.1 (hm5C), 258.1 → 126.1 (m5C) and 244.1 → 112.1 (rC). All quantifications were performed by converting the peak area from the LC-MS/MS to moles using the standard curve obtained from pure nucleoside standards. Then, the percentages of m5C and hm5C to C were calculated and compared across different samples. All modifications are quantified using external calibration curves rather than labeled internal standards.
Dot-blot assay
DNA samples were applied to an Amersham Hybond-N+ membrane, optimized for nucleic acid binding (GE Healthcare), in serial dilutions. After UV cross-linking three times in a Stratagene Stratalinker 2400 UV Crosslinker, the membrane was stained with 0.04% methylene blue in 0.5 M sodium acetate. Then, the membrane was washed with 1× phosphate-buffered saline, 0.1% Tween (PBST), blocked with 5% of bovine serum albumin (BSA) in PBST, and then incubated with 5% of BSA in 1 × PBST containing specific antibody (1:500) overnight at 4 °C. After washing three times with 1 × PBST, horseradish peroxidase (HRP)–conjugated secondary antibody (1:20,000) in 5% of BSA was used. The membrane was visualized by ECL Western Blotting Detection Kit (Thermo Fisher Scientific).
TET2-CS expression and purification
N-terminally FLAG-tagged TET2-CS, the crystal structure variant of the enzyme (1129–1936 Δ1481–1843), was purified from insect cells as previously described (46). Briefly, the construct, with an N-terminal FLAG tag, was subcloned into a pFastBac1 vector. After generation of baculovirus, Sf9 cells were infected and expression was performed for 24 h. Cells from a 1 L culture were collected and resuspended in lysis buffer [50 mM HEPES (pH 7.5), 300 mM NaCl, and 0.2% (v/v) NP-40] containing complete, EDTA-free Protease Inhibitor Cocktail (Roche, 1 tablet/10 ml). Cells were lysed by three passes through a microfluidizer at 15,000 psi. The lysate was cleared by centrifugation at 20,000g for 30 min. The supernatant was then passed two or three times over a 500-μl or 1-ml packed column of anti-FLAG M2 affinity resin (Sigma), prepared according to the manufacturer's instructions. The column was washed three times with 10 ml of wash buffer [50 mM HEPES (pH 7.5), 150 mM NaCl, and 15% (v/v) glycerol]. One column volume of elution buffer [wash buffer containing 100 μg/ml 3× FLAG peptide (Sigma)] was used to elute the bound protein by incubating on the column for 10 min, and serial elution was collected until no more protein was detected by the Bio-Rad Protein Assay. The three most concentrated fractions were pooled, aliquoted, and stored at −80 °C.
In vitro activity assays
The DNA or tRNA substrate was diluted to 100 nM or 200 nM, respectively, in reaction buffer [50 mM HEPES (pH 6.5), 100 mM NaCl, 1 mM alpha-ketoglutaric acid, 1 mM dithiothreitol, and 2 mM sodium ascorbate]. Denaturation of tRNAs was accomplished by heating the tRNA diluted in the appropriate volume of water at 80 °C for 5 min before immediately adding ice-cold reaction buffer. Fresh ammonium iron (II) sulfate was added to each reaction to a final concentration of 75 μM, and the purified enzyme was added lastly to the reactions at a final volume of 25 μl. The reactions were incubated at 37 °C for 1 h. DNA reactions were quenched by the addition of a premixed quenching solution [25 μl of H2O, 25 μl Oligo Binding Buffer (Zymo), and 100 μl of ethanol]. DNA oligonucleotide products were purified using the Zymo Oligo Clean & Concentrator kit and eluted in 10 μl of Millipore water. tRNA reactions were quenched by the addition of a premixed quenching and small-RNA extraction solution [25 μl RNA binding buffer (Zymo), 100 μl of ethanol]. Small-RNA (<200 nt) products were purified using the Zymo RNA Clean & Concentrator kit modified for small RNAs and eluted in 10 μl of Millipore water. DNA and RNA products were analyzed by LC-MS/MS.
In vitro luciferase translation assays
In vitro translation assays were accomplished using the Flexi Rabbit Reticulocyte Lysate System (Promega L4960). Translation reactions were supplemented with 20 ng/ul of luciferase mRNA (Promega L4561) and with varying amounts of purified small RNAs or tRNAs. Using this in vitro translation system with purified tRNAs, we can avoid the effects from other RNA species such as rRNA. Assembled reactions were incubated at 30 °C. Reactions were tested for functional luciferase production using the standard luciferase assay kit with a luminometer (Promega L4960).
Protein synthesis assay
The rate of global protein synthesis was determined using puromycin to label nascent peptides as described previously (47). Briefly, cells were labeled by adding 1 μM puromycin to the medium and incubating 1 h. The cells were collected and washed twice with PBS. Then, the cells were lysed by radioimmunoprecipitation assay buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol, and freshly added protease inhibitor). Samples were then analyzed by SDS-PAGE followed by Western blot using an anti-puromycin antibody (Sigma-Aldrich, MABE343), and GAPDH was used as loading control.
Data availability
All other data are available from the corresponding author upon reasonable request. This session contains all data availability information in this study.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
H. S. and R. J. O. prepared samples and conducted the LC-MS/MS experiments. H. S. and R. J. O. conducted the in vitro translation assays. M. C. O performed DNA extraction and LC-MS/MS. K. F. L. conceived the research and wrote the manuscript together with R. J. O., M. C. O., and R. K. R. K. supervised the in vitro demethylation assay. M. Y. L. constructed TET-CS–inducible HEK 293T cells. U. G. purified TET-CS proteins. All authors contributed to data analysis, writing, and editing of the manuscript.
Funding and additional information
This work was supported by the National Institutes of Health (R35GM133721 to K. F. L.; R01-GM118501 to R. M. K.; T32GM132039 to M. C. O.; and F31GM139325 to R. J. O.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Karin Musier-Forsyth
Footnotes
This article contains supporting information.
Supplementary data
Figures S1 to S6
References
- 1.Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 2.Li E., Zhang Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 4.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C.X., Zhang K. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeNizio J.E., Liu M.Y., Leddin E.M., Cisneros G.A., Kohli R.M. Selectivity and promiscuity in TET-mediated oxidation of 5-methylcytosine in DNA and RNA. Biochemistry. 2019;58:411–421. doi: 10.1021/acs.biochem.8b00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delatte B., Wang F., Ngoc L.V., Collignon E., Bonvin E., Deplus R., Calonne E., Hassabi B., Putmans P., Awe S., Wetzel C., Kreher J., Soin R., Creppe C., Limbach P.A. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 9.Shen Q.C., Zhang Q., Shi Y., Shi Q.Z., Jiang Y.Y., Gu Y., Li Z.Q., Li X., Zhao K., Wang C.M., Li N., Cao X.T. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature. 2018;554:123–127. doi: 10.1038/nature25434. [DOI] [PubMed] [Google Scholar]
- 10.Chen X., Li A., Sun B.F., Yang Y., Han Y.N., Yuan X., Chen R.X., Wei W.S., Liu Y.C., Gao C.C., Chen Y.S., Zhang M.M., Ma X.D., Liu Z.W., Luo J.H. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019;21:978–990. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y., Wang L., Han X., Yang W.L., Zhang M., Ma H.L., Sun B.F., Li A., Xia J., Chen J., Heng J., Wu B., Chen Y.S., Xu J.W., Yang X. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell. 2019;75:1188–1202. doi: 10.1016/j.molcel.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 12.Trixl L., Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA. 2019;10 doi: 10.1002/wrna.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vare V.Y., Eruysal E.R., Narendran A., Sarachan K.L., Agris P.F. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. 2017;7:29. doi: 10.3390/biom7010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shanmugam R., Fierer J., Kaiser S., Helm M., Jurkowski T.P., Jeltsch A. Cytosine methylation of tRNA-Asp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov. 2015;1:15010. doi: 10.1038/celldisc.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaefer M., Pollex T., Hanna K., Tuorto F., Meusburger M., Helm M., Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., Frye M., Helm M., Stoecklin G., Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 17.Tuorto F., Herbst F., Alerasool N., Bender S., Popp O., Federico G., Reitter S., Liebers R., Stoecklin G., Grone H.J., Dittmar G., Glimm H., Lyko F. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015;34:2350–2362. doi: 10.15252/embj.201591382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trixl L., Amort T., Wille A., Zinni M., Ebner S., Hechenberger C., Eichin F., Gabriel H., Schoberleitner I., Huang A., Piatti P., Nat R., Troppmair J., Lusser A. RNA cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cell Mol. Life Sci. 2018;75:1483–1497. doi: 10.1007/s00018-017-2700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P., Lukk M., Lombard P., Treps L., Popis M., Kellner S., Holter S.M., Garrett L., Wurst W., Becker L. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haag S., Sloan K.E., Ranjan N., Warda A.S., Kretschmer J., Blessing C., Hubner B., Seikowski J., Dennerlein S., Rehling P., Rodnina M.V., Hobartner C., Bohnsack M.T. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35:2104–2119. doi: 10.15252/embj.201694885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano S., Suzuki T., Kawarada L., Iwata H., Asano K., Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met) Nat. Chem. Biol. 2016;12:546–551. doi: 10.1038/nchembio.2099. [DOI] [PubMed] [Google Scholar]
- 22.Van Haute L., Dietmann S., Kremer L., Hussain S., Pearce S.F., Powell C.A., Rorbach J., Lantaff R., Blanco S., Sauer S., Kotzaeridou U., Hoffmann G.F., Memari Y., Kolb-Kokocinski A., Durbin R. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 2016;7:12039. doi: 10.1038/ncomms12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haag S., Warda A.S., Kretschmer J., Gunnigmann M.A., Hobartner C., Bohnsack M.T. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA. 2015;21:1532–1543. doi: 10.1261/rna.051524.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motorin Y., Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 25.Squires J.E., Preiss T. Function and detection of 5-methylcytosine in eukaryotic RNA. Epigenomics. 2010;2:709–715. doi: 10.2217/epi.10.47. [DOI] [PubMed] [Google Scholar]
- 26.Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A., Helm M., Bujnicki J.M. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long T., Li J., Li H., Zhou M., Zhou X.L., Liu R.J., Wang E.D. Sequence-specific and shape-selective RNA recognition by the human RNA 5-methylcytosine methyltransferase NSun6. J. Biol. Chem. 2016;291:24293–24303. doi: 10.1074/jbc.M116.742569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain S., Tuorto F., Menon S., Blanco S., Cox C., Flores J.V., Watt S., Kudo N.R., Lyko F., Frye M. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol. Cell Biol. 2013;33:1561–1570. doi: 10.1128/MCB.01523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbasi-Moheb L., Mertel S., Gonsior M., Nouri-Vahid L., Kahrizi K., Cirak S., Wieczorek D., Motazacker M.M., Esmaeeli-Nieh S., Cremer K., Weissmann R., Tzschach A., Garshasbi M., Abedini S.S., Najmabadi H. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012;90:847–855. doi: 10.1016/j.ajhg.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawarada L., Suzuki T., Ohira T., Hirata S., Miyauchi K., Suzuki T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017;45:7401–7415. doi: 10.1093/nar/gkx354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takemoto C., Spremulli L.L., Benkowski L.A., Ueda T., Yokogawa T., Watanabe K. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009;37:1616–1627. doi: 10.1093/nar/gkp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber S.M., van Delft P., Tanpure A., Miska E.A., Balasubramanian S. 2'-O-Methyl-5-hydroxymethylcytidine: a second oxidative derivative of 5-methylcytidine in RNA. J. Am. Chem. Soc. 2017;139:1766–1769. doi: 10.1021/jacs.6b12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He C., Sidoli S., Warneford-Thomson R., Tatomer D.C., Wilusz J.E., Garcia B.A., Bonasio R. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell. 2016;64:416–430. doi: 10.1016/j.molcel.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu L.L., Li Z., Cheng J.D., Rao Q.H., Gong W., Liu M.J., Shi Y.J.G., Zhu J.Y., Wang P., Xu Y.H. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Khandelia P., Yap K., Makeyev E.V. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:12799–12804. doi: 10.1073/pnas.1103532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nedialkova D.D., Leidel S.A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arioka Y., Watanabe A., Saito K., Yamada Y. Activation-induced cytidine deaminase alters the subcellular localization of Tet family proteins. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takano A., Endo T., Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005;309:140–142. doi: 10.1126/science.1113346. [DOI] [PubMed] [Google Scholar]
- 39.Shaheen H.H., Hopper A.K. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11290–11295. doi: 10.1073/pnas.0503836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaheen H.H., Horetsky R.L., Kimball S.R., Murthi A., Jefferson L.S., Hopper A.K. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8845–8850. doi: 10.1073/pnas.0700765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q., Liu X., Gao W., Li P., Hou J., Li J., Wong J. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked beta-N-acetylglucosamine transferase (OGT) J. Biol. Chem. 2014;289:5986–5996. doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan C.T., Pang Y.L., Deng W., Babu I.R., Dyavaiah M., Begley T.J., Dedon P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song C.X., He C. Potential functional roles of DNA demethylation intermediates. Trends Biochem. Sci. 2013;38:480–484. doi: 10.1016/j.tibs.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hon G.C., Song C.X., Du T., Jin F., Selvaraj S., Lee A.Y., Yen C.A., Ye Z., Mao S.Q., Wang B.A., Kuan S., Edsall L.E., Zhao B.S., Xu G.L., He C. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell. 2014;56:286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu F., Clark W., Luo G., Wang X., Fu Y., Wei J., Wang X., Hao Z., Dai Q., Zheng G., Ma H., Han D., Evans M., Klungland A., Pan T. ALKBH1-Mediated tRNA demethylation regulates translation. Cell. 2016;167:816–828.e816. doi: 10.1016/j.cell.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu M.Y., DeNizio J.E., Kohli R.M. Quantification of oxidized 5-methylcytosine bases and TET enzyme activity. Methods Enzymol. 2016;573:365–385. doi: 10.1016/bs.mie.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sondalle S.B., Longerich S., Ogawa L.M., Sung P., Baserga S.J. Fanconi anemia protein FANCI functions in ribosome biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2561–2570. doi: 10.1073/pnas.1811557116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S6
Data Availability Statement
All other data are available from the corresponding author upon reasonable request. This session contains all data availability information in this study.