Abstract
Meningiomas (MNs), arising from the arachnoid/meningeal layer, are nonresponsive to chemotherapies, with ∼50% showing loss of the Neurofibromatosis 2 (NF2) tumor suppressor gene. Previously, we established NF2 loss activates mechanistic target of rapamycin complex 1 (mTORC1) and mechanistic target of rapamycin complex 2 (mTORC2) signaling, leading to clinical trials for NF2 and MN. Recently our omics studies identified activated ephrin (EPH) receptor and Src family kinases upon NF2 loss. Here, we report increased expression of several ligands in NF2-null human arachnoidal cells (ACs) and the MN cell line Ben-Men-1, particularly neuregulin-1/heregulin (NRG1), and confirm increased NRG1 secretion and activation of V-ERB-B avian erythroblastic leukemia viral oncogene homolog 3 (ERBB3) receptor kinase. Conditioned-medium from NF2-null ACs or exogenous NRG1 stimulated ERBB3, EPHA2, and mTORC1/2 signaling, suggesting pathway crosstalk. NF2-null cells treated with an ERBB3-neutralizing antibody partially downregulated mTOR pathway activation but showed no effect on viability. mTORC1/2 inhibitor treatment decreased NRG1 expression and downregulated ERBB3 while re-activating pAkt T308, suggesting a mechanism independent of NRG1–ERBB3 but likely involving activation of another upstream receptor kinase. Transcriptomics after mTORC1/2 inhibition confirmed decreased ERBB3/ERBB4 while revealing increased expression of insulin-like growth factor receptor 1 (IGF1R). Drug treatment co-targeting mTORC1/2 and IGF1R/insulin receptor attenuated pAkt T308 and showed synergistic effects on viability. Our findings indicate potential autocrine signaling where NF2 loss leads to secretion/activation of NRG1-ERBB3 signaling. mTORC1/2 inhibition downregulates NRG1-ERBB3, while upregulating pAkt T308 through an adaptive response involving IGF1R/insulin receptor and co-targeting these pathways may prove effective for treatment of NF2-deficient MN.
Keywords: signaling, NF2, brain tumor, meningioma, insulin-like growth factor (IGF) receptor 1, mammalian target of rapamycin (mTOR), tumor suppressor gene, dual mTORC1/mTORC2 inhibition, Akt PKB, NRG1-ERBB3
Abbreviations: AC, arachnoid cell; APJ, apelin receptor; CM, concentrated medium; DRC, dose–response curve; EGFR, epidermal growth factor receptor; EPH receptor, erythropoietin-producing hepatocellular receptor; ERBB, V-ERB-B avian erythroblastic leukemia viral oncogene homolog; FOXO, Forkhead box protein O; IC50, inhibitory concentration 50%; IGF1R/IR, insulin-like growth factor receptor 1/insulin receptor; IRS, insulin receptor substrate; MN, meningioma; MR, maximum response; mTOR, mechanistic/mammalian target of rapamycin; mTORC1/mTORC2, mTOR complex 1/mTOR complex 2; NF2, Neurofibromatosis 2; NRG1, neuregulin-1/heregulin; PDK1, phosphoinositide-dependent kinase-1; PI3K, Phosphoinositide 3-kinase; qPCR, quantitative RT-PCR; RTK, receptor tyrosine kinase; SGK1, serum-/glucocorticoid-responsive kinase-1; TGFA, transforming growth factor-alpha; WHO, World Health Organization
The disease neurofibromatosis 2 (NF2) is characterized by multiple nervous system tumors, including bilateral vestibular schwannomas and intracranial meningiomas (MNs), and is caused by mutations in the NF2 gene (1, 2, 3). Separately, sporadic MNs are the most common primary intracranial tumors in adults, with ∼50% having biallelic, somatic inactivation of NF2. Benign MNs (World Health Organization grade I) are most common; however, they often cause severe neurologic morbidity and mortality because of compression of adjacent brain or spinal cord. Atypical (World Health Organization grade II) or anaplastic (World Health Organization grade III) MNs display more aggressive clinical behavior with rapid growth and increased recurrence rates (4, 5). With nonresponse to chemotherapies, the current standard of care for MN is maximal surgical resection, whereas radiation is reserved for recurrent or aggressive tumors. MNs that progress despite surgery and radiation show high morbidity and mortality (1, 6). Therefore, effective noninvasive therapies are much needed for both NF2-associated and sporadic MNs.
NF2 encodes the tumor suppressor protein merlin, which has been implicated in a wide range of mitogenic signaling pathways, including receptor tyrosine kinases (RTKs) (7), Rac/p21-activated kinase (8, 9), mammalian/mechanistic target of rapamycin complex 1 (mTORC1) (10, 11), and Hippo (12, 13) pathways. A nuclear function for merlin through regulation of E3 ubiquitin ligase Cullin-RING E3 ubiquitin ligase-4 (DDB1 and CUL4-associated factor-1) is also reported (14). Thus, merlin likely regulates these various implicated pathways in a cell context–dependent manner. Our studies employing isogenic human arachnoidal cell lines (ACs, cell of origin for MN) expressing or lacking NF2, generated with CRISPR-Cas9 genome editing, as well as primary and immortalized MN cells, established that NF2 loss leads to aberrant activation of mTORC1 and mTORC2 signaling (10, 15, 16, 17). This finding led to completed clinical trials with the mTORC1 inhibitor RAD001 for NF2 patients (18, 19, 20) and ongoing clinical trials using ATP-competitive mTOR kinase inhibitor AZD2014 (vistusertib) for NF2-associated and sporadic MN. Treating NF2 patients with the mTORC1 inhibitor RAD001 revealed cytostatic effects of delayed growth or stabilization of schwannomas and MN without tumor shrinkage (18, 19, 20), and the results of AZD2014 therapy remain under investigation.
To identify other relevant drug targets for NF2, we recently performed large-scale kinome and transcriptome analyses of our AC and MN cell models, which showed increased activation and expression of several EPH receptor family tyrosine kinases, Src family kinase members and c-KIT, which are all targets of dasatinib (21). In follow-up studies, we confirmed the expression/activation of EPH receptor family tyrosine kinases as well as downstream Src family kinases in NF2-null MN and schwannoma models and reported that combining a dual mTORC1/mTORC2 inhibitor, AZD2014 or INK128 (also called TAK-228), with dasatinib is synergistic in these models (22, 23). Here, extending analyses of transcriptomic data from our previous studies, we have identified several ligands, including NRG1, which encodes neuregulin-1. Although mTOR activation upon NF2 loss is well established, the upstream regulation of this activation and downstream consequences of mTOR activation remain unclear. Therefore, we set out to examine the role of neuregulin-1/heregulin (NRG1) in the context of NF2 loss. Our findings reveal that NF2 loss leads to NRG1 secretion that in turn activates the V-ERB-B avian erythroblastic leukemia viral oncogene homolog 3 (ERBB3) receptor kinase in an autocrine fashion. In addition, secreted and exogenous NRG1 activates downstream mTOR signaling and EPHA2. Interestingly, mTORC1/mTORC2 inhibition in our MN cellular models disrupts the NRG1-ERBB3 signaling while increasing pAkt T308, but not pAkt S473, because of an adaptive response that likely involves upregulation of insulin-like growth factor receptor 1 (IGF1R) expression and activation. Our results further show that combined inhibition of mTOR and IGF1R signaling is synergistic in MN cells and provide a compelling basis for in vivo testing in animal models of NF2.
Results
High throughput transcriptome analyses reveal increased expression of ligands in NF2-deficient cells
We recently carried out high throughput kinome and transcriptome analyses along with drug screening in NF2-null MN cellular models including CRISPR-modified human arachnoid cells (ACs) and an immortalized human MN line Ben-Men-1 (21, 22). Analysis of the RNA-sequencing (RNA-seq) dataset revealed significant up regulation of several genes including NRG1, HBEGF, APLN, and TGFA (21) that encode respective ligands NRG1, capable of binding ERBB3/HER3 and ERBB4/HER4 receptors (24); heparin-binding EGF-like growth factor which binds epidermal growth factor receptor (EGFR) (also known as ERBB1) and ERBB4 receptors (24); apelin, a ligand for apelin receptor APJ (25, 26); and transforming growth factor-alpha, which binds EGFR (21, 24) (Table 1). Using quantitative RT-PCR (qPCR), we confirmed increased expression of NRG1, HBEGF, and APLN in NF2-null(−) CRISPR-modified arachnoidal cells (AC-CRISPR) cells and MN line Ben-Men-1 compared with NF2-expressing(+) ACs (Fig. 1A), with no significant difference in TGFA by qPCR (data not shown). Interestingly, previous reports have implicated overexpression of epidermal growth factor receptor family tyrosine kinases EGFR and ERBB2/HER2 in NF2-associated schwannomas and MN (27, 28). Further, in our previous study employing shRNA-mediated kinome screening in Ben-Men-1 cells, in which we reported serum/glucocorticoid-regulated kinase 1 (SGK1) as a regulator of mTORC1 signaling, another top candidate to emerge was the ERBB3 receptor (15). We therefore chose to focus our studies on NRG1-ERBB3 signaling.
Table 1.
Ligand/growth factor genes showing increased expression in NF2-null AC and MN cells
| Gene ID | Protein name |
NF2(−) ACs versus NF2(+) ACsa |
Ben-Men-1 versus NF2(+) ACsb |
||
|---|---|---|---|---|---|
| log2FC | p-valuec | log2FC | p-valuec | ||
| NRG1 | Neuregulin/heregulin | 4.47 | 6.36 × 10−6 | 6.28 | 1.45 × 10−8 |
| HBEGF | Heparin-binding EGF-like growth factor | 3.00 | 2.00 × 10−4 | 6.76 | 8.33 × 10−11 |
| APLN | Apelin | 2.42 | 1.61 × 10−2 | 5.88 | 1.84 × 10−10 |
| TGFA | Transforming growth factor, alpha | 1.89 | 5.62 × 10−2 | 3.26 | 4.67 × 10−6 |
AC, arachnoid cell; MN, meningioma.
Data from genetically matched (isogenic) NF2(−) versus NF2(+) AC lines is summarized from previous report (21).
Genetically unmatched lines.
p-value is Bonferroni adjusted; log2FC, log2 fold change.
Figure 1.
NF2-null cells show increased NRG1 expression and secretion.A, quantitative RT-PCR (qPCR) for human NRG1 (hNRG1), hHBEGF, and hAPLN shows increased expression in NF2-null(−) ACs and Ben-Men-1 cells compared with NF2(+) cells. Data are expressed as relative fold change versus NF2(+) ACs, ±SD. Column scatter plots were generated using GraphPad Prism 8. B, immunoblotting of NF2/merlin in NF2-null ACs and Ben-Men-1 along with GAPDH (loading control). C, concentrated media (48 h collection, 48 h-CM) from NF2-null(−) ACs (upper and lower panel) and Ben-Men-1 (lower panel) show secreted NRG1 (secNRG1) compared with 48 h-CM from NF2(+) ACs. Media freshly added and then immediately collected (0 h-CM) serves as control (upper panel). Nonspecific band serves as loading control (lower panel, gray arrowhead). D, treatment of NF2(+) ACs with conditioned media from NF2(−) ACs activates pERBB3 Y1197, pEPHA2 S897, mTORC1 (pS6 S240/4 readout) and mTORC2 (pNDRG1 T346 readout) versus NF2(+) AC media. E, Incubation of NF2-null ACs and Ben-Men-1 cells in serum-free medium (∼20 h) followed by NRG1stimulation (5 nM, 30 min) leads to activation of pERBB3 Y1197, pEPHA2 S897, pAkt S473 (mTORC2 readout), and pS6K T389 (mTORC1 readout). For E and F, ImageJ/Fiji quantitation shows phosphorylated relative to total protein, with control lanes normalized to 1. AC, arachnoid cell; MN, meningioma; mTORC1, mechanistic target of rapamycin complex 1.
NF2-deficient AC and MN cells secrete NRG1 ligand that activates the ERBB3 receptor and downstream signaling
We next examined whether NF2-deficient AC cells and Ben-Men-1 cells (Fig. 1B) secrete NRG1 ligand. Media from NF2(+) and NF2(−) AC-CRISPR cells were collected at 48 h following a change to serum-free medium and then concentrated by centrifugal filtration. Immunoblotting revealed NRG1 secretion in 48 h-concentrated medium (48 h-CM) from NF2(−) AC cells and Ben-Men-1, whereas 48 h-CM from NF2(+) ACs or 0 h-CM from NF2(−) ACs showed no signal for secreted NRG1 (Fig. 1C). As a confirmation, we also performed qPCR analysis in Ben-Men-1 cells where NF2 had been re-introduced by lentiviral transduction and observed that re-expression of NF2 (NF2-CSCW2) demonstrated a significant decrease in NRG1 expression compared with empty vector (V-CSCW2) (Fig. S1).
Next, in conditioned medium experiments, we incubated NF2(+) ACs with medium from NF2(+) or NF2(−) ACs. In cells treated with NF2(−) 48 h-CM, we observed increased phosphorylation of ERBB3 Y1197 (pERBB3) receptor compared with 48 h-CM from NF2(+) ACs. We also found increased phosphorylation of the receptor EPHA2 S897 (pEPHA2) as well as activated mTOR signaling, as shown by increase in mTORC1 pathway readout pS6 S240/44 and mTORC2-SGK1 pathway readout pNDRG1 T346 (Fig. 1D). We confirmed these results using exogenous NRG1 to stimulate NF2-null ACs and observed not only activation of pERBB3 but also pEPHA2, pS6K T389 (mTORC1 readout), and pAkt S473 (another mTORC2 readout) (Fig. 1E). These data suggest that loss of NF2 in AC or MN cells leads to secretion of NRG1 ligand and activation of ERBB3 receptor in an autocrine fashion. In addition, upregulation of NRG1-ERBB3 signaling can cross-talk to EPHA2 receptor as well as activate downstream mTORC1/2 signaling.
Treatment with an ERBB3 neutralizing antibody downregulates activated ERBB3 and downstream signaling in NF2-deficient cells
To further understand the basal level of activation of ERBB3, EPHA2, and mTORC1/2 signaling upon NF2 loss, we treated our NF2-null ACs with the multi-ERBB inhibitor lapatinib or the EGFR-specific inhibitor erlotinib under NRG1-stimulated or unstimulated conditions. In NRG1-stimulated cells, lapatinib treatment led to inhibition of pERBB3, as expected, along with downregulation of pEPHA2 receptor and mTORC1/2 pathway readouts pS6K T389 and pAkt S473, respectively, whereas erlotinib treatment showed no effect. Conversely, in unstimulated cells, while lapatinib treatment blocked basal pERBB3, neither lapatinib nor erlotinib treatment attenuated basally activated pEPHA2 or mTORC1/2 signaling (Fig. 2A). We next treated our cells with MM-121/seribantumab (provided by Merrimack Pharmaceuticals), a human monoclonal antibody that specifically binds ERBB3 and prevents NRG1 ligand binding (29, 30). Immunoblotting demonstrated that treatment of NF2-null AC-CRISPR and Ben-Men-1 cells with MM-121 inhibited the basally activated and NRG1-stimulated pERBB3 Y1197, both at 2 h and 24 h time points (Fig. 2B). Further, similar to lapatinib, NRG1-stimulated cells treated with MM-121 also showed attenuated pEPHA2 S897, pAkt S473 (mTORC2 readout), and pS6K T389 (mTORC1 readout) at 2 h and 24 h treatment, as well as decreased pS6 S240/44 (mTORC1) in NF2-null ACs at 24 h and Ben-Men-1 at 2 h and 24 h (Fig. 2C). Moreover, when examining basally activated downstream signaling, we found that unlike lapatinib, treatment with MM-121 was capable of inhibiting pEPHA2 and mTORC1 signaling (pS6K and pS6) at 24 h in NF2-null ACs. It should be noted that, consistent with our previous reports, activated pAkt S473 was below detectable level in serum-deprived NF2-null ACs. In Ben-Men-1 cells, MM-121 treatment also downregulated basal pAkt (mTORC2), pS6K, and pS6 (mTORC1) at 2 h and 24 h, but had no effect on pEPHA2 (Fig. 2D). Here, our results confirm the signaling cross-talk seen in our conditioned media and NRG1-stimulation experiments and suggest that activated NRG1-ERBB3 signaling may be partially responsible for activated EPHA2 and mTORC1/2 pathways in NF2-null AC and Ben-Men-1 cells. We also show that direct inhibition of NRG1 ligand binding to ERBB3 is more effective than lapatinib to downregulate the basally activated signaling pathways.
Figure 2.
NRG1-ERBB3 signaling activates EPH-RTK and mTOR signaling pathways in NF2-deficient cells.A, NF2-null(−) AC-CRISPR cells show activated pERBB3 Y1197, pEPHA2 S897, pAkt S473 (mTORC2 readout), and pS6K T389 (mTORC1 readout) signatures compared to NF2-expressing(+) ACs, which were further increased by exogenous NRG1 stimulation (30 min, 5 nM). Following overnight serum deprivation, cells were co-treated for 2 h with lapatinib (500 nM), erlotinib (500 nM) or DMSO, with (+) or without (−) NRG1 stimulation in the final 30 min. Co-treatment with multi-ERBB inhibitor lapatinib attenuated the NRG1-stimulated downstream signaling, but EGFR-specific erlotinib did not. B, treatment of NF2-null ACs (top panel) or Ben-Men-1 cells (bottom panel) for 2 h or 24 h with ERBB3-specific neutralizing antibody MM-121 (0.5 μM or 1 μM) blocked activated pERBB3 Y1197 in unstimulated and stimulated cells. C and D, treatment of NF2-null ACs or Ben-Men-1 cells for 2 h or 24 h with MM-121 (0.5 μM) blocked NRG1-stimulated pEPHA2 S897, pAkt S473 and pS6K T389 (C). In addition, MM-121 downregulated basally activate pEPHA2 S897, pS6K T389 at 24 h treatment in NF2-null ACs, and pAkt S473, pS6K T389 at 2 h and 24 h in Ben-Men-1 cells (D). ImageJ/Fiji quantitation shows phosphorylated relative to total protein, with control lanes normalized to 1. AC-CRISPR, CRISPR-modified arachnoidal cells; EPH-RTK, EPH receptor family tyrosine kinases; ERBB3, V-ERB-B avian erythroblastic leukemia viral oncogene homolog 3; mTOR, mechanistic/mammalian target of rapamycin; mTORC1, mechanistic target of rapamycin complex 1.
We next examined the effect of MM-121 treatment on cell growth in NF2-null AC-CRISPR and Ben-Men-1 cells. Dose–response curves (DRCs) showed a slight effect in NF2-null ACs with a maximum response (MR) at 10 μM of 34.4% inhibition (65.6% viable cells remaining). In Ben-Men-1 cells, MM-121 treatment only led to an MR at 10 μM of 12.3% inhibition (87.7% viable cells remaining), and calculated IC50, defined as 50% viable cells remaining compared to vehicle control, could not be determined in either cell line (Fig. S2). Taken together, our data demonstrate that direct inhibition of ERBB3 receptor by preventing NRG1 ligand binding can lead to downregulation of ERBB3 as well as EPHA2 and mTORC1/2 signaling. However, our viability assays indicate that specific inhibition of NRG1-ERBB3 signaling in NF2-null AC and Ben-Men-1 cells is not sufficient to affect cellular growth.
Dual mTORC1/2 inhibition negatively regulates NRG1-ERBB3 signaling and induces activation PDK1-AKT signaling
We previously established that NF2 loss in AC and MN cells leads to constitutive activation of mTORC1/2 signaling (10, 15, 17). Recent studies in breast cancer cells demonstrated that treatment with a dual mTORC1/2 inhibitor AZD8055 led to increased expression and activation of several RTKs including the ERBB receptor family (ERBB1-4), IGF1R, and insulin receptor (IR), resulting in phosphoinositide-dependent kinase-1 (PDK1)–dependent activation/phosphorylation of Akt T308 (31). We therefore raised the question of whether mTOR pathway activation in NF2-null cells could similarly regulate NRG1-ERBB3 signaling. However, qPCR analysis revealed that treatment of NF2-null ACs and Ben-Men-1 with mTORC1/2 inhibitor, AZD2014 or INK128/TAK-228, as well as mTORC1-specific rapamycin led to decreased expression of NRG1 (Fig. 3A). Immunoblotting also showed attenuation of pERBB3 Y1197 with AZD2014 or INK128, as well as rapamycin treatment (Fig. 3, B–C). Further, similar to reports in breast cancer cells, we observed increased pAkt T308 with INK128 treatment, while pAkt S473 and pS6 remained inhibited. However, unlike breast cancer cells, this was independent of NRG1-ERBB3 signaling because co-treatment with INK128 combined with ERBB3 inhibitor MM-121 also showed increased pAkt T308, similar to INK128 alone (Fig. 3C). Transcriptome analyses of our NF2-null ACs after treatment with INK128 showed a decrease in NRG1, ERBB3, and ERBB4, suggesting that NRG1-ERBB pathway is downregulated at the transcription level by INK128 treatment (Table 2).
Figure 3.
mTORC1 inhibition downregulates NRG1-ERBB3 signaling.A, quantitation of NRG1 qPCR in NF2-null(−) ACs and Ben-Men-1 cells treated with mTORC1-specific rapamycin or dual mTORC1/2 inhibitors is shown. Data are expressed as relative fold change compared with DMSO-treated cells, ± SD. Each data point represents three technical replicates with biological replicate numbers (n) and p-value shown. Column scatter plots were generated using GraphPad Prism 8. Drug treatment time and doses are described in B and C. B, immunoblotting of NF2-null ACs and Ben-Men-1, treated for 24 h with rapamycin (20 nM) or mTORC1/2 inhibitor AZD2014 (300 nM), shows decrease in activated ERBB3 (pERBB3 Y1197) along with respective mTORC1/2 pathway readouts pAkt S473 (mTORC2) and pS6K T389, pS6 S240/44 (mTORC1). C, immunoblotting of NF2-null ACs and Ben-Men-1 treated for 24 h with mTORC1/2 inhibitor INK128 shows decreased pERBB3 and pAkt S473 (mTORC2 readout). However, INK128 treatment (alone or combined with ERBB3-specific inhibitor MM-121) revealed upregulation of PDK1-dependent pAkt T308, while still inhibiting pAkt S473. All treatment times were 24 h (A–C). ImageJ/Fiji quantitation shows phosphorylated relative to total protein, with control lanes normalized to 1. ERBB3, V-ERB-B avian erythroblastic leukemia viral oncogene homolog 3; mTORC1, mechanistic target of rapamycin complex 1; PDK1, phosphoinositide-dependent kinase-1.
Table 2.
Transcriptome changes in INK128-treated NF2-null AC-CRISPR cells
| Symbol | Ensemble ID | log2FC | FC | p value | BH adj p value |
|---|---|---|---|---|---|
| IRS2 | ENSG00000185950 | 1.45 | 2.73 | 4.57 × 10−12 | 3.59 × 10−10 |
| IGF1R | ENSG00000140443 | 0.79 | 1.72 | 2.77 × 10−10 | 1.05 × 10−08 |
| INSR | ENSG00000171105 | 0.56 | 1.47 | 1.06 × 10−03 | 3.22 × 10−03 |
| IGF2R | ENSG00000197081 | 0.43 | 1.34 | 4.23 × 10−04 | 1.48 × 10−03 |
| ERBB3 | ENSG00000065361 | −0.74 | −1.67 | 5.29 × 10−05 | 2.55 × 10−04 |
| ERBB4 | ENSG00000178568 | −1.58 | −2.99 | 9.59 × 10−09 | 1.92 × 10−07 |
| MTOR | ENSG00000198793 | −0.34 | −1.27 | 2.31 × 10−04 | 8.91 × 10−04 |
| NRG1 | ENSG00000157168 | −0.48 | −1.39 | 6.10 × 10−02 | ns |
AC-CRISPR, CRISPR-modified arachnoidal cells; BH adj, Benjamini-Hochberg adjusted; FC, fold change; ns, not significant.
Interestingly, RNA-seq analyses also showed a significant increase in IRS2 and IGF1R in NF2-null ACs treated with INK128 (Table 2). Treatment of two independent NF2-null AC-CRISPR clones and Ben-Men-1 cells with INK128 at 2 h and 24 h revealed increased expression of the IGF1R receptor by immunoblotting, confirming our RNA-seq results (Fig. 4A). Further, using an antibody that recognizes both pIGF1R Y1135/36 and pIR Y1150/51, we also found increased levels of pIGF1R/pIR as well as increased pAkt T308, but no activation of pAkt S473 after 24 h in NF2-null ACs and Ben-Men-1 cells (Fig. 4B). Treatment with INK128 at both 24 h and 48 h showed similar inhibition of pAkt S473 in our cells (data not shown). RTKs, including IGF1R/IR, are clearly established to activate downstream phosphoinositide 3-kinase (PI3K)/PDK1–dependent recruitment and phosphorylation of Akt T308 at the plasma membrane (32). We examined whether INK128 treatment might induce a similar signaling mechanism in NF2-null cells. Co-treatment of our NF2-null AC and Ben-Men-1 cells with INK128 combined with IGF1R inhibitor BMS-754807 downregulated both pIGF1R and pAkt T308 (Fig. 4, B–C), whereas co-treatment of INK128 with lapatinib or erlotinib had no effect on pAkt T308 levels (Fig. 4C). In addition, inhibition of both mTORC1/2 and IGF1R also downregulated pFOXO1/3a, a functional downstream phospho-target of Akt, compared with INK128 alone. (Fig. 4D). Together, these data suggest that mTOR kinase inhibition disrupts and downregulates NRG1-ERBB3 signaling, while inducing an adaptive response of PDK1-dependent pAkt T308 activation that involves upregulation of IGF1R in NF2-deficient AC and MN cells.
Figure 4.
mTOR kinase inhibition activates PDK1-dependent pAkt T308 through increased expression/activation of IGF1R/IR signaling.A, immunoblotting of two independent NF2-null (−) AC-CRISPR clones (#1, #2) and Ben-Men-1 cells treated with mTOR kinase inhibitor INK128 (200 nM, 2 h and 24 h) revealed increased expression of IGF1R and activation of PDK1-dependent pAkt T308 while still inhibiting pS6 S240/44 (mTORC1 readout) and pAkt S473 (mTORC2 readout). B, treatment of NF2(−) ACs and Ben-Men-1 with INK128 (200 nM, 24 h) led to increased phosphorylation of IGF1R/IR (antibody recognizes pIGF1R Y1135/36 and pIR Y1150/51). Co-treatment using INK128 and IGF1R inhibitor BMS-754807 (BMS-807, 3 μM, 24 h) downregulated pIGF1R/IR as well as pAkt T308 compared with INK128 alone. Readouts for mTORC1 (pS6 S240/44) and mTORC2 (pAkt S473) signaling are shown. C, co-treatment (24 h) of Ben-Men-1 cells with INK128 (200 nM) and lapatinib (500 nM) or erlotinib (500 nM) was unable to downregulated pAkt T308, unlike INK128 co-treatment with BMS-807 (3 μM). D, increased phosphorylation of the Akt downstream effector pFOXO1 T24/pFOXO3a T32 was observed upon INK128 treatment (200 nM, 24 h) which was downregulated by INK128 co-treatment with BMS-807 (3 μM). ImageJ/Fiji quantitation shows phosphorylated relative to total protein, with control lanes normalized to 1. PDK1, phosphoinositide-dependent kinase-1; mTOR, mechanistic/mammalian target of rapamycin; mTORC1, mechanistic target of rapamycin complex 1.
Co-treatment with INK128 and BMS-754807 reveals synergistic effects
We examined the effects of mTOR kinase inhibition and IGF1R inhibition, singly and combined, on cell viability in NF2-null cells including NF2-null ACs, two immortalized MN lines Ben-Men-1 and MN1-LF, and two primary MN lines MN646C and MN658 (Fig. S3). For single drug testing, in agreement with our previous reports using mTORC1/2 inhibitors (15, 22), cell viability assays in NF2-null cell lines demonstrated a reduction upon INK128 treatment. Moreover, INK128 treatment showed superior efficacy in all cell lines tested compared with BMS-754807 with IC50s in the nanomolar range for INK128 treatment compared with micromolar IC50 range for BMS-754807 (Fig. 5A, Table S1). Therefore, these data suggest a relative insensitivity to IGF1R inhibition alone in these cell lines.
Figure 5.
Dose–response testing of INK128 and BMS-754807 co-treatment in NF2-null cells.A, single drug dose–response curves (DRCs) are shown for NF2-null ACs and meningioma lines, including immortalized Ben-Men-1 and MN1-LF as well as two primary lines MN646C and MN658. DRCs demonstrate greater effectiveness of INK128 compared with BMS-754807 alone. Cell lines were treated (three replicates) with nine dosage points (1.5 nM–10 μM, 1:3 serial dilution) for each drug, and % cell viability is relative to DMSO vehicle treatment. B, Ben-Men-1 cells were treated (four replicates) with 10 dosage points (0.5 nM–10 μM, 1:3 serial dilution) for BMS-754807, either alone or in the presence of 400 nM INK128. C, MN1-LF cells were treated with BMS-754807 as in A, either alone or in the presence of INK128 (doses as indicated). In B and C, left-shifted DRCs revealed increased sensitivity of cells to IGFR1R/IR inhibition when co-treated with mTOR kinase inhibitor INK128 compared with BMS-754807 alone. IGF1R, insulin-like growth factor receptor 1; IR, insulin receptor; mTOR, mechanistic/mammalian target of rapamycin; NF2, neurofibromatosis 2.
Based on our RNA-seq and immunoblotting data showing upregulated IGF1R expression and activation in response to dual mTORC1/2 inhibition, we next performed co-treatment assays. Dose–response testing in Ben-Men-1 cells for BMS-754807 in the presence of 400 nM INK128 (BMS + 400 nM INK128) revealed a left-shifted DRC, with a decreased IC50 of 3.2 μM (MR, 77.7%) compared with 4.4 μM (MR, 75.7%) for BMS-754807 alone (Fig. 5B). We further extended the co-treatment dose–response testing in MN1-LF cells. DRCs for BMS-754807 co-treated with increasing doses of INK128 again revealed left-shifted DRCs suggesting increased sensitivity to IGF1R inhibition in a dose-dependent manner compared with BMS-754807 alone (Fig. 5C, Table S1).
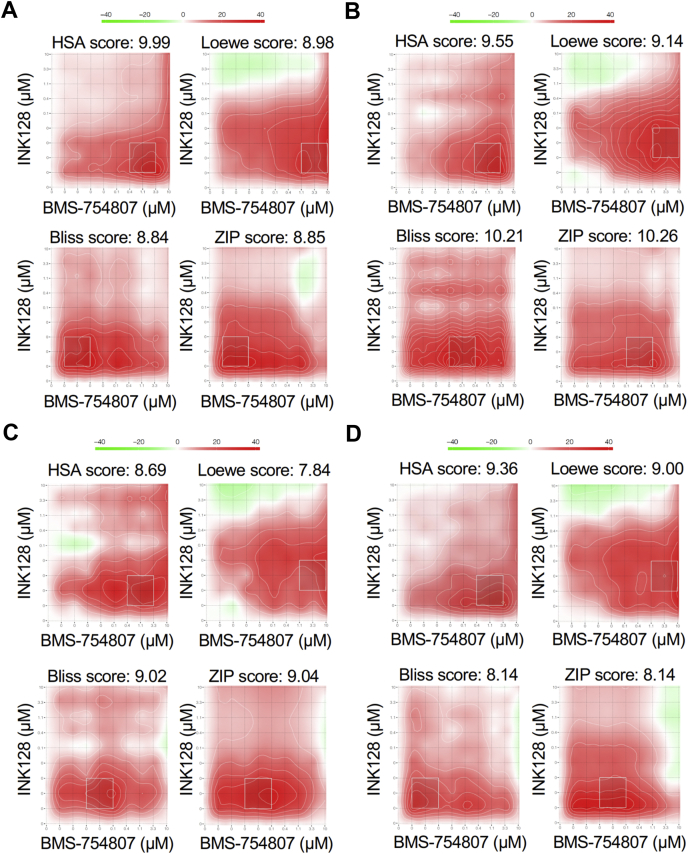
Expanding on these results, employing 10 × 10 dose matrix testing, we next asked whether co-treatment with INK128 and BMS-754807 has a synergistic effect. For drug synergy analysis, we utilized the web-based SynergyFinder application (33), which generates a synergy score by comparing drug treatment data to four different synergy reference models: (i) Highest Single Agent, (ii) Loewe Additivity, (iii) Bliss Independence, and (iv) Zero Interaction Potency. Co-treatment of Ben-Men-1 and MN1-LF cells as well as two human primary NF2-null MN lines, MN646C and MN658, with INK128 and BMS-754807 showed true synergistic effects, defined as a score >5.0 for all four reference models (34) (Fig. 6, Table 3). Taken together, cell viability assays strongly support our immunoblotting results and suggest a potential mechanism for drug synergy where mTOR kinase inhibition leads to IGF1R-PDK1-pAkt T308 pathway activation that in turn sensitizes NF2-null cells to IGF1R inhibition.
Figure 6.
Quantification of INK128 and BMS-754807 synergy in additional NF2-null meningioma cells.A–D, NF2-null immortalized lines, Ben-Men1 (A) and MN1-LF (B), as well as two primary lines MN646C (C) and MN658 (D) were co-treated in a 10 × 10 dose-matrix format (three replicates) with INK128 and BMS-754807 at 1.5 nM–10 μM (nine dosage points, threefold dilution series) of each drug and DMSO (vehicle). Data were calculated as percent viability at each treatment point relative to vehicle treated cells. Percent inhibition and synergy plots with scores for each reference model (HSA, Loewe, Bliss, and ZIP) were generated using SynergyFinder web application. White boxed regions indicate most synergistic areas, with scores summarized in Table S1. Synergy scoring scale is shown with red representing synergism and green representing antagonism (A–D). Bliss, Bliss independence; HSA, highest single agent; Loewe, Loewe additivity; ZIP, zero interaction potency.
Table 3.
SynergyFinder scores from NF2-null meningioma lines co-treated with INK128 and BMS-754807
| Cell line | Modela | SynergyFinder |
|
|---|---|---|---|
| Average score | Most synergistic area score | ||
| Ben-Men-1 (immortalized MN) | HSA | 9.99 | 24.79 |
| Ben-Men-1 (immortalized MN) | Loewe | 8.98 | 21.78 |
| Ben-Men-1 (immortalized MN) | Bliss | 8.84 | 21.98 |
| Ben-Men-1 (immortalized MN) | ZIP | 8.85 | 21.30 |
| MN1-LF (immortalized MN) | HSA | 9.55 | 25.76 |
| MN1-LF (immortalized MN) | Loewe | 9.14 | 21.27 |
| MN1-LF (immortalized MN) | Bliss | 10.21 | 22.52 |
| MN1-LF (immortalized MN) | ZIP | 10.26 | 22.76 |
| MN646 C (primary MN) | HSA | 8.69 | 18.78 |
| MN646 C (primary MN) | Loewe | 7.84 | 17.38 |
| MN646 C (primary MN) | Bliss | 9.02 | 22.64 |
| MN646 C (primary MN) | ZIP | 9.04 | 21.71 |
| MN658 (primary MN) | HSA | 9.36 | 21.29 |
| MN658 (primary MN) | Loewe | 9.00 | 19.87 |
| MN658 (primary MN) | Bliss | 8.14 | 20.92 |
| MN658 (primary MN) | ZIP | 8.14 | 20.86 |
MN, meningioma.
Synergy reference models: HSA, highest single agent; Loewe, Loewe additivity; Bliss, Bliss independence; ZIP, zero interaction potency.
Discussion
The ERBB family of RTKs (also known as HER family) include four distinct receptors, ERBB1 (EGFR/HER1), ERBB2 (neu/HER2), ERBB3 (HER3), and ERBB4 (HER4). They are often overexpressed, mutated, or amplified in human cancers, thus making them important therapeutic targets (35). Many extracellular ligands can bind ERBB receptors, initiating a cascade of biochemical and signaling events. Specifically, NRG1 and NRG2 can bind ERBB3 and ERBB4 receptors. Elevated NRG1 expression and activated ERBB3 are seen in many types of human cancers supporting the rationale to target the NRG1-ERBB3 axis (36, 37, 38, 39). More importantly, the NRG1 ligand and its target the ERBB family receptors have been implicated in schwannoma tumorigenesis (40, 41, 42) leading to preclinical testing of EGFR/ERBB2 kinase inhibitor lapatinib in a vestibular schwannoma model (27). A subsequent Phase II clinical trial of lapatinib for patients with NF2 and progressive vestibular schwannoma showed tumor regression and improvement of hearing in 4 of 17 patients treated (43). Based on these reports, we focused our study on elevated NRG1 expression, detected in our MN models by transcriptome analyses, to define the role of the NRG1-ERBB3 axis in MN with NF2 loss.
In addition to confirming the secretion of NRG1 and activation of downstream signaling by NRG1, our results here show that a multi-ERBB inhibitor lapatinib, but not EGFR-specific inhibitor erlotinib inhibits the NRG1-stimulated activation of ERBB3 (pERBB3), EPHA2 (pEPHA2), and mTOR (pAkt, pS6K) (Fig. 2), suggesting that NRG1-induced activation is not EGFR-dependent. Lapatinib however was unable to block basal activation of EPHA2 and mTOR signaling in NF2-null cells.
Interestingly, using a systems biology approach, a human monoclonal antibody seribantumab (MM-121), that inhibits NRG1-stimulated ERBB3 signaling with low nanomolar IC50 values compared with lapatinib and other ERBB inhibitors, was identified to be more effective in blocking ligand-induced activation of ERBB3 signaling network, which led to a Phase II clinical trial of human cancers (29, 30, 44). Therefore, we set out to test the ability of MM-121 to block NRG1-induced signaling and effectively inhibit cell proliferation in NF2-null MN cells. Our results show that MM-121 not only blocks NRG1-stimulated but also basal ERBB3 activation and downstream signaling upon NF2 loss; however, it is not effective in inhibiting proliferation of MN cells, suggesting that specific NRG1-ERBB3 signaling may not be a major player for MN growth. Our findings in fact show that NRG1 expression is regulated by mTORC1 as seen by a significant decrease in NRG1 expression after either mTORC1 inhibition by rapamycin or mTOR kinase inhibition (Fig. 3A), supporting the possibility that NRG1 expression and autocrine signaling could be partly because of mTORC1/mTORC2 activation upon NF2 loss (Fig. 7A).
Figure 7.
Proposed model for basal autocrine signaling and adaptive response to mTOR kinase inhibition in NF2-null cells.A, NF2/merlin loss activates mTOR signaling that in turn upregulates NRG1 transcription. Increased NRG1/neuregulin is secreted, which leads to increased activation of ERBB3 receptor signaling in an autocrine loop as well as potential cross-talk to EPH receptor and mTOR activation. B, treatment with dual mTORC1/2 inhibitor downregulates NRG1 expression and ERBB3 receptor activation, while upregulating PDK1-dependent pAkt T308 through a positive feedback loop involving IGF1R/IR signaling. ERBB3, V-ERB-B avian erythroblastic leukemia viral oncogene homolog 3; IGF1R, insulin-like growth factor receptor 1; IR, insulin receptor; mTOR, mechanistic/mammalian target of rapamycin; NF2, neurofibromatosis 2; NRG1, neuregulin-1/heregulin; PDK1, phosphoinositide-dependent kinase-1.
A previous study demonstrated that mTOR kinase inhibition of breast cancer cells for 24 h led to inhibition of pAkt S473 but activation of pAkt T308, along with downstream effectors of Akt, because of activation of several RTKs, including ERBB family members as well as IGF1R and IR. Treating breast cancer cell lines and in vivo xenograft models with a combination of a mTOR kinase inhibitor and a multi-ERBB inhibitor lapatinib abolished Akt signaling and resulted in cell death and tumor regression (31). Our results in MN cells similarly show that mTOR kinase inhibition persistently inhibits pAkt S473 while activating pAkt T308. Conversely, unlike breast cancer cells, mTOR inhibition downregulates NRG1 expression as well as pERBB3 activation, and treatment with MM-121 was unable to block activation of pAkt T308 induced by mTOR kinase inhibition (Fig. 3C), which was consistent with our RNA-seq results where we noted a decrease in ERBB3 and ERBB4 expression. Interestingly, upon mTOR kinase inhibition, we observed a significant increase in the expression of receptor genes IGF1R, INSR, and IGF2R, as well as IRS2, encoding insulin receptor substrate 2. It is interesting to note that insulin receptor substrate (IRS) proteins are recruited to active IGF/insulin receptors where they are phosphorylated, triggering PI3K-PDK1-Akt signaling (45). We confirmed that increased expression and activation of IGF1R/IR in our NF2-deficient cells is responsible for phosphorylation and activation of Akt at T308, a direct target site of PDK1. A recent report on melanoma revealed that long-term mTOR kinase inhibition leads to re-activation of both pAkt S473 and pAkt T308 and that induction of IGF1R/IR-dependent PI3K activation and Akt phosphorylation is mediated by an integrin/focal adhesion kinase/IGFR-dependent process (46). In our MN models, the activation of pAkt T308 by mTOR kinase inhibition, similar to melanoma cells, is mediated by IGF1R/IR; however, unlike melanoma, pAkt S473 remains fully inhibited by mTOR kinase inhibition in MN cells. Taken together, our results support the existence of cell-/context-dependent mechanisms for adaptive signaling observed upon mTOR kinase inhibition.
Treatment of NF2-associated MN and schwannomas with rapamycin analogs was found to be cytostatic (18, 19, 20). Further, mTORC1 inhibition with rapamycin and its analogs is known to relieve the negative feedback inhibition on IRS-1 (47, 48) and Grb10 (49, 50), as well as other negative regulation of mTORC2 independent of IRS-1 and Grb10 (51, 52), thus activating the PI3K-mTORC2-Akt prosurvival pathway. Therefore, mTOR kinase inhibitors were developed to overcome the limitations of rapamycin by effectively inhibiting both mTORC1 and mTORC2 (53). Our previous studies revealed mTORC2-SGK1 activation in addition to mTORC1 activation in NF2 tumors and demonstrated mTOR kinase inhibitors to be more effective than rapamycin (15, 17), which led to ongoing clinical trials with mTOR kinase inhibitor AZD2014 for MN. Our continued studies here show that mTOR activation in NF2-deficient cells leads to increased expression and secretion of NRG1 ligand in an autocrine fashion (Fig. 7A) and that mTOR kinase inhibition disrupts the NRG1-ERBB3 signaling. Nevertheless, inhibiting mTOR kinase induces an adaptive response involving IGFR/IR expression and activation in NF2-null MN cells (Fig. 7B), leading to activation of prosurvival pathways, including Akt T308 and Forkhead box protein O (FOXO) phosphorylation, which needs to be taken into account when considering combination treatment approaches. Importantly, our findings show that combining an IGFR inhibitor with an mTOR kinase inhibitor has synergistic effects, thus setting the stage for further in vivo studies and potential translation to the clinic for more effective treatment of NF2-associated tumors.
Experimental procedures
Cell lines and reagents
Human cell lines included an NF2-null immortalized MN line Ben-Men-1, NF2 AC-CRISPR, and two independent human primary MN for which cell line establishment and growth conditions have been previously described (15, 54). All primary cultures were collected following Massachusetts General Hospital Human Subjects protocols for tumor acquisition after informed consent. In addition, we generated an immortalized human MN line, MN1-LF from an independent NF2-null human primary MN cell line derived from a surgical resection. Immortalization methods and confirmation was carried out on a fee-for-service basis by Alstem, Inc. Briefly, 1 × 105 cells were transduced with lentivirus encoding SV40 large T antigen and a puromycin resistance gene. Cells were infected at a multiplicity of infection of 2. Following selection with 1.5 μg/ml puromycin for 3 days, cells underwent an additional three passages. PCR amplification of SV40 was performed to confirm immortalization followed by expansion and freezing of cells.
Reagents included exogenous heregulin/NRG1 (Sigma); inhibitor drugs lapatinib, erlotinib, INK128/TAK-228, and BMS-754807 (Selleck Chemicals); rapamycin (EMD Millipore); AZD2014 (obtained from AstraZeneca); and MM-121 (generously provided by Merrimack Pharmaceuticals). Drug treatment concentrations and times are described in the figure legends.
Re-expression of NF2 in Ben-Men-1 cells
Re-introduction of NF2 was performed using the full length cDNA coding sequence of NF2 (isoform 1, NCBI accession #NM_000268) cloned into the lentiviral vector CSCW2 followed by packaging as previously reported (16). For re-expression, Ben-Men-1 cells were transduced with either NF2-CSCW2 or V-CSCW2 at a multiplicity of infection of 50 along with 8 μg/ml polybrene by spin-infection as previously described (15). Cells were then expanded and harvested for RNA/cDNA synthesis for quantitative RT-PCR, and protein lysates were generated for immunoblotting.
Transcriptome/RNA-seq and analysis
Baseline transcriptome analyses between NF2(−) versus NF2(+) ACs and Ben-Men-1 were previously described (21). For analysis of transcriptome in posttreated cells, NF2-null AC-CRISPR cells were first treated for 24 h with 200 nM INK128 or DMSO control (final concentration of 0.1% on cells) followed by harvesting on ice by scraping in ice-cold PBS.
For transcriptome analyses, RNA was isolated from all MN-relevant lines using TRIzol reagent (15596026, ThermoFisher Scientific) according to the manufacturer's instructions. Pelleted cells were resuspended TRIzol reagent using microdounce homogenization then extracted with chloroform, followed by isopropanol precipitation of RNA from the aqueous phase and an 80% ethanol wash. RNA pellets were solubilized in 30 to 50 μl of RNase-free water (Ambion, AM9937). RNA quality was assessed using the Agilent Bioanalyzer Tapestation 2200 (Agilent Technologies).
For INK128 and DMSO-treated cell lines, mRNA libraries for RNA-seq were made in triplicate and sextuplicate, respectively, using TruSeq Stranded mRNA Library Preparation Kits (RS-122-2101, RS-122-2102, or RS-122-2103; Illumina). Libraries were analyzed using D1000 tape on Agilent Tapestation 2200 and/or by qPCR using the KAPA Library Quantification Kits (KK4854, KAPABiosystems) on a LightCycler480 (Roche Life Science). These libraries were sequenced on Illumina HiSeq2500 platform, generating paired-end sequencing reads of 75 bp. Quality checking of sequence reads was assessed by fastQC (v.0.10.1) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Libraries concentration was determined by Qubit, and quality assessment and average fragment length was determined by Agilent Tapestation. Equimolar 8- or 9-plex pools were run on a NextSeq 500 using a v2 high output 75 cycle kit.
Sequence reads were aligned to human reference genome Ensembl GRCh37 (v.75), using STAR (v. 2.5.2a) (55) with parameters ‘–outSAMunmapped Within –outFilterMultimapNmax 1 –outFilterMismatchNoverLmax 0.1 –alignIntronMin 21 –alignIntronMax 0 –alignEndsType Local –quantMode GeneCounts –twopassMode Basic’. In this step, STAR also generated gene level counts for all libraries relying on the gene annotation provided for Ensembl GRCh37 (v. 75). Quality checking of alignments was assessed by a custom script utilizing Picard Tools (http://broadinstitute.github.io/picard/), RNASeQC (56), RSeQC (57), and samTools (58). Differentially expressed genes in pair-wise comparisons were identified by edgeR's quasi-likelihood F test (v. 3.18.1) (59), which was run at the R platform (v. 3.4) on genes with greater than 10 counts across replicates per condition in pair-wise comparisons.
Quantitative RT-PCR
RNA extraction and cDNA synthesis were carried out as previously described (15). Quantitative RT-PCR was performed in three biological replicates (in triplicate) on a Roche Lightcycler 480 (software version 1.5.0. SP3) employing iQ-SYBER Green Supermix (Bio-Rad). Human primers included the following: for NRG1, one previously reported primer set (39) and a second primer set, hNRG1-F: 5ʹ-ATGTGTCTTCAGAGTCTCCCAT-3ʹ and hNRG1-R: 5ʹ-TGGACGTACTGTAGAAGCTGG-3ʹ (PrimerBank ID 408411a1, http://pga.mgh.harvard.edu/primerbank/index.html); for HBEGF, one previously reported primer set (60) and a second primer set, hHBEGF-F: 5ʹ-ATCGTGGGGCTTCTCATGTTT-3ʹ and hHBEGF-R: 5ʹ-TTAGTCATGCCCAACTTCACTTT-3ʹ (PrimerBank ID 194018480c1); for APLN, primer set hAPLN-F: 5ʹ- GTCTCCTCCATAGATTGGTCTGC-3ʹ and hAPLN-R: 5ʹ-GGAATCATCCAAACTACAGCCAG-3ʹ (PrimerBank ID 21314668a1); and for TGFA, one previously reported primer set (60) and a second primer set, hTGFA-F: 5ʹ-AGGTCCGAAAACACTGTGAGT-3ʹ and hTGFA-R: 5ʹ-AGCAAGCGGTTCTTCCCTTC-3ʹ (PrimerBank ID 345842399c1). Controls included human 18S: h18S-F, 5ʹ-ACCCGTTGAACCCCATTCGTGA-3ʹ and h18S-R, 5ʹ-GCCTCACTAAACCATCCAATCGG-3ʹ as well as human GAPDH: hGAPDH-F, 5ʹ- CCCCGGTTTCTATAAATTGAGC-3ʹ and hGAPDH-R, 5ʹ- CACCTTCCCCATGGTGTCT-3ʹ. Melt curves showed single peak specificity for each qRT-PCR primer set. Fold changes in gene expression were calculated using the comparative CT (threshold cycle) method, and expression levels were quantitated relative to control (normalized to 1.0). Column scatter plots were generated using Graphpad Prism 8, and data values are represented as mean ± SD. Within each group, Student t test was performed with a value of p < 0.05 considered significant.
Immunoblotting and antibodies
SDS-PAGE and immunoblotting were carried out as previously described (10). Cells were lysed with either radioimmunoprecipitation assay lysis buffer as previously described (10) or 1% Triton X-100 lysis buffer as described (46). Antibodies recognizing ERRB3 Y1197, ERBB3, EPHA2 S897, NDRG1 T346, Akt S473, Akt T308, Akt, p70S6K T389, p70S6K, ribosomal S6 S240/244, S6, IGF1R Y1135/36/IR Y1150/51, IGF1R, FOXO1/3a T24/T32, and FOXO3 were from Cell Signaling. Other antibodies included EPHA2 (Santa Cruz), secreted form of NRG1 (R&D Systems), NDRG1 (Abcam), and GAPDH (EMD Millipore). Anti-NF2/merlin polyclonal antibody has been previously described (61).
NRG1 secretion and conditioned media experiments
Experiments to examine secreted NRG1 were performed following previous reported methods with minor modifications (39). Briefly, AC-CRISPR or Ben-Men-1 cells were seeded at 1 × 106 cells/10 cm plate in full growth medium. The next day, cells were rinsed twice with PBS and once in serum-free DMEM (SF-DMEM), followed by addition of 10 ml SF-DMEM per plate. Cells were then incubated for 48 h. As a control, 10 ml of SF-DMEM was also added to cells and immediately collected (0 h). Next, 0 h- or 48 h-incubated medium was collected, briefly spun to remove debris, and then applied to an Amicon Ultra-15 3K filtration unit (EMD Millipore) followed by centrifugation at 3000g for ∼45 to 50 min. When total retentate volume was ∼700 μl, retentate was then transferred to an Amicon Ultra-0.5 ml 3K filtration unit (EMD Millipore), in two additions, and spun at 14,000 rpm until retentate reached a final volume of ∼200 μl. The resulting 0 h- or 48 h-concentrated media (0 h-CM or 48 h-CM) was collected, protease inhibitor cocktail (P8340, Sigma) was added (1 × final concentration), followed by immunoblotting of 40 μl for each sample.
For conditioned medium experiments, isogenic NF2-expressing and NF2-null AC-CRISPR cell lines were seeded in full growth medium. The next day, cells were rinsed twice with PBS followed by addition of SF-DMEM to each cell line, which served as the source of conditioned medium. Cells were then incubated for 48 h, and then conditioned medium was harvested. For treatment, NF2-expressing AC-CRISPR cells were seeded in full growth medium and incubated until ∼70% confluency. The growth medium was then removed, and cells were rinsed twice in PBS followed by 6 h treatment using respective conditioned medium harvested above. Treated cells were then lysed, followed by immunoblotting.
Cell viability and combination drug screening
Drug screening assays for all cell lines were carried out in a 384-well format using the CellTiter-Glo cell viability kit (Promega) according to the manufacturer's instructions. Relative luminescence units were measured using the EnVision 2103 Multilabel Reader (Perkin Elmer). For single drug DRCs, cells were seeded 24 h before drug treatment at 400 cells/well. Treatments using MM-121, INK128, and BMS-754807 were performed in full growth conditions for 72 h, and treatment dosage is outlined in the figure legends. Percent viability was calculated (relative luminescence units of drug treated versus vehicle treated), and DRCs and drug concentrations inhibiting cell growth by 50% (IC50) were calculated using GraphPad Prism 8 by nonlinear regression (curve fit) analysis with ± standard error of the mean determined (at least three replicates/cell line). For drug synergy testing, INK128 and BMS-754807 were arrayed in a standard 10 × 10 dose matrix (three replicates/cell line). Cells were seeded as above and treated for 72 h, and percent viability was calculated for each treatment dosage point. Synergy scores were generated using the web-based SynergyFinder application (https://synergyfinder.fimm.fi/) (33).
Data availability
All data described in this study are contained within the manuscript with the exception of the entire dataset for transcriptome changes in INK128-treated cells (described in Table 2), which is available upon request.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank Anna D. Ware and Breelyn Karno for technical help.
Author contributions
R. L. B. contributed to design of experiments, acquisition of data, analysis and interpretation of data, and drafting and revising the article; S. E. contributed to analysis and interpretation of data, and drafting and revising the article; L. W. contributed to acquisition of data; J. T. J., S. R. P. and J. F. G. contributed to interpretation of data, and drafting and revising the article; V. R. contributed to conception of study, design of experiments, interpretation of data, drafting and revising the article, and final approval of version to be published.
Funding and additional information
This study was supported by Children's Tumor Foundation-Synodos for NF2, NF Northeast, U.S. Army Medical Research Neurofibromatosis Program W81XWH1810547, National Institutes of Health R01 NS113854, and the S. Sydney DeYoung Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Eric Fearon
Footnotes
This article contains supporting information.
Supporting information
References
- 1.Blakeley J.O., Evans D.G., Adler J., Brackmann D., Chen R., Ferner R.E., Hanemann C.O., Harris G., Huson S.M., Jacob A., Kalamarides M., Karajannis M.A., Korf B.R., Mautner V.F., McClatchey A.I. Consensus recommendations for current treatments and accelerating clinical trials for patients with neurofibromatosis type 2. Am. J. Med. Genet. A. 2012;158A:24–41. doi: 10.1002/ajmg.a.34359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans D.G. Neurofibromatosis 2 [Bilateral acoustic neurofibromatosis, central neurofibromatosis, NF2, neurofibromatosis type II] Genet. Med. 2009;11:599–610. doi: 10.1097/GIM.0b013e3181ac9a27. [DOI] [PubMed] [Google Scholar]
- 3.Ruggieri M., Pratico A.D., Evans D.G. Diagnosis, management, and new therapeutic options in childhood neurofibromatosis type 2 and related forms. Semin. Pediatr. Neurol. 2015;22:240–258. doi: 10.1016/j.spen.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Claus E.B., Bondy M.L., Schildkraut J.M., Wiemels J.L., Wrensch M., Black P.M. Epidemiology of intracranial meningioma. Neurosurgery. 2005;57:1088–1095. doi: 10.1227/01.neu.0000188281.91351.b9. discussion 1088–1095. [DOI] [PubMed] [Google Scholar]
- 5.Kleihues P., Louis D.N., Scheithauer B.W., Rorke L.B., Reifenberger G., Burger P.C., Cavenee W.K. The WHO classification of tumors of the nervous system. J. Neuropathol. Exp. Neurol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. discussion 226–219. [DOI] [PubMed] [Google Scholar]
- 6.Apra C., Peyre M., Kalamarides M. Current treatment options for meningioma. Expert Rev. Neurother. 2018;18:241–249. doi: 10.1080/14737175.2018.1429920. [DOI] [PubMed] [Google Scholar]
- 7.McClatchey A.I., Fehon R.G. Merlin and the ERM proteins - regulators of receptor distribution and signaling at the cell cortex. Trends Cell Biol. 2009;19:198–206. doi: 10.1016/j.tcb.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw R.J., Paez J.G., Curto M., Morse Pruit W., Saotome I., O'Bryan J.P., Gupta V., Ratner N., Der C.J., Jacks T., McClatchey A.I. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev. Cell. 2001;1:63–72. doi: 10.1016/s1534-5807(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 9.Yi C., Wilker E.W., Yaffe M.B., Stemmer-Rachamimov A., Kissil J.L. Validation of the p21-activated kinases as targets for inhibition in neurofibromatosis type 2. Cancer Res. 2008;68:7932–7937. doi: 10.1158/0008-5472.CAN-08-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James M.F., Han S., Polizzano C., Plotkin S.R., Manning B.D., Stemmer-Rachamimov A.O., Gusella J.F., Ramesh V. NF2/Merlin is a novel negative regulator of mTOR complex 1 and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol. Cell Biol. 2009;29:4250–4261. doi: 10.1128/MCB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Lago M.A., Okada T., Murillo M.M., Socci N., Giancotti F.G. Loss of the tumor suppressor NF2/Merlin constitutively activates integrin-dependent mTORC1 signaling. Mol. Cell Biol. 2009;29:4235–4249. doi: 10.1128/MCB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 13.Yin F., Yu J., Zheng Y., Chen Q., Zhang N., Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W., You L., Cooper J., Schiavon G., Pepe-Caprio A., Zhou L., Ishii R., Giovannini M., Hanemann C.O., Long S.B., Erdjument-Bromage H., Zhou P., Tempst P., Giancotti F.G. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beauchamp R.L., James M.F., DeSouza P.A., Wagh V., Zhao W.N., Jordan J.T., Stemmer-Rachamimov A., Plotkin S.R., Gusella J.F., Haggarty S.J., Ramesh V. A high-throughput kinome screen reveals serum/glucocorticoid-regulated kinase 1 as a therapeutic target for NF2-deficient meningiomas. Oncotarget. 2015;6:16981–16997. doi: 10.18632/oncotarget.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James M.F., Lelke J.M., Maccollin M., Plotkin S.R., Stemmer-Rachamimov A.O., Ramesh V., Gusella J.F. Modeling NF2 with human arachnoidal and meningioma cell culture systems: NF2 silencing reflects the benign character of tumor growth. Neurobiol. Dis. 2008;29:278–292. doi: 10.1016/j.nbd.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James M.F., Stivison E., Beauchamp R., Han S., Li H., Wallace M.R., Gusella J.F., Stemmer-Rachamimov A.O., Ramesh V. Regulation of mTOR complex 2 signaling in neurofibromatosis 2-deficient target cell types. Mol. Cancer Res. 2012;10:649–659. doi: 10.1158/1541-7786.MCR-11-0425-T. [DOI] [PubMed] [Google Scholar]
- 18.Giovannini M., Bonne N.X., Vitte J., Chareyre F., Tanaka K., Adams R., Fisher L.M., Valeyrie-Allanore L., Wolkenstein P., Goutagny S., Kalamarides M. mTORC1 inhibition delays growth of neurofibromatosis type 2 schwannoma. Neuro Oncol. 2014;16:493–504. doi: 10.1093/neuonc/not242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goutagny S., Giovannini M., Kalamarides M. A 4-year phase II study of everolimus in NF2 patients with growing vestibular schwannomas. J. Neurooncol. 2017;133:443–445. doi: 10.1007/s11060-017-2447-3. [DOI] [PubMed] [Google Scholar]
- 20.Goutagny S., Raymond E., Esposito-Farese M., Trunet S., Mawrin C., Bernardeschi D., Larroque B., Sterkers O., Giovannini M., Kalamarides M. Phase II study of mTORC1 inhibition by everolimus in neurofibromatosis type 2 patients with growing vestibular schwannomas. J. Neurooncol. 2015;122:313–320. doi: 10.1007/s11060-014-1710-0. [DOI] [PubMed] [Google Scholar]
- 21.Synodos for NF2 Consortium. Allaway R., Angus S.P., Beauchamp R.L., Blakeley J.O., Bott M., Burns S.S., Carlstedt A., Chang L.S., Chen X., Clapp D.W., Desouza P.A., Erdin S., Fernandez-Valle C., Guinney J., Gusella J.F. Traditional and systems biology based drug discovery for the rare tumor syndrome neurofibromatosis type 2. PLoS One. 2018;13 doi: 10.1371/journal.pone.0197350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angus S.P., Oblinger J.L., Stuhlmiller T.J., DeSouza P.A., Beauchamp R.L., Witt L., Chen X., Jordan J.T., Gilbert T.S.K., Stemmer-Rachamimov A., Gusella J.F., Plotkin S.R., Haggarty S.J., Chang L.S., Johnson G.L. EPH receptor signaling as a novel therapeutic target in NF2-deficient meningioma. Neuro Oncol. 2018;20:1185–1196. doi: 10.1093/neuonc/noy046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sagers J.E., Beauchamp R.L., Zhang Y., Vasilijic S., Wu L., DeSouza P., Seist R., Zhou W., Xu L., Ramesh V., Stankovic K.M. Combination therapy with mTOR kinase inhibitor and dasatinib as a novel therapeutic strategy for vestibular schwannoma. Sci. Rep. 2020;10:4211. doi: 10.1038/s41598-020-60156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roskoski R., Jr. Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol. Res. 2019;139:395–411. doi: 10.1016/j.phrs.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Harford-Wright E., Andre-Gregoire G., Jacobs K.A., Treps L., Le Gonidec S., Leclair H.M., Gonzalez-Diest S., Roux Q., Guillonneau F., Loussouarn D., Oliver L., Vallette F.M., Foufelle F., Valet P., Davenport A.P. Pharmacological targeting of apelin impairs glioblastoma growth. Brain. 2017;140:2939–2954. doi: 10.1093/brain/awx253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y., Lv S.Y., Ye W., Zhang L. Apelin/APJ system and cancer. Clin. Chim. Acta. 2016;457:112–116. doi: 10.1016/j.cca.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Ammoun S., Cunliffe C.H., Allen J.C., Chiriboga L., Giancotti F.G., Zagzag D., Hanemann C.O., Karajannis M.A. ErbB/HER receptor activation and preclinical efficacy of lapatinib in vestibular schwannoma. Neuro Oncol. 2010;12:834–843. doi: 10.1093/neuonc/noq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osorio D.S., Hu J., Mitchell C., Allen J.C., Stanek J., Hagiwara M., Karajannis M.A. Effect of lapatinib on meningioma growth in adults with neurofibromatosis type 2. J. Neurooncol. 2018;139:749–755. doi: 10.1007/s11060-018-2922-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoeberl B., Faber A.C., Li D., Liang M.C., Crosby K., Onsum M., Burenkova O., Pace E., Walton Z., Nie L., Fulgham A., Song Y., Nielsen U.B., Engelman J.A., Wong K.K. An ErbB3 antibody, MM-121, is active in cancers with ligand-dependent activation. Cancer Res. 2010;70:2485–2494. doi: 10.1158/0008-5472.CAN-09-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoeberl B., Pace E.A., Fitzgerald J.B., Harms B.D., Xu L., Nie L., Linggi B., Kalra A., Paragas V., Bukhalid R., Grantcharova V., Kohli N., West K.A., Leszczyniecka M., Feldhaus M.J. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci. Signal. 2009;2:ra31. doi: 10.1126/scisignal.2000352. [DOI] [PubMed] [Google Scholar]
- 31.Rodrik-Outmezguine V.S., Chandarlapaty S., Pagano N.C., Poulikakos P.I., Scaltriti M., Moskatel E., Baselga J., Guichard S., Rosen N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., Abraham R.T. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ianevski A., Giri A.K., Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48:W488–W493. doi: 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malyutina A., Majumder M.M., Wang W., Pessia A., Heckman C.A., Tang J. Drug combination sensitivity scoring facilitates the discovery of synergistic and efficacious drug combinations in cancer. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Appert-Collin A., Hubert P., Cremel G., Bennasroune A. Role of ErbB receptors in cancer cell migration and invasion. Front. Pharmacol. 2015;6:283. doi: 10.3389/fphar.2015.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capparelli C., Purwin T.J., Heilman S.A., Chervoneva I., McCue P.A., Berger A.C., Davies M.A., Gershenwald J.E., Krepler C., Aplin A.E. ErbB3 targeting enhances the effects of MEK inhibitor in wild-type BRAF/NRAS melanoma. Cancer Res. 2018;78:5680–5693. doi: 10.1158/0008-5472.CAN-18-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capparelli C., Rosenbaum S., Berger A.C., Aplin A.E. Fibroblast-derived neuregulin 1 promotes compensatory ErbB3 receptor signaling in mutant BRAF melanoma. J. Biol. Chem. 2015;290:24267–24277. doi: 10.1074/jbc.M115.657270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capparelli C., Rosenbaum S., Berman-Booty L.D., Salhi A., Gaborit N., Zhan T., Chervoneva I., Roszik J., Woodman S.E., Davies M.A., Setiady Y.Y., Osman I., Yarden Y., Aplin A.E. ErbB3-ErbB2 complexes as a therapeutic target in a subset of wild-type BRAF/NRAS cutaneous melanomas. Cancer Res. 2015;75:3554–3567. doi: 10.1158/0008-5472.CAN-14-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson T.R., Lee D.Y., Berry L., Shames D.S., Settleman J. Neuregulin-1-mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer Cell. 2011;20:158–172. doi: 10.1016/j.ccr.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Doherty J.K., Ongkeko W., Crawley B., Andalibi A., Ryan A.F. ErbB and Nrg: potential molecular targets for vestibular schwannoma pharmacotherapy. Otol. Neurotol. 2008;29:50–57. doi: 10.1097/mao.0b013e31815d4429. [DOI] [PubMed] [Google Scholar]
- 41.Lallemand D., Manent J., Couvelard A., Watilliaux A., Siena M., Chareyre F., Lampin A., Niwa-Kawakita M., Kalamarides M., Giovannini M. Merlin regulates transmembrane receptor accumulation and signaling at the plasma membrane in primary mouse Schwann cells and in human schwannomas. Oncogene. 2009;28:854–865. doi: 10.1038/onc.2008.427. [DOI] [PubMed] [Google Scholar]
- 42.Stonecypher M.S., Chaudhury A.R., Byer S.J., Carroll S.L. Neuregulin growth factors and their ErbB receptors form a potential signaling network for schwannoma tumorigenesis. J. Neuropathol. Exp. Neurol. 2006;65:162–175. doi: 10.1097/01.jnen.0000199575.93794.2f. [DOI] [PubMed] [Google Scholar]
- 43.Karajannis M.A., Legault G., Hagiwara M., Ballas M.S., Brown K., Nusbaum A.O., Hochman T., Goldberg J.D., Koch K.M., Golfinos J.G., Roland J.T., Allen J.C. Phase II trial of lapatinib in adult and pediatric patients with neurofibromatosis type 2 and progressive vestibular schwannomas. Neuro Oncol. 2012;14:1163–1170. doi: 10.1093/neuonc/nos146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoeberl B., Kudla A., Masson K., Kalra A., Curley M., Finn G., Pace E., Harms B., Kim J., Kearns J., Fulgham A., Burenkova O., Grantcharova V., Yarar D., Paragas V. Systems biology driving drug development: from design to the clinical testing of the anti-ErbB3 antibody seribantumab (MM-121) NPJ Syst. Biol. Appl. 2017;3:16034. doi: 10.1038/npjsba.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boucher J., Kleinridders A., Kahn C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon S.O., Shin S., Karreth F.A., Buel G.R., Jedrychowski M.P., Plas D.R., Dedhar S., Gygi S.P., Roux P.P., Dephoure N., Blenis J. Focal adhesion- and IGF1R-dependent survival and migratory pathways mediate tumor resistance to mTORC1/2 inhibition. Mol. Cell. 2017;67:512–527. doi: 10.1016/j.molcel.2017.06.033. e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrington L.S., Findlay G.M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N.R., Cheng S., Shepherd P.R., Gout I., Downes C.P., Lamb R.F. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah O.J., Wang Z., Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 49.Hsu P.P., Kang S.A., Rameseder J., Zhang Y., Ottina K.A., Lim D., Peterson T.R., Choi Y., Gray N.S., Yaffe M.B., Marto J.A., Sabatini D.M. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Y., Yoon S.O., Poulogiannis G., Yang Q., Ma X.M., Villen J., Kubica N., Hoffman G.R., Cantley L.C., Gygi S.P., Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu P., Gan W., Inuzuka H., Lazorchak A.S., Gao D., Arojo O., Liu D., Wan L., Zhai B., Yu Y., Yuan M., Kim B.M., Shaik S., Menon S., Gygi S.P. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat. Cell Biol. 2013;15:1340–1350. doi: 10.1038/ncb2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo Y., Liu L., Wu Y., Singh K., Su B., Zhang N., Liu X., Shen Y., Huang S. Rapamycin inhibits mSin1 phosphorylation independently of mTORC1 and mTORC2. Oncotarget. 2015;6:4286–4298. doi: 10.18632/oncotarget.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thoreen C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y., Reichling L.J., Sim T., Sabatini D.M., Gray N.S. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puttmann S., Senner V., Braune S., Hillmann B., Exeler R., Rickert C.H., Paulus W. Establishment of a benign meningioma cell line by hTERT-mediated immortalization. Lab. Invest. 2005;85:1163–1171. doi: 10.1038/labinvest.3700307. [DOI] [PubMed] [Google Scholar]
- 55.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeLuca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L., Wang S., Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 58.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu K., Gualano R.C., Hibbs M.L., Anderson G.P., Bozinovski S. Epidermal growth factor receptor signaling to Erk1/2 and STATs control the intensity of the epithelial inflammatory responses to rhinovirus infection. J. Biol. Chem. 2008;283:9977–9985. doi: 10.1074/jbc.M710257200. [DOI] [PubMed] [Google Scholar]
- 61.Wiederhold T., Lee M.F., James M., Neujahr R., Smith N., Murthy A., Hartwig J., Gusella J.F., Ramesh V. Magicin, a novel cytoskeletal protein associates with the NF2 tumor suppressor merlin and Grb2. Oncogene. 2004;23:8815–8825. doi: 10.1038/sj.onc.1208110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in this study are contained within the manuscript with the exception of the entire dataset for transcriptome changes in INK128-treated cells (described in Table 2), which is available upon request.