Abstract
Spectral-domain optical coherence tomography (SD-OCT) is useful for visualizing retinal and ocular structures in vivo. In research, SD-OCT is a valuable tool to evaluate and characterize changes in a variety of retinal and ocular disease and injury models. In light induced retinal degeneration models, SD-OCT can be used to track thinning of the photoreceptor layer over time. In glaucoma models, SD-OCT can be used to monitor decreased retinal nerve fiber layer and total retinal thickness and to observe optic nerve cupping after inducing ocular hypertension. In diabetic rodents, SD-OCT has helped researchers observe decreased total retinal thickness as well as decreased thickness of specific retinal layers, particularly the retinal nerve fiber layer with disease progression. In mouse models of myopia, SD-OCT can be used to evaluate axial parameters, such as axial length changes. Advantages of SD-OCT include in vivo imaging of ocular structures, the ability to quantitatively track changes in ocular dimensions over time, and its rapid scanning speed and high resolution. Here, we detail the methods of SD-OCT and show examples of its use in our laboratory in models of retinal degeneration, glaucoma, diabetic retinopathy, and myopia. Methods include anesthesia, SD-OCT imaging, and processing of the images for thickness measurements.
Keywords: Optical coherence tomography, retina, retinal degeneration, glaucoma, diabetic retinopathy, myopia, rodent
SUMMARY:
Here, we describe the use of spectral-domain optical coherence tomography (SD-OCT) to visualize retinal and ocular structures in vivo in models of retinal degeneration, glaucoma, diabetic retinopathy, and myopia.
INTRODUCTION:
Spectral-domain optical coherence tomography (SD-OCT) is a precise, high-resolution imaging modality that allows clinicians and researchers to examine ocular structures non-invasively. This imaging technique is based on interferometry to capture three-dimensional retinal images in vivo on a micrometer scale1,2. It has become one of the most frequently used imaging modalities in vision research and in the clinic due to the easy detection and accuracy of pathological features such as structural defects and/or thinning of retinal layers and subretinal fluid3. In research using animal models of vision-related disorders, SD-OCT has provided essential noninvasive analyses of relationships between structure and function and their histopathological origins4. Due to its resolution (up to 2–3 microns, depending on the depth into the eye5), SD-OCT has the capability to detect even small changes in retinal layer thickness. This type of analysis can provide essential information for disease progression and assess the efficacy of neuroprotective methods and treatments for vision-related disorders.
SD-OCT is a non-invasive alternative to examining structure histologically, and the two have been shown to be correlated6. While SD-OCT does not reach cellular resolution, it does allow for longitudinal studies in animals. This is advantageous because disease progression can be tracked in individual animals over time as opposed to having to euthanize animals at specific time points. As imaging techniques continue to improve, SD-OCT technology will also progress, providing enhanced image quality as well as the capability to assess biological processes such as retinal blood vessel function in fine detail. Even since its advent in 1991, SD-OCT technology has seen huge advances in resolution, speed, and sensitivity7.
The present study utilizes an SD-OCT system (see table of materials) to quantify changes in retinal layers in rodent models of retinal degeneration, glaucoma, and diabetic retinopathy. The SD-OCT system used here is a Fourier-domain OCT-system that utilizes low-power, near-infrared light to acquire, process, and store depth-resolved images in real time. The SD-OCT system has extended depth-imaging capability in the 800-nanometers wavelength band, providing 8-millimeter depth and 4 micrometer resolution. In Fourier domain detection, the interference signal between scattered light from the tissue and a reference path is Fourier transformed to construct axial scans and or axial depth profiles of scattered intensity8. For our studies, the OCT beam is scanned over the desired retinal structure while serially acquiring axial scans. Typically, a scan pattern acquires the two-dimensional grid (B-Scans) as a collection of linear one-dimensional scan lines (A-Scans), which correspond to 2D cross-sectional images using a raster scan pattern. For studies focused on myopia in mice, this system is also used to measure dimensions of ocular structures (e.g. cornea thickness, lens thickness, vitreous chamber depth, and axial length).
Materials
| Name of Material/Equipment | Company | Catalog Number | Comments/Description |
|---|---|---|---|
| 1% tropicamide | Sandoz | Sandoz #6131403550; NDC-24208-585-59 | |
| 0.5% tetracaine | Alcon | NDC 0065-0741-12 | |
| Celluvisc gel | REFRESH® CELLUVISC® | #4554; NDC-0023-4554-30 | |
| saline | ADDIPAK | 200-39 | |
| MATLAB | Mathworks | mathematical modeling program | |
| System Envisu R4300 VHR 120V | Leica Bioptigen | 90-R4300-V1-120 | SD-OCT system |
| AIM-RAS G3 120V | Leica Bioptigen | 90-AIMRAS-G3-120 | Specialized platform to hold the OCT Scanner Head for mice |
| Mouse/Rat Kit | Leica Bioptigen | 90-KIT-M/R | Mouse/rat rodent alignment system |
| G3 Mouse Lens | Leica Bioptigen | 90-BORE-G3-M | |
| G3 Rat Lens | Leica Bioptigen | 90-BORE-G3-R | |
| G3 18mm Telecentric Lens | Leica Bioptigen | 90-BORE-G3-18 | |
| InVivoVue™ software | Leica Bioptigen | Specialized software that pairs with the Leica Bioptigen SD-OCT system | |
| heating pad | Fabrication | 11-1130 |
The current system allows users to design their own protocols, creating scans that can be tailored and selected based on the ocular structures of interest. The principal scans featured in these user defined protocols makes this imaging technique user-friendly. For image analyses, we have developed customized programming in a mathematic modeling program (see table of materials) SD-OCT is a powerful tool to non-invasively identify and quantify pathomorphological changes in ocular structures and monitor vision-related disease progression.
PROTOCOL:
All procedures described were approved by the Atlanta Veterans Affairs Institutional Animal Care and Use Committee and conformed to the National Institutes of Health guide for the care and use of laboratory animals (NIH Publications, 8th edition, updated 2011).
The SD-OCT system used to develop the protocol below is described in the table of materials. While some of the procedures are specific to this particular system, the overall approach can be adapted for other OCT devices and animal models. Further, in our lab, these protocols are commonly used in mice and rats; however, the overall approach can be adopted to different animal models and SD-OCT devices provided an individual has the correct lens and capabilities on their device.
1. Set up the optical coherence tomography equipment
-
1.1
Open the SD-OCT software (see table of materials).
-
1.2
Define who is taking the OCT, the study, and the treatment arm (if relevant). Name these categories in a way that will help researchers search for the desired scans later during data analysis.
-
1.2.1
In the “Patient/Exam” tab, click “Test Examiner”. Select the name of the examiner. (Use the “Setup Examiners & Physicians” button to add new examiners.)
-
1.2.2
Click “Study Name” to define the study. (Click the “Study” tab to add a new study or modify treatments in an existing study.) Click to the right of “Select Treatment Arm” to select a treatment arm.
-
1.2.1
-
1.3
Click the “Add Patient” button, which is used to add a new timepoint for an entire group. When the window appears, enter ID number, First name, and Last name. Select Male or Female. Enter the Date of Birth.
-
1.4
Click the “Add Exam” button to add the individual rats. To identify the rats, click on an exam. Click “Edit Exam”. Enter the ID number into the “Enter Notes” box. Click the “Save Changes” button.
-
1.5
Attach the proper lens to the device (Figure 1B), select the corresponding Configuration in the software, and dial in the associated reference arm position.
NOTE: The SD-OCT system described has customized lenses, preset scan patterns, and reference arm settings specific to the animal species and region of eye being imaged (retina or cornea, mouse or rat). Some of these details are specific to the SD-OCT system described (see table of materials). For example, not all devices offer manual adjustment of reference arm pathlength.
-
1.6
In the “Patient/Exam” tab, double click the highlighted exam to proceed to the “Imaging” tab and begin imaging or simply click the “Imaging” tab. If there is a default scan, right click to delete it.
-
1.7
Load a pre-set Scan Protocol by clicking the “Select a protocol from the list” button. (Or add individual scans.)
-
1.8
For rat models of glaucoma and diabetic retinopathy and mouse models of retinal degeneration, choose a pre-set that consists of four images: 2 OD and 2 OS scans. For mouse myopia, choose a pre-set that consists of 8 images: 4 OD and 4 OS scans.
NOTE: Pre-set imaging will be explained in more detail in Section 3. (This is something each laboratory makes for themselves or with the manufacturer during on-site installation).
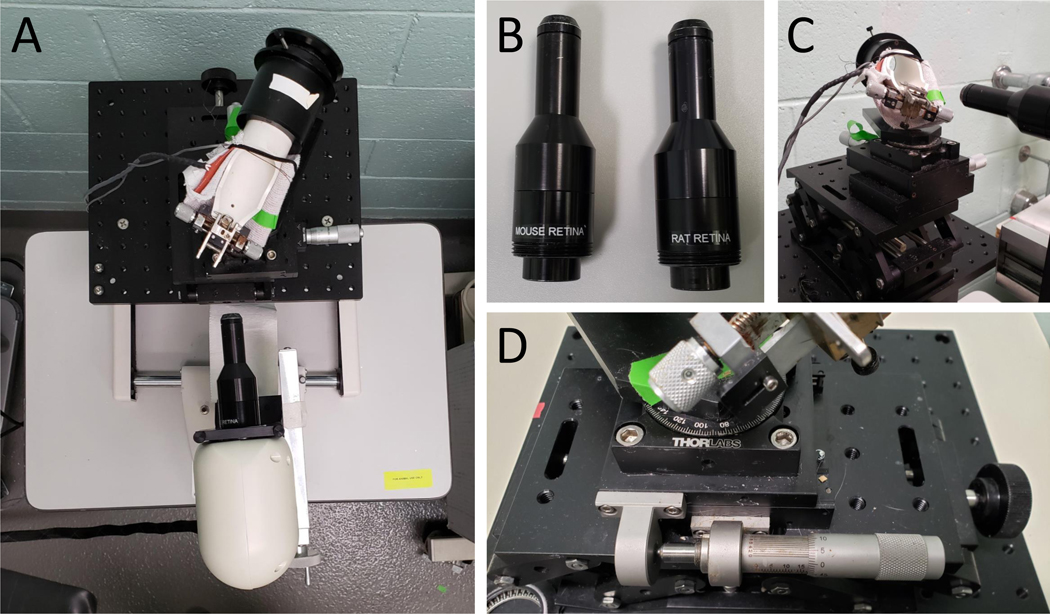
Figure 1: Set up of SD-OCT equipment.
(A) Picture of rodent alignment system and OCT scan head. (B) Picture of rat and mouse OCT lenses. (C) Picture of mouse rodent alignment system illustrating its ability to move in 3-dimensional space. (D) Close up of the rodent alignment system, specifically the knobs that control its movement.
2. Anesthetize the animal
-
2.1
Administer anesthetic.
-
2.1.1
Anesthetize rats with ketamine (60 mg/kg) and xylazine (7.5 mg/kg) via intraperitoneal injection.
-
2.1.2
Anesthetize mice with ketamine (80 mg/kg) and xylazine (16 mg/kg) via intraperitoneal injection.
-
2.1.3
Wait until animals are fully anesthetized and do not respond to toe pinch.
-
2.1.1
-
2.2
Administer pupil dilation drops (1% tropicamide). Wait for pupils to dilate before imaging.
NOTE: Dilation of pupils increases the field of view but is not a requirement. Local (corneal) anesthetic drops (0.5% tetracaine) to numb the eye should also be used if anything will be touching the eye (for example, if applying contact lenses or using a guide). A guide is a device that is placed over the scan head and helps beginners to line up the eye and the scan head.
-
2.3
After anesthetizing the rodent, place the rodent in a rodent alignment system that can rotate the animal in 3-dimensional space (Figure 1 A, C, & D).
NOTE: Currently, we use rodent alignment systems for mice and rats designed and sold with the SD-OCT device.
-
2.4
Apply liquid (e.g. saline or artificial tears) to keep the eyes lubricated. Ensure that the eye does not dry out during imaging so that the optical properties of the eye are maintained between scans (when the cornea is wet, the retina can be seen clearly).
-
2.4.1
Make sure to maintain moisture in the opposite eye as you are scanning the first eye so it does not dry out.
-
2.4.1
-
2.5
Use a delicate task wipe to wick away excess saline just before imaging, as too much or too little lubricant on the eye will affect the image quality.
NOTE: Lubricant eye gel can also be used. A contact lens can also be applied to ensure adequate moisture on the eye throughout the test. In our experience a contact lens did not provide a marked improvement in image quality, but contact lenses do help reduce the risk of the cornea drying during the imaging session.
3. Rodent OCT Imaging
-
3.1
Begin with one eye (OS or OD) and image the contralateral eye after.
-
3.1.1
Position the animal using the two rotational motions of the rodent alignment system, such that the gaze is horizontal and looking down the axis of the OCT Lens (Figure 1D).
-
3.1.2
Use the OCT in Free Run mode to orient the retina for data collection. Use the “Aim” mode (by clicking the “Aim” button) initially for a continuous display of both horizontal and vertical B-scans in real time.
-
3.1.3
Move the scan head closer to the eye until the retina is visible (as mouse and rat retina lenses are fixed-focus, moving the lens toward the eye focuses deeper into the retina). Then use the rodent alignment system to adjust the animal position up/down and swivel/twist to position the optic nerve head in the center, make the horizontal scan horizontal, and the vertical scan vertical (Figure 1A).
-
3.1.4
Adjust the working distance such that the retinal image is flat and not curved.
-
3.1.5
Adjust the reference arm position to keep the image near the top of the display window. Be careful not to push in too far or the eye image will flip back on itself.
-
3.1.1
3.2. Retinal Imaging
-
3.2.1
For glaucoma, retinal degeneration, and diabetic retinopathy models: Define a volume scan that consists of 1000 × 100 × 1 (A scans × B scans × repeated B scans) for averaging. In rats, take a volume scan that is 3 × 3 mm. In mice, take a 1.5 × 1.5 mm volume scan.
-
3.2.2
Center the optic nerve in the horizontal and the vertical access so that the volume scan is in the center. Take time to make sure the optic nerve head is at the center of the scan and straight along the nasal-temporal and superior-inferior axes (Figure 2). Scan and re-center to make sure it is exactly in the center, if needed. Repeat this scan as necessary until the optic nerve head is centered and aligned along both axes. Click the “Snapshot” button to take a photo.
NOTE: Some SD-OCT devices have the option of optically manipulating the curvature of the eye (e.g. the image is flattened) by adjusting the distance of the eye from the light source with the reference arm. We recommend flattening and centering the images when taking direct thickness measurements through the retinal layers to improve accuracy along the anterior-posterior direction.
-
3.2.3
Click the Save button to save the image.
-
3.2.4
Take a radial scan centered at the optic nerve head that is 1000 × 4 × 20 (A-scan × B-scan × repeated B-scans. Use repeated B scans to enhance image clarity of the eye or retina, which will help to interpret regions of the eye or layers of the retina during data analysis.
NOTE: Again, in rats this radial scan is 3 mm, while in mice the radial scan is 1.5 mm.
-
3.2.5
Save the image.
-
3.2.6
Repeat steps 3.1 through 3.2.5 in the contralateral eye.
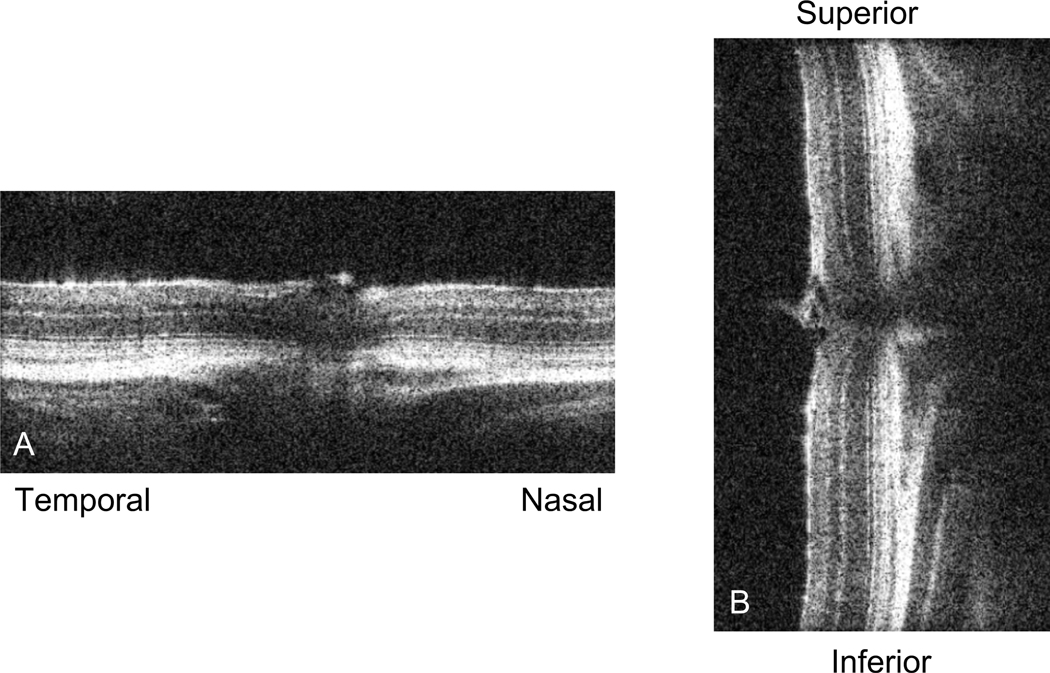
Figure 2: SD-OCT sample scan.
Picture of a live scan of the mouse retina just prior to taking a volume or radial scan. (A) depicts the nasal-temporal alignment, while (B) shows the superior-inferior alignment. Once the retinas in these two images are straight in their respective vertical or horizontal planes and the optic nerve is centered in both images, we proceed to acquire the SD-OCT image.
3.3. Axial Length Measurements
-
3.3.1
For projects that involve imaging the whole eye, such as mouse myopia, take three scans of the entire eye and one retina scan for each eye. Choose a pre-set that consists of a radial scan that is 500 × 20 × 1 and encompasses the full diameter of the eye.
NOTE: This setting provides an image of the entire length of the mouse eye from the cornea to the choroid.
-
3.3.2
Center the middle of the eye and retina in the field of view. Take three radial scans (whole eye scans): a linear B scan that is 1000 × 5 × 2 and two additional linear B scans of 1000 × 5 × 2 at the same location. Save the images.
-
3.3.3
Afterward, if desired, zoom in and take a volume or rectangular scan (retina scan) similar to the description in 3.2 that consists of 1000 × 20 A scans × B scans. Save the volume scan.
-
3.3.4
Repeat steps 3.3 through 3.3.3 in the contralateral eye.
Note: Axial length measurements are only possible on small eyes (mouse or smaller) since the imaging window of current systems is not large enough to capture a larger eye.
4. Post-imaging steps
-
4.1
Store saved data on a cloud, which is good practice for data management and allows for easy access for later analysis. Perform data analysis with custom software developed in a mathematical modeling program (see table of materials).
-
4.2
Remove the rodent from the rodent alignment system and give an intraperitoneal injection of atipamezole (1 mg/kg for rats and mice) to reverse the effects of the xylazine, so that the rodent will wake up more quickly and will be less likely to develop corneal ulcers.
-
4.3
Allow rodents to recover on a heating pad on low heat. Give additional saline drops as needed. Return rodents to their home cage when they have regained full ambulation.
-
4.4
Close program and turn off the OCT.
5. Post-processing of OCT images
-
5.1
Process the images using custom software developed in a mathematical modeling program to suit specific OCT needs, e.g. measure the thickness of areas of interest by manually marking the images.
-
5.2
Depending on the purpose of the image (mouse retina, rat retina, or myopia/axial length), use one of three different programs:
-
5.2.1
For processing the retina, select the OCT scans to load. First, define the center of the optic nerve head with a simple click.
-
5.2.2
Watch as the program generate vertical lines defining distances on either side of the optic nerve head. Note that in the rat retina, these lines are at 0.5 mm and 1.2 mm away from the center of the optic nerve head, for a total of 4 vertical lines representing the nasal-temporal and inferior-superior axes of the eye depending on the radial B scan currently analyzed.
NOTE: In the mouse retina, these vertical lines are at 0.25 mm and 0.5 mm from the optic nerve head center.
-
5.2.3
Delineate the following layers along each line:
The retinal nerve fiber layer (RNFL), inner plexiform layer (IPL), inner nuclear layer (INL), outer plexiform layer (OPL), outer nuclear layer (ONL), external limiting membrane (ELM), inner segments/outer segments (IS/OS), retinal pigment epithelium (RPE), and total retinal thickness.
NOTE: The radial scan does not typically have nasal/temporal and superior/inferior labels when it is opened. Scans may be created such that they have an n/t and s/I orientation, and those scans in particular are analyzed later.
-
5.2.4
After an image has been delineated and the program closed, export these measurements into a spreadsheet software for data analysis.
-
5.2.1
-
5.3
Use these length and thickness values from step 5 to make comparisons between groups, for example, determining if there are regional differences (n/t/s/i), or longitudinal changes.
-
5.4
For retinal measurements, first determine if there are any differences in the nasal-temporal and inferior-superior axis at the 0.5 mm and 1.2 mm distances.
NOTE: If differences in quadrants are not observed, the 0.5 mm and 1.2 mm measurements may be averaged together. This is a similar approach for the mouse retinal scans only at 0.25 mm and 0.5 mm.
-
5.53
For myopia studies, use this program to assess the ocular parameters along the optical axis of the eye. Open the mathematical modeling program. First, select an image to load.
-
5.3.1
After loading the image, manually mark each scan (radial and B scans). Mark the anterior and posterior edges of the cornea, lens, vitreous chamber, and retina, so that the program will calculate corneal thickness, lens thickness, anterior and vitreous chamber depth, total retinal thickness, total axial length.
-
3.2
After marking, exit the program which prompts a save menu. Save the delineated values in a spreadsheet software and average the three separate scans together.
-
5.3.1
REPRESENTATIVE RESULTS:
SD-OCT is considered successful if high quality images are obtained such that ocular dimensions can be reliably measured. Here, a variety of uses of SD-OCT are illustrated using models of retinal degeneration, glaucoma, diabetic retinopathy, and myopia.
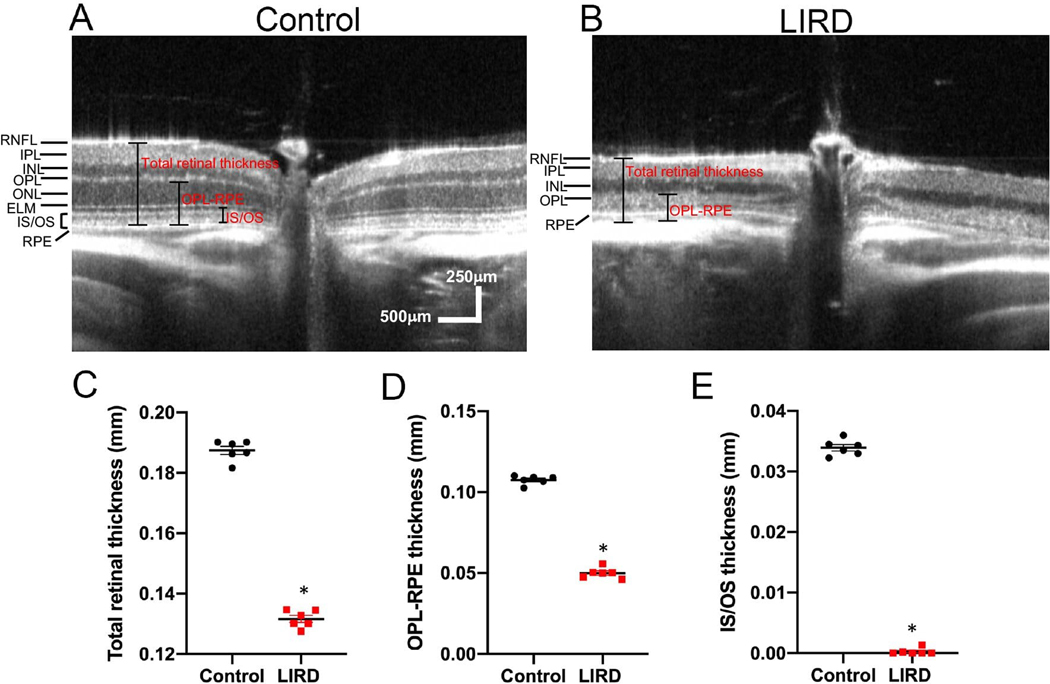
In a light-induced retinal degeneration (LIRD) model, exposure to bright light (10,000 lux) induces degeneration of photoreceptor cells in the retina9. Representative SD-OCT images reveal a thinner outer nuclear layer, which contains the photoreceptor cell bodies, in retinas from LIRD BALB/c mice compared with undamaged (control) mice (Figure 3A&B). After quantifying the retinal layer thickness, a significant difference between undamaged and LIRD mice was observed for total retinal thickness (Figure 3C), outer nuclear layer thickness (Figure 3D), and IS/OS thickness (Figure 3E).
Figure 3: Using SD-OCT to track thinning of the photoreceptor layer over time in a mouse model of retinal degeneration.
(A) Representative SD-OCT scan of an undamaged (control) retina from a BALB/c mouse. (B) Representative SD-OCT scan of a retina from a light-induced retinal degeneration (LIRD) BALB/c mouse. (C-E) Quantification of total retinal thickness (C), outer nuclear layer (ONL) thickness (D), and inner segment/outer segment (IS/OS) thickness (E) in undamaged and LIRD Balb/c mice. Mean ± SEM.
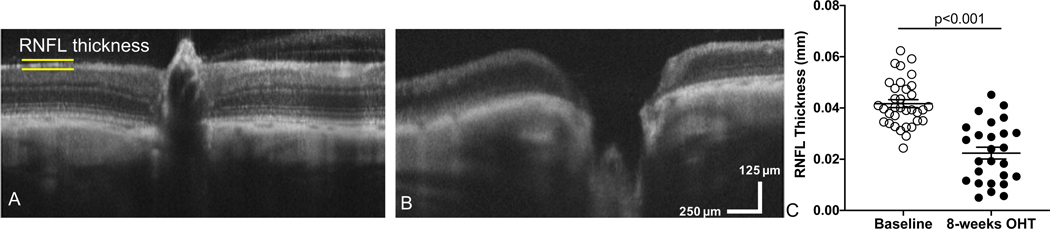
To experimentally model glaucomatous damage, we used a model of ocular hypertension (OHT)10. In brief, Brown Norway rats (n=35) received an injection of hypertonic saline into the limbus vein of one eye while the contralateral eye served as an internal control11. For glaucoma studies, we quantified retinal nerve fiber layer (RNFL) thickness. After 8 weeks of OHT, we observed distinct remodeling at the optic nerve head, including optic nerve cupping (Figure 4A&B). We then quantified RNFL thickness and found RNFL thinning after 8 weeks of OHT compared to baseline measurements (Figure 4C).
Figure 4: Using SD-OCT we measured a decrease in retinal nerve fiber layer thickness and observed optic nerve cupping after inducing ocular hypertension in a rat model of glaucoma.
(A) Representative SD-OCT scan of a retina and optic nerve head from a rat eye taken prior to inducing ocular hypertension (Baseline: OHT). (B) SD-OCT scan of the same rat retina after 8-weeks of OHT (experimental model of glaucoma). (C) Quantification of retinal nerve fiber layer (RNFL) thickness at baseline compared to OHT eyes. Mean ± SEM. This data has been modified from Feola et al.11
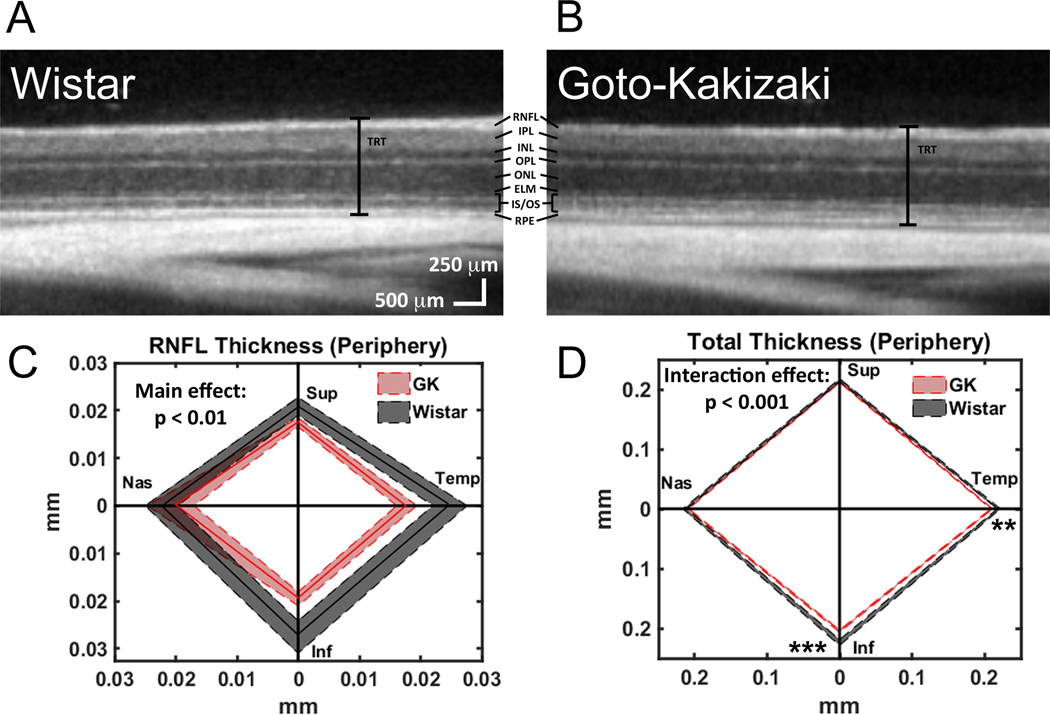
To model diabetic retinopathy, Goto-Kakizaki rats, a non-obese, polygenic model of diabetes that develops hyperglycemia as early as 2–3 weeks of age, were used12,13. Retinas from Goto-Kakizaki rats and Wistar rats (non-diabetic controls) were imaged using SD-OCT (Figure 5A&B). At 6 weeks of age, RNFL and total retinal thickness were reduced in Goto-Kakizaki rats compared with Wistar rats in the central retina (data not shown) and the peripheral retina (Figure 5C&D). The greatest differences were observed in the inferior and temporal quadrants of the retina (Figure 5C&D).
Figure 5: Using SD-OCT to observe decreased total retinal thickness as well as decreased thickness of specific retinal layers in a rat model of diabetes.
(A) Representative SD-OCT scan of a retina from a Wistar (Wild-type control) rat. (B) Representative SD-OCT scan of a retina from a Goto-Kakizaki (diabetic) rat. Retinal layers: retinal nerve fiber layer (RNFL), inner plexiform layer (IPL), inner nuclear layer (INL), outer plexiform layer (OPL), outer nuclear layer (ONL), external limiting membrane (ELM), inner segments/outer segments (IS/OS), retinal pigment epithelium (RPE), and total retinal thickness (TRT). (C-D) Quantification of RNFL (C) and total retinal thickness (D) in Wistar and Goto-Kakizaki retinas where the central line is the mean and the shaded area is the SEM for all four quadrants (Sup, Superior; Temp, Temporal; Inf, Inferior; Nas, Nasal) of the peripheral retina (1.2 mm from the optic nerve head). ** p < 0.01, *** p < 0.001. This figure has been modified from Allen et al.13
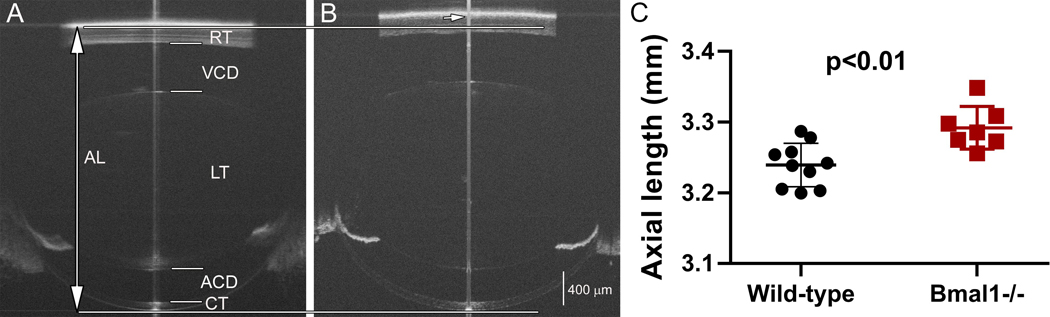
To evaluate mouse models for myopia, axial length was measured in Bmal1−/− mice. Bmal1 is a clock gene of interest because circadian rhythms may play a role in myopia development14,15. The axial length of the Bmal1−/− mouse eye (Figure 6B) is visibly longer than the wild-type eye (Figure 6A) in the OCT images. Quantification of the axial length confirms that Bmal1−/− mice have significantly longer axial lengths at 84 days of age (Figure 6C), showing that the lack of the clock gene contributes to myopia development.
Figure 6: Using SD-OCT to evaluate axial length in a mouse model of myopia.
Whole eye SD-OCT images of wild-type (A) and Bmal1−/− (B) mouse eyes at 84 days of age. The eyes of Bmal1−/− mice have significantly longer axial length than the wild-type eyes (C). AL: axial length; RT: retinal thickness; VCD: vitreous chamber depth; LT: lens thickness; ACD: anterior chamber depth; CT: corneal thickness. The long vertical line indicates axial length boundaries (top and bottom indicated by horizontal line) for the wild-type eye. Short arrow indicates the posterior axial length marking for the Bmal1−/− eye. Mean ± SEM. (Note: The central line down the middle of each image (A&B) is a vertical saturation artifact. It is typically used as a guide to center the eye, but if the scan is well aligned, it can be made to disappear.)
This protocol generated images of ocular structures in models of retinal degeneration, glaucoma, diabetic retinopathy, and myopia. Images were of sufficient quality such that ocular dimensions, including outer nuclear layer, retinal nerve fiber layer, total retinal thickness, and axial length, could be quantified. The results showed that significant differences in the dimensions of ocular structures could be observed in vivo using SD-OCT.
DISCUSSION:
High resolution imaging of ocular structures in vivo allows for the assessment of retinal and ocular changes over time. In this protocol, SD-OCT was demonstrated to capture differences in ocular structures in vivo in models of retinal degeneration, glaucoma, diabetic retinopathy, and myopia.
Critical steps, modifications, and troubleshooting
The most critical aspect when performing SD-OCT is obtaining a clear image of the retina or other ocular structure of interest. It is important to take time to make sure the retina is perfectly centered and has excellent clarity. Heavy breathing by the rodent can result in noisy images (the retina can actually be seen to wiggle on screen). This sometimes happens if an animal is not fully unconscious after anesthetic administration. In order to work around this issue, multiple B scans can be averaged to help visualize where the boundaries of the retinal layers are, and then the best single B scan image can be analyzed.
Another common mistake is that the eye is too dry or too wet. This can be checked easily by applying an additional drop of saline, wicking it away with a chem wipe, and assessing whether the image improved in clarity. A consideration to take into account when marking retinal layer thicknesses on SD-OCT images is how to mark the RNFL. While it is possible to differentiate between the RNFL and GCL on some rodent OCTs, often these two layers are indistinguishable. For consistency, we mark the entire RNFL region (RNFL + GCL, when visible) as the RNFL. Some studies report the RNFL and GCL as separate layers or combine the GCL and inner plexiform layer16–18, though this research was typically performed in humans, who have much larger eyes than rodents. Reporting of RNFL thickness is more typical in rodent studies11,13,19,20. Another important issue is that very slight changes in marking can cause a very large change, especially in myopia because of the small size of the structures being measured. For example, a 6 micron difference in measurement is equal to a diopter of change in refractive error21. Because slight changes make such a big difference in measurement, image clarity is critical.
Limitations of the technique
A limitation of this protocol, and of SD-OCT in general, is that clear ocular media is required for a good image. For example, corneal lesions, lens abnormalities, and cataracts can prevent users from obtaining clear images. This is a problem in diabetic retinopathy imaging, in particular, as cataracts commonly develop in diabetic rodents22. If the cataract or other ocular issue is small, it is sometimes possible to maneuver the scan head around it. For larger ocular media disruptions, retinal OCT images are impossible to obtain. These retinas could still be investigated using histology as retinal histology is not contingent on clear ocular media.
A further limitation is the fact that hyperreflective lesions, such as exudates and hemorrhages, as well as major retinal vessels, result in shadowing of the underlying retinal structures, and thereby details of the underlying morphology are lost. In a case exhibiting choroidal neovascular membrane and diabetic retinopathy/macular edema where the retinal thickness was over 400 μm, it was hard to discern the underlying pathology and choroid23. Additionally, SD-OCT can only be used to assess thickness at specific locations. SD-OCT also has a limited penetration depth for imaging the choroid and for imaging of whole eyes (the whole eye can be imaged in a mouse, but not in larger animals). Another limitation is that fluorescent or other markers cannot be used with SD-OCT as with scanning laser ophthalmoscopy (SLO). However, typical SLO devices do not allow for the visualization of retinal layers in cross-section with the same ease that is observed with SD-OCT. Finally, the resolution with SD-OCT is not perfect. However, it is much improved over the resolution available at the inception of SD-OCT and continues to improve over time.
Significance and future applications
In conclusion, the advantages and significance of the SD-OCT technique are that it allows for in vivo imaging of ocular structures and quantitative tracking of changes in ocular dimensions over time, and that it performs this imaging with rapid scanning speed. Because of the high resolution of SD-OCT, it can be used to detect subtle differences that are not observable with the naked eye (Figures 4&5). Further, SD-OCT is a useful tool to measure multiple parameters of the eye in a number of disease and injury models. In this protocol alone, SD-OCT was used to measure retinal thickness in models of retinal degeneration and diabetic retinopathy, retinal thickness and cupping in a glaucoma model, and axial length in a myopia model. SD-OCT can also be used to measure corneal curvature24, assess retinal changes after blast and traumatic brain injury19,25,26, identify pathology in age-related macular degeneration27, and monitor retinal health during and after ocular injections28 and retinal placement of prosthetic devices like subretinal implants29. It can be used in other animal models such as tree shrews30 and non-human primates31 as well. SD-OCT can also be used to localize retinal pathology based on quadrant (superior, inferior, nasal, temporal) and location (central vs. peripheral). The future SD-OCT devices will achieve even greater resolution. Additionally, OCT angiography is allowing for imaging of the retinal and choroidal microvasculature by utilizing the reflection of laser light off the surface of red blood cells as they move through the retinal vasculature32,33.
ACKNOWLEDGMENTS:
This work was supported by the Department of Veterans Affairs Rehab R&D Service Career Development Awards (CDA-1, RX002111; CDA-2; RX002928) to RSA, Merit Award (RX002615) and Research Career Scientist Award (RX003134) to MTP, Career Development Award (CDA-2, RX002342) to AJF, EY028859 to MTP, NEI Core Grant P30EY006360, Research to Prevent Blindness, and Foundation Fighting Blindness.
Footnotes
DISCLOSURES:
The authors have nothing to disclose.
REFERENCES
- 1.Wojtkowski M. et al. Ultrahigh-resolution, high-speed, Fourier domain optical coherence tomography and methods for dispersion compensation. Optics Express. 12 (11), 2404–2422, (2004). [DOI] [PubMed] [Google Scholar]
- 2.Nassif N. et al. In vivo high-resolution video-rate spectral-domain optical coherence tomography of the human retina and optic nerve. Optics Express. 12 (3), 367–376, (2004). [DOI] [PubMed] [Google Scholar]
- 3.Theelen T. & Teussink MM Inspection of the Human Retina by Optical Coherence Tomography. Methods in Molecular Biology. 1715 351–358, (2018). [DOI] [PubMed] [Google Scholar]
- 4.Nakazawa M, Hara A. & Ishiguro SI Optical Coherence Tomography of Animal Models of Retinitis Pigmentosa: From Animal Studies to Clinical Applications. Biomed Research International. 2019 8276140, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drexler W. et al. Ultrahigh-resolution ophthalmic optical coherence tomography. Nature Medicine. 7 (4), 502–507, (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.VanLeeuwen JE et al. Altered AMPA receptor expression with treadmill exercise in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-lesioned mouse model of basal ganglia injury. Journal of Neuroscience Research. 88 (3), 650–668, (2010). [DOI] [PubMed] [Google Scholar]
- 7.Tian J. et al. Performance evaluation of automated segmentation software on optical coherence tomography volume data. Journal of Biophotonics. 9 (5), 478–489, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraus MF et al. Motion correction in optical coherence tomography volumes on a per A-scan basis using orthogonal scan patterns. Biomedical Optics Express. 3 (6), 1182–1199, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boatright JH et al. Tool from ancient pharmacopoeia prevents vision loss. Molecular Vision. 12 1706–1714, (2006). [PubMed] [Google Scholar]
- 10.Morrison JC et al. A rat model of chronic pressure-induced optic nerve damage. Experimental Eye Research. 64 (1), 85–96, (1997). [DOI] [PubMed] [Google Scholar]
- 11.Feola AJ et al. Menopause exacerbates visual dysfunction in experimental glaucoma. Experimental Eye Research. 186 107706, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto Y, Kakizaki M. & Masaki N. Production of spontaneous diabetic rats by repetition of selective breeding. The Tohoku Journal of Experimental Medicine. 119 (1), 85–90, (1976). [DOI] [PubMed] [Google Scholar]
- 13.Allen RS et al. Retinal Deficits Precede Cognitive and Motor Deficits in a Rat Model of Type II Diabetes. Investigative Ophthalmolology & Visual Science. 60 (1), 123–133, (2019). [DOI] [PubMed] [Google Scholar]
- 14.Stone RA et al. Altered ocular parameters from circadian clock gene disruptions. PLoS One. 14 (6), e0217111, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakraborty R. et al. Circadian rhythms, refractive development, and myopia. Ophthalmic & Physiological Optics. 38 (3), 217–245, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies EC et al. Retinal ganglion cell layer volumetric assessment by spectral-domain optical coherence tomography in multiple sclerosis: application of a high-precision manual estimation technique. Journal of neuro-ophthalmology : the official journal of the North American Neuro-Ophthalmology Society. 31 (3), 260–264, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carnevali A. et al. Optical coherence tomography angiography analysis of retinal vascular plexuses and choriocapillaris in patients with type 1 diabetes without diabetic retinopathy. Acta Diabetologica. 54 (7), 695–702, (2017). [DOI] [PubMed] [Google Scholar]
- 18.Springelkamp H. et al. Population-based evaluation of retinal nerve fiber layer, retinal ganglion cell layer, and inner plexiform layer as a diagnostic tool for glaucoma. Investigative Ophthalmology & Visual Science. 55 (12), 8428–8438, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen RS et al. Long-Term Functional and Structural Consequences of Primary Blast Overpressure to the Eye. Journal of Neurotrauma. 35 (17), 2104–2116, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao D. et al. Age-related changes in the response of retinal structure, function and blood flow to pressure modification in rats. Scientific Reports. 8 (1), 2947, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmucker C. & Schaeffel F. A paraxial schematic eye model for the growing C57BL/6 mouse. Vision Research. 44 (16), 1857–1867, (2004). [DOI] [PubMed] [Google Scholar]
- 22.Aung MH, Kim MK, Olson DE, Thule PM & Pardue MT Early visual deficits in streptozotocin-induced diabetic long evans rats. Investigative Ophthalmology & Visual Science. 54 (2), 1370–1377, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puzyeyeva O. et al. High-Resolution Optical Coherence Tomography Retinal Imaging: A Case Series Illustrating Potential and Limitations. Journal of Ophthalmology. 2011 764183, doi: 10.1155/2011/764183, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu AS et al. Topography and pachymetry maps for mouse corneas using optical coherence tomography. Experimental Eye Research. 190 107868, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan K, Kecova H, Hernandez-Merino E, Kardon RH & Harper MM Retinal ganglion cell damage in an experimental rodent model of blast-mediated traumatic brain injury. Investigative Ophthalmology & Visual Science. 54 (5), 3440–3450, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harper MM et al. Blast-Mediated Traumatic Brain Injury Exacerbates Retinal Damage and Amyloidosis in the APPswePSENd19e Mouse Model of Alzheimer’s Disease. Investigative Ophthalmology Visual Science. 60 (7), 2716–2725, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M. et al. Advanced image processing for optical coherence tomographic angiography of macular diseases. Biomedical Optics Express. 6 (12), 4661–4675, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhlfriedel R. et al. Optimized Subretinal Injection Technique for Gene Therapy Approaches. Methods in Molecular Biology. 1834 405–412, (2019). [DOI] [PubMed] [Google Scholar]
- 29.Adekunle AN et al. Integration of Perforated Subretinal Prostheses With Retinal Tissue. Translational Vision Science & Technology. 4 (4), 5, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sajdak BS et al. Noninvasive imaging of the tree shrew eye: Wavefront analysis and retinal imaging with correlative histology. Experimental Eye Research. 185 107683, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominik Fischer M. et al. Detailed functional and structural characterization of a macular lesion in a rhesus macaque. Documenta Ophthalmologica. 125 (3), 179–194, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagag AM, Gao SS, Jia Y. & Huang D. Optical coherence tomography angiography: Technical principles and clinical applications in ophthalmology. Taiwan Journal of Ophthalmology. 7 (3), 115–129, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treister AD et al. Prevalence of Subclinical CNV and Choriocapillaris Nonperfusion in Fellow Eyes of Unilateral Exudative AMD on OCT Angiography. Translational Vision Science & Technology. 7 (5), 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]