Abstract
Chronic diabetic foot is a global burden affecting millions of people, and the chronicity of an ulcer is directly linked to the diverse bacterial burden and its biofilm mode of infection. The bacterial diversity of 100 chronic diabetic ulcer samples was profiled via traditional culturing method as well as metagenomic approach by sequencing the 16S rRNA V3 hyper‐variable region on Illumina Miseq Platform (Illumina, Inc., San Diego, CA). All the relevant clinical metadata, including duration of diabetes, grade of ulcer, presence of neuropathy, and glycaemic level, were noted and correlated with the microbiota. The occurrence and establishment of bacterial biofilm over chronic wound tissues was revealed by Fluorescent in situ Hybridization and Scanning Electron Microscopy. The biofilm‐forming ability of predominant bacterial isolates was studied via crystal violet assay and Confocal Laser Scanning Microscopy. The dominant phyla obtained from bacterial diversity analysis were Firmicutes, Proteobacteria, and Actinobacteria. The dominant aerobic pathogens identified by culture method are Pseudomonas, Proteus, Enterococcus, and Staphylococcus, whereas high‐throughput sequencing revealed heightened levels of Streptococcus and Corynebacterium along with 22 different obligate anaerobes. The biofilm occurrence in chronic diabetic ulcer infection is well analysed. Herein, we illustrate the comprehensive pattern of bacterial infection and identify the community composition of chronic wound pathogenic biofilm.
Keywords: bacterial biofilm, diabetic ulcer, metagenomics, polymicrobial infection
1. INTRODUCTION
Chronic wound infection is a major menace to millions of affected patients as well as health care systems. The main categories of chronic infections include diabetic ulcer, pressure sores, and venous insufficiency in which diabetic foot ulcer (DFU) scores the maximum chronicity and encumbrance. Nearly 24.4% of the total health care expenditure of the diabetic population is because of foot ulcer complications.1 Life‐time foot ulcer risk is about 25% for diabetic patients2 and accounts for two thirds of all non‐traumatic amputations.3 Unlike acute wounds, which heal within a predictable time period, diabetic ulcers do not show signs of healing even after 30 days of medication4 and become worse because of pathogenic microbial infection. Chronic ulcers display delayed healing due to various reasons, including low blood supply, uncontrolled inflammatory response, reduced reepithelialisation, and the presence of biofilm‐associated infections.5 The infection may be mono or polymicrobial, mostly polymicrobial with antimicrobial‐resistant bacteria organised as a complex biofilm community that acts as a major contributing factor to the chronicity of a non‐healing ulcer.
Polymicrobial communities associated with diabetic ulcer biofilm are refractory to conventional antibiotic therapy. According to a WHO report (2016), diabetes prevalence is rising rapidly in low‐ and middle‐income countries, and the related complications like non‐healing ulcers pose a severe burden to the economic and social life of affected patients. Such countries are solely dependent on traditional culturing techniques, which portray different diversity pattern and dominance information regarding the microbial load. The social epidemiology of diabetic foot in countries like India is entirely different from that of western countries. Several studies focusing on the microbial diversity of wound infections were reported frequently, but those centred on molecular diagnostic approach to sketch the comprehensive diversity pattern are meagre in such lower‐ and middle‐income countries. In this context, we aim to highlight the significance of both culturing and metagenomic approaches to provide a complete coverage of bacterial diversity of chronic diabetic ulcer and the nature of wound biofilms, as well as the diagnostic options to be used in the management of biofilm‐related infections.
2. METHODOLOGY
2.1. Patient recruitment and sample collection
In total, 100 patients with DFUs from the Govt. Medical College Hospital, Trivandrum, Kerala, India, were enrolled in this study after signing the informed consent protocol form in compliance with Institutional Human Ethics Committee (Reference number RGCB‐IEC No. IHEC/01/2013/11). The patients’ details, such as age, gender, duration of diabetes, age of wound, organ injured, grade of ulcer (based on Wagner's classification), glycaemic level (HbA1c), presence of neuropathy, and presence of vascularisation, were recorded. We did not interfere in the treatment procedures and antibiotic therapy.
Diabetic wound beds were debrided to remove superficial debris and were cleansed with sterile saline. Then, swabs obtained using the Levine technique were transported to the laboratory in Amies transport medium. Sharp debridement was performed with aseptic precautions as part of standard of care, and the tissue samples were collected in sterile bottles and immediately frozen at −80°C.
2.2. Swab culturing and 16S rRNA gene sequencing
The swab cultures were streaked onto 5% Sheep blood agar, Chocolate agar, and MacConkey agar. Pure colonies of morphologically different bacterial isolates were inoculated to Luria Bertani (LB) broth and incubated at 37°C for 18 to 24 hours. Cell lysates were prepared, and 16S rRNA gene was amplified using universal 16S rRNA primers.6 The amplified product was purified through USB Exosap‐IT (Affymetrix, Santa Clara, California) treatment. Then, the sequencing was performed using a Big Dye terminator cycle sequencing kit (Applied BioSystems, Foster City, California) and was resolved on an Applied BioSystems model 3100 automated DNA sequencing system (Applied BioSystems). The similarity and homology of the 16S rRNA partial gene sequences were analysed using BLAST search of the data bank of the National Center for Biotechnology Information (NCBI).
2.3. Metagenomic sequencing
Total DNA was isolated from each of the debridement tissue samples using the Wizard Genomic DNA Purification kit (Promega, Madison, Wisconsin), and the 16S rRNA gene (1500 bp) was amplified from each sample as described earlier. The samples were quantified using a Qubit 2.0 Fluorometer. The V3 hyper‐variable region was amplified from pooled PCR product using V3‐specific primers 314F‐5′CCTACGGGAGGCAGCAG3′ and 518R‐5′ATTACCGCGGCTGCTGG3′ with the following PCR condition: 98°C for 30 seconds, 30 cycles of 98°C for 10 seconds, 72°C for 30 seconds, and final extension at 72°C for 5 seconds. A second PCR was performed using Phusion Hot Start DNA Polymerase (New England Biolabs, Ipswich, Massachusetts) with a set of primers that has illumina‐indexed bar code sequences with PCR conditions of: 98°C for 30 seconds, 15 cycles of 98°C for 10 seconds, 72°C for 30 seconds, and extension at 72°C for 5 seconds followed by 4 °C hold, and the size selection was performed with a Pure link Gel extraction kit (Invitrogen, Carlsbad, California). Library validation was performed on an Agilent 2200 Tape Station Instrument. The library was then run on Illumina MiSeq platform utilising a 300‐cycle V2 Illumina MiSeq kit and custom primers for sequencing.
In total, 90% of the reads have a phred score greater than 30 (>Q30: error probability ≥0.001), and the GC content is in the range of 40% to 60%. The primers and spacers were trimmed, and the paired‐end reads were overlapped to assemble the V3 tag sequences using the ClustalO programme. After performing multiple filters, high‐quality paired‐end reads were aligned with each other with 0 mismatches, with an average contig length of ~135 to ~165 bp. After singleton removal, the PCR chimeras were removed using the UCHIME implemented in the tool USEARCH v7.0.1090.7 The pre‐processed consensus V3 sequences were clustered into operational taxonomic units (OTUs) using the Uclust programme (Similarity cut‐off = 0.97), and a representative sequence from each OTU was aligned against Greengenes core set of sequences using the PyNAST programme.8 Then, taxonomy classification was performed using the RDP classifier, and the phylum, class, order, family, genus, and species distribution for the sample was obtained. The Alpha diversity within the sample was computed by calculating Shannon, Chao1, and Observed species metrics, and the metric calculation was performed using QIIME software.
2.4. Fluorescent in situ hybridisation
In order to study the in situ distribution of wound biofilm, debridement tissues (5 samples of acute and chronic wounds each) fixed in 4% paraformaldehyde were cryosectioned in Cryotome (CM1850UV, Leica Instruments, Wetzlar, Germany) and embedded in poly‐l‐lysine (Sigma, St. Louis, Missouri)‐coated slides. After lysozyme treatment and dehydration using graded concentrations of ethanol, a hybridisation buffer (0.9 M NaCl, 20 mM Tris HCl, 0.01% SDS, and 10% Formamide) containing 50 pmol of cy3‐labelled EUB338 universal bacterial probe (5′‐GCTGCCTCCCGTAGGAGT‐3′) was applied to the slides and incubated at 46°C in a humid chamber for 90 minutes. The slides were dipped in wash buffer, washed with ice‐cold Milli Q, stained with DAPI (1.5 μg/mL), and proceeded for Confocal Laser Scanning Microscopy (Nikon, Melville, NY).
2.5. Scanning electron microscopy (SEM)
Debridement tissue samples fixed in 2.5% glutaraldehyde solution were coated with gold–platinum and visualised using Scanning Electron Microscopy (Oxford Instruments, Abingdon, UK).
2.6. Biofilm quantification
The predominant isolates obtained were inoculated in broth (Gram‐negative isolates in LB broth and Gram‐positive in Tryptic Soy Broth + 1% glucose) and incubated overnight at 30°C. A crystal violet assay was carried out in 96‐well microtitre plates (Nunc, Roskilde, Denmark) in triplicate, as described previously.9
2.7. Confocal laser scanning microscopy (CLSM)
Biofilm staining of Gram‐positive and Gram‐negative biofilm‐forming isolates with Syto9 (Invitrogen) was carried out as described previously.10, 11 The slides were then observed under 60× objective using a Nikon Eclipse Ti Confocal Laser scanning inverted microscope (Nikon). The excitation/emission wavelength for Syto9 was 488/525 nm. The measurement of biofilm thickness was performed using NIS‐Element AR software, version 4.00.04.
2.8. Statistical analysis
Statistical analysis by χ 2 test or Student's t test was applied to relate the microbial diversity and ulcer characteristics of the patient; P < 0.05 was considered significant.
3. RESULTS
3.1. Patient data
The details of clinical samples collected and the associated clinical factors are summarised in Table 1. Samples were collected from 100 patients, with mean age of 60 ± 10·074 years, and 75% were male; 81% of the samples were taken from patients with wound duration of more than 1 month and 32% with duration of more than 3 months to several years. The mean duration of diabetes among the subjects was 12 ± 7.5 years, and those with diabetes for more than 10 years was 62%. In total, 77.4% patients with diabetes for more than 10 years suffered from neuropathy. The mean HbA1c level was 7·99 ± 2·19, and 80% of patients had HbA1c level ≥ 6·5 and 18% with level ≥10. Grade 2 ulcers penetrating ligaments and muscles (51%) were more prevalent, and 18% were found to have osteomyelitis, that is, grade 3 ulcers (as per Wagner's classification).
Table 1.
Characteristics of patients and diabetic ulcer specimens
| Total no. of specimen | 100 |
| Age of subjects | 60 ± 10.074 y |
| Subjects with duration of wound ≥1 mo | 81% |
| Patients having neuropathy | 69% |
| HbA1c level | 7.99 ± 2.19 |
| Patients with HbA1c (>6.5) | 80% |
| Poor vascularisation | 24% |
| Duration of diabetes (≥10 y) | 62% |
| Grade 1 ulcera—Superficial ulcer, not involving underlying tissues | 20% |
| Grade 2 ulcer—Deep ulcer, penetrating ligaments and muscle | 51% |
| Grade 3 ulcer—Deep ulcer with cellulitis or abscess formation, often with osteomyelitis | 18% |
| Grade 4 ulcer—Localised gangrene | 11% |
Grade of ulcer based on Wagner's classification.
3.2. Bacterial diversity by standard culturing
Aerobic swab culturing of 100 DFU samples revealed that 85% of the infections are polymicrobial in nature. There was no particular pattern of coinfection, and the infections are highly diversified in nature. The proportions of different bacterial genera identified by swab culturing are shown in Figure 1. The major phyla obtained were Proteobacteria (87%), Firmicutes (49%), Actinobacteria (24%), and Bacteroidetes (2%). Pseudomonas sp. (43%) and Proteus sp. (34%) belonging to Gammaproteobacteria represented the highest number of occurrences, followed by Enterococcus sp. (30%) and Staphylococcus sp. (26%) belonging to Firmicutes. While 39% of the samples have only Gram‐negative bacteria, 13% have Gram‐positive bacteria, and 48% have both Gram‐positive and ‐negative bacteria. As part of the present study, Wohlfahrtiimonas chitiniclastica, a rare pathogen, was reported for the first time from an Asian country.12
Figure 1.
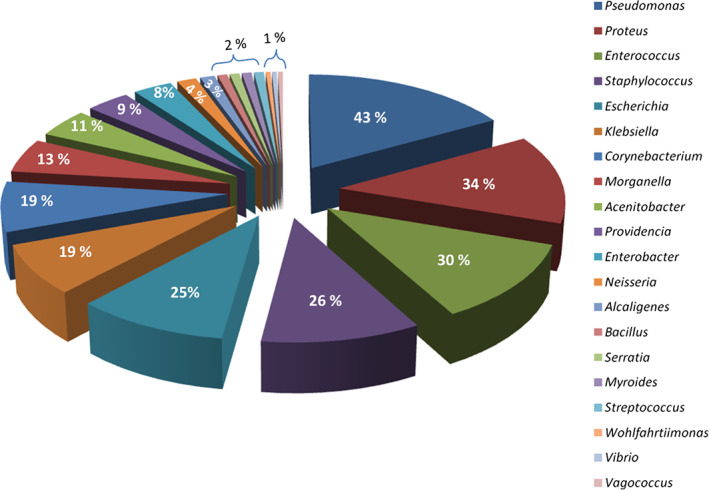
Pie chart depicting the bacterial profiling and its relative percentage obtained via swab culturing
3.3. Metagenomic analysis
In total, 3487 OTUs were identified from 542 641 reads, and 2106 OTUs were taken for further analysis after singleton removal. Raw datasets are submitted in the NCBI submission portal, SRA submission Id: SRX1453631/DU50. The heat map generated using MeV software depicted 54 different bacterial genera (Figure 2). This approach identified significantly more members at each taxonomic level when compared with the culture‐based identification. At the phylum level, Firmicutes constitute the highest proportion followed by Proteobacteria and Actinobacteria, while Bacteroidetes was comparatively lower followed by Acidobacteria, Gemmatimonadetes, and Chloroflexi. Streptococcus and Corynebacterium were found in high abundance along with the predominant ones identified via the standard culturing method. Other leading aerobic/facultative anaerobic genera identified were Helcococcus, Granulicatella, and Facklamia, and the obligate anaerobes that dominated were Finegoldia, Parvimonas, Peptostreptococcus, and Veillonella.
Figure 2.

Heat map showing the list of bacterial genera detected by the metagenomic approach. It depicts the relative percentage of 16S rRNA gene sequences assigned to each bacterial genus. Square colours shifted towards bright red indicate higher abundance
3.4. In situ analysis of bacterial biofilm
Confocal Laser Scanning Microscopy imaging demonstrated that bacteria colonising the chronic wound tissue sections appeared as large aggregates of discrete, multi‐cellular, biofilm communities (Figure 3A‐C). Three‐dimensional images were generated to visualise the biofilm pattern throughout the tissue sections. The presence of biofilm aggregates over chronic wound tissues is also confirmed by Scanning Electron Microscopy (Figure 3D‐F).
Figure 3.
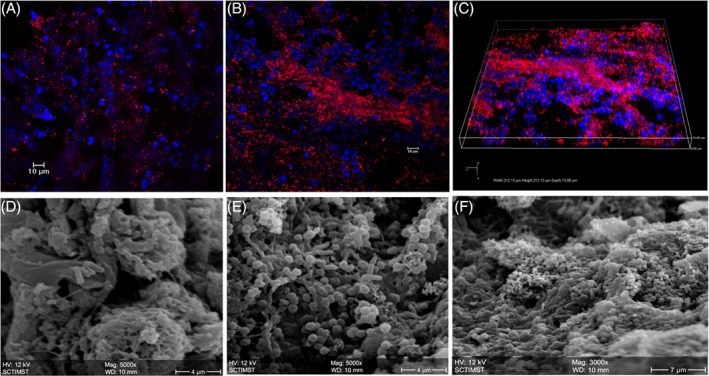
In situ visualisation of biofilm in wound tissue. Fluorescent in situ Hybridisation shows bacterial cells (red) attached to the host wound debridement tissue (blue ‐DAPI). A, Acute wound. B, Chronic wound with bacterial biofilm. C, Isometric view showing biofilm clusters attached to the chronic wound host tissue. Scanning Electron Microscopy (SEM) images of (D) acute wound with planktonic bacterial cells, 5000×; (E and F) Chronic wound with clustered bacterial biofilm, 5000× and 3000×, respectively
3.5. Biofilm formation of predominant bacteria
The majority of the predominant bacterial isolates were good biofilm producers. All the Proteus isolates (100%) intensely formed the biofilm, whereas other predominant genera, such as Enterococcus sp. (92%), Pseudomonas sp. (79%), and Staphylococcus sp. (72%), were also good biofilm formers (Figure 4). Biofilm assay demonstrated that Escherichia coli showed a low capacity to develop biofilm, without significant differences among the isolates. Biofilm development was maximum at 24 hours up to 48 hours of incubation, and after this period, cells detach, and the biofilm architecture becomes thinner. Confocal laser scanning analysis demonstrated the approximate thickness and distribution of biofilm‐forming bacterial isolates (Figure 5).
Figure 4.
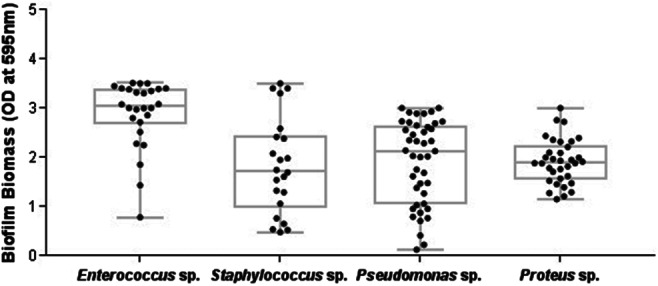
Biofilm formation of predominant isolates obtained from ulcer specimens. Each dot indicates the mean OD595 value from triplicates of each bacterial isolate. OD value <0 to 1 shows weak biofilm formers, 1 to 2 represents moderate biofilm formers, and ≥2 shows good/strong biofilm formers
Figure 5.
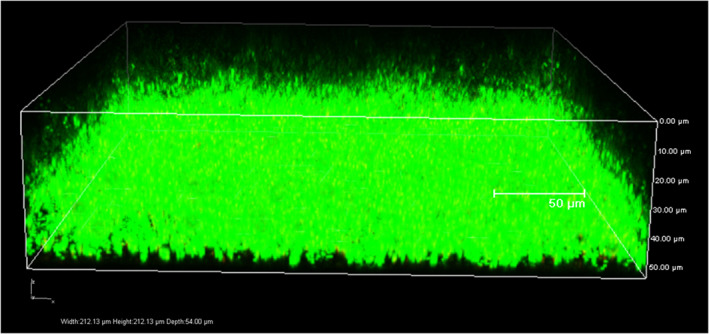
Confocal Laser scanning Microscopy analysis of biofilm formed by a Gram‐positive bacterium Enterococcus faecalis stained with Syto 9. 3D image of biofilm depicts biofilm thickness in μm
3.6. Statistical analysis
Statistical analysis revealed that the increasing duration of ulcers showed a significant incidence of members of the phylum Proteobacteria (P value 0.05) and a rising trend of polymicrobial infection (P value 0.003). The patient's grade of ulcer and the duration of diabetes were associated with a significant P value of 0.02.
4. DISCUSSION
Delayed wound healing and poor vascularisation associated with diabetes leads to severe amputations, and majority of the chronic wounds are found to be linked with biofilm infections.13 This creates a huge physiological as well as psychological impact on the patients and may lead to morbidity and increased mortality rates.14 Diverse bacterial load and its permanent establishment as biofilm create a barrier for the antibiotic therapy of chronic ulcer infections. Identifying the major culprits is of great concern as the traditional culturing techniques detect easily growing pathogens, like Staphylococcus, as the dominating ones. However, this will give a faulty picture of the microflora infecting the chronic ulcers. This does not mean that these pathogens are of mere importance, but this will conceal the role of other fastidious/non‐culturable bacteria in the polymicrobial infection leading to the chronicity of an ulcer and other related complications. Each pathogen in the polymicrobial biofilm can contribute to enhance the resistance, and the combined pathogenic effect will worsen the situation. Hence, the real depiction of the multispecies biofilm inhabiting the chronic ulcer must be well understood via next‐generation sequencing technologies.
The present study focused on delivering a comprehensive picture of the diabetic wound microbial ecology. The samples were taken from a government tertiary care hospital, and majority of the patients were from low‐ or middle‐income families. More male patients were hospitalised for DFU treatment than females. The age of the subjects does not show any influence on the bacterial population or its polymicrobial nature of infection. Most of the subjects had suffered a long duration of diabetes, and this may have influenced the grade as well as the chronicity of the ulcer. DFUs are typically associated with a prolonged diabetic condition, reduced multi‐organ efficiency, and poor vascularisation.15 The strong association between the duration of diabetes and foot complications was suggested by previous studies,16, 17 and the risk of amputation increases in patients suffering with diabetes for more than 10 years. In total, 70% of the patients had neuropathy, and this also plays a major role in ulcer chronicity, as previously reported.18 According to previous studies,19, 20 poor glycaemic control is another factor that plays a major role in retarding the healing process. In total, 80% of the ulcer patients had an HbA1c level > 6.5 at the time of hospitalisation, and the mean level was 7.99 ± 2.19, which also aggravates the healing condition.
In the present study, 85% of the infections were polymicrobial in nature, and this has proved that there is an increasing pattern of multispecies infection as the ulcer duration progresses. The predominant pathogens derived from the aerobic culturing were Pseudomonas, Proteus, Enterococcus, Staphylococcus, and E. coli. Previous studies have covered the culture analysis of diabetic ulcer specimens and reported Staphylococcus and the members of the Enterobacteriacea family as the most predominant ones.21, 22 The metagenomic approach showed heightened levels of Streptococcus and Corynebacterium along with the other predominant isolates obtained from the swab culturing method in this study. Pseudomonas, Staphylococcus, Enterococcus, and Streptococcus were considered to be the potential biofilm‐forming pathogens responsible for delayed wound healing as per the previous reports.23, 24 Other aerobic/facultative anaerobes like Helcococcus, Granulicatella, and Facklamia were also found in abundance via the metagenomic approach but were not detected by the culturing method. Helcococcus and Granulicatella are known to be slow growers and usually demonstrate satellitism around colonies of other bacteria like Staphylococcus. Hence, the bacterial culture results will be misinterpreted, and these bacteria are rarely reported previously. Besides, the current rapid microbial identification systems in use lack the database of such bacteria, which make their identification difficult. Although the pathogenesis of these bacterial genera was not well investigated, they were reported to be associated with wound infections, sepsis, and prosthetic joint infections.25, 26, 27 Bacteria like Aclanivorax, Balneimonas, Candidatus etc. reported in the present study were not identified as human inhabitants/pathogens so far, and hence, their pathogenesis is yet to be elucidated. The metagenomic approach makes the detection of microbes possible without environmental selection pressures inherent in the culturing process.
The metataxonomic analysis revealed 22 different genera of anaerobes in the diabetic ulcer specimens. A few groups have studied the anaerobic infection of diabetic ulcer, and anaerobes have been found to play a major role in delayed wound healing.28, 29 Smith et al30 have studied the new and recurring types of ulcer, and anaerobes were detected in nearly 87% of the samples. Finegoldia, Parvimonas, Peptostreptococcus, veillonella, Anaerococcus, Porphyromonas, Peptoniphilus, and Prevotella were the predominant anaerobic genera identified in the present study. Anaerobes are encased within the polymicrobial biofilm where oxygen can penetrate only up to few microns31 and hence were protected by acquiring an anaerobic condition within the open wounds. In clinical settings, not much importance is given to identify the anaerobic infection. Generally, clinicians provide broad‐spectrum antibiotics for ulcer infections, and metronidazole, the drug for treating anaerobic infections,32 is prescribed only in detected cases. This negligence may lead to a rising trend in anaerobic infections, and the treatment strategies adopted will not heal the wound promptly.
The diverse bacterial communities are found to adopt a biofilm mode of life in non‐healing ulcers. In our study, FISH‐CLSM and SEM imaging of the ulcer debridement samples showed aggregates of bacterial clusters on the wound surface, which helps to spot the abundance and pattern of biofilm over the infected tissue. James et al33 evaluated 50 chronic wounds and 16 acute wounds and microscopically confirmed the presence of biofilm in more than 60% of the chronic wounds. Similar previous studies have evaluated the presence of biofilm infections of S. aureus and Pseudomonas aeruginosa in various chronic wound specimens.34, 35 The predominant bacterial isolates were checked for biofilm‐forming ability in vitro, and the majority were found to be good biofilm formers and may exist as polymicrobial biofilm in the infected wound. Antibiotic administration may not affect the survival of good biofilm formers as well as the bacteria entrapped within the biofilm and will delay infection control. The concept of functionally equivalent pathogroup (FEP) populations suggested by36 also states that the non‐pathogenic species may live symbiotically and act synergistically and contribute to the chronicity of diabetic foot wounds.
The present investigation facilitates the understanding of polymicrobial communities associated with the diabetic ulcer, which gives a clear‐cut picture of wound ecology and routes for the better management of diabetic wounds. Along with other major aetiologies of diabetic ulcers, bacterial load and its biofilm mode of infection play a major role in ulcer chronicity. The metagenomic approach highlights the presence of viable but non‐culturable bacteria and obligate anaerobes, which may play a major role in pathogenicity, even though not much emphasis is given in the current treatment scenario. The ever‐rising pattern of diabetic ulcer infections globally drives the need for the development of better molecular diagnostic techniques for the surveillance of the bacterial community and its biofilm mode of infection for better patient management.
4.1. Metagenome data Submission
Raw datasets are submitted to the NCBI Sequence Read Archive under SRA accession number: SRX1453631, Bioproject: PRJNA304366.
ACKNOWLEDGEMENT
We thank Dr. A Sreekumar, Professor, Government Medical College Hospital, Trivandrum, Kerala, India, for permitting the collection of clinical samples. We also acknowledge Prof. M Radhakrishna Pillai, Director of Rajiv Gandhi Centre for Biotechnology, for providing the necessary facilities. The authors are thankful to Department of Biotechnology, Ministry of Science and Technology, Govt. of India for the funding provided. KS acknowledges KSCSTE, Govt. of Kerala, for the fellowship provided.
Suryaletha K, John J, Radhakrishnan MP, George S, Thomas S. Metataxonomic approach to decipher the polymicrobial burden in diabetic foot ulcer and its biofilm mode of infection. Int Wound J. 2018;15:473–481. 10.1111/iwj.12888
Funding information Department of Biotechnology , Ministry of Science and Technology, Grant/Award number: No.BT/PR15261/MED/29/994/2015
REFERENCES
- 1. Sargen MR, Hoffstad O, Margolis DJ. Geographic variation in Medicare spending and mortality for diabetic patients with foot ulcers and amputations. J Diabetes Complications. 2013;27(2):128‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lavery LA, Armstrong DG, Wunderlich RP, Tredwell J, Boulton AJM. Diabetic foot syndrome: evaluating the prevalence and incidence of foot pathology in Mexican Americans and non‐Hispanic whites from a diabetes disease management cohort. Diabetes Care. 2003;26(5):1435‐1438. [DOI] [PubMed] [Google Scholar]
- 3. Boulton AJM, Vileikyte L, Ragnarson‐Tennvall G, Apelqvist J. The global burden of diabetic foot disease. Lancet. 2005;366(9498):1719‐1724. [DOI] [PubMed] [Google Scholar]
- 4. Greenhalgh DG. Skin, soft tissue, and wound healing in the elderly. In: Yelon JA, Luchette FA, eds. Geriatric Trauma and Critical Care. New York, NY: Springer; 2014:37‐44. [Google Scholar]
- 5. Percival SL, Hill KE, Williams DW, Hooper SJ, Thomas DW, Costerton JW. A review of the scientific evidence for biofilms in wounds. Wound Repair Regen. 2012;20(5):647‐657. [DOI] [PubMed] [Google Scholar]
- 6. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697‐703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194‐2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069‐5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Merritt JH, Kadouri DE, O'Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol. 2005. Chapter 1:Unit 1B.1. 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dharmaprakash A, Mutt E, Jaleel A, Ramanathan S, Thomas S. Proteome profile of a pandemic Vibrio parahaemolyticus SC192 strain in the planktonic and biofilm condition. Biofouling. 2014;30(6):729‐739. [DOI] [PubMed] [Google Scholar]
- 11. Cerca N, Martins S, Sillankorva S, et al. Effects of growth in the presence of subinhibitory concentrations of dicloxacillin on Staphylococcus epidermidis and Staphylococcus haemolyticus biofilms. Appl Environ Microbiol. 2005;71(12):8677‐8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suryalatha K, John J, Thomas S. Wohlfahrtiimonas chitiniclastica‐associated osteomyelitis: a rare case report. Future Microbiol. 2015;10:1107‐1109. [DOI] [PubMed] [Google Scholar]
- 13. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin‐Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711‐745. [DOI] [PubMed] [Google Scholar]
- 14. Ismail K, Winkley K, Stahl D, Chalder T, Edmonds MA. Cohort study of people with diabetes and their first foot ulcer: the role of depression on mortality. Diabetes Care. 2007;30:1473‐1479. [DOI] [PubMed] [Google Scholar]
- 15. Brownrigg JRW, Griffin M, Hughes CO, et al. Influence of foot ulceration on cause‐specific mortality in patients with diabetes mellitus. J Vasc Surg. 2014;60:982‐986. [DOI] [PubMed] [Google Scholar]
- 16. Moss SE, Klein R, Klein BEK. The prevalence and incidence of lower extremity amputation in a diabetic population. Arch Intern Med. 1992;152:610‐616. [PubMed] [Google Scholar]
- 17. Lavery LA, Armstrong DG, Vela SA, Quebedeaux TL, Fleischli JG. Practical criteria for screening patients at high risk for diabetic foot ulceration. Arch Intern Med. 1998;158:157‐162. [DOI] [PubMed] [Google Scholar]
- 18. Grunfeld C. Diabetic foot ulcers: etiology, treatment, and prevention. Adv Intern Med. 1991;37:103‐132. [PubMed] [Google Scholar]
- 19. Al‐Rubeaan K, Al Derwish M, Ouizi S, Youssef AM, Subhani SN, Ibrahim HM, Alamri BN. Diabetic foot complications and their risk factors from a large retrospective cohort study. PLoS One 2015; 6:10(5):e0124446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boyko EJ, Ahroni JH, Stensel V, Forsberg RC, Davignon DR, Smith DGA. Prospective study of risk factors for diabetic foot ulcer. The Seattle diabetic foot study. Diabetes Care. 1999;22:1036‐1042. [DOI] [PubMed] [Google Scholar]
- 21. Bravo‐Molina A, Linares‐Palomino JP, Lozano‐Alonso S, Asensio‐GarcÃa R, Ros‐DÃe E, Hernández‐Quero J. Influence of wound scores and microbiology on the outcome of the diabetic foot syndrome. J Diabetes Complications. 2016;30:329‐334. [DOI] [PubMed] [Google Scholar]
- 22. Viswanathan V, Jasmine JJ, Snehalatha C, Ramachandran A. Prevalence of pathogens in diabetic foot infection in South Indian type 2 diabetic patients. J Assoc Physicians India. 2002;50:1013‐1016. [PubMed] [Google Scholar]
- 23. Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev. 2001;14(2):244‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dowd SE, Sun Y, Secor PR, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LaClaire L, Facklam R. Antimicrobial susceptibilities and clinical sources of Facklamia species. Antimicrob Agents Chemother. 2000;44(8):2130‐2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vergne A, Guerin F, Leinhard R, et al. Identification and clinical significance of Helcococcus kunzii in human samples. J Clin Microbiol. 2015;53(8):2703‐2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quénard F, Seng P, Lagier JC, Fenollar F, Stein A. Prosthetic joint infection caused by Granulicatella adiacens: a case series and review of literature. BMC Musculoskelet Disord. 2017;18:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aherrao N, Shahi SK, Dwivedi A, Kumar A, Gupta S, Singh SK. Detection of anaerobic infection in diabetic foot ulcer using PCR technique and the status of metronidazole therapy on treatment outcome. Wounds. 2012;24:283‐288. [PubMed] [Google Scholar]
- 29. Charles PGP, Uçkay I, Kressmann B, Emonet SP, Lipsky BA. The role of anaerobes in diabetic foot infections. Anaerobe. 2015;34:8‐13. [DOI] [PubMed] [Google Scholar]
- 30. Smith K, Collier A, Townsend EM, et al. One step closer to understanding the role of bacteria in diabetic foot ulcers: characterising the microbiome of ulcers. BMC Microbiol. 2016;16:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rasmussen K, Lewandowski Z. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol Bioeng. 1998;59:302‐309. [DOI] [PubMed] [Google Scholar]
- 32. Löfmark S, Edlund C, Nord CE. Metronidazole is still the drug of choice for treatment of anaerobic infections. Clin Infect Dis. 2010;50(suppl 1):S16‐S23. [DOI] [PubMed] [Google Scholar]
- 33. James GA, Swogger E, Wolcott R, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16:37‐44. [DOI] [PubMed] [Google Scholar]
- 34. Akiyama H, Huh WK, Fujii K, Yamasaki O, Oono T, Iwatsuki K. Confocal laser microscopic observation of glycocalyx production by Staphylococcus aureus in vitro . J Dermatol Sci. 2002;29:54‐61. [DOI] [PubMed] [Google Scholar]
- 35. Kirketerp‐Møller K, Jensen PA, Fazli M, et al. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol. 2008;46:2717‐2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One. 2008;3(10):e3326. [DOI] [PMC free article] [PubMed] [Google Scholar]


