Abstract
Hirschsprung disease (HSCR) is a heterogeneous group of neurocristopathy characterized by the absence of the enteric ganglia along a variable length of the intestine. Genetic defects play a major role in the pathogenesis of HSCR, whereas family studies of pathogenic variants in all the known genes (loci) only demonstrate incomplete penetrance and variable expressivity for unknown reasons. Here, we applied large-scale, quantitative proteomics of human colon tissues from 21 patients using isobaric tags for relative and absolute quantification. method followed by bioinformatics analysis. Selected findings were confirmed by parallel reaction monitoring verification. At last, the interesting differentially expressed proteins were confirmed by Western blot. A total of 5341 proteins in human colon tissues were identified. Among them, 664 proteins with >1.2-fold difference were identified in six groups: groups A1 and A2 pooled protein from the ganglionic and aganglionic colon of male, long-segment HSCR patients (n = 7); groups B1 and B2 pooled protein from the ganglionic and aganglionic colon of male, short-segment HSCR patients (n = 7); and groups C1 and C2 pooled protein from the ganglionic and aganglionic colon of female, short-segment HSCR patients (n = 7). Based on these analyses, 49 proteins from five pathways were selected for parallel reaction monitoring verification, including ribosome, endocytosis, spliceosome, oxidative phosphorylation, and cell adhesion. The downregulation of three neuron projection development genes ARF4, KIF5B, and RAB8A in the aganglionic part of the colon was verified in 15 paired colon samples using Western blot. The findings of this study will shed new light on the pathogenesis of HSCR and facilitate the development of therapeutic targets.
Keywords: Hirschsprung disease, iTRAQ, PRM, neuron projection development genes
Abbreviations: ENS, enteric nervous system; GO, gene ontology; HSCR, Hirschsprung disease; iTRAQ, isobaric tags for relative and absolute quantification; KEGG, Kyoto encyclopedia of genes and genomes; PRM, parallel reaction monitoring; WB, Western blot
Graphical Abstract
Highlights
-
•
Large-scale, quantitative proteomics of human colon tissues from Hirschsprung disease patients.
-
•
Parallel reaction monitoring, Western blotting, and immunohistochemical staining for validation.
-
•
Four pathways related to differentially expressed proteins: ribosome, endocytosis, spliceosome, and axon guidance.
-
•
Downregulation of ARF4, KIF5B, and RAB8A in the aganglionic (stenotic) colon segment.
In Brief
Quantitative proteomics revealed differentially expressed proteins in paired colon tissues from 21 patients diagnosed with Hirschsprung disease (HSCR). Hierarchical clustering suggests that the protein abundancy and expression pattern in normal intestinal tissues are closely related to gender. Gene ontology enrichment analysis revealed spliceosome, axon guidance, and extracellular exosome as the most related biological processes/molecular functions. Three neuron projection development molecules, ARF4, KIF5B, and RAB8A, were downregulated in the aganglionic segment which may be involved in the pathogenesis of HSCR.
Embryonic development is a complex and highly choreographed process, requiring exquisite temporal and spatial regulation (1). Specific signaling events, such as cell-fate progression, proliferation, differentiation, migration, metabolism, and DNA damage response, composing sets of coordinated functional genes occur to build a healthy embryo (2). Any disruption in the gene expression or regulation may interfere with early embryonic development and thus constitute a major disease risk for congenital malformations (3). Hirschsprung disease (HSCR, MIM 142623), also known as congenital aganglionosis, is characterized by the lack of enteric ganglia along the distal bowel, and when the gut fails to be innervated, it loses motility (4). HSCR patients often present with abdominal distension, vomiting, severe constipation, predisposition to bowel inflammation (enterocolitis), and even early death (5). HSCR patients can be classified into three main types according to the extent of aganglionosis: short-segment HSCR (S-HSCR, 80%), long-segment HSCR (L-HSCR, 15%), and total colonic aganglionosis (5%) (6). HSCR has an incidence of approximately 1 in every 5000 newborns of European ancestry and is twice as common in Asians, with a high heritability (>80%) and marked sex differences (male: female ratio, 4:1) (7). Although the disease occurs as an isolated disorder in approximately 70% of patients, it can also manifest with chromosomal abnormality, additional congenital anomalies, or recognized syndromes (8).
Genetic studies during the last decade have identified rare coding variants in 15 genes that together explain ∼10% of HSCR cases (9,10). The most frequent coding mutations occur in RET, which encodes a receptor tyrosine kinase that regulates the proliferation, differentiation, and migration of the enteric neural crest cells to enteric neurons (11). Other molecules participating in development of the enteric nervous system (ENS), such as GDNF (the RET receptor ligand), EDN3/EDNRB signaling pathway, transcription factors (SOX10, PHOX2B, ZEB2), SEMAs (axonal guidance molecules), L1CAM (cell adhesion molecule), and PROKR1/2 (coordinately acting with GDNF) were also verified to be encoded by susceptibility genes for HSCR (12, 13). In addition, many studies have also documented deficiencies in smooth-muscle proteins, extracellular matrix molecules, ion channels, and a variety of other important molecules in the HSCR colon. Nevertheless, family studies of all these pathogenic variants demonstrate incomplete penetrance and variable expressivity, the causes of which remain largely unexplained (14, 15, 16, 17).
Proteomics is an important technique to identify the key molecules and pathways in a variety of physiological and pathological processes and has been extensively used to characterize the molecular events occurring in numerous disease processes (18, 19, 20). Mass spectrometry (MS)-based two-dimensional gel electrophoresis has been applied to investigate the proteins that are relevant to the development of HSCR. However, previous studies were all greatly limited by the relatively low resolution of the technique itself (21, 22). Herein, we performed MS-based proteomics analysis with tandem mass tag (isobaric tags for relative and absolute quantification [iTRAQ]) labeling. Hierarchical clustering, Gene Ontology (GO) annotation, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) were utilized to analyze and classify the proteomic data. Interesting differential proteins were further validated by means of parallel reaction monitoring (PRM) in the stenotic (aganglionic) and normal (ganglionic) colon segment tissues from patients with HSCR. The downregulation of ARF4, KIF5B, and RAB8A was confirmed by Western blotting. Intense expression of all three proteins was observed in both the mucosal and myenteric plexuses of normal colon tissue by immunohistochemical staining. Our study explored for the first time proteomic changes in stenotic colon segment tissues from patients with HSCR and advanced the understanding of the etiology and pathogenesis of the disease.
Experimental Procedures
Human Subjects
In total, 21 children diagnosed with isolated HSCR (7 male S-HSCR: aged 2–10 months, mean=5.6 months; 7 male L-HSCR: aged 2–6 months, mean=3.0 months; 7 female S-HSCR: aged 2–11 months, mean=4.7 months) were recruited and studied here. All the intestinal tissues were collected during surgery, transported on ice, and stored at −80 °C for later use. Both the ganglionic and aganglionic segments were confirmed with immunohistochemical staining and the pathologist's examination. See details in supplemental Tables S1 and S2.
Protein Extraction
The One Step Animal Tissue Active Protein Extraction Kit (C500006, Sheng Gong, China) was used to extract the human colon tissue protein. For each sample, 50 mg human colon tissue was used to extract proteins. After 1 ml of precooled extraction reagent (with 1 μl of protease inhibitor, 5 μl of protease inhibitor, 1 μl of DTT, and 10 μl of PMSF) was added into each sample, the tissue was ground with a homogenizer and ultrasonically broken, 30 s each time, 8 to 10 times, with an interval of 1 min. After centrifuging for 15 min, the supernatant was transferred to a new precooled eppendorf tube for the next step of the filter aided sample preparation experiment.
Filter Aided Sample Preparation
As shown in the supplemental Table S1, the experimental samples included six groups, each of which was a mixed sample of seven patients with the same diagnosis. Briefly, protein from seven patients with the same diagnosis (14.3 μg for each patient) was mixed into 100 μg for each group. The total protein from each group was concentrated by a 3KD filter (UFC500396, Millipore, Thermo Fisher Scientific). Each 100 μg sample was added to 300 μl urea solution and centrifuged at 14,000g for 30 min. Then, 30 μl 10 mM DTT (297 μl UA + 3 μl 1 M DTT) was added and incubated at 37 °C for 1 h. Then, 15 μl 1 mM iodoacetamide was added, and the solution was kept in the dark for 20 min at room temperature (RT) and then centrifuged at 14,000g for 45 min. Next, 300 μl urea solution was added for washing three times, which was followed by three washes with 300 μl NH4HCO3. The total protein was dissolved in 100 μl 25 mM ammonium bicarbonate buffer and stored at −80 °C for the next experiment.
Protein Digestion and iTRAQ Labeling
Before the iTRAQ labeling experiments, 50 μg of protein, extracted as described above, were trypsinized. After an overnight digestion, iTRAQ labeling was performed using a 4-plex iTRAQ labeling kit (AB Sciex, catalog: 4466096) according to the manufacturer's protocol. The iTRAQ labeling was performed as shown in Table 1: 2 mixtures used as controls containing samples from all groups were labeled with iTRAQ reagent 114, A1 and B2 were labeled with iTRAQ reagent 115; A2 and C1 were labeled with iTRAQ reagent 116; C2 and B1 were labeled with iTRAQ reagent 117. The concentrated iTRAQ reagent-labeled digest samples from the six groups were next solubilized in 250 μl loading buffer A (20 mM ammonium formate, pH 10) and combined into one tube prior to fractionation.
Table 1.
The iTRAQ labeling details
| Type | L-HSCR male ganglionic | L-HSCR male aganglionic | S-HSCR male ganglionic | Mixture (pooled with A1, A2, B1, B2, C1, C2) | S-HSCR male aganglionic | S-HSCR female ganglionic | S-HSCR female aganglionic |
|---|---|---|---|---|---|---|---|
| Subgroup | A-1 | A-2 | B-1 | Mix | B-2 | C-1 | C-2 |
| iTRAQ labeling | 115 | 116 | 117 | 114∗2 | 115 | 116 | 117 |
HSCR, Hirschsprung disease; L-HSCR, long-segment HSCR; S-HSCR, short-segment HSCR.
High pH One-D Reversed-Phase Chromatography Fractionation
Fractionation of the peptide mixture was performed on a Dionex Ultimate 3000 HPLC system equipped with an UltiMate 3000 RS pump, an UltiMate 3000 RS column compartment and an UltiMate 3000 RS autosampler (Dionex). The column was initially equilibrated for 70 min in buffer A (20 mM ammonium formate, pH 10) as described above before sample injection. The tryptic digest was injected onto a Phenomenex column (Gemini-NX 3u C18 110A; 1502.00 mm) using a linear gradient increasing at a rate of 1% B per minute from 2 ± 45% mobile phase B (A: 20 mM HCOONH4 at pH 10; B: 20 mM HCOONH4, 80% CAN at pH 10) at a flow rate of 200 μl/min while monitoring a UV absorbance at 214 nm/280 nm. Fractions were collected every minute during the course of the run. Subsequently, all 48 collected fractions were mixed into 14 components, lyophilized, and kept at −80 °C before Nano-LC-MS/MS analysis.
HPLC/MS
The tryptic digests were injected into an UltiMate 3000 RSLCnano System (Dionex) and analyzed by a Q Exactive HF mass spectrometer (Thermo Scientific). The tryptic digests were suspended in buffer A (0.1% formic acid) and directly loaded onto an Acclaim PepMap100 C18 75 μm × 20 mm trap column (Thermo Fisher Scientific) for preconcentration and online desalting. Separation was then achieved using an EASY-Spray PepMap C18 75 μm × 150 mm column (Thermo Fisher Scientific), employing a linear gradient from 4 to 90% acetonitrile at 300 nl/min over 150 min. Q-Exactive HF MS System (Thermo Fisher Scientific) was operated in full MS/data-dependent acquisition MS/MS mode (data-dependent acquisition). The Orbitrap mass analyzer was used at a resolution of 60,000 to acquire full MS with an m/z range of 350 to 2000, incorporating a target automatic gain control value of 3E6 and maximum fill times of 20 ms. MS/MS fragments were measured at an Orbitrap resolution of 30,000 using an automatic gain control target of 5E5 and maximum fill times of 60 ms. Full-scan MS spectra in the m/z range of 350 to 2000 were acquired in the Orbitrap. Twenty of the most intense ions were isolated for MS/MS analysis. The raw data were processed using Proteome Discoverer (version 2.1.0.81, Thermo Fisher Scientific), searching with a database of human histone (www.uniprot.org, accessed February 2016, 91,974 sequences). Peptides were generated from a semitryptic digestion with up to four missed cleavages, carbamidomethylation of cysteines as a fixed modification, and oxidation of methionines as a variable modification. Precursor mass tolerance was 20 ppm, and product ions were searched at 0.05 Da tolerance. Peptide spectral matches were validated using a percolator based on q-values at a 1% false discovery rate. The dataset analyzed here has been deposited into the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (23) with a dataset identifier of PXD021292.
Development and Analytical Validation Targeted MS Assays/Measurements
The peptides passing the false discovery rate were exported to a text file and processed by PRM. The mass inclusion list involved mass, charge, and polarity. Lists of all peptides targeted in the PRM analyses are provided in supplemental Table S3. PRM methods were developed according to the Tier 3 level analyses. PRM method design and data analysis was performed using Skyline (ver. 20.2.0.286). Samples were analyzed by LC-MS/MS on a Dionex Ultimate 3000 nanoRSLC HPLC connected online to a Q Exactive HF Hybrid Quadrupole-Orbitrap mass spectrometer equipped with an EASY-Spray ion source (Thermo Scientific). Peptides were separated by direct injection over an EASY-Spray PepMap C18 column (2 μm; 15 cm × 75 μm, Thermo Fisher Scientific) for 150 min at 300 nl/min with a reverse phase linear ACN gradient from 4 to 90% solvent B (B: 80% ACN/0.1% formic acid/20% water, A: 2% ACN/0.05% formic acid/98% water). The mass spectrometer was configured for PRM acquisition. Six samples of each group were selected for PRM. The raw data were processed using Proteome Discoverer (version 2.1.0.81, Thermo Fisher Scientific), searching with a database of human histone (www.uniprot.org, accessed February 2016, 91,974 sequences). The proteome discovery results analyzed here have been deposited into the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with a dataset identifier of PXD021292.
PRM Data Analysis
The resulting MS data were processed using Skyline (version 20.2.0.286; AB Sciex). Peptide settings: enzyme was set as trypsin [KR/P], max missed cleavage set as 0, the peptide length was set as 7 to 25, and fixed modification was set to carbamidomethyl on Cys. Transition settings: precursor charges were set as 2, 3, ion charges were set as 1, ion types were set as b, y. The product ions were set as from ion 3 to last ion, and the ion match tolerance was set as 0.02 Da. Peptides were quantified by extracting the peak areas of accurate fragment ions, and they were then integrated across the peptides' elution profiles. For each peptide, transition peak areas were normalized by the average of the sum of the transition peak areas for all the peptides across the runs. The sum of the intensities of all detected peptides was used to compare the differences. If a peptide was not detected in the sample or the spectrum was not good, one sample was removed, and at least five samples were left for statistical analysis.
Experimental Design and Statistical Rationale
The colon tissue protein of 21 patients were divided into six groups, each of which was a mixed sample of seven patients with the same diagnosis (male S-HSCR, male L-HSCR, and female S-HSCR). The total protein from each group was digested, and iTRAQ labeling was used to perform quantitative proteomics. A mixture containing samples from all groups were used as controls to normalize the protein level in two batches. In results of quantitative analysis, we used at least two unique peptides to identify proteins and the abundances from each channel were used to calculate the mean ratios of identified proteins. We then set the ratio threshold at > 1.2 for protein upregulation and at < 0.83 (1/1.2) for protein downregulation.
Proteins with a fold change larger than 1.2 and a p value <0.05 were defined as significantly differentially expressed. Significantly changed proteins were screened by volcano plot analysis which is detected by Student’s t-test. Histograms of protein fold changes in proteomic data were used to demonstrate normal data distribution. Six individuals’ samples from group A1, A2, B1, B2, C1, and C2 were randomly chosen for PRM verification. Statistical analysis of PRM was performed using GraphPad Prism 6. PRM data were analyzed by an unpaired, two-tailed t-test. Statistical significance was considered at p value <0.05. All data were expressed as mean ± standard errors.
Western Blot (WB) Analysis
Total protein was extracted from human colon tissues using a Sangon kit (Sangon Biotech, Shanghai, China). Equal amounts of denatured protein were separated with 10% SDS-PAGE (Sangon Biotech) and then transferred to a nitrocellulose (NC) membrane (Millipore Corporation). The NC membranes were blocked with 10% skimmed milk for 1 h at RT and incubated with primary antibodies (anti-ARF4, 1:1000 dilution, ab171746, Abcam; anti-KIF5B, 1:1000 dilution, ab167429, Abcam; anti-RAB8A, 1:3000 dilution, ab188574, Abcam; and anti-β-actin, 1:1000 dilution, #3700, CST) at 4 °C overnight. After rinsing, the NC membranes were further incubated with the appropriate secondary antibody for 2 h at RT. An ECL reagent (GE Healthcare) was applied to visualize the band signals. The grayscale values of the bands were normalized to the values of the corresponding β-actin band to determine the expression levels of the proteins.
Immunohistochemistry
Immunohistochemical studies were performed on 3 μm sections obtained from paraffin-embedded blocks using the biotin–streptavidin complex method. Tissue sections were deparaffinized in xylene (four times, 20 min each), rehydrated with graded ethanol (100%, 90%, 80%, and 70%), and rinsed in distilled water (5 min). Antigen retrieval was performed with Tris/EDTA at pH 9.0 and 140 °C for 2 min. After treatment with 3% H2O2 (Beijing Yili) in methanol for 15 min at 37 °C to block endogenous peroxidases, the sections were blocked with 5% bovine serum albumin in PBS and incubated with the primary antibodies anti-ARF4 (1:500 dilution, ab171746, Abcam), anti-KIF5B (1:100 dilution, ab167429, Abcam), and anti-RAB8A (1:700 dilution, ab188574, Abcam) overnight at 4 °C. After washing with 0.1 M PBST; pH 7.4; (5 min, 3 times), antibodies were detected using the Polink-2 Plus polymer horseradish peroxidase detection system for goat primary antibodies (ZSGB-BIO) according to the manufacturer's instructions. After washing with PBS, the reaction was visualized with a 3,3′-diaminobenzidine tetrahydrochloride kit (Dako). All samples were counterstained with Harris hematoxylin, differentiated using hydrochloric acid in ethanol, and blued using ammonia water. Finally, the slides were dehydrated in graded alcohols, cleared with dimethylbenzene, and mounted with neutral gum. An equivalent volume of PBS instead of primary antibodies was used for negative controls. Tissue sections from human kidney, brain, and stomach served as positive controls for the primary anti-ARF4 antibody, anti-KIF5B antibody, and anti-RAB8A antibody, respectively. The stained tissue samples were assessed by two independent pathology researchers. Images were captured using a Nikon E800 digital microscope.
Results
Overview of the Workflow Used to Discover and Verify HSCR-Related Proteins
Our experimental strategy is illustrated in Figure 1. For the discovery step, the proteins generated from the ganglionic and aganglionic part of the male, L-HSCR (n = 7), male, S-HSCR (n = 7), and female, S-HSCR (n = 7) groups were analyzed by iTRAQ quantification. Proteins with >1.2-fold difference among the six groups were identified. For the verification step, a targeted PRM study was set up based on the information found in the prior study to validate quantitative findings from the iTRAQ discovery study. Finally, downregulation of three neuron projection development molecules, ARF4, KIF5B, and RAB8A, was verified in 15 paired colon samples using Western blotting.
Fig. 1.
The workflow of the experimental procedures. An overview of the workflow used for the identification of differentially expressed proteins in six groups of 21 children diagnosed with isolated HSCR. (A1: pooled ganglionic part of male L-HSCR patients (n = 7); A2: pooled aganglionic part of male L-HSCR patients (n = 7), B1: pooled ganglionic part of male S-HSCR (n = 7), B1: pooled aganglionic part of male S-HSCR (n = 7), C1: pooled ganglionic part of female S-HSCR (n = 7), C2: pooled aganglionic part of female S-HSCR (n = 7). Colon tissues of six groups were collected. Proteins were extracted and digested by trypsin. Peptides from each individual sample and from pooled internal reference samples were randomly distributed and labeled across two iTRAQ experiments. Following labeling, samples were pooled and analyzed by tandem mass spectrometry. The data were identified using Proteome Discoverer. Bioinformatics analysis was applied based on the quantified results, and the selected proteins of interest were verified using PRM experiment. Furthermore, WB was used to confirm the HSCR-related proteins. HSCR, Hirschsprung disease; L-HSCR, long-segment HSCR; PRM, parallel reaction monitoring; S-HSCR, short-segment HSCR; WB, Western blot.
Identification of Differentially Expressed Proteins Using iTRAQ Labeling and LC-MS/MS
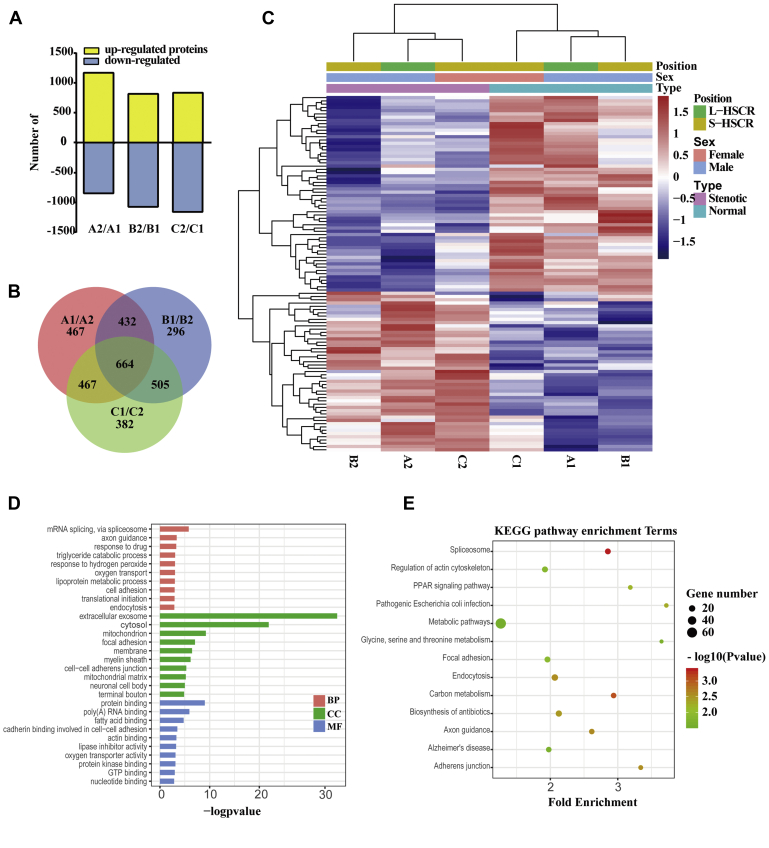
A total of 5341 proteins were identified by iTRAQ coupled 2D LC-MS/MS analysis in human colon tissues. Among them, 4907 proteins were detected in batch 1; 4792 proteins were detected in batch 2; and 4358 detected in both batches (supplemental Fig. S1). The detailed information including protein quantification date and average iTRAQ ratio for these identified proteins is shown in supplemental Tables S4–S6. Proteins with >1.2-fold difference in the male L-HSCR, male S-HSCR, and female S-HSCR patients were analyzed. In the male L-HSCR group, in the aganglionic part compared with the ganglionic part, 1169 proteins were upregulated, and 846 proteins were downregulated. In the male S-HSCR group, in the aganglionic part compared with the ganglionic part, 816 proteins were upregulated, and 1073 proteins were downregulated. In the female S-HSCR group, in the aganglionic part compared with the ganglionic part, 834 proteins were upregulated, and 1160 proteins were downregulated (Fig. 2A). Altogether, 664 common proteins displayed significantly altered levels with fold cut-off of ≥1.2 between A1 and A2, B1 and B2, and C1 and C2 (Fig. 2B and supplemental Table S7).
Fig. 2.
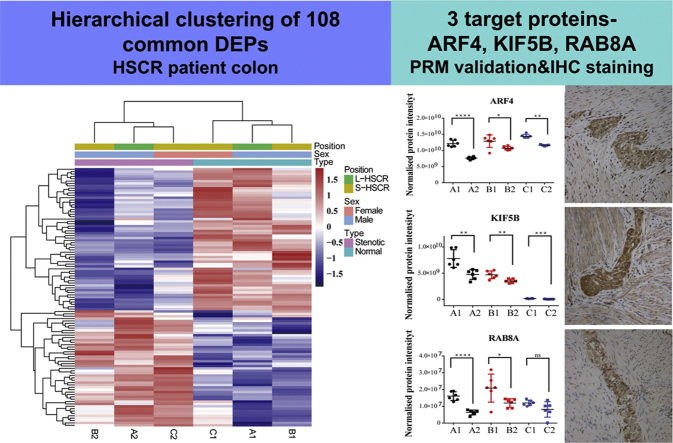
Bioinformatics analysis based on the sequencing results. A, the upregulated and downregulated proteins with >1.2-fold difference in the male L-HSCR, male S-HSCR, and female S-HSCR patients. B, Venn-diagram showing overlap of all differentially expressed proteins among the ganglionic and aganglionic part of three groups (A1 and A2: male L-HSCR; B1 and B2: male S-HSCR; and C1 and C2: female S-HSCR). C, hierarchical clustering of the 108 common differentially expressed proteins. The bar color represents a logarithmic scale from −1.5 to 1.5. D, gene Ontology enrichment results of the cellular components, biological processes, and molecular functions. The p value (-Log10) of each term is shown on the horizontal axis, and the vertical axis denotes the term name. E, reactome enrichment results are displayed through a bubble chart. The horizontal axis shows the rich factor of each pathway. The number of genes involved in each pathway is reflected by the size of the bubbles, whereas the color represents each p value. BP, biological processes; CC, cellular components; HSCR, Hirschsprung disease; KEGG, Kyoto Encyclopedia of Genes and Genomes; L-HSCR, long-segment HSCR; MF, molecular functions; S-HSCR, short-segment HSCR.
Bioinformatics Analysis
One hundred and eight proteins that were significantly changed in all six groups were selected for cluster analysis (Fig. 2C). The results showed that A1, B1, and C1 clustered to one group, and A2, B2, and C2 clustered to another group. These results indicated that there was some common difference between the ganglionic and the aganglionic part in HSCR patients. Six hundred sixty-four common proteins with >1.2-fold differences were chosen for further bioinformatics analysis based on the DAVID database (https://david.ncifcrf.gov). GO analysis includes biological process, molecular function, and cellular component. The top 10 items were chosen to draw Figure 2D. Pathway analysis was applied to classify proteins according to the pathway in the KEGG database; the top 13 pathways were selected to draw Figure 2E. The protein–protein interaction (PPI) was analyzed based on the STRING database (https://string-db.org), and the PPI result with all 664 proteins is shown in supplemental Fig. S2. The PPI network of 304 proteins was obtained by setting confidence value at 0.9, and then, Cytoscape 3.7.1 was used to adjust the PPI map. Finally, the PPI network of 117 key proteins was obtained by select degree ≥ 5 (Fig. 3).
Fig. 3.
Protein–protein interaction (PPI) networks analysis of the differentially expressed proteins. The networks analysis of the 117 key differentially expressed proteins was carried out using STRING software (https://string-db.org/). The confidence value is set at 0.9, and the degree ≥5. An edge was drawn with six color lines as shown (see details in the legend).
Validation of Differentially Expressed Proteins by PRM
Next, to verify the differentially expressed proteins, trypsin-digested proteins of the six groups were selected for PRM. According to the analysis results from KEGG and David, four pathways related to the differentially expressed proteins were selected: ribosome, endocytosis, spliceosome, and oxidative phosphorylation. In addition, we also selected cell adhesion molecules, which were previously known to be related to HSCR, and the total came to 49 proteins. The profile of the target peptide analyzed by Skyline was shown in supplemental Table S8. The PRM results of 25 proteins were consistent with iTRAQ. Among them, nine nucleoside-triphosphatase activity–related proteins, DDX5, ATP2A2, TUBB6, EIF4A1, NSF, RAB8A, ARF4, RAC2, and KIF5B, were confirmed to be decreased in all aganglionic segments of the three patient groups. Partial results are shown in Figure 4: in the aganglionic part of the three groups, the expression levels of quantitative peptide “QDLPNAMAISEMTDK” and sum of all detected peptides of ARF4 were decreased (Fig. 4, A–B); the sum of all detected peptides of RAB8A, KIF5B, ATP2A2, and RAC2 were almost all significantly decreased (Fig. 4, C–F). These results are consistent with the iTRAQ results, suggesting that the activity of nucleoside-triphosphatase was decreased in the aganglionic part of the colon in HSCR patients.
Fig. 4.
The relative protein levels of five nucleoside-triphosphatase activity-related proteins measured using PRM. A1: ganglionic part of male L-HSCR; A2: aganglionic part of male L-HSCR. B1: ganglionic part of male S-HSCR; B2: aganglionic part of male S-HSCR. C1: ganglionic part of female S-HSCR; C2: aganglionic part of female S-HSCR. Data represent mean ± SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001. The aganglionic part was compared with the ganglionic part; the p values were calculated using an unpaired t test. A, in the A group, the ARF4 peptide QDLPNAMAISEMTDK intensity of A1 and A2 were 6.427E8 ± 3.431E7 (n = 6) and 3.328E8 ± 1.403E7 (n = 6), p < 0.0001; in the B group, the ARF4 peptide QDLPNAMAISEMTDK intensity of B1 and B2 were 7.235E8 ± 5.877E7 (n = 6) and 4.832E8 ± 4.310E7 (n = 6), p = 0.0080; in the C group, the ARF4 peptide QDLPNAMAISEMTDK intensity of C1 and C2 were 7.435E8 ± 4.452E7 (n = 6) and 3.784E8 ± 7.738E7 (n = 6), p = 0.0022. B, in the A group, sum of all detected peptide intensities in ARF4 of A1 and A2 were 1.215E10 ± 4.241E8 (n = 6) and 7.602E9 ± 1.678E8 (n = 6), p < 0.0001; in the B group, sum of all detected peptide intensities in ARF4 of B1 and B2 were 1.288E10 ± 7.956E8 (n = 6) and 1.068E10 ± 2.301E8 (n = 6), p = 0.0243; in the C group, sum of all detected peptide intensities in ARF4 of C1 and C2 were 1.442E10 ± 2.400E8 (n = 6) and 1.160E10 ± 1.000E8 (n = 5), p = 0.0086. C, in the A group, sum of all detected peptide intensities in RAB8A of A1 and A2 were 1.607E7 ± 1.070E6 (n = 6) and 6.333E6 ± 589071 (n = 6), p < 0.0001; in the B group, sum of all detected peptide intensities in RAB8A of B1 and B2 were 2.086E7 ± 3.409E6 (n = 6) and 1.199E7 ± 1.029E6 (n = 6), p = 0.0319; in the C group, sum of all detected peptide intensities in RAB8A of C1 and C2 were 1.200E7 ± 682642 (n = 6) and 8.135E6 ± 1.878E6 (n = 6), p = 0.0818. D, in the A group, sum of all detected peptide intensities in KIF5B of A1 and A2 were 7.795E9 ± 6.902E8 (n = 6) and 4.763E9 ± 3.846E8 (n = 6), p = 0.0033; in the B group, sum of all detected peptide intensities in KIF5B of B1 and B2 were 4.693E9 ± 3.017E8 (n = 6) and 3.513E9 ± 1.534E8 (n = 6), p = 0.0059; in the C group, sum of all detected peptide intensities in KIF5B of C1 and C2 were 1.693E8 ± 1.735E7 (n = 6) and 5.742E7 ± 1.075E7 (n = 5), p = 0.0006. E, in the A group, sum of all detected peptide intensities in ATP2A2 of A1 and A2 were 6.842E9 ± 1.913E8 (n = 6) and 3.972E9 ± 9.854E7 (n = 6), p < 0.0001; in the B group, sum of all detected peptide intensities in ATP2A2 of B1 and B2 were 4.810E9 ± 5.061E8 (n = 6) and 3.565E9 ± 1.181E8 (n = 6), p = 0.0376; in the C group, sum of all detected peptide intensities in ATP2A2 of C1 and C2 were 7.778E9 ± 3.011E8 (n = 6) and 6.642E9 ± 1.639E8 (n = 5), p = 0.0123. F, in the A group, sum of all detected peptide intensities in RAC2 of A1 and A2 were 2.707E9 ± 1.072E8 (n = 6) and 1.840E9 ± 9.096E7 (n = 6), p = 0.0001; in the B group, sum of all detected peptide intensities in RAC2 of B1 and B2 were 4.610E9 ± 4.172E8 (n = 6) and 3.498E9 ± 2.521E8 (n = 6), p = 0.0458; in the C group, sum of all detected peptide intensities in RAC2 of C1 and C2 were 2.443E9 ± 1.642E8 (n = 6) and 1.710E9 ± 2.632E8 (n = 6), p = 0.0397. HSCR, Hirschsprung disease; L-HSCR, long-segment HSCR; S-HSCR, short-segment HSCR.
Immunohistochemistry Staining of ARF4, KIF5B, and RAB8A in the Normal Colon Specimens
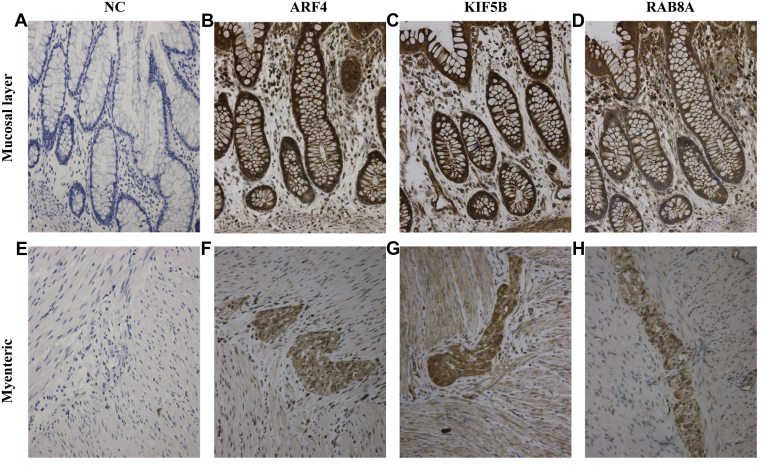
Among the 25 proteins verified by PRM, three key proteins, ARF4, KIF5B, and RAB8A, in the PPI analysis were classified as neuron projection development proteins in the STRING database. To further understand their possible pathological roles in the disease, we then used immunohistochemical staining to localize ARF4, KIF5B, and RAB8A in the colon tissue. Intense expression of all three neuron projection development proteins was observed in both the mucosal and myenteric plexuses of normal colon tissue. Omission of primary antibodies served as negative controls. No immunostaining was detected in the group of negative controls (Fig. 5).
Fig. 5.
Immunohistochemistry staining of ARF4, KIF5B, and RAB8A in the normal colonic sections. Intense expression of ARF4, KIF5B, and RAB8A were observed in both the mucosal (B, C and D) and myenteric (F, G and H) plexuses in the normal colon tissues. No immunostaining was detected in the group of negative controls (A and E). Representative colon sections are shown. Bars: 200 μm. NC, negative control.
Protein Expression Levels of ARF4, KIF5B, and RAB8A
Finally, to confirm the alterations in ARF4, KIF5B, and RAB8A at the protein level, we performed WB analysis. In total, 15 paired samples were examined, five each from the A, B, and C groups. Our results showed that the protein expression level was markedly decreased in the aganglionic segments compared with that of the ganglionic segments (p < 0.05) (Fig. 6). Equal amounts of loading were confirmed by β-actin staining of the stripped membranes.
Fig. 6.
WB confirmed downregulated expression of ARF4, KIF5B, and RAB8A in the aganglionic segments of HSCR patients. Representative staining bands are shown (A, ARF4; B, KIF5B, C, RAB8A). Equal amounts of loading were confirmed by β-actin staining. Values are given as the mean ± SD (D). G, ganglionic segment; A, aganglionic segment. HSCR, Hirschsprung disease; WB, Western blot.
Discussion
We here present a quantitative proteomic landscape of the most prevalent subtype of human HSCR, with comprehensive PRM validation, WB verification, and subsequent morphologic examination through immunohistochemical staining. Using colon tissues from male patients affected by L- and S-HSCR, as well as from female patients diagnosed with S-HSCR, robust proteomics and bioinformatics tools were utilized to explore shared and subtype-specific altered proteomes present in different gender- and phenotype-associated HSCR, providing insights into the biology of this complex human disease with variable expressivity. Among the six groups, 664 common proteins with >1.2-fold difference were identified and chosen for further investigation. GO analysis indicated that the differentially expressed proteins mainly function in spliceosome (mRNA splicing), axon guidance, extracellular exosome, cytosol, and protein binding. While using KEGG, 13 pathways were identified to be significantly enriched with association signals at the p < 0.05 level, including spliceosome, carbon metabolism, axon guidance, endocytosis, and adherens junction. These signaling pathways are significantly affected by different proteins, and these findings may provide valuable information for future studies of the molecular mechanism underlying development of the human ENS.
Altogether, 108 proteins with consistent trends of change among all the subgroups were detected and subjected to hierarchical clustering analysis. A clear distinction between the normal (ganglionic—A1, B1, and C1) and all stenotic (aganglionic—A2, B2, and C2) colon segment tissues was observed. The protein expression pattern of normal colon tissues in male, S-HSCR patients (B1) is more similar to that in male, L-HSCR patients (A1) than in female, S-HSCR patients (C1). Similarly, the protein expression pattern of the stenotic colon tissues in male, L-HSCR patients (A2) was unexpectedly more similar to that of female, S-HSCR patients (C2) than that of male, S-HSCR patients (B2), suggesting that (1) the protein abundancy and expression pattern in normal intestinal tissue seems to be quite different between different genders, and they are more similar in males compared with females and vice versa; (2) the threshold for HSCR to be manifested in females is probably significantly higher than that in males and consequently defines a “rule” that a similar pattern (and extent) of abnormal protein expression will present in males with L-HSCR (A2) as in females with S-HSCR (C2). A study with a bigger sample size and an in-depth analysis would be warranted for a better search for the etiology underlying the marked sex differences in HSCR.
Of 25 PRM validated proteins that were consistent with the iTRAQ result, three of them (ARF4, KIF5B, and RAB8A) are classified as neuron projection development–related proteins by the STRING database, serving as good candidates for being involved in the developmental defects of the ENS. The subsequent WB experiments confirmed their downregulation within the stenotic segment of colon in the patients, and a broad, intense expression in the normal colon tissue was observed, including both the submucosal and myenteric plexuses. ARF4 is a 20-kDa cellular protein that was initially identified and characterized as a member of the human ADP-ribosylation factor gene family. As the most divergent member of the human ADP-ribosylation factors, ARF4 is involved in controlling a multitude of cellular functions, including the assembly and dynamics of the microtubule and actin cytoskeletons, endomembrane trafficking, lipid metabolism, mitochondrial architecture, and other pathways in cell biology (24, 25). Global knockout of Arf4 in mice was demonstrated to be embryonic lethal, with mutant animals showing severe defects in ciliary assembly. Pearring et al. (26) found that knockout of Arf4 at postnatal day 2 (P2) resulted in elevated mortality by P10. Survivors at P10 were smaller than their littermates, and necropsy revealed a smaller pancreas, yellowish feces in the lower intestine, and abnormal microvilli in the visceral endoderm. KIF5B, one of the ∼40 Kinesin superfamily proteins identified in mammals to date, was reported to act as a molecular motor for RNA-containing particles in the nervous system (27). RNA transport is an important and fundamental event for local protein synthesis, especially in neurons. The process of dendritic protein synthesis was discovered to often accompany plasticity and participate in long-lasting changes in synaptic strength (28); such changes are key to establishing the ENS. RAB8A belongs to the RAS superfamily of small GTP/GDP-binding proteins with an average size of 200 amino acids. This protein shares 97% and 96% similarity with the dog RAB8 and mouse MEL proteins, respectively, and contains four GTP/GDP-binding sites that are present in all the RAS proteins, playing vital roles in the transport of proteins from the endoplasmic reticulum to the Golgi and the plasma membrane (29, 30). As a phosphorylation substrate of LRRK2, RAB8A was demonstrated to be involved in the repair of damaged endomembranes in macrophages (31), mediating endolysosomal trafficking alterations and causing familial Parkinson's disease (32), and coordinating endothelial cell tubulogenesis by controlling polarized membrane trafficking events (33). As a whole, whether and how the dysfunction of these molecules disrupts neuron projection development and contributes to a higher HSCR risk remains to be elucidated.
Proteins are a set of diverse and complicated molecules that contribute to the immense complexity of a biological system. Except for the huge bunch of functionally distinct proteins, biodiversity and the expanded-spectrum of human traits also comes from posttranscriptional modification, which cannot be predicted from the nucleic acid sequence. Hence, quantitative proteomic profiling might provide a more realistic picture of the functional aberrations that occur in defective cells or tissues and lead to the discovery of novel modules or pathways that might participate in the development of the organ involved in the disease.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (23) with the dataset identifier PXD021292.
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval
This study was approved by the medical ethics committee of the Capital Institute of Pediatrics (SHERLL 2013039) and was carried out according to the Declaration of Helsinki protocols.
Acknowledgments
We are grateful to the many affected individuals and their families whose cooperation made this study possible.
Funding and additional information
This work was made possible by a grant from the National Natural Science Foundation of China (81971397), the Public Service Development and Reform Pilot Project of Beijing Medical Research Institute (BMR2019-11), and the Research Foundation of the Capital Institute of Pediatrics (PY-2018-02) to Q. Z. B. B. was supported by the National Natural Science Foundation of China (81901167). Z. Z. was supported by the National Natural Science Foundation of China (81700451). Q. L. was supported by the Beijing Municipal Administration of Hospitals Incubating Program (PX2020054). Q. J. was supported by the grant from the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (CAMS-I2M) and the National Natural Science Foundation of China (81771620, 82070532).
Author contributions
Q. Z. and L.-H. W. prepared the manuscript. Q. L., Z. Z., H. W., and L. L. conducted sample acquisition and collected clinical information. Q. Z., B.-L. B., and D. L. extracted the protein, performed protein digestion, iTRAQ labeling, chromatography fractionation, HPLC/MS, data analysis, and PRM validation. L.-H. W. performed the WB experiment. L.-H. W. and P. X. conducted immunohistochemical staining. Q. Z. and Q. J. supervised the study and edited the manuscript.
Footnotes
This article contains supporting information.
Supporting Information
Supplemental Figures S1–S2 and Tables S1–S2
Supplemental Table S3
Supplemental Table S4
Supplemental Table S5
Supplemental Table S6
Supplemental Table S7
Supplemental Table S8
References
- 1.Timme-Laragy A.R., Hahn M.E., Hansen J.M., Rastogi A., Roy M.A. Redox stress and signaling during vertebrate embryonic development: regulation and responses. Semin. Cell Dev. Biol. 2018;80:17–28. doi: 10.1016/j.semcdb.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Theunissen T.W., Jaenisch R. Mechanisms of gene regulation in human embryos and pluripotent stem cells. Development. 2017;144:4496–4509. doi: 10.1242/dev.157404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh A.J., Ramsey S.A., Filtz T.M., Kioussi C. Differential gene regulatory networks in development and disease. Cell Mol. Life Sci. 2018;75:1013–1025. doi: 10.1007/s00018-017-2679-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler-Tjaden N.E., Trainor P.A. The developmental etiology and pathogenesis of Hirschsprung disease. Transl. Res. 2013;162:1–15. doi: 10.1016/j.trsl.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gosain A., Brinkman A.S. Hirschsprung's associated enterocolitis. Curr. Opin. Pediatr. 2015;27:364–369. doi: 10.1097/MOP.0000000000000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luzon-Toro B., Villalba-Benito L., Torroglosa A., Fernandez R.M., Antinolo G., Borrego S. What is new about the genetic background of Hirschsprung disease? Clin. Genet. 2020;97:114–124. doi: 10.1111/cge.13615. [DOI] [PubMed] [Google Scholar]
- 7.Amiel J., Sproat-Emison E., Garcia-Barcelo M., Lantieri F., Burzynski G., Borrego S., Pelet A., Arnold S., Miao X., Griseri P., Brooks A.S., Antinolo G., de Pontual L., Clement-Ziza M., Munnich A. Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- 8.Jaroy E.G., Acosta-Jimenez L., Hotta R., Goldstein A.M., Emblem R., Klungland A., Ougland R. “Too much guts and not enough brains”: (epi)genetic mechanisms and future therapies of Hirschsprung disease - a review. Clin. Epigenetics. 2019;11:135. doi: 10.1186/s13148-019-0718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alves M.M., Sribudiani Y., Brouwer R.W., Amiel J., Antiñolo G., Borrego S., Ceccherini I., Chakravarti A., Fernández R.M., Garcia-Barcelo M.M., Griseri P., Lyonnet S., Tam P.K., van Ijcken W.F., Eggen B.J. Contribution of rare and common variants determine complex diseases-Hirschsprung disease as a model. Dev. Biol. 2013;382:320–329. doi: 10.1016/j.ydbio.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Tilghman J.M., Ling A.Y., Turner T.N., Sosa M.X., Krumm N., Chatterjee S., Kapoor A., Coe B.P., Nguyen K.H., Gupta N., Gabriel S., Eichler E.E., Berrios C., Chakravarti A. Molecular genetic anatomy and risk profile of Hirschsprung's disease. N. Engl. J. Med. 2019;380:1421–1432. doi: 10.1056/NEJMoa1706594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emison E.S., Garcia-Barcelo M., Grice E.A., Lantieri F., Amiel J., Burzynski G., Fernandez R.M., Hao L., Kashuk C., West K., Miao X., Tam P.K., Griseri P., Ceccherini I., Pelet A. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am. J. Hum. Genet. 2010;87:60–74. doi: 10.1016/j.ajhg.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Q., Arnold S., Heanue T., Kilambi K.P., Doan B., Kapoor A., Ling A.Y., Sosa M.X., Guy M., Jiang Q., Burzynski G., West K., Bessling S., Griseri P., Amiel J. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability. Am. J. Hum. Genet. 2015;96:581–596. doi: 10.1016/j.ajhg.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phusantisampan T., Sangkhathat S., Phongdara A., Chiengkriwate P., Patrapinyokul S., Mahasirimongkol S. Association of genetic polymorphisms in the RET-protooncogene and NRG1 with Hirschsprung disease in Thai patients. J. Hum. Genet. 2012;57:286–293. doi: 10.1038/jhg.2012.18. [DOI] [PubMed] [Google Scholar]
- 14.O'Donnell A.M., Nakamura H., Tomuschat C., Marayati N.F., Puri P. Altered expression of KCNG3 and KCNG4 in Hirschsprung's disease. Pediatr. Surg. Int. 2019;35:193–197. doi: 10.1007/s00383-018-4394-2. [DOI] [PubMed] [Google Scholar]
- 15.O'Donnell A.M., Coyle D., Puri P. Deficiency of platelet-derived growth factor receptor-alpha-positive cells in Hirschsprung's disease colon. World J. Gastroenterol. 2016;22:3335–3340. doi: 10.3748/wjg.v22.i12.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura H., O'Donnell A.M., Tomuschat C., Coyle D., Puri P. Altered expression of caveolin-1 in the colon of patients with Hirschsprung's disease. Pediatr. Surg. Int. 2019;35:929–934. doi: 10.1007/s00383-019-04505-1. [DOI] [PubMed] [Google Scholar]
- 17.Gunadi, Kalim A.S., Liana E., Fauzi A.R., Sirait D.N., Afandy D., Kencana S.M.S., Purnomo E., Iskandar K., Makhmudi A. Aberrant UBR4 expressions in Hirschsprung disease patients. BMC Pediatr. 2019;19:493. doi: 10.1186/s12887-019-1879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djomehri S.I., Gonzalez M.E., da Veiga Leprevost F., Tekula S.R., Chang H.Y., White M.J., Cimino-Mathews A., Burman B., Basrur V., Argani P., Nesvizhskii A.I., Kleer C.G. Quantitative proteomic landscape of metaplastic breast carcinoma pathological subtypes and their relationship to triple-negative tumors. Nat. Commun. 2020;11:1723. doi: 10.1038/s41467-020-15283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Readhead B., Haure-Mirande J.V., Funk C.C., Richards M.A., Shannon P., Haroutunian V., Sano M., Liang W.S., Beckmann N.D., Price N.D., Reiman E.M., Schadt E.E., Ehrlich M.E., Gandy S., Dudley J.T. Multiscale analysis of independent Alzheimer's cohorts finds disruption of molecular, genetic, and clinical networks by human Herpesvirus. Neuron. 2018;99:64–82.e67. doi: 10.1016/j.neuron.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W., Zhang X., Wang W., Sun R., Liu B., Ma Y., Zhang W., Ma L., Jin Y., Yang S. Quantitative proteomics analysis of mitochondrial proteins in lung adenocarcinomas and normal lung tissue using iTRAQ and tandem mass spectrometry. Am. J. Transl. Res. 2017;9:3918–3934. [PMC free article] [PubMed] [Google Scholar]
- 21.Gao H., He X., Wu M., Zhang Z., Wang D., Lv L., Su Z., Huang Y. Proteomic analysis of differentially expressed proteins between stenotic and normal colon segment tissues derived from patients with Hirschsprung's disease. Protein J. 2011;30:138–142. doi: 10.1007/s10930-011-9314-4. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S.C., Chen F., Jiang K.L., Yuan Z.W., Wang W.L. Comparative proteomic profiles of the normal and aganglionic hindgut in human Hirschsprung disease. Pediatr. Res. 2014;75:754–761. doi: 10.1038/pr.2014.33. [DOI] [PubMed] [Google Scholar]
- 23.Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F., Hermjakob H., Zhu Y. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher S., Kuna D., Caspary T., Kahn R.A., Sztul E. ARF family GTPases with links to cilia. Am. J. Physiol. Cell Physiol. 2020;319:C404–C418. doi: 10.1152/ajpcell.00188.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monaco L., Murtagh J.J., Newman K.B., Tsai S.C., Moss J., Vaughan M. Selective amplification of an mRNA and related pseudogene for a human ADP-ribosylation factor, a guanine nucleotide-dependent protein activator of cholera toxin. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2206–2210. doi: 10.1073/pnas.87.6.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearring J.N., San Agustin J.T., Lobanova E.S., Gabriel C.J., Lieu E.C., Monis W.J., Stuck M.W., Strittmatter L., Jaber S.M., Arshavsky V.Y., Pazour G.J. Loss of Arf4 causes severe degeneration of the exocrine pancreas but not cystic kidney disease or retinal degeneration. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 28.Raymond C.R., Thompson V.L., Tate W.P., Abraham W.C. Metabotropic glutamate receptors trigger homosynaptic protein synthesis to prolong long-term potentiation. J. Neurosci. 2000;20:969–976. doi: 10.1523/JNEUROSCI.20-03-00969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cromm P.M., Spiegel J., Kuchler P., Dietrich L., Kriegesmann J., Wendt M., Goody R.S., Waldmann H., Grossmann T.N. Protease-resistant and cell-permeable double-stapled peptides targeting the Rab8a GTPase. ACS Chem. Biol. 2016;11:2375–2382. doi: 10.1021/acschembio.6b00386. [DOI] [PubMed] [Google Scholar]
- 30.Burtey A., Wagner M., Hodneland E., Skaftnesmo K.O., Schoelermann J., Mondragon I.R., Espedal H., Golebiewska A., Niclou S.P., Bjerkvig R., Kögel T., Gerdes H.H. Intercellular transfer of transferrin receptor by a contact-, Rab8-dependent mechanism involving tunneling nanotubes. FASEB J. 2015;29:4695–4712. doi: 10.1096/fj.14-268615. [DOI] [PubMed] [Google Scholar]
- 31.Herbst S., Campbell P., Harvey J., Bernard E.M., Papayannopoulos V., Wood N.W., Morris H.R., Gutierrez M.G. LRRK2 activation controls the repair of damaged endomembranes in macrophages. EMBO J. 2020;39 doi: 10.15252/embj.2020104494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivero-Rios P., Romo-Lozano M., Fernandez B., Fdez E., Hilfiker S. Distinct roles for RAB10 and RAB29 in pathogenic LRRK2-mediated endolysosomal trafficking alterations. Cells. 2020;9:1719. doi: 10.3390/cells9071719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norden P.R., Sun Z., Davis G.E. Control of endothelial tubulogenesis by Rab and Ral GTPases, and apical targeting of caveolin-1-labeled vacuoles. PLoS One. 2020;15 doi: 10.1371/journal.pone.0235116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures S1–S2 and Tables S1–S2
Supplemental Table S3
Supplemental Table S4
Supplemental Table S5
Supplemental Table S6
Supplemental Table S7
Supplemental Table S8
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (23) with the dataset identifier PXD021292.