Abstract
Reliable measurement of ligand binding to cell surface receptors is of outstanding biological and pharmacological importance. Resonance energy transfer–based assays are powerful approaches to achieve this goal, but the currently available methods are hindered by the necessity of receptor tagging, which can potentially alter ligand binding properties. Therefore, we developed a tag-free system to measure ligand‒receptor interactions in live cells using the Gaussia luciferase (GLuc) as a bioluminescence resonance energy transfer donor. GLuc is as small as the commonly applied Nanoluciferase but has enhanced brightness, and its proper substrate is the frequently used coelenterazine. In our assay, bystander bioluminescence resonance energy transfer is detected between a GLuc-based extracellular surface biosensor and fluorescent ligands bound to their unmodified receptors. The broad spectrum of applications includes equilibrium and kinetic ligand binding measurements for both labeled and competitive unlabeled ligands, and the assay can be utilized for different classes of plasma membrane receptors. Furthermore, the assay is suitable for high-throughput screening, as evidenced by the identification of novel α1 adrenergic receptor ligands. Our data demonstrate that GLuc-based biosensors provide a simple, sensitive, and cost-efficient platform for drug characterization and development.
Keywords: bioluminescence resonance energy transfer (BRET), G protein–coupled receptor (GPCR), cell surface receptor, signal transduction, molecular pharmacology, Gaussia luciferase (GLuc), NanoLuciferase (NanoLuc), ligand binding
Abbreviations: α1AAR, α1A adrenergic receptor; β2AR, β2 adrenergic receptor; AT1R, AT1 angiotensin receptor; BRET, bioluminescence resonance energy transfer; D1R, D1 dopamine receptor; DN-Dyn, dominant negative form of dynamin2A; EGFR, epidermal growth factor receptor; GLuc, Gaussia luciferase; GPCR, G protein–coupled receptor; NanoLuc, NanoLuciferase; PIP2, phosphatidylinositol 4,5-bisphosphate; PM, plasma membrane; SRE, serum response element; TAMRA‒AngII, red fluorophore–conjugated angiotensin II; TfR, transferrin receptor
The superfamily of plasma membrane (PM) receptors consists of a diverse range of signaling proteins, such as G protein–coupled receptors (GPCRs), tyrosine kinase receptors, enzyme-linked receptors, nutrient receptors, or ion channels. In addition to their important role in sensing environmental signals, their altered function is commonly reported in a wide array of pathological conditions. Accordingly, most of our currently prescribed drugs target cell surface receptors (1). A growing number of evidences indicates that characteristics of drug‒receptor interaction may profoundly shape the pharmacological outcome. In addition to affinity and efficacy, kinetic ligand parameters were also shown to be decisive factors for the clinical action of drugs. Association and dissociation rate constants (kon and koff) of drugs, by defining ligand residence time, have been linked to various drug properties, such as duration of action, efficacy, or occurrence of side effects (2, 3, 4, 5, 6, 7). Therefore, the proper characterization of drug‒receptor interactions represents a major task for drug development. Although various techniques with different advantageous properties are available, they all have serious drawbacks (8). For example, traditional radioligand binding measurements are ponderous and burdensome, especially in case of kinetic binding measurements, and the strict rules required for handling radioactive compounds limit their use. In contrast, fluorescence-based approaches are not hindered by the practical limitations surrounding the use of radioactivity but often suffer from low signal-to-noise ratio due to the autofluorescence of biological samples. By overcoming these limitations, resonance energy transfer–based measurements have revolutionized the application of fluorescent ligands in binding measurements (9, 10, 11, 12, 13). In these assays, resonance energy transfer is detected between a fluorescent or luminescent donor attached to a receptor and a fluorophore conjugated to a receptor ligand. Since the efficiency of resonance energy transfer strongly depends on the molecular proximity, a condition that is met during ligand binding, the signal-to-noise ratio is greatly enhanced. Time-resolved Förster resonance energy transfer (FRET) and bioluminescence resonance energy transfer (BRET) have been successfully used to detect ligand binding of various receptors for equilibrium and kinetic measurements as well. However, these methods require the covalent labeling of the target protein, which may substantially alter the receptor function. Furthermore, the high cost of some substrates may limit their use in high-throughput applications. Therefore, there is a continuous need for new and surpassing assays to assess ligand‒receptor binding. Here we report a novel BRET-based approach to measure ligand binding of cell surface receptors. The method is cost-effective, does not need receptor modification, and is also applicable in high-throughput drug screenings.
Results
Gaussia luciferase is a small and bright luciferase
Since N-terminal receptor tagging with naturally nonsecreted and high-molecular-weight luciferases (such as Renilla luciferase) induces endoplasmic retention and impaired PM expression of several receptors, it was not possible to study receptor‒ligand binding with BRET for a long time (8). This limitation has been overcome by the development of the small luciferase NanoLuciferase (NanoLuc) (14), a variant of the naturally secreted luciferase of the deep-sea shrimp Oplophorus gracilirostris, which has made BRET suitable to detect ligand binding of NanoLuc-tagged receptors. However, the high cost of furimazine, the proper substrate of NanoLuc, seriously hinders the widespread application, especially in high-throughput experiments. Therefore, we aimed to establish a system using an alternative coelenterazine-utilizing luciferase that is also secreted naturally; thus, it may not induce incorrect folding of proteins tagged on the extracellular side. The luciferase of the marine copepod Gaussia princeps (Gaussia luciferase, GLuc) has the same molecular weight as NanoLuc (19 kDa), but its appropriate substrate is a natural substance, the native coelenterazine (Fig. 1A) (15). Since GLuc is rapidly inactivated, we used a mutant of GLuc (GLucM23) (16), which emits light substantially longer and was shown to be even 10-fold brighter than the wildtype enzyme. First, we compared the properties of GLuc and NanoLuc with constructs that label the extracellular surface of the PM (GLuc‒PM and NanoLuc‒PM) by fusing them to a transmembrane domain. We also created versions of the constructs that were intracellularly tagged with Venus to perform expression-normalized comparisons (Fig. 1B). Using bioluminescent and fluorescent image acquisition, we verified that all constructs had proper PM localization (Fig. 1C). As shown in Figure 1, D and E, GLuc was found to have extreme brightness, was even brighter than NanoLuc, and both luciferases produced long-lasting (glow-type) luminescence (Fig. 1D). We tested the effect of different substrates in multiple concentrations. In agreement with previous studies, GLuc was the brightest when native coelenterazine was used (15, 16). The GLuc-emitted luminescence was already sufficiently detectable in the presence of 5 μM coelenterazine; therefore, this concentration was used in the experiments. Similar to previous reports (17, 18), we found that NanoLuc efficiently utilizes some other coelenterazine derivatives as well, such as 2-deoxycoelenterazine (coelenterazine h). Although coelenterazine h was shown to be inferior as a substrate of NanoLuc compared with furimazine, it still induced sufficient brightness to perform BRET measurements in our setup. We compared the emission spectra of GLuc and NanoLuc, and the emission of the former was right-shifted by approximately 20 nm (Fig. 1F), which may be beneficial for the excitation of red-emitting acceptors.
Figure 1.
GLuc is a bright alternative to NanoLuc.A, general characteristics of NanoLuc and GLuc. B, schematic structures of the used constructs. C, bioluminescent and fluorescent images show that the PM-targeted constructs have proper PM localization. Representative images from three independent experiments; the scale bars represent 10 μm. D, temporal decay of bioluminescence of NanoLuc and GLuc. Furimazine (fur; used in 1:200 dilution, stock concentration was not provided by Promega, the suggested dilution in protocol #TM439 is 1:500) or native coelenterazine (coel; 50 μM) was added as substrates, as indicated, and luminescence was continuously measured for 60 min, n = 3, data are mean ± SD. τNanoLuc = 12.2 min and τGLuc = 9.5 min assessed with one phase decay curve fitting. E, peak luminescence of GLuc and NanoLuc upon treatment with different substrates in distinct concentrations: native coelenterazine (coel; 50, 15, 5, 1.5, and 0.5 μM), coelenterazine h (coel h; 50, 15, 5, 1.5, and 0.5 μM), furimazine (fur; 1:200) and DeepBlueC (DBC; 5 μM). In D and E, the experiments were performed with the GLuc–PM–Venus and NanoLuc–PM–Venus constructs, and the measured luminescence was normalized to Venus fluorescence. Scatter dot plots with bars are shown, data are mean + SD. ∗∗∗∗p < 0.0001, one-way ANOVA, Tukey post hoc test, n = 3. F, emission spectra of GLuc and NanoLuc. GLuc–PM and NanoLuc–PM constructs were used for the measurements. Substrates were native coelenterazine (coel; 5 μM) for GLuc and coelenterazine h (coelenterazine h; 5 μM) or furimazine (fur; 1:200) for NanoLuc. Data are means of three independent experiments. GLuc, Gaussia luciferase; NanoLuc, NanoLuciferase; PM, plasma membrane.
GLuc is suitable for ligand binding applications
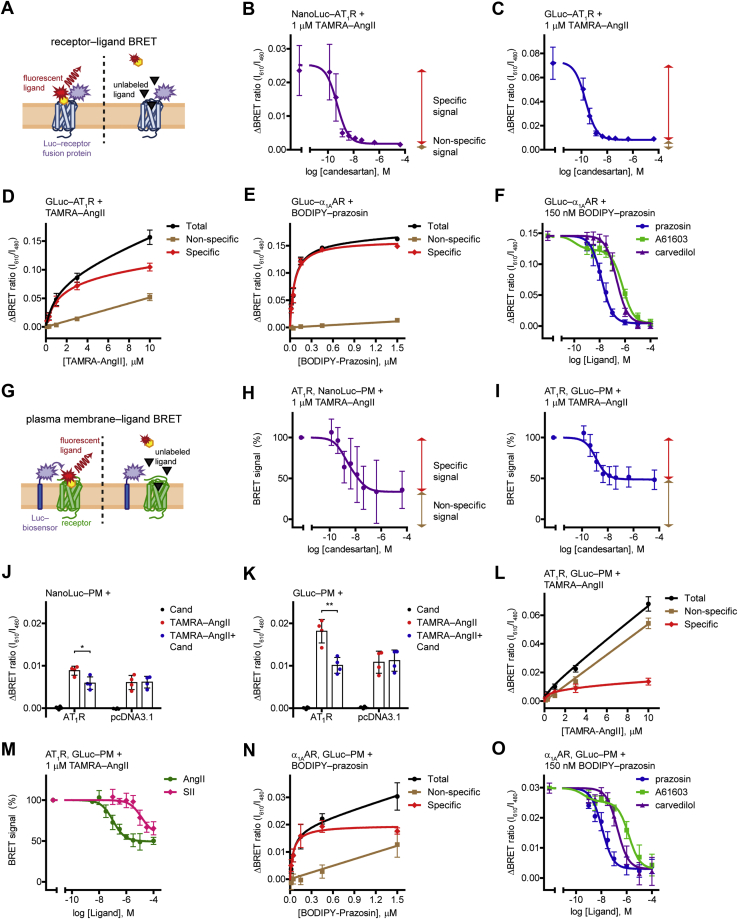
To test the performance of GLuc in BRET assays, we measured receptor‒ligand BRET in a similar manner as it was described in a previous study with NanoLuc (Fig. 2A) (12). We expressed GLuc- or NanoLuc-tagged AT1 angiotensin receptor (AT1R) in HEK 293T cells as BRET donors and treated them with red fluorophore–conjugated angiotensin II (TAMRA‒AngII) as BRET acceptor for 2 h at room temperature to reach equilibrium binding (Fig. 2, B and C). In both cases, the binding of TAMRA‒AngII to the tagged receptors resulted in an increase of the BRET signal, as the molecular proximity between the donor and the acceptor caused resonance energy transfer. The increase of the BRET ratio was higher with GLuc‒AT1R, for which a possible explanation could be the greater overlap between the excitation spectrum of TAMRA and the emission spectrum of GLuc than that of NanoLuc. The specificity of the signal was verified by competitive ligand binding measurements. The competitive AT1R antagonist candesartan prevented a great portion of the BRET signal, proving that this part of the signal originated from specific interaction between TAMRA‒AngII and AT1R. The remaining nonspecific signal reflects random collisions between the donor and acceptor molecules (bystander BRET), whose amplitude is linearly proportional to the acceptor concentration. Accordingly, increasing concentrations of TAMRA‒AngII elevated the nonspecific signal linearly, whereas the specific binding was saturable (Fig. 2D). To avoid possible receptor internalization–related changes in the signal, we always overexpressed an internalization inhibitor protein, the dominant negative form of dynamin2A (DN-Dyn). We confirmed that its application does not disturb the ligand binding properties of the receptor (Fig. S1, A and B). We also tested GLuc in receptor‒ligand BRET measurements for another GPCR, the α1A adrenergic receptor (α1AAR) (Fig. 2, E and F). The green-emitting fluorescent BODIPY FL‒prazosin was applied as the labeled ligand, the α1AAR antagonist prazosin and carvedilol or the α1AAR agonist A61603 were used as unlabeled ligands. Again, the ligand binding of the tagged receptor could be detected with GLuc. The tracer ligand was displaced by all α1AAR ligands, representing that binding of any orthosteric ligand can be measured.
Figure 2.
Measurement of ligand binding of tagged or unmodified receptors in equilibrium.A, schematic representation of the receptor–ligand BRET measurements. Binding of a fluorescent ligand to its luciferase (Luc)-tagged receptor results in the molecular proximity of the BRET donor and acceptor molecules, which promotes resonance energy transfer and increase of the BRET signal. A competitor unlabeled ligand prevents the receptor binding of the fluorescent ligand, thus the specific BRET signal does not develop. B and C, competitive binding curves of NanoLuc–AT1R (B) and GLuc–AT1R (C). One micromolar TAMRA–AngII and increasing concentrations of candesartan were applied. Red and brown two-headed arrows indicate the specific (candesartan-sensitive) and the nonspecific (candesartan-insensitive) signal, respectively. Two site competitive binding curves were fitted, B: IC50_Hi = 520 pM, IC50_Lo = 1 nM; n = 4. C: IC50_Hi = 207 pM, IC50_Lo = 1.3 nM; n = 3. D, concentration dependency of TAMRA–AngII binding to GLuc–AT1R. The BRET ratio was measured in GLuc–AT1R–expressing cells upon treatment of increasing concentrations of TAMRA–AngII. Nonspecific signal was assessed by cotreatment of 30 μM candesartan. Specific signal was calculated by subtracting the nonspecific signal from the total signal. Two site-specific, total, and nonspecific binding curves were fitted (KD_Hi = 1.016 μM and KD_Lo = 26.42 μM), n = 4. E, concentration dependency of BODIPY FL–prazosin binding to GLuc–ɑ1AAR. Total, specific, and nonspecific one site binding curves were fitted (KD = 61.13 nM), n = 4. F, competitive binding curves of GLuc–ɑ1AAR. BODIPY FL–prazosin, 150 nM, was used as the labeled ligand; prazosin, A61603, and carvedilol were added in increasing concentrations. One site competitive binding curves were fitted on the points of the ɑ1AAR antagonists (prazosin, Ki = 4.17 nM and carvedilol, Ki = 62.7 nM); two site competitive binding curve was fitted on the points of the ɑ1AAR agonist A61603 (Ki_Hi = 32.5 pM, Ki_Lo = 187 nM), n = 6. G, schematic representation of the PM–ligand BRET measurements. Cells are cotransfected with untagged receptors and cell surface–targeted luciferase (Luc-biosensor). Fluorescent ligand binding to the receptor leads to its enrichment at the PM, which results in the increase of bystander BRET between the luciferase and the fluorescent ligand. Binding of unlabeled ligand to the receptor prevents the signal. H and I, competitive binding curves of untagged AT1R obtained with NanoLuc–PM (H) or GLuc–PM (I). Similar ligand treatments and curve fitting were used as in B and C; data are presented as the percentage of the ΔBRET ratio induced by TAMRA–AngII in the absence of candesartan. H: 100% = 0.0062 ± 0.0017, IC50_Hi = 1.29 nM, IC50_Lo = 19.6 nM; n = 4. I: 100% = 0.0168 ± 0.004, IC50_Hi = 1.24 nM, IC50_Lo = 248 μM; n = 4. J and K, BRET change upon TAMRA–AngII treatment with NanoLuc–PM (J) or GLuc–PM (K) in the presence or absence of receptor coexpression. Cells were cotransfected with AT1R or pcDNA3.1, 1 μM TAMRA–AngII, 30 μM candesartan, or vehicle treatments were applied, as indicated, n = 4. ∗p = 0.0173, ∗∗p = 0.0029, unpaired, two-tailed t test. L, concentration dependency of TAMRA–AngII binding to AT1R measured with GLuc–PM. Similar ligand treatments and curve fitting were used as in D (KD_Hi = 287 nM and KD_Lo = 18.46 μM), n = 4. M, AT1R binding of agonists with distinct functional effects assessed with GLuc–PM. Two site competitive binding curves were fitted, IC50_Hi = 105 nM for AngII and IC50_Hi = 1.28 μM for SII, IC50_Lo fit values were ambiguous, n = 3. N, concentration dependency of BODIPY FL–prazosin binding to ɑ1AAR measured with GLuc–PM. Total, specific, and nonspecific one site binding curves were fitted (KD = 46.63 nM), n = 4. O, competitive binding curves of prazosin, A61603, and carvedilol to ɑ1AAR assessed with GLuc–PM. Similar curve fitting was applied as in F. Ki = 2.72 nM was for prazosin, Ki = 43.8 nM was for carvedilol, Ki_Hi = 94.8 pM and Ki_Lo = 348.8 nM was for A61603, n = 4. Scatter dot plots with mean + SD are shown in J and K, and data are mean ± SD in the other panels. Measurements were performed after 2-h incubation with the indicated compounds at room temperature. BRET, bioluminescence resonance energy transfer; GLuc, Gaussia luciferase; PM, plasma membrane; TAMRA‒AngII, red fluorophore–conjugated angiotensin II.
NanoLuc‒PM and GLuc‒PM biosensors allow the detection of ligand binding of unmodified GPCRs
It must be emphasized that the phenomenon of bystander BRET not only denotes a background signal but can also be beneficially exploited. Bystander BRET measurements are widely used to detect enrichment of a protein in a particular compartment, where a compartment-targeted molecule and the protein of interest are labeled with BRET partners. The labeling can even be indirect by tagging an interaction partner of the protein, which has the advantage that no molecular modification of the protein under investigation is required. Using bystander BRET, cellular redistribution of proteins, such as receptor internalization and β-arrestin recruitment, or lipid levels in the PM were previously successfully monitored (19, 20, 21, 22, 23). In these experiments, the acceptor accumulation in the donor’s proximity results in the elevation of the BRET signal. We applied a strategy based on similar principles to assess ligand binding of unmodified receptors. Instead of measuring BRET between luciferase-tagged receptors and their fluorescent ligands (“receptor‒ligand BRET”), we detected bystander BRET between luciferase-labeled PM and fluorescent ligands bound to their receptors (“PM‒ligand BRET”) (Fig. 2G). We coexpressed untagged AT1R with GLuc‒PM or NanoLuc‒PM constructs that label the extracellular side of the PM (Fig. 2, H and I). Treatment of cells with TAMRA‒AngII induced an increase of the BRET signal between the PM-targeted donor and TAMRA‒AngII. This signal originated from two types of bystander BRET. A significant portion of the signal could be prevented by the AT1R antagonist candesartan, showing that this signal reflects the specific interaction between TAMRA‒AngII and untagged AT1R, which caused the enrichment of TAMRA‒AngII at the PM. Accordingly, this signal was absent when no receptor was expressed (Fig. 2, J and K). The candesartan-insensitive part of the signal was caused by the other (nonspecific) type of bystander BRET (Fig. 2L), which is due to random collisions between PM–tagged luciferases and unbound acceptor-conjugated ligands. This candesartan-insensitive signal is similar and equally high as in receptor‒ligand BRET measurement (see Fig. 2, D and L). PM‒ligand BRET, in comparison with receptor‒ligand BRET, showed lower amplitude of the specific signal (ΔBRET values of the different setups are shown in Fig. 2, B, C, J, and K). This was in agreement with the fact that resonance energy transfer is greatly sensitive to the distance between donor and acceptor molecules, which is larger when the PM is labeled and not the target receptor. Remarkably, the half-maximal inhibitory concentration of candesartan for the high-affinity binding site (IC50_Hi) was significantly lower in the case of the receptor‒ligand BRET than that of the PM‒ligand BRET (Fig. S1, C and D). Accordingly, the dissociation constant of TAMRA‒AngII for the high-affinity binding site (KD_Hi) of the untagged receptor was smaller than the KD_Hi for the GLuc-labeled receptor (Fig. 2, D and L), demonstrating that receptor tagging altered the ligand binding properties of the receptor. These results are in good agreement with previous studies showing altered ligand binding properties of AT1R upon N-terminal modification (24, 25) and suggest that, although receptor‒ligand BRET is a sensitive approach to detect ligand binding, the results obtained with this method should be interpreted cautiously. Since we achieved larger BRET signal changes with GLuc than with NanoLuc in our system (compare Fig. 2, H with I and Fig. 2, J with K), only GLuc BRET measurements were performed in the further experiments. In principle, a ligand binding assay should be able to detect the binding of any orthosteric ligands regardless of their functional effects. In accordance, we were able to determine the binding of both the high-affinity full agonist angiotensin II (AngII) and the low-affinity β-arrestin‒biased agonist [Sar1,Ile4,Ile8]-angiotensin II (SII) (26) with our assay (Fig. 2M).
We also measured the ligand binding of α1AAR with the PM‒ligand BRET setup (Fig. 2, N and O). With this approach, we were able to measure the competition binding of all three unlabeled ligands, and no significant shift of competition binding curves was observed compared with GLuc‒α1AAR, suggesting that the effect of N-terminal receptor tagging on receptor conformation may vary between GPCRs.
We tested if the amplitude of the specific signal correlates with the amount of the receptor DNA transfected (Fig. S2). We found a positive correlation for both AT1R and α1AAR, suggesting that a high level of receptor expression is advantageous for good signal-to-noise separation.
PM‒ligand BRET using GLuc‒PM as a general approach for ligand binding measurement of multiple families of cell surface receptors
Theoretically, our PM‒ligand BRET system could be utilized for ligand binding detection of any cell surface receptors, if an appropriate fluorescent ligand is available. The approach is quick and easy, as it requires only the coexpression of the GLuc‒PM extracellular surface biosensor and the unmodified receptor, and no further protein fusion procedure is needed.
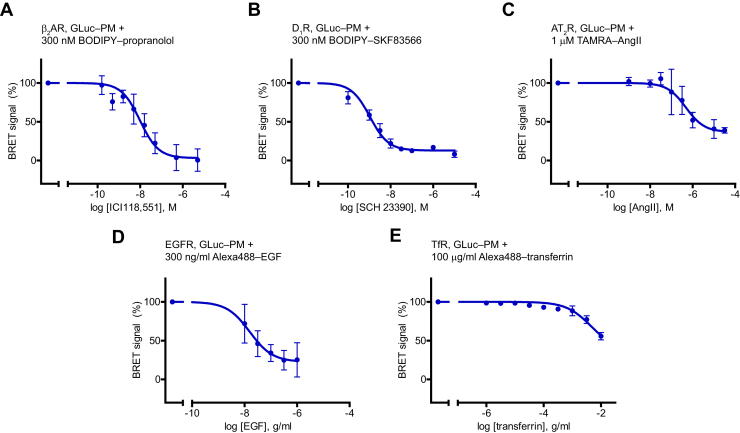
First, we adjusted our system to other GPCRs. We could detect the binding of BODIPY FL‒propranolol to the β2 adrenergic receptor (β2AR). Of interest, we found a consistent increase of the BRET ratio at low concentrations of the β2AR inverse agonist ICI118,552 instead of the expected drop in the signal (Fig. S3). We speculated that the 2-h incubation with the drug at room temperature may induce cell responses, for example, receptor externalization, which could influence the ligand binding results; therefore, we performed incubations on ice. Accordingly, we got regular competition binding curves (Fig. 3A). We could also successfully examine the ligand binding of the D1 dopamine receptor (D1R) or the AT2 angiotensin receptor (AT2R) (Fig. 3, B and C). A study of the latter with our assay could be especially useful in drug screening applications, because AT2R cannot be investigated with receptor signaling assays, as it does not signal in heterologous expression systems (27). It is interesting that AT2R had a TAMRA‒AngII affinity approximately two orders of magnitude higher than that of AT1R (Fig. S4), which explains the observed relatively higher IC50 of AngII to AT2R.
Figure 3.
Application of GLuc–PM to assess ligand binding of multiple cell surface receptor classes. Competitive binding curves of ICI118,551 to β2AR (A, n = 5), of SCH 23390 to D1R (B, n = 3), of angiotensin II (AngII) to AT2R (C, n = 5), of epidermal growth factor (EGF) to EGFR (D, n = 4), and of transferrin to TfR (E, n = 3). In A–E, cells were cotransfected GLuc–PM with the examined receptor, and the indicated labeled ligands were used (300 nM BODIPY FL‒(S)-propranolol, 300 nM BODIPY FL‒SKF83566, 1 μM TAMRA‒AngII, 300 ng/ml biotinylated EGF conjugated to Alexa Fluor 488 streptavidin, or 100 μg/ml Alexa Fluor 488–conjugated human transferrin). Two-hour incubation was made on ice for β2AR and D1R, and at room temperature for the other receptors. For TfR, the BRET ratio was normalized to treatment of transferrin without fluorescent ligand. Data are mean ± SD, expressed as the percentage of the ΔBRET ratio induced by the treatment with fluorescent ligand without unlabeled ligand (0.0142 ± 0.07 (A), 0.0262 ± 0.0023 (B), 0.0119 ± 0.0016 (C), 0.0186 ± 0.0057 (D), and 0.6519 ± 0.0344 (E) values were 100%). One site competitive binding curves were fitted. IC50 values were 9.84 nM (A), 1.07 nM (B), 479 nM (C), 15.8 pg/l (D), and 4.54 μg/l (E). β2AR, β2 adrenergic receptor; AT2R, AT2 angiotensin receptor; GLuc, Gaussia luciferase; PM, plasma membrane; TAMRA‒AngII, red fluorophore–conjugated angiotensin II.
We also performed experiments with non-GPCR cell surface receptors. We examined the epidermal growth factor receptor (EGFR), a prototypical tyrosine kinase receptor (Fig. 3D), and the transferrin receptor (TfR), as an example of cell surface nutrient receptors (Fig. 3E). In both cases, we could detect the specific binding of the Alexa488-conjugated ligands to their receptors, which was prevented by unlabeled ligands. These results show that PM‒ligand BRET using GLuc is a versatile tool for ligand binding measurement of cell surface receptors.
Real-time kinetic detection of ligand binding using GLuc BRET
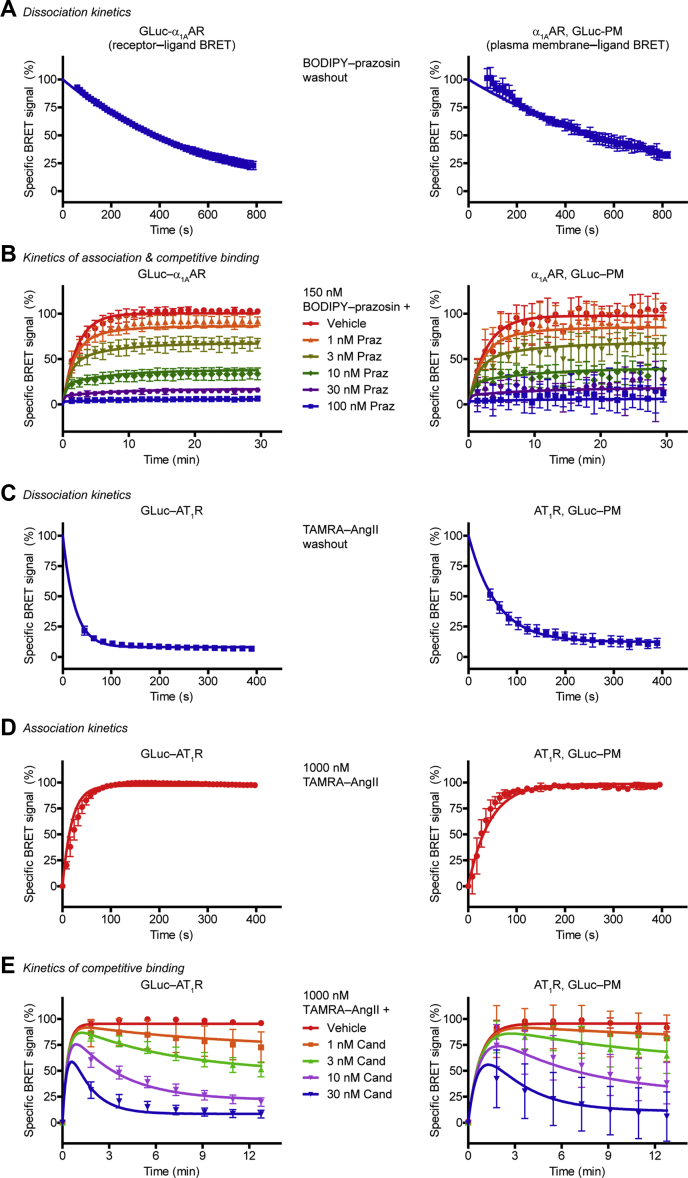
A major advantage of resonance energy transfer–based ligand binding assays is the ability to detect ligand binding in real time and high temporal resolution, allowing simple determination of kinetic ligand parameters. We performed kinetic experiments on α1AAR in live cells with both GLuc BRET setups (receptor‒and PM‒ligand BRET). We measured the dissociation rate constant (koff) of the fluorescent ligand after its washout: the medium was replaced and supplemented with unlabeled ligand to prevent rebinding (Fig. 4A). We could follow the ligand dissociation in both setups. Next, we monitored the association of the fluorescent ligand to the receptor with real-time measurements (Fig. 4B) and determined the association rate constant (kon). Kinetic ligand parameters of unlabeled ligands can be assessed with the help of the Motulsky–Mahan equation (28). The koff and kon values of the unlabeled ligand can be calculated by measuring the ligand binding after simultaneous treatment of the labeled ligand and the unlabeled ligand in different concentrations. With the GLuc BRET system we could successfully determine these parameters of prazosin for α1AAR (Fig. 4B).
Figure 4.
Measurement of kinetic ligand parameters using GLuc.Left panels, kinetic receptor–ligand BRET measurements; right panels, kinetic PM–ligand BRET measurements. A, dissociation kinetics of BODIPY FL–prazosin. After 15-min treatment with 150 nM BODIPY FL–prazosin, the ligand was washed out, and the decline of the BRET ratio was followed. Prazosin was added to prevent rebinding. One phase exponential decay curves were fitted, koff of BODIPY FL–prazosin was 1.84 × 10−3 s−1 for GLuc–ɑ1AAR, and koff was 1.33 × 10−3 s−1 for ɑ1AAR assessed with GLuc–PM, n = 3. B, kinetics of association and competitive ligand binding to ɑ1AAR. Cells were treated with 150 nM BODIPY FL–prazosin and increasing concentrations of prazosin (Praz) simultaneously. kon of BODIPY FL–prazosin was 3.86 × 104 s−1M−1 for GLuc–ɑ1AAR, and kon was 3.22 × 104 s−1 for ɑ1AAR assessed with GLuc–PM. Prazosin kon and koff values were 2.38 × 106 s−1M−1 and 3.51 × 10−3 s−1 for GLuc–ɑ1AAR, while they were 1.7 × 106 s−1M−1 and 2.55 × 10−3 s−1, respectively, for ɑ1AAR determined with GLuc–PM, n = 5. C, dissociation kinetics of TAMRA–AngII. Cells were treated with 1 μM TAMRA–AngII for 15 min, then TAMRA–AngII was washed out and BRET was measured in real time. Candesartan was applied to prevent rebinding. One phase exponential decay curves were fitted, koff of TAMRA–AngII was 0.04046 s−1 for GLuc–AT1R, and koff was 0.01731 s−1 for AT1R assessed with GLuc–PM, n = 3. D, association kinetics of TAMRA–AngII (1 μM). One phase association curves were fitted to calculate kon, kon of TAMRA–AngII was 1.15 × 104 s−1M−1 for GLuc–AT1R, and kon was 5.27 × 103 s−1M−1 for AT1R measured with GLuc–PM, n = 3. E, kinetics of competitive binding to AT1R. Cells were simultaneously treated with 1 μM TAMRA–AngII and candesartan (Cand) in the indicated concentrations, and BRET was followed in real time. Candesartan kon and koff values were 5.4 × 105 s−1M−1 and 1.23 × 10−3 s−1 for GLuc–AT1R, while they were 2.78 × 105 s−1M−1 and 8.33 × 10−4 s−1, respectively, for AT1R assessed with GLuc–PM, n = 5. Data are mean ± SD. α1AAR, α1A adrenergic receptor; BRET, bioluminescence resonance energy transfer; GLuc, Gaussia luciferase; PM, plasma membrane; TAMRA‒AngII, red fluorophore–conjugated angiotensin II.
We also performed similar studies with AT1R. TAMRA‒AngII binds AT1R with two different affinities; however, the proportion of high- and low-affinity states shows a temporal change upon agonist binding because of the ternary complex formation with effectors. For the sake of simplicity, we fitted one site binding curves on the measured points. The koff values of TAMRA‒AngII differed between receptor‒ligand BRET and PM‒ligand BRET setups (Fig. 4C), in agreement with the observation that the N-terminal tagging of AT1R alters its binding properties. Thereafter, we assessed the kon values of TAMRA‒AngII (Fig. 4D) and calculated kon and koff for the unlabeled ligand candesartan in kinetic competitive ligand binding measurements (Fig. 4E).
Screening of receptor ligands with GLuc BRET
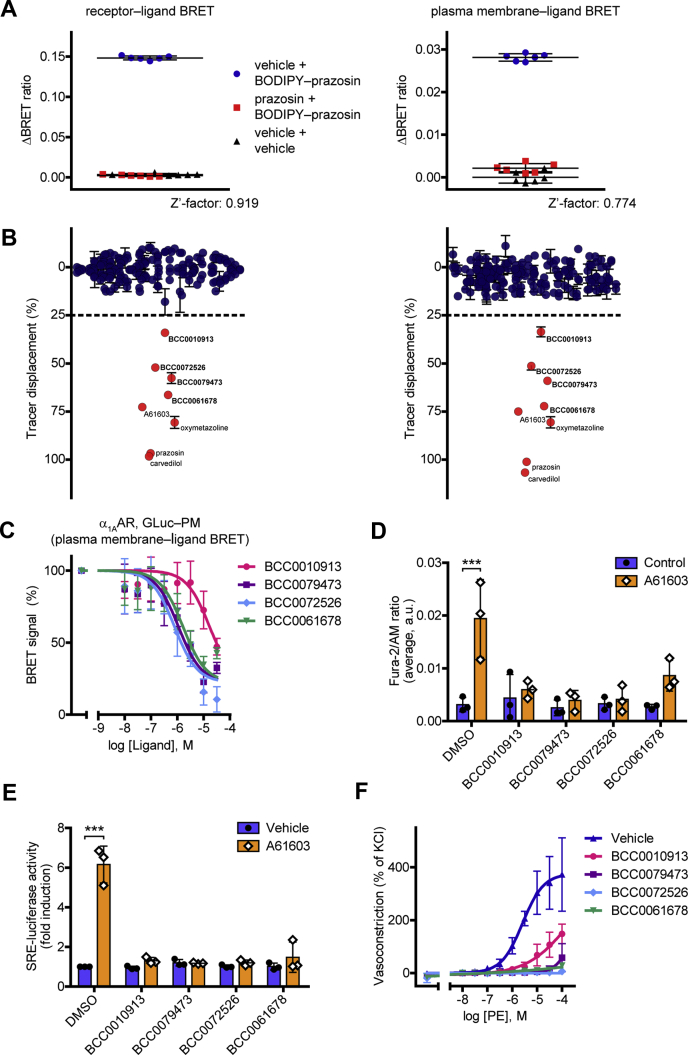
We tested whether our system could be applied for receptor ligand screening. As a model receptor, we chose the α1AAR, a major pharmacological target in the treatment of high or low blood pressure and benign prostate hyperplasia (29). In preliminary experiments, we compared the signal variance in equilibrium and kinetic binding measurements. From this viewpoint, the latter seemed to be more advantageous because of the possibility to apply baseline correction. To statistically characterize the suitability of our system for high-throughput screenings, we calculated the Z’-factor statistical parameter both for receptor‒ligand and PM‒ligand BRET (Fig. 5A) (30). It was 0.919 and 0.774, respectively, proving that our system could also be applied in high-throughput screenings. Thereafter, we made a test screening with a compound library of 180 compounds (Fig. 5B and Table S1). As positive controls, four known α1AAR ligands were also tested. We defined those compounds as hits that induced at least 25% decrease of the signal (induced 25% displacement of the tracer ligand). Four compounds were found as hits (Fig. S5), which were previously not known to bind to α1AAR. The hits were further characterized. Their KD values were assessed using both ligand binding setups (Fig. 5C and Fig. S6A). Their functional effects on α1AAR, a receptor known to trigger phospholipase C–mediated phosphatidylinositol 4,5-bisphosphate (PIP2) hydrolysis and concomitant calcium signaling, were also investigated. All four hits were able to prevent the cytosolic calcium release induced by the α1AAR agonist A61603 (Fig. 5D). Similarly, the hits abolished the A61603-induced serum response element (SRE)-luciferase activation, a reporter of mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling and serum response factor activity (Fig. 5E). Consistently with these results, they shifted the A61603 concentration‒response curve of PIP2 cleavage to the right, representing their competitive α1AAR antagonistic property (Fig. S6B). Moreover, they alone had no effect on the vascular tone of mouse aortic rings but antagonized the vasoconstrictor effect of phenylephrine, a well-known α1 adrenergic receptor agonist (Fig. 5F). These results demonstrate that our GLuc-based assay could be utilized as a powerful novel tool for drug screening.
Figure 5.
Receptor ligand screening with GLuc BRET.A, statistical validation of the suitability of receptor–ligand BRET and PM–ligand BRET for high-throughput screenings. After 20-min prazosin (10 μM) or dimethyl sulfoxide pretreatment, 150 nM BODIPY FL–prazosin was added to the cells. BRET was normalized to vehicle-treatment. Z’-factor values were 0.919 (GLuc–ɑ1AAR) and 0.774 (ɑ1AAR with GLuc–PM), respectively. Results of a representative experiment with six technical replicates are shown, n = 3. Data are mean ± SD. B, screening for ɑ1AAR ligands. Left panel, receptor–ligand BRET (GLuc–ɑ1AAR); right panel, PM–ligand BRET (ɑ1AAR with GLuc–PM). A 180-compound library was tested, and A61603, oxymetazoline, prazosin, and carvedilol were used as positive controls. After 20-min unlabeled ligand (10 μM) pretreatment, the displacement of 150 nM BODIPY FL–prazosin was assessed. The screening was performed in duplicate, data are mean ± range. C, competitive ligand binding of the hits using the PM–ligand BRET. Increasing concentrations of hits and 375 nM BODIPY FL–prazosin were added to the cells, n = 4. Data are mean ± SD. One site competitive ligand binding curves were fitted, the Ki values were 1.845 μM (BCC0010913), 121.6 nM (BCC0079473), 86.12 nM (BCC0072526), and 180.4 nM (BCC0061678). D, effects of the hits on calcium signaling of ɑ1AAR. Cytosolic calcium measurements were performed in Fura-2/AM-loaded cells that overexpressed ɑ1AAR. The cells were first treated with the hits (10 μM) or vehicle for 1 min, then 100 pM A61603 was used as a stimulus for 3 min. Averages of the signals are shown, data are mean ± SD, n = 3. The compounds did not induce significant calcium signal but prevented the A61603 effect (two-way ANOVA with Tukey post hoc test, ∗∗∗p = 0.0001). E, functional activity of the hits assessed with the SRE-luciferase reporter. After 20-min pretreatment with vehicle or the tested compound (30 μM), 2-h costimulation with 1 nM A61603 or vehicle was applied. All the hits were antagonists, as they did not induce SRE-luciferase activity but prevented the A61603 effect, n = 3. Scatter dot plots with bars are shown, data are mean + SD (two-way ANOVA with Tukey post hoc test, performed on the nonnormalized data, ∗∗∗p = 0.0003). F, the identified new ɑ1AAR ligands antagonize the phenylephrine-induced vasoconstriction. Myography measurements were performed on murine aortic rings. Data are mean ± SD. Concentration–response curves were fitted. No significant effect of hit treatment on resting tension was found (one-way ANOVA with Tukey post hoc test), n = 3. α1AAR, α1A adrenergic receptor; BRET, bioluminescence resonance energy transfer; GLuc, Gaussia luciferase; PM, plasma membrane; SRE, serum response element.
Discussion
Over the last 2 decades, resonance energy transfer–based methods are emerging in pharmacological studies, which is particularly true for ligand binding investigations (8, 13). Time-resolved FRET or BRET (using NanoLuc) was successfully adapted to study various receptors, including GPCRs and receptor tyrosine kinases (9, 11, 12, 31, 32, 33, 34). Here we described a novel ligand binding assay that has the merits of previous resonance energy transfer–based methods and overcomes many of their limitations. Like other resonance energy transfer–based approaches, our system has remarkably low signal variance, is simple and fast to perform as it lacks laborious experimental steps, and is harmless to health. In addition, resonance energy transfer–based systems allow ligand binding kinetics to be monitored in real time with exceptional temporal resolution. The importance of knowledge of ligand binding kinetics cannot be emphasized enough, as several studies have shown that the in vivo action of drugs correlates much better with kinetic ligand properties, such as kon, koff, or ligand residence time, than with ligand parameters assessed in equilibrium. Moreover, our approach is cost-effective, since it applies the extremely bright Gaussia luciferase that utilizes the native coelenterazine as a substrate. It should be noted that inexpensive substrates are available for NanoLuc as well, for example, coelenterazine h, but the brightness produced is significantly lower.
Previous resonance energy transfer–based approaches suffered from the need for molecular biological modification of the protein of interest, since the generation of properly functioning fusion proteins sometimes requires tremendous effort and the potential risk of altered function cannot be completely ruled out even with cautious design and application of appropriate controls. Accordingly, several cases of tag- or even linker-induced functional changes for multiple receptors have been described, such as endoplasmic reticulum retention, altered ligand affinity, or modified effector coupling (12, 24, 25, 35, 36). These examples may be just the tip of the iceberg, as malfunctioning constructs are not likely to be published. To examine ligand binding of unlabeled receptors, we developed a bystander BRET-based approach. Although we experienced a notable drop in the signal amplitude, the signal deviation remained low; thus, the separation between signal and background remained high. We believe that this price is significantly outweighed by the benefit of being able to investigate unmodified receptors.
Since only the unmodified receptor and the cell surface–targeted bioluminescent sensor have to be expressed in a cell line, our assay can be readily applied to study any cell surface receptor when a fluorescent ligand is available. We adapted our system for various GPCRs and non-GPCRs, including AT1R, AT2R, α1AAR, β2AR, D1R, EGFR, and TfR. We prefer GLuc to NanoLuc as a bioluminescent donor because of the higher effect size in our system, but NanoLuc can also be used based on similar logic. In addition to these proteins, we always coexpressed dominant-negative dynamin to prevent possible interference caused by internalization. We found a positive correlation between the amount of receptor DNA transfected and the amplitude of the specific BRET signal. Therefore, it must be noted that the assay is more suitable for measuring ligand binding of highly expressed receptors, and endogenous receptors with low copy number may not reach the detection limit of the method. The system allows the determination of any ligand binding properties and is particularly useful for kinetic ligand binding measurements as the assay allows real-time detection. Theoretically, the signal of the PM‒ligand BRET setup may be influenced by the change in distance between the fluorescent ligand-bound receptor and the GLuc‒PM biosensor. It is known that subtle changes of the bystander BRET between luciferase-tagged receptors and PM–targeted fluorescent proteins can also reflect the lateral movement of the receptor between PM compartments (20, 37). However, we found no indication that this phenomenon would contribute significantly to the measured signal in the ligand association experiments with AT1R or α1AAR. In addition, this issue is unlikely in dissociation experiments, because alterations in the localization of the unlabeled receptor are unseen by our assay after dissociation of the fluorescent ligand.
In high-throughput applications, ligand binding assays offer several advantages over approaches for assessing signaling pathways. Receptors are known to activate multiple signaling pathways, which are not equally modulated by certain ligands (biased agonists) (38, 39, 40). In order to achieve the highest possible hit rate with functional assays, multiple signaling pathways must be simultaneously examined with distinct assays, and different assay conditions are required for agonists and antagonists. This means that a compound needs to be screened in several setups, significantly increasing the cost and the time of the screening. These disadvantages are absent in ligand binding measurements; a single setup is sufficient to perform a receptor ligand screen. We proved that our assay is suitable for high-throughput screenings. As a demonstration, we screened a compound library of 180 molecules for α1AAR ligands with our system. We found four previously unknown α1AAR ligands, which acted as α1AAR antagonists in multiple functional assays. The relatively high hit rate is consistent with the observation that a large number of drugs exert off-target effects through α1AAR, causing orthostatic hypotension and vertigo (41).
In summary, we have developed a novel ligand binding assay that allows the determination of ligand binding with low signal variance and high temporal resolution, providing a powerful tool for drug characterization and development.
Experimental procedures
Materials
TAMRA‒AngII was purchased from AnaSpec. BODIPY FL‒prazosin, biotinylated EGF conjugated to Alexa Fluor 488 streptavidin, and Alexa Fluor 488–conjugated human transferrin were bought from Thermo Scientific. BODIPY FL‒SKF83566 and BODIPY FL‒(S)-propranolol were from HelloBio. Candesartan and SCH 23390 were purchased from Tocris. [Sar1,Ile4,Ile8]-angiotensin II was bought from Bachem. The compound library consisting of randomly selected 180 molecules was from BioAscent. Native coelenterazine and coelenterazine h were from Regis Technologies. Furimazine and the Luciferase Assay System were from Promega; DeepBlueC (coelenterazine 400a) was from Perkin-Elmer. Fura-2/AM was from Molecular Probes. ICI118,552 EGF, transferrin, angiotensin II, A61603, prazosin, and otherwise not stated materials were from Sigma-Aldrich.
Plasmid constructs
The plasmid constructs coding the rat AT1R, α1AAR, β2AR, EGFR, and the PM PIP2 BRET biosensor (L10‒Venus‒T2A‒PLCδ1-PH‒SLuc) were described (23, 42, 43). SRE-luciferase, containing the SRE promoter region fused with the coding sequence of firefly luciferase, was from Promega. The cDNA of human D1R and hemagglutinin‒dynamin2A‒K44A (DN-Dyn) were kind gifts from Dr Marc G. Caron and Dr Kazuhisa Nakayama, respectively. The coding sequence of untagged rat AT2R without the untranslated regions was polymerase chain reaction (PCR) amplified and inserted into pEYFP-N1 by replacing YFP. The C-terminal GFP-tag of TfR‒GFP (kindly provided by Dr Tamás Balla) was replaced by hemagglutinin-tag to create TfR‒hemagglutinin. To generate GLuc‒PM, the DNA sequence of Gaussia luciferase signal peptide followed by GLucM23 (humanized GLuc harboring the K50E, M60L, V113D, M127I, and G184D mutations (16)) fused to the transmembrane domain of platelet derived growth factor (PM) with a flexible (GGGSGGGSRSGGGSGGGSGGGS) linker was gBlock synthetized and was inserted into pcDNA3.1 vector. To create NanoLuc‒PM, the coding sequence of NanoLuc N-terminally fused with the secretion signal peptide of interleukin-6 was gBlock synthetized and the signal peptide-fused GLucM23 was replaced with that in GLuc‒PM. N-terminally GLuc- or NanoLuc-tagged AT1Rs harboring a short linker (GGGSGGGS) were generated by PCR amplification of the sequence of rat AT1R to replace PM in GLuc‒PM and NanoLuc‒PM, respectively. A similar strategy was used to generate GLuc‒α1AAR. To create GLuc‒PM‒Venus and NanoLuc‒PM‒Venus, GLuc‒PM and NanoLuc‒PM were PCR amplified without stop codons and were inserted into pVenus-N1 vector before the sequence of monomeric Venus.
Cell culture and transfection
HEK 293T cells (from American Type Culture Collection: CRL-3216) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 100 IU/ml penicillin/streptomycin. To visualize the cellular localization of the probes, the cells were plated on coverslips on the day before transfection, and the adherent cells were transfected with the calcium phosphate precipitation method: for each well (24-well plate), 1 μg plasmid DNA was diluted in 45 μl distilled water, to this 5 μl 2.5 M CaCl2 was rapidly added, then 50 μl 2× HEBS (Hepes-buffered salt solution, containing 274 mM NaCl, 15 mM glucose, 42 mM Hepes, 10 mM KCl, 1.4 mM Na2HPO4, pH 7.1) was added slowly, and the solution was added dropwise to the culture medium (1 ml) of cells. The amounts and volumes for calcium phosphate precipitation were proportionally increased or decreased for wells with bigger or smaller surfaces. For cytosolic calcium measurements, the cells were plated on IBIDI μ-Slide 8-well plates and a similar procedure was applied. Otherwise, the transfections were performed in suspension using the calcium phosphate precipitation method, and the cells were plated on white 96-well plates. Mixture volumes were multiplied by the number of wells transfected, plasmid DNAs were diluted in 9 μl distillated water, 1 μl 2.5 M CaCl2, and 10 μl 2× HEBS per well, as described above, and the transfection solution was added dropwise to HEK 293T cells suspended in supplemented DMEM (8 × 104 cells in 200 μl of medium per well). A volume of 200 μl of the mixture was seeded on each well. For receptor‒ligand BRET measurements, the plasmid DNA of luciferase-tagged receptor and DN-Dyn were transfected in 1:4 ratio (0.05 and 0.2 μg per well). In the PM‒ligand BRET measurements, the receptor:luciferase‒PM:DN-Dyn plasmid DNA ratio was 20:1:5 (0.2, 0.01, and 0.05 μg per well), unless otherwise stated.
Bioluminescent and fluorescent image acquisition
For live cell image acquisition, coverslips were held in Chamlide magnetic chambers in Hanks' balanced salt solution media. Images were collected with a Nikon Ti2 inverted microscope with a Hamamatsu ORCA Flash 4 V3 sCMOS camera using a 100× objective. Five micromolar coelenterazine h and 5 μM native coelenterazine were used as substrates for NanoLuc and GLuc, respectively. Luminescence was detected by applying 10-s exposure time with 4× averaging. Fluorescence was recorded by excitating at 488 nm laser, and emission was detected using a Quad band filter. The images were individually processed, looking up tables were adjusted in each image distinctively to achieve high contrast.
Luciferase activity measurements
The emission spectra of GLuc‒PM and NanoLuc‒PM were detected using a Thermo Scientific Varioskan Flash multimode plate reader (spectral scanning mode using a monochromator). Since the peak total luminescence of GLuc surpassed the detection limit of the Varioskan Flash plate reader, fluorescence and luminescence intensities of GLuc‒PM‒Venus or NanoLuc‒PM‒Venus were measured with a BMG Labtech CLARIOstar microplate reader. Venus fluorescence was assessed by exciting at 497/15 nm and measuring emission at 540/20 nm. Total luminescence was measured without filter (gain 1100, integration time 0.2 s).
Bioluminescence resonance energy transfer measurements
All measurements were performed on adherent cells. On the day after transfection, the cells were washed and the medium was changed to modified Krebs–Ringer medium (120 mM NaCl, 10 mM glucose, 10 mM Na-Hepes, 4.7 mM KCl, 1.2 mM CaCl2, 0.7 mM MgSO4, pH 7.4). Equilibrium binding studies were performed after 2-h incubation of labeled and unlabeled ligands at room temperature. Incubations were made on ice for β2AR and D1R binding measurements. Luminescence was measured using a Varioskan Flash multimode plate reader at 27 °C. For NanoBRET measurements, light intensities were measured with 460/20- and 610/60-nm bandpass filters after addition of 5 μM coelenterazine h. GLuc BRET measurements were performed after treatment with 5 μM native coelenterazine, TAMRA‒AngII binding was monitored using 480/20- and 610/60-nm filters, whereas 480/20- and 530/20-nm filters were applied in BRET measurements with green-emitting (BODIPY FL- or Alexa Fluor 488–conjugated) fluorescent ligands.
To assess Koff of fluorescent ligands, the ligands were preincubated with the cells for 15 min. Thereafter, the cells were washed and the medium was replaced with media containing coelenterazine and unlabeled ligand (10 μM candesartan for AT1R, 10 μM prazosin for α1AAR) to prevent rebinding. To determine the maximal signal, control cells were treated with labeled ligand and coelenterazine, but no unlabeled ligand was added. Basal BRET was assessed in cells treated with no labeled ligand. For analysis, basal BRET was always subtracted. For Kon measurements of fluorescent ligands, cells were treated with coelenterazine, baseline BRET was monitored for 3 min, and then the cells were treated with fluorescent ligands. Kinetic parameters of unlabeled ligands were measured similarly. After baseline BRET assessment, the cells were treated simultaneously with fluorescent ligand and increasing concentrations of unlabeled ligand.
PM PIP2 levels were monitored using a PIP2 BRET biosensor (L10–Venus–T2A–PLCδ1-PH–SLuc) (23). Bystander BRET was measured between Super Renilla luciferase-tagged PLCδ1-PH (the PIP2-binding domain of PLCδ1) and PM–targeted Venus in cells coexpressing PIP2 BRET biosensor and α1AAR. The BRET ratio decreased upon treatment with α1AAR agonist, which reflects the phospholipase C–mediated PIP2 cleavage. A 5-min pretreatment with the hit compounds (10 μM) was applied, then the cells were stimulated with increasing concentrations of α1AAR agonist A61603. First, the agonist-induced BRET change was determined (the BRET ratio after pretreatment was subtracted from the average of the BRET ratios measured in the first 5 min after stimulation), then the BRET signal was expressed in the percent of the 10 μM A61603-induced BRET change in vehicle-pretreated cells.
The screening was performed using a compound library harboring 180 compounds (Table S1). The compounds were randomly recruited from the BioAscent library of approximately 1.2 × 106 molecules. To obtain a better signal-to-noise ratio, 1.75 times more cells (1.4 × 105) were seeded per well on 96-well plates. Outer wells of plates were not used to rule out the edge effect; only the inner 60 wells were measured. Before the measurements, the cells were pretreated with the compounds for 20 min in 10 μM (which was diluted to 6 μM after substrate and ligand treatment). After addition of coelenterazine, baseline BRET was determined for 5 min, then the cells were stimulated with 150 nM BODIPY FL–prazosin or vehicle. The same experimental procedure was used for Z’-factor assessment, BRET was monitored in six wells per condition on 96-well plates, and the Z’-factor was calculated as previously described (30).
Cytosolic calcium concentration measurements
Adherent HEK 293T cells overexpressing α1AAR were treated with 200 μM sulfinpyrazone and loaded with the ratiometric fluorescent dye Fura-2/AM (2 μM, 45 min). Single-cell calcium measurements were performed using an inverted microscope (Axio Observer, Zeiss) equipped with a 40× oil immersion objective (Fluar, Zeiss) and a Cascade II camera (Photometrics). Excitation wavelengths were set by a random access monochromator connected to a xenon arc lamp (DeltaRAM, Photon Technology International). For ratiometric measurements of Fura-2 excitation wavelengths of 340 and 380 nm were selected combined with a 505-nm dichroic filter and a 525/36-nm emission filter set. Data acquisition was performed by the MetaFluor (Molecular Devices) software. Images were acquired every 5 s. After a control period (1 min), cells were treated with vehicle (dimethyl sulfoxide) or test compound (10 μM), then the cells were stimulated with 100 pM A61603 for 3 min. Image analysis was performed with ImageJ software; the ratio of fluorescent intensities measured at 340 and 380 nm excitations was taken. About 150 to 200 regions of interest representing individual cells from each experiment and each treatment were measured, and fluorometric ratios were normalized to the control period.
SRE-luciferase activity measurement
HEK 293T cells were transiently transfected with SRE-luciferase and receptor plasmids in suspension and were seeded on 96-well white plates. On the next day, the cells were serum starved for 2 h by replacing the media to DMEM containing 1% penicillin/streptomycin and 1% bovine serum albumin. Then the medium was changed to modified Krebs–Ringer medium, and the cells were pretreated with vehicle or the investigated compounds (30 μM) for 20 min, then 2-h costimulation with 1 nM A61603 or vehicle was applied at 37 °C. Then the cells were plated on ice and were lysed with passive lysis buffer (Promega). After 20-min rocking at room temperature, the Luciferase Assay System (Promega) substrate was added and luminescence was measured with a Thermo Scientific Varioskan Flash multimode plate reader at 27 °C.
Wire myography
Myography experiments were performed as described (44). Male C57BL/6 mice were terminated by cervical dislocation, and the vascular system was perfused through the left ventricle and a right atrial slit with Kreb’s solution containing 119 mM NaCl, 4.7 mM KCl, 2.5 mM CaCl2, 1.17 mM MgSO4, 20 mM NaHCO3, 1.18 mM KH2PO4, 0.027 mM EDTA, and 10 mM glucose (pH 7.4). Abdominal aortas were removed and placed into Kreb’s solution. Aortic rings (3–4 mm), five segments from each animal, were mounted onto a multichamber isometric myograph system (Danish Myo Technology). The organ chambers were filled with Kreb’s solution kept at 37 °C and bubbled with carbogen gas (5% CO2 and 95% O2). Recording of isometric tension was performed with the Powerlab data acquisition system and the LabChart evaluation program (ADInstruments). The segments were allowed to equilibrate for 30 to 40 min, and the resting tension was adjusted to 10 mN. First, contractions were elicited by hyperkalemic (124 mM) solution for 1 min. Endothelial integrity was tested with the vasodilatory response by acetylcholine (100 nM) after phenylephrine precontraction (100 nM). Before compound treatments, a 3-min KCl reference contraction was induced, then resting tension was adjusted again with washing steps. Treatments with the hit compounds (10 μM) were applied for 15 min. Next, phenylephrine was administered in cumulative concentrations (10 nM–100 μM). Vasoconstrictor responses were expressed as percentages of the 3-min KCl-induced reference contraction. All experimental procedures were reviewed and approved by the Animal Care Committee of Semmelweis University (Budapest, Hungary).
Mathematical and statistical analysis
Data plotting, statistical analysis, and curve fittings were performed with GraphPad Prism 9 software. One or two site-specific binding curves were fitted on the specific signal points of equilibrium binding measurements. With the use of the determined KD values, total (one site or two site) and nonspecific binding were also fitted. One or two site competition binding curves were fitted in the case of equilibrium competitive ligand binding measurements. Ki values were calculated with the Cheng–Prusoff equation (45). To determine labeled ligand–receptor Koff in measurements of Figure 4, one phase exponential decay curves were fitted. Thereafter, the “association kinetic (one ligand concentration)” equation was used to calculate labeled ligand–receptor Kon with constrained Koff and Bmax (maximum specific binding, determined in equilibrium binding measurements). Koff and Kon values of the unlabeled ligand–receptor interaction were determined with the “Kinetics of competitive binding” (Motulsky–Mahan) equation (28). To calculate τ, one phase association curves were fitted. The results of curve fits for ligand binding experiments in the main figures are summarized in Table S2.
For statistical comparison, unpaired two-tailed t test and one-way or two-way ANOVA with Tukey post hoc test were applied.
Data availability
GLuc‒PM and NanoLuc‒PM biosensors will be available from Addgene after publication.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article..
Supporting information
This article contains supporting information.
Acknowledgments
We are grateful to the Nikon Center of Excellence (Nikon-KOKI Imaging Center of the Institute of Experimental Medicine, Budapest) for providing us the Nikon Ti2 inverted microscope and to Dr László Barna for the fruitful discussions. We are thankful for the technical assistance of Ilona Oláh and Eszter Halász.
Author contributions
A. D. T., G. T., and L. H. conceptualized the work; A. D. T., D. G., and S. P. performed the BRET and luminometric measurements; G. T.. performed the calcium measurements; E. S. -K. performed the myography experiments. All authors analyzed the results, wrote the manuscript, and approved the final version of the manuscript.
Funding and additional information
This work was supported by the Hungarian National Research, Development and Innovation Fund (grants NKFI K116954 and NVKP_16-1-2016-0039) and the New National Excellence Program of the Ministry of Human Capacities (ÚNKP-17-3-III-SE-23, ÚNKP-20-3-II-SE-33).
Edited by Henrik Dohlman
Supporting information
References
- 1.Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G., Karlsson A., Al-Lazikani B., Hersey A., Oprea T.I., Overington J.P. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Copeland R.A., Pompliano D.L., Meek T.D. Drug–target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 3.de Witte W.E.A., Danhof M., van der Graaf P.H., de Lange E.C.M. In vivo target residence time and kinetic selectivity: The association rate constant as determinant. Trends Pharmacol. Sci. 2016;37:831–842. doi: 10.1016/j.tips.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Klein Herenbrink C., Sykes D.A., Donthamsetti P., Canals M., Coudrat T., Shonberg J., Scammells P.J., Capuano B., Sexton P.M., Charlton S.J., Javitch J.A., Christopoulos A., Lane J.R. The role of kinetic context in apparent biased agonism at GPCRs. Nat. Commun. 2016;7:10842. doi: 10.1038/ncomms10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Copeland R.A. The drug–target residence time model: A 10-year retrospective. Nat. Rev. Drug Discov. 2016;15:87–95. doi: 10.1038/nrd.2015.18. [DOI] [PubMed] [Google Scholar]
- 6.Sykes D.A., Moore H., Stott L., Holliday N., Javitch J.A., Lane J.R., Charlton S.J. Extrapyramidal side effects of antipsychotics are linked to their association kinetics at dopamine D2 receptors. Nat. Commun. 2017;8:763. doi: 10.1038/s41467-017-00716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vauquelin G. Effects of target binding kinetics on in vivo drug efficacy: k off, k on and rebinding. Br. J. Pharmacol. 2016;173:2319–2334. doi: 10.1111/bph.13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoddart L.A., Kilpatrick L.E., Hill S.J. NanoBRET approaches to study ligand binding to GPCRs and RTKs. Trends Pharmacol. Sci. 2018;39:136–147. doi: 10.1016/j.tips.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Castro M., Nikolaev V.O., Palm D., Lohse M.J., Vilardaga J.-P. Turn-on switch in parathyroid hormone receptor by a two-step parathyroid hormone binding mechanism. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16084–16089. doi: 10.1073/pnas.0503942102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohse M.J., Nuber S., Hoffmann C. Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacol. Rev. 2012;64:299–336. doi: 10.1124/pr.110.004309. [DOI] [PubMed] [Google Scholar]
- 11.Emami-Nemini A., Roux T., Leblay M., Bourrier E., Lamarque L., Trinquet E., Lohse M.J. Time-resolved fluorescence ligand binding for G protein–coupled receptors. Nat. Protoc. 2013;8:1307–1320. doi: 10.1038/nprot.2013.073. [DOI] [PubMed] [Google Scholar]
- 12.Stoddart L.A., Johnstone E.K.M., Wheal A.J., Goulding J., Robers M.B., Machleidt T., Wood K.V., Hill S.J., Pfleger K.D.G. Application of BRET to monitor ligand binding to GPCRs. Nat. Methods. 2015;12:661–663. doi: 10.1038/nmeth.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoddart L.A., White C.W., Nguyen K., Hill S.J., Pfleger K.D.G. Fluorescence- and bioluminescence-based approaches to study GPCR ligand binding. Br. J. Pharmacol. 2016;173:3028–3037. doi: 10.1111/bph.13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall M.P., Unch J., Binkowski B.F., Valley M.P., Butler B.L., Wood M.G., Otto P., Zimmerman K., Vidugiris G., Machleidt T., Robers M.B., Benink H.A., Eggers C.T., Slater M.R., Meisenheimer P.L. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaskova Z.M., Tsarkova A.S., Yampolsky I.V. 1001 lights: Luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev. 2016;45:6048–6077. doi: 10.1039/c6cs00296j. [DOI] [PubMed] [Google Scholar]
- 16.Lindberg E., Mizukami S., Ibata K., Fukano T., Miyawaki A., Kikuchi K. Development of cell-impermeable coelenterazine derivatives. Chem. Sci. 2013;4:4395. doi: 10.1002/chem.201302002. [DOI] [PubMed] [Google Scholar]
- 17.Inouye S., Sato J., Sahara-Miura Y., Yoshida S., Kurakata H., Hosoya T. C6-Deoxy coelenterazine analogues as an efficient substrate for glow luminescence reaction of nanoKAZ: The mutated catalytic 19kDa component of Oplophorus luciferase. Biochem. Biophys. Res. Commun. 2013;437:23–28. doi: 10.1016/j.bbrc.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Yano H., Cai N.S., Javitch J.A., Ferré S. Luciferase complementation based-detection of G-protein-coupled receptor activity. Biotechniques. 2018;65:9–14. doi: 10.2144/btn-2018-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan T.-H., Liu Q., Li C., Wu G., Lambert N.A. Sensitive and high resolution localization and tracking of membrane proteins in live cells with BRET. Traffic. 2012;13:1450–1456. doi: 10.1111/j.1600-0854.2012.01401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balla A., Tóth D.J., Soltész-Katona E., Szakadáti G., Erdélyi L.S., Várnai P., Hunyady L. Mapping of the localization of type 1 angiotensin receptor in membrane microdomains using bioluminescence resonance energy transfer-based sensors. J. Biol. Chem. 2012;287:9090–9099. doi: 10.1074/jbc.M111.293944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyombolai P., Tóth A.D., Tímár D., Turu G., Hunyady L. Mutations in the “DRY” motif of the CB1 cannabinoid receptor result in biased receptor variants. J. Mol. Endocrinol. 2015;54:75–89. doi: 10.1530/JME-14-0219. [DOI] [PubMed] [Google Scholar]
- 22.Namkung Y., Le Gouill C., Lukashova V., Kobayashi H., Hogue M., Khoury E., Song M., Bouvier M., Laporte S.A. Monitoring G protein-coupled receptor and β-arrestin trafficking in live cells using enhanced bystander BRET. Nat. Commun. 2016;7:12178. doi: 10.1038/ncomms12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tóth J.T., Gulyás G., Tóth D.J., Balla A., Hammond G.R.V., Hunyady L., Balla T., Várnai P. BRET-monitoring of the dynamic changes of inositol lipid pools in living cells reveals a PKC-dependent PtdIns4P increase upon EGF and M3 receptor activation. Biochim. Biophys. Acta. 2016;1861:177–187. doi: 10.1016/j.bbalip.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H., Unal H., Desnoyer R., Han G.W., Patel N., Katritch V., Karnik S.S., Cherezov V., Stevens R.C. Structural basis for ligand recognition and functional selectivity at angiotensin receptor. J. Biol. Chem. 2015;290:29127–29139. doi: 10.1074/jbc.M115.689000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wingler L.M., McMahon C., Staus D.P., Lefkowitz R.J., Kruse A.C. Distinctive activation mechanism for angiotensin receptor revealed by a synthetic nanobody. Cell. 2019;176:479–490.e12. doi: 10.1016/j.cell.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szakadáti G., Tóth A.D., Oláh I., Erdélyi L.S., Balla T., Várnai P., Hunyady L., Balla A. Investigation of the fate of type I angiotensin receptor after biased activation. Mol. Pharmacol. 2015;87:972–981. doi: 10.1124/mol.114.097030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H., Han G.W., Batyuk A., Ishchenko A., White K.L., Patel N., Sadybekov A., Zamlynny B., Rudd M.T., Hollenstein K., Tolstikova A., White T.A., Hunter M.S., Weierstall U., Liu W. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017;544:327–332. doi: 10.1038/nature22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motulsky H.J., Mahan L.C. The kinetics of competitive radioligand binding predicted by the law of mass action. Mol. Pharmacol. 1984;25:1–9. [PubMed] [Google Scholar]
- 29.Alexander S.P.H., Christopoulos A., Davenport A.P., Kelly E., Mathie A., Peters J.A., Veale E.L., Armstrong J.F., Faccenda E., Harding S.D., Pawson A.J., Sharman J.L., Southan C., Davies J.A., CGTP Collaborators The concise guide to pharmacology 2019/20: G protein-coupled receptors. Br. J. Pharmacol. 2019;176 Suppl:S21–S141. doi: 10.1111/bph.14748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 31.Christiansen E., Hudson B.D., Hansen A.H., Milligan G., Ulven T. Development and characterization of a potent free fatty acid receptor 1 (FFA1) fluorescent tracer. J. Med. Chem. 2016;59:4849–4858. doi: 10.1021/acs.jmedchem.6b00202. [DOI] [PubMed] [Google Scholar]
- 32.Kilpatrick L.E., Friedman-Ohana R., Alcobia D.C., Riching K., Peach C.J., Wheal A.J., Briddon S.J., Robers M.B., Zimmerman K., Machleidt T., Wood K.V., Woolard J., Hill S.J. Real-time analysis of the binding of fluorescent VEGF165a to VEGFR2 in living cells: Effect of receptor tyrosine kinase inhibitors and fate of internalized agonist-receptor complexes. Biochem. Pharmacol. 2017;136:62–75. doi: 10.1016/j.bcp.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mocking T.A.M., Verweij E.W.E., Vischer H.F., Leurs R. Homogeneous, real-time NanoBRET binding assays for the histamine H3 and H4 receptors on living cells. Mol. Pharmacol. 2018;94:1371–1381. doi: 10.1124/mol.118.113373. [DOI] [PubMed] [Google Scholar]
- 34.Kozielewicz P., Bowin C.-F., Turku A., Schulte G. A NanoBRET-based binding assay for smoothened allows real-time analysis of ligand binding and distinction of two binding sites for BODIPY-cyclopamine. Mol. Pharmacol. 2020;97:23–34. doi: 10.1124/mol.119.118158. [DOI] [PubMed] [Google Scholar]
- 35.Brothers S.P., Janovick J.A., Conn P.M. Unexpected effects of epitope and chimeric tags on gonadotropin-releasing hormone receptors: Implications for understanding the molecular etiology of hypogonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 2003;88:6107–6112. doi: 10.1210/jc.2003-031047. [DOI] [PubMed] [Google Scholar]
- 36.Al-Sabah S., Adi L., Bünemann M., Krasel C. The effect of cell surface expression and linker sequence on the recruitment of arrestin to the GIP receptor. Front. Pharmacol. 2020;11:1271. doi: 10.3389/fphar.2020.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tóth D.J., Tóth J.T., Gulyás G., Balla A., Balla T., Hunyady L., Várnai P. Acute depletion of plasma membrane phosphatidylinositol 4,5-bisphosphate impairs specific steps in endocytosis of the G-protein-coupled receptor. J. Cell Sci. 2012;125:2185–2197. doi: 10.1242/jcs.097279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schröder R., Schmidt J., Blättermann S., Peters L., Janssen N., Grundmann M., Seemann W., Kaufel D., Merten N., Drewke C., Gomeza J., Milligan G., Mohr K., Kostenis E. Applying label-free dynamic mass redistribution technology to frame signaling of G protein–coupled receptors noninvasively in living cells. Nat. Protoc. 2011;6:1748–1760. doi: 10.1038/nprot.2011.386. [DOI] [PubMed] [Google Scholar]
- 39.Luttrell L.M., Maudsley S., Bohn L.M. Fulfilling the promise of “biased” G protein–coupled receptor agonism. Mol. Pharmacol. 2015;88:579–588. doi: 10.1124/mol.115.099630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wacker D., Stevens R.C., Roth B.L. How ligands illuminate GPCR molecular pharmacology. Cell. 2017;170:414–427. doi: 10.1016/j.cell.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evers A., Klabunde T. Structure-based drug discovery using GPCR homology modeling: Successful virtual screening for antagonists of the Alpha1A adrenergic receptor. J. Med. Chem. 2005;48:1088–1097. doi: 10.1021/jm0491804. [DOI] [PubMed] [Google Scholar]
- 42.Tóth A.D., Prokop S., Gyombolai P., Várnai P., Balla A., Gurevich V.V., Hunyady L., Turu G. Heterologous phosphorylation–induced formation of a stability lock permits regulation of inactive receptors by β-arrestins. J. Biol. Chem. 2018;293:876–892. doi: 10.1074/jbc.M117.813139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tóth A.D., Gyombolai P., Szalai B., Várnai P., Turu G., Hunyady L. Angiotensin type 1A receptor regulates β-arrestin binding of the β2-adrenergic receptor via heterodimerization. Mol. Cell. Endocrinol. 2017;442:113–124. doi: 10.1016/j.mce.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 44.Szekeres M., Nádasy G.L., Turu G., Soltész-Katona E., Tóth Z.E., Balla A., Catt K.J., Hunyady L. Angiotensin II induces vascular endocannabinoid release, which attenuates its vasoconstrictor effect via CB1 cannabinoid receptors. J. Biol. Chem. 2012;287:31540–31550. doi: 10.1074/jbc.M112.346296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Y., Prusoff W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GLuc‒PM and NanoLuc‒PM biosensors will be available from Addgene after publication.