Influenza poses a significant burden on society and health care systems. Although antivirals are an integral tool in effective influenza management, the potential for the emergence of antiviral-resistant viruses can lead to uncertainty and hesitation among front-line prescribers and policy makers.
KEYWORDS: antiviral agents, drug resistance evolution, influenza
SUMMARY
Influenza poses a significant burden on society and health care systems. Although antivirals are an integral tool in effective influenza management, the potential for the emergence of antiviral-resistant viruses can lead to uncertainty and hesitation among front-line prescribers and policy makers. Here, we provide an overview of influenza antiviral resistance in context, exploring the key concepts underlying its development and clinical impact. Due to the acute nature of influenza in immunocompetent patients, resistant viruses that develop during antiviral treatment of a single patient (“treatment-emergent resistance”) are usually cleared in a relatively short time, with no impact on future antiviral efficacy. In addition, although available data are limited by small numbers of patients, they show that antiviral treatment still provides clinical benefit to the patient within whom resistance emerges. In contrast, the sustained community transmission of resistant variants in the absence of treatment (“acquired resistance”) is of greater concern and can potentially render front-line antivirals ineffective. Importantly, however, resistant viruses are usually associated with reduced fitness such that their widespread transmission is relatively rare. Influenza antivirals are an essential part of effective influenza management due to their ability to reduce the risk of complications and death in infected patients. Although antiviral resistance should be taken seriously and requires continuous careful monitoring, it is not comparable to antibiotic resistance in bacteria, which can become permanent and widespread, with far-reaching medical consequences. The benefits of antiviral treatment far outweigh concerns of potential resistance, which in the vast majority of cases does not have a significant clinical impact.
INTRODUCTION
Each year, seasonal influenza viruses infect approximately 1 billion people globally, leading to millions of hospitalizations and up to 650,000 excess deaths (1, 2). This remarkable and ongoing burden of influenza exists despite readily available low-cost vaccines and effective antivirals and will likely exert an increasingly significant toll on health care systems with the continued impact of coronavirus disease 2019 (COVID-19) (over 95 million severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2] infections and more than 2 million deaths worldwide as of January 2021) (3). Although vaccination is undoubtedly the most desirable option for influenza prevention, it is associated with variable rates of protection due to suboptimal uptake, mismatches with circulating influenza virus strains, long production times in chicken eggs, and within-season waning of effectiveness (4–6). It is therefore clear that effective approaches are needed for influenza treatment. Antivirals target the virus rather than offering only symptomatic relief and as such are key to the control and management of influenza and the reduction of morbidity and mortality (7–10). The utility of antivirals becomes clearer when juxtaposed to the huge societal and economic impact of the current COVID-19 pandemic, which is to a large extent driven by the lack of vaccine and antiviral options in a highly susceptible human population. Several antivirals are approved for the treatment of influenza; these are summarized in Table 1 along with the populations and settings where clinical efficacy has been demonstrated in a randomized controlled trial setting.
TABLE 1.
Summary of widely approved influenza antiviralsa
| Influenza antiviral | Approval | Mechanism of action | Administration route | Standard adult treatment regimen | Yr of first approval in any country | Patient population(s) with demonstrated efficacy from RCT data (reference[s]) |
|---|---|---|---|---|---|---|
| Amantadine | Worldwide (including the EU and U.S.); note that it is no longer used due to widespread resistance | M2 ion channel inhibitor | Oral | Once daily for ≥10 days (U.S.) | 1966 | Otherwise healthy (72), prophylaxis (73) |
| Rimantadine | Worldwide (excluding the EU); note that it is no longer used due to widespread resistance | M2 ion channel inhibitor | Oral | Twice daily for 7 days | 1993 | Otherwise healthy (74), children (75), prophylaxis (73) |
| Zanamivir | Worldwide (including the EU and U.S.) | Neuraminidase inhibitor | Inhalation or intravenous | Twice daily for 5 days (inhalation) or twice-daily infusion for 5–10 days (intravenous) | 1999 | Otherwise healthy (76), children ≥5 yrs of age (77), prophylaxis (78, 79) |
| Oseltamivir | Worldwide (including the EU and U.S.) | Neuraminidase inhibitor | Oral | Twice daily for 5 days | 1999 | Otherwise healthy (78, 80), high risk of complications (53), children ≥1 yr of age (51), prophylaxis (81) |
| Peramivir | Worldwide (including the EU and U.S.) | Neuraminidase inhibitor | Intravenous infusion | Single infusion over 15 min (minimum) | 2014 | Otherwise healthy (82) |
| Laninamivir | Japan only | Neuraminidase inhibitor | Inhalation | Single dose | 2010 | Otherwise healthy (83), children (≤9 yrs of age) (84), prophylaxis (85) |
| Baloxavir | Worldwide (including the EU and U.S.) | Endonuclease inhibitor | Oral | Single dose | 2018 | Otherwise healthy (49), high risk of complications (53), children ≥1 yr of age (54), prophylaxis (86) |
| Favipiravir | Japan and China (approved for novel pandemic or multiresistant strains only) | RNA-dependent RNA polymerase inhibitor | Oral | Twice daily for 5 days | 2014 | Otherwise healthy (87) |
RCT, randomized controlled trial; EU, European Union.
Although the body of evidence from clinical trials and real-world data demonstrate that antivirals are an integral tool in influenza clinical management, the possibility of antiviral resistance can lead to uncertainty and reservations among front-line prescribers and policy makers, which may in turn contribute to the huge morbidity and mortality burden of influenza (5). To help understand the risks and impact of antiviral resistance for patients and populations, we present the underlying concepts of antiviral resistance in influenza, place them in the context of antibiotic resistance in bacteria, and discuss what these core ideas mean for both prescribers and patients.
THE EVOLUTION OF DRUG RESISTANCE
Natural Selection at Play
The emergence and spread of drug resistance among viruses (as well as bacteria, fungi, and parasites) are governed by the long-established rules of Darwinian natural selection (11–13). Like most RNA viruses, influenza virus is subject to high rates of evolutionary change due to a combination of rapid mutation (approximately 1 error per virus genome, per replication) and rapid replication (14, 15). Hence, a diverse virus population is generated during every individual influenza virus infection (16). Some of the myriad mutations generated in the viral genome will affect key aspects of the virus phenotype, including the ability to circumvent preexisting antibodies or confer antiviral resistance, although the vast majority will impact virus function in a detrimental manner and hence be removed by purifying (i.e., negative) natural selection. However, if a viral variant arises that confers drug resistance at a time when the virus is exposed to that drug, this variant will be at a selective advantage and so will have the opportunity to leave more progeny than drug-sensitive strains (17). It is therefore no surprise that antiviral resistance has been described for all known antiviral treatments (17) and that this emergence can arise rapidly within individual patients (18). The rate of antiviral resistance can vary between influenza virus types, with lower resistance rates typically being observed for influenza B viruses than for influenza A virus subtypes (e.g., the World Health Organization [WHO] reports a neuraminidase [NA] inhibitor [NAI] resistance rate among currently circulating strains of 0% for influenza B virus, compared with 0.80% for A/H1N1pdm09 [19]). The reasons for this are not fully understood but may reflect marginally lower antiviral inhibition (oseltamivir and baloxavir 50% inhibitory concentration [IC50]/50% effective concentration [EC50] values are higher for influenza B viruses than for influenza A viruses) and, therefore, the selection pressures conferred by the drugs on influenza B viruses (20). As discussed below, however, of equal importance to how and at what rate influenza antiviral resistance is generated is whether the resistant variant retains fitness within the host and is transmitted efficiently between individuals in the population at large. This may also differ between influenza A and B viruses.
Reduced Susceptibility versus Resistance
Although the term “resistance” is typically used to describe influenza viral variants that are less well inhibited by an antiviral, they are in reality more accurately described as having “reduced susceptibility.” This is because susceptibility to an antiviral is not binary: mutations do not normally result in complete antiviral resistance but rather reduce antiviral susceptibility to various degrees, from a negligible effect through to “complete” resistance that cannot be overcome clinically (21, 22). Laboratory-based phenotypic assays typically determine the in vitro drug concentrations needed to inhibit influenza virus enzyme activity or viral replication, classifying viruses as having reduced or highly reduced susceptibility (23). The clinical utility of these assays is limited, however, as the potential correlation of these values with the clinical impact of any change on drug effectiveness is not well understood. Although it is possible to compare average antiviral exposure levels in plasma from patients receiving treatment with the in vitro EC50 values of a virus with reduced susceptibility to better understand the likely inhibitory effect and clinical outcome, this does not take into account all relevant variables. For example, if the antiviral resistance viruses represent only a minor component of the viral population (for example, the variant develops late in infection), the majority of viruses still remain fully antiviral susceptible (see “Clinical Impact of Resistance,” below). In addition, measurements of drug distribution in the lung require invasive techniques such as bronchoalveolar lavage (24), making it difficult to accurately correlate local drug concentrations in respiratory tract cells where the virus is present with plasma levels or in vitro EC50 values. As such, although the thresholds used by laboratories to define susceptibility can be a useful reference, they have an unclear clinical correlation. Genotypic assays are also used to determine the presence or absence of key amino acid substitutions in the influenza virus genome that are known to confer reduced or highly reduced susceptibility to antivirals (see below). For simplicity, and because “resistance” is often used to mean “reduced susceptibility,” we use the term “resistance” throughout this article; nevertheless, an appreciation of the points made above remains important whenever the term “resistance” is used.
Mechanisms of Resistance
Amino acid substitutions that confer resistance have been described for all three classes of influenza antivirals: the M2 ion channel inhibitors (adamantanes), the NAIs, and the polymerase inhibitors. For the M2 ion channel inhibitors, the S31N substitution (i.e., a serine-to-asparagine substitution at residue 31 in the M2 protein) is the most common resistance mutation, disrupting drug binding by altering the hydrogen-bonding network in the ion channel pore (25). S31N substitutions are found in virtually all currently circulating influenza A viruses such that the adamantanes are no longer appropriate for use to treat influenza (26).
In the case of the NAIs, amino acid substitutions that alter the shape of the NA enzymatic site, in turn reducing antiviral binding to NA, confer resistance and are often influenza virus type and subtype specific, and the same substitution may have a differential impact on the binding of different NAIs. For example, in A/H1N1 viruses, the most common amino acid substitution associated with resistance is H275Y, which confers reduced susceptibility to oseltamivir and peramivir but does not alter zanamivir or laninamivir inhibition. In H3N2 viruses, two different substitutions, E119V and R292K, result in resistance to multiple NAIs and are most commonly detected under NAI pressure (27). For the endonuclease inhibitor baloxavir, the I38T amino acid substitution in the polymerase acidic (PA) subunit (viral PA protein) is the most commonly described resistance mutation and occurs predominantly in A/H3N2 viruses (22). It acts by reducing van der Waals contacts and thereby decreasing the affinity between the drug and the endonuclease component of the polymerase complex (22). It is important to stress that although rates of resistance to NAIs (<1%) and baloxavir (<0.1%) in currently circulating strains from routine population surveillance are extremely low, resistance to the adamantane class remains at effectively 100% (19). Therefore, a greater range of antivirals with different mechanisms of action will ensure that alternative treatment options exist in the event that resistance develops against any given antiviral. This is a very active area and includes the development of novel M2 ion channel inhibitors with activity against viruses with the S31N substitution (28, 29) or that carry V27A, another clinically relevant adamantane-resistant variant in M2 (30, 31). Pimodivir is an example of a novel polymerase inhibitor (targeting the polymerase basic 2 subunit [PB2] protein) that has been undergoing evaluation in phase III clinical trials. However, a preplanned interim analysis led to its discontinuation as it did not offer any benefit in combination with standard-of-care treatment in hospitalized influenza patients (32). Favipiravir is a purine nucleoside that has received conditional marketing approval for pandemic use in Japan (33). Favipiravir-resistant variants have not been detected in clinical trials, but an in vitro serial passage study selected for variants that conferred a 30-fold reduction in susceptibility (34).
Antiviral Resistance Should Not Be Confused with Antibiotic Resistance
A multitude of recent initiatives and campaigns have highlighted the serious and growing global threat of resistance to a broad range of antimicrobial agents (12, 35). These messages have targeted front-line prescribers to raise awareness of the impact of the indiscriminate and inappropriate use of antibiotics. But what is often not well understood is that antibiotic resistance is a rather different phenomenon from influenza antiviral resistance, with different implications and risks in discrete populations.
As bacteria are part of the healthy flora of the human body, the indiscriminate and inappropriate use of antibiotics significantly increases the risk of antibiotic resistance developing among commensal host bacteria, which can become permanent and widespread and have a number of potentially detrimental host effects (36–38). In contrast to infections with bacteria or viruses that are retained in the body for many years (for example, chronic herpes simplex virus or cytomegalovirus infections), influenza virus causes an acute infection that is completely cleared in immunocompetent patients within 1 to 3 weeks of infection dependent on age (see below). This rapid clearance has two important implications: (i) the use of an influenza antiviral in a misdiagnosed (i.e., noninfluenza) patient will have no bearing on resistance development as these drugs are highly specific, and (ii) even if a resistant viral variant emerges in a patient with an acute influenza infection, the infection will be cleared by the immune system and will not impact the clinical progression or choice of therapy for future influenza infections so long as drug-sensitive strains remain dominant among globally circulating viruses.
Another key difference between antibiotics and influenza antivirals is that the former are often broad spectrum, which can lead to resistance developing in commensal bacteria in addition to the pathogenic bacteria being targeted (39). However, the influenza antivirals currently used were designed to be highly specific for key conserved regions of proteins that exist only in influenza viruses such that they have no activity against noninfluenza viruses. Influenza antiviral resistance and antibiotic resistance therefore have very different origins and clinical implications. Consequently, clinicians should ensure that their decisions are specific to the setting, disease, and drug, rather than adopting a “one-size-fits-all” approach to anti-infective therapies.
CLINICAL IMPACT OF ANTIVIRAL RESISTANCE
Acute versus Chronic Viral Infections
In chronic viral infections such as those caused by human immunodeficiency virus or some hepatitis B and C virus infections, the host immune response is unable to clear the virus even if antiviral treatment greatly suppresses viral replication (40, 41). Therefore, if antiviral resistance develops, the clinical benefit is lost, and the resistant virus can become fixed in the population even if treatment is stopped: this limits the future utility of that antiviral, or potentially an entire class of antivirals, for that patient (40–42). Importantly, however, these concerns do not apply to acute infections such as influenza, where all viruses (resistant or sensitive) are cleared within a few weeks in most immunocompetent patients (41). In some immunocompromised patients, the likelihood of influenza antiviral resistance developing is greater due to prolonged viral shedding for weeks or even months and the often extended antiviral treatment durations (43, 44). However, this risk may vary depending on the degree of immunosuppression experienced by the patient and may be reduced with combination therapy using antivirals with different modes of action (45–47).
Treatment-Emergent versus Acquired Resistance
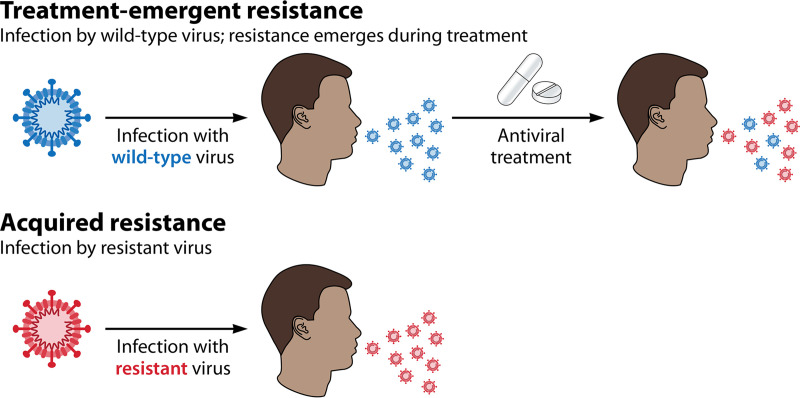
As noted above, the likelihood that a resistant virus can spread in a population is of critical importance. It is therefore important to distinguish between resistance that arises in an individual patient with no onward transmission and resistance that spreads in the population. We define “treatment-emergent” resistance as that arising during antiviral treatment of a single patient who was initially infected with a wild-type (i.e., drug-sensitive) virus. In contrast, “acquired” resistance refers to a circulating virus that already has antiviral resistance at the point of infecting a new host (Fig. 1). Importantly, rates of acquired resistance (such as those observed in routine influenza surveillance) are often lower than those of treatment-emergent resistance (such as those observed in a clinical trial), where all patients are subjected to the selection pressures imposed by antiviral treatment (e.g., the rate of acquired resistance to baloxavir is currently <0.1% in WHO population sampling surveillance [19, 48], compared with a treatment-emergent resistance rate of 9.7% from the CAPSTONE-1 clinical trial [49]). This suggests that evolving antiviral resistance normally imparts a fitness cost on the virus so that it is not widely transmitted in the population, although there are some notable exceptions (see Fitness and Transmission of Resistant Viruses, below). The detection of treatment-emergent resistance therefore provides no indication of the potential long-term fitness and transmissibility of the resistant virus, whereas the detection of high levels of acquired resistance implies that resistant viruses are fit (in the absence of drug) and able to transmit within a population.
FIG 1.
Two types of influenza antiviral resistance can be distinguished. Resistance to influenza antivirals is referred to as either “treatment emergent” or “acquired.” Treatment-emergent resistance refers to that which emerges de novo in an individual patient in response to the selection pressure of antiviral treatment, for example, during a clinical trial. In contrast, acquired resistance occurs without the selection pressure of antiviral treatment because the virus is already resistant before it is transmitted to the new host.
Clinical Impact of Resistance
From the perspective of the prescribing physician and the patient, it is important to understand the clinical impact of influenza antiviral resistance should it develop. When it occurs, treatment-emergent resistance usually develops on days 3 to 4 postinfection at the earliest. However, this can result in a transient increase in viral titers, particularly in young children, although levels of replication are substantially lower than those earlier in the infection and may last for only 1 to 2 days before the immune system clears the virus (50).
Although large-scale clinical studies have been conducted, drawing reliable conclusions on the clinical impact of treatment-emergent resistance is often difficult because this occurs in a relatively small number of patients (49, 51, 52). A recent example was seen with the newly licensed polymerase inhibitor baloxavir. In one study, the evolution of treatment-emergent antiviral resistance corresponded to an ∼12-h lengthening of symptom duration (50), whereas in a second study, treatment-emergent resistance was associated with an ∼11-h-shorter duration of symptoms than in those in whom resistance had not developed (53). Importantly, even in the study where an extension of symptom duration was observed, patients with (treatment-emergent) resistant viruses retained much of the benefit of treatment compared with those who received placebo. A similar picture is observed when looking at the possible clinical impact of treatment-emergent resistance in pediatric clinical studies, although patient numbers are small, and some studies lack control or comparator arms. Treatment-emergent resistance was detected in 18/77 (23.4%) children in a single-arm study of baloxavir, corresponding to an ∼36-h-longer time to illness alleviation and an ∼9-h-longer time to fever resolution than in patients without resistance (52). Comparable rates of treatment-emergent resistance (∼19%) were reported in two separate phase III studies of baloxavir in children (54, 55), with clinical benefit being observed regardless of viral variants; specifically, the time to alleviation of symptoms was comparable between baloxavir-treated patients who developed resistant viruses and the oseltamivir-treated population (54).
Outside randomized clinical trials, real-world surveillance studies have shown that while treatment-emergent resistance can have a minor effect in delaying clearance of residual virus, it has no impact on symptom duration or resolution (56). In contrast, acquired resistance can have an effect on the clinical outcome of treatment, as expected if the entire virus population is resistant to the antiviral from the onset of infection. This has been documented in the case of infection with oseltamivir-resistant viruses circulating in 2007 to 2009, where fever resolution did not differ from the case where patients had received no treatment at all (57, 58). However, it is important to note that these viruses retained sensitivity to an alternative neuraminidase inhibitor, zanamivir. Hence, resistance to one antiviral does not necessarily mean resistance to all antivirals of the same class.
It is also important to note that there is no rationale for the belief that antiviral resistance would affect the safety profile of the respective influenza antiviral, and no evidence of an altered safety profile exists among published studies for any antiviral.
FITNESS AND TRANSMISSION OF RESISTANT VIRUSES
As noted above, resistant viral variants often have a decreased ability to transmit between individuals and hence are of reduced fitness relative to wild-type drug-sensitive viruses, all other things being equal (22, 59). This, coupled with the fact that resistance will usually develop later in an infection and after any interhost transmission event, provides a simple biological explanation for why acquired resistance is expected to be relatively rare. In addition, as there is often a major population bottleneck as the virus transmits from one host to another, with most new infections being initiated by a very small number of virions, any low-frequency resistance mutation could easily be lost from the population simply by chance sampling (60). Despite this, even with reduced viral fitness, person-to-person transmission of resistant viruses can occur in closed settings such as households (61). Indeed, such short-lived, sporadic transmission has been documented for both oseltamivir- and baloxavir-resistant viruses (46, 51, 62, 63).
Of far greater concern, however, is the sustained interhost transmission of resistant viruses in the absence of drug treatment pressure, potentially leading to widespread global circulation. Notably, such a phenomenon has occurred with both subtypes of seasonal influenza virus in humans, A/H1N1 and A/H3N2, in which strains resistant to the M2 ion channel inhibitors (amantadine and rimantadine) emerged a number of years ago and remain in global circulation today, rendering these drugs ineffective in the treatment of influenza (64). In the case of A/H3N2, it has been suggested that the global spread of amantadine resistance was due to genetic “hitchhiking” with fitness-enhancing mutations impacting other aspects of virus biology and located elsewhere in the viral genome (65). An additional example of the global circulation of a resistant virus was the emergence in 2007 to 2008 of an oseltamivir-resistant influenza A/H1N1 virus in Europe (66) that was able to become the dominant circulating strain due to “permissive” mutations that helped overcome fitness defects associated with the H275Y resistance mutation (67). However, this strain was dominant for only 2 years before being naturally outcompeted from circulation by the newer oseltamivir-susceptible, but antigenically novel, pandemic A/H1N1pdm09 virus in 2009 that remains in circulation today (64).
Since 2009, there have been only a small number of reports of localized circulation of A/H1N1pdm09 oseltamivir-resistant viruses (68, 69), and these have not become widespread on a regional, national, or global level. In addition, there have been no reports of local clusters or widespread circulation of viruses resistant to other antiviral drugs such as baloxavir (polymerase acidic endonuclease inhibitor) or the NAIs zanamivir and laninamivir (50). Despite the relative rarity of these events, the ongoing potential for the global spread of resistance to front-line influenza antivirals is continuously monitored. In addition, although antiviral resistance mutations clearly face positive selection pressure due to antiviral use, it is important to note that their widespread circulation cannot be attributed solely to greater antiviral usage. For example, the oseltamivir-resistant A/H1N1 virus described was first reported at high rates in Europe against a backdrop of very low antiviral use (66), while resistance to the M2 ion channel inhibitors was detected in countries with both high and low antiviral use, with suggestions that the resistance mutation S31N arose at least 11 times independently (64). In addition, a surveillance network study suggested that despite high usage of NAIs throughout Japan from 2003 to 2007, the frequency of resistant mutations circulating in this period remained low (70). Therefore, it is likely that although greater antiviral usage can increase the risk of such resistance variants circulating in the community, historically, it has not been the major contributing factor.
MONITORING RESISTANCE
Rates of Treatment-Emergent Resistance
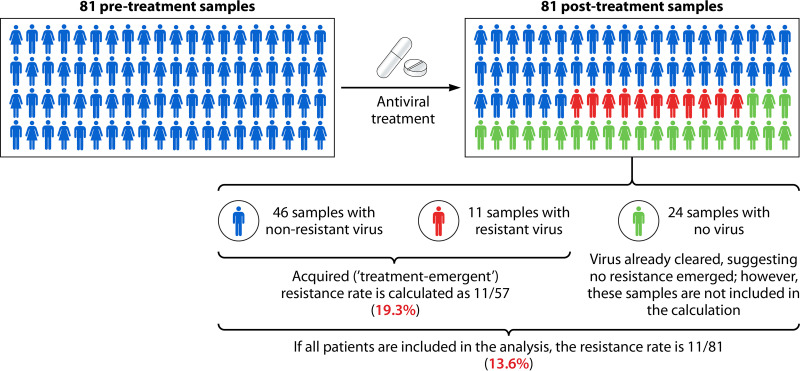
Estimates of rates of treatment-emergent resistance are based on the number of patients who develop resistant viruses as a consequence of antiviral treatment pressure and are usually measured using clinical trial data (50, 52, 54). For this calculation, paired patient samples are collected (one before and one after treatment) to enable laboratory analysis to determine whether a resistance-conferring mutation arose posttreatment. Importantly, only those patients in whom virus is still detectable after treatment (usually days 3 to 5) are included, with those who clear the virus before the second sampling time point being excluded. This likely leads to an overestimation of treatment-emergent resistance rates in clinical trials because patients who have cleared the virus by the second sampling point, and are not included in the overall denominator, are unlikely to have developed resistant virus (as it would have delayed clearance). For example, in the miniSTONE-2 trial, the reported resistance rate was 19.3%, based on 11 patient samples with resistant virus out of 57 paired samples (54). If all patients were part of the denominator, including those without a second virus sample (81 in total), then the resistance rate would be 13.6% (11 out of 81) (Fig. 2). More efficacious antiviral drugs will result in fewer samples with detectable virus posttreatment, further diminishing the denominator and accentuating the bias. As a result, it remains important to be aware of rates of treatment-emergent resistance calculated in different ways: (i) using only patients with paired samples and (ii) using the denominator of all patients treated. It is also generally important to separate pre- and posttreatment samples when calculating overall resistance rates in a population to ensure that rates of acquired and treatment-emergent resistance are not conflated.
FIG 2.
Currently used approaches for calculating antiviral resistance can result in overestimation of resistance rates. Rates of treatment-emergent resistance are estimated from clinical trial data and include only patients with detectable virus within the posttreatment sample. This means that resistance rates will be overestimated, as in most cases, the patients without detectable posttreatment virus will be excluded from the denominator but would most likely not have developed resistant virus. This is illustrated in the example from the miniSTONE-2 trial (54).
Global Prevalence of Antiviral Resistance
Rates of resistance, either treatment emergent or acquired, are regularly reported through the WHO Global Influenza Surveillance and Response System, which coordinates national organizations to assess antiviral resistance among circulating strains. These data are derived from viruses typically collected from untreated patients and thus provide an indication of the likelihood of antiviral resistance that may be observed in patients who acquire influenza in the community. A recent WHO global update on currently circulating influenza viruses (2019–2020 influenza season) reported a low frequency of reduced susceptibility to the neuraminidase inhibitors (<1%) or baloxavir (<0.1%) (19), suggesting that transmission of resistant strains should not be a major concern when considering antiviral treatment. To aid in antiviral treatment decision-making, primary prescribers should be made aware of the latest updates on resistance rates in their respective regions.
CONCLUSIONS
Influenza antivirals are currently the only virus-targeting treatments available and hence are an invaluable control measure for the optimal management of influenza. They play an important role in shortening the illness duration and reducing the risk of complications and death in high-risk individuals and hospitalized patients. As has been argued with respect to SARS-CoV-2, frequent mutations are in reality a routine element of RNA virus evolution (71). The occurrence of resistance to influenza antivirals is therefore a reality that we have to live with. Unlike antibiotic-resistant bacteria, resistant viruses are cleared from immunocompetent patients in a relatively short time and do not impact the future utility of the same antiviral upon subsequent infections if drug-sensitive strains still dominate in the population as a whole. Similarly, the benefit of the antiviral is retained in the majority of patients who develop treatment-emergent resistant viruses, and there are no safety concerns associated with resistance. Close-contact transmission of resistant viruses can occur, but if these remain rare, as is currently the case, they will be of little concern from a community public health perspective. However, evidence of the sustained transmission of these viruses needs to be closely monitored as it can render antivirals ineffective. Fortunately, global and national influenza networks are in place to monitor the frequency of acquired resistance and relay the information to public health bodies to inform appropriate clinical management. Importantly, the benefit of antiviral treatment for a disease that affects up to 1 billion people and causes hundreds of thousands of deaths each year vastly outweighs the concerns of resistance. With the long-term cocirculation of influenza virus and SARS-CoV-2 being a clear possibility, we argue that now is the time to acknowledge the complementary role that antivirals can play with vaccines in relieving the burden of influenza on our health care systems, which may free up capacity to effectively manage other respiratory pathogens.
ACKNOWLEDGMENTS
E.C.H. is funded by an Australian Research Council Australian Laureate fellowship (FL170100022). Third-party medical writing assistance, under the direction of the authors, was provided by John Bett of Gardiner-Caldwell Communications and was funded by F. Hoffmann-La Roche Ltd. Baloxavir and oseltamivir are licensed by F. Hoffmann-La Roche Ltd.
E.C.H. has no conflicts of interest to declare. A.C.H. and Z.D. are employees of F. Hoffmann-La Roche. B.C. is an employee of Roche Products Ltd. J.S.O. has received lecture and consultancy fees from Roche. P.A.P. reports personal fees (consultancy) from F. Hoffmann-La Roche and grants from Shionogi outside the submitted work.
Biographies

Edward C. Holmes is an Australian Research Council Australian Laureate Fellow with concurrent Professorial appointments in the School of Life and Environmental Sciences and the School of Medical Sciences, The University of Sydney, which he joined in 2012. He received his undergraduate degree from the University of London (1986) and his Ph.D. from the University of Cambridge (1990). Between 1993 and 2004, he held various positions at the University of Oxford, including University Lecturer in Evolutionary Biology and Fellow of New College. His research focuses on the emergence, evolution, and spread of RNA viruses, with special emphasis on revealing the genetic and epidemiological processes that underpin viral emergence, the molecular epidemiology of important human and animal pathogens (including influenza, dengue, Ebola, Zika, and SARS-CoV-2 viruses), understanding the nature of global virus diversity, and the major mechanisms of virus evolution. He was elected a Fellow of the Australian Academy of Science (FAA) in 2015 and a Fellow of the Royal Society (FRS) in 2017. He has published over 600 research articles.

Aeron C. Hurt is the Principal Global Medical Director, Influenza, at Roche, Basel, Switzerland. He was previously Head of the Antiviral Susceptibility Analysis Surveillance Unit and Research Group at the WHO Collaborating Centre for Reference and Research on Influenza, Melbourne, Australia. He has had a career-long interest in influenza antivirals and regularly acted as an external Temporary Advisor to the WHO. He has completed a number of short-term consultancies for the Vietnamese Ministry of Health, the WHO, and the Australian Government related to influenza surveillance, epidemiology, and laboratory analysis. He has published over 170 peer-reviewed research and review papers in the field of influenza, including articles in Science, Nature, the New England Journal of Medicine, Nature Medicine, and Lancet Infectious Diseases. He completed a Ph.D. in influenza virology at Monash University and holds an honorary Principal Fellow appointment, as Associate Professor, at the University of Melbourne.

Zuzana Dobbie is a Principal Scientific Communications Director for Infectious Diseases at Roche. She received her Ph.D. in Human Genetics from the University of Basel, Switzerland, and led a research group focusing on cancer genetics at the University Hospital, Basel. As a senior healthcare leader with 20 years of experience in the biotechnology, pharmaceutical, and diagnostics industries, her passion is scientific communications: translating complex scientific topics into simple and engaging narratives and ensuring that these reach the targeted audiences.

Barry Clinch is Global Head, Late-Stage Clinical Science for Influenza and Infectious Disease, at Roche, having over 20 years of industry experience that includes over a decade in infectious disease at Roche. During his time at Roche, he has worked on a variety of infectious disease programs, including a therapeutic vaccine for human papillomavirus (HPV), helping to shape Roche’s reentry into antibiotic development and more recently Roche’s response to SARS-CoV-2. The main part of his work has been the development of antivirals for respiratory viruses, particularly influenza, including both oseltamivir and baloxavir.

John S. Oxford was trained as a virologist by Dr. Geoffrey Schild and Sir Charles Stuart-Harris at Lodge Moor Hospital in Sheffield. He was then appointed Lecturer in Medical Microbiology in Sheffield before further training with Dr. Robert Webster and Dr. Graeme Laver in Canberra, Australia. Professor Oxford returned to London to research at the National Institute for Medical Research Mill Hill and thereafter at the National Institute for Biological Standards and Control. He was appointed Professor of Virology at the London Hospital Medical College and is now Emeritus Professor. Along with Leslie Collier, Professor Oxford authored the successful Oxford University Press book Human Virology (now in its 6th edition) and has also published over 400 peer-reviewed papers. He has been appointed Fellow of the Royal College of Physicians, Edinburgh, and the Royal College of Biology, as well as having been awarded a D.Sc. from the University of Kingston. A particular research interest has been in the genetics and origins of the 1918 pandemic influenza virus with Jeffery Taubenberger. Professor Oxford’s other accolades include being voted “science communicator of the year” and being among the 1,000 most influential Londoners after the 2009 influenza pandemic.

Pedro A. Piedra is Professor in the Department of Molecular Virology and Microbiology, and Pediatrics, and Director of the Respiratory Viral Diagnostic Laboratory at Baylor College of Medicine, Houston, TX. His academic career is devoted to translational research, with an overall goal of reducing the respiratory virus illness burden in infants, children, and the community through successful vaccination and antiviral programs. He has acted as principal investigator on numerous clinical trials studying viral infection and pathogenesis, immunology, vaccine safety and efficacy, and herd protection. He is author/coauthor of more than 220 articles, book chapters, and invited articles. He serves on NIH grant review committees and is a reviewer for numerous journals. He completed a 4-year term in the Vaccines and Related Biological Products Advisory Committee of the FDA (1 February 2012 to 31 January 2016) and has served as an advisor to the World Health Organization on respiratory syncytial virus.
REFERENCES
- 1.WHO. 2019. Global influenza strategy 2019-2030. WHO, Geneva, Switzerland. https://apps.who.int/iris/bitstream/handle/10665/311184/9789241515320-eng.pdf?sequence=18&isAllowed=y. Accessed 19 January 2021. [Google Scholar]
- 2.Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS, Global Seasonal Influenza-Associated Mortality Collaborator Network . 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johns Hopkins University. 2020. COVID-19 dashboard. Johns Hopkins University, Baltimore, MD. https://coronavirus.jhu.edu/map.html. Accessed 19 January 2021. [Google Scholar]
- 4.Schmid P, Rauber D, Betsch C, Lidolt G, Denker M-L. 2017. Barriers of influenza vaccination intention and behavior—a systematic review of influenza vaccine hesitancy, 2005-2016. PLoS One 12:e0170550. doi: 10.1371/journal.pone.0170550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Principi N, Camilloni B, Alunno A, Polinori I, Argentiero A, Esposito S. 2019. Drugs for influenza treatment: is there significant news? Front Med (Lausanne) 6:109. doi: 10.3389/fmed.2019.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng S, Chiu SS, Chan ELY, Kwan MYW, Wong JSC, Leung CW, Chung Lau Y, Sullivan SG, Malik Peiris JS, Cowling BJ. 2018. Effectiveness of influenza vaccination on influenza-associated hospitalisations over time among children in Hong Kong: a test-negative case-control study. Lancet Respir Med 6:925–934. doi: 10.1016/S2213-2600(18)30419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurt AC. 2019. Antiviral therapy for the next influenza pandemic. Trop Med Infect Dis 4:67. doi: 10.3390/tropicalmed4020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oxford JS, Oxford JR. 16 May 2020. Antiviral drugs. In eLS. John Wiley & Sons Ltd, Chichester, United Kingdom. doi: 10.1002/9780470015902.a0000410.pub3. [DOI] [Google Scholar]
- 9.De Clercq E, Li G. 2016. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bean B. 1992. Antiviral therapy: current concepts and practices. Clin Microbiol Rev 5:146–182. doi: 10.1128/cmr.5.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, Nisar MA, Alvi RF, Aslam MA, Qamar MU, Salamat MKF, Baloch Z. 2018. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist 11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moxon ER. 2011. Darwin, microbes and evolution by natural selection. Adv Exp Med Biol 697:77–86. doi: 10.1007/978-1-4419-7185-2_6. [DOI] [PubMed] [Google Scholar]
- 14.Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao W, Li X, Goraya MU, Wang S, Chen J-L. 2017. Evolution of influenza A virus by mutation and re-assortment. Int J Mol Sci 18:1650. doi: 10.3390/ijms18081650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wille M, Holmes EC. 2020. The ecology and evolution of influenza viruses. Cold Spring Harb Perspect Med 10:a038489. doi: 10.1101/cshperspect.a038489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nijhuis M, van Maarseveen NM, Boucher CAB. 2009. Antiviral resistance and impact on viral replication capacity: evolution of viruses under antiviral pressure occurs in three phases. Handb Exp Pharmacol 189:299–320. doi: 10.1007/978-3-540-79086-0_11. [DOI] [PubMed] [Google Scholar]
- 18.Rong L, Dahari H, Ribeiro RM, Perelson AS. 2010. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med 2:30–32. doi: 10.1126/scitranslmed.3000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO. 2020. Recommended composition of influenza virus vaccines for use in the 2021 Southern Hemisphere influenza season. WHO, Geneva, Switzerland. https://www.who.int/influenza/vaccines/virus/recommendations/202009_recommendation.pdf. Accessed 19 January 2021. [Google Scholar]
- 20.Zaraket H, Hurt AC, Clinch B, Barr I, Lee N. 2021. Burden of influenza B virus infection and considerations for clinical management. Antiviral Res 185:104970. doi: 10.1016/j.antiviral.2020.104970. [DOI] [PubMed] [Google Scholar]
- 21.Ikematsu H, Kawai N, Chong Y, Bando T, Iwaki N, Kashiwagi S. 2019. In vitro neuraminidase inhibitory concentration (IC50) of four neuraminidase inhibitors in the Japanese 2017-18 season: comparison with the 2010-11 to 2016-17 seasons. J Infect Chemother 25:649–652. doi: 10.1016/j.jiac.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Omoto S, Speranzini V, Hashimoto T, Noshi T, Yamaguchi H, Kawai M, Kawaguchi K, Uehara T, Shishido T, Naito A, Cusack S. 2018. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci Rep 8:9633. doi: 10.1038/s41598-018-27890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takashita E, Daniels RS, Fujisaki S, Gregory V, Gubareva LV, Huang W, Hurt AC, Lackenby A, Nguyen HT, Pereyaslov D, Roe M, Samaan M, Subbarao K, Tse H, Wang D, Yen HL, Zhang W, Meijer A. 2020. Global update on the susceptibilities of human influenza viruses to neuraminidase inhibitors and the cap-dependent endonuclease inhibitor baloxavir, 2017-2018. Antiviral Res 175:104718. doi: 10.1016/j.antiviral.2020.104718. [DOI] [PubMed] [Google Scholar]
- 24.Davies BE. 2010. Pharmacokinetics of oseltamivir: an oral antiviral for the treatment and prophylaxis of influenza in diverse populations. J Antimicrob Chemother 65(Suppl 2):ii5–ii10. doi: 10.1093/jac/dkq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomaston JL, DeGrado WF. 2016. Crystal structure of the drug-resistant S31N influenza M2 proton channel. Protein Sci 25:1551–1554. doi: 10.1002/pro.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong G, Peng C, Luo J, Wang C, Han L, Wu B, Ji G, He H. 2015. Adamantane-resistant influenza A viruses in the world (1902-2013): frequency and distribution of M2 gene mutations. PLoS One 10:e0119115. doi: 10.1371/journal.pone.0119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moscona A. 2005. Oseltamivir resistance—disabling our influenza defenses. N Engl J Med 353:2633–2636. doi: 10.1056/NEJMp058291. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Ma C, Wang J, Jo H, Canturk B, Fiorin G, Pinto LH, Lamb RA, Klein ML, DeGrado WF. 2013. Discovery of novel dual inhibitors of the wild-type and the most prevalent drug-resistant mutant, S31N, of the M2 proton channel from influenza A virus. J Med Chem 56:2804–2812. doi: 10.1021/jm301538e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon NA, McGuire KL, Wallentine SK, Mohl GA, Lynch JD, Harrison RG, Busath DD. 2017. Divalent copper complexes as influenza A M2 inhibitors. Antiviral Res 147:100–106. doi: 10.1016/j.antiviral.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Rey-Carrizo M, Gazzarrini S, Llabres S, Frigole-Vivas M, Juarez-Jimenez J, Font-Bardia M, Naesens L, Moroni A, Luque FJ, Vazquez S. 2015. New polycyclic dual inhibitors of the wild type and the V27A mutant M2 channel of the influenza A virus with unexpected binding mode. Eur J Med Chem 96:318–329. doi: 10.1016/j.ejmech.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 31.Barniol-Xicota M, Gazzarrini S, Torres E, Hu Y, Wang J, Naesens L, Moroni A, Vazquez S. 2017. Slow but steady wins the race: dissimilarities among new dual inhibitors of the wild-type and the V27A mutant M2 channels of influenza A virus. J Med Chem 60:3727–3738. doi: 10.1021/acs.jmedchem.6b01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.PR Newswire. 2020. Janssen to discontinue pimodivir influenza development program. PR Newswire, New York, NY. https://www.prnewswire.com/news-releases/janssen-to-discontinue-pimodivir-influenza-development-program-301122958.html. Accessed 19 January 2021. [Google Scholar]
- 33.Furuta Y, Komeno T, Nakamura T. 2017. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci 93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldhill DH, Te Velthuis AJW, Fletcher RA, Langat P, Zambon M, Lackenby A, Barclay WS. 2018. The mechanism of resistance to favipiravir in influenza. Proc Natl Acad Sci U S A 115:11613–11618. doi: 10.1073/pnas.1811345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.WHO. 2018. Antimicrobial resistance. WHO, Geneva, Switzerland. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed 19 January 2021. [Google Scholar]
- 36.Becattini S, Taur Y, Pamer EG. 2016. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Bijnen EME, Paget J, de Lange-de Klerk ESM, den Heijer CDJ, Versporten A, Stobberingh EE, Goossens H, Schellevis FG, APRES Study Team . 2015. Antibiotic exposure and other risk factors for antimicrobial resistance in nasal commensal Staphylococcus aureus: an ecological study in 8 European countries. PLoS One 10:e0135094. doi: 10.1371/journal.pone.0135094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fjalstad JW, Esaiassen E, Juvet LK, van den Anker JN, Klingenberg C. 2018. Antibiotic therapy in neonates and impact on gut microbiota and antibiotic resistance development: a systematic review. J Antimicrob Chemother 73:569–580. doi: 10.1093/jac/dkx426. [DOI] [PubMed] [Google Scholar]
- 39.Paharik AE, Schreiber HL, IV, Spaulding CN, Dodson KW, Hultgren SJ. 2017. Narrowing the spectrum: the new frontier of precision antimicrobials. Genome Med 9:110. doi: 10.1186/s13073-017-0504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pantaleo G, Graziosi C, Fauci AS. 1993. The immunopathogenesis of human immunodeficiency virus infection. N Engl J Med 328:327–335. doi: 10.1056/NEJM199302043280508. [DOI] [PubMed] [Google Scholar]
- 41.Mason S, Devincenzo JP, Toovey S, Wu JZ, Whitley RJ. 2018. Comparison of antiviral resistance across acute and chronic viral infections. Antiviral Res 158:103–112. doi: 10.1016/j.antiviral.2018.07.020. [DOI] [PubMed] [Google Scholar]
- 42.Lampejo T. 2020. Influenza and antiviral resistance: an overview. Eur J Clin Microbiol Infect Dis 39:1201–1208. doi: 10.1007/s10096-020-03840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hurt AC, Chotpitayasunondh T, Cox NJ, Daniels R, Fry AM, Gubareva LV, Hayden FG, Hui DS, Hungnes O, Lackenby A, Lim W, Meijer A, Penn C, Tashiro M, Uyeki TM, Zambon M, WHO Consultation on Pandemic Influenza A (H1N1) 2009 Virus Resistance to Antivirals . 2012. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis 12:240–248. doi: 10.1016/S1473-3099(11)70318-8. [DOI] [PubMed] [Google Scholar]
- 44.Rogers MB, Song T, Sebra R, Greenbaum BD, Hamelin ME, Fitch A, Twaddle A, Cui L, Holmes EC, Boivin G, Ghedin E. 2015. Intrahost dynamics of antiviral resistance in influenza A virus reflect complex patterns of segment linkage, reassortment, and natural selection. mBio 6:e02464-14. doi: 10.1128/mBio.02464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitha E, Krivan G, Jacobs F, Nagler A, Alrabaa S, Mykietiuk A, Kenwright A, Le Pogam S, Clinch B, Vareikiene L. 2019. Safety, resistance, and efficacy results from a phase IIIb study of conventional- and double-dose oseltamivir regimens for treatment of influenza in immunocompromised patients. Infect Dis Ther 8:613–626. doi: 10.1007/s40121-019-00271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiso M, Mitamura K, Sakai-Tagawa Y, Shiraishi K, Kawakami C, Kimura K, Hayden FG, Sugaya N, Kawaoka Y. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759–765. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- 47.Lumby CK, Zhao L, Oporto M, Best T, Tutill H, Shah D, Veys P, Williams R, Worth A, Illingworth CJR, Breuer J. 2020. Favipiravir and zanamivir cleared infection with influenza B in a severely immunocompromised child. Clin Infect Dis 71:e191–e194. doi: 10.1093/cid/ciaa023. [DOI] [PubMed] [Google Scholar]
- 48.WHO. 2020. Recommended composition of influenza virus vaccines for use in the 2020-2021 Northern Hemisphere influenza season. WHO, Geneva, Switzerland. https://www.who.int/influenza/vaccines/virus/recommendations/202002_recommendation.pdf. Accessed 19 January 2021. [Google Scholar]
- 49.Hayden FG, Sugaya N, Hirotsu N, Lee N, de Jong MD, Hurt AC, Ishida T, Sekino H, Yamada K, Portsmouth S, Kawaguchi K, Shishido T, Arai M, Tsuchiya K, Uehara T, Watanabe A, Baloxavir Marboxil Investigators Group . 2018. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N Engl J Med 379:913–923. doi: 10.1056/NEJMoa1716197. [DOI] [PubMed] [Google Scholar]
- 50.Uehara T, Hayden FG, Kawaguchi K, Omoto S, Hurt AC, De Jong MD, Hirotsu N, Sugaya N, Lee N, Baba K, Shishido T, Tsuchiya K, Portsmouth S, Kida H. 2020. Treatment-emergent influenza variant viruses with reduced baloxavir susceptibility: impact on clinical and virologic outcomes in uncomplicated influenza. J Infect Dis 221:346–355. doi: 10.1093/infdis/jiz244. [DOI] [PubMed] [Google Scholar]
- 51.Whitley RJ, Hayden FG, Reisinger KS, Young N, Dutkowski R, Ipe D, Mills RG, Ward P. 2001. Oral oseltamivir treatment of influenza in children. Pediatr Infect Dis J 20:127–133. doi: 10.1097/00006454-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 52.Hirotsu N, Sakaguchi H, Sato C, Ishibashi T, Baba K, Omoto S, Shishido T, Tsuchiya K, Hayden FG, Uehara T, Watanabe A. 2020. Baloxavir marboxil in Japanese pediatric patients with influenza: safety and clinical and virologic outcomes. Clin Infect Dis 71:971–981. doi: 10.1093/cid/ciz908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ison MG, Portsmouth S, Yoshida Y, Shishido T, Mitchener M, Tsuchiya K, Uehara T, Hayden FG. 2020. Early treatment with baloxavir marboxil in high-risk adolescent and adult outpatients with uncomplicated influenza (CAPSTONE-2): a randomised, placebo-controlled, phase 3 trial. Lancet Infect Dis 20:1204–1214. doi: 10.1016/S1473-3099(20)30004-9. [DOI] [PubMed] [Google Scholar]
- 54.Baker J, Block SL, Matharu B, Burleigh Macutkiewicz L, Wildum S, Dimonaco S, Collinson N, Clinch B, Piedra PA. 2020. Baloxavir marboxil single-dose treatment in influenza-infected children: a randomized, double-blind, active controlled phase 3 safety and efficacy trial (miniSTONE-2). Pediatr Infect Dis J 39:700–705. doi: 10.1097/INF.0000000000002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokoyama T, Sakaguchi H, Ishibashi T, Shishido T, Piedra PA, Sato C, Tsuchiya K, Uehara T. 2020. Baloxavir marboxil 2% granules in Japanese children with influenza: an open-label phase 3 study. Pediatr Infect Dis J 39:706–712. doi: 10.1097/INF.0000000000002748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lina B, Boucher C, Osterhaus A, Monto AS, Schutten M, Whitley RJ, Nguyen-Van-Tam JS. 2018. Five years of monitoring for the emergence of oseltamivir resistance in patients with influenza A infections in the Influenza Resistance Information Study. Influenza Other Respir Viruses 12:267–278. doi: 10.1111/irv.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuzaki Y, Mizuta K, Aoki Y, Suto A, Abiko C, Sanjoh K, Sugawara K, Takashita E, Itagaki T, Katsushima Y, Ujike M, Obuchi M, Odagiri T, Tashiro M. 2010. A two-year survey of the oseltamivir-resistant influenza A(H1N1) virus in Yamagata, Japan and the clinical effectiveness of oseltamivir and zanamivir. Virol J 7:53. doi: 10.1186/1743-422X-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawai N, Ikematsu H, Hirotsu N, Maeda T, Kawashima T, Tanaka O, Yamauchi S, Kawamura K, Matsuura S, Nishimura M, Iwaki N, Kashiwagi S. 2009. Clinical effectiveness of oseltamivir and zanamivir for treatment of influenza A virus subtype H1N1 with the H274Y mutation: a Japanese, multicenter study of the 2007-2008 and 2008-2009 influenza seasons. Clin Infect Dis 49:1828–1835. doi: 10.1086/648424. [DOI] [PubMed] [Google Scholar]
- 59.Irwin KK, Renzette N, Kowalik TF, Jensen JD. 2016. Antiviral drug resistance as an adaptive process. Virus Evol 2:vew014. doi: 10.1093/ve/vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCrone JT, Lauring AS. 2018. Genetic bottlenecks in intraspecies virus transmission. Curr Opin Virol 28:20–25. doi: 10.1016/j.coviro.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelso A, Hurt AC. 2012. The ongoing battle against influenza. Drug-resistant influenza viruses: why fitness matters. Nat Med 18:1470–1471. doi: 10.1038/nm.2954. [DOI] [PubMed] [Google Scholar]
- 62.Takashita E, Ichikawa M, Morita H, Ogawa R, Fujisaki S, Shirakura M, Miura H, Nakamura K, Kishida N, Kuwahara T, Sugawara H, Sato A, Akimoto M, Mitamura K, Abe T, Yamazaki M, Watanabe S, Hasegawa H, Odagiri T. 2019. Human-to-human transmission of influenza A(H3N2) virus with reduced susceptibility to baloxavir, Japan, February 2019. Emerg Infect Dis 25:2108–2111. doi: 10.3201/eid2511.190757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghedin E, Holmes EC, DePasse JV, Pinilla LT, Fitch A, Hamelin ME, Papenburg J, Boivin G. 2012. Presence of oseltamivir-resistant pandemic A/H1N1 minor variants before drug therapy with subsequent selection and transmission. J Infect Dis 206:1504–1511. doi: 10.1093/infdis/jis571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hussain M, Galvin HD, Haw TY, Nutsford AN, Husain M. 2017. Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist 10:121–134. doi: 10.2147/IDR.S105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simonsen L, Viboud C, Grenfell BT, Dushoff J, Jennings L, Smit M, Macken C, Hata M, Gog J, Miller MA, Holmes EC. 2007. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- 66.Meijer A, Lackenby A, Hungnes O, Lina B, van-der-Werf S, Schweiger B, Opp M, Paget J, van-de-Kassteele J, Hay A, Zambon M, European Influenza Surveillance Scheme . 2009. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerg Infect Dis 15:552–560. doi: 10.3201/eid1504.181280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bloom JD, Gong LI, Baltimore D. 2010. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takashita E, Kiso M, Fujisaki S, Yokoyama M, Nakamura K, Shirakura M, Sato H, Odagiri T, Kawaoka Y, Tashiro M. 2015. Characterization of a large cluster of influenza A(H1N1)pdm09 viruses cross-resistant to oseltamivir and peramivir during the 2013–2014 influenza season in Japan. Antimicrob Agents Chemother 59:2607–2617. doi: 10.1128/AAC.04836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hurt AC, Hardie K, Wilson NJ, Deng YM, Osbourn M, Leang SK, Lee RT, Iannello P, Gehrig N, Shaw R, Wark P, Caldwell N, Givney RC, Xue L, Maurer-Stroh S, Dwyer DE, Wang B, Smith DW, Levy A, Booy R, Dixit R, Merritt T, Kelso A, Dalton C, Durrheim D, Barr IG. 2012. Characteristics of a widespread community cluster of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza in Australia. J Infect Dis 206:148–157. doi: 10.1093/infdis/jis337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tashiro M, McKimm-Breschkin JL, Saito T, Klimov A, Macken C, Zambon M, Hayden FG, Neuraminidase Inhibitor Susceptibility Network . 2009. Surveillance for neuraminidase-inhibitor-resistant influenza viruses in Japan, 1996-2007. Antivir Ther 14:751–761. doi: 10.3851/IMP1194. [DOI] [PubMed] [Google Scholar]
- 71.Grubaugh ND, Petrone ME, Holmes EC. 2020. We shouldn’t worry when a virus mutates during disease outbreaks. Nat Microbiol 5:529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayden FG, Hall WJ, Douglas RG, Jr. 1980. Therapeutic effects of aerosolized amantadine in naturally acquired infection due to influenza A virus. J Infect Dis 141:535–542. doi: 10.1093/infdis/141.5.535. [DOI] [PubMed] [Google Scholar]
- 73.Dolin R, Reichman RC, Madore HP, Maynard R, Linton PN, Webber-Jones J. 1982. A controlled trial of amantadine and rimantadine in the prophylaxis of influenza A infection. N Engl J Med 307:580–584. doi: 10.1056/NEJM198209023071002. [DOI] [PubMed] [Google Scholar]
- 74.Hayden FG, Monto AS. 1986. Oral rimantadine hydrochloride therapy of influenza A virus H3N2 subtype infection in adults. Antimicrob Agents Chemother 29:339–341. doi: 10.1128/aac.29.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hall CB, Dolin R, Gala CL, Markovitz DM, Zhang YQ, Madore PH, Disney FA, Talpey WB, Green JL, Francis AB, Pichichero ME. 1987. Children with influenza A infection: treatment with rimantadine. Pediatrics 80:275–282. [PubMed] [Google Scholar]
- 76.Hayden FG, Osterhaus AD, Treanor JJ, Fleming DM, Aoki FY, Nicholson KG, Bohnen AM, Hirst HM, Keene O, Wightman K. 1997. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenzavirus infections. GG167 Influenza Study Group. N Engl J Med 337:874–880. doi: 10.1056/NEJM199709253371302. [DOI] [PubMed] [Google Scholar]
- 77.Hedrick JA, Barzilai A, Behre U, Henderson FW, Hammond J, Reilly L, Keene O. 2000. Zanamivir for treatment of symptomatic influenza A and B infection in children five to twelve years of age: a randomized controlled trial. Pediatr Infect Dis J 19:410–417. doi: 10.1097/00006454-200005000-00005. [DOI] [PubMed] [Google Scholar]
- 78.Treanor JJ, Hayden FG, Vrooman PS, Barbarash R, Bettis R, Riff D, Singh S, Kinnersley N, Ward P, Mills RG. 2000. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. US Oral Neuraminidase Study Group. JAMA 283:1016–1024. doi: 10.1001/jama.283.8.1016. [DOI] [PubMed] [Google Scholar]
- 79.Monto AS, Pichichero ME, Blanckenberg SJ, Ruuskanen O, Cooper C, Fleming DM, Kerr C. 2002. Zanamivir prophylaxis: an effective strategy for the prevention of influenza types A and B within households. J Infect Dis 186:1582–1588. doi: 10.1086/345722. [DOI] [PubMed] [Google Scholar]
- 80.Nicholson KG, Aoki FY, Osterhaus AD, Trottier S, Carewicz O, Mercier CH, Rode A, Kinnersley N, Ward P. 2000. Efficacy and safety of oseltamivir in treatment of acute influenza: a randomised controlled trial. Neuraminidase Inhibitor Flu Treatment Investigator Group. Lancet 355:1845–1850. doi: 10.1016/s0140-6736(00)02288-1. [DOI] [PubMed] [Google Scholar]
- 81.Welliver R, Monto AS, Carewicz O, Schatteman E, Hassman M, Hedrick J, Jackson HC, Huson L, Ward P, Oxford JS, Oseltamivir Post Exposure Prophylaxis Investigator Group . 2001. Effectiveness of oseltamivir in preventing influenza in household contacts: a randomized controlled trial. JAMA 285:748–754. doi: 10.1001/jama.285.6.748. [DOI] [PubMed] [Google Scholar]
- 82.Kohno S, Kida H, Mizuguchi M, Shimada J, S-021812 Clinical Study Group . 2010. Efficacy and safety of intravenous peramivir for treatment of seasonal influenza virus infection. Antimicrob Agents Chemother 54:4568–4574. doi: 10.1128/AAC.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watanabe A, Chang S-C, Kim MJ, Chu DW-S, Ohashi Y, MARVEL Study Group . 2010. Long-acting neuraminidase inhibitor laninamivir octanoate versus oseltamivir for treatment of influenza: a double-blind, randomized, noninferiority clinical trial. Clin Infect Dis 51:1167–1175. doi: 10.1086/656802. [DOI] [PubMed] [Google Scholar]
- 84.Sugaya N, Ohashi Y. 2010. Long-acting neuraminidase inhibitor laninamivir octanoate (CS-8958) versus oseltamivir as treatment for children with influenza virus infection. Antimicrob Agents Chemother 54:2575–2582. doi: 10.1128/AAC.01755-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kashiwagi S, Watanabe A, Ikematsu H, Awamura S, Okamoto T, Uemori M, Ishida K. 2013. Laninamivir octanoate for post-exposure prophylaxis of influenza in household contacts: a randomized double blind placebo controlled trial. J Infect Chemother 19:740–749. doi: 10.1007/s10156-013-0622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ikematsu H, Hayden FG, Kawaguchi K, Kinoshita M, de Jong MD, Lee N, Takashima S, Noshi T, Tsuchiya K, Uehara T. 2020. Baloxavir marboxil for prophylaxis against influenza in household contacts. N Engl J Med 383:309–320. doi: 10.1056/NEJMoa1915341. [DOI] [PubMed] [Google Scholar]
- 87.Toyama Chemical Co. 2017. Avigan Japanese label. Toyama Chemical Co, Tokyo, Japan. https://www.cdc.gov.tw/File/Get/ht8jUiB_MI-aKnlwstwzvw. Accessed 19 January 2021. [Google Scholar]