Abstract
Wound infection plays an important role in the development of chronicity, delaying wound healing. This study aimed to identify the bacterial pathogens present in infected wounds and characterise their resistance profile to the most common antibiotics used in therapy. Three hundred and twelve wound swab samples were collected from 213 patients and analysed for the identification of microorganisms and for the determination of their antibiotic susceptibility. Patients with diverse type of wounds were included in this retrospective study, carried out from March to September 2012. A total of 28 species were isolated from 217 infected wounds. The most common bacterial species detected was Staphylococcus aureus (37%), followed by Pseudomonas aeruginosa (17%), Proteus mirabilis (10%), Escherichia coli (6%) and Corynebacterium spp. (5%). Polymicrobial infection was found in 59 (27·1%) of the samples and was mainly constituted with two species. The most common association was S. aureus/P. aeruginosa. All Gram‐positives were susceptible to vancomycin and linezolid. Gram‐negatives showed quite high resistance to the majority of antibiotics, being amikacin the most active against these bacteria. This study is mostly oriented to health care practitioners who deal with wound management, making them aware about the importance of wound infection and helping them to choose the adequate treatment options to control microbial infection in wounds.
Keywords: Antibiotic susceptibility, Gram‐positive and Gram‐negative bacteria, Resistance pattern, Wound infection
Introduction
Wound care constitutes an important part of routine care given by health professionals to the community population 1. An effective management of wounds, especially chronic wounds, in the health care setting can have an impact in the population health, reducing morbidity and improving function and quality of life.
Wounds presented by patients vary from one setting to another, ranging from acute surgical wounds, traumatic wounds such as those that occur following an accident, burn wounds or chronic wounds such as diabetic foot, leg and pressure ulcers. All wounds are contaminated with microorganisms that are part of the saprophytic microflora of the skin and the type and quantity of these microorganisms vary from one wound to another 2. Some important factors such as origin, body location, size and duration of the wound should be taken into account in the wound management because of their impact on wound colonisation and infection 3. Microbial colonisation of wounds is characterised by the presence of multiplying microorganisms on the surface of a wound, but with no immune response from the host 4, 5 and with no associated clinical signs and symptoms. Differently, wound infection depends on the pathogenicity and virulence of the microorganisms and on the immune competency of the host and it is determined by the presence of clinical signs of infection such as erythema, pain, tenderness, heat, oedema, cellulites and abscess/pus 6, 7. Therefore, wound infection results in active disease that is likely to delay the wound healing process 8. Moreover, despite these common criteria to identify wound infection, clinicians should be aware that each wound type may present different clinical signs of infection. Thus, the presence of microorganisms per se is not indicative of wound infection 9. However, the probability that a critical microbial load may directly contribute to the non healing outcome in both acute and chronic wounds has been considered and evidence has been shown 5, 10. Other studies in other polymicrobial chronic infections suggest that the presence of specific pathogens is more important that the bacterial burden 11, 12. Generally, after the clinical diagnosis of infection is made, culture is recommended to identify the causative organisms and guide antibiotic therapy.
In this study, we investigated the bacterial profile and assessed their antimicrobial susceptibility pattern of infected wounds presented by patients from who swab samples were collected and analysed by Microbiology laboratory of Pescara Hospital, in central Italy, during a 6‐month period. The aim was to identify the bacterial species present within the wounds and mainly detect the resistance profile to the most common antibiotics used in therapy.
Patients and methods
This retrospective analysis was conducted by reviewing records of wound swab samples that arrived at the Microbiology laboratory of the ‘Santo Spirito’ Hospital of Pescara in Italy, from March to September 2012. Wounds from diverse aetiologies (predominantly leg ulcers, diabetic foot ulcers and pressure ulcers; surgical wounds were excluded), location and duration progress (acute and chronic) were considered in this study. Information about the gender and age of the patients was also provided. The wounds were sampled for microbiological analysis prior to any administration of antibiotic. In total, 312 wound samples were collected from 213 patients. Some patients had more than one wound.
Sample collection
After superficial precleansing of wounds with physiologic saline, each specimen was collected by rotating a sterile, premoistened swab (Nuova Aptaca SRL, Canelli, Italy) across the wound surface of a 1 cm2 area in a zig‐zag motion, from the centre to the outside of the wound. Then, the swab was placed in the tube containing the transport medium (Nuova Aptaca SRL) and sent to the Microbiology laboratory of the hospital for further culture analysis.
Microbiological analysis
Each swab was plated onto four media: blood agar, McConkey agar, mannitol salt agar and Sabouraud agar (SA). All plates except SA plates were incubated aerobically at 37°C for 24 hours. SA plates were also incubated aerobically at 30°C for 48 hours. Any growth was identified by morphologic aspects of the colonies and followed by biochemical identification using the automated Vitek 2 system (bioMérieux, Marcy l'Etoile, France). The antibiotic susceptibility pattern for each bacterial species previously identified was determined using the Vitek 2 susceptibility testing cards for Gram‐positives and Gram‐negatives. These procedures followed the manufacturer's recommendations.
Results
Overall, 312 wound samples were collected from 213 patients, 109 (51·2%) were female, and their age ranged from 20 to 100 years. Of all patients, 28 (13·5%) presented at least one relapse and 6 (2·9%) had two or more wounds during the 6‐month period considered in the study.
In 95 (30·5%) of the wounds only saprophytic bacterial flora was found and there were no signs of infection. The remaining wounds, 217 (69·5%), were considered infected and one or more microbial species with clinical importance were isolated from them. A total of 28 different microbial species were isolated; 44·2% were Gram‐positive and 55·8% were Gram‐negative. The most common bacterial species detected was Staphylococcus aureus (37%), followed by Pseudomonas aeruginosa (17%), Proteus mirabilis (10%), Escherichia coli (6%) and Corynebacterium spp. (5%) (Figure 1A). The most representative species of Enterococcus was Enterococcus cloacae.
Figure 1.
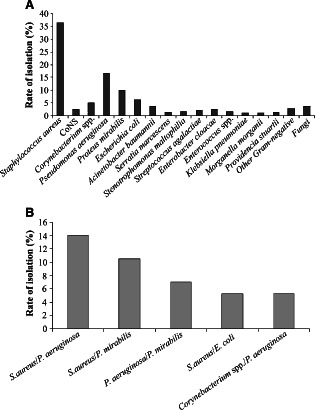
(A) Percentage of microorganisms isolated from 217 swab samples collected from patients with infected wounds. The ‘other Gram‐negative’ includes Alcaligenes faecalis, Citrobacter amalonaticus, Citrobacter koseri, Klebsiella oxytoca, Kocuria kristinae and Pseudomonas stutzeri. ‘Fungi’ comprises Candida albicans, Candida parapsilosis and Aspergillus niger. (B) Percentage of the most common bacterial associations found in the infected wounds analysed.
The presence of only one species isolated from each sample was the most frequent (72·8%). Polymicrobial infection was found in 59 (27·2%) of the infected wounds and was mainly constituted by two species; three species were the maximum number of species isolated per sample and represented only 3·4% of the total polymicrobial infections. The predominant species found in polymicrobial infections was S. aureus, P. aeruginosa and P. mirabilis. The most common association was S. aureus/P. aeruginosa (Figure 1B).
The antibiotic resistance pattern of the Gram‐positive isolates is shown in Table 1. S. aureus and coagulase‐negative staphylococci differ significantly in their resistance rate to oxicillin, being 21·8% and 85·7%, respectively. Corynebacterium spp. showed total resistance to oxacillin, as well as to penicillin G, amoxicillin/clavulanic acid, clindamycin, moxifloxacin and levofloxacin. In general, these species showed higher resistance to the majority of the antibiotics tested. The most active agents against all the Gram‐positive bacteria tested (with no resistance found) were vancomycin, teicoplanin, linezolid and daptomycin.
Table 1.
Antibiotic resistance pattern of Gram‐positive bacteria isolated from patients with infected wounds from March to September 2012 in the Microbiology laboratory of Pescara Hospital
| Microbial species isolated (No.) | Drugs tested No. (%) of resistance | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | AC | OX | G | V | T | CL | ER | MX | LX | PX | TS | TC | L | TG | FA | D | |
| Staphylococcus aureus (n = 101) | 72 (71·2) | 32 (31·6) | 22 (21·8) | 28 (27·7) | 0 (0) | 0 (0) | 16 (15·8) | 42 (41·6) | 41 (40·6) | 46 (45·5) | 45 (44·5) | 6 (5·9) | 8 (7·9) | 0 (0) | 0 (0) | 1 (0·99) | 2 (1·9) |
| CoNS (n = 7) | 5 (7·14) | 5 (71·4) | 6 (85·7) | 6 (85·7) | 0 (0) | 2 (28·6) | 3 (42·8) | 5 (71·4) | 5 (71·4) | 1 (14·3) | 5 (71·4) | 1 (14·3) | 2 (28·6) | 0 (0) | 0 (0) | 2 (28·6) | 0 (0) |
| Corynebacterium spp. (n = 14) | 14 (100) | 14 (100) | 14 (100) | 13 (92·8) | 0 (0) | 0 (0) | 14 (100) | 9 (64·3) | 14 (100) | 14 (100) | 10 (71·4) | 10 (71·4) | 11 (78·6) | 0 (0) | 1 (7·1) | – | 0 (0) |
CoNS, coagulase‐negative staphylococci; P, penicillin G; AC, amoxycillin/clavulanic acid; OX, oxacillin; G, gentamicin; V, vancomycin; T, teicoplanin; CL, clindamycin; ER, erythromycin; MX, moxifloxacin; LX, levofloxacin; PX, prulifloxacin; TS, trimethoprim/sulfamethoxazole; TC, tetracycline; L, linezolid; TG, tigecycline; FA, fosfomycin; D, daptomycin.
Additionally, in Table 2, the antibiotic resistance pattern of the three most common Gram‐negatives isolated from the wounds is shown. The most effective antibiotic against all these Gram‐negatives was amikacin; P. mirabilis and E. coli were fully susceptible to this antibiotic; however, 28·3% of P. aeruginosa showed resistance to it. The isolates of P. aeruginosa showed 100% of resistance to ampicillin, amoxicillin/clavulanic acid, ertapenem and trimethoprim/sulfamethoxazole. Moreover, P. mirabilis and E. coli also showed high resistance to ampicillin. Meropenem, ertapenem and fosfomycin were 100% active against E. coli.
Table 2.
Antibiotic resistance pattern of the most common Gram‐negative bacteria isolated from patients with infected wounds from March to September 2012 in the Microbiology laboratory of Pescara Hospital
| Microbial species isolated (No.) | Drugs tested No. (%) of resistance | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | AC | PB | CT | CZ | CP | M | E | G | AK | CX | TS | FA | |
| Pseudomonas aeruginosa (n = 46) | 46 (100) | 46 (100) | 24 (52·2) | 44 (95·6) | 23 (50) | 17 (36·9) | 14 (30·4) | 46 (100) | 18 (39·1) | 13 (28·3) | 21 (45·6) | 46 (100) | – |
| Proteus mirabilis (n = 28) | 25 (89·3) | 14 (50) | 1 (3·6) | 12 (42·8) | 8 (28·6) | 3 (10·7) | 1 (3·6) | 2 (7·1) | 15 (53·6) | 0 (0) | 19 (67·8) | 19 (67·8) | 11 (39·3) |
| Escherichia coli (n = 17) | 16 (94·1) | 8 (47·1) | 2 (11·8) | 4 (23·5) | 1 (5·9) | 2 (11·8) | 0 (0) | 0 (0) | 2 (11·8) | 0 (0) | 9 (52·9) | 11 (64·7) | 0 (0) |
A, ampicillin; AC, amoxycillin/clavulanic acid; PB, piperacillin/tazobactam; CT, cefotaxime; CZ, ceftazidime; CP, cefepime; M, meropenem; E, ertapenem; G, gentamicin; AK, amikacin; CX, ciprofloxacin; TS, trimethoprim/sulfamethoxazole; FA, fosfomycin.
Discussion
To prevent or reduce wound infection is a goal shared by health care practitioners in charge of wound management and care; however, when infection is already established, wound management practices should be specifically addressed and become more challenging and demanding. Antibiotic treatment is recommended but, previously, an antibiotic susceptibility test should be performed.
In this study, 28 microbial species were isolated from wounds with signs of infection. The majority of the wounds were colonised with a single bacterial species. The most common isolate was S. aureus, which was also reported in many other studies to be the predominant microorganism (40–60% of the total microorganisms) isolated from different types of wounds 13, 14, 15, 16, 17, 18. P. aeruginosa was the Gram‐negative more detected, which is also in agreement with other reports 15, 16, 19, 20, 21. It is well documented that bacteria such as S. aureus and P. aeruginosa produce very destructive virulence factors, responsible for maintaining infection and delay healing in chronic wounds. S. aureus causes clinically relevant infections mostly because of its virulence factors such as coagulase, catalase, clumping‐factor A and leucocidines 22. Similarly, the production of an elastase by P. aeruginosa has been associated to its pathogenicity in the wound environment 23. Thus, our results confirm the usual most prevalent microorganisms found in infected wounds. However, the role that each specific pathogen plays in both no healing and infected chronic wounds is not yet very defined; it is mostly based on hypotheses 5, 9. Beyond the presence of pathogens, it has been considered to be of paramount importance the presence of specific bacterial combinations and interactions in both acute and chronic wounds 24. In our study, only 27·2% of the wounds displayed polymicrobial infections. It is known that interspecies interactions consist mostly in bacterial synergy that enhances survival, therefore hampering the infection eradication. Moreover, microorganisms have the ability to establish themselves and proliferate as a biofilm, both in monomicrobial and polymicrobial biofilms, which are often considered to be a further complication that has a significant contribution to the lack of successful antibiotic treatment 25. Because of this problem, researchers are seeking for new alternative therapies useful to enhance wound healing, such as laser therapy 26.
The bacterial isolates were examined for their susceptibility pattern to the most commonly used antibiotics in therapy. Despite increasing concerns about antibiotic‐resistant bacteria, appropriate use of systemic antibiotics is still recommended where there is clear evidence of infection 9, 27, 28. The resistance to oxacillin is particularly important because it can give us the percentage of methicillin‐resistant Staphylococcus aureus (MRSA); in our study, a relevant percentage (21·8%) of S. aureus was oxacillin resistant. S. aureus has always been a major source of infection in acute soft‐tissue wounds, but MRSA has only been an infecting organism in a small fraction of the total. Nevertheless, MRSA is becoming a more common wound pathogen 29. The occurrence of MRSA presents two problems: the first is associated to the chronic wound being a source of other MRSA nosocomial infections and the second is related to the impact of MRSA on the chronic wound itself, that is, who have chronic wounds growing MRSA and have an increased risk of suffering a bacteremia by MRSA 29. Among the Gram‐positive bacteria, all isolates were susceptible to vancomycin and to linezolid; despite these two antibiotics are largely used, no resistance was found. Regarding the antibiotic resistance of Gram‐negative, the most common isolates, and in particular P. aeruginosa, showed a relatively high resistance to the majority of the antibiotics. Multidrug‐resistant isolates of P. aeruginosa, that is, fully resistant simultaneously to ampicillin, amoxicillin/clavulanic acid, ertapenem and trimethoprim/sulfamethoxazole, are of major concern. Additionally, the results indicate that P. aeruginosa is tending toward a high level of resistance to carbapenems and third‐generation cephalosporins. Similar evidence was reported by Nicoletti et al. 30 in a study regarding diverse severe infections. P. mirabilis and E. coli showed, however, a low‐resistance profile compared to P. aeruginosa.
An important limitation of this study must be mentioned. It is evident from the results stated that only aerobic/facultative microorganisms were investigated, as the procedures followed by the laboratory of the hospital did not perform the isolation of anaerobic bacteria from the swab samples collected from the wounds. Potential pathogenic anaerobes are frequently found in wounds 14, 31, 32. Bowler and Davies 13 reported that together with the aerobic and facultative population, the diversity of anaerobic bacteria in leg ulcers is considerable and their presence was particularly obvious in infected leg ulcers in respect to non infected ones. Despite the already demonstrated role and importance of anaerobes in wound infection, the techniques to isolate them still not being performed in many clinical laboratories in these days; the reason seems to be that the culture of anaerobes is more time‐consuming, labour‐intensive and expensive and, thus, too demanding. This is certainly a drawback in the wound management that should be addressed in order that the best treatment will be suggested. Therefore, it is very likely that the percentage of polymicrobial infection that we found is biased. Gjødsbøl et al. 15 performed a longitudinal study on chronic ulcers and reported that none of these ulcers was colonised with only one single bacterial species, but with two or more, being the average number of six species per ulcer.
Moreover, it seems to be opportune to state that the use of wound swab sampling has been questioned by some researchers 33 on the basis that it only permits to identify the microorganisms present in the surface, neglecting the ones present on the deeper tissue; thus, the sample might be lacking the correct information of the colonising organisms. However, that statement has been opposed by other studies 11, 34 that compared different methods to collect wounds samples. In these studies, they concluded that the wound swab sampling, if appropriate microbiologic culture techniques are used, can be an effective method to isolate the microorganisms present. In fact, all microbial contaminants (both aerobic and anaerobic) are originated from exogenous sources to the wound and, consequently, the microorganisms disseminated into deeper tissue must also be comprised in the superficial tissue of the wound.
The successful management of bacteria in a wound is of great importance; however, it is still a complex issue. Therefore, our study evaluates the current situation in a particular geographic area, which is mostly helpful to the clinicians and microbiologists involved because it can make them aware of the real circumstances that they are dealing with presently. Knowing the prevalent type of microorganisms present in infected wounds and their resistance pattern is clearly pertinent to choose the adequate treatment. The data presented here together with the discussion carried out can be useful to improve the management of wound infection.
ACKNOWLEDGEMENTS
The authors thank Dr Emanuela Di Campli, Soraya Di Bartolomeo and Eleonora Di Zio for their technical advice. The authors are also grateful to all members of the Microbiology laboratory of ‘Spirito Santo’ Hospital of Pescara.
References
- 1. Meaume S, Keriheul JC, Fromantin I, Téot L. Workload and prevalence of open wounds in the community: French Vulnus initiative. J Wound Care 2012;21:62–6. [DOI] [PubMed] [Google Scholar]
- 2. Cooper R, Lawrence JC. The isolation and identification of bacteria from wounds. J Wound Care 1996;5:335–40. [DOI] [PubMed] [Google Scholar]
- 3. White RJ, Cooper R, Kingsley A. Wound colonization and infection: the role of topical antimicrobials. Br J Nurs 2001;10:563–78. [DOI] [PubMed] [Google Scholar]
- 4. Edwards R, Harding KG. Bacteria and wound healing. Curr Opin Infect Dis 2004;17:91–6. [DOI] [PubMed] [Google Scholar]
- 5. Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 2001;14:244–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. European Wound Management Association (EWMA) . Position document: identifying criteria for wound infection. London: MEP Ltd, 2005. [Google Scholar]
- 7. Collier M. Recognition and management of wound infections. 2004. URL http://www.worldwidewounds.com/2004/january/Collier/Management‐of‐Wound‐infections.html [accessed on 12 October 2012]
- 8. Beldon P. Recognising wound infection. Nurs Times 2001;97:3–4. [PubMed] [Google Scholar]
- 9. Howell‐Jones RS, Wilson MJ, Hill KE, Howard AJ, Price PE, Thomas DW. A review of the microbiology, antibiotic usage and resistance in chronic skin wounds. J Antimicrob Chemother 2005;55:143–9. [DOI] [PubMed] [Google Scholar]
- 10. Serralta VW, Harrison‐Balestra C, Cazzaniga AL, Davis SC, Mertz PM. Lifestyles of bacteria in wounds: presence of biofilms? Wounds 2001;13:29–34. [Google Scholar]
- 11. Davies CE, Hill KE, Newcombe RG, Stephens P, Wilson MJ, Harding KG, Thomas DW. A prospective study of the microbiology of chronic venous leg ulcers to reevaluate the clinical predictive value of tissue biopsies and swabs. Wound Repair Regen 2007;15:17–22. [DOI] [PubMed] [Google Scholar]
- 12. Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 2008;3:e3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bowler PG, Davies BJ. The microbiology of infected and noninfected leg ulcers. Int J Dermatol 1999;38:573–8. [DOI] [PubMed] [Google Scholar]
- 14. Brook I, Frazier EH. Aerobic and anaerobic microbiology of chronic venous ulcers. Int J Dermatol 1998;37:426–8. [DOI] [PubMed] [Google Scholar]
- 15. Gjødsbøl K, Christensen JJ, Karlsmark T, Jørgensen B, Klein BM, Krogfelt KA. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 2006;3:225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davies CE, Hill KE, Wilson MJ, Stephens P, Hill CM, Harding KG, Thomas DW. Use of 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis for analysis of the microfloras of healing and nonhealing chronic venous leg ulcers. J Clin Microbiol 2004;42:3549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Urbancic‐Rovan V, Gubina M. Bacteria in superficial diabetic foot ulcers. Diabet Med 2000;17:814–5. [DOI] [PubMed] [Google Scholar]
- 18. Körber A, Schmid EN, Buer J, Klode J, Schadendorf D, Dissemond J. Bacterial colonization of chronic leg ulcers: current results compared with data 5 years ago in a specialized dermatology department. J Eur Acad Dermatol Venereol 2010;24:1017–25. [DOI] [PubMed] [Google Scholar]
- 19. Halbert AR, Stacey MC, Rohr JB, Jopp‐McKay A. The effect of bacterial colonization on venous leg ulcer healing. Australas J Dermatol 1992;33:75–80. [DOI] [PubMed] [Google Scholar]
- 20. Madsen SM, Westh H, Danielsen L, Rosdahl VT. Bacterial colonization and healing of venous leg ulcers. APMIS 1996;104:895–9. [DOI] [PubMed] [Google Scholar]
- 21. Burmølle M, Thomsen TR, Fazli M, Dige I, Christensen L, Homøe P, Tvede M, Nyvad B, Tolker‐Nielsen T, Givskov M, Moser C, Kirketerp‐Møller K, Johansen HK, Høiby N, Jensen PØ, Sørensen SJ, Bjarnsholt T. Biofilms in chronic infections – a matter of opportunity –monospecies biofilms in multispecies infections. FEMS Immunol Med Microbiol 2010;59:324–36. [DOI] [PubMed] [Google Scholar]
- 22. Dissemond J. Methicillin resistant Staphylococcus aureus (MRSA): diagnostic, clinical relevance and therapy. J Dtsch Dermatol Ges 2009;6:544–51. [DOI] [PubMed] [Google Scholar]
- 23. Schmidtchen A, Holst E, Tapper H, Bjorck L. Elastase‐producing Pseudomonas aeruginosa degrade plasma proteins and extracellular products of human skin and fibroblasts, and inhibit fibroblast growth. Microb Pathog 2003;34:47–55. [DOI] [PubMed] [Google Scholar]
- 24. Percival SL, Thomas JG, Williams DW. Biofilms and bacterial imbalances in chronic wounds: anti‐Koch. Int Wound J 2010;7:169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kirketerp‐Møller K, Jensen PØ, Fazli M, Madsen KG, Pedersen J, Moser C, Tolker‐Nielsen T, Høiby N, Givskov M, Bjarnsholt T. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol 2008;46:2717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baffoni M, Bessa LJ, Grande R, Di Giulio M, Mongelli M, Ciarelli A, Cellini L. Laser irradiation effect on Staphylococcus aureus and Pseudomonas aeruginosa biofilms isolated from venous leg ulcer. Int Wound J 2012;9:517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. National Institute for Clinical Excellence . Type 2 diabetes. Prevention and management of foot problems. Clinical guideline 10. London: National Institute for Clinical Excellence, 2004. [Google Scholar]
- 28. European Wound Management Association (EWMA) . Position document: management of wound infection. London: MEP Ltd, 2006. [Google Scholar]
- 29. Demling RH, Waterhouse B. The increasing problem of wound bacterial burden and infection in acute and chronic soft‐tissue wounds caused by methicillin‐resistant Staphylococcus aureus . J Burns Wounds 2007;7:86–98. [PMC free article] [PubMed] [Google Scholar]
- 30. Nicoletti G, Schito G, Fadda G, Boros S, Nicolosi D, Marchese A, Spanu T, Pantosti A, Monaco M, Rezza G, Cassone A, Garaci E. Bacterial isolates from severe infections and their antibiotic susceptibility pattern in Italy: a nationwide study in the hospital setting. J Chemother 2006;18:589–602. [DOI] [PubMed] [Google Scholar]
- 31. Ge Y, MacDonald D, Hait H, Lipsky B, Zasloff M, Holroyd K. Microbiological profile of infected diabetic foot ulcers. Diabet Med 2002;19:1032–4. [DOI] [PubMed] [Google Scholar]
- 32. Trengove NJ, Stacey MC, McGechie DF, Mata S. Qualitative bacteriology and leg ulcer healing. J Wound Care 1996;5:277–80. [DOI] [PubMed] [Google Scholar]
- 33. Bjarnsholt T, Kirketerp‐Møller K, Jensen PØ, Madsen KG, Phipps R, Krogfelt K, Høiby N, Givskov M. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen 2008;16:2–10. [DOI] [PubMed] [Google Scholar]
- 34. Cooper RA, Ameen H, Price P, McCulloch DA, Harding KG. A clinical investigation into the microbiological status of “locally infected” leg ulcers. Int Wound J 2009;6:453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]


