The bacterial pathogen Pseudomonas aeruginosa causes devastating infections in immunocompromised hosts, including chronic lung infections in cystic fibrosis patients. To combat infection, the host’s immune system produces the antimicrobial oxidants hypochlorous acid (HOCl) and hypothiocyanous acid (HOSCN). Little is known about how P. aeruginosa responds to and survives attack from these oxidants. To address this, we carried out two approaches: a mutant screen and transcriptional study. We identified the P. aeruginosa transcriptional regulator, RclR, which responds specifically to HOCl and HOSCN stress and is essential for protection against both oxidants. We uncovered a link between the P. aeruginosa transcriptional response to these oxidants and physiological processes associated with pathogenicity, including antibiotic resistance and the type 3 secretion system.
KEYWORDS: Pseudomonas aeruginosa, cystic fibrosis, efflux pumps, hypochlorous acid, hypothiocyanous acid, oxidative stress, peroxiredoxin
ABSTRACT
Pseudomonas aeruginosa is a significant nosocomial pathogen and is associated with lung infections in cystic fibrosis (CF). Once established, P. aeruginosa infections persist and are rarely eradicated despite host immune cells producing antimicrobial oxidants, including hypochlorous acid (HOCl) and hypothiocyanous acid (HOSCN). There is limited knowledge as to how P. aeruginosa senses, responds to, and protects itself against HOCl and HOSCN and the contribution of such responses to its success as a CF pathogen. To investigate the P. aeruginosa response to these oxidants, we screened 707 transposon mutants, with mutations in regulatory genes, for altered growth following HOCl exposure. We identified regulators of antibiotic resistance, methionine biosynthesis, catabolite repression, and PA14_07340, the homologue of the Escherichia coli HOCl-sensor RclR (30% identical), which are required for protection against HOCl. We have shown that RclR (PA14_07340) protects specifically against HOCl and HOSCN stress and responds to both oxidants by upregulating the expression of a putative peroxiredoxin, rclX (PA14_07355). Transcriptional analysis revealed that while there was specificity in the response to HOCl (231 genes upregulated) and HOSCN (105 genes upregulated), there was considerable overlap, with 74 genes upregulated by both oxidants. These included genes encoding the type 3 secretion system, sulfur and taurine transport, and the MexEF-OprN efflux pump. RclR coordinates part of the response to both oxidants, including upregulation of pyocyanin biosynthesis genes, and, in the presence of HOSCN, downregulation of chaperone genes. These data indicate that the P. aeruginosa response to HOCl and HOSCN is multifaceted, with RclR playing an essential role.
IMPORTANCE The bacterial pathogen Pseudomonas aeruginosa causes devastating infections in immunocompromised hosts, including chronic lung infections in cystic fibrosis patients. To combat infection, the host’s immune system produces the antimicrobial oxidants hypochlorous acid (HOCl) and hypothiocyanous acid (HOSCN). Little is known about how P. aeruginosa responds to and survives attack from these oxidants. To address this, we carried out two approaches: a mutant screen and transcriptional study. We identified the P. aeruginosa transcriptional regulator, RclR, which responds specifically to HOCl and HOSCN stress and is essential for protection against both oxidants. We uncovered a link between the P. aeruginosa transcriptional response to these oxidants and physiological processes associated with pathogenicity, including antibiotic resistance and the type 3 secretion system.
INTRODUCTION
Pseudomonas aeruginosa is a leading cause of nosocomial infections that are life-threatening and difficult to treat (1). As an ESKAPE pathogen, it is one of the bacteria that poses the greatest public health threat (2). Furthermore, P. aeruginosa is the major pathogen associated with chronic lung infections in patients suffering from cystic fibrosis (CF) (3, 4). This genetic disease is caused by mutations in the cystic fibrosis transmembrane conductance regulator, which results in abnormal ion and water transport across epithelial membranes and leads to dehydrated airways and the establishment of respiratory infections (5). The immune response to infection is characterized by persistent neutrophil-dominated inflammation that is ineffective at clearing infection and results in progressive lung tissue damage (3, 4).
Host cells defend themselves against invading pathogens by the production of oxidants during the innate immune response, including the hypohalous acids: hypochlorous acid (HOCl), the active ingredient in bleach, and hypothiocyanous acid (HOSCN) (6). HOCl is a potent oxidant produced by neutrophils (7). The neutrophil oxidative burst that follows phagocytosis involves the reduction of O2 to superoxide (O2−) by the enzyme NADPH oxidase and the dismutation of O2− to hydrogen peroxide (H2O2) (7). The heme enzyme myeloperoxidase (MPO) catalyzes the formation of HOCl from the reaction of H2O2 with chloride (Cl−) (7). MPO can also mediate H2O2 oxidation of other halides, including the pseudohalide thiocyanate (SCN−), to HOSCN. However, its primary physiological substrate in neutrophils is proposed to be HOCl, as the concentration of Cl− in plasma (100 to 140 mM) is much greater than that of SCN− (20 to 100 μM) (6, 8). The physiological concentration of HOCl is unknown due to the instability of this compound (6). At the epithelial cell surface of the lungs, HOSCN is produced through the concerted action of the dual oxidase (DUOX) and lactoperoxidase (LPO) (9). The epithelial cell DUOX releases H2O2 into the airway surface liquid, where LPO catalyzes the reaction of H2O2 with SCN− to form HOSCN (9). HOCl can target proteins, lipids, DNA, and RNA; the most reactive targets of HOCl are thiol (S-H) groups of cysteine and methionine (7). Oxidation of cysteine by HOCl results in the formation of sulfenic acid, which can react with other cysteine thiols to form reversible disulfide bonds, and oxidation of methionine by HOCl results in the formation of methionine sulfoxide (6). Similar to HOCl, HOSCN reacts fastest with thiol groups of cysteine forming sulfenic acids or disulfide bonds (6). However, HOCl is a stronger oxidant than HOSCN, with rate constants for cysteine thiol groups of 3.2 × 107 M−1 s−1 and 7.8 × 104 M−1 s−1, respectively (6).
Despite HOCl and HOSCN having greater antibacterial activities than H2O2, studies of the bacterial mechanisms used to protect against oxidative stress have, until recent years, focused on the less potent oxidants, H2O2 and O2− (6, 10). However, over the last 7 years, emerging studies into the bacterial response mechanisms to HOCl have identified thiol-based redox-sensing transcriptional regulators (11, 12). They include the Bacillus subtilis transcriptional factors OhrR, HypR, and PerR and the Escherichia coli transcriptional factors NemR, HypT, and RclR (13–17). HypT and RclR are the first described bacterial transcriptional regulators that are specific to sensing HOCl (16, 17); the others respond to a variety of stresses, including reactive electrophile species and organic hydroperoxides (11, 14, 15, 18–24). The identification of these HOCl-sensing regulators revealed that bacteria have specific mechanisms for responding to and protecting against HOCl stress and do not rely solely on mechanisms used against other oxidants. Virtually nothing is known about bacterial responses to HOSCN, apart from a study from Groitl et al. that investigated the transcriptional response of P. aeruginosa PA14 to HOSCN as well as to HOCl and another hypohalous acid, hypobromous acid (HOBr) (25). A key finding of this study was that all three oxidants caused protein aggregation and P. aeruginosa responded by increasing polyphosphate levels, which protected against protein aggregation and aided survival (25).
However, there is still very little known about how the clinically relevant pathogen P. aeruginosa senses, responds to, and protects against HOCl or HOSCN. Due to P. aeruginosa encountering these innate immune cell-derived oxidants in a variety of infection environments, including the neutrophilic environment of the CF lung, it is important to increase our knowledge of these specific oxidative stress responses.
Here, we have used a combination of mutant screening and transcriptomics to identify genes and systems used by P. aeruginosa to survive HOCl and HOSCN challenge. Our major finding is the identification of the P. aeruginosa RclR system that is specifically required for protection against both HOCl and HOSCN stress, in addition to discovering regulators with known roles in antibiotic resistance and methionine and carbon metabolism as having protective roles against HOCl.
RESULTS
Screening transposon mutants of P. aeruginosa regulatory genes for altered susceptibility to HOCl.
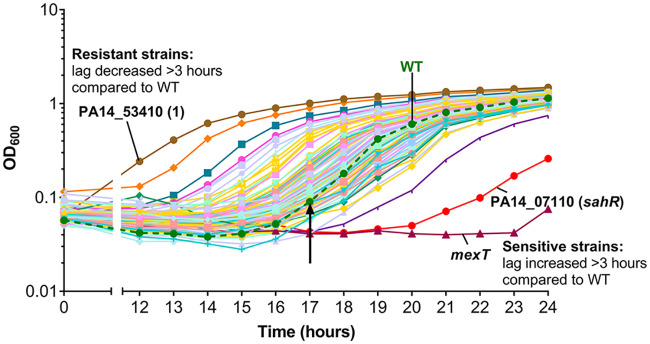
We reasoned that targeting regulatory systems would be an effective way to uncover processes used by P. aeruginosa to protect against HOCl stress. To address this, we used mutants from the PA14 nonredundant transposon insertion mutant library (26), which contains 5,850 mutants from 4,596 genes (out of the 5,977 annotated genes) in the PA14 chromosome. To compile a subset of available mutants from this library, in genes classed as having confirmed or predicted regulatory roles, we used gene ontology information from the Pseudomonas genome database (27). A search for mutants in transcriptional regulator, two-component system, catalase, Ser/Thr kinase, sigma factor, and other regulatory genes resulted in a subset of 707 mutants (see Table S1 in the supplemental material). These 707 mutants were screened for altered susceptibility compared to the wild type (WT) when grown in the presence of 4.4 mM HOCl over 24 h. The most sensitive strains were identified as those that consistently failed to grow during the time of the assay or had an increased lag of >3 h compared to the WT. Resistant strains were identified as those that consistently had a decreased lag of >3 h compared to the WT. Figure 1 shows an example of a 96-well screening plate with HOCl-sensitive and HOCl-resistant mutants labeled. Identified HOCl-sensitive and HOCl-resistant mutants were rescreened, and those consistently displaying altered HOCl susceptibility are shown in Fig. S1. Statistical analysis of these data revealed 16 mutants that were significantly HOCl sensitive and 13 mutants that were significantly HOCl resistant compared to the WT at set time points (Tables 1 and 2).
FIG 1.
Identification of PA14 regulatory gene mutants with altered susceptibility to HOCl. Shown is an example screening plate with HOCl-sensitive (mexT and PA14_07110 [sahR]) and HOCl-resistant (PA14_53410 [1]) mutants labeled. Strains were grown in a 96-well format in LB medium with 4.4 mM HOCl, and optical density (OD600) was recorded as a measure of growth. Mutant strains that had an increased lag of >3 h compared to the WT were identified as HOCl sensitive, and strains with a decreased lag of >3 h compared to the WT were identified as HOCl resistant. The arrow represents the end of the WT lag phase. Strains grown in the absence of HOCl showed minor variations in growth, but none of the strains selected as sensitive or resistant (Table 1 and 2) had growth defects in LB only (data not shown).
TABLE 1.
HOCl-sensitive regulatory gene mutants
| PA14 locus tag and gene namea | PAO1 locus tag | Protein description |
P value for mutant OD600 vs WT OD600 at set time pointsb
|
||
|---|---|---|---|---|---|
| 20 h | 22 h | 24 h | |||
| PA14_07110 sahR | PA0547 | ArsR family transcriptional regulator | 0.0420 | 0.0003 | <0.0001 |
| PA14_08780 rpoC | PA4269 | DNA-directed RNA polymerase β chain | 0.0280 | 0.0002 | <0.0001 |
| PA14_13660 tpbA | PA3885 | Protein tyrosine phosphatase | 0.0522 | 0.0006 | 0.0005 |
| PA14_17900 metR | PA3587 | LysR family transcriptional regulator | 0.0342 | 0.0002 | <0.0001 |
| PA14_20730 flgM (1) | PA3351 | Flagellar biosynthesis anti-sigma-28 factor | 0.0288 | 0.0002 | <0.0001 |
| PA14_20730 flgM (2) | PA3351 | Flagellar biosynthesis anti-sigma-28 factor | 0.0299 | 0.0001 | <0.0001 |
| PA14_22370 (2) | PA3233 | Hypothetical protein, cyclic nucleotide-binding domain | 0.0274 | 0.0002 | <0.0001 |
| PA14_29740 | PA2656 | Two-component sensor | 0.2349 | 0.0427 | 0.0214 |
| PA14_32410 mexT | PA2492 | LysR family transcriptional regulator of multidrug efflux | 0.0370 | 0.0002 | <0.0001 |
| PA14_45950 rsaL | PA1431 | Regulatory protein | 0.1666 | 0.0367 | 0.0385 |
| PA14_52570 rsmA | PA0905 | Posttranscriptional global regulator | 0.0340 | 0.0003 | <0.0001 |
| PA14_56620 pyeR (2) | PA4354 | ArsR family transcriptional regulator | 0.0396 | 0.0010 | 0.0068 |
| PA14_60860 nfxB | PA4600 | TetR family transcriptional regulator of multidrug efflux | 0.0515 | 0.0049 | 0.0235 |
| PA14_62490 dksA | PA4723 | RNA polymerase-binding transcription factor | 0.0088 | 0.0006 | 0.0018 |
| PA14_68680 envZ (3) | PA5199 | Two-component sensor | 0.0494 | 0.0082 | 0.0714 |
| PA14_70570 recGc | PA5345 | ATP-dependent DNA helicase | 0.0053 | <0.0001 | <0.0001 |
Mutants were identified as HOCl sensitive by visual inspection (increased lag of >3 h compared to the WT) and statistical analysis of growth. The numbers in parentheses next to some of the gene mutants correspond to genes that have more than one mutant strain in the transposon library (Table S1).
Statistical analysis performed on two biological replicates. P values were determined by one-way analysis of variance with Dunnett’s multiple-comparison test.
recG is not a regulatory mutant per se but is on the same operon as oxyR, which encodes the H2O2-sensing transcriptional regulator; the transposon mutant in oxyR was not recoverable.
TABLE 2.
HOCl-resistant regulatory gene mutants
| PA14 locus tag and gene namea | PAO1 locus tag | Description |
P value for mutant OD600 vs WT OD600 at set time pointsb
|
|||
|---|---|---|---|---|---|---|
| 12 h | 14 h | 16 h | 18 h | |||
| PA14_06950 (1) | PA0533 | LuxR family transcriptional regulator | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| PA14_09760 | PA4185 | GntR family transcriptional regulator | 0.9538 | 0.0098 | 0.0170 | 0.2154 |
| PA14_10800 ampR | PA4109 | LysR family transcriptional regulator of β-lactamase | <0.0001 | <0.0001 | <0.0001 | 0.0003 |
| PA14_27900 | PA2802 | GntR family transcriptional regulator | 0.1181 | 0.0040 | 0.0040 | 0.0442 |
| PA14_38040 cmrA | PA2047 | AraC family transcriptional regulator | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| PA14_44490 anr | PA1544 | Transcriptional regulator of anaerobic metabolism | <0.0001 | <0.0001 | <0.0001 | 0.0053 |
| PA14_50180 fleR (1) | PA1099 | Two-component response regulator of flagellar biosynthesis | 0.9990 | 0.2376 | 0.0471 | 0.3638 |
| PA14_50200 fleS (2) | PA1098 | Two-component sensor of flagellar biosynthesis | 0.0610 | <0.0001 | 0.0017 | 0.0481 |
| PA14_56620 pyeR (1) | PA4354 | ArsR family transcriptional regulator | <0.0001 | <0.0001 | <0.0001 | 0.0001 |
| PA14_62530 cbrA (1) | PA4725 | Two-component sensor of catabolic pathways | <0.0001 | <0.0001 | <0.0001 | 0.0398 |
| PA14_62530 cbrA (2) | PA4725 | Two-component sensor of catabolic pathways | 0.8620 | 0.0482 | 0.0405 | 0.3687 |
| PA14_62540 cbrB | PA4726 | Two-component response regulator of catabolic pathways | 0.0049 | 0.0003 | 0.0140 | 0.2098 |
| PA14_68680 envZ (1) | PA5199 | Two-component sensor | <0.0001 | <0.0001 | <0.0001 | 0.0015 |
Mutants were identified as HOCl resistant by visual inspection (decreased lag of >3 h compared to the WT) and statistical analysis of growth. The numbers in parentheses next to some of the gene mutants correspond to genes that have more than one mutant strain in the transposon library (Table S1).
Statistical analysis performed on two biological replicates. P values were determined by one-way analysis of variance with Dunnett’s multiple-comparison test.
Links between antimicrobial resistance and HOCl susceptibility: the MexEF-OprN multidrug efflux pump protects P. aeruginosa from HOCl.
A mutant in mexT was unable to grow in the presence of HOCl (Fig. 2A). MexT is a LysR-type regulator that positively regulates expression of its adjacent operon mexEF-oprN, which encodes a multidrug efflux pump (28, 29). MexT regulates a number of genes (30, 31), in addition to the mexEF-oprN operon; thus, to determine whether loss of the efflux pump is associated with HOCl sensitivity, transposon mutants of all three efflux pump genes were tested for altered HOCl susceptibility. They displayed increased sensitivity in the order of mexE > mexF > oprN (Fig. 2A). For all three mutant strains, the transposon is inserted in the middle of the gene. This suggests that the MexEF-OprN efflux pump provides P. aeruginosa with protection against HOCl. MexT positively regulates 12 other genes (30) in addition to mexEF-oprN and pyeR (mentioned below); mutants in 8 of these were available for screening, and 4 displayed HOCl sensitivity (PA14_41990, PA14_39060, PA14_27755, and PA14_27770), although none were as sensitive as mexE (Table S2).
FIG 2.
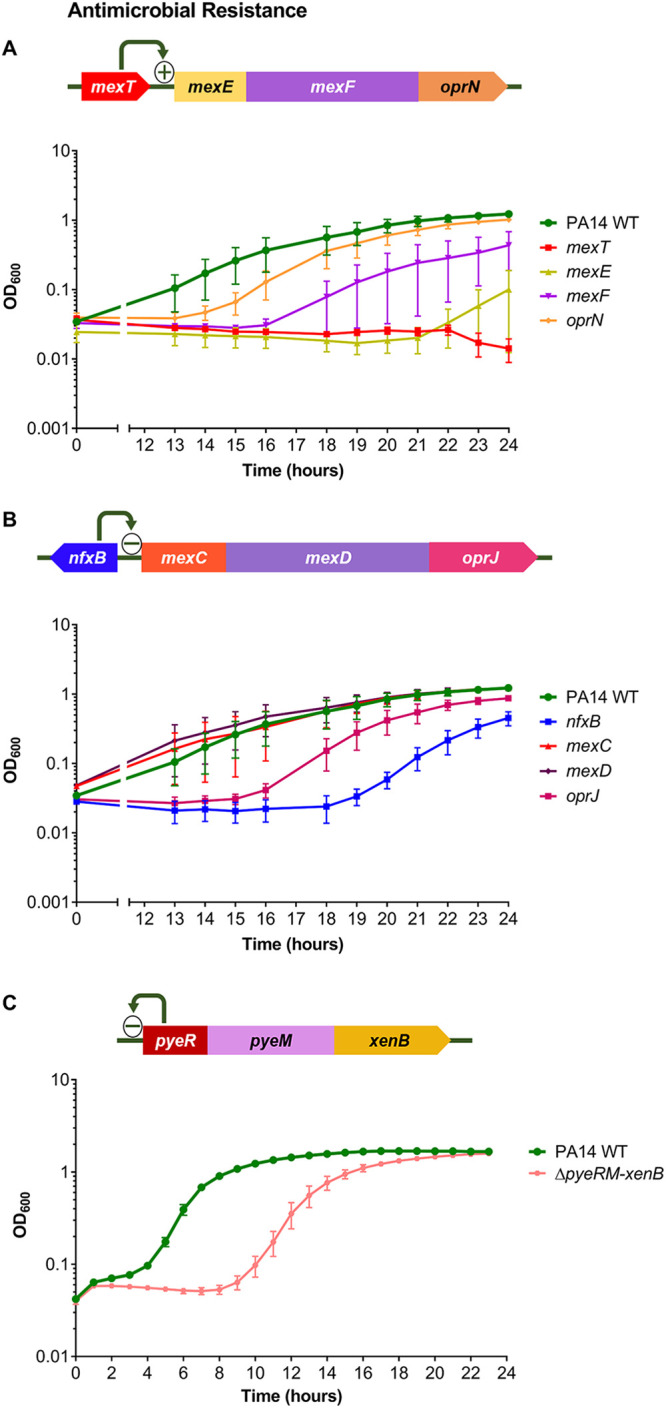
Mutants of genes involved in antimicrobial resistance displayed altered susceptibility to HOCl. (A) Gene arrangement of mexT and its adjacent mexEF-oprN operon. Growth of PA14 WT, mexT, mexE, mexF, and oprN strains in the presence of 4.4 mM HOCl. (B) Gene arrangement of nfxB and its adjacent mexCD-oprJ operon. Growth of PA14 WT, nfxB, mexC, mexD, and oprJ strains in the presence of 4.4 mM HOCl. For mexC and mexD, 2 transposon mutants of each gene were available and showed similar phenotypes; therefore, only 1 mutant of each is displayed. (C) Gene arrangement of the pyeRM-xenB operon; PyeR autorepresses the pyeRM-xenB operon (33). Growth of PA14 WT and ΔpyeRM-xenB strains in the presence of 5.1 mM HOCl. Graphs display the means and standard errors of the means for three independent experiments (A and B) and three biological replicates (C), representative of three independent experiments. The plus and minus signs in the gene arrangement diagrams represent positive and negative regulators, respectively. All mutant strains displayed statistically significant altered susceptibility to HOCl compared with the WT at different time points, apart from oprN, mexC, mexD, and oprJ (P < 0.05; one-way analysis of variance with Dunnett’s multiple-comparison test). Strains grown in the absence of HOCl showed no growth defects (data not shown). All transposon mutants and the in-frame deletion were confirmed by PCR.
Another multidrug efflux pump regulator, NfxB, was required for protection against HOCl, as an nfxB mutant had increased HOCl sensitivity (Fig. 2B). NfxB is a negative regulator of the mexCD-oprJ multidrug efflux operon (32), and transposon mutants in these genes were tested; only oprJ had altered susceptibility, displaying HOCl sensitivity (Fig. 2B).
MexT regulates the pyeRM-xenB operon, in which pyeR encodes an ArsR family repressor, pyeM encodes a major facilitator superfamily (MFS) transporter, and xenB encodes an uncharacterized xenobiotic reductase (33). Transposon mutants in pyeR showed variable susceptibility to HOCl (Fig. S1); therefore, we constructed a whole-operon in-frame ΔpyeRM-xenB deletion mutant, which displayed HOCl sensitivity (Fig. 2C). This suggests a role for this MexT-regulated operon in protecting P. aeruginosa against HOCl.
Metabolic regulators are involved in protection against HOCl.
A mutant in metR that encodes a regulator homologous (40% identity) to E. coli MetR, a regulator of methionine biosynthesis genes (34), was HOCl sensitive (Fig. 3A). A mutant in PA14_07110 (sahR) was also HOCl sensitive (Fig. 3A). This gene encodes an ortholog of SahR from Desulfovibrio spp., which is a transcriptional regulator of the S-adenosylmethionine cycle genes that are involved in methionine recycling (35).
FIG 3.
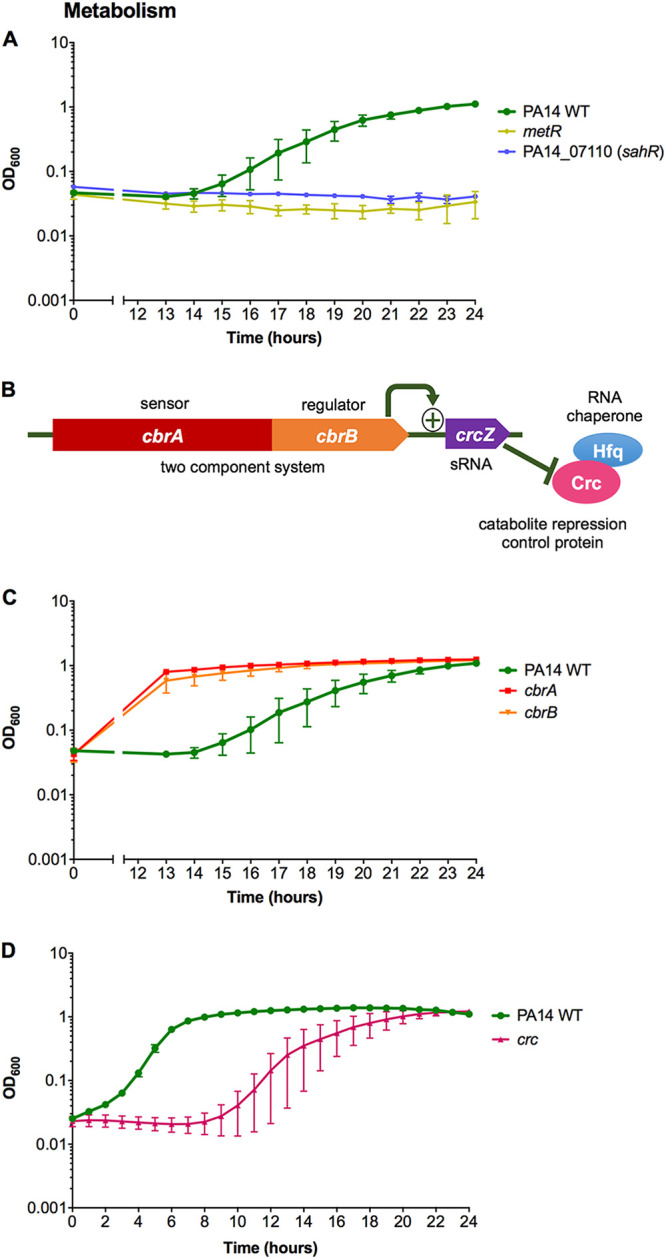
Mutants of genes involved in metabolism displayed altered susceptibility to HOCl. (A) Growth of PA14 WT, metR, and PA14_07110 (sahR) strains in the presence of 4.4 mM HOCl. (B) Gene arrangement of the two-component system cbrAB that regulates the expression of the sRNA CrcZ, which relieves Crc/Hfq-mediated repression of target mRNAs. (C and D) Growth of PA14 WT, cbrA, and cbrB strains (C) and PA14 WT and crc strains (D) in the presence of 4.4 mM HOCl. For cbrA, 2 transposon mutants were available and showed similar phenotypes; therefore, only 1 mutant is displayed. Graphs display the means and standard errors of the means for three independent experiments (A and C) and three biological replicates (D), representative of three independent experiments. The plus sign in the gene arrangement diagram represents a positive regulator. All mutant strains displayed statistically significant altered susceptibility to HOCl compared with the WT at different time points (P < 0.05; one-way analysis of variance with Dunnett’s multiple-comparison test). Strains grown in the absence of HOCl showed no growth defects (data not shown). All transposon mutants were confirmed by PCR.
The CbrA/B two-component system is involved in controlling carbon and nitrogen metabolism and carbon catabolite repression (36–38). CbrA/B acts through activation of the small RNA (sRNA) CrcZ, which sequesters the RNA chaperone Hfq and the catabolite repression control protein Crc from mRNA target genes, relieving catabolite repression (38–40) (Fig. 3B). Mutants of cbrAB were HOCl resistant (Fig. 3C), and subsequently the crc transposon mutant was tested and found to be HOCl sensitive (Fig. 3D). These phenotypes correlate with the opposing regulatory roles of CbrA/B and Crc.
Regulators with putative roles in the oxidative stress response are required for protection against HOCl.
A mutant in the algH gene, which encodes a hypothetical protein that has been crystallized and predicted to be a potential redox sensor, was HOCl sensitive (Fig. 4A) (41). algH is part of a 4-gene operon that includes gshB, which encodes a glutathione synthetase; a gshB mutant was tested, and it displayed HOCl sensitivity (Fig. 4A).
FIG 4.
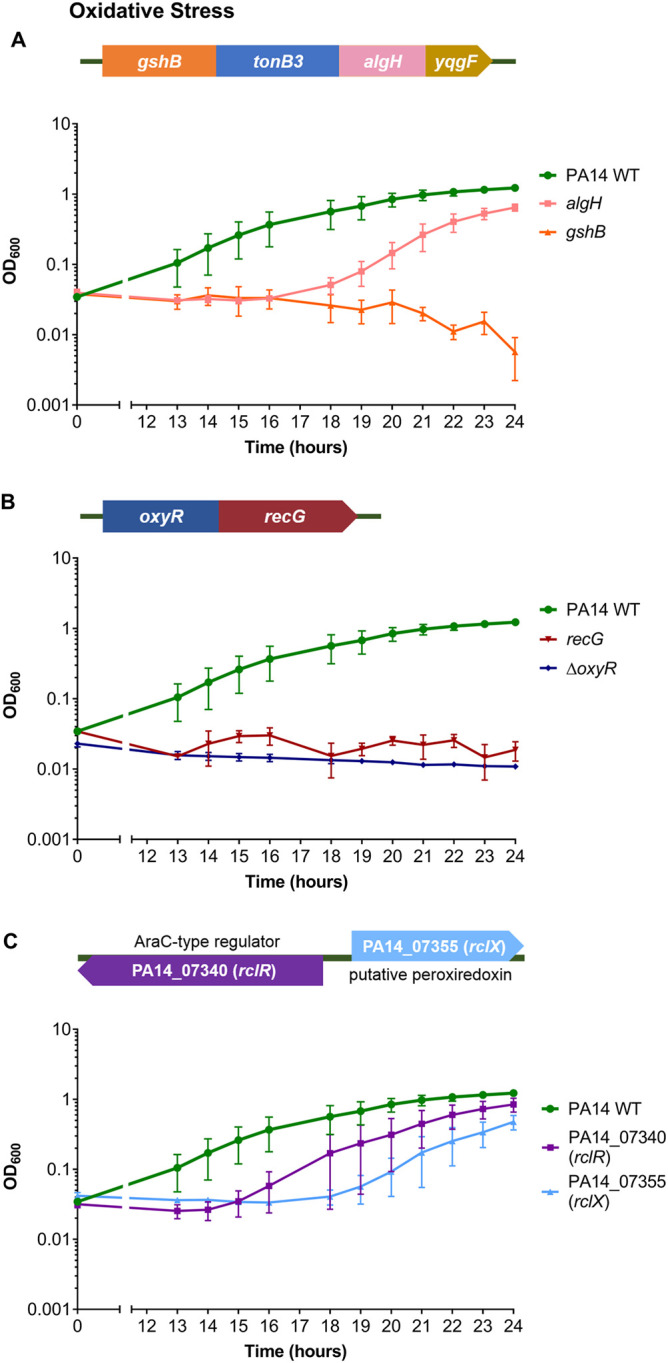
Mutants of genes involved in oxidative stress defense displayed altered susceptibility to HOCl. (A) Gene arrangement of the predicted 4-gene operon, gshB-tonB3-algH-yqgF. Growth of PA14 WT, algH, and gshB strains in the presence of 4.4 mM HOCl. PA14 transposon mutants were not available for yqgF but were for tonB3; however, they did not display altered HOCl susceptibility (data not shown). (B) Gene arrangement of the oxyR-recG operon. Growth of PA14 WT, recG, and ΔoxyR strains in the presence of 4.4 mM HOCl. (C) Gene arrangement of PA14_07340 (rclR) and PA14_07355 (rclX). Growth of PA14 WT, PA14_07340 (rclR), and PA14_07355 (rclX) strains in the presence of 4.4 mM HOCl. Graphs display the means and standard errors of the means for three independent experiments (two independent experiments only for recG). All mutant strains displayed statistically significant altered susceptibility to HOCl compared with the WT at different time points (P < 0.05; one-way analysis of variance with Dunnett’s multiple-comparison test). Strains grown in the absence of HOCl showed no growth defects (data not shown). All transposon mutants and the in-frame deletion were confirmed by PCR.
A mutant in oxyR encoding an H2O2-sensing transcriptional regulator (42) was nonviable; however, a mutant was available for its adjacent gene, a putative DNA repair enzyme, recG (42), and was HOCl sensitive (Fig. 4B). The sensitivity of the recG mutant to HOCl was not due to disruption of oxyR, as reverse transcription-PCR (RT-PCR) confirmed the oxyR transcript was present in the recG mutant (data not shown). We constructed an in-frame deletion of oxyR, which displayed HOCl sensitivity (Fig. 4B). This suggests that the oxyR-recG operon is involved in protection against HOCl as well as H2O2 (42).
P. aeruginosa mutants of homologues of other transcriptional regulators that respond to HOCl in bacteria, NemR (PA14_36300), HypT (PA14_71640), and OhrR (PA14_27230), did not display consistent HOCl-sensitive or -resistant phenotypes in the screen (Fig. S2). However, a mutant in PA14_07340, the P. aeruginosa homologue of the E. coli reactive chlorine-specific transcriptional regulator, RclR (17), was identified as HOCl sensitive (Fig. 4C). The rclR mutant was one of three strains from the screen that showed consistent, reproducible sensitivity to HOCl and displayed significant altered growth in the presence of the oxidant when rescreened (Fig. 4C) but did not show significant altered growth at the time points analyzed in the initial screen (Table 1). P. aeruginosa rclR (PA14_07340) is adjacent to a single-gene operon, PA14_07355, that encodes a putative peroxiredoxin enzyme of the alkylhydroperoxidase (AhpD) family (Fig. 4C). A mutant in PA14_07355 was tested and displayed HOCl sensitivity (Fig. 4C).
P. aeruginosa RclR is specifically required for protection against HOCl and HOSCN.
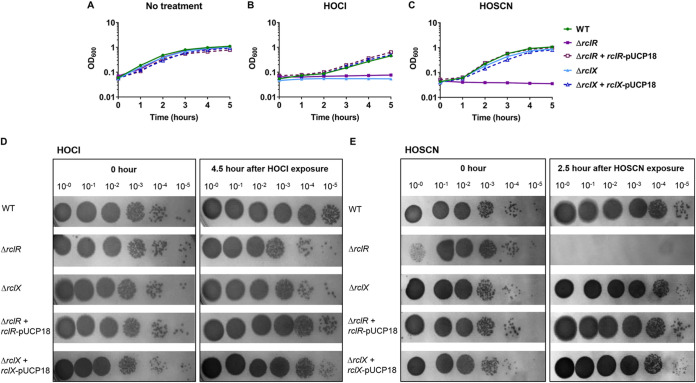
To confirm the role of P. aeruginosa RclR in protection against HOCl, in-frame deletion mutants of rclR and the putative peroxiredoxin gene, which we named rclX, were constructed. The susceptibility of the mutants to HOCl was retested; ΔrclR and ΔrclX strains grew similarly to the WT in the absence of oxidant (Fig. 5A), but neither mutant grew in the presence of HOCl over the time course of the experiment (Fig. 5B). Complementation of ΔrclR and ΔrclX strains with rclR-pUCP18 and rclX-pUCP18, respectively, restored growth in the presence of HOCl to that of the WT (Fig. 5B). To test the specificity of RclR, mutants were exposed to other reactive oxygen species (H2O2, the O2− generator methyl viologen, and tert-butyl hydroperoxide [TBH]), reactive electrophilic species (N-ethylmaleimide [NEM], methylglyoxal, and diamide), and the nitric oxide generator diethylamine NONOate (DEANO). These reactive species did not alter the growth of the ΔrclR strain compared to the WT, indicating that P. aeruginosa RclR is not required for protection against these compounds (Fig. S3). Due to HOSCN being another physiologically relevant, thiol-reactive oxidant produced by the immune system (9), we were interested in determining whether ΔrclR and ΔrclX strains had altered sensitivity to this oxidant. Growth of ΔrclR and ΔrclX strains in the presence of 0.8 mM HOSCN revealed that the ΔrclR strain was sensitive to HOSCN, whereas the ΔrclX strain was not at this concentration (Fig. 5C). Complementation of the ΔrclR strain with rclR-pUCP18 restored growth in the presence of HOSCN (Fig. 5C). These data were supported by viability assays, which showed HOCl inhibited growth of ΔrclR and ΔrclX strains (Fig. 5D), and HOSCN was bactericidal toward the ΔrclR strain (Fig. 5E). When the HOSCN concentration was increased to 1 mM, the ΔrclX as well as the ΔrclR strain displayed sensitivity to this oxidant (Fig. S4). Therefore, both RclR and RclX are required for protection against HOCl and HOSCN.
FIG 5.
Mutants of rclR and its adjacent gene rclX are sensitive to HOCl, and rclR is sensitive to the epithelium-derived oxidant HOSCN. (A to C) Growth of PA14 WT, ΔrclR, ΔrclR + rclR-pUCP18, ΔrclX, and ΔrclX + rclX-pUCP18 strains in LB medium (A), LB medium with 4.4 mM HOCl (B), or LB medium with 0.8 mM HOSCN (C). Graphs display the means and standard errors of the means for three biological replicates, representative of three independent experiments. For HOSCN sensitivity assays, a control reaction of glucose oxidase, glucose, and potassium thiocyanate was carried out, which resulted in the production of H2O2 only; the ΔrclR strain was not sensitive under these conditions, confirming sensitivity is due to HOSCN production (data not shown). The addition of the empty pUCP18 vector to ΔrclR and ΔrclX strains had no effect on the susceptibility of these mutants to HOCl or HOSCN (data not shown). (D and E) Viability CFU assays. Mid-exponential-phase cultures of PA14 WT, ΔrclR, ΔrclX, ΔrclR + rclR-pUCP18, and ΔrclX + rclX-pUCP18 strains were incubated in LB medium containing 4.4 mM HOCl (D) or 0.8 mM HOSCN (E). Samples were taken immediately after oxidant exposure and after 4.5 h (HOCl) or 2.5 h (HOSCN), diluted in PBS, spot titered onto LBA, and incubated at 37°C overnight or until CFU could be visualized. (E) For undiluted (10−0) ΔrclR samples taken immediately after HOSCN exposure (0 h), fewer viable cells are visualized because HOSCN acted immediately, whereas in the other dilution factors HOSCN was diluted down and, therefore, did not affect growth. Images are representative of three independent experiments.
The sensitivity assays thus far were performed on planktonic cells in Luria-Bertani (LB) medium; to determine whether the findings were consistent when cells were grown under conditions relevant to the CF lung environment, sensitivity assays for ΔrclR and ΔrclX mutants were repeated in ASM (artificial sputum medium), which mimics the CF sputum (43, 44). Both ΔrclR and ΔrclX strains displayed HOCl and HOSCN sensitivity when grown in ASM (Fig. S5), confirming the findings of the LB assays. Additionally, during chronic CF lung infections, P. aeruginosa persists in biofilms. To determine whether P. aeruginosa mutants display similar HOCl and HOSCN susceptibility when forming biofilms, ΔrclR and ΔrclX mutants were grown in a suspended ASM biofilm assay in the absence or presence of HOCl and HOSCN (adapted from reference 44). The WT, ΔrclR, and ΔrclX strains formed biofilms in ASM in the absence of the oxidants (Fig. S6). However, in the presence of 3.5 mM HOCl and 0.53 mM HOSCN, both ΔrclR and ΔrclX strains were unable to grow and appeared sensitive to these oxidants compared to the WT, which was able to form biofilms at these concentrations (Fig. S6). These data confirm the relevance of the impact of HOCl and HOSCN on P. aeruginosa in a more realistic CF environment.
RclR regulates induction of rclX expression in response to HOCl and HOSCN stress.
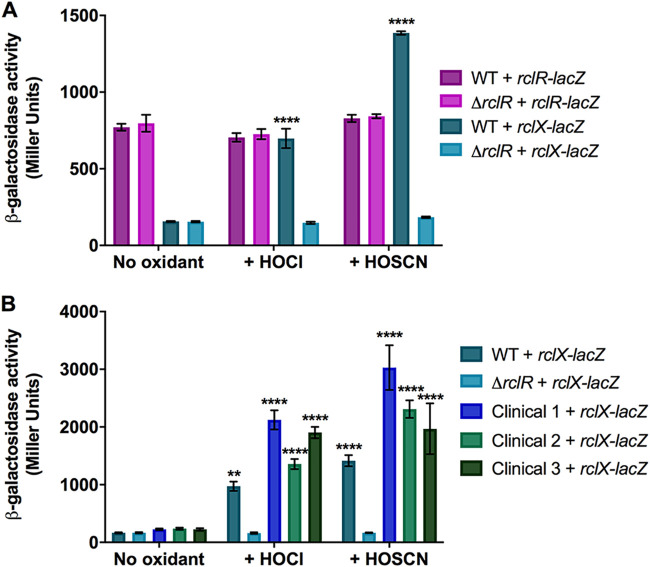
To determine whether RclR regulates expression of itself and/or rclX under HOCl and HOSCN stress, transcriptional rclR-lacZ and rclX-lacZ fusions were constructed and introduced into the WT and ΔrclR strains for in vivo β-galactosidase assays. The activity of rclX-lacZ increased following HOCl and HOSCN exposure in the WT strain but not in the ΔrclR strain, suggesting that expression of rclX increases following HOCl and HOSCN stress dependent on RclR regulation (Fig. 6A). No difference in the activity of rclR-lacZ was observed between the WT and ΔrclR strains in the absence or presence of HOCl and HOSCN, suggesting RclR does not autoregulate its own expression and is constitutively expressed (Fig. 6A). We next examined if the activation of rclX in response to these oxidants occurred in clinical CF isolates of P. aeruginosa. The activity of rclX-lacZ in three CF isolates was measured in the absence or presence of HOCl and HOSCN. The rclX gene was significantly induced in response to both oxidants in all three clinical isolates compared to untreated controls (Fig. 6B). This confirmed that rclX induction appears to be an important adaptive response to these oxidants across P. aeruginosa laboratory and clinical strains.
FIG 6.
HOCl and HOSCN stress induces RclR-dependent regulation of rclX expression in PA14 P. aeruginosa and clinical CF isolates. (A) Activity of rclR-lacZ and rclX-lacZ transcriptional fusions in the PA14 WT and ΔrclR strains in the absence or presence of 2.2 mM HOCl or 0.8 mM HOSCN. (B) Activity of the rclX-lacZ transcriptional fusions in WT, ΔrclR, and clinical CF isolates 1, 2, and 3 in the absence or presence of 2.2 mM HOCl or 0.8 mM HOSCN. Strains were grown aerobically in LB medium until mid-exponential phase, when HOCl, HOSCN, or LB only was added, and samples were taken for β-galactosidase activity measurements at 30 min. The concentrations of oxidants used caused a 30- to 120-min lag in growth of exponential-phase cultures. PA14 WT and ΔrclR strains carrying the empty pMP220 vector expressed low β-galactosidase activity (∼100 Miller units) under all of the treatment conditions (data not shown). Data show the means and standard errors of the means for three biological replicates, representative of three independent experiments (six biological replicates for untreated strains [B]). Statistical analysis was performed using one-way analysis of variance with Tukey’s multiple-comparison post hoc test. **, P < 0.01; ****, P < 0.0001; compared to the same strain untreated (no oxidant).
To investigate the possible function of RclX, we performed homology modeling that fitted the RclX sequence with 32.1% sequence identity to the AhpD-like protein, Lpg0406, from Legionella pneumophila, which belongs to the carboxymuconolactone decarboxylase family (45) (Fig. S7). The predicted structure was a homohexamer (Fig. S7A), formed from monomers with an AhpD-like fold containing six α-helices and a conserved catalytic CXXC motif (Fig. S7B). RclX was sequence aligned with Lpg0406, PA0269, another AhpD-like protein from P. aeruginosa (46), and MtAhpD, from Mycobacterium tuberculosis (47–49) (Fig. S7C). Four out of the five catalytic residues of MtAhpD responsible for peroxidase activity (47, 48) were conserved in RclX (Fig. S7C), suggesting a role for RclX in oxidant detoxification.
Transcriptional changes in PA14 WT and ΔrclR strains in response to HOCl and HOSCN exposure.
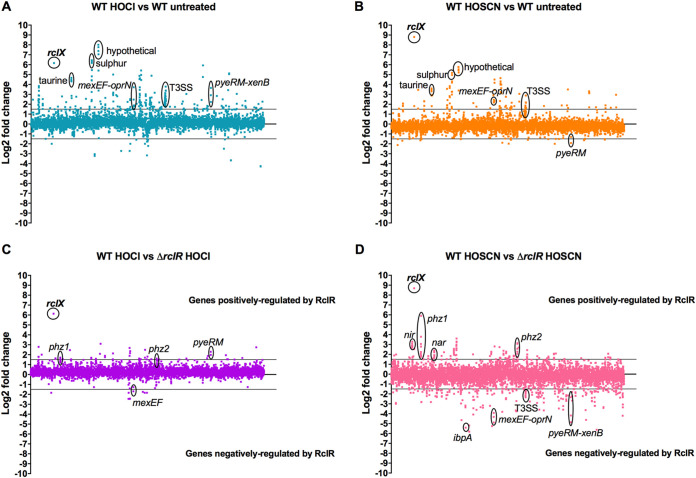
The response of P. aeruginosa to HOCl and HOSCN was further characterized by measuring changes in gene expression in the WT and ΔrclR strains. PA14 WT and ΔrclR exponential-phase cultures were exposed to 2.2 mM HOCl or 0.8 mM HOSCN, and gene expression was analyzed by transcriptome sequencing (RNA-seq). Genes with a log2 fold change of >1.5 were considered upregulated, and genes with a log2 fold change of <−1.5 were considered downregulated (Fig. 7). All transcriptomic data are presented in Data Set S1 and are accessible through the Gene Expression Omnibus (GEO) database under accession number GSE124385. For identification of genes whose expression is altered in the presence of HOCl or HOSCN, expression was compared in treated WT strains versus untreated WT strains. HOCl upregulated 231 genes and downregulated 20 genes in the WT strain (Fig. 7A). HOSCN upregulated 105 genes and downregulated 16 genes in the WT strain (Fig. 7B). The importance of rclX in the P. aeruginosa response to HOCl and HOSCN was confirmed, as it was strongly upregulated with a >6 and >8 log2 fold change, respectively, and this was consistent with the gene fusion expression data (Fig. 6 and 7A and B). These data have been validated by quantitative RT-PCR (qRT-PCR) for a subset of the most highly upregulated and downregulated genes (including rclX) whose patterns of expression were confirmed (5 upregulated and 1 downregulated) (Fig. S8). For the identification of genes whose expression is dependent on RclR, expression was compared in treated WT strains versus treated ΔrclR strains. Therefore, in Fig. 7C and D, genes displaying increased expression indicate genes positively regulated by RclR, and genes displaying decreased expression indicate genes negatively regulated by RclR. In the presence of HOCl, RclR upregulated 42 genes and downregulated 7 genes (Fig. 7C), and in the presence of HOSCN, RclR upregulated 132 genes and downregulated 213 genes (Fig. 7D). Regulation of rclX by RclR was confirmed, as it was the gene most strongly upregulated by RclR in the presence of HOCl and HOSCN (Fig. 7C and D). Expression of rclR was not altered following exposure to HOCl or HOSCN, agreeing with our previous gene fusion data (Fig. 6A and Data Set S1). Furthermore, the genes of homologues of other HOCl-responsive transcriptional regulators from E. coli and B. subtilis, NemR (PA14_36300), HypT (PA14_71640), and OhrR (PA14_27230), did not have altered expression in response to HOCl or HOSCN (Data Set S1).
FIG 7.
Relative changes in gene expression in PA14 WT and ΔrclR strains in the presence of HOCl or HOSCN. Log2 fold change of gene expression was plotted against the gene locus tag in order of genomic location. (A) Effect of HOCl on P. aeruginosa gene expression, WT plus HOCl versus WT untreated. (B) Effect of HOSCN on P. aeruginosa gene expression, WT plus HOSCN versus WT untreated. (C) Identification of genes regulated by RclR in the presence of HOCl, WT plus HOCl versus ΔrclR strain plus HOCl. (D) Identification of genes regulated by RclR in the presence of HOSCN, WT plus HOSCN versus ΔrclR strain plus HOSCN. Lines indicate cutoff values at 1.5 or −1.5 log2 fold change. (A and B) Highlighted upregulated genes include rclX, hypothetical operon (PA14_21570-PA14_21580-PA14_21590-PA14_21600), sulfur transport genes (ssuD-PA14_19570-ssuB), taurine transport genes (PA14_12920-PA14_12940-PA14_12960), drug efflux pump operon mexEF-oprN, pyeRM-xenB operon (HOCl), and T3SS genes. Downregulated genes include pyeRM (HOSCN). (C and D) In these plots, positive log2 fold change values indicate genes positively regulated by RclR, and negative log2 fold change values indicate genes negatively regulated by RclR. Highlighted genes positively regulated include rclX, pyocyanin biosynthesis operons phz1 and phz2, denitrification nir and nar operons (HOSCN), and pyeRM (HOCl). Negatively regulated genes include pyeRM-xenB (HOSCN), mexEF-oprN, T3SS genes (HOSCN), and heat shock protein ibpA (HOSCN).
The upregulated and downregulated genes were examined for overlap between the response to both oxidants and were categorized into functional groups based on assigned gene ontologies in the Pseudomonas genome database (27) (Fig. S9). Of the 105 genes upregulated in response to HOSCN, 74 (70%) were also upregulated in response to HOCl, indicating similarities between the bacterial responses to these two oxidants (Fig. S9A). The functional groups that displayed the largest percentage of genes upregulated in response to HOCl and HOSCN were noncoding RNA, protein secretion/export apparatus, and antibiotic resistance and susceptibility (Fig. S9B). Of the 132 RclR-regulated genes whose expression was upregulated in the presence of HOSCN, only 12 (9%) of those were also upregulated in the presence of HOCl (Fig. S9C), and of the 213 RclR-regulated genes downregulated following HOSCN exposure, only 2 (1%) were downregulated after HOCl exposure (Fig. S9D). For RclR-regulated genes, the functional group with the highest percentage of genes upregulated following HOCl and HOSCN exposure was secreted factors (Fig. S9E). RclR downregulated a large number of genes in the presence of HOSCN, particularly those in the chaperone and heat shock protein functional groups (Fig. S9F). The observed regulatory effects might be a direct consequence of loss of RclR-mediated regulation or a result of secondary pathways impacted by rclR deletion.
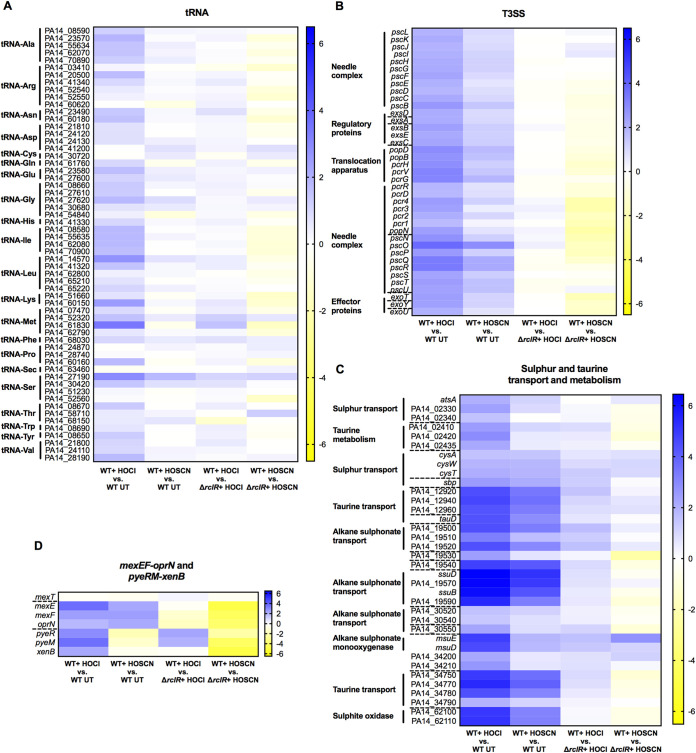
tRNA, type 3 secretion system (T3SS), sulfur and taurine metabolism, and MexT-regulated efflux pump genes show increased expression following exposure to HOCl and HOSCN.
The expression of functionally categorized genes was displayed in heat maps to demonstrate differences in gene expression in response to HOCl (first column) or HOSCN (second column) and in which upregulated genes are displayed in blue and downregulated in yellow (Fig. 8 and 9). Genes that are positively or negatively regulated by RclR are indicated by blue or yellow, respectively, in the presence of HOCl (third column) or HOSCN (fourth column) (Fig. 8 and 9).
FIG 8.
Heat maps displaying expression of tRNA, T3SS, sulfur and taurine transport and metabolism, and mexEF-oprN efflux pump and pyeRM-xenB genes in PA14 WT and ΔrclR strains after HOCl or HOSCN exposure. (A) tRNAs. (B) T3SS. (C) Sulfur and taurine transport and metabolism. (D) mexEF-oprN and pyeRM-xenB. Expression of genes is color coordinated from 6.5 log2 fold change (blue) to −6.5 log2 fold change (yellow). The first column indicates HOCl-responsive genes (log2 fold change of WT plus HOCl versus WT untreated [UT]), and the second column indicates HOSCN-responsive genes (log2 fold change of WT plus HOSCN versus WT UT). The third column indicates RclR-regulated genes under HOCl stress (log2 fold change of WT plus HOCl versus ΔrclR strain plus HOCl), and the fourth column indicates RclR-regulated genes under HOSCN stress (log2 fold change of WT plus HOSCN versus ΔrclR strain plus HOSCN). In these columns, genes that are positively or negatively regulated are indicated by blue or yellow, respectively. Broken lines indicate the start and end of operons.
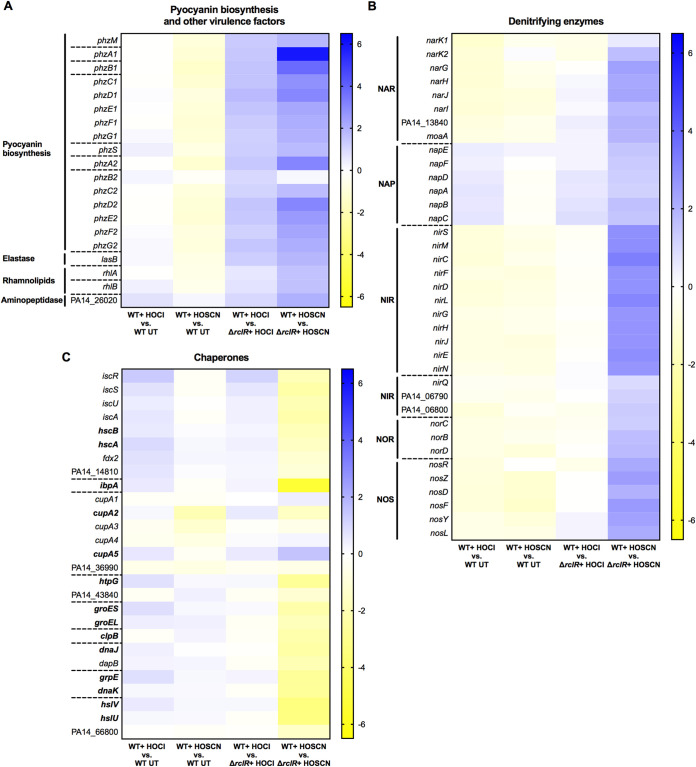
FIG 9.
Heat maps displaying expression of pyocyanin biosynthesis, denitrifying enzyme, and chaperone and heat shock genes in PA14 WT and ΔrclR strains after HOCl or HOSCN exposure. (A) Pyocyanin biosynthesis and other virulence factors. (B) Denitrifying enzymes. (C) Chaperones (genes in boldface indicate the chaperones). The expression of genes is color coordinated from 6.5 log2 fold change (blue) to −6.5 log2 fold change (yellow). The first column indicates HOCl-responsive genes (log2 fold change of WT plus HOCl versus WT UT), and the second column indicates HOSCN-responsive genes (log2 fold change of WT plus HOSCN versus WT UT). The third column indicates RclR-regulated genes under HOCl stress (log2 fold change of WT plus HOCl versus ΔrclR strain plus HOCl), and the fourth column indicates RclR-regulated genes under HOSCN stress (log2 fold change of WT plus HOSCN versus ΔrclR strain plus HOSCN). In these columns, genes that are positively or negatively regulated are indicated by blue or yellow, respectively. Broken lines indicate the start and end of operons.
The genes of noncoding RNAs were upregulated in response to HOCl and HOSCN; HOCl increased the expression of tRNAs (Fig. 8A), whereas HOSCN upregulated 5S and 16S rRNAs (Data Set S1). An rRNA depletion step had been carried out prior to RNA-seq using the highly efficient Ribo-Zero kit (50), and less than 2% rRNA was recovered; therefore, the physiological importance of this induction is unclear. All of the genes that are involved in biogenesis and regulation of the T3SS (51) were induced in the WT in response to both HOCl and HOSCN, while RclR appeared to downregulate a number of the T3SS genes in the presence of HOSCN (Fig. 8B and Data Set S1). Around 40% of the genes in the transport of small molecules category that were upregulated in the WT response to HOCl and HOSCN were involved in sulfur or taurine metabolism (Fig. 8C and Fig. S9B and Data Set S1). The mexEF-oprN operon, which encodes the efflux pump that we found plays a role in protection against HOCl (Fig. 2A), was upregulated in response to HOCl and HOSCN exposure (Fig. 8D and Data Set S1). The mexT regulator did not have altered gene expression (Fig. 8D and Data Set S1). Surprisingly, these data indicated that RclR downregulates the expression of the mexEF-oprN operon in the presence of HOCl and HOSCN (Fig. 8D and Data Set S1). The MexT-regulated operon, pyeRM-xenB, which is also involved in protection against HOCl (Fig. 2C), was upregulated in response to HOCl but was downregulated in response to HOSCN, and this appeared to be dependent on RclR (Fig. 8D and Data Set S1).
RclR regulates expression of pyocyanin biosynthesis genes in the presence of HOCl and HOSCN and denitrification and chaperone genes in the presence of HOSCN.
Heat maps of functionally categorized genes with altered expression in the ΔrclR deletion mutant included phz, denitrification, and chaperone genes. The phz genes are involved in the biosynthesis of the virulence factor pyocyanin (52). The overall WT response to HOCl and HOSCN had no and little effect, respectively, on phz expression (Fig. 9A and Data Set S1). However, the ΔrclR mutant displayed lower phz expression levels after HOCl and HOSCN exposure than the WT; thus, when values of the treated WT were divided by those for the treated ΔrclR strain, the results indicate that RclR positively regulates phz gene expression in the presence of both oxidants (Fig. 9A and Data Set S1). In the absence of these oxidants, there was no difference between pyocyanin production in WT and ΔrclR strains (data not shown). Similarly, the virulence factors elastase (lasB) and rhamnolipids (rhlA and rhlB) and an aminopeptidase (PA14_26020) did not display altered expression in response to HOCl or HOSCN in the WT but were upregulated by RclR in the presence of both oxidants (Fig. 9A and Data Set S1). In the WT strain, HOCl and HOSCN had little effect on the expression of genes required for anaerobic dissimilatory denitrification (Fig. 9B and Data Set S1). However, in the presence of HOSCN, but not HOCl, RclR positively regulated the expression of 6 operons encoding the denitrifying enzymes: nitrate reductase (NAR), periplasmic nitrate reductase (NAP), nitrite reductase (NIR), nitric oxide reductase (NOR), and nitrous oxide reductase (NOS) (Fig. 9B and Data Set S1). HOCl and HOSCN had little effect on the expression of chaperone and heat shock genes in the WT strain (Fig. 9C and Data Set S1). RclR did not regulate the expression of these genes in the presence of HOCl, yet in the presence of HOSCN, RclR negatively regulated the expression of 13 chaperone and heat shock genes, hcsB, hcsA, ibpA, cupA2, htpG, groES, groEL, clpB, dnaJ, grpE, dnaK, hslV, and hslU, and positively regulated the expression of the chaperone gene cupA5 (Fig. 9C and Data Set S1).
DISCUSSION
A major mechanism employed by the innate immune response to attack infecting bacteria, such as during lung infections, is through the production and release of the antimicrobial oxidants HOCl and HOSCN by neutrophils and the airway epithelium, respectively (7, 9).
In this study, we investigated how P. aeruginosa adapts to and protects itself against HOCl and HOSCN. This is of direct relevance to CF infections due to evidence of a compromised immune response in CF patients, including impaired HOCl and HOSCN formation (53–57), and evidence for HOSCN production being protective of lung function (58).
Our mutant screen identified regulators with known or putative roles in antibiotic resistance, methionine biosynthesis, catabolite repression, and the antioxidant response that are required for protecting P. aeruginosa against HOCl (Fig. 2, 3, and 4). We revealed that the RclR transcriptional regulator, the P. aeruginosa homologue of the E. coli RclR HOCl-specific sensor (17), is required specifically for protection of P. aeruginosa against HOCl and HOSCN and responds to the presence of these oxidants through activating expression of its adjacent gene, rclX (Fig. 5, 6, and 7).
E. coli RclR (RclREc) also induces the expression of its adjacent operon, rclABC, in the presence of HOCl (17). However, unlike rclREc (17), we found P. aeruginosa rclR (rclRPa) is constitutively expressed, not upregulated by HOCl (or HOSCN), and does not regulate its own expression (Fig. 6A). The rclX gene regulated by RclRPa encodes a putative peroxiredoxin, is one of the most upregulated genes in response to HOCl and HOSCN exposure, and is required for protection against both oxidants (Fig. 5, 6, and 7; see also Fig. S4 in the supplemental material). RclX is not homologous to any of the genes in the RclREc-regulated rclABC operon (17), indicating differing approaches to protection against HOCl between these Gram-negative bacteria. Homology modeling and multiple-sequence alignment support RclX being an AhpD-like protein, with 4 conserved catalytic residues, including the CXXC motif (Fig. S7), which is required for peroxidase activity in the functionally characterized AhpD from M. tuberculosis (47, 48). Therefore, it is probable RclX acts as a detoxification enzyme in the protective response to HOCl and HOSCN. The sensitivity of ΔrclR and ΔrclX mutants to HOCl and HOSCN, when grown in ASM in both planktonic and biofilm cultures (Fig. S5 and S6), together with HOCl- and HOSCN-dependent expression of rclX in clinical CF isolates of P. aeruginosa (Fig. 6B), indicates that the RclR-mediated response is important in clinical strains and may play a role during infection.
There was considerable overlap in the transcriptional response of P. aeruginosa to HOCl and HOSCN (Fig. S9A and B). RclR regulated expression of a larger number of genes in the presence of HOSCN than HOCl (Fig. S9C and D). HOSCN caused RclR-dependent upregulation of pyocyanin biosynthesis genes and denitrification genes and downregulation of chaperone and heat shock genes (Fig. 9). Three other transcriptional regulators that are HOCl responsive in other bacteria, NemR (PA14_36300), HypT (PA14_71640), and OhrR (PA14_27230) (13, 15, 16), did not have altered expression in response to HOCl or HOSCN (Data Set S1). In a previous transcriptome study, Groitl et al. reported that P. aeruginosa nemR was induced by HOCl and HOSCN and P. aeruginosa hypT was induced by HOCl (25). Nevertheless, we demonstrate consistencies in our results, as nemR, hypT, and ohrR were not HOCl responsive and mutants in these genes were not HOCl sensitive (Fig. S2). The reasons for these differences between the Groitl et al. study and ours are unclear, but they may be due to variations in the growth conditions used, in particular the media used and the carbon sources metabolized.
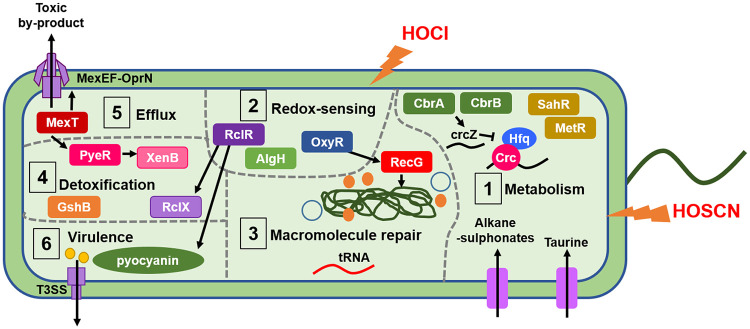
Our study revealed that the bacterial response to HOCl and HOSCN is multifaceted and incorporates an array of genes from different systems, including metabolism, redox sensing, macromolecule repair and detoxification, export of toxic compounds, and virulence. Figure 10 summarizes the physiological processes described here as having putative roles in protecting P. aeruginosa against HOCl and HOSCN in vitro and, we hypothesize, in the CF lung during infection. Sulfur transporter and metabolism genes were upregulated following HOCl and HOSCN exposure, and methionine metabolism regulators SahR and MetR appear required for survival against HOCl (Fig. 3A and 8C). During sulfur starvation, bacteria are able to use alkane sulfonates and taurine as a sulfur source (59–62). Therefore, it is plausible that P. aeruginosa requires increased uptake of alternative sulfur sources and careful control of methionine metabolism to maintain levels of sulfur-containing compounds that are under oxidative attack from HOCl and HOSCN. Upregulation of sulfur transport and metabolism, and methionine and cysteine biosynthesis genes, in response to HOCl stress has been reported in other bacteria (12, 63–65). We found the catabolite repression control system (38) plays a role in protection against HOCl; this indicates that appropriate regulation of carbon metabolism is important for survival against HOCl (Fig. 3B to D). RclR upregulated the expression of denitrification genes in the presence of HOSCN (Fig. 9B). As our experiments were carried out aerobically, we would expect these Anr-regulated genes to be downregulated (66); this suggests there is a physiological advantage to expressing these pathways aerobically in the presence of HOSCN.
FIG 10.
Model of the putative defense mechanisms used by P. aeruginosa in response to HOCl and HOSCN stress. In step 1, P. aeruginosa responds to HOCl and HOSCN by upregulation of sulfur transport and metabolism genes; this may facilitate increased uptake of alternative sulfur sources, including taurine, to replenish depleted sulfur levels caused by HOCl and HOSCN oxidative damage. Methionine biosynthesis and metabolism regulators MetR and SahR appear required to protect against HOCl, which we postulate is a response to oxidative disruption of the metabolism of the sulfur-containing amino acid methionine and the cellular need to maintain its levels. Catabolite repression by the Crc protein appears required for protection against HOCl, possibly indicating that optimal regulation of metabolic flexibility aids defense against HOCl. In step 2, the transcriptional regulator RclR, the H2O2 sensor OxyR (42), and the hypothetical protein AlgH (41) all have putative redox-sensing capabilities and were identified as playing a role in protection against HOCl. In step 3, macromolecular repair mechanisms are implicated in the response to HOCl and HOSCN. Upregulation of tRNAs in response to HOCl may occur to replace oxidatively damaged tRNA. The DNA repair enzyme RecG is required for protection against HOCl. In step 4, putative bacterial detoxification enzymes are involved in the response to HOCl and HOSCN. The PyeR-regulated oxidoreductase XenB is induced in response to HOCl and is required for protection, and the glutathione synthetase GshB is required for protection against HOCl. The peroxiredoxin RclX is positively regulated by RclR in response to HOCl and HOSCN and required for protection against both oxidants. In step 5, upregulation of the MexT-regulated mexEF-oprN operon, encoding a multidrug efflux pump (28, 29), occurs in response to HOCl and HOSCN and is required for protection against HOCl. This highlights a link between oxidative stress and antibiotic resistance and indicates a role for the efflux pump in expelling toxic by-products of these oxidants. In step 6, RclR is required for survival against HOCl and HOSCN and regulates a number of genes in the presence of both oxidants, including upregulation of the phz genes, which synthesize the virulence factor pyocyanin (52). The genes responsible for the biogenesis and regulation of another virulence mechanism, the T3SS (51), are upregulated in response to HOCl and HOSCN. Induction of pyocyanin and the T3SS may occur as a mechanism to counterattack the innate immune cell-derived HOCl and HOSCN-mediated oxidative attack on the bacteria.
Genes involved in repairing macromolecules and the detoxification of oxidants were found to be important in the defense against HOCl and HOSCN. The H2O2 sensor OxyR and DNA repair enzyme RecG (42) were required for protection against HOCl (Fig. 4B). Noncoding tRNAs were upregulated in response to HOCl (Fig. 8A), perhaps to replace oxidatively damaged tRNA. Surprisingly, tRNA-Cys expression was unchanged in the presence of HOCl. RclR downregulated the expression of 13 chaperone and heat shock genes and upregulated the expression of the chaperone gene cupA5 in the presence of HOSCN but not HOCl (Fig. 9C). In the WT strain, expression of the chaperone genes was not markedly altered in the presence of HOCl or HOSCN (Fig. 9C). In contrast, Groitl et al. found that HOCl and HOBr induced transcription of chaperone and heat shock genes and, together with HOSCN, increased production of the chaperone polyphosphate, which protects P. aeruginosa against protein unfolding and aggregation caused by the oxidants (25). As previously mentioned, these differences may be due to variations in methodological approaches used in our and the Groitl et al. studies, including the growth media and oxidant concentrations.
We found detoxification enzymes, in addition to RclX, were required for survival against HOCl, including the pyeRM-xenB operon, which encodes the oxidoreductase XenB (33) (Fig. 2C). The glutathione synthetase GshB was required for protection against HOCl (Fig. 4A); this enzyme is involved in synthesizing glutathione, which maintains the redox potential of the cell in response to oxidative stress (12, 67). Previous transcriptome assays have highlighted the importance of repair and detoxifying enzymes in the response of bacteria to HOCl stress (63–65, 68–71).
A striking finding of this work is a connection between the protective response to HOCl and HOSCN stress and antibiotic resistance. MexT and its regulated multidrug efflux pump, MexEF-OprN, were required for survival against HOCl (Fig. 2A). Additionally, the mexEF-oprN operon was upregulated in response to both HOCl and HOSCN (Fig. 8D), consistent with recent findings (25). While the fluoroquinolone-, chloramphenicol-, and trimethoprim-exporting MexEF-OprN efflux pump is normally quiescent in WT cells or not expressed in strains with an inactive chromosomal copy of mexT, it is highly induced in nfxC-type phenotypic mutants (mexT, mexS, or mvaT) in a MexT-dependent manner (72–73). This leads to increased resistance to fluoroquinolones and chloramphenicol and hypersensitivity to β-lactams (73). Of direct relevance to CF lung infections is the repeated observation that mutations in mexT occur in persistently infecting P. aeruginosa, both loss-of-function and gain-of-function mutations (nfxC type) (74–77).
MexT has been shown previously to be a redox-responsive regulator modulating the response to electrophilic and nitrosative stress (78, 79). Our data extend the role of MexT and MexEF-OprN to protection of P. aeruginosa against HOCl and HOSCN stress and suggest that the cell has a pleiotropic need for induction of the MexEF-OprN efflux pump in response to a wide range of oxidative stress events. MexEF-OprN can expel products other than antibiotics, and increased expression of mexEF-oprN is associated with decreased quorum sensing due to the efflux of quorum-sensing molecules, specifically the Pseudomonas quinolone signal precursors 4-hydroxy-2-heptylquinoline and kynurenine (80, 81). Therefore, one can propose that MexEF-OprN might additionally expel toxic by-products of HOCl and HOSCN reactions.
In conclusion, our findings have identified a wide range of different genes involved in the bacterial response to HOCl and HOSCN stress and provide the foundation from which further exploration of these mechanisms in protecting P. aeruginosa from these oxidants in the context of infection can be conducted.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains, plasmids, and primers used in this study are listed in Table S1 in the supplemental material. Three anonymized clinical isolates of P. aeruginosa from airway secretions of infected CF patients were obtained from the Royal Brompton Hospital. PCR products and plasmids were sequenced by GENEWIZ, Inc. Bacteria were routinely grown aerobically at 37°C, with shaking at 200 rpm or 700 rpm when in 96-well plates, in LB medium (5 g/liter NaCl, 5 g/liter yeast, and 10 g/liter tryptone) or on LBA plates (LB and 1.5% [wt/vol] agar) supplemented with appropriate antibiotics. P. aeruginosa strains were isolated on Pseudomonas isolation agar (PIA) (Sigma) supplemented with 20% (vol/vol) glycerol. ASM was prepared as described by Kirchner et al.; however, all individual amino acids, apart from l-tryptophan, were replaced with Casamino Acids, as described by Sriramulu et al. (43, 44). The components of ASM per liter were 4 g low-molecular-weight salmon sperm DNA, 5 g mucin from porcine stomach (type II), 4.75 g Casamino Acids, 0.25 g l-tryptophan, 5 g NaCl, 2.2 g KCl, 5.9 mg diethylenetriaminepentaacetic acid (DTPA), and 5 ml egg yolk emulsion. The pH of ASM was adjusted to 6.9, filter sterilized (44), and stored at 4°C for up to 1 month. After filtration, the ASM slowly turns cloudy due to precipitation of the salts from the media, but this does not impact its quality. Pyocyanin was extracted from 20-h cultures grown in LB, and the concentration was expressed in micrograms per milliliter, as described previously (82).
Construction of plasmids and mutant strains.
Details of genes were obtained from The Pseudomonas Genome Database (27). In-frame deletions were constructed as previously described (83). Briefly, for the construction of deletion mutants of PA14 rclR, rclX, pyeRM-xenB operon, and oxyR, an ∼500-bp upstream and ∼500-bp downstream fragment of the genes was amplified by PCR using primer pairs 1F and 2R as well as 3F and 4R (Table S1). Subsequent overlap PCR was performed using primers 1F and 4R (Table S1) to fuse the upstream and downstream fragments together to form a mutator fragment that was purified and ligated into the pCR-Blunt cloning vector (Invitrogen) for sequence verification using M13F and M13R primers. The rclR, rclX, and pyeRM-xenB mutator fragments were cloned into the BamHI site of the pKNG101 suicide vector, and the oxyR mutator fragment was cloned into the XbaI site of pKNG101; all were transformed into the donor CC118λpir E. coli strain and plated onto LBA supplemented with 50 μg/ml streptomycin. The rclR-pKNG101, rclX-pKNG101, pyeRM-xenB-pKNG101, and oxyR-pKNG101 plasmids from the donor strain were introduced into the PA14 recipient strain by conjugation. The DH5α E. coli strain containing the helper plasmid pRK2013 was grown on LBA supplemented with 50 μg/ml kanamycin, and the recipient PA14 strain was grown on LBA. Colonies of the donor, helper, and recipient strains were mixed together on LBA and incubated overnight. PA14 with the rclR-pKNG101, rclX-pKNG101, pyeRM-xenB-pKNG101, and oxyR-pKNG101 plasmids integrated site specifically into the chromosome as single crossovers were isolated on PIA with 2,000 μg/ml streptomycin. Subsequently, for sucrose counterselection, isolated colonies were streaked onto LBA without NaCl and with 10% (wt/vol) sucrose and incubated at room temperature for 48 h to obtain double-crossover unmarked in-frame deletion mutants. For oxyR deletion, 100 U of catalase from bovine liver per ml was added to the media. This is due to ΔoxyR mutants being unable to survive in LB, as components within the media autoxidize to generate ∼1.2 μM H2O2 min−1 (84). Colonies were isolated on LBA, and deletion of rclR, rclX, pyeRM-xenB, and oxyR was confirmed by PCR and sequencing using 5F and 6R primers (Table S1). Subsequent initial overnight growth of the ΔoxyR strain on LBA or in LB broth included 100 U/ml catalase. Complement plasmids rclR-pUCP18 and rclX-pUCP18 were constructed by PCR amplification of the promoter region and open reading frame of PA14 rclR and rclX using primers listed in Table S1 and subsequent cloning into the HindIII/XbaI sites of the pUCP18 shuttle vector. The plasmids were transformed into DH5α E. coli and grown on LBA with 100 μg/ml ampicillin prior to sequence verification using M13F and M13R primers (Table S1) and transformation into PA14 ΔrclR or ΔrclX mutant strains. The rclR-lacZ and rclX-lacZ plasmids were constructed by PCR amplification of the upstream promoter DNA (117 bp) of rclR and rclX using primers listed in Table S1 and ligation into pCR-Blunt (Invitrogen) for sequence verification using M13F and M13R primers. The promoter DNA of rclX and rclR was cloned into the EcoRI/PstI sites of the transcriptional fusion promoter-probe vector pMP220 (containing the lacZ gene), transformed into DH5α E. coli, and grown on LBA with 25 μg/ml tetracycline. Plasmids were sequence verified using primer Mid-PrclR or Mid-PrclX with Mid-lacZ (Table S1) prior to conjugation into PA14 WT and ΔrclR strains. Additionally, the rclX-lacZ plasmid was conjugated into three clinical CF isolates.
Preparation of HOCl and HOSCN solutions.
Sodium hypochlorite solution with 10 to 15% available chlorine (Sigma) was diluted directly into LB or ASM to prepare the final HOCl concentrations required for assays. The concentration of HOCl was determined by diluting sodium hypochlorite into 40 mM potassium phosphate buffer (pH 7.5) with 10 mM sodium hydroxide and measuring the absorbance at 292 nm (ɛ, 350 M−1 cm−1) (25, 85). HOSCN was generated enzymatically by combining glucose, glucose oxidase (GO), potassium thiocyanate (KSCN), and LPO. GO catalyzes the reaction of glucose with O2 to form H2O2, which reacts with KSCN, catalyzed by LPO, to form HOSCN. Stock concentrations of the enzymes, 200 U/ml GO (Sigma) and 1,000 U/ml LPO (Sigma), were prepared in 50% (vol/vol) phosphate-buffered saline (PBS) and 50% (vol/vol) glycerol solution and stored at –20°C. Stock concentrations of 4% (wt/vol) glucose and 750 mM KSCN (8 M; Fluka) were prepared in H2O and stored at 4°C. Varying the amount of glucose altered the amount of HOSCN produced, and a final sublethal concentration of 0.01% glucose was selected for all assays performed in LB (apart from Fig. S4, for which a final amount of 0.02% glucose was used). The final concentrations of LPO, GO, and KSCN were based on those used in reference 86. For the assays, all components were diluted directly into LB or ASM to final concentrations of 0.01% glucose (0.02% glucose for Fig. S4), 0.5 U/ml GO, 0.75 mM KSCN, and 3 U/ml LPO. A control assay without LPO that would only result in H2O2 generation (0.01% glucose, 0.5 U/ml GO, and 0.75 mM KSCN) was performed to examine the effect of H2O2 (data not shown). The concentration of HOSCN was determined by reaction with 5-thio-2-nitrobenzoic acid (TNB), as described previously (25, 87). Briefly, 10 mM 5,5′-dithiobis (2-nitrobenzoic acid) (Thermo Scientific), prepared in 40 mM potassium phosphate buffer (pH 7.5), was hydrolyzed by the addition of sodium hydroxide to generate TNB. The TNB concentration was determined by measuring absorbance at 412 nm (ɛ, 14,150 M−1 cm−1). HOSCN (0.01% glucose [0.02% glucose for Fig. S4], 0.5 U/ml GO, 0.75 mM KSCN, and 3 U/ml LPO) prepared in 40 mM potassium phosphate buffer (pH 7.5) was diluted 1:1 in TNB, and the loss of absorption at 412 nm was measured after 20 min. The concentration of HOSCN was determined from the concentration of TNB consumed during the reaction (stoichiometry, 2:1 [TNB:HOSCN]). A concentration of 0.8 mM HOSCN was produced from 0.01% glucose and 1 mM HOSCN from 0.02% glucose. For the ASM assays, 0.8 mM HOSCN was produced, as described above, which was then diluted in ASM to give the final HOSCN concentrations used.
HOCl and HOSCN susceptibility and viability assays.
A total of 707 strains with mutations in genes with regulatory functions were selected from the PA14 nonredundant single-transposon mutant library (26) (Table S1). The strains were grown overnight in LB medium with 15 μg/ml gentamicin, alongside a PA14 WT control grown in LB medium, in 2.2 ml 96-well deep-well plates (VWR). For the HOCl screen, overnight cultures were subcultured 1:20 into LB medium with 15 μg/ml gentamicin in 96-well microtiter plates (Falcon) and grown for 3 h. Cultures were subcultured again at 1:20 (optical density [OD], 0.05 ± 0.03) in LB medium or LB medium containing 4.4 mM HOCl in 96-well microtiter plates (Falcon) and grown for up to 24 h, with the OD at 600 nm (OD600) recorded as a measure of growth at hourly time points. LB medium was used for HOCl screening to facilitate growth of the large number of mutant strains, some of which may require nutrient-rich broth. The concentration of HOCl used was selected from a preliminary experiment, which identified 4.4 mM HOCl as being sublethal to PA14 WT, causing a lag in growth but not complete inhibition (data not shown). However, for the assays that tested the HOCl susceptibility of ΔpyeRM-xenB, 5.1 mM HOCl was used. For PA14 WT and ΔrclR and ΔrclX mutant strains, and ΔrclR plus rclR-pUCP18 and ΔrclX plus rclX-pUCP18 complemented strains, overnight cultures were prepared by growing the strains in LB medium or LB medium with 500 μg/ml carbenicillin for the pUCP18 complemented strains. Overnight cultures were subcultured 1:20 in LB medium and grown for 3 h prior to subculturing again at 1:20 (OD, 0.05 ± 0.02) into LB medium or LB medium with 4.4 mM HOCl or 0.8 mM HOSCN. We monitored growth by recording the OD600. The concentration of HOSCN was chosen from initial growth curve analyses, which identified 0.8 mM and 1 mM HOSCN as subinhibitory for ΔrclR and ΔrclX strains, respectively (data not shown). However, as the higher concentration was lethal to the ΔrclR strain, the lower concentration of 0.8 mM was used for all experiments performed in LB, apart from Fig. S4. For the HOCl and HOSCN susceptibility assays in ASM, the PA14 WT and ΔrclR and ΔrclX mutant strains were grown as described above. The only difference was the ASM was filtered again prior to adding HOCl at a final concentration of 3.1 mM and adding components to make 0.8 mM HOSCN, which was diluted to a final concentration of 0.53 mM. Lower concentrations were required in ASM, as bacteria had increased susceptibility to both oxidants in this medium. For HOCl and HOSCN viability assays, cultures were sampled at 0 h and 2.5 h or 4.5 h after HOSCN or HOCl exposure, respectively, 10-fold serial dilutions in PBS were prepared, and dilutions were drop plated on LBA. For other chemical susceptibility assays, PA14 WT, ΔrclR, and ΔrclX strains were grown as described above but in LB medium containing 0.125 mM NEM, 5 mM diamide, 2.5 mM methylglyoxal, 5 mM H2O2, 0.5 mM TBH, 1 mM methyl viologen, or 10 mM DEANO. The concentrations of chemicals used were determined from preliminary experiments that tested a range of concentrations to identify those that caused a lag, but not inhibition, in the growth of PA14 WT (data not shown). All chemicals were from Sigma, apart from TBH, which was from EMD Millipore. For ASM biofilm assays, we adapted the method from reference 44. Overnight cultures of PA14 WT, ΔrclR, and ΔrclX strains were subcultured 1:50 in LB and grown for 3 h prior to subculturing again at 1:50 into 2 ml ASM without or with HOCl (3.1 or 3.5 mM) and HOSCN (0.43 mM or 0.53 mM) in a 24-well tissue culture-treated plate (Falcon), which was incubated at 37°C, with low-speed shaking, for 3 days to allow biofilm formation and then left at room temperature, static, for a further 4 days before visualization.
RNA extraction and RNA-seq analysis.
Exponential-phase PA14 WT and ΔrclR cultures were subcultured 1:50 in LB (OD, 0.05 ± 0.02) and grown for 2.5 h prior to adding HOCl to a final concentration of 2.2 mM, HOSCN to a final concentration of 0.8 mM, or no treatment (LB only). LB medium was used for consistency with the HOCl screen. The concentrations of oxidants used were chosen due to causing a 30- to 120-min lag in growth of exponential-phase cultures. Treated and untreated WT and ΔrclR cultures were incubated for a further 20 min, and then cells were collected and centrifuged at 13,000 × g for 5 min. The pellets were washed twice with PBS, and RNAlater (Ambion) was added to preserve the RNA. Two biological replicates of WT and ΔrclR strains from independent experiments, treated with HOCl or HOSCN or left untreated, were collected for RNA purification. Enzymatic lysis and proteinase K digestion of bacteria followed by RNA purification were carried out using the RNeasy Protect Bacteria minikit (Qiagen). DNase treatment of RNA samples was performed using the TURBO DNA-free kit (Ambion). Removal of rRNA, preparation of cDNA libraries, and sequencing using the Illumina NextSeq 500 system and a 75-bp read length were performed by Vertis Biotechnologie AG. Reads were aligned to the P. aeruginosa UCBPP-PA14 complete genome (NCBI accession number NC_008463.1) using bowtie2, and expression values in reads per kilobase per million mapped reads (RPKM) were calculated using FeatureCounts. Normalization of the RPKM values against the OD600 of the samples prior to RNA extraction was performed, and the values of the replicates were averaged. The log2 fold change of the RPKM values of treated WT compared to untreated WT or treated WT compared to treated ΔrclR strain was calculated. Genes with a log2 fold change of >1.5 and <−1.5 were considered differentially expressed.
Quantitative real-time PCR.
Cultures of PA14 WT were grown to mid-exponential phase in LB and treated with HOCl or HOSCN for 20 min prior to RNA extraction; this was performed as described for RNA-seq. The cDNA library was prepared using the high-capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR was set up with Fast SYBR green mix (Invitrogen) and using the Applied Biosystems 7500 fast real-time PCR instrument. Expression ratios were calculated by comparing the expression of each gene in treated PA14 WT cultures to untreated PA14 WT cultures by the ΔΔCT method (88) and normalized to expression of rpoD (RNA polymerase sigma factor), which shows unchanged expression levels under the conditions tested. Primers used for qRT-PCR are listed in Table S1.
β-Galactosidase assays.
PA14 WT and ΔrclR strains containing rclR-lacZ and rclX-lacZ plasmids, and clinical CF isolates 1, 2, and 3 containing the rclX-lacZ plasmid, were grown in LB medium with 100 μg/ml tetracycline overnight. Cultures were subcultured 1:50 (OD, 0.05 ± 0.02) in LB medium in 2.2-ml 96-well deep-well plates (VWR) and grown to exponential phase (3 h) and then were left untreated or treated with 2.2 mM HOCl or 0.8 mM HOSCN. Cultures were collected 30 min after treatment with HOCl or HOSCN. β-Galactosidase activity was assayed using the modified version (89) of the Miller method (90).
Homology modeling and sequence analysis.
The predicted structure of RclX was modeled with SWISS-MODEL (91), and illustrations of protein structures were prepared with EzMol (92). Amino acid sequences were aligned using Clustal Omega (93) and visualized with ESPript 3.0 (94).
Data availability.
The transcriptomic data of our RNA-seq experiments have been deposited in the NCBI’s GEO database (95) under accession number GSE124385.
Supplementary Material
ACKNOWLEDGMENTS
We thank Emily Calamita for her technical support in the construction of the ΔpyeRM-xenB mutant.
This work was funded by a Medical Research Council Doctoral Training Grant and was also supported by the Cystic Fibrosis Trust Strategic Research Center via a grant titled “Personalized Approach to Pseudomonas aeruginosa.”
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Moradali MF, Ghods S, Rehm BH. 2017. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 3.Williams HD, Davies JC. 2012. Basic science for the chest physician: Pseudomonas aeruginosa and the cystic fibrosis airway. Thorax 67:465–467. doi: 10.1136/thoraxjnl-2011-201498. [DOI] [PubMed] [Google Scholar]
- 4.Smith WD, Bardin E, Cameron L, Edmondson CL, Farrant KV, Martin I, Murphy RA, Soren O, Turnbull AR, Wierre-Gore N, Alton EW, Bundy JG, Bush A, Connett GJ, Faust SN, Filloux A, Freemont PS, Jones AL, Takats Z, Webb JS, Williams HD, Davies JC. 2017. Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol Lett 364:fnx121. doi: 10.1093/femsle/fnx121. [DOI] [PubMed] [Google Scholar]
- 5.Cant N, Pollock N, Ford RC. 2014. CFTR structure and cystic fibrosis. Int J Biochem Cell Biol 52:15–25. doi: 10.1016/j.biocel.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Pattison DI, Davies MJ, Hawkins CL. 2012. Reactions and reactivity of myeloperoxidase-derived oxidants: differential biological effects of hypochlorous and hypothiocyanous acids. Free Radic Res 46:975–995. doi: 10.3109/10715762.2012.667566. [DOI] [PubMed] [Google Scholar]
- 7.Winterbourn CC, Kettle AJ, Hampton MB. 2016. Reactive oxygen species and neutrophil function. Annu Rev Biochem 85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- 8.van Dalen CJ, Whitehouse MW, Winterbourn CC, Kettle AJ. 1997. Thiocyanate and chloride as competing substrates for myeloperoxidase. Biochem J 327:487–492. doi: 10.1042/bj3270487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer H. 2009. Mechanisms and function of DUOX in epithelia of the lung. Antioxid Redox Signal 11:2453–2465. doi: 10.1089/ars.2009.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillion M, Antelmann H. 2015. Thiol-based redox switches in prokaryotes. Biol Chem 396:415–444. doi: 10.1515/hsz-2015-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray MJ, Wholey WY, Jakob U. 2013. Bacterial responses to reactive chlorine species. Annu Rev Microbiol 67:141–160. doi: 10.1146/annurev-micro-102912-142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi BK, Gronau K, Mader U, Hessling B, Becher D, Antelmann H. 2011. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics 10:M111.009506. doi: 10.1074/mcp.M111.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palm GJ, Chi BK, Waack P, Gronau K, Becher D, Albrecht D, Hinrichs W, Read RJ, Antelmann H. 2012. Structural insights into the redox-switch mechanism of the MarR/DUF24-type regulator HypR. Nucleic Acids Res 40:4178–4192. doi: 10.1093/nar/gkr1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray MJ, Wholey WY, Parker BW, Kim M, Jakob U. 2013. NemR is a bleach-sensing transcription factor. J Biol Chem 288:13789–13798. doi: 10.1074/jbc.M113.454421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebendorfer KM, Drazic A, Le Y, Gundlach J, Bepperling A, Kastenmuller A, Ganzinger KA, Braun N, Franzmann TM, Winter J. 2012. Identification of a hypochlorite-specific transcription factor from Escherichia coli. J Biol Chem 287:6892–6903. doi: 10.1074/jbc.M111.287219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker BW, Schwessinger EA, Jakob U, Gray MJ. 2013. The RclR protein is a reactive chlorine-specific transcription factor in Escherichia coli. J Biol Chem 288:32574–32584. doi: 10.1074/jbc.M113.503516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JW, Soonsanga S, Helmann JD. 2007. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A 104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol 183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuangthong M, Herbig AF, Bsat N, Helmann JD. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J Bacteriol 184:3276–3286. doi: 10.1128/jb.184.12.3276-3286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JW, Helmann JD. 2006. The PerR transcription factor senses H2O2 by metal-catalyzed histidine oxidation. Nature 440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 22.Umezawa Y, Shimada T, Kori A, Yamada K, Ishihama A. 2008. The uncharacterized transcription factor YdhM is the regulator of the nemA gene, encoding N-ethylmaleimide reductase. J Bacteriol 190:5890–5897. doi: 10.1128/JB.00459-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C, Shin J, Park C. 2013. Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coli K-12. Mol Microbiol 88:395–412. doi: 10.1111/mmi.12192. [DOI] [PubMed] [Google Scholar]
- 24.Ozyamak E, de Almeida C, de Moura AP, Miller S, Booth IR. 2013. Integrated stress response of Escherichia coli to methylglyoxal: transcriptional readthrough from the nemRA operon enhances protection through increased expression of glyoxalase I. Mol Microbiol 88:936–950. doi: 10.1111/mmi.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groitl B, Dahl JU, Schroeder JW, Jakob U. 2017. Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol Microbiol 106:335–350. doi: 10.1111/mmi.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty LK, Pechere JC. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol 23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 29.Kohler T, Epp SF, Curty LK, Pechere JC. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol 181:6300–6305. doi: 10.1128/JB.181.20.6300-6305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian ZX, Fargier E, Mac Aogain M, Adams C, Wang YP, O'Gara F. 2009. Transcriptome profiling defines a novel regulon modulated by the LysR-type transcriptional regulator MexT in Pseudomonas aeruginosa. Nucleic Acids Res 37:7546–7559. doi: 10.1093/nar/gkp828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian ZX, Mac Aogain M, O'Connor HF, Fargier E, Mooij MJ, Adams C, Wang YP, O'Gara F. 2009. MexT modulates virulence determinants in Pseudomonas aeruginosa independent of the MexEF-OprN efflux pump. Microb Pathog 47:237–241. doi: 10.1016/j.micpath.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li XZ, Nishino T. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol 21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 33.Mac Aogain M, Mooij MJ, McCarthy RR, Plower E, Wang YP, Tian ZX, Dobson A, Morrissey J, Adams C, O'Gara F. 2012. The non-classical ArsR-family repressor PyeR (PA4354) modulates biofilm formation in Pseudomonas aeruginosa. Microbiology 158:2598–2609. doi: 10.1099/mic.0.058636-0. [DOI] [PubMed] [Google Scholar]
- 34.Urbanowski ML, Stauffer LT, Plamann LS, Stauffer GV. 1987. A new methionine locus, metR, that encodes a trans-acting protein required for activation of metE and metH in Escherichia coli and Salmonella typhimurium. J Bacteriol 169:1391–1397. doi: 10.1128/jb.169.4.1391-1397.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novichkov PS, Li X, Kuehl JV, Deutschbauer AM, Arkin AP, Price MN, Rodionov DA. 2014. Control of methionine metabolism by the SahR transcriptional regulator in Proteobacteria. Environ Microbiol 16:1–8. doi: 10.1111/1462-2920.12273. [DOI] [PubMed] [Google Scholar]
- 36.Nishijyo T, Haas D, Itoh Y. 2001. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol Microbiol 40:917–931. doi: 10.1046/j.1365-2958.2001.02435.x. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Lu CD. 2007. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J Bacteriol 189:5413–5420. doi: 10.1128/JB.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonnleitner E, Abdou L, Haas D. 2009. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 106:21866–21871. doi: 10.1073/pnas.pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.La Rosa R, Behrends V, Williams HD, Bundy JG, Rojo F. 2016. Influence of the Crc regulator on the hierarchical use of carbon sources from a complete medium in Pseudomonas. Environ Microbiol 18:807–818. doi: 10.1111/1462-2920.13126. [DOI] [PubMed] [Google Scholar]
- 40.Pei XY, Dendooven T, Sonnleitner E, Chen S, Blasi U, Luisi BF. 2019. Architectural principles for Hfq/Crc-mediated regulation of gene expression. Elife 8:e43158. doi: 10.7554/eLife.43158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urbauer JL, Cowley AB, Broussard HP, Niedermaier HT, Bieber Urbauer RJ. 2015. Solution structure and properties of AlgH from Pseudomonas aeruginosa. Proteins 83:1137–1150. doi: 10.1002/prot.24811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol 182:4533–4544. doi: 10.1128/jb.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sriramulu DD, Lunsdorf H, Lam JS, Romling U. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol 54:667–676. doi: 10.1099/jmm.0.45969-0. [DOI] [PubMed] [Google Scholar]
- 44.Kirchner S, Fothergill JL, Wright EA, James CE, Mowat E, Winstanley C. 2012. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J Vis Exp 64:e3857. doi: 10.3791/3857:e3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, Hu Y, Yang B, Gong X, Zhang N, Niu L, Wu Y, Ge H. 2015. Structure of lpg0406, a carboxymuconolactone decarboxylase family protein possibly involved in antioxidative response from Legionella pneumophila. Protein Sci 24:2070–2075. doi: 10.1002/pro.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clarke TE, Romanov V, Chirgadze YN, Klomsiri C, Kisselman G, Wu-Brown J, Poole LB, Pai EF, Chirgadze NY. 2011. Crystal structure of alkyl hydroperoxidase D like protein PA0269 from Pseudomonas aeruginosa: homology of the AhpD-like structural family. BMC Struct Biol 11:27. doi: 10.1186/1472-6807-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koshkin A, Nunn CM, Djordjevic S, Ortiz de Montellano PR. 2003. The mechanism of Mycobacterium tuberculosis alkylhydroperoxidase AhpD as defined by mutagenesis, crystallography, and kinetics. J Biol Chem 278:29502–29508. doi: 10.1074/jbc.M303747200. [DOI] [PubMed] [Google Scholar]
- 48.Nunn CM, Djordjevic S, Hillas PJ, Nishida CR, Ortiz de Montellano PR. 2002. The crystal structure of Mycobacterium tuberculosis alkylhydroperoxidase AhpD, a potential target for antitubercular drug design. J Biol Chem 277:20033–20040. doi: 10.1074/jbc.M200864200. [DOI] [PubMed] [Google Scholar]
- 49.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 50.Petrova OE, Garcia-Alcalde F, Zampaloni C, Sauer K. 2017. Comparative evaluation of rRNA depletion procedures for the improved analysis of bacterial biofilm and mixed pathogen culture transcriptomes. Sci Rep 7:41114. doi: 10.1038/srep41114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fragoso MA, Fernandez V, Forteza R, Randell SH, Salathe M, Conner GE. 2004. Transcellular thiocyanate transport by human airway epithelia. J Physiol 561:183–194. doi: 10.1113/jphysiol.2004.071548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moskwa P, Lorentzen D, Excoffon KJ, Zabner J, McCray PB, Jr, Nauseef WM, Dupuy C, Banfi B. 2007. A novel host defense system of airways is defective in cystic fibrosis. Am J Respir Crit Care Med 175:174–183. doi: 10.1164/rccm.200607-1029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conner GE, Wijkstrom-Frei C, Randell SH, Fernandez VE, Salathe M. 2007. The lactoperoxidase system links anion transport to host defense in cystic fibrosis. FEBS Lett 581:271–278. doi: 10.1016/j.febslet.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Painter RG, Bonvillain RW, Valentine VG, Lombard GA, LaPlace SG, Nauseef WM, Wang G. 2008. The role of chloride anion and CFTR in killing of Pseudomonas aeruginosa by normal and CF neutrophils. J Leukoc Biol 83:1345–1353. doi: 10.1189/jlb.0907658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Painter RG, Valentine VG, Lanson NA, Jr, Leidal K, Zhang Q, Lombard G, Thompson C, Viswanathan A, Nauseef WM, Wang G, Wang G. 2006. CFTR expression in human neutrophils and the phagolysosomal chlorination defect in cystic fibrosis. Biochemistry 45:10260–10269. doi: 10.1021/bi060490t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorentzen D, Durairaj L, Pezzulo AA, Nakano Y, Launspach J, Stoltz DA, Zamba G, McCray PB, Jr, Zabner J, Welsh MJ, Nauseef WM, Banfi B. 2011. Concentration of the antibacterial precursor thiocyanate in cystic fibrosis airway secretions. Free Radic Biol Med 50:1144–1150. doi: 10.1016/j.freeradbiomed.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Ploeg JR, Iwanicka-Nowicka R, Bykowski T, Hryniewicz MM, Leisinger T. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J Biol Chem 274:29358–29365. doi: 10.1074/jbc.274.41.29358. [DOI] [PubMed] [Google Scholar]
- 60.van der Ploeg JR, Weiss MA, Saller E, Nashimoto H, Saito N, Kertesz MA, Leisinger T. 1996. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J Bacteriol 178:5438–5446. doi: 10.1128/jb.178.18.5438-5446.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kertesz MA. 1996. Desulfonation of aliphatic sulfonates by Pseudomonas aeruginosa PAO. FEMS Microbiol Lett 137:221–225. doi: 10.1111/j.1574-6968.1996.tb08109.x. [DOI] [PubMed] [Google Scholar]
- 62.Kertesz MA, Schmidt-Larbig K, Wuest T. 1999. A novel reduced flavin mononucleotide-dependent methanesulfonate sulfonatase encoded by the sulfur-regulated msu operon of Pseudomonas aeruginosa. J Bacteriol 181:1464–1473. doi: 10.1128/JB.181.5.1464-1473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Small DA, Chang W, Toghrol F, Bentley WE. 2007. Toxicogenomic analysis of sodium hypochlorite antimicrobial mechanisms in Pseudomonas aeruginosa. Appl Microbiol Biotechnol 74:176–185. doi: 10.1007/s00253-006-0644-7. [DOI] [PubMed] [Google Scholar]
- 64.Wang S, Phillippy AM, Deng K, Rui X, Li Z, Tortorello ML, Zhang W. 2010. Transcriptomic responses of Salmonella enterica serovars Enteritidis and Typhimurium to chlorine-based oxidative stress. Appl Environ Microbiol 76:5013–5024. doi: 10.1128/AEM.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S, Deng K, Zaremba S, Deng X, Lin C, Wang Q, Tortorello ML, Zhang W. 2009. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl Environ Microbiol 75:6110–6123. doi: 10.1128/AEM.00914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams HD, Zlosnik JE, Ryall B. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv Microb Physiol 52:1–71. doi: 10.1016/S0065-2911(06)52001-6. [DOI] [PubMed] [Google Scholar]
- 67.Wongsaroj L, Saninjuk K, Romsang A, Duang-Nkern J, Trinachartvanit W, Vattanaviboon P, Mongkolsuk S. 2018. Pseudomonas aeruginosa glutathione biosynthesis genes play multiple roles in stress protection, bacterial virulence and biofilm formation. PLoS One 13:e0205815. doi: 10.1371/journal.pone.0205815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bodet C, Sahr T, Dupuy M, Buchrieser C, Hechard Y. 2012. Legionella pneumophila transcriptional response to chlorine treatment. Water Res 46:808–816. doi: 10.1016/j.watres.2011.11.059. [DOI] [PubMed] [Google Scholar]
- 69.Ceragioli M, Mols M, Moezelaar R, Ghelardi E, Senesi S, Abee T. 2010. Comparative transcriptomic and phenotypic analysis of the responses of Bacillus cereus to various disinfectant treatments. Appl Environ Microbiol 76:3352–3360. doi: 10.1128/AEM.03003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jang HJ, Nde C, Toghrol F, Bentley WE. 2009. Global transcriptome analysis of the Mycobacterium bovis BCG response to sodium hypochlorite. Appl Microbiol Biotechnol 85:127–140. doi: 10.1007/s00253-009-2208-0. [DOI] [PubMed] [Google Scholar]
- 71.Peeters E, Sass A, Mahenthiralingam E, Nelis H, Coenye T. 2010. Transcriptional response of Burkholderia cenocepacia J2315 sessile cells to treatments with high doses of hydrogen peroxide and sodium hypochlorite. BMC Genomics 11:90. doi: 10.1186/1471-2164-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Llanes C, Kohler T, Patry I, Dehecq B, van Delden C, Plesiat P. 2011. Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob Agents Chemother 55:5676–5684. doi: 10.1128/AAC.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uwate M, Ichise YK, Shirai A, Omasa T, Nakae T, Maseda H. 2013. Two routes of MexS-MexT-mediated regulation of MexEF-OprN and MexAB-OprM efflux pump expression in Pseudomonas aeruginosa. Microbiol Immunol 57:263–272. doi: 10.1111/1348-0421.12032. [DOI] [PubMed] [Google Scholar]
- 74.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Diaz Caballero J, Clark ST, Coburn B, Zhang Y, Wang PW, Donaldson SL, Tullis DE, Yau YC, Waters VJ, Hwang DM, Guttman DS. 2015. Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung. mBio 6:e00981-15. doi: 10.1128/mBio.00981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Mansfeld R, de Been M, Paganelli F, Yang L, Bonten M, Willems R. 2016. Within-host evolution of the Dutch high-prevalent Pseudomonas aeruginosa clone ST406 during chronic colonization of a patient with cystic fibrosis. PLoS One 11:e0158106. doi: 10.1371/journal.pone.0158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sherrard LJ, Tai AS, Wee BA, Ramsay KA, Kidd TJ, Ben Zakour NL, Whiley DM, Beatson SA, Bell SC. 2017. Within-host whole genome analysis of an antibiotic resistant Pseudomonas aeruginosa strain sub-type in cystic fibrosis. PLoS One 12:e0172179. doi: 10.1371/journal.pone.0172179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fargier E, Mac Aogain M, Mooij MJ, Woods DF, Morrissey JP, Dobson AD, Adams C, O'Gara F. 2012. MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J Bacteriol 194:3502–3511. doi: 10.1128/JB.06632-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fetar H, Gilmour C, Klinoski R, Daigle DM, Dean CR, Poole K. 2011. mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob Agents Chemother 55:508–514. doi: 10.1128/AAC.00830-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamarche MG, Deziel E. 2011. MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS One 6:e24310. doi: 10.1371/journal.pone.0024310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olivares J, Alvarez-Ortega C, Linares JF, Rojo F, Kohler T, Martinez JL. 2012. Overproduction of the multidrug efflux pump MexEF-OprN does not impair Pseudomonas aeruginosa fitness in competition tests, but produces specific changes in bacterial regulatory networks. Environ Microbiol 14:1968–1981. doi: 10.1111/j.1462-2920.2012.02727.x. [DOI] [PubMed] [Google Scholar]
- 82.Essar DW, Eberly L, Hadero A, Crawford IP. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol 172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muhl D, Filloux A. 2014. Site-directed mutagenesis and gene deletion using reverse genetics. Methods Mol Biol 1149:521–539. doi: 10.1007/978-1-4939-0473-0_40. [DOI] [PubMed] [Google Scholar]
- 84.Hassett DJ, Alsabbagh E, Parvatiyar K, Howell ML, Wilmott RW, Ochsner UA. 2000. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J Bacteriol 182:4557–4563. doi: 10.1128/jb.182.16.4557-4563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morris JC. 1966. The acid ionization constant of HOCl from 5 to 35°. J Phys Chem 70:3798–3805. doi: 10.1021/j100884a007. [DOI] [Google Scholar]
- 86.Sermon J, Wevers EM, Jansen L, De Spiegeleer P, Vanoirbeek K, Aertsen A, Michiels CW. 2005. CorA affects tolerance of Escherichia coli and Salmonella enterica serovar Typhimurium to the lactoperoxidase enzyme system but not to other forms of oxidative stress. Appl Environ Microbiol 71:6515–6523. doi: 10.1128/AEM.71.11.6515-6523.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nagy P, Jameson GN, Winterbourn CC. 2009. Kinetics and mechanisms of the reaction of hypothiocyanous acid with 5-thio-2-nitrobenzoic acid and reduced glutathione. Chem Res Toxicol 22:1833–1840. doi: 10.1021/tx900249d. [DOI] [PubMed] [Google Scholar]
- 88.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Griffith KL, Wolf RE, Jr.. 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun 290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- 90.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 91.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modeling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reynolds CR, Islam SA, Sternberg MJE. 2018. EzMol: a web server wizard for the rapid visualization and image production of protein and nucleic acid structures. J Mol Biol 430:2244–2248. doi: 10.1016/j.jmb.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic data of our RNA-seq experiments have been deposited in the NCBI’s GEO database (95) under accession number GSE124385.