Abstract
This study outlines the potential of a novel therapeutic dressing for the management of chronic wounds. The dressing incorporates polyphosphate, a non toxic compound with a number of beneficial characteristics in terms of wound healing, in a foam matrix. The aim of this study was to identify the potential of polyphosphate incorporated in the foam dressing to sequester the activity of matrix metalloproteinases (MMPs) and proteases derived from Pseudomonas aeruginosa. Methods used included gelatin zymography and milk‐casein agar plate analysis. Results have shown that this dressing is effectively capable of reducing the levels of MMP‐2 and MMP‐9 in both their active and latent forms using an in vitro model. The dressing also demonstrated the compound's potential in the regulation of P. aeruginosa derived proteases.
Keywords: Biofilm, Enzymes, MMP, Proteases, Wound healing
Introduction
Cutaneous wound healing is a complex process which involves a careful balance between tissue synthesis, deposition and breakdown. A number of cell types and molecular mediators are involved in the process, which work synergistically to achieve rapid restoration of tissue architecture and function 1. If this balance is disrupted, however, healing is hampered leading the wound into a state of chronicity 2, 3. Such instances include chronic ulcers in which excessive tissue degradation prevails and keloid scars are instigated by excessive extracellular matrix production 4. Proteases including the matrix metalloproteinases (MMPs) and their inhibitors, particularly the natural inhibitors, tissue inhibitors of metalloproteinases (TIMPs), play a vital role in this equilibrium and are important factors of the healing process 2. A number of cell types including keratinocytes, fibroblasts and endothelial cells contribute to the proteolytic environment within the wound bed. Whilst fibroblasts produce MMPs, invading neutrophils and macrophages produce elastase and collagenase 5, 6, 7. MMPs are of particular interest in terms of cutaneous wound healing as a number of MMPs have been shown to be upregulated within the tissues of chronic wounds that fail to heal adequately. Indeed, research has showed that the expression of various MMPs are upregulated during wound repair and regeneration, with mRNAs encoding specific MMPs being upregulated in wounded skin as compared with normal uninjured skin. For example, a study by Lund et al. 8 demonstrated via a murine wound model that MMP‐2, ‐9, ‐3, ‐13, ‐11, 12 and ‐14 were all upregulated in terms of mRNA expression in skin wounds when compared with normal control skin 8. However, a proteinase inhibitor, galardin (an inhibitor of plasminogen), was tested for its action in the wound healing process and was found to inhibit the healing capacity of the skin, an indirect effect caused by an inability of fibroblasts to direct their way through the fibrin rich wound bed via the actions of proteases 8.
Given the extensive evidence that suggests excessive MMP levels as a central pathological feature of chronic wounds, much research interest has been aimed at redressing this proteolytic imbalance and is an important goal of medical material science 9. Whilst many wound dressings are aimed at promoting healing through the absorption of exudates and reduction in bioburden through antimicrobial actions 10, a number of dressings are now available that are aimed at sequestering MMP levels in the wound bed. Such dressings modulate the biological molecules including growth factors and MMPs involved in wound healing 11.
A limited number of studies have demonstrated the ability of polyphosphate dressings to sequester various proteinases within the wound bed. Unbound polyphosphate has been shown to promote healing of chronic wounds though the inhibition of proteases, in turn maintaining a robust extracellular matrix 12. A number of polyphosphates have been used including trimetaphosphate and hexametaphosphate. Such polyphosphates may be used externally in wound dressings or may be administered enterally and have been shown to be effective against pepsin, collagenase, elastase and hyaluronidase and are active at a broad pH range of 2–6. A number of enzyme dose–response assays showed that polyphosphates are active against wound‐associated proteases in a concentration‐dependant manner 12. Such dressings may prove to be more cost effective for the patient and health care services alike, when compared to other high technology approaches such as skin substitutes used to balance tissue degeneration 13.
The aim of this study was to examine the potential MMP modulating capacity of a foam dressing containing polyphosphate compared with a commercially available sterile, freeze‐dried dressing with a collagen/oxidised regenerated cellulose (ORC) composition, a soft adherent foam dressing with Technology Lipido‐Colloid (TLC) with the healing accelerator nano‐oligosaccharide factor (NOSF) and a standard polyurethane foam dressing. Given the evidence that suggests an elevated level of MMP‐2 and MMP‐9 within the chronic wound milieu 14, this study has focussed specifically on the ability of the selected dressings to sequester these MMPs.
Methods
Chemicals and reagents
All chemicals and reagents were purchased from Sigma Aldrich (Dorset, UK) unless otherwise stated.
Ethical approval
Approval for this study was granted by the Ethics Committee, University of Liverpool and informed consent was obtained from the owners of the animals participating in this study.
Dressings
Dressings used in this study included a standard foam dressing (Smith and Nephew, Hull, UK), a collagen/ORC dressing (Johnson and Johnson, New Brunswick, NJ), a TLC‐NOSF dressing (Urgo Medical, Loughborough, UK) and a foam dressing containing polyphosphate (Advanced Medical Solutions Plc, Winsford, Cheshire, UK).
MMP‐2 isolation
Equine dermal tissue was obtained either at post mortem of horses which had been euthanised for medical reasons or from tissue debrided during wound management procedures, in which case consent was given by the owner. All ethical considerations were ensured. Tissue sections were washed twice in 1× Hank's balanced salt solution (HBSS) (Gibco, Invitrogen, Paisley, UK) and minced into 3–5 mm2 pieces and placed into 25 cm2 culture flasks. Fibroblasts were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, Invitrogen) supplemented with 10% foetal calf serum (FCS), 20 mM HEPES buffer, 100 µg/ml gentamicin and 0·5 µg/ml amphotericin B (Fungizone). The cultures were incubated in 5% CO2 in air at 37°C. Following a culture period of 5–10 days, cells were examined microscopically (Nikon Eclipse, Melville, NY, USA) for outgrowth and confluency. When confluent, cells were passaged in a ratio of 1:4 with 0·05% trypsin/EDTA and cells of passage 3–8 were used experimentally. Once cells regained confluency the media was removed and the cell layer washed with HBSS to remove all traces of FCS. Cells were then cultured in serum‐free DMEM with 20 mM HEPES buffer and 1% penicillin–streptomycin (Invitrogen) to confluency (approximately 5 days). The media was removed and cells were centrifuged at a speed of 164 g for 5 minutes. The supernatant media was removed and stored at −80°C ready for enzyme purification and the pellet was discarded appropriately.
MMP‐9 isolation
Equine peripheral blood was collected by venepuncture, anticoagulated with heparin and refrigerated for 30 minutes. Monocytes were isolated using Histopaque solution according to the manufacturer's instructions. In brief, the anticoagulated blood was layered on top of the Histopaque at a 1:1 ratio in 50 ml centrifuge tubes. Blood samples were centrifuged at 400 g for 30 minutes to separate the monocyte layer from the blood plasma and red blood cells. Differential migration resulted in the clearly identifiable monocyte cell layer which was removed and successively washed in 1× HBSS. The monocytes were pelleted and stored in 0·5 ml 1× HBSS. Cells were lysed with 1 ml 0·1% triton X‐100 per pellet and freeze‐thawed four times to release the enzyme. The lysed cells were stored at −80°C ready for enzyme purification.
MMP‐2 and MMP‐9 purification using gelatin sepharose chromatography
A gelatin sepharose (GE Healthcare, Little Chalfont, UK) chromatography column was prepared using a 20 ml disposable syringe (without needle) with a small amount of glass wool applied to the bottom and overlaid with 5 ml gelatin sepharose solution. A separate column was prepared for MMP‐2 and for MMP‐9. The column was washed twice with equilibration buffer; 0·05 M Tris base, 0·5 M NaCl (Fisher Bioreagents, Fisher Scientific, Loughborough, UK), 0·005 M CaCl2, 0·05% Brij, 0·02% sodium azide, pH 7·6. The fibroblast spent media or lysed cell suspensions were passed through the appropriate column and discarded. The column was washed with 5× 5 ml equilibration buffer. The gelatin sepharose‐bound enzyme was eluted with 10 ml 10% dimethyl sulphoxide in 80% elution buffer (0·05 M Tris–HCl, 1 M NaCl, 0·005 M CaCl2, 0·05% Brij 35%, 0·02% sodium azide) and collected in 15 ml centrifuge tubes. The eluent was then dialysed overnight in MMP dialysis buffer (0·05 M Tris–HCl, 0·005 M CaCl2, 0·05% Brij 35, 0·02% sodium azide) at 4°C. Dialysed aliquots were then stored at −80°C until required for dressing incubation experiments. The columns were rehydrated with equilibration buffer, sealed and stored at 4°C for further use.
Dressing interaction studies using purified MMPs
In order to assess the ability of each dressing to sequester MMP‐2 and MMP‐9 activity an in vitro model was used, as adapted from the study by Walker et al. 15. Dressing samples weighing 0·01 g were prepared and incubated with 250 µl of either MMP‐2 or MMP‐9 in triplicate for each of the four dressings in 24‐well culture plates. Separate samples were incubated for 0 (1 minute), 3, 6, 24 and 48 hours. At each corresponding time point 500 µl 1× sterile phosphate buffered saline (PBS) was added to each well of the plate, to stop the action of the dressing and to aid in the elution of the enzyme solution from the dressing samples, and the enzyme samples were recovered and the supernatant stored at −80°C until further analysis of enzyme activity using gelatin zymography.
Dressing interaction studies using Pseudomonas aeruginosa conditioned media
Bacterial isolation and identification
Pseudomonas aeruginosa isolates were obtained from chronic equine wounds using an optimised swab method and stored in Stuart's transport medium (Oxoid, Basingstoke, UK). Individual colonies were isolated on tryptone soy agar (TSA) (LabM, Bury, UK) plates and colonies were confirmed as P. aeruginosa on 5% blood agar plates and MacConkey agar (LabM), followed by Gram staining and analysis under oil immersion. Oxidase tests and API identification strips (API 20 E, Biomerieux, Basingstoke, UK) were used for final identification. An ATCC type strain of P. aeruginosa (ATCC 27853) was used as a control.
P. aeruginosa culture
Sterile Mueller Hinton broth (MHB) (LabM) was inoculated in 30 ml portions with each of the corresponding P. aeruginosa isolates and cultured by shaking in a shaking incubator set at 37°C for 24 hours in 75 cm3 culture flasks. Cultures were harvested and centrifuged at 121 g for 10 minutes to pellet the cells. Conditioned culture supernatant was filtered through 0·45 µm sterile filters.
In order to assess the ability of each dressing to sequester P. aeruginosa derived protease activity in vitro, 0·01 g dressing samples were prepared and incubated with 250 µl of each corresponding P. aeruginosa conditioned media in triplicate for each of the four dressings in 24‐well culture plates. Separate samples were incubated for 0 (1 minute), 3, 6, 24 and 48 hours. At each corresponding time point 500 µl sterile PBS was added to each well of the plate and the enzyme samples were recovered and the supernatant stored at −80°C until further analysis of enzyme activity. Dressing interaction studies were also performed with a combination of MMP‐2 and P. aeruginosa conditioned media: 125 µl of MMP‐2 was added to the plate wells with 50 µl conditioned P. aeruginosa media. Samples were treated as previously described following incubation, and supernatants were harvested ready for analysis via the milk‐casein agar plate method.
Milk‐casein agar plate inoculation
Whilst the milk‐casein agar plate method for detecting proteolytic activity is only semi‐quantitative, it provides an initial visual representation of the levels of general proteolytic activity of different bacterial isolates over time, prior to further investigation. This method is commonly used for the screening of general proteases produced by both Gram negative and Gram positive bacteria, as reviewed by Kasana, Salwan and Yadav 16. Whilst some studies have measured the proteolytic activity of various microorganisms from the proteolytic halo zone formed around colonies from microorganisms isolated from various settings 17, 18, 19, 20, 21, in this study the method was standardised using wells cut into the agar plates into which an aliquot of conditioned bacterial media from a culture sample normalised to a set optical density (OD) was placed. This method has been used previously to detect the presence of bacterial proteolytic activity in the clinical setting 22, 23.
Milk‐casein agar was prepared with 25 g skim milk powder (BD, Oxford, UK), 2·5 g casein (BDH, VWR, Lutterworth, UK), 1·25 g yeast extract (Oxoid, Basingstoke, UK), 0·5 g d‐glucose (BDH, Oxford, UK) and 6·25 g No. 1 agar (LabM). Agar was sterilised by autoclaving and poured to equal levels into 9 cm round petri dishes. A sterile 8 mm punch biopsy was used to bore single wells in each of the milk plates. Samples recovered from the dressing incubations with P. aeruginosa conditioned cell‐free media were used to inoculate milk‐casein agar plates. A 100 µl aliquot of each sample was applied to the 8 mm wells made in milk plates in triplicate. Plates were incubated for 24 and 48 hours. Following incubation, the caseinolytic area was measured; two measurements were made in mm at right angles to each other and an average measurement made, minus the 8 mm well (see Figure 1). For calculations of percentage decrease in caseinolytic activity, the average measurement of the clear caseinolytic zone (in mm; minus the 8 mm well) for each sample was given as a percentage of the average measurement of the clear caseinolytic zone (mm) (minus the 8 mm well) for the wells containing conditioned media which was previously incubated without any of the test dressings (control wells). Percentage decrease in the caseinolytic zone, as compared with the control wells, was then calculated.
Figure 1.
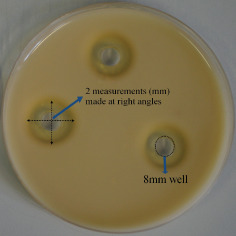
Method of calculation of caseinolytic zone using the milk‐casein agar plate method.
Dressing interaction studies with P. aeruginosa live cultures
P. aeruginosa cultures, both wound‐isolates and ATCC type strain, were prepared as previously described. Live cultures were standardised to an OD of 0·85 at 600 nm using a spectrophotometer. Aliquots of 100 µl of each individual culture were applied, in triplicate, to 8 mm wells in milk‐casein agar plates, prepared as previously described. About 0·01 g portions of each dressing were overlaid over the cultures in the corresponding wells. Plates were incubated at 37°C for 24 and 48 hours. Caseinolytic zones were measured as described above and averages taken.
Treatment of samples for gelatin zymography
Samples were diluted with non reducing Laemmli sample buffer (Bio‐Rad, Hemel Hempstead, UK) at a ratio of 1:5 and distilled water to an overall 1:5 dilution of the sample. Samples were then incubated in a 37°C water bath for 1 hour to activate the enzyme prior to application to the gel. A Miniprotean II gel system (Bio‐Rad) was used for zymography gels. A 7·5% acrylamide/bis‐acrylamide (Severn Biotech Ltd., Kidderminster, UK) resolving gel copolymerised with a 1% gelatin solution (EIA grade reagent gelatin) (Bio‐Rad) was prepared to create a 0·25% gelatin zymogram in a pH 8·8 resolving buffer (1·5 M Tris/HCl). A volume of 3·2 ml of this was cast in between ethanol washed glass gel plates and overlain with isobutanol to level the gel. The gel was left to set. Once set, the resolving gel was layered with a stacking gel made up with a pH 6·8 buffer (0·5 M Tris/HCl) and was left to set with a 10 lane comb inserted on top of the gel. The gels were placed in a tank filled with running buffer and 20 µl portions of each sample were loaded into the gel and subjected to electrophoresis at 200 V for 1 hour or until the lane marker had run off the end of the gel. Mark‐12 high molecular weight lane markers (Invitrogen, LC5677, unstained standards) were run on each gel. Following electrophoresis, gels were carefully removed from the glass plates and the lane markers were removed and placed in 0·02% Coomassie brilliant blue stain (BDH) for 15–20 minutes. The gels were placed in 2·5% Triton X‐100 and agitated on a plate shaker for 1 hour to remove all traces of SDS. The gels were washed thoroughly in distilled water and placed in MMP incubation buffer/gelatin refolding buffer overnight. After incubation, the gels were stained for 1 hour in 0·5% Coomassie stain and then destained in a solution of diluted methanol/acetic acid until clear bands of digested gelatin were visible against a darkly stained blue background. Gels were visualised and imaged using Genesnap software. The gels were dried in cellophane sheets using a Hoeffer gel drying dock.
Gel quantification
Gel images were captured and analysed using the GeneGenius Gel Documentation System (Syngene, Cambridge, UK). The relative activity value (RAV) was calculated using the following equation:
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of conditioned P. aeruginosa media
A 15% acrylamide mini gel was prepared using 3·2 ml resolving gel in a 1·5 M Tris/HCl buffer of pH 8·8. Once set this gel was overlaid with a stacking gel made up in a 0·5 M Tris/HCl buffer, pH 6·8 in a Miniprotean gel system (Bio‐Rad). Samples were prepared by adding 13 µl original sample, 2 µl DTT and 5 µl non reducing sample buffer with marker dye (Thermo Scientific). Samples were denatured by heating for 10 minutes in a heat block pre‐set to 80°C. The samples were vortexed briefly. A 20 µl aliquot of each sample was added to each corresponding lane of the gel. The gel tank was filled with a 1:20 dilution of NUPAGE MES SDS running buffer (Invitrogen) in distilled water. The gel was run at 15 V for approximately 2 hours or until the lane marker ran to the bottom of the gel. The gels were carefully removed from the glass plates and placed in 0·02% Coomassie brilliant blue stain for 2 hours with agitation. Gels were then destained in a solution of diluted methanol/acetic acid until dark bands appeared against a stain‐free background.
Results
Dressing incubations with MMP‐2 and MMP‐9
Following incubation at 37°C with all of the dressings tested, the residual proteinase activity was assayed at 0 (1 minute), 1, 3, 6, 24 and 48 hours to evaluate the capacity for each dressing to inactivate both purified MMP‐2 and MMP‐9. All of the dressings tested were able to reduce the levels of active and latent enzymes when compared to the control (no dressing) from 1 minute onwards. The standard foam dressing was less effective than the other dressings, yet was still able to actively reduce MMP levels. This could be attributed to the dressing's active fluid management system. Samples incubated with the Collagen/ORC dressing demonstrated complete inhibition of both MMP‐2 and MMP‐9 in their latent (Figures 2 and 3) and active forms (Figures 4 and 5) from 1 hour onwards. The foam dressing containing polyphosphate consistently outperformed both the standard foam dressing and the TLC‐NOSF dressing in terms of the sequestration of the active and latent forms of MMP‐2 and MMP‐9. Samples treated with the foam dressing containing polyphosphate demonstrated a time‐dependant reduction in MMP activity, decreasing MMP‐2 and MMP‐9 by more than 50% by 48 hours in both the active and latent forms.
Figure 2.
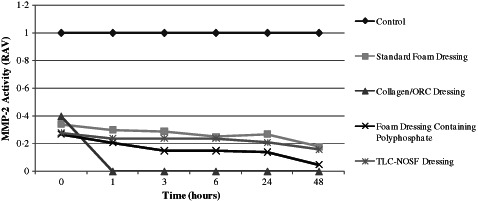
Action of each of the test dressings against the activity of matrix metalloproteinase (MMP)‐2 in the latent form. Dressings were incubated with MMP‐2 (n = 3) for 0, 1, 3, 6, 24 and 48 hours.
Figure 3.
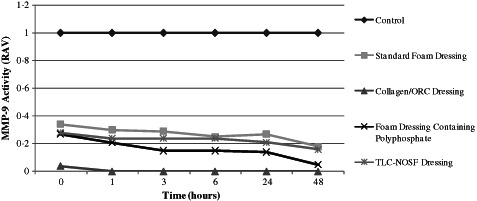
Action of each dressing against the activity of matrix metalloproteinase (MMP)‐9 in the latent form. Dressings were incubated with MMP‐9 (n = 3) for 0, 1, 3, 6, 24 and 48 hours.
Figure 4.
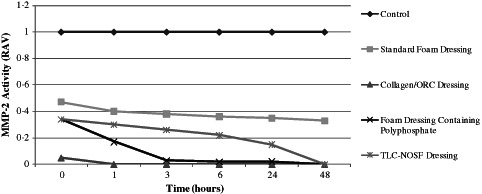
Action of each dressing against the activity of matrix metalloproteinase (MMP)‐2 in the active form. Dressings were incubated with MMP‐2 (n = 3) for 0, 1, 3, 6, 24 and 48 hours.
Figure 5.
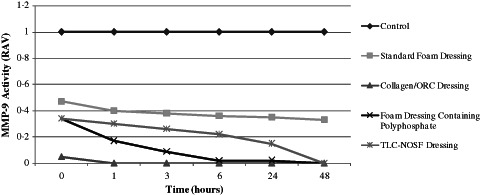
Action of each dressing against the activity of matrix metalloproteinase (MMP)‐9 in the active form. Dressings were incubated with MMP‐9 (n = 3) for 0, 1, 3, 6, 24 and 48 hours.
Incubations with MMP‐2 and MMP‐9 showed that the foam dressing containing polyphosphate sequestered significantly more latent enzyme than the TLC‐NOSF dressing at 3 and 48 hours (P < 0·005). The performance of the foam dressing containing polyphosphate was significantly greater when compared to the standard foam dressing following 1, 3, 6, 24 and 48 hours incubation with the enzymes (P < 0·001) (see Figures 6 and 7).
Figure 6.

Example zymogram of samples from dressing incubations with matrix metalloproteinase (MMP)‐2. The 72 kDa bands are indicative of latent MMP‐2 whereas the 64 kDa bands are indicative of active MMP‐2. Lanes 1–3: 0 hours, standard foam dressing; Lanes 4–6: 0 hours, Collagen/ORC dressing; Lanes 7–9: 0 hours, foam dressing containing polyphosphate; Lane 10: 0 hours, Control MMP‐2 (no dressing treatment).
Figure 7.

Example zymogram of samples from dressing incubations with matrix metalloproteinase (MMP)‐2. The 72 kDa bands are indicative of latent MMP‐2 whereas the 64 kDa bands are indicative of active MMP‐2. Lane 1: 3 hours, Control MMP‐2 (no dressing treatment); Lanes 2–4: 3 hours, standard foam dressing; Lanes 5–7: 3 hours, Collagen/ORC dressing; Lanes 8–10: 3 hours, foam dressing containing polyphosphate.
The foam dressing containing polyphosphate significantly reduced the active enzymes (both MMP‐2 and MMP‐9) when compared to the TLC‐NOSF dressing at 1, 3, 6 and 24 hours (P < 0·005). Likewise, there was a significant reduction (P < 0·001) in active MMP‐2 and MMP‐9 when tested against the standard foam dressing at 1, 3, 6, 24 and 48 hours.
Dressing interaction studies using P. aeruginosa conditioned media
Each of the test dressings was incubated as described for the MMP incubations, with 250 µl filtered 24 hour conditioned culture media from one ATCC type strain and two chronic wound‐isolated strains of P. aeruginosa. Milk‐casein agar plate evaluation demonstrated a reduction in caseinolytic activity following dressing incubation with each of the dressings when compared with the bacterial conditioned media alone. The percentage decrease in caseinolytic zone area showed that the foam dressing containing polyphosphate sequestered P. aeruginosa derived proteases to a greater extent when compared to the standard foam dressing and the TLC‐NOSF dressing after 24 hours (Figure 8) when incubated with the ATCC type strain derived conditioned media. In contrast, the proteolytic activity of the ATCC P. aeruginosa conditioned media incubated with the TLC‐NOSF dressing and the standard foam dressing returned after 24 hours. The Collagen/ORC dressing had the greatest effect against the proteases of this strain, reducing the proteolytic activity from 1 hour onwards when compared in terms of percentage decrease with the other dressing types. The foam dressing containing polyphosphate showed optimum activity at each time point against the wound‐isolated strain with high biofilm‐forming capacity, when compared to each of the other three dressings.
Figure 8.
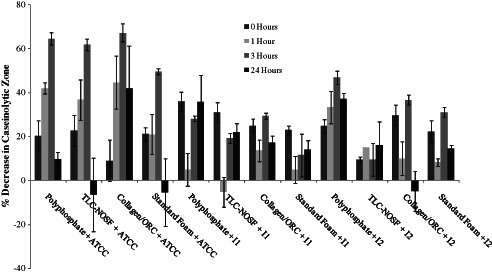
Percentage decrease in caseinolytic zone of milk‐casein agar when incubated with each dressing and protease‐rich conditioned media of an ATCC type strain and 2 chronic wound‐isolated Pseudomonas aeruginosa. Milk‐casein agar plates were incubated (n = 3) for 0, 1, 3, 6 and 24 hours. I1: P. aeruginosa Wound strain 1 (low biofilm potential); I2: P. aeruginosa Wound strain 2 (high biofilm potential). ‘Polyphosphate’ refers to the foam dressing containing polyphosphate.
The activity of the Collagen/ORC dressing peaked following 3 hours incubation with the conditioned media of each strain but this dressing demonstrated the greatest activity against the ATCC P. aeruginosa reference strain derived conditioned media when compared to the chronic wound isolated P. aeruginosa cultures. Results showed that following 24 hours incubation, the foam dressing containing polyphosphate was more active at reducing caseinolytic zones from the high biofilm‐forming P. aeruginosa isolate derived proteases than the Collagen/ORC dressing. The foam dressing containing polyphosphate was also significantly better at reducing proteolytic activity than the TLC‐NOSF dressing after 3 hours of incubation with the conditioned media from the high biofilm‐forming isolate (P < 0·05). SDS‐PAGE analysis was conducted to identify the major proteases produced by P. aeruginosa isolates in vitro (see Figure 9).
Figure 9.

Example SDS‐PAGE showing the major protein bands produced by Pseudomonas aeruginosa chronic equine wound isolates at 4 hours (Lanes 2 and 3), 8 hours (Lanes 4 and 5), 12 hours (Lanes 6 and 7) and 24 hours (Lanes 8 and 9) after MHB inoculation. Lane 1: Protein markers; Lanes 2, 4, 6 and 8: Low biofilm‐forming isolate; Lanes 3, 5, 7 and 9: High biofilm‐forming isolate. The second bands from the top of the gel may represent the 33 kDa protein, mature elastase whereas the top bands seen on the gel may represent the 51 kDa proelastase or the 53 kDa preproenzyme of elastase (McIver, Kessler & Ohman, 1991). However, for confirmation further protein analysis would be required.
Dressing incubation with live P. aeruginosa cultures
Each of the dressings were tested for their potential ability in sequestering the proteases secreted into the media of a number of live P. aeruginosa cultures. Results showed that after 24 hours incubation the foam dressing containing polyphosphate reduced the caseinolytic zones of milk‐casein agar plates inoculated with each of the clinical P. aeruginosa isolates when compared with the test plates with no dressing and compared to each of the other dressings (see Figure 10).
Figure 10.
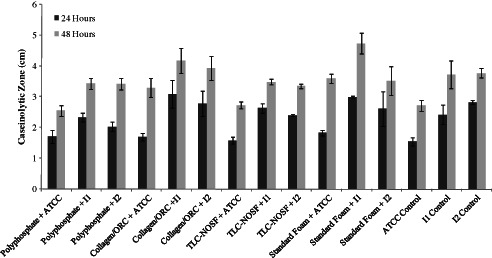
Caseinolytic zone measurements (average of 2 measurements) (n = 3) of milk‐casein agar plates inoculated with Pseudomonas aeruginosa live cultures, overlaid with each of the four dressings. Plates were incubated for 24 hours and 48 hours and measurements made at each time point. ATCC, P. aeruginosa reference strain; I1, P. aeruginosa Wound strain 1 (low biofilm potential); I2, P. aeruginosa Wound strain 2 (high biofilm potential). ‘Polyphosphate’ refers to the foam dressing containing polyphosphate.
Dressing incubations with a combination of purified MMP‐2 and P. aeruginosa conditioned media
Each of the dressings were tested for their potential ability in sequestering the proteases of P. aeruginosa conditioned media in combination incubations with purified MMP‐2 (see Figures 11 and 12). Results showed that the foam dressing containing polyphosphate reduced the activity of both MMP‐2 and the P. aeruginosa derived protease steadily over a 6‐hour period. However, following further incubation, the activity of the bacterial derived protease increased. This also occured in the other dressing incubations, apart from the Collagen/ORC dressing which maintained its activity over the 24‐hour period. This may be due to the limited effectiveness of dressings over extended time periods or may be as a result of proteases being released back into the surrounding supernatant from the dressing samples. The foam dressing containing polyphosphate performed better than the standard foam dressing throughout the incubation period.
Figure 11.
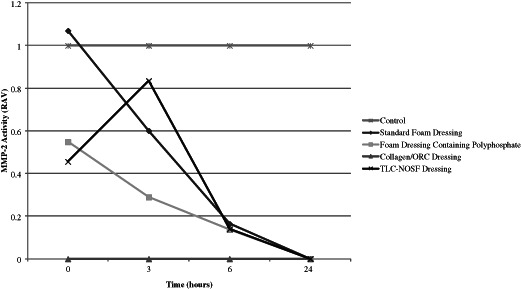
Action of each dressing against the activity of matrix metalloproteinase (MMP‐2). Dressings were incubated with MMP‐2 (n = 3) for 0, 3, 6 and 24 hours.
Figure 12.
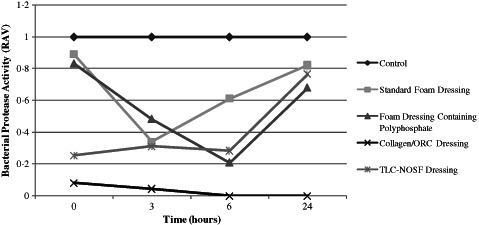
Action of each dressing against the activity of Pseudomonas aeruginosa‐derived protease. Dressings were incubated with protease‐rich conditioned P. aeruginosa media (n = 3) for 0, 3, 6 and 24 hours.
Each of the dressings were incubated over a period of 24 hours as described previously. Dressings were incubated with a combination of MMP‐2 (125 µl) and P. aeruginosa protease‐rich conditioned media (50 µl). Results from the zymography analysis showed that the dressings had an immediate action in terms of sequestration of the active MMP‐2 enzyme when in combination with P. aeruginosa derived protease. Whether this is a direct synergistic effect of the presence of bacterial protease remains to be elucidated. Similarly, the level of P. aeruginosa protease activity decreased rapidly throughout the incubation period for all dressings tested. However, when incubated with the standard foam dressing the activity of P. aeruginosa derived protease increased after 3 hours before reaching an activity that was over 80% of the control after 24 hours incubation. This also occured when samples were incubated with both the foam dressing containing polyphosphate and the TLC‐NOSF dressing but only after a 6‐hour incubation period. The reason for this increase is not known, but may suggest that the dressings are only able to hamper the activity of these enzymes for a limited period. The Collagen/ORC dressing was effectively able to reduce the activities of both MMP‐2 and P. aeruginosa derived protease following 0 (1 minute), 3, 6 and 24 hours incubation when tested in this in vitro model.
Discussion
All chronic wounds are characterised by an elevated inflammatory response, resulting in increases in protease activity and abundance and decreased growth factor activity 2. MMPs are able to break down peptide links of growth factors thus indirectly inhibiting matrix remodelling 10. This dual biochemical imbalance in wound tissues is, in part, responsible for the failing of healing in such wounds, given the net loss of tissue 2. MMPs have also been implicated in the pathogenesis of cancer metastasis, as a result of increased tissue breakdown and remodelling during invasive tumour growth and angiogenesis 24. Indeed, raised blood plasma levels of MMP‐3 and MMP‐9 have been found in patients with medullary carcinoma 24.
It has been consistently reported that elevated and potentially harmful levels of proteases, particularly the metalloproteases MMP‐2 and MMP‐9 are present in chronic wound fluids in a range of mammalian models 2, 8, 25. It is likely that the sequestration of these harmful levels of MMPs within the wound bed could enhance healing and encourage matrix reformation. Whilst it may not be possible, and would not be necessary, to completely remove MMPs from the wound environment or to completely inhibit their action, their activity may be contained sufficiently.
A number of dressings have been developed with the aim to retain MMPs within the wound bed or to sequester protease activity, including silver dressings 15, collagen‐containing dressings which act as an alternative substrate for MMPs 2 and dressings which act to absorb and retain proteases within the matrix of the dressing whilst immobilising bacteria 26. Whilst collagen is useful in wound healing as a mechanical scaffold to promote fibroblast migration 13 and can enhance the metabolic activity of newly formed granulation tissue, regenerated cellulose is able to effectively reduce levels of proteases. Another type of dressing incorporates a compound aimed at wound closure primarily through MMP inhibition, namely NOSF. Polyphosphates have been used for their ability to chelate alginate, a product of biofilm‐forming bacteria. Molecularly dehydrated polyphosphates have been added to wound dressings to reduce the alginate present in infected wounds and they have proved more successful than organic surfactants in terms of their toxicity to the skin. Furthermore, other chelating agents require stabilisation of pH to ensure cytoprotection, which is not always possible during wound management.
Ribeiro et al. 1 described a chitosan hydrogel dressing which incorporated polyphosphate as an anticoagulant along with silver as an antibacterial agent. Whilst many research studies have focussed on the anticoagulant properties of polyphosphates in wound dressings, the potential of this substance in protease sequestration is emerging. As polyphosphates are negatively charged, they associate with and bind cations, including zinc ions and so prove useful in the sequestration of metalloproteinases. Furthermore, polyphosphate has also been shown to have antibacterial properties, although this has been insufficiently studied experimentally.
In this study, it has been demonstrated that a foam dressing containing polyphosphate was able to sequester both MMP‐2 and MMP‐9 in their active and latent states as visualised using gelatin zymography. The results from this study also showed that the Collagen/ORC dressing effectively inhibited the activity of both MMP‐9 and MMP‐2, corresponding well to published data on the efficacy of this dressing 27. The Collagen/ORC dressing functions to actively bind and inactivate proteases involved in wound healing including MMPs, plasmin and neutrophil elastase 27. As in other studies, this dressing sustained its activity throughout the 48 hour test period. This coincides with the dressing change time period recommended for this type of dressing 27. The collagen component of this dressing acts as an alternative substrate, binding the proteases and thus inactivating them. The Collagen/ORC active agent is negatively charged, and is able to physically bind positively charged molecules including metal ions important for MMP structure and stability.
This study represents significant data relating to the efficacy of a foam dressing containing polyphosphate in the sequestration of MMP‐2 and MMP‐9: the dressing was effectively able to reduce MMP‐2 and MMP‐9 activity throughout the 48‐hour test period, with optimal activity observed after 6 hours of incubation with these enzymes. Furthermore, the foam dressing containing polyphosphate outperformed the standard foam dressing and the TLC‐NOSF dressing in terms of reducing the levels of active MMP‐2 and ‐9 over the entire incubation period and until 24 hours incubation, respectively. Future studies should aim to assess the capacity of the foam dressing containing polyphosphate to sequester other important proteases involved in wound healing, including neutrophil elastase and also any effect of this dressing on the levels of relevant protease inhibitors.
The foam dressing containing polyphosphate also demonstrated potential in the inactivation of P. aeruginosa derived proteases, both alone and in combination with MMP‐2. Moreover, the foam dressing containing polyphosphate was more effective in the inhibition of proteases derived from a P. aeruginosa chronic wound‐derived isolate with high biofilm‐forming capacity when compared to a type strain and a low biofilm‐forming isolate. This may be important in relation to chronic wounds in which high levels of bacterial bioburden exist, particularly in the biofilm state. There is very little evidence in the literature to suggest a role for therapeutic wound dressings in inhibition of bacterial derived proteases despite evidence suggesting that these proteases play a vital pathological role in wound chronicity. Whilst the methods used in this study for the analysis of protease activity are semi‐quantitative, the study demonstrates the potential of a novel foam dressing containing polyphosphate to inhibit both host MMPs‐2 and ‐9 and also those exogenous proteases found within the wound bed. In future experimentation, fully quantitative methods should be used to confirm the levels of specific bacterial derived proteases; substrate assays can be utilised to assess specific protease activity. For instance, the elastin Congo red assay can be used for the detection of P. aeruginosa elastase. Fluorescent substrate assays can also be used for quantitative analysis of proteolytic activity. Furthermore, other bacterial proteases should be considered in future studies. For example, Staphylococcus aureus produces a number of proteases (including the serine protease, V8 protease) which are considered important virulence factors within the wound milieu. Different combinations of host and bacterial proteases could be used to assess the ability of the foam dressing containing polyphosphate to sequester those proteases found within the chronic wound. Although the study of the mechanism(s) of action of the polyphosphate foam dressing was beyond the scope of this study, the effect seen could relate to the inhibition of biofilm formation through the binding of zinc which is also the key pathway in the inhibition of metalloproteinases. Furthermore, chelation of Zn2+ specifically prevents biofilm formation by Staphylococcus epidermidis and methicillin‐resistant S. aureus (MRSA) 28. The therapeutic inhibition of zinc molecules within the wound bed may prove useful in inhibiting both biofilm formation 28 and in the inactivation of proteases, both bacterial and host‐derived.
Further investigation into the potential of the foam dressing containing polyphosphate to sequester proteases should also consider dressing absorption; proteases which are free within the dressing once absorbed may still be active and thus would be free to migrate back into the wound bed. It is important to investigate the mechanism of action of the dressing in terms of reduction of protease activity and to address the questions of whether proteases are bound within the dressing and whether they are permanently inactivated. In Figures 2, 3, 4, 5, 11 and 12, there is evidence of protease activity returning over time following incubation of the proteases with the test dressing samples. This may be indicative of an initial decrease in protease activity due to dressing absorption; in future experimentation it may be useful to pre‐wet the dressing samples so as to exclude absorption as an influential factor.
In terms of transposing the data presented to the human chronic wound, the equine model is an ideal model in which to study the naturally occurring human chronic wound 25, 29. The pathophysiology of the equine chronic wound is very similar to that of the human chronic wound. For example, the equine indolent wound is similar in appearance to human venous leg ulcers; they produce little granulation tissue and are often inflamed and infected 25. The equine wounds used to isolate bacteria used in this study were naturally occurring chronic wounds. Furthermore, gelatinase activity in equine tissue homogenates shows that the major bands of activity in equine granulation tissue correspond to human MMP‐2 and MMP‐9 and proteolytic bands have been shown to be less prominent in normal tissue homogenates compared with granulation tissue homogenates 25. Whilst no in vitro model can mimic the true clinical environment in terms of the levels of proteolytic activity, future experimentation could utilise wound fluids collected from acute and chronic wounds to test the ability of wound dressings to sequester various proteases.
To conclude, this study has presented evidence to suggest a role for polyphosphate when incorporated into a foam wound dressing for the management of chronic wounds with elevated protease activity. A foam dressing incorporating polyphosphate has been shown to reduce the activity of both host MMPs‐2 and ‐9 and P. aeruginosa derived proteinases via an in vitro model.
Acknowledgement
This work was funded by an educational grant from Advanced Medical Solutions Ltd.
References
- 1. Ribeiro MP, Epspiga A, Silva D, Baptista P, Henriques J, Ferreira C, Silva JC, Borges JP, Pires E, Chaves P. Development of a new chitosan hydrogel for wound dressing. Wound Repair Regen 2009;17:817–24. [DOI] [PubMed] [Google Scholar]
- 2. Cullen B, Smith R, Mcculloch E, Silcock D, Morrison L. Mechanism of action of PROMOGRAN, a protease modulating matrix, for the treatment of diabetic foot ulcers. Wound Repair Regen 2002;10:16–25. [DOI] [PubMed] [Google Scholar]
- 3. Wright JB, Lam K, Buret AG, Olson ME, Burrell RE. Early healing events in a porcine model of contaminated wounds: effects of nanocrystalline silver on matrix metalloproteinases, cell apoptosis, and healing. Wound Repair Regen 2002;10:141–51. [DOI] [PubMed] [Google Scholar]
- 4. Ravanti L, Kahari V. Matrix metalloproteinases in wound repair (review). Int J Mol Med 2000;6:391–407. [PubMed] [Google Scholar]
- 5. Trengove NJ, Stacey MC, Macauley S, Bennett N, Gibson J, Burslem F, Murphy G, Schultz G. Analysis of the acute and chronic wound environments: the role of proteases and their inhibitors. Wound Repair Regen 1999;7:442–52. [DOI] [PubMed] [Google Scholar]
- 6. Tarnuzzer RW, Schultz GS. Biochemical analysis of acute and chronic wound environments. Wound Repair Regen 1996;4:321–5. [DOI] [PubMed] [Google Scholar]
- 7. McCarty SM, Cochrane CA, Clegg PD, Percival SL. The role of endogenous and exogenous enzymes in chronic wounds: a focus on the implications of aberrant levels of both host and bacterial proteases in wound healing. Wound Repair Regen 2012;20:125–36. [DOI] [PubMed] [Google Scholar]
- 8. Lund LR, Romer J, Bugge TH, Nielsen BS, Frandsen TL, Degen JL, Stephens RW, Danø K. Functional overlap between two classes of matrix‐degrading proteases in wound healing. EMBO J 1999;18:4645–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edwards JV, Howley PS. Human neutrophil elastase and collagenase sequestration with phosphorylated cotton wound dressings. J Biomed Mater Res A 2007;83:446–54. [DOI] [PubMed] [Google Scholar]
- 10. Timmons J. Evaluating a new foam dressing with a healing accelerator. Wounds UK 2010;6:88–92. [Google Scholar]
- 11. Veves A, Sheehan P, Pham HT. A randomized, controlled trial of Promogran (a collagen/oxidized regenerated cellulose dressing) vs standard treatment in the management of diabetic foot ulcers. Arch Surg 2002;137:822–7. [DOI] [PubMed] [Google Scholar]
- 12. Richardson JC, Dettmar PW, Allen RL, Coyle CP. Chronic wound treatment. Google Patents, 2009.
- 13. Hart J, Silcock D, Gunnigle S, Cullen B, Light ND, Watt PW. The role of oxidised regenerated cellulose/collagen in wound repair: effects in vitro on fibroblast biology and in vivo in a model of compromised healing. Int J Biochem Cell Biol 2002;34:1557–70. [DOI] [PubMed] [Google Scholar]
- 14. Wysocki AB, Staiano‐Coico L, Grinnell F. Wound fluid from chronic leg ulcers contains elevated levels of metalloproteinases MMP‐2 and MMP‐9. J Invest Dermatol 1993;101:64–8. [DOI] [PubMed] [Google Scholar]
- 15. Walker M, Bowler PG, Cochrane CA. In vitro studies to show sequestration of matrix metalloproteinases by silver‐containing wound care products. Ostomy Wound Manage 2007;53:18–25. [PubMed] [Google Scholar]
- 16. Kasana RC, Salwan R, Yadav SK. Microbial proteases: Detection, production, and genetic improvement. Crit Rev Microbiol 2011;37:262–76. [DOI] [PubMed] [Google Scholar]
- 17. Sokol PA, Ohman DE, Iglewski BH. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa . J Clin Microbiol 1979;9:538–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Howe TR, Iglewski BH. Isolation and characterization of alkaline protease‐deficient mutants of Pseudomonas aeruginosa in vitro and in a mouse eye model. Infect Immun 1984;43:1058–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nicas TI, Iglewski BH. Isolation and characterization of transposon‐induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect Immun 1984;45:470–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wery N, Gerike U, Sharman A, Chaudhuri JB, Hough DW, Danson M. Use of a packed‐column bioreactor for isolation of diverse protease‐producing bacteria from Antarctic soil. J Appl Environ Microbiol 2003;69:1457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ponmurugan P. Proteolytic Activity of Bacillus cereus Under in vitro Condition. J Biol Sci 2007;7:65–7. [Google Scholar]
- 22. Palolahti M, Lauharanta J, Stephens RW, Kuusela P, Vaheri A. Proteolytic activity in leg ulcer exudate. Exp Dermatol 1993;2:29–37. [DOI] [PubMed] [Google Scholar]
- 23. Wladyka B, Bista M, Sabat AJ, Bonar E, Grzeszczuk S, Hryniewicz W, Dubin A. A novel member of the thermolysin family, cloning and biochemical characterization of metalloprotease from Staphylococcus pseudintermedius. Acta Biochim Pol 2008;55:525–36. [PubMed] [Google Scholar]
- 24. Komorowski J, Pasieka Z, Jankiewicz‐Wika J, Stepien H. Matrix metalloproteinases, tissue inhibitors of matrix metalloproteinases and angiogenic cytokines in peripheral blood of patients with thyroid cancer. Thyroid 2002;12:655–62. [DOI] [PubMed] [Google Scholar]
- 25. Cochrane CA. Models in vivo of wound healing in the horse and the role of growth factors. Vet Dermatol 1997;8:259–72. [DOI] [PubMed] [Google Scholar]
- 26. Walker M, Hobot JA, Newman GR, Bowler PG. Scanning electron microscopic examination of bacterial immobilisation in a carboxymethyl cellulose (Aquacel®) and alginate dressings. Biomaterials 2003;24:883–90. [DOI] [PubMed] [Google Scholar]
- 27. Cullen B, Watt PW, Lundqvist C, Silcock D, Schmidt RJ, Bogan D, Light ND. The role of oxidised regenerated cellulose/collagen in chronic wound repair and its potential mechanism of action. Int J Biochem Cell Biol 2002;34:1544–56. [DOI] [PubMed] [Google Scholar]
- 28. Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. A zinc‐dependent adhesion molecule is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA 2008;105:19456–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cochrane CA, Pain R, Knottenbelt DC. In‐vitro wound contraction in the horse: differences between body and limb wounds. Wounds UK 2003;15:175–81. [Google Scholar]


