Abstract
The DNA replication stress-induced checkpoint activated through the TopBP1-ATR axis is important for maintaining genomic stability. However, the regulation of TopBP1 in DNA-damage responses remains unclear. In this study, we identify the deubiquitinating enzyme (DUB) USP13 as an important regulator of TopBP1. Mechanistically, USP13 binds to TopBP1 and stabilizes TopBP1 by deubiquitination. Depletion of USP13 impedes ATR activation and hypersensitizes cells to replication stress-inducing agents. Furthermore, high USP13 expression enhances the replication stress response, promotes cancer cell chemoresistance, and is correlated with poor prognosis of cancer patients. Overall, these findings suggest that USP13 is a novel deubiquitinating enzyme for TopBP1 and coordinates the replication stress response.
Keywords: USP13, TopBP1, replication, DNA damage
1. Introduction
DNA replication is an essential process for propagating genetic information. Imperfect DNA replication leads to the accumulation of genomic instability and is a major driver of tumorigenesis [1]. To maintain the accuracy of DNA replication, replication forks must be protected from a variety of structural disruptions such as DNA lesions, transcription complexes, or DNA-RNA complexes [2–4]. Numerous mechanisms help to minimize error frequencies in replication. Stalled replication fork restart is one such mechanism for maintaining genomic stability in response to replication stress. During DNA replication, exo- or endogenous carcinogens generate several types of DNA damage which lead to stalled replication forks. These stalled replication forks generate single stranded DNA (ssDNA) that is sensed by the RPA complex, which in turn serves as a platform for recruiting the replication repair protein machinery such as Rad17 complex, Rad9-Hus1-Rad1 (9-1-1), and TopBP1. Recruited TopBP1 directly interacts with and activates ATR. Activated ATR phosphorylates substrates including RPA and Chk1 and regulates downstream stress responses such as cell cycle arrest, cell death when DNA damage repair fails, or replication fork restart after damage resolution [5–9]. It has previously been reported that TopBP1 is tightly regulated by post-translational-modification (PTM) following DNA damage, such as acetylation [10], phosphorylation [11], or ubiquitination [12]. Ubiquitination is the most common PTM regulating protein degradation. TopBP1 is ubiquitinated by the HECT domain containing ligase hHYD in unstressed cells. While X ray-irradiation is capable of diminishing the levels of ubiquitinated TopBP1, the responsible proteins and mechanism leading to decreased TopBP1 ubiquitination following DNA damage remain elusive.
Ubiquitin-specific-protein 13 (USP13) is a deubiquitinase belonging to the USP subclass of the deubiquitinating enzyme (DUB) superfamily. USPs can remove ubiquitin chains from its substrates to inhibit protein degradation. USP13 has been reported to be involved in diverse cellular processes such as autophagy, endoplasmic reticulum-associated degradation (ERAD), and DNA repair by deubiquitinating substrates BECN1, STING, SKP2, RAP80 and USP10 [13–18]. Despite its role in multiple cellular processes, the role of USP13 in tumorigenesis remains unclear. Some findings suggest that USP13 may promote tumor initiation or progression. For example, USP13 is overexpressed and promotes protein metabolism by deubiquitinating ATP citrate lyase and oxoglutarate dehydrogenase in ovarian cancer [19]. In addition, USP13 stabilizes microphthalmia-associated transcription factor and promotes cell invasion in melanoma [20]; and, USP13 inhibition decreases proliferation of glioma stem cells by inducing c-Myc degradation [21]. On the other hand, some studies suggest USP13 might act as a tumor suppressor. In oral squamous cell carcinoma and breast cancer USP13 inhibits tumorigenesis by deubiquitinating PTEN [22–24]. It has also been reported that USP13 promotes interferon signaling by stabilizing STAT1 [25]. USP13 also deubiquitinates USP10 and stabilizes its substrate TP53 [18]. Thus, the role of USP13 in tumor is most likely context-dependent.
Here, we show that USP13 is involved in the replication stress response. USP13 deubiquitinates and stabilizes TopBP1, which subsequently promotes ATR and checkpoint activation. This function of USP13 is important for regulating cancer cell survival in response to several anticancer agents.
2. Materials and Methods
2.1. Cell culture and inhibitor
HEK293T (CRL-3216), SK-OV-3 (HTB-77) and U2OS (HTB-96) cell lines were purchased from ATCC and cultured in Dulbecco’s Modified Eagle’s Medium or McCoy’s 5A supplemented with 10% fetal bovine serum (FBS) at 37 °C in 5% (v/v) CO2. The following inhibitors were used: MG132 (Sigma: C2211), Chloroquine (Sigma: C6628), and spautin-1 (Selleckchem: S7888 ).
2.2. Plasmids and antibodies
HA-FLAG-USP family vectors and HA-FLAG-USP13 were purchased from Addgene. USP13 site mutants were generated by site-directed mutagenesis (Stratagene). GFP-TopBP1 was a gift from Prof. Wojciech Niedzwiedz (Institute of Cancer Research, London, UK). The following antibodies were used: anti-USP13 (GTX118595, dilution: 1:500), anti-phospho RPA32 (S33) (Bethyl Laboratories: A300-246A-M, 1: 2000), anti-RPA32 (Bethyl Laboratories: A300-244A, 1:5000, Santa Cruz Biotechnology: sc-56770, 1: 2000), anti-γH2AX (EMD Millipore Crop: 2884537, 1: 1000, Bethyl Laboratories: A300-081A, 1: 1000), anti-phospho Chk1 (S345) (CST: 2348, 1:1000), anti-ATR (CST: 13934, 1: 1000), anti-TopBP1 (Bethyl Laboratories: A300-111A, 1: 1000), anti-Rad17 (Santa Cruz: sc-17761, 1: 1000), anti-ATRIP (CST: 2737, 1: 1000), anti-β-actin (Sigma: A2228, 1: 2000), anti-HA (Sigma: H6908, 1: 2000), anti-FLAG (Sigma: F1804, 1: 2000, F7425, 1: 2000), anti-GFP (Santa Cruz: sc-9996, 1: 1000), anti-UB (Santa Cruz: sc-8017, 1: 1000), anti-BrdU antibody (BD Bioscience: 347580, Abcam: ab6326).
2.3. sgRNAs and shRNAs
For CRISPR/Cas9 knockout of USP13 cells, the following sgRNAs were used. sgUSP13 #1: 5-GACCTGGGCACGCGGATCGT-3; sgUSP13 #2: 5-GCATGGAGGCGGCAACCAACA-3. All USP13 shRNAs were used as previous reports (17). These sgRNA and shRNA vectors were used for lentiviral infection.
2.4. DNA transfection, virus packaging and lentiviral infection
All DNA transfections were performed using TransIT-X2 (MIRUS Bio). Lentiviruses for infection were packaged in HEK293T cells using targeted shRNA, sgRNA, with pMD2.G and pSPAX2 vectors, respectively. Media containing lentivirus was collected forty-eight hours after transfection. Cells were inoculated with harvested virus and 8μg/mL polybrene.
2.5. Immunofluorescence
U2OS cells were cultured on coverslips 24 h before experiments. Cells were fixed with methanol: acetone (1:1) at −20 °C for 20 min, followed by washing two times with PBS, and blocked with 5% goat serum for 30 min. The cells were then incubated with primary antibodies diluted in PBS containing 1% BSA overnight at 4 °C, followed by secondary antibodies incubated at room temperature for 1 h. After washing for two times, cells were counter-stained by DAPI at room temperature for 5 min mounted with anti-fade solution, and visualized using a Nikon eclipse 80i Fluorescence microscope and foci number were counted by Image-Xpress confocal High-content imaging system (Molecular device).
2.6. Immunoprecipitation and immunoblotting
For immunoprecipitation, cells were lysed with NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40) for 10 min. Whole cell lysates were centrifuged at 12,000 rpm for 10 min. Supernatants were incubated with ANTI-FLAG® M2 Affinity Gel (Sigma: A2220) at 4 °C for 2 hours or overnight. After incubation, beads were washed three times with NETN buffer. The bound proteins on the beads were heated and eluted with 2x Laemmli buffer. For immunoblotting, cells were lysed in NETN buffer for 10 min. Whole cell lysates were centrifuged at 12,000 rpm for 10 min. Supernatants were heated in 2x Laemmli buffer and loaded to SDS-PAGE, transferred to PVDF membrane, and immunoblotting with the indicated antibodies.
2.7. Ubiquitination assay
For the denaturing ubiquitination assay, cells were collected and pellets were washed once in PBS. Cells were lysed with Urea lysis buffer (8 M Urea, 0.1 M NaH2PO4, 300 mM NaCl and 0.01 M Tris (pH 8.0)). Lysates were briefly sonicated to shear DNA and incubated with Ni-NTA magnetic agarose (Thermo Scientific: #78605) for 2 h at RT. Beads were washed five times with Urea wash buffer (8 M Urea, 0.1 M NaH2PO4, 300 mM NaCl and 0.01 M Tris (pH 8.0)). Input and beads were boiled in loading buffer. For ubiquitination assay, control cells or USP13 knockout cells were lysed in SDS buffer (62.5 mM Tris-HCl (PH 6.8), 2% SDS, 10% glycerol, 20 mM NEM and 1 mM iodoacetamide) and boiled for 15 min then diluted 10 times with NETN buffer containing protease inhibitors. The cell lysates were subjected to immunoprecipitation with the indicated antibodies, and immunoblotting with the indicated antibodies.
2.8. Colony formation assay
500 cells were plated in triplicates in each well of 6 well plates. 16 hours later, cells were treated with hydroxyurea (HU, mM), camptothecin (CPT, nM), ultra violet (UV, J/m2), or Fluorouracil (5-FU, μM) and incubated for 10 – 14 days at 37 °C to allow colony formation. Colonies were stained with Giemsa and counted. Results were normalized to plating efficiencies. The graphs represent mean ± S.D., two-tailed, unpaired t-test. n = 3 independent experiments (*. P < 0.05; **. P < 0.01; ***. P < 0.001).
2.9. DNA fiber assay
To assess restart efficiency of stalled replication forks, cells were labeled with IdU 25 μM for 20 min and then washed twice with media before being treated with HU 4 mM for 2 h. After washing with media, cells were recovered in fresh medium with CldU 200 μM for indicated time point. Cells were trypsinized and resuspended in PBS to a concentration of 2.5×105 cells/ml and diluted 1:40 with unlabeled cells at the same concentration. 5 μl of cells was mixed with 15 μl of lysis buffer (200 mM Tris-HCl, pH 7.4, 50 mM EDTA and 0.5% SDS) on a clean glass slide. After 8 min incubation, the slides were tilted at 15° to horizontal, allowing the lysate to slowly flow down along the slide. The slides were then air-dried, fixed in 3:1 methanol/acetic acid and stored at 4 °C overnight. The slides were treated with 2.5 M HCl for 1 h, neutralized in 0.1 M Na2B4O7, pH 8.5, and rinsed three times in PBST (PBS with 0.1% Tween-20). The slides were then blocked in blocking buffer (PBST containing 1% BSA) for 20 min and incubated with anti-BrdU antibody (BD Bioscience: 347580, Abcam: ab6326) in blocking buffer at 37 °C for 1 h. After washing, secondary antibodies were diluted in PBS containing 1% BSA and incubated with cell at room temperature for 1 h. After incubation, the slides were washed once with low-salt TBST (36 mM Tris-HCl pH8.0, 0.5 M NaCl, 0.5% Tween-20) and 3 times with PBST. After washing, cells were mounted with anti-fade solution and visualized using a Nikon eclipse 80i Fluorescence microscope. All fiber lengths were measured using Image J.
2.10. Flow cytometry analysis
For cell cycle or sub G1 analysis, USP13 proficient or deficient cells were treated with 2 mM HU. After 24 h, cells were washed, replaced with fresh media, and harvested by trypsinization at the indicated time. Cells were fixed in 70% ice-cold ethanol, treated with RNase A, stained with propidium iodide and analyzed on an Attune Nxt Flow cytometer (Thermo Fisher). For staining, USP13 proficient or deficient cells were fixed with 70% ice-cold ethanol then permeabilized with 0.25% Triton in PBS. Cells were incubated with indicated primary antibody for 3 h at RT, followed by secondary antibody for 1 h at RT, with PBS wash for two times in between. Cells were treated with RNase A, stained with propidium iodide and analyzed on an Attune Nxt Flow cytometer (Thermo Fisher), and data was analyzed with Flow Jo.
2.11. In vitro deubiquitination assay
For the preparation of ubiquitinated TopBP1 as the substrate for the in vitro deubiquitination assay, HEK293T cells were transfected with the FLAG-TopBP1. Proteins were purified from the cell extracts with FLAG conjugated beads in lysis buffer included 20 mM NEM and 1 mM iodoacetamide and then FLAG-TopBP1 is eluted in elution buffer (50 mM Tris-Hcl buffer pH 8.0 with 3x FLAG peptide). The recombinant GST-USP13 WT and GST-USP13 CA were expressed in BL21 cells and purified following standard protocol. For the deubiquitination assay in vitro, ubiquitinated proteins were incubated with recombinant USP13 in a deubiquitination buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol) for overnight at room temperature, and immunoblotting with the indicated antibodies.
2.12. Survival analysis
Survival analysis was performed on Ovarian cancer (n=655) and Gastric cancer (n=631) datasets from KMplotter (https://kmplot.com/analysis/). Overall survival was analyzed against expression of USP13 (probes 226902_at and 227788_at), split at median. Survival hazard was estimated by Kaplan-Meier method and statistical significance was tested by Gehan-Breslow-Wilcoxin test. The data was downloaded, replotted and statistically analyzed by Graphpad Prism 8.0.
2.13. Genetic variation of USP13 in cancer patients
Genetic variation of USP13 from TCGA cohorts were analyzed from cBioPortal (www.cBioPortal.org) [26, 27]. This portal collected NGS data from The Cancer Genome Atlas (TCGA) PanCancer Atlas Studies (32 studies, 10967 samples).
2.14. Statistics and reproducibility
Data in bar and line graphs are presented as mean ± SD of at least three independent experiments. All western blot assays shown here were successfully repeated at least three times.
3. Results
3.1. USP13 regulates TopBP1 expression level via proteasome dependent degradation
TopBP1 is an important protein for activating ATR following replication stress. To study TopBP1 regulation, we screened a library of USP superfamily members to identify TopBP1 deubiquitinating enzymes. As shown in Fig. 1A and Fig. S1A, we found that overexpression of USP13 significantly increased TopBP1 protein levels. To confirm the role of USP13 in regulating levels of TopBP1 protein, we assessed TopBP1 in cells overexpressing USP13 alone. As shown in Fig. 1B and Fig. S1B, USP13 overexpression increased TopBP1 protein level but not TopBP1 mRNA level. The levels of other key factors in the replication stress pathway such as ATR and Rad17 did not change. Furthermore, we confirmed that TopBP1 was reduced in USP13 deficient cells (Fig. 1C). Consistent with these findings, cells treated with the USP13 inhibitor spautin-1 downregulated TopBP1 expression (Fig. 1D). These results indicate that USP13 positively regulates TopBP1 protein levels. Importantly, TopBP1 levels could be restored in USP13 knockout (KO) cells by treatment with the proteasome inhibitor MG132 but not with the lysosomotropic agent chloroquine (CQ) (Fig. 1E). To examine whether USP13 regulates TopBP1 stability, we repeated our experiments in the presence of the protein synthesis inhibitor cycloheximide and found that TopBP1 protein stability was significantly decreased in both USP13 depleted cells and spautin-1 treated cells compared to control cells (Fig. 1F–G). Taken together, these results suggest that USP13 regulates TopBP1 stability in a proteasome-dependent manner and that USP13 is a potential DUB for TopBP1.
Fig. 1. USP13 regulates TopBP1 expression level via proteasome degradation.
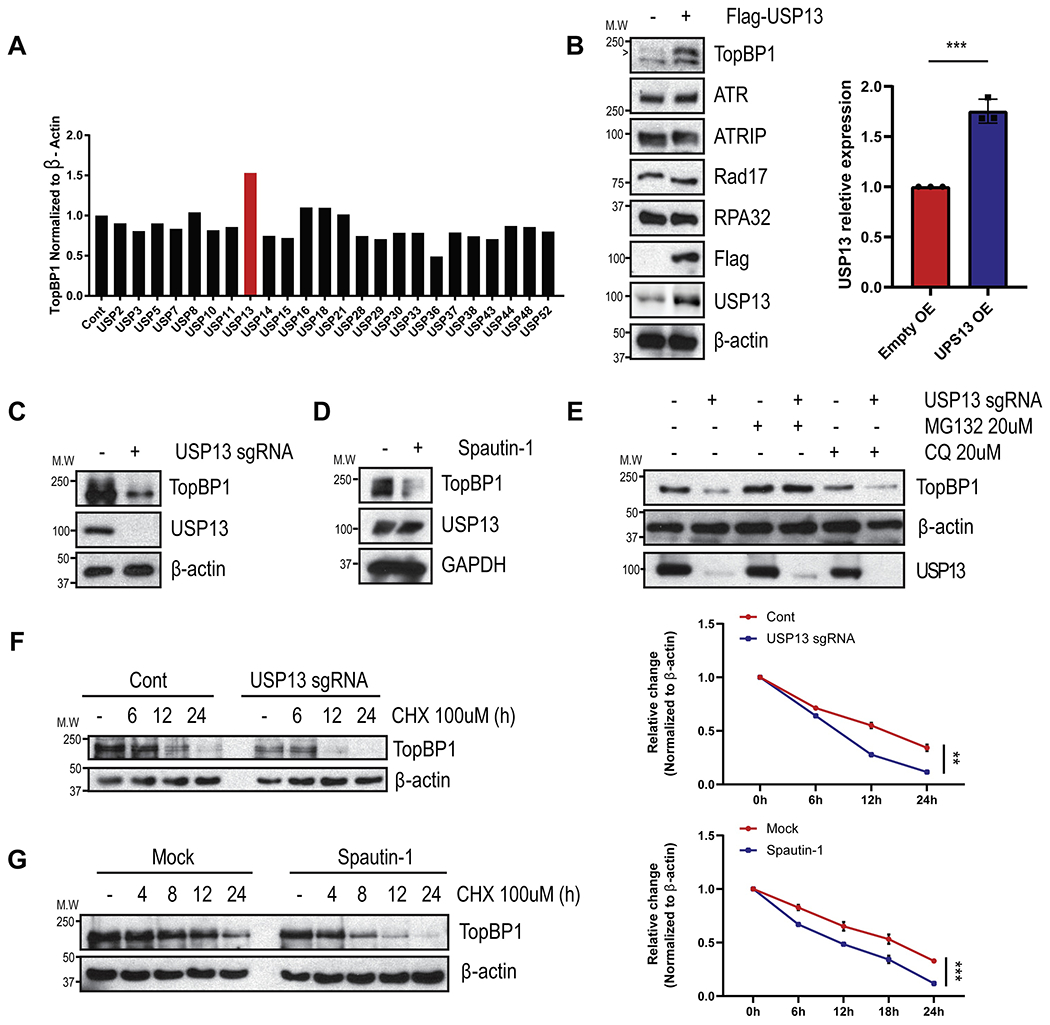
A. 293T cells were transfected with USP family cDNAs (Addgene) and screened for TopBP1 protein levels. B-D. TopBP1 protein level were evaluated by immunoblotting upon USP13-overexpressed (B), knockout in 293T cells (C), or spautin-1 (1 μM, 24 h) treatment in SK-OV-3 cells (D). E. Cells were treated with MG132 (20 uM) or CQ (20 uM) for 24 h in 293T cells. Each sample was immunoblotted with indicated antibodies. F-G. Cells were treated with cyclohexamide and TopBP1 protein levels were examined by immunoblotting in USP13 deficient 293T cells (F) or spautin-1 (1 μM, 24 h) treated SK-OV-3 cells (G). Each sample was immunoblotted with indicated antibodies. TopBP1 protein levels were measured by image J and visualized. The graphs represent mean ± S.D., two-tailed, unpaired t-test. n = 3-4 independent experiments.
3.2. USP13 deubiquitinates TopBP1
To test whether USP13 is a deubiquitinase for TopBP1, we first examined whether USP13 interacts with TopBP1. We found that endogenous TopBP1 co-immunoprecipitated with FLAG-USP13 (Fig. 2A). Because USP13 is a deubiquitinase, we checked whether USP13 affected ubiquitination of TopBP1. As shown in Fig. 2B, overexpression of wildtype (WT) USP13 (USP13-WT) decreased TopBP1 ubiquitination. Overexpression of a catalytically inactive (C345A) mutant (CA) was unable to reduce TopBP1 ubiquitination (Fig. 2B). USP13 deficiency dramatically elevated the ubiquitination level of TopBP1 (Fig. 2C). Furthermore, TopBP1 deubiquitination could be rescued in USP13 knockout cells by restoring with USP13 WT but not with the CA mutant (Fig. 2D). To further confirm this, we performed in vitro deubiquitinase assays and we found USP13 WT deubiquitinated TopBP1. The USP13 CA mutant failed to deubiquitinate TopBP1 (Fig. 2E). Collectively, these results suggest that USP13 regulates TopBP1 protein level through its deubiquitinase activity.
Fig. 2. USP13 deubiquitinates TopBP1.
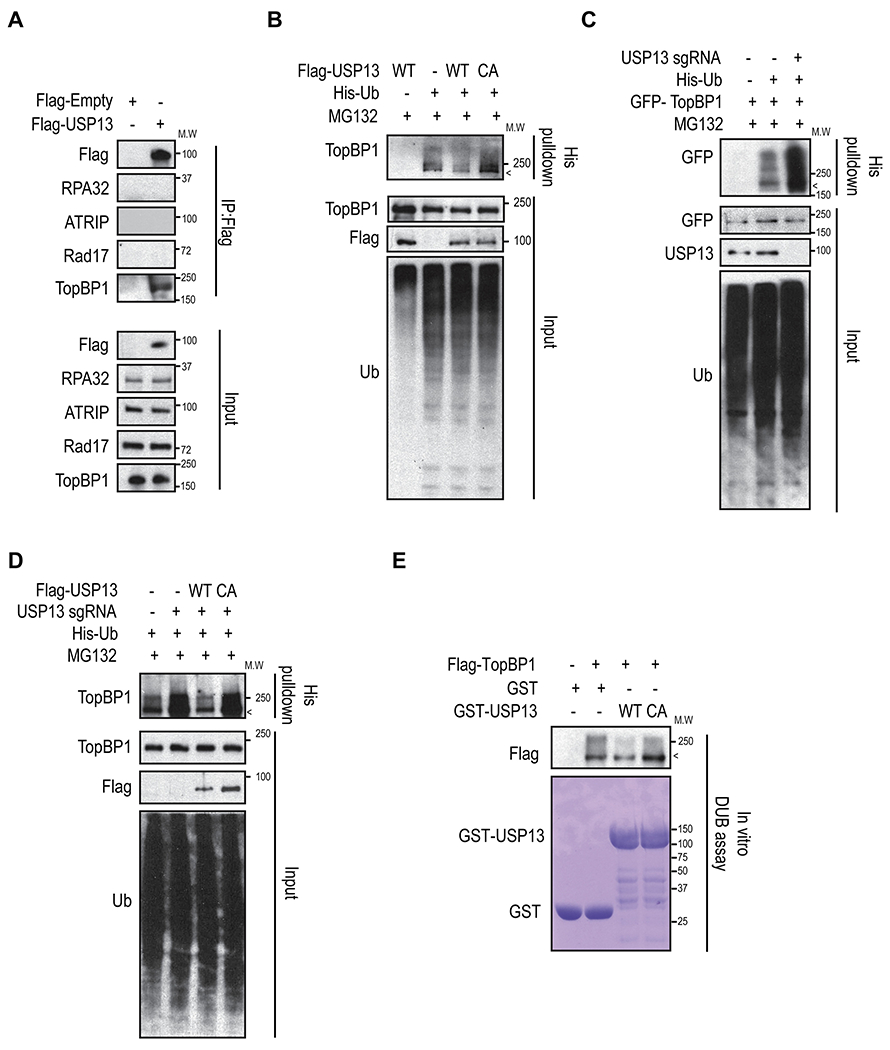
A. Whole cell lysates from FLAG-USP13 overexpressed 293T cells were used for immunoprecipitation. B. 293T cells were transfected with USP13 WT or CA mutant and His-Ub for 48h then ubiquitination assay were performed. C. Cells were transfected with GFP-TopBP1 and His-Ub in USP13 proficient or deficient 293T cells for 48h, and ubiquitination assay were performed. D. Ubiquitination assays were performed with USP13 WT, and CA mutant reconstituted USP13 knockout 293T cells. Each sample was immunoblotted with indicated antibodies. E. Ubiquitinated FLAG-TopBP1 was incubated with purified GST-USP13 WT or USP13 CA and then blotted with the indicated antibodies. Non-ubiquitinated TopBP1 proteins were indicated by <.
3.3. USP13 regulates the ATR signaling pathway via TopBP1
Since our results showed that USP13 regulates TopBP1 protein levels by deubiquitination, we hypothesized that USP13 is involved in ATR axis signal transduction under stressed conditions. To test this hypothesis, we checked RPA32 phosphorylation at serine 33 (p-RPA32 (S33)), which is a marker for ATR activation. As shown in Fig. 3A–B and Fig. S2, depletion of USP13 diminished p-RPA32 (S33) staining after hydroxyurea (HU) treatment. p-RPA32 (S33) foci were restored by reconstitution with USP13 WT but not with USP13 CA mutant. Furthermore, we found that Chk1 phosphorylation on Serine 345 was decreased in USP13 deficient cells or spautin-1 treated cells (Fig. 3C–D). Next, we wondered whether USP13 deficiency impedes activation of ATR signaling via TopBP1. To test this, we ectopically expressed TopBP1 in USP13 knockout cells and checked p-RPA32 (S33). As shown in Fig. 3E, reconstituting TopBP1 restores phosphorylation of RPA32 (S33). Taken together, these results suggest that USP13 is important for proper activation of the ATR pathway and the replication stress response.
Fig. 3. USP13 is involved in the ATR signaling pathway.
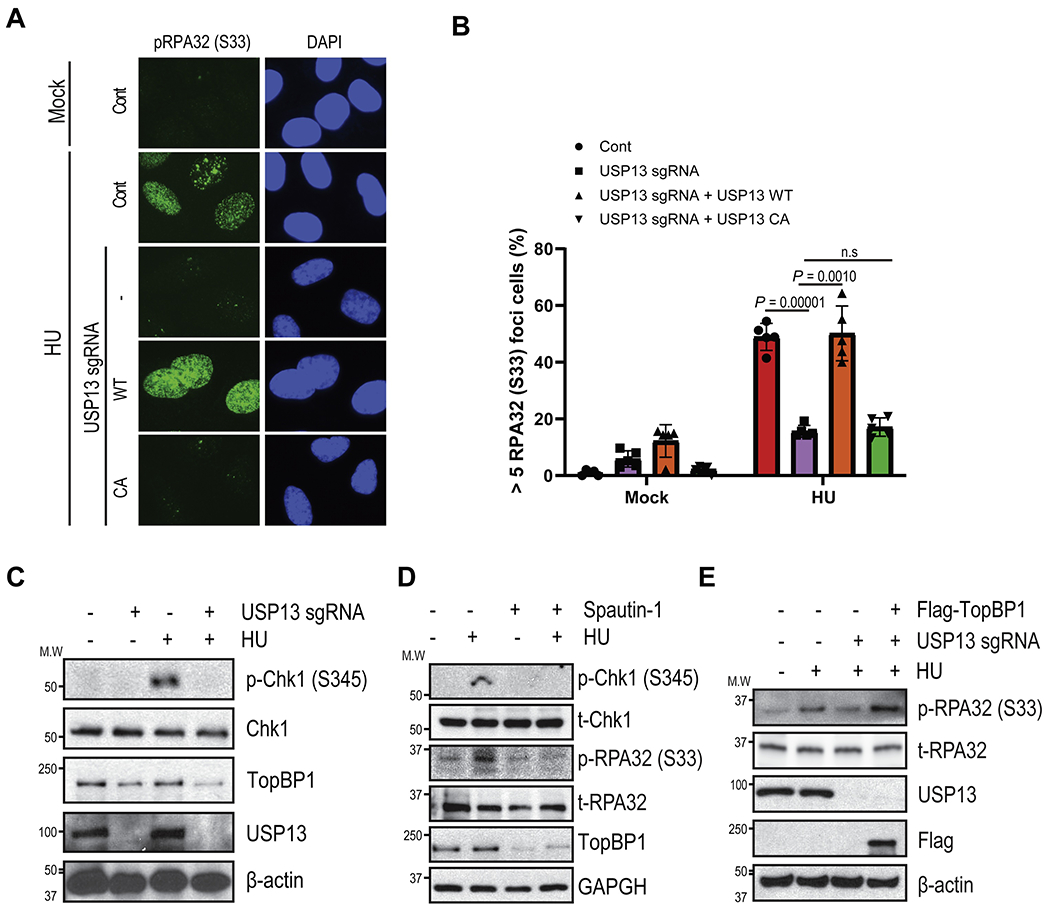
A-B. phospho-RPA32 (S33) was determined after 1h HU treatment by immunofluorescence in USP13 proficient or deficient U2OS cells. C-D. Phospho- Chk1 and RPA32 levels were determined in USP13 proficient, deficient (C) or spautin-1 treated SK-OV-3 cells (D) by immunoblotting. E. Cells were reconstituted with TopBP1 in USP13 knockout cells then Phospho- Chk1 and RPA32 levels were determined. Each sample was immunoblotted with indicated antibodies. The graphs represent mean ± S.D., two-tailed, unpaired t-test. n = 3 independent experiments. Foci numbers were counted by Image-Xpress confocal High-content imaging system (Molecular device).
3.4. USP13 is important for the replication checkpoint activation
The TopBP1-ATR pathway plays an important role in DNA-damage checkpoint activation and recovery from stalled replication forks. Since Chk1 phosphorylation is abrogated by USP13 deficiency, we tested whether USP13 is involved in cell cycle checkpoint activation following replication stress. Cell cycle profiles showed that USP13 deficient U2OS cells had a slightly increased G1-phase population, while USP13 inhibitor treated SK-OV3 ovarian cancer cell lines showed a slight increase in the S-phase population under unstressed conditions. Upon HU treatment, control cells were arrested in S phase, while USP13 deficient or USP13 inhibitor treated cells were not affected (Fig. 4A–B, Fig. S3A–B). This result indicates that USP13 deficiency or inhibition prevents cell cycle arrest in S phase in response to replication stress. We then assessed checkpoint activation using IdU/CIdU incorporation assay. As shown in Fig. 4C–F, the IdU positive cell population was similar between USP13 proficient and USP13 deficient cells (Fig. 4D). However, depletion of USP13 increased the CIdU uptake cell population under stressed condition (Fig. 4E–F). These data strongly suggest that USP13 deficiency prevents activation of the cell cycle checkpoint following replication stress. Furthermore, we examined DNA synthesis using DNA fiber assays. USP13 deficient cells showed a slight increase in their synthesized DNA fiber lengths compared to USP13 proficient cells under unstressed conditions (Fig. 4G, Fig. S3C, D). USP13 deficient cells also exhibited significantly increased replication strand lengths under replication stress (Fig. 4H, Fig. S3C, E). This is consistent with defective checkpoint activation [28]. The increased replicated strand lengths were reversed by reconstitution with USP13 WT, but not with the USP13 CA mutant (Fig. 4G–H). Taken together, our results suggest that USP13 is important for cell cycle checkpoint activation under replication stress by regulating the TopBP1-ATR signaling pathway.
Fig. 4. USP13 is important for DNA-damage checkpoint activation.
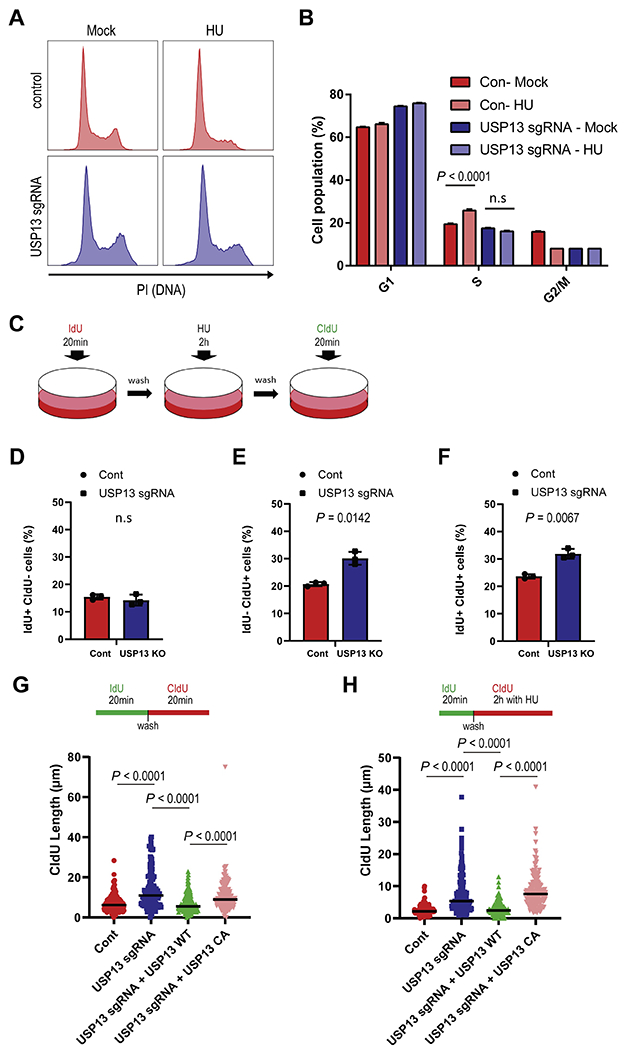
A-B. Cell cycle distribution was determined in USP13 proficient or deficient U2OS cells by flow cytometry (A). Population fractions were analyzed by Modfit (B). C-F. Schematic representation of IdU/CIdU incorporation assay (C). Labeled cells were counted by flow cytometry IdU+ CIdU− (D), IdU− CIdU+ (E), and IdU+ CIdU+ (F) in USP13 proficient or deficient U2OS cells. G-H. Cells were labeled with IdU and CldU for indicated time points. Fork speeds (G), and DNA synthesis (H) were determined by measuring DNA fiber length in USP13 proficient or deficient U2OS cells. All fiber lengths were measured using Image J. The graphs represent mean ± S.D., two-tailed, unpaired t-test. n = 3 independent experiments.
3.5. USP13 maintains genomic stability
Since our data show that USP13 deficient cells are defective in the replication stress response, we analyzed spontaneous and induced DNA damage in USP13 proficient or deficient cells with or without replication stress. As shown in Fig. 5A, the DNA-damage marker γH2AX signals increased in USP13 deficient cells under both unstressed and stressed conditions. To further investigate this, we tested whether USP13 deficiency sensitizes cells to replication stress inducing agents. Loss or inhibition of USP13 caused hypersensitivity to HU, camptothecin (CPT), ultraviolet (UV), and Fluorouracil (5-FU) treatment (Fig. 5B–I). On the other hand, overexpression of USP13 WT, but not the CA mutant, increased resistance to replication stress inducing agents (Fig. S4A–D). We next explored the potential role of USP13 in cancer development and prognosis. First, we analyzed the type and frequency of genetic variation in USP13 reported in the TCGA database (http://www.cBioPortal.org). This analysis included data from 10967 samples of 32 different tumor types. We found that the USP13 was frequently amplified, overexpressed and mutated in several cancer tissues. Importantly, increased USP13 copy number is positively correlated with TopBP1 protein levels (Fig. S4E–F), consistent with our conclusion. Next, we performed Kaplan–Meier survival analysis on several cancer types using median USP13 expression level. The survival of patients with high expression levels of USP13 was significantly shorter compared to patients with low USP13 expression levels in ovarian (Fig. 5J–K) and gastric cancer (Fig. S4G–H). Taken together, our results indicate that USP13 plays an important role in the replication stress response and may be associated with clinical outcome of patients with cancer.
Fig. 5. USP13 maintains genomic stability.
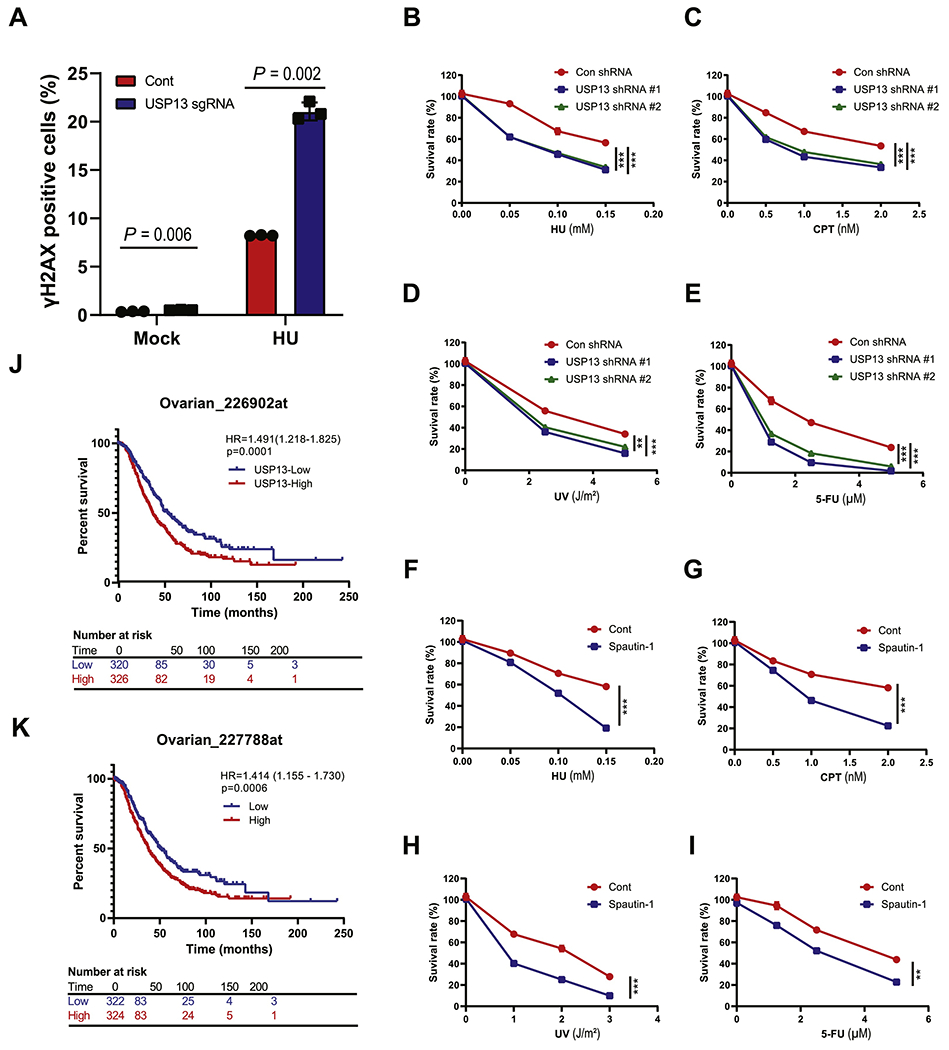
A. U2OS cells were treated with or without HU (2 mM, 24 h), and then genomic stability was examined by measuring γH2AX. The γH2AX positive population was analyzed by Flow Jo. B-E. USP13 proficient or deficient SK-OV-3 cells were plated and treated with hydroxyurea (HU, mM) (B), camptothecin (CPT, nM) (C), ultraviolet (UV, J m−2) (D), and fluorouracil (5-FU, μM) (E). F-1. SK–OV-3 cells were plated with or without spautin-1 (0.5 μM) and treated with hydroxyurea (HU, mM) (F), camptothecin (CPT, nM) (G), ultraviolet (UV, J/m−2) (H), and fluorouracil (5-FU, μM) (I). After 14 days, colony numbers were counted. J-K. Survival analysis produced from ovarian cancer specimens by KM-plot. The graphs represent mean ± S.D., two-tailed, unpaired t-test. n = 3 independent experiments.
4. Discussion
Here, we show that USP13 is a novel deubiquitinating enzyme for TopBP1 and promotes ATR activation under replication related stress. This work identifies a mechanism of TopBP1 regulation and sheds light on the regulation of replication stress-related DNA-damage responses TopBP1 was previously found to be ubiquitinated by the HECT-domain ubiquitin ligase hHyd (12). However, how TopBP1 deubiquitination is regulated was not elucidated clearly before this work.
Our study shows that USP13 interacts with TopBP1. USP13 then stabilizes TopBP1 by deubiquitination (Fig. 2). Depletion of USP13 hypersensitizes cells to replication stress inducing agents, prevents activation of the ATR pathway and the S phase checkpoint, and leads to the accumulation of spontaneous DNA damage (Fig. 4–5). We found that USP13 is frequently amplified and mutated in several types of human cancers (Fig. S4E–F). Furthermore, high USP13 expression is associated with poor survival (Fig. 5J–K, Fig. S4G–H). Collectively, our results demonstrate that USP13 is a novel deubiquinitating enzyme for TopBP1 and our study provides evidence for a relationship between USP13 and the replication stress response (Fig. 6).
Fig. 6. A model for TopB1 stability regulated by USP13.
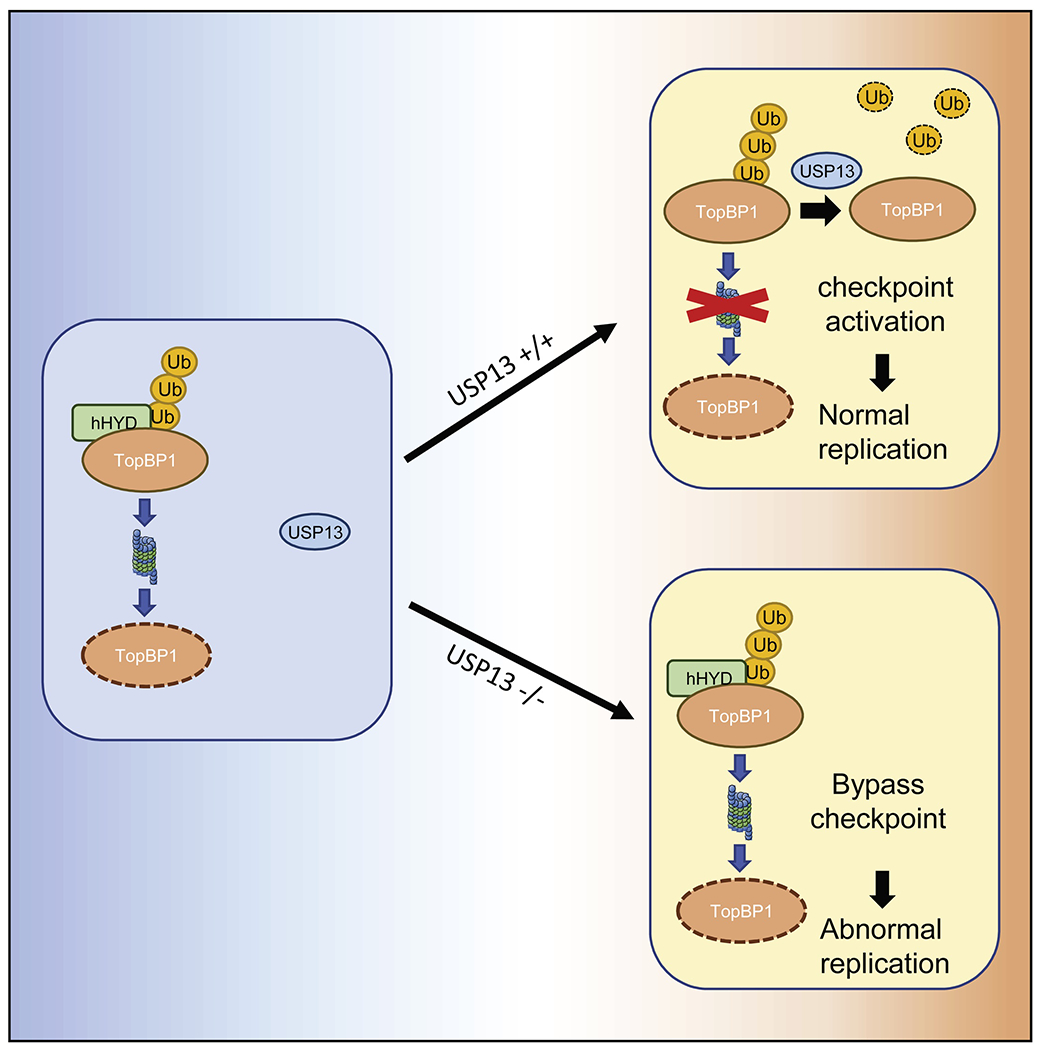
USP13 deubiquitinates TopBP1. The deubiquitinated TopBP1 is stabilized and then activates ATR- TopBP1 axis pathway.
While TopBP1 is amplified in multiple cancers and plays a role in chemotherapy resistance, it has been challenging to develop an inhibitor targeting TopBP1 because it lacks enzymatic activity. Here we show that USP13 is a deubiquitinase that stabilizes TopBP1 and represents a potential target for cancer therapeutics. Furthermore, we show that the USP13 inhibitor spautin-1 decreases survival from replication stress inducing drugs in ovarian cancer cell lines. As mentioned above, DUBs play important roles in many different biological processes including cell cycle control, DNA-damage responses, and transcription factor regulation. Development of highly specific USP13 inhibitors may benefit cancer patients receiving chemotherapy; and, the combination of replication stress inducing therapy combined with USP13 inhibition may be a new candidate therapeutic approach.
Supplementary Material
Hightlight.
USP13 is a novel deubiquinitating enzyme for TopBP1
TopBP1 deubiquitination and stabilization is regulated by USP13 in DNA replication stress responses
The mechanism of TopBP1 regulation and sheds light on the regulation of replication stress-related DNA-damage responses
Acknowledgements
We thank to Wojciech Niedzwiedz for GFP-TopBP1 plasmids.
Funding
This work was supported by the National Institutes of Health [RO1CA203971 to Z.L., S.H.K]; and the Mayo Foundation.
Abbreviations
- DUB
Deubiquitinating enzyme
- ssDNA
Single stranded DNA
- 9-1-1
Rad9-Hus1-Rad1
- PTM
Post-translational-modification
- USP13
Ubiquitin-specific-protein 13
- ERAD
Endoplasmic reticulum-associated degradation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors have declared that no competing interest exists.
References
- [1].Anindya R, Single-stranded DNA damage: Protecting the single-stranded DNA from chemical attack, DNA Repair (Amst), 87 (2020) 102804. [DOI] [PubMed] [Google Scholar]
- [2].Fedeles BI, Essigmann JM, Impact of DNA lesion repair, replication and formation on the mutational spectra of environmental carcinogens: Aflatoxin B1 as a case study, DNA Repair (Amst), 71 (2018) 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lans H, Hoeijmakers JHJ, Vermeulen W, Marteijn JA, The DNA damage response to transcription stress, Nat Rev Mol Cell Biol, 20 (2019) 766–784. [DOI] [PubMed] [Google Scholar]
- [4].Wells JP, White J, Stirling PC, R Loops and Their Composite Cancer Connections, Trends Cancer, 5 (2019) 619–631. [DOI] [PubMed] [Google Scholar]
- [5].Saldivar JC, Cortez D, Cimprich KA, The essential kinase ATR: ensuring faithful duplication of a challenging genome, Nat Rev Mol Cell Biol, 18 (2017) 622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cortez D, Preventing replication fork collapse to maintain genome integrity, DNA Repair (Amst), 32 (2015) 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zeman MK, Cimprich KA, Causes and consequences of replication stress, Nat Cell Biol, 16 (2014) 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fanning E, Klimovich V, Nager AR, A dynamic model for replication protein A (RPA) function in DNA processing pathways, Nucleic Acids Res, 34 (2006) 4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zou L, Elledge SJ, Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes, Science, 300 (2003) 1542–1548. [DOI] [PubMed] [Google Scholar]
- [10].Liu T, Lin YH, Leng W, Jung SY, Zhang H, Deng M, Evans D, Li Y, Luo K, Qin B, Qin J, Yuan J, Lou Z, A divergent role of the SIRT1-TopBP1 axis in regulating metabolic checkpoint and DNA damage checkpoint, Mol Cell, 56 (2014) 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bigot N, Day M, Baldock RA, Watts FZ, Oliver AW, Pearl LH, Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint, Elife, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Honda Y, Tojo M, Matsuzaki K, Anan T, Matsumoto M, Ando M, Saya H, Nakao M, Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response, J Biol Chem, 277 (2002) 3599–3605. [DOI] [PubMed] [Google Scholar]
- [13].Xie W, Jin S, Cui J, The NEDD4-USP13 axis facilitates autophagy via deubiquitinating PIK3C3, Autophagy, (2020) 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Scortegagna M, Subtil T, Qi J, Kim H, Zhao W, Gu W, Kluger H, Ronai ZA, USP13 enzyme regulates Siah2 ligase stability and activity via noncatalytic ubiquitin-binding domains, J Biol Chem, 286 (2011) 27333–27341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu Y, Soetandyo N, Lee JG, Liu L, Xu Y, Clemons WM Jr., Ye Y, USP13 antagonizes gp78 to maintain functionality of a chaperone in ER-associated degradation, Elife, 3 (2014)e01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sun H, Zhang Q, Jing YY, Zhang M, Wang HY, Cai Z, Liuyu T, Zhang ZD, Xiong TC, Wu Y, Zhu QY, Yao J, Shu HB, Lin D, Zhong B, USP13 negatively regulates antiviral responses by deubiquitinating STING, Nat Commun, 8 (2017) 15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li Y, Luo K, Yin Y, Wu C, Deng M, Li L, Chen Y, Nowsheen S, Lou Z, Yuan J, USP13 regulates the RAP80-BRCA1 complex dependent DNA damage response, Nat Commun, 8(2017)15752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Li Y, Hao Y, Choi A, Ke H, Ma D, Yuan J, Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13, Cell, 147 (2011) 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Han C, Yang L, Choi HH, Baddour J, Achreja A, Liu Y, Li Y, Li J, Wan G, Huang C, Ji G, Zhang X, Nagrath D, Lu X, Amplification of USP13 drives ovarian cancer metabolism, Nat Commun, 7 (2016) 13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhao X, Fiske B, Kawakami A, Li J, Fisher DE, Regulation of MITF stability by the USP13 deubiquitinase, Nat Commun, 2 (2011) 414. [DOI] [PubMed] [Google Scholar]
- [21].Fang X, Zhou W, Wu Q, Huang Z, Shi Y, Yang K, Chen C, Xie Q, Mack SC, Wang X, Carcaboso AM, Sloan AE, Ouyang G, McLendon RE, Bian XW, Rich JN, Bao S, Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination, J Exp Med, 214 (2017) 245–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Qu Z, Zhang R, Su M, Liu W, USP13 serves as a tumor suppressor via the PTEN/AKT pathway in oral squamous cell carcinoma, Cancer Manag Res, 11 (2019) 9175–9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xiang S, Fang J, Wang S, Deng B, Zhu L, MicroRNA135b regulates the stability of PTEN and promotes glycolysis by targeting USP13 in human colorectal cancers, Oncol Rep, 33 (2015) 1342–1348. [DOI] [PubMed] [Google Scholar]
- [24].Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, Wang M, Chen D, Sun Y, Hung MC, Chen J, Ma L, Deubiquitylation and stabilization of PTEN by USP13, Nat Cell Biol, 15 (2013) 1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yeh HM, Yu CY, Yang HC, Ko SH, Liao CL, Lin YL, Ubiquitin-specific protease 13 regulates IFN signaling by stabilizing STAT1, J Immunol, 191 (2013) 3328–3336. [DOI] [PubMed] [Google Scholar]
- [26].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, Cancer Discov, 2 (2012) 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal, Sci Signal, 6 (2013) pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y, The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses, Mol Cell Biol, 27 (2007) 5806–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


