Abstract
Citizen science (CS) currently refers to the participation of non-scientist volunteers in any discipline of conventional scientific research. Over the last two decades, nature-based CS has flourished due to innovative technology, novel devices, and widespread digital platforms used to collect and classify species occurrence data. For scientists, CS offers a low-cost approach of collecting species occurrence information at large spatial scales that otherwise would be prohibitively expensive. We examined the trends and gaps linked to the use of CS as a source of data for species distribution models (SDMs), in order to propose guidelines and highlight solutions. We conducted a quantitative literature review of 207 peer-reviewed articles to measure how the representation of different taxa, regions, and data types have changed in SDM publications since the 2010s. Our review shows that the number of papers using CS for SDMs has increased at approximately double the rate of the overall number of SDM papers. However, disparities in taxonomic and geographic coverage remain in studies using CS. Western Europe and North America were the regions with the most coverage (73%). Papers on birds (49%) and mammals (19.3%) outnumbered other taxa. Among invertebrates, flying insects including Lepidoptera, Odonata and Hymenoptera received the most attention. Discrepancies between research interest and availability of data were as especially important for amphibians, reptiles and fishes. Compared to studies on animal taxa, papers on plants using CS data remain rare. Although the aims and scope of papers are diverse, species conservation remained the central theme of SDM using CS data. We present examples of the use of CS and highlight recommendations to motivate further research, such as combining multiple data sources and promoting local and traditional knowledge. We hope our findings will strengthen citizen-researchers partnerships to better inform SDMs, especially for less-studied taxa and regions. Researchers stand to benefit from the large quantity of data available from CS sources to improve global predictions of species distributions.
Introduction
Species distribution models have become a widely used tool in ecology and have tackled diverse scientific issues at different spatial and temporal scales in recent years [1–3]. Understanding the association between the occurrence of species and environmental conditions is a first step in addressing questions about species distributions, abundances and habitat preferences [4, 5]. In fact, knowledge on species distributions is paramount in order to develop biodiversity conservation and management strategies [6]. Current global-scale issues such as climate and land-use changes have also increased the need to be able to predict the distribution of migratory or invasive species across a landscape. The fundamental theory behind species distribution models (SDMs, hereafter) assumes that the presence of a species in a given location depends on the environment, which implies that ecologists are able to estimate past, current, or future species distributions based on the environmental characteristics of unsurveyed locations [3, 5, 7, 8]. Specifically, SDMs link information about the presence of a species to the environmental variables of their known locations, and apply statistical models to predict the spatial distribution of species [4, 5, 9]. Consequently, three major components can be identified in any framework for SDMs: species presence data, landscape or environmental data, and a statistical model that links the first two components [4, 5, 10]. Species distribution models are widely used in both fundamental science and applied sciences in biogeography, evolution, dispersal, migration, species invasion, meta-population, conservation, and climate change [3]. For example, SDMs have shown their value to assess species invasions [11], to predict spatial changes in response to climate change or land-use changes [12–14], or to assess the suitability of possible conservation areas [15–17].
Modelling the distribution of species usually requires a large amount of information collected over multiple years of standardized fieldwork [18–20]. However, the long-term collection of broad-scale information on a wide range of species is prohibitively expensive [21]. Yet, for some taxa, an impressive volume of data collected using mostly non-standardized protocols is currently available on online portals, through efforts collectively labelled as citizen science (CS, hereafter). Globally, a huge variety of CS programs are currently being implemented involving a wide range of taxa [22]. Nevertheless, CS data are still challenging to analyze due to the intrinsic issues of non-standardized protocols that can affect the credibility and quality of the data [23, 24].
Issues within CS datasets arise from the large number of observations that vary in quality when used as a source for research [25, 26]. Previous studies have tackled the different sources of error and bias in CS data [27–29]. Firstly, CS datasets are typically biased towards human population centers, areas that are easy to access, protected areas, or regions frequented by active observers [30–32]. These problems lead to disparities in effort between over-sampled and under-sampled areas [19, 29, 30, 33–35]. Secondly, geographical coverage of CS data can be biased towards well-financed and more industrialized countries, mainly in North America and Europe [25, 32, 36]. These two regions contribute substantially more data than any other region in the Global Biodiversity Information Facility (GBIF) database [37–41]. Consequently, a large proportion of samples occur in a restricted geographical extent, controlled by administrative borders. This results in a non-representative sample of species’ distribution. Thirdly, over time, the observation and reporting protocols can change. For example, the Audubon Christmas Bird Count at its start in 1900 aimed at offering an alternative to hunting on Christmas Day morning, with a loose survey protocol. The date for conducting the count became flexible over the years. For example, it was conducted during a window of 7 days in 1940 [42]. The sampling window then expanded to 12 days in 1966 [43] and the protocol was further standardized to collect a snapshot of wintering birds around Christmas time [44]. The survey period expanded again in 2000, this time to 23 days [45]. Unfortunately, changes in survey protocols were often poorly documented [46, 47]. Fourthly, CS observations can be taxonomically biased because volunteers are usually attracted to large and common species, to species that are brightly colored and easy to detect, and to more charismatic groups [28, 40, 47, 48]. This taxonomical disparity results in more information on relatively well-known groups than for under-reported groups. Finally, another source of variation in CS programs includes the variation in skill and expertise among observers, primarily due to the participation of a wide range of volunteers [49, 50]. The quality of observations depends on the ability of observers to correctly detect and identify species. This inter-observer sampling variation increases for species that are harder to identify [49, 51, 52]. Bias and precision associated with each of these five sources of variation can influence predictions of future trends. A major challenge is to account for these issues in species distribution models [53, 54].
Despite these issues regarding data quality, the use of CS has increased in recent years in different fields of study [55, 56]. For instance, CS is used in astronomy to classify galaxy images or to search for signals in radio data, and in atmospheric sciences to record the quality of air, soil, and water [55, 57, 58]. However, the main application of CS is in conservation and ecology to monitor species occurrence [39, 59]. Past reviews have focused on how CS contributes to biodiversity monitoring [39], global change [60], and conservation biology [61]. However, considering the increasing prevalence of CS in ecological studies, it is critical to gain a better understanding of how this data source contributes to peer-reviewed research and to what extent CS can fill gaps in under-represented species and locations. Furthermore, the degree to which scientists already use CS data to build SDMs is not well documented. Understanding the contributions of different forms of CS that provide data for SDMs should help to better allocate research efforts in the future and outline specific strategies to increase the usefulness of CS.
The main objective of this review was to quantify the variation and gaps in the use of CS as an input for modelling species distribution. To achieve this objective, we assessed the current strengths in the use of CS in SDMs and identified partiality and under-use relative to taxa, regions, and data acquisition methods. We formulated three research questions: (1) What is the trend in the use of CS data for SDMs over the last decade? We expected the number of papers using CS data to have increased at a faster rate than the SDM field as a whole given the increasing contribution of citizen science in different field studies; (2) Is there variation across regions, taxa, and types of data used? We anticipated that because volunteers behave differently according to the region and group of interest, the set of papers would reflect clear preferences towards regions that are easy to access and groups that are visually appealing to volunteers. However, it is expected that these preferences will gradually fade due to the growing diversification of initiatives and platforms worldwide over the last decade; (3) What are the information gaps and how can research needs be met in the near future? We expected that new approaches of collecting data for sensitive species and under-sampled locations play an important role for filling research gaps.
Materials and methods
Paper selection
We used the Scopus search engine to conduct a literature review of peer-reviewed papers focusing on species distribution models that used citizen science. Our search spanned a period of 10 years, considering papers from 2010, when the “citizen science” term was widely accepted by several authors [58, 62, 63], until 17 October 2019. We searched for papers using the following combination of keywords: (“citizen science” OR “public participation” OR “community monitoring program” OR “participatory monitoring”) AND (“species distribution model” OR “predictive model” OR “distribution map” OR “invasive species” OR “occupancy model” OR “occurrence” OR “migration” OR “climate change”). The integration of citizen science data and advances in occupancy modelling allows researchers to build species distribution models that account for imperfect detection probability. Indeed, several authors encourage their use for a variety of species [59, 64, 65]. For our review, papers using occupancy models were included only when they were used for mapping species distribution.
Data collection
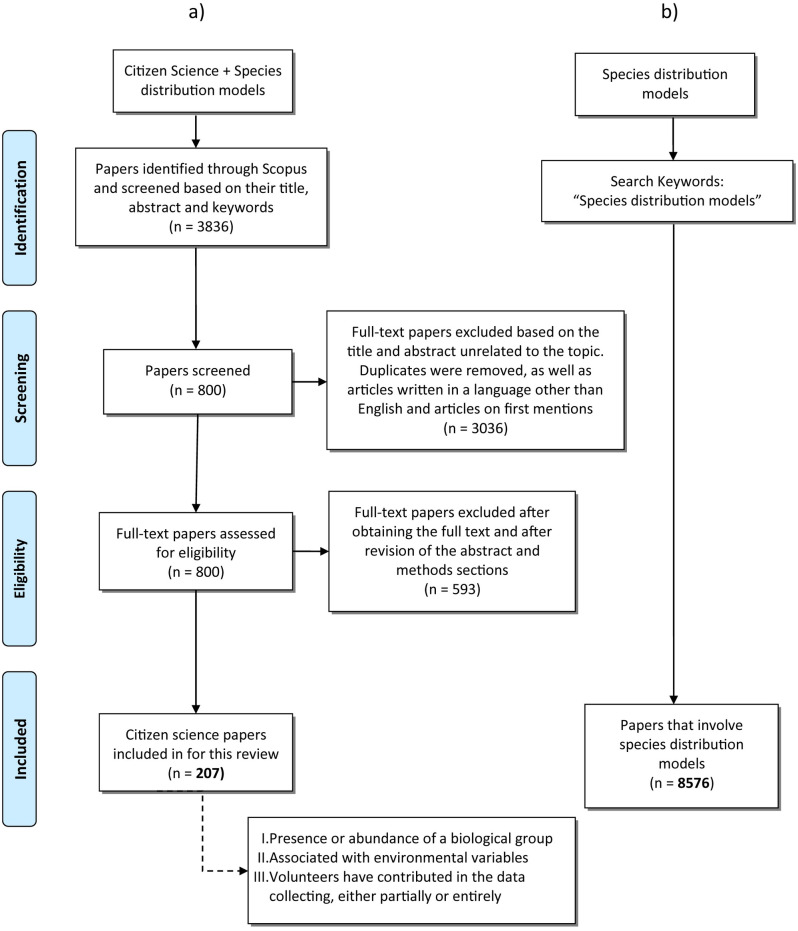
From the first Scopus search, a total of 3,836 papers were screened based on their title, abstract and keywords (Fig 1a). Those not related with either citizen science (CS) or species distribution models (SDMs) were dismissed. The remaining 800 papers were reduced after a further revision of abstracts and methodologies. We excluded papers written in languages other than English (n = 4), all review papers, and also papers using data gathered by volunteers but without applications of SDMs (e.g., first report of a species or new occurrence data). To consider a given paper as relevant for our review, each of the following three conditions had to be met: the data included the presence or abundance of a biological group, the data were collected by volunteers (either partially or entirely), and a statistical method was applied to assess relationships with environmental data (Fig 1a). Our search resulted in 207 papers from peer-reviewed journals. These papers formed the basis of the analyses presented herein. Some papers that used CS as a data source for SDM are not in the Scopus database and were certainly missed. Nevertheless, we consider that this database is a representative sample that covers most of the peer-reviewed journals of world science at present within the SDM field in the last decade. Details and extracted information about all papers included in our review are listed in (S1 Table). This list may not reflect the full influence of CS programs in SDMs, but only their contribution to published articles.
Fig 1. Flow chart of paper selection for a) the citizen science (CS) papers and b) for the entire species distributions models (SDMs) field.
All 207 papers in a) are listed in S1 Table. From: Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred Reporting Iterns for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med 6(7): e1000097. doi:10.1371/journal.pmed1000097 For more information, visit www.prisma-statement.org.
From each paper, the following information was extracted: (1) year of publication; (2) focal taxa; (3) source of data or platform used (if any); (4) country and region where the data was taken; (5) scope or central objective of the study—when appearing in the title, abstract, or keywords, including species conservation, land-use changes, biogeography, habitat suitability, population trends climate change, invasive species or migration; (6) data type used (presence-only, presence-absence, or abundance); (7) statistical approach used, and 8) the method of collecting CS data (opportunistic data, count data, community-based monitoring, historical records, local ecological knowledge, or trained volunteers). In order to assess the contribution of CS to SDMs over the last decade, we compiled papers within Scopus by using the keywords “species distribution models” to obtain the number of papers in this field (Fig 1b). This search engine seeks matches within the title, abstract, keywords, and indexed keywords and returned 8576 papers.
Data analyses
Contribution of citizen science to species distribution models
We tested for differences in the rate of increase of CS-SDM papers and the overall number of SDM papers using generalized linear models (GLM) with a Poisson distribution that included an interaction term between the year and type of paper (CS-SDM vs SDMs). We expected papers using CS data to have increased at a faster rate than the SDM field as a whole.
Taxonomic groups
In order to assess the representation of CS within the biological groups, each paper was categorized within the following taxonomic groups: invertebrates, plants and fungi (including bryophytes and lichens), fish, reptiles, amphibians, mammals, and birds. We used a chi-square test to first compare the number of CS papers with data on each group observed to the numbers expected based on the proportion of species in each group according to the Catalogue of Life [66] (accession date April 2020). A second chi-square test was used to compare the number of occurrences for each taxa contained in the GBIF dataset (accession date 23 November 2020). To compare the observed (CS) and expected proportions for each of the five taxonomic groups, we constructed a logistic regression model excluding the intercept to estimate the logit of the probability that a taxa t appears in a CS study:
| (1) |
We then calculated a Z-score from the difference between βt and the logit of Et, the expected proportion for that taxa, scaled by the standard error of βt:
| (2) |
We obtained a two-tailed p-value for the null hypothesis that the observed proportion was equal to the expected proportion by comparing Zt to the standard normal distribution. We excluded papers that focused on more than one taxonomic group to meet assumptions of statistical independence of observations. For invertebrates, only the taxonomic orders represented in our CS set of papers were analyzed (Lepidoptera, Odonata, Hymenoptera, Coleoptera and Mollusca). The remaining invertebrate groups were not represented or were in very low numbers in our review (e.g., spiders and hemiptera).
Geographic regions
Papers were individually classified into country and continent of origin of the CS data, including Africa, Asia, Eastern Europe, Western Europe, Oceania, North America, Central America, and South America. To assess if these regions were over or under-sampled in the CS papers set, we used a one-sample chi-square test to compare the number of CS papers in each region to the number expected based on the proportion of the Earth’s land area covered by each region (from http://www.worlddata.info; accessed 29.11.19). Then, we compared these observed and expected proportions for each of these geographic regions. Using the same strategy as above, we constructed a logistic regression model that excluded the intercept to estimate the logit of the probability that the regiony appears in a CS study:
| (3) |
We then calculated a Z-score from the difference between βy and the logit of Ey, the expected proportion for that region, scaled by the standard error of βy:
| (4) |
We compared the Zy against the standard normal distribution. Papers that focused on more than one region were excluded to meet assumptions of statistical independence of the observations. All statistical analyses were performed in R version 3.5.0 [67].
Results and discussion
Year of publication
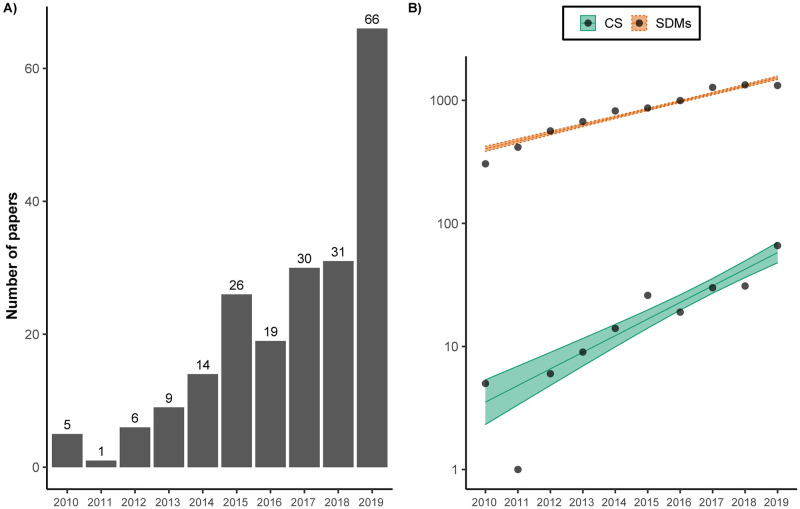
As we expected, our analysis indicates that the use of CS data in the peer-reviewed SDM literature has increased in frequency over the past 10 years (Fig 2a). Numerous authors have indicated the increase in publications using different types of CS data [55, 56, 60, 68, 69], but also the growing rate of SDMs in publications [70, 71]. Our analysis shows that the use of CS in SDMs has grown approximately twice as fast as the number of papers using SDMs in general (publication increases of 16% for SDMs and 36% for CS on average per year). In addition, given its peak in 2019 with 66 papers (Fig 2a), the next few years may extend the rapid growth of the use of CS in SDMs.
Fig 2.
(a) Annual number of papers that have used species distribution models (SDMs) with citizen science (CS) data; (b) generalized linear model with Poisson distribution of the total papers using SDMs (blue) and the papers using CS data (red) across the 10-year period covered by our review (difference in slopes: -0.16, Z = -5.4, P < 0.001), resulting in publication growth of 16% for SDMs and 36% for CS on average per year.
Taxonomic groups
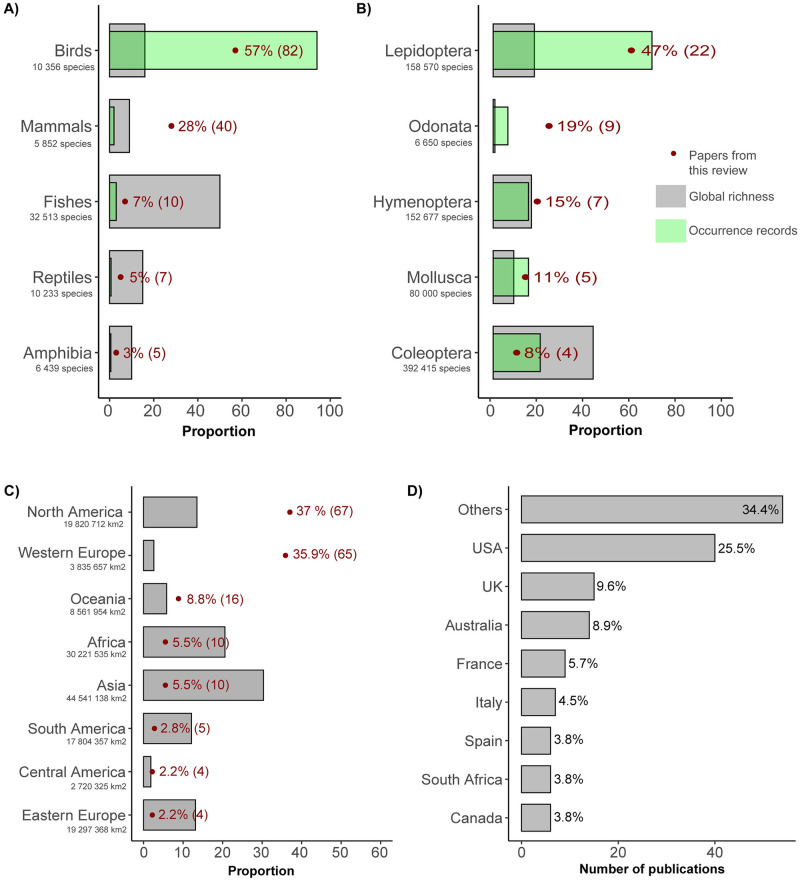
Following our predictions for the more visually appealing species, there were marked variations among taxonomic groups, with birds (n = 83; 41%), invertebrates (n = 47; 23,3%), and mammals (n = 39; 19.3%) being the main taxa studied. Reptiles (n = 12; 5.9%), fish (n = 11; 5.4%) and amphibians (n = 10; 4.9%) received less attention (Fig 3a). This taxonomic preference towards bird species was previously noted by other authors [39, 55, 60], and is reflected in occurrence records from GBIF (94%; Fig 3a).
Fig 3. Proportion of citizen science (CS) papers from this review (red point) relative to the proportion of global richness in the Catalogue of Life (grey bars) by taxa and number of global occurrence in GBIF (green bars), for (a) vertebrates and (b) invertebrates; (c) proportion of CS papers by data collection region (red dotted lines) relative to each region’s fraction of the Earth’s land area (grey bars); and (d) proportion of CS papers by country.
Compared to their global richness, vertebrate groups were unequally represented in the CS papers (χ2 = 262.77, df = 4, p < 0.001). Specifically, birds (Zbirds = 11.6; p < 0.001) and mammals (Zmammals = 6.8; p < 0.001) were over-represented, whereas amphibians (Zamphibians = -2.3; p = 0.02), reptiles (Zreptiles = -8.1; p < 0.001) and fish (Zfish = -7.7; p < 0.001) were under-represented in CS papers compared to their estimated global richness in the Catalogue of Life database (Fig 3). These differences were mostly driven by the interest of the volunteers collecting the data, which results in over-representation of charismatic taxa relative to groups generating less interest [72]. However, compared to the global occurrence records (number of occurrences per group instead of richness), mammals (Zmammals = 15.9; p < 0.001), reptiles (Zreptiles = 11.34; p < 0.001), amphibians (Zamphibians = 4.1; p < 0.001) and fish (Zfish = 2.7; p = 0.006) were over-represented in CS papers. In contrast, birds were largely under-represented compared to their occurrence records (Zbirds = -13.9; p < 0.001) since nearly all occurrence records were concentrated within this group (94% in Fig 3a). Such results suggest a great imbalance between research interests and availability of CS data, especially for amphibians, reptiles and fishes, which are over-represented in publications as compared to their occurrence data but under-represented compared to their estimated global species richness. Thus, there is a need to increase data acquisition on a wider range of species by volunteers on these specific groups containing several lesser-known species, with poorly defined distributions and conservation status. In the case of birds, reducing structured sampling efforts in areas already well-covered by volunteers may help redirect limited monitoring resources to sample less populated areas or to target more cryptic species. Filling such information gaps may also require local and traditional knowledge or community-based monitoring approaches, especially if research knowledge gaps also meet local concerns.
We found differences between the observed proportion of invertebrates in our CS data set and the proportion expected based on global richness (χ2 = 93.08, df = 4, p < 0.001). Lepidoptera (Zlepidoptera = 4.4; p < 0.001) and Odonata (Zodonata = 5.9; p < 0.001) were over-sampled relative to their global richness (Fig 3b). Only Coleoptera (Zcoleoptera = -4.1; p < 0.001) were under-sampled. The proportion of papers on Mollusca (Zmollusca = 0.49; p = 0.62) and Hymenoptera (Zhymenoptera = -0.29; p = 0.77) did not differ from the proportion expected from global species richness. The remaining invertebrate orders in the Catalogue of Life database did not occur or were in very low numbers in the set of CS papers studied. Considering occurrences records, proportions between our number of CS papers and the GBIF global dataset did not differ: Lepidoptera (Zlepidoptera = -0.5; p = 0.58), Mollusca (Zmollusca = 0.08; p = 0.93), Coleoptera (Zcoleoptera = -1.03; p = 0.3), Odonata (Zodonata = 1.9; p = 0.051) and Hymenoptera (Zhymenoptera = 1.01 p = 0.3). Such results suggest a better balance between research interests and availability of CS data for invertebrates than for vertebrates.
The plant and fungi group included papers involving vascular plants (n = 14), fungi (n = 3), lichens (n = 1), and bryophytes (n = 1; S1 Table). Considering the known number of species in each group according to the Catalogue of Life database (vascular plants: 348,000 species; fungus: 140,000 species; bryophytes 16,000 species), plant taxonomic groups were remarkably under-represented in CS papers (n = 13; 8% of all groups in this review). The major obstacle could be that identifying plants up to species level in the field is sometimes complex, even for expert botanists [73, 74]. Plant identification is time consuming for several families, requires significant botanical skills, and can be frustrating for non-expert volunteers [74]. In addition, there is not as strong a tradition for botanists in sharing observations using online portals, compared to animal databases. Nonetheless, plant initiatives seem to be highly attractive to the general public. Millions of observations are produced and stored in broad databases such as GBIF, iNaturalist, and in particular botanic platforms such as Pl@ntNet [75], Project Bud Burst [76], or Plant Watch Canada [77]. Several authors recommend using this information collected from volunteers for the early detection and control of invasive species [78, 79], or to improve the performance of models [79–81]. Nevertheless, our review confirms a notable under-use of plant, fungi, lichen and bryophyte public databases in the last decade in papers that model the distribution of species, as compared to the interest that they generate in CS programs.
Geographic coverage
Our review identified strong geographic biases in CS sampling efforts (χ2 = 1374.5, df = 7, p < 0.001). While Western Europe (ZWesternEurope = 20.5; p < 0.001) and North America (ZNorthAmerica = 7.7; p < 0.001) were over-sampled relative to their fraction of the planet’s land area, Africa (ZAfrica = -4.4; p < 0.001), Asia (ZAsia = -6.1; p < 0.001), South America (ZSouthAmerica = -3.6; p < 0.001) and Eastern Europe (ZEasternEurope = -3.8; p < 0.001) were under-sampled (Fig 3c). Oceania (ZOceania = 1.6; p = 0.11) and Central America (ZCentralAmerica = 0.3; p = 0.77) were sampled proportionally to their area. At the country level, most of the papers using CS data were from USA (n = 40; 25.5%), the UK (n = 15; 9.5%), Australia (n = 14; 8.9%), France (n = 9; 5.7%), Italy (n = 7; 4.5%), South Africa, Spain and Canada (n = 6; 3.8%; Fig 3d).
Such a strong geographic inclination toward Europe and North America has already been indicated by several authors [39, 41, 82]. Others also revealed the same pattern of CS being predominantly conducted in Europe and North America, but with a greater number of studies in South and Central America [16, 32] than reported in our study. This geographical disparity of CS-based papers among regions is likely influenced by three factors. First, North America and Europe host more developed countries, which have more funding available for research [48], and consequently tend to publish more. Some of these countries have traditional national platforms such as the National Biodiversity Gateway (NBN) in the United Kingdom (containing around 127 million records), the Atlas of Living Australia (ALA; containing 87,179,824 records on 19th April 2020), or the Sweden Species Gateway (containing around 60,000 species). Individual country platforms share characteristics associated with successful CS programs that contributed more to global biodiversity monitoring. These platforms receive important support by national governments and are linked to well-funded institutions with active involvement of academic researchers [39]. These factors explain why the expansion of CS platforms in developing countries might be limited by the availability of necessary infrastructures [39]. Secondly, this geographic pattern is consistent with the tradition of CS, which emerged in North America and then spread globally, primarily driven by some iconic platforms and surveys such as the Christmas Bird Count, eBird, and Project BudBurst [76]. Lastly, in regions with fewer papers using CS data, sharing biodiversity data remains difficult due to a lack of a tradition of open-access databases, but also to language barriers [39, 83]. A limitation of our study is that papers in languages other than English were not included in our review. These papers represent approximately 15% of papers within Scopus [84]. Thus, we acknowledge that restricting our review to English papers may have reduced our coverage of certain areas such as Arab countries, Latin America, or Asia [82]. In addition to language barriers and geographic location, national security concerns and economy also creates spatial variations in the coverage of global databases [82].
Source of data
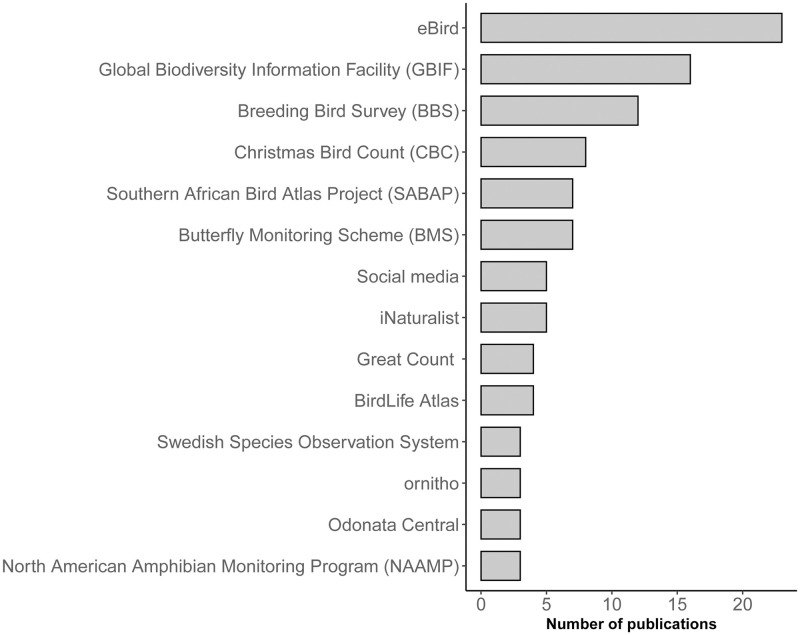
A wide variety of sources were identified, most being taxa-specific (e.g. birds or flying insects), whereas platforms that included various taxa were used to a lesser extent. The main reason for the predominance of bird CS papers was the use of three widespread networks of birders: the eBird project, the Breeding Bird Survey (BBS), and the Southern African Bird Atlas Project (SABAP, Fig 4). For insects, the Butterfly Monitoring Scheme (BMS) was the most widely used. Even if GBIF was the second most used source of information, this portal aggregates global biodiversity information from a variety of sources [39], including other CS portals listed in Fig 4. Indeed, the major GBIF contributor is eBird [39, 82]. For that reason, the GBIF database cannot be dissociated from other sources of CS in Fig 4.
Fig 4. Sources of information used in the papers included in the literature review (n = 207).
Only databases with three or more papers are shown. Great Count: world-wide surveys targeting birds and mammals.
Study scope
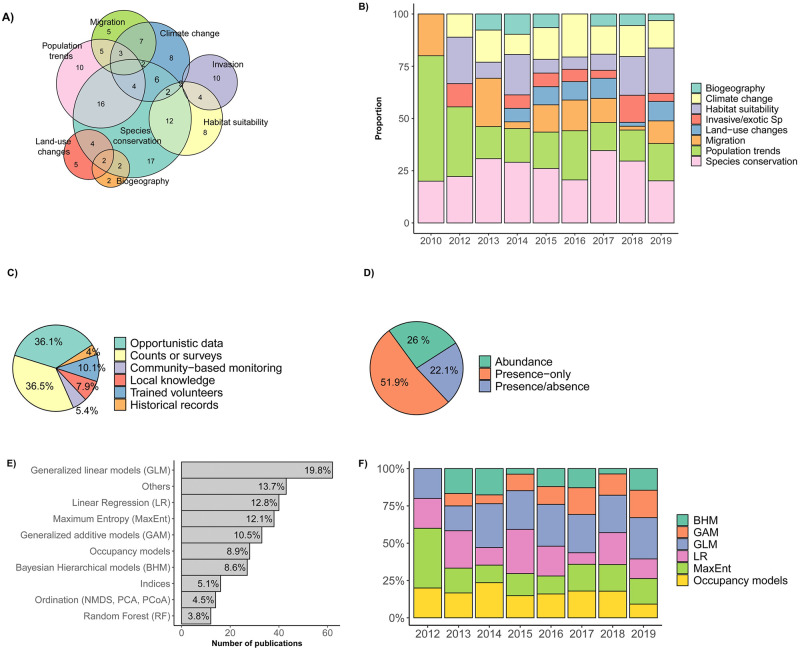
Although the scope and geographical coverage varied greatly among SDM papers using CS data (Fig 5a), most papers addressed issues related to species conservation (n = 95; 25.5%), followed by population trends (n = 67; 17.9%), habitat suitability (n = 56; 15%), and climate change (n = 52; 13.9%) (Fig 5a). Furthermore, species conservation was the central aim of these studies throughout the last decade, particularly in recent years (Fig 5b). The same pattern was observed for SDMs in tropical regions [1]. Most species conservation papers using CS documented species of conservation concern, rare species, or poorly-studied regions [34, 85].
Fig 5. Papers that have analyzed species distribution models (SDMs) using citizen science (CS) data in our literature review illustrating (a) study scope illustrated by a Venn diagram (intersections containing a single study not shown); (b) study scope per year; (c) method of collecting citizen science data; (d) data type used; (e) statistical approach and (f) statistical approach per year.
All details for the 207 papers are cited in S1 Table.
Method of collecting citizen science data
Several methods of collecting CS data were reported in the papers reviewed. The predominant method of collection consisted of count surveys (n = 101; 36.5%). In contrast to opportunistic data collection, count surveys differ in the structure of the methodology used, and may involve transect counts, point counts, or censuses. The second most used was opportunistic data collection which accounted for 36.1% (n = 100) of the papers (Fig 5c). With the growing popularity of online databases compiling occurrence data, the predominance of opportunistic data collection is not surprising [86]. Opportunistic data can be collected in many forms, including crowdsourcing databases or single random observations stored on online portals. Historical databases store information on past occurrence data usually collected opportunistically but generally found in naturalist diaries, letters or newspapers [87]. Our review found only 11 papers in the last decade that included historical databases (3.9% of the papers reviewed) (Fig 5c). Among other methods of collection, CS with trained volunteers only comprised a small proportion (n = 28; 10,1%) of the papers reviewed, highlighting that training volunteers is not a barrier to publishing. Nevertheless, projects with trained volunteers are more likely to be published than projects without training [60]. Data collection based on local knowledge (n = 22; 7,9%) and community-based monitoring (CBM) (n = 15; 5,4%) were rarely used (Fig 5c). Local and traditional knowledge of indigenous communities could improve SDMs. Indeed, by combining multiple types (or system) of knowledges, the collaboration between researchers and indigenous communities may help to expand the understanding of distribution boundaries, habitat and environmental associations, incorporate long-term observations, find local solutions for conservation actions and support the maintenance of local languages and culture. Additionally, engaging local participation may incorporate spatial guidance for gathering species records, increase the quantity of the data collected and expand the number of taxa covered by incorporating more observers [88, 89]. However, to fully benefit from this data collection method, researchers must be familiar with social science methods. Researchers may encounter difficulties in cross-cultural interactions, including language communication barriers and the reticence of the communities to share information about their environment [90]. Despite such difficulties, both scientist and communities can benefit from building on the interest and concerns of local community members when applying local knowledge and CBM [81]. Usually, the full potential of CBM programs is expressed when local communities participate actively during the entire scientific program, from the conceptual design and interpretation of results to the formulation of conclusions [88, 91]. Such cases have rarely occurred in the last decade for SDMs, probably because these programs are typically designed to monitor environmental factors rather than to collect species occurrence.
Data type
Citizen science data usually consist of presence-only data because they are easier to collect and require less effort than any other type of data. In our literature review, 120 out of 207 (52%) papers used PO data, 60 (26%) used abundance data and 51 (22%) used presence-absence data (Fig 5d). Twenty-two papers used two types of data (see S1 Table) and one paper used all three data types [92]. Several authors have highlighted the limitations of presence-only data, which confound information about habitat preferences and availability, have a strong spatial bias with more effort in sampling certain areas than others, and ignore environmental conditions associated with species occurrence [19, 27, 93]. Despite such restrictions, prominent modelling approaches dealing with presence-only data in the SDM literature can provide a performance comparable to models with presence-absence data [90, 94]. Some of those techniques include MaxEnt [95–97] especially for rare species or remote areas with low data points, spatial point-process models [98–100], or generalized additive models (GAMs) [101]. Furthermore, when absence data is not available in presence-only models, an alternative approach to account for potential sampling bias is to simulate pseudo-absences by including background information about non-occupied environments [19, 53, 102]. This is usually achieved by treating random point locations as absences in the same numbers as the presence data set [103–105]. Methods that generate simulated pseudo-absences instead of presence-only are an open research field for SDMs [104, 106, 107], highlighted in our review for 27 (13%) papers. After the generation of pseudo-absences, both the presence and pseudo-absence data can be analyzed using standard analyses for presence-absence data [108, 109], which is appropriate for a wide range of SDMs [110–112]. Despite these recent advances in presence-only data, there were high proportions of presence-absence and abundance data in the papers reviewed (n = 111; 48% in total; Fig 5d). Presence-absence data allow the comparison of a species’ occupancy between different areas or time periods [27], but is generally less common in CS data. Recent developments of various occupancy model types, accounting for imperfect detection probability [93, 113, 114], contributed to the increasing use of presence-absence data to infer the spatial distribution of species. Hence, this development would explain the increasing use of PA data obtained from CS databases. Abundance (AB) data occurred in similar proportions to PA in the set of studies reviewed (n = 60, 26%). Information on the number of individuals is essential to detect changes in population sizes [27]. Presence-absence (PA) and AB data can both be obtained from checklists, point-counts, or transect surveys by volunteers [93].
Statistical approach
The statistical approaches used in the papers reviewed were diverse, including linear regression approaches (LR; n = 40; 12.8%), maximum entropy (MaxEnt; n = 38, 12.4%), generalized linear models (GLM; n = 62; 19.8%), occupancy models (n = 28; 8.9%), and generalized additive models (GAM; n = 33; 10.5%; Fig 5e). Presence-only (PO) data were most frequently analyzed with MaxEnt (n = 36), whereas PA data were most frequently analyzed with occupancy models and GLM (n = 17 and 12, respectively). Abundance (AB) data were most often analyzed with GLM (n = 24). The proportion of use of the statistical approaches in the CS papers we reviewed did not seem to change between 2010 and 2019, with the exception of Bayesian hierarchical models (BHM) and GAMs appearing in papers from 2013 onward (Fig 5f).
Multiple data sources
Of the 207 articles reviewed, 81 (39.1%) used multiple sources of data, merging data from CS recorded by the public with professional data collected by experts. In 29 of these cases, authors compared results of volunteers’ observations with those obtained by professional scientists and only three studies revealed mismatches, particularly for species abundance [51, 115, 116]. The integration of CS and professional data has been a growing trend in recent years [105, 117, 118] and shows promise to improve inferences and the predictive ability of models, as well as to fill knowledge gaps for under-studied areas or poorly studied species [105, 117–119]. This approach of combining data benefits from robust survey schemes and expands the geographic and taxonomic coverage using unstructured opportunistic schemes. However, integrating highly heterogeneous data types such as large unstructured presence-only data and standardized abundance surveys, is still challenging for modelling purposes.
Conclusions
In this review, we examined the trends and information gaps in the use of citizen science (CS) data for species distribution models (SDMs) in peer-reviewed papers over the last decade. Citizen science already makes substantial contributions to the field of SDMs and this trend will probably continue. We presented examples of the use of CS and highlighted recommendations to motivate further research, such as combining multiple data sources and promoting local and traditional knowledge.
The reviewed citizen science papers considered a wide range of taxa, regions, and countries, from numerous biomes and landscape forms. However, taxonomic and geographic unevenness of CS projects for SDMs still remain [39, 48, 120]. It is imperative to better cover a wide range of taxonomic diversity to optimize the use of SDMs for species conservation. Accounting for these disparities in CS is crucial to adequately cover spatial and temporal scales, and strategically deploying formal surveys in areas or for species not covered by volunteers can be a key to better predict species distribution. Despite its huge contribution, the potential of citizen science can be maximized only if its value is recognized and data are analyzed rigorously. Therefore, we strongly encourage that researchers use as well as actively contribute to citizen science because they might have a major impact over the entire community of observers. The active participation of researchers in citizen science platforms (e.g. validating species identifications) can not only increase the amount of accessible data, but also increase the interest of local participants in countries where little information is currently available on the distribution of certain species.
Supporting information
(DOCX)
(DOC)
Acknowledgments
Earlier versions of the manuscript benefited from comments by A. Nolin.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
This project was made possible with funding provided by the Natural Sciences and Engineering Research Council (NSERC) of Canada – Université du Québec en Abitibi-Témiscamingue (UQAT) industrial research chair on northern biodiversity in a mining context.
References
- 1.Cayuela L, Golicher DJ, Newton AC, Kolb M, de Alburquerque FS, Arets E, et al. Species distribution modeling in the tropics: problems, potentialities, and the role of biological data for effective species conservation. Trop Conserv Sci. 2009; 2: 319–352. 10.1177/194008290900200304 [DOI] [Google Scholar]
- 2.Allouche O, Tsoar A, Kadmon R. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006; 43:1223–32. 10.1111/j.1365-2664.2006.01214.x [DOI] [Google Scholar]
- 3.Guisan A, Thuiller W. Predicting species distribution: offering more than simple habitat models. Ecol Lett. 2005; 8: 993–1009. 10.1111/j.1461-0248.2005.00792.x [DOI] [PubMed] [Google Scholar]
- 4.Austin MP. Spatial prediction of species distribution: an interface between ecological theory and statistical modelling. Ecological Modelling. 2002; 157: 101–118. 10.1016/S0304-3800(02)00205-3 [DOI] [Google Scholar]
- 5.Elith J, Leathwick JR. Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst. 2009; 40: 677–697. 10.1146/annurev.ecolsys.110308.120159 [DOI] [Google Scholar]
- 6.van Maes D, Isaac NJB, Harrower CA, Collen B, Roy DB, Van Strien AJ. The use of opportunistic data for IUCN Red List assessments. Biological Journal of the Linnean Society. 2015; 115:690–706. 10.1111/bij.12530 [DOI] [Google Scholar]
- 7.Pearson RG. Species’ distribution modeling for conservation educators and practitioners. Synthesis American Museum of Natural History. 2007; 50:54–89. [Google Scholar]
- 8.Guillera-Arroita G, Lahoz-Monfort JJ, Elith J, Gordon A, Kujala H, Lentini PE, et al. Is my species distribution model fit for purpose? Matching data and models to applications. Global Ecology and Biogeography. 2015; 24: 276–292. 10.1111/geb.12268 [DOI] [Google Scholar]
- 9.Guisan A, Zimmermann NE. Predictive habitat distribution models in ecology. Ecological modelling. 2000; 135: 147–186. 10.1016/s0304-3800(00)00354-9 [DOI] [Google Scholar]
- 10.Austin M. Species distribution models and ecological theory: a critical assessment and some possible new approaches. Ecological modelling. 2007; 200:1–19. 10.1016/j.ecolmodel.2006.07.005 [DOI] [Google Scholar]
- 11.van Wilgen NJ, Roura-Pascual N, Richardson DM. A quantitative climate-match score for risk-assessment screening of reptile and amphibian introductions. Environmental Management. 2009; 44: 590–607. 10.1007/s00267-009-9311-y [DOI] [PubMed] [Google Scholar]
- 12.Randin CF, Engler R, Normand S, Zappa M, Zimmermann NE, Pearman PB, et al. Climate change and plant distribution: local models predict high-elevation persistence. Glob Chang Biol. 2009; 15: 1557–1569. 10.1111/j.1365-2486.2008.01766.x [DOI] [Google Scholar]
- 13.Wilson RJ, Davies ZG, Thomas CD. Modelling the effect of habitat fragmentation on range expansion in a butterfly. Proceedings of the Royal Society B: Biological Sciences. 2009; 276: 1421–1427. 10.1098/rspb.2008.0724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McRae BH, Schumaker NH, McKane RB, Busing RT, Solomon AM, Burdick CA. A multi-model framework for simulating wildlife population response to land-use and climate change. Ecological modelling. 2008; 219: 77–91. 10.1016/j.ecolmodel.2008.08.001 [DOI] [Google Scholar]
- 15.Hauser WR, Heise-Pavlov SR. Can incidental sighting data be used to elucidate habitat preferences and areas of suitable habitat for a cryptic species? Integrative zoology. 2017; 12: 186–197. 10.1111/1749-4877.12227 [DOI] [PubMed] [Google Scholar]
- 16.Fois M, Cuena-Lombraña A, Fenu G, Bacchetta G. Using species distribution models at local scale to guide the search of poorly known species: Review, methodological issues and future directions. Ecological Modelling. 2018; 385: 124–132. 10.1016/j.ecolmodel.2018.07.018 [DOI] [Google Scholar]
- 17.Tye CA, McCleery RA, Fletcher RJ, Greene DU, Butryn RS. Evaluating citizen vs. professional data for modelling distributions of a rare squirrel. J Appl Ecol. 2016; 54: 628–637. 10.1111/1365-2664.12682 [DOI] [Google Scholar]
- 18.Bled F, Nichols JD, Altwegg R. Dynamic occupancy models for analyzing species’ range dynamics across large geographic scales. Ecol Evol. 2013; 3(15): 4896–4909. 10.1002/ece3.858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips SJ, Dudik M, Elith J, Graham CH, Lehmann A, Leathwick J, et al. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecological applications. 2009; 19: 181–197. 10.1890/07-2153.1 [DOI] [PubMed] [Google Scholar]
- 20.Stockwell DRB, Peterson AT. Effects of sample size on accuracy of species distribution models. Ecol Modell. 2002; 148: 1–13. 10.1016/s0304-3800(01)00388-x [DOI] [Google Scholar]
- 21.Peterson AT, Navarro-Sigüenza AG, Benítez-Díaz H. The need for continued scientific collecting; a geographic analysis of Mexican bird specimens. Ibis (Lond 1859). 2008; 140: 288–294. 10.1111/j.1474-919x.1998.tb04391.x [DOI] [Google Scholar]
- 22.van Strien AJ, van Swaay CAM, Termaat T. Opportunistic citizen science data of animal species produce reliable estimates of distribution trends if analysed with occupancy models. Journal of Applied Ecology. 2013; 50: 1450–1458. 10.1111/1365-2664.12158 [DOI] [Google Scholar]
- 23.Tulloch AI, Possingham HP, Joseph LN, Szabo J, Martin TG. Realising the full potential of citizen science monitoring programs. Biological Conservation. 2013; 165: 128–138. 10.1016/j.biocon.2013.05.025 [DOI] [Google Scholar]
- 24.Cooper CB, Shirk J, Zuckerberg B. The invisible prevalence of citizen science in global research: Migratory birds and climate change. PloS One. 2014; 9 (9). 10.1371/journal.pone.0106508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmeller DS, Henry PY, Julliard R, Gruber B, Clobert J, Dziock F, et al. Advantages of volunteer-based biodiversity monitoring in Europe. Conservation biology. 2009; 23: 307–316. 10.1111/j.1523-1739.2008.01125.x [DOI] [PubMed] [Google Scholar]
- 26.Hutchinson, RA, He L, Emerson SC. Species distribution modeling of citizen science data as a classification problem with class-conditional noise. In Proceedings of Thirty-First AAAI Conference on Artificial Intelligence. 2017
- 27.Bird TJ, Bates AE, Lefcheck JS, Hill NA, Thomson RJ, Edgar GJ, et al. Statistical solutions for error and bias in global citizen science datasets. Biological Conservation. 2014; 173: 144–154. 10.1016/j.biocon.2013.07.037 [DOI] [Google Scholar]
- 28.Ward DF. Understanding sampling and taxonomic biases recorded by citizen scientists. Journal of insect conservation. 2014; 18: 753–756. 10.1007/s10841-014-9676-y [DOI] [Google Scholar]
- 29.Hugo S, Altwegg R. The second Southern African Bird Atlas Project: causes and consequences of geographical sampling bias. Ecology and evolution. 2017; 7: 6839–6849. 10.1002/ece3.3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy S, Davalos LM. Geographical sampling bias and its implications for conservation priorities in Africa. J Biogeogr. 2003; 30: 1719–1727. 10.1046/j.1365-2699.2003.00946.x [DOI] [Google Scholar]
- 31.Botts EA; Erasmus BF; Alexander GJ. Geographic sampling bias in the South African Frog Atlas Project: implications for conservation planning. Biodiversity and Conservation 2011; 20: 119–139. 10.1007/s10531-010-9950-6 [DOI] [Google Scholar]
- 32.Martin LJ, Blossey B, Ellis E. Mapping where ecologists work: biases in the global distribution of terrestrial ecological observations. Front Ecol Environ. 2012; 10: 195–201. 10.1890/110154 [DOI] [Google Scholar]
- 33.Collins SD, Abbott JC, McIntyre NE. Quantifying the degree of bias from using county-scale data in species distribution modeling: Can increasing sample size or using county-averaged environmental data reduce distributional overprediction? Ecology and evolution. 2017; 7: 6012–6022. 10.1002/ece3.3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiménez-Valverde A, Peña-Aguilera P, Barve V, Burguillo-Madrid L. Photo-sharing platforms key for characterising niche and distribution in poorly studied taxa. Insect Conservation and Diversity. 2019; 12: 389–403. 10.1111/icad.12351 [DOI] [Google Scholar]
- 35.Ruete A. Displaying bias in sampling effort of data accessed from biodiversity databases using ignorance maps. Biodivers Data J. 2015; 1–15. 10.3897/BDJ.3.e5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Gabbas A, Dormann CF. Improved species-occurrence predictions in data-poor regions: using large-scale data and bias correction with down-weighted Poisson regression and Maxent. Ecography. 2018; 41: 1161–1172. 10.1111/ecog.03149 [DOI] [Google Scholar]
- 37.Beck J, Ballesteros-Mejia L, Nagel P, Kitching IJ. Online solutions and the ‘Wallacean shortfall’: what does GBIF contribute to our knowledge of species’ ranges? Divers Distrib. 2013; 19: 1043–1050. 10.1111/ddi.12083 [DOI] [Google Scholar]
- 38.Peterson AT, Soberón J, Krishtalka L. A global perspective on decadal challenges and priorities in biodiversity informatics. BMC Ecol. 2015; 15: 15. 10.1186/s12898-015-0046-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandler M, See L, Copas K, Bonde AM, López BC, Danielsen F, et al. Contribution of citizen science towards international biodiversity monitoring. Biological Conservation. 2017; 213: 280–294. 10.1016/j.biocon.2016.09.004 [DOI] [Google Scholar]
- 40.Amano T, Lamming JDL, Sutherland WJ. Spatial gaps in global biodiversity information and the role of citizen science. Bioscience. 2016; 66: 393–400. 10.1093/biosci/biw022 [DOI] [Google Scholar]
- 41.Pocock MJ, Chandler M, Bonney R, Thornhill I, Albin A, August T, et al. A vision for global biodiversity monitoring with citizen science. Advances in Ecological Research. 2018; 59: 169–223. [Google Scholar]
- 42.National Audubon Society. Forty-first Christmas Bird Count. Audubon Magazine’ Supplement: 1941; 74–148 [Google Scholar]
- 43.Robbins C. Sixty-sixth Christmas Bird Count. 241. Southern Dorchester County, Md. Audubon Field Notes. 1966; 20:180.
- 44.Niven DK, Butcher GS, Bancroft GT. Northward shifts in early winter abundance. Am Birds. 2010; 63: 10–15. [Google Scholar]
- 45.National Audubon Society. Alphabetical index and future National Audubon Society—The Christmas Bird Count Dates. 1999: 5. [Google Scholar]
- 46.Szabo J.K.; Vesk P.A.; Baxter P.W.; Possingham H.P. Regional avian species declines estimated from volunteer-collected long-term data using List Length Analysis. Ecological Applications 2010, 20, 2157–2169. 10.1890/09-0877.1 [DOI] [PubMed] [Google Scholar]
- 47.Boakes E, Gliozzo G, Seymour V, Harvey M, Smith C, Roy DB, et al. Patterns of contribution to citizen science biodiversity projects increase understanding of volunteers’ recording behaviour. Scientific reports. 2016; 6: 33051. 10.1038/srep33051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newbold T. Applications and limitations of museum data for conservation and ecology, with particular attention to species distribution models. Prog Phys Geogr. 2010; 34: 3–22. 10.1177/0309133309355630 [DOI] [Google Scholar]
- 49.Fitzpatrick MC, Preisser EL, Ellison AM, Elkinton JS. Observer bias and the detection of low-density populations. Ecological Applications. 2009; 19: 1673–1679. 10.1890/09-0265.1 [DOI] [PubMed] [Google Scholar]
- 50.Cox TE, Philippoff J, Baumgartner E, Smith CM. Expert variability provides perspective on the strengths and weaknesses of citizen-driven intertidal monitoring program. Ecological Applications. 2012; 22: 1201–1212. 10.1890/11-1614.1 [DOI] [PubMed] [Google Scholar]
- 51.Kamp J, Oppel S, Heldbjerg H, Nyegaard T, Donald PF. Unstructured citizen science data fail to detect long-term population declines of common birds in Denmark. Diversity and Distributions. 2016; 22: 1024–1035. 10.1111/ddi.12463 [DOI] [Google Scholar]
- 52.Kelling S, Johnston A, Hochachka WM, Iliff M, Fink D, Gerbracht J, et al. Can observation skills of citizen scientists be estimated using species accumulation curves? PLoS One. 2015; 10: e0139600. 10.1371/journal.pone.0139600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hertzog LR, Besnard A, Jay-Robert P. Field validation shows bias-corrected pseudo-absence selection is the best method for predictive species-distribution modelling. Diversity and distributions. 2014; 20: 1403–1413. 10.1111/ddi.12249 [DOI] [Google Scholar]
- 54.Fithian W, Elith J, Hastie T, Keith DA. Bias correction in species distribution models: pooling survey and collection data for multiple species. Methods in Ecology and Evolution. 2015; 6: 424–438. 10.1111/2041-210X.12242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Follett R, Strezov V. An analysis of citizen science based research: usage and publication patterns. PloS One. 2015; 10: 1–14. 10.1371/journal.pone.0143687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kullenberg C, Kasperowski D. What is citizen science?–A scientometric meta-analysis. PloS One. 2016; 11:e0147152. 10.1371/journal.pone.0147152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grey F. Citizen cyberscience: the new age of the amateur. CERN Courier. 2011. [Google Scholar]
- 58.Silvertown J. A new dawn for citizen science. Trends in ecology & evolution. 2009; 24: 467–471. 10.1016/j.tree.2009.03.017 [DOI] [PubMed] [Google Scholar]
- 59.Brown ED, Williams BK. The potential for citizen science to produce reliable and useful information in ecology. Conservation Biology. 2018; 33: 561–569. 10.1111/cobi.13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theobald EJ, Ettinger AK, Burgess HK, DeBey LB, Schmidt NR, Froehlich HE, et al. Global change and local solutions: Tapping the unrealized potential of citizen science for biodiversity research. Biol Conserv. 2015; 181: 236–244. 10.1016/j.biocon.2014.10.021 [DOI] [Google Scholar]
- 61.McKinley DC, Miller-Rushing AJ, Ballard HL, Bonney R, Brown H, Cook-Patton SC, et al. Citizen science can improve conservation science, natural resource management, and environmental protection. Biol Conserv. 2017; 208: 15–28. [Google Scholar]
- 62.Bonney R, Cooper CB, Dickinson J, Kelling S, Phillips TB, Rosenberg KV., et al. Citizen Science: a developing tool for expanding science knowledge and scientific literacy. BioScience. 2009; 59: 977–984. 10.1525/bio.2009.59.11.9 [DOI] [Google Scholar]
- 63.Cohn JP. Citizen science: Can volunteers do real research? Bioscience. 2008; 58: 192–7. 10.1641/b580303 [DOI] [Google Scholar]
- 64.Altwegg R, Wheeler M, Erni B (2008) Climate and the range dynamics of species with imperfect detection. Biol Lett 2008; 4: 581–584. 10.1098/rsbl.2008.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kéry M, Royle JA, Schmid H, Schaub M, Volet B, Häfliger G, et al. Site-occupancy distribution modeling to correct population-trend estimates derived from opportunistic observations. Conserv. Biol. 2010; 24: 1388–1397. 10.1111/j.1523-1739.2010.01479.x [DOI] [PubMed] [Google Scholar]
- 66.Bisby, F. A., Roskov, Y. R., Orrell, T. M., Nicolson, D., Paglinawan, L. E., Bailly, N., et al. 2011. Species 2000 & ITIS Catalogue of Life: 2019 Annual Checklist. 2019
- 67.Team, R.C. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. 2018.
- 68.Ries L, Oberhauser K. A citizen army for science: Quantifying the contributions of citizen scientists to our understanding of monarch butterfly biology. Bioscience. 2015; 65: 419–430. 10.1093/biosci/biv011 [DOI] [Google Scholar]
- 69.Welvaert M, Caley P. Citizen surveillance for environmental monitoring: combining the efforts of citizen science and crowdsourcing in a quantitative data framework. SpringerPlus. 2016; 5: 1890. 10.1186/s40064-016-3583-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lobo JM, Jiménez-Valverde A, Hortal J. The uncertain nature of absences and their importance in species distribution modelling. Ecography. 2010; 33: 103–114. 10.1111/j.1600-0587.2009.06039.x [DOI] [Google Scholar]
- 71.Guisan A, Tingley R, Baumgartner JB, Naujokaitis-Lewis I, Sutcliffe PR, Tulloch AI, et al. Predicting species distributions for conservation decisions. Ecol Lett. 2013; 16: 1424–1435. 10.1111/ele.12189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cerrano C, Milanese M, Ponti M. Diving for science-science for diving: volunteer scuba divers support science and conservation in the Mediterranean Sea. Aquat Conserv. 2017; 27: 303–323. 10.1002/aqc.2663 [DOI] [Google Scholar]
- 73.Champ J, Lorieul T, Bonnet P, Maghnaoui N, Sereno C, Dessup T, et al. Categorizing plant images at the variety level: Did you say fine-grained? Pattern Recognition Letters. 2016; 81: 71–79. 10.1016/j.patrec.2016.05.022 [DOI] [Google Scholar]
- 74.Wäldchen J, Mäder P. Plant species identification using computer vision techniques: A systematic literature review. Archives of Computational Methods in Engineering. 2018; 25: 507–543. 10.1007/s11831-016-9206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Joly A, Bonnet P, Goëau H, Barbe J, Selmi S, Champ J, et al. A look inside the Pl@ ntNet experience. Multimedia Systems. 2016; 22: 751–766. [Google Scholar]
- 76.Havens K, Henderson S. Citizen science takes root. American Scientist. 2013; 101: 378–385. 10.1511/2013.104.378 [DOI] [Google Scholar]
- 77.Plant Watch Canada. What is Plant Watch? Available online www.naturewatch.ca/plantwatch (accessed on 18 Mai 2020).
- 78.de Sá NC, Marchante H, Marchante E, Cabral JA, Honrado JP, Vicente JR. Can citizen science data guide the surveillance of invasive plants? A model-based test with Acacia trees in Portugal. Biological Invasions. 2019; 21: 2127–2141. 10.1007/s10530-019-01962-6 [DOI] [Google Scholar]
- 79.Nimis PL, Pittao E, Altobelli A, De Pascalis F, Laganis J, Martellos S. Mapping invasive plants with citizen science. A case study from Trieste (NE Italy). Plant Biosystems. 2018: 1–10. 10.1080/11263504.2018.1536085 [DOI] [Google Scholar]
- 80.Crall AW, Jarnevich CS, Young NE, Panke BJ, Renz M, Stohlgren TJ. Citizen science contributes to our knowledge of invasive plant species distributions. Biol Invasions. 2015; 17: 2415–2427. 10.1007/s10530-015-0885-4 [DOI] [Google Scholar]
- 81.Dyderski MK, Paź S, Frelich LE, Jagodziński AM. How much does climate change threaten European forest tree species distributions? Glob. Chang. Biol. 2018; 24: 1150–1163. 10.1111/gcb.13925 [DOI] [PubMed] [Google Scholar]
- 82.Amano T, Sutherland WJ. Four barriers to the global understanding of biodiversity conservation: wealth, language, geographical location and security. Proceedings of the Royal Society B: Biological Sciences. 2013; 280: 20122649. 10.1098/rspb.2012.2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hobern D, Baptiste B, Copas K, Guralnick R, Hahn A, van Huis E, et al. Connecting data and expertise: a new alliance for biodiversity knowledge. Biodiversity data journal. 2019; 7:e33679. 10.3897/BDJ.7.e33679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Moya-Anegón F.; Chinchilla-Rodríguez Z.; Vargas-Quesada B.; Corera-Álvarez E.; Muñoz-Fernández F.; González-Molina A.; et al. Coverage analysis of Scopus: A journal metric approach. Scientometrics 2007, 73, 53–78. 10.1007/s11192-007-1681-4 [DOI] [Google Scholar]
- 85.Richardson SJ, Clayton R, Rance BD, Broadbent H, McGlone MS, Wilmshurst JM. Small wetlands are critical for safeguarding rare and threatened plant species. Applied Vegetation Science. 2015; 18: 230–241. 10.1111/avsc.12144 [DOI] [Google Scholar]
- 86.Martínez-Minaya J, Cameletti M, Conesa D, Pennino MG. Species distribution modeling: A statistical review with focus in spatio-temporal issues. Stoch Environ Res Risk Assess. 2018; 7: 1–18. 10.1007/s00477-018-1548-7 [DOI] [Google Scholar]
- 87.Boshoff AF and Kerley G I. Historical mammal distribution data: how reliable are written records?–South Afr. J. Sci. 2010; 106: 26–33. 10.4102/sajs.v106i1/2.116 [DOI] [Google Scholar]
- 88.Skroblin A, Carboon T, Bidu G, Chapman N, Miller M, Taylor K, et al. Including Indigenous knowledge in species distribution modelling for increased ecological insights. Conservation Biology. 2019. 10.1111/cobi.13373 [DOI] [PubMed] [Google Scholar]
- 89.Mistry J and Berardi A. Bridging indigenous and scientific knowledge. Science 2016; 352: 1274–1275. 10.1126/science.aaf1160 [DOI] [PubMed] [Google Scholar]
- 90.Wang Y and Stone L. Understanding the connections between species distribution models for presence-background data. Theoretical Ecology 2019; 12: 73–88. 10.1007/s12080-018-0389-9 [DOI] [Google Scholar]
- 91.Bélisle AC, Asselin H, LeBlanc P, Gauthier S. Local knowledge in ecological modeling. Ecology and Society. 2018; 23(2). 10.5751/es-09949-230214 [DOI] [Google Scholar]
- 92.Pagel J, Anderson BJ, O’Hara RB, Cramer W, Fox R, Jeltsch F, et al. Quantifying range-wide variation in population trends from local abundance surveys and widespread opportunistic occurrence records. Methods in Ecology and Evolution. 2014; 5: 751–760. 10.1111/2041-210X.12221 [DOI] [Google Scholar]
- 93.Guillera-Arroita G. Modelling of species distributions, range dynamics and communities under imperfect detection: advances, challenges and opportunities. Ecography. 2017; 40: 281–295. 10.1111/ecog.02445 [DOI] [Google Scholar]
- 94.Mateo RG, Croat TB, Felicísimo ÁM, Munoz J. Profile or group discriminative techniques? Generating reliable species distribution models using pseudo-absences and target-group absences from natural history collections. Diversity and Distributions 2010; 16: 84–94. 10.1111/j.1472-4642.2009.00617.x [DOI] [Google Scholar]
- 95.Phillips SJ, Dudík M, Schapire RE. A maximum entropy approach to species distribution modeling. In Proceedings of Proceedings of the twenty-first international conference on Machine learning 2204: 655–692.
- 96.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological modelling 2006; 190: 231–259. 10.1016/j.ecolmodel.2005.03.026 [DOI] [Google Scholar]
- 97.West AM, Kumar S, Brown CS, Stohlgren TJ, Bromberg J. Field validation of an invasive species Maxent model. Ecological Informatics 2016; 36: 126–134. 10.1016/j.ecoinf.2016.11.001 [DOI] [Google Scholar]
- 98.Warton DI, Shepherd LC. Poisson point process models solve the “pseudo-absence problem” for presence-only data in ecology. The Annals of Applied Statistics 2010; 4: 1383–1402. 10.1214/10-aoas331corr [DOI] [Google Scholar]
- 99.Warton DI, Renner IW, Ramp D. Model-based control of observer bias for the analysis of presence-only data in ecology. PloS one 2013; 8: e79168. 10.1371/journal.pone.0079168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Renner IW, Warton DI. Equivalence of MAXENT and Poisson point process models for species distribution modeling in ecology. Biometrics 2013; 69: 274–281. 10.1111/j.1541-0420.2012.01824.x [DOI] [PubMed] [Google Scholar]
- 101.Grüss A, Drexler MD, Chancellor E, Ainsworth CH, Gleason JS, Tirpak JM, et al. Representing species distributions in spatially-explicit ecosystem models from presence-only data. Fisheries Research 2019; 210: 89–105. 10.1016/j.fishres.2018.10.011 [DOI] [Google Scholar]
- 102.Engler R, Guisan A, Rechsteiner L. An improved approach for predicting the distribution of rare and endangered species from occurrence and pseudo-absence data. Journal of applied ecology 2004; 41: 263–274. 10.1111/j.0021-8901.2004.00881.x [DOI] [Google Scholar]
- 103.Ranc N, Santini L, Rondinini C, Boitani L, Poitevin F, Angerbjörn A, et al. Performance tradeoffs in target-group bias correction for species distribution models. Ecography 2017; 40: 1076–1087. 10.1111/ecog.02414 [DOI] [Google Scholar]
- 104.Barbet-Massin M, Jiguet F, Albert CH, Thuiller W. Selecting pseudo-absences for species distribution models: how, where and how many? Methods in ecology and evolution 2012; 3: 327–338. 10.1111/j.2041-210x.2011.00172.x [DOI] [Google Scholar]
- 105.Miller DA, Pacifici K, Sanderlin JS, Reich BJ. The recent past and promising future for data integration methods to estimate species’ distributions. Methods in Ecology and Evolution 2019; 10: 22–37. 10.1111/2041-210x.13110 [DOI] [Google Scholar]
- 106.Iturbide M, Bedia J, Herrera S, del Hierro O, Pinto M, Gutiérrez JM. A framework for species distribution modelling with improved pseudo-absence generation. Ecological Modelling 2015; 312: 166–174. 10.1016/j.ecolmodel.2015.05.018 [DOI] [Google Scholar]
- 107.VanDerWal J, Shoo LP, Graham C, Williams SE. Selecting pseudo-absence data for presence-only distribution modeling: how far should you stray from what you know? Ecological modelling 2009; 220: 589–594. 10.1016/j.ecolmodel.2008.11.010 [DOI] [Google Scholar]
- 108.Pearce JL, Boyce MS. Modelling distribution and abundance with presence-only data. J. Appl. Ecol. 2006; 43: 405–412. 10.1111/j.1365-2664.2005.01112.x [DOI] [Google Scholar]
- 109.Elith J, Graham CH, Anderson RP, Dudík M, Ferrier S, Guisan A, et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006; 29: 129–151. [Google Scholar]
- 110.Dorazio RM, Gotelli NJ, Ellison AM. Modern methods of estimating biodiversity from presence-absence surveys. Biodiversity loss in a changing planet 2011: 277–302. 10.5772/23881 [DOI] [Google Scholar]
- 111.Mouton AM, De Baets B, Goethals PL. Ecological relevance of performance criteria for species distribution models. Ecological modelling 2010; 221: 1995–2002. 10.1016/j.ecolmodel.2010.04.017 [DOI] [Google Scholar]
- 112.Brotons L, Thuiller W, Araújo MB, Hirzel AH. Presence-absence versus presence-only modelling methods for predicting bird habitat suitability. Ecography 2004; 27: 437–448. 10.1111/j.0906-7590.2004.03764.x [DOI] [Google Scholar]
- 113.MacKenzie DI, Nichols JD, Royle JA, Pollock KH, Bailey L, Hines JE. Occupancy estimation and modeling: inferring patterns and dynamics of species occurrence; Elsevier: 2017. [Google Scholar]
- 114.Royle JA, Dorazio RM. Hierarchical modeling and inference in ecology: the analysis of data from populations, metapopulations and communities: Elsevier; 2008. [Google Scholar]
- 115.Snäll T, Kindvall O, Nilsson J, Pärt T. Evaluating citizen-based presence data for bird monitoring. Biological conservation 2011; 144: 804–810. 10.1016/j.biocon.2010.11.010 [DOI] [Google Scholar]
- 116.Cantú-Salazar L, Gaston KJ. Species richness and representation in protected areas of the Western hemisphere: discrepancies between checklists and range maps. Divers Distrib. 2013; 19: 782–793. 10.1111/ddi.12034 [DOI] [Google Scholar]
- 117.Fletcher RJ Jr, Hefley TJ, Robertson EP, Zuckerberg B, McCleery RA, Dorazio RM. A practical guide for combining data to model species distributions. Ecology. 2019: e02710. 10.1002/ecy.2710 [DOI] [PubMed] [Google Scholar]
- 118.Isaac NJ, Jarzyna MA, Keil P, Dambly LI, Boersch-Supan PH, Browning E, et al. Data Integration for Large-Scale Models of Species Distributions. Trends in ecology & evolution. 2019; 35: 56–67. 10.1016/j.tree.2019.08.006 [DOI] [PubMed] [Google Scholar]
- 119.Pacifici K, Reich BJ, Miller DA, Pease BS. Resolving misaligned spatial data with integrated species distribution models. Ecology. 2019: e02709. 10.1002/ecy.2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Devictor V, Whittaker RJ, Beltrame C. Beyond scarcity: citizen science programmes as useful tools for conservation biogeography. Diversity and distributions. 2010; 16: 354–362. 10.1111/j.1472-4642.2009.00615.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOC)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.