ABSTRACT
Long non-coding RNAs (ncRNAs) are major regulators of gene expression and cell fate. The INK4 locus encodes the tumour suppressor proteins p15INK4b, p16INK4a and p14ARF required for cell cycle arrest and whose expression increases during senescence. ANRIL is a ncRNA antisense to the p15 gene. In proliferative cells, ANRIL prevents senescence by repressing INK4 genes through the recruitment of Polycomb-group proteins. In models of replicative and RASval12 oncogene-induced senescence (OIS), the expression of ANRIL and Polycomb proteins decreases, thus allowing INK4 derepression. Here, we found in a model of RAF1 OIS that ANRIL expression rather increases, due in particular to an increased stability. This led us to search for circular ANRIL isoforms, as circular RNAs are rather stable species. We found that the expression of two circular ANRIL increases in several OIS models (RAF1, MEK1 and BRAF). In proliferative cells, they repress p15 expression, while in RAF1 OIS, they promote full induction of p15, p16 and p14ARF expression. Further analysis of one of these circular ANRIL shows that it interacts with Polycomb proteins and decreases EZH2 Polycomb protein localization and H3K27me3 at the p15 and p16 promoters, respectively. We propose that changes in the ratio between Polycomb proteins and circular ANRIL isoforms allow these isoforms to switch from repressors of p15 gene to activators of all INK4 genes in RAF1 OIS. Our data reveal that regulation of ANRIL expression depends on the senescence inducer and underline the importance of circular ANRIL in the regulation of INK4 gene expression and senescence.
KEYWORDS: Non-coding RNAs, circular RNAs, INK4 locus, oncogene-induced senescence, gene expression regulation, Polycomb proteins
Introduction
Long non-coding RNAs (lncRNAs) play an important role in the control of gene expression. They are usually involved in the local targeting of chromatin-modifying enzymes [1]. Although there are many examples of lncRNAs functioning in trans, they usually regulate genes in cis, meaning close to where they are produced. One of the best described lncRNA, which targets chromatin-modifying enzymes to chromatin, is the ANRIL ncRNA [2]. ANRIL is produced from the tumour suppressor INK4 locus, which contains two protein-coding genes, CDKN2A and CDKN2B. The CDKN2B gene encodes the p15INK4b (p15) cyclin-dependent kinase (CDK) inhibitor. The CDKN2A gene encodes two proteins, the p16INK4a (p16) CDK inhibitor and p14ARF, by a different reading frame using an alternate first exon located 20 kb upstream [3]. ANRIL is actually transcribed antisense to the p15/CDKN2B gene [4].
ANRIL has been extensively studied given that it has been identified as a risk locus in many human diseases from Single Nucleotide Polymorphism (SNP) association by Genome Wide Association Studies (GWAS) [5,6]. Notably, a 53 kb region associated with increased risk of coronary artery diseases (CAD) is located in the second half of the ANRIL gene [5]. Increased or reduced ANRIL expression has been reported in diverse diseases, including cancers, diabetes or cardiovascular diseases, whose risk is linked with ageing [7]. Notably, ANRIL is overexpressed in many cancers and promote cell proliferation of cancer cell lines from diverse origins, including colon carcinoma [8,9], oesophageal carcinoma and bladder cancers [10,11]. Moreover, ANRIL regulates inflammatory responses and is a strong candidate for genetic susceptibility to many inflammatory diseases [12].
The INK4 locus plays a major role in the control of cellular senescence [13], which is characterized by a stable proliferation arrest. Indeed, the two major pathways that mediate senescence commitment are the p53 and Rb tumour suppressor pathways, which are both regulated by genes encoded by the INK4 locus [14]. Upon senescence induction, INK4 gene expression is activated. The p15 and p16 CDK inhibitors activate the Rb pathway by inhibiting its phosphorylation by Cyclin/CDK, while p14ARF represses HDM2 function, allowing activation of the p53-p21 pathway. Both events result in cell cycle arrest, which is made irreversible by subsequent events.
Senescence was first described as a stable cell proliferation arrest after a defined number of divisions of normal cells in culture, referred to as replicative senescence. It is triggered by telomere shortening in the absence of telomerase activity and the incomplete replication of the very end of chromosomes [15]. It is now clear that senescence is a cell fate induced upon various kinds of stresses, such as oncogene activation, and characterized by a potent and permanent cell proliferation arrest, as well as a number of associated changes, such as chromatin reorganization and the setting up of a specific genetic program [16]. Senescence is considered as a major tumour suppressor pathway, through the elimination of cells with oncogenic activation, not repairable DNA damages or uncontrolled genetic instability [17].
ANRIL prevents senescence induction by repressing the expression of the p15/CDKN2B and p16/CDKN2A genes at the INK4 locus in proliferative cells by locally recruiting the repressive Polycomb complexes [18,19]. Polycomb-group proteins are involved in transcriptional gene silencing and heterochromatin formation [20,21] and mainly form two multimolecular complexes, PRC1 and PRC2. PRC2 contains the EZH2 histone methyl transferase, which methylates K27 of histone H3. PRC1 specifically recognizes methylated H3K27 and mediates transcription inhibition, at least in part through histone H2AK119 monoubiquitinylation [22]. In proliferative cells, ANRIL is able to recruit these two complexes in cis to the INK4 locus, allowing its transcriptional silencing [18,19]. ANRIL has also been shown to regulate gene expression in trans but the molecular mechanisms involved are still not clear [23,24].
During replicative and oncogene-induced senescence (OIS), ChIP experiments indicate that the recruitment of Polycomb-group proteins and H3K27 methylation decrease at the INK4 locus [25–28]. Although other mechanisms have been reported, such as the general decrease of Polycomb-group protein expression [25–29], a senescence-associated inhibition of ANRIL expression was proposed to be involved in this process. Indeed, ANRIL expression was found to decrease in MEFs undergoing replicative senescence [19], and in a model of OIS induced by oncogenic RAS [18].
Circular RNAs (circRNAs) are ncRNAs produced by back-splicing, which, by the splicing of a downstream donor splice site to an upstream acceptor splice site leads to the production of a circular RNA molecule [30]. CircRNAs are naturally resistant to degradation by exonucleases and hence, they are more stable than linear RNA molecules [31,32]. Many exonic circRNAs have been shown to be predominantly cytoplasmic [32,33], while intronic circRNAs are nuclear [34]. CircRNA expression is upregulated during cellular differentiation while their parental gene expression is mostly unchanged [35–38]. CircRNAs are also misregulated in cancers [39]. Recently, emerging functions of circRNAs have been described, especially linked to the regulation of transcription, splicing, protein interaction dynamics or microRNA sponging [40]. Many different isoforms of ANRIL have been identified [23], including circular ANRIL isoforms [41–43], whose expression correlates with anti-proliferative properties and CAD protection [42].
In a recent study, we performed genome-wide analyses of strand-specific RNA expression in WI38 human primary fibroblasts undergoing OIS upon activation of RAF1 oncogene [44]. In this experimental setting, INK4 genes are activated, and their activation is known to be important for the associated cell cycle arrest and other senescence features [45–47]. Here, we observed in this model of OIS and other models of OIS an increased ANRIL expression, indicating that repression of ANRIL expression is not general during senescence induction and is not required for activation of INK4 genes. We further identified several circular ANRIL isoforms whose expression increases in RAF1-induced senescence. They repress p15 gene expression in proliferative cells, while they are important for full induction of INK4 genes in RAF1-induced senescence. Our results indicate that ANRIL switches from a negative role on p15 to a positive role on INK4 gene expression during RAF1-induced senescence progression.
Materials and methods
Cell culture
WI38 hTERT RAF1-ER, IMR90 hTERT RASval12-ER and BJ hTERT BRAF V600E cells were a kind gift from Dr C. Mann (CEA, France) [45,48,49]. IMR90 hTERT MEK1-ER cells [50] were a kind gift from Dr M. Djabali. WI38-, IMR- and BJ-derived cells were grown in MEM supplemented with L-glutamine, non-essential amino acids, sodium pyruvate, penicillin–streptomycin and 10% foetal bovine serum in normoxic (5% O2) culture conditions. For senescence induction in WI38 hTERT RAF1-ER and IMR90 hTERT MEK1-ER, cells were treated with, respectively, 20 nM or 100 nM of 4-HT (Sigma) for 3 days. IMR90 hTERT RASval12-ER cells were treated with 100 nM 4-HT for 8 days (cells were dividing once or twice before they completely stopped proliferating). BJ hTERT BRAF V600E cells were treated with 1 µg/mL doxycycline for 3 days. For replicative senescence, human embryonic fibroblasts WI38 were grown until they entered senescence after 63 population doublings (cells were harvested 4 weeks after their last division). Proliferative WI38 were harvested at 49 population doublings. During long kinetics of RAF1-induced senescence, the medium (including 4-HT) was changed every 3 days. In parallel to senescent cells, because 4-HT was diluted in ethanol, proliferative cells were supplemented with the same volume of ethanol used to treat senescent cells with 4-HT. For transcription inhibition, cells were treated with 1 µM flavopiridol (Sigma) for the indicated times. siRNA transfection was performed using the Dharmafect 4 reagent (Dharmacon) according to the manufacturer’s recommendations, except that 100 nM of siRNA were used and then diluted twice 24 h later by adding the same volume of medium. Cells were harvested 72 h following transfection. For the transfection of RAF1-induced senescent cells, cells were treated 72 h with 4-HT, then transfected and cultured without 4-HT for 72 h. For samples treated with control siRNAs (which do not target any sequence in the genome), either one single control siRNA (cont3 or scramble) or a pool of 8 control siRNAs (thus diluting each siRNA control to avoid potential off-target effects of each of them (pool sicont)) was transfected. siRNA sequences are available in Table S5.
Chromatin immunoprecipitations
ChIP was performed as previously described [46] except that Nuclear lysis buffer was diluted twice before use and chromatin samples were diluted 5 times in dilution buffer.
Antibodies
Antibody to the GAPDH protein (MAB374) was purchased from Millipore. Antibodies to the p15 (sc-612) and p300 (sc-585) proteins were purchased from Santa Cruz. Antibodies to SUZ12 (Ab12073), H3 (Ab1791), H3K27Ac (Ab4729) and H3K27me3 (Ab6002) epitopes were purchased from Abcam. The antibodies to EZH2 (AC22, #3147) and to RNA pol II (A304-405A) were purchased from Cell Signalling and Bethyl, respectively.
Analysis of RNA-seq datasets
RNA-seq experiments, normalization and calculation of RNA-seq ratios between senescent and proliferative samples within specific regions were previously described in [44]. For the analysis of spliced reads, the numbers of aligned reads in the ANRIL locus or in all gene loci (28,445; from RefSeq database) were calculated. Genes were only analysed if 10 or more reads were aligned on the gene for each sample and each replicate (15,027 genes). For the same regions, the number of aligned reads, which did not align consecutively on the genome, were calculated and referred to as spliced reads. The percentage of spliced reads was calculated by computing the ratio of the number of spliced reads over the number of total aligned reads for each region. The start and the end of the gap for each spliced sequence were analysed for the ANRIL locus (Table S1).
For back-spliced read analysis (Table S1), a new pipeline was developed including three different detection tools in combination (CircExplorer2, CircRNA finder and FindCirc) to detect all potential circular RNA reads, and an annotation step of the identified back-spliced junctions (Delmas et al., manuscript in preparation).
For RNA-seq following depletion of circANRIL e16-e5, 5–10 μg of total RNA, extracted as described below in (RNA extraction and Reverse transcription), was submitted to EMBL-GeneCore, Heidelberg, Germany. Two replicates of each sample were sequenced. We used strand-specific total RNA-seq method, relying on UTP incorporation in the second cDNA strand. RNA-seq samples were sequenced using Illumina NextSeq 500 sequencer, paired-end, 80-bp reads. The quality of each raw sequencing file (fastq) was verified with FastQC [51]. Files were aligned to the reference human genome (hg38) in paired-end mode with STAR Version 2.5.2 and processed (sorting and indexing) with samtools [52]. Raw reads were counted, per gene_id, using HT-seq Version 0.6.1 [53] on the NCBI refseq annotation gtf file from UCSC in a strand-specific mode with default parameters. Genes with a mean of less than one read count for all the samples were eliminated. Differential analysis was performed with DESeq2 Bioconductor R package, Version 1.22.1 with default parameters but using normalization by the total read number per sample. Genes of interest were selected when |log2FoldChange| was higher than 1 and adjusted p-value lower than 0.05. For comparison between the genes regulated by circANRIL e16-e5 and genes regulated in senescence, proliferation and senescence RNA-seq datasets were processed exactly the same way by HT-seq and DESeq2.
EdU and hoechst staining
EdU and Hoechst staining were performed as previously described [44].
RNA extraction, RT, qPCR and outward-facing PCR
Total RNA was extracted from cells using the MasterPure RNA Purification Kit from Epicentre following the manufacturer’s protocol, except that 5 µL of 50 µg/µL proteinase K was used for cellular lysis. Alternatively, TRIzol was used. After total nucleic acid precipitation, we proceeded to removal of contaminating DNA using a mixture of DNase I and Baseline Zero DNase, supplemented with Riboguard RNAse inhibitor, for 45 min at 37°C. Usually, 500 ng of total RNA was used for reverse transcription (RT) using the Superscript III Reverse Transcriptase (Invitrogen). A minus RT reaction was performed for each sample and analysed by qPCR for the expression of GAPDH (using GAPDH e9 primers) to verify the absence of DNA contamination. qPCR was performed using the SYBR premix Ex Taq from Takara and the Biorad CFX thermocycler. GAPDH expression (measured using GAPDH e9 primers) was used for normalization between samples in each RT-qPCR experiment.
Outward-facing PCR reactions were performed on 6 ng of random primed cDNA using the GoTaq G2 DNA polymerase (Promega) following the manufacturer’s recommendations in the Techne TC-3000 PCR thermal cycler (initial denaturation of 2 min 95°C, followed by 35 cycles of 30 sec 95°C, 30 sec 58°C, 3 min 72°C; final extension of 5 min 72°C). Primers in exon 5 or exon 6 were used since exon5-exon6 junction was the most abundant junction found in RAF1-induced senescence in our RNA-seq datasets (Table S1). The detection of circular isoforms was also tested using outward-facing primers in exon 16 (because it was found in back-spliced junctions with exon 5 from different studies [41–43]) and in exon 1 (a control since the first exon of a transcript cannot be included in circular RNA molecules because it does not possess splicing signals at its 5ʹ end) (Fig. S6). PCR products were then run on a 1% agarose gel and visualized using GelRed Nucleic Acid Gel Stain (Biotium). Only well-separated bands corresponding to the main PCR products obtained were cut out from the gel and PCR products were extracted from gel slices using GenElute Agarose Spin Columns (Sigma-Aldrich). The PCR products were then ethanol precipitated, resuspended in 10 µL of water and sent to Eurofins MWG for sequencing using one of the primers used for the PCR reaction.
Many products were detected using the outward-facing primers in exons 5, 6 and 16, likely representing circular ANRIL species. Note that, although the exon 16-containing species were identified using outward-facing primers in exon 16, these species were not identified using outward-facing primers in exon 5 or exon 6. This is probably due to the fact that the most abundant PCR products obtained using outward-facing primers in exon 5 or exon 6 correspond to small isoforms. Nevertheless, many bands corresponding to larger isoforms were also detected using outward-facing primers in exon 5 or exon 6, but as these were not properly separated from one another, it was impossible to specifically extract them from the gel.
Sequences of primers are available in Table S5.
RNase R treatment
A 3.5 µg total RNA was incubated for 5 min at 65°C and immediately placed on ice. Total RNA was then treated for 15 min at 37°C with 10 U RNase R from Epicentre in a 20 μl reaction volume. A parallel reaction was performed without adding RNase R. 180 μl of TE buffer (10 mM Tris and 1 mM EDTA) was added to each reaction. Purification and precipitation of RNA were performed using the MasterPure RNA Purification Kit from Epicentre by adding 200 μl of 2X T and C Lysis Solution and following the manufacturer’s protocol. A 500 ng of untreated total RNA from the parallel reaction and an equal volume of the RNase R-treated RNA sample were used for random primed RT with Superscript III Reverse Transcriptase.
Labelling and capture of nascent RNA
Nascent RNA was labelled and captured using the Click-iT Nascent RNA Capture Kit from Life Technologies following the manufacturer’s protocol. Cells were incubated with EU (5-Ethynyl Uridine), an analogue of uridine, at a concentration of 0.2 mM for 30 min or 1 h or at a concentration of 0.5 mM for 10 min, as specified in the figure legends. A 1 µg of total RNA was used for the click reaction.
In vitro RNA synthesis for rescue experiments
A 10 µg of plasmid (sequences can be found in Table S5) was digested overnight using PvuII for scramble and SmaI for circANRIL. Linearized plasmids were used as template for in vitro transcription using T7 RNA polymerase (Promega) and a ribonucleotide set (NEB) following the manufacturer’s recommendations for 2 h at 37°C. Given that in vitro transcription using T7 RNA polymerase can introduce untemplated nucleotides at the 5ʹ and 3ʹ ends of the synthesized RNA, a linear RNA already containing the e16-e5 backspliced junction was produced, in order to avoid introducing mutations in this specific junction. Thus, a linear RNA containing the 3ʹ end of exon 7 followed by the entire exon 16-exon 5-exon 6 sequence followed by the 5ʹ end of exon 7 (Fig. S9B) was produced, so that untemplated nucleotides eventually introduced by T7 RNA polymerase are located in the middle of exon 7. In vitro transcribed RNAs were incubated for 1 h at 37°C after addition of 10 units DNase I and 10 units DNase Zero (Epicentre). The RNAs were then ethanol precipitated and resuspended in water.
For two of the rescue experiments (out of four independent experiments), the in vitro transcribed RNAs were directly used for ligation at this step. However, because we found that the ligation efficiency was very low, we modified the protocol to improve this ligation efficiency. For the other rescue experiments, in vitro transcribed RNAs were then dephosphorylated using 2 units Alkaline Phosphatase (FastAP, Thermo Scientific) for 30 min at 37°C following the manufacturer’s recommendations. De-phosphorylated RNAs were purified by phenol/chloroform extraction followed by ethanol precipitation, re-phosphorylated using 10 units T4 PNK (Promega) for 30 min at 37°C and ethanol precipitated. Around 500 ng of RNA was ligated overnight at 16°C using 20 units of T4 RNA ligase 1 (NEB) in the presence of 10% PEG 8000 to favour intramolecular ligations.
Ligated RNAs were then purified using phenol/chloroform extraction followed by ethanol precipitation and then digested using 10 U RNase R from Epicentre as described in the RNase R treatment paragraph. The size of the in vitro transcribed RNAs was checked on gel (Fig. S9C). Between 24 h and 72 h following siRNA transfection, 0.036 µg of in vitro synthesized circular RNAs were transfected in WI38 RAF1-ER cells, using Dharmafect 4 (Dharmacon) according to the manufacturer’s recommendations. Overexpression of the in vitro transcribed RNAs was checked by analysing the RNAs purified from transfected cells by RT-qPCR in each experiment (Fig. S9B).
Upon ligase treatment, ligation of the two halves of exon 7 (producing the entire exon 7) reconstitutes a circularized RNA containing the exact e16-e5 junction and nearly identical to the isoform we identified in senescent cells. Ligation efficiency was checked before (Fig. S9C) and after transfection in senescent cells (data not shown). Although the circularization was effective, it was not very efficient and variable from one experiment to the other. Thus, for circANRIL e16-e5 overexpression, the e16-e5 backspliced junction was overexpressed on average 1300 times (Fig. S9B) whereas the circularized product only 19 times (data not shown). As such, most of the overexpressed circANRIL e16-e5 RNAs were linear in transfected cells, but contain the e16-e5 backspliced junction within its natural context and may thus mimic the function of circANRIL e16-e5 natural RNAs, at least to some extent.
In vitro RNA synthesis for pull-down experiments
A 10 µg of plasmid (Table S5) was digested overnight using PvuII for scramble and SmaI for circANRIL. Linearized plasmids were used as template for in vitro transcription using T7 RNA polymerase (Promega) and biotin RNA labelling mix (Sigma Aldrich) following the manufacturer’s recommendations for 2 h at 37°C. In vitro transcribed RNAs were incubated for 1 h at 37°C after addition of 10 units DNase I and 10 units DNase Zero (Epicentre). The RNAs were then ethanol precipitated and resuspended in water and dephosphorylated using 2units Alkaline Phosphatase (FastAP, Thermo Scientific) for 30 min at 37°C following the manufacturer’s recommendations. De-phosphorylated RNAs were purified by phenol/chloroform extraction followed by ethanol precipitation, re-phosphorylated using 10 units T4 PNK (Promega) for 30 min at 37°C and ethanol precipitated. Around 500 ng of RNAs was ligated overnight at 16°C using 20 units of T4 RNA ligase 1 (NEB) in the presence of 10% PEG 8000 to favour intramolecular ligations. Ligated RNAs were then purified using phenol/chloroform extraction followed by ethanol precipitation. For four of the pull-down experiments (out of eight independent experiments), we used the MasterPure RNA purification kit from Epicentre instead of phenol/chloroform extraction to purify RNA after the dephosphorylation and ligation steps.
Biotinylated RNA pull-down
A 30 µL of HeLa nuclear extract (purchased from Computer Cell Culture Centre, prepared according to the classical Dignam protocol) were diluted 10 times in binding buffer (20 mM Tris pH 8.0, 100 mM NaCl, 5 mM MgCl2, 0.4% NP40) and supplemented with protease inhibitors (Roche), 60 µg of yeast tRNA (Invitrogen) and 8 U of DNase I (Epicentre). These diluted nuclear extracts were pre-cleared using 25 µL of previously blocked Streptavidin Sepharose beads (GE Healthcare) for 2 h at 4°C. Blocking was achieved by incubating the beads with 1 mg.mL−1 of Ultrapure BSA and 0.5 mg.mL−1 of salmon sperm DNA (Invitrogen) overnight at 4°C. The precleared nuclear extracts were recovered, supplemented with 2 µL Riboguard (Epicentre) and incubated for 30 min at room temperature with 500 ng of in vitro synthesized RNAs. 10 µL of blocked beads were then added and incubation was pursued for another 30 min at room temperature. After 4 washes in binding buffer, beads were resuspended in 20 µL binding buffer and 10 µL 4X Laemmli sample buffer (Biorad) supplemented with b-mercaptoethanol. Samples were boiled for 5 min at 95°C before being loaded on a 3–8% NuPAGE Tris-Acetate gel (Invitrogen). Western blots were quantified using ImageJ. Linearity of the EZH2 and SUZ12 western blots was controlled by serial dilutions of an input.
RNA FISH
The protocol used for RNA FISH experiments has been described elsewhere [46] except that probes were denatured for 7 min at 75°C and additionally pre-incubated 10 min at 37°C. Red-dUTP (02N34-050, Abbot Molecular) labelled probes were generated by nick translation using the WI2-48,403-G248P84444H2 fosmid. All images were taken on a fluorescence microscope DM5000 B Leica with a Retiga R3 camera controlled by the Metamorph 7.7 software using x40 objective. Different wavelength probes were used (DAPI (360 nm, 470 nm) and Texas Red (596 nm, 612 nm)). Images were quantified using Columbus, the integrated software of the Operetta automated high-content screening microscope (PerkinElmer).
Accession numbers
RNA-seq data are available at GEO (GSE 143957).
Results
ANRIL expression increases in a model of senescence induced by oncogenic RAF1
To characterize transcriptional changes in cells undergoing oncogene-induced senescence, we have recently performed strand-specific RNA-seq experiments using either proliferative WI38 hTERT RAF1-ER cells (derived from normal human fibroblasts and immortalized by ectopic hTERT expression) or the same cells made senescent by the activation of the RAF1 oncogene [44]. This cell line contains a fusion protein RAF1-ER (oestrogen receptor), which allows the induction of senescence after the addition of 4-hydroxy-tamoxifen (4-HT) [45]. In this model, all cells stop dividing after 3 days of 4-HT treatment and present senescence characteristics, such as SAHF formation or the induction of anti-proliferative genes including the INK4 genes [45,46]. Analysis of RNA-seq data at the INK4 locus, which encodes three major mediators of senescence (p15, p16 and p14ARF), shows that p15, p16 and p14ARF-mRNAs are strongly induced in senescence with p15 being particularly activated (Fig. S1A), as previously shown by RT-qPCR experiments [46]. We also observed that expression of ANRIL, which is antisense to the p15-encoding gene and involved in its repression in proliferative cells [18,19,54], increases upon senescence induction (Fig. 1A-B and S1B). This observation is surprising given that in the generally accepted model, ANRIL expression decreases in senescent cells, leading to the activation of genes from the INK4 locus [18,19].
Figure 1.
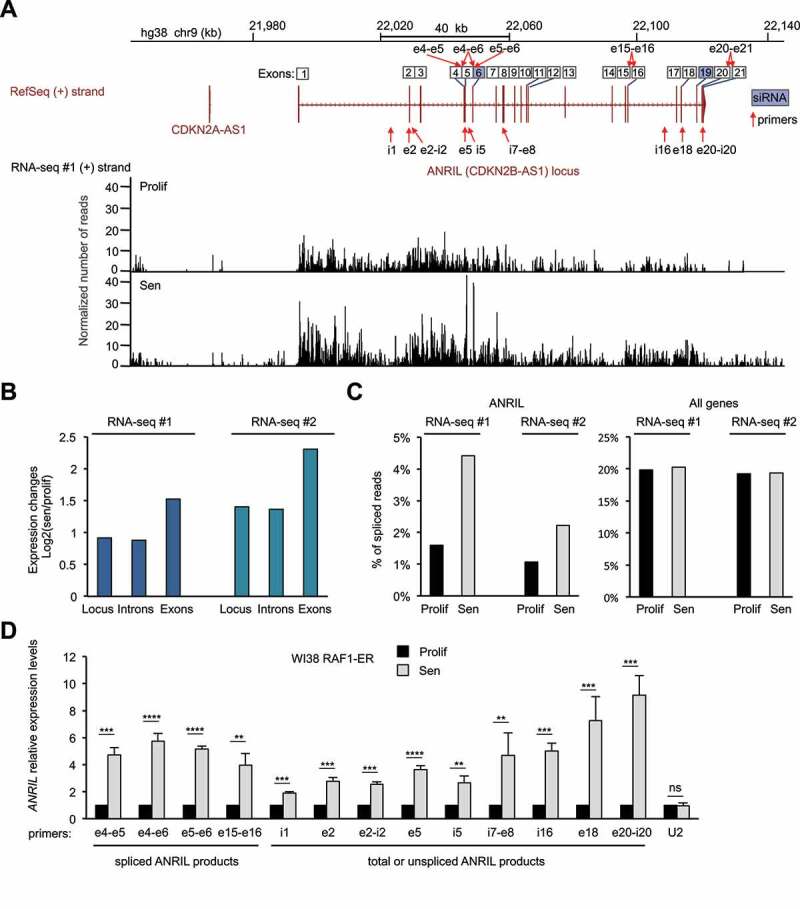
ANRIL expression increases in a model of RAF1 oncogene-induced senescence
(A) RNA-seq data (from the 1st replicate (RNA-seq#1)) showing expression of the (+) strand at the ANRIL gene, in WI38 hTERT RAF1-ER cells either proliferative (Prolif) or induced to senescence (Sen) by 4-HT treatment, as indicated. Strand-specific RNA-seq tracks show the number of reads per base normalized by the total number of aligned reads multiplied by 100 million. The ANRIL locus is also shown with all the annotated exons from the various transcript variants from the RefSeq database. The primers and siRNAs targeting ANRIL, used in this study, are shown with red arrows and blue squares, respectively. Primers detecting spliced products are shown on top of the exon numbers while primers detecting total (spliced + unspliced) or unspliced ANRIL are shown below the locus. (B) Quantification of the RNA-seq signal from the 1st (RNA-seq #1) and 2nd (RNA-seq #2) replicates at the ANRIL locus, exons or introns. For each analysed region, the mean per base of the normalized number of aligned reads was computed. The ratio of this number in senescence to the number in proliferation was then calculated (log2). (C) Analysis of strand-specific RNA-seq datasets for spliced reads relative to the total number of reads in proliferation or senescence for the ANRIL gene or all expressed genes (15,027 genes analysed as described in the Methods section, showing the median value of the whole population). (D) WI38 hTERT RAF1-ER cells were induced to senescence or maintained proliferative, as indicated. Seventy-two hours later, total RNA was extracted, and ANRIL expression was measured by RT-qPCR using the indicated primers. U2 ncRNA was measured as a control whose expression does not change in RAF1-induced senescence. The means and standard deviations from four independent experiments are shown, relative to GAPDH and normalized to 1 in proliferative cells for each experiment (before calculating the mean between experiments, to compare the variations in expression changes between the different regions). Significant differences are indicated with asterisks (*: p value < 0.05, ** to ****: p values < 10−2, 10−3 and 10−4, respectively; two-sided paired Student’s t-test on log2 values). i = introns, e = exons, ns = not significant.
Close examination of RNA-seq data further shows that the expression of exons tends to increase more than that of introns, suggesting that the percentage of spliced forms of ANRIL increases in senescence (Fig. 1B). We, therefore, calculated from our two RNA-seq replicates the percentage of spliced reads relative to the total number of reads at the ANRIL gene. This percentage is very low in proliferative cells (about 1%), whereas it increases by more than 2 folds in senescent cells (Fig. 1C). This increase is very specific to ANRIL because the percentage of spliced reads considering all genes is stable during senescence induction (Fig. 1C). Thus, these data suggest that spliced mature ANRIL transcripts specifically increase in senescence. We next analysed in detail the ANRIL spliced junctions found in the RNA-seq experiments. For simplicity, as ANRIL has many transcript variants annotated in the reference genome (RefSeq hg38), we numbered the exons based on the position of all the exons of these different variants and used this nomenclature throughout this study to name primers and siRNAs (ANRIL locus, Fig. S2). We found that the most abundant splicing event of ANRIL in senescence occurs between the exons 5 and 6 (Table S1). Although small ANRIL isoforms ending at exon 13 are described in the reference genome (Fig. S2), we found only one read containing a spliced junction with exon 13 in a single RNA-seq replicate (data not shown). In contrast, several reads were found connecting the downstream exons in both RNA-seq replicates, suggesting that the small ANRIL isoforms are largely under-represented compared to large ANRIL isoforms ending at exon 21 in WI38 cells (Table S1). We also observed that ANRIL expression increases more in its second part than in its first part (Fig. S3A-C).
To validate these results, we induced WI38 hTERT RAF1-ER cells to senesce or not. Senescence was controlled by the lack of EdU incorporation and SAHF formation (Fig. S4). We first confirmed the increase of ANRIL expression at the individual cell level by RNA FISH (Fig. S5). We then recovered total RNAs and confirmed the increase of ANRIL expression at the cell population level by Reverse Transcription followed by qPCR (RT-qPCR) at different locations across ANRIL using primers detecting only the spliced ANRIL (in exon-exon junctions), the unspliced ANRIL (within introns or intron-exon junctions), or both spliced and unspliced ANRIL referred to as total ANRIL (within exons) (Fig. 1D). We also found that ANRIL spliced isoforms increase more than total or unspliced ANRIL measured in the first part of the gene (from i1 to i5) (Fig. 1D). Moreover, the second half of total or unspliced ANRIL transcripts (measured in i7-e8 to e20-i20) increases more than its first part (Fig. 1D), consistent with our RNA-seq analysis (Fig. S3).
Taken together, these data indicate that in WI38 cells undergoing RAF1-induced senescence, ANRIL expression increases, and show a complex regulation among its transcript variants.
ANRIL stability increases in RAF1-induced senescence
To investigate how ANRIL expression increases in RAF1-induced senescence, we monitored transcription by capture of nascent transcripts. We found that p15 and p16 transcription increases in senescence (Fig. 2A). Such increase is comparable to the increase of their steady-state mRNA levels (see total RNA levels as compared to nascent RNA levels). In contrast, ANRIL transcription increased only weakly, compared to ANRIL RNA expression (measured in the exon 5, Fig. 2A). This result suggests that the stability of ANRIL RNA might also be affected by senescence progression. We thus monitored ANRIL half-life in proliferative and senescent cells following transcription inhibition [55–57]. As controls, we found that the stability of p15 or p16 mRNAs is not affected during senescence induction (Fig. 2B). In contrast, ANRIL stability is increased, with its half-life of about 2 h in proliferative cells (as previously described [58]) increasing to more than 5 h in senescent cells. Thus, Fig. 2 data indicate that the increase in ANRIL expression in WI38 cells undergoing RAF1-induced senescence is due to both an increase in its transcription and an increase in its stability.
Figure 2.
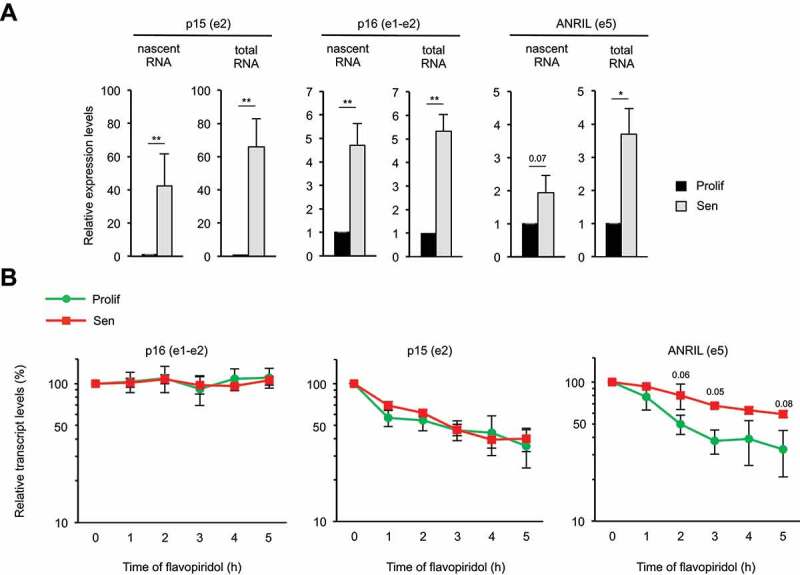
ANRIL is stabilized in senescence
(A) WI38 hTERT RAF1-ER cells were induced to senescence or maintained proliferative, as indicated. Seventy-two hours later, cells were treated with EU for 1 h, total RNAs were extracted and EU labelled nascent RNAs were purified using the Click-iT technology. Total and nascent p15, p16 and ANRIL levels were measured by RT-qPCR using the primers indicated in parentheses. The means and standard deviations from three independent experiments are shown, relative to GAPDH (e9) and normalized to 1 in proliferative cells for each experiment. Significant differences are indicated with asterisks (*: p value < 0.05, ** to ****: p values < 10−2, 10−3 and 10−4, respectively; two-sided paired Student’s t-test on log2 values) or by the number of the p value when it is between 0.05 and 0.1. (B) WI38 hTERT RAF1-ER cells were induced to senescence or maintained proliferative, as indicated. Seventy-two hours later, cells were treated with flavopiridol to inhibit transcription for the indicated times. p15, p16 and ANRIL expression was monitored by RT-qPCR using the indicated primers at the indicated times following flavopiridol addition. The levels of RNA were normalized to those of GAPDH (e9) and then normalized to 100% at 0 time point for each experiment. The means and standard deviations from three independent experiments are shown (logarithmic scale). Significant differences were calculated as in (A); not significant p values are not indicated.
Circular ANRIL RNAs are strongly induced in senescence
Many circular ANRIL isoforms containing the exon 5 have been identified [41–43]. Circular RNAs are more stable than linear RNAs [31,32,38], and hence, we hypothesized that the increase in ANRIL stability could be due to an increase in circular ANRIL species containing the exon 5. To identify circular forms of ANRIL in RAF1-induced senescence, we performed random primed RT followed by PCR using outward-facing primers in different exons (Fig. S6). After purification and sequencing of the PCR products, we identified four circular ANRIL species (Fig. S6), which were previously reported [41–43]. We detected back-splicing from exon 6 to exon 5, from exon 7 to exon 5, and from exon 16 to exon 5. These data indicate that circular ANRIL species produced by back-splicing at exon 5 are readily detected in RAF1-induced senescent WI38 cells.
Interestingly, circANRIL e7-e5 and circANRIL e16-e5 are strongly induced in senescence, measured by RT-qPCR using a reverse primer in exon 5 associated with a primer spanning the back-spliced junction (Fig. 3A-B). However, we did not pursue analysis of circANRIL e6-e5 isoform because we were not able to specifically detect this back-spliced junction by RT-qPCR (data not shown).
Figure 3.
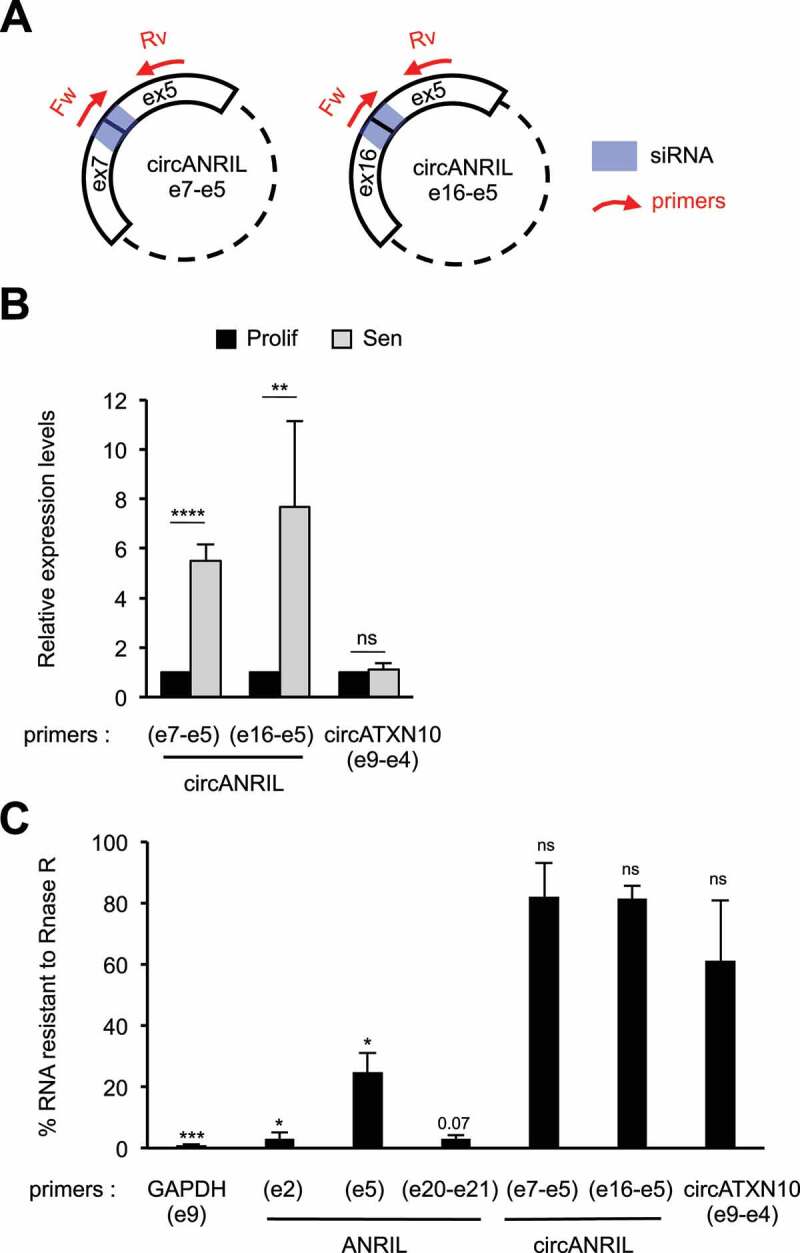
Circular ANRIL species are strongly induced in senescent cells
(A) Schematization of the circular ANRIL isoforms containing the specific back-spliced junctions that we identified in our study (Fig. S6). The primers and siRNAs targeting these back-spliced junctions are shown in red and blue, respectively. Note that although these primers give a unique PCR product, they can detect all circular species containing these specific back-spliced junctions, irrespective of the number or identity of spliced exons or introns located between exon 5 and the reversed spliced exon. (B) Total RNA was extracted from proliferative or senescent WI38 hTERT RAF1-ER cells. The expression of circular ANRIL species was measured by RT-qPCR using the indicated primers. CircATXN10 was measured as a control whose expression does not change in RAF1-induced senescence. The levels of RNA were normalized to those of GAPDH (e9) and then normalized to 1 in proliferative cells for each experiment. The means and standard deviations from four independent experiments are shown. Significant differences are indicated with asterisks (*: p value < 0.05, ** to ****: p values < 10−2, 10−3 and 10−4, respectively; two-sided paired Student’s t-test on log2 values), the number of the p value is indicated when it is between 0.05 and 0.1; ns: not significant. (C) Total RNAs from senescent WI38 hTERT RAF1-ER cells were treated or not with RNase R to digest all linear RNAs, and analysed by RT-qPCR. Percentages of RNAs resistant to RNase R treatment were calculated relative to the levels of these RNAs measured in untreated samples. Primers designed for e20-e21 junction detect both spliced and unspliced products, the intron 20 being very small (93 nt). Means and standard deviations from three independent experiments are shown (only two for ANRIL e20-e21, circANRIL e7-e5 and circANRIL e16-e5 primers). Significant differences are indicated as in (B).
To assess the proportion of circular ANRIL compared to total ANRIL, we first analysed splicing events in RNA-seq data. We detected 11 reads corresponding to back-spliced junctions, compared to 48 reads containing the e5-e6 junction (present in both linear and circular ANRIL) in senescent cells (Table S1), suggesting that about 25% of spliced ANRIL are present within circular species in senescent cells. To confirm this result, we used RNAse R, an exonuclease, which specifically degrades linear RNAs [59]. As expected, both circular forms of ANRIL analysed above (circANRIL e7-e5 and circANRIL e16-e5) are largely resistant to RNAse R degradation, whereas virtually all GAPDH mRNA is degraded by RNAse R (Fig. 3C). Strikingly, about 25% of ANRIL exon 5, which is included within all circular ANRIL species we identified (Fig. S6), is resistant to RNAse R degradation in senescent cells. In contrast, ANRIL exon 2 and exon 20-exon 21 regions are almost completely degraded by RNase R treatment, suggesting that these regions are mostly included in linear species. Note that exon 20-exon 21 junction degradation is consistent with the fact that the last exon of a transcript cannot circularize as it does not possess splicing signals at its 3ʹ end. These data suggest that around 25% of ANRIL exon 5 is included within circular species in senescent cells. Altogether, these data indicate that significant amounts of circular ANRIL species are produced during RAF1-induced senescence progression of WI38 fibroblasts.
Oncogenic RAF1-, MEK1- or RAS-induced senescence leads to a specific regulation of ANRIL expression
Although cell cycle arrest is complete at 3 days after 4-HT addition in this RAF1-induced senescence model [45], we wondered whether the changes in gene expression that we observed remain stable after 3 days of senescence induction. By performing kinetics of senescence induction up to 15 days, we measured the expression of total (measured in ANRIL exon 5) and circular ANRIL transcripts, p21 mRNA (which encodes another Cyclin/CDK inhibitor, p21) and INK4 mRNAs (p15, p16 and p14ARF). Their expression increases up to 6–9 days following 4-HT addition (Fig. 4A). After this time point, their levels remain stable up to 15 days of 4-HT treatment (Fig. 4A), indicating that ANRIL RNAs, as INK4 mRNAs, are not transiently induced in response to RAF1 oncogenic stress but are rather stably associated with the senescent state.
Figure 4.
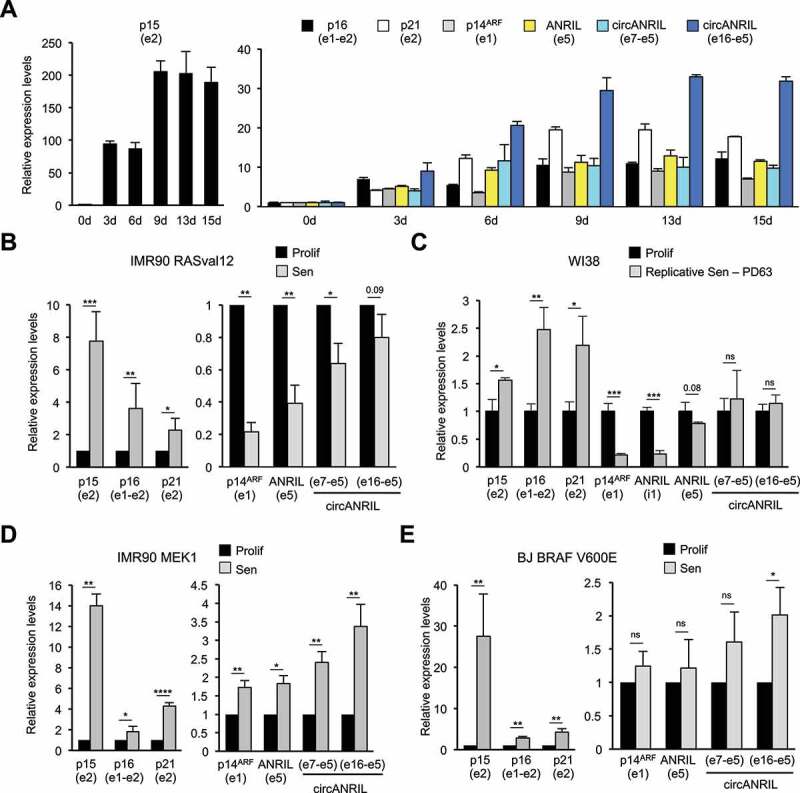
ANRIL expression follows p14ARF expression in different models of OIS
(A) Long kinetics of RAF1-induced senescence. WI38 hTERT RAF1-ER cells were treated with 4-HT for the indicated days (d). RNA expression was analysed by RT-qPCR using the primers indicated in parentheses. The means and standard deviations from the PCR triplicates of one representative experiment out of 2 are shown, relative to GAPDH and normalized to 1 in proliferative cells. (B) IMR90 hTERT RASval12-ER cells were induced to senescence by 4-HT treatment (Sen) or maintained proliferative (Prolif) for 8 days. Total RNA was extracted, and RNA expression was measured by RT-qPCR using the primers indicated in parentheses. The means and standard deviations from four independent experiments are shown, relative to GAPDH and normalized to 1 in proliferative cells for each experiment. Significant differences are indicated with asterisks (*: p value < 0.05, ** to ****: p values < 10−2, 10−3 and 10−4, respectively; two-sided paired Student’s t-test on log2 values); the number of the p value is indicated when it is between 0.05 and 0.1; ns: not significant. (C) WI38 cells were grown until replicative senescence (Sen, 63 population doublings (PD)) or arrested while proliferating (Prolif). RT-qPCR experiments were performed as in (B) from three independent experiments, except that data were normalized to 1 after calculating the means and two-sided unpaired Student’s t-tests were applied. (D) IMR90 hTERT ΔMEK1-ER cells were induced to senescence by 4-HT addition or maintained proliferative for 3 days. RT-qPCR experiments were performed as in (B) from three independent experiments. (E) BJ hTERT BRAF V600E cells were induced to senescence by doxycycline addition or maintained proliferative for 3 days. RT-qPCR experiments were performed as in (B) from three independent experiments.
Conversely, a decrease in ANRIL expression was reported in a RASval12 model of OIS and in replicative senescence [18,19]. Therefore, we asked whether increased ANRIL expression is specific to RAF1-induced senescence of WI38 cells or whether we did not analyse the same ANRIL transcript isoforms. Using the same primers (measuring total ANRIL in the exon 5 or circANRIL isoforms), we monitored ANRIL expression in an IMR90 cell line expressing an ER-RASval12 fusion allowing the induction of senescence following 4-HT treatment. We confirmed that, as previously published in WI38 cells induced in senescence by activated H-RasG12V [18], total ANRIL expression decreased about 3 fold in activated RAS-induced senescence (Fig. 4B). Circular ANRIL expression also decreased although to a lesser extent (Fig. 4B).
We also monitored ANRIL expression in WI38 cells undergoing replicative senescence. Although unspliced ANRIL decreases and total ANRIL expression slightly decreases, consistently with previous observation [19], circular ANRIL levels remain stable (Fig. 4C). From these experiments, we conclude that ANRIL expression is differentially regulated in senescence with respect to senescence inducers.
To investigate whether increased ANRIL expression is restricted to RAF1-induced senescence, we used other in vitro models of OIS. We induced senescence in IMR90 hTERT ΔMEK1-ER cells by a 3-day treatment with 4-HT [50] or in BJ BRAF V600E cells by a 3-day treatment with doxycycline [48], and monitored the expression of ANRIL. In IMR90 hTERT ΔMEK1-ER cells, the expression of total ANRIL and circular ANRIL increases with senescence (Fig. 4D). In BJ BRAF V600E cells, whereas total ANRIL levels do not change much, the expression of circANRIL e16-e5 is significantly induced in senescence and, although it is not significant, the expression of circANRIL e7-e5 is induced in all three experiments (Fig. 4E). These results indicate that the increase in total or circular ANRIL expression is not specific to RAF1-induced senescence nor to WI38 cells.
Notably, changes in ANRIL expression (up-regulation or down-regulation) in these various models of senescence parallels that of p14ARF, consistent with the fact that they are produced from divergent promoters (Fig. 4). By contrast, p15, p16 and p21 gene expression always increases (Fig. 4). In RAF1-induced senescence, short kinetics of senescence induction indicate that ANRIL induction occurs concomitantly with p14ARF induction or even slightly earlier, and after the induction of p15 and p16 (Fig. S7A). Finally, in all the senescence models we tested, we observed that circular ANRIL expression increases more or decreases less than total ANRIL expression, suggesting either that ANRIL circularization is more efficient or that circular ANRIL is stabilized in senescent cells (Fig. 4).
Taken together, these data suggest that ANRIL regulation by RAF1 is linked to the senescence process per se and not to RAF1 activation. To further demonstrate this point, we inhibited endogenous RAF1 in parental WI38 cells. This does not affect ANRIL production (Fig. S7B), suggesting that ANRIL is not directly targeted by RAF1-dependent pathways. Moreover, we prevented senescence induction using previously characterized siRNAs against p16 and p21 mRNAs [45]. Depletion of p16 and p21 abolishes the activation of both total and circular ANRIL expression induced by RAF1 activation in WI38 cells (Fig. S7C), therefore demonstrating that it is linked to the process of senescence.
ANRIL participates to INK4-protein-coding gene activation in RAF1-induced senescence
Next, we investigated the role of ANRIL and its circular RNA species in RAF1-induced senescence. To test whether they could regulate the expression of the gene to which ANRIL is an antisense, the p15-encoding gene, we first transfected senescent cells with an siRNA targeting exon 6 of ANRIL. This siRNA efficiently decreases total ANRIL expression measured in exon 5 as well as the expression of the two circular ANRIL species analysed above (Fig. 5A). Surprisingly, considering the published role of ANRIL in repressing p15-encoding gene expression in proliferative cells, ANRIL depletion in senescent cells leads to a decrease in p15-encoding gene expression (Fig. 5A and S8A). This was confirmed at the protein level by western blotting (Fig. S8B). This result is not due to an off-target effect of the siRNA used in these experiments, because it was confirmed using an siRNA targeting exon 19 of ANRIL associated to another control siRNA (Fig. S8C).
Figure 5.
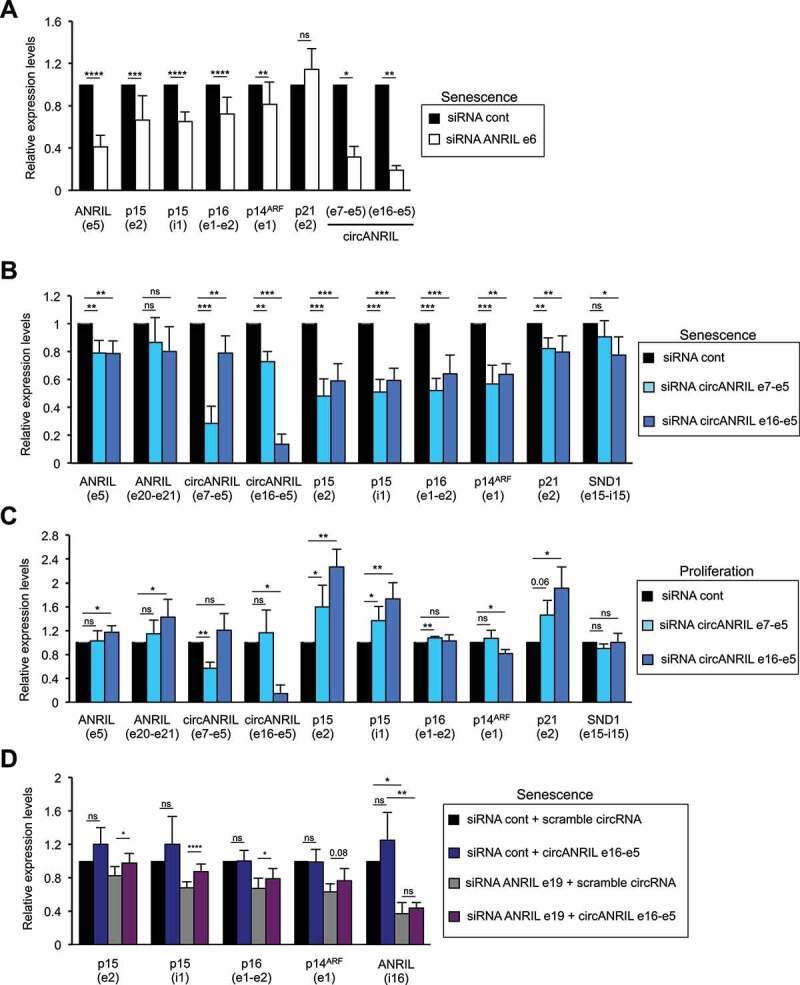
Circular ANRIL species participate in INK4 gene regulation in proliferative or senescent cells
(A) Senescent WI38 hTERT RAF1-ER cells were transfected with the indicated siRNA. Seventy-two hours later, total RNA was extracted. ANRIL, p15, p16, p14ARF and p21 mRNA expression was measured by RT-qPCR using the primers indicated in parentheses, calculated relative to GAPDH and normalized to 1 for the control siRNA in each experiment. The means and standard deviations from 3 to 14 independent experiments are shown, depending on the primers. Significant differences are indicated with asterisks (*: p value < 0.05, ** to ****: p values < 10−2, 10−3 and 10−4, respectively, two-sided paired Student’s t-test on log2 values); the number of the p value is indicated when it is between 0.05 and 0.1; ns: not significant. (B) Same as in (A), except that the indicated siRNAs were used. (C) Proliferative WI38 hTERT RAF1-ER cells were transfected using the indicated siRNAs and analysed as in (A). (D) Senescent WI38 hTERT RAF1-ER cells were transfected with the indicated siRNAs. 24 to 72 h later, the cells were transfected with the indicated in vitro synthesized circular RNAs. 24 h later, total RNA was extracted and analysed by RT-qPCR using the primers indicated in parentheses. The means and standard deviations from four independent experiments are shown. Significant differences are indicated as in (A).
The p15-encoding gene is located within the INK4 locus, which also contains the CDKN2A gene encoding proteins important for senescence induction: the p16 Cyclin/CDK inhibitor as well as by a distinct promoter, p14ARF, an upstream activator of the p53 pathway. We thus analysed the role of ANRIL on the expression of the p16 and p14ARF mRNAs in senescence. Depletion of ANRIL decreases p16 and p14ARF but not p21 (Fig. 5A and S8C), indicating that in senescent cells, ANRIL specifically increases the expression of the senescence-associated genes at the INK4 locus.
Circular ANRIL species mediate INK4 gene regulation in proliferative and RAF1 induced-senescent cells
Circular ANRIL species increase in RAF1 induced-senescent cells and siRNAs targeting exon 6 or exon 19 of ANRIL, both strongly affect circular ANRIL expression (Fig. 5A and S8C). We, therefore, hypothesized that circular ANRIL isoforms could play a role in INK4 gene regulation. To test this hypothesis, we used siRNAs targeting exon 7-exon 5 or exon 16-exon 5 back-spliced junctions (represented in Fig. 3A), which strongly and specifically affect the expression of circANRIL e7-e5 and circANRIL e16-e5, respectively (Fig. 5B-C). Importantly, they only weakly affect ANRIL exon 5, consistent with our estimation that approximately 25% of exon 5 is present within the diverse circular ANRIL isoforms and thus, each individual circular species accounts for only a small fraction of total exon 5. Moreover, as expected, they do not affect the expression of linear ANRIL species (measured in exon 20-exon 21 junction, which, as shown in Fig. 3C, is present within linear species) (Fig. 5B-C).
Depletion of both circular isoforms in RAF1-induced senescence decreased the expression of p15, p16 and p14ARF INK4 genes whereas it had only a weak effect, if any, on the expression of p21 and SND1 (as a control) genes (Fig. 5B). Similar results were obtained when using another siRNA against circANRIL e16-e5 and a control scrambled siRNA (Fig. S9A). Most importantly, transfection of in vitro produced RNAs containing the e16-e5 backspliced junction (see Fig. S9B-C and materials and methods for details of its preparation) reverses the effect of siRNAs decreasing circANRIL e16-e5 in RAF1-induced senescence (Fig. 5D and S8C), ruling out off-target effects of siRNAs.
In proliferative cells, depletion of circANRIL e16-e5 and circANRIL e7-e5 both increase p15 expression, while p16, p14ARF and SND1 expression is largely unaffected (Fig. 5C). These results, which are consistent with the known function of ANRIL in repressing the INK4 gene locus in proliferative cells [18,19], suggest that circular isoforms of ANRIL participate in the repression of the p15 gene in proliferative cells. Unexpectedly, the depletion of the two circular ANRIL species also increases the expression of p21 mRNA, suggesting that they can repress anti-proliferative genes in trans (Fig. 5C).
Thus, these data indicate that circular species of ANRIL are responsible, at least in part, for the repression of p15 expression in proliferative WI38 fibroblasts, as well as for the full induction of all INK4 genes during RAF1-induced senescence progression.
Altogether, these data suggest that at least two circular ANRIL isoforms shift from being repressors of p15 expression in proliferative cells to activators of INK4 genes in RAF1-induced senescent cells.
CircANRIL e16-e5 regulates H3K27 modifications at the p16 gene promoter in RAF1-induced senescence
Next, we investigated how circular ANRIL species positively regulate INK4 gene expression in RAF1-induced senescent cells. The observation that circANRIL e7-e5 or e16-e5 RNAs favour p15 intron expression (Fig. 5B-D and S9A) suggests that p15 activation occurs transcriptionally. To test this possibility, we monitored nascent p15 and p16 RNAs and found that their expression is decreased upon total ANRIL or circANRIL e7-e5 or e16-e5 depletion, indicating that they control transcription of INK4 genes in RAF1-induced senescence (Fig. S10A). Furthermore, we found that depleting circANRIL e16-e5 leads to a decrease in RNA pol II recruitment at the p15 and p16 promoters (Figure S10B). Of note, we did not analyse p14ARF promoter because it is also the promoter of ANRIL, which could affect the results.
We next analysed the consequences of depleting circANRIL e16-e5 in RAF1-induced senescent cells on chromatin marks known to regulate INK4 gene expression. Among them, H3K27 methylation and its counteracting modification, H3K27 acetylation, have both been involved in INK4 gene regulation [26,27]. We did not find any significant effect on H3K27 modifications at the p15 promoter (Fig. 6A). These results may be biased by the fact that the p15 gene is antisense and thus included within the ANRIL gene whose expression is increased in senescence. However, the presence of H3K27ac is decreased whereas the presence of H3K27me3 is increased at the p16 promoter upon circANRIL e16-e5 depletion in RAF1-induced senescent cells (Fig. 6A). Thus, circANRIL e16-e5 regulates H3K27 modifications, at least at the p16 promoter.
Figure 6.
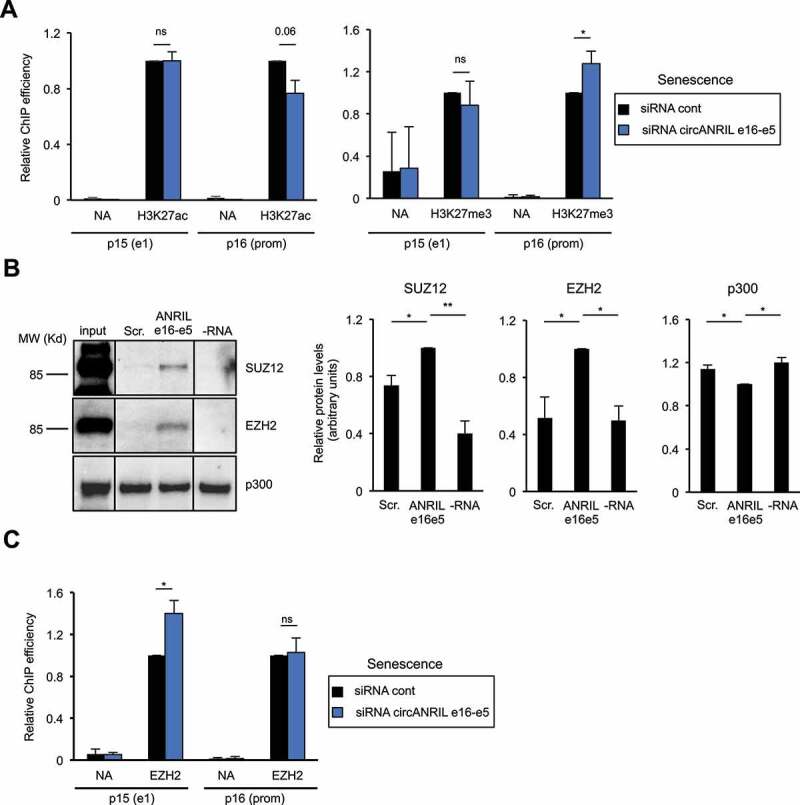
Circular ANRIL e16-e5 regulates H3K27me3 at the p16 promoter in senescent cells
(A) Senescent WI38 hTERT RAF1-ER cells were transfected with the circANRIL e16-e5 siRNA. Three days later, cells were harvested and subjected to a ChIP analysis using H3K27ac (left panel) or H3K27me3 (right panel) or total histone H3 antibody or no antibody (NA) as a control. The amount of p15 e1, p16 promoter (prom) or GAPDH e9 (used as a positive control since it also detects pseudogenes with significant H3K27ac and H3K27me3 levels) sequences was quantified by qPCR. The percentage of input of p15 and p16 sequences was calculated relative to 1 for the control siRNA sample following standardization with total nucleosome occupancy (H3 ChIP values) and GAPDH e9. The means and standard deviations from three independent experiments are shown. Significant differences are indicated by an asterisk (p value < 0.05, two-sided paired Student’s t-test on log2 values); the number of the p value is indicated when it is between 0.05 and 0.1; ns: not significant. (B) In vitro transcribed biotinylated RNAs either containing the ANRIL e16-e5 spliced junction or a scrambled sequence (Scr) were incubated with HeLa nuclear extracts. Biotinylated RNAs were then recovered using streptavidin beads and co-precipitated proteins were analysed by western blotting using SUZ12 or EZH2 antibodies, as indicated. Typical western blots are shown. Lines indicate unnecessary intervening lanes that have been spliced out. The histograms show the means and standard deviations of the means (SDOM) from eight independent experiments (for SUZ12 and EZH2) or five independent experiments (for p300) excluding outliers. Significant differences are indicated with asterisks (*: p value < 0.05, **: p values < 10−2, two-sided paired Student’s t-test on log2 values).C) Same as in (A), except that an anti-EZH2 antibody was used and only GAPDH e9 but not total nucleosome occupancy was used for standardization.
To test whether this effect could be direct, and given that in vitro produced RNAs containing the circANRIL e16-e5 junction can reverse the effect of siRNAs on INK4 gene expression (Fig. 5C), we checked whether enzymes regulating H3K27 modifications could interact with these in vitro produced RNAs. H3K27 tri-methylation is mediated by the histone methyltransferase EZH2, which is the catalytic subunit of the PRC2 Polycomb complex, whereas H3K27 acetylation is mediated by CBP/p300 [22]. We incubated in vitro produced biotinylated RNAs containing the circANRIL e16-e5 junction with nuclear extracts and recovered interacting proteins using streptavidin beads. No specific binding of p300 to circANRIL e16-e5 junction-containing RNAs is observed, p300 being pulled-down by streptavidin beads even in the absence of biotinylated RNA (Fig. 6B). In contrast, SUZ12 and EZH2 proteins are more efficiently recovered using RNAs containing the circANRIL e16-e5 junction than using control scramble RNAs (Fig. 6B). Actually, depending on the experiment, from 1.2% to 13% of the input EZH2 or SUZ12 are recovered with in vitro produced RNAs containing the circANRIL e16-e5 junction. Thus, these data indicate that RNAs containing the ANRIL e16-e5 backspliced junction can specifically interact with the SUZ12 and EZH2 Polycomb-group proteins.
The PRC2 complex containing EZH2 is directly recruited to the INK4 locus [27,28]. We thus investigated whether circANRIL e16-e5 regulates EZH2 recruitment to the INK4 locus. In RAF1-induced senescence, circANRIL e16-e5 depletion increases EZH2 recruitment at the p15 promoter whereas no effect is observed at the p16 promoter (Fig. 6C). These data suggest that circANRIL e16-e5 regulates the recruitment of EZH2 to the p15 promoter at the INK4 locus during RAF1-induced senescence, resulting in changes of H3K27me3 methylation at the p16 promoter, probably because of chromatin folding [60].
Altogether, these data indicate that some circular species of ANRIL favour the transcription of p15- and p16-encoding genes in RAF1-induced senescence probably by sequestering EZH2 and preventing its recruitment to some regions of the INK4 locus, therefore decreasing the presence of the repressive H3K27 trimethylation epigenetic mark.
CircANRIL e16-e5 is important for the genetic program of senescence
Given the importance of INK4 genes in the control of cell proliferation and senescence induction, we next analysed whether circular ANRIL species are important for these processes. Depletion of total ANRIL, circANRIL e16-e5 and e7-e5 in proliferative cells leads to a decrease in their proliferating capacity measured by reduced EdU staining (Fig. S11), in agreement with the increased expression of p15 and p21 (Fig. 5C). Depletion of these RNAs in senescent cells, alone or in combination with p16 and/or p21 inhibition does not increase the number of cells able to proliferate and does not decrease the presence of SAHF measured by DAPI heterogeneity (data not shown), suggesting that it is not sufficient to revert senescence. Given that trans effects of ANRIL on gene expression were reported [23], we analysed whether it could be important for the genetic program associated with senescence besides INK4 gene regulation. We thus performed RNA-seq experiments following depletion of circANRIL e16-e5 in senescence and performed a differential gene expression analysis. We found 296 up-regulated genes and 294 down-regulated genes by more than two folds (see Table S2 for the list of regulated genes). A gene ontology analysis for these up- and down-regulated genes is shown in Tables S3 and S4, respectively. Of note, with respect to senescence features, genes induced upon circANRIL e16-e5 depletion are enriched in inflammation-linked genes and include three genes from the SASP (genes coding for IL6, IL15RA and FGF), whereas genes down-regulated are linked to regulation of cell proliferation.
To test whether circANRIL e16-e5 could be important for the genetic program associated with senescence, we crossed these lists with genes differentially regulated upon senescence induction in the same cells [44]. Strikingly, amongst the 296 genes repressed by circANRIL e16-e5 (i.e., upregulated upon circANRIL e16-e5 depletion), 115 are also repressed in senescence. This is much more than expected by chance, a difference, which is highly significant (53 expected, p-value 1.85*10−21, chi2 test). Similarly, 78 genes activated by circANRIL e16-e5 (downregulated upon circANRIL e16-e5 depletion) are also activated in senescence, again significantly different from what randomly expected (26 expected, p-value: 2.81*10−27). These data indicate that circANRIL e16-e5 is important for the genetic program associated with RAF1-induced senescence.
Discussion
Here, we show that, contrary to the proposed model based on the analyses of ANRIL expression changes in Rasval12-induced and replicative senescence [18,19], senescence is not always associated with a decrease in the expression of ANRIL. Instead, we observe in various models of OIS that ANRIL expression increases during senescence (Fig. 1 and 4). This indicates that, depending on the inducer of senescence, the cell response at the INK4 locus is different, at least for ANRIL and p14ARF expression. Indeed, although RAS activation is upstream RAF1 and MEK1 activation in the ERK MAP kinase hyperactivation pathway [61], RAF1-induced senescence differs from RASval12-induced senescence. In contrast to RAF1-induced senescence, senescence following RASval12 activation requires replication stress, ROS activation and a proliferation burst before entering senescence [45]. The flattened cell morphology of RAS-induced senescent cells also differs from RAF1-induced senescent cells, which adopt a fusiform cell morphology [45]. In addition to the activation of the RAF-MEK-ERK pathway, RAS activation was found to activate other pathways, which could explain the differences observed between RAF1- and RASval12-induced senescent phenotypes [61].
In addition to changes in ANRIL expression, our data uncover a complex regulation among its transcript variants, which we did not fully investigate in this manuscript. Obtaining a complete picture of the whole spectrum of ANRIL variant expression changes associated with senescence is warranted and would require further investigation. In this manuscript, we focused on two circular ANRIL species, given that the function of circular isoforms of ANRIL was not previously investigated during senescence induction.
Our data also indicate that a decrease in ANRIL expression is not absolutely required for p15 induction during senescence. Indeed, p15 is strongly induced during RAF1-, BRAF V600E- and MEK1-induced senescence, while a decrease in ANRIL expression does not occur (Fig. 1 and 4). This indicates that the relief of p15 silencing by Polycomb complexes could be mediated by other mechanisms than the decrease of ANRIL expression, such as the decrease of Polycomb-group protein expression [25–29]. Moreover, recent data demonstrate the critical role played by the MSK kinase, which, by phosphorylating H3S28, relieves Polycomb-mediated repression at the INK4 locus in OIS [62].
In agreement with the previously reported function of ANRIL on INK4 gene expression in proliferative cells [18,19], we found in our cell model that circular ANRIL represses p15 expression in proliferative cells. However, we further describe that ANRIL participates in the activation of INK4 genes during RAF1-induced senescence (Fig. 5). Other studies support the role of ANRIL in the activation of the expression of the INK4 locus. As such, inhibiting ANRIL expression has been reported to slightly decrease p15 expression in colon cancer cells [8]. Moreover, deletion of the mouse region orthologous to the human chromosome 9p21 CAD risk locus, which is localized in the second half of the ANRIL human gene [5], leads to reduced INK4 gene expression [63]. Thus, the fact that ANRIL can participate in the full activation of INK4 gene expression [8,63], as we show here during RAF1-induced senescence, could provide the basis for data showing a positive correlation between ANRIL and protein-coding INK4 gene expression in normal and pathological tissues [4,41,64–70].
Here, we also uncover that circular ANRIL expression can be regulated during a physiological cell response, i.e. senescence induced by the activation of oncogenes. Indeed, we identified two circular ANRIL species, circANRIL e7-e5 and e16-e5, whose expression increases in RAF1-induced senescence as well as in MEK1- or BRAF V600E-induced senescence (Fig. 3 and 4). This increase in circular ANRIL isoforms, at least in the RAF1-induced model of senescence, likely explains the increase of total ANRIL stability measured in exon 5 (Fig. 2B). RNA circularization can be regulated by the binding of specific factors to linear RNAs [35,71], but also by the rate of transcription elongation [38]. ANRIL circularization could thus be differentially controlled between proliferative and senescent cells by specific interactions with back-splicing regulators. However, it is tempting to speculate that increased production of circular ANRIL species is due to a change in the transcription elongation rate at the ANRIL locus during RAF1-induced senescence. Indeed, we have previously shown that transcription elongation rate is increased at some loci in RAF1-induced senescence [44].
Most importantly, we demonstrate that these circular isoforms of ANRIL play an important function and can actually recapitulate ANRIL functions on INK4 gene expression regulation as previously shown for ANRIL in proliferative cells [18,19], and in RAF1-induced senescence (this study). Indeed, we found that these circular forms of ANRIL repress p15-encoding gene expression in proliferative cells and activate INK4 gene expression in RAF1-induced senescent cells (Fig. 5). Such a role of circANRIL isoforms in RAF1-induced senescence could also be shared in other models of OIS, such as in MEK1- and BRAF V600E-induced senescence, in which we show that the expression of circANRIL isoforms also increases. Note that we were not able to demonstrate whether linear ANRIL species can also mediate ANRIL function. Indeed, siRNAs targeting the first or the last spliced junction, which cannot be included in circular species, were inefficient (data not shown). Moreover, the formation of circular RNAs is largely post-transcriptional [38] and can occur after alternative splicing of their precursor linear RNAs [40,72]. Targeting the ANRIL exon 19, which is not included in the circular species identified here but decreases the level of all circular ANRIL species tested (Fig. S8C), indicates that ANRIL circularization occurs post-transcriptionally. As such, we were unable to specifically inactivate linear RNAs without affecting the circular isoforms because linear RNAs are actually precursors of the circular isoforms.
A key question is what could be the mechanisms by which circular ANRIL isoforms regulate INK4 gene transcription and switch from being repressors of p15 to activators of all INK4 genes during RAF1-induced senescence progression?
In proliferative cells, circANRIL isoforms could participate, together with other species of ANRIL [18], in the recruitment of Polycomb proteins at its site of transcription to repress the expression of the p15-encoding gene. Indeed, circANRIL e16-e5 can interact with Polycomb proteins (Fig. 6B). In proliferative cells, this mechanism seems to be restricted to the p15-encoding gene amongst INK4 genes, since p16- and p14ARF-encoding genes are not regulated by circANRIL species (Fig. 5C). This specificity could be related to the fact that ANRIL is antisense to p15 but not to the other INK4 genes and is consistent with previous observations for total ANRIL [18].
In RAF1-induced senescence, transfection of circANRIL e16-e5 produced in vitro reverses the effect of siRNAs suggesting that circANRIL e16-e5 can function in trans to regulate INK4 gene expression (Fig. 5D). Moreover, we found that it inhibits the binding of EZH2 to the p15 promoter (Fig. 6C). One hypothesis is that circANRIL e16-e5 competes with linear ANRIL isoforms localized at their transcription site for recruitment of PRC2 at this target site. Indeed, linear ANRIL isoforms carrying the e1-e2 junction have been shown to participate in PRC2 recruitment on the p15 gene [18], which is located on the antisense strand within the intron 1 of the ANRIL gene. On the p16 promoter, which is outside the ANRIL gene and where PRC2 recruitment is not affected by circANRIL e16-e5 depletion, H3K27 marks could be regulated by circANRIL by chromatin folding between the p15 and p16 promoters within the INK4 locus [60]. Of note, this mechanism could operate on other loci besides the INK4 locus, in particular on genes down-regulated upon circANRIL e16-e5 depletion. Such a model of competition for binding to an RNA binding protein is similar to what has been proposed for circANRIL function on ribosomal RNA maturation in atherosclerosis [42].
How then does ANRIL shift from being a repressor to an activator of INK4 gene expression during senescence progression? We propose that this is linked to the senescence-associated change in the relative ratio of Polycomb-group proteins and circular ANRIL species, resulting from the increase in circular ANRIL expression (this study) but also from the very strong decrease of Polycomb-group protein expression [25–29], including EZH2 [25,27,73]. In proliferative cells, the excess of Polycomb relative to ANRIL would ensure that all ANRIL species present at the INK4 locus locally recruit Polycomb proteins. Upon senescence induction, an increased amount of circular ANRIL species would sequester the residual Polycomb-group proteins outside of the INK4 locus and prevent them from being recruited to the INK4 locus. For example, this could occur while the excess of circular ANRIL RNAs is taken in charge by the RNA export machinery to the cytoplasm. Here, we thus propose a mechanism, relying on changes in the ratio between a non-coding RNA and its effector, by which a non-coding RNA can shift from being a negative to a positive regulator of the same genes upon commitment into a specific cell fate.
Supplementary Material
Acknowledgments
This work was supported by grants from the Institut National du Cancer (INCa, PLBio program), the Fondation ARC pour la Recherche sur le Cancer (programme ARC), the Fondation Toulouse Cancer Santé and the Ligue Nationale Contre le Cancer (LNCC, équipe labélisée) to DT and from the Fondation de France to LM. EN is employed by INSERM. LM was supported by fellowships from the LNCC, the Fondation ARC and the Fondation de France. MA was supported by a fellowship from the Fondation pour la Recherche Médicale (FRM), SL was supported by studentships from the Fondation ARC and JO by a studentship from the French Ministry of Research. We thank Malek Djabali and Jacques Côté for constructive discussions throughout the study, Céline Vallot and Claire Rougeulle for sharing their expertise on nuclear RNA FISH and Olivier Sordet for critically reading the manuscript.
Funding Statement
This work was supported by the Fondation ARC pour la Recherche sur le Cancer [PGA120150202320]; Fondation de France [185742_ 00087515]; Institut National du Cancer [DA N°2012-123]; Fondation Toulouse Cancer Santé [DRUGSPEED 2018-2]; Ligue Nationale Contre le Cancer [EL-190350].
Abbreviations
- ncRNAs,
non-coding RNAs
- OIS,
oncogene-induced senescence
- ANRIL,
antisense non-coding RNA in the INK4 locus
- CDK,
cyclin-dependent kinase
- CAD,
coronary artery diseases
- circRNAs,
circular RNAs
- SAHF,
senescence–associated heterochromatin foci
- PRC,
Polycomb repressive complex
- SASP,
- senescence-associated secretory phenotype
- ER,
estrogen receptor
- hTERT,
human telomerase reverse transcriptase
- 4-HT,
4-hydroxy-tamoxifen
- RNA pol II,
RNA polymearse II
- EdU,
5-ethynyl-2’-deoxyuridine
- EU,
5-ethynyl uridine
- RT-qPCR,
reverse transcription-quantitative PCR
- ChIP,
chromatin immunoprecipitation
- FISH,
fluorescence in situ hybridization
- RNA-seq,
RNA sequencing
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental material
Supplemental data for this article can be accessed here.
References
- [1].Magistri M, Faghihi MA, St Laurent G., et al.. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 2012;28(8):389–396. 3rd et al. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Aguilo F. Zhou MM and Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71(16):5365–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7(9):667–677. [DOI] [PubMed] [Google Scholar]
- [4].Pasmant E, Laurendeau I, Heron D, et al.. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67(8):3963–3969. DOI: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- [5].Broadbent HM, Peden JF, Lorkowski S, et al.. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17(6):806–814. DOI: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- [6].Pasmant E, Sabbagh A, Vidaud M, et al.. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. Faseb J. 2011;25(2):444–448. DOI: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- [7].Congrains A, Kamide K, Ohishi M, et al.. ANRIL: molecular mechanisms and implications in human health. Int J Mol Sci. 2013;14(1):1278–1292. DOI: 10.3390/ijms14011278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Naemura M, Tsunoda T, Inoue Y, et al.. ANRIL regulates the proliferation of human colorectal cancer cells in both two- and three-dimensional culture. Mol Cell Biochem. 2016;412(1–2):141–146. DOI: 10.1007/s11010-015-2618-5. [DOI] [PubMed] [Google Scholar]
- [9].Wan G, Mathur R, Hu X, et al.. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013;25(5):1086–1095. DOI: 10.1016/j.cellsig.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen D, Zhang Z, Mao C, et al. ANRIL inhibits p15(INK4b) through the TGFbeta1 signaling pathway in human esophageal squamous cell carcinoma. Cell Immunol. 2014;289:91–96. [DOI] [PubMed] [Google Scholar]
- [11].Zhu H, Li X, Song Y, et al.. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem Biophys Res Commun. 2015;467(2):223–228. DOI: 10.1016/j.bbrc.2015.10.002. [DOI] [PubMed] [Google Scholar]
- [12].Zhou X, Han X, Wittfeldt A, et al.. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-kappaB pathway. RNA Biol. 2016;13(1):98–108. DOI: 10.1080/15476286.2015.1122164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Simboeck E, Ribeiro JD, Teichmann S, et al.. Epigenetics and senescence: learning from the INK4-ARF locus. Biochem Pharmacol. 2011;82(10):1361–1370. DOI: 10.1016/j.bcp.2011.07.084. [DOI] [PubMed] [Google Scholar]
- [14].Campisi J, d’Adda Di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740. [DOI] [PubMed] [Google Scholar]
- [15].Bodnar AG, Ouellette M, Frolkis M, et al.. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–352. DOI: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- [16].Salama R, Sadaie M, Hoare M, et al.. Cellular senescence and its effector programs. Genes Dev. 2014;28(2):99–114. DOI: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kuilman T, Michaloglou C, Mooi WJ, et al.. The essence of senescence. Genes Dev. 2010;24(22):2463–2479. DOI: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kotake Y, Nakagawa T, Kitagawa K, et al.. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. DOI: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yap KL, Li S, Munoz-Cabello AM, et al.. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. DOI: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12(2):123–135. [DOI] [PubMed] [Google Scholar]
- [21].Masui O, RN HE. A and protein actors in X-chromosome inactivation. Cold Spring Harb Symp Quant Biol. 2006;71:419–428. [DOI] [PubMed] [Google Scholar]
- [22].Aranda S. Mas G and Di Croce L. Regulation of gene transcription by Polycomb proteins. Sci Adv. 2015;1(11):e1500737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Holdt LM, Hoffmann S, Sass K, et al.. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9(7):e1003588. DOI: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sato K, Nakagawa H, Tajima A, et al.. ANRIL is implicated in the regulation of nucleus and potential transcriptional target of E2F1. Oncol Rep. 2010;24(3):701–707. DOI: 10.3892/or_00000910. [DOI] [PubMed] [Google Scholar]
- [25].Agger K, Cloos PA, Rudkjaer L, et al.. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23(10):1171–1176. DOI: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Agherbi H, Gaussmann-Wenger A, Verthuy C, et al.. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS One. 2009;4(5):e5622. DOI: 10.1371/journal.pone.0005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bracken AP, Kleine-Kohlbrecher D, Dietrich N, et al.. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21(5):525–530. DOI: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kotake Y, Cao R, Viatour P, et al.. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 2007;21(1):49–54. DOI: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gil J, Bernard D, Martinez D, et al.. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6(1):67–72. DOI: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- [30].Wilusz JE. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 2017;14(8):1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Enuka Y, Lauriola M, Feldman ME, et al.. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44(3):1370–1383. DOI: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jeck WR, Sorrentino JA, Wang K, et al.. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. DOI: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Salzman J, Gawad C, Wang PL, et al.. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. DOI: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang Y, Zhang XO, Chen T, et al.. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. DOI: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- [35].Conn SJ, Pillman KA, Toubia J, et al.. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. DOI: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- [36].Rybak-Wolf A, Stottmeister C, Glazar P, et al.. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. DOI: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- [37].You X, Vlatkovic I, Babic A, et al.. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–610. DOI: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang Y, Xue W, Li X, et al.. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15(3):611–624. DOI: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- [39].Qu S, Zhong Y, Shang R, et al.. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. DOI: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li X, Yang L, Chen LL, et al.. Functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. [DOI] [PubMed] [Google Scholar]
- [41].Burd CE, Jeck WR, Liu Y, et al.. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. DOI: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Holdt LM, Stahringer A, Sass K, et al.. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7(1):12429. DOI: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sarkar D, Oghabian A, Bodiyabadu PK, et al. Multiple isoforms of ANRIL in melanoma cells: structural complexity suggests variations in processing. Int J Mol Sci. 2017;18(7):1378. DOI: 10.3390/ijms18071378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Muniz L, Deb MK, Aguirrebengoa M, et al.. Control of gene expression in senescence through transcriptional read-through of convergent protein-coding genes. Cell Reports. 2017;21(9):2433–2446. DOI: 10.1016/j.celrep.2017.11.006. [DOI] [PubMed] [Google Scholar]
- [45].Jeanblanc M, Ragu S, Gey C, et al.. Parallel pathways in RAF-induced senescence and conditions for its reversion. Oncogene. 2012;31(25):3072–3085. DOI: 10.1038/onc.2011.481. [DOI] [PubMed] [Google Scholar]
- [46].Lazorthes S, Vallot C, Briois S, et al.. A vlincRNA participates in senescence maintenance by relieving H2AZ-mediated repression at the INK4 locus. Nat Commun. 2015;6(1):5971. DOI: 10.1038/ncomms6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhu J, Woods D, McMahon M, et al.. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12(19):2997–3007. DOI: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Carvalho C, L’Hote V, Courbeyrette R, et al. Glucocorticoids delay RAF-induced senescence promoted by EGR1. J Cell Sci. 2019;132.132(16):jcs230748. DOI:10.1242/jcs.230748. [DOI] [PubMed] [Google Scholar]
- [49].Young AR, Narita M, Ferreira M, et al.. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23(7):798–803. DOI: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Collado M, Gil J, Efeyan A, et al.. Tumour biology: senescence in premalignant tumours. Nature. 2005;436(7051):642. DOI: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- [51].Wingett SW, FastQ Screen AS. A tool for multi-genome mapping and quality control. F1000Res. 2018;7:1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li H, Handsaker B, Wysoker A, et al.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. DOI: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Anders S. Pyl PT and Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yu W, Gius D, Onyango P, et al.. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–206. DOI: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bensaude O. Inhibiting eukaryotic transcription: which compound to choose?. How to Evaluate Its Activity? Transcription. 2011;2:103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in Vivo. J Biol Chem. 2001;276(34):31793–31799. [DOI] [PubMed] [Google Scholar]
- [57].Rahl PB, Lin CY, Seila AC, et al.. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. DOI: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tani H, Mizutani R, Salam KA, et al.. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Research. 2012;22(5):947–956. DOI: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Panda AC, Gorospe M. Detection and analysis of circular RNAs by RT-PCR. Bio Protoc. 2018;8(6):e2775. DOI: 10.21769/BioProtoc.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hirosue A, Ishihara K, Tokunaga K, et al.. Quantitative assessment of higher-order chromatin structure of the INK4/ARF locus in human senescent cells. Aging Cell. 2012;11(3):553–556. DOI: 10.1111/j.1474-9726.2012.00809.x. [DOI] [PubMed] [Google Scholar]
- [61].Rajalingam K, Schreck R, Rapp UR, et al.. Ras oncogenes and their downstream targets. Biochim Biophys Acta. 2007;1773(8):1177–1195. DOI: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- [62].Culerrier R, Carraz M, Mann C, et al.. MSK1 triggers the expression of the INK4AB/ARF locus in oncogene-induced senescence. Mol Biol Cell. 2016;27(17):2726–2734. DOI: 10.1091/mbc.e15-11-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Visel A, Zhu Y, May D, et al.. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464(7287):409–412. DOI: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cunnington MS, Santibanez Koref M, Mayosi BM, et al.. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6(4):e1000899. DOI: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Drak Alsibai K, Vacher S, Meseure D, et al. High positive correlations between ANRIL and p16-CDKN2A/p15-CDKN2B/p14-ARF gene cluster overexpression in multi-tumor types suggest deregulated activation of an ANRIL-ARF bidirectional promoter. Noncoding RNA. 2019;5(3):44. DOI: 10.3390/ncrna5030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Folkersen L, Kyriakou T, Goel A, et al. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of Eight New ANRIL Splice Variants. PLoS One. 2009;4:e7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Holdt LM, Beutner F, Scholz M, et al.. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30(3):620–627. DOI: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- [68].Jarinova O, Stewart AF, Roberts R, et al.. Functional analysis of the chromosome 9p21. 3 Coronary Artery Disease Risk Locus. Arterioscler Thromb Vasc Biol. 2009;29(10):1671–1677. DOI: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- [69].Liu Y, Sanoff HK, Cho H, et al.. Expression of p16 INK4a in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8(4):439–448. DOI: 10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Liu Y, Sanoff HK, Cho H, et al.. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4(4):e5027. DOI: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Li X, Liu C-X, Xue W, et al.. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Molecular Cell. 2017;67(2):214–227 e217. DOI: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- [72].Wilusz JE. A 360 degrees view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9(4):e1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ito T, Teo YV, Evans SA, et al.. Regulation of cellular senescence by polycomb chromatin modifiers through distinct DNA damage- and histone methylation-dependent pathways. Cell Rep. 2018;22(13):3480–3492. DOI: 10.1016/j.celrep.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


