Abstract
Barbara McClintock first hypothesized that interspecific hybridization could provide a “genomic shock” that leads to the mobilization of transposable elements (TEs). This hypothesis is based on the idea that regulation of TE movement is potentially disrupted in hybrids. However, the handful of studies testing this hypothesis have yielded mixed results. Here, we set out to identify if hybridization can increase transposition rate and facilitate colonization of TEs in Saccharomyces cerevisiae × Saccharomyces uvarum interspecific yeast hybrids. Saccharomyces cerevisiae have a small number of active long terminal repeat retrotransposons (Ty elements), whereas their distant relative S. uvarum have lost the Ty elements active in S. cerevisiae. Although the regulation system of Ty elements is known in S. cerevisiae, it is unclear how Ty elements are regulated in other Saccharomyces species, and what mechanisms contributed to the loss of most classes of Ty elements in S. uvarum. Therefore, we first assessed whether TEs could insert in the S. uvarum sub-genome of a S. cerevisiae × S. uvarum hybrid. We induced transposition to occur in these hybrids and developed a sequencing technique to show that Ty elements insert readily and nonrandomly in the S. uvarum genome. We then used an in vivo reporter construct to directly measure transposition rate in hybrids, demonstrating that hybridization itself does not alter rate of mobilization. However, we surprisingly show that species-specific mitochondrial inheritance can change transposition rate by an order of magnitude. Overall, our results provide evidence that hybridization can potentially facilitate the introduction of TEs across species boundaries and alter transposition via mitochondrial transmission, but that this does not lead to unrestrained proliferation of TEs suggested by the genomic shock theory.
Keywords: hybridization, transposable elements, transposition rate, Ty element, Saccharomyces
Significance
When two different species mate and produce hybrid offspring, the union of the two genomes may result in transposable element (TE) mobilization due to the disruption of TE regulation in the hybrid. We test the hypothesis that TE mobilization is increased in hybrids, as has been supported by some studies, but rejected by others. We find no evidence for increased mobilization of TEs in hybrid yeast, however, we show that which species you inherit your mitochondria from does change transposition rate.
Introduction
Transposable elements (TEs) are mobile, repetitive genetic elements that have colonized nearly every organism across the tree of life. TEs self-encode machinery to either replicate or excise themselves from one genomic location and re-insert at another genomic location, which can disrupt genes or gene expression and promote chromosomal rearrangements through ectopic recombination. Due to the high potential of fitness costs of these mutations, most organisms have evolved host defense systems to regulate TEs (Rebollo et al. 2012). However, although experiments and population genetics show that the average effect of TE insertions is deleterious, individual transposition events may be neutral or even advantageous (Wilke et al. 1992; González and Petrov 2009; Stoebel and Dorman 2010; Van’t Hof et al. 2016; Hope et al. 2017; Li et al. 2018; Esnault et al. 2019; Niu et al. 2019). Far from their historical status of “junk DNA,” TEs are now known to contribute to a variety of processes including telomere maintenance (Pardue and DeBaryshe 2011), centromere structure (Casola et al. 2008; Carbone et al. 2012; Gao et al. 2015; Kursel and Malik 2016; Jangam et al. 2017), sex chromosome evolution (Bachtrog 2003; Ellison and Bachtrog 2013; Dechaud et al. 2019), regulation of gene expression, evolution of genome size, karyotype, and genomic organization across the tree of life (Petrov 2002; Jiang et al. 2004; Gregory and Johnston 2008; Pellicer et al. 2014; Schubert and Vu 2016; Kapusta et al. 2017; Bourque et al. 2018; Thybert et al. 2018).
The type and number of TEs in a genome vary between populations and species, as do the regulatory systems organisms use to suppress TEs (Bourque et al. 2018). In her Nobel prize lecture in 1983, Barbara McClintock hypothesized that hybridization between different populations or species could act as a “genomic shock” that initiates TE mobilization that could lead to the formation of new species (McClintock 1984). This idea revolves in part around the idea that hybridization could cause a de-repression of TE regulation, perhaps by mismatch of the repression system in the hybrid genome. Evidence supporting this hypothesis is mixed. Initial excitement centered on the hybrid dysgenesis system in Drosophila melanogaster, where an intraspecific cross between a strain carrying the P-element transposon to a strain without P-elements produced sterile offspring (Kidwell et al. 1977; Bingham et al. 1982; Kidwell 1983; Rose and Doolittle 1983; Bucheton et al. 1984). However, attempts to test this model of transposon induced speciation across other species of Drosophila demonstrated this applied in certain crosses but not others (Coyne 1985, 1986, 1989; Hey 1988; Lozovskaya et al. 1990; Labrador et al. 1999; Kelleher et al. 2012). Studies in the Arabidopsis species complex are similarly mixed, with evidence that crosses between Arabidopsis thaliana and Arabidopsis arenosa lead to an upregulation of the retrotransposon ATHILA, the level of which is linked to hybrid inviability (Josefsson et al. 2006); but crosses between A. thaliana and A. lyrata show no change in expression of TEs in interspecific hybrids (Göbel et al. 2018). Iconic studies in desert sunflowers revealed that three independent hybrid species formed by crosses of Helianthus annuus and Helianthus petiolaris had elevated copy number of long terminal repeat (LTR) retrotransposons compared with their parent species (Ungerer et al. 2006, 2009; Staton et al. 2009). However, contemporary crosses of the same Helianthus parental species did not lead to large scale proliferation of TEs, although the TEs remain transcriptionally active (Kawakami et al. 2011; Ungerer and Kawakami 2013; Renaut et al. 2014). From all of these studies, there is evidence that hybridization in some cases can lead to a misregulation of the TE repression system and potential proliferation of TEs, but it remains unclear how widespread this phenomenon is and what factors contribute to this process.
In this study, we use Saccharomyces cerevisiae × Saccharomyces uvarum interspecific hybrids as a system to explore the hypotheses that hybridization can lead to an increase in transposition of TEs, and that hybridization could provide an avenue for colonization of a genome by new TEs. Saccharomyces cerevisiae has been used as a model to understand retrotransposition for decades. Saccharomyces cerevisiae TEs are made up of LTR retrotransposons which fall into six families, Ty1, Ty2, Ty3, Ty3_1p, Ty4, and Ty5 (Kim et al. 1998; Carr et al. 2012). Ty elements make up a small fraction of the genome (<5%), with a total of approximately 50 full-length Ty elements and over 400 solo LTRs in the S. cerevisiae reference genome (Kim et al. 1998; Carr et al. 2012). Ty1 is the most abundant and well-studied Ty element, representing almost 70% of the full length TEs in the reference genome, with its closely related family Ty2 making up a further 25%. Ty1 preferentially integrates near genes transcribed by RNA Polymerase III through an association between integrase and Pol III-complexes (Mularoni et al. 2012). The other families are rare; Ty3 and Ty4 are thought to be active families (Hansen and Sandmeyer 1990; Hug and Feldmann 1996; Nelson et al. 2017), and no intact copies of Ty3_1p or Ty5 are known (Voytas and Boeke 1992; Carr et al. 2012).
Ty content and copy number vary across strains and species (Liti et al. 2005, 2009; Bleykasten-Grosshans et al. 2013), with Ty elements inherited vertically and horizontally (Liti et al. 2005; Carr et al. 2012; Bergman 2018; Czaja et al. 2020), and certain Ty families lost. For example, S. uvarum, a cold-tolerant species 20 million years divergent from S. cerevisiae, has no full-length Ty elements with the exception of the Ty4-like Tsu4 (which likely evolved from the Ty4/Tsu4 superfamily which gave rise to the Ty4 element in the S. cerevisiae/S. paradoxus lineage) (Neuvéglise et al. 2002; Liti et al. 2005; Bergman 2018). Although there are no intact copies of Ty1 elements in the S. uvarum reference genome assembly, there are a number of Ty1 and Ty2 solo LTRs, indicative of past retrotransposition events (Scannell et al. 2011).
Saccharomyces are particularly interesting because the clade has recently lost RNAi regulation of TEs (Drinnenberg et al. 2009). Instead, S. cerevisiae and S. paradoxus Ty1 is regulated through a novel mechanism, copy number control (CNC) (Garfinkel et al. 2003, 2016; Saha et al. 2015; Ahn et al. 2017). A truncated form of the Ty-encoded Gag capsid protein (p22) disrupts virus-like particle assembly in a dose-dependent manner, allowing high levels of retrotransposition when few Ty1 elements are present and inhibiting transposition as copy number increases (Garfinkel et al. 2005; Saha et al. 2015). However, re-introducing the proteins Dicer and Argonaute of Naumovozyma castellii to S. cerevisiae can restore RNAi, and are sufficient to silence endogenous Ty retrotransposition (Drinnenberg et al. 2009). Saccharomyces uvarum and some strains of its close relative S. eubayanus are the only Saccharomyces species to still retain Dicer (Wolfe et al. 2015), but how this may contribute to Ty regulation is unclear. CNC is not well understood for Ty elements besides Ty1, nor is it known how CNC functions in other species of Saccharomyces outside of S. cerevisiae and S. paradoxus (Moore et al. 2004; Czaja et al. 2020).
Here, we use Ty-specific sequencing and transposition assays in lab-created interspecific hybrids to understand how hybridization impacts Ty mobilization. We show that hybridization does not lead to an increase in transposition rate or proliferation of Ty1 elements in hybrids. However, we do document variation in transposition rate in hybrids that is mediated through a curious phenomenon of mitochondrial inheritance, such that hybrids with S. uvarum mitochondria have a lower rate of transposition than hybrids with S. cerevisiae mitochondria.
Materials and Methods
Strains and Plasmids Used
Strains YMD119 and YMD120 are haploid S. cerevisiae strains of GRF167 background (YMD119: MATα ura3-167; YMD120: MATα ura3-167). YMD119 is a high-Ty strain created by repeated induced transposition of Ty1, whereas YMD120 has a Ty1 profile similar to S288C (Scheifele et al. 2009). These strains were crossed to YMD366, a S. uvarum lab strain of background CBS7001, to create hybrids YMD130, and YMD129, respectively. Strains yCSH141 (MATα his3d200 ura3-167, Ty1his3AI-242 [chrXII]) and yCSH142 (MATα his3d200 ura3-167, Ty1his3AI-273 [chrII]) carry an integrated, marked Ty1 element for use in transposition assays (gifts from Mary Bryk, see Bryk et al. 1997). yCSH141 and yCSH142 were crossed to yCSH143 (MATa his3-del200 ura3-52) to create S. cerevisiae diploids yCSH144 and yCSH145, and to yCSH189 (MATahoΔ::KAN lys2-1his3Δ::Hyg) to create S. cerevisiae × S. uvarum hybrids (yCSH192, 193, 195–198) for transposition assays. Strain yCSH182 (MATa dcr1Δ::KanMX hoΔ::NatMX) was provided by Chris Hittinger. yCSH182 was modified to knockout HIS3, yCSH187 (MATa dcr1Δ::KanMX hoΔ::NatMX his3Δ::Hyg). yCSH187 was crossed to yCSH141 to create hybrid yCSH671 with a S. uvarum dcr1 knockout for transposition assays. Strains yCSH215, yCSH216, and yCSH217 are ρ0 (mtDNA absent) versions of yCSH141, yCSH142, and yCSH189, respectively, which were created via passage on ethidium bromide. yCSH215 was crossed to yCSH189 to form hybrids yCSH218-220; yCSH216 was crossed to yCSH189 to form hybrids yCSH221–223; yCSH217 was crossed to yCSH141 to form hybrids yCSH224–226; and yCSH217 was crossed to yCSH142 to create hybrids yCSH227–229. These hybrids were used in transposition assays to test the role of mitochondrial inheritance. The Ty1his3AI plasmid was a gift from David Garfinkel, as used in Curcio and Garfinkel (1991) (see supplementary table S1, Supplementary Material online, for a list of all strains used, crossing information, and their purpose in the study).
Survey of S. uvarum Ty Elements
We downloaded sequencing reads for 54 S. uvarum isolates (Almeida et al. 2014), and used several programs for TE detection. We used the program deviaTE (Weilguny and Kofler 2019) with a custom Ty element consensus library based on the library utilized in Carr et al. (2012) and supplemented with Tsu4 sequence from (Neuvéglise et al. 2002), as per Bergman (2018). We used default parameters, and the option –rpm to normalize sequencing coverage for comparison across samples. We summarized the coverage by average coverage, percent of the query with coverage >0, and percent of the query with coverage >25 (supplementary table S2, Supplementary Material online). We also employed RetroSeq version 1.41 (Keane et al. 2013) on a subset of these samples (samples with paired-end read sequencing data) to call novel insertions in the S. uvarum genome. Each call was manually inspected using Integrative Genomics Viewer (Robinson et al. 2011).
TySeq Library Creation and Sequencing
DNA was extracted using the Hoffman–Winston protocol (Hoffman and Winston 1987), cleaned using the Zymo Clean and Concentrate kit (Zymo Research, Irving, CA), and quantified on the Qubit fluorometer. To identify Ty elements, we took a sequencing based approach modified from previous methods (van Opijnen et al. 2009; Mularoni et al. 2012), which we call TySeq. The library preparation was based on previously described methods (Wetmore et al. 2015; Sanchez et al. 2019), modified as described here (supplementary fig. S1, Supplementary Material online, for detailed protocol, supplementary table S3, Supplementary Material online, for primers). About 1 µg of genomic DNA was sheared to an average size of 800 bp using a Covaris machine with default settings. The sheared DNA fragments were blunt ended, and A-tails were added to the fragments to ligate the Illumina adapter sequences.
We used a nested PCR approach, in which we first attempted to amplify full-length Ty1 and Ty2 elements using custom primers designed to target sequences interior to Ty1 and Ty2 elements, avoiding the LTR sequences (see supplementary table S3, Supplementary Material online, for primers used, supplementary fig. S10, Supplementary Material online), and custom indexed primers that target the Illumina adapter sequence were used to enrich for genomic DNA with Ty1 and Ty2 insertion sites. We designed a single primer, Ty1_3prime2R (supplementary table S3, Supplementary Material online), which is 27 bp long, and identically matches the sequence of about half of the annotated Ty1 elements in the S. cerevisiae S228C reference genome (supplementary fig. S10, Supplementary Material online). The primer differs from other Ty1 and Ty2 elements at four sites. We tested the Ty1_3prime2R primer via PCR for known genomic Ty1 and Ty2 elements, and the Ty1his3AI reporter construct utilized in later experiments and confirm that it successfully amplifies sequence from these elements despite sequence differences.
The second PCR used the product from PCR#1 with the same indexed primer that binds the Illumina adapter, and a second primer that binds the Ty1 and Ty2 LTR and adds the second Illumina adapter (supplementary fig. S1, Supplementary Material online). The resulting libraries were quantified on a Qubit and run on a 6%TBE gel to assess library size. Libraries were sequenced on an Illumina NextSeq 500 (150 bp PE) using a custom R1 sequencing primer (LTRseqF) that binds the Ty1 and Ty2 LTR. Due to the low complexity of the libraries, libraries were never allowed to exceed 10–15% of a sequencing run.
TySeq of induced transposition with the marked Ty1 was produced as above, except using a primer that binds to HIS3 instead of Ty1 (see supplementary table S3, Supplementary Material online, for primers used). Strain CSH153 was transformed with the Ty1his3AI plasmid and crossed to S. uvarum strain CSH6 to create strain CSH177. Biological replicates of CSH177 were grown overnight in C-URA media to maintain the plasmid, then a small number of cells were used to inoculate 48 replicates of 1 ml C-URA + 2% galactose, which was grown for 2 days at 20 °C. Replicates were then pooled together and plated on C-HIS plates. Plates were scraped and pooled together to be used for DNA library preparation.
TySeq Sequencing Analysis
We took a stringent approach to filtering TySeq reads for alignment. First, R1 reads were cropped to 27 bp in length using trimmomatic v0.32 (Bolger et al. 2014) and aligned to a Ty element reference genome, which contained all annotated LTR and Ty elements in the S. cerevisiae S288C reference genome (obtained from SGD, last updated January 13, 2015), using bwa aln (Li and Durbin 2009). Only reads mapping to this Ty reference genome were used in later steps. To better understand which elements we were sequencing, we identified all unique 27 bp reads and identified LTR and Ty elements with 100% match of the reads (no gaps, mismatches, full 27 bp matching) using blast+ version 2.2.29 blastn “blastn-short” (Altschul et al. 1990; Camacho et al. 2009) with the same database of all annotated solo LTR and Ty elements from S. cerevisiae used above (obtained from SGD, last updated January 13, 2015). We identified reads mapping to all annotated Ty1 and Ty2 elements (supplementary table S4, Supplementary Material online), which suggests that we are capturing a diversity of Ty elements in the genome. We find an overrepresentation of the sequence “ATTATCTCAACATTCACCCATTTCTC” in our sequencing in all samples, which matches a subset of Ty1 elements including the Ty1 element on the Ty1his3AI plasmid. We note this is in line with an increased number of Ty1 elements derived from the Ty1his3AI plasmid in strain YMD130. The most common sequences and the Ty elements they match are included in supplementary table S4, Supplementary Material online.
We then subset all 150 bp reads to only reads that mapped to the Ty reference genome using seqtk subseq (https://github.com/lh3/seqtk). These full-length R1 reads then had the first 27 bp cropped using trimmomatic to remove the LTR-specific sequence from the read. A second filtering step was taken to remove all reads mapping to Ty elements using the same approach as above. This step may remove a percentage of real inserts, due to the nature of Ty element insertions to occur nested within other Ty elements. However, due to the sequencing design, a portion of the reads are expected to derive from full-length Ty element, so filtering these reads out aids in unique read mapping. Finally, reads not mapping to Ty elements were aligned to the reference genome, sacCer3 or Sbay.ultrascaf (Scannell et al. 2011). Only positions with a read depth of 50 reads were considered likely insertions. All potential inserts were visually inspected using Integrative Genomics Viewer (Robinson et al. 2011) and we confirmed a subset of the insertions using PCR. Genome coverage in 25 bp intervals was assessed using igvtools count (Robinson et al. 2011). Overlap of Ty elements between different samples was assessed using bedtools “window,” and proximity to sequence features was assessed using bedtools “closest” (Quinlan and Hall 2010).
Transposition Rate Assays
Transposition rate was measured in strains with an integrated Ty1 tester Ty1his3AI as has been previously described (Curcio and Garfinkel 1991; Bryk et al. 1997; Dunham et al. 2015). A strain was grown overnight, then cell count was assessed by hemacytometer. Approximately 2500 cells were diluted in 10 ml of YPD then inoculated in 100 µl volume in a 96-well plate, such that there were less than 500 cells per well. The plate was sealed with a breathable membrane and incubated without shaking at 20 °C for 4 days. All exterior wells were discarded. C-HIS plates were prepared for the assay by drying via blotting with sterile Watson filter paper or incubation in a 30-incubator for 2 days. Three wells were titered on YPD plates to assess population size and the remaining wells entire contents were individually, independently spotted onto very dry C-HIS plates and left to incubate at 30 °C for 3 days. Patches were scored as zero or nonzero. Each assay examined on average 57 patches, with at least two biological replicates. Transposition rate was scored via a maximum-likelihood method (Lea and Coulson 1949).
Whole Genome Sequencing of Selected Hybrids
Based on results from transposition assays, four strains were selected for whole genome sequencing (yCSH195, yCSH198, yCSH193, yCSH196). Strains were grown up overnight, and a portion of each was used to start new transposition assays. The remaining cells had DNA extracted using the Hoffman Winston protocol followed by library preparation using the Illumina Nextera library kit. The samples were sequenced on an Illumina NextSeq 500 and reads were aligned to a concatenated reference genome of S. cerevisiae and S. uvarum (Scannell et al. 2011) using bwa mem and default parameters (Li and Durbin 2009). Read depth was assessed using igvtools (Robinson et al. 2011) and normalized to account for average genome wide coverage. Read depth per homolog was used to detect copy number change in the hybrid.
Plate Reader Assay
We used a BioTek Synergy H1 plate reader to assay growth rate by measuring OD600 every 15 min at 25 °C with agitation over the course of 60 h. Three replicates of each strain (CSH218, 219, 221, 222, 224, 225, 227, 228) were grown in rich media (YPD), and three replicates of each strain were grown in media with glycerol as the sole carbon source (YPG).
Statistical Analyses
Statistics were conducted using R packages “dplyr,” “FSA,” and “car.”
Results
Variation in Ty Element Content in Isolates of S. uvarum
Characterization of the CBS7001 lab strain of S. uvarum determined that S. uvarum was devoid of full-length Ty elements with the exception of Tsu4 (Bon et al. 2000; Neuvéglise et al. 2002; Liti et al. 2005; Scannell et al. 2011). We conducted a bioinformatics-based survey of 54 worldwide isolates from natural and fermentation conditions (Almeida et al. 2014) to identify if the characterization of CBS7001 was representative of the species as a whole. We largely confirm S. uvarum to be missing full-length Ty elements, with the exception of Tsu4, which was present in almost every isolate surveyed (supplementary table S2, Supplementary Material online). Several strains have potential full-length Ty2 elements and partial Ty1 and Ty2 elements, and some of these strains have introgressions derived from S. eubayanus, S. kudriavzevii, and S. cerevisiae. Given the history of hybridization in many of these strains, we sought to identify if hybridization could provide a possible mechanism for Ty elements to insert in a species’ genome.
TySeq, a Sequencing Method for Detecting De Novo Transposable Element Insertions
Detecting TEs in sequencing data is notoriously difficult. Their repetitive nature and large size (e.g., the Ty1 is approximately 6 kb) present major challenges to genome assembly, and traditional alignment pipelines will miss new insertions due to their absence in the reference genome. There have been many advances in the computational detection of TEs using short read sequencing data (Ewing 2015; Rishishwar et al. 2017), and long-read sequencing will likely represent the new gold standard for TE annotation (Disdero and Filée 2017; Bergman 2018; Kutter et al. 2018; Shahid and Slotkin 2020). However, there is still a wide range of false positives and false negatives associated with computational methods, and long-read sequencing is currently more expensive and less high-throughput than short read methods. We therefore present a method, TySeq, adapted from previous methods (van Opijnen et al. 2009; Mularoni et al. 2012), which can identify novel or nonreference Ty1 element insertions. Although we apply this to Ty1 and Ty2 elements in Saccharomyces specifically, it is easily adapted to support the detection of other TEs in other organisms.
Briefly, we created a sequencing library quite similar to traditional whole genome sequencing library methods with small modifications (supplementary fig. S1, Supplementary Material online). We started with a sheared genomic library of 800 bp, large enough to span the LTR region of Ty elements and capture flanking genomic sequence. We created a biased library by using primers that amplify DNA fragments which contain a full-length Ty1 or Ty2 element (supplementary table S3, Supplementary Material online). We then used a custom sequencing primer that sequences off the LTR, capturing the flanking genomic region. These reads can be mapped back to a reference genome, thus identifying locations of new, nonreference, and reference TE insertions.
We applied TySeq to S. cerevisiae × S. uvarum hybrid strains to demonstrate proof of principle (fig. 1, supplementary figs. S2–S4, Supplementary Material online). We identified 52 putative Ty1 and Ty2 elements (read depth of 50+ reads supporting, supplementary table S5, Supplementary Material online) in the S. cerevisiae sub-genome of a hybrid strain (hereafter, “control hybrid”). Although the strain background differs from the S. cerevisiae reference genome, we find a similar number of Ty1 and Ty2 elements present. We additionally utilized a “high-Ty” hybrid, in which the S. cerevisiae portion of the genome carries a higher load of Ty1 elements derived from repeated induction of transposition using a synthetic construct (Scheifele et al. 2009). We identified 71 putative Ty1 and Ty2 elements (read depth of 50+ reads supporting, supplementary table S5, Supplementary Material online) in the S. cerevisiae sub-genome of this high-Ty hybrid. We then created a synthetic mixed population (90% control hybrid, 10% high-Ty hybrid) to test the sensitivity of our TySeq protocol in detecting low frequency Ty insertions. We detected 87 Ty1 and Ty2 elements in the synthetic mixed sample, largely recapitulating Ty elements derived from both the control hybrid (49/52 elements detected at a read depth of 50+ reads) and high-Ty hybrid (69/72 elements detected at read depth of 50+ reads), indicating we can detect most Ty elements which are only present in 10% of a population. We detected in 8/87 in the mixed sample but not in either control or high-Ty strain and 5/87 were present in control and/or high-Ty strain but not in mixed sample. The majority of these cases are the result of presence of an element with 50 or more reads in one sample, with reads between 1–49 read depth in the other sample(s) (supplementary table S5, Supplementary Material online). However, we should note that particular Ty elements are overrepresented in TySeq results, and it is difficult to uncouple whether this reflects an amplification bias or a biological basis (see Materials and Methods; supplementary table S4, Supplementary Material online). This may account for some of the variability we see between samples.
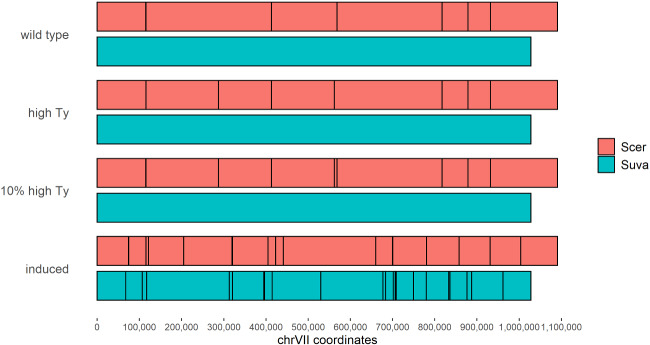
Fig. 1.
Using TySeq to identify Ty elements in S. cerevisiae × S. uvarum hybrids. Ty elements detected with TySeq are shown as black lines across chrVII for the S. cerevisiae (pink) and S. uvarum (blue) portions of a hybrid genome. Ty elements are shown for control (YMD129), high-Ty (YMD130), a mixed sample of 90% YMD129 and 10% YMD130, and a pool of His+ colonies obtained from induced transposition. No Ty elements were detected in the S. uvarum portion of the hybrid genome except when transposition was artificially induced (these insertions are plotted using S. uvarum genome coordinates). For whole genome figures, see supplementary figs. S2–S4, S6, and S7, Supplementary Material online. For coordinates of insertions, see supplementary tables S5–S7, Supplementary Material online.
We did not identify Ty1 or Ty2 elements in the S. uvarum sub-genome of these hybrid strains, consistent with the previously identified absence of full length Ty1 or Ty2 elements in the reference genome of S. uvarum (fig. 1, supplementary figs. S2–S4, Supplementary Material online). This furthermore suggests that new insertions do not occur early in the outgrowth of the colony from a single hybrid zygote.
We next sought to identify if we could induce transposition and detect novel insertions in a hybrid genome, and in particular, if insertions would occur in the S. uvarum sub-genome. We used a marked Ty1 element, Ty1his3AI on a plasmid under galactose induced expression (Curcio and Garfinkel 1991). This construct has a full-length Ty1 element with a HIS3 reporter gene interrupted with an artificial intron. Upon transposition, the intron is spliced out, restoring functionality to HIS3 and allowing detection of transposition events by growth on media lacking histidine (supplementary fig. S5, Supplementary Material online). We sequenced two replicates of a pool of His+ colonies and detected 23,693 and 31,083 reads mapping to the S. cerevisiae sub-genome, and 33,427 and 45,272 reads mapping to the S. uvarum sub-genome. We identified 93 and 122 insertions in the S. cerevisiae sub-genome respectively (with 50+ reads, supplementary table S6, Supplementary Material online, fig. 1, supplementary figs. S6 and S7, Supplementary Material online), with many of these sites differing from those identified in the control and high-Ty hybrid. A similar number of insertions were identified in the S. uvarum sub-genome, with 121 and 109 insertions detected, respectively (fig. 1, supplementary figs. S6, S7, and table S7, Supplementary Material online). These results suggest that Ty1 is equally likely to insert into either S. cerevisiae or S. uvarum genomes.
In S. cerevisiae, Ty1 elements preferentially insert near PolIII transcribed genes, like tRNAs (Mularoni et al. 2012). Here, we show that in the two replicates, 83.68% and 88.55% of reads that map to the S. uvarum genome are within 2 kb of an annotated tRNA gene. This is similar to the 93.6% reported for S. cerevisiae (Mularoni et al. 2012), suggesting the insertion preference for Ty1 is conserved despite 20 Myr divergence between the two species. The discrepancy between S. cerevisiae and S. uvarum might be due in part to differences in annotation between the two species reference genomes (there are fewer tRNA genes annotated in the S. uvarum reference). Our results thus show that Ty1 elements can insert in the S. uvarum genome and suggest that hybridization may be a mechanism through which TEs could hop from one species genome to another. However, backcrossing to S. uvarum following hybridization would be necessary for the establishment of Ty1 in the S. uvarum species, and further work is needed to explore this mechanism generally.
Variable Transposition Rate in Hybrids
We then directly measured transposition rate in S. cerevisiae × S. uvarum hybrids to test the hypothesis that transposition is increased in interspecific hybrids. We used S. cerevisiae strains which have a marked Ty1 element, Ty1his3AI, integrated on chrII and chrXII, respectively (supplementary table S1 and fig. S5, Supplementary Material online). These marked S. cerevisiae strains were crossed to an unmarked S. cerevisiae strain to create diploids, and to an unmarked S. uvarum strain to make hybrids. Transposition rate was scored via the fluctuation method (Lea and Coulson 1949). Briefly, a small number of cells were inoculated into independent cultures and allowed to grow for 4 days at 20 °C. Each culture was individually spotted on to selective media (agar plates lacking histidine), and then each patch was scored for the presence or absence of His+ colonies. Transposition rate was scored as the natural log of the number of patches with no His+ colonies divided by the population size of the culture.
Transposition rate is dependent on the location of the marked Ty1 element, and can depend upon ploidy, where diploids may have a lower rate of transposition compared with haploids due to MATa/α repression (Elder et al. 1981; Herskowitz 1988; Garfinkel et al. 2005). We first repeated transposition assays in marked S. cerevisiae haploids and recapitulate previously published results, that S. cerevisiae haploid Ty1his3AI strains have transposition rates of 10−6–10−7 per generation (Curcio and Garfinkel 1991, 1992; Bryk et al. 1997). We furthermore recapitulate results of similar haploid and diploid rates (table 1) (Garfinkel et al. 2005).
Table 1.
Variable Transposition Rate across Hybrids
| Strain Number | Ploidy, Species | Location of Marked Ty | Transposition Ratea (SE, replicate trials) |
|---|---|---|---|
| CSH141 | Haploid S. cerevisiae | chrXII | 1.6 × 10−7 (Bryk et al. 1997) |
| CSH142 | Haploid S. cerevisiae | chrII | 1.5 × 10−7 (Bryk et al. 1997) |
| CSH144 | Diploid S. cerevisiae | chrXII | 1.48 × 10−7 (NA, 1) |
| CSH145 | Diploid S. cerevisiae | chrII | 7.91 × 10−8 (3.76 × 10−8, 2) |
| CSH192 | Diploid hybrid | chrXII | 1.05 × 10−7 (4.60 × 10−9, 3) |
| CSH194 | Diploid hybrid | chrII | 4.22 × 10−8 (6.30 × 10−9, 2) |
| CSH195 | Diploid hybrid | chrXII | 0 (0, 2) |
| CSH196 | Diploid hybrid | chrXII | 5.08 × 10−8 (1.45 × 10-8, 2) |
| CSH193 | Diploid hybrid | chrII | 5.68 × 10−8 (3.17 × 10-8, 2) |
| CSH197 | Diploid hybrid | chrII | 4.53 × 10−8 (1.38 × 10−8, 3) |
| CSH198 | Diploid hybrid | chrII | 5.73 × 10−9 (1.12 × 10−9, 2) |
The rate of His+ prototroph formation per cell per generation, as determined by the maximum-likelihood method of Lea and Coulson (1949).
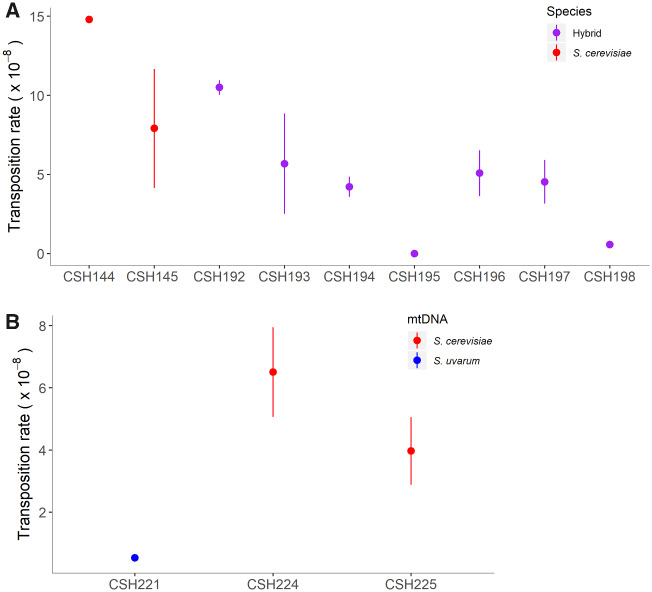
We tested transposition rate in seven independent hybrid crosses (fig. 2A, table 1, supplementary table S1, Supplementary Material online). We clearly show that hybridization does not increase transposition rate in the genetic background tested, with the highest rate of transposition observed in hybrids at approximately 1.05 × 10−7 (±4.60 × 10−9), similar to rates in haploid S. cerevisiae, ranging to undetectably low levels of transposition (scored as a rate of 0).
Fig. 2.
Variable transposition rate in hybrids. (A) Transposition rate in S. cerevisiae diploids (red) and in interspecific hybrids (purple) (see table 1 for transposition rate, error, and replicates). (B) Transposition rate in hybrids from controlled crosses, with S. uvarum mtDNA (blue) or S. cerevisiae mtDNA (red) (see table 2 for transposition rate, error, and replicates).
We tested the hypothesis that the maintenance of one of the RNAi genes, Dicer (DCR1), in S. uvarum may be responsible for the absence of most Ty elements in that species. DCR1 is absent in S. cerevisiae, so hybrids would normally have only the single S. uvarum copy of DCR1. We created a hybrid with a S. uvarum dcr1 knockout. If DCR1 mediates transposition rate, we would expect that dcr1 hybrids would have an increased transposition rate. Instead, we found the rate in these hybrids to be 5.44 × 10−8 (±5.26 × 10−9), similar to the rate observed in hybrids with an intact copy of S. uvarum DCR1 (table 1).
Although we did not identify increased transposition in hybrids, we did identify significant variation in transposition rate between hybrids (one-way ANOVA, F6 = 7.16, P = 0.0049). Hybrids should be isogenic within a cross, and between crosses should only be differentiated by the marked Ty1 element residing on chrII or chrXII. Differences in transposition rate between independent hybrid matings could result from copy number variation resulting from genomic instability following hybridization, a point mutation or insertion/deletion that occurred during the grow up of the culture for the transposition assay, or differential mitochondrial inheritance.
To identify the causal variants contributing to transposition rate variation in these hybrids, we selected strains that exhibited a low transposition rate (yCSH195, yCSH198), and strains with a diploid-like transposition rate (yCSH193, yCSH196) for whole genome sequencing. We identified a loss of part of the S. cerevisiae copy of chrXII in yCSH195, which resulted in the loss of the marked Ty1, hence the observed rate of 0 (supplementary fig. S8, Supplementary Material online). We did not identify any other copy number variants, point mutations, or insertion/deletions in the remaining strains; however, we observed that the other hybrid with low transposition rate (yCSH198) inherited the S. uvarum mitochondrial genome (mtDNA), whereas the other strains (yCSH193, yCSH196) inherited the S. cerevisiae mtDNA. mtDNA is inherited from one parent (uniparental inheritance) in almost all sexual eukaryotes (Birky 1995, 2001), including the Saccharomyces yeasts. Previous work has observed a transmission bias in S. cerevisiae × S. uvarum hybrids, which typically inherit the S. cerevisiae mtDNA, although there are a variety of genetic and environmental factors that contribute to mtDNA inheritance such as temperature and carbon source (Marinoni et al. 1999; Lee et al. 2008; Hsu and Chou 2017; Hewitt et al. 2020). Mitotype can affect a number of phenotypes, such as temperature tolerance in yeast hybrids (Baker et al. 2019; Li et al. 2019; Hewitt et al. 2020), but to our knowledge has not been previously implicated in transposition.
Saccharomyces uvarum mtDNA Decreases Transposition Rate in S. cerevisiae × S. uvarum hybrids
We set out to test the hypothesis that mitotype can influence transposition rate in hybrids by creating a set of crosses with controlled mtDNA inheritance. We induced strains of S. cerevisiae and S. uvarum to lose their mtDNA (denoted as ρ0) through passage on ethidium bromide, then crossed these ρ0 strains to the corresponding species with mtDNA intact. We conducted transposition assays in these newly created hybrids and demonstrate that the inheritance of S. uvarum mtDNA results in a significantly lower transposition rate (Welch’s two-sample t-test, P = 0.0039; fig. 2B, table 2). A series of growth curves on fermentable and nonfermentable carbon sources illustrates that S. uvarum mtDNA is still functioning in respiration, although results in a slightly slower growth rate than the identical strain with S. cerevisiae mtDNA (supplementary fig. S9, Supplementary Material online).
Table 2.
Saccharomyces uvarum mtDNA Decreases Hybrid Transposition Rate by an Order of Magnitude
| Strain Number | Ploidy, Species | mtDNA | Transposition Ratea (SE, replicate trials) |
|---|---|---|---|
| CSH218 | Diploid hybrid | S. uvarum | 0 (0, 2) |
| CSH221 | Diploid hybrid | S. uvarum | 5.28 × 10−9 (1.09 × 10−9, 3) |
| CSH224 | Diploid hybrid | S. cerevisiae | 6.51 × 10−8 (1.44 × 10−8, 3) |
| CSH225 | Diploid hybrid | S. cerevisiae | 3.97 × 10−8 (1.09 × 10−8, 3) |
The rate of His+ prototroph formation per cell per generation, as determined by the maximum-likelihood method of Lea and Coulson (1949).
Discussion
In summary, we combined a modified sequencing strategy, TySeq, with in vivo transposition rate assays to test the hypothesis that TE mobilization may be increased in interspecific hybrids. Using an integrated, marked Ty element construct to quantify transposition rate, we identified significant variation in transposition rate among strains that we expected to be isogenic. We show that mitochondrial inheritance can explain this variation, with S. uvarum mtDNA decreasing transposition rate in hybrids by an order of magnitude. Thus, although we reject the hypothesis that hybridization increases TE mobilization, we demonstrate hybridization can impact transposition rate in novel ways.
Intrinsic and Extrinsic Variables That Affect Transposable Element Movement
There is considerable variation in TE content across species and between populations, and many extrinsic and intrinsic factors that mediate transposition rate. Both the rate and distribution of TEs are governed by their overall deleterious effect (Charlesworth and Langley 1989). All organisms have evolved defenses to limit TE movement, although these systems vary across species and include zinc-finger proteins, small RNA-based silencing strategies, DNA methylation, and chromatin modifications (Rebollo et al. 2012). TE elements and their host defense systems continue to evolve, which in turn changes transposition rate. For example, Kofler et al. (2018) utilized experimental evolution to observe the evolution of a P-element invasion in populations of naïve D. simulans, documenting the emergence over time of P-element-specific piRNAs that curbed the spread of the P-element. In S. cerevisiae and S. paradoxus, recent work discovered two variants of the Ty1 element segregating in populations of wild and human-associated strains that determine rates of Ty mobility (Czaja et al. 2020). Strains with the canonical Ty1 element show reduced mobility of canonical Ty1 whereas strains with the divergent Ty1’ (and lack of genomic canonical Ty1) show increased mobility of canonical Ty1. This is presumably a result of the TE defense system (CNC) being Ty specific, such that Ty1’ CNC cannot control the mobility of Ty1.
One important caveat to our study is that transposition rate is dependent on genetic background, and more specifically, the existing Ty content present in a strain (Garfinkel et al. 2005; Czaja et al. 2020). Transposition of Ty1 is higher in S. cerevisiae and S. paradoxus strain backgrounds in which canonical Ty1 elements are absent or very low in copy number. The GRF167 S. cerevisiae strain background utilized in this study is likely “restrictive” for Ty mobilization due to the high copy number of Ty1 elements, and thus, testing transposition rate in interspecific hybrids derived from other “permissive” strain backgrounds is needed. A recent study examining Ty content across natural and experimentally evolved hybrids in the S. paradoxus species complex also found no evidence for increased Ty mobilization in hybrids (Hénault et al. 2020). Changes in Ty copy number in some experimentally evolved lines were observed but were not associated with evolutionary divergence between hybrid parents, and instead were highly genotype specific. These results further support prior work that transposition rate depends on genetic background.
Here, we find that mitochondrial inheritance in hybrids significantly changes transposition rate, the first study to document this connection. A mechanism of how mtDNA is influencing transposition is unclear, although mitochondria function in a huge variety of processes beyond generating cellular energy (Malina et al. 2018; Dujon 2020; Hose et al. 2020). Evidence from Drosophila, silkworm, and mice suggests that piRNA biogenesis, and thereby regulation of TEs, in germ cells is mediated through mitochondrial bound proteins and mitochondrial membrane metabolism/signaling (Pane et al. 2007; Watanabe et al. 2011; Nishida et al. 2018). The unique pattern of mtDNA inheritance and large numbers of nuclear-encoded mitochondrial genes contribute to mito-nuclear incompatibilities that underlie some speciation events (Lee et al. 2008; Gershoni et al. 2009; Chou and Leu 2010; Burton and Barreto 2012; Crespi and Nosil 2013) and human diseases (Duchen and Szabadkai 2010; Vafai and Mootha 2012). Moreover, species-specific inheritance of mtDNA in hybrids results in a strong environmentally dependent allele preference for one species’ alleles or the other (Hewitt et al. 2020). Perhaps this species-specific allele expression results in the suppression of S. cerevisiae encoded Ty elements in a hybrid with S. uvarum mtDNA, causing the observed lower rates of transposition.
Temperature also seems to play a mediating role in mitochondrial inheritance, mitochondria function, and TE movement. Mitochondria have been repeatedly implicated in adaptation to different temperatures (e.g., the “mitochondrial climatic adaptation hypothesis”) (Mishmar et al. 2003; Ballard and Whitlock 2004; Ruiz-Pesini et al. 2004; Wallace 2007; Dowling 2014; Camus et al. 2017). For example, in hybrids between thermotolerant S. cerevisiae and cryotolerant S. uvarum or S. eubayanus, S. cerevisiae mtDNA confers growth at high temperatures, whereas S. uvarum or S. eubayanus mtDNA confers growth at low temperatures (Baker et al. 2019; Li et al. 2019; Hewitt et al. 2020). An Australian cline of D. melanogaster showed thermal performance associated with each mitotype corresponds with its latitudinal prevalence (Camus et al. 2017). Intriguingly, TEs were shown to play a significant role in adaptation to the climatic variables in this same D. melanogaster cline (González et al. 2008, 2010). Recently Kofler et al. (2018) used experimental evolution of D. simulans at cold and warm temperatures and showed that temperature drastically impacts the rate at which a TE can spread in a population. In S. cerevisiae, rates of transposition are estimated to be 100 fold higher at temperatures 15–20 °C than at the normal lab conditions of 30 °C (Paquin and Williamson 1984; Garfinkel et al. 2005). All transposition assays were conducted at the standard 20 °C in this study, but future work could explore how temperature impacts transposition rate in non S. cerevisiae species, particularly the cold tolerant S. uvarum and S. eubayanus. If transposition rate is increased at cold temperatures, reduced transposition rate may be an evolutionary response to curb TE mobilization in cryotolerant species. This is certainly an intriguing area for further study.
The Role of Transposable Elements in Evolution
In recent years, we have witnessed a shift from viewing TEs as solely parasitic genetic elements, to appreciating the myriad ways in which TEs impact eukaryotic evolution. In our own work in laboratory evolution experiments, we have shown that Ty elements are often breakpoints for adaptive copy number variants and that insertions can cause adaptive gain and loss of function mutations (Dunham et al. 2002; Gresham et al. 2008; Hope et al. 2017). Intriguingly, we have previously observed fewer copy number variants in S. uvarum than S. cerevisiae evolved populations, perhaps related to their paucity of repetitive elements to facilitate such mutational events (Smukowski Heil et al. 2017, 2019). Copy number events, and in particular chromosome rearrangements can cause inviability between crosses (e.g., chromosomal speciation) (Hou et al. 2014), which may represent more relevant paths in which TEs may impact speciation (Serrato-Capuchina and Matute 2018). Although the evidence that TE mobilization in hybrids can facilitate speciation is limited, there remains much to be explored regarding evolution of host-TE dynamics between closely related species.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Marcus Annable for help in shearing TySeq libraries, Mary Bryk and Chris Hittinger for sharing strains, and David Garfinkel for sharing the Ty1his3AI plasmid. They also thank three anonymous reviewers for feedback on the manuscript. This work was supported by the National Science Foundation (1516330 to M.J.D.). C.S.H. was supported in part by the NIH/NHGRI Genome Training (T32 HG00035). During this work, M.J.D. was a Senior Fellow in the Genetic Networks program at the Canadian Institute for Advanced Research and a Rita Allen Foundation Scholar. M.J.D. was supported in part by a Faculty Scholars grant from the Howard Hughes Medical Institute.
Data Availability
All strains are available upon request. Sequencing data are deposited under BioProject ID PRJNA639117.
Literature Cited
- Ahn HW, Tucker JM, Arribere JA, Garfinkel DJ.. 2017. Ribosome biogenesis modulates Ty1 copy number control in Saccharomyces cerevisiae. Genetics 207(4):1441–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida P, et al. 2014. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 5:4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J. Mol. Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. 2003. Accumulation of Spock and Worf, two novel non-LTR retrotransposons, on the neo-Y chromosome of Drosophila miranda. Mol. Biol. Evol. 20(2):173–181. [DOI] [PubMed] [Google Scholar]
- Baker EP, et al. 2019. Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 5(1):eaav1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JWO, Whitlock MC.. 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13(4):729–744. [DOI] [PubMed] [Google Scholar]
- Bergman CM. 2018. Horizontal transfer and proliferation of Tsu4 in Saccharomyces paradoxus. Mob. DNA 9(18):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham PM, Kidwell MG, Rubin GM.. 1982. The molecular basis of P-M hybrid dysgenesis: the role of the P element, a P-strain-specific transposon family. Cell 29(3):995–1004. [DOI] [PubMed] [Google Scholar]
- Birky CW. 1995. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl. Acad. Sci. U.S.A. 92(25):11331–11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky CW. 2001. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35(1):125–148. [DOI] [PubMed] [Google Scholar]
- Bleykasten-Grosshans C, Friedrich A, Schacherer J.. 2013. Genome-wide analysis of intraspecific transposon diversity in yeast. BMC Genomics 14(1):399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon E, et al. 2000. Genomic exploration of the hemiascomycetous yeasts: 5. Saccharomyces bayanus var. uvarum. FEBS Lett. 487(1):37–41. [DOI] [PubMed] [Google Scholar]
- Bourque G, et al. 2018. Ten things you should know about transposable elements. Genome Biol. 19(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M, et al. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11(2):255–269. [DOI] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ.. 1984. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell 38(1):153–163. [DOI] [PubMed] [Google Scholar]
- Burton RS, Barreto FS.. 2012. A disproportionate role for mtDNA in Dobzhansky-Muller incompatibilities? Mol. Ecol. 21(20):4942–4957. [DOI] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus MF, Wolff JN, Sgrò CM, Dowling DK.. 2017. Experimental support that natural selection has shaped the latitudinal distribution of mitochondrial haplotypes in Australian Drosophila melanogaster. Mol. Biol. Evol. 34(10):2600–2612. [DOI] [PubMed] [Google Scholar]
- Carbone L, et al. 2012. Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol. Evol. 4(7):648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr M, Bensasson D, Bergman CM.. 2012. Evolutionary genomics of transposable elements in Saccharomyces cerevisiae. PLoS One 7(11):e50978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C, Hucks D, Feschotte C.. 2008. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol. Biol. Evol. 25(1):29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH.. 1989. The population genetics of Drosophila transposable elements. Annu. Rev. Genet. 23(1):251–287. [DOI] [PubMed] [Google Scholar]
- Chou J-Y, Leu J-Y.. 2010. Speciation through cytonuclear incompatibility: insights from yeast and implications for higher eukaryotes. BioEssays 32(5):401–411. [DOI] [PubMed] [Google Scholar]
- Coyne JA. 1985. Genetic studies of three sibling species of Drosophila with relationship to theories of speciation. Genet. Res. 46(2):169–192. [DOI] [PubMed] [Google Scholar]
- Coyne JA. 1986. Meiotic segregation and male recombination in interspecific hybrids of Drosophila. Genetics 114(2):485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA. 1989. Mutation rates in hybrids between sibling species of Drosophila. Heredity. 63(2):155–162. [DOI] [PubMed] [Google Scholar]
- Crespi B, Nosil P.. 2013. Conflictual speciation: species formation via genomic conflict. Trends Ecol. Evol. 28(1):48–57. [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Garfinkel DJ.. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 88(3):936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Garfinkel DJ.. 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell Biol. 12(6):2813–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja W, Bensasson D, Ahn HW, Garfinkel DJ, Bergman CM.. 2020. Evolution of Ty1 copy number control in yeast by horizontal transfer and recombination. PLoS Genet. 16(2):e1008632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechaud C, Volff J-N, Schartl M, Naville M.. 2019. Sex and the TEs: transposable elements in sexual development and function in animals. Mob. DNA. 10:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disdero E, Filée J.. 2017. LoRTE: detecting transposon-induced genomic variants using low coverage PacBio long read sequences. Mob. DNA. 8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DK. 2014. Evolutionary perspectives on the links between mitochondrial genotype and disease phenotype. Biochim. Biophys. Acta 1840(4):1393–1403. [DOI] [PubMed] [Google Scholar]
- Drinnenberg IA, et al. 2009. RNAi in budding yeast. Science 326(5952):544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen MR, Szabadkai G.. 2010. Roles of mitochondria in human disease. Essays Biochem. 47:115–137. [DOI] [PubMed] [Google Scholar]
- Dujon B. 2020. Mitochondrial genetics revisited. Yeast 37(2):191–205. [DOI] [PubMed] [Google Scholar]
- Dunham MJ, et al. 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 99(25):16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MJ, Gartenberg M, Brown GW.. 2015. Methods in yeast genetics and genomics, 2015 edition: a CSHL course manual. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. [Google Scholar]
- Elder RT, et al. 1981. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. II. Recombination and expression of Ty1 and adjacent sequences. Cold Spring Harb. Symp. Quant. Biol. 45(0):581–591. [DOI] [PubMed] [Google Scholar]
- Ellison CE, Bachtrog D.. 2013. Dosage compensation via transposable element mediated rewiring of a regulatory network. Science 342(6160):846–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Lee M, Ham C, Levin HL.. 2019. Transposable element insertions in fission yeast drive adaptation to environmental stress. Genome Res. 29(1):85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD. 2015. Transposable element detection from whole genome sequence data. Mob. DNA 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Jiang N, Wing RA, Jiang J, Jackson SA.. 2015. Transposons play an important role in the evolution and diversification of centromeres among closely related species. Front. Plant Sci. 6:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel DJ, Nyswaner K, Wang J, Cho J-Y.. 2003. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165(1):83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel DJ, Nyswaner KM, Stefanisko KM, Chang C, Moore SP.. 2005. Ty1 copy number dynamics in Saccharomyces. Genetics 169(4):1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel DJ, et al. 2016. A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces. Curr. Genet. 62(2):321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni M, Templeton AR, Mishmar D.. 2009. Mitochondrial bioenergetics as a major motive force of speciation. BioEssays 31(6):642–650. [DOI] [PubMed] [Google Scholar]
- Göbel U, et al. 2018. Robustness of transposable element regulation but no genomic shock observed in interspecific Arabidopsis hybrids. Genome Biol. Evol. 10(6):1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA.. 2008. High rate of recent transposable element–induced adaptation in Drosophila melanogaster. PLoS Biol. 6(10):e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Petrov DA.. 2009. The adaptive role of transposable elements in the Drosophila genome. Gene 448(2):124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Karasov TL, Messer PW, Petrov DA.. 2010. Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PLoS Genet. 6(4):e1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR, Johnston JS.. 2008. Genome size diversity in the family Drosophilidae. Heredity 101(3):228–238. [DOI] [PubMed] [Google Scholar]
- Gresham D, et al. 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4(12):e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LJ, Sandmeyer SB.. 1990. Characterization of a transpositionally active Ty3 element and identification of the Ty3 integrase protein. J. Virol. 64(6):2599–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hénault M, Marsit S, Charron G, Landry CR.. 2020. The effect of hybridization on transposable element accumulation in an undomesticated fungal species. bioRxiv 2020.06.17.157636. 10.1101/2020.06.17.157636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. 1988. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52(4):536–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SK, et al. 2020. Plasticity of mitochondrial DNA inheritance and its impact on nuclear gene transcription in yeast hybrids. Microorganisms 8(4):494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. 1988. Speciation via hybrid dysgenesis: negative evidence from the Drosophila affinis subgroup. Genetica 78(2):97–103. [Google Scholar]
- Hoffman CS, Winston F.. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57(2–3):267–272. [DOI] [PubMed] [Google Scholar]
- Hope EA, et al. 2017. Experimental evolution reveals favored adaptive routes to cell aggregation in yeast. Genetics 206(2):1153–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hose J, et al. 2020. The genetic basis of aneuploidy tolerance in wild yeast. eLife. 9:e52063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Friedrich A, de Montigny J, Schacherer J.. 2014. Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Curr. Biol. 24(10):1153–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-Y, Chou J-Y.. 2017. Environmental factors can influence mitochondrial inheritance in the Saccharomyces yeast hybrids. PLoS One 12(1):e0169953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug AM, Feldmann H.. 1996. Yeast retrotransposon Ty4: the majority of the rare transcripts lack a U3-R sequence. Nucleic Acids Res. 24(12):2338–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangam D, Feschotte C, Betrán E.. 2017. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet. 33(11):817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR.. 2004. Pack-MULE transposable elements mediate gene evolution in plants. Nature 431(7008):569–573. [DOI] [PubMed] [Google Scholar]
- Josefsson C, Dilkes B, Comai L.. 2006. Parent-dependent loss of gene silencing during interspecies hybridization. Curr. Biol. 16(13):1322–1328. [DOI] [PubMed] [Google Scholar]
- Kapusta A, Suh A, Feschotte C.. 2017. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. U.S.A. 114(8):E1460–E1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Dhakal P, Katterhenry AN, Heatherington CA, Ungerer MC.. 2011. Transposable element proliferation and genome expansion are rare in contemporary sunflower hybrid populations despite widespread transcriptional activity of LTR retrotransposons. Genome Biol. Evol. 3:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Wong K, Adams DJ.. 2013. RetroSeq: transposable element discovery from next-generation sequencing data. Bioinforma 29(3):389–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Edelman NB, Barbash DA.. 2012. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 10(11):e1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. 1983. Evolution of hybrid dysgenesis determinants in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 80(6):1655–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA.. 1977. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86(4):813–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF.. 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8(5):464–478. [DOI] [PubMed] [Google Scholar]
- Kofler R, Senti K-A, Nolte V, Tobler R, Schlötterer C.. 2018. Molecular dissection of a natural transposable element invasion. Genome Res. 28(6):824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel LE, Malik HS.. 2016. Centromeres. Curr. Biol. 26(12):R487–R490. [DOI] [PubMed] [Google Scholar]
- Kutter C, Jern P, Suh A.. 2018. Bridging gaps in transposable element research with single-molecule and single-cell technologies. Mob. DNA 9:34. [Google Scholar]
- Labrador M, Farré M, Utzet F, Fontdevila A.. 1999. Interspecific hybridization increases transposition rates of Osvaldo. Mol. Biol. Evol. 16(7):931–937. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA.. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49(3):264–285. [DOI] [PubMed] [Google Scholar]
- Lee H-Y, et al. 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135(6):1065–1073. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-W, et al. 2018. Transposable elements contribute to the adaptation of Arabidopsis thaliana. Genome Biol. Evol. 10(8):2140–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Peris D, Hittinger CT, Sia EA, Fay JC.. 2019. Mitochondria-encoded genes contribute to evolution of heat and cold tolerance in yeast. Sci. Adv. 5(1):eaav1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Peruffo A, James SA, Roberts IN, Louis EJ.. 2005. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast 22(3):177–192. [DOI] [PubMed] [Google Scholar]
- Liti G, et al. 2009. Population genomics of domestic and wild yeasts. Nature 458(7236):337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovskaya ER, Scheinker VS, Evgen’ev MB.. 1990. A hybrid dysgenesis syndrome in Drosophila virilis. Genetics 126(3):619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina C, Larsson C, Nielsen J.. 2018. Yeast mitochondria: an overview of mitochondrial biology and the potential of mitochondrial systems biology. FEMS Yeast Res. 18:foy040. [DOI] [PubMed] [Google Scholar]
- Marinoni G, et al. 1999. Horizontal transfer of genetic material among Saccharomyces yeasts. J. Bacteriol. 181(20):6488–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1984. The significance of responses of the genome to challenge. Science 226(4676):792–801. [DOI] [PubMed] [Google Scholar]
- Mishmar D, et al. 2003. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. U.S.A. 100(1):171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SP, et al. 2004. Analysis of a Ty1-less variant of Saccharomyces paradoxus: the gain and loss of Ty1 elements. Yeast 21(8):649–660. [DOI] [PubMed] [Google Scholar]
- Mularoni L, et al. 2012. Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res. 22(4):693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MG, Linheiro RS, Bergman CM.. 2017. McClintock: an integrated pipeline for detecting transposable element insertions in whole-genome shotgun sequencing data. Genes Genomes Genet. 7:2763–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvéglise C, Feldmann H, Bon E, Gaillardin C, Casaregola S.. 2002. Genomic evolution of the long terminal repeat retrotransposons in hemiascomycetous yeasts. Genome Res. 12:930–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, et al. 2018. Hierarchical roles of mitochondrial Papi and Zucchini in Bombyx germline piRNA biogenesis. Nature 555(7695):260–264. [DOI] [PubMed] [Google Scholar]
- Niu X-M, et al. 2019. Transposable elements drive rapid phenotypic variation in Capsella rubella. Proc. Natl. Acad. Sci. U.S.A. 116(14):6908–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opijnen T. V, Bodi KL, Camilli A.. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6(10):767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T.. 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12(6):851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin CE, Williamson VM.. 1984. Temperature effects on the rate of Ty transposition. Science 226(4670):53–55. [DOI] [PubMed] [Google Scholar]
- Pardue M-L, DeBaryshe PG.. 2011. Retrotransposons that maintain chromosome ends. Proc. Natl. Acad. Sci. U.S.A. 108(51):20317–20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer J, Kelly LJ, Leitch IJ, Zomlefer WB, Fay MF.. 2014. A universe of dwarfs and giants: genome size and chromosome evolution in the monocot family Melanthiaceae. New Phytol. 201(4):1484–1497. [DOI] [PubMed] [Google Scholar]
- Petrov DA. 2002. Mutational equilibrium model of genome size evolution. Theor. Popul. Biol. 61(4):531–544. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinforma 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL.. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 46(1):21–42. [DOI] [PubMed] [Google Scholar]
- Renaut S, Rowe HC, Ungerer MC, Rieseberg LH.. 2014. Genomics of homoploid hybrid speciation: diversity and transcriptional activity of long terminal repeat retrotransposons in hybrid sunflowers. Philos. Trans. R. Soc. B Biol. Sci. 369:20130345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishishwar L, Mariño-Ramírez L, Jordan IK.. 2017. Benchmarking computational tools for polymorphic transposable element detection. Brief Bioinform. 18(6):908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. 2011. Integrative genomics viewer. Nat Biotechnol. 29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MR, Doolittle WF.. 1983. Molecular biological mechanisms of speciation. Science 220(4593):157–162. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC.. 2004. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303(5655):223–226. [DOI] [PubMed] [Google Scholar]
- Saha A, et al. 2015. A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J. Virol. 89(7):3922–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MR, et al. 2019. Transposon insertional mutagenesis in Saccharomyces uvarum reveals trans-acting effects influencing species-dependent essential genes. Genome Res. 29(3):396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, et al. 2011. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. Genes Genomes Genet. 1:11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifele LZ, Cost GJ, Zupancic ML, Caputo EM, Boeke JD.. 2009. Retrotransposon overdose and genome integrity. Proc. Natl. Acad. Sci. U.S.A. 106(33):13927–13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I, Vu GTH.. 2016. Genome stability and evolution: attempting a holistic view. Trends Plant Sci. 21(9):749–757. [DOI] [PubMed] [Google Scholar]
- Serrato-Capuchina A, Matute DR.. 2018. The role of transposable elements in speciation. Genes 9(5):254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid S, Slotkin RK.. 2020. The current revolution in transposable element biology enabled by long reads. Curr. Opin. Plant Biol. 54:49–56. [DOI] [PubMed] [Google Scholar]
- Smukowski Heil CS, et al. 2017. Loss of heterozygosity drives adaptation in hybrid yeast. Mol. Biol. Evol. 34(7):1596–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukowski Heil CS, et al. 2019. Temperature preference can bias parental genome retention during hybrid evolution. PLoS Genet. 15(9):e1008383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staton SE, Ungerer MC, Moore RC.. 2009. The genomic organization of Ty3/gypsy-like retrotransposons in Helianthus (Asteraceae) homoploid hybrid species. Am. J. Bot. 96(9):1646–1655. [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Dorman CJ.. 2010. The effect of mobile element IS10 on experimental regulatory evolution in Escherichia coli. Mol. Biol. Evol. 27(9):2105–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thybert D, et al. 2018. Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. Genome Res. 28(4):448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Kawakami T.. 2013. Transcriptional dynamics of LTR retrotransposons in early generation and ancient sunflower hybrids. Genome Biol. Evol. 5(2):329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Strakosh SC, Stimpson KM.. 2009. Proliferation of Ty3/gypsy-like retrotransposons in hybrid sunflower taxa inferred from phylogenetic data. BMC Biol. 7(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Strakosh SC, Zhen Y.. 2006. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 16(20):R872–R873. [DOI] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK.. 2012. Mitochondrial disorders as windows into an ancient organelle. Nature 491(7424):374–383. [DOI] [PubMed] [Google Scholar]
- Van’t Hof AE, et al. 2016. The industrial melanism mutation in British peppered moths is a transposable element. Nature 534(7605):102–105. [DOI] [PubMed] [Google Scholar]
- Voytas DF, Boeke JD.. 1992. Yeast retrotransposon revealed. Nature 358(6389):717. [DOI] [PubMed] [Google Scholar]
- Wallace DC. 2007. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76(1):781–821. [DOI] [PubMed] [Google Scholar]
- Watanabe T, et al. 2011. MitoPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell. 20(3):364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilguny L, Kofler R.. 2019. DeviaTE: assembly‐free analysis and visualization of mobile genetic element composition. Mol Ecol Resour. 19(5):1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore KM, et al. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6(3):e00306–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CM, Maimer E, Adams J.. 1992. The population biology and evolutionary significance of Ty elements in Saccharomyces cerevisiae. Genetica 86(1–3):155–173. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, et al. 2015. Clade- and species-specific features of genome evolution in the Saccharomycetaceae. FEMS Yeast Res. 15(5):fov035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains are available upon request. Sequencing data are deposited under BioProject ID PRJNA639117.